- 1School of Agriculture, Meiji University, Kawasaki, Japan
- 2Graduate School of Agriculture, Meiji University, Kawasaki, Japan
- 3Department of Chemical and Environmental of Engineering, Malaysia-Japan International Institute of Technology, Universiti Teknologi Malaysia, Kuala Lumpur, Malaysia
- 4Department of Biotechnology, Graduate School of Agricultural and Life Sciences, The University of Tokyo, Tokyo, Japan
The conversion of tropical peat swamp forests to oil palm plantations has become a focal point in relation to global change. However, it is difficult to understand the ecological consequences of this conversion because little is known about how the microorganisms in these ecosystems respond to land-use conversion. Therefore, in this study, we assessed the microbial community structures of tropical peatland under two land uses, peat swamp forest and oil palm plantation (OP), to investigate how changes in local environmental conditions due to the conversion from forest to OP may have affected the microbial communities. For each land use, the microbial communities were assessed at three depths (0–5 cm, 20–25 cm, and 40–50 cm) using meta-16S amplicon analysis with Illumina Miseq. We found that the microbial communities under both land uses were dominated by anaerobes and fermenters, such as Acidobacteriota, Proteobacteria and Actinobacteria, which accounted for 80%–90% of the total abundance. There were strong similarities between the microbial communities in the 0–5 cm forest samples and the OP samples at all depths. However, the microbial communities in the 20–25 cm and 40–50 cm forest samples were different from the other samples. The differences in the deeper forest samples were likely related to water table and peat quality. CO2 fluxes from the forest were significantly higher than from OP, with mean fluxes of 190 ± 66.92 and 57.86 ± 33.66 mg m−2 h−1, respectively (p < 0.05). However, there were no differences in either CH4 or N2O fluxes between the forest and OP. We detected nine microbial taxa that characterized the differences in the microbial communities between the forest and OP (p > 0.05); Bathyarchaeia, Dadabacteriales, Syntrophobacter, and Subgroup_13 were significantly more abundant in the forest, especially in the deeper peat layers, whereas Acidobacter, Bryobacter, 11–24, Leptospirillum, and WPS-2 were significantly more abundant in the OP (p > 0.05). This study helps improve our understanding of the differences in microbial community structures between peat swamp forests and OP systems.
1 Introduction
Tropical peatlands are different from most northern peatlands in that they are typically forested and rich in carbon both above and below ground (Dargie et al., 2017). There are various kinds of tropical peatland forest systems characterized by differences in peat thickness, hydrology, and nutrient supply (Ribeiro et al., 2021). Southeast Asian peatlands, the second-largest tropical peatlands, are predominantly domed, nutrient-poor ombrotrophic bogs that receive nutrients solely from precipitation due to their raised shape, with relatively narrow flooded minerotrophic margins close to rivers (Newbery et al., 1999). Southeast Asian peatlands store about 69 Gt of carbon and absorb about 2.6 t CO2 per hectare per annum (Miettinen and Liew, 2010). However, these peatlands have experienced high rates of deforestation and have rapidly declined from 76% (11.9 Mha) of swamp forest cover in 1990 to 29% (4.6 Mha) in 2015, with the majority of the forest cover being converted to plantations and secondary vegetation (Miettinen et al., 2016). In Malaysia, there are approximately 2.7 Mha of peatlands, with about 1.7 Mha in Malaysian Borneo (Melling, 2016) and 1.0 Mha in peninsular Malaysia, out of which more than 1 Mha have already been converted to oil palm, and the total converted area is expected to rise to 2.3 Mha by 2030 (Miettinen et al., 2016). This conversion from a multi-layered heterogeneous canopy tropical peatland forest system into a monocrop open–cover oil palm system can severely impact soil moisture content, nutrient availability, organic matter quality, and above- and below-ground biodiversity (Luskin and Potts, 2011; Posa et al., 2011; Tonks et al., 2016; Too et al., 2018; Dhandapani et al., 2019b, 2020; Azizan et al., 2021).
Peatland microorganisms play an important role in the formation of peat, directly influencing the carbon turnover and nutrient mineralization required to support high primary production, which affects the overall function and sustainability of the ecosystem (Andersen et al., 2013). Substrate availability and redox potential vary with peat depth, while moisture and oxygen levels can influence the structure and diversity of microbial communities in peatlands (Birnbaum et al., 2022; Zhao et al., 2022). Microbial communities in deeper peat layers are essential for driving the carbon and nitrogen cycles throughout the peat profile, for example, carrying out anaerobic ecosystem functions like methanogenesis (Wendlandt et al., 2010) and ammonia oxidation (Hu et al., 2011; Oshiki et al., 2016). A study by Dhandapani et al. (2020) found that microbial diversity is highest in intermediate depths due to the co-existence of oxic and anoxic layers, with a subsequent decrease in diversity in deeper peat layers.
Forested swamp ecosystems typically have high carbon content, nutrient-poor, acidic and water-logged conditions, and contain a variety of soil microbial communities (Kanokratana et al., 2011; Dhandapani et al., 2019b; Dom et al., 2021). Recent advances in high-throughput sequencing, such as next-generation sequencing (NGS) and bioinformatics, have enabled higher-resolution assessments of the microbial compositions in such complex tropical peatland ecosystems (Jackson et al., 2008; Kanokratana et al., 2011; Kwon et al., 2013; Tripathi et al., 2016; Too et al., 2018; Dom et al., 2021). These assessments include studies on how differences in swamp forest system characteristics affect microbial community structure, including characteristics such as high acidity (Jackson et al., 2008; Dom et al., 2021), nutrient gradients (Sjögersten et al., 2011), peat dome variation (Dom et al., 2021), as well as the influences of different trees or vegetation types (Too et al., 2018; Girkin et al., 2020a,b). However, there have been no microbial studies related to land-use change in tropical peatlands using NGS, although there have been a few studies on microbial composition in drained tropical peatlands using methods like Polymerase chain reaction/denaturing gradient gel electrophoresis (PCR-DGGE) and enzyme activities (Nurulita et al., 2016), chloroform fumigation (Könönen et al., 2018) and phenotypic structure by phospholipid fatty acid (PLFA; Dhandapani et al., 2020). Therefore, this study aimed to utilize advanced high-throughput sequencing methods to compare microbial communities in peat swamp forests and drained oil palm plantations to assess how changes in microbial communities may affect biogeochemical cycling. We hypothesized (1) that shifts in microbial communities due to changes in land use are associated with water table, nutrient availability and GHG emissions, (2) that microbial communities in peat swamp forests differ significantly with depth due to changes in moisture content, oxygen content, redox potential, and substrate availability, and (3) that in oil palm plantations, microbial communities do not change significantly with depth, because the OP peat quality is less varied with depth due to a low water table, peat subsidence, and higher long-term decomposition rates.
2 Materials and methods
2.1 Study area
This study was conducted from August to September 2019 (end of the dry season to early in the wet season) on sites under two land uses: (1) peat swamp forest (3°26′20.29 N” 101°20′02.28″ E) and (2) oil palm plantation (3°25′29.92″ N 101°20′10.47″ E; Figure 1). The forest sites are located in Raja Musa Peat Swamp Forest Reserve (PSFR), Malaysia, which is part of the North Selangor Peat Swamp Forest (NSPSF), comprising 23,486 hectares out of the total 81,304 hectare NSPSF area.

Figure 1. Sampling locations for the two land uses (A) and surface cover conditions in the peat swamp forest (B), and oil palm plantation (C).
The Raja Musa PSFR is an ombrotrophic peatland that was partially logged in the 1980s and is considered to be a secondary forest that has a high water table and is mainly flooded during the wet season (Dhandapani et al., 2019b; Azizan et al., 2021). Currently, more than 50% of Raja Musa PSFR is a mix of high- (trees more than 25-m tall) and medium-height (trees 15–25-m tall) forest with 50%–80% forest density (Selangor State Forestry Department, 2014). The dominant tree species in most of Raja Musa PSFR are Kempas (Koompassia malaccensis), Kedondong (Santiria spp.), and Kelat (Syzgium spp.).
The oil palm plantation sites (hereafter called OP) are part of a mature smallholder plantation on a former peat swamp forest, i.e., 10–15-year-old oil palms in first crop rotations, located adjacent to the Raja Musa PSFR (Figure 1). The OP trees were 8–12-m tall and had been under drainage and fertilization since the cultivation started. The main fertilizers used in the OP sites were urea-based fertilizers, applied at rates of 50–kg/palm/year (based on observation during the fieldwork and personal communication by the smallholder).
2.2 Sampling design and field data collection
Three sites were established within each land use (forest and OP), with distances between the sites of approximately 300 m for the forest sites and 700 m for the OP sites. Peat samples were collected from three depths (0–5 cm, 20–25 cm, and 40–50 cm) at each site using a peat auger (Royal Eijkelkamp, Giesbeek, The Netherlands). The peat auger was washed with ddH2O and carefully sterilized with ethanol before each sample collection. The peat samples were carefully placed in sterilized 50-ml falcon tubes using a sterilized spatula. A total of nine samples were collected from each land use (forest and OP) for microbial and nutrient analysis.
Gas sampling was performed with two-part static chambers consisting of a blue PVC base collar (30-cm height × 15-cm radius) and a top chamber (Petersen et al., 2012). At each site, three replicate collars were inserted 5-cm deep into an un-vegetated area of the peat, with approximately 10–15 m distance between each collar. The collars were installed on 1 August 2019, 3 weeks prior to the first flux measurements, and remained in the same position throughout the entire study period. The detachable top chamber was sealed to the collar during each gas flux measurement. Gas samples (30 mL) were taken at 0, 10, and 20 min after sealing the top chamber, using a syringe to withdraw the gas from a sampling port fitted to the top of the chamber, and transferred into an evacuated vial. Gas fluxes were measured at three-day intervals for 1 month (540 gas samples). On each measurement date, gas sampling was conducted between 10:00 and 12:00 at the forest sites and between 13:00 and 16:00 at the OP sites. All peat and gas samples were kept in an ice box away from direct sunlight prior to transport to the local laboratory (MJIIT, Malaysia). In the local laboratory, the peat and gas samples were stored at −80°C and 4°C, respectively, until transport to Japan on 29 September 2019, for further analysis.
In-situ field data measurements were carried out for one and a half months (8 August–26 September 2019). Data loggers (Em50, Decagon Devices, United States) were installed at one forest site and two OP sites to measure soil temperature (5 and 30-cm), soil volumetric water content (VWC; 5 and 30-cm), and water table (WT). Manual WT measurements were also conducted at the other sampling sites. The WT was measured by installing perforated PVC pipes (15-cm diameter × 2-m height) and using the following equation: WT (cm) = H1–H2, where H1 is the height from the top of the pipe to the water surface inside the pipe, and H2 is the height from the top of the pipe to the ground surface outside the pipe. An average reading was obtained from measurements taken at three different points around the circumference of the pipe.
