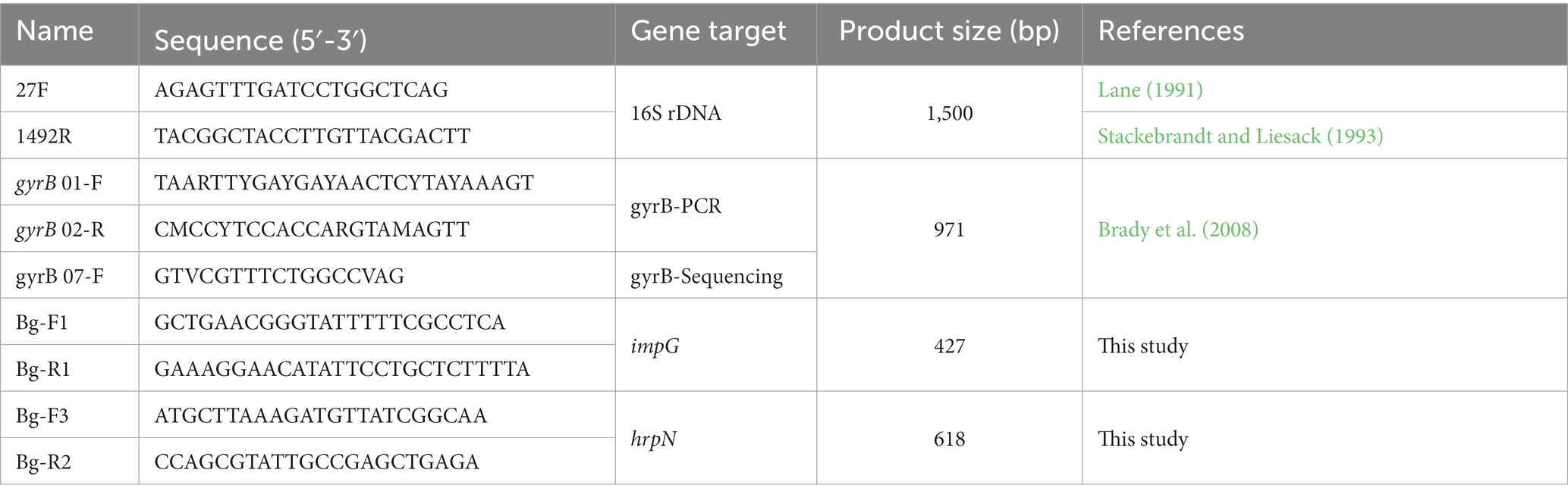
- 1Department of Entomology and Plant Pathology, College of Agricultural Technology, University College of Agriculture and Natural Resources, University of Tehran, Tehran, Iran
- 2Centre for Research in Bioscience, Faculty of Health and Life Sciences, University of the West of England, Bristol, United Kingdom
- 3Department of Plant Protection, Sari Agricultural Science and Natural Resources University, Sari, Iran
Chestnut-leaved oak (Quercus castaneifolia) and oriental beech (Fagus orientalis) are among the major tree species in the Hyrcanian forests. Brenneria goodwinii was identified as the causal agent of necrotic lesions and stem bleeding on affected oak trees in different countries. Oak and oriental beech trees with bleeding symptoms were observed in a few forest sites in northern Iran. The objectives of the present study were to identify and characterize the causal agents of bark canker in oak and oriental beech trees and develop a primer set for specific detection, using polymerase chain reaction (PCR), of Brenneria goodwinii strains. A total of 31 and 20 samples from oak and oriental beech trees, respectively, with stem bleeding and bark canker symptoms were collected from Golestan and Mazandaran forests in northern Iran in 2020–2021. Bacterial strains displaying a green metallic sheen on EMB-agar medium were isolated from symptomatic oak (105 strains) and oriental beech samples (32 strains), while 31 and 20 strains were also isolated from healthy oak and oriental beech, respectively. Pathogenicity tests indicated that 51 and 25 strains isolated from oak and oriental beech, respectively were able to induce a necrotic area on oak acorns 15 days following inoculation. Moreover, four and two representative strains inoculated on oak and oriental beech twigs, respectively induced necrosis on all inoculated green twigs 1 month after inoculation. The sequences of the 16S rRNA and gyrB genes of representative strains isolated from and proved pathogenic on oak and oriental beech trees were 100% and over 99% similar to B. goodwinii LMG 26270T, respectively, which revealed the strains belong to B. goodwinii species. The primer pair BgF3/R2, which was designed to target the hrpN gene, was proven to be specific in the detection of B. goodwinii strains. The primer pair amplified a 618-bp DNA fragment from strains of B. goodwinii only and not from strains belonging to Rahnella, Gibbsiella, Lonsdalea, and the other Brenneria species among several other pathogenic bacteria tested. No fragment was amplified from DNA extracted from healthy trees or seedlings in PCR using this primer pair.
Introduction
Chestnut-leaved oak (Quercus castaneifolia) and oriental beech (Fagus orientalis) are among the main tree species in the Hyrcanian or Caspian forests in northern Iran, with a history that dates back to 25–50 million years (Alavi et al., 2020).
Forest decline, described as loss in tree vigor and increased mortality due initially to climate change events (e.g., climate warming and drought) and accompanied by the contribution of pathogens and pests (Manion, 1991), is becoming widespread in some local stands (Sangüesa-Barreda et al., 2015). Several bacterial species including Gibbsiella quercinecans (Brady et al., 2010), Brenneria goodwinii (Denman et al., 2012), Brenneria roseae (Brady et al., 2014a), Rahnella victoriana (Brady et al., 2014b), Bacillus pumilus and Stenotrophomonas maltophilia (Ahmadi et al., 2019), and “Brenneria izadpanahii” (Bakhshi Ganje et al., 2021) have been identified as the causal agents of necrotic lesions and stem bleeding on oak trees with decline symptoms. Although several bacterial species were found to be associated with acute oak decline (AOD) (Denman et al., 2012), the most frequently encountered pathogens are G. quercinecans (Brady et al., 2010) and B. goodwinii (Denman et al., 2012). The latter is supposed to be the most damaging among AOD agents (Doonan et al., 2019). B. goodwinii was isolated from different Quercus species in the United Kingdom (UK) (Denman et al., 2012), Latvia (Zalkalns and Celma, 2021), Spain (González and Ciordia, 2020), Iran (Bakhshi Ganje et al., 2020), Switzerland (Ruffner et al., 2020), and Poland (Tkaczyk et al., 2021). Additionally, it was associated with acute decline symptoms on hornbeam (Carpinus betulus) trees in Iran (Moradi-Amirabad et al., 2019).
Diagnostic approaches commonly employed for the identification of B. goodwinii species consist of traditional (phenotypic biochemical and nutritional) and pathogenicity tests and sequencing of housekeeping genes including gyrB, infB, rpoB, and atpD (Denman et al., 2012; Bakhshi Ganje et al., 2020), which are both time- and labor-consuming procedures. Recently, a multiplex TaqMan polymerase chain reaction (PCR) assay was developed for simultaneous detection of four bacterial species involved in the AOD syndrome in the UK, including B. goodwinii, G. quercinecans, R. victoriana, and Lonsdalea britannica (Crampton et al., 2020). Nonetheless, a reliable molecular method for the rapid, accurate, and culture-independent detection and identification of B. goodwinii strains in infected plant samples is still lacking.
Stem bleeding and bark canker of oak and oriental beech trees, similar to those reported in bacterial canker, were observed in the Hyrcanian or Caspian forests in northern Iran during the summer months of 2020–2021 in the forests of Golestan and Mazandaran provinces. The objectives of the present study were to identify and characterize the causal agents of bark canker in oak and oriental beech trees by bacterial isolation and prove pathogenicity on oak and oriental beech seedlings and oak acorns, phenotypic characterization, and phylogenetic analyses based on 16S rRNA and gyrB genes. As no species-specific primers have thus far been designed for the detection of B. goodwinii strains, attempts were made to devise and prove the reliability of a primer set for specific detection, using PCR, of B. goodwinii strains from different hosts.
