
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. For. Glob. Change, 14 March 2025
Sec. Forest Management
Volume 8 - 2025 | https://doi.org/10.3389/ffgc.2025.1549531
Introduction: Accurate biomass estimation is crucial for quantifying forest carbon storage and guiding sustainable management. In this study, we developed four biomass modeling systems for natural white birch (Betula platyphylla Sukaczev) in northeastern China using field data from 148 trees.
Methods: The data included diameter at breast height (DBH), tree height (H), crown dimensions, and biomass components (stem, branch, foliage, and root biomass), as well as soil and climate variables. We employed Seemingly Unrelated Regression (SUR) and mixed-effects models (SURM) to account for component correlations and spatial variability.
Results: The base model (SURba), using only the DBH variable, explained 89-96% of the biomass variance (RMSE%: 1.34-19.94%). The second model (SURbio) incorporated H for stem/branch biomass and crown length (CL) for foliage, improving the predictions of stem, branch, and foliage biomass (R2 increased by 1.69–4.86%; RMSE% decreased by 10.76-59.04%). Next, the SURba-abio and SURbio-abio models integrated abiotic factors, including soil organic carbon content (SOC), mean annual precipitation (MAP), degree-days above 18°C (DD18), and soil bulk density (BD). Both models showed improvement, with the abiotic factor model SURba-abio performing similarly to the biotic factor model SURbio (ΔR2 < 4.36%), while the SURbio-abio model performed the best. Subsequently, random effects were introduced at the sampling point (Forestry Bureau) level, developing seemingly unrelated mixed-effects models (SURMba, SURMbio, SURMba-abio, SURMbio-abio), which improved model fitting and prediction accuracy. The gap between the SURMba-abio model (with abiotic factors) and the SURMbio-abio model (including both biotic and abiotic factors) was minimal (ΔR2 < 2.80%). The random effects model stabilized when calibrated with aboveground biomass measurements from four trees.
Discussion: In conclusion, these models provide an effective approach for estimating the biomass of natural white birch in northeastern China. In the absence of biotic factors, the SURba-abio and SURMba-abio models serve as reliable alternatives, emphasizing the importance of abiotic factors in biomass estimation and offering a practical solution for predicting birch biomass.
Accurate forest biomass estimation serves as a foundational pillar for quantifying terrestrial carbon stocks, evaluating ecosystem productivity, and informing climate change mitigation frameworks (Dutcă et al., 2020; Testolin et al., 2023). Natural forests, particularly in northern and central China, serve as critical carbon sinks and biodiversity reservoirs. Their carbon storage significantly influences the regional ecological balance and efforts to mitigate climate change (Wei et al., 2014; Sun and Liu, 2019). Among the dominant species in these forests, Betula platyphylla Sukaczev (white birch) emerges as an ecologically pivotal species due to its extensive geographic distribution, rapid growth rates, and adaptive plasticity—traits that collectively enhance soil stabilization and carbon sequestration (Wang et al., 2018; Geng et al., 2022; Liu et al., 2023). However, accurate biomass estimation in natural forests remains challenging due to structural complexity—heterogeneous tree densities, multi-layered canopies, and diverse species compositions—which traditional single-variable models (e.g., DBH, height) fail to capture (Saatchi et al., 2011). Furthermore, the high cost and operational difficulties associated with ground surveys in natural forests pose significant barriers to large-scale biomass assessments (Holly et al., 2007). Although remote sensing offers scalable monitoring solutions, it struggles with species-specific biomass differentiation and belowground carbon quantification (Litton et al., 2007; Fassnacht et al., 2014). These limitations underscore the necessity for integrative approaches that synergize in-situ measurements with environmental covariates to advance accuracy and scalability.
Recent advances advocate combining biotic factors (e.g., DBH, crown dimensions) with abiotic variables (e.g., soil organic carbon, climate indices) to improve model accuracy (Poorter et al., 2008). Notably, extensive evidence highlights the critical role of climatic and edaphic variables in influencing tree growth and biomass distribution (Fu et al., 2017; Chen et al., 2021; Wang et al., 2023; Li et al., 2024). For example, climatic factors such as the mean annual temperature and the precipitation strongly influence the biomass distribution across different forest components (Uscanga et al., 2023; Yang et al., 2024). Soil properties, including the soil organic carbon (SOC) content and bulk density (BD), directly affect root system development and, consequently, the distribution of total biomass (Ola et al., 2018). They are closely related to forest biomass (Lal, 2018; Petaja et al., 2023). While prior studies have integrated biotic and abiotic variables for white birch biomass estimation, a systematic framework addressing spatial heterogeneity and incorporating hard-to-measure biotic variables remains underdeveloped in natural forest contexts.
Statistical modeling techniques have improved to meet these challenges. For example, during model application, even small deviations can be exaggerated at the stand or even regional scales. Mixed-effects models (MEMs) have been widely used in ecological research and biomass estimation because of their ability to account for both fixed and random effects, effectively capturing spatial variations and reducing biases from local environmental differences or data inconsistencies (Bates et al., 2014; Xie et al., 2023). The efficacy of these models in large-scale biomass estimation, particularly in carbon stock assessments, by improving model adaptability through sampling correction and prediction error reduction. Furthermore, the use of MEMs can reduce the need for direct underground biomass measurements by improving the prediction of aboveground biomass, thereby lowering costs and enhancing overall model precision (Bates et al., 2014). Despite their theoretical advantages, mixed-effects models (MEMs) face practical limitations hindering broad application. These include dependency on specialized statistical software for parameter estimation and the need for resource-intensive sampling protocols to calibrate their enhanced predictive accuracy. To mitigate these limitations, this study synergizes mixed-effects models (MEMs) with readily available abiotic covariates (e.g., bioclimatic indices, edaphic properties) to investigate the feasibility of achieving robust biomass predictions under reduced sampling intensities. Therefore, the focus of this study is efficient and accurate biomass estimation for natural white birch to further advance regional forest carbon stock assessments and climate change prediction and provide a reliable theoretical foundation and technical support for forest management, carbon trading market analyses, and ecological restoration. The specific steps are as follows: (1) develop a seemingly unrelated additive biomass model (SUR) that incorporates abiotic factors and additional biotic factors (such as topography, soil, and climate factors) to achieve high prediction accuracy; (2) analyze the influence of the abiotic factors included in the biomass model on the biomass components of natural white birch, compare the variability associated with the abiotic factors and additional biotic factors, and explore whether abiotic factors can be substituted for more difficult-to-obtain biotic factors; and (3) introduce random effects from sampling points (Forestry Bureau) to establish the SURM biomass model, which improves predictive accuracy for large-scale biomass estimation, finally, the predictive capability of the random-effects model is evaluated through aboveground biomass calibration.
In this study, we collected data from 148 natural white birch trees managed by 12 Forestry Bureaus in northeastern China, encompassing diameter at breast height (DBH), tree height (H), crown length (CL), crown width (CW) and topographic factors, such as elevation, slope, and slope direction. All trees were destructively divided into stems, branches, roots, and foliage in order to gain access to the various tissues and organs and to the above- and below-ground biomass. Stems were cut into 1-meter-long differentiated segments and then weighed. At the end of each stem segment, we collected a disc approximately 3 cm thick. The living crown of the tree is divided into three layers (upper, middle, and lower), and all the living branches in each layer are cut and weighed. One or two average-sized branches were taken from each layer, and the branch and foliage were weighed separately. Branch and foliage separation was used to calculate the specific weight of branch and foliage and to extrapolate the branch and foliage biomass of other branches. Roots (≥ 5 mm in diameter) were removed from the soil and weighed separately, classified into three diameter groups: large roots (≥ 5 cm), medium roots (2–5 cm), and small roots (< 2 cm). Fine roots (< 5 mm) were excluded from biomass measurements due to the labor-intensive collection process and their small biomass. After measuring the fresh weight of each organ in the field, the dry-to-fresh weight ratio was determined in the laboratory to calculate the dry biomass of the stem, branch, foliage, and root. Approximately 100 grams of wood samples and 50 grams of bark samples from the discs were collected. Branch (50–100 g) and foliage (50–100 g) samples were taken from average-sized branches selected from three canopy layers. Around 100 g of roots were sampled from each group. In the laboratory, all subsamples were chopped into small pieces and dried at 85°C until a constant weight was achieved. The dry biomass of the stem (sum of wood and bark), branch, foliage, and root was calculated by multiplying the fresh weight of each part by its corresponding fresh-to-dry weight ratio.
The geographic locations of the sampled trees and corresponding statistics are shown in Figure 1 and Table 1. The allometric growth relationships between the biomass of the different components of natural birch and the tree variables (DBH, H, CL, and CW) are shown in Figure 2. On the basis of the geographic coordinates of the sampling points, 10 soil factors, such as the soil bulk weight, cation exchange capacity, and organic carbon content, as well as 17 climatic factors, such as the mean annual temperature, mean annual precipitation, and relative humidity, were obtained for the sample trees. Meteorological data for all years corresponding to the age of each sample tree were extracted on the basis of the age of each tree via ClimateAP (Wang et al., 2017). Climatic data were aligned with individual tree age and investigation year to capture growth-phase climatic influences. For structural components (stem, branch, and root biomass), mean annual climate values were applied. Foliage biomass exclusively utilized harvest-year climatic data due to its non-cumulative nature. The soil data were extracted from the SoilGrids250 m system on the basis of latitude and longitude (Hengl et al., 2017). Eleven soil variables were weighted and averaged by depth of the soil profile (0–5, 5–15, 15–30, 30–60, and 60–100 cm) for use in the present study. The aim of this study was to comprehensively analyze the relationships between biotic and abiotic factors and the biomass of natural birch and to construct a natural birch biomass model with the addition of abiotic factors. The basic statistical information for the abiotic factors is shown in Table 2.
Figure 1. Geographic distribution of natural white birch (Betula platyphylla) sampling sites in northeastern China. Blue dots indicate individual tree locations.
