
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. For. Glob. Change , 27 October 2023
Sec. Pests, Pathogens and Invasions
Volume 6 - 2023 | https://doi.org/10.3389/ffgc.2023.1240804
This article is part of the Research Topic "Forentomics": forest pest and pathogen biology, ecology, and management using omics View all 10 articles
Termites play an important role as decomposers of organic matter in forests by utilizing their gut symbionts and associated carbohydrate-active enzymes (CAZymes) to digest wood materials. However, there is a limited understanding of the entire repertoire of CAZymes and their evolution in termite genomes. Here we identified the gene families of CAZymes in publicly available termite genomes and analyzed the evolution of abundant gene families. We found that 79 CAZyme gene families from the carbohydrate-binding module and four CAZyme classes, including glycosyl transferase (GT), glycoside hydrolase (GH), auxiliary activity (AA) and carbohydrate esterase (CE), were present in termites with minor variations across termite species except for a few gene families. The gene trees of the large and conserved gene families have several groups of genes from all species, and each group encodes enzymes with complete corresponding domains. Three gene families, namely GT1, GH1 and AA3, exhibited significant variations in gene numbers and experienced several losses and a few duplications, which might be related to their rich gut symbionts and newly gained functions. Furthermore, the overall expression of CAZymes appears to have a caste- and tissue-specific pattern, reflecting a division of labor in termite colonies. Overall, these results reveal a likely stable CAZyme repertoire in termites and pave the way for further research on the functional contribution of termites to wood digestion.
Termites are important decomposers of organic matter. They can digest recalcitrant plant materials predominantly composed of lignocellulose, contributing to the decomposition of more than half of the dead wood in tropical and subtropical forests (Griffiths et al., 2019; Wu et al., 2021). In addition to their essential role in nutritional cycling, they also can influence soil moisture in tropical forests via their mounds (Ashton et al., 2019). However, due to their wood digestion ability, termites cause damage to wooden constructions and crops, with an estimated 40 billion United States dollars of global annual economic losses (Rust and Su, 2012; Kalleshwaraswamy et al., 2022).
Originating from a wood-feeding ancestor, they can efficiently digest recalcitrant lignocellulose, which relies on a large number of carbohydrate digestion enzymes produced by their associated symbionts. Because of their diversified feeding habits, termites harbor diverse gut microbiota, producing abundant CAZymes to adapt to their feeding preferences (Arora et al., 2022). In lower termites the digestion ability mainly relies on the protists and associated bacteria in their guts. While in higher termites their digestion ability depends on their symbiotic bacteria (Brune, 2014; Brune and Dietrich, 2015; Arora et al., 2022) because of the loss of the protists during their evolution (Bucek et al., 2019). These symbiotic microbes can possess a diverse repertoire of carbohydrate-active enzymes (CAZymes), including plant cell wall digestion enzymes. CAZymes are generally classified into five classes: glycoside hydrolases (GHs), glycosyl transferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs) and auxiliary activities (AAs). The termite symbionts possess a wide array of gene sets encoding active functional enzymes from these classes (Tartar et al., 2009; Marynowska et al., 2017; Hervé et al., 2020; Arora et al., 2022). Although the cellulolytic symbionts have been extensively investigated in termite digestion, the CAZyme genes originating from termites have gained traction since the discovery of the first cellulase in termites (Watanabe et al., 1998).
In addition to their symbionts, termites have specific CAZyme genes contributing to lignocellulose digestion. The combined efforts of both termite enzymes and those of their symbionts allow efficient digestion of wooden materials (Poulsen et al., 2014; Arora et al., 2022). Among them, endo-β-1,4-glucanases and β-glucosidases have been widely studied in cellulose hydrolysis. Both genes have multiple copies in each available termite genome (Tokuda, 2019) and were expressed mainly in salivary glands and gut with different expression patterns in different species (Tokuda et al., 2004; Fujita et al., 2008). For example, an endo-β-1,4-glucanase was specifically expressed in the salivary glands of lower termites, while it was mainly expressed in the midguts of higher termites (Tokuda et al., 2004). Similarly, a digestive β-glucosidase was explicitly expressed in the salivary glands of a lower termite Neotermes koshunensis (Shiraki) and the salivary glands and midguts of a higher termite Nasutitermes takasagoensis (Tokuda et al., 2002, 2009). Furthermore, due to the division of labor within the reproductive caste system, different termite castes showed different enzymatic activity. For instance, in a lower termite, Hodotermopsis sjostesti, the expression of endo-β-1,4-glucanase was higher in workers than soldiers (Fujita et al., 2008). In addition, other CAZyme genes were also characterized in termite species, such as chitin metabolism-related lytic polysaccharide monooxygenases (LPMOs) and auxiliary activity (AA) 15 in Coptotermes gestroi (Cairo et al., 2020).
Both well-studied gene families, endo-β-1,4-glucanases and β-glucosidases, have ancient origins (Davison and Blaxter, 2005; Shelomi et al., 2020; He et al., 2022). However, some of these genes have also gained functions other than digestion during termite evolution. A fascinating example is a GH1 β-glucosidase gene that is specifically expressed in the accessory glands of female ovaries (Shigenobu et al., 2022) and suppresses the production of new female reproductives in the colony (Korb et al., 2009; Zhang et al., 2012). In addition, another GH1 gene has been implicated in the recognition of termite eggs (Matsuura et al., 2009), although the gene is still not characterized.