Materials and methods
Sampling and isolation
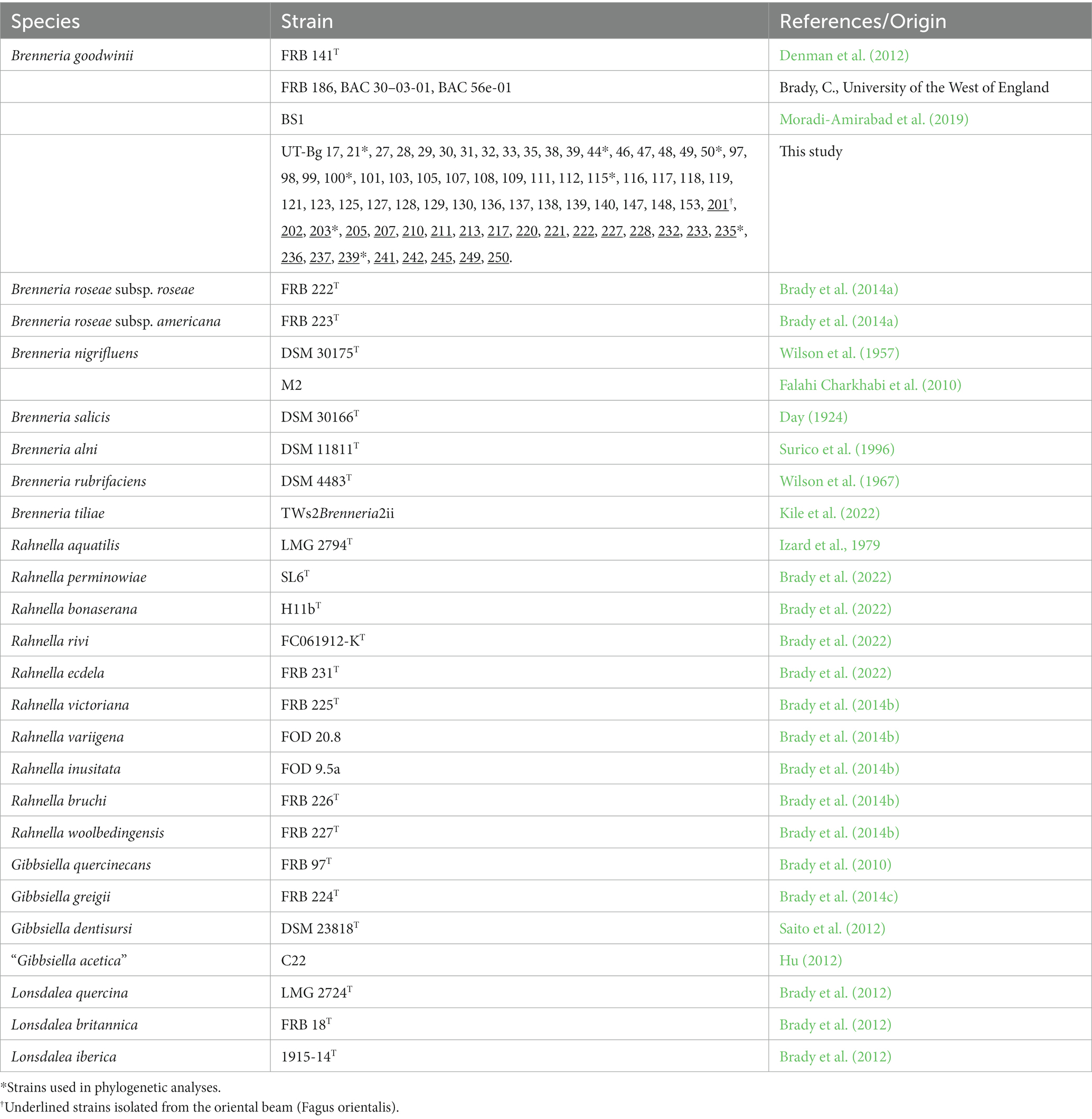
Samples of the outer bark, inner bark, and sapwood tissues from cankered sites on the trunk and main branches of oak (Q. castaneifolia) and oriental beech (F. orientalis) trees were collected from Gorgan, Golestan province and Sari, Mazandaran province, respectively, in the summer of 2020–2021. Five symptomless samples were collected from the healthy trees of each host. Moreover, three samples were collected from two- and three-year-old healthy oak seedlings in the greenhouse. Tissues were washed under tap water for 5 min, then surface sterilized in 70% ethanol for 30 s and rinsed three times in sterile distilled water (SDW). The symptomatic tissues were ground to small pieces in SDW using a mortar and pestle and a loopful of resulting suspensions streaked on plates of EMB medium (Merck, Darmstadt, Germany) containing 1% glycerol and 0.5% yeast extract and incubated at 28°C. Bacterial colonies were isolated after 48–72 h and purified by streaking on nutrient agar (NA) (Merck, Germany, Darmstadt) plates. The strains were stored in 15% glycerol stocks at −80°C. Bacterial strains included in this study are listed in Table 1.
Pathogenicity assay
Pathogenicity testing of all strains, including 105 and 32 strains isolated from oak and oriental beech symptomatic trees, was performed on chestnut-leaved oak acorns by injecting 10 μL of a bacterial suspension of each isolate prepared from an overnight culture on NA in SDW with an optical density of 0.1 at 600 nm using a disposable insulin syringe with needle (González and Ciordia, 2020). B. goodwinii FRB 141T, G. quercinecans LMG 25500T, and R. victoriana FRB 225T were used as positive controls and SDW as the negative control, respectively. Symptom progression was observed for 15 days. Pathogenicity assays were performed in two replicates each containing three acorns per strain.
Moreover, pathogenicity of representative strains (UT-Bg 21, 44, 100, 115, 235, and 245) isolated from oak and oriental beech trees was tested by inoculating two- and three-year-old oak and oriental beech seedlings. Seedlings were kept in a greenhouse under conditions of 16 h light at 28°C and 8 h dark at 24°C with 75% relative humidity. The surface of the young green twigs was first disinfected using 95% ethanol for 1 min. The bacterial suspensions of the representative strains with OD600 = 0.1 were prepared in SDW and injected into twigs using sterile syringes. Inoculated twigs were covered with parafilm, and symptoms were monitored 2 months post-inoculation. To fulfill Koch’s postulates, bacterial re-isolation from inoculated acorns, and twigs were performed 15 and 30 days post-inoculation on EMB-agar, respectively. Representative strains were inoculated on twigs in three replicates. Re-isolated bacteria were identified by colony morphology on EMB-agar and PCR using specific primers.
Phenotypic characterization
Ten strains isolated from each host were subjected to the following diagnostic tests: Gram reaction using 3% KOH (Suslow et al., 1982), levan formation from sucrose, oxidase reaction, starch hydrolysis, and several other phenotypic tests according to Schaad et al. (2001). Utilization of carbon sources was assayed using the basal medium of Ayers et al. (1919) supplemented with filter-sterilized carbohydrate sources at 0.25% final concentration. Cultures of Brenneria nigrifluens M2 (Falahi Charkhabi et al., 2010), B. goodwinii FRB 141T, G. quercinecans LMG2 5500T, and R. victoriana FRB 225T were also used as reference strains in all tests.
Molecular characterization
Genomic DNA was extracted from overnight cultures on NA using the alkalic lysis procedure (Niemann et al., 1997). PCR was performed in a final volume of 20 μL containing 2x Taq DNA Polymerase Master Mix (Ampliqon A/S, Odense, Denmark), 10 pmol of each primer, and 2 μL of template DNA. The reaction was performed in a thermocycler (Mastercycler®, Eppendorf, Hamburg, Germany). PCR amplification of the 16S rRNA gene of the five strains (UT-Bg 21, 44, 100, 235, and 239) was performed using the primer pair 27F/1492R (Lane, 1991; Stackebrandt and Liesack, 1993; Table 2). The PCR program used consisted of an initial denaturation at 94°C for 4 min, followed by 30 cycles of denaturing at 94°C for 1 min, annealing at 59°C for 1 min, and extension at 72°C for 2.5 min. The final extension was performed at 72°C for 10 min.
A 1050 bp fragment of the housekeeping gene gyrB (DNA gyrase subunit β) of each of the eight strains including UT-Bg 21, 44, 50, 100, 115, 203, 235, and 239 was amplified and sequenced using the primers and conditions previously published (Brady et al., 2008; Table 2).