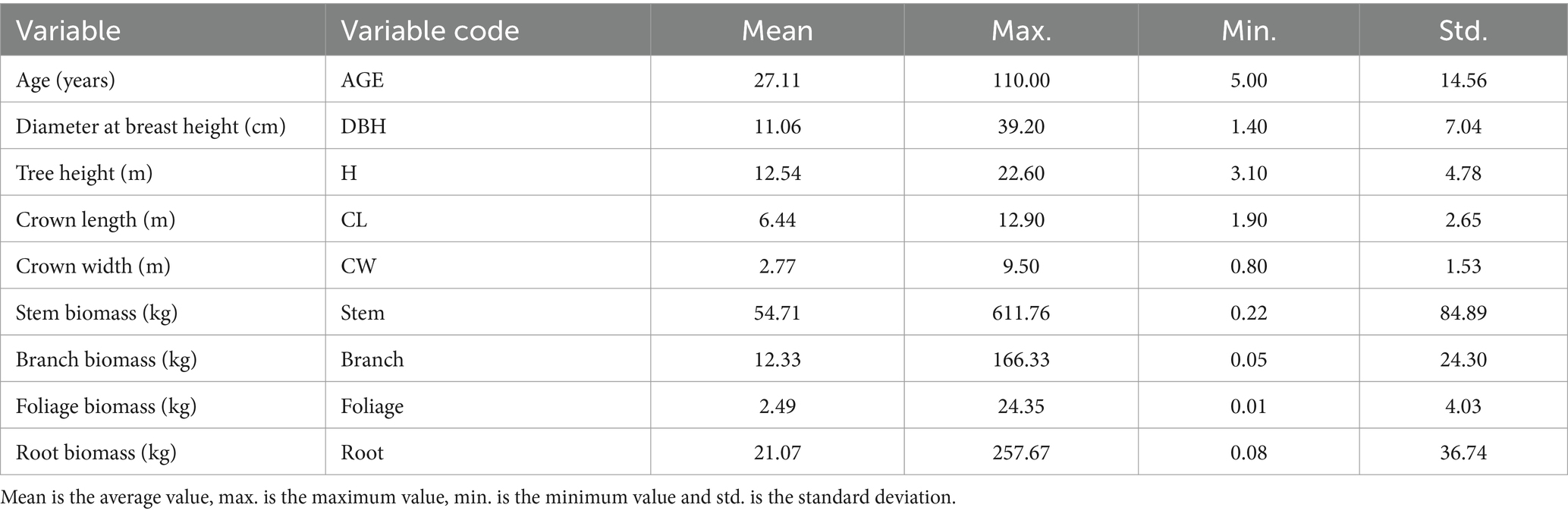
Table 1. Basic statistical information for tree parameters and the biomass of natural white birch trees.
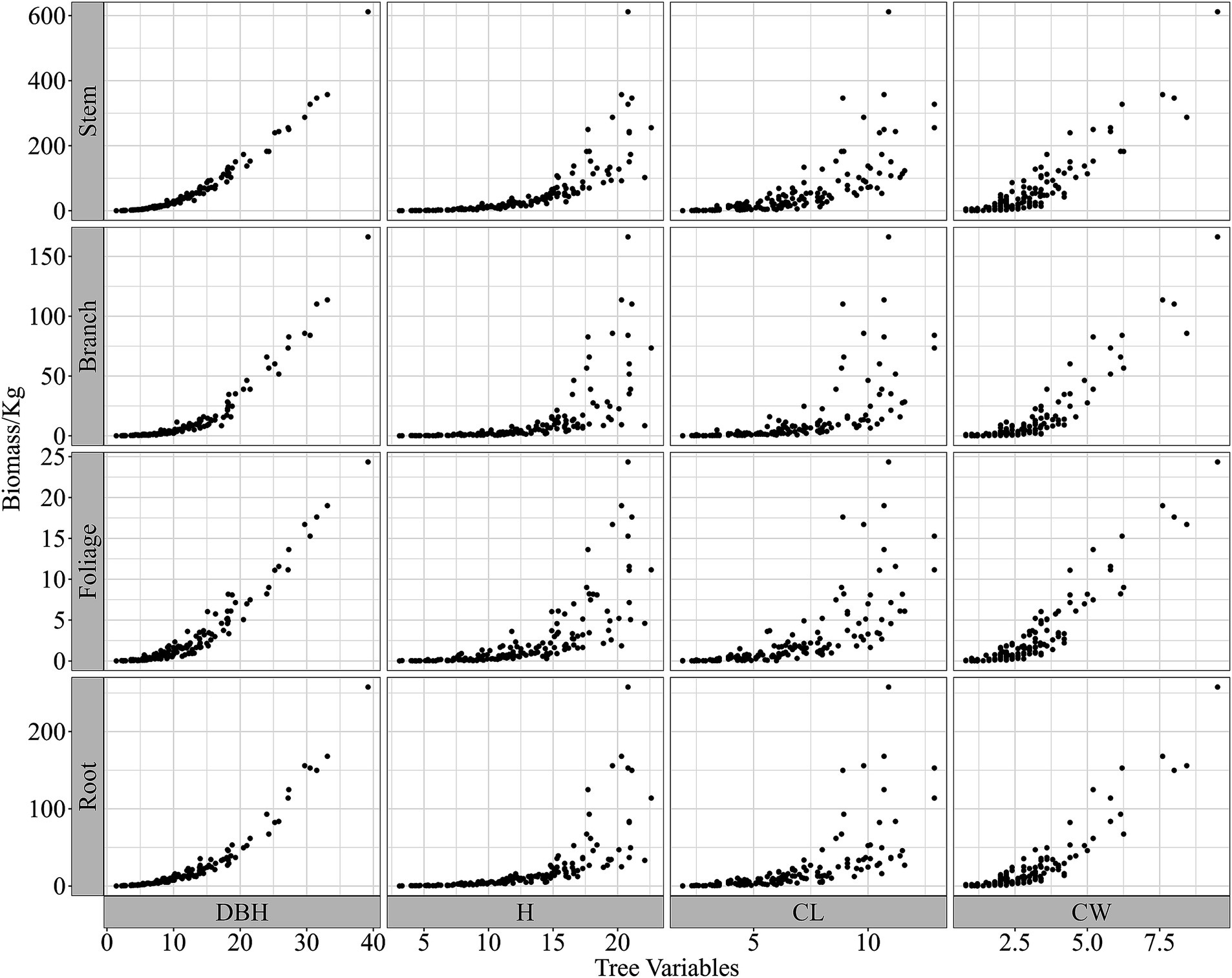
Figure 2. Allometric relationships between biomass components (stem, branch, foliage, root) and tree variables for natural white birch: diameter at breast height (DBH, cm), tree height (H, m), crown length (CL, m), and crown width (CW, m).
Power functions are commonly used to describe the relationships between tree attributes and component biomass (Dong et al., 2014). The error structure of a model can be additive or multiplicative, with likelihood analysis used to determine the more appropriate form (Xiao et al., 2011). Maintaining model additivity—a fundamental requirement where total biomass equals the sum of its constituent compartments—necessitated implementing a seemingly unrelated regression (SUR) method. This approach preserves the mass-balance relationship between aboveground (foliage, branch, stem) and belowground biomass components through simultaneous equation estimation (Bi et al., 2015; Dong et al., 2018; Xie et al., 2023). Machine learning algorithms were excluded based on sample size constraints (n = 148) and interpretability requirements. Model selection diagnostics demonstrated statistical superiority of multiplicative error structures (ΔAICc>2) (Dong et al., 2014), establishing log-linear regression as the foundational modeling approach.
Additionally, many studies have shown that adding tree height and canopy variables to biomass models, alongside diameter at breast height (DBH), can improve prediction accuracy (Zhao et al., 2015; Bronisz and Mehtätalo, 2020). Furthermore, abiotic factors are known to affect biomass (Chen et al., 2021). In this study, we consider both biotic and abiotic factors when modeling the biomass of four components of natural birch. Three types of models are constructed via an additive approach. The first model (base model: SURba, Equation 1) uses DBH as the sole predictor, as it is the key predictor and easy to measure. The second model (a model incorporating additional biotic factors: SURbio, Equation 2) builds on SURba by incorporating single-tree factors, such as tree height or canopy variables. Finally, abiotic factors are added to the SURba and SURbio models to create two additional models, SURba-abio and SURbio-abio (Equation 3). The specific forms of these models are as follows:
where , , , , and represent the stem biomass, branch biomass, foliage biomass, root biomass, and diameter at breast height, respectively; represents the estimated parameters; represents the covariates, i.e., H, CL, and CW for single trees; and represents the abiotic factors (soil and climate variables). The best covariates were determined by comparing the modeling effects of different covariates; is the error term, and .
Equations 1–3 can be converted into matrix form, as noted by Xie et al. (2023). In addition, to separate the effects of each variable on the biomass model, hierarchical partitioning (HP) analysis was used (Chevan and Sutherland, 1991). HP analysis was performed via the “hier.part” package in R software (Nally and Walsh, 2004).
To address spatial heterogeneity across 12 Forestry Bureau jurisdictions, we incorporated a bureau-level random effect into the model framework. This specification takes into account differences in biomass-allometry relationships that happen at different sites for trees that are dendrometrically similar. This makes the estimation more accurate (Ou et al., 2016; Xie et al., 2023). Therefore, on the basis of the SUR modeling system, the random effect of the sampling point characteristics (Forestry Bureau) was introduced to construct a seemingly unrelated regression mixed-effects (SURM) system. That is, the SURM modeling system includes four variants: SURMba, SURMbio, SURMba-abio, and SURMbio-abio. They are the models of SURba, SURbio, SURba-bio, and SURbio-abio after adding random effects, respectively. The form of the SURM model is as follows:
where is the n -order fixed-effects design matrix ( is the number of fixed-effects parameters for each submodel); is the vector of dimensional fixed-effects parameters; is the n -order random-effects design matrix ( is the number of random-effects parameters for each submodel); and is the vector of dimensional random-effects parameters; the other variables are as defined above. Equation 4 can be converted to matrix form, as described by Xie et al. (2023). For the log-transformed linear SURM model, the parameters were estimated via the restricted maximum likelihood (REML) method via the lme function in the nlme package in R4.1.2 software.
