- 1Colorado Parks and Wildlife, Fort Collins, CO, United States
- 2Colorado Cooperative Fish and Wildlife Research Unit, Department of Fish, Wildlife and Conservation Biology, U.S. Geological Survey, Colorado State University, Fort Collins, CO, United States
- 3Genomic Variation Laboratory, Department of Animal Science, University of California, Davis, Davis, CA, United States
Introduction: Myxobolus cerebralis, the parasite responsible for salmonid whirling disease, was unintentionally introduced to and became established in Colorado in the 1990s. Mortality of young-of-year fish due to infection by M. cerebralis resulted in recruitment failure and subsequent significant declines in Rainbow Trout (Oncorhynchus mykiss) populations. The complex multistage lifecycle of M. cerebralis makes it difficult to eradicate and manage, and hatchery control strategies do not work in the wild. A viable method that has been utilized for wild populations is enhancing host resistance. Myxobolus cerebralis resistant Rainbow Trout were discovered at a hatchery in Germany and subsequently incorporated into Colorado's brood stock program. Since 2004, M. cerebralis resistant strains have been stocked into all major Colorado coldwater drainages to re-establish Rainbow Trout populations after whirling disease-related declines, with documented survival and reproduction of stocked disease resistant fish.
Methods and results: Genetic population assignment tests (via putatively neutral microsatellite markers) were used to monitor the stocked populations and indicated that, after only a few years, many of the individuals in these populations unexpectedly assigned to genetic strains that were historically susceptible to M. cerebralis. To further investigate the genetic composition of these fish, a single nucleotide polymorphism (SNP) panel was used to determine the percent genetic composition of resistant strain in these individuals. Microsatellites and SNPs provided similar results, indicating a low percentage of ancestry from the resistant strain in these fish, but they continued to survive exposure to M. cerebralis, suggesting that these individuals possessed genetic loci necessary for resistance. Finally, a quantitative trait locus (QTL) region (termed WDRES-9) was used to identify individuals with alleles associated with disease resistance. Implementation of the WDRES-9 QTL test allowed for more accurate determination of M. cerebralis resistant individuals within wild populations and better described their variability in resistance.
Discussion: Overall, reintroductions and genetic monitoring required a suite of tools to understand the effects of M. cerebralis exposure on the genetic resistance of wild fish populations over time.
1 Introduction
Infectious diseases have major negative biological, ecological and economic implications for both fish production and aquatic ecosystems and pathogen control or eradication is a major priority for managers (Chapman et al., 2021; Fraslin et al., 2020; Das and Sahoo, 2014). Several management options are available to hatchery facilities to reduce or eliminate infectious pathogens, such as antibiotic treatment, vaccination, and probiotics, among others (Das and Sahoo, 2014). Hatcheries can also manipulate their facilities to eliminate pathogens using techniques such as dewatering and changing water sources (Wagner, 2002; Hedrick et al., 1998) but most of these methods are cost prohibitive. Additionally, these hatchery management options are not possible or appropriate for wild and native fisheries. Small-scale modifications, such as removal of depositional backwaters, can be used to reduce secondary host or pathogen habitat availability locally (Thompson, 2011). Although possible, large-scale environmental manipulations are difficult to implement in the wild (Nehring et al., 2018), and other strategies need to be considered. One viable option is using host innate disease resistance. Disease resistance in fishes is a well-established concept and thought to be an adaptive genetically controlled mechanism (Fraslin et al., 2020) that could be used to re-establish populations in situations in which the pathogen cannot be eliminated (Hedrick et al., 1998; Fraslin et al., 2020).
Whirling disease, caused by the parasite Myxobolus cerebralis, has caused catastrophic declines in many important fisheries in the United States intermountain west beginning in the 1980s (Vincent, 1996; Nehring and Walker, 1996). The life cycle includes the primary host, a salmonid, and a secondary host tubificid worm, Tubifex tubifex. Clinical signs of disease in salmonids include deformities of the cranium, operculum, lower jaw and spine, black tail, and erratic whirling swimming behavior. The complex multistage life cycle of M. cerebralis makes it difficult to eliminate in the wild once it is established. Due to the difficulty of eliminating the parasite in the wild, host resistance was thought to be one of the most viable options available to disrupt the parasite life cycle (El-Matbouli et al., 1999; Kerans et al., 1999; Beauchamp et al., 2002; Schisler et al., 2000; Wagner et al., 2006; Fetherman et al., 2011, 2012; Nehring et al., 2013, 2016; Avila et al., 2018). Multiple attempts were made to use resistant lineages of the secondary T. tubifex worm host (Beauchamp et al., 2005; Nehring et al., 2013), but efforts to shift community composition, as well as introduction efforts, were met with limited success (Clapp, 2009; Winkelman and Gigliotti, 2014). Serendipitously, a whirling disease resistant Rainbow Trout Oncorhynchus mykiss strain was discovered in a German hatchery that presumably developed resistance to whirling disease through continuous exposure to M. cerebralis for over a century (El-Matbouli et al., 2002). The German Rainbow Trout strain (referred to hereafter as GR) is more resistant to whirling disease than many other Rainbow Trout strains found in North America (Hedrick et al., 2003). However, due to the long history of domestication of the GR (roughly 100 years; El-Matbouli et al., 2002; Hedrick et al., 2003), it was thought that their survival in the wild would be lower than other Rainbow Trout historically stocked in Colorado (Schisler et al., 2006), and this was supported by laboratory (Fetherman et al., 2011) and field experiments (Fetherman et al., 2020). To overcome the effects of domestication, increase wild survival, and maintain resistance to M. cerebralis, Colorado developed a breeding program to cross the GR with other Rainbow Trout strains (Schisler et al., 2006; Fetherman et al., 2011, 2012; Avila et al., 2018, 2022). The resulting Rainbow Trout offspring are M. cerebralis-resistant (Schisler et al., 2006; Fetherman et al., 2011, 2012) and have been stocked into all major Colorado coldwater drainages since 2004.
Given their performance during controlled lab exposure experiments (Schisler et al., 2006; Fetherman et al., 2011, 2012), offspring of GR crosses were expected to exhibit increased survival upon exposure to M. cerebralis in the wild. However, the successful reestablishment of Rainbow Trout in rivers where the parasite had become established was dependent not only upon the survival of stocked fish, but also their ability to reproduce and incorporate the genetic basis of whirling disease resistance into wild populations. Monitoring and research efforts to assess both survival and reproduction in the wild have been ongoing (Fetherman et al., 2014; Avila et al., 2018, 2023), and included efforts to assess the genetic composition of the populations over time, especially the incorporation of whirling disease resistant genetics into wild populations (Fetherman et al., 2014, 2018, 2020; Fetherman and Schisler, 2014). Our overall objective in this manuscript was to utilize the multiple genetic tools available at the time of sampling to understand genetic patterns of M. cerebralis resistance in wild Rainbow Trout populations in the Colorado and Gunnison rivers due to the reintroduction of M. cerebralis-resistant Rainbow Trout strains.
2 Methods
The upper Colorado River is located in Grand County, Colorado with the primary study sites located 1.6 km downstream of Windy Gap Reservoir (Figure 1). Flow ranges from a mean of 2.2 to 22.5 cm. Temperature in the upper Colorado River ranges from 3.4 to 16.2°C and has a mean annual temperature of 10.7°C (Fetherman et al., 2014). The lower Gunnison River is located in Montrose and Delta counties, Colorado (Figure 2), flowing from Crystal dam downstream through the Black Canyon of the Gunnison National Park and Gunnison Gorge National Conservation Area. Flow varies from 0.48 to 58 cm with an annual mean of 20 cm (U.S. Geological Survey [USGS], 2016).