2.3 Peat physicochemical properties and GHG fluxes
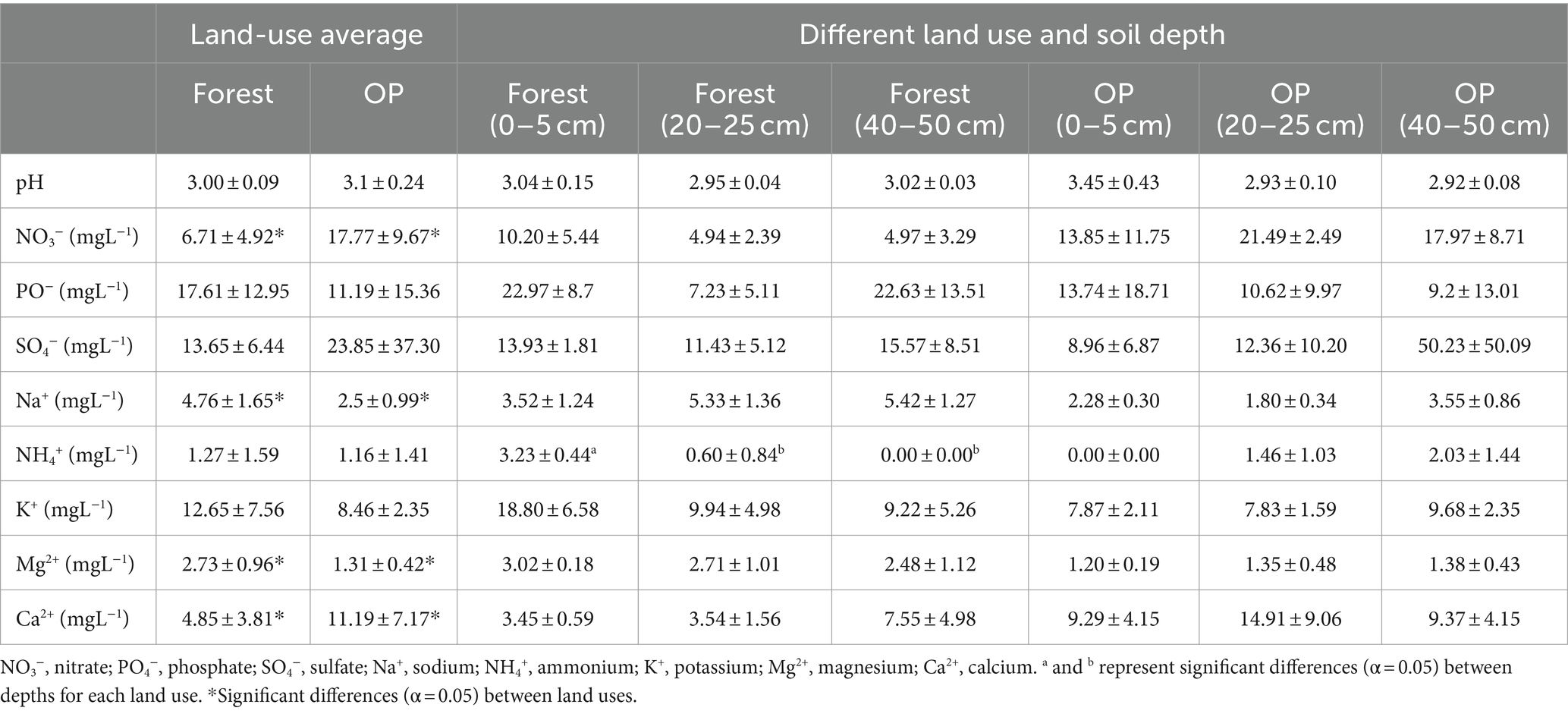
The pH and nutrient availabilities of each peat sample were measured as follows. The peat samples were air-dried for 24 h, then sieved and ground. Then, 10 mL of ultra-pure deionized water was added to approximately 1 g of each peat sample, and the mixture was shaken for 24 h. The pH of each sample was measured using an MM60R Water Quality Checker (DKK-TOA, Tokyo, Japan). Then the samples were filtered by syringe filter (0.2 μm), and the sample extracts were analyzed for the following nutrient availabilities using an Ion analyzer IA 300 (DKK-TOA, Tokyo, Japan) fitted with an ICA-200AS autosampler: NO3−, PO4−, SO4−, Na+, NH4+, K+, Mg2+, and Ca2+.
All gas samples were analyzed by gas chromatography (Agilent Technologies 6890, California, SC, United States) using a flame ionization detector (FID) for CH4 and CO2 and an electron capture detector (ECD) for N2O. The ppm data were converted to gas concentration values in mg m−3 using the ideal derivative gas law equation, W = MVP/RT. This equation was derived from PV = nRT, where P is pressure (1 atm), V is volume (ppm), n is the number of moles of gas, R is the universal gas constant (0.082 ATM L mol−1 K−1), and T is temperature in degrees kelvin (K). Hence, n = W/M, where W is the weight of the gas (mg m−3) and M is the molar mass of the gas (g mol−1). The gas emission from each chamber on each sampling occasion was then calculated using the linear regression equation F = Δc/Δt × h (Levy et al., 2011), where F is the gas flux (mg m−2 h−1), Δc/Δt is the change in gas concentration inside the chamber during the sampling period and h is the height inside the chamber above the peat surface (m).
A mixed-model ANOVA was used (1) to examine differences in the peat physicochemical properties between different depths and land uses and (2) to compare the physicochemical properties, in-situ environmental variables and GHG emission data between the land uses. All statistical analyses were conducted in Datatab,1 and the significance value was set at p < 0.05 throughout the study.
2.4 Metabarcoding protocols and analyses
2.4.1 Extraction of eDNA
DNA was extracted from each peat sample (3 sites × 3 depths for each land use) using the ISOIL for Beads Beating DNA Extraction Kit (NIPPON GENE CO., LTD), following the manufacturer’s instructions. Then, the three DNA samples for each depth at each site were combined into a single pooled sample. Each pooled DNA sample was subjected to sequencing analysis using a Next Generation Sequencing GENEWIZ (Genomics Japan Corp., Tokyo, Japan).
2.4.2 Library preparation and Illumina sequencing
The DNA concentration of each pooled sample was quantified using an Equalbit dsDNA High Sensitivity Assay Kit. Then, 20–30 ng of DNA was used for PCR amplification using a panel of proprietary primers (designed by GENEWIZ) aimed at relatively conserved regions bordering the V3 and V4 hypervariable regions of bacteria 16S and Archaea 16S rDNA. Also, the samples were prepared for NGS sequencing by adding a linker with an index to the end of the 16S rDNA PCR products. The library was purified with magnetic beads, and then the concentration was measured by a microplate reader, with the fragment size assessed by agarose gel electrophoresis. The library was quantified to 10 nM, and PE250/FE300 paired-end sequencing was performed using Illumina MiSeq/Novaseq (Illumina, San Diego, CA, United States).
2.4.3 Sequencing data processing
There were 331,014–405,206 raw sequences in the forest peat samples and 256,740–278,922 raw sequences in the OP peat samples. Quality control was performed by Cutadapt (v1.9.1) and QIIME (1.9.1), while chimera sequence removal was performed by VSEARCH (1.9.6). Then, sequences with low-quality scores (<20) were removed before sequences with a final base-pair length of more than 200 were processed. The number of retained sequences for the forest peat samples ranged from 151,274–185,509 with an average length of 441.9 bp, while for the OP peat samples, there were 117,553–130,377 sequences with an average length of 441.3 bp. Hence, a total of 502,704 and 371,591 high-quality gene sequences were obtained for community analyses of the three pooled forest peat samples and three pooled OP peat samples, respectively. All optimized sequences were clustered into operational taxonomic units (OTUs) with a 97% similarity using VEARCH (1.9.6). Representative sequences of each OTU were classified to different levels (phylum, class, order, family, genus, and species) against SILVA database v.138 to obtain the community composition of each sample using the Ribosomal Database Project (RDP) classifier v.2.2 (Cole et al., 2014).
2.4.4 Sequencing data analysis
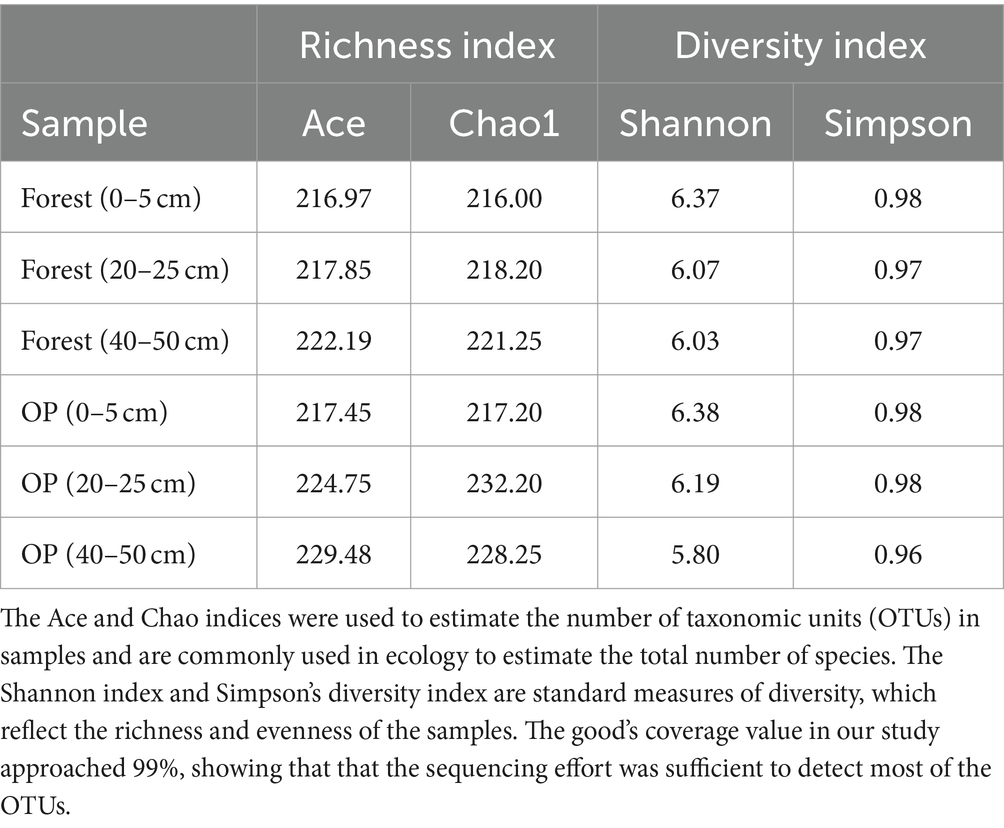
Sequences were rarefied to 80,000 reads (Supplementary Data 1) prior to the calculation of alpha and beta-diversity statistics. Alpha diversity indices (e.g., Shannon, Simpson, Chao1, and ACE) were generated for each sample based on the OTU table using QIIME (1.9.6), which reflected the species richness and evenness at different depths of each land use. In addition, box plots of the Shannon and Chao1 alpha diversity indices were generated for each land use using R (v3.3.1). The differences in Alpha diversity indexes between the land uses were assessed using the Wilcoxon rank-sum test (p < 0.05). Non-metric multidimensional scaling (NMDS) with 999 permutations was performed for beta diversity based on unweighted UniFrac and Bray–Curtis dissimilarity across the samples. A stress value of less than 0.2 indicated that the NMDS analysis was reliable. A one-way analysis of similarity (ANOSIM) was performed to determine differences in microbial communities between the land uses (Clarke et al., 2008), after which intra-group and inter-group beta distance boxplot diagrams were generated. The NMDS and ANOSIM were produced using R (v3.3.1). Linear discriminant analysis effect size (LefSe) was performed to rank the most abundant taxa (modules) in each of the forest and OP samples. A size-effect threshold of 2.0 on the logarithmic LDA score was used for s for discriminative functional biomarkers (OTUs). LefSe was performed online using the Galaxy workflow framework. Furthermore, metastats gap analysis, together with Fisher’s exact test, was used to identify differences in microbial community abundance (genus level) between the two land-use groups.
3 Results
3.1 Peat properties, in-situ environmental variables and GHG fluxes
The physicochemical properties of the peat samples for each depth and land use are listed in Table 1. The forest peat acidity was similar at all depths, but in the OP, the deeper layers were more acidic. There was little effect of depth on nutrient availability, except in the forest where available NH4+ at 0–5 cm depth was significantly higher than at the other depths (p < 0.05). While the available SO4− in OP at 40–50 cm depth was higher than other depths, the large variation between samples meant that the difference between depths was not statistically significant. Comparison of the pooled data across all depths for each land use showed that the OP had significantly more available NO3− and Ca2+, but significantly lower Na+ and Mg2+ (p < 0.05).
The in-situ temperature and moisture content (VWC) of the peat under each land use, as well as the GHG fluxes, are shown in Figure 2. In the forest, there were no significant differences in soil moisture or soil temperature between the 5-cm and 30-cm depths, but in OP, the temperature at 30-cm depth was 2–3°C higher than at 5-cm (p < 0.05). There was no significant difference in temperatures between depths in OP. There was also no difference in WT between the two land uses, although the WT level measurements in the forest were more variable.