Mixed effects models can yield two types of predictions (Bronisz and Mehtätalo, 2020; Bronisz et al., 2021). The first type relies only on fixed effects, and the random effects calibration sample size is assumed to be 0. The second type combines both fixed and random effects, reflecting the case in which the available sample size for random effects calibration is greater than zero. For the SURM biomass model, the random-effects parameters for the model can be calculated from submodels for which there are existing measurements of the response variable, and then the random-effects parameters for submodels corresponding to observations of the nonresponse variable can be calculated on the basis of the correlation between the random-effects parameters, i.e., allowing for the use of biomass measurements for some subsets of the model and calibrating all five subsets of the model’s random-effects parameters on the basis of the variance–covariance matrix of the random-effects parameter vectors (Bronisz and Mehtätalo, 2020; Xie et al., 2021; Xie et al., 2023). Because the acquisition of biomass data is time-consuming, especially for belowground biomass, stem, branch, and foliage biomass measurements were used to calibrate the random-effects parameters of the SURM model.
A random sampling method was used to select sample trees in the Forestry Bureau to correct the random effects parameters of the SURM model on the basis of stem, branch, and foliage biomass measurements. Random effects parameters were calculated on the basis of the estimated empirical best linear unbiased predictor (EBLUP) (Mehtätalo and Lappi, 2020; Xie et al., 2021). The formula for calculating the vector of random effects parameters for a given sample site is as follows:
where is the random effects parameter for the kth Forestry Bureau; is the matrix obtained by removing the columns corresponding to the random-effects parameters in the model for the unobserved response variable from the matrix; is the matrix obtained by removing the rows and columns corresponding to the unobserved response variable from the matrix, with the superscript denoting the vector transpose; , and are obtained from the fitting process of the model; and the and matrices are the matrices of all the response variables and predictor variables constructed separately, including observed and unobserved variables. The matrix is computed from the partial derivatives of the model function with respect to its stochastic parameters, as follows: , where , is the size of Forestry Bureau k, and is the number of random-effects parameters.
The models were fitted using all the data, and the goodness of fit of the models was evaluated on the basis of two metrics: the coefficient of determination (R2, Equation 6), root mean square error (RMSE%, Equation 7) and mean absolute percentage error (MAPE%, Equation 8). Model evaluation prioritized the R2 over its adjusted counterpart to provide a clearer understanding of the model’s effectiveness. The leave-one-out cross-validation (LOOCV) protocol was systematically implemented to mitigate overfitting potential during model training. The predictive ability of the SUR and SURM models was evaluated via the LOOCV (Xie et al., 2023). On the basis of the values predicted via the LOOCV method at the sample level, the MAPE% value was calculated to evaluate the predictive ability of the models. The formula is as follows:
where is the size of the sample tree. , and are the observation, antilogarithmic fitted value and antilogarithmic LOOCV prediction for the ith subcomponent of biomass, respectively, and is the mean of the biomass subcomponent observations.
A fundamental biomass modeling system based on biotic and abiotic factors (SUR modeling system) was developed using all the data, and the parameter estimates and fitting effects are shown in Table 3. The SURba model accounted for 89–96% of natural white birch biomass variance (RMSE%: 1.34–19.94%). Its extension SURbio integrated dendrometric covariates (H for stem/branch biomass, CL for foliage biomass), yielding component-specific improvements: R2 increased by 1.69–4.86% and RMSE% reductions of 10.76–59.04%. On the other hand, biotic covariates (H, CL, crown width) showed no predictive value for root biomass (parameters lacked statistical significance, p > 0.05), justifying their exclusion from the belowground model.
Conversely, the SURba-abio model, constructed by incorporating abiotic factors (organic carbon content (SOC) for the stem biomass model, mean annual precipitation (MAP) for the branch biomass model, degree-days above 18°C (DD18) for the foliage biomass model, and soil bulk density (BD) for the root biomass model), also improved the branch and foliage biomass models, with an increase in of approximately 0.23 to 2.16% and a decrease in RMSE% of 1.93 to 13.92%. The SURbio-abio model demonstrated optimal performance when both biotic factors (e.g., tree height and crown length) and abiotic factors were included, resulting in an increase in of approximately 3.49 to 5.39% and a decrease in RMSE% of 12.92 to 59.09%. Overall, the model that incorporated additional biotic factors, such as tree height and crown length, as well as abiotic factors, such as soil and climate variables, exhibited superior predictive performance.
Figure 3 presents the MAPE% of biomass components for different tree diameter classes on the basis of LOOCV method. The results indicate that the inclusion of additional biotic factors significantly enhanced the prediction accuracy of stem biomass, with greater efficacy observed for the prediction of the stem biomass and total biomass of larger trees (DBH ≥ 15 cm) than for those of smaller trees. Abiotic factors, such as climate and soil factors, were particularly effective for improving the prediction accuracy for the branch, foliage, root, and total biomass of smaller trees (DBH < 15 cm). The model incorporating both biotic and abiotic factors exhibited the lowest prediction bias for stem and branch biomass, whereas for foliage biomass prediction for smaller trees (DBH < 15 cm), the model with abiotic factors alone demonstrated the lowest prediction bias. Nevertheless, it is evident that the base models still exhibited substantial prediction bias for branch and foliage biomass.
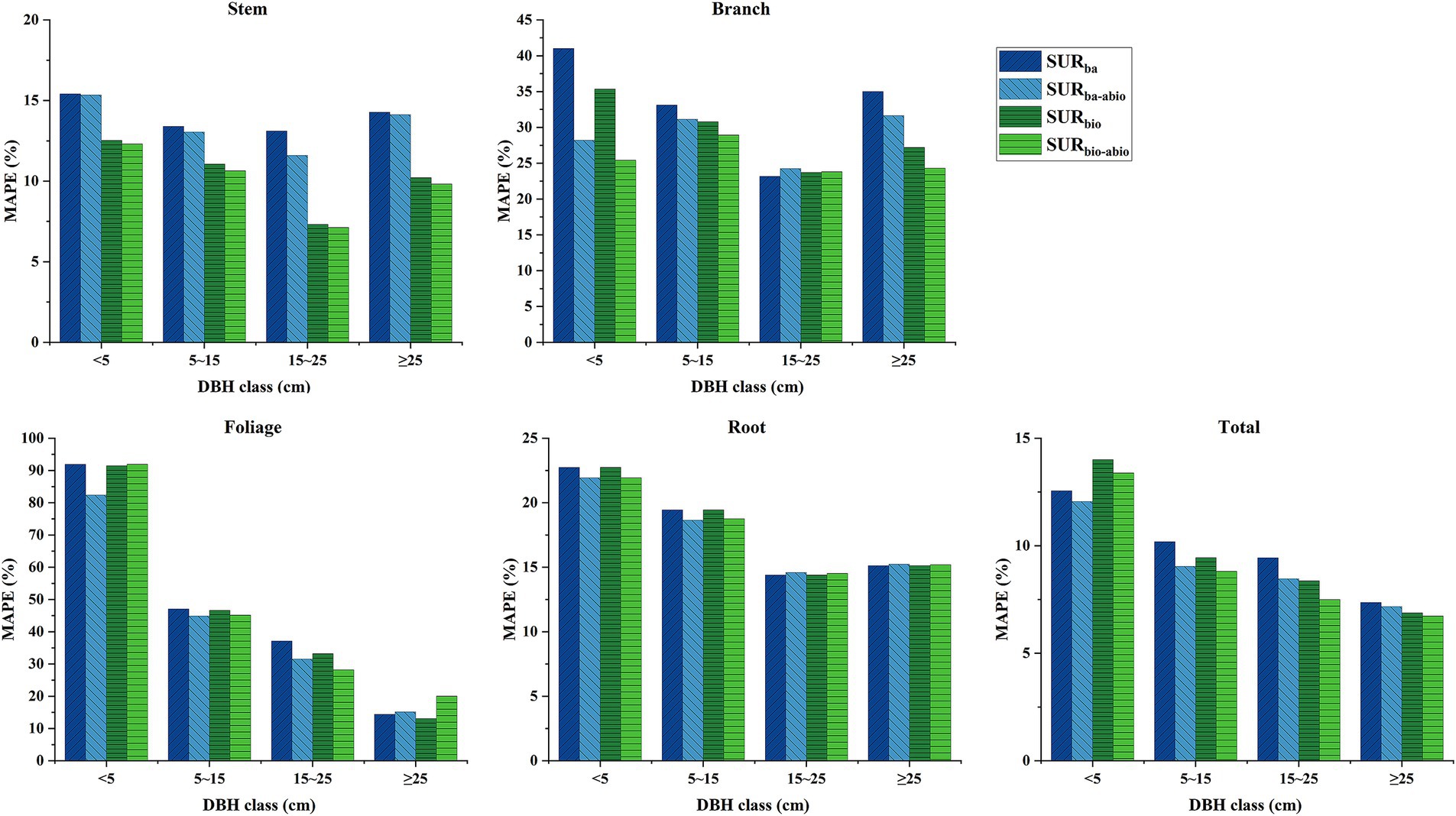
Figure 3. Mean absolute percentage error (MAPE%) of SUR model predictions for biomass components across DBH classes, evaluated via leave-one-out cross-validation (LOOCV). Models include: SURba (DBH only), SURbio (DBH + H/CL), SURba-abio (DBH + abiotic factors), and SURbio-abio (DBH + H/CL + abiotic factors). Abiotic factors: soil organic carbon (SOC) for stem, mean annual precipitation (MAP) for branche, degree-days above 18°C (DD18) for foliage, and soil bulk density (BD) for root.
