
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Fungal Biol. , 10 August 2022
Sec. Fungal Biotechnology
Volume 3 - 2022 | https://doi.org/10.3389/ffunb.2022.944234
 Taryn A. Miller†
Taryn A. Miller† Debbie A. Hudson†
Debbie A. Hudson† Richard D. Johnson
Richard D. Johnson Jaspreet S. Singh
Jaspreet S. Singh Wade J. Mace
Wade J. Mace Natasha T. Forester
Natasha T. Forester Paul H. Maclean
Paul H. Maclean Christine R. Voisey
Christine R. Voisey Linda J. Johnson*
Linda J. Johnson*Epichloë festucae var. lolii and Epichloë sp. LpTG-3 are filamentous fungal endophytes of perennial ryegrass (Lolium perenne) that have a substantial impact on New Zealand’s agricultural economy by conferring biotic advantages to the host grass. Overall, Epichloë endophytes contribute NZ$200 million to the economy annually, with strain AR37 estimated to contribute NZ$3.6 billion to the New Zealand economy over a 20-year period. This strain produces secondary metabolites, including epoxyjanthitrems, which are a class of indole diterpenes, associated with the observed effects of AR37 on livestock and insect pests. Until very recently, AR37 was intractable to genetic modification but this has changed with the application of CRISPR-Cas9 based gene editing techniques. In this paper, gene inactivation by CRISPR-Cas9 was used to deconvolute the genetic basis for epoxyjanthitrem biosynthesis, including creating an AR37 strain that has been edited to remove the biosynthesis of all indole diterpenes. We show that gene editing of Epichloë can be achieved without off-target events or introduction of foreign DNA (footprint-less) through an AMA1-based plasmid that simultaneously expresses the CRISPR-Cas9 system and selectable marker. Genetic modification events in these transformants were investigated through genome sequencing and in planta chemistry.
Epichloë endophytes have been successfully commercialised as a seed-borne technology for pastoral agriculture to confer host persistence from invertebrate and vertebrate grazing pressures in New Zealand, and to a lesser extent, Australia and the USA (Johnson et al., 2013a). The symbiotic association that these filamentous fungi form with Pooideae grasses such as perennial ryegrass (Lolium perenne) are particularly crucial for conferring insect bioprotection in New Zealand farming systems (Caradus and Johnson, 2019; Johnson and Caradus, 2019). A widely adopted Epichloë strain for perennial ryegrass pastures in New Zealand is AR37 (Lolium perenne taxonomic group 3, LpTG3). This endophyte strain provides protection against Argentine stem weevil (Listronotus bonariensis), pasture mealy bug (Balanococcus poae), porina (Wiseana spp), black beetle (Heteronychus arator), and root aphid (Aploneura lentisci). Of the known secondary metabolite pathways in Epichloë, AR37 has a non-functional peramine pathway and a functional indole diterpene (epoxyjanthitrems) pathway, with both the ergot alkaloid and loline pathways absent (Johnson et al., 2013a).
Indole diterpenes are a class of secondary metabolite compounds comprised of a cyclic diterpene-derived skeleton joined to an indole moiety that are produced by filamentous fungi from the phylum Ascomycota e.g., Penicillium, Aspergillus, Claviceps, and Epichloë. Structural and therefore bioactive diversity is brought about through differences in stereochemistry, prenylation, hydroxylation, epoxidation, methylation, and oxidation patterns (Kozák et al., 2019). All species that produce indole diterpenes contain a core set of four genes that code enzymes involved in the production of the first stable intermediate; IdtG (geranylgeranyl pyrophosphate synthase) converts isopentenyl diphosphate (IPP) and/or dimethylallyl pyrophosphate (DMAPP) and/or geranyl pyrophosphate (GPP) and/or farnesyl pyrophosphate (FPP) into geranylgeranyl pyrophosphate (GGPP). IdtC (a geranylgeranyl transferase) catalyses the indole condensation of GGPP with indole-3-glycerol phosphate (IGP) to produce 3-geranylgeranyl indole (3-GGI). IdtM (a FAD dependent epoxidase) either single or double epoxidates, depending on species, the 3-GGI, which is then finally cyclised by IdtB (a cyclase) via either a mechanism of Markovnikov’s or anti-Markovnikov’s rule, again species dependent. In Epichloë, all indole diterpene pathways are double epoxidated and cyclised through the Markovnikov’s mechanism to produce the first stable intermediate paspaline (Parker and Scott, 2004).
In Epichloë, the indole diterpene pathway produces two well characterised classes of decorated indole diterpenes, the lolitrems and the epoxyjanthitrems. These two classes of indole diterpenes are produced by a core set of genes as well as a small further subset of unique genes and are typically only expressed in planta (Young et al., 2009; Ludlow et al., 2019; Finch et al., 2020). The lolitrems are produced in a range of Epichloë species, with diversification through non-functional genes or the presence or absence of genes (Young et al., 2009), with lolitrem B responsible for ryegrass staggers, a neurological disorder of grazing livestock (Fletcher and Harvey, 1981). This pathway consists of an interconnected network of compounds, rather than a linear pathway, synthesised by 11 genes in total grouped into 3 clusters interspaced with AT-rich regions in a single sub-telomeric loci (Young et al., 2006; Saikia et al., 2012). Further investigation has been conducted into identifying the exact pathway intermediates between paspaline and lolitrem B, including structure, pathway placement, tremorgenicity, and insect bioactivity, e.g. lolitrems A, B, and F have long-term tremors while lolitrem E is non-tremorgenic, to build up an understanding of this important bioactive secondary metabolite pathway (Saikia et al., 2008; Saikia et al., 2012). In contrast, epoxyjanthitrems are produced in a restricted group of strains within the taxonomic grouping LpTG-3 (Ludlow et al., 2019) (Linda Johnson, unpublished data). These compounds are associated with mild ryegrass staggers but more importantly bioactivity against porina (Hennessy et al., 2016; Finch et al., 2020). Recently, a single loci consisting of 16 genes associated into 4 clusters was identified as potentially being involved in epoxyjanthitrem biosynthesis through bioinformatic analysis of the genome of Epichloë sp. LpTG-3 strain NEA12 (Ludlow et al., 2019). Clusters 1 and 2 were analogous to the first two clusters in the lolitrem pathway (Young et al., 2009), cluster 3 was novel and linked to epoxyjanthitrem biosynthesis through RNAi knock-down of jtmD (idtD) followed by in planta chemistry, and cluster 4 was also novel but no additional experimental evidence was collected. A network pathway was then proposed based on the jtmD RNAi chemistry with proposed gene functions, and indole diterpene structures from this study (Ludlow et al., 2019). In addition, the chemical structures of all five epoxyjanthitrem compounds have now been elucidated (Finch et al., 2020).
Traditionally, genetic modification of Epichloë has involved the use of homologous recombination in conjunction with protoplast mediated transformation to either knock-out or knock-in genes. Sexual species such as the model E. festucae strain Fl1, are genetically tractable to these techniques allowing a thorough investigation into the genes involved in secondary metabolite biosynthesis as well as the Epichloë-ryegrass symbiotic association (Fleetwood et al., 2007; Eaton et al., 2008; Tanaka et al., 2008; Charlton et al., 2012; Saikia et al., 2012; Johnson et al., 2013b; Tanaka et al., 2013; Berry et al., 2015; Johnson et al., 2015; Voisey et al., 2016; Rahnama et al., 2018; Scott et al., 2018; Hassing et al., 2019; Rahnama et al., 2019; Lukito et al., 2020; Noorifar et al., 2021; Johnson et al., 2021; Hassing et al., 2022). However, in general, these techniques have been less effective on asexual strains such as AR37 and AR1, making molecular investigation into these strains near impossible (Panaccione et al., 2001) (Linda Johnson, unpublished).
The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas (CRISPR Associated protein) system is a naturally derived immune system originally discovered from bacteria and archaea, and is divided into two Classes, I and II, each further classified by sub-class I-VI. For example, Cas9 is a class II sub-class II CRISPR/Cas enzyme system (Jinek et al., 2012; Makarova et al., 2018). By 2013 CRISPR-Cas had been adapted as a novel gene editing technique in a range of systems e.g., mammals, plants, insects, and fungi (Jiang et al., 2021). More recently, Song et al. (2018) developed a sophisticated CRISPR-Cas9 delivery system that has been successfully used in Aspergillus via an autonomously replicating plasmid ANEp8_Cas9_LIC that simultaneously expresses the Cas9 protein, associated guide, and selectable marker for CRISPR genetic modification (Kun et al., 2021). CRISPR has been successfully implemented in other Epichloë species through alternative expression systems (Florea et al., 2021; Wang et al., 2021). The CRISPR-Cas system used by Wang et al. (2021) was based on a single plasmid harbouring a chimeric RNA guide and the cas9 gene under a fungal promoter, however without the autonomously replicating AMA sequence present in the vector delivery system used by Song et al. (2018). In contrast the approach taken by Florea et al. (2021) was based on Cas9-sgRNA (single guide and tracrRNA) ribonucleoprotein complexes (RNPs).
In this paper, we used a modification of the CRISPR-Cas9 autonomously replicating plasmid, developed by Song et al. (2018), to inactivate genes in the epoxyjanthitrem pathway of AR37 in order to i) investigate a novel method of genetic modification of AR37, ii) investigate the footprint-less and off-target nature of this specific CRISPR-Cas9 system, iii) compare the epoxyjanthitrem genes in AR37 with those in NEA12 (Ludlow et al., 2019) through bioinformatic analysis of the genome, iv) confirm involvement of ltmM (hence forth idtM), jtmO (hence forth idtO), jtm02 (hence forth idtA), and ltmF (hence forth idtF) in epoxyjanthitrem synthesis, and v) provide further validation of the proposed epoxyjanthitrem pathway, which we have determined differs to the previously published version by Ludlow (2019).
