
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Fungal Biol., 30 May 2022
Sec. Fungi-Animal Interactions
Volume 3 - 2022 | https://doi.org/10.3389/ffunb.2022.911366
This article is part of the Research TopicInsights in Fungi-Animal Interactions 2021View all 5 articles
Fungi rely on major signaling pathways such as the MAPK (Mitogen-Activated Protein Kinase) signaling pathways to regulate their responses to fluctuating environmental conditions, which is vital for fungi to persist in the environment. The cosmopolitan Metarhizium fungi have multiple lifestyles and remarkable stress tolerance. Some species, especially M. robertsii, are emerging models for investigating the mechanisms underlying ecological adaptation in fungi. Here we review recently identified new downstream branches of the MAPK cascades in M. robertsii, which controls asexual production (conidiation), insect infection and selection of carbon and nitrogen nutrients. The Myb transcription factor RNS1 appears to be a central regulator that channels information from the Fus3- and Slt2-MAPK cascade to activate insect infection and conidiation, respectively. Another hub regulator is the transcription factor AFTF1 that transduces signals from the Fus3-MAPK and the membrane protein Mr-OPY2 for optimal formation of the infection structures on the host cuticle. Homologs of these newly identified regulators are found in other Metarhizium species and many non-Metarhizium fungi, indicating that these new downstream signaling branches of the MAPK cascades could be widespread.
Metarhizium is a genus in the Clavicipitaceae family, and it represents a continuum of species and strains that are extraordinarily versatile (St Leger and Wang, 2020). Some Metarhizium species have multiple lifestyles and they are pathogens, but most are also saprophytes, rhizosphere colonizers and beneficial root endophytes, with the ability to switch between these different lifestyles (Moonjely and Bidochka, 2019). Although M. viride and M. granulomatis are pathogenic to reptiles, most Metarhizium species are entomopathogenic, including M. anisopliae, M. robertsii, M. brunneum, M. globosum, M. acridum, M. majus, M. flavoviride, M. frigidum, M. rileyi, M. pingshaense, M. lepidiotae, and M. guizhouense (St Leger and Wang, 2020). The Nobel Prize winner Elie Metchnickoff initiated trials of a Metarhizium fungus for control of the wheat cockchafer Anisoplia austriaca in 1879 (Lord, 2005). Currently, Metarhizium fungi are being used worldwide as environmentally friendly alternatives to chemical insecticides to control agricultural and forest pests, and vectors of diseases (Zhao et al., 2016). Detailed mechanistic knowledge of Metarhizium pathogenicity to insects, conidiation and conidial tolerance to abiotic stresses is needed for mycoinsecticide improvement.
Insect infection by the Metarhizium fungi is initiated when conidia are attached to the insect cuticle and produce germ tubes that differentiate into infection structures called appressoria. The appressoria produce infection pegs, which penetrate the cuticle via a combination of mechanical pressure and cuticle degradation by diverse enzymes. In the insect hemocoel, the fungi undergo dimorphism from hyphae to yeast-like cells (i.e., blastospores), and the insect is killed by a combination of fungal growth and toxins. Finally, the mycelium grown in the hemocoel reemerges from the dead insect, resulting in a muscardine cadaver covered with dark green conidia. The formation of appressorium by Metarhizium is similar to plant pathogenic fungi such as Magnaporthe oryzae and Colletotrichum lagenarium (Kojima et al., 2002; Zhao et al., 2007). The protein kinase A and hydrophobins are functionally conserved during appressorial formation between M. oryzae and Metarhizium (Fang et al., 2009). Metarhizium fungi and mammal pathogenic fungi also have many parallels with regard to pathogenesis such as degrading proteinaceous integuments and evasion of the host innate immune system (Scully and Bidochka, 2006; Ortiz-Urquiza and Keyhani, 2013). Therefore, Metarhizium species can be used as representatives to study broad themes of fungal pathogenicity.
In the first dozen years of this century, robust genetic manipulation technologies including Agrobacterium tumefaciens-mediated fungal transformation and gene deletion based on homologous recombination had been developed for the Metarhizium fungi (Fang et al., 2006; Wang and St Leger, 2006; Xu et al., 2014; Su et al., 2018), and the genomes of Metarhizium species, M. robertsii, and M. acridum, were sequenced (Gao et al., 2011; Hu et al., 2014). This has been significantly speeding up the research on molecular mechanisms underlying insect infection and development by the Metarhizium fungi. Several groups in the field have been interested in regulators that control the insect infection and conidiation by M. robertsii, M. acridum, and M. rileyi. Many major regulators, which had been previously functionally characterized in other fungi including plant and mammal pathogenic fungi and even non-pathogenic fungi such as Saccharomyces cerevisiae and Neurospora crassa, were investigated in Metarhizium. These include a variety of transcription factors (Huang et al., 2015; Wei et al., 2017; Song et al., 2019), epigenetic regulators (Fan et al., 2017; Li et al., 2017; Wang et al., 2017; Lai et al., 2020), and several major signaling pathways such as cAMP-PKA, MAPK and GPCR (Fang et al., 2009; Jin et al., 2012, 2014; Liu et al., 2012; Chen et al., 2016; Song et al., 2016; Wang et al., 2018; Shang et al., 2021; Yu et al., 2021). However, these imitative studies recently revealed several new regulators of insect infection and conidiation by M. robertsii, which have not been functionally characterized in other fungi (Guo et al., 2017; Meng et al., 2021a; Zhang et al., 2021). Most noticeably, several new components constitute new downstream signaling branches of the MAPK signaling pathways in M. robertsii, which will be reviewed in this minireview.