The mixed-effects model parameter estimates and fitting effects are shown in Table 4. The SURMba model effectively predicted the variation in natural birch biomass, with an accuracy of approximately 93 to 98%, and the RMSE% ranged from 1.04 to 12.54%; the SURMbio and SURMba-abio models effectively predicted the variation in natural birch biomass, with an accuracy of approximately 94 to 99%, and the RMSE% ranged from 0.82 to 8.49%; and the SURMbio-abio model effectively predicted the variation in natural birch biomass, with an accuracy of approximately 96 to 99%, and the RMSE% ranged from 0.82 to 8.02%. The white birch biomass was associated with approximately 96 to 99% of the variance, with the RMSE% ranging from 0.82 to 8.02%. The models all improved after the introduction of sampling point (Forestry Bureau)-level random effects; notably, increased by approximately 0.03% ~ 5.00%, and the RMSE% of each component biomass model decreased by 1.69% ~ 37.12%. On the other hand, the performance of the SURMba-abio model differed very little from that of the SURMbio and SURMbio-abio models and was better than that of the SURMbio model in terms of branch and root biomass fitting.
The predictive ability of the mixed-effects model with random-effects correction was assessed via independent data through the LOOCV method to evaluate the performance of the SURM model at the Forestry Bureau level. Measurements of aboveground biomass (such as stem, branch, and foliage biomass) from 1 to 6 randomly selected trees within the Forestry Bureau were used as response variables to calibrate the random effects for each biomass model. The MAPE% of the biomass components was calculated on the basis of the correlation between the residuals of the SURM model and the associations among the random effects parameters. As illustrated in Figure 4, the prediction accuracy of the corrected SURM model improved with increasing number of sampled trees, with this model outperforming the pure fixed-effects SURM model (with a correction sample size of zero). Moreover, the MAPE% values for each biomass component stabilized once the sample size reached four trees, indicating that the introduction of random effects enhanced the predictive accuracy for each component.
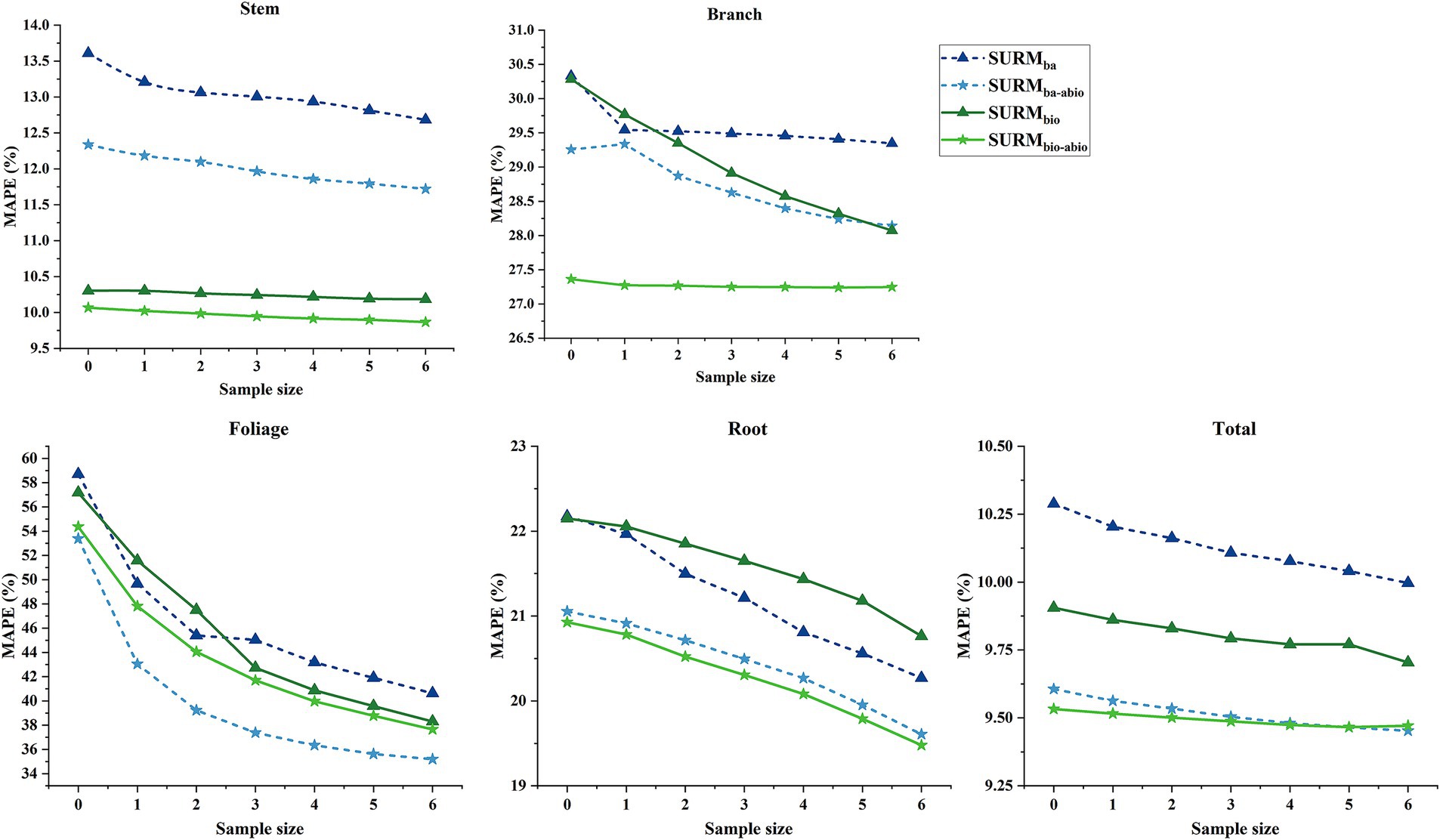
Figure 4. MAPE% of SUR mixed-effects models (SURM) for biomass predictions across sample groups. SURM variants: SURMba (SURba + random effects), SURMbio (SURbio + random effects), SURMba-abio (SURba-abio + random effects), and SURMbio-abio (SURbio-abio + random effects).
From the perspective of MAPE% values, the prediction bias for stem, root, and total biomass was relatively low (with MAPE% values ranging from 9.87 to 22.92%), whereas the bias for branch and foliage biomass remained comparatively high (with MAPE% values ranging from 27.23 to 58.69%). Overall, when predicting independent data, the SURM model incorporating additional individual tree factors and abiotic factors (SURMbio-abio) exhibited superior performance for predicting stem, branch, and root biomass compared with the other models (Figure 4). The SURM model that included only abiotic factors (SURMba-abio) demonstrated certain advantages in predicting foliage biomass and performed comparably to the SURMbio-abio model for total biomass prediction.
To further evaluate the impact of correcting for random effects using only aboveground biomass given the prediction bias across different tree diameter classes, the measured aboveground biomass values from four randomly selected trees were used to correct the random effects in the SURM models. The predictive performance of the models was assessed by comparing the MAPE% for each biomass component. As illustrated in Figure 4, the SURMbio-abio model yielded the lowest prediction error for stem biomass, indicating that the joint inclusion of additional biotic and abiotic factors provided the highest prediction accuracy for stem biomass. The SURMba-abio model displayed the lowest prediction bias for foliage, root, and total biomass for smaller trees (DBH < 15 cm), highlighting the significant effects of climate and soil on the biomass of young natural birch trees, as corroborated by the SUR model predictions. Furthermore, while the SURMba-abio model demonstrated slightly worse performance than the SURMbio model in terms of the prediction of stem biomass, it performed comparably or even better for other components and total biomass. These findings suggest that for predicting the biomass of natural birch, a model incorporating only abiotic factors may serve as a viable alternative to models that include biotic factors, such as tree height and crown length, which are more challenging to obtain.
From the model parameter estimates in Tables 3, 4, the parameter estimates of the abiotic factor (LnSOC) were negative, and the parameter estimates of LnMAP, LnDD18 and LnBD were positive, indicating that the organic carbon content (SOC) was negatively correlated with the natural white birch stem biomass; additionally, the mean annual precipitation (MAP), the degree-days above 18°C (DD18), and the soil bulk density (BD) were negatively correlated with the branch biomass, foliage biomass and root biomass. The relative importance of the variables analyzed via HP is shown in Figure 5. DBH contributed the most to the modeling of the biomass of each component of natural white birch, followed by H, CL, and the abiotic factors (SOC, MAP, DD18, and BD). Among them, abiotic factors had a certain effect on branch, foliage and root biomass, with a relative importance of as high as 12.10–23.72%; additionally, the stem biomass was less affected by the abiotic factors, with a relative importance of 2.16–2.53%. Moreover, HP analyses of SURbio-abio and SURba-abio revealed that abiotic factors (SOC, MAP, DD18, and BD) account for a portion of the prediction errors even after the introduction of variables such as H and CL; thus, introducing abiotic factors into the natural birch biomass model is necessary. The elevated predictive uncertainty (MAPE%: 25–66%) observed in branch and foliage biomass estimation of juvenile trees (DBH < 15 cm). This uncertainty remained even with abiotic indices (e.g., DD18), indicating their limited capacity to capture such biological complexity.
