- 1Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Japan
- 2Bioproduction Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Sapporo, Japan
- 3Computational Bio Big-Data Open Innovation Laboratory (CBBD-OIL), National Institute of Advanced Industrial Science and Technology (AIST), Tokyo, Japan
Peptidyl compounds produced by filamentous fungi, which are nonribosomal peptides (NRPs) and ribosomally synthesized and post-translationally modified peptides (RiPPs), are rich sources of bioactive compounds with a wide variety of structures. Some of these peptidyl compounds are useful as pharmaceuticals and pesticides. However, for industrial use, their low production often becomes an obstacle, and various approaches have been challenged to overcome this weakness. In this article, we summarize the successful attempts to increase the production of NRPs and RiPPs in filamentous fungi and present our perspectives on how to improve it further.
1 Introduction
Fungal natural products or secondary metabolites are valuable sources for pharmaceutical compounds, such as antibiotics and anticancer drugs, because they have a wide range of structures with unexpected unique bonds and modifications, which are often difficult to chemically synthesize. Penicillin, an antibiotic, and cyclosporin, an immunosuppressant, are examples of clinically used bioactive compounds produced by filamentous fungi. Macrocyclic peptides, such as penicillin, are a major group of fungal natural products, and they are further classified by their biosynthetic pathways into nonribosomal peptides (NRPs), and ribosomally synthesized and post-translationally modified peptides (RiPPs). The former is not synthesized via ribosomes, but several amino acid residues are concatenated and cyclized by a large enzyme of thousands or more kilodaltons called nonribosomal peptide synthetase (NRPS), which has domains of adenylation, peptidyl carrier protein, condensation, and thioesterase. On the other hand, the backbone peptides for RiPPs are encoded in their corresponding precursor genes, which are translated by ribosomes in the same manner as ordinary proteins and further modified through such as methylation, prenylation, and cyclization by oxydation.
NRPs and RiPPs are expected to be designed and modified by altering the genes responsible for their synthesis; this aspect makes them more attractive targets for drug discovery. However, there are still many difficulties in making fungal NRPs and RiPPs available in the industry. One of the main issues is their low production, especially in liquid media. It is not unusual that a fungal cyclic peptide cannot be produced in more than 0.1 mg/L yield, even though the corresponding biosynthetic genes are overexpressed. Here, we summarize the current advances in the production of fungal peptidyl compounds and discuss perspectives to overcome their low production based on recent achievements.
2 Production of nonribosomal peptides in filamentous fungi
Currently, 180 NRPS pathways in Ascomycota are registered in MIBiG (Kautsar et al., 2020), the database of natural compound biosynthetic gene clusters. Within them, there are several valuable NRPs that are used as pharmaceutical agents, such as penicillin and cyclosporin, or that have the potential to be used as such (Keller, 2019) (Figure 1A). The production of these NRPs has been improved for industrial use, and random mutagenesis is one of the most frequently applied techniques. For example, penicillin production increased by more than three orders of magnitude from that of the original producer, Penicillium spp. wild-type strain, by repetition of random mutagenesis (Fierro et al., 2022). Throughout the repeated random mutagenesis process, both the copy number of penicillin-biosynthetic genes increased, and metabolic reprogramming for more efficient secondary metabolite production occurred (McLean et al., 2015). Specifically, major industrial Penicillium strains for penicillin production originated from a naturally-isolated wild-type Penicillium chrysogenum strain NRRL1951 (later renamed Penicillium rubens) (Houbraken et al., 2011). NRRL1951 was then subjected to repeated mutagenesis, and after 15 generations of mutation, the Wisconsin54-1255 mutant strain with improved penicillin production was developed (Van den Berg, 2011). Wisconsin54-1255 has been used in laboratories for research, and by pharmaceutical companies for development of industrial strains by further random mutagenesis. Thus, most industrial strains are derivatives of Wisconsin54-1255. In addition to mutant development, the liquid fermentation conditions have been optimized. Fed-batch culture was applied aseptically in stainless steel tank reactors of 30,000-100,000-gallon capacity. As a result, 40-50 g/L penicillin yields have been attained to date (Elander, 2003).
