
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Fungal Biol. , 24 October 2022
Sec. Fungal Pathogenesis
Volume 3 - 2022 | https://doi.org/10.3389/ffunb.2022.1010782
This article is part of the Research Topic Emergence of Antifungal Resistance: an international call for prevention View all 5 articles
 Pilar Escribano1,2
Pilar Escribano1,2 Jesús Guinea1,2,3*
Jesús Guinea1,2,3*Candida parapsilosis is a leading cause of invasive candidiasis in southern Europe, Latin America and Asia. C. parapsilosis has been mostly considered susceptible to triazoles, but fluconazole resistance is on the rise in some countries. The main mechanism related to fluconazole resistance is the presence of ERG11p substitutions, dominated by the Y132F amino acid substitution. Isolates harbouring this substitution mimic C. auris given that they may cause hospital outbreaks, become endemic, and emerge simultaneously in distant areas around the world. At the moment, Spain is experiencing a brusque emergence of fluconazole resistance in C. parapsilosis; isolates harbouring the Y132F substitution were detected for the first time in 2019. A recent study on Candida spp isolates from blood cultures collected in 16 hospitals located in the Madrid metropolitan area (2019 to 2021) reported that fluconazole resistance in C. parapsilosis reached as high as 13.6%. Resistance rates rose significantly during those three years: 3.8% in 2019, 5.7% in 2020, and 29.1% in 2021; resistant isolates harboured either the dominant Y132F substitution (a single clone found in four hospitals) or G458S (another clone found in a fifth hospital). The COVID-19 pandemic may have increased the number of candidaemia cases. The reason for such an increase might be a consequence of uncontrolled intra-hospital patient-to-patient transmission in some hospitals, as an increase not only in C. parapsilosis candidaemia episodes but also in the spread of clonal fluconazole-resistant isolates might have occurred in other hospitals during the pandemic period. Patients affected with fluconazole-resistant C. parapsilosis harbouring the Y132F substitution presented a mortality rate ranging from 9% to 78%, were mainly admitted to intensive care wards but did not have differential risk factors compared to those infected by susceptible isolates. With scarce exceptions, few patients (≤20%) infected with fluconazole-resistant isolates had previously received fluconazole, thus supporting the fact that, although fluconazole might have been a key factor to promote resistance, the main driver promoting the spread of fluconazole-resistant isolates was patient-to-patient transmission.
Candida parapsilosis is a leading cause of invasive candidiasis, particularly in southern Europe, Latin America and Asia (Nucci et al., 2010; Guinea et al., 2014; Guo et al., 2021). A population-based study conducted in Spain showed that C. parapsilosis accounted for over 25% of all candidaemia isolates collected (Guinea, 2014). A single-hospital study confirmed a similar and steady-over-time proportion of episodes of candidaemia caused by C. parapsilosis in patients admitted to a hospital located in Madrid from 2007 to 2019; the same observation was recently confirmed in a multi-centre study involving 16 hospitals located in the same city (Diaz-Garcia et al., 2021b, Díaz-García et al., 2022b). The reasons why C. parapsilosis is a frequent cause of candidaemia in some geographic areas is still unknown. Since C. parapsilosis represented 27% of isolates from blood cultures but only 2.6% from intra-abdominal samples (Diaz-Garcia et al., 2021a), the species seems to be prone to cause catheter-related candidaemia (Puig-Asensio et al., 2014).
C. parapsilosis has been mostly considered susceptible to fluconazole and newer triazoles, however, the number of reported fluconazole-resistant isolates has been increasing simultaneously in different countries in the last few years. Azole resistance in C. parapsilosis is a matter of concern given the intrinsic diminished susceptibility of the species to echinocandins (Diaz-Garcia et al., 2021b). The environmental trait of C. parapsilosis, its high ability to form biofilms, and its potential to promote fluconazole resistance may explain the spread of fluconazole-resistant isolates across some hospitals, to the point of becoming endemic and a public health problem.
The present review covers the state-of-the-art of the emerging threat concerning fluconazole resistance in C. parapsilosis. The topics revised include an overview of the fluconazole resistance rates in C. parapsilosis, the dominant underlying mechanisms of resistance, the spread and tracking of resistant isolates, and the clinical impact of resistance. We performed a literature search in PubMed using the following keywords: “Candida”, “parapsilosis”, “fluconazole”, and “resistance”. Of the studies found (n=884), for the current review, we selected those reporting fluconazole antifungal susceptibility data, characterization of resistance mechanisms, and genotyping (n=38).
C. parapsilosis has been historically reported as susceptible to fluconazole; however, several reports have warned of the recent increasing rate of fluconazole resistance in some geographic regions. A study conducted on isolates collected worldwide between 1997 and 2016 reported a fluconazole resistance rate of 3.9% (Pfaller et al., 2019). Likewise, a population-based study conducted in Spain in 2010 and 2011 reported a low rate of fluconazole resistance (2.5%) in that species (Guinea et al., 2014). Other studies reported similar rates in Italy (3.6%) (Prigitano et al., 2016), Portugal (4%) (Faria-Ramos et al., 2014), and Denmark (6%) (Arendrup et al., 2011).
However, multi-centre studies may overshadow local epidemiology. For example, high azole resistance rates have been reported in single-centre studies recently conducted in France (9.2%) (Fekkar et al., 2021), Turkey (26.4%) (Arastehfar et al., 2020a), Italy (33%) (Mesini et al., 2020), Saudi Arabia (33%) (Aldardeer et al., 2020), Mexico (54%) (Corzo-Leon et al., 2021), Brazil (67.9%) (Thomaz et al., 2021), and South Africa (78%) (Magobo et al., 2020). Local epidemiology among hospitals located in the same area may differ. For example, fluconazole-resistant C. parapsilosis was not detected in patients admitted to the Gregorio Marañón hospital (Madrid, Spain), whereas other hospitals located in the same region reported disparate resistance rates (Diaz-Garcia et al., 2021a, Diaz-Garcia et al., 2021b, Diaz-Garcia et al., 2022b). Such notable differences among centres can be the consequence of infection control policies, prior use of azoles, and the fluconazole-resistant clones spreading across hospitals.
Fluconazole prevents fungal Candida spp cell growth by inhibiting lanosterol 14-α demethylase (ERG11p), a protein encoded by the ERG11 gene, which leads to a blockade of ergosterol synthesis, an essential component of fungal cell membranes (Grossman et al., 2015). Fluconazole resistance mechanisms are well known in C. albicans and their understanding has been helpful to bridge the knowledge gap in species such as C. parapsilosis.
Fluconazole resistance mechanisms may work alone or simultaneously. A major mechanism of resistance in C. albicans involves the ERG11 gene, in which the presence of some substitutions may lead to either a reduced affinity of ERG11p for the drug (Sanglard and Odds, 2002) or ERG11 gene up-regulation when mutations occur in the UPC2 gene encoding the transcriptional regulator of sterol biosynthesis genes (Schneider and Morschhauser, 2015). Some substitutions in the ERG3 gene (encoding a sterol-desaturase) may lead to loss of function of the enzyme, promote the accumulation of deleterious sterols, and allow the fungus to survive in the presence of fluconazole; however, these mechanisms are much less frequent (Morio et al., 2012). The other main mechanism involves the presence of mutations in the TAC1 (transcriptional activator of CDR genes) and MRR1 (transcription factor and multidrug resistance regulator) genes, which lead to overexpression of CDR and MDR1, respectively, resulting in the pumping of fluconazole out of the cell (Morschhauser et al., 2007; Liu and Myers, 2017).
