
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
OPINION article
Front. Fungal Biol. , 01 December 2021
Sec. Fungal Physiology and Metabolism
Volume 2 - 2021 | https://doi.org/10.3389/ffunb.2021.743070
This article is part of the Research Topic The use of metabolic engineering techniques to increase the productivity of primary and secondary metabolites within filamentous fungi View all 5 articles
Filamentous fungi are multicellular eukaryotic microorganisms that primarily obtain energy via the degradation of macromolecules in their environment. The production of many of the hydrolytic enzymes required for feeding is prolific and the capacity has been exploited for the mass production of industrially relevant enzymes such as amylase, protease, and lipase. Fungi can also produce large quantities of chemicals. In 1917, Aspergillus niger was found to have the capacity to produce citrate, a primary metabolite, that is now ubiquitously used as an acidulent (Cairns et al., 2018). In 1928, Penicillium rubens was found to produce penicillin, a secondary metabolite exhibiting antibacterial activity (Browne et al., 2013). In the intervening decades, filamentous fungi have been intensively researched to identify new chemicals, of which many are widely used as pharmaceuticals (e.g., cyclosporin and lovastatin) (Boruta, 2018), pigments (e.g., Monascus pigment) (Liu et al., 2018), platform chemicals (e.g., itaconic acid) (Huang et al., 2021), and cosmetics (e.g., kojic acid) (Terabayashi et al., 2010).
Industry has used a variety of microbial hosts including bacteria (e.g., Escherichia coli, Streptomyces sp., Corynebacterium sp.), yeasts (e.g., Saccharomyces cerevisiae, Pichia pastoris, Yarrowia lipolytica), and filamentous fungi to produce valuable metabolites. Among them, filamentous fungi have considerable merits that are ability to degrade polymers, availability in solid-state cultures, and suitability to produce key primary and secondary metabolites (Abe et al., 2006; Machida et al., 2008).
In recent years, next-generation DNA sequencing (NGS) technology has been widely used for genome and transcriptomic analysis (Goodwin et al., 2016). Noteworthy, NGS devices enabled high-throughput sequencing at a much lower cost as compared to conventional Sanger sequencing (Reuter et al., 2015). The impact of post-sequencing analytical tools to precisely annotate genes cannot be understated (Gene Ontology Consortium, 2021). For example, industrial strains of filamentous fungi other than type strains have been subjected to genome sequencing by NGS and the specific genes involved in the biosynthesis of many target metabolites can now be predicted in industrial strains. Thus, technology based on genomic information has in fact become applicable to industrial strains. Furthermore, enhancing industrial productivities of target metabolites by metabolic engineering can be attained.
However, there are still several limitations to industrial utilization. Based on author's experimental experience of filamentous fungi Aspergillus sp., for example, it is difficult to obtain high-density liquid cultures because of pellet formation (Miyazawa et al., 2016). Additionally, laborious experiments are required for their genetic manipulation compared to other host microorganisms. For several strains, single spore isolation is necessary for mutant construction due to their multinuclear sporulation characteristics (Maruyama et al., 2002). High-throughput screening is also difficult owing to large colony formation. The time taken to construct mutants via recombination is longer because several days are required for spore formation using agar culture. Therefore, research aiming to overcome these drawbacks has been performed for many years. Several progresses have been reported and are briefly introduced at the next two sections.
In filamentous fungi, the integration of transforming DNA by the non-homologous end joining (NHEJ) pathway occur at a much higher frequency than by the homologous recombination (HR) pathway (Takahashi et al., 2004). Thus, DNA fragments taken up by nuclei end up being integrated at random locations in the genome rather than at specific, targeted locations. Since most strategies for strain improvement require targeted integration of DNA, NHEJ is a hindrance. However, in 2004 and 2006, genetic factors involved in the NHEJ pathway were identified in Neurospora crassa (Ninomiya et al., 2004; Ishibashi et al., 2006). Following this identification, mutation of NHEJ-involved genes such as ku70, ku80, and ligD that were conserved among many fungal species created strains whereby subsequent DNA integrations occurred mostly by HR rather than NHEJ. This increased efficiency of HR (exceeding 90% in some filamentous fungi) vastly improved strain breeding pipelines (Takahashi et al., 2006; Mizutani et al., 2008).