Escherichia coli strains Top10 and One Shot™ ccdB survival™ 2 T1R competent cells (Invitrogen Corp., Carlsbad, California, USA) were grown at 37°C at 180 rpm in Luria–Bertani broths or 1.5% (w/v) agar plates (made to manufactures specifications) supplemented where necessary with ampicillin (100 µg/mL).
Cultures of Epichloë were maintained on 1.5% (w/v) potato dextrose agar (PDA) plates (Difco, Sparks, Maryland, USA) (made to manufactures specifications) supplemented where necessary with hygromycin (150 µg/mL) and grown at 22°C at 180 rpm.
The ANEp8_Cas9_LIC plasmid (Song et al., 2018) (Figure S1A and Table S1) was constructed through the introduction of a 38-bp ligation independent cloning (LIC) site, centred with a SwaI restriction site, into the ANEp8_Cas9 plasmid described in Song et al. The ANEp8_Cas9_gRNA plasmid (obtained from Concordia University) (Figure S1B and Table S1) was constructed through the introduction of a gRNA cassette, via the LIC method, into ANEp8_Cas9_LIC shown in Song et al. - Figure S5 We adapted the ANEp8_Cas9_LIC plasmid to contain a hygromycin cassette in place of the PyrG gene for selection. The ANEp8_Cas9_LIC plasmid was initially digested with NotI to liberate a 5.3 kb fragment containing the AMA1 cassette (purified by gel extraction) and a 10.3 kb fragment (purified by gel extraction) containing the Cas9 and PyrG genes. Subsequent digestion of the 10.3 kb fragment with KpnI liberated a 9 kb Cas9 cassette (purified by gel extraction) containing the Cas9 gene and removal of PyrG. To amplify the hygromycin resistance cassette (2.9 kb), primer PCR was performed on pDONR221-Hyg template with restriction enzyme adapted primers KpnI TtrpC Hyg DONR R and NotI PgpdA Hyg pDONR F (Table S1), with the resulting product being digested with KpnI and NotI prior to ligation. The Cas9 (NotI/KpnI digested) cassette and the Hygromycin resistance (NotI/KpnI digested) cassette were ligated with T4 ligase (Invitrogen Corp., Carlsbad, California, USA) at 16°C overnight, creating the Cas9Hyg plasmid. The Cas9Hyg plasmid was re-digested with NotI to linearise and alkaline phosphatase treated (purified by gel extraction) before its T4 ligation with the AMA1 (NotI digested) cassette, creating the Cas9HygAMA plasmid. The addition of the gRNA cassette containing the mock sgRNA SapI protospacer was achieved through fusion PCR and LIC. The plasmid ANEp8-Cas9-gRNA was used as a template in the gRNA cassette construction which was assembled by fusion PCR of the two DNA fragments using the primers: SapI site CRISPR Fw P1 and Rev LIC2 as well as SapI site CRISPR Rev P1 and Fw LIC2 (Table S1). The first fragment containing the tRNA promoter was amplified from ANEp8-Cas9-gRNA plasmid and the 20 bp protospacer sequence containing the SapI sites was added at the 3’-end through PCR using an overlapping sequence (Table S1). The second fragment was amplified from ANEp8-Cas9-gRNA plasmid with the 20 bp SapI protospacer sequence and tRNA terminator sequence added at 5’and 3’ends respectively via PCR (Table S1). In the fusion, PCR 1ng of each of the gel extracted PCR fragments were used as template in a 25 μL reaction and performed with Phusion DNA polymerase (New England Biolabs (NEB, Ipswich, Massachusetts, USA). The resulting PCR product was inserted into the Cas9HygAMA plasmid through LIC via its SwaI restriction enzyme site creating Cas9HygAMASapI. A ccdB lethal cassette was cloned between the two SapI sites to aid in the efficiency of future protospacer cloning. To amplify the ccdB lethal cassette sequence (2 kb), primer PCR was performed on the split marker plasmid pDONR-SM1 template with SapI restriction enzyme adapted primers SapI ccdB F and SapI ccdB R (Table S1). The resulting product was digested with SapI prior to its ligation with Cas9HygAMASapI (SapI digested) creating the Cas9HygAMAccdB (Figure S1C and Table S1) plasmid.
The AR37 epoxyjanthitrem genes idtM, idtD, idtO, idtA, and idtF were screened for CRISPR-Cas9 target sites N(21)GG, with the 3′ PAM sequence (NGG) using Geneious (2019.1.1) (Biomatters Ltd, Auckland, New Zealand). Two protospacer sequences targeting each gene (with no predicted off target-sites) were selected (Table 1) (Doench et al., 2014; Xie et al., 2014).
The ANEp8-Cas9-LIC (Figure S1A and Table S1) plasmid was used as host plasmid to harbour the gRNA cassette by using the LIC method (Song et al., 2018). The idtM and idtD gRNA cassette used for plasmid construction was amplified with a pair of end primers to link with LIC sequence sites at both sides (idtM g32 Rev P1/idtM g32 Fw P1 = g32, idtM g38 Rev P1/idtM g38 Fw P1 = g38, idtD g63 Rev P1/idtD g63 Fw P1 = g63, and idtD g148 Rev P1/idtD g148 Fw P1 = g148) (Table S1) to generate complementary single-strand overhangs between ANEp8-Cas9 plasmid and the idtM/idtD gRNA cassette insert. The SwaI linearised ANEp8-Cas9 and the idtM/idtD gRNA cassette DNA (ending with LIC tails) were treated by T4 DNA polymerase in the presence of dGTP and dCTP, respectively. The 20 μL reaction mixture contained 0.2 pmol of DNA, 0.8 μL of 100 mM dithiothreitol, 2 μL of 25 mM dGTP or dCTP, and 3 U of T4 DNA polymerase in NEB buffer 2.1. The reaction was carried out at 22°C for 30 mins followed by enzyme inactivation by heating at 75°C for 20 mins. The insert and plasmid were mixed in a 3:1 molar ratio. To achieve annealing, the mixture was first heated at 60°C for 5 mins and then gradually decreased to 4°C (reduce 0.1°C per second). The annealed products were transformed into E.coli Top10 (Invitrogen Corp., Carlsbad, California, USA) competent cells to generate plasmids Cas9HygAMA-idtMg32 (Figure S1D), Cas9HygAMA-idtMg38, Cas9HygAMA-idtDg63, and Cas9HygAMA-idtDg148 (Table S1). Transformants were pre-screened by PCR using PrimeSTAR GXL polymerase (Takara Bio Inc, USA) with the gRNA screen F and gRNA screen R primers (Table S1). ZymoPure large plasmid prep kit (Zymo Research, Orange, California, USA) was used to obtain the plasmids and these were checked by sequencing using the gRNA screen F and gRNA screen R primers (Table S1).
Primers were designed to contain 20 bp guide protospacer sequences for the idtO (idtO g119 Top/idtO g119 Btm = g119 and idtO g144 Top/idtO g144 Btm = g144), idtA (idtA g101 Top/idtA g101 Btm = g101 and idtA g64 Top/idtA g64 Btm = g64), and idtF (idtF g86 Top/idtF g86 Btm = g86 and idtF g119 Top/idtF g119 Btm = g11) (Table S1) genes, with the appropriate SapI overhang at each end for cloning into the Cas9HygAMAccdB (Figure S1C) plasmid. Each protospacer was generated by annealing 15ng of the forward and reverse oligonucleotides in annealing buffer (10 mM Tris-HCl pH 8, 50 mM NaCl, 1 mM EDTA, pH 8). The following thermocycler program was used for annealing: 5 mins at 95°C, 20 sec at 95°C, a decrease of 0.5°C/cycle for 140 cycles, 1 mins at 25°C. The annealed oligonucleotides were ligated with the digested Cas9HygAMAccdB (SapI) plasmid using T4 ligase (Invitrogen Corp., Carlsbad, California, USA) at 20°C for 15 mins. The annealed products were transformed into E.coli Top10 competent cells to generate Cas9HygAMA-idtOg119 and Cas9HygAMA-idtOg144 for the idtO gene, Cas9HygAMA-idtAg64 and Cas9HygAMA-idtAg101 for the idtA gene, and Cas9HygAMA-idtFg86 and Cas9HygAMA-idtFg119 for the idtF gene (Table S1). Transformants were screened by colony PCR using Sapphire Amp Fast PCR master mix (Takara Bio Inc, USA) with the idtO/idtA/idtF gRNA Top anneal oligonucleotide and the reverse gRNA-specific primer gRNA Screen R (Table S1). Sequencing to confirm the correct gRNAs was achieved by primer PCR using the gRNA screen F and gRNA screen R primers (Table S1).
Genomic DNA for PCR screening was isolated from Epichloë mycelium using the Fungal Bacterial DNA mini kit (Zymo Research, Orange, California, USA). Genomic DNA was extracted from AR37 idtM, idtD, and idtA CRISPR gene edited strains using modified small scale Byrd et al. (1990) method: where the chloroform:phenol extraction steps were replaced by the Fungal Bacterial DNA mini kit (Zymo Research, Orange, California, USA). Plasmid DNA was isolated and purified from E. coli cultures using the ZymoPure large Plasmid Prep kit (Zymo Research, Orange, California, USA). Plasmids and transformants were sequenced at Massey Genome Service (Palmerston North, New Zealand) using the ABI 3730 and the ABI 3500xl DNA Analyser, with data analysed using Geneious (2019.1.1) software. The genomic DNA was sequenced at Massey Genome Service (Palmerston North, New Zealand) by single lane of 250 bp paired-end sequence on an Illumina MiSeq. The reads were dynamically trimmed using the SolexaQA package to their longest fragment such that the base call error rates did not exceed a P value of 0.05, and paired end reads of less than 100 bp discarded.
