- 1College of Biological and Agricultural Sciences, Honghe University, Mengzi, Yunnan, China
- 2College of Resources and Environment, Yunnan Agricultural University, Kunming, Yunnan, China
The negative impacts of continuous cropping and long-term single crop planting on soil quality significantly restrict the high yield cultivation of perennial orchards. Intercropping can facilitate continuous cropping and improve the quality of the soil environment. However, it is still unclear whether the interplanting of faba bean in perennial orchards will increase the concentration of soil nutrients, change the composition of the soil microbial community, and increase the abundance of carbon (C) and nitrogen (N) cycling microorganisms. We interplanted faba beans in a perennial pomegranate orchard, and used sequencing and qPCR technology to study the effects on soil microbial diversity and C and N cycling genes. The results indicated that the interplanting of faba bean significantly increased the total N concentration by 28.6%, total phosphorus(P) concentration by 73.0% and available P concentration by 103.4%. The composition and structure of the soil microbial community were significantly changed, and the bacteria significantly enriched were Gaiellales and Rhizobiales at the order level and Nitrosomonadaceae at the family level. The fungi significantly enriched were Pezizomycetes at the class level, Pezizales and Sordariales at the order level, Ascodesmidaceae and Ophiocordycipitaceae at the family level, Cephaliophora, Parachaetomium, and Purpureocillium at the genus level, and Lilacinum, Lavendulum, Carinthiacum, Tropica, Chaetomium, and Delphinoides at the species level. The copy numbers of cbbL and nifH genes in soil were significantly increased by 79.9 and 168.5%, respectively. Changes in major nutrient elements explained 71.2% of the variance at the family level for bacteria and 46.0% of the variance at the family level for fungi. These results provided a scientific basis for the improvement of soil environmental quality and soil microorganisms by interplanting cash crops in perennial orchards.
Highlights
• Faba bean interplanting significantly increased the total N, total P, and available P concentrations in soil.
• Faba bean interplanting significantly changed the soil microbial community structure but had no significant effect on microbial diversity.
• Faba bean interplanting significantly increased the copy numbers of cbbL genes and nifH genes in soil.
1 Introduction
Pomegranate (Punica granatum L.) is a perennial deciduous shrub or small tree and is an important economic fruit tree in China. Pomegranate fruit is widely used for food, medical purposes, and organic synthesis, and has high economic value (Marathe et al., 2017). To meet increasing demand for the fruit, continuous cropping in recent years has led to soil hardening, decline of organic matter, decrease of soil microbial diversity, and aggravation of soil-borne diseases (Panneerselvam et al., 2020). Studies have shown that soil microecological imbalance, the accumulation of harmful microorganisms, and degradation of beneficial microbial populations are key problems preventing soil continuous cropping in pomegranate orchards (Kumar et al., 2020).
Soil microorganisms connect above-ground and subsurface ecosystems and help regulate ecosystem functions (De Corato, 2020; Vishwakarma et al., 2020). Long-term monoculture can decrease soil microbial diversity, decrease beneficial microbial population, and increase the harmful microbial population (Zou et al., 2019; Jin et al., 2020). Intercropping of gramineae and legumes can increase the quantity and population structure of soil microorganisms. Wheat-faba bean intercropping increased rhizosphere microbial activity, improved microbial diversity, and changed microbial community function (Lv et al., 2020). Intercropping of peanut and maize improved the number of species of N-fixing microorganisms in soil, changed their overall composition and structure, and changed the rhizosphere nutrient utilization status (Li et al., 2020). Changes in soil microbial activity can greatly affect soil N content. The amounts of N conversion function genes, such as nifH, amoA, nirK, and nirS, are commonly used to quantify the potential microbial activity of a specific N transformation process (Hao et al., 2022; Zhao et al., 2022; Frey et al., 2023). The nifH gene is carried by N-fixing bacteria and encodes ferritin subunits of N-fixing enzymes that catalyze biological N fixation, which increases N in soil and N uptake and utilization by plants (Burén et al., 2020). CO2 assimilation is the process by which atmospheric CO2 is fixed by autotrophic or heterotrophic microorganisms, including photosynthetic bacteria, chemoautotrophic bacteria, and algae in soil. The main C fixation pathway is the Calvin-Bassham-Benson cycle, and the key enzyme in this pathway is CO2-fixing enzyme ribulose diphosphate carboxylase/oxygenase. The majority of photoautotrophs and chemoautotrophs use this pathway to fix C, including most ammoniating bacteria involved in the N conversion process, and the key functional gene of this pathway is cbbL (Xu et al., 2019; Shi et al., 2023; Yao et al., 2023). Expression of functional genes (AOB, nirK) was increased in the intercropping system of alfalfa (Medicago sativa L.) and Siberian wildrye (Elymus sibiricus L.) (Mei et al., 2010; Qisong et al., 2018; Liao et al., 2020; Wang et al., 2020, 2022; Wu et al., 2020). The intercropping of grain/legumes may enhance symbiotic N fixation by increasing biological N fixation (Gong et al., 2019). The abundance of nifH was higher with interplanting than with monoculture (Chen et al., 2018). The addition of legumes to a cucumber intercropping system significantly improved the N fixation ability of soil, increased soil nifH gene abundance by 33.7%, and increased soil microbial biomass compared with monoculture (Gro et al., 2022).
Based on previous work described above, we proposed to improve soil nutrient availability and microbial community structure by interplanting faba bean in a pomegranate orchards. Faba bean is a widely grown leguminous crop in this area and has high economic value. However, it is unclear how the main nutrient elements of soil change, the response of soil microbial community structure, and the response of functional genes of soil N cycle and C cycle when faba beans are interplanted in pomegranate orchards. Therefore, the purpose of this study was to (1) study the effects of interplanting faba beans on the soil physicochemical properties of pomegranate orchards; (2) study the response of the soil microbial community in a pomegranate orchard after with faba bean inter-plantation; and (3) determine the effects of interplanting faba bean on functional genes of soil C and N cycling in pomegranate orchards. The results provide theoretical support to improve soil fertility and the microbial community structure by interplanting faba bean in pomegranate orchards.
2 Materials and methods
2.1 Description of the experimental site
2.1.1 Experimental location
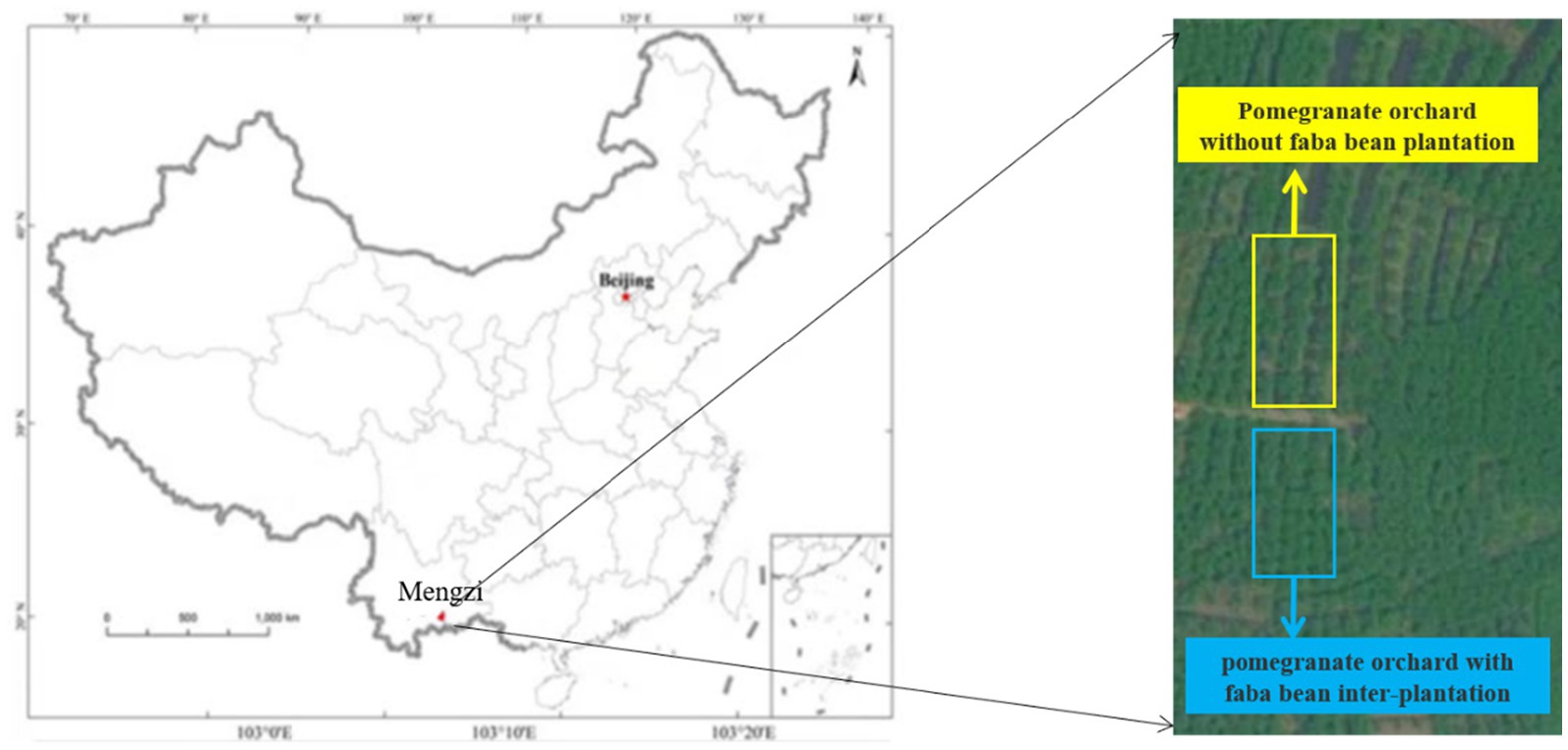
The field plot experiment was carried out in a pomegranate orchard in Wanmu, Xinansuo Town, Mengzi City, Yunnan Province, China (103.4291772E; 23.3710138 N) (Figure 1). Yunnan Mengzi city has subtropical low latitude plateau climate, annual average temperature of 18.6°C, extreme maximum temperature of 33.8°C, extreme minimum temperature of 2.9°C, frost-free period of 337 days, and annual rainfall of 815.8 mm.