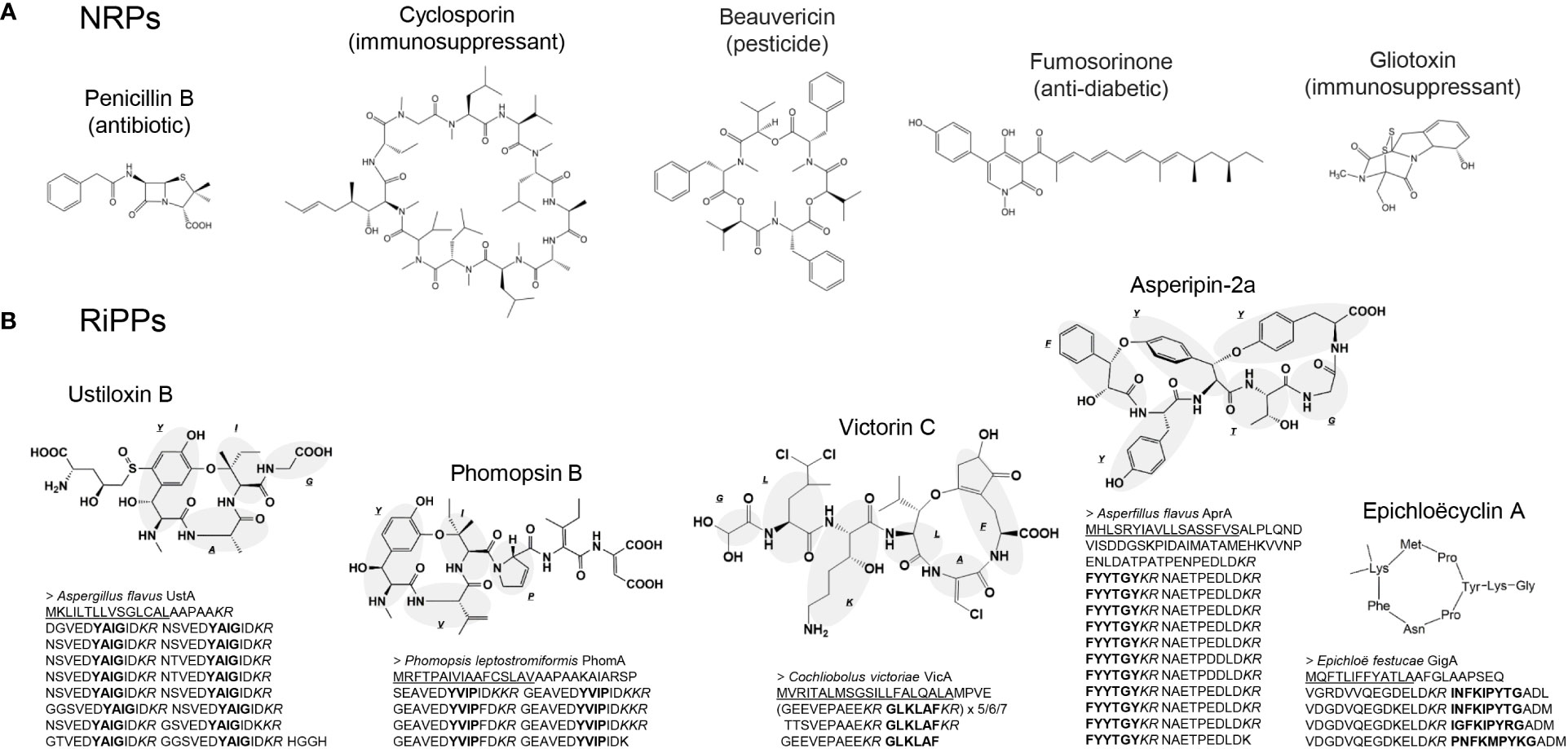
Figure 1 Examples of (A) nonribosomal peptides (NRPs) and (B) ribosomally synthesized and post-translationally modified peptides (RiPPs) produced by filamentous fungi. In (A), the major known bioactivities are described for each compound. In (B), the precursor protein sequences are shown below the corresponding compounds, where the endoplasmic reticulum signal peptide, core peptides, and Kex2 cleavage sites are underlined, bolded, and italicized, respectively.
However, there are three severe drawbacks to applying random mutagenesis to filamentous fungi: large colony size, slow growth, and the requirement of homokaryon isolation (Tamano and Yoshimi, 2021). Therefore, an approach involving knockout of the targeted gene(s) by recombination and subsequent functional studies, called reverse genetics, is more suitable for improving the production of fungal natural products than random mutagenesis. In this section, we describe what has been accomplished to increase NRP production yield by reverse genetics.
2.1 Overexpression of BGC transcription factor and amino-acid-biosynthesis positive regulator genes
A primitive NRP cyclic peptide synthesized by NRPS is usually further modified by other tailoring enzymes as the final NRP. The genes for these enzymes generally accumulate in the chromosome of the producer strain as a biosynthetic gene cluster (BGC) in the same manner as other fungal secondary metabolic genes. A transcription factor gene typically exists within the BGC where it positively regulates the expression of other genes (Keller, 2019). Hence, overexpression of the transcription factor gene by recombination increases the expression of other genes in the BGC, resulting in increase of the production of the corresponding compound. Many NRPs have been reported to be produced at higher yields by overexpressing transcription factor genes (Wang et al., 2021). In the case of the NRP FR901469 produced by an unidentified fungal sp. No. 11243 (Matsui et al., 2015), the production yield was enhanced by repetitive recombination. During the first recombination, the transcription factor gene frbF in the BGC was overexpressed, resulting in a 3.4-fold increase in FR901469 production (Matsui et al., 2017a). At the second recombination, another gene, cpcA, located outside the cluster, was overexpressed in addition to the frbF overexpression. Homologs of CpcA have been reported to positively regulate amino acid biosynthesis in Neurospora crassa (Paluh et al., 1988) and Saccharomyces cerevisiae (Tian et al., 2007). As a result, FR901469 production further increased by 1.8-fold (Matsui et al., 2017b). Prior to cpcA overexpression, Matsui et al. (2017b) found that the addition of some types of amino acids to the culture medium increased FR901469 production. Thus, increased production by cpcA overexpression is consistent with previous research.
2.2 Overexpression of global regulator genes
Other strategies can be applied in cases of BGCs without transcription factor genes. A global regulatory unit that epigenetically governs gene expression throughout the genome is known to exist in filamentous fungi. It is called the velvet complex, and the LaeA protein in this complex has histone methyltransferase activity. When histones binding to the chromosome are subjected to methylation by LaeA, DNA-histone complexes are dissociated, leading to an increase in the accessibility of RNA polymerases to DNA. As a result, the transcription of the genes located there is facilitated. Some successful cases are reported that genes in the BGC of an NRP were expressed by laeA overexpression, increasing the corresponding NRP production up to 25%, 20%, and 126%, respectively, higher than the parental strains (Kosalková et al., 2009; Martín, 2017; Yin et al., 2021).