Protoplasts of Epichloë were prepared as described in Fleetwood et al. (2007). AR37 wild type protoplasts were transformed as described in Fleetwood et al. (2007) with 300 fmol of the appropriate Cas9HygAMAgRNA plasmid, or for idtA/idtF inactivated gene edits, AR37 inactivated idtA protoplasts were transformed with the corresponding idtF Cas9HygAMAgRNA plasmid (Table S1). Transformants were selected on RG Media (PD with 0.8 M sucrose pH 6.5) containing hygromycin (150 µg/mL). To obtain clonal isolates, the resulting transformants were purified by sub-culturing two times as described by Young et al. (2005) and screened by PCR for CRISPR gene edits as detailed in Table S1, with gene editing events confirmed by sequencing.
For each guide, a single transformant predicted to result in early translational termination due to a frame shift in the predicted functional domain (i.e., non-functional transcript) was selected for inoculation in preparation for in planta chemistry. Endophyte-free seedlings were inoculated using the method of Latch and Christensen (1985) as follows: Perennial ryegrass (Lolium perenne cv. Samson) were inoculated with Epichloë AR37 wild type or CRISPR-Cas9 gene edited strains. Seedlings were grown in proprietary potting mixture in 45 mm pots under glasshouse conditions for 6 weeks and assessed for endophyte infection by immunoblotting (Simpson et al., 2012). Each CRISPR guide edit was successfully infected into at least 6 plant genotypes. Plants were allowed to mature for a minimum of a further 3 months under glasshouse conditions before being sampled for chemical analysis.
Pseudo-stems, the basal section of the tiller, were harvested from each endophyte-infected plant. Samples were harvested into liquid nitrogen and then transferred to a freeze drier (Freezone Plus12, Labconco Corporation, Kansas City, MI, USA). Once lyophilized the samples were ground and homogenised with a bead mill (Omni Bead Ruptor 24, Omni International Inc., Kennesaw, GA, USA) in a 7 mL vial using a ¼ inch zirconium bead (30 seconds at 4.5 m/s). Sub-samples (50 mg) were extracted with 1 mL of the prepared extraction solvent (80% v/v methanol with 0.54 ng/mL ergotamine, 0.202 ng/ml festuclavine, and 1.7 ng/mL homoperamine as internal standards) in 2 mL plastic vials for 1 hour by end-over-end rotation (30 Hz) in the dark. After centrifuging (5000 g, 5 min), the supernatant was transferred to 2 mL amber HPLC vials for analysis. Along with the samples, duplicate reference samples of AR37 (for quantifying the epoxyjanthitrems) were similarly extracted and analysed.
Samples were analysed according to Berry et al. (2022). For the epoxyjanthitrems, due to the instability of epoxyjanthitrems as pure compounds, the peak areas and known concentrations of the reference samples (AR37) were used to determine response factors, which were used to subsequently quantify each epoxyjanthitrem compound in the samples. All values are an average of up to two guides, with each guide in at least 6 plant genotypes, per CRISPR gene edit (Table 1). Table S2 shows the mass spectrometer parameters specific to each compound. For each indole diterpene a one-way ANOVA was used to compare the means of the inactivated genes to the AR37 wild type using MiniTab v19.1.1 (Minitab LLC, State College, PA, USA). A Fisher pairwise comparison was used to show significantly different means.
Samples were analysed according to Johnson et al. (2015) with modification of the linear gradient profile (eluent A, aqueous 0.1% formic acid and eluent B, acetonitrile with 0.1% formic acid); time 0 mins (T0) at 5% B and held for 2 mins, T10 at 35% B, T16 at 95% B and held for 2 mins, followed by equilibration to initial conditions over the following 6 mins. Detection and quantitation were achieved using a LTQxl (Thermo Fisher Scientific, Waltham, MA, USA) in ESI positive ion mode collecting MS1 spectra in the range 300 – 1300 m/z. The raw data was interrogated using LCQuan v5.0 (Thermo Scientific Inc, San Jose, CA, USA). The epichloëcyclins specific to AR37 were detected as peaks of 562.3 and 570.3 m/z at 8.8 and 7.5 mins respectively. No attempt was made to quantify the epichloëcylins, as detection alone was sufficient to confirm infection with AR37.
Genome for AR37 wild type and the CRISPR gene inactivated strains (multiple strains per guide) (Table S3) were assembled using MaSuRCA version 3.4.1 (Zimin et al., 2013) from reads previously generated by Illumina HiSeq and PacBio platforms. The trimmed MiSeq reads from each of the gene edited strains were aligned to the AR37 genome and the ANEp8-Cas9-LIC1 host plasmid sequence using the “mem” algorithm of BWA version 0.7.17-r118 (Li, 2013). The read alignments were then converted into the binary alignment (BAM) format using samtools version 1.8 (Li et al., 2009) for viewing in the Integrative Genomics Viewer (IGV) and for plasmid detection. Fuzznuc version 6.6.0.0 (EMBOSS) was used to find potential off-target sites in the AR37 genome by searching for each of the CRISPR target (protospacer and PAM) sequences with up to 5 mismatches in the first 21 bases. Up to 300 bases either side of the identified potential off target sites were visually inspected using IGV software. The view function of samtools version 1.8 (Li et al., 2009) was used to reveal any plasmid sequence integration into the genomes of the edits by counting the number of sequencing reads aligning to the plasmid (Table S3). Samples with more reads mapping to the plasmid than the control samples were confirmed visually using the IGV, whereas samples with less reads mapping to the plasmid than the control samples were considered ‘footprint-less’. Control DNA was generated by using an AR37 strain which had previously been genome sequenced twice, with and without spiking with plasmid, ANEp8-Cas9-LIC1, with the unspiked strain yielding no detectable vector contamination.
Given the structural similarities between janthitrems produced by Penicillium janthinellum (Nicholson et al., 2015) and epoxyjanthitrems produced by Epichloë sp. LpTG-3 strain AR37 (Tapper and Lane, 2004), the nucleotide sequence of P. janthinellum janD (accession KF280651:44,433-45,819) was used as the query input sequence for blastn and blastx homology searches within Epichloë sp. LpTG-3 strain AR37 genome assembly contigs. A hit was found by blastx with 52% protein level identity and 83% coverage. The corresponding gene was subsequently found within the same genetic scaffold containing known indole diterpene gene clusters by further blastn analyses using larger sequence regions surrounding the janD homologue following long-read DNA sequencing. In keeping with agreed naming conventions for indole diterpene genes of Epichloë sp., the janD equivalent gene was designated idtD. Coding sequences for the genes of this scaffold were determined using transcript information generated previously (Forester, unpublished) and the 19 proteins were subjected to blastp analysis for functional prediction. This led to the identification of four putative indole diterpene gene clusters containing 19 genes overall (Figure 1): cluster 1 and cluster 2 were homologous to lolitrem B cluster 1 and cluster 2 respectively (Young et al., 2006), cluster 3 containing the janD homolog (Nicholson et al., 2015), and cluster 4 containing genes of known indole diterpene biosynthesis. Another study recently reported on epoxyjanthitrems production in another Epichloë sp. LpTG-3 strain, NEA12, highlighting four genes (jtmO, jtmD, jtm01, and jtm02) relevant to the biosynthetic pathway (Ludlow et al., 2019). Through genomic sequence comparison an identical genomic sequence to AR37, was shown for the indole diterpene gene cluster NEA12 (Figure 1). Genbank accession numbers for the idt gene clusters (and features therein) were assigned as ON500678 for AR37 and ON500679 for NEA12. A selected number of AR37 idt genes were then chosen for functional characterisation through CRISPR gene inactivation.
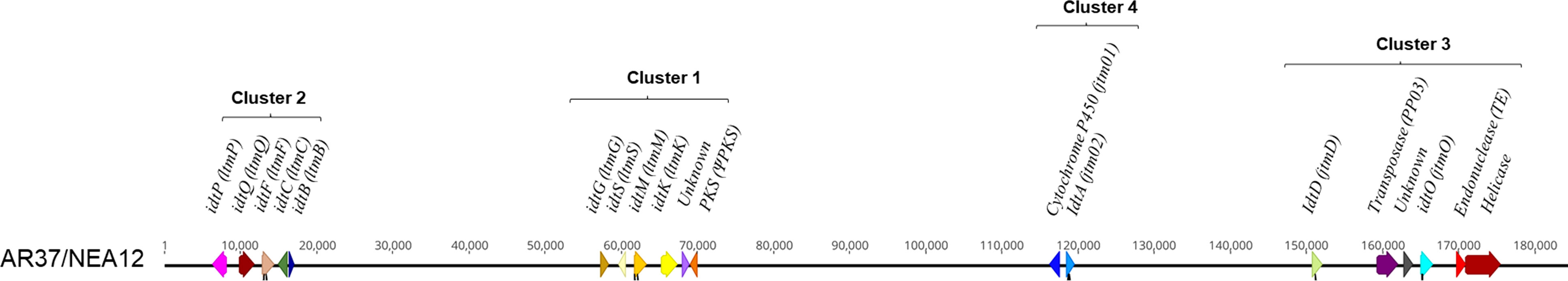
Figure 1 A comparison of syntenic analyses between AR37 and NEA12 epoxyjanthitrem biosynthetic clusters. Simplified gene location for epoxyjanthitrem, a group of indole diterpenes in Epichloë sp. LpTG-3 strain AR37 and NEA12. Identification and comparison of the genetic region containing the gene clusters proposed to be involved in the production of epoxyjanthitrem compounds in AR37 and NEA12. Genes are depicted as coloured arrows with direction indicating the direction of the gene transcription, size proportional to the gene size, and colour indicating gene homology. Either the given name or function is labelled above the gene, with gene names in brackets referring to corresponding genes in Ludlow et al. (2019). Black lines under the arrows indicate the guide locations. The row of numbers are the base pairs across the region.