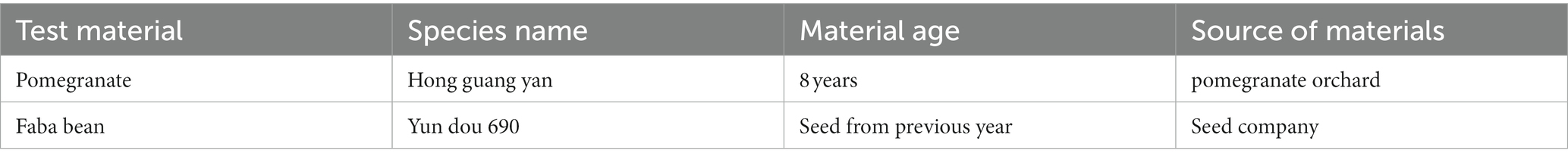
The test materials are described in Table 1.
2.2 Experimental design and implementation
The pomegranate orchard had been planted for 8 years, with fertilization performed each year, between the time of leaf germination and fruit harvest (Between April and October). Compound fertilizer was applied twice a year (application amount: 600 kg/ha), and urea was applied once a year (application amount: 600 kg/ha). Trace element fertilizer was applied twice a year (application amount: 300 kg/ha). The experiment was carried out after the pomegranate fruit harvest. During the experiment, neither the pomegranate orchard with faba bean nor those without faba bean were fertilized.
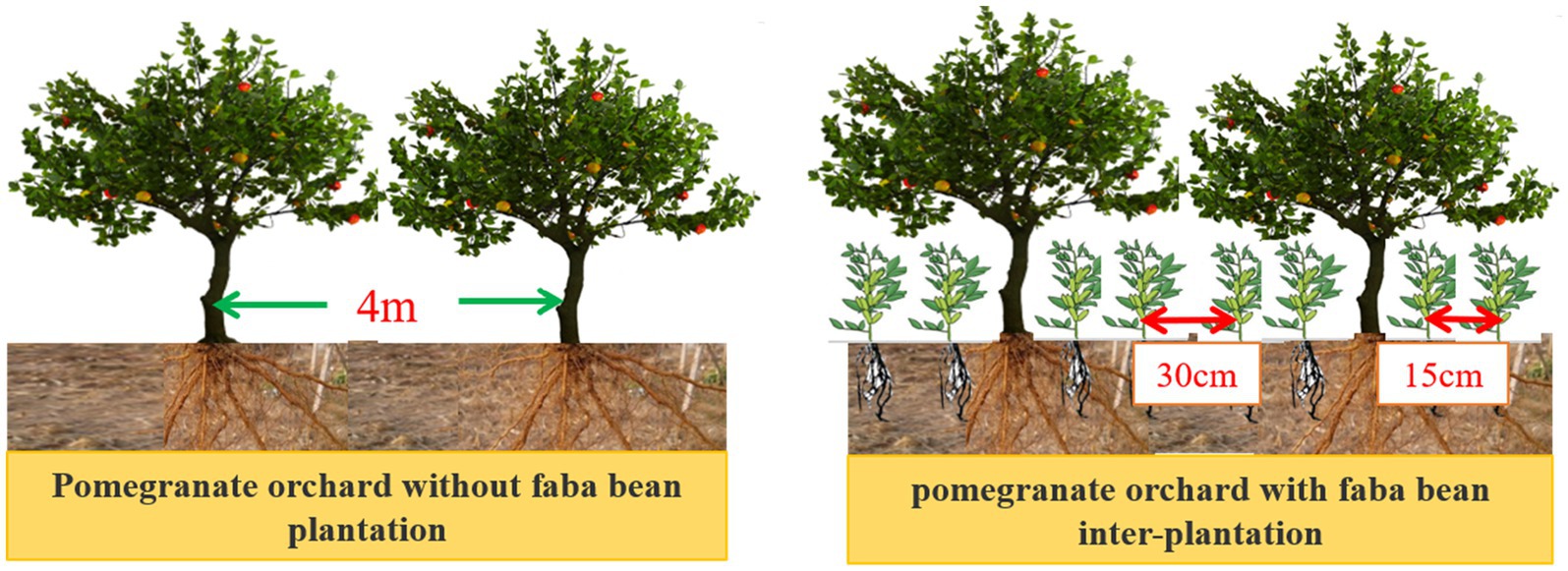
The 2,640 m2 pomegranate orchard was divided evenly into two halves, with one half planted with faba bean and the other without faba bean. With faba bean inter-plantation and without faba bean plantation are divided into two treatments, and each experimental group was divided into four repeat plots, each with an area of about 330 m2. During the experiment, the faba bean seed was sown on September 25, 2021 after harvesting the pomegranate fruit. Sowing was done with 16 rows of seeds between pomegranate trees, double rows of 15 cm × 15 cm, with rows of 30 cm (Figure 2). In the growing period, irrigation was carried out during the blooming and filling stages. Regular weeding and removal of aphids and leaf miner flies were performed for both treatments. Fresh pods were picked when the beans were ripe and the pod shells were tender green. The field trial ended on January 23, 2022.
2.3 Sample collection
The soil samples around the pomegranate trunk were collected from the plot without faba beans. For the plant with interplanting of faba beans, the soil between the planting of faba bean plants around the pomegranate trunk was collected. A soil drill was used to collect 0–30 cm of soil at each sampling site. Each soil sample is a mixture of soil samples from multiple sampling sites. Soil samples were stored in a sterile tube and stored at −80°C prior to soil microbial detection. The remaining soil samples were naturally air-dried, ground, and sifted through 1 and 0.25 mm mesh for determination of soil physical and chemical properties, respectively.
2.4 Soil physicochemical properties
Soil physicochemical properties of pH, available N, available P, total N, total P, and soil organic matter were measured as described previously (Zhang et al., 2022).
2.5 Soil bacterial and fungal community analysis
Extraction of soil DNA was performed using a FastDNA® Spin Kit (6560–200, MP Biomedicals, United States) according to the manufacturer’s instructions. ABI GeneAmp® 9700 PCR thermocycler (ABI, CA, United States) was used to amplify the bacterial 16S rRNA hypervariable region V3-V4 and the fungal ITS rRNA hypervariable region ITS1. Purified amplicons were pooled in equimolar amounts and paired-end sequenced on an Illumina Miseq PE300 platform (Illumina, San Diego, United States) according to standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). The detailed procedures are presented in Supplementary Data. The raw sequence data were uploaded to the NCBI sequence, Read Archive (SRA) with accession numbers PRJNA995182 and PRJNA995172.
2.6 Quantification of C and N cycling genes by qPCR
Primers AmoA-1F/AmoA-2R, PolyF/PolyR, and K2F/V2R were used for the amplification of the bacterial amoA, nifH, and cbbL genes, respectively (Rotthauwe et al., 1997; Nanba et al., 2004; Xu et al., 2019). Cloning of target genes, production of standard curves, real-time quantitative PCR experimental materials and instruments, PCR reaction conditions, and other calculations were as described (Zhang et al., 2022).
2.7 Data analysis
The bioinformatic analysis of the sequencing results was carried out according to previous methods (Zhang et al., 2022). The detailed procedures are presented in Supplementary Data. One-way ANOVA (p < 0.05) was performed among the different treatments with SPSS Statistics 16.0 (IBM, United States), and a value of p < 0.05 was considered a significant difference among different treatments. The figures were generated using OriginPro 2022b software (Origin Lab Corporation, Northampton, MA, United States).