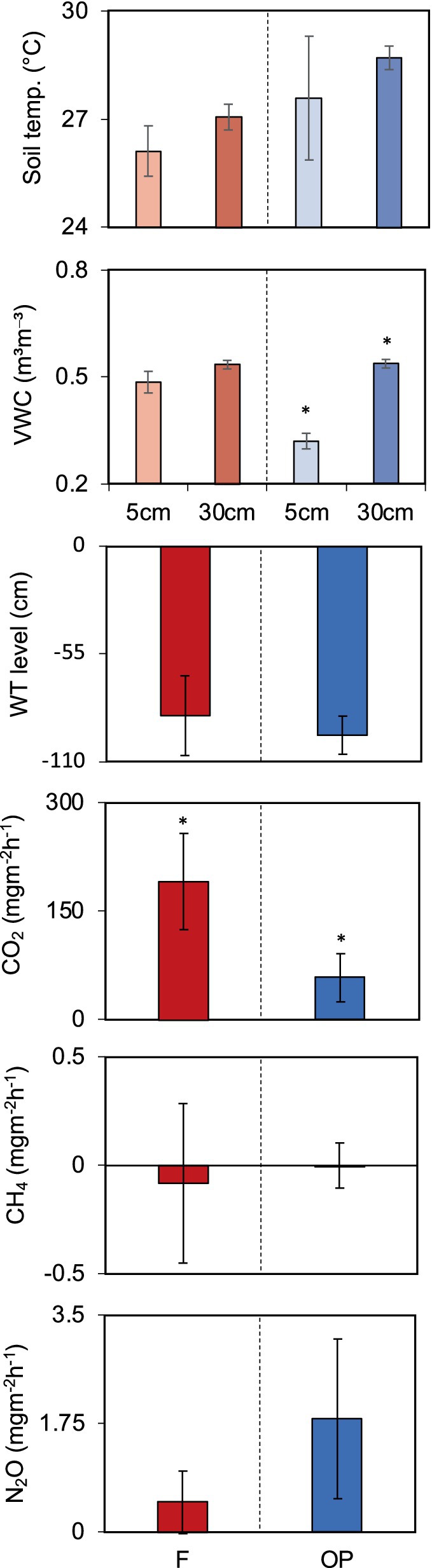
Figure 2. In-situ peat conditions, water table and GHG fluxes at the forest (F) and oil palm plantation (OP). VWC: volumetric water content (soil moisture); WT, water table. * represents significantly different (p < 0.05).
The CO2 fluxes from the forest were significantly higher than from OP, with mean fluxes of 190 ± 66.92 and 57.86 ± 33.66 mg m−2 h−1, respectively (p < 0.05). Notably, 90% of the measured CO2 fluxes at the forest were at least 20% higher than the OP fluxes on the same day (Supplementary Data 2). Although there were no significant differences in CH4 and N2O fluxes between the forest and OP sites, the CH4 fluxes were more variable in the forest (−0.91 to 0.54 mg m−2 h−1) than in the OP (−0.13 to 0.26 mg m−2 h−1), whereas the N2O fluxes were more variable in the OP (0.56–4.09 mg m−2 h−1) than in the forest (0.02–1.65 mg m−2 h−1; Figure 2).
3.2 Analysis of 16S-rRNA gene sequencing results
A total of 247 OTUs were obtained at a sequence-similarity level of 97%, with more than 50% of the OTUs being taxonomically unclassified at the genus level. The rarefaction curves (Wang et al., 2007) for the OTUs detected in this study showed that the quantity of observed OTUs increased as the sequencing depth increased (Supplementary Data 1). The ends of the rarefaction curves tapered off with increasing numbers of sequences per sample, as is commonly observed with sequencing data. The detected bacteria were classified into 22 phyla, 30 classes, 51 orders, 55 families, and 59 genera, while the archaea were classified into three phyla, three classes, four orders, four families, and five genera.
3.3 Relative abundance of microbial communities and differences between land uses
High-quality sequences were identified for 25 microbial phyla, 29 orders, and 30 genera based on their relative abundance, and these were compared across the different land uses and sampling depths (Figure 3). Overall, the four most dominant phyla, each accounting for more than 3% of the mean relative abundance of all microbial communities, were Acidobacteriota, Proteobacteria, Thermoplasmatota, and Actinobacteriota. The most abundant phyla at 0–5 cm and 20–25 cm depths of both sites were Acidobacteriota (also known as Acidobacteria), but at 40–50 cm depth, Proteobacteria were equally abundant as Acidobacteriota. The most important Proteobacteria classes in this study were Alphaproteobacteria and Gammaproteobacteria, and these were most abundant in the deeper peat samples (Supplementary Data 3). Thermoplasmatota were present at both sites but were only recorded at the phylum level. The proportions of Actinobacteriota were similar at all depths of both sites.
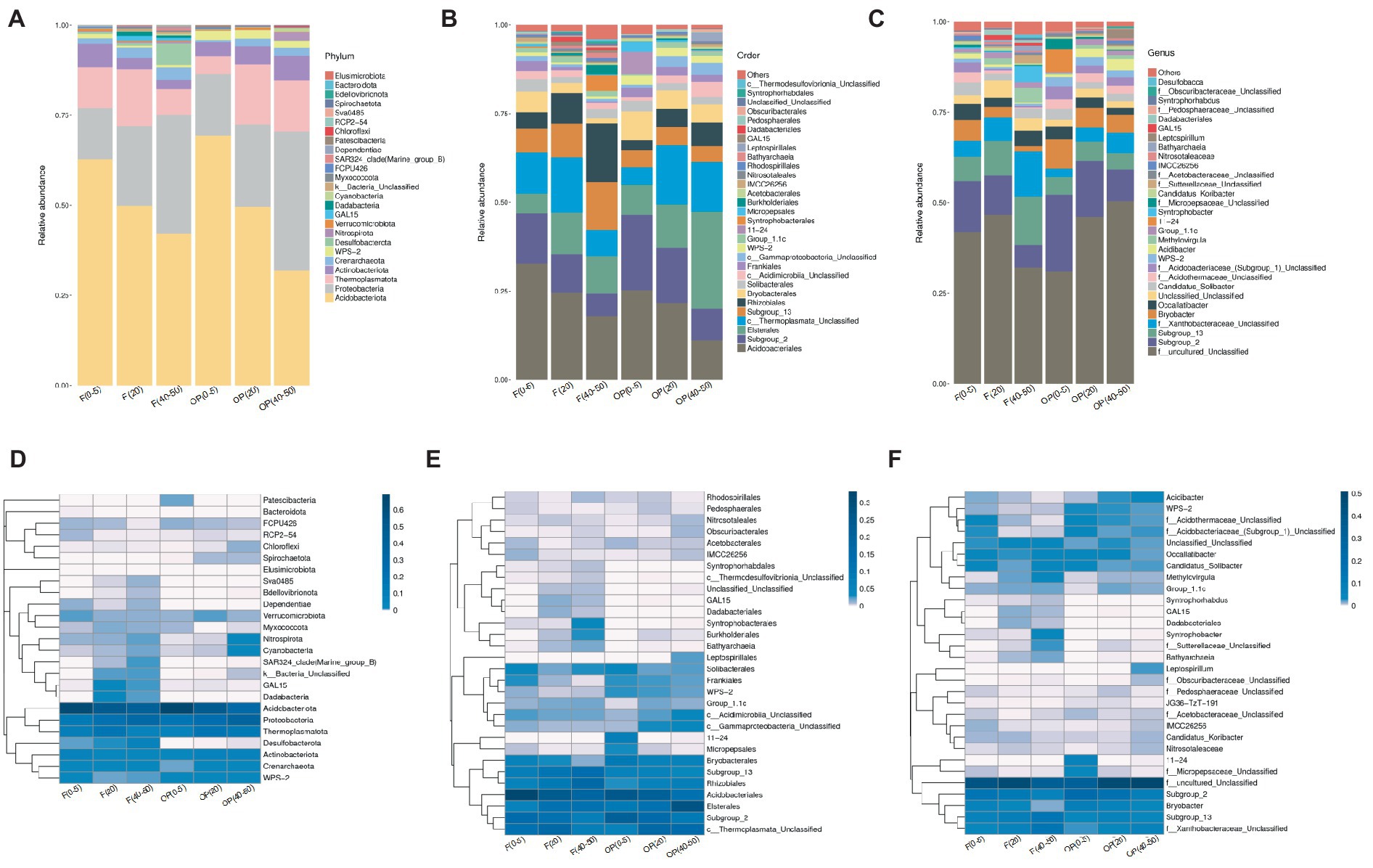
Figure 3. Relative abundances of microbial communities at phylum (A), order (B), and genus (C) levels in the forest (F) and OP peat profiles. Taxa clustering is shown in the heatmap at phylum (D), order (E), and genus (F). * c__ and f__ indicate class and family levels, respectively.
Cyanobacteria and Nitrospirota phyla were closely clustered and mainly found in the deepest peat sample layers in both the forest and OP (Figures 3A,D). Some taxa were only found in major abundance in one of the sites. For example, while Desulfobacterota (phylum) were found in all forest layers, with the highest abundance at 40–50 cm, they were virtually absent in all OP samples (relative abundance <0.01%). Similarly, Syntrophobacterales, (order), Burkholderiales (order), Syntrophobacter (genus), and Sutterellaceae (family) were only present at 40–50 cm in the forest. Other taxa were only found at the OP. WPS-2 (order and genus levels) was abundant at all OP depths, but virtually absent in the forest. Also, the 11–24 (order and genus levels), micropepsales (order) and micropepsaceae (family) were only found at 0–5 cm in the OP, while Leptospirillum (genus) was only present at OP 40–50 cm, while Acidibacter (genus) was present at both OP 20–25 cm and 40–50 cm.
FDR metastats gap analysis shows differences in microbial relative abundance (at genus level) between the forest and OP. The relative abundance of Subgroup_13 (member of phylum Acidobacteriota) and Syntrophobacter (member of phylum Desulfobacterota) genera were significantly higher in the forest than in the OP (Figure 4, p < 0.05). In contrast, Acidibacter (member of phylum Proteobacteria, class Gammaproteobacteria), Bryobacter (member of phylum Acidobacteriota), and WSP-2 (member of phylum WPS-2) genera had significantly greater relative abundance in the OP than in the forest (Figure 4, p < 0.05).
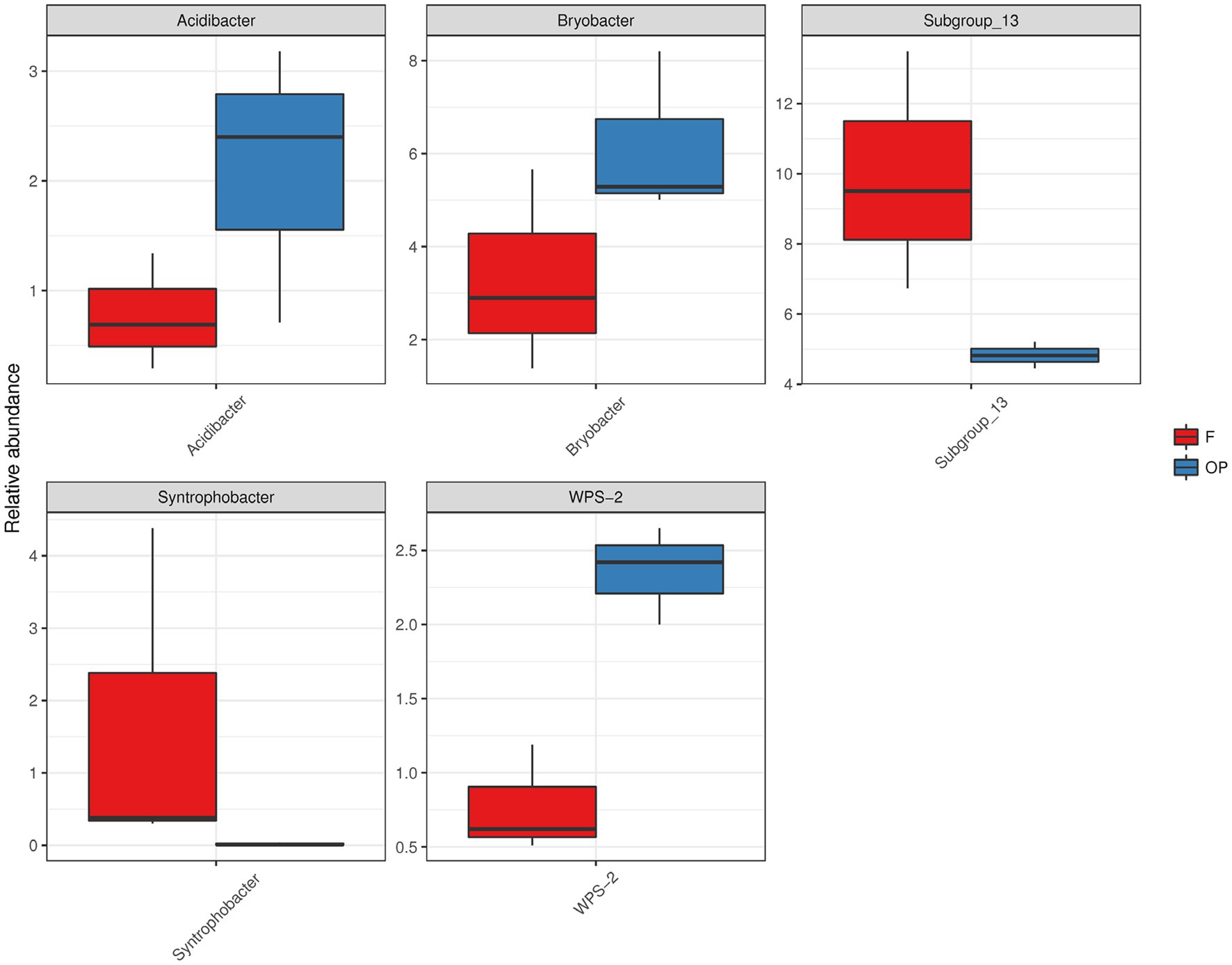
Figure 4. Relative abundance distributions of the five genera with the largest differences between the forest and OP (p < 0.05). The names of the five strains are shown on the X-axis, with the relative abundance shown on the Y-axis.