The dominant mechanism of fluconazole resistance in C. parapsilosis is the presence of ERG11p substitutions (Table 1). Y132F is the dominant amino acid substitution and has also been described in C. albicans, C. tropicalis, and C. auris (Morio et al., 2010; Jiang et al., 2013; Healey et al., 2018). C. parapsilosis isolates harbouring the Y132F ERG11p substitution have been recently reported in several countries (Table 2 and Figure 1). The first isolate was detected in Turkey in 2004 followed by other countries, with Spain being the last to be added to the list in 2019. The number of affected countries is expected to increase in the near future.
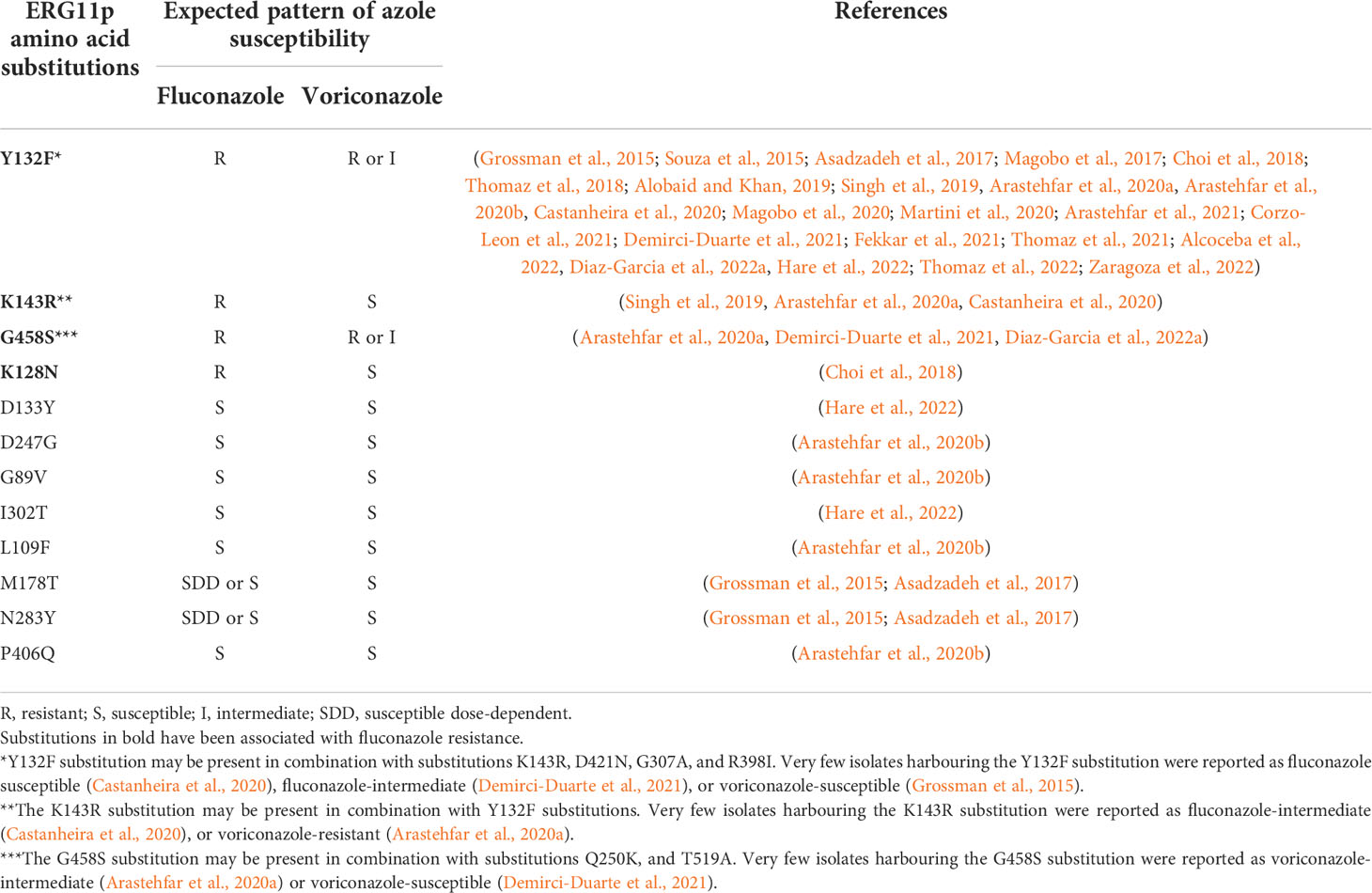
Table 1 ERG11p substitutions reported in C. parapsilosis isolates and their fluconazole resistance profile.
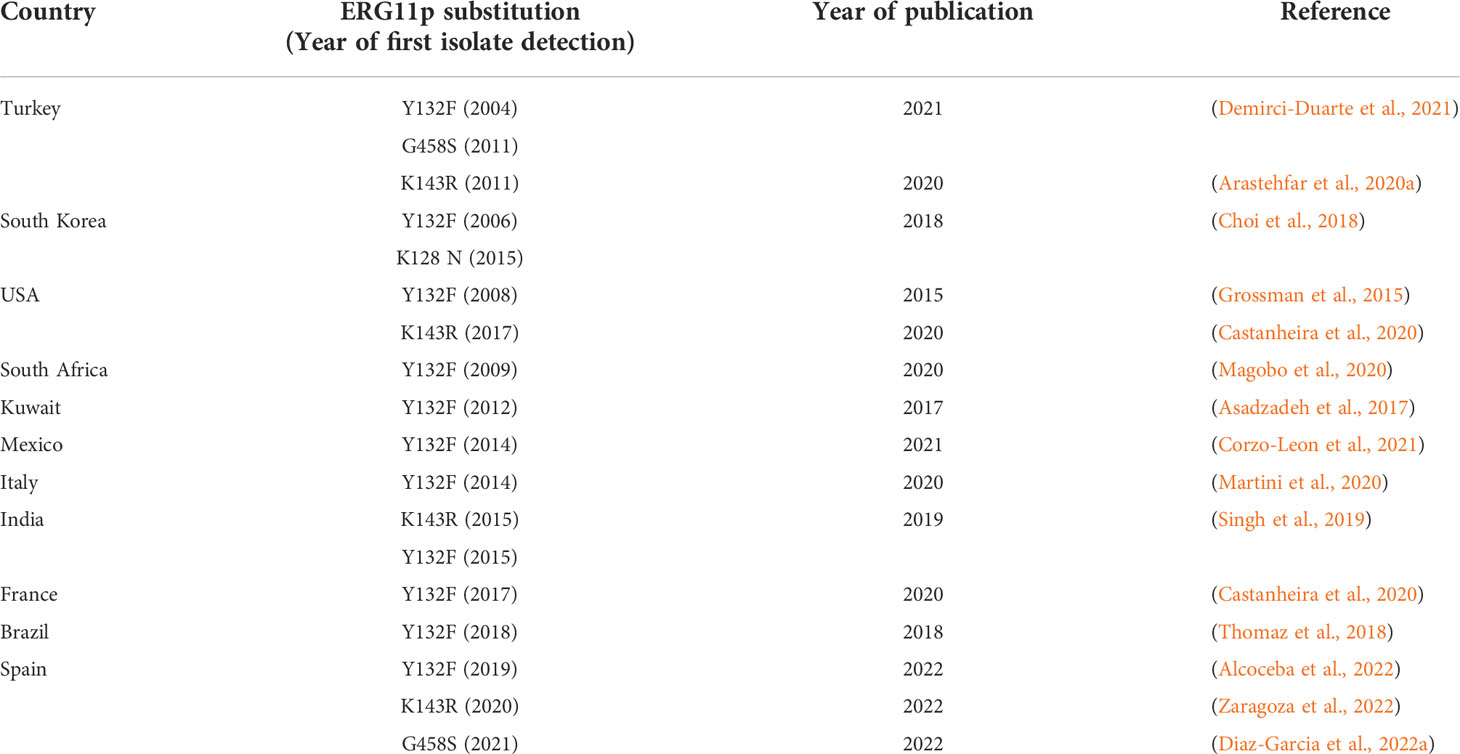
Table 2 Studies in which fluconazole-resistant C. parapsilosis isolates harbouring Y132F, K143R, G458S, and K128N ERG11p substitutions were reported for the first time (year of detection) in the countries affected.