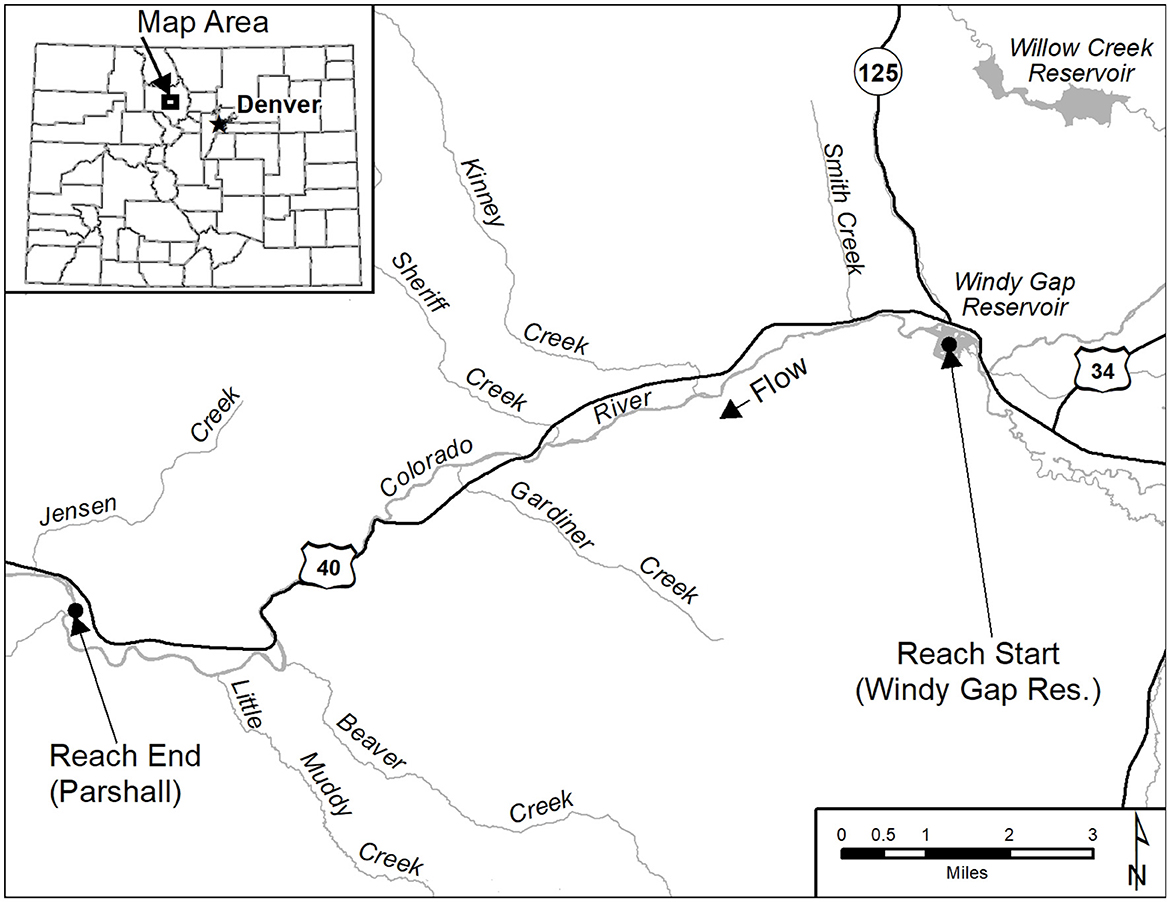
Figure 1. The upper Colorado River study site. The 23.42 km study site was located between Windy Gap Reservoir and Parshall, CO. Start and end points are shown with black dots.
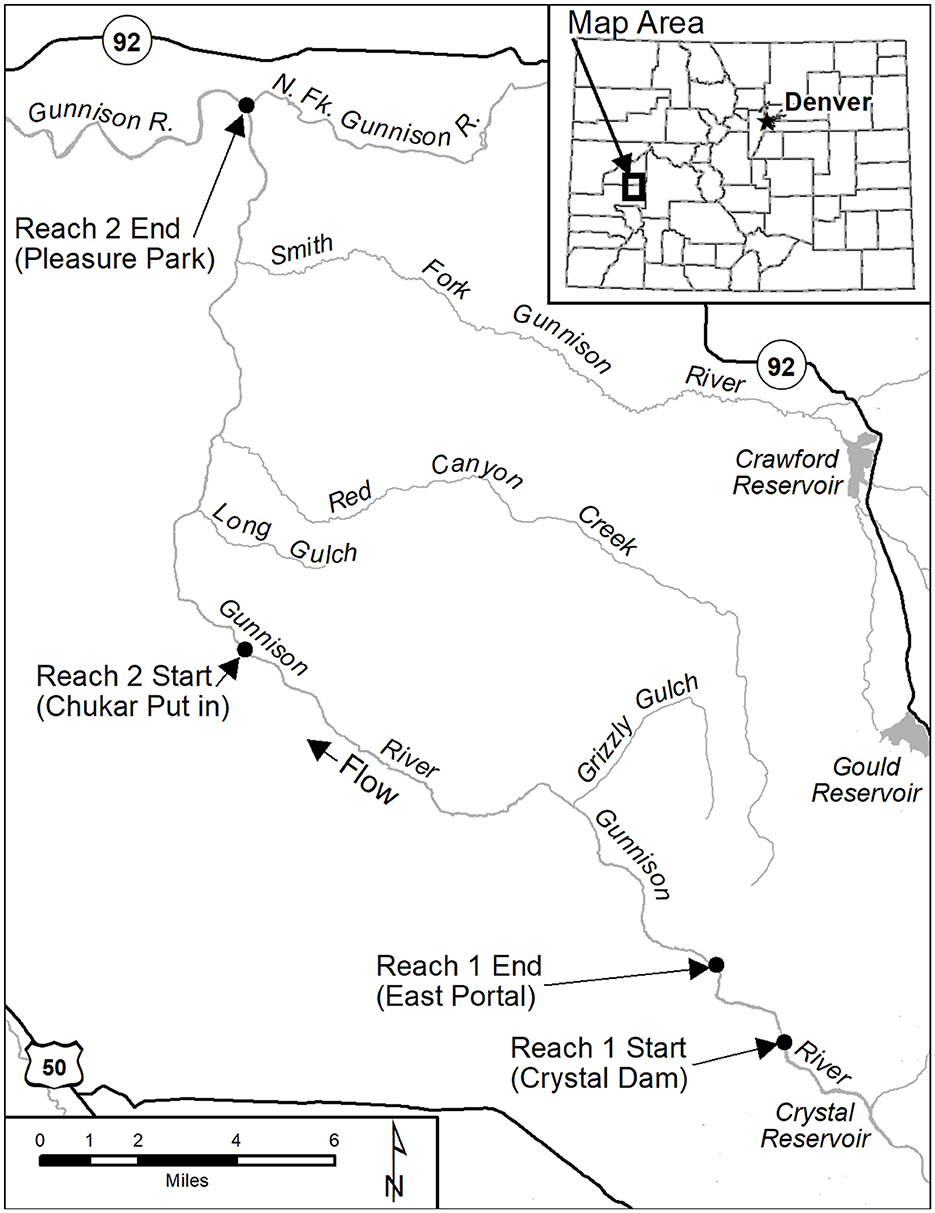
Figure 2. The Gunnison River study sites are located northwest of the Black Canyon of the Gunnison National Park, Colorado. The first study site was 4151.69 m long and starts below Crystal Reservoir. The second study site was 21.5 km long and located within the Gunnison Gorge Wilderness Area. Start and end points are shown with black dots.
2.1 Disease history in the Colorado and Gunnison rivers
Prior to the introduction of whirling disease, a thriving, self-sustaining Rainbow Trout population existed in the upper Colorado River, with adult Rainbow Trout outnumbering adult Brown Trout Salmo trutta 2:1 (Nehring, 2006). In 1981, Colorado Parks and Wildlife (CPW) started taking wild Rainbow Trout eggs from the stretch of river located below Windy Gap Reservoir to establish a wild Rainbow Trout brood stock known as the Colorado River Rainbow (CRR). The CRR was stocked widely throughout Colorado to establish many naturally reproducing wild Rainbow Trout fisheries (Walker and Nehring, 1995). M. cerebralis was unintentionally introduced to the upper Colorado River in the 1980s when privately reared Rainbow Trout previously exposed to M. cerebralis were stocked into three private water bodies located upstream of Windy Gap Reservoir. Fish below Windy Gap Reservoir tested positive for M. cerebralis in 1988, and a subsequent decline in the younger age classes of Rainbow Trout was observed in the early 1990s (Nehring, 2006). Although several possible reasons for the losses were investigated (Walker and Nehring, 1995; Schisler et al., 1999a,b, 2000), whirling disease was determined to be the primary cause for the disappearance of younger age classes (Nehring and Thompson, 2001). Despite repeated stocking efforts with CRRs (average of 11,634 stocked annually), Rainbow Trout abundances eventually dropped to lower than 90% of those observed prior to the establishment of M. cerebralis (Nehring, 2006). A similar drop in Brown Trout populations was not observed (Nehring and Thompson, 2001; Nehring, 2006) because they are more resistant to M. cerebralis than Rainbow Trout, having evolved with M. cerebralis, in their native, European home ranges (Hoffman, 1970; Hedrick et al., 1999, 2003).
Whirling disease was introduced to the Gunnison River basin above Blue Mesa, Morrow Point, and Crystal reservoirs in 1987 through fish stocked into Meridian Lake by a private hatchery that subsequently tested positive for M. cerebralis. Fish in the East River below Meridian Lake began showing signs of disease and tested positive in 1994. Initial introduction to the lower Gunnison River, where our study occurred, likely happened in 1993 when an uncontrolled surface spill of water over Crystal Dam carried the parasite over the dam. The lower Gunnison River below Crystal Dam had not been stocked with fish of any kind since the late 1970s, eliminating stocking as a potential disease introduction route (Nehring, 2006). The Rainbow Trout population in the lower Gunnison River collapsed, declining by more than 90% in the years following M. cerebralis introduction (Nehring and Thompson, 2001).