The evolutionarily conserved MAPK cascades function as key signal transducers that channel information via protein phosphorylation/dephosphorylation cycles (Rispail et al., 2009). A MAPK cascade usually contains three kinases ([MAPKKKs (Mitogen-activated protein kinase kinase kinase), MAPKKs (Mitogen-activated protein kinase kinase) and MAPKs]) that are activated sequentially. The activated MAPKs then phosphorylate downstream regulators such as transcription factors, resulting in specific output responses. In fungi, four MAPK cascades have been documented, including the complete Fus3-, Hog1-, and Slt2-MAPK cascades, and the Ime2-MAPK cascade where MAPKKK and MAPKK have not been documented (Xu et al., 2017).
Gene expansion and loss have resulted in significant diversity in the MAPK cascades in the fungal kingdom. A macroevolutionary analysis showed that a complete loss of the MAPK cascades was found in 17 microsporidia, some of which are insect pathogens (Xu et al., 2017). However, in four filamentous insect pathogenic fungi (Metarhizium, Beauveria, Cordyceps and Ophiocordyceps), Fus3-, Hog1-, and Slt2-MAPK cascades are all identified (Zheng et al., 2011; Xu et al., 2017). In another widely investigated insect pathogenic fungus Beauveria bassiana, duplication of the MAPK genes occurred (Tong and Feng, 2019). In contrast, a genome analysis identified complete Fus3-, Hog1-, and Slt2-MAPK cascades in nine Metarhizium species including the early diverging species M. album and M. rileyi, and no gene duplication and loss occurred in these species (Table 1).
All 10 components in the MAPK cascades have been systematically characterized in M. robertsii (Chen et al., 2016), and a few components in the cascades were also investigated in M. acridum and M. rileyi (Jin et al., 2012, 2014; Song et al., 2016; Wang et al., 2018). No obvious phenotypic changes were found to result from the deletion of the gene encoding Ime2-MAPK in M. robertsii (Chen et al., 2016). In M. robertsii, the Fus3-MAPK cascade regulates conidiation, and the three gene deletion mutants in this cascade all showed reduced conidial yields and impaired conidial pigmentation. The Fus3-MAPK cascade is indispensable for cuticle penetration, and the mutants cannot produce appressoria on the insect cuticle. However, this cascade is not involved in hemocoel colonization. The Fus3-MAPK cascade is also indispensable for tolerance to the hyperosmotic stress because the hyphal tips of the mutants swollen and burst under this stress. The Fus3-MAPK cascade is also involved in the tolerance to cell wall disturbing agent Congo Red (Chen et al., 2016). The Fus3-MAPK cascade is generally functionally conserved in the Metarhizium genus, but diversifications have also occurred. As with M. robertsii, the Fus3-MAPK (MaMK1) is also necessary for appressorium formation and following cuticle penetration, and is not involved in hemocoel colonization (Jin et al., 2014). In contrast to M. robertsii, the Fus3-MAPK is not involved in conidiation in M. acridum (Jin et al., 2014). In addition, the Fus3-MAPK is necessary for M. acridum to reemerge from the dead insects, which however is not found in M. robertsii (Jin et al., 2014; Chen et al., 2016).
The Hog1-MAPK is important for tolerance to hyperosmolarity in M. robertsii, M. acridum and M. rileyi. In contrast to M. robertsii, MaHog1 is also important for oxidative stress conferred by H2O2 in M. acridum (Jin et al., 2012). Hog1-MAPK is involved in cuticle penetration and hemocoel colonization in M. robertsii, M. acridum and M. rileyi (Jin et al., 2012; Chen et al., 2016; Song et al., 2016; Wang et al., 2018). The Hog1-MAPK cascade positively regulates conidiation in M. robertsii and M. rileyi, and their mutants all showed reduced conidial yields and altered conidial pigmentation (Chen et al., 2016; Song et al., 2016). In M. robertsii, the mutant of Hog1-MAPK showed significantly lower conidial yield than the mutants of the MAPKK and MAPKKK in the cascade, which could be due to the cross-talk that occurs between the Slt2-MAPK and Hog1-MAPK cascade during conidiation, with the Slt2-MAPK phosphorylating the Hog1-MAPK (Chen et al., 2016). In M. robertsii, the Hog1-MAPK regulates conidiation via control of the central regulatory pathway for conidiation, which mainly contains three sequentially controlled transcription factors (BrlA, AbaA and WetA) and is conserved in the Ascomycete fungi. The Pks1 gene cluster for the conidial pigmentation is also controlled by the Hog1-MAPK via the central regulatory pathway (Zeng et al., 2017).