LefSe analysis was performed to rank the most important features of the microbial communities in the forest and OP. As shown in Figure 5A, 27 microbial clades (taxa) had statistically significant LDA scores of >2 (Figure 5B). Notably, 14 of these enriched microbial taxa were in the forest, and 13 were in the OP. At the genus and family levels, the most differentially abundant bacterial taxa in the forest were Dadabacteriales (family), Syntrophobacter (genus), and Subgroup_13 (genus), while in the OP it was Leptospirillum (genus), 11–24 (genus), and WPS-2 (genus). In the archaea (kingdom), the representative taxon in the forest was Bathyarchaeia (genus).
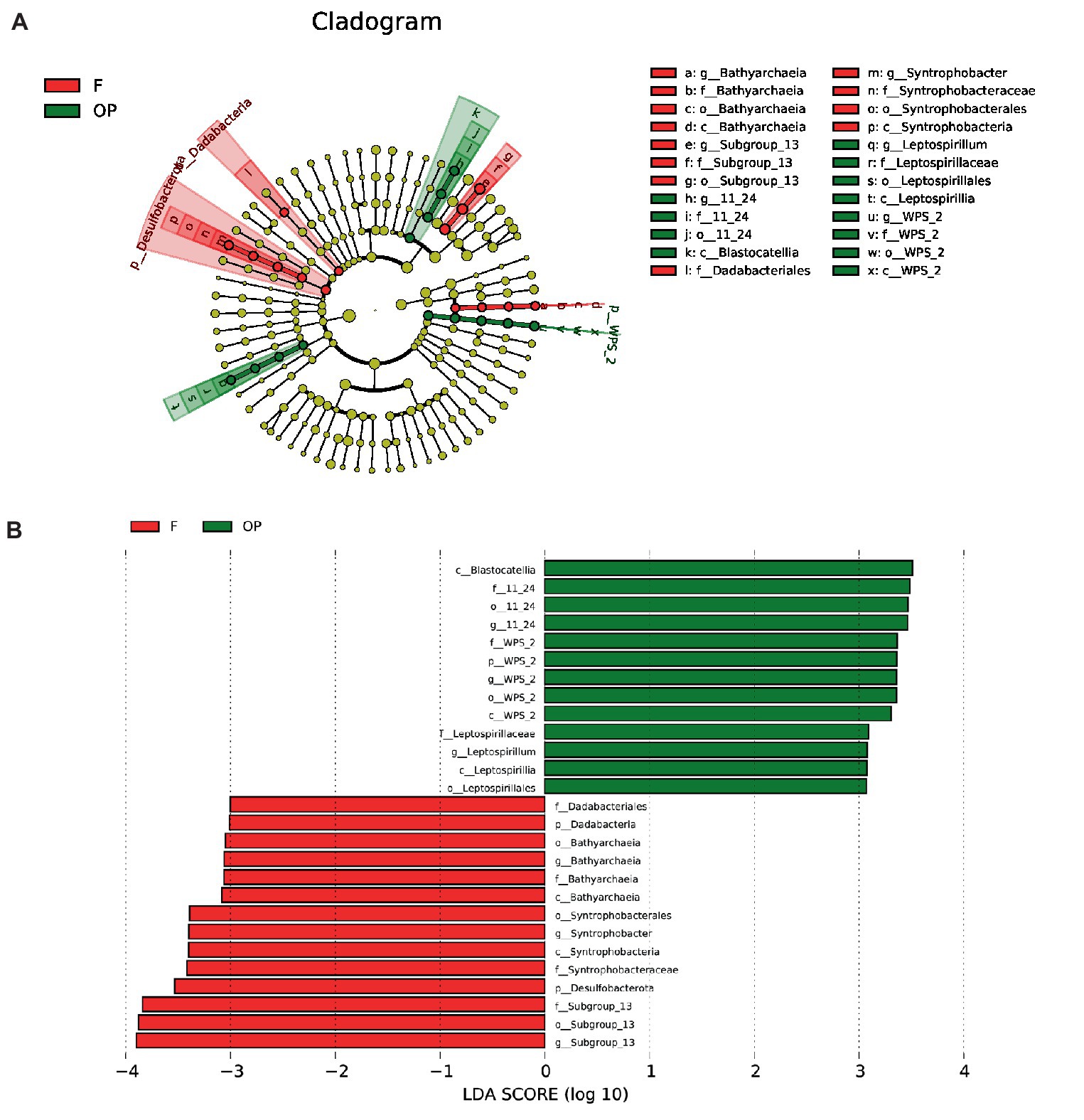
Figure 5. Linear discriminant analysis effect size (LEfSe) analysis to identify differences in the abundant taxa of the forest (F) and OP. (A) Cladogram representing statistically significant differences in microbial clades between the forest and OP identified by Wilcoxon rank-sum test. Circles in the diagram are shaded with either red (F), green (OP), or yellow (no significant difference between them represents the abundances of the taxa in the respective groups). (B) Microbial groups in the forest and OP with LDA score > 2.
3.4 Diversities and dissimilarities in microbial communities
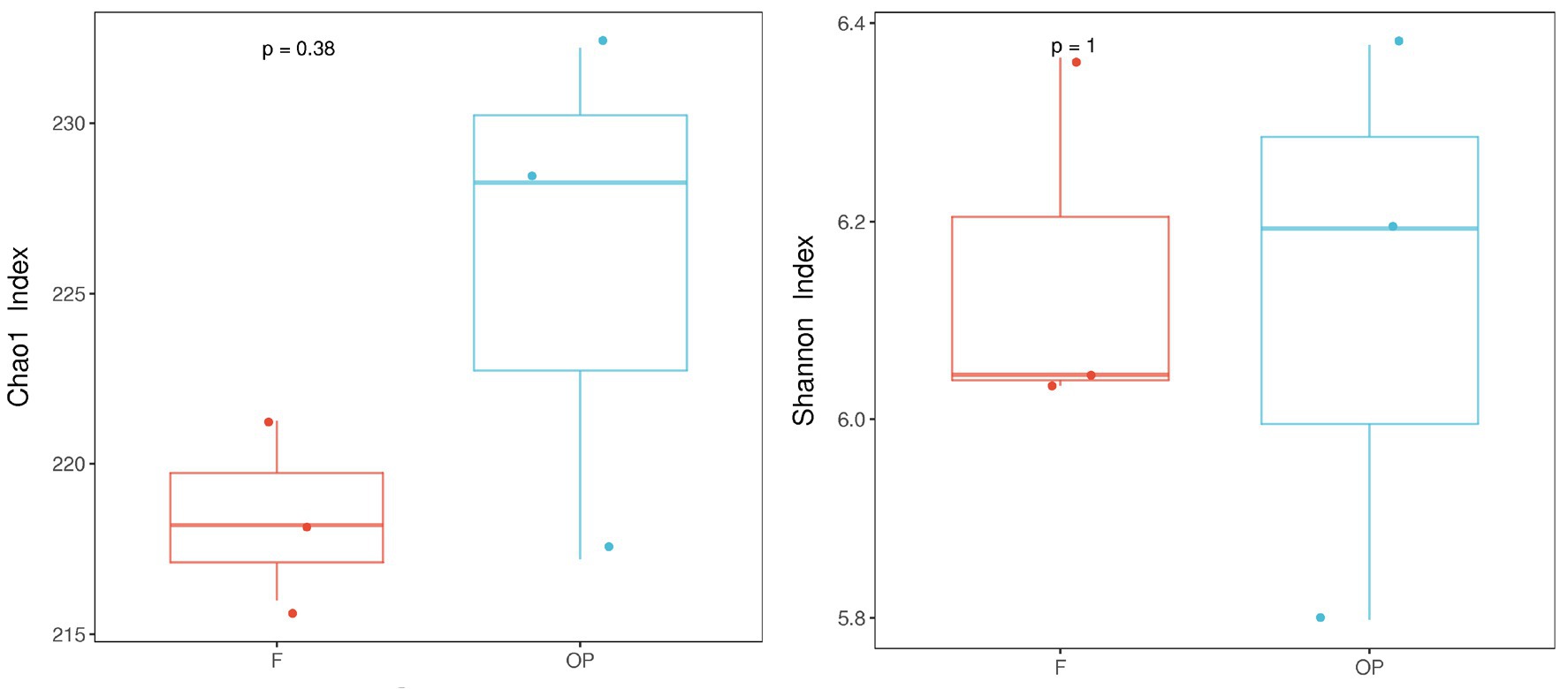
Alpha-diversity indices (Ace, Chao 1, Simpson, and Shannon; Figure 6; Table 2) and beta-diversity indices were calculated for the forest and OP peats. Alpha diversity did not vary significantly with peat depth for either land use (Table 2). Also, there was no significant difference in richness index (Chao 1; 218.48 ± 2.64, 225.88 ± 7.77) or diversity index (Shannon; 6.15 ± 0.18 and 6.12 ± 0.29) between the forest and OP peats (Figure 6). Beta diversity was used to analyze the spatial and depth changes in species composition in order to assess any differences between the two land uses. Although there were no statistically significant differences in the inter-group or intra-group beta distances between the microbial communities of the forest and OP groups (Figure 7), the NMDS analysis revealed dissimilarity in the microbial communities at different depths and sites (Figure 8). The microbial communities at all depths in the OP peat showed strong similarity to the 0–5 cm layer of the forest. The microbial communities in the 20–25 cm and 40–50 cm layers of the forest were closely related to each other but were different from the 0–5 cm layer and all OP layers.
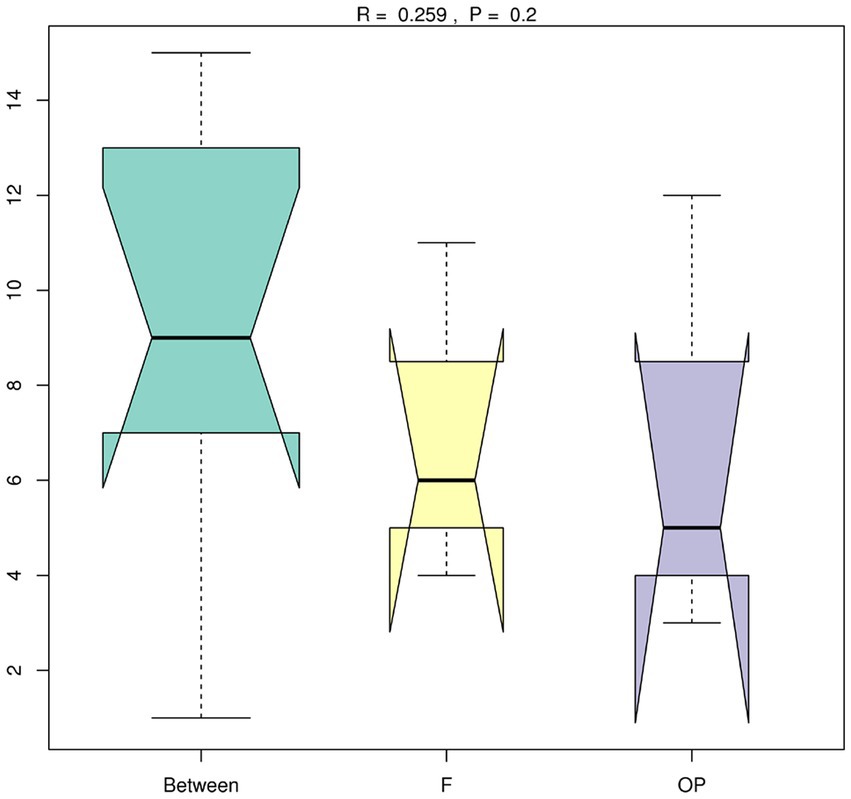
Figure 7. Box plot of inter-group and intra-group Beta distances for the forest (F) and OP (ANOSIM analysis). The x-axis represents the grouping, and the y-axis represents the distance calculated by Unweighted_unifrac. R-value range (−1, 1): an R-value ≤ 0 represents no significant difference between the inter-group and intra-group, while an R-value of >0 indicates that inter-group differences are greater than intra-group differences. The p-value represents the confidence level of the statistical analysis; p < 0.05 reflects a statistically significant difference. Boxes represent the interquartile range (IQR) between the first and third quartiles (25th and 75th percentiles, respectively), and the horizontal line inside the box defines the median. Whiskers represent the lowest and highest values within 1.5 times the IQR from the first and third quartiles, respectively.