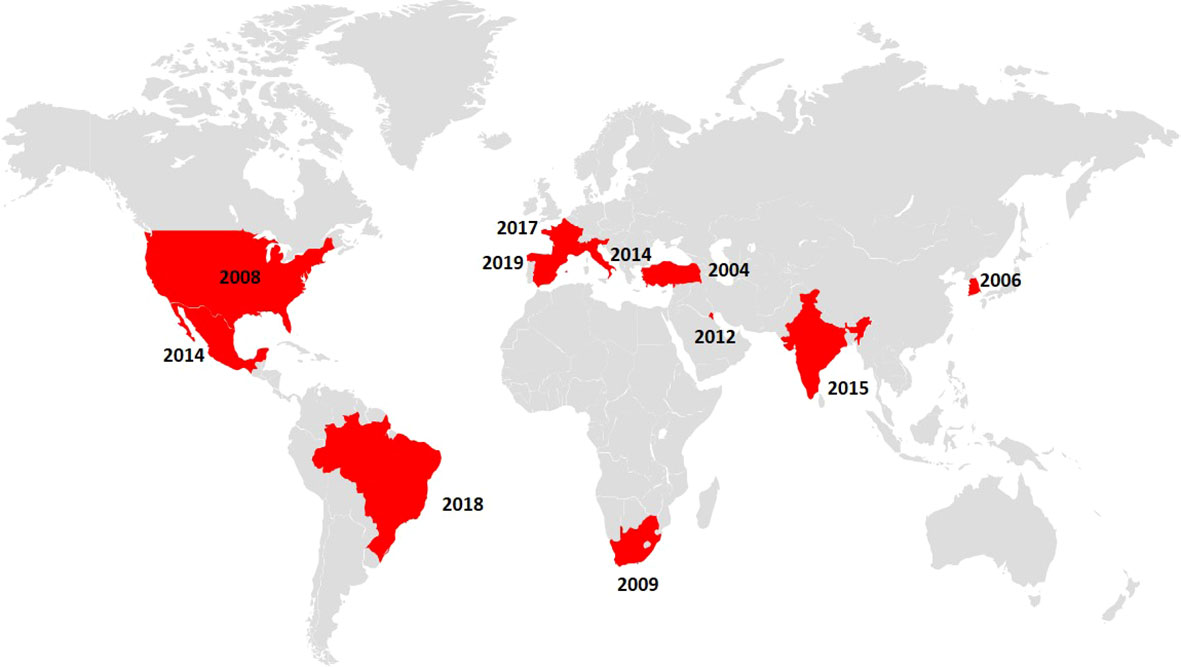
Figure 1 Map indicating the countries in which fluconazole-resistant C. parapsilosis isolates harbouring the Y132F ERG11p substitution were reported (depicted in red) alongside the year of detection of the first isolate.
The presence of G458S, K128N, and K143R ERG11p substitutions has been also associated with fluconazole resistance (Table 1). Substitutions can sometimes be found in combination; among others, the R398I substitution, as well as silent mutations, have been previously described in both susceptible and resistant isolates. Therefore, the role of these “accompanying” or “compensatory” mutations is uncertain and may represent mere polymorphisms, not necessarily associated with resistance (Table 1) (Grossman et al., 2015; Singh et al., 2019).
The profile of azole resistance is influenced by the type of amino acid substitution, although the data available is very limited. Isolates harbouring ERG11p substitutions are fully susceptible to amphotericin, micafungin and anidulafungin, and ibrexafungerp also (Diaz-Garcia et al., 2022b). Isolates harbouring the Y132F substitution are fluconazole-resistant and the vast majority are either voriconazole-resistant or voriconazole-intermediate (Table 1); one study reported that some isolates could also be non-wild type to posaconazole or isavuconazole (Diaz-Garcia et al., 2022a). Isolates harbouring the G458S substitution are fluconazole-resistant and mostly voriconazole-resistant (Table 1); one study reported that all isolates were non-wild type to posaconazole or isavuconazole, thus suggesting a pan-azole resistance phenotype (Table 1) (Diaz-Garcia et al., 2022a). Isolates with the K143R substitution are fluconazole-resistant and mostly voriconazole-susceptible and posaconazole and isavuconazole wild type (Table 1). Finally, isolates with the K128N substitution were reported in a single study as fluconazole-resistant and voriconazole-susceptible (Table 1).
Some fluconazole-resistant C. parapsilosis isolates presented a wild-type ERG11 gene sequence (Arastehfar et al., 2020a, Arastehfar et al., 2020b), thus suggesting alternative mechanisms of resistance. At this point in time, such alternative mechanisms play an uncertain role in azole resistance. Missense mutations in the transcriptional regulators TAC1 and MRR1 do not necessarily correlate with the overexpression of CDR1 and MDR1 (Berkow et al., 2015; Grossman et al., 2015; Asadzadeh et al., 2021). The genome plasticity and aneuploidy of C. parapsilosis can promote cell growth in the presence of azoles and have been suggested as a resistance mechanism (Yang et al., 2021).
Candida spp can commonly cause outbreaks of candidaemia in patients admitted to intensive care units and post-surgical units (Guducuoglu et al., 2016; Guinea et al., 2021). C. parapsilosis outbreaks have been reported in adult/neonatal intensive care units, frequently associated with patient-to-patient transmission (Clark et al., 2004; Diab-Elschahawi et al., 2012). In the event of an outbreak, the study of infecting isolates and environmental isolates, collected from the vicinity of the patient, is needed to abate the outbreak.
