
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Fungal Biol. , 10 January 2023
Sec. Fungal Physiology and Metabolism
Volume 3 - 2022 | https://doi.org/10.3389/ffunb.2022.1061841
This article is part of the Research Topic Aspergillus and Fusarium beyond the toxins: Enzyme potential from genes to applications. View all articles
 Ami Koizumi1
Ami Koizumi1 Ken Miyazawa1,2
Ken Miyazawa1,2 Makoto Ogata3
Makoto Ogata3 Yuzuru Takahashi4
Yuzuru Takahashi4 Shigekazu Yano4
Shigekazu Yano4 Akira Yoshimi5,6,7
Akira Yoshimi5,6,7 Motoaki Sano8
Motoaki Sano8 Masafumi Hidaka9
Masafumi Hidaka9 Takanori Nihira10
Takanori Nihira10 Hiroyuki Nakai10
Hiroyuki Nakai10 Satoshi Kimura11
Satoshi Kimura11 Tadahisa Iwata11
Tadahisa Iwata11 Keietsu Abe1,5*
Keietsu Abe1,5*Aspergillus fungi contain α-1,3-glucan with a low proportion of α-1,4-glucan as a major cell wall polysaccharide. Glycosylphosphatidylinositol (GPI)-anchored α-amylases are conserved in Aspergillus fungi. The GPI-anchored α-amylase AmyD in Aspergillus nidulans has been reported to directly suppress the biosynthesis of cell wall α-1,3-glucan but not to degrade it in vivo. However, the detailed mechanism of cell wall α-1,3-glucan biosynthesis regulation by AmyD remains unclear. Here we focused on AoAgtA, which is encoded by the Aspergillus oryzae agtA gene, an ortholog of the A. nidulans amyD gene. Similar to findings in A. nidulans, agtA overexpression in A. oryzae grown in submerged culture decreased the amount of cell wall α-1,3-glucan and led to the formation of smaller hyphal pellets in comparison with the wild-type strain. We analyzed the enzymatic properties of recombinant (r)AoAgtA produced in Pichia pastoris and found that it degraded soluble starch, but not linear bacterial α-1,3-glucan. Furthermore, rAoAgtA cleaved 3-α-maltotetraosylglucose with a structure similar to the predicted boundary structure between the α-1,3-glucan main chain and a short spacer composed of α-1,4-linked glucose residues in cell wall α-1,3-glucan. Interestingly, rAoAgtA randomly cleaved only the α-1,4-glycosidic bonds of 3-α-maltotetraosylglucose, indicating that AoAgtA may cleave the spacer in cell wall α-1,3-glucan. Consistent with this hypothesis, heterologous overexpression of agtA in A. nidulans decreased the molecular weight of cell wall α-1,3-glucan. These in vitro and in vivo properties of AoAgtA suggest that GPI-anchored α-amylases can degrade the spacer α-1,4-glycosidic linkages in cell wall α-1,3-glucan before its insolubilization, and this spacer cleavage decreases the molecular weight of cell wall α-1,3-glucan in vivo.
The cell wall of fungi is composed mainly of polysaccharides; it protects cells from stresses and maintains cell morphology (Latgé, 2010; Yoshimi et al., 2016). Aspergillus cell wall is composed of α-1,3-glucan (with a low proportion of α-1,4-glucan), β-1,3-glucan (with β-1,6-branches), galactomannan and chitin (Latgé, 2010; Yoshimi et al., 2016). In pathogenic fungi, α-1,3-glucan conceals cell wall β-1,3-glucan and chitin and consequently prevents recognition by the host immune system (Rappleye et al., 2004; Rappleye et al., 2007; Fujikawa et al., 2009; Fujikawa et al., 2012; Beauvais et al., 2013). α-1,3-Glucan is also an adhesion factor in hyphal aggregation (Yoshimi et al., 2013; Miyazawa et al., 2016; Zhang et al., 2017; Miyazawa et al., 2018).
Two α-1,3-glucan synthase genes (agsA, agsB) are known in the model fungus Aspergillus nidulans, with agsB functioning mainly during vegetative hyphal growth (Yoshimi et al., 2013). In A. nidulans, the amyD and amyG genes are located near the agsB locus and are predicted to encode glycosylphosphatidylinositol (GPI)-anchored α-amylase and intracellular α-amylase, respectively (He et al., 2014). The cluster of these three genes (agsB–amyD–amyG) is conserved among Aspergillus fungi except for Aspergillus fumigatus (He et al., 2014).
The mechanism of cell wall α-1,3-glucan biosynthesis was first predicted in the fission yeast Schizosaccharomyces pombe (Grün et al., 2005). In the cytoplasm, an α-1,3-glucan synthase Ags1 transfers glucose units from uridine diphosphate glucose to the non-reducing end of the primer maltooligosaccharide and thus forms an α-1,3-glucan subunit, which is then exported by the same enzyme to the extracellular space (Grün et al., 2005). The extracellular domain of Ags1 is thought to connect two exported α-1,3-glucan subunits by transglycosylation (Grün et al., 2005). In S. pombe, a spacer oligosaccharide (≈12 residues), which is derived from the primer oligosaccharide, is present between two α-1,3-glucan chains (≈120 residues each) (Grün et al., 2005). We previously estimated the mechanism of cell wall α-1,3-glucan biosynthesis in A. nidulans with reference to the speculative mechanism in S. pombe (Yoshimi et al., 2017; Miyazawa et al., 2018; Miyazawa et al., 2020; Figure 1). Cell wall α-1,3-glucan is mainly synthesized by AgsB, which is composed of the extracellular, intracellular, and multitransmembrane domains (Yoshimi et al., 2017; Miyazawa et al., 2020). The primer maltooligosaccharides, which are likely required for α-1,3-glucan biosynthesis, are predicted to be produced by AmyG (He et al., 2014; Miyazawa et al., 2018; Miyazawa et al., 2020). Kazim et al. (2021) reported that, under submerged culture conditions, amyG disruption in A. nidulans causes the formation of dispersed hyphae and decreases pellet size and α-1,3-glucan content in the cell wall in comparison with the parental strain; these changes can be reverted by adding maltose or maltotriose to the culture media, suggesting that AmyG provides maltooligosaccharides for α-1,3-glucan biosynthesis. Chemical analyses of cell wall α-1,3-glucan in A. nidulans (Miyazawa et al., 2018) and Aspergillus wentii (Choma et al., 2013) revealed concatenation of a subunit consisting of about 200 α-1,3-linked glucose residues and a spacer of several 1,4-linked glucose residues. The anomer of the glucose residues of the spacer in Aspergillus fungi has not been analyzed, but it is presumed to be α-type (Latgé, 2010).
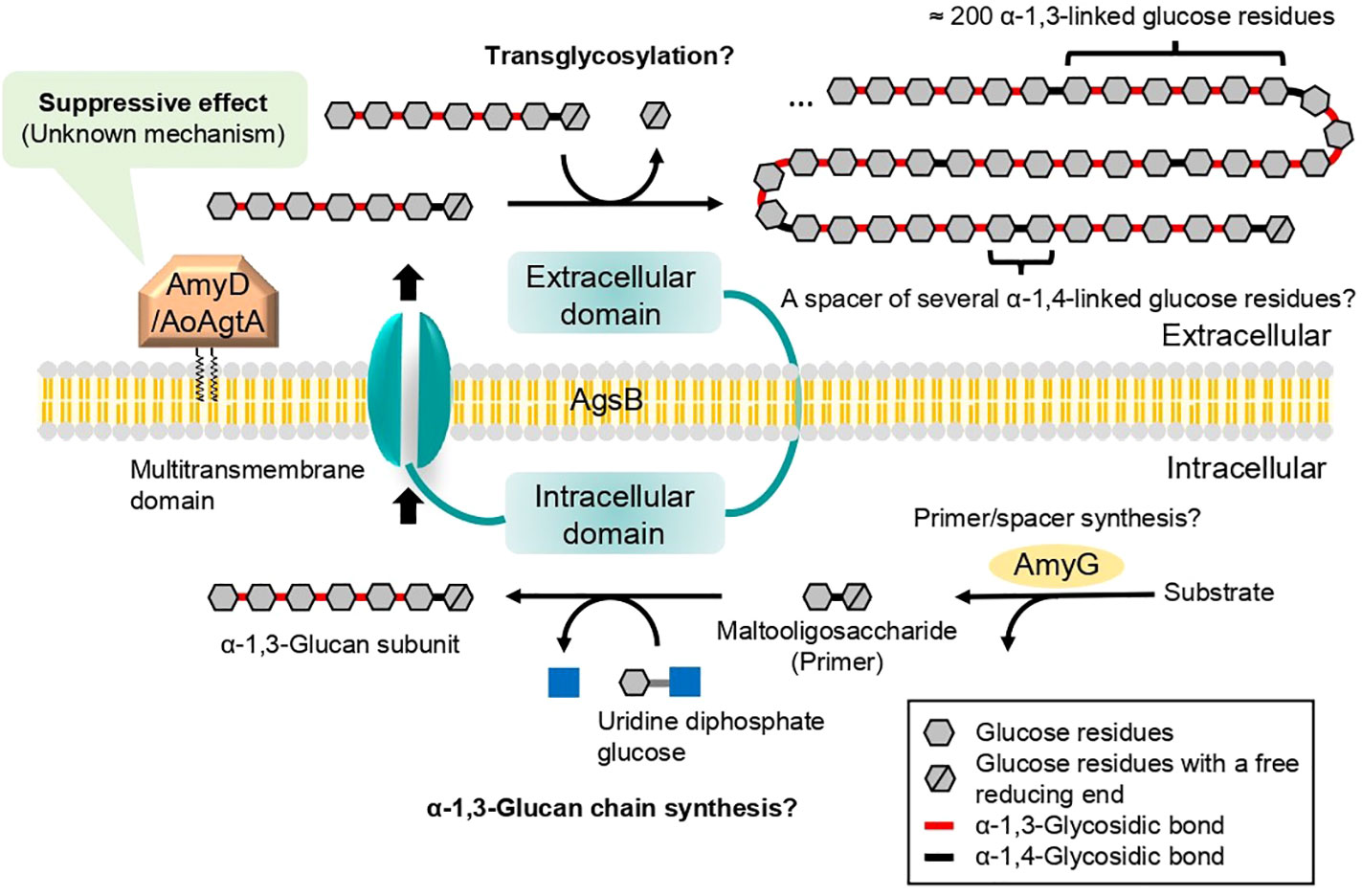
Figure 1 Speculative model for biosynthesis of cell wall α-1,3-glucan in A. nidulans and A. oryzae. Cell wall α-1,3-glucan is mainly synthesized by AgsB, which is composed of extracellular, intracellular, and multitransmembrane domains. Maltooligosaccharide is likely required as a primer for α-1,3-glucan biosynthesis and may be produced by intracellular α-amylase AmyG. The α-1,3-glucan chains may be synthesized by the intracellular domain of AgsB, and the resulting α-1,3-glucan subunits may be exported extracellularly by the multitransmembrane domain. The α-1,3-glucan subunits may be linked by transglycosylation catalyzed by the extracellular domain of AgsB to complete α-1,3-glucan maturation. Glycosylphosphatidylinositol-anchored α-amylase AmyD/AoAgtA regulates cell wall α-1,3-glucan biosynthesis, but the underlying mechanism remains unclear. Question marks indicate steps that are not firmly established.