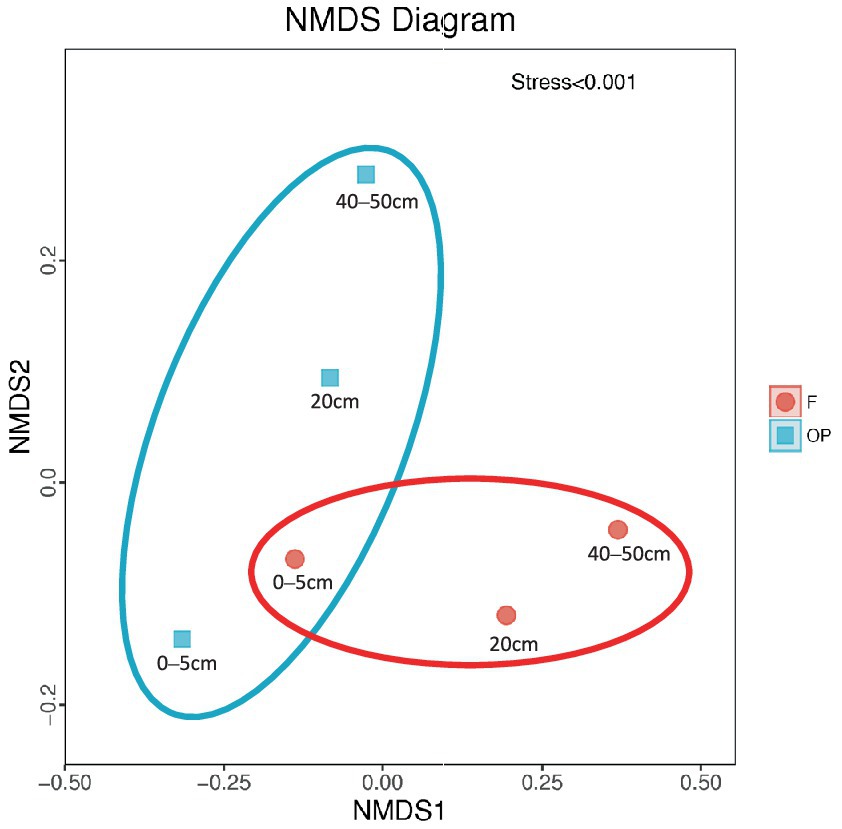
Figure 8. Non-metric multidimensional scaling (NMDS) analysis. Each point on the graph represents one sample, and each color represents a different land use group. The distance between points represents the level of difference. A stress level lower than 0.2 indicates that the NMDS analysis is reliable. The closer the proximity of the samples in the graph, the higher their similarity.
4 Discussion
Although, in recent years, there have been some microbial studies in tropical peatlands (Jackson et al., 2008; Tripathi et al., 2016; Too et al., 2018; Dhandapani et al., 2020; Dom et al., 2021), there is still much less than the amount of research on northern peatlands. Therefore, in this study, we used 16S-rRNA gene Illumina HiSeq sequencing to investigate microbial community composition in a tropical peatland in peninsular Malaysia, comparing the peat profiles of a peat swamp forest with that of an adjacent oil palm plantation (OP). While the peat swamp forest and OP soil physicochemical properties were similar to those in other tropical peatland studies (Sjögersten et al., 2011; Swails et al., 2018; Dhandapani et al., 2019a), there were some differences between the peat swamp forest and OP (Table 1). For example, while peatlands are generally nitrate-limited, the drainage and application of fertilizers in the OP likely contributed to the significantly higher NO3− and Ca2+ in the OP peat, as well as the lower Na+ and Mg2+ (p < 0.05). However, there was little variation in depth in either land use.
4.1 Microbial abundance in peat swamp forest and oil palm plantation
The major operational taxonomic units (OTUs) identified in this study are associated with bacterial communities of Acidobacteriota, Proteobacteria, Actinobacteria, and the archaeal communities of Thermoplasmata and Crenarchaeota. The most dominant phyla were Acidobacteriota and Proteobacteria which appear widely in most peatland ecosystems (Williams and Crawford, 1983; Kanokratana et al., 2011; Lin et al., 2012; Tsitko et al., 2014; Tripathi et al., 2016; Urbanová and Bárta, 2016; Espenberg et al., 2018; Too et al., 2018; Dom et al., 2021; Chen et al., 2022). Acidobacteriota has a diverse set of transporters for absorbing various substrates and can adapt to organic-rich oligotrophic conditions with low nutrients and pH (Lin et al., 2011; Trivedi et al., 2016; Zhang et al., 2016). On the other hand, Proteobacteria are known to be associated with high organic C soils (Lin et al., 2011) and play an important role in the carbon, sulfur and nitrogen cycles (Kersters et al., 2006). The ratio of Acidobacteriota to Proteobacteria has been used as an indicator for different types of peatlands (Urbanová and Bárta, 2016) and also for the nutrient status in soil ecosystems (Smit et al., 2001). In our study, the ratio of Acidobacteriota to Proteobacteria decreased with depth in both land uses, suggesting relatively low nutrient availability but high organic carbon in the deeper peat layers.
In the current study, Acidobacteriota comprised 30%–60% of the relative abundance in both the peat swamp forest and OP (Figures 3A,D). Acidobacteriota roles in soils include biopolymer decomposition and participation in global carbon, iron (Fe), and hydrogen (H) and sulfur cycling, but this list of functional capabilities is far from complete and is attributed to only a few sub-groups of this phylum (Ivanova et al., 2020). Metagenome data analysis has shown that Acidobacteriota, particularly members of sub-divisions SD1, SD3, and SD4, dominate the microbiota in oxic and anoxic soil layers of many wetlands (Dedysh et al., 2011). The major orders found in this study, such as Acidobacteriales, Bryobacterales, and 11–24, are affiliated with SD1, SD3, and SD4, respectively. Members of Acidobacteriota SD1 and SD3 are characterized as acidophilic (acidotolerant), mesophilic, psychrotolerant, and chemo-heterotrophic bacteria that use a variety of sugars and polysaccharides and have a wide range of hydrolytic abilities (Dedysh and Yilmaz, 2018). Other studies have suggested that cultivated Acidobacteriota of SD1 and SD3 are potentially aerobes or facultative anaerobes (Dedysh et al., 2006; Kulichevskaya et al., 2010; Dedysh et al., 2017) and may able to dissimilate inorganic and/or organic sulfur compounds (Hausmann et al., 2018; Woodcroft et al., 2018). Furthermore, members of Acidobacteriota SD4 have been characterized as aerobic, mesophilic or thermophilic, chemo-heterotrophic bacteria that specialize in the breakdown of complex proteinaceous substances (Huber et al., 2017).
In the current study, Proteobacteria comprised 23–26% of the microbial relative abundance in both the peat swamp forest and OP, which is similar to the findings in other tropical peatland studies, Southeast Asian peat (Too et al., 2018; Dom et al., 2021) and Central American peat (Troxler et al., 2012). The dominant classes in the current study were Alphaproteobacteria and Gammaproteobacteria (Supplementary Data 3), which are typical heterotrophic bacteria that play important roles in the degradation of organic nitrogen (Elliott et al., 2015; Truu et al., 2020; Prasitwuttisak et al., 2022). The order Rhizobiales from Alphaproteobacteria was more abundant in the peat swamp forest than in OP (Figure 3B), and it has been reported to be associated with the nitrogen cycle (Prasitwuttisak et al., 2022). Within the same order, Methylovirgula spp., which belongs to the family Beijerinckiaceae, was most prominent in the 40–50 cm layer of the peat swamp forest (Figure 3F). They have been categorized as methanotrophic and methylotrophic aerobic bacteria in a genus that is obligately acidophilic and mesophilic (Marín and Arahal, 2014). Its presence in the deepest sample layer of the peat swamp forest peat suggests it utilizes the carbon substrate while decomposing fallen woody materials and plant debris (Warneke et al., 1999) and is also potentially involved in CH4 oxidation.
The Actinobacteria phylum is involved in the degradation and mineralization of plant and humic materials in soil (Lewin et al., 2016) and is associated with the decomposition of organic compounds such as cellulose in peat (Pankratov et al., 2006). In the current study, Actinobacteria was relatively abundant at 0–5 cm in the peat swamp forest and at all depths in the OP (Figure 3A), suggesting that these are the locations where more intense peat decomposition occurs. A study by Zhou et al. (2017) also found a relatively high abundance of Actinobacteria in a rubber plantation tropical soil.
Two phyla of archaea were found in this study, Thermoplasmata and Crenarchaeota. In the peat swamp forest, there was a higher relative abundance of Thermoplasmata in the 0–5 cm and 20–25 cm layers, whereas in OP, they were more abundant in the 20–25 cm and 40–50 cm layers (Figure 3A). However, the order of Thermoplasmata found in this study was classified as uncultured. Other studies have identified members of Thermoplasmata in coniferous forest soils (Truu et al., 2020) and boreal acidic forest soils (Kemnitz et al., 2007), and they have been associated with Fe and sulfur (S) cycling (Jin et al., 2017), and with methanogenesis (Iino et al., 2013). The other archaea phyla found in the current study, Crenarchaeota, have typically been found in deeper peat layers of tropical peatlands (Dom et al., 2021), subtropical peatlands (Espenberg et al., 2018) and boreal peatlands (Bomberg et al., 2010; Sun et al., 2014) and they are thought to be crucial for carbon cycling in deeper peat layers (Lin et al., 2014). In the current study, the Crenarchaeota phylum was also most abundant in the 20–25 cm and 40–50 cm layers of both the peat swamp forest and OP. The major classes found in this study were Nitrososphaeria and Bathyarchaeia (currently reclassified into the phylum Bathyarchaeota; Supplementary Data 3).
4.2 Land use and depth affect microbial community structure, GHG emission and peat properties
There were significant differences in microbial composition at different depths in the forest peat, as has been previously reported in earlier studies conducted in the same North Selangor Peat Swamp Forest (Jackson et al., 2008; Too et al., 2018). NMDS analysis demonstrated differences in microbial community composition between the peat swamp forest and OP sites and between depths (Figure 8), suggesting that the compositions of the microbial communities at each land use site and depth may be related to WT, and the availability and quality of labile carbon (Könönen et al., 2018). The microbial community structure at 0–5 cm in the peat swamp forest was similar to that in all three sampled peat layers in OP, suggesting similarly high availability of labile carbon and similar peat decomposition processes (Yule and Gomez, 2009; Kwon et al., 2013). The different microbial community structures found at 20–25 cm and 40–50 cm depths of the peat swamp forest may relate to anoxic conditions in deeper peat that are known to accumulate recalcitrant carbon (Cooper et al., 2019).
Contrary to our expectation, there were no statistical differences in overall microbial diversity and evenness between land uses (Figures 6, 7). However, we detected nine microbial taxa that characterized differences in the microbial communities between the peat swamp forest and OP (p > 0.05). Taxa of Syntrophobacter, Dadabacteriales, Bathyarchaeia, and Subgroup_13 were significantly more abundant in the peat swamp forest, whereas Bryobacter, 11–24, Acidobacter, Leptospirillum, and WPS-2 were more abundant in the OP (Figures 4, 5, p > 0.05).