The amplified fragments were electrophoresed in 0.5X TBE buffer (tris-borate) and visualized by staining with ethidium bromide. The conserved genes were sequenced by Microsynth Co. (Balgach, Switzerland) using the Sanger method, and the obtained sequences were compared with those deposited in GenBank. Alignments were performed in ClustalW (Thompson et al., 1994), and phylogenetic trees were constructed using MEGA X (Kumar et al., 2018) with the maximum likelihood method based on the lowest value of the Bayesian information criterion (BIC). The robustness of the branches was determined by bootstrap analysis using 1000 replicates. Xenorhabdus nematophila ATCC 19061T (DSM 3370T) was used as an outgroup.
Primer designing
The genome sequence of B. goodwinii FRB 141T (GenBank accession no. NZ_CP014137.1) was aligned against genomes of the other B. goodwinii strains including B. goodwinii FRB 171 (NZ_MJLY00000000.1) and B. goodwinii OBR1 (CGIG01000001.1). Genes with more than 95% similarity were selected and aligned with those of ‘Affinibrenneria salicis’ strain L3-3HA, Brenneria corticis CFCC 11842, ‘B. izadpanahii’ Iran 50, B. nigrifluens ATCC 13028, B. nigrifluens LMG 2694T, Brenneria populi EniD312, B. roseae subsp. roseae LMG 27714T, B. roseae subsp. americana LMG 27715T, Brenneria rubrifaciens 6D370, Brenneria salicis ATCC 15712T, and Brenneria sp. hezel4-2-4. Potential genes with less than 85% similarity were screened, and candidate genes were aligned with those of Brenneria (taxid:71655), Gibbsiella (taxid:929812), Rahnella (taxid:34037), Lonsdalea (taxid:1082702), Dickeya (taxid:204037), Pantoea (taxid:53335), Serratia (taxid:613), Pectobacterium (taxid:122277), Enterobacterales (taxid:91347), Xanthomonas (taxid:338), and Pseudomonas (taxid:286), as well as NCBI BioProject PRJNA342025. Finally, the gene encoding protein IpmG of the type VI secretion system and the gene encoding HrpN of the type III secretion system in B. goodwinii strains FRB 141T, FRB 171, and OBR1 were selected for designing the primers. Two primer pairs were designed and evaluated for PCR detection of B. goodwinii: BgF1/R1, which amplified a 427 bp fragment of impG gene and BgF3/R2 that amplified a 618 bp fragment of hrpN as the target (Table 2). Primers were synthesized by the Metabion Company (Bavaria, Germany).
Primer specificity
The two primer pairs BgF1/R1 and BgF3/R2 were evaluated for their specificity in targeting the specific genes in B. goodwinii. PCR was performed in 20 μL-reaction mixtures containing 2x Taq DNA Polymerase Master Mix (Ampliqon A/S, Odense, Denmark), 10 pmol of each primer, and 2 μL of template DNA. The PCR program consisted of an initial denaturation step of 5 min at 94°C, followed by 28 cycles of 1 min at 94°C, 1 min at 56°C, and 45 s at 72°C, and a final extension step of 5 min at 72°C. B. goodwinii FRB 141T and B. goodwinii BS1 (Moradi-Amirabad et al., 2019) were used as positive controls. SDW and strains of the other species and representative species of Brenneria, Rahnella, Gibbsiella, and Lonsdalea inciting decline on other forest trees (Table 1), plus strains of some distantly related or unrelated genera, including Enterobacter ludwigii A21, Serratia marcescens A80, Pectobacterium carotovorum ATCC 15713T, Xanthomonas translucens pv. undulosa ICMP11055, X. translucens pv. cerealis ICMP 5752, Pantoea agglomerans NK16, Agrobacterium radiobacter PK, Pseudomonas viridiflava ATCC 13223T, P. viridiflava FBF52, Pseudomonas sp. A44, Pseudomonas tolaasii T7, and Pseudomonas amygdali pv. lachrymans GSPB 82a, A77: Klebsiella oxytoca A77, Enterobacter ludwigii A21, Bacillus sp. A11, and Lysinibacillus sp. A31, were all used as negative controls. Bacterial strains isolated from symptomless oak trees and seedlings were also used in the specificity assay.
In planta detection of Brenneria Goodwinii
For direct detection of B. goodwinii in infected tissues, oak symptomatic samples collected from Golestan and Mazandaran forests as well as oriental beech artificially inoculated plants were used. Inoculation was performed as described above. Three weeks after inoculation, 0.5 g of artificially infected oriental beech twig tissues was cut and homogenized with a mortar pestle in 4-ml SDW and incubated at room temperature for 1 h. Moreover, oak symptomatic samples were homogenized with a mortar pestle in phosphate-buffered saline buffer and incubated at room temperature for 1 h. DNA was extracted from the suspension using the alkalic lysis procedure (Niemann et al., 1997), 2 μL of which was used in the species-specific PCR as described above. Twigs injected with SDW were used as a negative control.
Primer sensitivity
To determine the sensitivity and limit of detection of the designed primer pair, DNA was extracted from serial 10-fold dilutions of B. goodwinii, with suspensions starting with OD600 = 0.1, by the alkalic lysis procedure (Niemann et al., 1997), and 2 μL was used as templates in the PCR tests as described above.
Results
Symptoms, sampling, and isolation
In the summer of 2020–2021, 31 samples and 20 samples from decline-affected oak and oriental beech trees were collected in different areas of the Golestan and Mazandaran forests, respectively. The most characteristic symptoms were tissue necrosis, canker, and bleeding of a dark sticky fluid from vertical cracks in the bark of the trunk and scaffold branches of the infected trees (Figure 1). A total of 105 and 32 strains with a green metallic sheen on EMB-agar medium were isolated from the symptomatic oak and oriental beech samples, respectively. Thirty-one strains were also isolated from symptomless oak and oriental beech trees. Additionally, 15 strains were isolated from two- and three-year-old oak seedlings in the greenhouse and used in some tests.
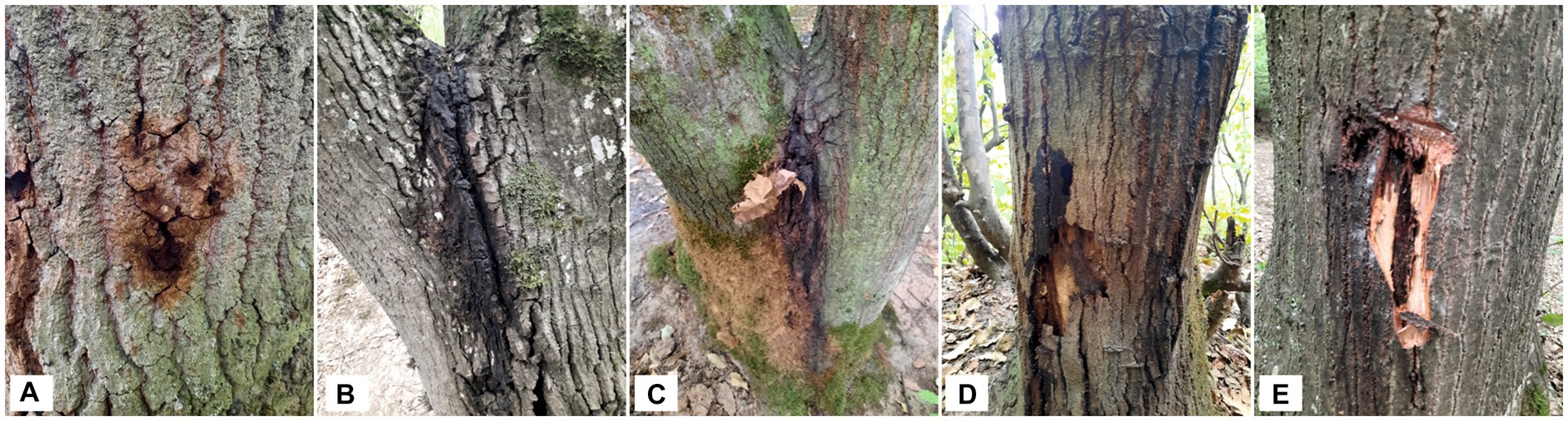
Figure 1. Symptoms of acute decline of chestnut-leaved oak (Quercus castaneifolia) in Iran, canker lesions, stem bleeding of a dark sticky fluid from vertical cracks (A–C), and necrotic underlying tissue lesions in the inner bark (D–E).