AgtA of Aspergillus oryzae (AoAgtA) and proteins that are encoded by the orthologues of A. oryzae agtA, which are defined here as “Agt proteins”, are expected to be α-amylases of the glycoside hydrolase family 13 (GH13) and contain a C-terminal GPI-anchor site. He et al. (2014) reported that overexpression of A. nidulans amyD, orthologous to A. oryzae agtA, decreases the amount of cell wall α-1,3-glucan, whereas amyD disruption increases it, suggesting that amyD suppresses α-1,3-glucan biosynthesis. He et al. (2017) showed that the amount of cell wall α-1,3-glucan was lower in the amyD-overexpressing strain than in the parental A. nidulans strain at all time points examined, whereas in the α-1,3-glucanase overexpressing strains it was similar to that in the parental strain at the beginning of the time course and became lower from the middle of the culture. These results suggest that the mechanism of the decrease in the amount of cell wall α-1,3-glucan by AmyD differs from that by α-1,3-glucanase. We recently performed in vivo functional analysis of amyD in A. nidulans and found that not only the amount but also the molecular weight (MW) of cell wall α-1,3-glucan was decreased by amyD overexpression (Miyazawa et al., 2022). Aspergillus niger AgtA (AnAgtA) cannot use α-1,3-glucan derived from A. nidulans as a substrate and shows remarkable transglycosylation activity to produce maltooligosaccharides with a degree of polymerization (DP) of at least 28 (van der Kaaij et al., 2007). Overall, the mechanisms underlying the decrease in the amount and MW of α-1,3-glucan by Agt proteins are still unknown.
We have been developing A. oryzae strains with dispersed hyphae and high levels of enzyme production, which lack α-1,3-glucan and extracellular matrix galactosaminogalactan (Miyazawa et al., 2016; Miyazawa et al., 2019; Ichikawa et al., 2022). Although hyphal pellet formation of filamentous fungi under submerged culture conditions has been controlled by genetic approaches and culture conditions (Miyazawa et al., 2020), the options for controlling it could be expanded by adding approaches based on the catalytic properties of enzymes involved in cell wall α-1,3-glucan biosynthesis. Therefore, understanding the mechanism how AoAgtA regulates α-1,3-glucan biosynthesis would contribute to further development of phenotype control of A. oryzae from the viewpoint of hyphal pellet formation. There are three α-1,3-glucan synthase genes in A. oryzae (agsA, agsB, agsC), and agsB deletion leads to the loss of cell wall α-1,3-glucan (Zhang et al., 2017).
The aim of the present study was to understand the function of AoAgtA, which is involved in cell wall α-1,3-glucan biosynthesis and influences hyphal pellet formation in A. oryzae. We characterized an agtA-overexpressing (agtAOE) strain of A. oryzae and analyzed the enzymatic properties of recombinant (r)AoAgtA for maltooligosaccharides and their derivatives. By in vivo analysis of A. nidulans strains heterologously overexpressing agtA, we showed that AoAgtA decreases the MW of cell wall α-1,3-glucan. We discussed the enzymatic properties and the involvement of AoAgtA in cell wall α-1,3-glucan biosynthesis.
The signal peptide sequence was predicted by SignalP-5.0 Server (https://services.healthtech.dtu.dk/service.php?SignalP-5.0). The GPI anchor site was predicted by GPI Modification Site Prediction (https://mendel.imp.ac.at/gpi/gpi_server.html). The structure of AoAgtA was predicted using AlphaFold2 (Jumper et al., 2021; Tunyasuvunakool et al., 2021).
p-Nitrophenyl α-maltopentaoside (Mal5-α-pNP) was synthesized in our laboratory (Usui and Murata, 1988). Maltooligosaccharides were kindly supplied by Kikkoman Corporation (Noda, Japan). Nigerooligosaccharides were prepared by two methods: (i) partial acid degradation of linear α-1,3-glucan produced by GtfJ, a glucosyltransferase from Streptococcus salivarius ATCC 25975 (Puanglek et al., 2016), according to Czerwonka et al. (2019). Briefly, bacterial α-1,3-glucan (250 mg) was suspended in 1 mL of 0.1 M H2SO4 and incubated at 100°C for 1 h; (ii) enzymatic synthesis using α-1,3-glucoside phosphorylase (Nihira et al., 2014). Dextran from Leuconostoc mesenteroides (average MW 9,000–11,000) was purchased from Merck (Darmstadt, Germany). Pullulan was kindly supplied by Dr. Tasuku Nakajima (Tohoku University) and nigeran by Dr. Keiko Uechi (University of the Ryukyus). Pustulan was purchased from Calbiochem (San Diego, CA, United States) and treated with alcohol as in Hattori et al. (2013). Laminaran from Eisenia bicyclis was purchased from Tokyo Chemical Industry (Tokyo, Japan). All other commonly used chemicals were obtained from commercial sources.
The A. oryzae and A. nidulans strains are listed in Table 1. Escherichia coli DH5α was used for plasmid amplification. Pichia pastoris SMD1168H (Thermo Fisher Scientific, Waltham, MA, United States) was used for protein expression.
Primers are listed in Supplementary Table 1. An agtA overexpression plasmid, pNEN142-agtA, was constructed as follows. The agtA gene was amplified by polymerase chain reaction (PCR) with the agtA-Fw-NotI and agtA-Rv-NotI primers (designed with a NotI restriction site each) and wild-type A. oryzae genomic deoxyribonucleic acid (DNA) as a template (Supplementary Figure 1A). The PCR product was ligated into the NotI site of the pNEN142 vector (Minetoki et al., 2003) with an In-Fusion HD Cloning Kit (Takara Bio, Kusatsu, Japan); this vector contains the improved promoter of the A. oryzae enoA gene. The pNEN142-agtA plasmid was transformed into wild-type A. oryzae (Supplementary Figure 1B). The agtAOE transformants were selected on standard Czapek–Dox (CD) medium (Miyazawa et al., 2019); the niaD gene was used as a selectable marker. The transformants were subjected to single sporing as described previously (Hara et al., 2002). Integration of the agtA overexpression cassette was confirmed by PCR (Supplementary Figure 1C).
The agtA-disruption (ΔagtA) strain was created as described previously (Tamano et al., 2007). The agtA gene disruption cassette was generated by fusion PCR using an Expand High Fidelity PCR System (F. Hoffmann-La Roche, Basel, Switzerland). The 5′- and 3′-arms of agtA were amplified from wild-type A. oryzae genomic DNA with the primers agtA-LU and agtA-LL+adeA for the 5′-arm, and agtA-RU+adeA and agtA-RL for the 3′-arm (Supplementary Figure 2A). adeA was amplified with the primer pair agtA-AU and agtA-AL (Supplementary Figure 2A). A mixture of the 5′-flanking amplicon:adeA:3′-flanking amplicon at a 1:3:1 molar ratio was used as a template. The PCR products were used for a second PCR round with the primer pair agtA-LU and agtA-RL to fuse the 5′ and 3′ regions of the target gene at each end of the adeA gene (Supplementary Figure 2A). The amplified fragment was transformed into wild-type A. oryzae (Supplementary Figure 2B). The transformants were subjected to two consecutive rounds of single sporing. Replacement of the agtA gene was confirmed by PCR (Supplementary Figure 2C).
Conidia (final concentration, 1 × 105/mL) of A. oryzae strains were inoculated into 50 mL of YPD medium (1% yeast extract, 2% peptone, and 2% glucose) in 200-mL Erlenmeyer flasks and rotated at 120 rpm at 30°C for 24 h. The mean diameter of hyphal pellets was determined by measuring 10 randomly selected pellets under a stereomicroscope (M125; Leica Microsystems, Wetzlar, Germany). Conidia (final concentration, 5 × 105/mL) of A. nidulans strains were inoculated into 50 mL of CD medium (Miyazawa et al., 2018) in 200-mL Erlenmeyer flasks and rotated at 160 rpm at 37°C for 24 h.
Strains of A. oryzae were inoculated into YPD medium and cultured as described in the Analysis of growth characteristics subsection. Strains of A. nidulans were inoculated into 200 mL of CD medium in 500-mL Erlenmeyer flasks and rotated at 160 rpm at 37°C for 24 h. Mycelia were collected by filtration through Miracloth (Merck), washed with distilled water, lyophilized, and pulverized in a Mixer Mill MM 400 (Retsch, Haan, Germany). The resulting A. nidulans powder (0.5 g) was suspended in chloroform–methanol (3:1 vol/vol) and delipidized as described previously (Miyazawa et al., 2022). The pulverized A. oryzae mycelia (0.25–1 g) or delipidized A. nidulans mycelia were suspended in 0.1 M sodium phosphate buffer (pH 7.0). Cell wall components were fractionated by hot-water and alkali treatment (Yoshimi et al., 2013); the fractionation resulted in hot-water-soluble, alkali-soluble (AS), and alkali-insoluble fractions. The AS fraction was further separated into a fraction soluble in water at neutral pH (AS1) and an insoluble fraction (AS2). The AS2 fraction was used as α-1,3-glucan; this fraction was hydrolyzed and its glucose content was quantified as described previously (Yoshimi et al., 2013).
An agtA-expression plasmid, pPICZα B-agtA, was constructed as follows. The gene encoding AoAgtA without the C-terminus (amino acids (aa) 1–518; whole protein, 549 aa) to prevent GPI-anchoring was amplified with the primers agtA-Fw-NdeI (designed with a NdeI restriction site) and agtA-Rv-SmaI (designed with a SmaI restriction site) from wild-type A. oryzae complementary DNA (cDNA) as a template. The PCR product was digested with NdeI and SmaI and ligated into the NdeI–SmaI site of the pIVEX 2.3d vector (F. Hoffmann-La Roche). Then the AoAgtA coding region without the 1st–23th aa (putative signal peptide) and with a PGGGS linker and a (His)6 tag at the C-terminus was amplified with the primers agtA-Fw-PstI (designed with a PstI restriction site) and agtA-Rv-XbaI (designed with a XbaI restriction site); pIVEX 2.3d-agtA was used as a template. The PCR product was digested with PstI and XbaI and ligated into the PstI–XbaI site of the pPICZα B vector (Thermo Fisher Scientific), which encodes a secretion signal of Saccharomyces cerevisiae α-factor. The pPICZα B-agtA plasmid was linearized by SacI and integrated into the chromosomal DNA of P. pastoris SMD1168H competent cells with an EasySelect Pichia Expression Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The agtA-expressing transformants were cultured on YPDS (1% yeast extract, 2% peptone, 2% glucose, and 1 M sorbitol) agar plates containing zeocin (100 µg/mL) at 30°C for 2 days.
Buffered glycerol-complex (BMGY) medium and buffered methanol-complex (BMMY) medium were prepared according to the instructions of the EasySelect Pichia Expression Kit. The rAoAgtA-expressing P. pastoris strain was inoculated into 25 mL of BMGY medium in a 100-mL Erlenmeyer flask and rotated at 160 rpm at 30°C until OD600 4–6. The culture broth was centrifuged at 3,000 × g for 5 min. The collected cells were resuspended into 100 mL BMMY medium in a 500-mL Erlenmeyer flask and rotated at 200 rpm at 30°C for 6 days. To induce and maintain the expression of rAoAgtA, methanol (final concentration, 0.5% vol/vol) was added to culture broth every 24 h. The culture broth was centrifuged at 3,000 × g for 5 min, and the supernatant was dialyzed against 10 mM Tris-HCl buffer (pH 8.0). The supernatant (25 mL) was applied to a Ni Sepharose 6 Fast Flow column (1.8 × 5 cm; Cytiva, Marlborough, MA, United States) equilibrated in 20 mM Tris-HCl buffer (pH 8.0). The column was washed with the same buffer, and bound enzyme was eluted with 250 mM imidazole in the same buffer with pH readjusted to 8.0. The eluant was concentrated and buffer was replaced with 10 mM Tris-HCl buffer (pH 8.0) in Amicon Ultra-15 (nominal MW limit, 10,000) (Merck); the enzyme was stored in this buffer at -80°C for future reactions. At each stage, protein content was determined by Bradford method with bovine serum albumin as a standard, and rAoAgtA activity was determined as in the Recombinant AoAgtA activity measurement subsection. To check protein purity and determine the MW of the purified protein, it was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli, 1970), and the gels were stained with Coomassie Brilliant Blue R-250; the MW standards (10,000–250,000) were Precision Plus Protein Kaleidoscope Standards (Bio-Rad Laboratories, Hercules, CA, United States). Purified protein (4 μg) was deglycosylated with 500 U of Endoglycosidase H (Endo H) (New England Biolabs, Ipswich, MA, United States) according to the manufacturer’s instructions.