With the advent of omics technologies, including genomics and transcriptomics, the CAZyme genes have been comprehensively investigated in several termite species (Yuki et al., 2008; Tartar et al., 2009; Zhang et al., 2012; Poulsen et al., 2014; Korb et al., 2015; Geng et al., 2018; Shigenobu et al., 2022). However, a systematic analysis of the evolution of the CAZyme gene families is still lacking. Currently genomes of 5 termite species belonging to four of the seven termite families are publicly available, including Zootermopsis nevadensis in Archotermopsidae, Cryptotermes secundus in Kalotermitidae, Reticulitermes speratus and Coptotermes formosanus in Rhinotermitidae, Macrotermes natalensis in termitidae. The former four termite species are wood-feeding lower termites, whereas M. natalensis is a fungus cultivating higher termites. In this study, we take advantage of these publicly available genomes to compare the digestion enzyme repertoire in five termite species to reveal the duplications and losses of CAZymes of termite origin during termite evolution.
The genomes, corresponding proteomes and GFF annotations were obtained from publicly available resources. Specifically, the data of Zootermopsis nevadensis and Cryptotermes secundus were retrieved from the NCBI RefSeq database; the data of Coptotermes formosanus was obtained from the NCBI genome assembly database (Itakura et al., 2020); the data of Macrotermes natalensis and Reticulitermes speratus were sourced from previously published datasets (Poulsen et al., 2014; Shigenobu et al., 2022).
To predict and annotate the CAZyme genes in the termite genomes, the longest isoform of each gene in Z. nevadensis and C. secundus were extracted by the function “retrieve_longest_isoforms” in the R package orthologr (Drost et al., 2015). The other three genomes have one protein for each gene in their proteomes. The completeness of proteomes was assessed by BUSCO with insecta_odb10 dataset before CAZyme gene prediction (Manni et al., 2021). Subsequently, the CAZyme genes in the termite genomes were annotated by the standalone tool run_dbCAN (Zhang et al., 2018), which uses three programs, including HMMER, diamond and eCAMI. The genes were considered to be confidently annotated if the protein had consistent annotations from at least two prediction programs. Subsequently, the proteins were mapped to the genomes for predicting unannotated CAZyme genes in all genomes by miniprot (Li, 2023), and the newly annotated genes were manually curated, and their corresponding proteins were subjected to CAZyme prediction as well. The confidently predicted CAZyme proteins of the genes were queried against the non-redundant protein database in NCBI to identify the origin of the predicted genes by following a method we described previously, and last common ancestor (LCA) of up to the top 10 best targets for each query was inferred using the ete3 toolkit (Huerta-Cepas et al., 2016). Additionally, the domains of the annotated proteins were identified by searching the InterPro database with InterProScan (Jones et al., 2014).
For each gene family, the corresponding proteins of the studied species were retrieved and aligned by using MAFFT with L-INS-I (Katoh and Standley, 2013) and muscle (Edgar, 2004). After further refining with RASCAL (Thompson et al., 2003) and scoring by normd (Thompson et al., 2001), the alignment with the highest normd score was subjected to phylogeny construction. IQ-TREE (Minh et al., 2020) was employed to construct phylogenetic trees with model selection and 1,000 ultrafast bootstrap replicates.
We used Notung, which takes a non-dated species tree and the constructed gene trees as input to infer gene duplication and loss for large gene families. The phylogenetic relationships of the five termite species were inferred from a previously published study (Bucek et al., 2019). The duplication and losses were inferred with the rearrange model for reconciliation at a 90% threshold to reduce the penalty of low supported branches (Durand et al., 2005).
For gene families with large duplications, specifically GT1, GH1 and AA3, we further inferred their duplication mode by using duplicate_gene_classifier in MCScanx (Wang et al., 2012) for each species. The duplicate genes of each species were classified into four modes: whole genome/segmental duplications (match genes in syntenic blocks), tandem duplications (continuous repeats), proximal duplications (not adjacent but in nearby chromosomal/scaffold regions at a maximum distance of 10 genes), and dispersed duplications (other modes than segmental, tandem, and proximal). The similarity of the duplications was determined by TBtools (Chen C. et al., 2020). The chromosomal location of these genes and their corresponding collinear blocks were also inferred in MCScanx. The results were visualized using circos (Krzywinski et al., 2009).
To gain preliminary insights into the expression patterns of the identified gene families, we analyzed the expression of identified CAZyme genes in R. speratus using publicly available expression data (Shigenobu et al., 2022). The data contain two body parts, head and body (throax+abdomen), from three castes: workers, soldiers and reproductives. Raw data were filtered with a minimum of 1 count-per-million in at least three samples, then log-transformed for visualization using ggplot2 (Wickham, 2016).
Four of the five CAZyme classes, including GT, GH, AA, and CE, are presented in all termite species with varying numbers. To sum up all the identified CAZyme genes, we found that R. speratus has the highest total CAZyme gene number (287), followed by C. secundus (277), Z. nevadensis (269) and C. formosanus (266), whereas M. natalensis has the lowest gene number (235) (Figure 1). Among them, 17 genes were newly annotated by homology-based mapping to the genomes (Supplementary Table S1) except RsGH9a, which was reported in the previous genome but without annotation. The most abundant class is GT, which accounts for approximately half of the identified genes. GH is the second largest class followed by AA, CBM (Carbohydrate-Binding Module) and CE.