Whirling disease resistant Rainbow Trout were first stocked in the lower Gunnison River in 2004 and upper Colorado River in 2006 (Schisler and Fetherman, 2010), with subsequent yearly stocking occurring in both rivers (average of 19,60 and 23,652 stocked annually into the Gunnison and Colorado rivers, respectively). Both rivers were stocked with the first filial (F1) generation of crosses between the GR and CRR, which were effectively 50% GR and 50% CRR (50:50 GRxCRR) and exhibited resistance characteristics similar to those of the GR strain (Schisler et al., 2006; Fetherman et al., 2012). In addition, between 2006 and 2013, the East Portal was stocked and managed as a potential wild brood source for GRxCRR. The F1 fish were capable of attaining critical swimming velocities similar to those of the CRR strain (Fetherman et al., 2011) and were therefore the best candidate for reestablishing Rainbow Trout populations in large river systems with high water velocity. Larger individuals (≥ 150 mm total length) were initially used for stocking events because they were less susceptible to M. cerebralis infection (Ryce et al., 2005) and Brown Trout predation, but survival and recruitment were low (Fetherman et al., 2014). To reduce hatchery related behavioral conditioning and increase survival, fry were stocked. Compared to larger fish, fry survived and recruited into the population at a higher rate in Colorado's large river systems (Fetherman et al., 2014; Fetherman and Schisler, 2016). Although this study is focused on the incorporation of disease resistance following these initial reintroductions with GRxCRR offspring, Colorado Parks and Wildlife continues to stock fry from various resistant strains to maintain resistance and increase Rainbow Trout abundance.
2.2 Sample collection
Fin clips have been collected from Rainbow Trout populations in the Colorado and Gunnison rivers during population surveys since 2006. Population surveys were completed using raft and backpackmark-recapture or removal electrofishing efforts (Fetherman et al., 2014). Fin clips consisted of a 1 cm squared piece of fin collected from the upper caudle fin with scissors, placed into a 2 ml vial with 95% ETOH, and archived in a freezer by date and location of collection. Over 3,500 fin clips have been collected from several rivers across Colorado from 2007–2023 to investigate M. cerebralis resistance. Samples for this study were haphazardly selected to be genotyped from the upper Colorado River and Gunnison River. The data set used here consists of 1,765 genetic samples collected from the upper Colorado River and the Gunnison River, Colorado (Table 1) and samples were selected in years following the initial resistant Rainbow Trout stocking and to determine changes in genetic resistance characteristics of these populations over time. In the Colorado River, the study reach extended 23.42 km from Windy Gap dam on the upstream end to Parshall, Colorado on the downstream end (Figure 1). The Gunnison River contained two study reaches and Rainbow Trout populations of interest: one in the Gunnison Gorge and the other in the East Portal in Black Canyon of the Gunnison National Park. The East Portal study reach included the 4.2 km stretch between Crystal Dam to the Gunnison Diversion Dam (Figure 2). The Gunnison Gorge reach included the 21.5 km stretch beginning at the Chukar access point below the Black Canyon of the Gunnison National Park and extended to Pleasure Park (Figure 2). In the Colorado River and Gunnison Gorge stretch of the Gunnison River where few wild Rainbow Trout remained after the introduction of M. cerebralis, it was thought that GR alleles and disease resistance would rapidly be incorporated into the remnant populations. In contrast, fewer losses occurred in the East Portal Rainbow Trout population because there is less worm habitat available for the intermediate Tubifex host in this tailwater stretch and no upstream source of infection. Therefore, it was thought that incorporation of GR alleles and disease resistance would take longer, given that both survival and reproductive success could differ between remnant wild fish and stocked disease resistant fish and may interact in unpredictable ways.
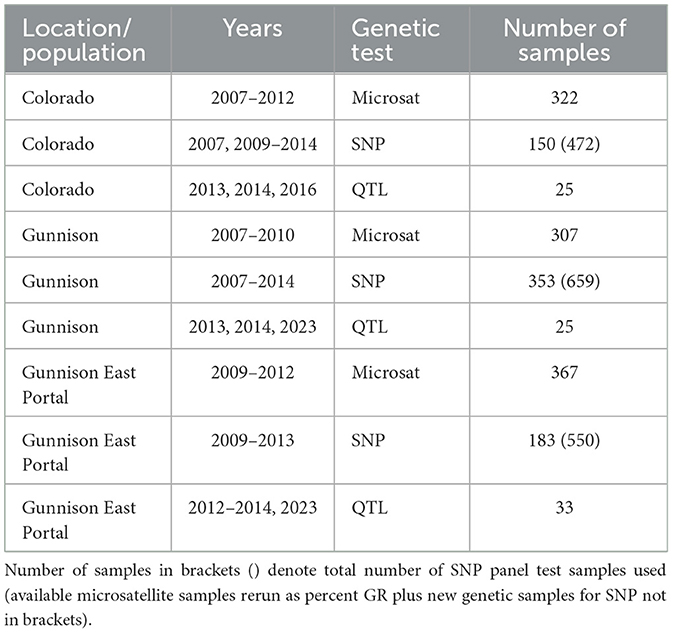
Table 1. Total number of genetic samples that were processed for each genetic test from each location/population and corresponding years.
Laboratory analysis: All genetic samples were sent to the Genomic Variation Laboratory (GVL) at the University of California, Davis for processing and genetic testing. There were three genetic analysis methods used between 2007 and 2023. The first was genetic population assignment, the details of which are described in Fetherman et al. (2014). Briefly, 18 microsatellite markers were genotyped at the GVL and software programs NewHybrids (Nielsen et al., 2001) and Structure 2.3.3 (Pritchard et al., 2000) were used in tandem and compared to distinguish individual assignment to strain, filial generation cross, or backcross (e.g., pure CRR strain, pure GR strain, GRxCRR, backcross of GRxCRR with GR or CRR). The second used genetic population assignment and estimation of the amount (percent) German Rainbow (hereafter referred to as percent GR test) within a fish. A single nucleotide polymorphism (SNP) panel, made of 46 SNPs, was developed by using restriction site associated DNA sequences (RAD-seq) at the GVL. Individuals were genotyped using Integrated Fluidic Circuits (IFC) on a Standard BioTools BioMark platform (South San Francisco, CA). Software program NewHybrids (Nielsen et al., 2001) was used to classify crosses (e.g., F1, F2, B1, B2, pure CRR, pure GR) and program Structure 2.3.3 (Pritchard et al., 2000) was used to determine percent GR ancestry (Fetherman and Schisler, 2013, 2016). The SNP panel did not target specific resistance loci, but rather identified putatively neutral loci with substantial allele frequency differences between pure GR and pure CRR strains. All SNPs were evaluated for neutrality relative to the known resistance markers and it was determined that they were unlikely to be influenced by the quantitative trait loci (QTL) described below (nearest genetic distance of 14 cm from the QTL). However, because the SNPs were developed prior to the completion of the Rainbow Trout genome mapping project, they were not mapped to determine relative distance to other potential survival loci upon which selection could be acting, and doing so was outside the scope of this study. Power of microsatellite and SNP panels were simulated to evaluate the accuracy of hybrid classes and allele frequencies associated with that class and were compared to known samples (Fetherman and Schilser, 2017). The final genetic method used a quantitative trait locus (QTL), known as the WDRES-9 QTL, to determine if fish have a major-effect M. cerebralis-resistant allele. Full description of the QTL is outlined in Baerwald et al. (2011). Briefly, the WDRES-9 QTL is located on chromosome Omy9 and explains a large amount of the phenotypic variance (50–86%) for M. cerebralis resistance in GRxCRR F2 families (Baerwald et al., 2011). The WDRES-9 QTL is in close proximity to the microsatellite marker BX310634, and individuals that are homozygous at this QTL marker for the resistance allele (RR) developed fewer myxospores than individuals that were heterozygous (RS) or did not have the resistant alleles (homozygous non-resistant; SS) when exposed to M. cerebralis (Baerwald et al., 2011; Fetherman et al., 2014, 2020).
2.3 Analysis
Data visualization and general statistics were done in program R (version 4.4.0; R Core Team, 2024) with packages ggplot2 (Wickham, 2016) and dplyr (Wickham et al., 2023). A total of 996 genetic samples were used for genetic assignment with three possible assignments: CRR, GRxCRR, backcrosses, and unknown. Genetic assignment results are presented as the proportion of fish assigned to one of the three categories for each year sampled in each of the three populations. A total of 1,681 genetic samples were used for the percent GR test, which ranged between 0 and 100% GR (Table 2). Comparing expected genetic percentages against known assignments of pure strains and filial generation crosses and backcrosses, showed that an individual 50:50 GRxCRR fish had a percent GR that ranged between 0.40 and 0.60 (Supplementary Figure 1). Backcrossing a 50:50 GRxCRR with a GR or a CRR resulted in an individual with percent GR ranging between 0.60 and 0.80 and 0.20 and 0.40, respectively. Pure GR fish had a percent GR very close to 1.00, and pure CRR fish had a percent GR below 0.20 (Supplementary Figure 1, Fetherman and Schisler, 2017). When examining samples collected from wild populations, we assumed that higher levels of GR ancestry (>20%) may indicate increased likelihood of M. cerebralis-resistance and a lower number of myxospores when exposed to M. cerebralis (Fetherman et al., 2012). Results are presented as the proportion of fish tested that fell into one or the other of these bins for each year sampled in each of the three populations of interest. A total of 83 genetic samples were used for the WDRES-9 QTL analysis. Results of the WDRES-9 QTL could fall into one of three categories: homozygous resistant (RR), homozygous susceptible (SS), and heterozygous resistant (RS). WDRES-9 QTL results are presented as the proportion of fish tested that fell into one of these three categories for each year sampled in each of the three populations of interest.