So far, the Slt2-MAPK cascade has only been characterized in M. robertsii. Slt2-MAPK is involved in cuticle penetration and hemocoel colonization. The Slt2-MAPK lost its ability to produce appressorium, but appressorial formation was only delayed in the mutants of MAPKKK and MAPKK in this cascade. Although the mutant of the Slt2-MAPK cannot produce appressorium, it is still able to infect insects, and this could be due to the upregulation of cuticle-degrading proteases in the mutant, which could facilitate cuticle penetration. As with the Fus3- and Hog1-MAPK cascade, the Slt2-MAPK cascade also positively regulates conidiation; in addition to the reductions in conidial yield and alteration of conidial pigmentation, the colonies of the three mutants in this cascade showed severe sectorization. As describe above, Slt2-MAPK is required for Hog1-MAPK phosphorylation during conidiation. Similar to the Slt2-MAPK cascade in many other fungi, the Slt2-MAPK cascade regulates the tolerance to cell wall disturbing agent Congo Red in M. robertsii. This cascade is also involved in tolerance to hyperosmotic stress (Chen et al., 2016).
Comparison of the gene expression of the wild-type (WT) strain with the MAPK mutants using RNA-Seq analysis revealed many genes regulated by the MAPKs in M. robertsii (Chen et al., 2016). Further characterization of MAPK-regulated genes has identified several new and important components in the MAPK signaling pathways, which provide significant insights into pathogenesis and conidiation in M. robertsii (Guo et al., 2017; Meng et al., 2019, 2021a,b).
During cuticle penetration, the transcription factor AFTF1 with a Zn2Cys6 fungal type DNA binding domain is positively regulated by the Fus3-MAPK cacade. Compared to the WT strain, appressorial formation by the deletion mutant of Aftf1, and to a lesser extent the strain Aftf1OE with Aftf1 overexpressed was delayed, and their pathogenicity was accordingly reduced. Therefore, a precisely modulated optimal level of the AFTF1 protein is needed to choreograph appressorial formation. The expression of Aftf1 is induced by the transcription factor STE12 (named as MrSt12 in M. robertsii); MrSt12 achieves this by physically interacting with the cis-acting element (ATGAAACA) in the promoter of Aftf1. As with the transcription factor STE12 in many other fungi, the activity of MrSt12 is directly phosphorylated by the Fus3-MAPK for activation. In M. robertsii, the deletion mutant of MrSt12 is unbale to form appressorium and is thus nonpathogenic, suggesting that MrSt12 also regulates genes that are not controlled by AFTF1 (Meng et al., 2019). The transcription factor STE12 in M. acridum and M. rileyi have similar functions, and they regulate appressorium formation as the deletion mutants cannot form mature appressoria (Wei et al., 2017; Lin et al., 2021). Therefore, AFTF1 is a new component in the Fus3-MAPK/STE12 signaling branch that regulates cuticle penetration. Homologs of AFTF1 are found in almost all Metarhizium species, and in the plant pathogenic fungi such as Fusarium oxysporum (EWY87630) and M. oryze (XP_003715433), suggesting that the Fus3-MAPK/STE12/AFTF1 signaling branch could be widespread.
Conversely, the membrane protein Mr-OPY2 negatively regulates Aftf1 expression to ensure that the expression level of Aftf1 is not too high for optimum effect (Guo et al., 2017). Upregulation of the Mr-OPY2 protein is required to repress the expression of Aftf1 and initiate appressorium formation for cuticle penetration, whereas reduced production of Mr-OPY2 elicits saprophytic growth and conidiation. The precise regulation of the Mr-OPY2 protein production is achieved via alternative transcription start sites. During saprophytic growth, only a single long transcript is expressed with small upstream open reading frames in its 5′ untranslated region, which inhibits translation of Mr-OPY2. Increased production of the Mr-OPY2 protein on the insect cuticle is achieved by expression of a transcript variant lacking a small upstream open reading frame, which can be highly translated. In addition to regulation of Aftf1, Mr-OPY2 also regulates tolerance to high osmotic stress by controlling the Hog1-MAPK and Fus3-MAPK cascade through the adaptor protein STE50. Mr-OPY2 physically contacts the STE50, which in turn interacts with the MAPKKKs in the Hog1- and Fus3-MAPK cascade. In the yeast Saccharomyces cerevisiae, three binding sites (CR-A, CR-B, and CR-D) in the OPY2 protein were found to interact with STE50; CR-A binds to STE50 constitutively to transmit signals to both the Hog1- and Fus3-MAPK cascade, while CR-B binds to STE50 under glucose-rich conditions to transmit the signal preferentially to the Hog1-MAPK cascade (Yamamoto et al., 2010). However, Mr-OPY2 does not have the CR-A, CR-B and CR-D sites, so the how Mr-OPY2 interacts with STE50 in different conditions remains to be determined. Since the high osmotic stress did not upregulate the Mr-OPY2 protein, a low level of this protein may be sufficient for stress resistance. Mr-OPY2 was the first OPY2 protein characterized in the filamentous fungal pathogens, and a recent study on its homolog in the plant pathogenic fungus Magnaporthe oryzae showed that the OPY2 protein is generally functionally conserved among the insect and plant pathogenic fungi (Cai et al., 2022).