Expression of CRISPR-Cas9 through Cas9HygAMAccdB containing either guide 32 or guide 38 designed to target idtM in AR37, successfully resulted in 18% inactivation rate (from 16 transformants) or 68% inactivation rate (from 16 transformants) respectively (Table 1). One transformant from each guide was taken forward for further investigation; a single C insertion in transformant #12 for guide 32 or a single A deletion in transformant #8 for guide 38 (Figure 2 and Table 1). In planta chemistry confirmed that both these idtM gene inactivated strains did not produce any indole diterpenes (Table 2A), with the endophyte presence confirmed through the detection of epichloëcyclins (Johnson et al., 2015), a characterised endophyte metabolite.
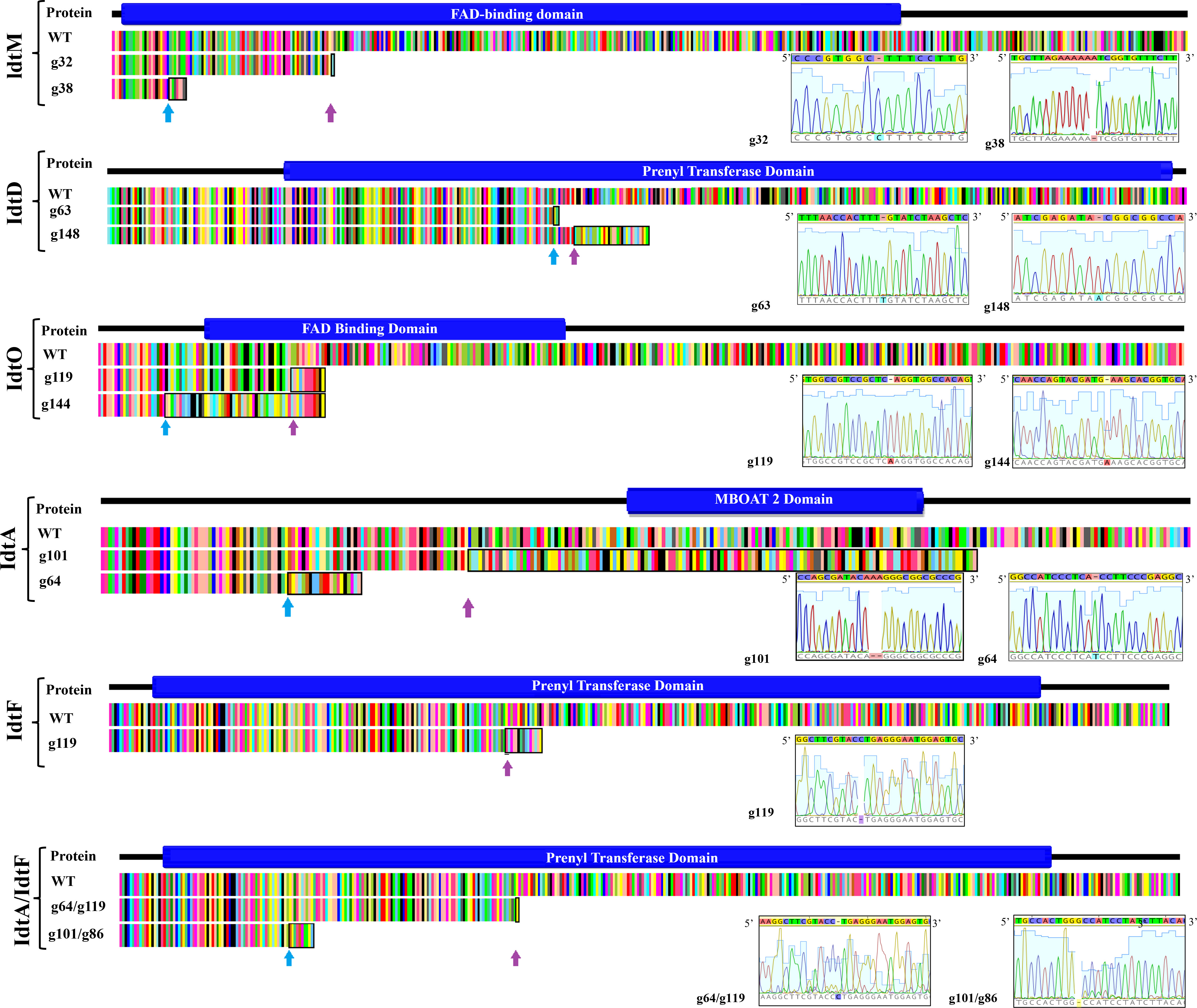
Figure 2 Resultant polypeptide and corresponding transcript of CRISPR gene edited indole diterpene genes in the proposed epoxyjanthitrem pathway. Pictorial depiction of the resultant polypeptide and corresponding transcript due to CRISPR gene inactivation of the indole diterpene genes idtM, idtD, idtO, idtA, idtF, and idtA/idtF when compared to the AR37 wild type transcript (WT) and the functional domain. The functional domain was predicted using the online software IntroPro Scan. The amino acid sequence of the AR37 wild type and CRISPR gene edited strains are colour coordinated by amino acid type with the indel site indicated in the CRISPR gene edited strains by a coloured arrow. The black box highlights the non-homologous amino acid sequence resulting from the indel. Description of the gene, guide, transformant number, indel, and frameshift is provided on the left-hand side of the figure and/or in the inserted table, with sequencing data provided in inserted chromatogram.
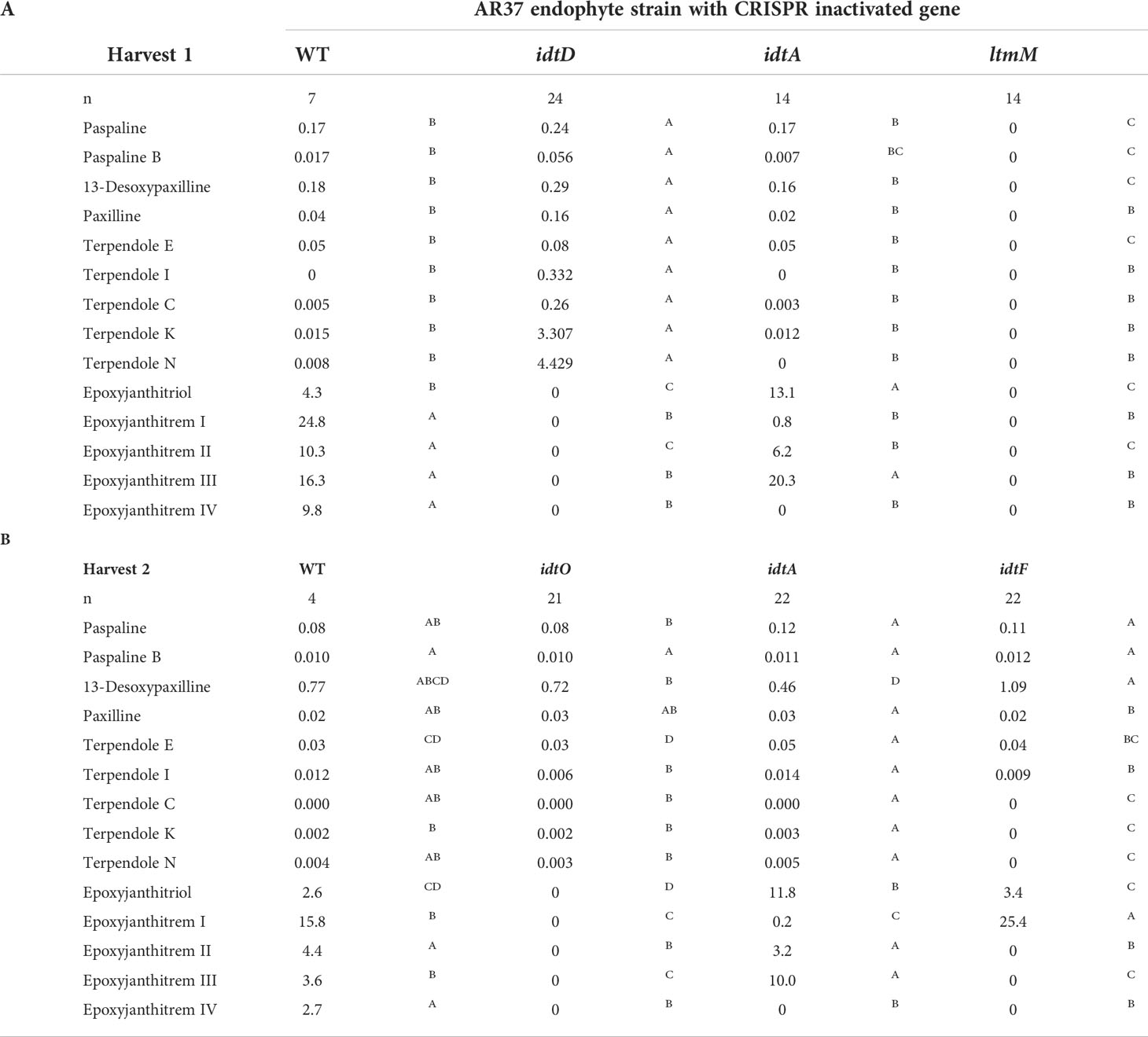
Table 2 Levels of indole diterpenes in AR37 wild type (WT) and AR37 strains with biosynthetic genes inactivated through CRISPR gene editing.