3 Results
3.1 Effects of interplanting faba bean on basic soil properties
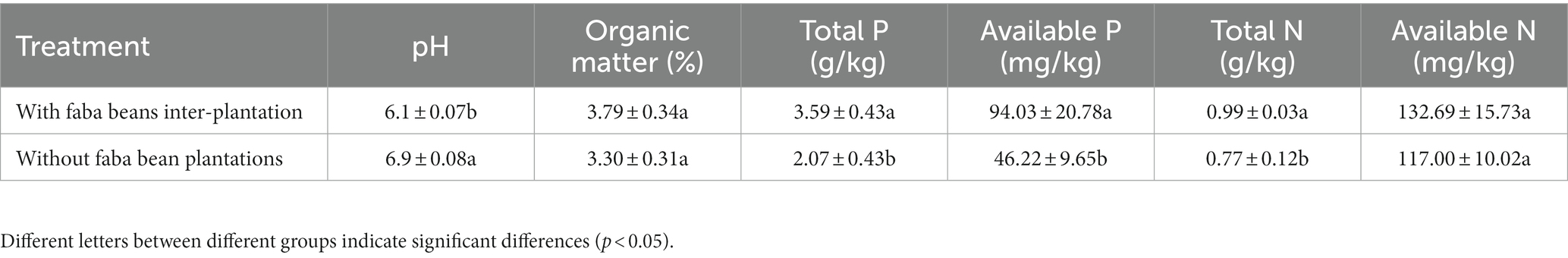
The concentrations of soil total P, organic matter, available P, and total N were increased by the interplanting of faba bean, and the soil pH was decreased. The total P concentration was increased by 73.0%, the available P concentration was increased by 103.4%, and the total N concentration was increased by 28.6% after interplanting (Table 2).
3.2 Response of soil microorganisms diversity to pomegranate/faba bean intercropping
3.2.1 Changes in soil microbial diversity
The Illumina MiSeq platform was used to conduct high-throughput sequencing of the V3-V4 region of the 16S rRNA gene of soil bacteria to determine changes in the soil bacterial community in response to interplanting faba bean. Overall, we obtained a total of 563,897 optimized sequences from eight soil samples, four from each treatment group. A total of 38 phyla, 112 classes, 226 orders, 414 families, 755 genera, 1,450 species, and 3,661 OTUs were included in the sample annotation analysis. High-throughput sequencing of ITS rRNA gene ITS1 region of soil fungi was performed to determine changes of the soil fungal community in response to interplanting faba bean. In total, we obtained a total of 510,559 optimized sequences from the eight soil samples. The sample annotation analysis included 10 phyla, 35 classes, 91 orders, 207 families, 411 genera, 623 species, and 1,617 OTUs. The Shannon and Chao indices of soil bacteria and fungi increased slightly, but the differences were not significant, compared with that without faba bean plantation (Figures 3A,B).
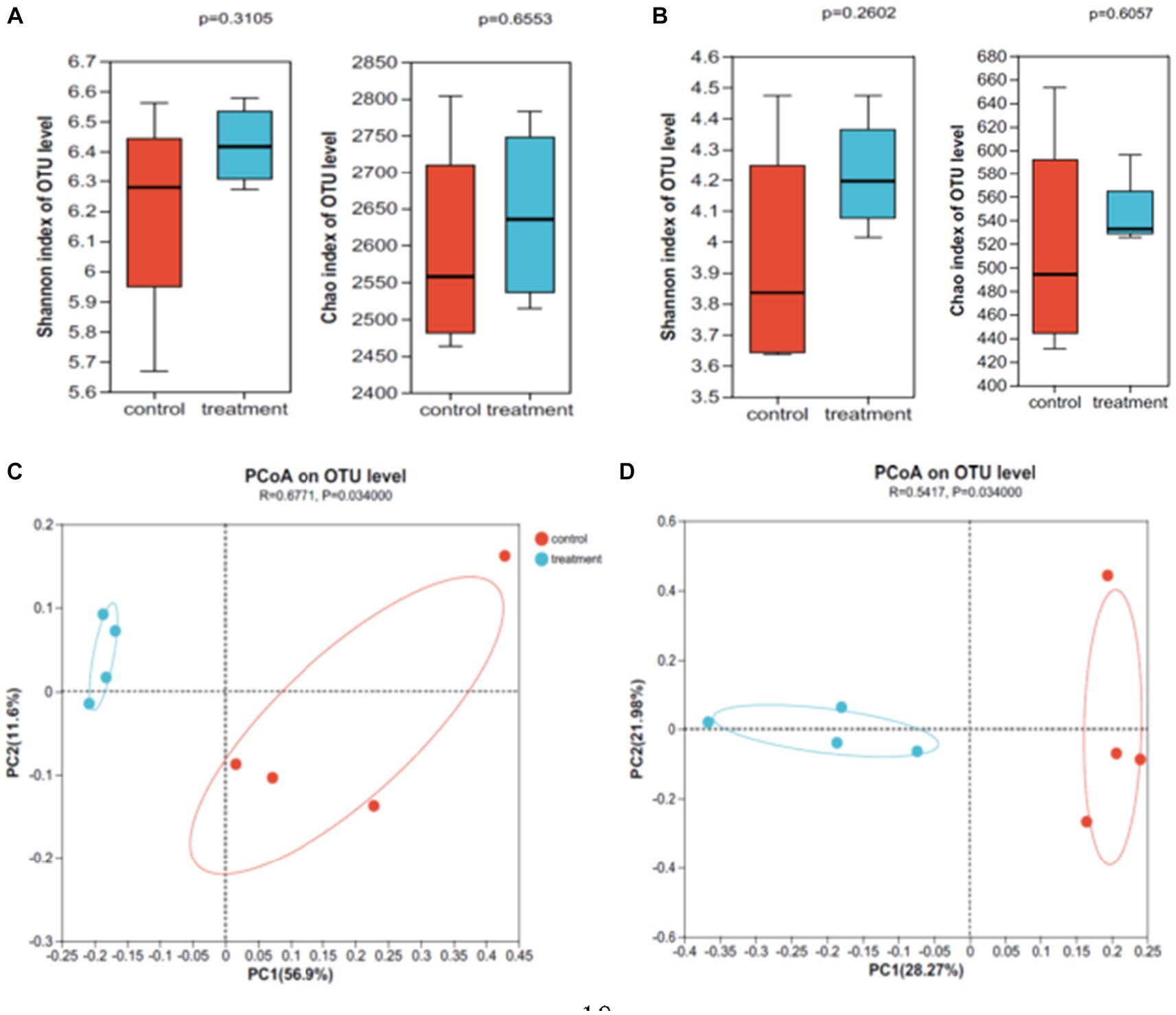
Figure 3. Soil microbial diversity (A,C: bacteria; B,D: fungi; control: without faba bean plantation, treatment: with faba bean inter-plantation; the same below).
The Braye Curtis distances were visualized by PCoA to assess differences in the microbial community structure between treatments. The results showed significant differences in soil bacteria and fungi between with faba bean inter-plantation and without faba bean plantation (R = 0.6771, p = 0.034; R = 0.5417, p = 0.034). In Figures 3C,D, with faba bean inter-plantation is plotted on the left side of the Y axis and without faba bean plantation is plotted on the right side of the Y axis. The separation of the two treatments indicates that the interplanting had a significant impact on both soil bacteria (Figure 3C) and fungi (Figure 3D). For soil bacteria, PC1 and PC2 accounted for 68.5% of the difference. For soil fungi, PC1 and PC2 together explained 50.2% of the difference.
3.2.2 Changes in soil microbial community composition
With faba bean inter-plantation resulted in a change in the composition of soil dominant flora. The distribution at the phylum level is shown in Figure 4. For soil bacteria, these sequences are divided into 39 phyla, as listed in Supplementary Table S1. Without faba bean plantation, the average proportion of Actinobacteriota in all soil samples reached 32.3%, accounting for the largest proportion of all bacteria in the phylum level. Other dominant phyla were Chloroflexi (20.3%), Proteobacteria (18.2%), Acidobacteriota (9.4%), Gemmatimonadota (7.3%), Firmicutes (4.3%), Myxococcota (2.3%), Bacteroidota (1.5%), Nitrospirota (1.0%), Methylomirabilota (0.6%), Verrucomicrobiota (0.6%), and Cyanobacteria (0.3%). with faba bean inter-plantation, the same bacteria were detected, but with some small differences in the abundances. The proportion of Actinobacteriota in all soil samples was still the highest, with an average of 35%. Other dominant phyla were Chloroflexi (17.5%), Proteobacteria (19.2%), Acidobacteriota (9.4%), Gemmatimonadota (3.9%), Firmicutes (4.3%), Myxococcota (2.3%), Bacteroidota (1.5%), Nitrospirota (1.0%), Methylomirabilota (1.3%), Cyanobacteria (0.9%), and Verrucomicrobiota (0.2%).