Another novel transcription factor RNS1 (regulation of nutrient selection 1) is also directly regulated by the Fus3-MAPK cascade, but shows no direct relationship with the transcription factor AFTF1 (Meng et al., 2021a). RNS1 is a Myb transcription factor containing a SANT domain (pfam00249). On the insect cuticle, the Fus3-MAPK physically contacts and phosphorylates the threonine at position 215 (Thr-215) and the serine at position 226 (Ser-226) in the RNS1 protein, and this facilitates the entry of RNS1 into nuclei. The phosphorylated RNS1 binds to the target DNA motif (ACCAGAC) in its own promoter to self-induce expression. Abundant phosphorylated RNS1 protein activates the expression of the genes for degrading cuticular proteins, chitin, and lipids, and this provides nutrients for hyphal growth and facilitates the entry of the fungus into the insect hemocoel for subsequent colonization (Meng et al., 2021a).
In addition to regulation of hydrolysis of the proteins, chitin, and lipids in the insect cuticle, using the same mechanisms described above on the insect cuticle, the Fus3-MAPK/RNS1 signaling branch also activates the genes for utilizing common complex and less-favored nitrogen and carbon sources (casein, colloid chitin, and hydrocarbons) that are not derived from insects. This is repressed by favored organic carbon and nitrogen nutrients, including glucose and glutamine. Therefore, the Fus3-MAPK/RNS1 branch controls fungal nitrogen and carbon metabolism and is implicated in the entomopathogenicity of M. robertsii. Homologs of RNS1 are found in other Metarhizium species and many other non-Metarhizium Ascomycete fungi including the saprophytes Neurospora crassa and Aspergillus fungi, and other insect pathogens, plant or mammal pathogens, suggesting that the Fus3-MAPK/RNS1 cascade could be a common mechanism in the phylum of the Ascomycota (Meng et al., 2021a).
Utilization of environmental carbon and nitrogen sources is a key physiological process for fungi to obtain the needed building blocks to sustain life and grow (Luo et al., 2014). Fungi have evolved sophisticated regulatory mechanisms to enable them to respond rapidly to fluctuating availability of carbon and nitrogen nutrients in the environment. In addition to the Fus3-MAPK/RNS1 signaling branch found in M. robertsii, two other regulatory mechanisms have been widely documented in fungi, which ensure that the genes for utilization of less-favored carbon or nitrogen sources are expressed only in the absence of the favored nutrients so that fungi preferentially utilize favored carbon (e.g., glucose) and nitrogen (e.g., ammonium and glutamine) sources. One mechanism is the nitrogen metabolite repression (NMR). The regulatory protein NmrA deactivates the GATA transcription factor AREA in the presence of favored nitrogen, whereas in the absence of favored nitrogen sources, NAD+ binds to NmrA, which in turn dissociates from AREA, thereby allowing AREA to activate expression of the genes for utilizing less-favored nitrogen sources (Wong et al., 2008). The other mechanism is called the carbon catabolite repression (CCR). Dependent on the repressor CreA, CCR ensures glucose is preferentially utilized by preventing the expression of the genes for utilizing less-favored carbon sources (Beattie et al., 2017). The CCR and NMR are functionally conserved in fungi, including several Metarhizium species (Song et al., 2019; Lai et al., 2020; Meng et al., 2021a). However, the Fus3-MAPK/RNS1 signaling branch acts independently from CCR and NMR, and other regulators of fungal carbon and nitrogen metabolism, including Tps1, Snf1, and TOR kinase (Meng et al., 2021a).