The same plasmid system was used to target idtD in AR37 through the expression of either guide 63 which resulted in a 62% inactivation rate (from 8 transformants) or guide 148 which resulted in a 100% inactivation rate (from 1 transformant) (Table 1). The two transformants taken forward where #113 which had a single T insertion and transformant #9 which had a single A insertion, respectively for each of the guides (Figure 2 and Table 1). For both these idtD gene inactivated strains, in planta chemistry detected no epoxyjanthitrem compounds, but did detect paspaline, paspaline B, 13-desoxypaxilline, paxilline, and terpendole compounds E, I, C, N, and K at increased levels when compared to the AR37 wild type strain (Table 2A).
When Cas9HygAMAccdB expressed guide 119 and guide 144, which both target idtO in AR37, a 10% inactivation rate (from 10 transformants) and 100% inactivation rate (from 7 transformants) was achieved for each guide, respectively (Table 1). Both transformants taken forward, #1 for guide 119 and #8 for guide 144, had an A insertion (Figure 2 and Table 1). The in planta chemistry again detected no epoxyjanthitrem compounds. However, in contrast to the idtD inactivated strains, paspaline, paspaline B, 13-desoxypaxilline, paxilline, and terpendole compounds E, I, C, N, and K were detected at similar to that if the AR37 wild type strain (Table 2B).
The idtA gene in AR37 was targeted through expression of guide 64 or guide 101 using the Cas9HygAMAccdB expression system and resulted in a 15% inactivation rate (from 20 transformants) or 33% inactivation rate (from 9 transformants) respectively (Table 1). Transformant #19-9 containing a single T insertion from guide 64 and transformant #3-4 containing a double A deletion from guide 101 were taken forward for further investigation (Figure 2 and Table 1). For both these idtA gene inactivated strains, in planta chemistry showed that epoxyjanthitrems I and IV were eliminated or reduced, respectively, while the levels of epoxyjanthitrems II and III were equivalent and epoxyjanthitriol was increased, in comparison to the AR37 wild type strain. The detected levels of paspaline, paspaline B, 13-desoxypaxilline, paxilline, and terpendole compounds E, I, C, N, and K were comparable to those of the AR37 wild type strain (Table 2A).
In AR37, idtF was targeted with the same plasmid system through the expression of guide 119 resulting in a 75% inactivation rate (from 4 transformants) (Table 1), with transformant #4 containing a C deletion taken forward in this study (Figure 2 and Table 1). In planta chemistry detected, equivalent levels of epoxyjanthitriol and increased levels of epoxyjanthitrem I compared to AR37 wild type strain, while epoxyjanthitrems II, III, and IV were eliminated. The levels of paspaline, paspaline B, 13-desoxypaxilline, paxilline, and terpendole I were comparable to the AR37 wild type strain, while terpendoles C, K, and N were eliminated (Table 2B).
Finally, double CRISPR gene inactivated strains were created using idtA inactivated protoplasts; IdtAg64 strain with idtF guide 119 or IdtA101 strain with idtF guide 86. The gene inactivation rates were 75% (from 4 transformants) and 33% (from 9 transformants) for the two idtF guides respectively (Table 1). The transformants taken forward were #4 containing a C insertion and transformant #8 containing a G deletion for idtF guides, respectively (Figure 2 and Table 1). In planta chemistry showed levels of epoxyjanthitriol were increased while levels of epoxyjanthitrem I were heavily reduced. Again, epoxyjanthitrems II, III, and IV were eliminated along with terpendoles C, K, and N. Detected levels of paspaline, paspaline B, 13-desoxypaxilline, paxilline, and terpendole I were equivalent to the AR37 wild type strain (Table 2B).
Illumina genome sequencing was performed for all idtM, idtD, and idtA CRISPR gene edited strains as well as the AR37 wild type protoplast cell line, with genome sequencing of the remaining gene edited strains still currently underway for idtO, idtF, and idtA/idtF. Bioinformatic analysis of these sequenced genomes against a AR37 PacBio/Illumina consensus sequence concluded that fragments of the CRISPR plasmid (containing the guide, Cas9 gene, and hygromycin resistance gene) had not integrated into the genome for the selected transformants analysed (Table S3). Note that two reads mapped to vector sequence in whole genome reads from idtM strain transformed with guide 38, but both reads were subsequently ruled out as integrated vector sequence. In one case, this was because only one of the pair-end reads mapped to the vector, of which the alignment was to several vectors (not used in this study), whereas the other read aligned to sequence of bird origin, suggesting cross contamination during the sequencing process.
Although zero off-target sites were a criterion for protospacer design in the Geneious software, we additionally performed bioinformatic analysis on the same sequenced genomes to investigate possible off-target effects. To date, no off-target events have been detected using the search parameters employed, therefore we conclude that only the intended CRISPR gene edits are present.
For the first time, CRISPR-Cas9 has been successfully used in the commercial AR37 strain to validate the epoxyjanthitrem pathway. Through the gene inactivation of idtM, idtD, idtO, idtA, idtF and idtA/F, we have confirmed the role of each of these genes in epoxyjanthitrem biosynthesis as well as pathway intermediates at each step (Figure 3). In addition, this research has established that CRISPR-Cas9 can be used to carry out footprint-less gene inactivation to manipulate the secondary metabolite pathways of commercial Epichloë endophyte strains to provide greater chemical diversity, retaining or improving bioactivity against pests whilst eliminating animal toxicity.
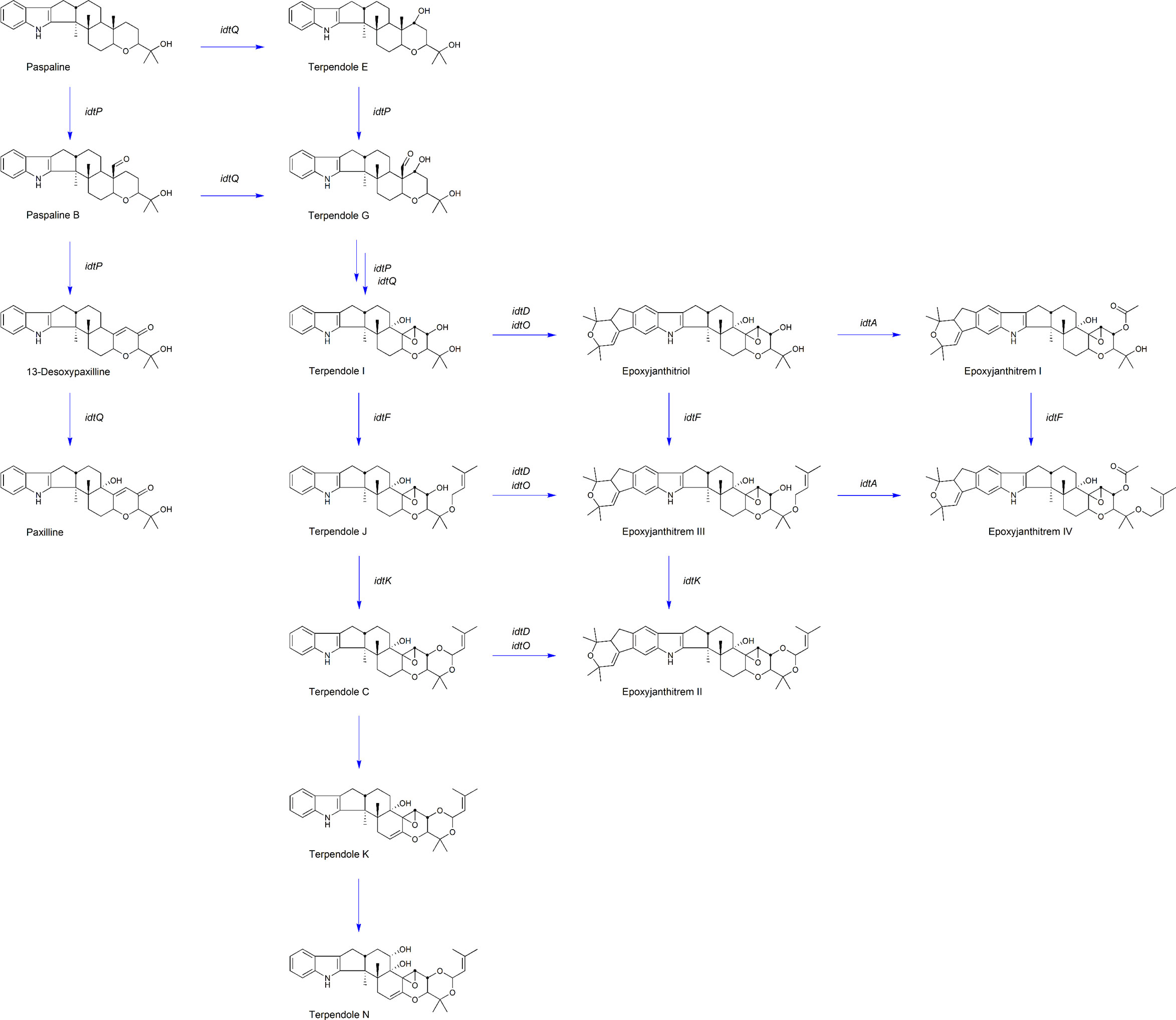
Figure 3 AR37 epoxyjanthitrem biosynthetic pathway. Key compounds proposed in the epoxyjanthitrem biosynthetic pathway in AR37. The compound name is labelled below the structure, with the corresponding gene involved in the compound production labelled next to the associated arrow.
When idtM, one of the key enzymes involved in the first committed step in the epoxyjanthitrem pathway was inactivated, all indole diterpene compounds were eliminated (Table 2A). As idtM is part of the core indole diterpene gene set (Parker and Scott, 2004), this definitively links AR37 epoxyjanthitrem production to this pathway and fits the results from previous structural elucidation studies by Finch et al. (2020). Importantly, idtM gene inactivation eliminated all indole diterpenes, which are the only known bioactive compounds identified in AR37 to date.