The genus Syntrophobacter belonging to the phylum Desulfobacterota, had significantly greater relative abundance in the peat swamp forest than in OP (Figures 4, 5), especially at 40–50 cm depth (Figure 3F). Syntrophobacter is known to oxidize organic matter incompletely and produce acetate as the end-product, which is the most common intermediate in anaerobic decomposition of organic matter and is an essential substrate for methanogens to produce CH4 (Liu et al., 2018). However, since no methanogens were found in any forest sample, we hypothesize that this member of sulfate-reducing bacteria may use all the products of primary fermentation and oxidize them to CO2 coupled to sulfate reduction (alternative anaerobic degradation processes; Muyzer and Stams, 2008; Plugge et al., 2011). This could explain the low CH4 production coupled with significantly higher CO2 emissions in the peat swamp forest (p < 0.05; Figure 2). Furthermore, the absence of Syntrophobacter in OP may indicate that this taxon favored anaerobic, acidic, and sulfate-depleted habitats (Pankratov et al., 2011; Too et al., 2018), which could also explain their higher abundance in the forest peat, which had low SO4— (Table 1). In the current study, there was also a significantly high abundance of the family Dadabacteriales in the peat swamp forest (Figure 5). Dadabacteriales belong to the phylum Dadabacteria and are characterized as sulfate-reducing bacteria. They have been detected in marine ecosystems (Graham and Tully, 2020). To our knowledge, this is the first study that has recorded Dadabacteriales in a peatland ecosystem. This taxon together with Syntrophobacter could be the key drivers in sulfate recycling in peat swamp ecosystems.
The archaeal genus Bathyarchaeia, formerly known as the Miscellaneous Crenarchaeotal Group (MCG) but now assigned to the phylum Bathyarchaeota based on the Genome Taxonomy Database (Rinke et al., 2021; Hou et al., 2022), has previously been detected in peatland ecosystems (Xiang et al., 2017; Truu et al., 2020; Prasitwuttisak et al., 2022). Several studies have suggested that Bathyarchaeia are organotrophic and are able to utilize lignin as an energy source for acetogenesis (Yu et al., 2018), leading to the degradation of a variety of organic carbon compounds such as acetate and complex hydrocarbons (He et al., 2016). Also, some members of Bathyarchaeia may be able to undergo anaerobic methane production (Evans et al., 2015). The higher abundance of Bathyarchaeia in the peat swamp forest than in OP (Figure 5, p < 0.05), especially in the deeper sample layers (Figure 3F), suggests a preference for anaerobic water-logged peat with recalcitrant organic material (Xiang et al., 2017). Also, the absence of Bathyarchaeia in the OP suggests that the displacement of this taxon could be related to changes in environmental factors such as WT and organic matter content.
Three taxa belonging to the phylum Acidobacteriota characterize the differences between the peat swamp forest and OP. Subgroup_13 was significantly more abundant in the peat swamp forest, while Bryobacter and 11–24 were significantly more abundant in OP (Figures 4, 5). Subgroup_13, belonging to SD3 in the Acidobacteriae class, is associated with carbon degradation, especially hemicellulose degradation (Ivanova et al., 2020). Bryobacter belonging to SD3 Acidobacteriota from the order Bryobacterales, is characterized by the ability to grow under anoxic conditions and to reduce Fe and NO3−. They are active in plant-derived polymer degradation (such as cellulose) and in N and Fe cycling (Küsel et al., 2008; Eichorst et al., 2011). The significant abundance of Bryobacter in OP suggests greater peat degradation and active N transformation pathways in the peat under this land use. This result could explain the high NO3− (Table 1) and the low C/N measured at the same sites in an earlier study (Azizan et al., 2021). Another study that found differences in relative abundances of Bryobacter depending on soil carbon was a study by Eichorst et al. (2011) in Michigan, United States, who found higher proportions of Bryobacter in arable soils with lower carbon than in grassland soils with higher carbon.
The third taxon, 11–24 from the class Blastocatellia, belongs to SD4 Acidobacteriota and was also present in OP, but their function remains unknown. Blastocatellia has been characterized as an aerobic, mesophilic or thermophilic, chemo-heterotrophic bacteria specializing in the breakdown of complex proteinaceous substances (Huber et al., 2017). It has been reported to be associated with habitats that are dry and low in organic matter (Ivanova et al., 2020). Therefore, since 11–24 belongs to the class Blastocatellia, it suggests that this taxon may be associated with aerobic heterotrophic metabolism.
Another taxon that had a different abundance in the peat swamp forest and OP is the genus Acidibacter from class Gammaproteobacteria of the phylum Proteobacteria. They were significantly more abundant in OP than in the peat swamp forest (Figure 4). They are acidophilic bacteria that have the capacity to employ ferric iron (Fe3+) as an alternate electron acceptor to oxygen when growing in anaerobic or microaerobic environments (Falagán et al., 2014). Previous studies have reported Acidibacter in a pit lake at an abandoned metal mine (Falagán et al., 2014) and in acid mine drainage from an abandoned gold mine (Bonilla et al., 2018).
The genus Leptospirillum, from the class of Leptospirallales belonging to phylum Nitrospirota, also had significantly different abundance between the peat swamp forest and OP. This genus has been reported to have the ability to live in strongly acidic environments (acidophilic bacteria), and they are also known to be aerobic Fe oxidizers (González-Toril et al., 2003) and nitrogen fixers (Méndez-García et al., 2015). Some studies have reported that Leptospirillum is always found together with highly active sulfur-oxidizing species in conditions with large amounts of sulfide minerals (Acosta et al., 2017; Bulaev et al., 2017). Other studies have reported that Leptospirillum is mostly found in limited N environments such as acid mine drainage (Parro and Moreno-Paz, 2004; Méndez-García et al., 2015). However, in the current study, this taxon was only significantly abundant at the 40–50 cm depth of the N-rich OP (Figures 3F, 5). Therefore, we suspect that the abundance of this taxon in this location may also be associated with the much higher SO4− concentrations (50.23 ± 50.09 mgL−1) recorded in the 40–50 cm OP layer than in the other layers or the forest peat (Table 1).
WPS-2 has been detected in cold, moss-associated acidic peat environments, including in temperate and Arctic peatlands, where their abundances correlate well with the abundance of methanotrophs (Holland-Moritz et al., 2018; Woodcroft et al., 2018; Sheremet et al., 2020; Kolton et al., 2022). Cold, moss-associated WPS-2 is thought likely to be involved with anoxygenic phototrophs that are capable of carbon fixation via RuBisCo and utilizes byproducts of photorespiration (Holland-Moritz et al., 2018; Kolton et al., 2022). However, in this study, the high abundance of WPS-2 in the aerobic conditions at the OP (Figures 4, 5) could suggest a different potential ecological function of this taxon. Cross-referencing of analysis (Single- and Metagenome-amplified genomes) suggests that WPS-2 have a predominantly aerobic organoheterotrophic lifestyle, possibly based on parasitizing or scavenging amino acids, nucleotides, and complex oligopeptides, together with a lithotrophic capacity for thiosulfate (Sheremet et al., 2020). However, since the function of WPS-2 in tropical peatlands remains unclear, further studies are needed to understand this versatile taxon.
5 Conclusion
This study helps to improve our understanding of the differences in microbial community composition between peat swamp forest and oil palm plantation systems. Although there were no statistically significant differences in overall microbial diversity and evenness between the two different land uses, there were differences in the abundances of nine specific microbial taxa between the peat swamp forest and the OP. The absence of Syntrophobacter and Bathyarchaeia in the OP suggests that these taxa may be disappearing from converted swamp forests, which could serve as an indicator of swamp forest conversion. Nevertheless, we recommend that further studies on microbial ecology should be conducted in converted and recovering tropical peatlands to confirm this. Also, we suggest that more attention be paid to the organic layers in studies of the belowground microbiome (e.g., using meta-transcriptomic technologies) during periods of agricultural conversion in tropical peatlands (e.g., clearance burning, early-stage oil palm planting), particularly with regard to S, C, N and phosphorus cycling since these processes are fundamental to the maintenance of this type of ecosystem. In addition, we think that further research is needed for specific taxa such as WPS-2 because there is no information about this taxon in tropical peatlands ecosystems.
Data availability statement
Sequencing data were submitted to NCBI Sequence Read Archive database SRX19834842, SRX19834841, SRX19834840, SRX19834838, and SRX19834837 under the Bioproject number PRJNA950583.
Author contributions
SNFA: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. SM: Supervision, Writing – review & editing. IM: Supervision, Writing – review & editing. NY: Data curation, Formal analysis, Writing – review & editing. SS: Methodology, Writing – review & editing. HH: Supervision, Validation, Writing – review & editing. KN: Funding acquisition, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the International Collaborative Research Promotion Project (MU-OSRI-ICRPP2016-209 and MU-OSRI-ICRPP2017-102) Meiji University, and JSPS Grant-in-Aid for Challenging Research (Pioneering) (22K19230).
Acknowledgments
The authors thank the Selangor State Forestry Department for granting site access. Special thanks to Azri and Yunus (forest rangers) for field assistance and to members of Memobio Ikohza (Malaysia–Japan International Institute of Technology, Malaysia) for participating in the sample collection. We thank Kotaro Seki and Yuki Sunakawa for their guidance on using the gas chromatography and ion analyzer equipment, respectively.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2024.1305491/full#supplementary-material
Footnotes
References
Acosta, M., Galleguillos, P., Guajardo, M., and Demergasso, C. (2017). “Microbial survey on industrial bioleaching heap by high-throughput 16s sequencing and metagenomics analysis” in State Phenomena, 262 SSP. eds. S. Hedrich, K. Rübberdt, F. Glombitza, W. Sand, A. Schippers, M. Vera Véliz, and S. Willscher (Zurich) vol. 262 (Trans Tech Publications Ltd), 219–223.
Andersen, R., Chapman, S. J., and Artz, R. R. E. (2013). Microbial communities in natural and disturbed peatlands: a review. Soil Biol. Biochem. 57, 979–994. doi: 10.1016/j.soilbio.2012.10.003
Azizan, S. N. F., Goto, Y., Doi, T., Kamardan, M. I. F., Hara, H., McTaggart, I., et al. (2021). Comparing GHG emissions from drained oil palm and recovering tropical peatland forests in Malaysia. Water 13:3372. doi: 10.3390/W13233372
Birnbaum, C., Wood, J., Lilleskov, E., Lamit, L. J., Shannon, J., Brewer, M., et al. (2022). Degradation reduces microbial richness and alters microbial functions in an Australian peatland. Microb. Ecol. 1, 1–17. doi: 10.1007/S00248-022-02071-Z/FIGURES/6
Bomberg, M., Montonen, L., and Timonen, S. (2010). Anaerobic Eury- and Crenarchaeota inhabit Ectomycorrhizas of boreal Forest scots pine. Eur. J. Soil Biol. 46, 356–364. doi: 10.1016/j.ejsobi.2010.09.002
Bonilla, J. O., Kurth, D. G., Cid, F. D., Ulacco, J. H., Gil, R. A., and Villegas, L. B. (2018). Prokaryotic and eukaryotic community structure affected by the presence of an acid mine drainage from an abandoned gold mine. Extremophiles 22, 699–711. doi: 10.1007/s00792-018-1030-y
Bulaev, A., Belyi, A., Panyushkina, A., Solopova, N., and Pivovarova, T. (2017). “Microbial population of industrial biooxidation reactors” in State Phenomena, 262 SSP. eds. S. Hedrich, K. Rübberdt, F. Glombitza, W. Sand, A. Schippers, M. Vera Véliz, and S. Willscher (Zurich) vol. 262 (Trans Tech Publications Ltd.), 48–52.