As described above, the Metarhizium fungi have the central conidiation regulatory pathway containing the transcription factors BrlA, AbaA and WetA. As with other ascomycete fungi, in M. robertsii the BrlA gene has two overlapping transcripts BrlAα and BrlAβ; they have the same major ORF, but the 5′ UTR of BrlAβ is 835 bp longer than the one of BrlAα. Multiple upstream ORFs (uORFs) are in the 5′ UTR of BrlAβ, which could inhibit the translation of the downstream major ORF (Guo et al., 2017; Meng et al., 2021b). During conidiation, the Slt2-MAPK phosphorylates the serine at the position 306 in RNS1. The phosphorylated RNS1 self-induces by binding the BM2 motif (CCCAGAC) in its own promoter, which in turn binds to the BM2 motif in the promoter of the BrlA gene and induces the expression of the transcript BlrAα. Since no uORFs are in the 5′ UTR of BrlAα, the major ORF can be highly translated. The BrlA protein in turn activates the expression of AbaA and WetA for optimal conidiation (Meng et al., 2021b).
The aerial hyphae of the deletion mutant of BrlA cannot differentiate to produce conidiophores (Zeng et al., 2017), whereas the deletion mutant of Rns1 can produce conidia though its conidial yield is reduced, suggesting that RNS1 only regulates the expression of the transcript BrlAα and the transcript BrlAβ could also function during conidiation. The contributions of BrlAα and BrlAβ to conidiation remain to be clarified. In addition to conidiation, deletion of the gene encoding Slt2-MAPK also reduced the tolerance to hyperosmotic stress and to the cell wall-disturbing agent Congo red; but deletion of Rns1 had no impact on the tolerance of M. robertsii to hyperosmotic stress and cell wall-disturbing agent. Therefore, the Slt2-MAPK cascade has other downstream branches than the Slt2-MAPK/RNS1.
The Metarhizium fungi have multiple lifestyles and remarkable stress tolerance (Słaba et al., 2013; Moonjely and Bidochka, 2019), resulting in their cosmopolitan distribution. The species M. robertsii is an emerging model to investigate the mechanisms underlying ecological adaptation in fungi. Major signaling pathways such as the conserved MAPK cascades play key roles in fungal sense and response to fluctuating environmental conditions, which is vital for fungi to persist in environment. In recent years, several novel downstream signaling branches of the MAPK cascades have been discovered in M. robertsii, which regulate conidiation and nutrient selection and entomopathogenicity (Figure 1). The Myb transcription factor RNS1 appears to be a central regulator that controls fungal responses to multiple environmental cues. In response to less-favored carbon and nitrogen sources, RNS1 phosphorylated by the Fus3-MAPK activates genes for the utilization of these complex nutrients, which is implicated in entomopathogenicity. The Slt2-MAPK phosphorylates RNS1 during conidiation to activate the central regulatory pathway to initiate asexual reproduction. The transcription factor AFTF1 also appears to act as a hub regulator that channels information from the Fus3-MAPK and the membrane protein Mr-OPY2 for optimal formation of the infection structure appressorium on the host cuticle. As with the components in the MAPK cascades, homologs of RNS1 and AFTF1 are found in other Metarhizium species and many non-Metarhizium fungi, indicating that these new downstream signaling branches of the MAPK cascades could be widespread.
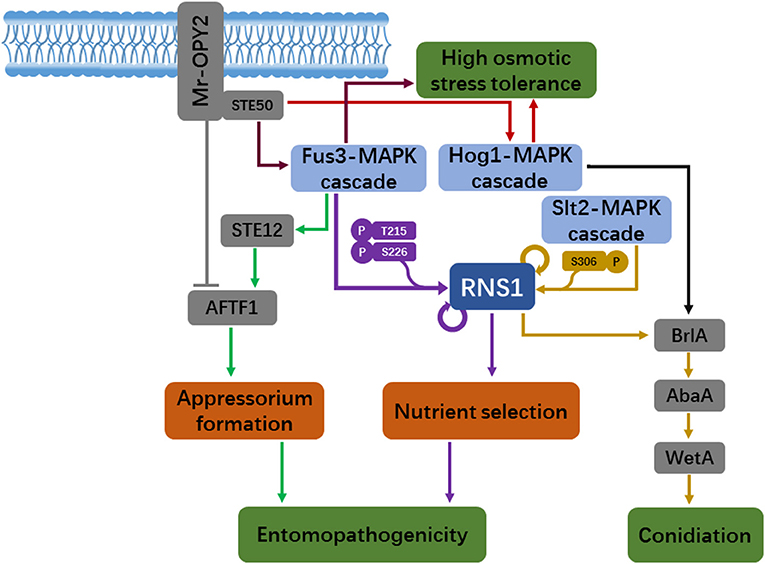
Figure 1. A schematic model of new downstream signaling branches of the MAPK cascades, which regulate asexual reproduction, nutrient selection and insect infection in the fungus Metarhizium robertsii. Mr-OPY2-mediated regulation of the MAPK signaling pathways and the transcript factor AFTF1 for appressorial formation and osmotic stress tolerance is also shown. Arrows: induction; Dashes: repression.
WF conceived the idea and wrote the manuscript. DT and XT wrote the manuscript. All authors contributed to the article and approved the submitted version.