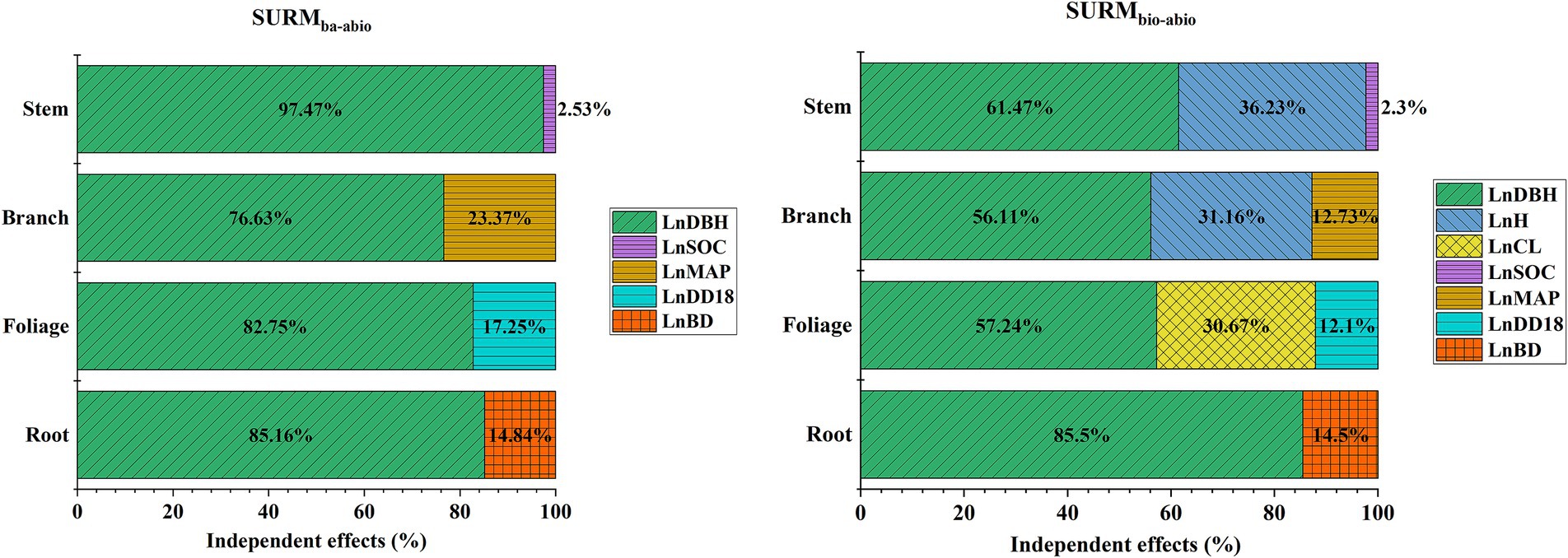
Figure 5. Relative contributions of biotic (DBH, H, CL) and abiotic (SOC, MAP, DD18, BD) factors to SURMba-abio and SURMbio-abio models, analyzed via hierarchical partitioning (HP). Variables are log-transformed (LnDBH: diameter at breast height; LnH: height; LnCL: crown length; LnSOC: soil organic carbon; LnMAP: mean annual precipitation; LnDD18: degree-days above 18°C; LnBD: bulk density).
This study developed an integrated biomass model for natural white birch that synthesizes tree measurement attributes with soil (SOC, BD) and climatic (MAP, DD18) predictors. The incorporation of multifactorial drivers significantly improved model precision, particularly for stem and foliage biomass. Spatial heterogeneity across administrative jurisdictions was explicitly addressed through a mixed-effects framework incorporating bureau-level random intercepts, which enhanced prediction robustness. The mixed-effects framework incorporated bureau-level random intercepts to explicitly address spatial heterogeneity across administrative jurisdictions, thereby enhancing predictive robustness through systematic variance partitioning. In terms of natural birch biomass prediction ability, these abiotic factors can effectively replace or even be more useful than more difficult-to-obtain biotic variables, such as tree height and crown length (Tables 3, 4; Figures 3, 4, 6). Notably, the influence of abiotic variables (e.g., climate and soil parameters) on forest biomass is very important, as observed in previous studies (Bennett et al., 2020; Chen et al., 2021). This may be a result of the interactive relations with biotic variables and the consequent confounding effects associated with abiotic variables. Analyses of models developed under traditional conditions, such as those of static climates or stable sites, suggest that unavoidable errors (significant differences in soil physicochemical properties and climate) may occur when nonenvironmentally adapted models are used at large geographic or temporal scales. Currently, high-resolution layers of global abiotic factors facilitate the large-scale extraction of environmental features from sample plots (Fick and Hijmans, 2017; Hengl et al., 2017; Wang et al., 2017), which largely solves the problem of measuring the cost of environmental factors. In this context, deciphering the influence of abiotic factors on the modeling of natural birch biomass is necessary.
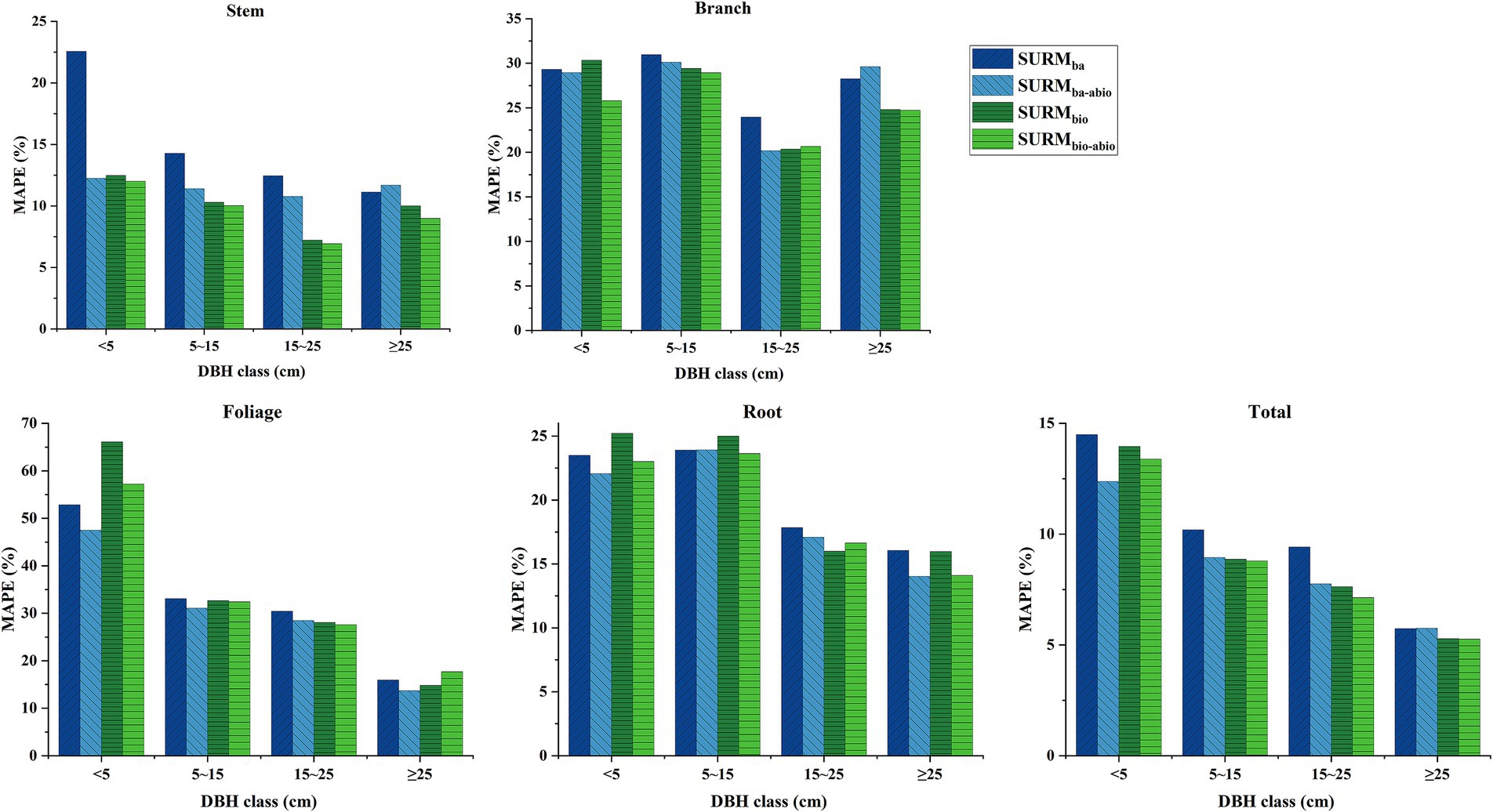
Figure 6. MAPE% of biomass component predictions by SURM models across DBH classes, calibrated with four trees per sampling site.
Prior to the development of environmentally adapted biomass models, SUR biomass models (SURba and SURbio) containing only DBH and introducing H and CL and corresponding SUR biomass mixed-effects models (SURMba and SURMbio) were constructed. Our results suggest that the allometric relationships between the biomass of natural birch fractions and various factors are plastic rather than fixed and that the W-DBH relationship of each component is constrained to varying degrees by other biological factors. Biomass models that include both biotic and abiotic factors (SURba-abio, SURbio-abio, SURMba-abio, and SURMbio-abio) can account for more variability in the observed biomass data of each component than the less flexible generalized biomass models that include only DBH and H (Bi et al., 2004; Dong et al., 2014). Interestingly, when we compared the prediction results of the SURba-abio and SURbio models and the prediction results of the SURba-abio and SURMba-abio models, the SURba-abio and SURMba-abio models performed only slightly worse than SURbio in terms of the prediction of stem biomass and yielded results similar to or even better than the SURbio and SURMbio models in terms of the prediction accuracy of the other components and total biomass. These findings suggest that a model with only abiotic factors can be an effective alternative to models with additional biotic factors, such as H and CL, which are more difficult to obtain for predicting natural birch biomass. This is one of the main goals of our study: constructing biomass prediction models that are convenient and widely applicable.