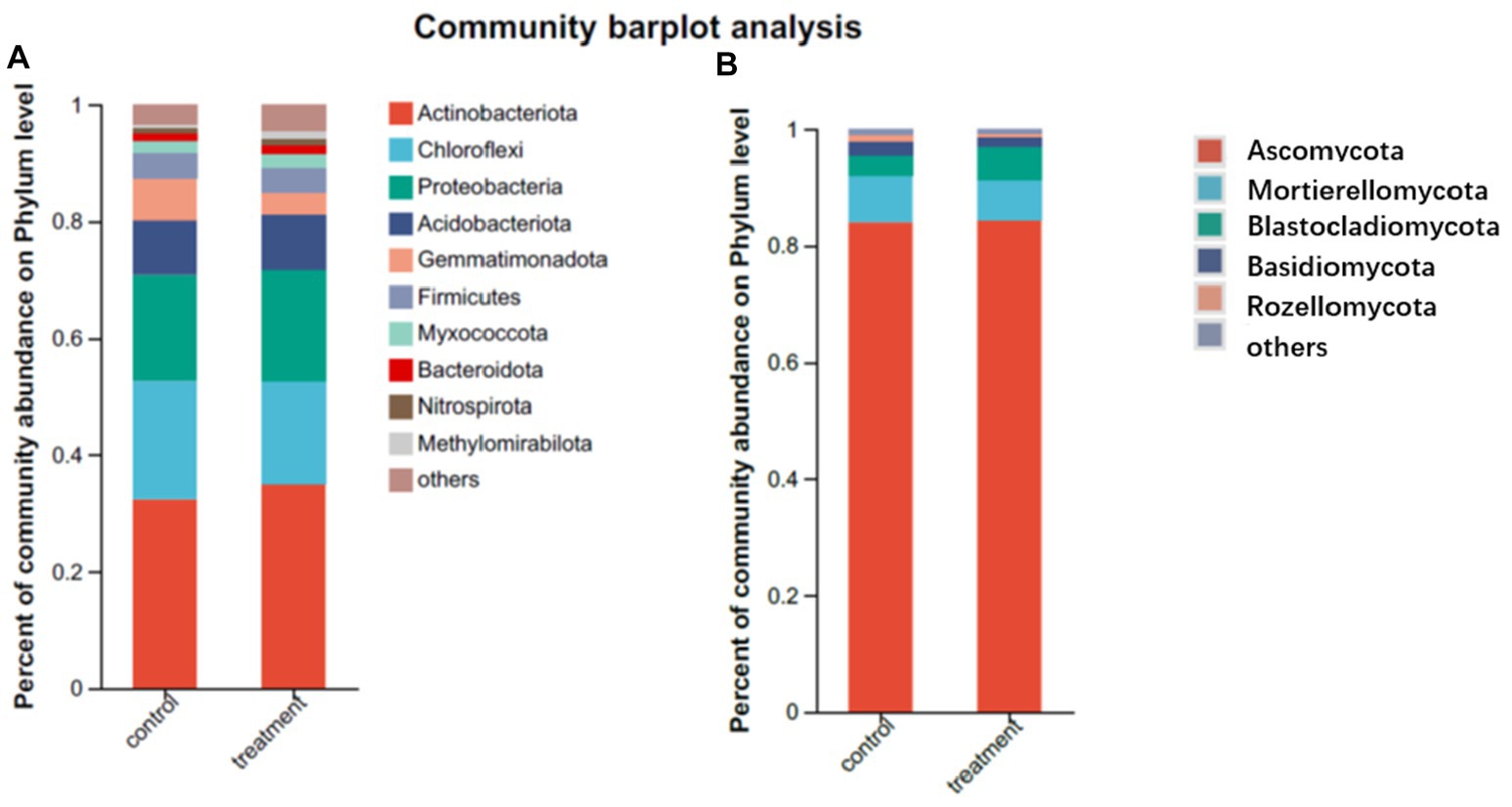
Figure 4. Composition of the rhizosphere soil microbial community of a pomegranate orchard with and without interplanting (A: bacteria; B: fungi).
For soil fungi, the obtained sequences were assigned to 10 phyla, as shown in Supplementary Table S1. Without faba bean plantations, the proportion of Ascomycota in soil fungi was the highest, reaching 84% on average. Other dominant phyla were Mortierellomycota (8.1%), Blastocladiomycota (3.4%), Basidiomycota (2.3%), Rozellomycota (1.2%), Chytridiomycota (0.6%), and Glomeromycota (0.3%). With interplanting of faba beans, the same fungi were detected, but there were slight changes in their relative abundances. The proportion of Ascomycota (84.4%) in soil fungi remained the highest. The other dominant phyla were Mortierellomycota (6.9%), Blastocladiomycota (5.6%), Basidiomycota (1.7%), Rozellomycota (0.5%), Chytridiomycota (0.5%), and Glomeromycota (0. 2%).
3.2.3 Microbial analysis of soil significance difference
LEfSe is software to discover high-dimensional biomarkers and revealing genomic features. The non-parametric factorial Kruskal-Wallis sum-rank test was used to detect features with significant abundance differences and find groups with significant differences in abundance. Finally, LEfSe based on linear discriminant analysis (LDA) was used to estimate the influence of the abundance of each species on the differential effect.
Linear discriminant analysis scores of 3.5 and higher were confirmed by LEfSe (Figure 5). Without faba bean plantation, the significantly enriched bacteria were Gemmatimonadota at the phylum level. At the class level, Gemmatimonadetes, Ktedonobacteriae, and Acidobacteriae were enriched. At the order level, Bacillales, Streptomycetales, Micromonosporales, Gemmatimonadales, and Ktedonobacteriales were enriched. At the family level, Micromonosporaceae, Gemmatimonadaceae, Streptomycetaceae, Ktedonobacteraceae, and Burkholderiaceae were enriched. At the genus level, Streptomyces, Hamadaea, Sphingomonas, Gemmatimonas, Caballeronia, Ktedonobacter, Calditerricola, and Micromonospora were enriched. With the interplanting of faba beans, the significantly enriched bacteria were Rhizobiales and Microtrichales at the order level. At the family level, Nitrosomonadaceae and Gaiellaceae were significantly enriched. At the genus level, Gaiella and Thiobacter were enriched.
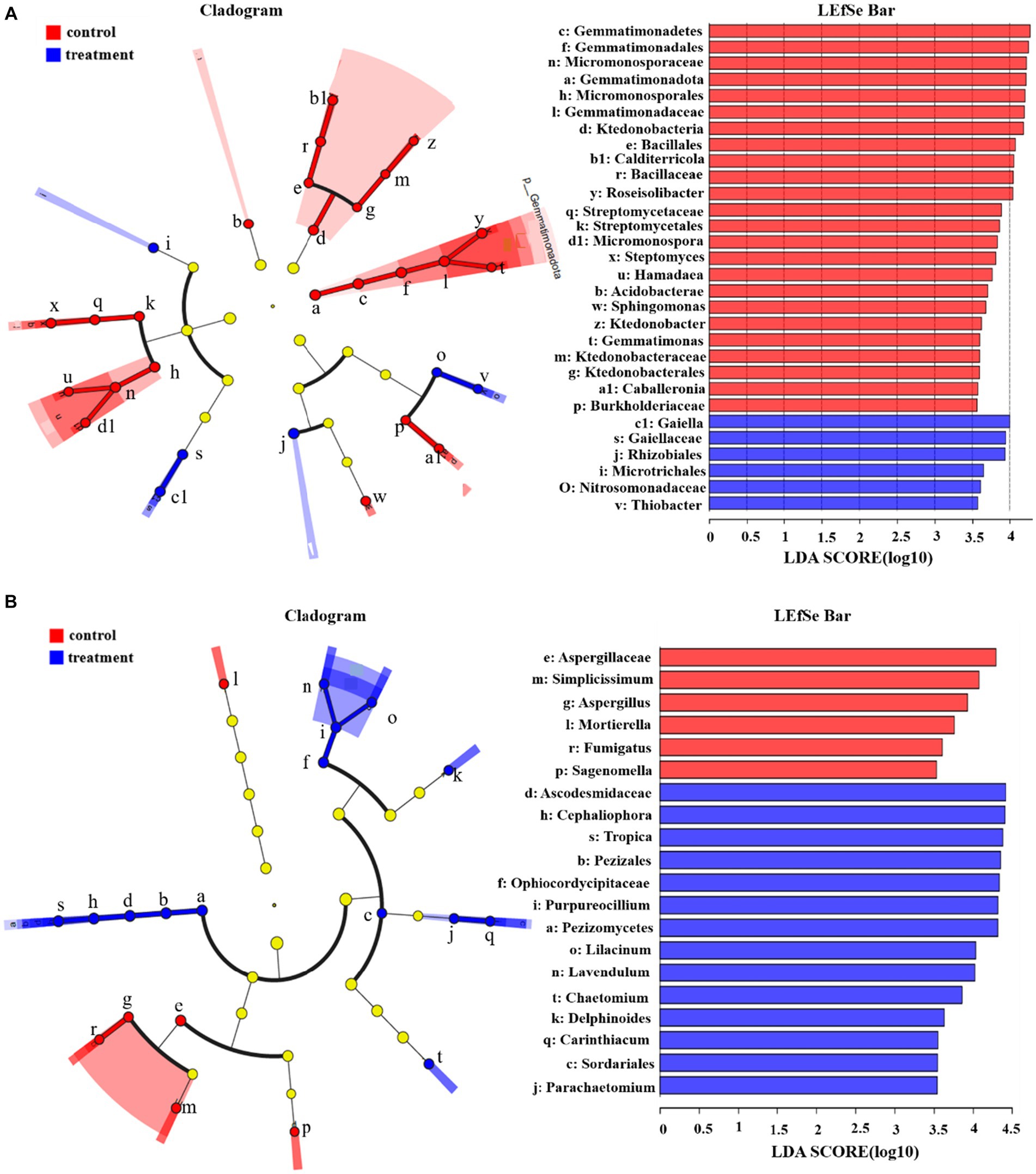
Figure 5. LEfSe diagram of soil microorganisms (A: bacteria; B: fungi; Only taxa meeting a linear discriminant analysis significance threshold of >3.5 are shown; different color nodes indicate microbial groups significantly enriched in the corresponding group. Light yellow nodes indicate that the microbiome does not differ significantly among different groups.).