2.3 Increasing copy number of BGC
In addition to modifying gene regulation, increasing gene copy number is also effective. Increasing the copy number of penicillin BGC by recombination caused an increase in penicillin production (Nijland et al., 2010). Interestingly, the industrial strain of Penicillium rubens used for mass production, which was bred via random mutagenesis, has been proven to have multiple penicillin BGCs (Fierro et al., 1995; Van den Berg, 2011). The same strategy of BGC copy number increase could be applied to heterologous production, which is discussed below.
2.4 Optimizing cell physiology and metabolism
Further, optimizing the cell physiology and metabolic pathways involved in NRP biosynthesis is a good approach for improving the production of secondary metabolites. Let us explain using penicillin as an example. Penicillin is synthesized by three enzymes: δ-(l-α-aminoadipyl)-L-cysteinyl-D-valine synthetase (ACVS), isopenicillin N synthase (IPNS), and isopenicillin N acyltransferase (IAT). ACVS is an NRPS and concatenates three amino acids, L-aminoadipic acid, L-valine, and L-cysteine, and then cyclizes the peptide chain. IPNS then forms the β-lactam structure in the intermediate, and IAT confers a phenyl residue to generate the final product. ACVS and IPNS work in the cytosol, whereas IAT works in the peroxisome.
The first successful case to increase penicillin production was the removal of autophagy function from the producer cells. Since autophagosomes possibly degrade enzymes involved in penicillin biosynthesis, the gene atg1 involved in autophagy was deleted in P. chrysogenum (Bartoszewska et al., 2011). Penicillin production increased by 1.4-fold (37%). The second case was the relocation of ACVS to the peroxisome, where the final reaction by IAT was performed. AcvA (a homolog of ACVS) was tagged with peroxisomal targeting signal 1 (PTS1, tripeptide composed of serine, lysine, and leucine) at the C-terminus, increasing penicillin production 3.2-fold (Herr and Fischer, 2014). The third case involved increasing the number of peroxisomes. Overexpression of Pex11-family genes has been reported to increase the number of peroxisomes in mammalian and fungal cells (Marshall et al., 1996; Schrader et al., 1998). With pex11 overexpression in Penicillium sp., the peroxisome number increased, and penicillin production also increased 2-fold (Kiel et al., 2005). The fourth case involved overexpression of the transporter gene. From the transcriptome analysis under penicillin production-facilitating culture conditions, it was found that the penT gene encoding a major facilitator superfamily transporter was upregulated, and based on the homology search, it was deduced to be involved in the transport of the penicillin precursor from the cytosol to the peroxisome. Thus, penT was overexpressed, and penicillin production increased 2-fold (Yang et al., 2012).
2.5 Heterologous production
If it is technically difficult to modify genes in the original producer strains, strains grow very slowly, or protoplasts cannot be sufficiently prepared for transformation, a promising strategy to increase NRP production involves the heterologous expression of the corresponding BGCs in other host strains, where convenient genetic tools are applicable (Alberti et al., 2017; Vassaux et al., 2019). When heterologously expressing a target BGC, it is better to replace the original promoter sequences in the BGC with those of constitutively highly expressed genes in the host strain. This is because the original promoters are related to secondary metabolism, which is often regulated at low levels of expression. Indeed, when the PKS-NRPS metabolite tenellin produced by Beauveria bassiana was challenged for heterologous production in Aspergillus oryzae, all promoters in the BGC were replaced with those of an A. oryzae amylase gene, which is highly expressed in maltose-supplemented media. As a result, a yield of 243 mg/L was attained, which was 5-fold higher than that of the original strain (Heneghan et al., 2010). Enniatin production is another successful example. The gene esyn1, which encodes NRPS involved in enniatin biosynthesis in Fusarium sp., was heterologously expressed in Aspergillus niger using the Tet-on inducible promoter system. This resulted in about 950-fold increase from the initial yield (Richter et al., 2014). Importantly, such BGCs were integrated to chromosomal loci, such as the pyrG locus, where genes were highly expressed. High expression in the heterologous host by locus selection can be expected, based on the epigenetic effect of chromosomal gene expression (Palmer and Keller, 2010). In addition, finding such loci at several positions on a chromosome is useful when considering the introduction of several copies of BGC to the heterologous host. Currently, because the BGCs of filamentous fungal NRPs generally reach dozens of kilobases in length, their transfer to other strains requires much labor and a longer period. However, if the original strain is difficult to genetically modify, heterologous production to improve NRP production is a challenge.