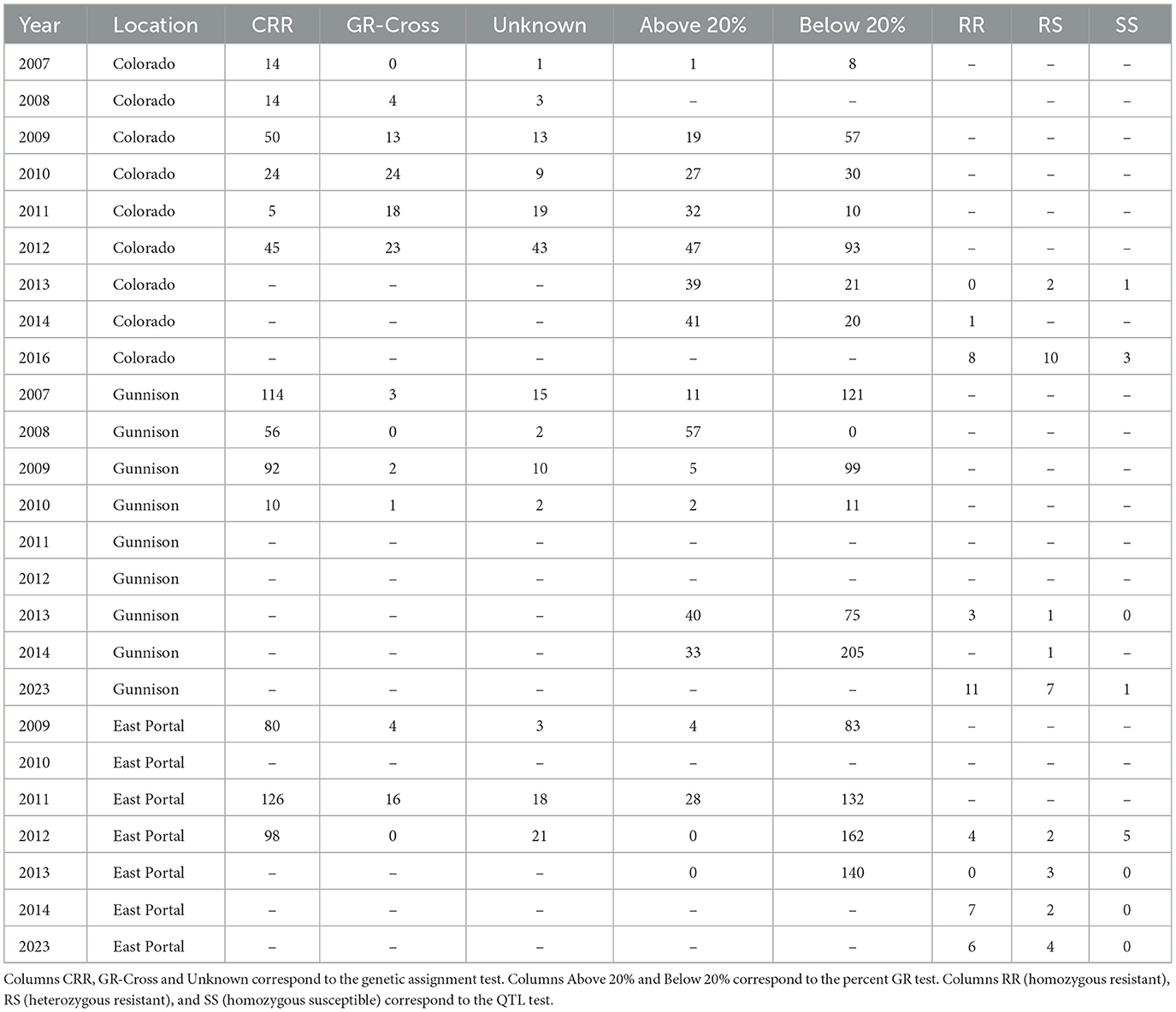
Table 2. Results for numbers of fish falling into each of the categories or bins from each genetic test by year and location.
3 Results
3.1 Microsatellites
The number of genetic samples analyzed within each year varied by location, with between 15 and 111 samples analyzed from the upper Colorado River between 2007 and 2010, 13 and 132 samples from the Gunnison River between 2007 and 2010, and 87 and 160 samples from the East Portal of the Gunnison River between 2009 and 2012 (Table 2).
In the upper Colorado River population, there was a lag between when the first GR × CRR fish were stocked and when the assignment test showed an increase in GR-cross fish, suggesting it took some time for the fish to establish, and recruit to and start to reproduce. However, the number of fish assigned as CRR decreased and fish assigned GR increased from 2007 through 2011, as expected (Figure 3A). The increase in GR genetics after stocking indicated that GR-cross fish were surviving exposure to M. cerebralis and natural reproduction was increasing M. cerebralis resistance within the population. We also observed this trend of admixture and increasing GR-cross fish within years in the upper Colorado River. As an example, data collected from fry within 2014 shows that in July 2014, 69% of the fry collected from the Colorado River were assigned as CRR, whereas in October 2014, only 3% of the fry assigned as CRR. These results suggest that GR-cross individuals were surviving exposure to M. cerebralis and contributing to future generations of this population. The number of fish classified as unknown increased during these years as well, but the NewHybrid results could have been affected by the proximity of the BX310634 microsatellite to the resistance QTL. The increasing number of fish classified as unknown was one impetus for developing the SNP panel that allowed us to account for variability in genetic structure through quantification of percent GR in each fish.
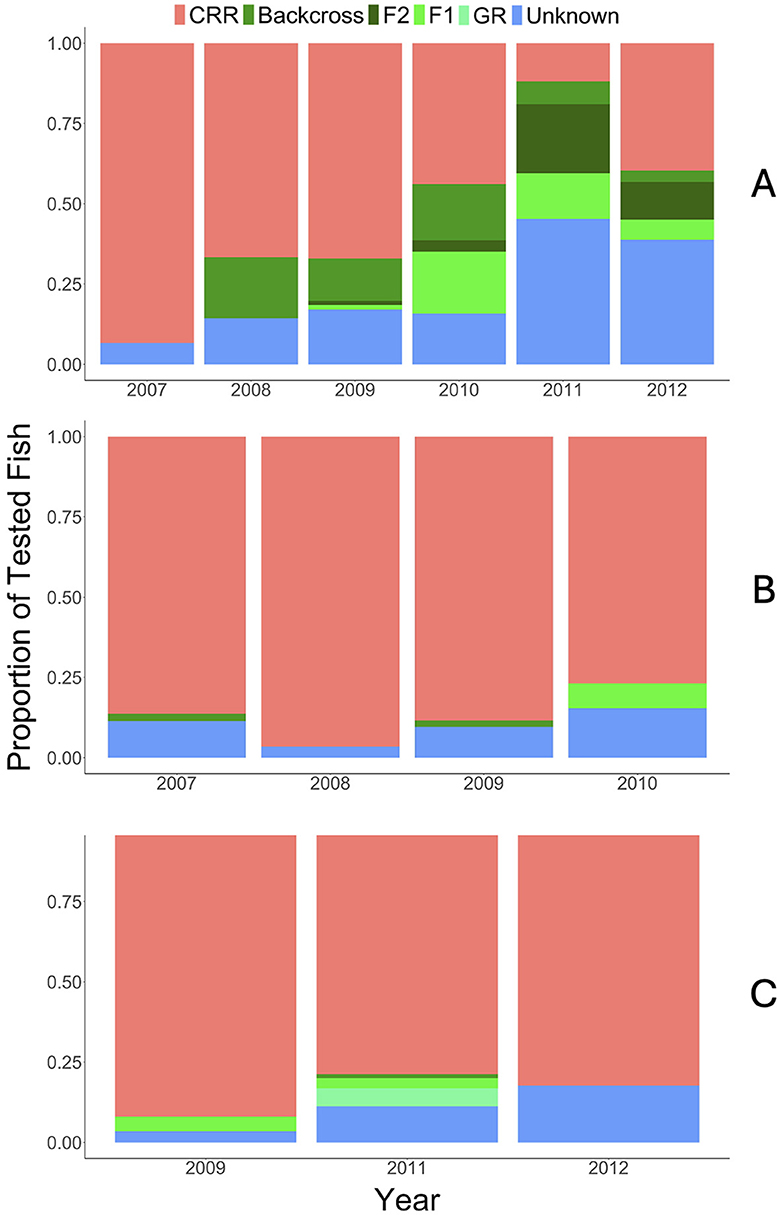
Figure 3. Resulting genetic assignment (CRR, red; Backcross, green; F2, dark green; F1, neon green; GR, light green; Unknown, blue) for each location/population (A) upper Colorado River, (B) Gunnison River (Gunnison Gorge through Pleasure Park), and (C) East Portal of the Gunnison River. Y-axis represents proportion of tested fish and x-axis represents year for each location.