Genotyping of isolates using highly discriminative tools may be useful to unravel the outbreak source and support infection control investigations. This approach might be particularly useful in hospital wards with a high number of candidaemia episodes. A variety of methods are available for genotyping, such as Southern blot hybridization with specific probes, electrophoretic karyotyping, amplified fragment length polymorphism (AFLP) analysis, restriction fragment length polymorphism (RFLP) analysis, randomly amplified polymorphism DNA (RAPD) analysis, multilocus sequence typing, and microsatellites, however, there is no ‘gold standard’ as yet (Marcos-Zambrano et al., 2014). Microsatellites are an attractive method and species-specific panels have been developed for C. parapsilosis (Lasker et al., 2006; Sabino et al., 2010; Diab-Elschahawi et al., 2012).
We have been genotyping Candida spp isolates from blood cultures for the last 20 years; some genotypes may be found only in a given patient (singleton genotypes) whereas others may be found in two or more patients (clusters) (Guinea et al., 2020). The presence of clusters could suggest a common source of infection or patient-to-patient transmission and cause infections in the form of outbreaks. Clusters may go unnoticed and are only unveiled by blindly genotyping consecutive isolates causing candidaemia (Escribano et al., 2013). A high number of clusters could indicate high patient-to-patient transmission in hospital wards with a high incidence of candidaemia; as a matter of fact, the implementation of prevention campaigns regarding catheter-related infections in our hospital correlated with a decrease in both the number of candidaemia episodes and the number of C. albicans and C. parapsilosis clusters (Escribano et al., 2018).
We previously observed that the presence of C. albicans and C. parapsilosis clusters was not infrequent and both species fulfilled a species-specific pattern: while C. albicans clusters involved a limited number of patients and were limited in time, C. parapsilosis clusters involved a higher number of patients, could become endemic in the unit, and persist in the ward for years (Guinea et al., 2021). Not all clusters involved patients with an epidemiological relationship; some patients involved in a given cluster could be admitted to different hospital wards at the same hospital or even different hospitals (Escribano et al., 2018; Guinea et al., 2020). The reason for these “unexplained” clusters is unclear but they may represent genotypes prone to being widespread across different geographic regions. When isolates are tagged by a phenotypic characteristic, such as the presence of fluconazole resistance, isolate genotyping may be helpful to track them down and understand the dynamics of their transmission, as well as understanding the clones implicated.
Several studies have shown the ability of C. parapsilosis isolates mainly harbouring substitution Y132F ERG11p to cause hospital outbreaks (Grossman et al., 2015; Souza et al., 2015; Pinhati et al., 2016; Choi et al., 2018; Thomaz et al., 2018; Singh et al., 2019; Magobo et al., 2020; Martini et al., 2020; Arastehfar et al., 2021; Corzo-Leon et al., 2021; Fekkar et al., 2021; Thomaz et al., 2021). The clonal spread of fluconazole-resistant isolates is not surprising and follows the previously described pattern of C. parapsilosis isolates in the clinical setting. Some clusters harbouring the Y132F substitution became endemic in the hospital environment (Choi et al., 2018; Thomaz et al., 2018; Martini et al., 2020; Alcoceba et al., 2022; Ramos-Martinez et al., 2022). Sometimes, different C. parapsilosis genotypes harbouring the Y132F substitution coexisted within a particular hospital (Magobo et al., 2017; Choi et al., 2018; Arastehfar et al., 2021; Corzo-Leon et al., 2021; Thomaz et al., 2021). Conversely, a particular clone could be found in different hospitals (Grossman et al., 2015; Magobo et al., 2017; Zhang et al., 2020, Diaz-Garcia et al., 2022a, Thomaz et al., 2022). These observations may simply reflect transfers of patients among hospitals of a given region and the active spread of isolates across hospitals (Chow et al., 2018). Isolates harbouring the Y132F substitution mimic C. auris given that they may cause hospital outbreaks, become endemic, and emerge simultaneously in distant areas around the world.
Studies reporting isolates harbouring the remaining relevant ERG11p substitutions (K134R, G458S, and K128N) are very limited. One study reported a number of isolates harbouring the K134R substitution in patients admitted to Indian hospitals. These isolates followed a pattern similar to that found in isolates harbouring the Y132F substitution: different clones could be found in a given hospital, whereas a clone could be found in different hospitals (Singh et al., 2019). We recently conducted a surveillance study in the Madrid metropolitan area in which we reported the presence of isolates harbouring the G458S substitution in a single hospital (Diaz-Garcia et al., 2022a). Finally, to date, isolates harbouring the K128N substitution have not been reported as a cause of outbreaks (Choi et al., 2018).
A major limitation of previously reported studies lies in the fact that they were mostly conducted retrospectively. Furthermore, environmental isolates were only genotyped in a single study conducted in Brazil, which proved the presence of resistant isolates in the vicinity of the patients (Thomaz et al., 2021). Although limited, available data suggests that the niche of fluconazole-resistant C. parapsilosis isolates may be in the environment of the infected patients.
Spain is currently experiencing a brusque emergence of fluconazole resistance in C. parapsilosis. The rate of fluconazole resistance in C. parapsilosis reported in the CANDIPOP study conducted in Spain in 2010 and 2011 was 2.5% (Guinea et al., 2014). Ten years later, the presence of fluconazole-resistant C. parapsilosis isolates harbouring the Y132F substitution was reported for the first time in Spain, in isolates collected in 2019 in the Son Espases hospital, a public 750-bed tertiary referral hospital located in the Balearic Islands (Alcoceba et al., 2022). The fact that the first resistant isolate (fluconazole plastic strip MIC > 4 mg/L) was detected in October 2015 (isolates collected from 2015 to April 2019 were unfortunately unavailable) suggested that the onset of the outbreak was in 2015 rather than in 2019. Since then, the number of patients with fluconazole-resistant C. parapsilosis isolates has been on the rise and fluconazole resistance rates in C. parapsilosis overall (83.7%) or isolates from blood cultures (70%) were extremely high.
The Son Espases hospital study represented a textbook example of a hospital severely hit by fluconazole-resistant C. parapsilosis (Alcoceba et al., 2022). The first resistant isolate showed up a long time ago, then its offspring adapted to the hospital environment to the point of becoming endemic. Such a spread may have been a consequence of delayed detection of resistant isolates; for this reason, prospective and long-lasting antifungal resistance surveillance studies are helpful in terms of allowing the early detection of resistant isolates and monitoring the resistance trend. In January 2019, we initiated the prospective monitoring of antifungal resistance in Candida spp isolates collected from blood cultures and intra-abdominal samples of patients admitted to any of the 16 hospitals located in the Madrid metropolitan area (CANDIMAD study). Data on 2,107 Candida spp isolates (1,895 patients) collected from 2019 to 2021 in the CANDIMAD study were recently reported (Diaz-Garcia et al., 2022b), including a separate analysis of C. parapsilosis (Diaz-Garcia et al., 2022a). Fluconazole resistance was higher in blood cultures than in intra-abdominal samples (9.1% versus 8.2%; P>0.05), especially for C. parapsilosis (16.6% versus 3.6%, P<0.05). A total of 13.6% of C. parapsilosis isolates were fluconazole-resistant and sourced mostly from blood cultures (94%). Moreover, overall resistance rates in C. parapsilosis were rising during these three years: 3.8% in 2019, 5.7% in 2020, and 29.1% in 2021 (P<0.05). C. parapsilosis resistant isolates involved patients admitted to five hospitals and were detected for the first time in May 2019, September 2020, October 2020, February 2021, and August 2021, at each hospital. Hospitals were affected to a different extent; whereas the fluconazole resistance rate in some hospitals was 0%, it reached up to 37.7% in others. All fluconazole-resistant isolates harboured either the Y132F (n=43/48) or G458S (n=5/48) substitution in ERG11p. Isolates harbouring the Y132F substitution were found in four hospitals, whereas isolates harbouring the G458S substitution were found in the fifth hospital.