The mixed-effects models (SURMba, SURMbio, SURMba-abio, and SURMbio-abio) constructed with random-effect levels at the Forestry Bureau outperformed the SUR model, indicating that the mixed-effects model can improve the accuracy of biomass estimation (Xie et al., 2021; Xie et al., 2023). LOOCV verified the prediction accuracy of the SURM models for independent data (Figures 4, 6). Mixed-effects models are widely used in forestry modeling because of their ability to represent data hierarchies; additionally, they are more accurate than general regression models. However, they are typically used in single models and rarely used in joint estimation with multiple equations (Maltamo et al., 2012; Xie et al., 2023). In terms of calibrating SURM models, when one measure of at least one response variable is available, the correlation of the random effects parameter can be used to predict the sample effects for all subcomponents of the SURM models (Bronisz and Mehtätalo, 2020; Xie et al., 2021). Given that obtaining biomass data requires harvesting trees for measurement, this calibration method is important for the practical application of the SURM biomass model; calibrating the model using response variable data can significantly reduce the needs for time, labor, and financial resources (Xie et al., 2021; Xie et al., 2023). When calibrated via measurements of all component biomasses, the random effects in the component-specific biomass model are associated with other random effects given the correlations among random effect parameters, and the best prediction accuracy is achieved by using all subcomponent biomasses to calibrate the random effects for prediction (Bronisz et al., 2021; Xie et al., 2023). Notwithstanding their predictive advantages, mixed-effects models necessitate labor-intensive calibration sampling to achieve optimal performance (Zuur et al., 2009), imposing substantial cost burdens that constrain operational scalability. Belowground biomass quantification presents disproportionate methodological challenges and financial expenditures compared to aboveground compartment measurements (stem, branch, foliage), particularly regarding root system excavation and processing (Xie et al., 2023). This methodological constraint prompted us to use only aboveground biomass measurements to calculate the random effects for enhanced prediction. Crucially, our calibration experiments demonstrated that sampling intensity can be optimized to four trees per administrative unit while maintaining prediction fidelity, as validated through leave-one-bureau-out cross-validation (Figure 3). The MAPE% value for each biomass component decreased with increasing number of calibration samples, indicating that using only aboveground biomass measurements in the calibration prediction test is reliable and valid. This is because there is a high correlation between aboveground and belowground biomass for most tree species (Mehtätalo and Lappi, 2020). Overall, calibration via aboveground biomass measurements produced robust model predictions.
In this study, three biotic factors, DBH, H, and CL, were selected as independent variables to construct a modeling system for natural birch biomass. Among them, diameter at breast height is the most reliable predictor of biomass because of it is easy to measure and highly correlated with other tree dimensions (Dong et al., 2020). In addition, tree height (H) and crown length (CL), as additional biotic factors, improved the prediction accuracy of the biomass model for stems, branches and leaves. Zhao et al. also reported that in addition to diameter at breast height, tree height, crown length and crown width had important effects on biomass modeling for various subcomponents (Zhao et al., 2015; Xie et al., 2021; Xie et al., 2023). Recent studies have shown that the enhancement effect of component-specific biomass modeling for some species is not significant when only tree height is used as a covariate (Dong et al., 2015; Kusmana et al., 2018). In this study, only tree height was included as the best covariate for stem and branch biomass modeling in the biomass SURba model with DBH, a finding that is biologically significant because of the presence of stems and branches throughout the tree structure. For the foliage biomass model, crown length was more important than the other variables, suggesting that the relationship between crown length and crown biomass is stronger than the relationship between tree height and crown biomass. For tree root biomass, no additional biological factors were introduced as covariates. Although the introduction of covariates has been shown to improve the modeling of aboveground biomass, the effect of covariates on root biomass may be relatively small (Kusmana et al., 2018; Xie et al., 2021; Xie et al., 2023). Figure 3 shows that the inclusion of four abiotic factors (SOC, MAP, DD18, and BD) improved the prediction accuracy of the biomass of each component for natural white birch trees of different diameters; in particular, the inclusion of four abiotic factors was most effective for predicting the biomass of branches, foliage and root biomass of trees in the small diameter class (DBH < 15 cm) than the inclusion of additional biotic factors (H and CL).
The soil characteristic variables (SOC and BD) were introduced into the stem biomass and root biomass models based on previous findings of the effects of soil on forest biomass (Becknell and Powers, 2014; Caleño-Ruíz et al., 2023). SOC is an extremely important component of soil that is not only closely or positively correlated with soil fertility but also strongly affects the Earth’s carbon cycle and is negatively correlated with natural birch stem biomass (Tables 3, 4). This may be due to complementary resource utilization among coexisting tree species. In natural forests, there is a significant interaction effect between soil fertility and aboveground biomass, with forests having different mycorrhizal compositions depending on soil fertility. Aboveground biomass in natural forests usually increases with species richness, whereas this positive diversity effect diminishes with increasing soil fertility (Ma et al., 2023). Stem biomass is an important component of aboveground biomass, and it contributes to the negative correlation between SOC and stem biomass. BD is also a key soil indicator that is positively correlated with tree root biomass (Tables 3, 4). Changes in soil fertility are often accompanied by changes in soil structure and bulk density, and high soil fertility tends correspond to low BD; however, studies have shown that tree biodiversity has a positive effect on below-ground biomass (Xu et al., 2019), which is consistent with the negative correlation between SOC and aboveground biomass (Li et al., 2018; Mao et al., 2023). It has been shown that an increase in the soil nutrient concentration should result in increased plant biomass; however, this phenomenon only seems to apply to small trees (Li et al., 2017). Radial growth responsivity to temperature and drought stressors exhibits progressive attenuation across ontogenetic stages (Drake et al., 2010). Such ontogenetic plasticity in environmental response necessitates the integration of bioclimatic drivers into allometric models (McDowell et al., 2020), particularly for long-term biomass projections.
The climate factor MAP was incorporated into the branch biomass model as a measure of the mean annual precipitation at the sampling sites. Our results were consistent with the theoretical expectation that dendritic biomass increases with MAP (Tables 3, 4). Natural white birch is a cold-temperate deciduous broad-leaved tree species that is light loving, shade tolerant, cold tolerant, highly adaptable to different soil conditions, and shallow rooted, preferring moist environments. Some studies have shown that the biomass of boreal conifers tends to increase in areas with low precipitation, whereas the biomass of cold temperate deciduous broadleaf trees tends to increase in areas with high precipitation (Obata et al., 2023). A reduction in precipitation decreases biomass, abundance, and mean individual weight for some species but increases biomass, abundance, and mean individual weight for others. Compared with those with deep roots, species with strong resource-acquiring traits (e.g., shallow-rooted species) displayed stronger positive responses to precipitation changes in terms of biomass and mean individual weight (Zhang et al., 2020). DD18, as a temperature indicator, was positively correlated with foliage biomass, which was consistent with theoretical expectations. Specifically, temperature changes drive trees to utilize light energy during photosynthetic uptake to increase aboveground biomass (Černý et al., 2020). The linear relationship between the photosynthetic radiation absorbed by different tree species and tree biomass has also been confirmed in previous studies (Teklehaimanot, 2004).
The integration of abiotic predictors (SOC, MAP, DD18, BD) significantly enhanced predictive accuracy for natural birch biomass, particularly for branch, foliage, and root biomass in juvenile trees (DBH <15 cm). Crucially, abiotic covariates function as complementary rather than substitutive parameters to biotic measurements. Comparative analysis revealed comparable performance between models utilizing only DBH with abiotic variables (SURba-abio, SURMba-abio) and those incorporating biotic factors (SURbio, SURMbio) for branch and foliage biomass (ΔMAPE% <6.14). However, stem biomass predictions exhibited marked divergence (ΔMAPE% = 18.12), underscoring the persistent importance of dendrometric variables like tree height (Figure 6). This evidence delineates a context-dependent optimization framework: abiotic proxies demonstrate operational utility where biotic data acquisition proves logistically constrained (e.g., remote monitoring), or for biomass components (foliage, roots) with stronger environmental regulation (Schimel et al., 2015). This is particularly relevant in large-scale or remote applications in which data on tree height and canopy characteristics may be difficult to obtain. As Gustafson et al. (2010) emphasized, the use of soil and climate data in biomass modeling provides a reliable foundation for forest biomass estimation across different geographic regions. With the increasing availability of high-resolution global datasets, such as WorldClim and SoilGrids250m, researchers can now efficiently integrate localized environmental data into biomass models, thereby increasing the predictive accuracy without the need for intensive field measurements. These global datasets provide spatially detailed climate and soil information that improves model adaptability across diverse environmental conditions (Fick and Hijmans, 2017; Hengl et al., 2017).
This study highlights the effectiveness of integrating biotic and abiotic factors in SURM models to estimate the biomass of natural white birch. By combining traditional tree measurements with environmental variables such as SOC, BD, MAP, and DD18, the model enhances accuracy and adaptability across different forest environments. This modeling framework demonstrates operational scalability for large-scale forest biomass estimation. By leveraging existing national forest inventory (NFI) permanent plots and management survey data, high-accuracy predictions can be achieved through diameter-at-breast-height (DBH) measurements combined with globally accessible abiotic covariates from WorldClim (climate) and SoilGrids250m (edaphic). The streamlined protocol substantially lowers survey costs through minimized reliance on labor-intensive dendrometric variables (e.g., H and CL). On the other hand, Hierarchical variance partitioning revealed distinct environmental mediation patterns: abiotic drivers explained 12.10–23.37% of variability in branch, foliage, and root biomass, but minimally influenced stem biomass (2.30–2.53%) (Figure 5). The negative correlation between soil organic carbon (SOC) and stem biomass (Tables 3, 4) likely reflects nutrient competition dynamics in high-fertility soils—a pattern consistent with recent findings in temperate forests (Ma et al., 2023). These results necessitate interpreting abiotic coefficients as ecological process mediators rather than direct predictive parameters, particularly for woody biomass components.