Pathogenicity tests
All strains were inoculated on oak acorns, among which 51 strains isolated from oak and 25 strains isolated from oriental beech were able to induce a necrotic area 15 days following inoculation (Figures 2H–KM–Q). Furthermore, B. goodwinii FRB 141T, G. quercinecans LMG 25500T, and R. victoriana FRB 225T also induced necrotic areas on oak acorns. Four representative strains induced necrosis on all inoculated oak green twigs, 1 month after inoculation under greenhouse conditions (Figures 2B–D). Moreover, two strains inoculated on oriental beech caused necrosis on green twigs and die-back (Figure 2F). Re-isolation from the inoculated six acorn samples, four oak, and three oriental beech twigs was performed on EMB and yielded a profuse number of colonies with a green metallic sheen similar to the inoculated ones. Moreover, the identification of re-isolated strains was confirmed by PCR using the developed specific primers. No symptoms were observed on test acorns and twigs injected with SDW (Figures 2A,E,G,L).

Figure 2. Pathogenicity assays of Brenneria goodwinii strains isolated from chestnut leaved oak (Quercus castaneifolia) and oriental beech (Fagus orientalis) on the oak and oriental beech seedlings and oak acorns, 30 and 15 days after inoculation, respectively. (A,E,G,L) Negative control infiltrated using sterile deionized water; oak seedlings inoculated using strains UT-Bg 21 (B), UT-Bg 100 (C), UT-Bg 115 (D); oriental beech seedling inoculated using strain UT-Bg 245 (F), acorn inoculated using strains UT-Bg 46 (H), UT-Bg 98 (I), UT-Bg 222 (J), UT-Bg 235 (K), UT-Bg 115 (M), UT-Bg 21 (N), UT-Bg 44 (O), UT-Bg 239 (P,Q).
Phenotypic characteristics
Twenty strains isolated from the affected trees were Gram-negative. Colonies on nutrient agar were cream-colored, circular, convex, and smooth with entire margins and produced a green metallic sheen on EMB-Agar. Strains were negative in tests for oxidase, starch, Tween 20 hydrolysis, and the production of levan. Acid was produced from arabinose, fructose, D-lactose, trehalose, mannitol, and sorbitol. Strains utilized histidine, alanine, and serine but not arginine, lysine, ornithine, and tryptophan as sole carbon sources. The phenotypic characters of isolated strains were identical with B. goodwinii LMG 26270T, which indicated the strains belong to B. goodwinii species.
Sequence analysis
Sequences of 976 bp and 718 bp fragments of the 16S rRNA and gyrB genes, respectively, of representative strains isolated from oak and oriental beech revealed similarity levels of 100% and over 99% to B. goodwinii LMG 26270T, respectively, which revealed the strains belong to B. goodwinii species. The sequences were deposited in the NCBI GenBank database under accession numbers ON398765 to ON398769 for 16S rRNA (UT-Bg 21, 44, 100, 235, and 239), and ON412785 to ON412792 for gyrB (UT-Bg 21, 44, 50, 100, 115, 203, 235, and 239).
In the phylogenetic tree (Figure 3A) inferred from the 16S rRNA gene sequences of strains UT-Bg 21, 44, 100, 235, and 239, the strains clustered within the type strain of B. goodwinii supported with bootstrap values of 100%. In the phylogenetic tree constructed from the gyrB sequences of strains UT-Bg 21, 44, 50, 100, 115, 203, 235, and 239, all strains clustered with B. goodwinii LMG 26270T and B. goodwinii FRB 171 with high bootstrap support (Figure 3B). Based on phenotypic characters and phylogenetic analysis, the strains belong to B. goodwinii species.
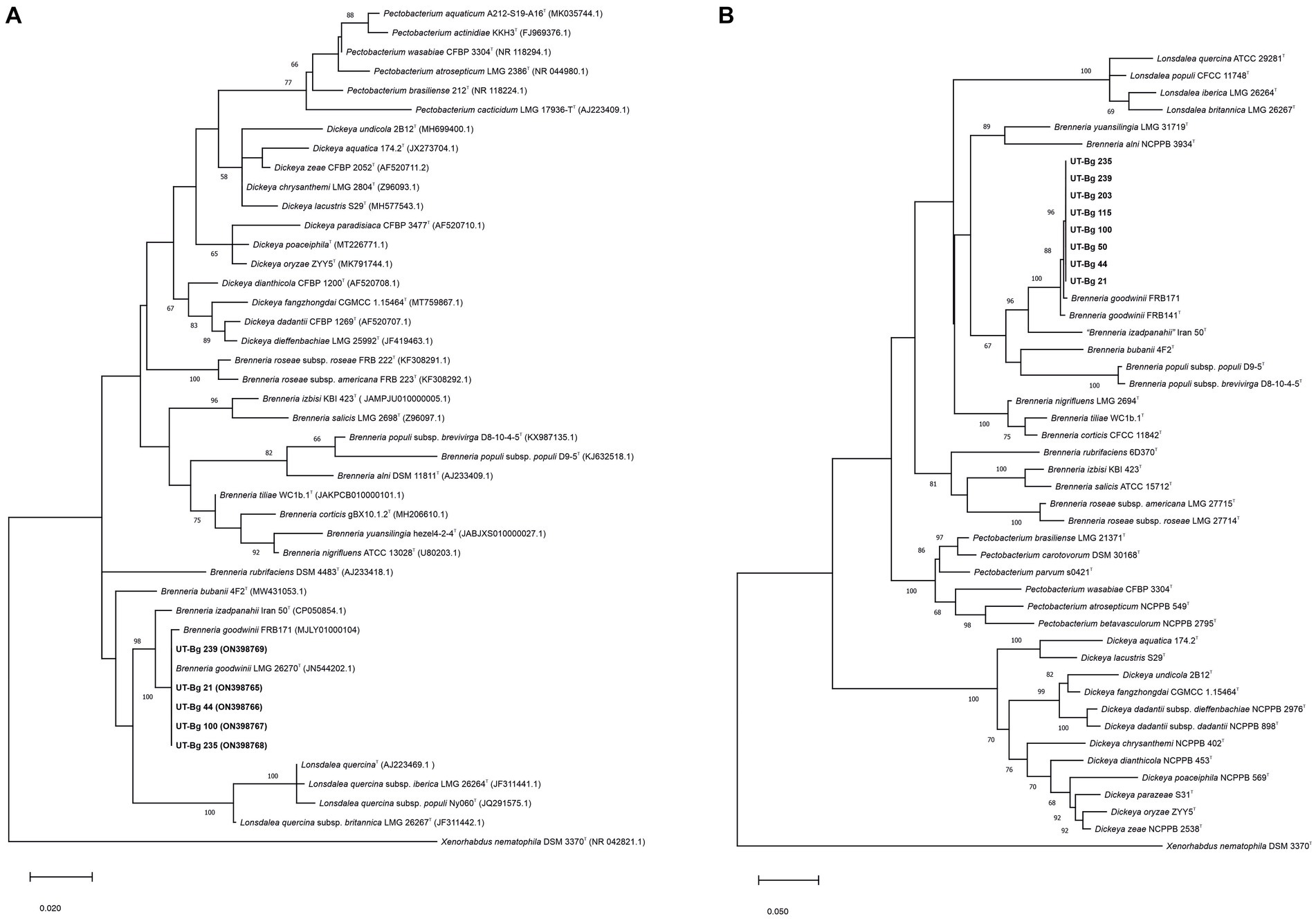
Figure 3. (A) Maximum likelihood phylogenetic tree based on partial sequence of 16S rRNA gene showing the taxonomic position of Brenneria goodwinii strains isolated from oak (UT-Bg 21, UT-Bg 44, UT-Bg 100) and oriental beech (UT-Bg 235 and UT-Bg 239). The tree was constructed using the Hasegawa-Kishino-Yano (HKY+ G+ I) based on the lowest Bayesian information criterion (BIC) score. Bootstrap values calculated for 1,000 replications are indicated. Xenorhabdus nematophila ATCC 19061T (DSM 3370T) was used as the outgroup. (B) Maximum likelihood phylogenetic tree based on partial sequence of gyrB gene showing the taxonomic position of Brenneria goodwinii strains isolated from oak (UT-Bg 21, UT-Bg 44, UT-Bg 50, UT-Bg 100, and UT-Bg 115) and oriental beech (UT-Bg 203, UT-Bg 235, and UT-Bg 239). The tree was constructed using the general time reversible (GTR) with a gamma distribution and invariant sites (GTR + G + I) based on the lowest BIC score. Bootstrap values calculated for 1,000 replications are indicated. Xenorhabdus nematophila ATCC 19061T (DSM 3370T) was used as the outgroup.