This work was supported by the National Natural Science Foundation of China Grants 31872021 and 32172470.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Beattie, S. R., Mark, K. M. K., Thammahong, A., Ries, L. N. A., Dhingra, S., Caffrey-Carr, A. K., et al. (2017). Filamentous fungal carbon catabolite repression supports metabolic plasticity and stress responses essential for disease progression. PLoS Pathog. 13, e1006340. doi: 10.1371/journal.ppat.1006340
Cai, Y. Y., Wang, J. Y., Wu, X. Y., Liang, S., Zhu, X. M., Li, L., et al. (2022). MoOpy2 is essential for fungal development, pathogenicity, and autophagy in Magnaporthe oryzae. Environ. Microbiol. 24, 1653–1671. doi: 10.1111/1462-2920.15949
Chen, X., Xu, C., Qian, Y., Liu, R., Zhang, Q., Zeng, G., et al. (2016). MAPK cascade-mediated regulation of pathogenicity, conidiation and tolerance to abiotic stresses in the entomopathogenic fungus Metarhizium robertsii. Environ. Microbiol. 18, 1048–1062. doi: 10.1111/1462-2920.13198
Fan, A., Mi, W., Liu, Z., Zeng, G., Zhang, P., Hu, Y., et al. (2017). Deletion of a histone acetyltransferase leads to the pleiotropic activation of natural products in Metarhizium robertsii. Org. Lett. 19, 1686–1689. doi: 10.1021/acs.orglett.7b00476
Fang, W., Pava-Ripoll, M., Wang, S., and St Leger, R. J. (2009). Protein kinase A regulates production of virulence determinants by the entomopathogenic fungus, Metarhizium anisopliae. Fungal Genet. Biol. 46, 277–285. doi: 10.1016/j.fgb.2008.12.001
Fang, W., Pei, Y., and Bidochka, M. J. (2006). Transformation of Metarhizium anisopliae mediated by Agrobacterium tumefaciens. Can. J. Microbiol. 52, 623–626. doi: 10.1139/w06-014
Gao, Q., Jin, K., Ying, S. H., Zhang, Y., Xiao, G., Shang, Y., et al. (2011). Genome sequencing and comparative transcriptomics of the model entomopathogenic fungi Metarhizium anisopliae and M. acridum. PLoS Genet. 7, e1001264. doi: 10.1371/journal.pgen.1001264
Guo, N., Qian, Y., Zhang, Q., Chen, X., Zeng, G., Zhang, X., et al. (2017). Alternative transcription start site selection in Mr-OPY2 controls lifestyle transitions in the fungus Metarhizium robertsii. Nat. Commun. 8, 1565. doi: 10.1038/s41467-017-01756-1
Hu, X., Xiao, G., Zheng, P., Shang, Y., Su, Y., Zhang, X., et al. (2014). Trajectory and genomic determinants of fungal-pathogen speciation and host adaptation. Proc. Natl. Acad. Sci. U.S.A. 111, 16796–16801. doi: 10.1073/pnas.1412662111
Huang, W., Shang, Y., Chen, P., Gao, Q., and Wang, C. (2015). MrpacC regulates sporulation, insect cuticle penetration and immune evasion in Metarhizium robertsii. Environ. Microbiol. 17, 994–1008. doi: 10.1111/1462-2920.12451
Jin, K., Han, L., and Xia, Y. (2014). MaMk1, a FUS3/KSS1-type mitogen-activated protein kinase gene, is required for appressorium formation, and insect cuticle penetration of the entomopathogenic fungus Metarhizium acridum. J. Invertebr. Pathol. 115, 68–75. doi: 10.1016/j.jip.2013.10.014
Jin, K., Ming, Y., and Xia, Y. X. (2012). MaHog1, a Hog1-type mitogen-activated protein kinase gene, contributes to stress tolerance and virulence of the entomopathogenic fungus Metarhizium acridum. Microbiology 158, 2987–2996. doi: 10.1099/mic.0.059469-0
Kojima, K., Kikuchi, T., Takano, Y., Oshiro, E., and Okuno, T. (2002). The mitogen-activated protein kinase gene MAF1 is essential for the early differentiation phase of appressorium formation in Colletotrichum lagenarium. Mol. Plant-Microbe. 15, 1268–1276. doi: 10.1094/MPMI.2002.15.12.1268
Lai, Y., Cao, X., Chen, J., Wang, L., Wei, G., and Wang, S. (2020). Coordinated regulation of infection-related morphogenesis by the KMT2-Cre1-Hyd4 regulatory pathway to facilitate fungal infection. Sci. Adv. 6, eaaz1659. doi: 10.1126/sciadv.aaz1659
Li, W., Wang, Y., Zhu, J., Wang, Z., Tang, G., and Huang, B. (2017). Differential DNA methylation may contribute to temporal and spatial regulation of gene expression and the development of mycelia and conidia in entomopathogenic fungus Metarhizium robertsii. Fungal Biol. 121, 293–303. doi: 10.1016/j.funbio.2017.01.002