The same analysis was performed for fungi, with LDA scores of 3.5 or higher confirmed by LEfSe. Without the planting of faba beans, the soil fungi that showed differences were Aspergillaceae at the family level and Aspergillus at the genus level. At the species level, Simplicissimum, Mortierella, Sagenomella, and Fumigatus were enriched. With faba bean planting, the differentially dominant fungi were Pezizomycetes at the class level, Pezizales and Sordariales at the order level, Acodesmidaceae and Ophiocordycipitaceae at the family level, Cephaliophora, Parachaetomium and Purpureocillium at the genus level, and Lilacinum, Lavendulum, Carinthiacum, Tropica, Chaetomium, and Delphinoides at the species level.
3.3 Relationship between soil properties and microbial community
Redundancy analysis (RDA) was used to analyze the relationships between environmental factors and the soil microbial community structure (Figure 6). The results showed that the eigenvalues of the level of soil bacteria in axis I and axis II were 59.39 and 11.84%, respectively, and the first two axes retained 71.23% of the total variance of soil bacteria, which was mainly determined by axis I. The eigenvalues of the level of soil fungi in axis I and axis II were 32.04 and 13.97%, respectively. The first two axes retained 46.01% of the total variance of soil bacteria, which was mainly determined by axis I.
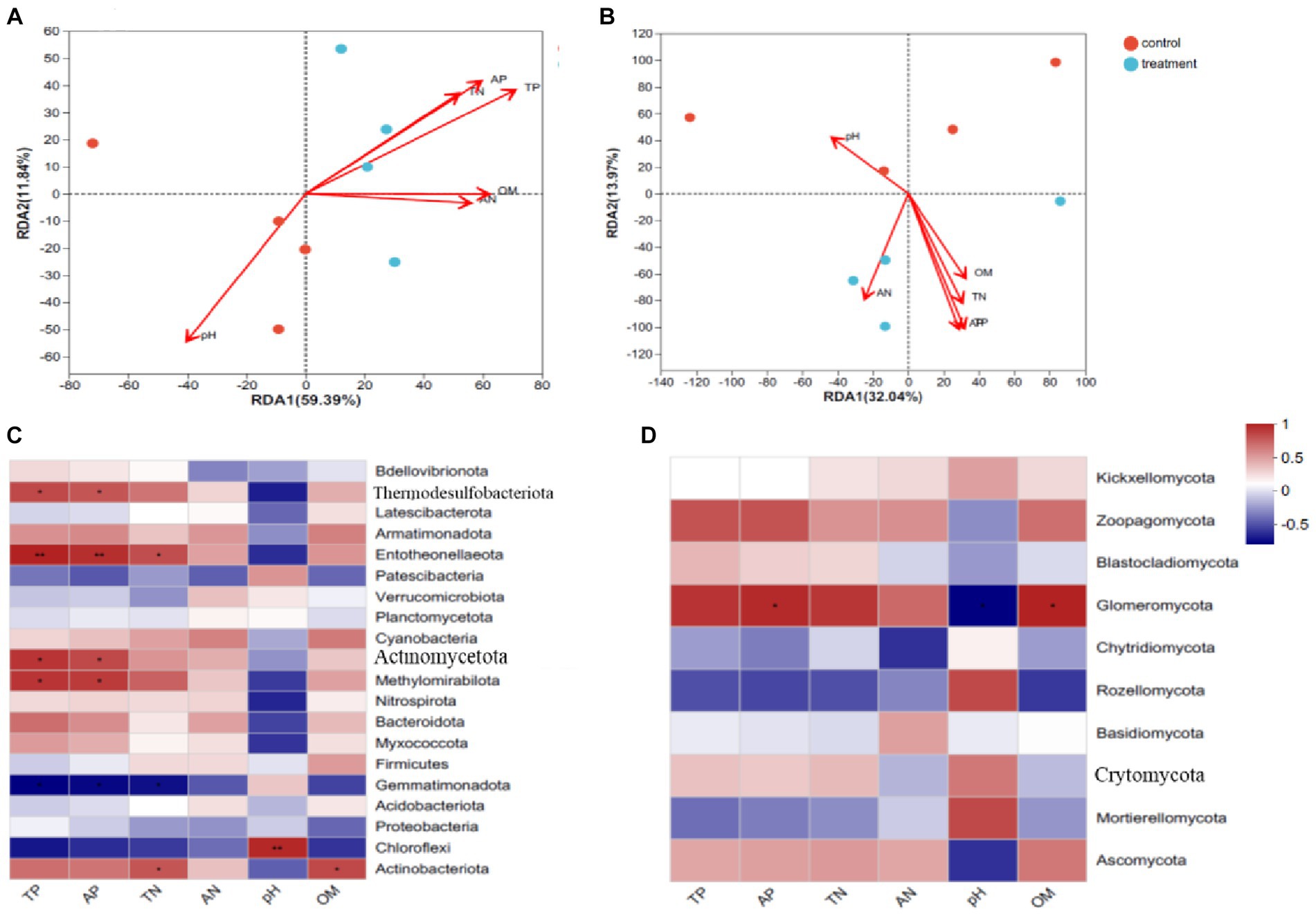
Figure 6. RDA revealed the relationships of soil microbial community of with soil physicochemical properties. The heatmaps show the correlation of soil properties with the dominant bacteria and fungi phyla. *Indicates a significant correlation between soil properties and microbes (A,C: bacteria; B,D: fungi).
The soil environmental factors were positively correlated with bacteria in the soil samples from interplanted faba bean soil, and negatively correlated with bacteria in the control treatment. For soil bacteria, the influence of environmental factors on bacterial community was greater than that of total P, available N, and organic matter. The effect of pH on the interplanted soil samples was negative, and the effect on the control soil samples was positive. For soil fungi, environmental factors (available N, total N, available P, total P, and organic matter) were positively correlated with interplanted faba bean soil samples, and negatively correlated without interplanted faba bean soil samples. The effects of total P and available P on fungal community were greater than those of total N, available N, and organic matter. The effect of pH on the interplanted faba bean soil samples was negative, and the effect on the control soil samples was positive.
There were several changes in soil bacteria. Entotheonellaeota was positively correlated with soil total P, available P, and total N. Chloroflexi exhibited a significant positive correlation with soil pH. Merhylomirabilota, Themodesulfobacteriota, and Actinomycetota were positively correlated with total P and available P. Actinobacteriota showed a significant positive correlation with soil pH and soil organic matter. Gemmatimonadota was negatively correlated with total P, available P, and total N. Among soil fungi, Glomeromycota was positively correlated with soil available P and organic matter.
3.4 Response of soil N and C cycling genes to pomegranate/faba bean intercropping
The copy numbers of amoA, nifH, and cbbL genes were increased by interplanting faba bean. There was a significant change in the copy number of the cbbL gene with interplanting, with an increase of 79.9%, from 4.4904E+08 copies/g of soil to 8.0778E+08 copies/g of soil (p < 0.05). There was also a significant increase in the copy number of the nifH gene with interplanting, from 1.4819E+08 copies/g of soil to 5.5185E+07 copies/g of soil, an increase of 168.5% (p < 0.05; Figure 7).