Ludlow et al. (2019) identified four clusters predicted to be involved in epoxyjanthitrem biosynthesis, and used RNAi to knock-down idtD, from cluster 3, resulting in a reduction of epoxyjanthitrem compounds. However, this current study conclusively links idtD to epoxyjanthitrem synthesis through an idtD CRISPR gene inactivation that removed all epoxyjanthitrem compounds (Table 2A). In addition, idtD encodes a protein which is predicted to be a prenyltransferase (Ludlow et al., 2019) and structural elucidation has confirmed that all five epoxyjanthitrem compounds contain a double prenylation on ring A (Finch et al., 2020). Overall, this supports the role of IdtD in the synthesis of all epoxyjanthitrem compounds i.e., the initial gene involved in epoxyjanthitrem biosynthesis (Figure 3). Ludlow (2019) only investigated the levels of paxilline, which increased in the idtD knock-down and was also observed in this study (4-fold increase) (Table 2A). However, we additionally show that terpendole compounds are also produced by AR37, where they are barely detectable in the AR37 wild type, likely due to the rapid conversion of these compounds towards epoxyjanthitrems. The terpendoles are elevated to a greater extent (56-fold for terpendole C) when idtD is inactivated (Table 2A). The idtD gene inactivation therefore provides further confirmation and clarification of the terpendoles as intermediates of the epoxyjanthitrem biosynthesis pathway (Figure 3).
Another gene identified in the same cluster as idtD by Ludlow et al. (2019)was idtO, predicted to encode a P450 monooxygenase (Figure 1). This was not an unexpected finding given that prenyltransferases and P450 monooxygenases are often associated together both in terms of the genome location as well as functionality in Epichloëe.g., idtF/idtK and idtE/idtJ (Young et al., 2006; Young et al., 2009). When CRISPR was used to inactivate this gene, all epoxyjanthitrem compounds were removed (Table 2B) showing that IdtO, like IdtD, is also involved in the initial synthesis of all epoxyjanthitrem compounds i.e., IdtD adds the two prenyl groups and IdtO cyclises these to form the ring structure. This is the first evidence presented that confirms idtO involvement in epoxyjanthitrem biosynthesis (Figure 3). However, the indole diterpene profile of the intermediate indole diterpene compounds upstream of the epoxyjanthitrem compounds for the idtO gene edit differs to the idtD gene edit (Table 2A and Table 2B). These results imply that the product of IdtD (predicted to be a diprenylated compound) is exerting a uncompetitive feedback inhibition where the product of one gene inhibits the upstream gene product stopping the terpendoles from accumulating (Guo et al., 2018) (Figure 3). Attempts thus far have failed to detect the predicted compound.
In addition to idtD and idtO (located in cluster 3), idtA (located in cluster 4) (Figure 1) (Ludlow et al., 2019) was also inactivated using CRISPR. This resulted in the reduction of epoxyjanthitrem I, the removal of epoxyjanthitrem IV, the continued production of epoxyjanthitrems II and III, and the elevation of epoxyjanthitriol, in comparison to the AR37 wild type (Table 2A). This is the first evidence that confirms the involvement of cluster 4 in epoxyjanthitrem synthesis as well as the specific involvement of idtA in the direct synthesis of epoxyjanthitrem I and IV (Figure 3). It is hypothesised that the low levels of epoxyjanthitrem I may be due to promiscuous enzymatic activity of Epichloë and/or Lolium derived enzymes. This promiscuity is not routinely observed in other Epichloë indole diterpene pathways e.g., the lolitrem B pathway, due to the much lower levels of indole diterpene compounds present in those pathways in relation to the epoxyjanthitrem pathway i.e., promiscuous activity below levels of detection. Again, the predicted functionality of the protein encoded by idtA as an acetyltransferase (Ludlow et al., 2019) corresponds to the structures of the synthesised epoxyjanthitrem I and IV e.g., the only epoxyjanthitrem compounds with an added acetyl group on carbon 43 (Finch et al., 2020). However, acetyltransferases are common in indole diterpene pathways in other species (Parker and Scott, 2004; Scott et al., 2013).
As seen in the idtD inactivated strains (Table 2A), removal of a gene involved in epoxyjanthitrem biosynthesis can result in changes to levels of upstream intermediate indole diterpene compounds. This would explain the changes in the amount of epoxyjanthitrems II and III as well as the accumulation of epoxyjanthitriol in the idtA inactivated strains even though idtA is not directly involved in synthesis of those compounds (Table 2B). By dramatically reducing the amount of epoxyjanthitrem I in the idtA inactivated strains this should reduce the tremorgenic effect of AR37, however there will be an associated reduction in insect bioactivity (Finch et al., 2020). The tremorgenic and bioactive properties of the remaining indole diterpene intermediate compounds would need to be investigated to get a complete understanding of the effect that changing the epoxyjanthitrem ratio would have on grazing livestock and invertebrate pests.
Finally, idtF CRISPR inactivation eliminated epoxyjanthitrems II, III, and IV, while maintaining production of epoxyjanthitriol and epoxyjanthitrem I (Table 2B). IdtF is therefore involved in the epoxyjanthitrem pathway as well as the lolitrem B pathway, as previously reported, specifically in the direct synthesis of epoxyjanthitrems II, III, and IV in the epoxyjanthitrem pathway, which again corresponds to the function of IdtF as a prenyltransferase (Saikia et al., 2012; Schardl et al., 2013) (Figure 3).
In addition, when idtF is inactivated in an idtA inactivated strain, a hybrid indole diterpene profile is observed. In this case, epoxyjanthitriol was the major compound produced with a minimal amount of epoxyjanthitrem I, as observed in idtA, and the elimination of epoxyjanthitrems II, III, and IV as seen in idtF (Table 2B). This further confirms the roles of idtA and idtF as an acetyl transferase and prenyltransferase, respectively (Figure 3).
Bioinformatic analysis of the epoxyjanthitrem gene cluster loci discovered in AR37 in this study and the epoxyjanthitrem gene cluster loci from Ludlow et al. (2019) re-analysed in this study, showed that the loci are identical within and between the clusters (Figure 1). In addition, both pathways predict an interconnected network of epoxyjanthitrem compounds, rather than a linear model, increasing compound diversity and therefore bioactivity, which is indicative of Epichloë indole diterpene pathways (Young et al., 2009; Ludlow et al., 2019) (Figure 3). In Ludlow et al. (2019), only paxilline and the four epoxyjanthitrems (I-IV) were detected and the proposed pathway was based on this limited data set. In contrast, the current study gathered data on a broader range of indole diterpene metabolites, including the terpendoles (Table 2A and Table 2B). The result of the removal of IdtD activity (namely the higher accumulation of terpendoles with low accumulation of paxilline) implies that the biosynthetic pathway progresses via the terpendoles, as seen in the biosynthetic pathway of lolitrem B, with terpendole I shown to be the first compound on which IdtD would act (Young et al., 2009). The biosynthetic pathway then simultaneously branches according to which enzyme interacts with successive products. Biosynthesis of epoxyjanthitrems via terpendole I also explains the presence of epoxyjanthitriol (Finch et al., 2020) that is absent in Ludlow’s (2019) pathway and which is analogous to lolitriol in the lolitrem B pathway (Schardl et al., 2013). Lastly, structural elucidation has confirmed that all epoxyjanthitrem compounds have α-stereochemistry at C-10, such as through the terpendoles, removing the need to invert the stereochemistry of the β-paxitriol precursor (Ludlow et al., 2019; Finch et al., 2020) (Figure 3).
The CRISPR-Cas9 delivery system developed by Song et al. (2018) has now been successfully used in four different filamentous fungal species; Aspergillus niger (Song et al., 2018; Kun et al., 2021), Venturia (Rocafort et al., 2022), Dothistroma septosporum (McCarthy et al., 2022), and Epichloë sp. LpTG-3 strain AR37 (this study) to inactivate a range of genes. These gene edits have been confirmed either through amplicon sequencing, high-resolution melting (HRM) curve, southern hybridisation, or genome sequencing. Aspergillus is a genetically tractable species and when this CRISPR-Cas9 system was developed and optimised for this specific species, the gene inactivation rate was highly efficient (75%-100%) although was dependent on the guide and gene targeted (Song et al., 2018; Kun et al., 2021). A similar gene inactivation rate was observed in the Dothistroma study (90%-100%), which was again gene and guide dependent (McCarthy et al., 2022). In contrast, Venturia is a genetically intractable species and when this CRISPR-Cas9 system (not developed or optimised for this specific species) was used, the gene inactivation rate dropped to 0%-16.7%, dependent on the guide. However this was the first time that targeted gene editing had been achieved in Venturia (Rocafort et al., 2022). In the current study, the gene inactivation rate of the genetically intractable Epichloë sp. LpTG-3 AR37 strain fell within the Aspergillus and Venturia ranges of 15%-100%, depending on the guide and gene targeted. The difference in gene inactivation rate in the different species could be attributed to the guide used and gene targeted as well as the Cas9 gene being codon optimised to Aspergillus and/or the species natural non-homologous end joining (NHEJ) efficacy rate i.e., high fidelity of NHEJ means lower mutation frequency. The Aspergillus studies, Venturia study, and Dothistroma study all used Geneious to select the guide, and also found that the guide genetic modification efficacy prediction did not correlate to the experimental rate (Song et al., 2018; Kun et al., 2021; Rocafort et al., 2022).