The high-performance liquid chromatography (HPLC) analysis was carried out with a Jasco Intelligent System Liquid Chromatograph (Jasco, Hachioji, Japan) under Conditions 1 and 2. Under Condition 1, a Unison UK-C18 column (4.6 × 250 mm; Imtakt, Kyoto, Japan) was used and p-nitrophenyl α-maltooligoside (PNM) was detected at 300 nm. The bound material was eluted with 20% methanol at a flow rate of 1.0 mL/min at 40°C. Under Condition 2, a Shodex HILICpak VG-50 4E column (4.6 × 250 mm; Showa Denko, Tokyo, Japan) was used and maltooligosaccharide was detected with a refractive index detector. The bound material was eluted with 65% acetonitrile at a flow rate of 0.7 mL/min at 40°C. 3-α-Maltooligosylglucose that presents at low level was analyzed under Condition 3: the HPLC analysis was carried out with a Hitachi Elite LaChrom HPLC System (Hitachi, Tokyo, Japan) on the same column as under Condition 2, and the detection was performed with an Alltech 3300 ELSD (Buchi Labortechnik, Flawil, Switzerland), an evaporative light scattering detector that is more sensitive than a refractive index detector. The bound material was eluted with 65% acetonitrile at a flow rate of 0.5 mL/min at 40°C. When the products of substrate degradation by rAoAgtA were analyzed by HPLC, data analysis was performed as follows. Products formed from the initial substrate at an early stage (up to approximately 15% degradation) were analyzed. Under Condition 1, the amount of each p-nitrophenylated product was calculated by multiplying the ratio of each peak area to the total peak area in the chromatogram by the concentration of the initial substrate. Under Conditions 2 and 3, the amount of each product was calculated from each peak area on the chromatogram and the calibration curve for each oligosaccharide.
A mixture (20 μL) containing 1 mM Mal5-α-pNP and an appropriate amount of rAoAgtA in 50 mM sodium acetate (Na-Ac) buffer (pH 5.5) was incubated at 40°C for 10 min. Aliquots (2 μL) were taken at 2-min intervals, and the reaction was immediately stopped with 40 μL of methanol. Then H2O (158 µL) was added to each aliquot, and the samples were subjected to HPLC analysis under Condition 1. The release velocity of p-nitrophenyl α-maltoside (Mal2-α-pNP) was determined as the slope of the time-course plot of Mal2-α-pNP amount. One unit of rAoAgtA activity was defined as the amount of enzyme required to liberate 1 μmol of Mal2-α-pNP from Mal5-α-pNP per minute.
Corn starch, potato starch, soluble starch, dextran, pullulan, bacterial α-1,3-glucan, nigeran, cellulose, pustulan, and laminaran were tested as rAoAgtA substrates. A mixture (12 μL) containing 2 mg/mL (0.2%) each substrate and 24 mU/mL purified rAoAgtA in 50 mM Na-Ac buffer (pH 5.5) was incubated at 40°C for 4 h. Then, H2O (138 μL) was added, and the reaction was immediately stopped by boiling for 5 min. The amount of reducing sugar generated from each substrate was measured by bicinchoninic acid method (Doner and Irwin, 1992; Utsumi et al., 2009). The bicinchoninic acid working reagent was prepared according to Utsumi et al. (2009), and 100 μL was added to a 100-μL aliquot of each boiled reaction mixture. The sample was incubated at 80°C for 40 min and then at room temperature for 15 min, and the absorbance at 560 nm was measured with a Multiscan Spectrum spectrophotometer (Thermo Fisher Scientific). One unit of enzyme activity was defined as the amount of enzyme required to liberate 1 μmol of reducing sugars (D-glucose conversion) from the substrate per minute. The detection limit was 1 mU/mL.
Maltooligosaccharides (DP 2–8, Mal2–8) and PNMs (DP 2–8, Mal2–8-α-pNP) were used to determine the modes of enzymatic bond cleavage by rAoAgtA. PNMs were synthesized from Mal5-α-pNP by transglycosylation catalyzed by rAoAgtA as described in Supplementary Material, and their structures were evaluated by 1H nuclear magnetic resonance (NMR) analysis in D2O (Supplementary Figure 3). To test maltooligosaccharides, a mixture (40 μL) containing 25 mM each substrate and 9.5 mU/mL purified rAoAgtA in 50 mM Na-Ac buffer (pH 5.5) was incubated at 40°C for 20 min. An aliquot (10 μL) was taken, and the reaction was immediately stopped with 65 μL of acetonitrile. Then H2O (25 µL) was added to the aliquot, and the sample was subjected to HPLC analysis under Condition 2. The frequency of rAoAgtA-catalyzed cleavages of glycosidic linkages was calculated from the amount of each product.
To test PNMs, a mixture (20 μL) containing 1.6 mM each substrate and 9.5 mU/mL purified rAoAgtA in 50 mM Na-Ac buffer (pH 5.5) was incubated at 40°C for 5 min. Aliquots (2 μL) were taken at 1-min intervals, and the reaction was immediately stopped with 40 μL of methanol. Then H2O (158 µL) was added to each aliquot, and the samples were subjected to HPLC analysis under Condition 1. The release velocity of each PNM from the initial substrate was determined as the slope of the time-course plot of the amount of that PNM. The frequency of rAoAgtA-catalyzed cleavages of glycosidic linkages was calculated from the release velocities of different PNMs.
Seven substrate concentrations ([S]) for Mal5-α-pNP (1.6–100 mM), six each for Mal6-α-pNP and Mal7-α-pNP (1.6–50 mM), and five for Mal8-α-pNP (1.6–25 mM) were used. The degradation reactions of these PNMs, including hydrolysis and transglycosylation, catalyzed by rAoAgtA and HPLC analyses were performed as described in the Analysis of the modes of bond cleavage subsection. The initial velocity (v) of substrate degradation was determined as the slope of the time-course plot of the total amount of all PNMs liberated from each initial substrate. The kinetic parameters of the Michaelis–Menten equation were evaluated by Hanes–Woolf plots ([S]/v versus [S]) and the least-squares method. The initial concentration of rAoAgtA in the reaction solution was 1.83 × 10-4 mM.
Mal5-α-pNP and various sugars (nigerose, nigerotriose, and glucose) were prepared at a final concentration of 1.6 mM and 16 mM, respectively. The reactions and HPLC analyses were performed as described in the Analysis of the modes of bond cleavage subsection, and the release velocity of Mal2-α-pNP was calculated.
A mixture (530 μL) containing Mal5-α-pNP (50 mg, 100 mM), nigerose (100 mg, 560 mM), and 180 mU/mL rAoAgtA in dialyzed culture supernatant in 50 mM Na-Ac buffer (pH 5.5) was incubated at 40°C for 24 h. The reaction was stopped by adding 11 mL of methanol. The reaction mixture was concentrated, lyophilized, dissolved in a small amount of H2O, and then applied to an ODS column (1.3 × 50 cm; Yamazen, Osaka, Japan) equilibrated with H2O at a flow rate of 3.0 mL/min to remove PNMs. The eluate (800 mL) was collected as a single fraction, concentrated, and lyophilized. The partially purified products were dissolved in a small amount of H2O, and then applied to a Toyopearl HW-40S column (3.5 × 62 cm; Tosoh, Tokyo, Japan) equilibrated with H2O at a flow rate of 0.5 mL/min. The eluate was collected in 2-mL fractions (300 mL in total). Each fraction was analyzed by HPLC under Condition 3, and the fractions containing 3-α-maltotetraosylglucose (Mal4α1,3Glc), 3-α-maltotriosylglucose (Mal3α1,3Glc), 3-α-maltosylglucose (Mal2α1,3Glc), and nigerose were concentrated and then lyophilized. Fractions containing products of insufficient purification were rechromatographed under the same conditions. Finally, Mal4α1,3Glc (7.3 mg, yield 16.7% based on Mal5-α-pNP), Mal3α1,3Glc (3.1 mg, 8.8%), Mal2α1,3Glc (10.8 mg, 40.7%), and nigerose (65.7 mg) were obtained.
The structures of the synthesized products were evaluated by 1H and 13C NMR analysis in D2O (Supplementary Tables 2, 3); 500 MHz 1H NMR spectra and 125 MHz 13C NMR spectra were recorded using a Bruker Avance Neo-500 NMR spectrometer (Bruker, Billerica, MA, United States). Matrix assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectra were acquired using an Autoflex Speed spectrometer (Bruker). MALDI-TOF mass analysis of 3-α-maltooligosylglucose showed m/z 527.167 [M + Na]+ (calcd for C18H32NaO16, 527.159), 689.210 [M + Na]+ (calcd for C24H42NaO21, 689.212), and 851.392 [M + Na]+ (calcd for C30H52NaO26, 851.264).
A mixture (12 μL) containing 25 mM Mal4α1,3Glc and 180 mU/mL purified rAoAgtA in 50 mM Na-Ac buffer (pH 5.5) was incubated at 40°C for 30 min. An aliquot (3 µL) was taken, and the reaction was immediately stopped with 44 µL of acetonitrile. Then H2O (21 µL) was added to the aliquot, and the sample was subjected to HPLC analysis under Condition 3. The substrate-degradation velocity and the frequency of rAoAgtA-catalyzed cleavages of glycosidic linkages were determined from the amounts of degradation products. The substrates Mal5 and nigeropentaose were used for comparison. For Mal5, the reaction time was set to 8 min and degradation velocity was determined from the amount of Mal2 liberated.
The agtAOE strains were constructed by inserting the agtA overexpression cassette into the disrupted amyD locus. The pNEN142-agtA(-intron) and pAHdPT-agtA plasmids were first constructed (Supplementary Figure 4A). To remove the intron in the open reading frame of agtA, PCR was performed using PrimeSTAR Max DNA Polymerase (Takara Bio) and the pNEN142-agtA plasmid as a template. The product was transformed into E. coli to obtain the pNEN142-agtA(-intron) plasmid. To construct pAHdPT-agtA, a fragment containing the agtA open reading frame and agdA terminator was amplified from pNEN142-agtA(-intron). Primers AopyrG-IF-Right-Fw and Ptef1-tail-Rv were used in PCR with pAHdPT-amyD (Miyazawa et al., 2022) as a template. The two fragments were fused using the In-Fusion HD Cloning Kit according to the manufacturer’s instructions. The resulting plasmid pAHdPT-agtA was digested with SacI and transformed into the ΔamyD and agsBOEΔamyD strains (Supplementary Figure 4B). Candidate strains were selected on CD medium containing uridine, uracil, and 1.3 mg/mL 5-fluoroorotic acid and then cultured on CD medium containing uridine, uracil, and 800 µg/mL hygromycin. Correct insertion of the cassette was confirmed by PCR (Supplementary Figure 4C).