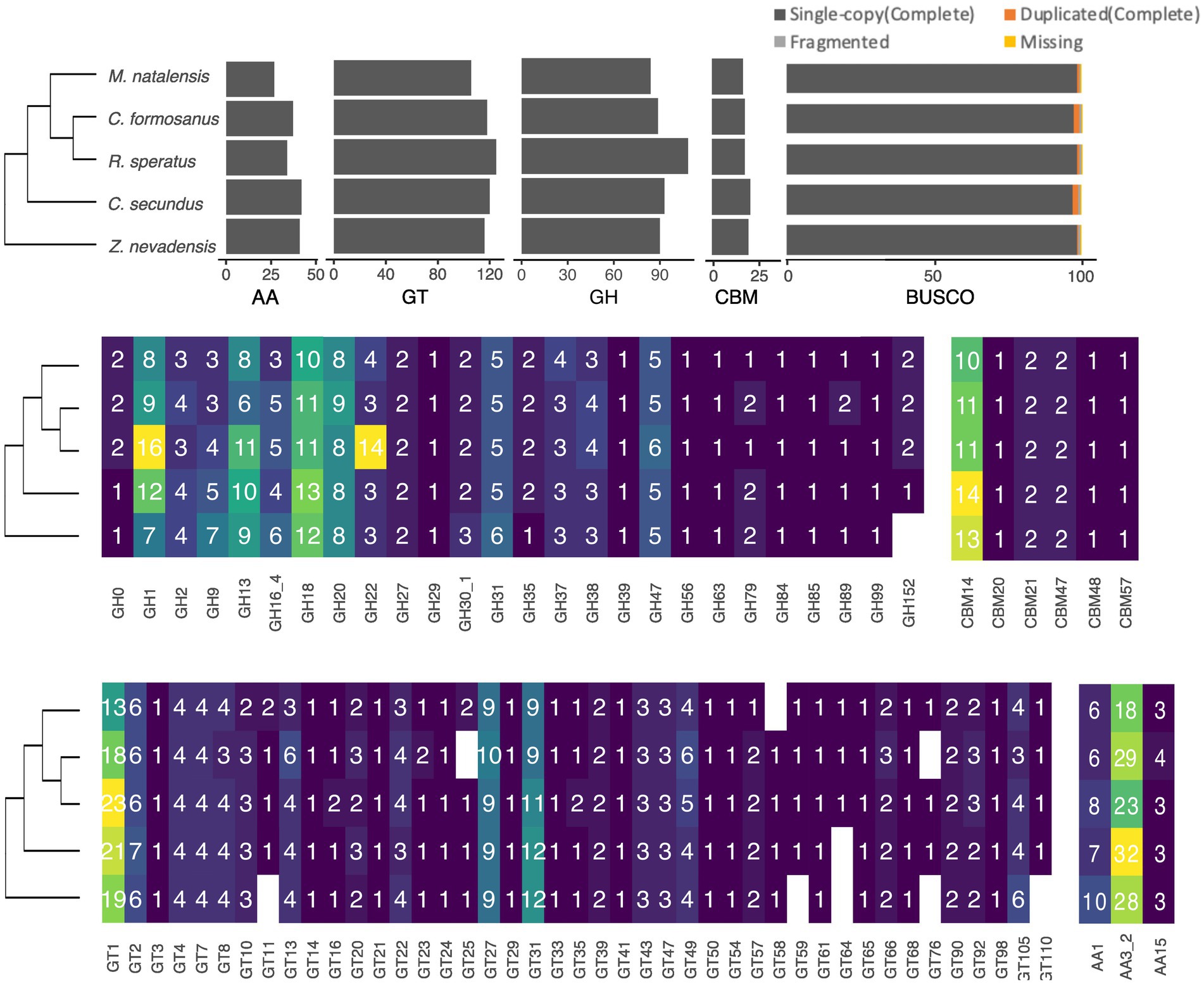
Figure 1. The identified gene numbers of CAZyme classes in different termite species based on a phylogeny inferred from published research (Bucek et al., 2019). At the top panel, the bar plots present the total gene numbers of four main CAZyme classes, including Auxiliary Activity (AA), Glycosyl Transferase (GT), Glycoside Hydrolase (GH), and Carbohydrate-Binding Module (CBM), as well as the BUSCO completeness of the genomes used in this study. The different gene numbers of GH gene families (middle-left panel), CBM (middle-right panel), GT (bottom-left panel), AA (bottom-right panel) are present along with the phylogeny of the top panel.