Percent GR did not increase in the Gunnison Gorge as quickly as in the Colorado River (Figure 3B). However, this was not unexpected for the East Portal population given that there were more wild Rainbow Trout in this stretch of the Gunnison River prior to stocking the GR-crosses. Within both Gunnison River populations, 75% or greater of the fish were assigned CRR between 2007 and 2012, and low numbers of fish were assigned GR or unknown, suggesting that admixture was limited or non-existent (Figures 3B, C). Assignments within a year support this in both the Gunnison River and the East Portal populations. In the Gunnison River in August 2014, 83% of the fry were assigned as CRR, increasing to 94% in October 2014, suggesting limited admixture. Admixture in the East Portal population appeared to be non-existent, with 100% of the fry collected in both July and November 2012 assigned as CRR.
3.2 Single nucleotide polymorphisms
The number of genetic samples analyzed within each year varied by location, with between 9 and 140 samples analyzed from the upper Colorado River between 2007 and 2014, 87 and 162 samples from the Gunnison River between 2007 and 2014, and 13 and 238 samples from the East Portal of the Gunnison River between 2009 and 2013 (Table 2).
Within the upper Colorado River population, the number of fish with >20% GR ancestry increased and fish with <20% GR ancestry decreased from 2007 through 2011 (Figure 4A). With the exception of 2012, which had a larger proportion of fish with <20% GR ancestry than years prior, the number of fish with <20% GR ancestry remained fairly stable after 2011. Percent GR ancestry also increased within years. As an example, in 2014, the percent GR ancestry of the fry collected averaged (± SD) 11 ± 15%, increasing to 47 ± 12% in October. The percent GR test did not produce any unknown fish in either river, resolving one of the issues experienced when using the prior genetic assignment test.
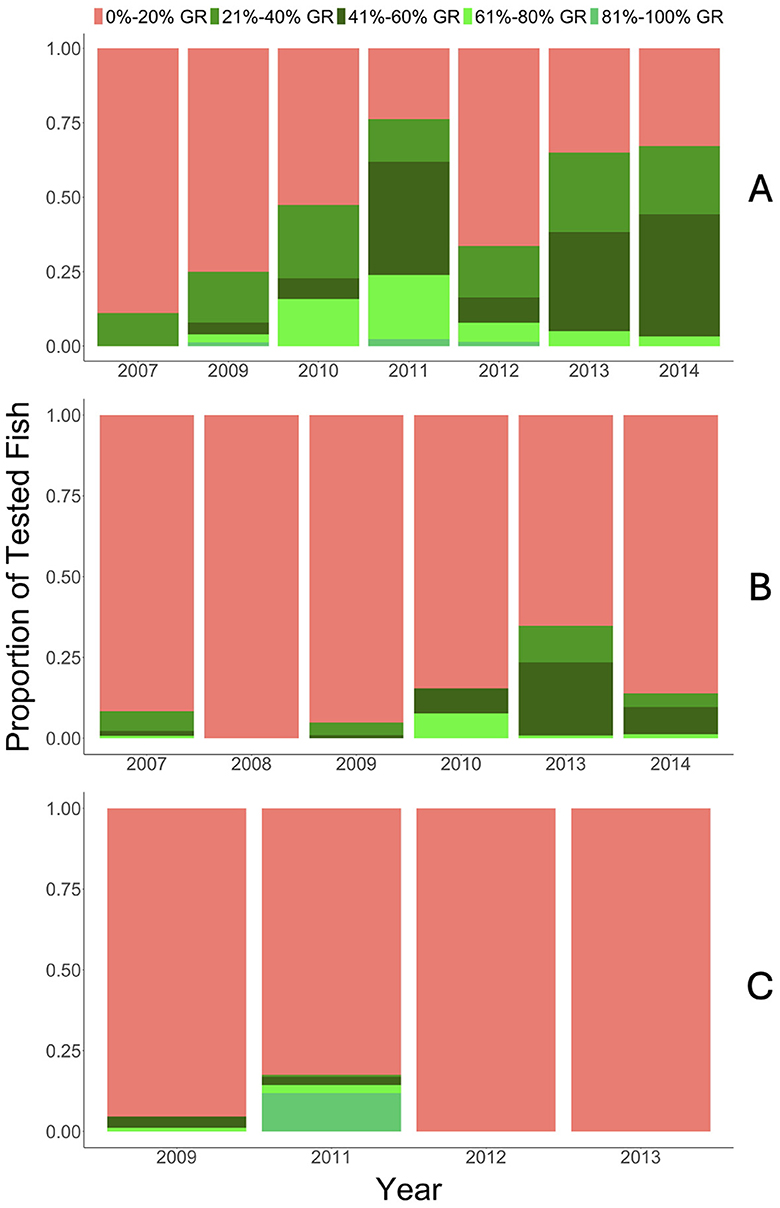
Figure 4. Resulting percent GR values (0%–20%, red; 21%–40%, medium green; 41%–60%, dark green; 61%–80%, neon green; 81%–100%, green) for each location/population (A) upper Colorado River, (B) Gunnison River (Gunnison Gorge through Pleasure Park), and (C) East Portal of the Gunnison River. Y-axis represents proportion of tested fish and x-axis represents year for each location.
Seventy-five percent or greater of the proportion of tested fish had <20% GR ancestry and a low proportion of fish tested had >20% GR ancestry within the Gunnison River populations between 2007 and 2014 (Figures 4B, C). The Gunnison Gorge population did show an increase in the proportion of fish with >20% GR ancestry between 2009 and 2012, but this increase was not as large as seen in the Colorado River where a similar lack of wild Rainbow Trout remained prior to stocking. Additionally, within a year, percent GR ancestry in fry was relatively low. For example, in 2014, the average percent GR ancestry in the fry collected was 7 ± 13% and remained low in October at 2 ± 6%. Similarly, in the East Portal in 2012, average percent GR ancestry was 2 ± 2% in July and 4 ± 3% in November. Percent GR results, especially in the East Portal of the Gunnison River, indicate that despite stocking only M. cerebralis-resistant Rainbow Trout, most fish genetically appeared to be pure CRR given the SNPs used to differentiate the genetic composition of the population.
3.3 WDRES-9 quantitative trait locus
The number of genetic samples analyzed within each year varied by location, with between 1 and 21 samples analyzed from the upper Colorado River in 2012 and 2023, 1 and 19 samples from the Gunnison River in 2012 and 2023, and 6 and 10 samples from the East Portal of the Gunnison River in 2012 and 2023 (Table 2).
Resulting genotypes of the proportion of tested fish for all populations indicated that most (66% or greater) of the fish had M. cerebralis-resistant alleles (i.e., were RR or RS; Figure 5). This result suggests that 1) selection may be acting most strongly upon the resistance QTL and other genes involved in resistance such that fish are RR or RS at this loci and 2) wild CRR genetics needed for survival in Colorado's large river systems (e.g., feeding behavior, swimming performance, predator avoidance, etc.) are maintained in individuals surviving, reproducing, and contributing to the spawning populations.
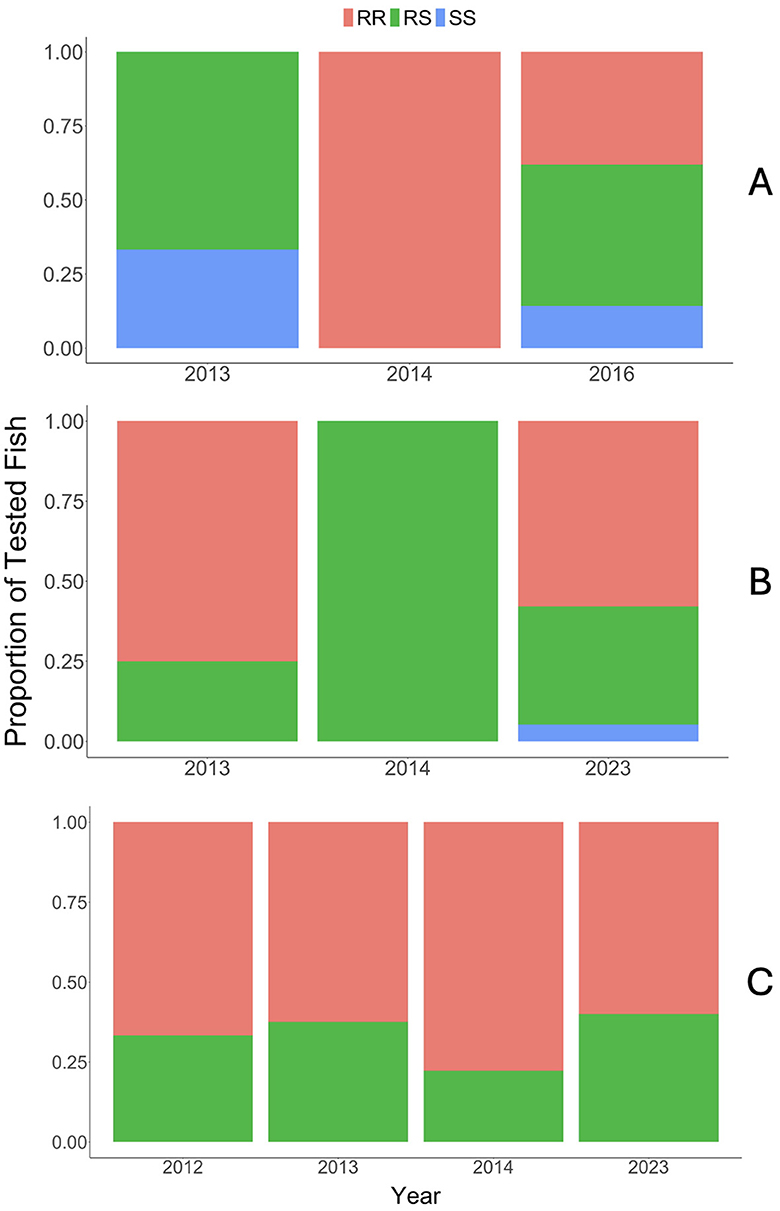
Figure 5. Resulting genotypes (RR, homozygous resistant, red; RS, heterozygous resistant, green; SS, homozygous susceptible, blue) from the QTL test for each location/population (A) upper Colorado River, (B) Gunnison River (Gunnison Gorge through Pleasure Park), and (C) East Portal of the Gunnison River. Y-axis represents proportion of tested fish and x-axis represents year for each location.