All fluconazole-resistant isolates collected at the Son Espases hospital grouped into 11 clonally related genotypes, with one genotype accounting for most isolates. Likewise, all fluconazole-resistant isolates collected in the Madrid surveillance study harbouring the Y132F substitution grouped into three clonally related genotypes; one genotype dominated and was the first detected, whereas isolates harbouring the G458S substitution grouped into another genotype. On balance, the fluconazole-resistant genotypes detected in the Balearic Islands and Madrid were different one from another, and different from those involving susceptible isolates. To date, two main observations can be drawn. First, clones harbouring the Y132F substitution in the two regions of the country were different and caused unrelated outbreaks. Second, the clone harbouring the G458S substitution in Madrid was different and emerged independently in another hospital. At the moment, we are tracking down fluconazole-resistant C. parapsilosis isolates from other Spanish regions and countries. Moreover, we are keeping a closer watch on the evolution of the resistant clones in the Madrid region.
The COVID-19 pandemic may have increased the number of cases of candidaemia (Nucci et al., 2021; Machado et al., 2022; Papadimitriou-Olivgeris et al., 2022; Seagle et al., 2022). This increase may be a consequence of either an increase in the number of at-risk patients admitted or higher patient-to-patient transmission, but this might be a hospital-dependent phenomenon. In our hospital, whereas the incidence of candidaemia was higher in patients with COVID-19 than without, genotyping demonstrated that the increase was not due to uncontrolled intra-hospital patient-to-patient transmission (Machado et al., 2022). Conversely, a study conducted in another Madrid hospital showed that C. parapsilosis candidaemia episodes increased significantly during the pandemic period, including the number of cases caused by a fluconazole-resistant C. parapsilosis genotype harbouring the Y132F ERG11p substitution and spreading at that time (Ramos-Martinez et al., 2022). In line with these observations, another study conducted in Greece showed that both the incidence of candidaemia and fluconazole resistance in C. parapsilosis increased during the pandemic; unfortunately, the isolates were not molecular characterized (Routsi et al., 2022). In the Madrid study, many resistant isolates showed up before the COVID-19 pandemic; however, their spread become more evident during the pandemic, suggesting that all triggers increasing the number of candidaemia cases may have been effective in terms of promoting the clonal spread of resistant isolates (Ramos-Martinez et al., 2022).
The clinical description of patients affected with fluconazole-resistant C. parapsilosis harbouring the Y132F ERG11p substitution is quite limited. Some studies compared patients infected with fluconazole-resistant isolates with those infected with susceptible ones and did not find any differences in terms of clinical presentation or risk factors (Fekkar et al., 2021; Alcoceba et al., 2022). The reports warn that a large number of patients affected with resistant isolates were admitted to intensive care wards (Thomaz et al., 2018; Corzo-Leon et al., 2021; Fekkar et al., 2021; Thomaz et al., 2021; Alcoceba et al., 2022; Ramos-Martinez et al., 2022). An interesting observation was the fact that, with an exception (Thomaz et al., 2018), few patients infected with fluconazole-resistant isolates had previously received fluconazole (≤20%) (Corzo-Leon et al., 2021; Fekkar et al., 2021; Alcoceba et al., 2022; Ramos-Martinez et al., 2022). This supports the fact that, fluconazole might have been a key factor in promoting resistance, however, the main driver promoting the spread of fluconazole-resistant isolates was patient-to-patient transmission. Patient mortality ranged from 9% to 78% (Thomaz et al., 2018; Corzo-Leon et al., 2021; Fekkar et al., 2021; Thomaz et al., 2021; Alcoceba et al., 2022; Ramos-Martinez et al., 2022).
This review demonstrates the importance of tracking, genotyping, and controlling the spread of fluconazole-resistant C. parapsilosis isolates. Future studies are warranted in order to assess the clinical impact of this emerging problem, the quality of hospital control policies to halt future spreading, as well as the development of techniques to allow a fast and accurate detection of isolates in routine clinical microbiology.
PE and JG wrote all sections of the manuscript. Both authors contributed to the article and approved the submitted version.
This work was supported by grants PI18/01155 and PI19/00074 from the Fondo de Investigación Sanitaria (FIS. Instituto de Salud Carlos III. Plan Nacional de I+D+I 2017-2020). The study was co-funded by the European Regional Development Fund (FEDER) ‘A way of making Europe.’ The funders had no role in the study design, data collection, analysis, decision to publish, or preparation/content of the manuscript. PE (CPI20/00015) is a recipient of a Miguel Servet contract supported by the FIS. JG is a permanent researcher contracted by Fundación para Investigación Sanitaria del Hospital Gregorio Marañón.
We are grateful to Helena Kruyer for editing assistance.