Among the identified 43 GT gene families, 36 are present in all species, while seven families are absent in one or two species (Figure 1). Notably, GT1 (13–23 copies), GT27 (9–10 copies) and GT31 (8–12 copies) are the largest GT families in termites followed by GT2, GT13, GT49, GT4, GT22, GT7, GT8 and GT105 with 4–6 copies; the remaining families have 1–3 copies. In the GH class, 26 gene families exist in all species except GH152 lacking in Z. nevadensis. Among these GH families, the abundant families are GH1 (7–16 copies), GH18 (10–13 copies), GH13 (6–11 copies) and GH20 (8–9 copies) followed by GH16, GH31, GH47 and GH9 with 3–9 copies (Figure 1). In addition, we found a considerable variation of gene numbers in GH22 among termites ranging from three copies in Z. nevadensis, C. secundus, and C. formosanus to 14 copies in R. speratus. Within the AA class, three families are present in all species, with AA3 being the most abundant (18–32 copies) followed by AA1 (6–10 copies) and AA15 (3–4 copies) (Figure 1). In the CE class, only one family, CE9, is present in termites with one copy in each species. Apart from these four classes, we observed seven groups of CBM in termites, with CBM14, which is also present in GH18, being the most abundant, ranging from 10 to 14 copies.
Most gene families in CAZyme classes have a few but relatively stable gene numbers across the five termite species. We selected the abundant or highly variable gene families in each class for further phylogenetic analysis. Therefore, we constructed the gene trees for the following gene families: GT1, GT27, GT31, GH1, GH9, GH13, GH18, GH20, GH22, AA1, AA3 and CBM14.
As the largest family in GT, GT1 genes were classified into 12 groups (Figure 2A). Group 12 has the highest gene numbers, representing approximately a quarter of the identified GT1 genes in termites; while the remaining groups contain 1–3 gene copies from each species. However, no identified GT1 genes of M. natalensis was included in Group 1, 10, 11, and 12. GT27 and GT31 could be categorized into 9 and 11 groups, respectively, (Supplementary Figure S1); most groups contain one copy from each species except a few GT31 groups that either lack genes from less than two species or have no more than two copies from certain species.
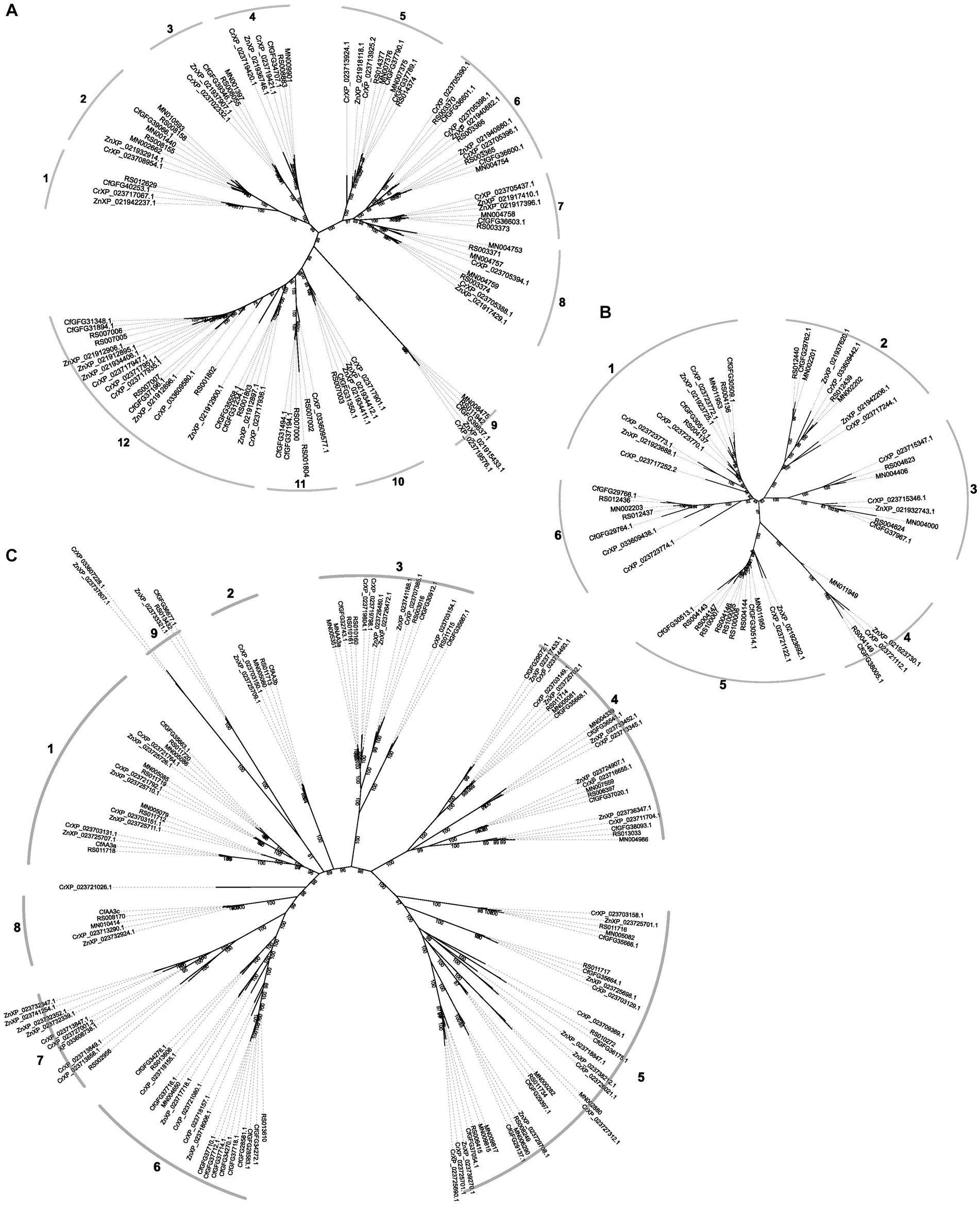
Figure 2. The gene trees of GT1 (A), GH1 (B), and AA3 (C) inferred from corresponding identified proteins with IQ-TREE. The ids in the phylogenies are the ids of identified genes for R. speratus (starting with RS) and Macrotermes natalensis (starting with MN), and labeled as species abbreviations (Z. nevadensis, Zn; C. secundus, Cr; C. formosanus, Cf) and related protein ids for the rest three species. The numbers in the gene trees represent bootstrap values for branch support; the numbers along the grey curves around the gene trees indicate the group numbers of each gene family.