The advantage of using genome sequencing to confirm gene edits in this study is that it allowed for bioinformatic analysis of the whole genome to investigate the footprint-less and off-target nature of this CRISPR-Cas9 delivery system. For the first time, it has been confirmed that CRISPR can be implemented in Epichloë in a footprint-less manner to inactivate genes without off-target effects. This study highlights the importance of genome sequencing to identify gene edited strains for downstream applications. Furthermore, since a nucleic acid template was not used to guide Cas9 and the repair mechanism was through NHEJ, gene edits generated with this delivery system are classified as site-directed nuclease 1 (SDN-1), which are not regulated as genetically modified organisms (GMO) in selected jurisdictions (Lema, 2021; Kawall, 2021).
For the first time, the genetically intractable commercial AR37 strain of Epichloë sp. LpTG-3 successfully underwent targeted genetic engineering using a CRISPR-Cas9 autonomously replicating system. The target genes were either previously associated with the Epichloë lolitrem B pathway (idtM, and idtF) or hypothesised to be associated with the epoxyjanthitrem pathway (idtD, idtO, and idtA) in AR37. The resultant CRISPR gene edits demonstrate that new biochemical variation can be created within the epoxyjanthitrem pathway with the accumulation of intermediate compounds and the removal of different epoxyjanthitrem compounds. The complete removal of the entire epoxyjanthitrem pathway through the precise gene inactivation of the idtM gene will be a useful tool for determining the potential for novel bioactivity of other secondary metabolite pathways that may be expressed by AR37. We have also confirmed the involvement of idtD, idtO, idtA, and idtF in the epoxyjanthitrem biosynthetic pathway based on the genetic dissection of these genes. This CRISPR-Cas9 system can now be applied to other Epichloë endophytes strains that remain genetically intractable to investigate genes of interest to advance Epichloë endophyte research. The manipulation of secondary metabolite pathways by CRISPR-Cas9 gene inactivation has the potential to deliver SDN-1 type endophytes for grasses with enhanced pest protection properties and reduced or no animal health and welfare issues.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ON500678, ON500679.
RJ, LJ, WM, CV, DH, and JS designed the research. DH, JS, WM, NF, and PM performed experiments and analyzed data. TM, LJ, RJ, and WM wrote the manuscript. LJ, RJ, and CV acquired funding. All authors contributed to the article and approved the submitted version.
Funding was provided through the Ministry of Business, Innovation and Employment (MBIE) Partnerships Investment Contract C10X1902 and with commercial partners Grasslanz Technology Ltd and PGG Wrightson Seeds Ltd under contract CV/IP/274.
Concordia University for providing the CRISR-Cas9 plasmid delivery system ANEp8_Cas9_LIC plasmid and ANEp8_Cas9 plasmid (Song et al., 2018). Sarah Finch, and Pranav Chettri for reviewing this manuscript.
The authors declare that this study received funding from Grasslanz Technology Ltd and PGG Wrightson Seeds Ltd. The funders were not involved in the study design, collection, analysis, interpretation of dat or the writing of this article. They were however involved in the decision to submit it for publication.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2022.944234/full#supplementary-material
Berry D., Lee K., Winter D., Mace W., Becker Y., Nagabhyru P., et al. (2022). Cross-species transcriptomics identifies core regulatory changes differentiating the asymptomatic asexual and virulent sexual life cycles of grass-symbiotic epichloë fungi. G3 12, jkac043. doi: 10.1093/g3journal/jkac043
Berry D., Takach J. E., Schardl C. L., Charlton N. D., Scott B., Young C. A. (2015). Disparate independent genetic events disrupt the secondary metabolism gene perA in certain symbiotic epichloë species. Appl. Environ. Microbiol. 81, 2797–2807. doi: 10.1128/AEM.03721-14
Byrd A. D., Schardl C. L., Songlin P. J., Mogen K. L., Siegel M. R. (1990). The β-tubulin gene of epichloë typhina from perennial ryegrass (Lolium perenne). Curr. Genet. 18, 347–354. doi: 10.1007/BF00318216
Caradus J. R., Johnson L. J. (2019). Improved adaptation of temperate grasses through mutualism with fungal endophytes. Endophyte. Biotechnol.: Potential. Agric. Pharmacol. 8, 85.
Charlton N. D., Shoji J.-Y., Ghimire S. R., Nakashima J., Craven K. D. (2012). Deletion of the fungal gene soft disrupts mutualistic symbiosis between the grass endophyte epichloë festucae and the host plant. Eukaryotic. Cell 11, 1463–1471. doi: 10.1128/EC.00191-12
Doench J. G., Hartenian E., Graham D. B., Tothova Z., Hegde M., Smith I., et al. (2014). Rational design of highly active sgRNAs for CRISPR-Cas9–mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267. doi: 10.1038/nbt.3026
Eaton C. J., Jourdain I., Foster S. J., Hyams J. S., Scott B. (2008). Functional analysis of a fungal endophyte stress-activated MAP kinase. Curr. Genet. 53, 163–174. doi: 10.1007/s00294-007-0174-6
Finch S. C., Prinsep M. R., Popay A. J., Wilkins A. L., Webb N. G., Bhattarai S., et al. (2020). Identification and structure elucidation of epoxyjanthitrems from lolium perenne infected with the endophytic fungus epichloë festucae var. lolii and determination of the tremorgenic and anti-insect activity of epoxyjanthitrem I. Toxins 12, 526. doi: 10.3390/toxins12080526
Fleetwood D. J., Scott B., Lane G. A., Tanaka A., Johnson R. D. (2007). A complex ergovaline gene cluster in epichloë endophytes of grasses. Appl. Environ. Microbiol. 73, 2571–2579. doi: 10.1128/AEM.00257-07
Fletcher L., Harvey I. (1981). An association of a lolium endophyte with ryegrass staggers. New Z. Veterinary. J. 29, 185–186. doi: 10.1080/00480169.1981.34839
Florea S., Jaromczyk J., Schardl C. L. (2021). Non-transgenic CRISPR-mmdiated knockout of entire ergot alkaloid gene clusters in slow-growing asexual polyploid fungi. Toxins 13, 153. doi: 10.3390/toxins13020153
Guo L., Wang P., Jaini R., Dudareva N., Chapple C., Morgan J. A. (2018). Dynamic modeling of subcellular phenylpropanoid metabolism in arabidopsis lignifying cells. Metab. Eng. 49, 36–46. doi: 10.1016/j.ymben.2018.07.003
Hassing B., Candy A., Eaton C. J., Fernandes T. R., Mesarich C. H., Di PIETRO A., et al. (2022). Localisation of phosphoinositides in the grass endophyte epichloë festucae and genetic and functional analysis of key components of their biosynthetic pathway in e. festucae symbiosis and fusarium oxysporum pathogenesis. Fungal Genet. Biol., 103669. PLoS ONE 14(2): e0209463. doi: 10.1016/j.fgb.2022.103669
Hassing B., Winter D., Becker Y., Mesarich C. H., Eaton C. J., Scott B. (2019). Analysis of epichloë festucae small secreted proteins in the interaction with lolium perenne. PloS One 14, e0209463. doi: 10.1371/journal.pone.0209463
Hennessy L. M., Popay A. J., Finch S. C., Clearwater M. J., Cave V. M. (2016). Temperature and plant genotype alter alkaloid concentrations in ryegrass infected with an epichloë endophyte and this affects an insect herbivore. Front. Plant Sci. 7, 1097. doi: 10.3389/fpls.2016.01097
Jiang C., Lv G., Tu Y., Cheng X., Duan Y., Zeng B., et al. (2021). Applications of CRISPR/Cas9 in the synthesis of secondary metabolites in filamentous fungi. Front. Microbiol. 12, 164. doi: 10.3389/fmicb.2021.638096
Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J. A., Charpentier E. (2012). A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Johnson L. J., Bonth De, Briggs L. R., Caradus J. R., Finch S. C., Fleetwood D. J., et al. (2013a). The exploitation of epichloae endophytes for agricultural benefit. Fungal Diversity 60, 171–188. doi: 10.1007/s13225-013-0239-4
Johnson l. J., Caradus J. R. (2019). The science required to deliver epichloë endophytes to commerce. Endophytes. Growing. World 343, 343–370. ISBN: 9781108471763.