Primer specificity
The designed primer pair BgF1/R1 amplified a 427-bp fragment from B. goodwinii FRB 141T and B. goodwinii BS1 strains. This primer pair amplified the desired fragment in two-thirds of B. goodwinii strains (data not shown). Contrarily, the primer pair BgF3/R2 amplified a 618 bp DNA fragment from 51 and 25 B. goodwinii strains isolated from affected oak and oriental beech, respectively, as well as FRB 141T (Denman et al., 2012), BS1 (Moradi-Amirabad et al., 2019), and FRB 186, BAC 30–03-01, and BAC 56e-01. None of the bacterial strains belonging to the genera Rahnella, Gibbsiella, Lonsdalea, or other Brenneria species inciting decline on other forest trees, as well as strains representing a number of the other plant pathogenic bacteria, were amplified by these primer pairs (Figure 4). DNA samples and crude extracts of healthy trees or seedlings used did not yield any amplified product in PCR with these primer sets (Figure 5).
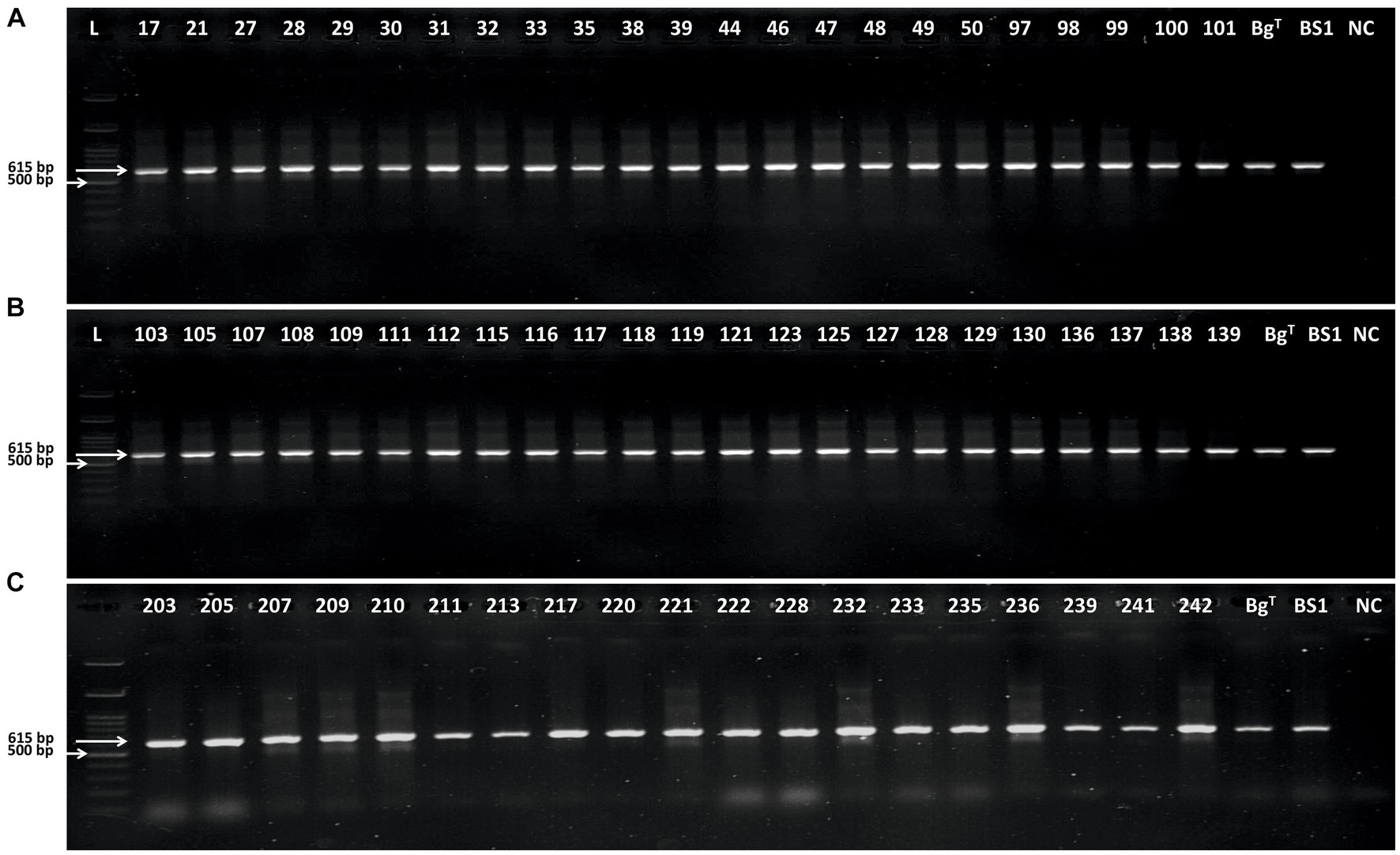
Figure 4. Gel electrophoresis analysis of polymerase chain reaction products amplified using BgF3/R2 primer pair. (A) L: 100 bp DNA Ladder Ready to Load (RT), Solis BioDyne, Tartu, Estonia;17–101: Brenneria goodwinii strains UT-Bg 17 to UT-Bg 101 isolated from oak; BgT: B. goodwinii FRB 141T; BS1: B. goodwinii BS1; and NC: negative control. (B): 103–129: B. goodwinii strains UT-Bg103 to UT-Bg 129 isolated from oak; BgT: B. goodwinii FRB 141T; BS1: B. goodwinii BS1; and NC: negative control. (C): 203–242: B. goodwinii strains UT-Bg203 to UT-Bg 242 isolated from oriental beech; BgT: B. goodwinii FRB 141T; BS1: B. goodwinii BS1; NC: negative control.