3 Production of ribosomally synthesized and post-translationally modified peptides in filamentous fungi
Fungal RiPPs were discovered relatively recently. In 2007, amatoxin (a strong food-poisoning toxin with RNA polymerase II inhibitory activity) and phallotoxin (F-actin stabilizer) produced by Amanita mushroom, were the first reported RiPPs in fungi, synthesized through ribosomes (Hallen et al., 2007). In filamentous fungi, ustiloxin B, which has strong inhibitory activity against tubulin assembly (Koiso et al., 1992), was first reported as a RiPP in Aspergillus flavus (Umemura et al., 2013; Umemura et al., 2014). Another RiPP class in fungi, later collectively called borosins, found in the basidiomycete Omphalotus olearius was reported in 2017 (Ramm et al., 2017). In this article, we focus on RiPPs produced by filamentous fungi or ascomycetes. The precursor protein for ustiloxin B is composed of a signal peptide to the endoplasmic reticulum at its N-terminus, followed by a 16-times repeated sequence that is separated by the recognition site of Kex2 protease into peptides containing the compound backbone (core) peptide. Proteins with these characteristics were later termed Kex2-processed repeat proteins (KEPs) (Marquer et al., 2019). The ustiloxin precursor gene is accompanied by two genes encoding the cyclic factor UstYa homolog (previously called DUF3328 domain-containing protein). RiPPs produced from KEPs as precursor proteins and cyclized by UstYa homolog(s) were named dikaritins (Ding et al., 2016). A bioinformatics survey of 1461 fungal genomes showed that KEPs are conserved among most fungal strains, with at least 838 different types, and 22% of KEP genes are accompanied by UstYa homolog genes (Umemura, 2020). Nevertheless, only five dikaritins, which are the sole examples of RiPPs in filamentous fungi or ascomycetes so far, have been reported: ustiloxin (Umemura et al., 2014), asperipin-2a (Nagano et al., 2016), phomopsin (Ding et al., 2016), epichloëcyclin (Johnson et al., 2015), and victorin (Kessler et al., 2020) (Figure 1B). The limited number of reported dikaritins and our experience suggests that most dikaritins are produced under limited conditions or in small quantities. We reported asperipin-2a and its biosynthetic genes from a reverse genetics approach (Nagano et al., 2016; Ye et al., 2019). However, a considerable amount of time and effort was required to obtain a sufficient amount of the compound for identification because asperipin-2a was produced only in a solid maize medium with a < 0.01 mg/kg yield in the original producer strain. The water-soluble properties of dikaritins make them further difficult to prepare because such compounds cannot be concentrated by water-insoluble organic solvents such as ethyl acetate and chloroform; in that sense, asperipin-2a was still better because it could be extracted by n-butanol, which is separated from the water layer. Therefore, increasing the production of dikaritins is crucial for obtaining sufficient amounts of compounds to identify absolute structures, and for industrial use.
Similar to NRPs, one of the most direct and effective ways to increase RiPP production is to overexpress the genes responsible for the biosynthesis of target compounds. If there is a transcription factor gene in the target RiPP BGC, it would be better to overexpress it to upregulate the entire BGC. This strategy was used in the case of ustiloxin; the yield of ustiloxin B increased 4.8 times by overexpressing ustR, a transcription factor that regulates the whole ustiloxin gene cluster composed of 15 genes (Umemura et al., 2014). In the case of asperipin-2a, we could not apply this strategy because there was no transcription factor gene, at least around the precursor gene. Instead, Ye et al. (2019) succeeded in obtaining 1 mg of asperipin-2a from 400 mL of the potato-maltose liquid medium by heterologous overexpression of the four genes responsible for asperipin-2a biosynthesis originating from A. flavus, in A. oryzae. The successful heterologous expression of ustiloxin and its intermediates in A. oryzae yielded sufficient amounts of compounds to identify their absolute structures (Ye et al., 2016). These two examples show that the heterologous expression strategy should work for the RiPP biosynthetic pathways. When attempting the heterologous production of fungal secondary metabolites, it is important to use a “clean” host, which produces a small number of secondary metabolites by itself, to reduce noisy background signals, observed such as in LC-MS chromatograms. In this regard, A. oryzae is one of the best host strains for the heterologous production of fungal natural compounds (Sakai et al., 2012).
Phomopsin (Ding et al., 2016) and victorin (Kessler et al., 2020), both of which are produced by plant pathogenic fungi and are widely known mycotoxins, were identified as RiPPs by deleting the corresponding biosynthetic genes in the producing strains. This was the same for epichloëcyclin (Johnson et al., 2015). Sogahata et al. (2021) demonstrated that three UstYa homologs in the biosynthesis of phomopsin are essential not only for cyclization but also for the desaturation of amino acid moieties; however, they did so not by heterologous expression, but by deleting the corresponding genes in the producing strain. These results and our experiences strongly imply that there are one or more hindrances to the heterologous production of fungal RiPPs, even though the corresponding genes are expressed. Heterologous production of ustiloxin and asperipin-2a, which were originally identified in A. flavus, was successful in A. oryzae, possibly because A. flavus and A. oryzae are genetically similar.
Another possible feature of dikaritins that improves their production is the highly repetitive sequence of their precursor proteins. On average, one dikaritin precursor protein contains a 5-fold repeated sequence (Umemura, 2020). We wondered why such highly repetitive sequences are retained ubiquitously in fungi because such structures should be unstable and costly to maintain in the genome. Recently, we reported that the repeat number in a dikaritin precursor protein contributes not only to compound production but also to the transcript level of the precursor protein itself (Umemura et al., 2022). With the increasing number of repeats in the ustiloxin precursor protein, compound production increased quadratically and not linearly. Surprisingly and importantly, the transcript level of the precursor gene increased linearly with an increasing number of repeats. This result suggests that the transcript of the precursor gene is stabilized via a highly repeated structure in a feedback manner by an unknown mechanism. As a result, compound production increased quadratically because the factor of repeat number was multiplied by the factor of the linearly increasing transcript level. We do not yet know how the repeated structure of the precursor gene or protein regulates its transcript level. The secondary structure of the highly repeated mRNA may be more stable than that of the unrepeated mRNA, although we could not find any meaningful secondary structure of such mRNAs by computational prediction. The translated protein product might function as a chaperone or RNA-binding protein to protect its transcript. Regardless of the mechanism, the repetitive structure of the ustiloxin precursor protein contributes to compound production, not just as abundant substrates but also as a stabilizer for its transcript. Although this phenomenon has been confirmed only in ustiloxin biosynthesis to date, when attempting to produce sufficient amounts of dikaritins, it might be better to maintain the repetitive structure or even increase the repeat number in dikaritin precursor genes. In addition, overexpression of the precursor protein gene, not the entire biosynthetic pathway, might also increase dikaritin yield.