Johnson L. J., Koulman A., Christensen M., Lane G. A., Fraser K., Forester N., et al. (2013b). An extracellular siderophore is required to maintain the mutualistic interaction of epichloë festucae with lolium perenne. PloS Pathog. 9. doi: 10.1371/journal.ppat.1003332
Johnson R. D., Lane G. A., Koulman A., Cao M., Fraser K., Fleetwood D. J., et al. (2015). A novel family of cyclic oligopeptides derived from ribosomal peptide synthesis of an in planta-induced gene, gigA, in epichloë endophytes of grasses. Fungal Genet. Biol. 85, 14–24. doi: 10.1016/j.fgb.2015.10.005
Johnson L. J., Bastías D. A., Caradus J. R., Chettri P., Forester N. T., Mace W. J., et al. (2021). Chapter 6 - The dynamic mechanisms underpinning symbiotic Epichloë–grass interactions: implications for sustainable and resilient agriculture, Editor(s): White James, Kumar Ajay, Droby Samir, Microbiome Stimulants for Crops, Woodhead Publishing, Elsevier, 2021, Pages 73–108. doi: 10.1016/B978-0-12-822122-8.00008-X
Kawall K. (2021). The generic risks and the potential of SDN-1 applications in crop plants. Plants 10, 2259. doi: 10.3390/plants10112259
Kozák L., Szilágyi Z., Tóth L., Pócsi I., Molnár I. (2019). Tremorgenic and neurotoxic paspaline-derived indole-diterpenes: biosynthetic diversity, threats and applications. Appl. Microbiol. Biotechnol. 103, 1599–1616. doi: 10.1007/s00253-018-09594-x
Kun R. S., Garrigues S., Di Falco M., Tsang A., De Vries R. P. (2021). The chimeric GaaR-XlnR transcription factor induces pectinolytic activities in the presence of d-xylose in aspergillus niger. Appl. Microbiol. Biotechnol. 105, 5553–5564. doi: 10.1007/s00253-021-11428-2
Latch G., Christensen M. (1985). Artificial infection of grasses with endophytes. Ann. Appl. Biol. 107, 17–24. doi: 10.1111/j.1744-7348.1985.tb01543.x
Lema M. (2021). Regulatory assessment of off-target changes and spurious DNA insertions in gene-edited organisms for agri-food use. J. Regul. Sci. 9, 1–15. doi: 10.21423/jrs-v09i1lema
Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li H. (2013). Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. preprint. arXiv. 1303, 3997. doi: 10.6084/M9.FIGSHARE.963153.V1
Ludlow E. J., Vassiliadis S., Ekanayake P. N., Hettiarachchige I. K., Reddy P., Sawbridge T. I., et al. (2019). Analysis of the indole diterpene gene cluster for biosynthesis of the epoxy-janthitrems in epichloë endophytes. Microorganisms 7, 560. doi: 10.3390/microorganisms7110560
Lukito Y., Lee K., Noorifar N., Green K. A., Winter D. J., Ram A., et al. (2020). Host infection by the grass-symbiotic fungus epichloë festucae requires catalytically active H3K9 and H3K36 methyltransferases. bioRxiv 2116–2131. doi: 10.1111/1462-2920.15370
Makarova K. S., Wolf Y. I., Koonin E. V. (2018). Classification and nomenclature of CRISPR-cas systems: where from here? CRISPR. J. 1, 325–336. doi: 10.1089/crispr.2018.0033
Mccarthy H. M., Tarallo M., Mesarich C. H., Mcdougal R. L., Bradshaw R. E. (2022). Targeted gene mutations in the forest pathogen dothistroma septosporum using CRISPR/Cas9. Plants 11, 1016. doi: 10.3390/plants11081016
Nicholson M. J., Eaton C. J., Stärkel C., Tapper B. A., Cox M. P., Scott B. (2015). Molecular cloning and functional analysis of gene clusters for the biosynthesis of indole-diterpenes in penicillium crustosum and p. janthinellum. Toxins 7, 2701–2722. doi: 10.3390/toxins7082701
Noorifar N., Savoian M. S., Ram A., Lukito Y., Hassing B., Weikert T. W., et al. (2021). Chitin deacetylases are required for epichloë festucae endophytic cell wall remodeling during establishment of a mutualistic symbiotic interaction with lolium perenne. Mol. Plant-Microbe Interact. 34, 1181–1192. doi: 10.1094/MPMI-12-20-0347-R
Panaccione D. G., Johnson R. D., Wang J., Young C. A., Damrongkool P., Scott B., et al. (2001). Elimination of ergovaline from a grass–neotyphodium endophyte symbiosis by genetic modification of the endophyte. Proc. Natl. Acad. Sci. 98, 12820–12825. doi: 10.1073/pnas.221198698
Parker E. J., Scott D. B. (2004). Handbook of industrial mycology - indole-diterpene biosynthesis in ascomycetous fungi (New York, NY, USA: Marcel Dekker).
Rahnama M., Johnson R., Voisey C., Simpson W., Fleetwood D. (2018). The global regulatory protein VelA is required for symbiosis between the endophytic fungus epichloë festucae and lolium perenne. Mol. Plant-Microbe Interact. 31, 591–604. doi: 10.1094/MPMI-11-17-0286-R
Rahnama M., Maclean P., Fleetwood D., Johnson R. (2019). The LaeA orthologue in epichloë festucae is required for symbiotic interaction with lolium perenne. Fungal Genet. Biol. 129, 74–85. doi: 10.1016/j.fgb.2019.05.001
Rocafort M., Arshed S., Hudson D., Sidhu J. S., Bowen J. K., Plummer K. M., et al. (2022). CRISPR-Cas9 gene editing and rapid detection of gene-edited mutants using high-resolution melting in the apple scab fungus, venturia inaequalis. Fungal Biol. 126, 35–46. doi: 10.1016/j.funbio.2021.10.001
Saikia S., Nicholson M. J., Young C., Parker E. J., Scott B. (2008). The genetic basis for indole-diterpene chemical diversity in filamentous fungi. Mycological. Res. 112, 184–199. doi: 10.1016/j.mycres.2007.06.015
Saikia S., Takemoto D., Tapper B. A., Lane G. A., Fraser K., Scott B. (2012). Functional analysis of an indole-diterpene gene cluster for lolitrem b biosynthesis in the grass endosymbiont epichloë festucae. FEBS Lett. 586, 2563–2569. doi: 10.1016/j.febslet.2012.06.035
Schardl C. L., Young C. A., Hesse U., Amyotte S. G., Andreeva K., Calie P. J., et al. (2013). Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the clavicipitaceae reveals dynamics of alkaloid loci. PloS Genet. 9, e1003323. doi: 10.1371/journal.pgen.1003323
Scott B., Green K., Berry D. (2018). The fine balance between mutualism and antagonism in the epichloë festucae–grass symbiotic interaction. Curr. Opin. Plant Biol. 44, 32–38. doi: 10.1016/j.pbi.2018.01.010
Scott B., Young C. A., Saikia S., Mcmillan L. K., Monahan B. J., Koulman A., et al. (2013). Deletion and gene expression analyses define the paxilline biosynthetic gene cluster in penicillium paxilli. Toxins 5, 1422–1446. doi: 10.3390/toxins5081422
Simpson W., Schmid J., Singh J., Faville M., Johnson R. (2012). A morphological change in the fungal symbiont neotyphodium lolii induces dwarfing in its host plant lolium perenne. Fungal Biol. 116, 234–240. doi: 10.1016/j.funbio.2011.11.006
Song L., Ouedraogo J.-P., Kolbusz M., Nguyen T. T. M., Tsang A. (2018). Efficient genome editing using tRNA promoter-driven CRISPR/Cas9 gRNA in aspergillus niger. PloS One 13, e0202868. doi: 10.1371/journal.pone.0202868
Tanaka A., Cartwright G. M., Saikia S., Kayano Y., Takemoto D., Kato M., et al. (2013). ProA, a transcriptional regulator of fungal fruiting body development, regulates leaf hyphal network development in the epichloë festucae–lolium perenne symbiosis. Mol. Microbiol. 90, 551–568. doi: 10.1111/mmi.12385
Tanaka A., Takemoto D., Hyon G. S., Park P., Scott B. (2008). NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between epichloe festucae and perennial ryegrass. Mol. Microbiol. 68, 1165–1178. doi: 10.1111/j.1365-2958.2008.06217.x
Tapper B., Lane G. (2004) in Proceedings of the 5th International Symposium on Neotyphodium/Grass interactions (University of Arkansas Press: Fayetteville, AR).
Voisey C. R., Christensen M. T., Johnson L. J., Forester N. T., Gagic M., Bryan G. T., et al. (2016). cAMP signaling regulates synchronised growth of symbiotic epichloë fungi with the host grass lolium perenne. Front. Plant Sci. 7, 1546. doi: 10.3389/fpls.2016.01546
Wang R., Luo S., Clarke B. B., Belanger F. C. (2021). The epichloë festucae antifungal protein efe-AfpA is also a possible effector protein required for the interaction of the fungus with its host grass festuca rubra subsp. rubra. Microorganisms 9, 140. doi: 10.3390/microorganisms9010140
Xie S., Shen B., Zhang C., Huang X., Zhang Y. (2014). sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS One 9, e100448. doi: 10.1371/journal.pone.0100448
Young C., Bryant M., Christensen M., Tapper B., Bryan G., Scott B. (2005). Molecular cloning and genetic analysis of a symbiosis-expressed gene cluster for lolitrem biosynthesis from a mutualistic endophyte of perennial ryegrass. Mol. Genet. Genomics 274, 13–29. doi: 10.1007/s00438-005-1130-0
Young C. A., Felitti S., Shields K., Spangenberg G., Johnson R. D., Bryan G. T., et al. (2006). A complex gene cluster for indole-diterpene biosynthesis in the grass endophyte neotyphodium lolii. Fungal Genet. Biol. 43, 679–693. doi: 10.1016/j.fgb.2006.04.004
Young C. A., Tapper B. A., May K., Moon C. D., Schardl C. L., Scott B. (2009). Indole-diterpene biosynthetic capability of epichloë endophytes as predicted by ltm gene analysis. Appl. Environ. Microbiol. 75, 2200–2211. doi: 10.1128/AEM.00953-08
Keywords: secondary metabolite, gene editing, endophyte, footprint-less, mass spectrometry, bioinformatics, off-targets, indole diterpene
Citation: Miller TA, Hudson DA, Johnson RD, Singh JS, Mace WJ, Forester NT, Maclean PH, Voisey CR and Johnson LJ (2022) Dissection of the epoxyjanthitrem pathway in Epichloë sp. LpTG-3 strain AR37 by CRISPR gene editing. Front. Fungal Biol. 3:944234. doi: 10.3389/ffunb.2022.944234
Received: 14 May 2022; Accepted: 18 July 2022;
Published: 10 August 2022.
Edited by:
Scott E. Baker, Pacific Northwest National Laboratory (DOE), United StatesReviewed by:
Jun-ichi Maruyama, The University of Tokyo, JapanCopyright © 2022 Miller, Hudson, Johnson, Singh, Mace, Forester, Maclean, Voisey and Johnson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Linda J. Johnson, TGluZGEuSm9obnNvbkBhZ3Jlc2VhcmNoLmNvLm56
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.