The MW of glucan in the AS2 fraction was determined by gel permeation chromatography according to Miyazawa et al. (2022). Polystyrene (MW, 13,900–3,850,000; Showa Denko) was used as a standard to calibrate the column.
The t-test or Welch’s test was used to compare paired samples, and Tukey’s test was used to compare multiple samples.
According to the NCBI database, the agtA (AO090003001498) gene in A. oryzae consists of 1,711 base pairs and encodes a 549-aa protein with a putative MW of 60,400. A comparison of the cDNA and genome sequence revealed one intron. The predicted signal peptide (1–23 aa) and a GPI anchor site (ω site, Ser 530) suggested that AoAgtA is a cell membrane and/more cell wall-bound protein.
The aa sequence of AoAgtA showed 44% identity with that of Taka-amylase A of A. oryzae (encoded by amyA, amyB, and amyC; hereinafter TAA) belonging to GH13. A comparison between the AlphaFold2-predicted model of AoAgtA and the TAA crystal structure (PDB, 2GVY) (Vujičić-Žagar and Dijkstra, 2006) showed that the overall structure, catalytic residues, and Ca2+-binding residues were highly conserved (Supplementary Figure 5).
We constructed the agtAOE and ΔagtA strains from wild-type A. oryzae (Supplementary Figures 1, 2). In submerged culture, the size of the hyphal pellets of the agtAOE strain was as small as that of the ΔagsAΔagsBΔagsC (ΔAG) strain lacking cell wall α-1,3-glucan (Figures 2A, B). We quantified the amount of glucose in the AS2 fraction obtained from lyophilized mycelia of each strain; this fraction contains mainly α-1,3-glucan. The proportion of glucose in the AS2 fraction from the agtAOE strain was reduced to 24% of that from the wild-type (Figure 2C; P < 0.01), whereas the size of hyphal pellets and the α-1,3-glucan content of ΔagtA strain were similar to those of the wild-type (Figure 2). These results suggest that AoAgtA negatively regulates cell wall α-1,3-glucan biosynthesis in A. oryzae.
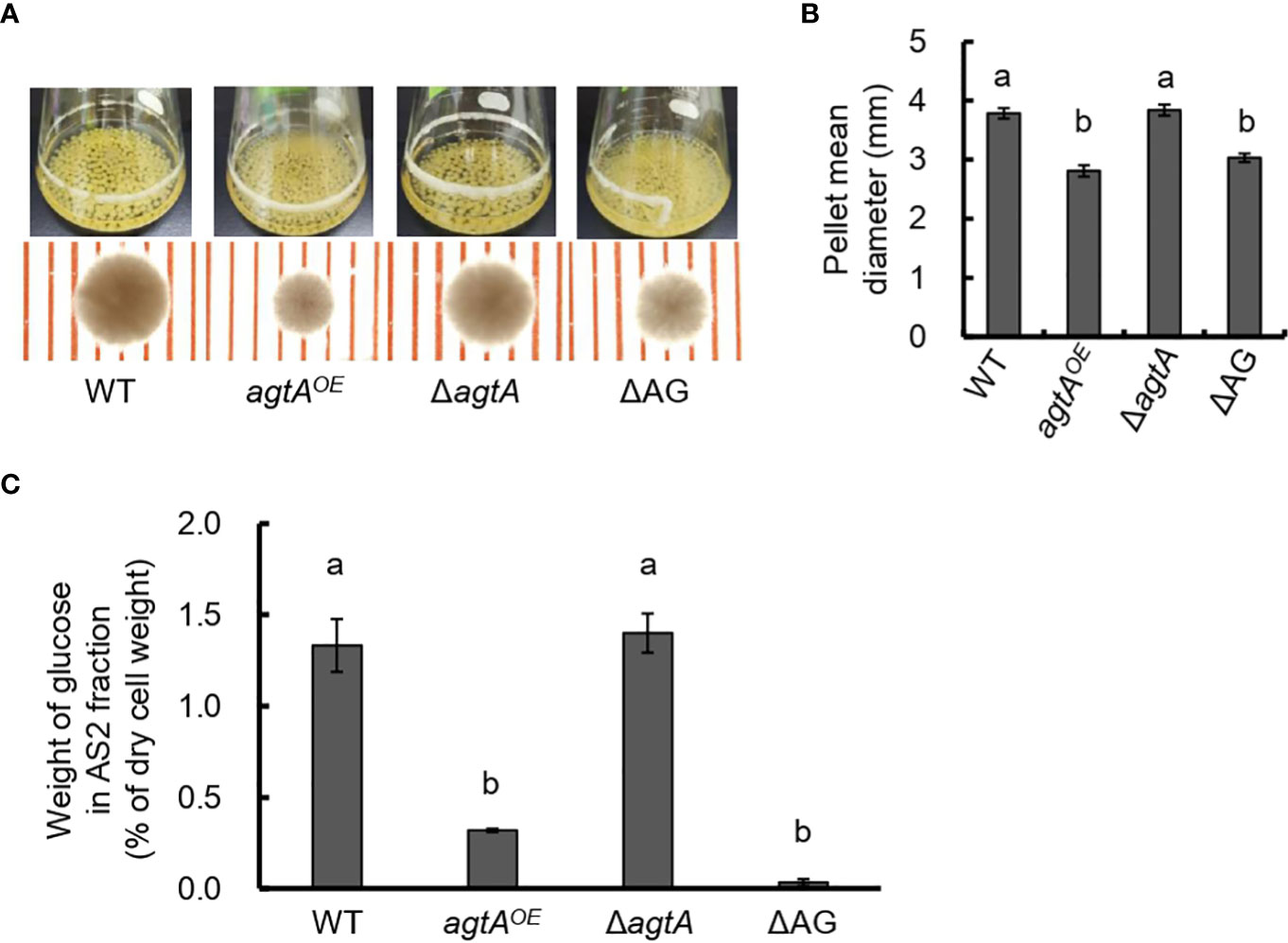
Figure 2 Analysis of the agtAOE strain of A. oryzae. (A) Growth characteristics of A. oryzae wild-type (WT), agtAOE, ΔagtA, and ΔAG strains in submerged culture. Conidia (1 × 105/mL) of each strain were inoculated into 50 mL of YPD medium and rotated at 120 rpm at 30°C for 24 h. Upper images, cultures in Erlenmeyer flasks; lower images, representative hyphal pellets under a stereomicroscope (scale = 1 mm). (B) The mean diameter of hyphal pellets. Error bars represent the standard error of the mean calculated from three replicates. Different letters above bars indicate significant difference by Tukey’s test (P < 0.01). (C) Amount of glucose in the alkali-soluble water-insoluble (AS2) fraction. Error bars represent the standard error of the mean calculated from three replicates. Different letters above bars indicate significant difference by Tukey’s test (P < 0.01).
Because agtA overexpression in A. oryzae led to a decrease in the amount of cell wall α-1,3-glucan and consequently to the formation of small hyphal pellets, we purified and characterized rAoAgtA produced in P. pastoris. In a single-step purification of (His)6-tagged rAoAgtA, 22% of enzymatic activity was recovered from culture supernatant on day 6 of culture; 5.2-fold purification was achieved (Table 2). Purified rAoAgtA showed a smeared band on SDS-PAGE with a MW of 73,200 (Figure 3). Removal of N-glycosylation by Endo H treatment decreased the apparent protein MW to 51,700 (Figure 3); the expected MW of rAoAgtA without the signal peptide and GPI anchor site but with the linker and the (His)6 tag at the C-terminus is 56,800. Therefore, the Endo H treatment revealed N-glycosylation of rAoAgtA.
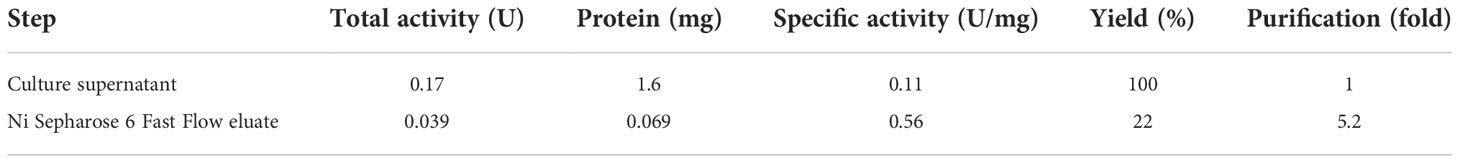
Table 2 Purification of rAoAgtA from the culture supernatant of the rAoAgtA-expressing P. pastoris strain.
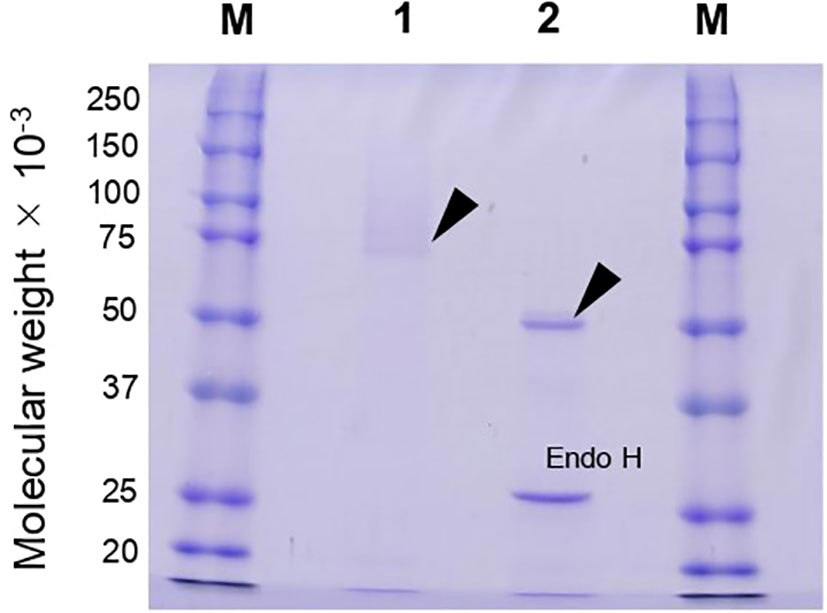
Figure 3 SDS-PAGE analysis of rAoAgtA produced in P. pastoris. Lanes M, Molecular weight markers; Lane 1, rAoAgtA purified on a Ni Sepharose 6 Fast Flow column; Lane 2, The same rAoAgtA deglycosylated with Endo H. Arrowheads indicate rAoAgtA bands. The band at 29,000 in lane 2 is Endo H.
To characterize the biochemical properties of rAoAgtA, we investigated optimal conditions and stability as described in Supplementary Material. The optimal temperature of rAoAgtA was 40°C (Supplementary Figure 6A). After 30-min incubation at 10–70°C, the protein retained >80% of enzymatic activity (i.e., was stable) at up to 30°C (Supplementary Figure 6B). The optimal pH of rAoAgtA was 5.5 (Supplementary Figure 6C). After 30-min incubation in each buffer at 4°C, the protein retained >80% enzymatic activity (i.e., was stable) at pH 4–9 (Supplementary Figure 6D).
Cu2+ and ethylenediaminetetraacetic acid (EDTA) reduced rAoAgtA activity to undetectable and 21 ± 6%, respectively (Supplementary Table 4). rAoAgtA activity was restored by adding Ca2+ after EDTA treatment (Supplementary Table 5). The predicted conservation of the Ca2+-binding residues between AoAgtA and TAA (Supplementary Figure 5E) is consistent with these results.