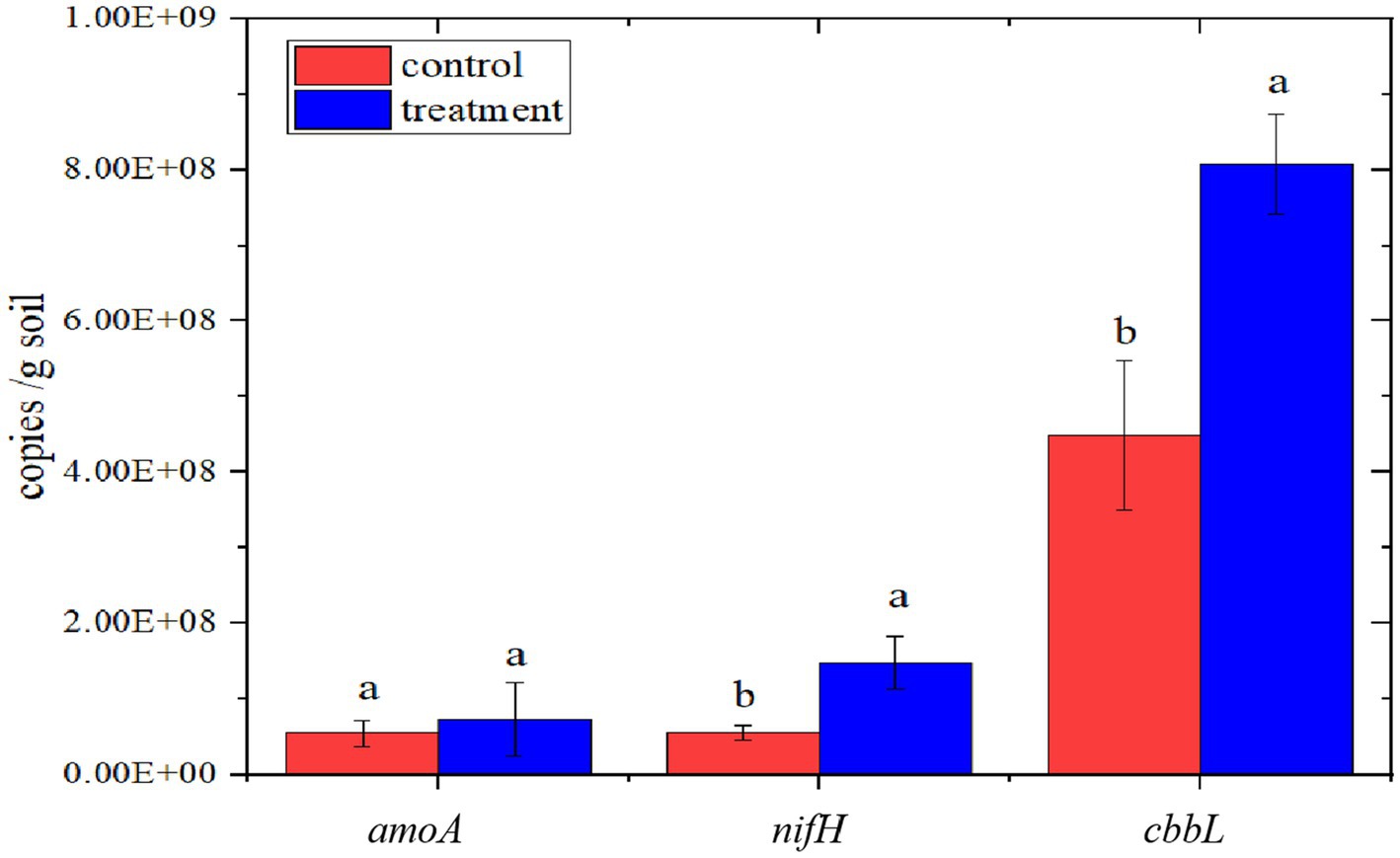
Figure 7. Copy numbers of soil C and N cycling genes. Different letters between different groups indicate significant differences (p < 0.05).
4 Discussion
4.1 Interplanting faba bean changed soil microbial community structure in pomegranate orchard
The results of our study indicated that intercropping of faba beans increased the Shannon and Chao1 indices of soil bacteria by 3.01 and 3.04%, respectively, and those of soil fungi by 7.0 and 5.22%, respectively (Figure 3). Plant diversity was previously shown to enrich soil microbial communities (Wu et al., 2020). Compared with monoculture, the α diversity, β diversity, and abundances of soil bacterial and fungal communities were increased under intercropping conditions. Previous work showed increased soil bacterial diversity with maize and peanut intercropping (Jin et al., 2020). The coexistence of different plants increased the complexity of soil microbial community and the diversity of soil microbial functions (Li et al., 2020). Foxtail millet and mung bean intercropping changed soil microbial diversity and foxtail millet rotation promoted soil bacterial diversity (Singh and Ahlawat, 2005). The microbial environment of foxtail millet rhizosphere soil was the best with rotation of foxtail millet and soybean (Arriagada et al., 2007). Continuous monocultures decrease the diversity of soil bacterial communities, and intercropping can increase the diversity of soil bacterial communities (Qiao et al., 2012).
Our results showed that faba bean inter-plantation changed the community structure of soil microorganisms (Figures 3C,D). The bacterial and fungal communities without faba bean plantation clustered together, while those with faba bean inter-plantation clustered together with significant differences. Soil microbial community has an important influence on soil ecosystem and the composition and function of soil microorganisms can change with the change of plant diversity (Curtright and Tiemann, 2021). Multi-dimensional scaling analysis (NMDS) and cluster analysis clearly showed clustering of the intercropping soil bacterial communities of maize and peanut, that differed from soil bacterial communities without intercropping (Chen et al., 2018).
Our results revealed different bacteria between soil with and without faba bean planting. With faba beans, the highest LDA scores were Rhizobiales and Nitrosomonadaceae, suggesting these are key bacteria for this treatment. The LDA score of Rhizobiales order was 4.0221, indicating significant enrichment. Nitrogen fixation is mainly carried out in the soil by N-fixing bacteria and archaea. Among N-fixing bacteria, Rhizobia can live in the soil as saprophytic bacteria or live as symbiotic bacteria in the root nodules of host legumes. Rhizobiales can also promote the dissolution and release of inorganic P in soil for a P solubilization effect (Cavalca et al., 2019; Zhao et al., 2022).
With faba bean inter-plantation, the soil differential dominant fungi were Pezizomycetes at the class level, Pezizales and Sordariales at the order level, and Ascodesmidaceae and Ophiocordycipitaceae at the family level. At the genus level, Cephaliophora, Parachaetomium, and Purpureocillium were enriched, and at the species level, Lilacinum, Lavendulum, Carinthiacum, Tropica, Chaetomium, and Delphinoides were enriched. The LDA values of Purpureocillium, Lilacinum, and Lavendulum were 4.2888, 4.0482, and 4.9407, respectively. These fungi can promote the dissolution of insoluble and difficult-to dissolve phosphate for P solublization (Baron et al., 2020; Girardi et al., 2022).
4.2 Interplanting faba bean changed the abundances of C and N cycling genes in the soil of pomegranate orchard
Interplanting of faba bean significantly increased the copy numbers of cbbL and nifH genes in soil by 79.9 and 168.5%, respectively (Figure 7). The expression level of nifH gene was significantly increased, indicating that interplanting of faba bean increased the number of C and N-fixing microorganisms in the soil of the pomegranate orchard. These changes should promote the fixation of C and N.
Fixation, nitrification, and denitrification of soil N are key biogeochemical processes in the soil N cycle, and each process is driven by associated microorganisms (Zhao et al., 2022). The nitrogenase nifH gene is a key marker gene (Yfantopoulos et al., 2022). The abundance of nifH genes in soil is influenced by plant species diversity. Crop rotation and shrub mix increased the copy number of the nifH gene and affected the soil microbial community structure through effects on soil P content (Hao et al., 2022). Short-term changes in the external environment can increase soil nifH gene abundance (Zhao et al., 2022). Crop diversification can affect the soil N cycle and its regulatory processes in terrestrial managed ecosystems and an increase of plant species can increase soil N fixation and denitrification genes to improve the accumulation of soil N (Yu, 2020; Hao et al., 2022). Changes in soil pH, ammonium N, and available P can significantly affect the soil bacterial community structure, including changes in the expression levels of soil N cycling genes (amoA, nirk, and nifH; Frey et al., 2023).
Fixed C microorganisms convert CO2 into organic matter in a process that is critical to the regeneration cycle of organic matter in the soil. The abundance and diversity of fixed C microbes can represent the potential of C fixation (Xu et al., 2019). The Calvin-Benson-Bassham cycle allows the absorption and conversion of C dioxide by light self-nourished organisms, and this cycle is regulated by Ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO), which is encoded by the cbbL gene (Yao et al., 2023). Melon/cowpea intercropping increased the contents of total organic C, total N and total ammonium in soil, increased the diversity of soil bacteria and the number of beneficial soil microorganisms, and increased the abundance of N cycle genes (Cuartero et al., 2022). Intercropping between Medicago sativa and Dactylis glomerata showed that intercropping altered soil N cycling community microorganisms and increased the abundance of N cycling functional genes (Zhao et al., 2017). Sugarcane/soybean intercropping changed the abundance of functional genes of soil C cycling function (Yu, 2020). Interplanting soybean in tea plantation could increase the abundance of functional genes related to soil C cycle (Wang et al., 2022).
Our study showed that the faba bean interplanting increased the concentration of soil organic matter, available P, total P, and total N, and decreased the soil pH value. Total P concentration was increased by 73% (p < 0.05), available P concentration was increased by 103.44% (p < 0.05) and total N concentration was increased by 28.6% (p < 0.05). Corn/faba bean intercropping releases a large number of protons into the rhizosphere through N fixation, resulting in a decrease in rhizosphere pH. During N fixation, faba beans absorb more cations than anions, and the roots release H+, which acidifies the soil (Chen et al., 2007). In soils, total N content is determined by processes of biological N fixation, soil organic N mineralization, nitrification, and denitrification, all of which are driven by soil microorganisms (Yfantopoulos et al., 2022; Zhao et al., 2022). Therefore, changes in soil microbial activity can have a large impact on soil N content. Grain/legumes intercropping may enhance symbiotic N fixation (Cen et al., 2023). The abundance of nifH genes in intercropping systems was higher than that in monoculture systems. The nifH gene encodes the ferritin subunit of the N-fixing enzyme that catalyzes biological N fixation. Nitrogen fixation can improve soil N availability and plant N status (Zeleke, 2019; Burén et al., 2020). Intercropping significantly increased the total N concentration and the abundance of nifH and amoA-AOB genes in soil planted with peanuts (Zhao et al., 2022).