In the GH class, GH1, GH9, and GH22 gene trees have clades with varying gene copies (up to 7) (Figure 2B; Supplementary Figure S2). In the gene trees of GH1 and GH22, most groups contain multiple genes from each species, while one group contains one gene from each species. The gene trees of GH13, GH18, and GH20 consist primarily of clades with one gene copy from each species, except a few groups that either lack genes from less than two species or have 2–3 copies (Supplementary Figure S3).
Similarly, in the AA class, the AA1 gene tree contains nine groups, of which eight groups have one gene from most species and one group contains only two gene copies of Z. nevadensis (Supplementary Figure S4). The gene tree of AA3 is more complex, where most groups and subgroups contain one gene copy from each species with some groups that lack genes from one or two species (Figure 2C). In contrast, several groups of AA3 have multiple copies (up to nine) from C. formosanus, C. secundus and Z. nevadensis.
In the CBM modules, most groups in the CBM14 gene tree contain only one copy from each species, although a few groups lack 1–2 species or have two copies from the same species (Supplementary Figure S4).
In the AA class, most AA1 proteins contain three domains: Multicopper oxidase C-terminal, Multicopper oxidase N-terminal, and Multicopper oxidase second cupredoxin domain. Five AA1 proteins in five gene groups and three species lack one or two of these domains (Supplementary Figure S5). Most AA3 proteins have both C-terminal and N-terminal domains of Glucose-methanol-choline oxidoreductase, except for two proteins that have Glucose-methanol-choline oxidoreductase N-terminal domain and a few proteins that have truncated domains (Supplementary Figure S6). However, two proteins from Z. nevadensis and C. secundus clustered together in a subgroup in the gene tree have dual C-terminal and N-terminal domains.
In the GH class, most GH1 proteins have a complete Glycoside hydrolase family 1 domain. However, six GH1s from Z. nevadensis, C. secundus, and C. formosanus spreading in groups 1, 2, 3, and 4 have two GH1 domains (Figure 3A). Additionally, five GH1s from C. formosanus, M. natalensis, and R. speratus in group 2 and 5 have truncated GH1 domain. For GH9 most proteins contain a single GH9 domain except for one gene from M. natalensis, which contains two domains with one being incomplete; a few proteins of Z. nevadensis in group 3 and a protein of R. speratus have incomplete GH9 domains (Supplementary Figure S7). In GH13 proteins from group 25 and group 8 have conserved domains (glycoside hydrolase, family 13, N-terminal, Alpha-amylase/branching enzyme, C-terminal all beta, glycosyl hydrolase, family 13, catalytic domain; glycogen debranching enzyme, N-terminal domain, glucanotransferase domain, central domain, C-terminal) in each gene from each species; whereas in GH13_15 most proteins have two domains (Alpha-amylase/branching enzyme, C-terminal all beta and Glycosyl hydrolase, family 13, catalytic domain) except for one protein that lacks one domain in each of C. secundus, R. speratus, and C. formosanus (Supplementary Figure S7). In GH13_17 most proteins in three groups have two domains (glycosyl hydrolase, family 13, catalytic domain; Solute carrier family 3 member 2, N-terminal domain), and proteins in one group have one domain (glycosyl hydrolase, family 13, catalytic domain). In GH20 all proteins contain a Glycoside hydrolase family 20, catalytic domain, and most proteins in groups 5–8 have an additional domain Beta-hexosaminidase, eukaryotic type, N-terminal (Supplementary Figure S8). In GH22 most proteins have a C-type lysozyme/alpha-lactalbumin family domain except for one gene from R. speratus and C. formosanus having a Destabilase domain (Supplementary Figure S8).