Chen, C. Y., Ling Lai, I., and Chang, S. C. (2022). Changes in soil microbial community and carbon flux regime across a subtropical montane peatland-to-Forest successional series in Taiwan. Forests 13:958. doi: 10.3390/F13060958
Clarke, K. R., Somerfield, P. J., and Gorley, R. N. (2008). Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J. Exp. Mar. Biol. Ecol. 366, 56–69. doi: 10.1016/j.jembe.2008.07.009
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Yanni Sun, C., et al. (2014). Ribosomal database project: data and tools for high throughput RRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Cooper, H. V., Vane, C. H., Evers, S., Aplin, P., Girkin, N. T., and Sjögersten, S. (2019). From peat swamp Forest to oil palm plantations: the stability of tropical peatland carbon. Geoderma 342, 109–117. doi: 10.1016/J.GEODERMA.2019.02.021
Dargie, G. C., Lewis, S. L., Lawson, I. T., Mitchard, E. T. A., Page, S. E., Bocko, Y. E., et al. (2017). Age, extent and carbon storage of the Central Congo Basin peatland complex. Nature 542, 86–90. doi: 10.1038/nature21048
Dedysh, S. N., Kulichevskaya, I. S., Huber, K. J., and Overmann, J. (2017). Defining the taxonomic status of described subdivision 3 Acidobacteria: proposal of Bryobacteraceae fam. Nov. Int. J. Syst. Evol. Microbiol. 67, 498–501. doi: 10.1099/ijsem.0.001687
Dedysh, S. N., Neufeld, J. D., and Dumont, M. G. (2011). Cultivating uncultured Bacteria from northern wetlands: Knowledge gained and remaining gaps. Front. Microbiol. 2:184. doi: 10.3389/fmicb.2011.00184
Dedysh, S. N., Pankratov, T. A., Belova, S. E., Kulichevskaya, I. S., and Liesack, W. (2006). Phylogenetic analysis and in situ identification of Bacteria community composition in an acidic Sphagnum peat bog. Appl. Environ. Microbiol. 72, 2110–2117. doi: 10.1128/AEM.72.3.2110-2117.2006
Dedysh, S. N., and Yilmaz, P. (2018). Refining the taxonomic structure of the phylum Acidobacteria. Int. J. Syst. Evol. Microbiol. 68, 3796–3806. doi: 10.1099/ijsem.0.003062
Dhandapani, S., Ritz, K., Evers, S., Cooper, H., Tonks, A., and Sjögersten, S. (2020). Land-use changes associated with oil palm plantations impact PLFA microbial phenotypic community structure throughout the depth of tropical peats. Wetlands 40, 2351–2366. doi: 10.1007/s13157-020-01342-0
Dhandapani, S., Ritz, K., Evers, S., and Sjögersten, S. (2019a). Environmental impacts as affected by different oil palm cropping Systems in Tropical Peatlands. Agric. Ecosyst. Environ. 276, 8–20. doi: 10.1016/j.agee.2019.02.012
Dhandapani, S., Ritz, K., Evers, S., Yule, C. M., and Sjögersten, S. (2019b). Are secondary forests second-rate? Comparing peatland greenhouse gas emissions, chemical and microbial community properties between primary and secondary forests in peninsular Malaysia. Sci. Total Environ. 655, 220–231. doi: 10.1016/j.scitotenv.2018.11.046
Dom, S. P., Ikenaga, M., Ling, S. Y., Lau, S. R., Midot, F., Yap, M. L., et al. (2021). Linking prokaryotic community composition to carbon biogeochemical cycling across a tropical peat dome in Sarawak, Malaysia. Sci. Rep. 11:6416. doi: 10.1038/s41598-021-81865-6
Eichorst, S. A., Kuske, C. R., and Schmidt, T. M. (2011). Influence of plant polymers on the distribution and cultivation of Bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 77, 586–596. doi: 10.1128/AEM.01080-10
Elliott, D. R., Caporn, S. J. M., Felix Nwaishi, R., Nilsson, H., and Sen, R. (2015). Bacterial and fungal communities in a degraded Ombrotrophic peatland undergoing natural and managed re-vegetation. PLoS One 10:e0124726. doi: 10.1371/JOURNAL.PONE.0124726
Espenberg, M., Truu, M., Mander, Ü., Kasak, K., Nõlvak, H., Ligi, T., et al. (2018). Differences in microbial community structure and nitrogen cycling in natural and drained tropical peatland soils. Sci. Rep. 8:4742. doi: 10.1038/s41598-018-23032-y
Evans, P. N., Parks, D. H., Chadwick, G. L., Robbins, S. J., Orphan, V. J., and Golding, S. D. et al. (2015). Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science 350, 434–438. doi: 10.1126/science.aac7745
Falagán, C., Barrie, D., and Johnson, A. (2014). Acidibacter Ferrireducens gen. Nov., Sp. Nov.: an acidophilic ferric Iron-reducing Gammaproteobacterium. Extremophiles 18, 1067–1073. doi: 10.1007/s00792-014-0684-3
Girkin, N. T., Dhandapani, S., Evers, S., Ostle, N., Turner, B. L., and Sjögersten, S. (2020a). Interactions between labile carbon, temperature and land use regulate carbon dioxide and methane production in tropical peat. Biogeochemistry 147, 87–97. doi: 10.1007/s10533-019-00632-y
Girkin, N. T., Lopes dos Santos, R. A., Vane, C. H., Ostle, N., Turner, B. L., and Sjögersten, S. (2020b). Peat properties, dominant vegetation type and microbial community structure in a tropical peatland. Wetlands 40, 1367–1377. doi: 10.1007/s13157-020-01287-4
González-Toril, E., Llobet-Brossa, E., Casamayor, E. O., Amann, R., and Amils, R. (2003). Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 69, 4853–4865. doi: 10.1128/AEM.69.8.4853-4865.2003
Graham, E. D., and Tully, B. J. (2020). Marine Dadabacteria exhibit genome streamlining and Phototrophy-driven niche partitioning. ISME J. 15, 1248–1256. doi: 10.1038/s41396-020-00834-5
Hausmann, B., Pelikan, C., Herbold, C. W., Köstlbacher, S., Albertsen, M., Eichorst, S. A., et al. (2018). Peatland Acidobacteria with a dissimilatory sulfur metabolism. ISME J. 12, 1729–1742. doi: 10.1038/s41396-018-0077-1
He, Y., Li, M., Perumal, V., Feng, X., Fang, J., Xie, J., et al. (2016). Genomic and enzymatic evidence for Acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat. Microbiol. 1:35. doi: 10.1038/nmicrobiol.2016.35
Holland-Moritz, H., Stuart, J., Lewis, L. R., Miller, S., Mack, M. C., McDaniel, S. F., et al. (2018). Novel bacterial lineages associated with boreal Moss species. Environ. Microbiol. 20, 2625–2638. doi: 10.1111/1462-2920.14288
Hou, J., Wang, Y., Zhu, P., Yang, N., Liang, L., Tiantian, Y., et al. (2022). Taxonomic and carbon metabolic diversification of Bathyarchaeia during its 4 co-evolution history with the early earth surface environment. Sci. Adv. 9:eadf5069. doi: 10.1101/2022.11.19.517056
Hu, B. L., Rush, D., van der Biezen, E., Zheng, P., van Mullekom, M., Schouten, S., et al. (2011). New anaerobic, ammonium-oxidizing community enriched from peat soil. Appl. Environ. Microbiol. 77, 966–971. doi: 10.1128/AEM.02402-10
Huber, K. J., Pascual, J., Foesel, B. U., and Overmann, J. (2017). “Blastocatellia” in Bergey’s Manual of Systematics of Archaea and Bacteria. Ed: WB Whitman: Hoboken, NJ, United States: John Wiley & Sons, Inc.
Iino, T., Tamaki, H., Tamazawa, S., Ueno, Y., Ohkuma, M., Suzuki, K. I., et al. (2013). Candidatus Methanogranum Caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. Nov. and Methanomassiliicoccales Ord. Nov., for a methanogenic lineage of the class Thermoplasmata. Microbes Environ. 28, 244–250. doi: 10.1264/JSME2.ME12189
Ivanova, A. A., Zhelezova, A. D., Chernov, T. I., and Dedysh, S. N. (2020). Linking ecology and systematics of Acidobacteria: distinct habitat preferences of the Acidobacteriia and Blastocatellia in tundra soils. PLoS One 15:e0230157. doi: 10.1371/journal.pone.0230157
Jackson, C. R., Liew, K. C., and Yule, C. M. (2008). Structural and functional changes with depth in microbial communities in a tropical Malaysian peat swamp Forest. Microb. Ecol. 57, 402–412. doi: 10.1007/s00248-008-9409-4
Jin, L., Lee, C. S., Ahn, C. Y., Lee, H. G., Lee, S., Shin, H. H., et al. (2017). Abundant Iron and sulfur oxidizers in the stratified sediment of a eutrophic freshwater reservoir with annual cyanobacterial blooms. Sci. Rep. 7, 1–10. doi: 10.1038/srep43814
Kanokratana, P., Uengwetwanit, T., Rattanachomsri, U., Bunterngsook, B., Nimchua, T., Tangphatsornruang, S., et al. (2011). Insights into the phylogeny and metabolic potential of a primary tropical peat swamp Forest microbial community by metagenomic analysis. Microb. Ecol. 61, 518–528. doi: 10.1007/s00248-010-9766-7
Kemnitz, D., Kolb, S., and Conrad, R. (2007). High abundance of Crenarchaeota in a temperate acidic Forest soil. FEMS Microbiol. Ecol. 60, 442–448. doi: 10.1111/J.1574-6941.2007.00310.X
Kersters, K., De Vos, P., Gillis, M., Swings, J., Vandamme, P., and Stackebrandt, E. (2006). “Introduction to the Proteobacteria” in The prokaryotes: Volume 5: Proteobacteria: Alpha and Beta subclasses. eds. M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (New York, NY: Springer New York), 3–37.
Kolton, M., Weston, D. J., Mayali, X., Weber, P. K., McFarlane, K. J., Pett-Ridge, J., et al. (2022). Defining the Sphagnum Core microbiome across the north American continent reveals a central role for Diazotrophic Methanotrophs in the nitrogen and carbon cycles of boreal peatland ecosystems. MBio 13:21. doi: 10.1128/mbio.03714-21
Könönen, M., Jauhiainen, J., Straková, P., Heinonsalo, J., Laiho, R., Kusin, K., et al. (2018). Deforested and drained tropical peatland sites show poorer peat substrate quality and lower microbial biomass and activity than unmanaged swamp Forest. Soil Biol. Biochem. 123, 229–241. doi: 10.1016/J.SOILBIO.2018.04.028
Kulichevskaya, I. S., Suzina, N. E., Liesack, W., and Dedysh, S. N. (2010). Bryobacter Aggregatus gen. Nov., Sp. Nov., a peat-inhabiting, aerobic chemo-Organotroph from subdivision 3 of the Acidobacteria. Int. J. Syst. Evol. Microbiol. 60, 301–306. doi: 10.1099/ijs.0.013250-0
Küsel, K., Blöthe, M., Schulz, D., Reiche, M., and Drake, H. L. (2008). Microbial reduction of Iron and Porewater biogeochemistry in acidic peatlands. Biogeosciences 5, 1537–1549. doi: 10.5194/bg-5-1537-2008
Kwon, M. J., Haraguchi, A., and Kang, H. (2013). Long-term water regime differentiates changes in decomposition and microbial properties in tropical peat soils exposed to the short-term drought. Soil Biol. Biochem. 60, 33–44. doi: 10.1016/j.soilbio.2013.01.023
Levy, P. E., Gray, A., Leeson, S. R., Gaiawyn, J., Kelly, M. P. C., Cooper, M. D. A., et al. (2011). Quantification of uncertainty in trace gas fluxes measured by the static chamber method. Eur. J. Soil Sci. 62, 811–821. doi: 10.1111/j.1365-2389.2011.01403.x
Lewin, G. R., Carlos, C., Chevrette, M. G., Horn, H. J., McDonald, B. R., Stankey, R. J., et al. (2016). Evolution and ecology of Actinobacteria and their bioenergy applications. Ann. Rev. Microbiol. 70, 235–254. doi: 10.1146/annurev-micro-102215-095748
Lin, X., Green, S., Tfaily, M. M., Prakash, O., Konstantinidis, K. T., Corbett, J. E., et al. (2012). Microbial community structure and activity linked to contrasting biogeochemical gradients in bog and fen environments of the glacial Lake Agassiz peatland. Appl. Environ. Microbiol. 78, 7023–7031. doi: 10.1128/AEM.01750-12
Lin, Y.-T., Jangid, K., Whitman, W. B., Coleman, D. C., and Chiu, C.-Y. (2011). Soil bacterial communities in native and regenerated Perhumid montane forests. Appl. Soil Ecol. 47, 111–118. doi: 10.1016/j.apsoil.2010.11.008
Lin, X., Malak, M., Tfaily, J., Megan, S., Patrick, C., and Kaitlin, E. (2014). “Microbial Community Stratification Linked to Utilization of Carbohydrates and Phosphorus Limitation in a Boreal Peatland at Marcell Experimental Forest, Minnesota, USA.” Applied and Environmental Microbiology 80, 3518–30. doi: 10.1128/AEM.00205-14
Liu, P., Pommerenke, B., and Conrad, R. (2018). Identification of Syntrophobacteraceae as major acetate-degrading sulfate reducing Bacteria in Italian Paddy soil. Environ. Microbiol. 20, 337–354. doi: 10.1111/1462-2920.14001
Luskin, M. S., and Potts, M. D. (2011). Microclimate and habitat heterogeneity through the oil palm lifecycle. Basic Appl Ecol 12, 540–551. doi: 10.1016/j.baae.2011.06.004
Marín, I., and Arahal, D. R. (2014). The family Beijerinckiaceae. Prokaryotes 9783642301971, 115–133. doi: 10.1007/978-3-642-30197-1_255
Melling, L. (2016). “Peatland in Malaysia” in Tropical peatland ecosystems. eds. M. Osaki and N. Tsuji (Tokyo: Springer Japan), 59–73.