4.3 The significance of interplanting faba bean to the production and management of pomegranate orchard
Interplanting legumes and other cash crops in a pomegranate orchard can effectively increase economic benefit, improve the soil quality of the pomegranate orchard, and alleviate the restrictions that arise from continuous cropping. Long-term cultivation of single crops can reduce soil microbial diversity and the numbers of beneficial microorganisms, resulting in an imbalance of soil microbial structure and the aggravation of soil-borne diseases (Kelderer et al., 2012; Urashima et al., 2012; Yang et al., 2012; Zhao et al., 2016). An increase of plant diversity can promote the increase of soil microbial diversity and decrease the abundance of soil pathogenic microorganisms (Qiao et al., 2012). In an intercropping system, crop root interaction can cause changes in soil microorganisms, and plant root exudates can attract more beneficial microorganisms to participate in nutrient cycling and inhibit the spread of soil-borne pathogens. In this study, the bacterial abundance decreased with the decrease of Proteobacteria abundance in the rhizosphere soil of a pomegranate orchard. Interplanting with faba bean increased the Shannon and Chao1 indices of both soil bacteria and fungi, significantly changed the soil microbial community structure, significantly enriched N-fixing and C-converting microorganisms, and significantly increased the copy numbers of cbbL and nifH genes in soil. These changes can alter the soil microbial community structure and improve soil C and N nutrition in orchards, and these results provide new ideas for the prevention and control of soil microbial structure imbalance and soil-borne diseases caused by long-term single continuous planting in orchards.
5 Conclusion
To study the response of soil microbial diversity and carbon and nitrogen cycling genes after interplanting, it is important to understand the effect of soil microbial reconstruction on soil fertility and microbial improvement after interplanting. The results of this study showed increased copy numbers of the soil carbon cycle cbbl gene and the nitrogen cycle nifH gene and increased concentrations of soil nitrogen and phosphorus after interplanting faba bean. These are changes that would allow a reduction in the amount of N and P fertilizer required. At the same time, there were significant changes of soil microbial community structure. This study confirmed the effect of interplanting faba bean on the improvement of soil carbon and nitrogen cycling microorganisms in a pomegranate orchard, and identified soil microbial community changes. These results provide a reference for the further popularization of this technology.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
DeZ: Writing – original draft, Supervision. DiZ: Data curation, Writing – review & editing. MW: Investigation, Writing – review & editing. XS: Data curation, Writing – review & editing. YC: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by the National Natural Science Foundation of China and the Science and Technology Program of Yunnan Province (grants 31860199 and 202001BA070001-190).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arriagada, C. A., Herrera, M. A., and Ocampo, J. A. (2007). Beneficial effect of saprobe and arbuscular mycorrhizal fungi on growth of Eucalyptus globulus co-cultured with Glycine max in soil contaminated with heavy metals. J. Environ. Manag. 84, 93–99. doi: 10.1016/j.jenvman.2006.05.005
Baron, N. C., Pollo, A. D. S., and Rigobelo, E. C. (2020). Purpureocillium lilacinum and Metarhizium marquandii as plant growth-promoting fungi. Peer 8:e9005. doi: 10.7717/peerj.9005
Burén, S., Jiménez-Vicente, E., Erasun, C. E., and Rubio, L. M. (2020). Biosynthesis of Nitrogenase cofactors. Chem. Rev. 120, 4921–4968. doi: 10.1021/acs.chemrev.9b00489
Cavalca, L., Zecchin, S., Zaccheo, P., Abbas, B., Rotiroti, M., Bonomi, T., et al. (2019). Exploring biodiversity and arsenic metabolism of microbiota inhabiting arsenic-rich Groundwaters in northern Italy. Front. Microbiol. 10:1480. doi: 10.3389/fmicb.2019.01480
Cen, Z., Zheng, Y., Guo, Y., Yang, S., and Dong, Y. (2023). Nitrogen fertilization in a Faba bean-wheat intercropping system can alleviate the autotoxic effects in Faba bean. Plants 12, 267–277. doi: 10.3390/plants12061232
Chen, J., Arafat, Y., Wu, L. K., Xiao, Z. G., Li, Q. S., Khan, M. A., et al. (2018). Shifts in soil microbial community, soil enzymes and crop yield under peanut/maize intercropping with reduced nitrogen levels. Appl. Soil Ecol. 124, 327–334. doi: 10.1016/j.apsoil.2017.11.010
Chen, Y. X., Zhang, F. D., Tang, L., Zheng, Y., Li, Y. J., Christie, P., et al. (2007). Wheat powdery mildew and foliar N concentrations as influenced by N fertilization and belowground interactions with intercropped faba bean. Plant Soil 291, 1–13. doi: 10.1007/s11104-006-9161-9
Cuartero, J., Pascual, J. A., Vivo, J. M., Özbolat, O., Sánchez-Navarro, V., Weiss, J., et al. (2022). Melon/cowpea intercropping pattern influenced the N and C soil cycling and the abundance of soil rare bacterial taxa. Front. Microbiol. 13, 545–567. doi: 10.3389/fmicb.2022.1004593
Curtright, A. J., and Tiemann, L. K. (2021). Intercropping increases soil extracellular enzyme activity: a meta-analysis. Agric. Ecosyst. Environ. 319, 107489–107567. doi: 10.1016/j.agee.2021.107489
De Corato, U. (2020). Soil microbiota manipulation and its role in suppressing soil-borne plant pathogens in organic farming systems under the light of microbiome-assisted strategies. Chem. Biolog. Technol. Agric. 7, 455–467. doi: 10.1186/s40538-020-00183-7
Frey, B., Moser, B., Tytgat, B., Zimmermann, S., Alberti, J., Biederman, L. A., et al. (2023). Long-term N-addition alters the community structure of functionally important N-cycling soil microorganisms across global grasslands. Soil Biol. Biochem. 176, 108887–107889. doi: 10.1016/j.soilbio.2022.108887
Girardi, N. S., Girardi, N. S., Sosa, A. L., Sosa, A. L., Etcheverry, M. G., Etcheverry, M. G., et al. (2022). Invitro characterization bioassays of the nematophagous fungus Purpureocillium lilacinum: evaluation on growth, extracellular enzymes, mycotoxins and survival in the surrounding agroecosystem of tomato. Fungal Biol. 126, 300–307. doi: 10.1016/j.funbio.2022.02.001
Gong, X., Li, C. J., Luo, J., Yang, Y., Zhang, Q. H., Yang, W. L., et al. (2019). Responses of rhizosphere soil properties, enzyme activities and microbial diversity to intercropping patterns on the loess plateau of China. Soil Tillage Res. 195, 104355–103356. doi: 10.1016/j.still.2019.104355
Gro, C., Hossen, S., Hartmann, H., Noll, M., and Borken, W. (2022). Biological nitrogen fixation and nif H gene abundance in deadwood of 13 different tree species. Biogeochemistry 161, 353–371. doi: 10.1007/s10533-022-00986-w
Hao, J., Feng, Y., Wang, X., Yu, Q., Zhang, F., Yang, G., et al. (2022). Soil microbial nitrogen-cycling gene abundances in response to crop diversification: a meta-analysis. Sci. Total Environ. 838:156621. doi: 10.1016/j.scitotenv.2022.156621
Jin, X., Shi, Y. J., Wu, F. Z., Pan, K., and Zhou, X. G. (2020). Intercropping of wheat changed cucumber rhizosphere bacterial community composition and inhibited cucumber fusarium wilt disease. Sci. Agric. 77, 5678–5689. doi: 10.1590/1678-992x-2019-0005
Kelderer, M., Manici, L. M., Caputo, F., and Thalheimer, M. (2012). Planting in the 'inter-row' to overcome replant disease in apple orchards: a study on the effectiveness of the practice based on microbial indicators. Plant Soil 357, 381–393. doi: 10.1007/s11104-012-1172-0
Kumar, A., Sharma, J., Munjal, V., Sakthivel, K., Thalor, S. K., Mondal, K. K., et al. (2020). Polyphasic phenotypic and genetic analysis reveals clonal nature of Xanthomonas axonopodis pv. Punicae causing pomegranate bacterial blight. Plant Pathol. 69, 347–359. doi: 10.1111/ppa.13128
Li, Y., Feng, J., Zheng, L., Huang, J., and Li, X. (2020). Intercropping with marigold promotes soil health and microbial structure to assist in mitigating tobacco bacterial wilt. J. Plant Pathol. 102, 731–742. doi: 10.1007/s42161-020-00490-w