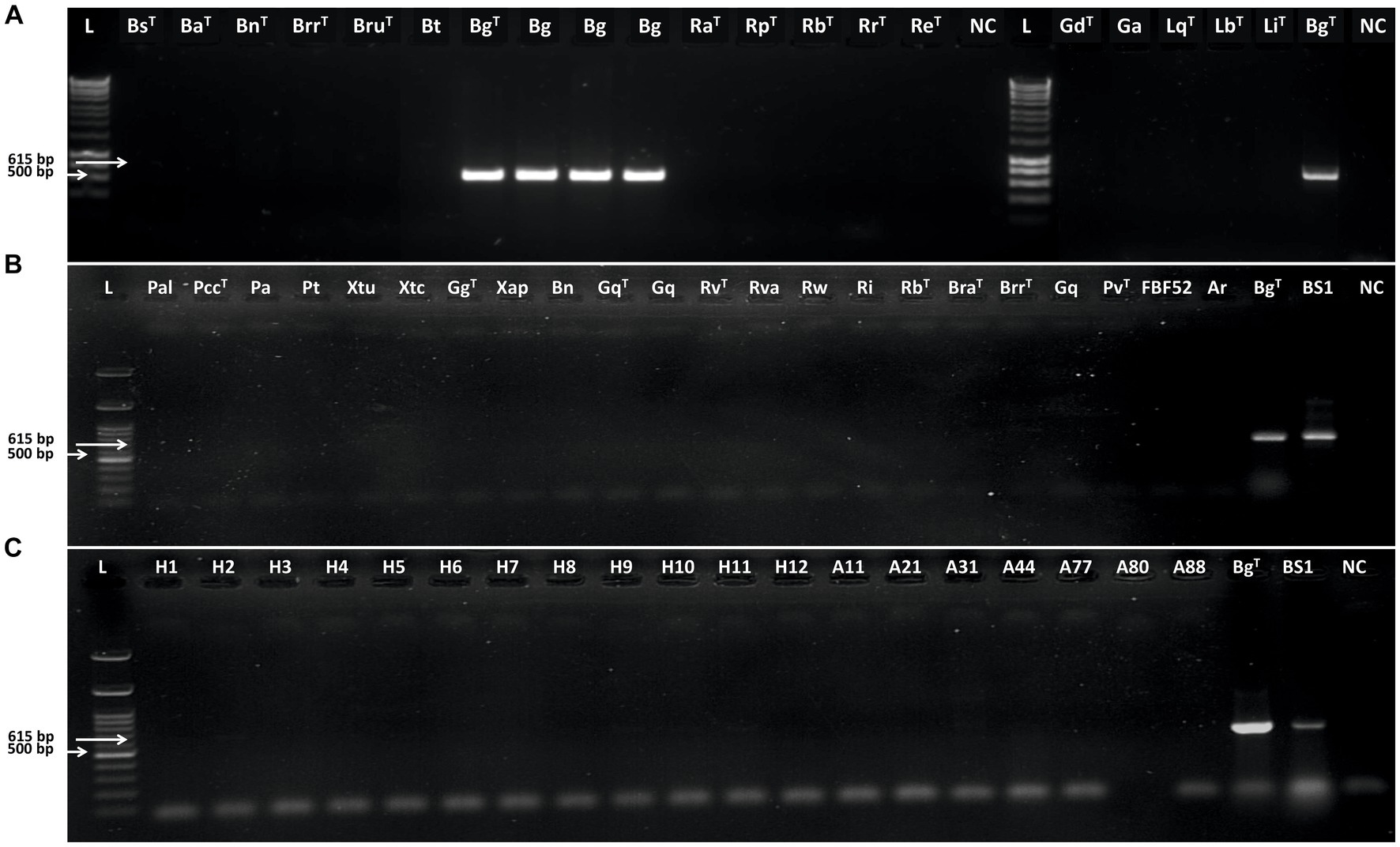
Figure 5. Gel electrophoresis analysis of polymerase chain reaction products amplified using BgF3/R2 primer pair. (A) L: Hyperladder 1 kb (Bioline); BsT: Brenneria salicis DSM 30166T, BaT: Brenneria alni DSM 11811T: Bn: Brenneria nigrifluens DSM 30175T; BrrT: Brenneria roseae ssp. roseae FRB 222T; BrT: Brenneria rubrifaciens DSM 4483T; Bt: Brenneria tiliae; TWs2II: Brenneria tiliae; BgT; Brenneria goodwinii FRB 14IT; Bg1: B. goodwinii FRB 186; Bg2: B. goodwinii BAC 30–03-01;Bg3: B. goodwinii BAC 56e-01; RaT: Rahnella aquatilis LMG 2794T; RpT: Rahnella perminowiae SL6T; RbT: Rahnella bonaserana H11bT; RrT: Rahnella rivi FC061912-KT; ReT: Rahnella ecdela FRB 231T; NC: Negative Control; L: Hyperladder 1 kb (Bioline); GdT: Gibbsiella dentisursi DSM 23818T; Ga: “Gibbsiella acetica” C22; LqT: Lonsdalea quercina LMG 2724T; LbT: Lonsdalea britannica FRB 18T; LiT: Lonsdalea iberica 1915-14T; BgT: B. goodwinii FRB 14IT; and NC: negative control. (B): L: 100 bp DNA Ladder Ready to Load (RT), Solis BioDyne, Tartu, Estonia; Pal: Pseudomonas amygdali pv. lachrymans; PccT: Pectobacterium carotovorum ATCC 15713; Pa: Pantoea agglomerans NK16; Pt: Pseudomonas tolaasii T7; Xtu: Xanthomonas translucens pv. undulosa ICMP 11055; Xtc: X. translucens pv. cerealis ICMP 5752; GgT: Gg: Gibbsiella greigii FRB 224T; Xap: X. arboricola pv. pruni; Bn: Brenneria nigrifluens M2; GqT: GqT: Gibbsiella quercinecans FRB 97T; Gq: G. quercinecans I2; RvT: Rahnella victoriana FRB 225T; Rva: Rahnella variigena FOD 20.8; RwT: Rahnella woolbedingensis FRB 227T; Ri: Rahnella inusitata FOD 9.5a; Rb: Rahnella bruchi FRB 226T; BraT: Brenneria roseae subsp. americana FRB 223T; BrrT: Brenneria roseae subsp. roseae FRB 222T; Gq: G. quercinecans KE1; Pseudomonas viridiflava ATCC 13223T; FBF52: P. viridiflava FBF52; Ar: Agrobacterium radiobacter PK, BgT: B. goodwinii FRB 141T; BS1: B. goodwinii BS1; and NC: negative control. (C): L: 100 bp DNA Ladder Ready to Load (RT), Solis BioDyne, Tartu, Estonia; H1-H12: Strains isolated from healthy trees and seedlings; A11: Bacillus sp. A11; A21: Enterobacter ludwigii A21; A31: Lysinibacillus sp. A31; A44: Pseudomonas putida A44; A77: Klebsiella sp. A77; A80: Serratia marcescens A80; A88: Stenotrophomonas maltophilia A88; B. goodwinii FRB 141T; BS1: B. goodwinii BS1; and NC: negative control.
In planta detection of Brenneria Goodwinii
B. goodwinii was successfully re-isolated from artificially infected oriental beech inoculated with UT-Bg 235 and 239 strains. As expected, a specific amplicon of 618 bp was obtained from all seven artificially infected oriental beech twigs three weeks post-inoculation. Furthermore, the desired fragment was amplified in four oak symptomatic samples. Twigs injected with SDW resulted in no symptoms and negative PCR reactions.
Primer sensitivity
The sensitivity threshold of BgF3/R2 primer pair in the detection of B. goodwinii, as estimated by a 10-fold dilution in SDW of cell cultures of the strain UT-Bg 100 from the initial suspension having OD600 = 0.1 (containing 1 × 1011 CFU/mL) on NA (Merck, Germany, Darmstadt), was approximately 1 × 106 CFU/mL (data not shown).
Discussion
Decline is described as decreasing health and vigor of forest trees incited by abiotic and biotic agents (Helms, 1998). Forest tree declines characterized by trunk bark necrosis and bleeding of a dark sticky fluid from the vertical cracks developed on the trunks have been increasing in prevalence and severity in the Alborz and Zagros forests of oak, walnut, and alder in Iran (Ahmadi et al., 2019; Bakhshi Ganje et al., 2020; Moradi-Amirabad and Khodakaramian, 2020; Allahverdipour et al., 2021; Bakhshi Ganje et al., 2021). In the present study, we characterized strains of B. goodwinii associated with oak and oriental beech trees; the latter was identified for the first time as a new host species impacted by B. goodwinii.
Pathogenicity assay of isolated strains was performed on oak acorns according to González and Ciordia (2020), which was conducted for stains associated with forest tree decline symptoms. As the isolated pathogens and symptoms in oak and oriental beach trees were the same, pathogenicity assays of all strains were performed on oak acorns, and the results indicated that B. goodwinii strains, regardless of the host species, are pathogenic on oak acorns. Moreover, the pathogenicity of representative strains on the corresponding plant hosts showed that B. goodwinii strains cause necrosis and die-back on the host twigs.
In this study, bacterial colonies with a green metallic sheen on EMB-agar were consistently isolated from the oak and oriental beech trees displaying canker and stem bleeding symptoms in northern Iran. The strains were identified as B. goodwinii on the basis of their phenotypic characteristics, pathogenicity features, and sequences of 16S rRNA and gyrB genes. All strains plus the type strain and four other known strains of B. goodwinii were also detected by PCR, using the newly developed primer pair BgF3/R2, which targets the hrpN gene of B. goodwinii.
The decline and bleeding stem canker of beech in Europe and the United States are mostly caused by Phytophthora × cambivora (Jung et al., 2005, 2017). The disease is characterized by dark-colored bleeding areas, which usually appear near the collar, and occasionally, bleeding may be observed on the trunk. In this study, B. goodwinii was identified as associated with brown spots, tissue necrosis, and canker on the trunk of oriental beech. To the best of our knowledge, this is the first bacterial disease of oriental beech.
B. goodwinii, as a key species involved in characteristic weeping symptoms and live bark degradation, has a type 3 secretion system (T3SS) and associated harpins and effectors that could manipulate the host. The other two prevalent bacterial species, G. quercinecans and R. victoriana (Brady et al., 2017; Denman et al., 2018), mainly with enzyme-degrading plant cell walls, enhance or facilitate necrosis (Doonan et al., 2019).