Lin, Y., Wang, J., Yang, K., Fan, L., Wang, Z., and Yin, Y. (2021). Regulation of conidiation, polarity growth, and pathogenicity by MrSte12 transcription factor in entomopathogenic fungus, Metarhizium rileyi. Fungal Genet. Biol. 155, 103612. doi: 10.1016/j.fgb.2021.103612
Liu, S., Peng, G., and Xia, Y. (2012). The adenylate cyclase gene MaAC is required for virulence and multi-stress tolerance of Metarhizium acridum. BMC Microbiol. 12, 163. doi: 10.1186/1471-2180-12-163
Lord, J. C. (2005). From Metchnikoff to Monsanto and beyond: the path of microbial control. J. Invert. Pathol. 89, 19–29. doi: 10.1016/j.jip.2005.04.006
Luo, Z., Qin, Y., Pei, Y., and Keyhani, N. O. (2014). Ablation of the creA regulator results in amino acid toxicity, temperature sensitivity, pleiotropic effects on cellular development and loss of virulence in the filamentous fungus Beauveria bassiana. Environ. Microbiol. 16, 1122–1136. doi: 10.1111/1462-2920.12352
Meng, Y., Tang, X., Bao, Y., Zhang, M., Tang, D., Zhang, X., et al. (2021b). Slt2-MAPK/RNS1 controls conidiation via direct regulation of the central regulatory pathway in the fungus Metarhizium robertsii. J. Fungi. 8, 26. doi: 10.3390/jof8010026
Meng, Y., Zhang, X., Guo, N., and Fang, W. (2019). MrSt12 implicated in the regulation of transcription factor AFTF1 by Fus3-MAPK during cuticle penetration by the entomopathogenic fungus Metarhizium robertsii. Fungal Genet. Biol. 131, 103244. doi: 10.1016/j.fgb.2019.103244
Meng, Y., Zhang, X., Tang, D., Chen, X., Zhang, D., Chen, J., et al. (2021a). Novel nitrogen and carbon metabolism regulatory cascade is implicated in entomopathogenicity of the fungus Metarhizium robertsii. mSystems 6, E0049921. doi: 10.1128/mSystems.00499-21
Moonjely, S., and Bidochka, M. J. (2019). Generalist and specialist Metarhizium insect pathogens retain ancestral ability to colonize plant roots. Fungal Ecol. 41, 209–217. doi: 10.1016/j.funeco.2019.06.004
Ortiz-Urquiza, A., and Keyhani, N. O. (2013). Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects 4, 357–374. doi: 10.3390/insects4030357
Rispail, N., Soanes, D. M., Ant, C., Czajkowski, R., Grünler, A., Huguet, R., et al. (2009). Comparative genomics of MAP kinase and calcium-calcineurin signalling components in plant and human pathogenic fungi. Fungal Genet. Biol. 46, 287–298. doi: 10.1016/j.fgb.2009.01.002
Scully, L. R., and Bidochka, M. J. (2006). Developing insect models for the study of current and emerging human pathogens. FEMS Microbiol. Lett. 263, 1–9. doi: 10.1111/j.1574-6968.2006.00388.x
Shang, J., Shang, Y., Tang, G., and Wang, C. (2021). Identification of a key G-protein coupled receptor in mediating appressorium formation and fungal virulence against insects. Sci. China Life Sci. 64, 466–477. doi: 10.1007/s11427-020-1763-1
Słaba, M., Bernat, P., Rózalska, S., Nykiel, J., and Długoński, J. (2013). Comparative study of metal induced phospholipid modifications in the heavy metal tolerant filamentous fungus Paecilomyces marquandii and implications for the fungal membrane integrity. Acta Biochim. Pol. 60, 695–700 doi: 10.18388/abp.2013_2043
Song, D., Shi, Y., Ji, H. Q., Xia, Y., and Peng, G. (2019). The MaCreA gene regulates normal conidiation and microcycle conidiation in Metarhizium acridum. Front. Microbiol. 10, 1946. doi: 10.3389/fmicb.2019.01946
Song, Z., Zhong, Q., Yin, Y., Shen, L., Li, Y., and Wang, Z. (2016). The high osmotic response and cell wall integrity pathways cooperate to regulate morphology, microsclerotia development, and virulence in Metarhizium rileyi. Sci. Rep. 6, 38765. doi: 10.1038/srep38765
St Leger, R. J., and Wang, J. B. (2020). Metarhizium: jack of all trades, master of many. Open Biol. 10, 200307. doi: 10.1098/rsob.200307
Su, Y., Wang, Z., Shao, C., Luo, Y., Wang, L., and Yin, Y. (2018). An efficient gene disruption method using a positive-negative split-selection marker and Agrobacterium tumefaciens-mediated transformation for Nomuraea rileyi. World J. Microbiol. Biotechnol. 34, 26. doi: 10.1007/s11274-018-2409-8