The constructed NHEJ-knockout strain can be utilized for metabolic engineering. For metabolic engineering, the pipeline for strain breeding generally includes deletion of a single or multiple genes and gene overexpression experiments. To remove unnecessary metabolic pathways to allow target metabolite overproduction, it is fundamental to inactivate the first reaction of unnecessary pathways by deleting a key enzyme-coding gene (Tamano, 2014). Target metabolite biosynthesis may also be facilitated by overexpression of a bottleneck enzyme-coding gene (Wong et al., 2021). In this case, the original promoter is changed to a promoter from a constitutively highly expressed gene. For gene deletion, target gene is replaced with a selectable marker by two HR events between chromosome and introduced DNA (e.g., double HR) (Tamano et al., 2015; Wendt et al., 2016); for gene overexpression, target gene promoter is replaced with a set of selectable marker and constitutive high-expression promoter by double HR (Tamano et al., 2013). Gene overexpression can alternatively be attained by transforming with a plasmid harboring the overexpression cassette (Kanamasa et al., 2001). In this case, a single HR event is sufficient for the cassette to be inserted into the chromosome.
Recently, the newly discovered editing technology, CRISPR/Cas9, has been applied to modify fungal genomes (reviewed in Jiang et al., 2021). In this case, gene disruption and overexpression occur via a targeted double strand DNA break followed by genome editing. For gene disruption, a selectable marker is introduced into the target gene; for gene overexpression, a DNA cassette composed of a selectable marker and constitutive high-expression promoter is introduced to a locus just upstream of the start codon of the gene.
Selectable marker genes must be available for mutant construction for metabolic engineering. They are categorized into three groups: [1] antifungal resistance, [2] auxotrophy restoration, and [3] nutrient-catabolizing. Five genes commonly used to confer resistance to different drugs (pyrithiamine, hygromycin, bleomycin, aureobasidin, and oligomycin) are ptrA, hph, ble, aur, and oliC31 (Gritz and Davies, 1983; Ward et al., 1986; Glumoff et al., 1989; Heidler and Radding, 1995; Kubodera et al., 2000). Three genes commonly used to restore auxotrophy are pyrG, niaD, and sC (van Hartingsveldt et al., 1987; Unkles et al., 1989; Yamada et al., 1997). amdS is the lone nutrient-catabolizing enzyme gene commonly used (Gomi et al., 1991). Some selectable markers can be “reused” to allow sequential engineering events through a process called counterselection. For example, the portion of a DNA cassette, pyrG, conferring pyrimidine prototrophy integrated into a chromosome by HR can be removed via DNA loop-out or by Cre recombinase coupled with loxP recognition sites under existence of 5-fluoroorotic acid (Zhang et al., 2017). Thus, using this marker-recycling system, multiple mutation events are possible in a single genome with essentially no limitation. By combining the use of these selectable markers, strains genetically engineered at multiple genomic locations can be constructed.
Additionally, high-expression promoters have to be utilized for gene overexpression. The translation elongation factor 1 (tef1)-encoding gene is constitutively expressed at high levels in Aspergillus oryzae (Kitamoto et al., 1998). Because this gene and promoter are highly conserved in distantly related fungi, the level of gene expression in heterologous hosts should be similar. Moreover, genes encoding glycolytic enzymes such as enolase are constitutively expressed at high levels. Promoters of these genes are usually utilized for gene overexpression. Recently, Jin et al. (2021) provided a comprehensive review of selectable markers and promoters commonly used to genetically engineer Aspergillus oryzae.