While the current framework exhibits robust predictive capabilities, future refinements could strengthen its ecological fidelity and operational versatility. Integrating temporal dynamics—including growth trajectories, soil nutrient fluxes, and climatic oscillations—would enhance responsiveness to environmental stochasticity under climate change scenarios. Expanding the covariate matrix to incorporate critical edaphic factors (e.g., nitrogen mineralization rates) and bioclimatic extremes (e.g., seasonal thermal thresholds) would enable more comprehensive characterization of biomass partitioning mechanisms across tree compartments. Ecophysiological trait integration (e.g., interspecific variation in wood density and leaf economics spectrum parameters) could facilitate cross-taxa generalization, while synergizing UAV-derived canopy structural indices (e.g., CHM-based vertical complexity metrics) with abiotic gradients may automate calibration workflows, reducing reliance on labor-intensive field measurements. Systematic validation across biomes—particularly in boreal and subtropical ecotones—would delineate model transferability boundaries and refine allometric scaling rules under divergent resource regimes. Operational optimization requires determining minimum calibration sample sizes through power analysis across forest structural types, thereby balancing prediction uncertainty with survey costs—a critical consideration for resource-limited regions. Although demonstrated for Betula platyphylla, model generalizability to phylogenetically distinct taxa (e.g., conifers) or tropical systems necessitates explicit validation given interspecific allometric divergence and ecosystem-specific gene–environment interactions.
This study developed SUR and SURM models for estimating the biomass of natural white birch in northeastern China by integrating biotic (DBH, H, CL) and abiotic factors (SOC, MAP, DD18, BD). The SURM framework, incorporating random effects at the Forestry Bureau level, significantly improved prediction accuracy (R2: 0.94–0.99), particularly for stem and foliage biomass, while demonstrating robust calibration using aboveground biomass measurements from four trees per sampling site. The inclusion of abiotic factors enhanced model generalizability, offering a scalable solution for regions where biotic variables like tree height or crown dimensions are logistically challenging to obtain. However, our findings also highlight several limitations and opportunities for future research. First, while abiotic factors (e.g., SOC, MAP) effectively compensated for the absence of biotic variables in branch and foliage biomass predictions, their substitution for dendrometric parameters in stem biomass estimation showed reduced efficacy (ΔMAPE% = 18.12). This suggests that abiotic proxies are context-dependent and may not universally replace biotic measurements, particularly for woody biomass components. Second, the reliance on global datasets such as SoilGrids250m and ClimateAP introduces uncertainties, as their coarse spatial resolution (~1 km) may inadequately capture microsite variations in heterogeneous natural forests. Future applications could benefit from localized soil-climate data to refine predictions. Additionally, while the model stabilized with four calibration trees, operational scalability in diverse ecological contexts—especially in regions with differing species compositions or disturbance regimes—requires further validation. Practically, this framework offers a cost-effective tool for large-scale carbon stock assessments. Integrating DBH with globally accessible abiotic covariates enables efficient resource allocation in remote areas. For instance, embedding the model into national forest inventories could streamline carbon trading analyses or reforestation planning under climate change. Adaptive calibration protocols tailored to local gradients (e.g., soil fertility) are recommended to enhance robustness. Future work should explore temporal dynamics (e.g., climate-growth feedbacks) and expand covariates (e.g., UAV-derived canopy metrics). Cross-biome validation, particularly in boreal and subtropical forests, will clarify transferability and strengthen global forest management applications.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
AM: Conceptualization, Formal analysis, Investigation, Methodology, Software, Visualization, Writing – original draft, Writing – review & editing. ZM: Methodology, Writing – original draft, Writing – review & editing. LX: Writing – review & editing. JT: Visualization, Writing – original draft. XZ: Writing – original draft. LD: Conceptualization, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research and/or publication of this article. This research was Supported by the National Key R&D Program of China (NO. 2022YFD2201001), the Heilongjiang Provincial Natural Science Foundation Project (NO. YQ2022C005), the Fundamental Research Funds for the Central Universities (NO. 2572023AW12).
The authors would like to thank the faculty and students of the Department of Forest Management, Northeast Forestry University (NEFU), China, who collected and provided the data for this study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2014). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01
Becknell, J., and Powers, J. (2014). Stand age and soils as drivers of plant functional traits and aboveground biomass in secondary tropical dry forest. Can. J. For. Res. 44, 604–613. doi: 10.1139/cjfr-2013-0331
Bennett, A., Penman, T., Arndt, S., Roxburgh, S., and Bennett, L. (2020). Climate more important than soils for predicting forest biomass at the continental scale. Ecography 43, 1692–1705. doi: 10.1111/ecog.05180
Bi, H., Murphy, S., Volkova, L., Weston, C., Fairman, T., Li, Y., et al. (2015). Additive biomass equations based on complete weighing of sample trees for open eucalypt forest species in South-Eastern Australia. For. Ecol. Manag. 349, 106–121. doi: 10.1016/j.foreco.2015.03.007
Bi, H., Turner, J., and Lambert, M. (2004). Additive biomass equations for native eucalypt forest trees of temperate Australia. Trees 18, 467–479. doi: 10.1007/s00468-004-0333-z
Bronisz, K., Bijak, S., Wojtan, R., Tomusiak, R., Bronisz, A., Baran, P., et al. (2021). Seemingly unrelated mixed-effects biomass models for black locust in West Poland. Forests 12:380. doi: 10.3390/f12030380
Bronisz, K., and Mehtätalo, L. (2020). Seemingly unrelated mixed-effects biomass models for young silver birch stands on post-agricultural lands. Forests 11:381. doi: 10.3390/f11040381
Caleño-Ruíz, B., Garzón-R, F., López Camacho, R., Pizano, C., Salinas, V., and González-M, R. (2023). Soil resources and functional trait trade-offs determine species biomass stocks and productivity in a tropical dry forest. Front. Forests Glob. Change 6:1028359. doi: 10.3389/ffgc.2023.1028359
Černý, J., Pokorny, R., Vejpustková, M., Srámek, V., and Bednář, P. (2020). Air temperature is the main driving factor of radiation use efficiency and carbon storage of mature Norway spruce stands under global climate change. Int. J. Biometeorol. 64, 1599–1611. doi: 10.1007/s00484-020-01941-w
Chen, R., Ran, J., Hu, W., Dong, L., Ji, M., Jia, X., et al. (2021). Effects of biotic and abiotic factors on forest biomass fractions. National Science. Review 8:nwab025. doi: 10.1093/nsr/nwab025
Chevan, A., and Sutherland, M. (1991). Hierarchical partitioning. Am. Stat. 45, 90–96. doi: 10.1080/00031305.1991.10475776
Dong, L.-H., Zhang, Z., Chen, X., and Li, W. (2020). Comparison of tree biomass modeling approaches for larch (Larix olgensis Henry) trees in Northeast China. Forests 11:202. doi: 10.3390/f11020202
Dong, L., Zhang, L., and Li, F. (2014). A compatible system of biomass equations for three conifer species in northeast, China. For. Ecol. Manag. 329, 306–317. doi: 10.1016/j.foreco.2014.05.050
Dong, L., Zhang, L., and Li, F. (2015). Developing additive systems of biomass equations for nine hardwood species in Northeast China. Trees. 29, 1149–1163. doi: 10.1007/s00468-015-1196-1
Dong, L.-H., Zhang, L., and Li, F. (2018). Additive biomass equations based on different Dendrometric variables for two dominant species (Larix gmelini Rupr. And Betula platyphylla Suk.) in natural forests in the eastern Daxing’an mountains, Northeast China. Forests 9:261. doi: 10.3390/f9050261
Drake, J., Raetz, L., Davis, S., and DeLucia, E. (2010). Hydraulic limitation not declining nitrogen availability causes the age-related photosynthetic decline in loblolly pine (Pinus taeda L.). Plant Cell Environ. 33, 1756–1766. doi: 10.1111/j.1365-3040.2010.02180.x
Dutcă, I., Mather, R., and Ioraș, F. (2020). Sampling trees to develop allometric biomass models: how does tree selection affect model prediction accuracy and precision? Ecol. Indic. 117:106553. doi: 10.1016/j.ecolind.2020.106553
Fassnacht, F. E., Hartig, F., Latifi, H., Berger, C., Hernández, J., Corvalán, P., et al. (2014). Importance of sample size, data type and prediction method for remote sensing-based estimations of aboveground forest biomass. Remote Sens. Environ. 154, 102–114. doi: 10.1016/j.rse.2014.07.028
Fick, S., and Hijmans, R. (2017). WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37, 4302–4315. doi: 10.1002/joc.5086
Fu, L., Sun, W., and Wang, G. (2017). A climate-sensitive aboveground biomass model for three larch species in northeastern and northern China. Trees 31, 557–573. doi: 10.1007/s00468-016-1490-6
Geng, W., Li, Y., Sun, D., Li, B., Zhang, P., Chang, H., et al. (2022). Prediction of the potential geographical distribution of Betula platyphylla Suk. In China under climate change scenarios. PLoS One 17:e0262540. doi: 10.1371/journal.pone.0262540
Gustafson, E., Shvidenko, A., Sturtevant, B., and Scheller, R. (2010). Predicting global change effects on forest biomass and composition in south-Central Siberia. Ecol. Appl. 20, 700–715. doi: 10.1890/08-1693.1
Hengl, T., Mendes de Jesus, J., Heuvelink, G., Gonzalez, M., Kilibarda, M., Blagotić, A., et al. (2017). SoilGrids250m: global gridded soil information based on machine learning. PLoS One 12:e0169748. doi: 10.1371/journal.pone.0169748
Holly, K. G., Sandra, B., John, O. N., and Jonathan, A. F. (2007). Monitoring and estimating tropical forest carbon stocks: making REDD a reality. Environ. Res. Lett. 2:045023 (13pp). doi: 10.1088/1748-9326/2/4/045023
Kusmana, C., Hidayat, T., Tiryana, T., and Rusdiana, O. (2018). Allometric models for above- and below-ground biomass of Sonneratia spp. Global Ecol. Conserv. 15:e00417. doi: 10.1016/j.gecco.2018.e00417
Lal, R. (2018). Digging deeper: a holistic perspective of factors affecting soil organic carbon sequestration in agroecosystems. Glob. Chang. Biol. 24, 3285–3301. doi: 10.1111/gcb.14054
Li, F., Shi, Z., Liu, S., Chen, M., Xu, G., Chen, J., et al. (2024). Stand structural diversity and edaphic properties regulate aboveground biomass of Abies fargesii var. faxoniana primary forest on the eastern Qinghai-Tibetan plateau. Eur. J. For. Res. 143, 1401–1410. doi: 10.1007/s10342-024-01697-7
Li, S., Su, J., Lang, X., Liu, W., and Guanglong, O. (2018). Positive relationship between species richness and aboveground biomass across forest strata in a primary Pinus kesiya forest. Sci. Rep. 8:2227. doi: 10.1038/s41598-018-20165-y
Li, Y., Tian, D., Yang, H., and Niu, S. (2017). Size-dependent nutrient limitation of tree growth from subtropical to cold temperate forests. Funct. Ecol. 32, 95–105. doi: 10.1111/1365-2435.12975
Litton, C. M., Raich, J. W., and Ryan, M. G. (2007). Carbon allocation in forest ecosystems. Glob. Chang. Biol. 13, 2089–2109. doi: 10.1111/j.1365-2486.2007.01420.x
Liu, J., Wang, X., Kou, Y., Zhao, W., and Liu, Q. (2023). Differences in the effects of broadleaf and coniferous trees on soil nematode communities and soil fertility across successional stages. Plant Soil 485, 197–212. doi: 10.1007/s11104-022-05677-x
Ma, J., Chen, L., Mi, X., Ren, H., Liu, X., Wang, Y., et al. (2023). The interactive effects of soil fertility and tree mycorrhizal association explain spatial variation of diversity–biomass relationships in a subtropical forest. J. Ecol. 111, 1037–1049. doi: 10.1111/1365-2745.14076
Maltamo, M., Mehtätalo, L., Vauhkonen, J., and Packalen, P. (2012). Predicting and calibrating tree attributes by means of airborne laser scanning and field measurements. Can. J. For. Res. 42, 1896–1907. doi: 10.1139/x2012-134
Mao, Z., Van der Plas, F., Corrales, A., Anderson-Teixeira, K., Bourg, N., Chu, C., et al. (2023). Scale-dependent diversity–biomass relationships can be driven by tree mycorrhizal association and soil fertility. Ecol. Monogr. 93:e1568. doi: 10.1002/ecm.1568
McDowell, N. G., Allen, C. D., Anderson-Teixeira, K., Aukema, B. H., Bond-Lamberty, B., Chini, L., et al. (2020). Pervasive shifts in forest dynamics in a changing world. Science 368:eaaz9463. doi: 10.1126/science.aaz9463
Mehtätalo, L., and Lappi, J., (2020). Biometry for forestry and environmental data: With examples in R. Chapman and Hall/CRC.