We assessed the ability of purified rAoAgtA to catalyze the degradation of various natural glucans by measuring the amounts of reducing sugars produced from the substrates. rAoAgtA showed a weak degradation activity (3.23 ± 0.28 mU/mL) only for soluble starch among glucans (corn starch, potato starch, soluble starch, dextran, pullulan, and nigeran) containing α-1,4-glycosidic bonds (Supplementary Table 6). Furthermore, rAoAgtA was unable to degrade bacterial α-1,3-glucan that contained only α-1,3-glycosidic bonds, as well as cellulose, pustulan, and laminaran (Supplementary Table 6). Since rAoAgtA did not degrade bacterial α-1,3-glucan and nigeran, which also contains α-1,3-glycosidic bonds, AoAgtA appears not to cleave the α-1,3-glycosidic bonds in cell wall α-1,3-glucan.
Since rAoAgtA degraded soluble starch, we evaluated its degradation activity and the modes of bond cleavage for 25 mM maltooligosaccharides (Mal2–8) and 1.6 mM their pNP-derivatives (Mal2–8-α-pNP). The cleavage positions and frequencies of glycosidic bonds degraded by rAoAgtA in each oligosaccharide substrate are shown in Figure 4. rAoAgtA showed degradation activity against maltooligosaccharide substrates with a DP of at least 5 (Mal5–8 and Mal5–8-α-pNP). HPLC chromatograms of the substrates and products of Mal5–8-α-pNP degradation by rAoAgtA are shown in Supplementary Figure 7. rAoAgtA catalyzed not only hydrolysis but also transglycosylation. For example, when the substrate was Mal5, the degradation products Mal2 and Mal3, and the transglycosylation product Mal8 were observed. Since the molar concentration of Mal2 was equal to the sum of molar concentrations of Mal3 and Mal8 (data not shown), we concluded that the cleavage position in Mal5 was the third glycosidic bond from the non-reducing end (Figure 4). In maltooligosaccharide substrates (DP of at least 5) other than Mal5 and Mal5-α-pNP, rAoAgtA randomly cleaved the internal α-1,4-glycosidic bonds (Figure 4). The mode of bond cleavage by rAoAgtA in maltooligosaccharide substrates with a DP of at least 5 was similar to that of TAA, which is a typical α-amylase (Nitta et al., 1971; Suganuma, 1983). On the other hand, a major difference was observed in that TAA can hydrolyze Mal2–4 (Nitta et al., 1971; Suganuma, 1983), whereas rAoAgtA cannot.
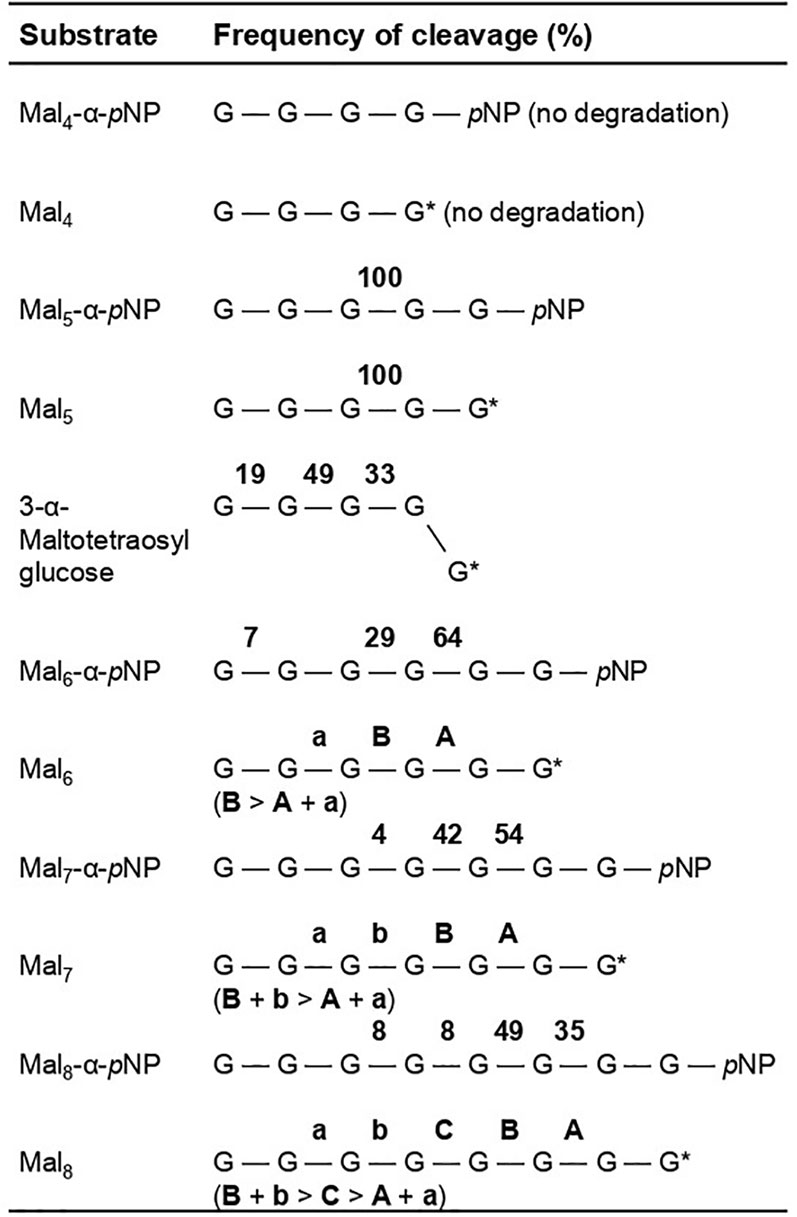
Figure 4 Frequency of site-specific cleavage during rAoAgtA-catalyzed degradation of maltooligosaccharides and their derivatives. The values in bold are the cleavage frequencies of glycosidic linkages. The cleavage positions that could not be distinguished are indicated as “a and A” or “b or B”. G, glucose residue; pNP, p-nitrophenly group; G*, glucose residue with a reducing end.
We conducted kinetic studies of the degradation of PNMs (Mal5–8-α-pNP) by rAoAgtA to further investigate its properties (Table 3 and Supplementary Figure 8). Ideally, the kinetics should be analyzed in the absence of transglycosylation, but we could not prevent transglycosylation. Therefore, we analyzed the kinetics in the presence of PNM hydrolysis and transglycosylation to the initial substrate, and obtained the apparent kinetic parameter values. The Km value of rAoAgtA was the lowest for Mal8-α-pNP (25.1 mM), and the kcat/Km value was the highest for Mal6-α-pNP (1.42 s-1mM-1). Other known α-amylases often have Km values of at most several mM and larger kcat/Km values than those of rAoAgtA for maltooligosaccharide substrates with a DP at least 5 (Nitta et al., 1971; Suganuma, 1983; Usui et al., 1992; Okada et al., 2000). Thus, the affinity and catalytic efficiency of rAoAgtA for maltooligosaccharide substrates were lower than those of other known α-amylases.
Nigerooligosaccharides can be considered as part of the structure of cell wall α-1,3-glucan of Aspergillus fungi. In the presence of rAoAgtA, 1.6 mM Mal5-α-pNP as a substrate, and 16 mM nigerose or nigerotriose, the release velocity of Mal2-α-pNP was 1.2 times and 1.4 times, respectively, that in the control without nigerooligosaccharides (Supplementary Figure 9; P < 0.01). Addition of glucose instead of nigerooligosaccharides had no effect (Supplementary Figure 9). This may be a result of substrate degradation due to the occurrence of transglycosylation with nigerooligosaccharides as acceptors. Therefore, we synthesized transglycosylation products using Mal5-α-pNP as a donor, nigerose as an acceptor, and an excess of rAoAgtA in dialyzed culture supernatant and obtained Mal4α1,3Glc, Mal3α1,3Glc, and Mal2α1,3Glc, in which maltooligosaccharide or glucose was transferred to the non-reducing end of nigerose with a formation of an α-1,4-glycosidic bond (Supplementary Tables 2, 3).
The 3-α-maltooligosylglucose motifs are present as part of the speculative cell wall α-1,3-glucan structure. We evaluated the behavior of rAoAgtA with 25 mM Mal4α1,3Glc and found that it cleaved Mal4α1,3Glc at the first, second, and third glycosidic bonds from the non-reducing end at the 19:49:33 ratio (Figure 4). The rAoAgtA substrate-degradation velocity for Mal4α1,3Glc was 0.133 ± 0.008 mM/min (mean ± standard deviation of three replicates), whereas rAoAgtA scarcely degraded nigeropentaose, which contains only α-1,3-glycosidic bonds. In Mal5, the third glycosidic bond from the non-reducing end was mainly cleaved by rAoAgtA, as described in the Analysis of the modes of bond cleavage subsection (Figure 4). The rAoAgtA substrate-degradation velocity for Mal5 was 0.320 ± 0.027 mM/min, and it was slower for Mal4α1,3Glc (0.42-fold, P < 0.01 in t-test) than for Mal5. The rAoAgtA activity for Mal4α1,3Glc showed two interesting characteristics: (i) a random cleavage mode of α-1,4-glycosidic bonds for Mal4α1,3Glc but not for Mal5; (ii) rAoAgtA catalyzed hydrolysis and self-transglycosylation with Mal5 but almost exclusively self-transglycosylation with Mal4α1,3Glc (data not shown). We found that rAoAgtA randomly cleaves the α-1,4-glycosidic bonds of Mal4α1,3Glc. Its structure can be considered as part of the structure of cell wall α-1,3-glucan in Aspergillus fungi (Miyazawa et al., 2018).
Since we have not established a method to measure the MW of the alkali-soluble glucan in the AS2 fractions derived from any A. oryzae strains, we introduced the agtA overexpression cassette into the ΔamyD and agsBOEΔamyD strains of A. nidulans to obtain the ΔamyD-agtAOE and agsBOEΔamyD-agtAOE strains (Supplementary Figure 4). In submerged culture, the ΔamyD strain formed tightly aggregated hyphal pellets, but the hyphae of the ΔamyD-agtAOE strain were almost fully dispersed (Figure 5). The latter result is consistent with that for the ΔamyD-amyDOE strain (Figure 5) reported by Miyazawa et al. (2022). The flask culture of the agsBOEΔamyD-agtAOE strain seemed cloudier than those of agsBOEΔamyD and agsBOEΔamyD-amyDOE strains, but there was only a slight difference in the hyphal pellets they formed (Figure 5). Overexpression of agtA had no marked effect on the phenotype of the agsBOEΔamyD strain.