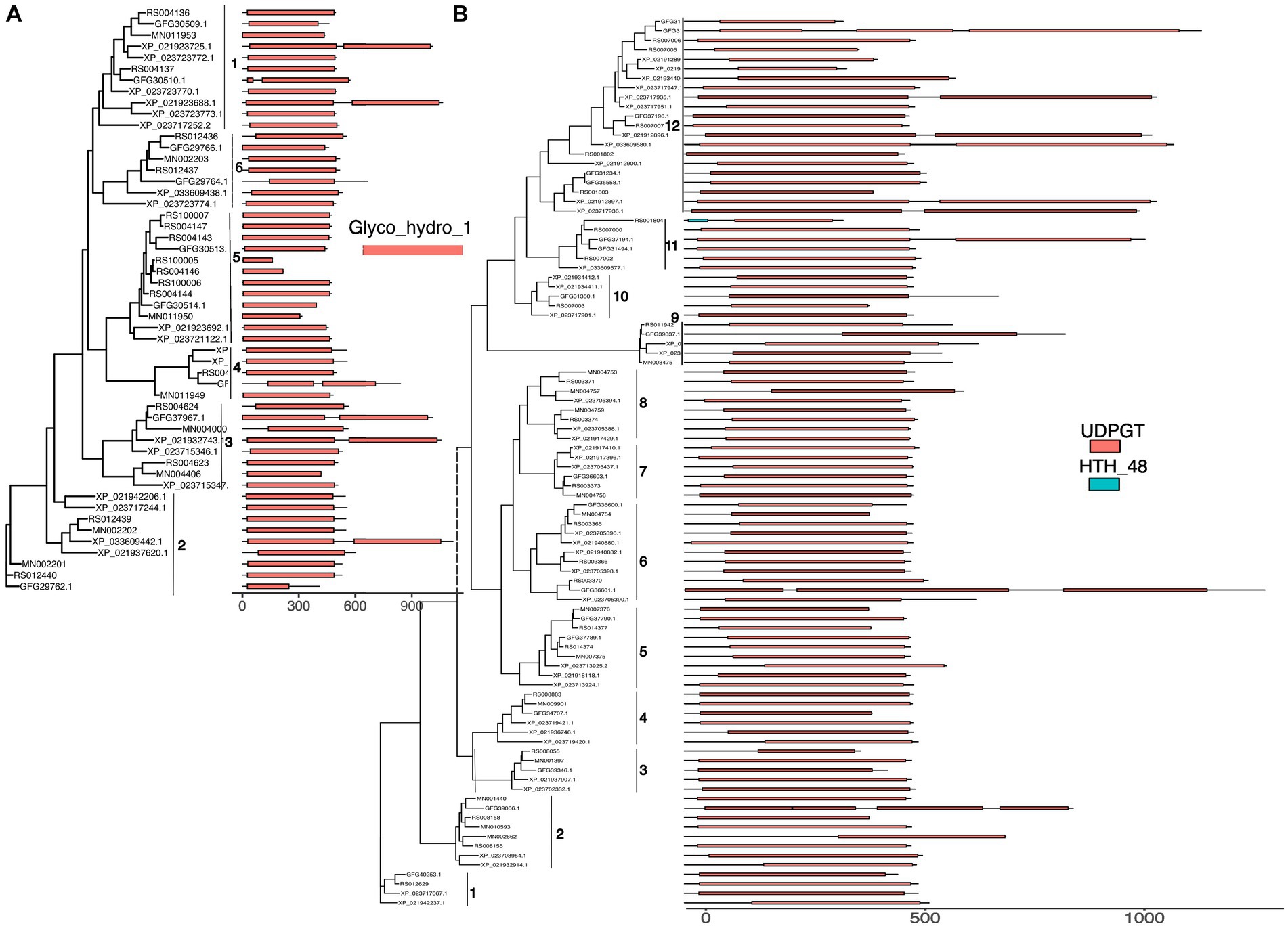
Figure 3. The domain structure (identified by searching the InterPro database with InterProScan, colored boxes) of GH1 (A) and GT1 (B) aligned to related gene trees from Figure 2. The x axis represents the length of the predicted proteins. Different numbers along the grey lines next to the gene phylogenies indicate the group numbers from Figure 2.
Similarly, most GT1 proteins have UDP-glucoronosyl and UDP-glucosyl transferase domains (Figure 3B). Five proteins in group 12 from Z. nevadensis and C. secundus and one protein in group 11 from C. formosanus have two domains, while each protein of C. formosanus in group 12, 6, and 2 have three domains; a few proteins in groups 3, 12, 11 have shorter domains than other members in the groups. All GT27 proteins have two domains, namely glycosyl transferase family 2 and Ricin-type beta-trefoil lectin domain, except for two proteins from C. formosanus and R. speratus in group 4, which lack the Ricin-type beta-trefoil lectin domain, and one protein from R. speratus in group 2 having N-terminal domain of galactosyltransferase instead of the GT2 domain (Supplementary Figure S9). In GT31 the proteins in groups 1–4 have one or two fringe-like domains, while the proteins in group 2 have a single chondroitin N-acetylgalactosaminyltransferase domain (Supplementary Figure S9). Additionally, the proteins in other groups of GT31 have one galactosyltransferase, except proteins in group 9, which have two galactosyltransferase domains. However, a protein of M. natalensis, MN006382-PA, has two domains, Bcl2−/adenovirus E1B nineteen kDa-interacting protein 2 and Divergent CRAL/TRIO domain.
We analyzed the duplications and losses of relatively large gene families to explore the evolution of identified gene families. Overall, the large gene families had large gene numbers in the common ancestor of the selected species with numerous losses and only a few duplications at most branches (Figure 4). Most duplications were found in the AA3 and GT1 gene families in Z. nevadensis, C. formosanus, and the ancestor of R. speratus and M. natalensis. In addition, five duplications of GH1 and AA3 were found in R. speratus and C. secundus, respectively. Moreover, most selected gene families had one duplication in the ancestor of R. speratus, M. natalensis, and C. formosanus.