JG has received funds for participating in educational activities organized on behalf of Pfizer, Gilead, and MSD; he has also received research funds from FIS, Gilead, Scynexis, F2G, and Cidara outside the submitted work.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be constructed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alcoceba E., Gomez A., Lara-Esbri P., Oliver A., Beltran A. F., Ayestaran I., et al. (2022). Fluconazole-resistant Candida parapsilosis clonally related genotypes: First report proving the presence of endemic isolates harbouring the Y132F ERG11 gene substitution in Spain. Clin. Microbiol. Infect. 28 (8), 1113–1119. doi: 10.1016/j.cmi.2022.02.025
Aldardeer N. F., Albar H., Al-Attas M., Eldali A., Qutub M., Hassanien A., et al. (2020). Antifungal resistance in patients with candidaemia: A retrospective cohort study. BMC Infect. Dis. 20 (1), 55. doi: 10.1186/s12879-019-4710-z
Alobaid K., Khan Z. (2019). Epidemiologic characteristics of adult candidemic patients in a secondary hospital in Kuwait: A retrospective study. J. Mycol Med. 29 (1), 35–38. doi: 10.1016/j.mycmed.2018.12.001
Arastehfar A., Daneshnia F., Hilmioglu-Polat S., Fang W., Yasar M., Polat F., et al. (2020a). First report of candidemia clonal outbreak caused by emerging fluconazole-resistant Candida parapsilosis isolates harboring Y132F and/or Y132F+K143R in Turkey. Antimicrob. Agents Chemother. 64 (10), e01001–e01020. doi: 10.1128/AAC.01001-20
Arastehfar A., Daneshnia F., Najafzadeh M. J., Hagen F., Mahmoudi S., Salehi M., et al. (2020b). Evaluation of molecular epidemiology, clinical characteristics, antifungal susceptibility profiles, and molecular mechanisms of antifungal resistance of Iranian Candida parapsilosis species complex blood isolates. Front. Cell Infect. Microbiol. 10, 206. doi: 10.3389/fcimb.2020.00206
Arastehfar A., Hilmioglu-Polat S., Daneshnia F., Pan W., Hafez A., Fang W., et al. (2021). Clonal candidemia outbreak by Candida parapsilosis carrying Y132F in Turkey: Evolution of a persisting challenge. Front. Cell Infect. Microbiol. 11, 676177. doi: 10.3389/fcimb.2021.676177
Arendrup M. C., Bruun B., Christensen J. J., Fuursted K., Johansen H. K., Kjaeldgaard P., et al. (2011). National surveillance of fungemia in denmark, (2004 to 2009). J. Clin. Microbiol. 49 (1), 325–334. doi: 10.1128/JCM.01811-10
Asadzadeh M., Ahmad S., Al-Sweih N., Khan Z. (2017). Epidemiology and molecular basis of resistance to fluconazole among clinical Candida parapsilosis isolates in Kuwait. Microb. Drug Resist. 23 (8), 966–972. doi: 10.1089/mdr.2016.0336
Asadzadeh M., Dashti M., Ahmad S., Alfouzan W., Alameer A. (2021). Whole-genome and targeted-amplicon sequencing of fluconazole-susceptible and -resistant Candida parapsilosis isolates from Kuwait reveals a previously undescribed N1132D polymorphism in CDR1. Antimicrob. Agents Chemother. 65 (2), e01633–e01620. doi: 10.1128/AAC.01633-20
Berkow E. L., Manigaba K., Parker J. E., Barker K. S., Kelly S. L., Rogers P. D. (2015). Multidrug transporters and alterations in sterol biosynthesis contribute to azole antifungal resistance in Candida parapsilosis. Antimicrob. Agents Chemother. 59 (10), 5942–5950. doi: 10.1128/AAC.01358-15
Castanheira M., Deshpande L. M., Messer S. A., Rhomberg P. R., Pfaller M. A. (2020). Analysis of global antifungal surveillance results reveals predominance of Erg11 Y132F alteration among azole-resistant Candida parapsilosis and Candida tropicalis and country-specific isolate dissemination. Int. J. Antimicrob. Agents 55 (1), 105799. doi: 10.1016/j.ijantimicag.2019.09.003
Choi Y. J., Kim Y. J., Yong D., Byun J. H., Kim T. S., Chang Y. S., et al. (2018). Fluconazole-resistant Candida parapsilosis bloodstream isolates with Y132F mutation in ERG11 gene, south Korea. Emerg. Infect. Dis. 24 (9), 1768–1770. doi: 10.3201/eid2409.180625
Chow N. A., Gade L., Tsay S. V., Forsberg K., Greenko J. A., Southwick K. L., et al. (2018). Multiple introductions and subsequent transmission of multidrug-resistant candida auris in the USA: A molecular epidemiological survey. Lancet Infect. Dis. 18 (12), 1377–1384. doi: 10.1016/S1473-3099(18)30597-8
Clark T. A., Slavinski S. A., Morgan J., Lott T., Arthington-Skaggs B. A., Brandt M. E., et al. (2004). Epidemiologic and molecular characterization of an outbreak of Candida parapsilosis bloodstream infections in a community hospital. J. Clin. Microbiol. 42 (10), 4468–4472. doi: 10.1128/JCM.42.10.4468-4472.2004
Corzo-Leon D. E., Peacock M., Rodriguez-Zulueta P., Salazar-Tamayo G. J., MacCallum D. M. (2021). General hospital outbreak of invasive candidiasis due to azole-resistant Candida parapsilosis associated with an Erg11 Y132F mutation. Med. Mycol 59 (7), 664–671. doi: 10.1093/mmy/myaa098
Demirci-Duarte S., Arikan-Akdagli S., Gulmez D. (2021). Species distribution, azole resistance and related molecular mechanisms in invasive Candida parapsilosis complex isolates: Increase in fluconazole resistance in 21 years. Mycoses 64 (8), 823–830. doi: 10.1111/myc.13296
Diab-Elschahawi M., Forstner C., Hagen F., Meis J. F., Lassnig A. M., Presterl E., et al. (2012). Microsatellite genotyping clarified conspicuous accumulation of Candida parapsilosis at a cardiothoracic surgery intensive care unit. J. Clin. Microbiol. 50 (11), 3422–3426. doi: 10.1128/JCM.01179-12
Diaz-Garcia J., Gomez A., Alcala L., Reigadas E., Sanchez-Carrillo C., Perez-Ayala A., et al. (2022a). Evidence of fluconazole-resistant Candida parapsilosis genotypes spreading across hospitals located in Madrid, Spain and harboring the Y132F ERG11p substitution. Antimicrob. Agents Chemother. 66 (8), e0071022. doi: 10.1128/aac.00710-22
Díaz-García J., Gómez A., Machado M., Alcalá L., Reigadas E., Sánchez-Carrillo C., et al. (2022b). Blood and intra-abdominal Candida spp from a multicentre study conducted in Madrid using EUCAST: Emergence of fluconazole resistance in C. parapsilosis, low echinocandin resistance, and absence of C. auris. J. Antimicrob. Chemother. dkac288. doi: 10.1093/jac/dkac288
Diaz-Garcia J., Mesquida A., Gomez A., Machado M., Martin-Rabadan P., Alcala L., et al. (2021a). Antifungal susceptibility testing identifies the abdominal cavity as a source of Candida glabrata-resistant isolates. Antimicrob. Agents Chemother. 65 (12), e0124921. doi: 10.1128/AAC.01249-21
Diaz-Garcia J., Mesquida A., Sanchez-Carrillo C., Reigadas E., Munoz P., Escribano P., et al. (2021b). Monitoring the epidemiology and antifungal resistance of yeasts causing fungemia in a tertiary care hospital in Madrid, Spain: Any relevant changes in the last 13 years? Antimicrob. Agents Chemother. 65 (4), e01827-20. doi: 10.1128/AAC.01827-20