4 Discussion
Our analysis of the WDRES-9 QTL suggests that the Rainbow Trout populations within the upper Colorado River and Gunnison River maintained the M. cerebralis resistance alleles despite showing low percent GR genetics in most of the individuals tested. Our results indicate that stocked M. cerebralis resistant Rainbow Trout reproduced in the wild and produced offspring that genetically appeared to be wild CRR fish but maintained M. cerebralis resistance. The WDRES-9 QTL gives a more nuanced understanding of the dynamics and retention of M. cerebralis resistance in wild Rainbow Trout. The development of the WDRES-9 QTL allowed for genotyping of the specific resistant alleles in M. cerebralis-resistant Rainbow Trout (Baerwald et al., 2011) and investigating the presence of the WDRES-9 QTL in individual fish started in Colorado in 2020. Laboratory results indicated that myxospore counts appeared correlated with alleles of a microsatellite marker in close proximity to the QTL. Individuals homozygous for the resistance-associated allele developed fewer myxospores than individuals that were heterozygous for the resistance-associated allele or did not have the allele (Fetherman et al., 2020). Presence of the resistance associated allele in the upper Colorado River, coupled with the across and within year assignment and percent GR ancestry, suggest that GR resistance was incorporated into the population by continued stocking and subsequent natural reproduction of the stocked GR-crosses. Conversely, in the Gunnison River population, there was little evidence of admixture, suggesting that the resistance allele may have already been present at low frequencies prior to the introduction of M. cerebralis.
Prior to the development of the QTL, genetic monitoring was initially done with genetic assignment because the number of potential outcrosses or backcrosses was low, and it was thought that genetic assignment would be an acceptable method to classify stocked Rainbow Trout based on previous laboratory research. In the mid-2000s when stocking of GRxCRR began, the estimated abundance of CRR was low in the upper Colorado and the Gunnison rivers, with a 90 to 100% reduction in abundance compared to the years prior to the introduction of M. cerebralis (Nehring, 2006). Therefore, we thought that stocked GR-cross fish would mainly reproduce with other stocked GR-cross fish due to the very low numbers of CRR in these systems. However, our results indicate that, although there was an increase in GR-cross fish in the Colorado River, there was also an increase in unknown and CRR fish that was unexpected. In the Gunnison River populations, there were also more CRR fish than we thought based on the low number of wild CRR and large numbers of GR-cross fish stocked in the Gunnison Gorge. In addition, the GR-cross fish did not appear to be reproducing and contributing to genetic resistance in the East Portal population. The percent GR test was developed and implemented to try to overcome some of the short falls of genetic assignment and did resolve unknowns obtained from the assignment test. Evaluations of the technique during development indicated that fish with higher percent GR developed lower myxospores compared to fish with lower percent GR (Fetherman and Schisler, 2013, 2016).
Stocking GRxCRR was expected to improve the survival, recruitment, and disease resistance in these systems by increasing the percent GR, and it was thought this would occur quickly. However, the overall inference from the percent GR test was the same as the assignment test. Fish in both systems had lower percent GR despite the large amounts of GR-cross fish stocked between 2004 and 2014 (upper Colorado River: 988,190 total GR-cross; Gunnison River: 689,342 total). One possible reason for the high proportion of individuals exhibiting a low percent of GR genetics was a mismatch of spawning times between the GRxCRR and CRR (pure GR typically spawn in the late fall in the hatchery). Differences in spawning time have been seen in Brown Trout, with stocked Brown Trout spawning earlier than wild Brown Trout (Stefanik and Sandheinrich, 1999). However, spring population sampling in both the Colorado River and the East Portal of the Gunnison River showed that the GR-cross fish were spawning at the same time as the wild Rainbow Trout. Another potential reason for the high proportion of individuals with low GR ancestry in the population could have been that GR-crosses spawn with CRR but only offspring that have wild CRR genetics survive due to the history of domestication in the GR-cross. However, higher CRR survival was thought to be unlikely given their history of susceptibility to M. cerebralis and the high myxospore counts developed by pure CRRs in the wild (Nehring and Thompson, 2001, 2003; Fetherman et al., 2014) and during laboratory exposure experiments (Schisler et al., 2006; Fetherman et al., 2012).
We feel that two other explanations for the patterns we observed were more likely. First, within the East Portal and Gunnison populations, the CRR may have developed resistance over 20 years of continuous exposure in the wild. Selective breeding in an aquaculture environment has helped Rainbow Trout develop Flavobacterium psychrophilum resistance in just a few generations (Leeds et al., 2010). We estimate that there could have been between five and eight generations between 1988 and 2006 since M. cerebralis has been a continuous stressor since the late 1980s (Nehring and Thompson, 2001; Nehring and Walker, 1996). Survival of exposure to M. cerebralis was expected to exert a high selective pressure, and because the WDRES-9 QTL was present in the CRR populations at lower frequencies (Baerwald et al., 2011) development of resistance could have been possible. This is especially true in the East Portal of the Gunnison River since the reduction in the wild CRR population was smaller than in other areas. Second, within the upper Colorado River population, fish reproducing in the wild required CRR traits to survive and recruit to the spawning population (e.g., to overcome competition, avoid predators), but also retained the specific M. cerebralis-resistance alleles needed to survive M. cerebralis exposure. Prior to the development of the WDRES-9 QTL it was estimated that there were 9 ± 5 loci involved in resistance to M. cerebralis and F1 crosses of the GR and CRR showed the highest resistance to the disease in terms of myxospores per fish (Fetherman et al., 2012), however, the SNP panel developed for percent GR did not specifically target M. cerebralis resistant alleles making it impossible to investigate this possibility using this genetic test. Our results from the Colorado River are similar to those obtained in the lab showing that fish with higher percent GR ancestry are surviving and reproducing within this population.
Natural selection likely played a role in the genetic structure that was observed when using assignment and percent GR. Fish needed two characteristics to survive: (1) resistance to M. cerebralis, which is provided by the WDRES-9 QTL and a few additional genes from the GR-cross fish; and (2) wild survival characteristics including feeding behavior, swimming performance, and predator avoidance, the genetic components for which came from the CRR. Selection for M. cerebralis resistance has been reported within Rainbow Trout populations (Miller and Vincent, 2008). The GR is the most resistant known Rainbow Trout strain and developed resistance to M. cerebralis by continuous exposure (~100 years) within a hatchery environment (El-Matbouli et al., 2002; Hedrick et al., 2003). The Harrison Lake Rainbow Trout population (Montana, USA) has shown some development of M. cerebralis resistance in the wild. Miller and Vincent (2008) showed the average histology scores for Harrison Lake Rainbow Trout declined over time and approached the resistant GR strain scores over a short period of time (1995 to 2003). The decline of histology scores suggests increased resistance and it was suggested that rapid natural selection was the process of increased M. cerebralis resistance in the Harrison Lake population. Our results show similar effects for rapid natural selection. The GR-crosses were created and selected for M. cerebralis resistance within the laboratory. Within a short amount of time, roughly 20 years, stocked populations of M. cerebralis-resistant fish were selected for both wild survival traits and maintenance of M. cerebralis resistance, and the fish with the correct combination of these two are the ones that are surviving and reproducing in these populations today.