Process of improving industrial strains by metabolic engineering, as an opinion of authors, is outlined in Figure 1. The first step of the process is to identify a wild-type strain that produces a valuable metabolite. Large-scale screening of filamentous fungal strains is necessary for the identification. However, it does not promise successful identification of such strains. Therefore, this step is most challenging. Next, as the second step, establishing adequate culture conditions is critical, especially defining the minimal medium composition because auxotrophic selectable markers may be used in subsequent strain improvement efforts. Similarly, as drug-resistant selectable markers may be considered for strain improvement, it is important to investigate drug sensitivity. The third step is NGS-based genome sequencing, gene annotation, and whole-genome transcriptional analysis. The latter transcriptional analysis can be particularly helpful if culture conditions conducive to product formation are established. The comparison of gene transcription after growth on metabolite production media vs. non-production media can contribute significantly to the accuracy of predicting genes involved in the biosynthesis of the target metabolite. The step requires NGS sequencing, and thus seems most expensive. The fourth step is to construct a genetic transformation system of the wild-type strain identified. Polyethylene glycol-mediated transformation protocol using protoplasts is very common and has been developed for a wide range of filamentous fungi (Tamano et al., 2007). This step requires trial and error to find an optimal condition of protoplast formation. Cell wall-lysing enzymes and the mixed composition must be optimized. Hence, it is labor-intensive. If protoplasts cannot be prepared in sufficient amounts to be used for transformation, a number of Agrobacterium-mediated transformation protocols have been developed (Takahashi et al., 2011). The fifth step is the construction of a basic strain for metabolic engineering. Briefly, genes ku70, ku80, and ligD responsible for NHEJ are extracted by homology search and then one of them is mutated by low-efficiency double HR with use of a drug-resistant selectable marker. The loss of NHEJ will allow acquisition of subsequent HR-mediated mutant strains at high-efficiency. After removing NHEJ capacity, a gene involved in essential metabolism is knocked out by double HR with another drug-resistant selectable marker. For example, pyrG gene, which partakes in the pyrimidine biosynthesis, is comparatively short and completely lethal when knocked out; therefore, it is a good candidate of knockout as well as an auxotrophic selectable marker (van Hartingsveldt et al., 1987). A marker-recycling system of Cre/loxP is then introduced to an appropriate locus on the chromosome with use of auxotrophic selectable marker enabled just before. At this point, the DNA fragment is designed as the Cre recombinase gene expression can be induced specifically under certain culture conditions. For instance, a xylanase gene promoter is placed ahead of the Cre-coding sequence; thus, Cre is only expressed when xylose is used as a carbon source in the culture (Zhang et al., 2017). After induction of the Cre/loxP system, an auxotrophic strain in which the NHEJ pathway is defective and the marker-recycling system reside is consequently constructed via counter-selection using 5-fluoroorotic acid. It is the basic strain for breeding. Lastly, as the sixth step, the metabolic engineers can now work directly to enhance target metabolite synthesis. A first common target is to identify and disable dispensable metabolic pathways that would consume primary feedstocks essential for target metabolite synthesis. The first reaction of a dispensable metabolic pathway competing with target metabolite biosynthesis is knocked out by deletion of the associated metabolic enzyme gene. Next, engineers focus on identifying “bottleneck” enzyme reactions that limit target metabolite biosynthesis predicted by analyzing transcriptomic and metabolomic data. One common solution to overcome a bottleneck is to overexpress the rate limiting enzyme-coding gene (Tamano, 2014). The relief of one bottleneck often uncovers secondary bottlenecks, which requires the process to be repeated. That is, repetition of workflow composed of prediction of bottleneck reaction via analysis of transcriptomic and metabolomic data, and subsequent cancellation of the bottleneck by metabolic engineering. This manner of productivity enhancement involving repeated workflow is referred to as the Design-Build-Test-Learn (DBTL) cycle (Vavricka et al., 2020).
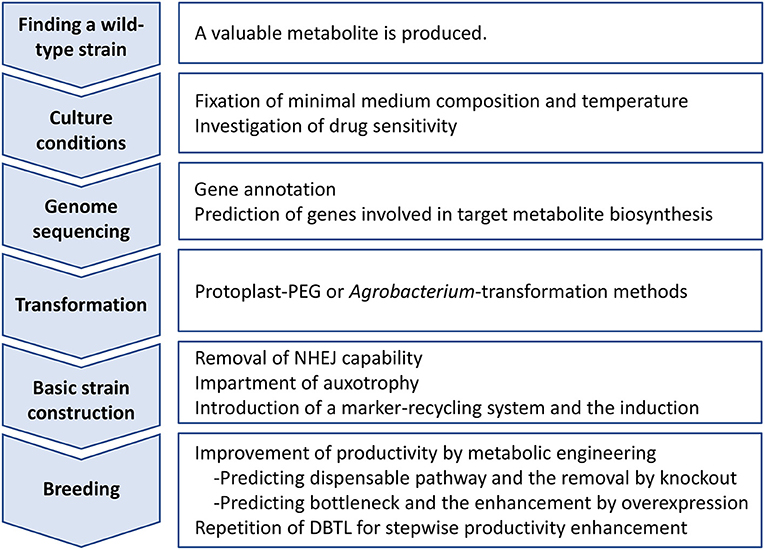
Figure 1. Outline of industrial strain improvement of filamentous fungi for enhancing productivity of a target metabolite. DBTL, Design-Build-Test-Learn cycle; NHEJ, non-homologous end joining; PEG, polyethylene glycol.