Escribano P., Rodriguez-Creixems M., Sanchez-Carrillo C., Munoz P., Bouza E., Guinea J. (2013). Endemic genotypes of Candida albicans causing fungemia are frequent in the hospital. J. Clin. Microbiol. 51 (7), 2118–2123. doi: 10.1128/JCM.00516-13
Escribano P., Sanchez-Carrillo C., Munoz P., Bouza E., Guinea J. (2018). Reduction in percentage of clusters of Candida albicans and Candida parapsilosis causing candidemia in a general hospital in Madrid, Spain. J. Clin. Microbiol. 56 (7), e00574–e00518. doi: 10.1128/JCM.00574-18
Faria-Ramos I., Neves-Maia J., Ricardo E., Santos-Antunes J., Silva A. T., Costa-de-Oliveira S., et al. (2014). Species distribution and in vitro antifungal susceptibility profiles of yeast isolates from invasive infections during a Portuguese multicenter survey. Eur. J. Clin. Microbiol. Infect. Dis. 33 (12), 2241–2247. doi: 10.1007/s10096-014-2194-8
Fekkar A., Blaize M., Bougle A., Normand A. C., Raoelina A., Kornblum D., et al. (2021). Hospital outbreak of fluconazole-resistant Candida parapsilosis: Arguments for clonal transmission and long-term persistence. Antimicrob. Agents Chemother. 65 (5), e02036–e02020. doi: 10.1128/AAC.02036-20
Grossman N. T., Pham C. D., Cleveland A. A., Lockhart S. R. (2015). Molecular mechanisms of fluconazole resistance in Candida parapsilosis isolates from a U.S. surveillance system. Antimicrob. Agents Chemother. 59 (2), 1030–1037. doi: 10.1128/AAC.04613-14
Guducuoglu H., Gultepe B., Otlu B., Bektas A., Yildirim O., Tuncer O., et al. (2016). Candida albicans outbreak associated with total parenteral nutrition in the neonatal unit. Indian J. Med. Microbiol. 34 (2), 202–207. doi: 10.4103/0255-0857.180303
Guinea J. (2014). Global trends in the distribution of Candida species causing candidemia. Clin. Microbiol. Infect. 20 (Suppl 6), 5–10. doi: 10.1111/1469-0691.12539
Guinea J., Arendrup M. C., Canton R., Canton E., Garcia-Rodriguez J., Gomez A., et al. (2020). Genotyping reveals high clonal diversity and widespread genotypes of Candida causing candidemia at distant geographical areas. Front. Cell Infect. Microbiol. 10, 166. doi: 10.3389/fcimb.2020.00166
Guinea J., Mezquita S., Gomez A., Padilla B., Zamora E., Sanchez-Luna M., et al. (2021). Whole genome sequencing confirms Candida albicans and Candida parapsilosis microsatellite sporadic and persistent clones causing outbreaks of candidemia in neonates. Med. Mycol 60 (1), myab068. doi: 10.1093/mmy/myab068
Guinea J., Zaragoza O., Escribano P., Martin-Mazuelos E., Peman J., Sanchez-Reus F., et al. (2014). Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob. Agents Chemother. 58 (3), 1529–1537. doi: 10.1128/AAC.02155-13
Guo J., Zhang M., Qiao D., Shen H., Wang L., Wang D., et al. (2021). Prevalence and antifungal susceptibility of Candida parapsilosis species complex in Eastern China: A 15-year retrospective study by ECIFIG. Front. Microbiol. 12, 644000. doi: 10.3389/fmicb.2021.644000
Hare R. K., Arastehfar A., Rosendahl S., Charsizadeh A., Daneshnia F., Eshaghi H., et al. (2022). Candidemia among hospitalized pediatric patients caused by several clonal lineages of Candida parapsilosis. J. Fungi (Basel) 8 (2), 183. doi: 10.3390/jof8020183
Healey K. R., Kordalewska M., Jimenez Ortigosa C., Singh A., Berrio I., Chowdhary A., et al. (2018). Limited ERG11 mutations identified in isolates of Candida auris directly contribute to reduced azole susceptibility. Antimicrob. Agents Chemother. 62 (10), e01427-18. doi: 10.1128/AAC.01427-18
Jiang C., Dong D., Yu B., Cai G., Wang X., Ji Y., et al. (2013). Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 68 (4), 778–785. doi: 10.1093/jac/dks481
Lasker B. A., Butler G., Lott T. J. (2006). Molecular genotyping of Candida parapsilosis group I clinical isolates by analysis of polymorphic microsatellite markers. J. Clin. Microbiol. 44 (3), 750–759. doi: 10.1128/JCM.44.3.750-759.2006
Liu Z., Myers L. C. (2017). Mediator tail module is required for Tac1-activated CDR1 expression and azole resistance in Candida albicans. Antimicrob. Agents Chemother. 61 (11), e01342–e01317. doi: 10.1128/AAC.01342-17
Machado M., Estevez A., Sanchez-Carrillo C., Guinea J., Escribano P., Alonso R., et al. (2022). Incidence of candidemia is higher in COVID-19 versus non-COVID-19 patients, but not driven by intrahospital transmission. J. Fungi (Basel) 8 (3), 305. doi: 10.3390/jof8030305
Magobo R. E., Lockhart S. R., Govender N. P. (2020). Fluconazole-resistant Candida parapsilosis strains with a Y132F substitution in the ERG11 gene causing invasive infections in a neonatal unit, south Africa. Mycoses 63 (5), 471–477. doi: 10.1111/myc.13070
Magobo R. E., Naicker S. D., Wadula J., Nchabeleng M., Coovadia Y., Hoosen A., et al. (2017). Detection of neonatal unit clusters of Candida parapsilosis fungaemia by microsatellite genotyping: Results from laboratory-based sentinel surveillance, south africa 2009-2010. Mycoses 60 (5), 320–327. doi: 10.1111/myc.12596
Marcos-Zambrano L. J., Escribano P., Bouza E., Guinea J. (2014). Use of molecular typing tools for the study of hospital outbreaks of candidemia. Rev. Iberoam Micol 31 (2), 97–103. doi: 10.1016/j.riam.2013.06.003
Martini C., Torelli R., de Groot T., De Carolis E., Morandotti G. A., De Angelis G., et al. (2020). Prevalence and clonal distribution of azole-resistant Candida parapsilosis isolates causing bloodstream infections in a Large Italian hospital. Front. Cell Infect. Microbiol. 10, 232. doi: 10.3389/fcimb.2020.00232
Mesini A., Mikulska M., Giacobbe D. R., Del Puente F., Gandolfo N., Codda G., et al. (2020). Changing epidemiology of candidaemia: Increase in fluconazole-resistant Candida parapsilosis. Mycoses 63 (4), 361–368. doi: 10.1111/myc.13050
Morio F., Loge C., Besse B., Hennequin C., Le Pape P. (2010). Screening for amino acid substitutions in the Candida albicans Erg11 protein of azole-susceptible and azole-resistant clinical isolates: New substitutions and a review of the literature. Diagn. Microbiol. Infect. Dis. 66 (4), 373–384. doi: 10.1016/j.diagmicrobio.2009.11.006
Morio F., Pagniez F., Lacroix C., Miegeville M., Le Pape P. (2012). Amino acid substitutions in the Candida albicans sterol {Delta}5,6-desaturase (Erg3p) confer azole resistance: Characterization of two novel mutants with impaired virulence. J. Antimicrob. Chemother. 67 (9), 2131–2138. doi: 10.1093/jac/dks186