Tong, S. M., and Feng, M. G. (2019). Insights into regulatory roles of MAPK-cascaded pathways in multiple stress responses and life cycles of insect and nematode mycopathogens. Appl. Microbiol. Biotechnol. 103, 577–587. doi: 10.1007/s00253-018-9516-1
Wang, C., and St Leger, R. J. (2006). A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc. Natl. Acad. Sci. U.S.A. 103, 6647–6652. doi: 10.1073/pnas.0601951103
Wang, Y., Wang, T., Qiao, L., Zhu, J., Fan, J., Zhang, T., et al. (2017). DNA methyltransferases contribute to the fungal development, stress tolerance and virulence of the entomopathogenic fungus Metarhizium robertsii. Appl. Microbiol. Biotechnol. 101, 4215–4226. doi: 10.1007/s00253-017-8197-5
Wang, Z., Song, Z., Zhong, Q., Du, F., and Yin, Y. (2018). Adaption to stress via Pbs2 during Metarhizium rileyi conidia and microsclerotia development. World J. Microbiol. Biotechnol. 34, 107. doi: 10.1007/s11274-018-2475-y
Wei, Q., Du, Y., Jin, K., and Xia, Y. (2017). The Ste12-like transcription factor MaSte12 is involved in pathogenicity by regulating the appressorium formation in the entomopathogenic fungus, Metarhizium acridum. Appl. Microbiol. Biotechnol. 101, 8571–8584. doi: 10.1007/s00253-017-8569-x
Wong, K. H., Hynes, M. J., and Davis, M. A. (2008). Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryotic Cell 7, 917–925. doi: 10.1128/EC.00076-08
Xu, C., Liu, R., Zhang, Q., Chen, X., Qian, Y., and Fang, W. (2017). The diversification of evolutionarily conserved MAPK cascades correlates with the evolution of fungal species and development of lifestyles. Genome Biol. Evol. 9, 311–322. doi: 10.1093/gbe/evw051
Xu, C., Zhang, X., Qian, Y., Chen, X., Liu, R., Zeng, G., et al. (2014). A high-throughput gene disruption methodology for the entomopathogenic fungus Metarhizium robertsii. PLoS ONE 9, e107657. doi: 10.1371/journal.pone.0107657
Yamamoto, K., Tatebayashi, K., Tanaka, K., and Saito, H. (2010). Dynamic control of yeast MAP kinase network by induced association and dissociation between the Ste50 scaffold and the Opy2 membrane anchor. Mol. Cell. 40, 87–98. doi: 10.1016/j.molcel.2010.09.011
Yu, D., Xie, R., Wang, Y., Xie, T., Xu, L., and Huang, B. (2021). The G-protein coupled receptor GPRK contributes to fungal development and full virulence in Metarhizium robertsii. J. Invertebr. Pathol. 183, 107627. doi: 10.1016/j.jip.2021.107627
Zeng, G., Chen, X., Zhang, X., Zhang, Q., Xu, C., Mi, W., et al. (2017). Genome-wide identification of pathogenicity, conidiation and colony sectorization genes in Metarhizium robertsii. Environ. Microbiol. 19, 3896–3908. doi: 10.1111/1462-2920.13777
Zhang, X., Meng, Y., Huang, Y., Zhang, D., and Fang, W. (2021). A novel cascade allows Metarhizium robertsii to distinguish cuticle and hemocoel microenvironments during infection of insects. PLoS Biol. 19, e3001360. doi: 10.1371/journal.pbio.3001360
Zhao, H., Lovett, B. R., and Fang, W. (2016). Genetically engineering entomopathogenic fungi. Adv. Genet. 94, 137–163. doi: 10.1016/bs.adgen.2015.11.001
Zhao, X., Mehrabi, R., and Xu, J. R. (2007). Mitogen-activated protein kinase pathways and fungal pathogenesis. Eukaryotic Cell 6, 1701–1714. doi: 10.1128/EC.00216-07
Keywords: Metarhizium, MAPK cascade, entomopathogenicity, conidiation, signaling pathway
Citation: Tang D, Tang X and Fang W (2022) New Downstream Signaling Branches of the Mitogen-Activated Protein Kinase Cascades Identified in the Insect Pathogenic and Plant Symbiotic Fungus Metarhizium robertsii. Front. Fungal Biol. 3:911366. doi: 10.3389/ffunb.2022.911366
Received: 02 April 2022; Accepted: 19 April 2022;
Published: 30 May 2022.
Edited by:
Chengshu Wang, Center for Excellence in Molecular Plant Sciences (CAS), ChinaReviewed by:
Brian Lovett, West Virginia University, United StatesCopyright © 2022 Tang, Tang and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weiguo Fang, d2ZhbmcxQHpqdS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.