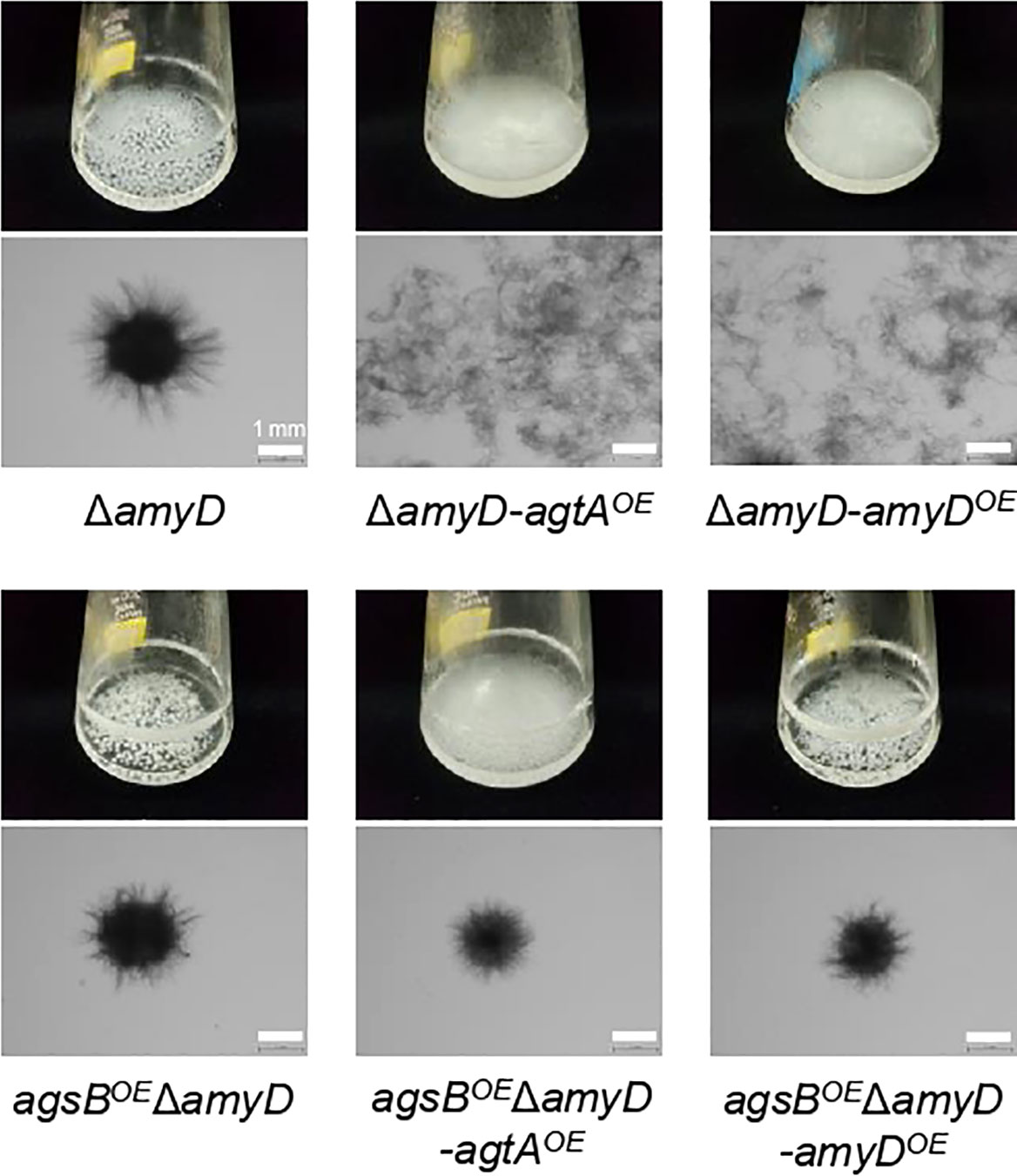
Figure 5 Growth characteristics of agtAOE strains of A. nidulans in submerged culture. Conidia (5 × 105/mL) of each strain were inoculated into 50 mL of standard Czapex–Dox (CD) medium and rotated at 160 rpm at 37°C for 24 h. Upper images, cultures in Erlenmeyer flasks; lower images, representative hyphae under a stereomicroscope. Scale bars, 1 mm.
We determined the amount of glucose and MW of glucan in the AS2 fractions from the agsBOEΔamyD and agsBOEΔamyD-agtAOE strains. The proportion of glucose in the AS2 fraction seemed to be slightly lower in agsBOEΔamyD-agtAOE than in agsBOEΔamyD (Figure 6A), but the difference was not statistically significant, presumably due to the masking effect of agsB overexpression. On the other hand, the number-average MW of glucan in the AS2 fraction from agsBOEΔamyD-agtAOE (95,700 ± 1,300) was significantly lower than that from agsBOEΔamyD (462,000 ± 38,000; Figure 6B; Table 4, P < 0.01 in Welch’s test). These results indicate that AoAgtA decreases the MW of cell wall α-1,3-glucan, similar to A. nidulans AmyD (Miyazawa et al., 2022).
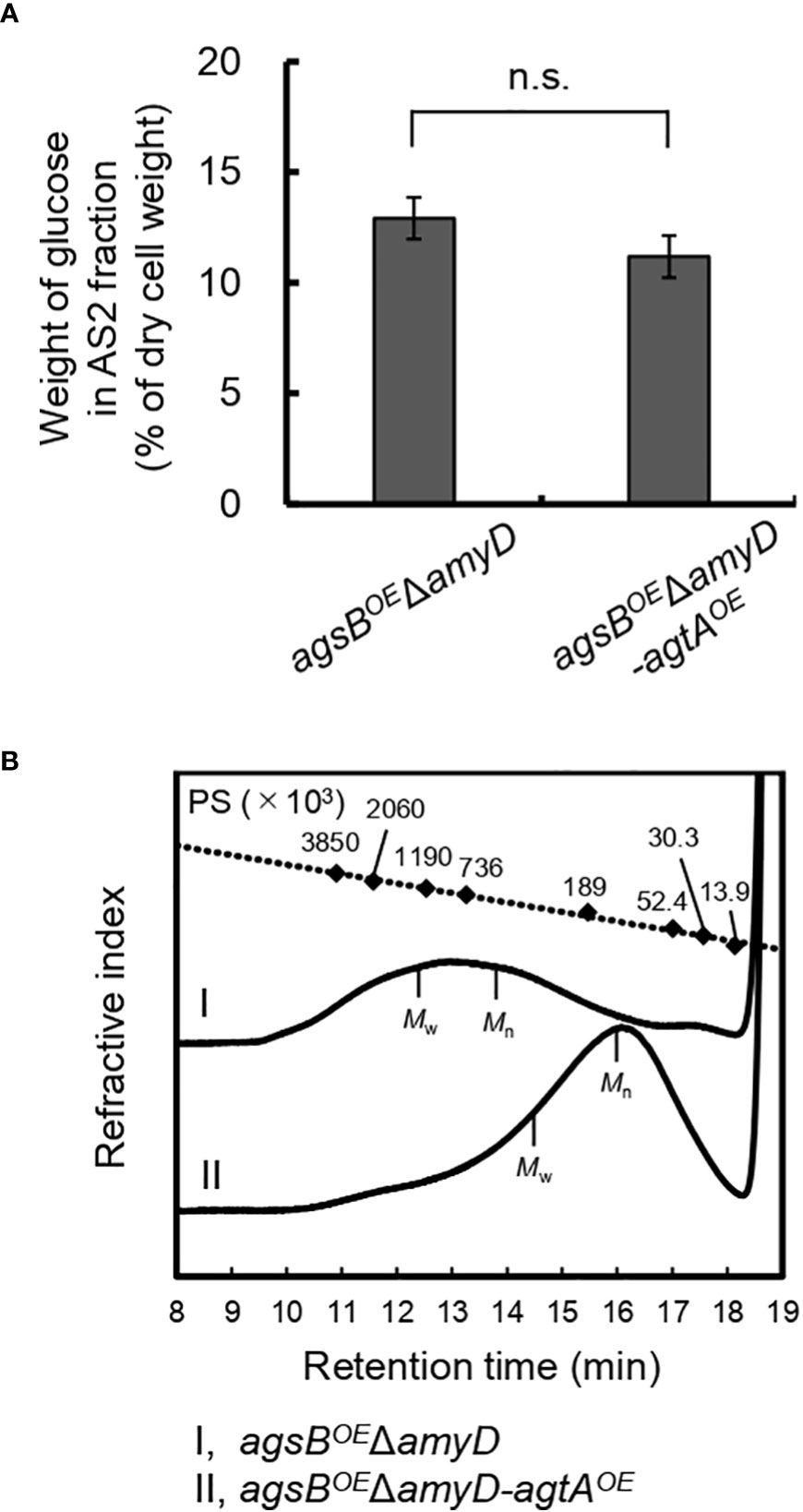
Figure 6 (A) Glucose content and (B) gel permeation chromatography elution profile of the AS2 fraction in the agsBOEΔamyD-agtAOE strain of A. nidulans. (A) Conidia (5 × 105/mL) of each strain were inoculated into CD medium and rotated at 160 rpm at 37°C for 24 h. Error bars represent the standard error of the mean calculated from three replicates. n.s., not significant in t-test (P ≥ 0.05). (B) The AS2 fraction from 24-h-cultured mycelia of each strain was dissolved in 20 mM LiCl/N,N-dimethylacetamide. The elution profile was monitored with a refractive index detector. Molecular weights of the glucan peaks were determined from a calibration curve of polystyrene (PS) standards (♦). Mw, weight-average molecular weight; Mn, number-average molecular weight.
Although Agt proteins encoded by the orthologous agtA genes in Aspergillus fungi are thought to be GPI-anchored α-amylases, their function may be related to the biosynthesis of cell wall α-1,3-glucan rather than to starch catabolism (van der Kaaij et al., 2007; He et al., 2014; He et al., 2017; Miyazawa et al., 2022). In the present study, similarly to amyD in A. nidulans (He et al., 2014; He et al., 2017; Miyazawa et al., 2022), agtA overexpression in A. oryzae decreased the amount of α-1,3-glucan in the cell wall (Figure 2), suggesting that AoAgtA suppresses cell wall α-1,3-glucan biosynthesis in A. oryzae. We also investigated the enzymatic properties of rAoAgtA to further understand its contribution to cell wall α-1,3-glucan biosynthesis.
Among several types of glucans, rAoAgtA degraded only soluble starch (Supplementary Table 6). Therefore, AoAgtA appeared to act on α-1,4-glycosidic linkages and may degrade the spacer composed of α-1,4-linked glucose residues that is thought to be incorporated into cell wall α-1,3-glucan (Figure 1) during its biosynthesis. We analyzed the modes of bond cleavage by rAoAgtA, including hydrolysis and transglycosylation, using maltooligosaccharide substrates. rAoAgtA showed an endo-type cleavage mode for substrates with a DP at least 5 (Figure 4). The weak activity of rAoAgtA on soluble starch and its substrate specificity for maltooligosaccharide substrates were consistent with the properties of AnAgtA of A. niger, which also has 4-α-glucanotransferase activity and is expected to belong to a new subgroup of GH13 (van der Kaaij et al., 2007). Although the length (DP) of the spacer has not been clarified, if AoAgtA cleaves the spacer, its DP should be at least 5, as suggested by the failure of rAoAgtA to degrade nigeran, a polysaccharide consisting of alternating α-1,3- and α-1,4-linked glucose residues (Supplementary Table 6), and by the modes of bond cleavage in maltooligosaccharide substrates (Figure 4). However, the kinetic studies of PNM degradation (Table 3) demonstrated an extremely low rAoAgtA activity for maltooligosaccharide substrates compared with other known α-amylases. The reason for such low activity may be a replacement of a His residue in Region I of the Agt proteins, which was also mentioned in the case of AnAgtA by van der Kaaij et al. (2007) (Supplementary Table 7). This residue is particularly conserved among the members of the α-amylase family (Svensson, 1994; Kuriki et al., 2006), and its mutation reportedly decreases enzyme activity and alters the type of reaction catalyzed by the enzyme (Nakamura et al., 1993; Svensson, 1994; Chang et al., 2003; Leemhuis et al., 2004). This His residue is changed to Asn in AoAgtA (Supplementary Table 7). The enzymatic characteristics of rAoAgtA might reflect the evolutionary divergence of the functions of Agt proteins from those of α-amylases highly active on starch.