Our continuous monitoring of M. cerebralis resistant Rainbow Trout allowed for the development and use of different genetic tools over time for observing the Rainbow Trout population genetic structure and determine that rapid natural selection for both wild survival and resistance traits is occurring in the wild. Moving forward, when managing populations of interest, it will be important to recognize the multiple selective pressures that shape population genetic structure, including the presence of pathogens, environmental and stochastic events, and anthropomorphic change. The Rainbow Trout that have been produced because of this selection within the East Portal of the Gunnison River are prime candidates for a wild Rainbow Trout broodstock based on their survival, reproduction, YOY recruitment, and parasite resistance. These Gunnison River Rainbow Trout (GRR) that originate from the East Portal, as well as crosses of the GRR and GR, are now widely stocked in Colorado and their performance is constantly evaluated as M. cerebralis continues to be a major issue for reestablished wild Rainbow Trout populations to overcome. Overall, stocking of M. cerebralis-resistant Rainbow Trout has led to an increase in abundance and recruitment numbers to Rainbow Trout populations that have been affected by the parasite.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, per Colorado Parks and Wildlife policies.
Ethics statement
The animal study was approved by the Institutional Animal Care and Use Committee at Colorado State University (Protocol Numbers: 10-1957A and 14-5112A). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
BA: Data curation, Formal analysis, Project administration, Writing – original draft, Writing – review & editing. EF: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. DW: Funding acquisition, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing. MB: Data curation, Formal analysis, Investigation, Methodology, Resources, Software, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Funding was provided by the Federal Aid in Sport Fish Restoration Program, project F-394, and by the Colorado Parks and Wildlife Aquatic Research Section, Sport Fish Research Studies.
Acknowledgments
We thank Barry Nehring, George Schisler, Jon Ewert, Dan Kowalski, Eric Gardunio, and Kevin Thompson for their help with field work and data collection. In addition we thank Alisha Goodbla, Emily Funk, and Andrea Schreier from the Genomic Variation Laboratory (GVL) at the University of California, Davis for sample genotyping. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffwsc.2025.1500903/full#supplementary-material
Supplementary Figure 1 | Median proportion German Rainbow (GR) genes identified using SNPs in fish known to be pure GR, pure Colorado River Rainbow (CRR), pure Harrison Lake Rainbow (HL), or crosses therein (black boxes; SD bars). The proportion GR expected for each strain or cross is represented by the gray circle.
References
Avila, B. W., Winkelman, D. L., and Fetherman, E. R. (2018). Survival of whirling-disease-resistant rainbow trout fry in the wild: a comparison of two strains. J. Aquat. Anim. Health 30, 280–290. doi: 10.1002/aah.10040
Avila, B. W., Winkelman, D. L., and Fetherman, E. R. (2022). Dual resistance to Flavobacterium psychrophilum and Myxobolus cerebralis in rainbow trout (Oncorhynchus mykiss, Walbaum). J. Fish Dis. 45, 801–813. doi: 10.1111/jfd.13605
Avila, B. W., Winkelman, D. L., and Fetherman, E. R. (2023). Biotic and abiotic factors affecting short-term survival of two age-0 Rainbow trout strains in Colorado streams. N. Am. J. Fish. Manage. 43, 1–8. doi: 10.1002/nafm.10895
Baerwald, M. R., Petersen, J. L., Hedrick, R. P., Schisler, G. J., and May, B. (2011). A major effect quantitative trait locus for whirling disease resistance identified in rainbow trout (Oncorhynchus mykiss). Heredity 106, 920–926. doi: 10.1038/hdy.2010.137
Beauchamp, K. A., Gay, M., Kelley, G. O., El-Matbouli, M., Kathman, R. D., Nehring, R. B., et al. (2002). Prevalence and susceptibility of infection to Myxobolus cerebralis, and genetic differences among populations of Tubifex tubifex. Dis. Aquat. Org. 51, 113–121. doi: 10.3354/dao051113
Beauchamp, K. A., Kelley, G. O., Nehring, R. B., and Hedrick, R. P. (2005). The severity of whirling disease among wild trout corresponds to the differences in genetic composition of Tubifex tubifex populations in central Colorado. J. Parasitol. 91, 53–60. doi: 10.1645/GE-327R
Chapman, J. M., Kelly, L. A., Teffer, A. K., Miller, K. M., and Cooke, S.J. (2021). Disease ecology of wild fish: opportunities and challenges for linking infection metrics with behaviour, condition, and survival. Can. J. Fish. Aquat. Sci. 78, 995–1007. doi: 10.1139/cjfas-2020-0315
Clapp, C. M. (2009). Investigating competition among Myxobolus cerebralis resistant and susceptible mitochondrial lineages of Tubifex tubifex and the potential for biological control of whirling disease (Masters thesis). Colorado State University, Fort Collins, CO, United States.
Das, S., and Sahoo, P. K. (2014). Markers for selection of disease resistance in fish: a review. Aquacult. Int. 22, 1793–1812. doi: 10.1007/s10499-014-9783-5
El-Matbouli, M., Gay, M., McDowell, T. S., Georgiadis, M. P., and Hedrick, R. P. (1999). The potential for using biological control technologies in the management of whirling disease,” in 5th Annual Whirling Disease Symposium: Research and Management Perspectives. Whirling Disease Symposium (Missoula, Montana), 191–195.
El-Matbouli, M. R., Hoffman, W., and Küppers, M. P. (2002). “Identification of a whirling disease resistant strain of Rainbow Trout in Germany,” in 8th Annual Whirling Disease Symposium: Putting a Fresh Spin on Whirling Disease (Denver, CO), 29–32.
Fetherman, E. R., Neuschwanger, B., Avila, B. W., and Riepe, T. B. (2020). “Sport fish research studies,” in Annual Report, ed. Anonymous (Fort Collins, CO: Colorado Parks and Wildlife, Aquatic Research Section), 77.
Fetherman, E. R., and Schisler, G. J. (2013). “Sport fish research studies,” in Federal Aid in Fish and Wildlife Restoration, Job Progress Report, ed. Anonymous (Fort Collins, CO: Colorado Parks and Wildlife, Aquatic Wildlife Research Section), 135.
Fetherman, E. R., and Schisler, G. J. (2014). “Sport fish research studies,” in Federal Aid in Fish and Wildlife Restoration, Job Progress Report, ed. Anonymous (Fort Collins, CO: Colorado Parks and Wildlife, Aquatic Wildlife Research Section), 82.
Fetherman, E. R., and Schisler, G. J. (2016). “Sport fish research studies,” in Federal Aid in Fish and Wildlife Restoration, Job Progress Report, ed. Anonymous (Fort Collins, CO: Colorado Parks and Wildlife, Aquatic Wildlife Research Section), 51.
Fetherman, E. R., and Schisler, G. J. (2017). “Sport fish research studies,” in Federal Aid in Fish and Wildlife Restoration, Job Progress Report, ed. Anonymous (Fort Collins, CO: Colorado Parks and Wildlife, Aquatic Wildlife Research Section), 38.
Fetherman, E. R., Schisler, G. J., and Avila, B. W. (2018). “Sport fish research studies,” in Federal Aid in Fish and Wildlife Restoration, Job Progress Report, ed. Anonymous (Fort Collins, CO: Colorado Parks and Wildlife, Aquatic Wildlife Research Section), 76.
Fetherman, E. R., Winkelman, D. L., Baerwald, M. R, and Schisler, G. J. (2014). Survival and reproduction of Myxobolus cerebralis-resistant Rainbow Trout introduced to the Colorado River and increased resistance of age-0 progeny. PLoS ONE, 9:e96954. doi: 10.1371/journal.pone.0096954
Fetherman, E. R., Winkelman, D. L., Schisler, G. J., and Antolin, M. F. (2012). Genetic basis of differences in myxospore count between whirling disease-resistant and susceptible strains of rainbow trout. Dis. Aquat. Org. 102, 97–106. doi: 10.3354/dao02543
Fetherman, E. R., Winkelman, D. L., Schisler, G. J., and Myrick, C. A. (2011). The effects of Myxobolus cerebralis on the physiological performance of whirling disease resistant and susceptible strains of rainbow trout. J. Aquat. Anim. Health 23, 169–177. doi: 10.1080/08997659.2011.630273
Fraslin, C., Quillet, E., Rochat, T., Dechamp, N., Bernardet, J-F., Collet, B., et al. (2020). Combining multiple approaches and models to dissect the genetic architecture of resistance to infections in fish. Front. Genet. 11:677. doi: 10.3389/fgene.2020.00677
Hedrick, R. P., Adkison, M. A., El-Matbouli, M., and MacConnell, E. (1998). Whirling disease: re-emergence among wild trout. Immunol. Rev. 166, 365–376. doi: 10.1111/j.1600-065X.1998.tb01276.x
Hedrick, R. P., McDowell, T. S., Gay, M., Marty, G. D., Georgiadis, M. P., and MacConnell, E. (1999). Comparative susceptibility of Rainbow Trout Oncorhynchus mykiss and Brown Trout Salmo trutta to Myxobolus cerebralis, the cause of salmonid whirling disease. Dis. Aquat. Org. 37, 173–183. doi: 10.3354/dao037173
Hedrick, R. P., McDowell, T. S., Marty, G. D., Fosgate, G. T., Mukkatira, K., Myklebust, K., et al. (2003). Susceptibility of two strains of Rainbow Trout (one with suspected resistance to whirling disease) to Myxobolus cerebralis infection. Dis. Aquat. Org. 55, 37–44. doi: 10.3354/dao055037
Hoffman, G. L. (1970). “Intercontinental and transcontinental dissemination and transfaunation of fish parasites with emphasis on whirling disease (Myxosoma cerebralis) and its effects on fish,” in Symposium on Diseases of Fisheries and Shellfishes, ed. S. F. Snieszko (Bethesda, MD: American Fisheries Society, Special Publication), 69–81.