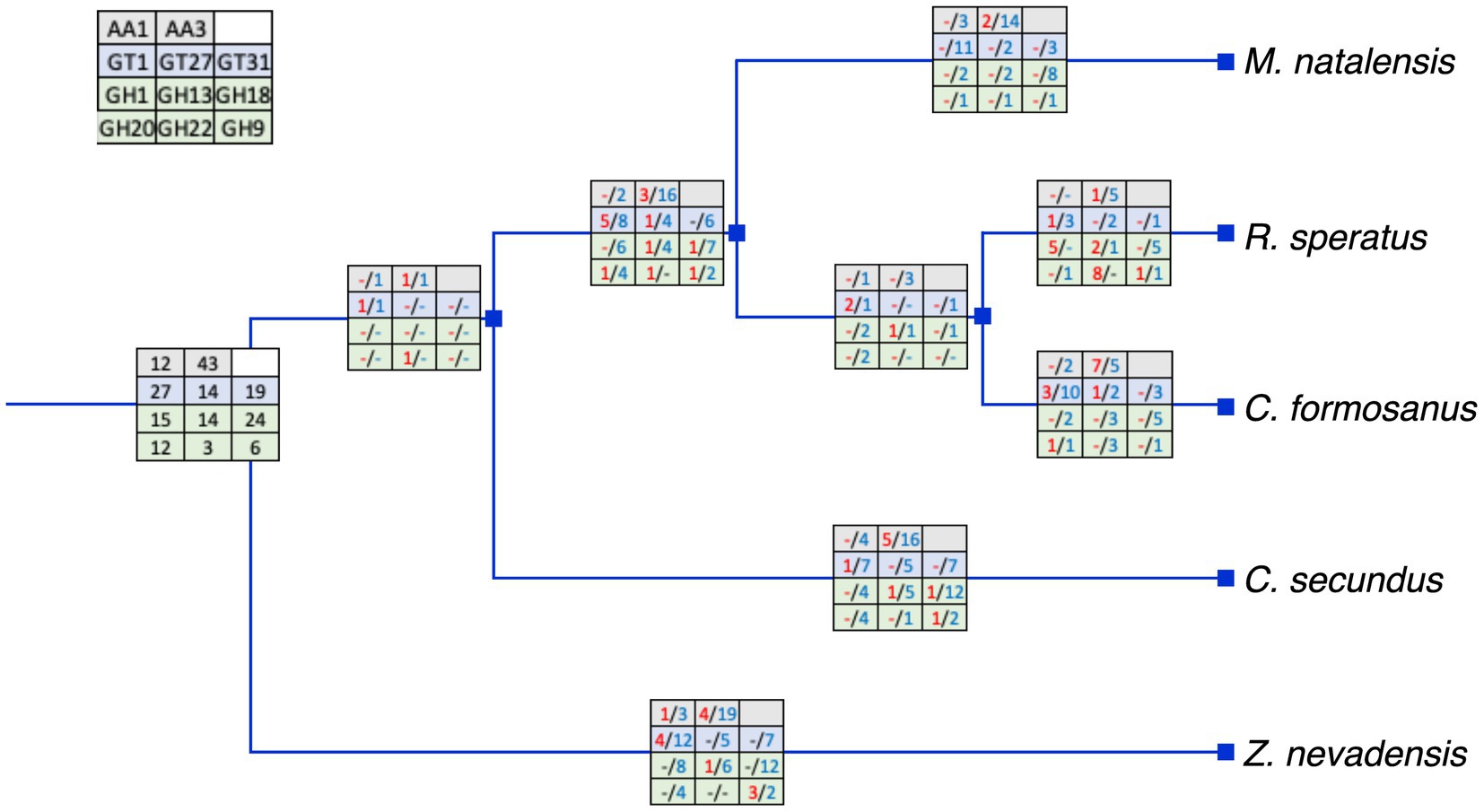
Figure 4. The inferred duplications and losses of selected gene families (left-up panel). Numbers in red represent duplications and numbers in blue represent losses. The duplications and losses were inferred by Notung with the species tree in Figure 1 and related gene trees in Figure 2. Represents no duplication or loss.
As most of the examined CAZyme gene families had limited gene duplications in termites, we analyzed the collinearity and duplication modes of the gene families that experienced several duplications, namely GT1, GH1, and AA3.
In the GT1 gene family, more than half of the genes were tandem duplications and a few were proximal duplications, located in three collinear blocks among all species (Figure 5A). Additionally, approximately five GT1s in each species were dispersed duplications. Most GH1 genes were tandemly duplicated and are located in a few collinear blocks among all species, especially in a few blocks on one contig of R. speratus and a collinear block between C. secundus, C. formosanus and R. speratus (Figure 5B). Interestingly, nearly half of AA3 in all species were tandemly duplicated (Figure 5C). These tandem duplications in M. natalensis, R. speratus, and Z. nevadensis are located in a collinear block, whereas the duplications in C. secundus and C. formosanus are dispersed in multiple contigs.