Genomic information coupled with new techniques to genetically engineer filamentous fungi has led to tremendous progress in strain improvement in recent years. Hereafter, the development of new techniques focused on increasing efficiency and moving to high throughput mutant construction is required. Moreover, the application of flux balance analysis based on metabolism modeling and other multi-omics analysis will allow engineers to more accurately identify bottleneck genes. Continued progress in development of new experimental microbiology and molecular biology techniques as well as bioinformatic tools related to metabolic engineering, is required to attain the efficient productivity improvements needed by society up until now and from now on.
KT drafted the opinion text. AY checked the draft. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Dr. Daren W. Brown (National Center for Agricultural Utilization Research, USDA, USA) for his critical reading of the manuscript as well as his helpful suggestions. We also thank Editage for English language editing.
Abe, K., Gomi, K., Hasegawa, F., and Machida, M. (2006). Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia 162, 143–153. doi: 10.1007/s11046-006-0049-2
Boruta, T. (2018). Uncovering the repertoire of fungal secondary metabolites: from Fleming's laboratory to the International Space Station. Bioengineered 9, 12–16. doi: 10.1080/21655979.2017.1341022
Browne, A. G. P., Fisher, M. C., and Henk, D. A. (2013). Species-specific PCR to describe local-scale distributions of four cryptic species in the Penicillium chrysogenum complex. Fungal Ecol. 6, 419–429. doi: 10.1016/j.funeco.2013.04.003
Cairns, T. C., Nai, C., and Meyer, V. (2018). How a fungus shapes biotechnology: 100 years of Aspergillus niger research. Fungal Biol. Biotechnol. 5:13. doi: 10.1186/s40694-018-0054-5
Gene Ontology Consortium. (2021). The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325–D334. doi: 10.1093/nar/gkaa1113
Glumoff, V., Käppeli, O., Fiechter, A., and Reiser, J. (1989). Genetic transformation of the filamentous yeast, Trichosporon cutaneum, using dominant selection markers. Gene 84, 311–318. doi: 10.1016/0378-1119(89)90505-2
Gomi, K., Kitamoto, K., and Kumagai, C. (1991). Cloning and molecular characterization of the acetamidase-encoding gene (amdS) from Aspergillus oryzae. Gene 108, 91–98. doi: 10.1016/0378-1119(91)90491-S
Goodwin, S., McPherson, J. D., and McCombie, W. R. (2016). Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 17, 333–351. doi: 10.1038/nrg.2016.49
Gritz, L., and Davies, J. (1983). Plasmid-encoded hygromycin B resistance: the sequence of hygromycin B phosphotransferase gene and its expression in Escherichia coli and Saccharomyces cerevisiae. Gene 25, 179–188. doi: 10.1016/0378-1119(83)90223-8
Heidler, S. A., and Radding, J. A. (1995). The AUR1 gene in Saccharomyces cerevisiae encodes dominant resistance to the antifungal agent aureobasidin A (LY295337). Antimicrob. Agents Chemother. 39, 2765–2769. doi: 10.1128/AAC.39.12.2765
Huang, X., Men, P., Tang, S., and Lu, X. (2021). Aspergillus terreus as an industrial filamentous fungus for pharmaceutical biotechnology. Curr. Opin. Biotechnol. 69, 273–280. doi: 10.1016/j.copbio.2021.02.004
Ishibashi, K., Suzuki, K., Ando, Y., Takakura, C., and Inoue, H. (2006). Nonhomologous chromosomal integration of foreign DNA is completely dependent on MUS-53 (human Lig4 homolog) in Neurospora. Proc. Natl. Acad. Sci. U. S. A. 103, 14871–14876. doi: 10.1073/pnas.0604477103