We consider that Agt proteins are specialized in cleaving α-1,4-linked oligosaccharides or glucan with α-1,3-glycosidic bonds on the side of the reducing end or both sides in vivo. As preparation of such predicted substrates is currently difficult, we examined the effect of nigerooligosaccharides on rAoAgtA catalytic activity and found that they increased Mal5-α-pNP-degradation (Supplementary Figure 9). AnAgtA can use nigerooligosaccharides with a small DP as acceptors for transglycosylation (van der Kaaij et al., 2007). We predicted that rAoAgtA also conducts transglycosylation with nigerooligosaccharides as acceptors. In fact, 3-α-maltooligosylglucose was synthesized by rAoAgtA transglycosylation activity with Mal5-α-pNP as a donor and nigerose as an acceptor (Supplementary Tables 2, 3). The transfer of maltooligosaccharide to nigerooligosaccharide with a formation of an α-1,4-glycosidic bond, which is catalyzed by rAoAgtA, has not been known for other α-amylases. The study of AnAgtA has not elucidated the chemical structures of the transglycosylation products with nigerooligosaccharides as acceptors (van der Kaaij et al., 2007). The degradation of Mal5-α-pNP catalyzed by rAoAgtA likely proceeds via a covalent glycosyl (mainly Mal3)–enzyme intermediate. We expect that such an intermediate would be more vulnerable to nucleophilic attack with oligosaccharides than with H2O.
The structure of 3-α-maltooligosylglucose containing both nigerooligosaccharide and maltooligosaccharide components is present as parts of the structure in the speculative cell wall α-1,3-glucan structure. In fact, rAoAgtA was able to cleave Mal4α1,3Glc (Figure 4). The activity of rAoAgtA with Mal4α1,3Glc was weak, with a slower substrate-degradation velocity than that for Mal5. The length of the maltooligosaccharide moiety of the Mal4α1,3Glc might be insufficient for the maximum activity of rAoAgtA, as mentioned above. Interestingly, we found that the rAoAgtA mode of bond cleavage in Mal4α1,3Glc clearly differed from that in Mal5 (Figure 4).
We here propose a hypothetical role of AoAgtA in cell wall α-1,3-glucan biosynthesis. rAoAgtA randomly cleaved only the α-1,4-glycosidic bonds of Mal4α1,3Glc (Figure 4), and the structure of Mal4α1,3Glc is similar to the predicted boundary structure between α-1,3-glucan main chain and the spacer in cell wall α-1,3-glucan synthesized by AgsB (Figure 7A). Miyazawa et al. (2022) suggested that AmyD of A. nidulans requires a GPI anchor to act on α-1,3-glucan in vivo and that AmyD reacts with cell wall α-1,3-glucan shortly after it is synthesized by α-1,3-glucan synthase on the plasma membrane. Therefore, we speculate that AoAgtA cleaves some of the spacers of cell wall α-1,3-glucan in the process of synthesis in vivo (Figure 7B). The random mode of α-1,4-glycosidic bond cleavage in Mal4α1,3Glc by rAoAgtA (Figure 4) supports the possibility that AoAgtA cleaves spacers in cell wall α-1,3-glucan. In fact, the MW of glucan in the AS2 fraction derived from the agtAOE strain of A. nidulans was significantly lower than in that from the parental stain (Figure 6B; Table 4). Thus, similar to AmyD (Miyazawa et al., 2022), AoAgtA decreased the MW of cell wall α-1,3-glucan in vivo. The MW of glucan in the AS2 fraction extracted from agsBOEΔamyD was not affected by purified rAoAgtA (data not shown), perhaps because mature cell wall α-1,3-glucan is packed by hydrogen bonds, and its spacers cannot be accessed by the enzyme. To prove that AoAgtA acts on cell wall α-1,3-glucan in vitro, it is necessary to establish a new evaluation system.
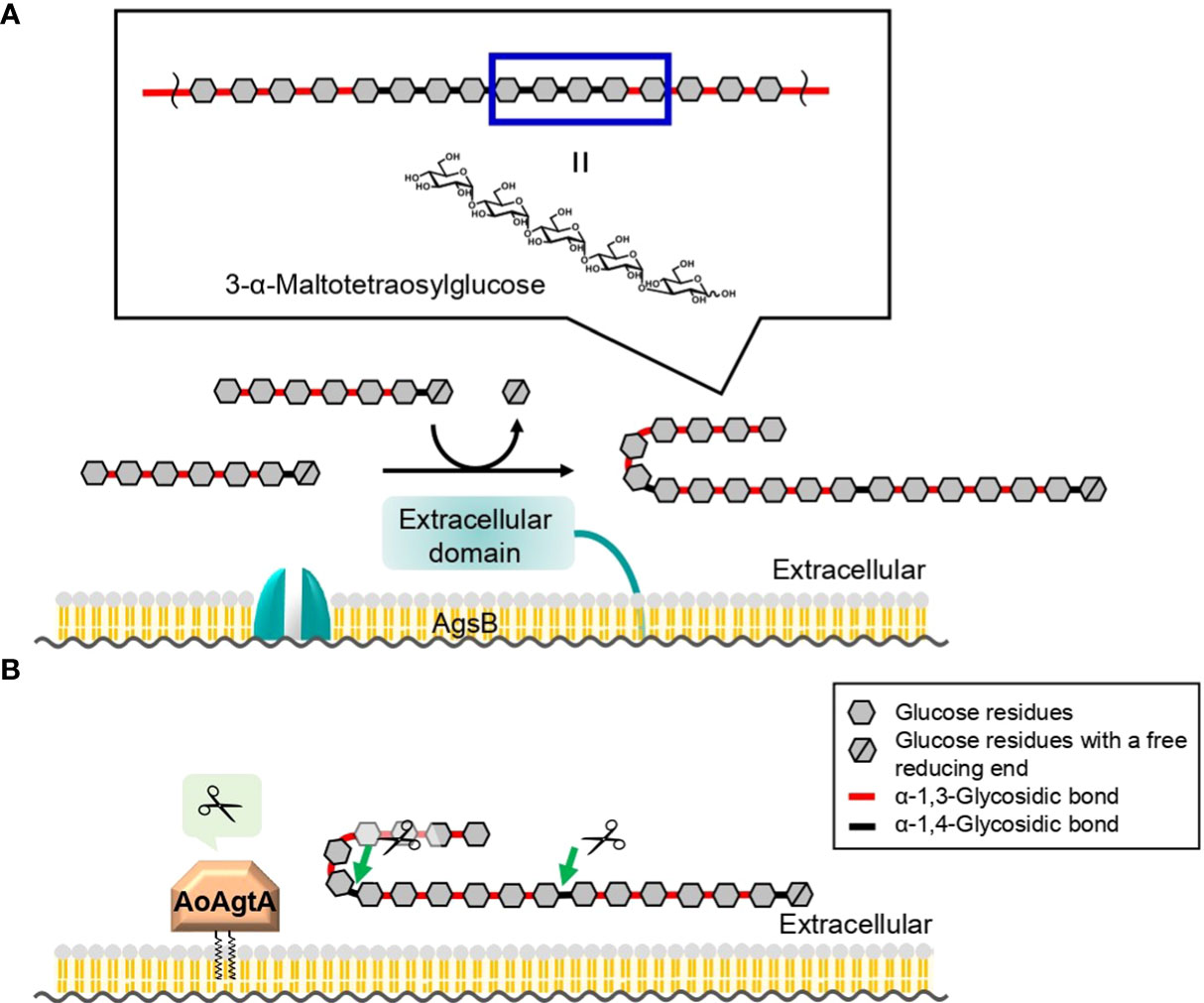
Figure 7 Predicted role of AoAgtA in cell wall α-1,3-glucan biosynthesis in A. oryzae. (A) During biosynthesis, α-1,3-glucan subunits may be interconnected by transglycosylation catalyzed by the extracellular domain of AgsB; the structure of 3-α-maltotetraosylglucose is consistent with the structure in which α-1,3-glucan subunits are linked. (B) We speculate that AoAgtA cleaves the spacers composed of α-1,4-linked glucose residues in cell wall α-1,3-glucan before its insolubilization.
In the present study, we showed that overexpression of the agtA gene in A. oryzae decreased the amount of α-1,3-glucan in the cell wall, suggesting that AoAgtA suppresses cell wall α-1,3-glucan biosynthesis. Analysis using the agtAOE strain of A. nidulans showed that AoAgtA decreases the MW of cell wall α-1,3-glucan. Purified rAoAgtA randomly cleaved the α-1,4-glycosidic bonds of Mal4α1,3Glc, which can be considered as part of the structure of cell wall α-1,3-glucan. We conclude that AoAgtA likely cleaves the spacers composed of α-1,4-linked glucose residues in cell wall α-1,3-glucan before its insolubilization. Taken together with the results of in vivo functional analysis of the amyD gene of A. nidulans (Miyazawa et al., 2022), the present study suggests that AoAgtA plays an important role in the biosynthesis of cell wall α-1,3-glucan.
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
AK, KM, AY, and KA conceived and designed the experiments. AK performed most experiments and analyzed the data. AK and MS constructed the A. oryzae mutants. AK, YT, SY, and MS constructed the P. pastoris strain and purified rAoAgtA. AK and MO performed enzymatic assays and chemical analyses of oligosaccharides. AK and KM constructed the A. nidulans mutants. TN and HN produced nigerooligosaccharides. SK and TI enzymatically produced bacterial α-1,3-glucan. MH performed structural analysis of AoAgtA. KA supervised this research and acquired funding. AK, KM, MO, AY, and KA wrote the paper. All authors contributed to the article and approved the submitted version.
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Nos. JP20H02895, JP26292037 (KA), JP18K05384 (AY), JP18J11870, and JP20K22773 (KM). This work was also supported by the Institute for Fermentation, Osaka (Grant No. L-2018–2–014) (KA), and New Energy and Industrial Technology Development Organization (NEDO), JPNP20011 (KA).