Kerans, B. L., Gangloff, M. M., and Vincent, E. R. (1999). “Spatial relations between whirling disease severity and tubificids,” in 5th Annual Whirling Disease Symposium: Research and Management Perspectives1999 Whirling Disease Symposium (Missoula, MT), 103–108.
Leeds, T. D., Silverstein, J. T., Weber, G. M., Vallejo, R. L., Palti, Y., Rexroad, C. E., et al. (2010). Response to selection for bacterial cold water disease resistance in Rainbow Trout. J. Anim. Sci. 88, 1936–1946. doi: 10.2527/jas.2009-2538
Miller, M. P., and Vincent, E. R. (2008). Rapid natural selection for resistance to an introduced parasite of rainbow trout. Evol. Appl. 1, 336–341. doi: 10.1111/j.1752-4571.2008.00018.x
Nehring, R. B., Alves, J., Nehring, J. B., and Felt, B. (2018). Elimination of Myxobolus cerebralis in Placer Creek, a native cutthroat trout stream in Colorado. J. Aquat. Anim. Health 30, 264–279. doi: 10.1002/aah.10039
Nehring, R. B., Hancock, B., Catanese, M., Stinson, M. E. T., and Winkelman, D. L. (2013). Reduced Myxobolus cerebralis actinospore production in a Colorado reservoir may be linked to changes in Tubifex tubifex population structure. J. Aquat. Anim. Health 25, 205–220. doi: 10.1080/08997659.2013.788581
Nehring, R. B., Schisler, G. J., Chiaramonte, L., Horton, A., and Poole, B. (2016). Accelerated deactivation of Myxobolus cerebralis myxospores by susceptible and non-susceptible Tubifex tubifex. Dis. Aquat. Org. 121, 37–47. doi: 10.3354/dao03025
Nehring, R. B., and Thompson, K. G. (2003). “Whirling disease risk assessment: the Colorado perspective,” in 9th Annual Whirling Disease Symposium: Managing the Risk (Seattle, Washington), 31–32.
Nehring, R. B., and Walker, P. G. (1996). Whirling disease in the wild: the new reality in the Intermountain West. Fisheries 21, 28–30.
Nehring, R. B. (2006). Colorado's Coldwater Fisheries: Whirling Disease Case Histories and Insights for Risk Management. Colorado Division of Wildlife Special Report, 79. Colorado Division of Wildlife, Fort Collins, CO.
Nehring, R. B., and Thompson, K. G. (2001). Impact Assessment of Some Physical and Biological Factors in the Whirling Disease Epizootic Among Wild Trout in Colorado. Colorado Division of Wildlife Special Report, 76. Colorado Division of Wildlife, Fort Collins, CO.
Nielsen, E. E., Hansen, M. M., and Bach, L. A. (2001). Looking for a needle in a haystack: discovery of indigenous Atlantic salmon (Salmo salar L.) in stocked populations. Conserv. Genet. 2, 219–232. doi: 10.1023/A:1012239029574
Pritchard, J. K., Stephens, M., and Donnely, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959. doi: 10.1093/genetics/155.2.945
R Core Team. (2024). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available at:https://www.r-project.org/ (accessed August 1, 2024).
Ryce, E. K. N., Zale, A. V., MacConnell, E., and Nelson, M. (2005). Effects of fish age versus size on the development of whirling disease in rainbow trout. Dis. Aquat. Org. 63, 69–76. doi: 10.3354/dao063069
Schisler, G. J., Bergersen, E. P., and Walker, P. G. (1999a). Evaluation of chronic gas supersaturation on growth, morbidity, and mortality of fingerling rainbow trout infected with Myxobolus cerebralis. N. Am. J. Aquacult. 61, 175–183. doi: 10.1577/1548-8454(1999)061<0175:EOCGSO>2.0.CO
Schisler, G. J., Bergersen, E. P., and Walker, P. G. (2000). Effects of multiple stressors on morbidity and mortality of fingerling rainbow trout infected with Myxobolus cerebralis. Trans. Am. Fish. Soc. 129, 859–865. doi: 10.1577/1548-8659(2000)129<0859:EOMSOM>2.3.CO
Schisler, G. J., and Fetherman, E. R. (2010). “Sport fish research studies,” in Federal Aid in Fish and Wildlife Restoration, Job Progress Report, ed. Anonymous (Fort Collins, CO: Colorado Parks and Wildlife, Aquatic Wildlife Research Section), 40.
Schisler, G. J., Myklebust, K. A., and Hedrick, R. P. (2006). Inheritance of Myxobolus cerebralis resistance among F1-generation crosses of whirling disease resistant and susceptible Rainbow Trout strains. J. Aquat. Anim. Health 18, 109–115. doi: 10.1577/H05-047.1
Schisler, G. J., Walker, P. G., Chittum, L. A., and Bergersen, E. P. (1999b). Gill ectoparasites of juvenile rainbow trout and brown trout in the upper Colorado, River. J. Aquat. Anim. Health 11, 170–174. doi: 10.1577/1548-8667(1999)011<0170:GEOJRT>2.0.CO
Stefanik, E. L., and Sandheinrich, M. B. (1999). Differences in spawning and emergence phenology between stocked and wild populations of Brown Trout in Southwestern Wisconsin streams. N. Am. J. Fish. Manage. 19, 1112–1116. doi: 10.1577/1548-8675(1999)019<1112:DISAEP>2.0.CO
Thompson, K. G. (2011). Evaluation of small-scale habitat manipulation to reduce the impact of the whirling disease parasite in streams. Aquat. Ecosyst. Health Manage. 14, 305–317. doi: 10.1080/14634988.2011.602276
U.S. Geological Survey [USGS] (2016). USGS 13200000 Mores Creek AB Robie Creek NR Arrowrock Dam ID, in USGS water data for the Nation: U.S. Geological Survey National Water Information System database. Available at: https://waterdata.usgs.gov/nwis/dv?referred_module=sw&site_no=13200000 (accessed December 16, 2016).
Vincent, E. R. (1996). Whirling disease and wild trout: the Montana experience. Fisheries 21, 32–34.
Wagner, E. J. (2002). Whirling disease prevention, control, and management: a review. Am. Fish. Soc. Symp. 29, 217–225.
Wagner, E. J., Wilson, C., Arndt, R., Goddard, P., Miller, M., Hodgson, A., et al. (2006). Evaluation of disease resistance of the Fish Lake-DeSmet, wounded man, and Harrison lake strains of rainbow trout exposed to Myxobolus cerebralis. J. Aquat. Anim. Health, 18, 128–135. doi: 10.1577/H05-039.1
Walker, P. G., and Nehring, R. B. (1995). An Investigation to Determine the Cause(s) of the Disappearance of Young Wild Rainbow Trout in the Upper Colorado River, in Middle Park, Colorado (Denver, CO: Colorado Division of Wildlife Report).
Wickham, H., François, R., Henry, L., Müller, K., and Vaughan, D. (2023). dplyr: A Grammar of Data Manipulation. R Package Version 1.1.4. Available at: https://github.com/tidyverse/dplyr; https://dplyr.tidyverse.org
Keywords: rainbow trout, whirling disease, genetics, resistance, quantitative trait locus
Citation: Avila BW, Fetherman ER, Winkelman DL and Baerwald MR (2025) Genetics of wild, whirling disease resistant rainbow trout populations in Colorado. Front. Freshw. Sci. 3:1500903. doi: 10.3389/ffwsc.2025.1500903
Received: 24 September 2024; Accepted: 02 January 2025;
Published: 03 March 2025.
Edited by:
Ioannis A. Giantsis, Aristotle University of Thessaloniki, GreeceReviewed by:
Doru Stelian Banaduc, Lucian Blaga University of Sibiu, RomaniaNicolas Pech, UMR 1467 Recover Aix-Marseille University - INRAE, France
Copyright At least a portion of this work is authored by Dana L. Winkelman on behalf of the U.S. Government and as regards Dr. Winkelman and the U.S. Government, is not subject to copyright protection in the United States. Foreign and other copyrights may apply. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brian W. Avila, YnJpYW4uYXZpbGFAc3RhdGUuY28udXM=
 Brian W. Avila
Brian W. Avila Eric R. Fetherman
Eric R. Fetherman Dana L. Winkelman
Dana L. Winkelman Melinda R. Baerwald3
Melinda R. Baerwald3