Li, Y., Li, Z., Chang, S. X., Cui, S., and Cai, Y. (2020). Residue retention promotes soil carbon accumulation in minimum tillage systems: implications for conservation agriculture. Sci. Total Environ. 740:140147. doi: 10.1016/j.scitotenv.2020.140147
Liao, H., Qin, F., Wang, K., Zhang, Y., Hao, X., Chen, W., et al. (2020). Long-term chemical fertilization-driving changes in soil autotrophic microbial community depresses soil CO (2) fixation in a Mollisol. Sci. Total Environ. 748:141317. doi: 10.1016/j.scitotenv.2020.141317
Lv, J. X., Dong, Y., Dong, K., Zhao, Q., Yang, Z. X., and Chen, L. (2020). Intercropping with wheat suppressed fusarium wilt in faba bean and modulated the composition of root exudates. Plant Soil 448, 153–164. doi: 10.1007/s11104-019-04413-2
Marathe, R. A., Sharma, J., Murkute, A. A., and Babu, K. D. (2017). Response of nutrient supplementation through organics on growth, yield and quality of pomegranate. Sci. Hortic. 214, 114–121. doi: 10.1016/j.scienta.2016.11.024
Mei, S. Y., Nan, Z. N., Tao, W. E., Li, Y. H., Shui, Y. J., and Xin, C. W. (2010). Influence of intercropping and intercropping plus rhizobial inoculation on microbial activity and community composition in rhizosphere of alfalfa (Medicago sativa L.) and Siberian wild rye (Elymus sibiricus L.). Fems microbiology. Ecology 70, 218–226. doi: 10.1111/j.1574-6941.2009.00752.x
Nanba, K., King, G. M., and Dunfield, K. (2004). Analysis of facultative Lithotroph distribution and diversity on volcanic deposits by use of the large subunit of ribulose 1, 5-bisphosphate carboxylase/oxygenase. Appl. Environ. Microbiol. 70, 2245–2253. doi: 10.1128/AEM.70.4.2245-2253.2004
Panneerselvam, P., Selvakumar, G., Ganeshamurthy, A. N., Mitra, D., and Senapati, A. (2020). Enhancing pomegranate (Punica granatum L.) plant health through the intervention of a Streptomyces consortium. Biocontrol Sci. Tech. 31, 430–442. doi: 10.1080/09583157.2020.1859095
Qiao, Y. J., Li, Z. Z., Wang, X., Zhu, B., and Zeng, Z. H. (2012). Effect of legume-cereal mixtures on the diversity of bacterial communities in the rhizosphere. Plant Soil Environ. 58, 174–180. doi: 10.17221/351/2011-PSE
Qisong, L., Jun, C., Linkun, W., Xiaomian, L., Na, L., and Yasir, A. (2018). Belowground interactions impact the soil bacterial community, soil fertility, and crop yield in maize/Peanut intercropping systems. Int. J. Mol. Sci. 19, 3345–3356. doi: 10.3390/ijms19020622
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The ammonia monooxygenase structural gene amo a as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712. doi: 10.1128/aem.63.12.4704-4712.1997
Shi, M., Li, J., Gao, R., Song, X., Wang, G., Gao, Y., et al. (2023). Contrasting effects of elevated CO2 on autotrophic prokaryotes with different CO2 fixation strategies in tea plantation soil. Biol. Fertil. Soils 59, 205–215. doi: 10.1007/s00374-023-01700-0
Singh, D. P., and Ahlawat, I. P. S. (2005). Greengram (Vigna radiata) and blackgram (V-Mungo) improvement in India past, present and future prospects. Indian J. Agric. Sci. 75, 243–250. doi: 10.1016/j.compag.2004.11.014
Urashima, Y., Sonoda, T., Fujita, Y., and Uragami, A. (2012). Application of PCR-denaturing-gradient gel electrophoresis (DGGE) method to examine microbial community structure in Asparagus fields with growth inhibition due to continuous cropping. Microbes Environ. 27, 43–48. doi: 10.1264/jsme2.ME11222
Vishwakarma, K., Kumar, N., Shandilya, C., Mohapatra, S., Bhayana, S., and Varma, A. (2020). Revisiting plant-microbe interactions and microbial consortia application for enhancing sustainable agriculture: a review. Front. Microbiol. 11:560406. doi: 10.3389/fmicb.2020.560406
Wang, T., Duan, Y., and Liu, G. (2022). Tea plantation intercropping green manure enhances soil functional microbial abundance and multifunctionality resistance to drying-rewetting cycles. Sci. Total Environ. 810, 151282–151292. doi: 10.1016/j.scitotenv.2021.151282
Wang, Y., Huang, Q., Liu, C., Ding, Y., and Zhao, Z. (2020). Mulching practices alter soil microbial functional diversity and benefit to soil quality in orchards on the loess plateau. J. Environ. Manag. 271:110985. doi: 10.1016/j.jenvman.2020.110985
Wu, H. M., Lin, M. H., Rensing, C., Qin, X. J., Zhang, S. K., Chen, J., et al. (2020). Plant-mediated rhizospheric interactions in intraspecific intercropping alleviate the replanting disease of Radix pseudostellariae. Plant Soil 454, 411–430. doi: 10.1007/s11104-020-04659-1
Xu, Y., Wang, T., Li, H., Ren, C., Chen, J., Yang, G., et al. (2019). Variations of soil nitrogen-fixing microorganism communities and nitrogen fractions in a Robinia pseudoacacia chronosequence on the loess plateau of China. Catena 174, 316–323. doi: 10.1016/j.catena.2018.11.009
Yang, J. I., Ruegger, P. M., Mckenry, M. V., Ole, B. J., James, B., and Yang, C. H. (2012). Correlations between root-associated microorganisms and peach replant disease symptoms in a California soil. PLoS One 7:e46420. doi: 10.1371/journal.pone.0046420
Yao, Y., Shen, X., Wang, L., Zhao, J., Gong, L., Wang, S., et al. (2023). Effects of tillage management on cbb L-carrying bacteria and soil organic carbon dynamics across aggregate size classes in the farmland of North China plain. Ecol. Indic. 150, 110213–114466. doi: 10.1016/j.ecolind.2023.110213
Yfantopoulos, D., Ntatsi, G., Gruda, N., Bilalis, D., and Savvas, D. (2022). Effects of the preceding crop on soil N availability, biological nitrogen fixation, and fresh pod yield of organically grown Faba bean (Vicia faba L.). Horticulturae 8, 5667–5676. doi: 10.3390/horticulturae8060496
Yu, Y. W. J. (2020). The soil carbon cycle determined by geo Chip 5.0 in sugarcane and soybean intercropping systems with reduced nitrogen input in South China. Appl. Soil Ecol. 155, 103653–103406. doi: 10.1016/j.apsoil.2020.103653
Zeleke, K. T. (2019). Aqua crop calibration and validation for Faba bean (Vicia faba L.) under different agronomic managements. Agronomy 9, 4423–4432. doi: 10.3390/agronomy9060320
Zhang, D., Lei, M., Wan, X., Guo, G., Zhao, X., and Liu, Y. (2022). Responses of diversity and arsenic-transforming functional genes of soil microorganisms to arsenic hyperaccumulator (Pteris vittata L.)/pomegranate (Punica granatum L.) intercropping. Sci. Total Environ. 850:157767. doi: 10.1016/j.scitotenv.2022.157767
Zhao, M., Jones, C. M., and Meijer, J. (2017). Intercropping affects genetic potential for inorganic nitrogen cycling by root-associated microorganisms in Medicago sativa and Dactylis glomerata. Appl. Soil Ecol. 119, 260–266. doi: 10.1016/j.apsoil.2017.06.040
Zhao, Y. P., Lin, S., Chu, L., Gao, J. T., Azeem, S., and Lin, W. (2016). Insight into structure dynamics of soil microbiota mediated by the richness of replanted Pseudostellaria heterophylla. Sci. Rep. 6:26175. doi: 10.1038/srep26175
Zhao, F., Sun, Z., Feng, L., Zhang, Y., Feng, C., Bai, W., et al. (2022). Biological N fixation but not mineral N fertilization enhances the accumulation of N in peanut soil in maize/peanut intercropping system. J. Agric. Food Res. 10, 100365–133355. doi: 10.1016/j.jafr.2022.100365
Keywords: intercropping, soil microbial community, N cycling genes, C cycling genes, pomegranate
Citation: Zhang D, Zhu D, Wang M, Shi X and Chen Y (2023) Responses of diversity and carbon and nitrogen cycling genes of soil microorganisms to pomegranate (Punica granatum L.)/faba bean (Vicia faba L.) intercropping. Front. For. Glob. Change. 6:1295732. doi: 10.3389/ffgc.2023.1295732
Edited by:
Andreas Schindlbacher, Austrian Research Center for Forests (BFW), AustriaReviewed by:
Mubasher Nasir, Northwest A&F University, ChinaAndrea Visca, Italian National Agency for New Technologies, Energy and Sustainable Economic Development (ENEA), Italy
Copyright © 2023 Zhang, Zhu, Wang, Shi and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongchuan Chen, Y2hlbnlvbmdjaHVhbjc3NEBzb2h1LmNvbQ==
 Degang Zhang1
Degang Zhang1 Yongchuan Chen
Yongchuan Chen