We are grateful to Kikkoman Corporation for providing maltooligosaccharides. We thank Professor Emeritus Tasuku Nakajima (Tohoku University) for providing pullulan and for critical discussion on the research, Dr. Keiko Uechi (University of the Ryukyus) for providing nigeran, and Dr. Daisuke Sugimori (Fukushima University) for lending equipment.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffunb.2022.1061841/full#supplementary-material
Beauvais A., Bozza S., Kniemeyer O., Formosa C., Balloy V., Henry C., et al. (2013). Deletion of the α-(1,3)-glucan synthase genes induces a restructuring of the conidial cell wall responsible for the avirulence of Aspergillus fumigatus. PloS Pathog. 9, e1003716. doi: 10.1371/journal.ppat.1003716
Chang C. T., Lo H. F., Chi M. C., Yao C. Y., Hsu W. H., Lin L. L. (2003). Identification of essential histidine residues in a recombinant α-amylase of thermophilic and alkaliphilic Bacillus sp. strain TS-23. Extremophiles 7, 505–509. doi: 10.1007/s00792-003-0341-8
Choma A., Wiater A., Komaniecka I., Paduch R., Pleszczyńska M., Szczodrak J. (2013). Chemical characterization of a water insoluble (1 → 3)-α-D-glucan from an alkaline extract of Aspergillus wentii. Carbohydr. Polym. 91, 603–608. doi: 10.1016/j.carbpol.2012.08.060
Czerwonka A., Wiater A., Komaniecka I., Adamczyk P., Rzeski W., Pleszczyńska M. (2019). Antitumour effect of glucooligosaccharides obtained via hydrolysis of α-(1 → 3)-glucan from Fomitopsis betulina. Mol. Biol. Rep. 46, 5977–5982. doi: 10.1007/s11033-019-05032-x
Doner L. W., Irwin P. L. (1992). Assay of reducing end-groups in oligosaccharide homologues with 2,2’-bicinchoninate. Anal. Biochem. 202, 50–53. doi: 10.1016/0003-2697(92)90204-k
Fujikawa T., Kuga Y., Yano S., Yoshimi A., Tachiki T., Abe K., et al. (2009). Dynamics of cell wall components of Magnaporthe grisea during infectious structure development. Mol. Microbiol. 73, 553–570. doi: 10.1111/j.1365-2958.2009.06786.x
Fujikawa T., Sakaguchi A., Nishizawa Y., Kouzai Y., Minami E., Yano S., et al. (2012). Surface α-1,3-glucan facilitates fungal stealth infection by interfering with innate immunity in plants. PloS Pathog. 8, e1002882. doi: 10.1371/journal.ppat.1002882
Grün C. H., Hochstenbach F., Humbel B. M., Verkleij A. J., Sietsma J. H., Klis F. M., et al. (2005). The structure of cell wall α-glucan from fission yeast. Glycobiology 15, 245–257. doi: 10.1093/glycob/cwi002
Hara S., Tsuji R. F., Hatamoto O., Masuda T. (2002). A simple method for enrichment of uninucleate conidia of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 66, 693–696. doi: 10.1271/bbb.66.693
Hattori T., Kato Y., Uno S., Usui T. (2013). Mode of action of a β-(1→6)-glucanase from Penicillium multicolor. Carbohydr. Res. 366, 6–16. doi: 10.1016/j.carres.2012.11.002
He X., Li S., Kaminskyj S. G. W. (2014). Characterization of Aspergillus nidulans α-glucan synthesis: roles for two synthases and two amylases. Mol. Microbiol. 91, 579–595. doi: 10.1111/mmi.12480
He X., Li S., Kaminskyj S. (2017). An amylase-like protein, AmyD, is the major negative regulator for α-glucan synthesis in Aspergillus nidulans during the asexual life cycle. Int. J. Mol. Sci. 18, 695. doi: 10.3390/ijms18040695
Ichikawa H., Miyazawa K., Komeiji K., Susukida S., Zhang S., Muto K., et al. (2022). Improved recombinant protein production in Aspergillus oryzae lacking both α-1,3-glucan and galactosaminogalactan in batch culture with a lab-scale bioreactor. J. Biosci. Bioeng. 133, 39–45. doi: 10.1016/j.jbiosc.2021.09.010
Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kazim A. R. S., Jiang Y., Li S., He X. (2021). Aspergillus nidulans AmyG functions as an intracellular α-amylase to promote α-glucan synthesis. Microbiol. Spectr. 9, e00644–e00621. doi: 10.1128/Spectrum.00644-21
Kuriki T., Takata H., Yanase M., Ohdan K., Fujii K., Terada Y., et al. (2006). The concept of the α-amylase family: a rational tool for interconverting glucanohydrolases/glucanotransferases, and their specificities. J. Appl. Glycosci. 53, 155–161. doi: 10.5458/jag.53.155
Laemmli U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. doi: 10.1038/227680a0
Latgé J. P. (2010). Tasting the fungal cell wall. Cell. Microbiol. 12, 863–872. doi: 10.1111/j.1462-5822.2010.01474.x
Leemhuis H., Wehmeier U. F., Dijkhuizen L. (2004). Single amino acid mutations interchange the reaction specificities of cyclodextrin glycosyltransferase and the acarbose-modifying enzyme acarviosyl transferase. Biochemistry 43, 13204–13213. doi: 10.1021/bi049015q
Minetoki T., Tsuboi H., Koda A., Ozeki K. (2003). Development of high expression system with the improved promoter using the cis-acting element in Aspergillus species. J. Biol. Macromol. 3, 89–96.
Miyazawa K., Yamashita T., Takeuchi A., Kamachi Y., Yoshimi A., Tashiro Y., et al. (2022). A glycosylphosphatidylinositol-anchored α-amylase encoded by amyD contributes to a decrease in the molecular mass of cell wall α-1,3-glucan in Aspergillus nidulans. Front. Fungal Biol. 2. doi: 10.3389/ffunb.2021.821946
Miyazawa K., Yoshimi A., Abe K. (2020). The mechanisms of hyphal pellet formation mediated by polysaccharides, α-1,3-glucan and galactosaminogalactan, in Aspergillus species. Fungal Biol. Biotechnol. 7, 10. doi: 10.1186/s40694-020-00101-4
Miyazawa K., Yoshimi A., Kasahara S., Sugahara A., Koizumi A., Yano S., et al. (2018). Molecular mass and localization of α-1,3-glucan in cell wall control the degree of hyphal aggregation in liquid culture of Aspergillus nidulans. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02623
Miyazawa K., Yoshimi A., Sano M., Tabata F., Sugahara A., Kasahara S., et al. (2019). Both galactosaminogalactan and α-1,3-glucan contribute to aggregation of Aspergillus oryzae hyphae in liquid culture. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.02090
Miyazawa K., Yoshimi A., Zhang S., Sano M., Nakayama M., Gomi K., et al. (2016). Increased enzyme production under liquid culture conditions in the industrial fungus Aspergillus oryzae by disruption of the genes encoding cell wall α-1,3-glucan synthase. Biosci. Biotechnol. Biochem. 80, 1853–1863. doi: 10.1080/09168451.2016.1209968
Mizutani O., Kudo Y., Saito A., Matsuura T., Inoue H., Abe K., et al. (2008). A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet. Biol. 45, 878–889. doi: 10.1016/j.fgb.2007.12.010
Nakamura A., Haga K., Yamane K. (1993). Three histidine residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of the replacement on pH dependence and transition-state stabilization. Biochemistry 32, 6624–6631. doi: 10.1021/bi00077a015
Nihira T., Nishimoto M., Nakai H., Ohtsubo K., Kitaoka M. (2014). Characterization of two α-1,3-glucoside phosphorylases from Clostridium phytofermentans. J. Appl. Glycosci. 61, 59–66. doi: 10.5458/jag.jag.JAG-2013_013
Nitta Y., Mizushima M., Hiromi K., Ono S. (1971). Influence of molecular structures of substrates and analogues on taka-amylase a catalyzed hydrolyses. i. effect of chain length of linear substrates. J. Biochem. 69, 567–576.
Okada M., Ogawa K., Usui T. (2000). Substrate specificity of human and bovine pancreatic α-amylases using end-blocked maltooligosaccharides. J. Appl. Glycosci. 47, 35–44. doi: 10.5458/jag.47.35
Puanglek S., Kimura S., Enomoto-Rogers Y., Kabe T., Yoshida M., Wada M., et al. (2016). In vitro synthesis of linear α-1,3-glucan and chemical modification to ester derivatives exhibiting outstanding thermal properties. Sci. Rep. 6, 30479. doi: 10.1038/srep30479
Rappleye C. A., Eissenberg L. G., Goldman W. E. (2007). Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc. Natl. Acad. Sci. U. S. A. 104, 1366–1370. doi: 10.1073/pnas.0609848104
Rappleye C. A., Engle J. T., Goldman W. E. (2004). RNA Interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol. Microbiol. 53, 153–165. doi: 10.1111/j.1365-2958.2004.04131.x
Suganuma T. (1983). Subsite structures and transglycosylation of taka-amylase a and Bacillus subtilis saccharifying α-amylase. J. Jpn. Soc Starch Sci. 30, 149–158. doi: 10.5458/jag1972.30.149
Svensson B. (1994). Protein engineering in the α-amylase family: Catalytic mechanism, substrate specificity, and stability. Plant Mol. Biol. 25, 141–157. doi: 10.1007/BF00023233
Tamano K., Satoh Y., Ishii T., Terabayashi Y., Ohtaki S., Sano M., et al. (2007). The β-1,3-exoglucanase gene exgA (exg1) of Aspergillus oryzae is required to catabolize extracellular glucan, and is induced in growth on a solid surface. Biosci. Biotechnol. Biochem. 71, 926–934. doi: 10.1271/bbb.60591
Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Žídek A., et al. (2021). Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596. doi: 10.1038/s41586-021-03828-1
Usui T., Murata T. (1988). Enzymatic synthesis of p-nitrophenyl α-maltopentaoside in an aqueous-methanol solvent system by maltotetraose-forming amylase: a substrate for human amylase in serum. J. Biochem. 103, 969–972. doi: 10.1093/oxfordjournals.jbchem.a122395
Usui T., Ogawa K., Nagai H., Matsui H. (1992). Enzymatic synthesis of p-nitrophenyl 45-O-β-D-galactosyl-α-maltopentaoside as a substrate for human α-amylases. Anal. Biochem. 202, 61–67. doi: 10.1016/0003-2697(92)90206-m
Utsumi Y., Yoshida M., Francisco P. B. J., Sawada T., Kitamura S., Nakamura Y. (2009). Quantitative assay method for starch branching enzyme with bicinchoninic acid by measuring the reducing terminals of glucans. J. Appl. Glycosci. 56, 215–222. doi: 10.5458/jag.56.215
van der Kaaij R. M., Yuan X. L., Franken A., Ram A. F. J., Punt P. J., van der Maarel M. J. E. C., et al. (2007). Two novel, putatively cell wall-associated and glycosylphosphatidylinositol-anchored α-glucanotransferase enzymes of Aspergillus niger. Eukaryot. Cell 6, 1178–1188. doi: 10.1128/EC.00354-06
Vujičić-Žagar A., Dijkstra B. W. (2006). Monoclinic crystal form of Aspergillus niger α-amylase in complex with maltose at 1.8 Å resolution. Acta Crystallogr. Sect. F: Struct. Biol. Cryst. Commun. 62, 716–721. doi: 10.1107/S1744309106024729
Yoshimi A., Miyazawa K., Abe K. (2016). Cell wall structure and biogenesis in Aspergillus species. Biosci. Biotechnol. Biochem. 80, 1700–1711. doi: 10.1080/09168451.2016.1177446
Yoshimi A., Miyazawa K., Abe K. (2017). Function and biosynthesis of cell wall α-1,3-glucan in fungi. J. Fungi 3, 63. doi: 10.3390/jof3040063
Yoshimi A., Sano M., Inaba A., Kokubun Y., Fujioka T., Mizutani O., et al. (2013). Functional analysis of the α-1,3-glucan synthase genes agsA and agsB in Aspergillus nidulans: AgsB is the major α-1,3-glucan synthase in this fungus. PloS One 8, e54893. doi: 10.1371/journal.pone.0054893
Keywords: α-amylase, cell wall, α-1,3-glucan, glycosylphosphatidylinositol-anchored protein, filamentous fungi, Aspergillus oryzae, oligosaccharide
Citation: Koizumi A, Miyazawa K, Ogata M, Takahashi Y, Yano S, Yoshimi A, Sano M, Hidaka M, Nihira T, Nakai H, Kimura S, Iwata T and Abe K (2023) Cleavage of α-1,4-glycosidic linkages by the glycosylphosphatidylinositol-anchored α-amylase AgtA decreases the molecular weight of cell wall α-1,3-glucan in Aspergillus oryzae. Front. Fungal Biol. 3:1061841. doi: 10.3389/ffunb.2022.1061841
Received: 05 October 2022; Accepted: 29 November 2022;
Published: 10 January 2023.
Edited by:
Tamás Papp, University of Szeged, HungaryReviewed by:
Miklós Takó, University of Szeged, HungaryCopyright © 2023 Koizumi, Miyazawa, Ogata, Takahashi, Yano, Yoshimi, Sano, Hidaka, Nihira, Nakai, Kimura, Iwata and Abe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Keietsu Abe, a2VpZXRzdS5hYmUuYjVAdG9ob2t1LmFjLmpw
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.