Although identification of B. goodwinii by sequencing housekeeping genes is a robust and reliable method, it is also laborious and time-consuming. Here, we indicated that PCR using specific primers is a valuable, efficient, and less labor-intensive method for direct and culture-independent detection of B. goodwinii in forest trees, as well as its rapid and precise identification in samples or cultures. The designed PCR requires only a commercial DNA Polymerase Master Mix that is not expensive. Therefore, this protocol is easy to use in different laboratories, including those located in low-income countries. Moreover, the target gene was detected in PCR with DNA extracted directly and culture-independently using the alkalic procedure that is rapid and cheap. Regarding the increase in tree diseases in Iran, the development of specific primers could facilitate rapid identification of the pathogen.
Pathogenicity/virulence genes like pth, hrp, and vir of several other phytopathogenic genera and species were the most commonly used targets for designing primers specific for detection and identification of the phytopathogenic bacteria at the species or subspecies level by PCR (López et al., 2003). For instance, for designing specific primers pel (pectate lyase) genes are used for detecting Pectobacterium and Dickeya species (Darrasse et al., 1994; Louws et al., 1999). Designing primers from conserved regions of pathogenicity gene clusters could avoid non-specific amplicon production from non-pathogenic species or other pathogens. In this study, we used the hrpN gene in detecting B. goodwinii strains in oak and oriental beech bark samples. HrpN, encoded by hrpN, induces the hypersensitive response (HR) in resistant hosts and non-host plants, implying that the hrp genes may also be involved in host specificity (Yang et al., 2002; Holeva et al., 2004). These results indicated that the hrpN primer pair has both a high degree of sensitivity and specificity in detecting B. goodwinii from the bacterial strains belonging to the genera Rahnella, Gibbsiella, Lonsdalea, or other Brenneria species inciting decline on other forest trees. Moreover, this primer pair was able to detect B. goodwinii from symptomatic tissues. Thus, hrpN was determined to be useful as an appropriate target for the specific detection of B. goodwinii directly from infected or diseased tissues.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
This research does not contain any studies with human participants or animals performed by any of the authors.
Author contributions
M-HA: Investigation, Validation, Writing – review & editing, Funding acquisition. NC: Validation, Writing – review & editing, Conceptualization, Formal analysis, Investigation, Methodology, Project administration, Software, Supervision, Visualization, Writing – original draft. CB: Methodology, Validation, Writing – review & editing. HR: Conceptualization, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was funded by University of Tehran (Grant numbers: 148605).
Acknowledgments
The authors thank Mohammad-Ali Mirhabibi and Mohammad Ashrafi for their help in performing the experiments.
Conflict of interest
The authors declare that they have no conflict of interest and the authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmadi, E., Kowsari, M., Azadfar, D., and Salehi Jouzani, G. (2019). Bacillus pumilus and Stenotrophomonas maltophilia as two potentially causative agents involved in Persian oak decline in Zagros forests (Iran). For. Pathol. 49:12541. doi: 10.1111/efp.12541
Alavi, S. J., Ahmadi, K., Dormann, C. F., Serra-Diaz, J., and Nouri, Z. (2020). Assessing the dominant height of oriental beech (Fagus orientalis L.) in relation to edaphic and physiographic variables in the Hyrcanian forests of Iran. Biotechnol. Agron. Soc. Environ. 24, 262–273.
Allahverdipour, T., Shahryari, F., and Falahi Charkhabi, N. (2021). Gibbsiella quercinecans and Brenneria roseae subsp. roseae associated to the canker disease of walnut trees in northwestern Iran. Eur. J. Plant Pathol. 161, 783–797. doi: 10.1007/s10658-021-02359-9
Ayers, S. H., Rupp, P., and Johnson, W. T. (1919). A study of the alkali-forming bacteria in milk. U. S. Dep. Agric. Bull. 782, 1–39. doi: 10.5962/bhl.title.108233
Bakhshi Ganje, M., Mackay, J., Nicolaisen, M., and Shams-Bakhsh, M. (2021). Comparative genomics, pangenome and phylogenomic analyses of Brenneria spp., delineation of Brenneria izadpanahii sp. nov. Phytopathology 111, 78–95. doi: 10.1094/PHYTO-04-20-0129-FI
Bakhshi Ganje, M., Shams-Bakhsh, M., Mackay, J., and Rahimian, H. (2020). Identification and characterization of bacterial strains associated with diseased oak trees in northern Iran. For. Pathol. 50:12571. doi: 10.1111/efp.12571
Brady, C., Arnold, D., McDonald, J. E., and Denman, S. (2017). Taxonomy and identification of bacteria associated with acute oak decline. World J. Microbiol. Biotechnol 33:1. doi: 10.1007/s11274-017-2296-4
Brady, C., Asselin, J. A., Beer, S., Brurberg, M. B., Crampton, B., Venter, S., et al. (2022). Emended description of the genus Rahnella. Int. J. Syst. Evol. Microbiol. 72: p.005190. doi: 10.1099/ijsem.0.005190
Brady, C. L., Cleenwerck, I., Denman, S., Venter, S. N., Rodríguez-Palenzuela, P., Coutinho, T. A., et al. (2012). Proposal to reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben et al. 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. iberica subsp. nov. and Lonsdalea quercina subsp. britannica subsp. nov., emendation of the description of the genus Brenneria, reclassification of Dickeya dieffenbachiae as Dickeya dadantii subsp. dieffenbachiae comb. nov., and emendation of the comb. nov., and emendation of the description of Dickeya dadantii. Int. J. Syst. Evol. Microbiol. 62, 1592–1602. doi: 10.1099/ijs.0.035055-0
Brady, C., Cleenwerck, I., Venter, S., Vancanneyt, M., Swings, J., and Coutinho, T. (2008). Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 31, 447–460. doi: 10.1016/j.syapm.2008.09.004
Brady, C., Denman, S., Kirk, S., Venter, S., Rodríguez-Palenzuela, P., and Coutinho, T. (2010). Description of Gibbsiella quercinecans gen. Nov., sp. nov., associated with acute oak decline. Syst. Appl. Microbiol. 33, 444–450. doi: 10.1016/j.syapm.2010.08.006
Brady, C., Hunter, G., Kirk, S., Arnold, D., and Denman, S. (2014a). Description of Brenneria roseae sp. nov. and two subspecies, Brenneria roseae subspecies roseae ssp. nov and Brenneria roseae subspecies Americana ssp. nov. isolated from symptomatic oak. Syst. Appl. Microbiol. 37, 396–401. doi: 10.1016/j.syapm.2014.04.005
Brady, C., Hunter, G., Kirk, S., Arnold, D., and Denman, S. (2014b). Rahnella victoriana sp. nov., Rahnella bruchi sp. nov., Rahnella woolbedingensis sp. nov., classification of Rahnella genomospecies 2 and 3 as Rahnella variigena sp. nov. and Rahnella inusitata sp. nov., respectively and emended description of the genus. Rahnella. Syst. Appl. Microbiol. 37, 545–552. doi: 10.1016/j.syapm.2014.09.001
Brady, C., Hunter, G., Kirk, S., Arnold, D., and Denman, S. (2014c). Gibbsiella greigii sp. nov., a novel species associated with oak decline in the USA. Syst. Appl. Microbiol. 37, 417–422. doi: 10.1016/j.syapm.2014.07.002
Crampton, B. G., Plummer, S. J., Kaczmarek, M., McDonald, J. E., and Denman, S. (2020). A multiplex real-time PCR assay enables simultaneous rapid detection and quantification of bacteria associated with acute oak decline. Plant Pathol. 69, 1301–1310. doi: 10.1111/ppa.13203
Darrasse, A., Priou, S., Kotoujansky, A., and Bertheau, Y. (1994). PCR and restriction fragment length polymorphism of a pel gene as a tool to identify Erwinia carotovora in relation to potato diseases. Appl. Environ. Microbiol. 60, 1437–1443. doi: 10.1128/aem.60.5.1437-1443.1994
Denman, S., Brady, C. L., Kirk, S., Cleenwerck, I., Venter, S. N., Coutinho, T. A., et al. (2012). Brenneria goodwinii sp. nov., associated with acute oak decline in the UK. Int. J. Syst. Evol. Microbiol. 62, 2451–2456. doi: 10.1099/ijs.0.037879-0
Denman, S., Doonan, J., Ransom-Jones, E., Broberg, M., Plummer, S., Kirk, S., et al. (2018). Microbiome and infectivity studies reveal complex polyspecies tree disease in acute oak decline. ISME J. 12, 386–399. doi: 10.1038/ismej.2017.170
Doonan, J., Denman, S., Pachebat, J. A., and McDonald, J. E. (2019). Genomic analysis of bacteria in the acute oak decline pathobiome. Microb. Genom. 5:e000240. doi: 10.1099/mgen.0.000240
Falahi Charkhabi, N., Shams-bakhsh, M., and Rahimian, H. (2010). Genetic diversity among Brenneria nigrifluens strains in Iran. Eur. J. Plant Pathol. 128, 303–310. doi: 10.1007/s10658-010-9667-0
González, A. J., and Ciordia, M. (2020). Brenneria goodwinii and Gibbsiella quercinecans isolated from weeping cankers on Quercus robur L. Spain. Eur. J. Plant Pathol. 156, 965–969. doi: 10.1007/s10658-019-01891-z
Helms, J. A. (1998). The dictionary of forestry. Bethesda, MD: The Society of American Foresters. 210.