Morschhauser J., Barker K. S., Liu T. T., Bla B. W. J., Homayouni R., Rogers P. D. (2007). The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PloS Pathog. 3 (11), e164. doi: 10.1371/journal.ppat.0030164
Nucci M., Barreiros G., Guimaraes L. F., Deriquehem V. A. S., Castineiras A. C., Nouer S. A. (2021). Increased incidence of candidemia in a tertiary care hospital with the COVID-19 pandemic. Mycoses 64 (2), 152–156. doi: 10.1111/myc.13225
Nucci M., Queiroz-Telles F., Tobon A. M., Restrepo A., Colombo A. L. (2010). Epidemiology of opportunistic fungal infections in Latin America. Clin. Infect. Dis. 51 (5), 561–570. doi: 10.1086/655683
Papadimitriou-Olivgeris M., Kolonitsiou F., Kefala S., Spiliopoulou A., Aretha D., Bartzavali C., et al. (2022). Increased incidence of candidemia in critically ill patients during the coronavirus disease 2019 (COVID-19) pandemic. Braz. J. Infect. Dis. 26 (2), 102353. doi: 10.1016/j.bjid.2022.102353
Pfaller M. A., Diekema D. J., Turnidge J. D., Castanheira M., Jones R. N. (2019). Twenty years of the SENTRY antifungal surveillance program: Results for Candida species from 1997-2016. Open Forum Infect. Dis. 6 (Suppl 1), S79–S94. doi: 10.1093/ofid/ofy358
Pinhati H. M., Casulari L. A., Souza A. C., Siqueira R. A., Damasceno C. M., Colombo A. L. (2016). Outbreak of candidemia caused by fluconazole resistant Candida parapsilosis strains in an intensive care unit. BMC Infect. Dis. 16 (1), 433. doi: 10.1186/s12879-016-1767-9
Prigitano A., Cavanna C., Passera M., Ossi C., Sala E., Lombardi G., et al. (2016). CAND-LO 2014-15 study: Changing epidemiology of candidemia in Lombardy (Italy). Infection 44 (6), 765–780. doi: 10.1007/s15010-016-0951-6
Puig-Asensio M., Padilla B., Garnacho-Montero J., Zaragoza O., Aguado J. M., Zaragoza R., et al. (2014). Epidemiology and predictive factors for early and late mortality in Candida bloodstream infections: A population-based surveillance in Spain. Clin. Microbiol. Infect. 20 (4), O245–O254. doi: 10.1111/1469-0691.12380
Ramos-Martinez A., Pintos-Pascual I., Guinea J., Gutierrez-Villanueva A., Gutierrez-Abreu E., Diaz-Garcia J., et al. (2022). Impact of the COVID-19 pandemic on the clinical profile of candidemia and the incidence of fungemia due to fluconazole-resistant Candida parapsilosis. J. Fungi (Basel) 8 (5), 451. doi: 10.3390/jof8050451
Routsi C., Meletiadis J., Charitidou E., Gkoufa A., Kokkoris S., Karageorgiou S., et al. (2022). Epidemiology of candidemia and fluconazole resistance in an ICU before and during the COVID-19 pandemic era. Antibiotics (Basel) 11 (6), 771.doi: 10.3390/antibiotics11060771
Sabino R., Sampaio P., Rosado L., Stevens D. A., Clemons K. V., Pais C. (2010). New polymorphic microsatellite markers able to distinguish among Candida parapsilosis sensu stricto isolates. J. Clin. Microbiol. 48 (5), 1677–1682. doi: 10.1128/JCM.02151-09
Sanglard D., Odds F. C. (2002). Resistance of Candida species to antifungal agents: Molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2 (2), 73–85. doi: 10.1016/S1473-3099(02)00181-0
Schneider S., Morschhauser J. (2015). Induction of Candida albicans drug resistance genes by hybrid zinc cluster transcription factors. Antimicrob. Agents Chemother. 59 (1), 558–569. doi: 10.1128/AAC.04448-14
Seagle E. E., Jackson B. R., Lockhart S. R., Georgacopoulos O., Nunnally N. S., Roland J., et al. (2022). The landscape of candidemia during the coronavirus disease 2019 (COVID-19) pandemic. Clin. Infect. Dis. 74 (5), 802–811. doi: 10.1093/cid/ciab562
Singh A., Singh P. K., de Groot T., Kumar A., Mathur P., Tarai B., et al. (2019). Emergence of clonal fluconazole-resistant Candida parapsilosis clinical isolates in a multicentre laboratory-based surveillance study in India. J. Antimicrob. Chemother. 74 (5), 1260–1268. doi: 10.1093/jac/dkz029
Souza A. C., Fuchs B. B., Pinhati H. M., Siqueira R. A., Hagen F., Meis J. F., et al. (2015). Candida parapsilosis resistance to fluconazole: Molecular mechanisms and In vivo impact in infected Galleria mellonella larvae. Antimicrob. Agents Chemother. 59 (10), 6581–6587. doi: 10.1128/AAC.01177-15
Thomaz D. Y., de Almeida J. N. Jr., Lima G. M. E., Nunes M. O., Camargo C. H., Grenfell R. C., et al. (2018). An azole-resistant Candida parapsilosis outbreak: Clonal persistence in the intensive care unit of a Brazilian teaching hospital. Front. Microbiol. 9, 2997. doi: 10.3389/fmicb.2018.02997
Thomaz D. Y., de Almeida J. N. Jr., Sejas O. N. E., Del Negro G. M. B., Carvalho G., Gimenes V. M. F., et al. (2021). Environmental clonal spread of azole-resistant Candida parapsilosis with Erg11-Y132F mutation causing a Large candidemia outbreak in a Brazilian cancer referral center. J. Fungi (Basel) 7 (4), 259. doi: 10.3390/jof7040259
Thomaz D. Y., Del Negro G. M. B., Ribeiro L. B., da Silva M., Carvalho G., Camargo C. H., et al. (2022). A Brazilian inter-hospital candidemia outbreak caused by fluconazole-resistant Candida parapsilosis in the COVID-19 era. J. Fungi (Basel) 8 (2), 100. doi: 10.3390/jof8020100
Yang F., Lu H., Wu H., Fang T., Berman J., Jiang Y. Y. (2021). Aneuploidy underlies tolerance and cross-tolerance to drugs in Candida parapsilosis. Microbiol. Spectr. 9 (2), e0050821. doi: 10.1128/Spectrum.00508-21
Zaragoza O., Alcázar-Fuoli L., Trevijano-Contador N., Torres-Cano A., Carballo-González C., Puig-Asensio M., et al. (2022). Global emergence of resistance to fluconazole and voriconazole in Candida parapsilosis in tertiary hospitals in Spain during the COVID-19 pandemic. medRXiv. doi: 10.1101/2022.06.06.22275514
Zhang L., Yu S. Y., Chen S. C., Xiao M., Kong F., Wang H., et al. (2020). Molecular characterization of Candida parapsilosis by microsatellite typing and emergence of clonal antifungal drug resistant strains in a multicenter surveillance in China. Front. Microbiol. 11, 1320. doi: 10.3389/fmicb.2020.01320
Keywords: Candida parapsilosis, fluconazole, resistance, ERG11, Y132F, G458S, K128N, K143R
Citation: Escribano P and Guinea J (2022) Fluconazole-resistant Candida parapsilosis: A new emerging threat in the fungi arena. Front. Fungal Bio. 3:1010782. doi: 10.3389/ffunb.2022.1010782
Received: 03 August 2022; Accepted: 06 October 2022;
Published: 24 October 2022.
Edited by:
João Nobrega De Almeida Júnior, Clinical Hospital, University of São Paulo, BrazilReviewed by:
Christopher Heath, Fiona Stanley Hospital, AustraliaCopyright © 2022 Escribano and Guinea. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jesús Guinea, amd1aW5lYW9ydGVnYUB5YWhvby5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.