4 Discussion
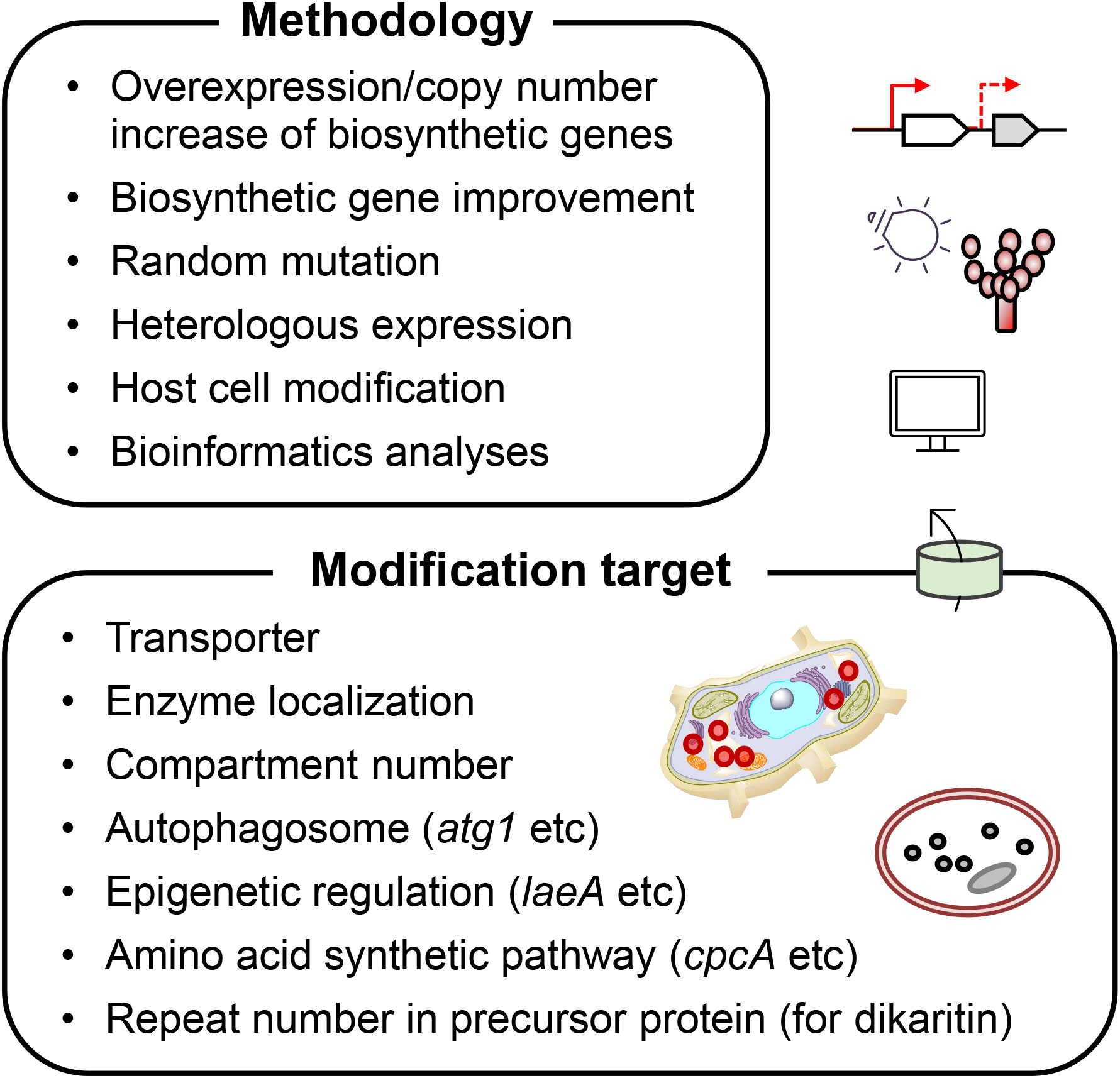
We described successful attempts to increase the production of NRPs and RiPPs in filamentous fungi through reverse genetics and heterologous production (Figure 2). Currently, there is still a large hindrance to introducing an entire BGC of NRP or RiPP into other host strains because it is generally large, approximately dozens of kilobases; preparing such a long gene, either by cloning or by artificial gene synthesis, is extremely difficult. Further, it is laborious to express biosynthetic genes for an NRP or RiPP in a heterologous manner when they are located at different loci in the genome, or when some of them are redundant. Genetic techniques to conjugate several DNA fragments instantly, or at least in a few steps, are anticipated to overcome this difficulty.
To modify host strains to improve target compound production, it is reasonable to optimize the localization of the corresponding biosynthetic enzymes in the cells. Some of the fungal peptidyl compounds reviewed by Skellam (2022) are synthesized in compartments. For example, IAT responsible for penicillin biosynthesis, localizes in peroxisomes (Meijer et al., 2010), and SimA functioning as cyclosporin synthetase, localizes in vacuoles (Hoppert et al., 2001). The prolyl oligopeptidase for maturation of amanitin (POPB) may localize in vacuoles (Luo et al., 2010). As mentioned in Section 2, penicillin production was improved using the localization information of biosynthetic enzymes. Probably, for other NRPs and dikaritins, we will be able to improve their production by optimizing the localization of biosynthetic enzymes in cells. To take this approach, the top priority is to elucidate when, where, and how the compounds are synthesized in cells and by which transporter(s) they are transported from one organelle to another; we are currently working on this.
Additionally, in NRPs containing non-proteogenic amino acids such as ornithine (Sheridan et al., 2015), metabolic modification to enhance biosynthesis of non-proteogenic amino acids is likely to increase NRP production. Moreover, for the enzymatic reaction, NRPS requires post-translational modification via 4’-phosphopantetheinylation. Therefore, improving 4’-phosphopantetheine biosynthesis by metabolic modification would be useful. Although no successful attempts have been reported to date, this strategy has the potential to be effective.