Holeva, M. C., Bell, K. S., Hyman, L. J., Avrova, A. O., Whisson, S. C., Birch, P. R., et al. (2004). Use of a pooled transposon mutation grid to demonstrate roles in disease development for Erwinia carotovora subsp. atroseptica putative type III secreted effector (DspE/A) and helper (HrpN) proteins. Mol. Plant Microbe Interact. 17, 943–950. doi: 10.1094/MPMI.2004.17.9.943
Izard, D., Gavini, F., Trinel, P. A., and Leclerc, H. (1979). Rahnella aquatilis, nouveau membre de la famille des Enterobacteriaceae. Ann. Microbiol. 130, 163–177.
Jung, T., Hudler, G. W., Jensen-Tracy, S. L., Griffiths, H. M., Fleischmann, F., and Osswald, W. (2005). Involvement of Phytophthora species in the decline of European beech in Europe and the USA. Mycologist 19, 159–166. doi: 10.1017/S0269-915X(05)00405-2
Jung, T., Jung, M. H., Cacciola, S. O., Cech, T., Bakonyi, J., Seress, D., et al. (2017). Multiple new cryptic pathogenic Phytophthora species from Fagaceae forests in Austria, Italy and Portugal. IMA Fungus 8, 219–244. doi: 10.5598/imafungus.2017.08.02.02
Kile, H., Arnold, D., Allainguilaume, J., Denman, S., and Brady, C. (2022). Brenneria tiliae sp. nov., isolated from symptomatic Tilia× moltkei and Tilia× europaea trees in the UK. Int. J. Syst. Evol. Microbiol. 72:5515. doi: 10.1099/ijsem.0.005515
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lane, D. J. (1991). “16S/23S rRNA sequencing” in Nucleic acid techniques in bacterial systematics. ed. E. G. M. Stackebrandt (New York: John Wiley & Sons, Inc.), 115–176.
López, M. M., Bertolini, E., Olmos, A., Caruso, P., Gorris, M. T., Llop, P., et al. (2003). Innovative tools for detection of plant pathogenic viruses and bacteria. Int. Microbiol. 6, 233–243. doi: 10.1007/s10123-003-0143-y
Louws, F. J., Rademaker, J. L. K., and de Bruijn, F. J. (1999). The three ds of PCR-based genomic analysis of phytobacteria: diversity, detection, and diagnosis. Annu. Rev. Phytopathol. 37, 81–125. doi: 10.1146/annurev.phyto.37.1.81
Moradi-Amirabad, Y., and Khodakaramian, G. (2020). First report of bleeding canker caused by sp. on Populus nigra in Iran. New Dis. Rep. 41:37. doi: 10.5197/j.2044-0588.2020.041.037
Moradi-Amirabad, Y., Rahimian, H., Babaeizad, V., and Denman, S. (2019). Brenneria spp. and Rahnella victoriana associated with acute oak decline symptoms on oak and hornbeam in Iran. For. Pathol. 49:12535. doi: 10.1111/efp.12535
Niemann, S., Pühler, A., Tichy, H. V., Simon, R., and Selbitschka, W. (1997). Evaluation of the resolving power of three different DNA fingerprinting methods to discriminate among isolates of a natural Rhizobium meliloti population. J. Appl. Microbiol. 82, 477–484. doi: 10.1046/j.1365-2672.1997.00141.x
Ruffner, B., Schneider, S., Meyer, J., Queloz, V., and Rigling, D. (2020). First report of acute oak decline disease of native and non-native oaks in Switzerland. New Dis. Rep. 41, 18–5197. doi: 10.5197/j.2044-0588.2020.041.018
Saito, M., Shinozaki-Kuwahara, N., and Takada, K. (2012). Gibbsiella dentisursi sp. nov., isolated from the bear oral cavity. Microbiol. Immunol. 56, 506–512. doi: 10.1111/j.1348-0421.2012.00464.x
Sangüesa-Barreda, G., Camarero, J. J., Oliva, J., Montes, F., and Gazol, A. (2015). Past logging, drought and pathogens interact and contribute to forest dieback. Agric. For. Meteorol. 208, 85–94. doi: 10.1016/j.agrformet.2015.04.011
Schaad, N. W., Jones, J. B., and Chun, W. (2001). Laboratory guide for the identification of plant pathogenic bacteria (No. Ed. 3). St. Paul, MN: American Phytopathological Society (APS Press).
Stackebrandt, E., and Liesack, W. (1993). “Nucleic acids and classification” in Handbook of new bacterial systematics. eds. M. Goodfellow and A. G. O'Donnell (London, England: Academic Press)
Surico, G., Mugnai, L., Pastorelli, R., Giovannetti, L., and Stead, D. E. (1996). Erwinia alni, a new species causing bark cankers of alder (Alnus miller) species. Int. J. Syst. Evol. Microbiol. 46, 720–726. doi: 10.1099/00207713-46-3-720
Suslow, T. V., Schroth, M. N., and Isaka, M. (1982). Application of a rapid method for gram differentiation of plant pathogenic and saprophytic bacteria without staining. Phytopathology 72:917. doi: 10.1094/Phyto-72-917
Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. doi: 10.1093/nar/22.22.4673
Tkaczyk, M., Celma, L., Ruņģis, D. E., and Bokuma, G. (2021). First report of Brenneria goodwinii and Gibbsiella quercinecans bacteria, detected on weaken oak trees in Poland. Balt. For. 27:563. doi: 10.46490/BF563
Wilson, E. E., Starr, M. P., and Berger, J. A. (1957). Bark canker, a bacterial disease of the Persian walnut tree. Phytopathology 47, 669–673.
Wilson, E. E., Zeitoun, F. M., and Fredrickson, D. L. (1967). Bacterial phloem canker, a new disease of Persian walnut trees. Phytopathology 57, 618–621.
Yang, C. H., Gavilanes-Ruiz, M., Okinaka, Y., Vedel, R., Berthuy, I., Boccara, M., et al. (2002). Hrp genes of Erwinia chrysanthemi 3937 are important virulence factors. Mol. Plant-Microbe Interact. 15, 472–480. doi: 10.1094/MPMI.2002.15.5.472
Keywords: emerging diseases, forest decline, Fagus orientalis , Quercus castaneifolia , Iran
Citation: Araeinejhad M-H, Charkhabi NF, Brady C and Rahimian H (2024) Reliable and specific detection and identification of Brenneria goodwinii, the causal agent of oak and oriental beech decline. Front. For. Glob. Change. 7:1325897. doi: 10.3389/ffgc.2024.1325897
Edited by:
Quan Lu, Chinese Academy of Forestry, ChinaReviewed by:
Diana Marčiulynienė, Lithuanian Research Centre for Agriculture and Forestry, LithuaniaYong Li, Chinese Academy of Forestry, China
Copyright © 2024 Araeinejhad, Charkhabi, Brady and Rahimian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nargues Falahi Charkhabi, ZmFsYWhpY2hhcmtoYWJpQHV0LmFjLmly
†These authors have contributed equally to this work
 Mohammad-Hossein Araeinejhad1†
Mohammad-Hossein Araeinejhad1† Nargues Falahi Charkhabi
Nargues Falahi Charkhabi Carrie Brady
Carrie Brady Heshmat Rahimian
Heshmat Rahimian