Jiang, C., Lv, G., Tu, Y., Cheng, X., Duan, Y., Zeng, B., et al. (2021). Applications of CRISPR/Cas9 in the synthesis of secondary metabolites in filamentous fungi. Front. Microbiol. 12:638096. doi: 10.3389/fmicb.2021.638096
Jin, F. J., Hu, S., Wang, B. T., and Jin, L. (2021). Advances in genetic engineering technology and its application in the industrial fungus Aspergillus oryzae. Front. Microbiol. 12:644404. doi: 10.3389/fmicb.2021.644404
Kanamasa, S., Takada, G., Kawaguchi, T., Sumitani, J., and Arai, M. (2001). Overexpression and purification of Aspergillus aculeatus beta-mannosidase and analysis of the integrated gene in Aspergillus oryzae. J. Biosci. Bioeng. 92, 131–137. doi: 10.1016/S1389-1723(01)80213-9
Kitamoto, N., Matsui, J., Kawai, Y., Kato, A., Yoshino, S., Ohmiya, K., et al. (1998). Utilization of the TEF1-alpha gene (TEF1) promoter for expression of polygalacturonase genes, pgaA and pgaB, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 50, 85–92. doi: 10.1007/s002530051260
Kubodera, T., Yamashita, N., and Nishimura, A. (2000). Pyrithiamine resistance gene (ptrA) of Aspergillus oryzae: cloning, characterization and application as a dominant selectable marker for transformation. Biosci. Biotechnol. Biochem. 64, 1416–1421. doi: 10.1271/bbb.64.1416
Liu, L., Zhao, J., Huang, Y., Xin, Q., and Wang, Z. (2018). Diversifying of chemical structure of native Monascus pigments. Front. Microbiol. 9:3143. doi: 10.3389/fmicb.2018.03143
Machida, M., Yamada, O., and Gomi, K. (2008). Genomics of Aspergillus oryzae: learning from the history of Koji mold and exploration of its future. DNA Res. 15, 173–183. doi: 10.1093/dnares/dsn020
Maruyama, J., Nakajima, H., and Kitamoto, K. (2002). Observation of EGFP-visualized nuclei and distribution of vacuoles in Aspergillus oryzae arpA null mutant. FEMS Microbiol. Lett. 206, 57–61. doi: 10.1111/j.1574-6968.2002.tb10986.x
Miyazawa, K., Yoshimi, A., Zhang, S., Sano, M., Nakayama, M., Gomi, K., et al. (2016). Increased enzyme production under liquid culture conditions in the industrial fungus Aspergillus oryzae by disruption of the genes encoding cell wall α-1,3-glucan synthase. Biosci. Biotechnol. Biochem. 80, 1853–1863. doi: 10.1080/09168451.2016.1209968
Mizutani, O., Kudo, Y., Saito, A., Matsuura, T., Inoue, H., Abe, K., et al. (2008). A defect of LigD (human Lig4 homolog) for nonhomologous end joining significantly improves efficiency of gene-targeting in Aspergillus oryzae. Fungal Genet. Biol. 45, 878–889. doi: 10.1016/j.fgb.2007.12.010
Ninomiya, Y., Suzuki, K., Ishii, C., and Inoue, H. (2004). Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc. Natl. Acad. Sci. U. S. A. 101, 12248–12253. doi: 10.1073/pnas.0402780101
Reuter, J. A., Spacek, D. V., and Snyder, M. P. (2015). High-throughput sequencing technologies. Mol. Cell 58, 586–597. doi: 10.1016/j.molcel.2015.05.004
Takahashi, T., Hatamoto, O., Koyama, Y., and Abe, K. (2004). Efficient gene disruption in the koji-mold Aspergillus sojae using a novel variation of the positive-negative method. Mol. Genet. Genomics 272, 344–352. doi: 10.1007/s00438-004-1062-0
Takahashi, T., Masuda, T., and Koyama, Y. (2006). Enhanced gene targeting frequency in ku70 and ku80 disruption mutants of Aspergillus sojae and Aspergillus oryzae. Mol. Genet. Genomics 275, 460–470. doi: 10.1007/s00438-006-0104-1
Takahashi, T., Mizutani, O., Shiraishi, Y., and Yamada, O. (2011). Development of an efficient gene-targeting system in Aspergillus luchuensis by deletion of the non-homologous end joining system. J. Biosci. Bioeng. 112, 529–534. doi: 10.1016/j.jbiosc.2011.08.007