We have not mentioned the bioinformatics approach in this article, but it is one of the central methodologies for the improvement of compound production. Using bioinformatics, we can narrow down target genes that are bottlenecks for compound production and detect biosynthetic core genes for target NRPs/RiPPs using biological omics data. Indeed, we identified ustiloxin biosynthetic genes using a bioinformatics algorithm, which analyzes genomic information combined with transcriptomic data obtained under compound-producing and non-producing culture conditions (Umemura et al., 2013). In addition to conventional homology search and statistical data analyses of biological large data, machine-learning technologies are rapidly developing in biological fields, which can predict protein structures (Jumper et al., 2021), cell-type-specific gene expression profiles (Newman et al., 2019), protein-protein interactions (Zubek et al., 2015), and points clustered from single-molecule localization microscopy data (Williamson et al., 2020). Using emerging informatics technology, we will be able to take this shortcut to genetically modify filamentous fungal strains to produce target compounds in high yields.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding author.
Author contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Acknowledgments
We thank Editage for English language editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alberti F., Foster G. D., Bailey A. M. (2017). Natural products from filamentous fungi and production by heterologous expression. Appl. Microbiol. Biotechnol. 101, 493–500. doi: 10.1007/s00253-016-8034-2
Bartoszewska M., Kiel J. A., Bovenberg R. A., Veenhuis M., van der Klei I. J. (2011). Autophagy deficiency promotes beta-lactam production in Penicillium chrysogenum. Appl. Environ. Microbiol. 77, 1413–1422. doi: 10.1128/AEM.01531-10
Ding W., Liu W.-Q., Jia Y., Li Y., Donk W. A., Zhang Q. (2016). Biosynthetic investigation of phomopsins reveals a widespread pathway for ribosomal natural products in ascomycetes. Proc. Natl. Acad. Sci. U. S. A. 113, 3521. doi: 10.1073/pnas.1522907113
Elander R. P. (2003). Industrial production of beta-lactam antibiotics. Appl. Microbiol. Biotechnol. 61, 385–392. doi: 10.1007/s00253-003-1274-y
Fierro F., Barredo J. L., Díez B., Gutierrez S., Fernández F. J., Martín J. F. (1995). The penicillin gene cluster is amplified in tandem repeats linked by conserved hexanucleotide sequences. Proc. Natl. Acad. Sci. U. S. A. 92, 6200–6204. doi: 10.1073/pnas.92.13.6200
Fierro F., Vaca I., Castillo N. I., García-Rico R. O., Chávez R. (2022). Penicillium chrysogenum, a vintage model with a cutting-edge profile in biotechnology. Microorganisms 6, 573. doi: 10.3390/microorganisms10030573
Hallen H. E., Luo H., Scott-Craig J. S., Walton J. D. (2007). Gene family encoding the major toxins of lethal Amanita mushrooms. Proc. Natl. Acad. Sci. U. S. A. 104, 19097. doi: 10.1073/pnas.0707340104
Heneghan M. N., Yakasai A. A., Halo L. M., Song Z., Bailey A. M., Simpson T. J., et al. (2010). First heterologous reconstruction of a complete functional fungal biosynthetic multigene cluster. Chembiochem 11, 1508–1512. doi: 10.1002/cbic.201000259
Herr A., Fischer R. (2014). Improvement of Aspergillus nidulans penicillin production by targeting AcvA to peroxisomes. Metab. Eng. 25, 131–139. doi: 10.1016/j.ymben.2014.07.002
Hoppert M., Gentzsch C., Schoergendorfer K. (2001). Structure and localization of cyclosporin synthetase, the key enzyme of cyclosporin biosynthesis in Tolypocladium inflatum. Arch. Microbiol. 176, 285–293. doi: 10.1007/s002030100324
Houbraken J., Frisvad J. C., Samson R. A. (2011). Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA. Fungus. 2, 87–95. doi: 10.5598/imafungus.2011.02.01.12
Johnson R. D., Lane G. A., Koulman A., Cao M., Fraser K., Fleetwood D. J., et al. (2015). A novel family of cyclic oligopeptides derived from ribosomal peptide synthesis of an in planta-induced gene, gigA, in Epichloë endophytes of grasses. Fungal Genet. Biol. 85, 14. doi: 10.1016/j.fgb.2015.10.005
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583. doi: 10.1038/s41586-021-03819-2
Kautsar S. A., Blin K., Shaw S., Navarro-Muñoz J.-C., Terlouw B. R., van der Hooft J. J. J., et al. (2020). MIBiG 2.0: a repository for biosynthetic gene clusters of known function. Nucleic Acid Res. 48, D454. doi: 10.1093/nar/gkz882
Keller N. P. (2019). Fungal secondary metabolism: Regulation, function and drug discovery. Nat. Rev. Microbiol. 17, 167–180. doi: 10.1038/s41579-018-0121-1
Kessler S. C., Zhang X., McDonald M. C., Gilchrist C. L. M., Lin Z., Rightmyer A., et al. (2020). Victorin, the host-selective cyclic peptide toxin from the oat pathogen Cochliobolus victoriae, is ribosomally encoded. Proc. Natl. Acad. Sci. U. S. A. 117, 24243. doi: 10.1073/pnas.2010573117