Méndez-García, C., Peláez, A. I., Mesa, V., Sánchez, J., Golyshina, O. V., and Ferrer, M. (2015). Microbial diversity and metabolic networks in acid mine drainage habitats. Front. Microbiol. 6:475. doi: 10.3389/fmicb.2015.00475
Miettinen, J., and Liew, S. C. (2010). Degradation and development of peatlands in peninsular Malaysia and in the islands of Sumatra and Borneo since 1990. Land Degrad. Dev. 21, 285–296. doi: 10.1002/ldr.976
Miettinen, J., Shi, C., and Liew, S. C. (2016). Land cover distribution in the peatlands of peninsular Malaysia, Sumatra and Borneo in 2015 with changes since 1990. Glob Ecol Conserv 6, 67–78. doi: 10.1016/J.GECCO.2016.02.004
Muyzer, G., and Stams, A. J. M. (2008). The ecology and biotechnology of Sulphate-reducing Bacteria. Nat. Rev. Microbiol. 6, 441–454. doi: 10.1038/nrmicro1892
Newbery, D. M.., Kennedy, D. N., Petol, G. H., Madani, L., and Ridsdale, C. E. (1999). Primary forest dynamics in lowland dipterocarp forest at Danum Valley, Sabah, Malaysia, and the role of the understorey. hil. Trans. R. Soc. Lond. B 354, 1763–1782. doi: 10.1098/rstb.1999.0519
Nurulita, Y., Adetutu, E. M., Kadali, K. K., Shahsavari, E., Zul, D., Taha, M., et al. (2016). Assessment of the influence of oil palm and rubber plantations in tropical peat swamp soils using microbial diversity and activity analysis. J Agric Chem Environ 5, 53–65. doi: 10.4236/jacen.2016.52006
Oshiki, M., Satoh, H., and Okabe, S. (2016). Ecology and physiology of anaerobic ammonium oxidizing Bacteria. Environ. Microbiol. 18, 2784–2796. doi: 10.1111/1462-2920.13134
Pankratov, T. A., Dedysh, S. N., and Zavarzin, G. A. (2006). The leading role of Actinobacteria in aerobic cellulose degradation in Sphagnum peat bogs. Dokl. Biol. Sci. 410, 428–430. doi: 10.1134/S0012496606050243
Pankratov, T. A., Ivanova, A. O., Dedysh, S. N., and Liesack, W. (2011). Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat. Environ. Microbiol. 13, 1800–1814. doi: 10.1111/j.1462-2920.2011.02491.x
Parro, V., and Moreno-Paz, M. (2004). Nitrogen fixation in Acidophile Iron-oxidizing Bacteria: the Nif regulon of Leptospirillum Ferrooxidans. Res. Microbiol. 155, 703–709. doi: 10.1016/j.resmic.2004.05.010
Petersen, S. O., Hoffmann, C. C., Schäfer, C. M., Blicher-Mathiesen, G., Elsgaard, L., Kristensen, K., et al. (2012). Annual emissions of CH 4 and N 2O, and ecosystem respiration, from eight organic soils in Western Denmark managed by agriculture. Biogeosciences 9, 403–422. doi: 10.5194/bg-9-403-2012
Plugge, C. M., Zhang, W., Scholten, J. C. M., and Stams, A. J. M. (2011). Metabolic flexibility of sulfate-reducing Bacteria. Front. Microbiol. 2:81. doi: 10.3389/fmicb.2011.00081
Posa, M. R., Wijedasa, L. S., and Corlett, R. T. (2011). Biodiversity and conservation of tropical peat swamp forests. BioScience 61, 49–57. doi: 10.1525/bio.2011.61.1.10
Prasitwuttisak, W., Hoshiko, Y., Maeda, T., Haraguchi, A., and Yanagawa, K. (2022). Microbial community structures and methanogenic functions in wetland peat soils. Microbes Environ. 37:ME22004. doi: 10.1264/JSME2.ME22004
Ribeiro, K., Pacheco, F. S., Ferreira, J. W., de Sousa-Neto, E. R., Hastie, A., Krieger, G. C., et al. (2021). Tropical peatlands and their contribution to the global carbon cycle and climate change. Glob. Chang. Biol. 27, 489–505. doi: 10.1111/gcb.15408
Rinke, C., Chuvochina, M., Mussig, A. J., Chaumeil, P. A., Davín, A. A., Waite, D. W., et al. (2021). A standardized archaeal taxonomy for the genome taxonomy database. Nat. Microbiol. 6, 946–959. doi: 10.1038/s41564-021-00918-8
Selangor State Forestry Department (2014). Integrated management plan for North Selangor peat swamp Forest 2014–2023. Vol. 1. Selangor, Malaysia:Selangor State Forestry Department.
Sheremet, A., Jones, G. M., Jarett, J., Bowers, R. M., Bedard, I., Culham, C., et al. (2020). Ecological and genomic analyses of candidate phylum WPS-2 Bacteria in an Unvegetated soil. Environ. Microbiol. 22, 3143–3157. doi: 10.1111/1462-2920.15054
Sjögersten, S., Cheesman, A. W., Lopez, O., and Turner, B. L. (2011). Biogeochemical processes along a nutrient gradient in a tropical Ombrotrophic peatland. Biogeochemistry 104, 147–163. doi: 10.1007/s10533-010-9493-7
Smit, E., Leeflang, P., Gommans, S., Van Den Broek, J., Van Mil, S., and Wernars, K. (2001). Diversity and seasonal fluctuations of the dominant members of the bacterial soil Community in a Wheat Field as determined by cultivation and molecular methods. Appl. Environ. Microbiol. 67, 2284–2291. doi: 10.1128/AEM.67.5.2284-2291.2001
Sun, H., Terhonen, E., Koskinen, K., Paulin, L., Kasanen, R., and Asiegbu, F. O. (2014). Bacterial diversity and community structure along different peat soils in boreal Forest. Appl. Soil Ecol. 74, 37–45. doi: 10.1016/j.apsoil.2013.09.010
Swails, E., Jaye, D., Verchot, L., Hergoualc’h, K., Schirrmann, M., Borchard, N., et al. (2018). Will CO2 emissions from drained tropical peatlands decline over time? Links between soil organic matter quality, nutrients, and C mineralization rates. Ecosystems 21, 868–885. doi: 10.1007/s10021-017-0190-4
Tonks, A. J., Aplin, P., Beriro, D. J., Cooper, H., Evers, S., Vane, C. H., et al. (2016). Impacts of conversion of tropical peat swamp Forest to oil palm plantation on peat organic chemistry, physical properties and carbon stocks. Geoderma 289, 36–45. doi: 10.1016/j.geoderma.2016.11.018
Too, C. C., Keller, A., Sickel, W., Lee, S. M., and Yule, C. M. (2018). Microbial community structure in a Malaysian tropical peat swamp Forest: the influence of tree species and depth. Front. Microbiol. 9:2859. doi: 10.3389/FMICB.2018.02859/BIBTEX
Tripathi, B. M., Woojin Song, J. W. F., Slik, R. S., Sukri, S. J., Dong, K., and Adams, J. M. (2016). Distinctive tropical Forest variants have unique soil microbial communities, but not always low microbial diversity. Front. Microbiol. 7:376. doi: 10.3389/fmicb.2016.00376
Trivedi, P., Delgado-Baquerizo, M., Anderson, I. C., and Singh, B. K. (2016). Response of soil properties and microbial communities to agriculture: implications for primary productivity and soil health indicators. Front. Plant Sci. 7:990. doi: 10.3389/fpls.2016.00990
Troxler, T. G., Ikenaga, M., Scinto, L., Boyer, J. N., Condit, R., Perez, R., et al. (2012). Patterns of soil Bacteria and canopy community structure related to tropical peatland development. Wetlands 32, 769–782. doi: 10.1007/s13157-012-0310-z
Truu, M., Nõlvak, H., Ostonen, I., Oopkaup, K., Maddison, M., Ligi, T., et al. (2020). Soil bacterial and archaeal communities and their potential to perform N-cycling processes in soils of boreal forests growing on well-drained peat. Front. Microbiol. 11:358. doi: 10.3389/fmicb.2020.591358
Tsitko, I., Lusa, M., Lehto, J., Parviainen, L., Ikonen, A. T. K., Lahdenperä, A.-M., et al. (2014). The variation of microbial communities in a depth profile of an acidic, nutrient-poor boreal bog in southwestern Finland. Open J Ecol 4, 832–859. doi: 10.4236/oje.2014.413071
Urbanová, Z., and Bárta, J. (2016). Effects of long-term drainage on microbial community composition vary between peatland types. Soil Biol. Biochem. 92, 16–26. doi: 10.1016/j.soilbio.2015.09.017
Wang, Q., Garrity, GM., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–7. doi: 10.1128/AEM.00062-07
Warneke, C., Karl, T., Judmaier, H., Hansel, A., Jordan, A., Lindinger, W., et al. (1999). Acetone, methanol, and other partially oxidized volatile organic emissions from dead plant matter by Abiological processes: significance for atmospheric HOx chemistry. Global Biogeochem. Cycles 13, 9–17. doi: 10.1029/98GB02428
Wendlandt, K.-D., Stottmeister, U., Helm, J., Soltmann, B., Jechorek, M., and Beck, M. (2010). The potential of methane-oxidizing Bacteria for applications in environmental biotechnology. Eng. Life Sci. 10, 87–102. doi: 10.1002/elsc.200900093
Williams, R. T., and Crawford, R. L. (1983). Microbial diversity of Minnesota peatlands. Microb. Ecol. 9, 201–214. doi: 10.1007/BF02097737
Woodcroft, B. J., Singleton, C. M., Boyd, J. A., Evans, P. N., Emerson, J. B., Zayed, A. A. F., et al. (2018). Genome-centric view of carbon processing in thawing permafrost. Nature 560, 49–54. doi: 10.1038/s41586-018-0338-1
Xiang, X., Wang, R., Wang, H., Gong, L., Man, B., and Ying, X. (2017). Distribution of Bathyarchaeota communities across different terrestrial settings and their potential ecological functions. Sci. Rep. 7:28. doi: 10.1038/srep45028
Yu, T., Weichao, W., Liang, W., Lever, M. A., Hinrichs, K. U., and Wang, F. (2018). Growth of sedimentary Bathyarchaeota on lignin as an energy source. Proc. Natl. Acad. Sci. U. S. A. 115, 6022–6027. doi: 10.1073/pnas.1718854115
Yule, C. M., and Gomez, L. N. (2009). Leaf litter decomposition in a tropical peat swamp Forest in peninsular Malaysia. Wetl. Ecol. Manag. 17, 231–241. doi: 10.1007/s11273-008-9103-9
Zhang, C., Liu, G., Xue, S., and Wang, G. (2016). Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the loess plateau. Soil Biol. Biochem. 97, 40–49. doi: 10.1016/j.soilbio.2016.02.013
Zhao, M., Wang, M., Zhao, Y., Nanlin, H., Qin, L., Ren, Z., et al. (2022). Soil microbial abundance was more affected by soil depth than the altitude in peatlands. Front. Microbiol. 13:8504. doi: 10.3389/FMICB.2022.1068540
Keywords: microbial communities and structure, 16 rRNA, land-use change, peat swamp forest, oil palm plantation
Citation: Azizan SNF, Murakami S, McTaggart I, Yusof N, Sha’arani S, Hara H and Noborio K (2024) Changes in specific microbial groups characterize the impact of land conversion to oil palm plantations on peat. Front. For. Glob. Change. 7:1305491. doi: 10.3389/ffgc.2024.1305491
Edited by:
Selva Dhandapani, Agri-Food and Biosciences Institute, IrelandReviewed by:
Louis Lamit, Syracuse University, United StatesGusti Anshari, Tanjungpura University, Indonesia
Copyright © 2024 Azizan, Murakami, McTaggart, Yusof, Sha’arani, Hara and Noborio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Siti Noor Fitriah Azizan, YXppemFuLnNuZml0cmlhaEBvdXRsb29rLmNvbQ==
 Siti Noor Fitriah Azizan
Siti Noor Fitriah Azizan Shuichiro Murakami1
Shuichiro Murakami1 Iain McTaggart
Iain McTaggart