Tamano, K. (2014). Enhancing microbial metabolite and enzyme production: current strategies and challenges. Front. Microbiol. 5:718. doi: 10.3389/fmicb.2014.00718
Tamano, K., Bruno, K. S., Karagiosis, S. A., Culley, D. E., Deng, S., Collett, J. R., et al. (2013). Increased production of fatty acids and triglycerides in Aspergillus oryzae by enhancing expressions of fatty acid synthesis-related genes. Appl. Microbiol. Biotechnol. 97, 269–281. doi: 10.1007/s00253-012-4193-y
Tamano, K., Bruno, K. S., Koike, H., Ishii, T., Miura, A., Umemura, M., et al. (2015). Increased production of free fatty acids in Aspergillus oryzae by disruption of a predicted acyl-CoA synthetase gene. Appl. Microbiol. Biotechnol. 99, 3103–3113. doi: 10.1007/s00253-014-6336-9
Tamano, K., Satoh, Y., Ishii, T., Terabayashi, Y., Ohtaki, S., Sano, M., et al. (2007). The beta-1,3-exoglucanase gene exgA (exg1) of Aspergillus oryzae is required to catabolize extracellular glucan, and is induced in growth on a solid surface. Biosci. Biotechnol. Biochem. 71, 926–934. doi: 10.1271/bbb.60591
Terabayashi, Y., Sano, M., Yamane, N., Marui, J., Tamano, K., Sagara, J., et al. (2010). Identification and characterization of genes responsible for biosynthesis of kojic acid, an industrially important compound from Aspergillus oryzae. Fungal Genet. Biol. 47, 953–961. doi: 10.1016/j.fgb.2010.08.014
Unkles, S. E., Campbell, E. I., Carrez, D., Grieve, C., Contreras, R., Fiers, W., et al. (1989). Transformation of Aspergillus niger with the homologous nitrate reductase gene. Gene 78, 157–166. doi: 10.1016/0378-1119(89)90323-5
van Hartingsveldt, W., Mattern, I. E., van Zeijl, C. M., Pouwels, P. H., and van den Hondel, C. A. (1987). Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Mol. Gen. Genet. 206, 71–75. doi: 10.1007/BF00326538
Vavricka, C. J., Hasunuma, T., and Kondo, A. (2020). Dynamic metabolomics for engineering biology: accelerating learning cycles for bioproduction. Trends Biotechnol. 38, 68–82. doi: 10.1016/j.tibtech.2019.07.009
Ward, M., Wilkinson, B., and Turner, G. (1986). Transformation of Aspergillus nidulans with a cloned, oligomycin-resistant ATP synthase subunit 9 gene. Mol. Gen. Genet. 202, 265–270. doi: 10.1007/BF00331648
Wendt, K. E., Ungerer, J., Cobb, R. E., Zhao, H., and Pakrasi, H. B. (2016). CRISPR/Cas9 mediated targeted mutagenesis of the fast growing cyanobacterium Synechococcus elongatus UTEX (2973). Microb. Cell Fact. 15:115. doi: 10.1186/s12934-016-0514-7
Wong, P. S., Tamano, K., and Aburatani, S. (2021). Improvement of free fatty acid secretory productivity in Aspergillus oryzae by comprehensive analysis on time-series gene expression. Front. Microbiol. 12:605095. doi: 10.3389/fmicb.2021.605095
Yamada, O., Lee, B. R., and Gomi, K. (1997). Transformation system for Aspergillus oryzae with double auxotrophic mutations, niaD and sC. Biosci. Biotechnol. Biochem. 61, 1367–1369. doi: 10.1271/bbb.61.1367
Keywords: filamentous fungi, metabolite, metabolic engineering, gene knockout, gene overexpression
Citation: Tamano K and Yoshimi A (2021) Metabolic Engineering Techniques to Increase the Productivity of Primary and Secondary Metabolites Within Filamentous Fungi. Front. Fungal Biol. 2:743070. doi: 10.3389/ffunb.2021.743070
Received: 17 July 2021; Accepted: 20 October 2021;
Published: 01 December 2021.
Edited by:
Katherine A. Borkovich, University of California, Riverside, United StatesReviewed by:
Tomasz Boruta, Lodz University of Technology, PolandCopyright © 2021 Tamano and Yoshimi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Koichi Tamano, dGFtYW5vLWtAYWlzdC5nby5qcA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.