Kiel J. A., van der Klei I. J., van den Berg M. A., Bovenberg R. A., Veenhuis M. (2005). Overproduction of a single protein, pc-Pex11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet. Biol. 42, 154–164. doi: 10.1016/j.fgb.2004.10.010
Koiso Y., Natori M., Iwasaki S., Sato S., Sonoda R., Fujita Y., et al. (1992). Ustiloxin: A phytotoxin and a mycotoxin from false smut balls on rice panicles. Tetrahedron. Lett. 33, 4157. doi: 10.1016/S0040-4039(00)74677-6
Kosalková K., García-Estrada C., Ullán R. V., Godio R. P., Feltrer R., Teijeira F., et al. (2009). The global regulator LaeA controls penicillin biosynthesis, pigmentation and sporulation, but not roquefortine c synthesis in Penicillium chrysogenum. Biochimie 91, 214–225. doi: 10.1016/j.biochi.2008.09.004
Luo H., Hallen-Adams H. E., Scott-Craig J. S., Walton J. D. (2010). Colocalization of amanitin and a candidate toxin-processing prolyl oligopeptidase in amanita basidiocarps. Eukaryot. Cell. 9, 1891. doi: 10.1128/EC.00161-10
Marquer M., Clemente H. S., Roux C., Savelli B., Frei N. (2019). Identification of new signalling peptides through a genome-wide survey of 250 fungal secretomes. BMC Genomics 20, 64. doi: 10.1186/s12864-018-5414-2
Marshall P. A., Dyer J. M., Quick M. E., Goodman J. M. (1996). Redox-sensitive homodimerization of Pex11p: A proposed mechanism to regulate peroxisomal division. J. Cell. Biol. 135, 123–137. doi: 10.1083/jcb.135.1.123
Martín J. F. (2017). Key role of LaeA and velvet complex proteins on expression of β-lactam and PR-toxin genes in Penicillium chrysogenum: Cross-talk regulation of secondary metabolite pathways. J. Ind. Microbiol. Biotechnol. 44, 525–535. doi: 10.1007/s10295-016-1830-y
Matsui M., Yokoyama T., Nemoto K., Kumagai T., Tamano K., Machida M., et al. (2017b). Further enhancement of FR901469 productivity by co-overexpression of cpcA, a cross-pathway control gene, and frbF in fungal sp. no. 11243. J. Biosci. Bioeng. 124, 8–14. doi: 10.1016/j.jbiosc.2017.02.015
Matsui M., Yokoyama T., Nemoto K., Kumagai T., Terai G., Arita M., et al. (2015). Genome sequence of fungal species No.11243, which produces the antifungal antibiotic FR901469. Genome Announc. 3, e00118–e00115. doi: 10.1128/genomeA.00118-15
Matsui M., Yokoyama T., Nemoto K., Kumagai T., Terai G., Tamano K., et al. (2017a). Identification of a putative FR901469 biosynthesis gene cluster in fungal sp. no. 11243 and enhancement of the productivity by overexpressing the transcription factor gene frbF. J. Biosci. Bioeng. 123, 147–153. doi: 10.1016/j.jbiosc.2016.08.007
McLean K. J., Hans M., Meijrink B., van Scheppingen W. B., Vollebregt A., Tee K. L., et al. (2015). Single-step fermentative production of the cholesterol-lowering drug pravastatin via reprogramming of Penicillium chrysogenum. Proc. Natl. Acad. Sci. U. S. A. 112, 2847–2852. doi: 10.1073/pnas.1419028112
Meijer W. H., Gidijala L., Fekken S., Kiel J. A., van den Berg M. A., Lascaris R., et al. (2010). Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum. Appl. Environ. Microbiol. 76, 5702–5709. doi: 10.1128/AEM.02327-09
Nagano N., Umemura M., Izumikawa M., Kawano J., Ishii T., Kikuchi M., et al. (2016). Class of cyclic ribosomal peptide synthetic genes in filamentous fungi. Fungal Genet. Biol. 86, 58. doi: 10.1016/j.fgb.2015.12.010
Newman A. M., Steen C. B., Liu C. L., Gentles A. J., Chaudhuri A. A., Scherer F., et al. (2019). Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37, 773. doi: 10.1038/s41587-019-0114-2
Nijland J. G., Ebbendorf B., Woszczynska M., Boer R., Bovenberg R. A., Driessen A. J. (2010). Nonlinear biosynthetic gene cluster dose effect on penicillin production by Penicillium chrysogenum. Appl. Environ. Microbiol. 76, 7109–7115. doi: 10.1128/AEM.01702-10
Palmer J. M., Keller N. P. (2010). Secondary metabolism in fungi: Does chromosomal location matter? Curr. Opin. Microbiol. 13, 431–436. doi: 10.1016/j.mib.2010.04.008
Paluh J. L., Orbach M. J., Legerton T. L., Yanofsky C. (1988). The cross-pathway control gene of Neurospora crassa, cpc-1, encodes a protein similar to GCN4 of yeast and the DNA-binding domain of the oncogene v-jun-encoded protein. Proc. Natl. Acad. Sci. U. S. A. 85, 3728–3732. doi: 10.1073/pnas.85.11.3728
Ramm S., Krawczyk B., Mühlenweg A., Poch A., Mösker E., Süssmuth R. D. (2017). A self-sacrificing n-methyltransferase is the precursor of the fungal natural product omphalotin. Angew. Chem. Int. Ed. 56, 9994. doi: 10.1002/anie.201703488
Richter L., Wanka F., Boecker S., Storm D., Kurt T., Vural Ö., et al. (2014). Engineering of Aspergillus niger for the production of secondary metabolites. Fungal Biol. Biotechnol. 1, 4. doi: 10.1186/s40694-014-0004-9
Sakai K., Kinoshita H., Nihira T. (2012). Heterologous expression system in Aspergillus oryzae for fungal biosynthetic gene clusters of secondary metabolites. Appl. Microbiol. Biotechnol. 93, 2011. doi: 10.1007/s00253-011-3657-9