Nally, R. M., and Walsh, C. J. (2004). Hierarchical partitioning public-domain software. Biodivers. Conserv. 13, 659–660. doi: 10.1023/B:BIOC.0000009515.11717.0b
Obata, A., Yoshida, T., and Hiura, T. (2023). Estimation of stand biomass and species-specific biomass in Japanese northern mixed forests in 1920–1930s: understanding environmental factors affecting carbon sequestration before recent climate change. Ecol. Indic. 154:110495. doi: 10.1016/j.ecolind.2023.110495
Ola, A., Schmidt, S., and Lovelock, C. E. (2018). The effect of heterogeneous soil bulk density on root growth of field-grown mangrove species. Plant Soil 432, 91–105. doi: 10.1007/s11104-018-3784-5
Ou, G., Wang, J., Xu, H., Chen, K., Zheng, H., Zhang, B., et al. (2016). Incorporating topographic factors in nonlinear mixed-effects models for aboveground biomass of natural Simao pine in Yunnan, China. J. For. Res. 27, 119–131. doi: 10.1007/s11676-015-0143-8
Petaja, G., Bardule, A., Zalmanis, J., Lazdina, D., Daugaviete, M., Skranda, I., et al. (2023). Changes in organic carbon stock in soil and whole tree biomass in afforested areas in Latvia. Plants-Basel 12. doi: 10.3390/plants12122264
Poorter, L., Wright, S. J., Paz, H., Ackerly, D. D., Condit, R., Ibarra-Manríquez, G., et al. (2008). Are functional traits good predictors of demographic rates? Evidence from five neotropical forests. Ecology 89, 1908–1920. doi: 10.1890/07-0207.1
Saatchi, S., Harris, N., Brown, S., Lefsky, M., Mitchard, E., Salas, W., et al. (2011). Benchmark map of forest carbon stocks in tropical regions across three continents. Proc. Natl. Acad. Sci. USA 108, 9899–9904. doi: 10.1073/pnas.1019576108
Schimel, D., Pavlick, R., Fisher, J. B., Asner, G. P., Saatchi, S., Townsend, P., et al. (2015). Observing terrestrial ecosystems and the carbon cycle from space. Glob. Chang. Biol. 21, 1762–1776. doi: 10.1111/gcb.12822
Sun, W., and Liu, X. (2019). Review on carbon storage estimation of forest ecosystem and applications in China. Forest Ecosyst. 7:4. doi: 10.1186/s40663-019-0210-2
Teklehaimanot, Z. (2004). Physiological plant ecology: ecophysiology and stress physiology of functional groups, 4th edn. Forestry 77, 365–366. doi: 10.1093/forestry/cph039
Testolin, R., Dalmonech, D., Marano, G., Bagnara, M., D'Andrea, E., Matteucci, G., et al. (2023). Simulating diverse forest management options in a changing climate on a Pinus nigra subsp. laricio plantation in southern Italy. Sci. Total Environ. 857:159361. doi: 10.1016/j.scitotenv.2022.159361
Uscanga, A., Bartlein, P. J., and Silva, L. C. R. (2023). Local and regional effects of land-use intensity on aboveground biomass and tree diversity in tropical montane cloud forests. Ecosystems 26, 1734–1752. doi: 10.1007/s10021-023-00861-1
Wang, Y., Miao, Z., Hao, Y., Dong, L.-H., and Li, F. (2023). Effects of biotic and abiotic factors on biomass conversion and expansion factors of natural White birch Forest (Betula platyphylla Suk.) in Northeast China. Forests 14:362. doi: 10.3390/f14020362
Wang, T., Wang, G., Innes, J., Seely, B., and Chen, B. (2017). ClimateAP: An application for dynamic local downscaling of historical and future climate data in Asia Pacific. Front. Agric. Sci. Eng. 4, 448–458. doi: 10.15302/J-FASE-2017172
Wang, X., Zhao, D., Liu, G., Yang, C., and Teskey, R. O. (2018). Additive tree biomass equations for Betula platyphylla Suk. Plantations in Northeast China. Ann. For. Sci. 75:60. doi: 10.1007/s13595-018-0738-2
Wei, Y., Yu, D., Lewis, B. J., Zhou, L., Zhou, W., Fang, X., et al. (2014). Forest carbon storage and tree carbon pool dynamics under natural forest protection program in northeastern China. Chin. Geogr. Sci. 24, 397–405. doi: 10.1007/s11769-014-0703-4
Xiao, X., White, E., Hooten, M., and Durham, S. (2011). On the use of log-transformation vs. nonlinear regression for analyzing biological power laws. Ecology 92, 1887–1894. doi: 10.2307/23034821
Xie, L., Fu, L., Widagdo, F., Dong, L.-H., and Li, F. (2021). Improving the accuracy of tree biomass estimations for three coniferous tree species in Northeast China. Trees 36, 451–469. doi: 10.1007/s00468-021-02220-w
Xie, L., Wang, T., Miao, Z., Hao, Y., Dong, L.-H., and Li, F. (2023). Considering random effects and sampling strategies improves individual compatible biomass models for mixed plantations of Larix olgensis and Fraxinus mandshurica in northeastern China. For. Ecol. Manag. 537:120934. doi: 10.1016/j.foreco.2023.120934
Xu, Y., Zhang, Y., Li, W., Liu, W., Gu, X., Guan, Z., et al. (2019). Effects of tree functional diversity and environmental gradients on belowground biomass in a natural old-growth forest ecosystem. Can. J. For. Res. 49, 1623–1632. doi: 10.1139/cjfr-2019-0254
Yang, Y., Bao, W., Hu, H., Wu, N., Li, F., Wang, Z., et al. (2024). Environmental factors drive latitudinal patterns of fine-root architectures of 96 xerophytic species in the dry valleys of Southwest China. Sci. Total Environ. 950:175352. doi: 10.1016/j.scitotenv.2024.175352
Zhang, B., Hautier, Y., Tan, X., You, C., Cadotte, M., Chu, C., et al. (2020). Species responses to changing precipitation depend on trait plasticity rather than trait means and intraspecific variation. Funct. Ecol. 34, 2622–2633. doi: 10.1111/1365-2435.13675
Zhao, D., Kane, M., Markewitz, D., and Clutter, M. (2015). Additive tree biomass equations for Midrotation loblolly pine plantations. For. Sci. 61. doi: 10.5849/forsci.14-193
Keywords: biomass estimation, SUR mixed-effects models, biotic and abiotic factors, natural white birch, SUR
Citation: Ma A, Miao Z, Xie L, Tian J, Zhao X and Dong L (2025) Integrating climate and soil factors enhances biomass estimation for natural white birch (Betula platyphylla Sukaczev). Front. For. Glob. Change. 8:1549531. doi: 10.3389/ffgc.2025.1549531
Received: 21 December 2024; Accepted: 27 February 2025;
Published: 14 March 2025.
Edited by:
Yashwant Singh Rawat, Federal Technical and Vocational Education and Training Institute (FTVETI), EthiopiaReviewed by:
Praveen Kumar, Dr. Yashwant Singh Parmar University of Horticulture and Forestry, IndiaCopyright © 2025 Ma, Miao, Xie, Tian, Zhao and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihu Dong, bGlodWRvbmdAbmVmdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.