Schrader M., Reuber B. E., Morrell J. C., Jimenez-Sanchez G., Obie C., Stroh T. A., et al. (1998). Expression of PEX11beta mediates peroxisome proliferation in the absence of extracellular stimuli. J. Biol. Chem. 273, 29607–29614. doi: 10.1074/jbc.273.45.29607
Sheridan K. J., Dolan S. K., Doyle S. (2015). Endogenous cross-talk of fungal metabolites. Front. Microbiol. 5. doi: 10.3389/fmicb.2014.00732
Skellam E. (2022). Subcellular localization of fungal specialized metabolites. Fungal Biol. Biotechnol. 9, 11. doi: 10.1186/s40694-022-00140-z
Sogahata K., Ozaki T., Igarashi Y., Naganuma Y., Liu C., Minami A., et al. (2021). Biosynthetic studies of phomopsins unveil posttranslational installation of dehydroamino acids by UstYa family proteins. Angew. Chem. Int. Ed. 60, 25729. doi: 10.1002/anie.202111076
Tamano K., Yoshimi A. (2021). Metabolic engineering techniques to increase the productivity of primary and secondary metabolites within filamentous fungi. Front. Fungal. Biol. 2. doi: 10.3389/ffunb.2021.743070
Tian C., Kasuga T., Sachs M. S., Glass N. L. (2007). Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot. Cell. 6, 1018–1029. doi: 10.1128/EC.00078-07
Umemura M. (2020). Peptides derived from Kex2-processed repeat proteins are widely distributed and highly diverse in the fungi kingdom. Fungal Biol. Biotechnol. 7, 11. doi: 10.1186/s40694-020-00100-5
Umemura M., Koike H., Nagano N., Ishii T., Kawano J., Yamane N., et al. (2013). MIDDAS-m: Motif-independent de novo detection of secondary metabolite gene clusters through the integration of genome sequencing and transcriptome data. PloS One 8, e84028. doi: 10.1371/journal.pone.0084028
Umemura M., Kuriiwa K., Linh V. D. (2022). Tandem repeats in precursor protein stabilize transcript levels and production levels of the fungal ribosomally synthesized and post-translationally modified peptide ustiloxin b. Fungal Genet. Biol. 160, 103691. doi: 10.1016/j.fgb.2022.103691
Umemura M., Nagano N., Koike H., Kawano J., Ishii T., Miyamura Y., et al. (2014). Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin b in Aspergillus flavus. Fungal Genet. Biol. 68, 23. doi: 10.1016/j.fgb.2014.04.011
Van den Berg M. A. (2011). Impact of the Penicillium chrysogenum genome on industrial production of metabolites. Appl. Microbiol. Biotechnol. 92, 45–53. doi: 10.1007/s00253-011-3476-z
Vassaux A., Meunier L., Vandenbol M., Baurain D., Fickers P., Jacques P., et al. (2019). Nonribosomal peptides in fungal cell factories: From genome mining to optimized heterologous production. Biotechnol. Adv. 37, 107449. doi: 10.1016/j.biotechadv.2019.107449
Wang W., Yu Y., Keller N. P., Wang P. (2021). Presence, mode of action, and application of pathway specific transcription factors in Aspergillus biosynthetic gene clusters. Int. J. Mol. Sci. 22, 8709. doi: 10.3390/ijms22168709
Williamson D. J., Burn G. L., Simoncelli S., Griffié J., Peters R., Davis D. M., et al. (2020). Machine learning for cluster analysis of localization microscopy data. Nat. Commun. 11, 1493. doi: 10.1038/s41467-020-15293-x
Yang J., Xu X., Liu G. (2012). Amplification of an MFS transporter encoding gene penT significantly stimulates penicillin production and enhances the sensitivity of Penicillium chrysogenum to phenylacetic acid. J. Genet. Genomics 39, 593–602. doi: 10.1016/j.jgg.2012.08.004
Ye Y., Minami A., Igarashi Y., Izumikawa M., Umemura M., Nagano N., et al. (2016). Unveiling the biosynthetic pathway of the ribosomally synthesized and post-translationally modified peptide ustiloxin b in filamentous fungi. Angew. Chem. Int. Ed. 55, 8072. doi: 10.1002/anie.201602611
Ye Y., Ozaki T., Umemura M., Liu C., Minami A., Oikawa H. (2019). Heterologous production of asperipin-2a: Proposal for sequential oxidative macrocyclization by a fungi-specific DUF3328 oxidase. Org. Biomol. Chem. 17, 39. doi: 10.1039/C8OB02824A
Yin M., Xiao D., Wang C., Zhang L., Dun B., Yue Q. (2021). The regulation of BbLaeA on the production of beauvericin and bassiatin in Beauveria bassiana. World J. Microbiol. Biotechnol. 38, 1. doi: 10.1007/s11274-021-03162-8
Keywords: nonribosomal peptide (NRP), ribosomally synthesized and post-translationally modified peptide (RiPP), cyclic peptide, secondary metabolite, heterologous expression
Citation: Umemura M and Tamano K (2022) How to improve the production of peptidyl compounds in filamentous fungi. Front. Fungal Biol. 3:1085624. doi: 10.3389/ffunb.2022.1085624
Received: 31 October 2022; Accepted: 02 December 2022;
Published: 22 December 2022.
Edited by:
Katherine A. Borkovich, University of California, Riverside, United StatesReviewed by:
Andy Bailey, University of Bristol, United KingdomCopyright © 2022 Umemura and Tamano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koichi Tamano, dGFtYW5vLWtAYWlzdC5nby5qcA==
 Maiko Umemura
Maiko Umemura Koichi Tamano
Koichi Tamano