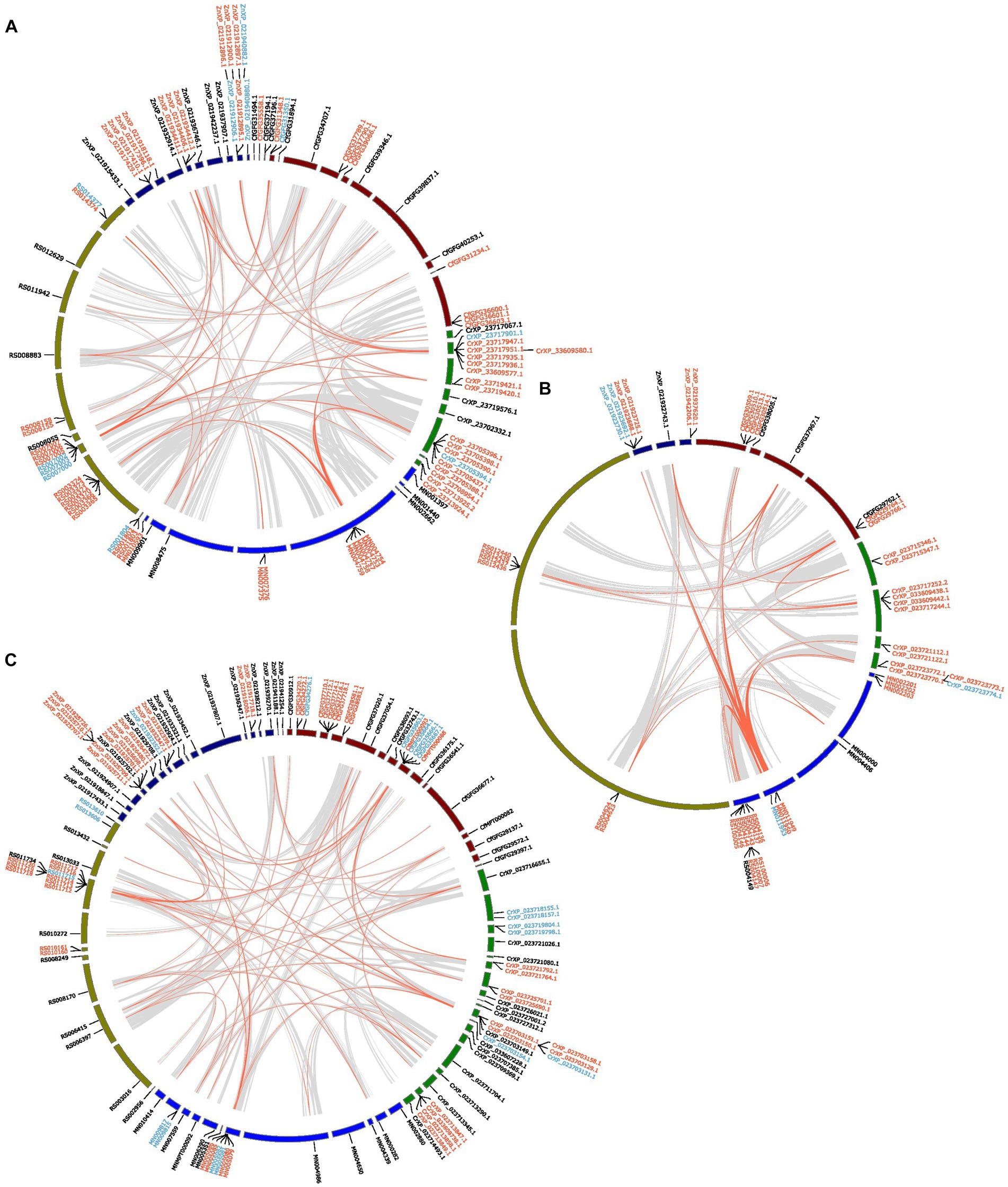
Figure 5. The inferred collinearity and duplication modes of GT1 (A), GH1 (B), AA3 (C) of five termite species. Gene ids in red represent tandem duplications, Gene ids in blue indicate proximal duplications, Gene ids in black represent dispersed duplications. The grey links show the collinear blocks between the scaffolds/contigs from different species that containing identified genes; the red links show the collinear blocks containing identified genes in the referred gene families. The collinearity analysis was performed at protein level.
The overall sequence similarity of the genes in the GT1, GH1, and AA3 gene families is 52.5%. The average sequence similarities for the dispersed, proximal, and tandem duplications of these three gene families are 46.89%, 55.27%, and 55.52%, respectively. Though the average similarities of different duplication types differ among these three gene families, the dispersed duplications have the lowest sequence similarity (AA3, 50.51%; GT1, 47.20%; GH1, 37.09%). In addition, the proximal duplications of GT1 and GH1 have slightly higher sequence similarities (GT1, 55.68%; GH1, 55.27%) than the corresponding tandem duplications (GT1, 51.85%; GH1, 55.52%). However, the tandem duplications of AA3 have a higher sequence similarity (62.15%) than its proximal duplications (54.50%).
Regarding the expression patterns of the identified CAZyme genes, we found an overall tissue- and caste-specific expression pattern but no noticeable sex difference except between the body parts of female and male reproductive (Supplementary Figure S6). Approximately half of the genes have generally low expression in the body parts of all castes. Among the expressed genes, 15 genes showed higher expression in the head than the body of different castes (Figure 6). Among them, a GH30_1 (RS014869) showed a specific expression in the head of workers and reproductives. Additionally, GH9 (RS012687), GH1 (RS004136), and GH16_4 (RS100018) had higher expression in the body of workers than the reproductives and soldiers. Another GH1 (RS004624) had a specific high expression in the body parts of female reproductives. In addition, we observed increased expression levels of three GH22 genes (RS014698, RS100022, and RS100023) in the body parts of soldiers.
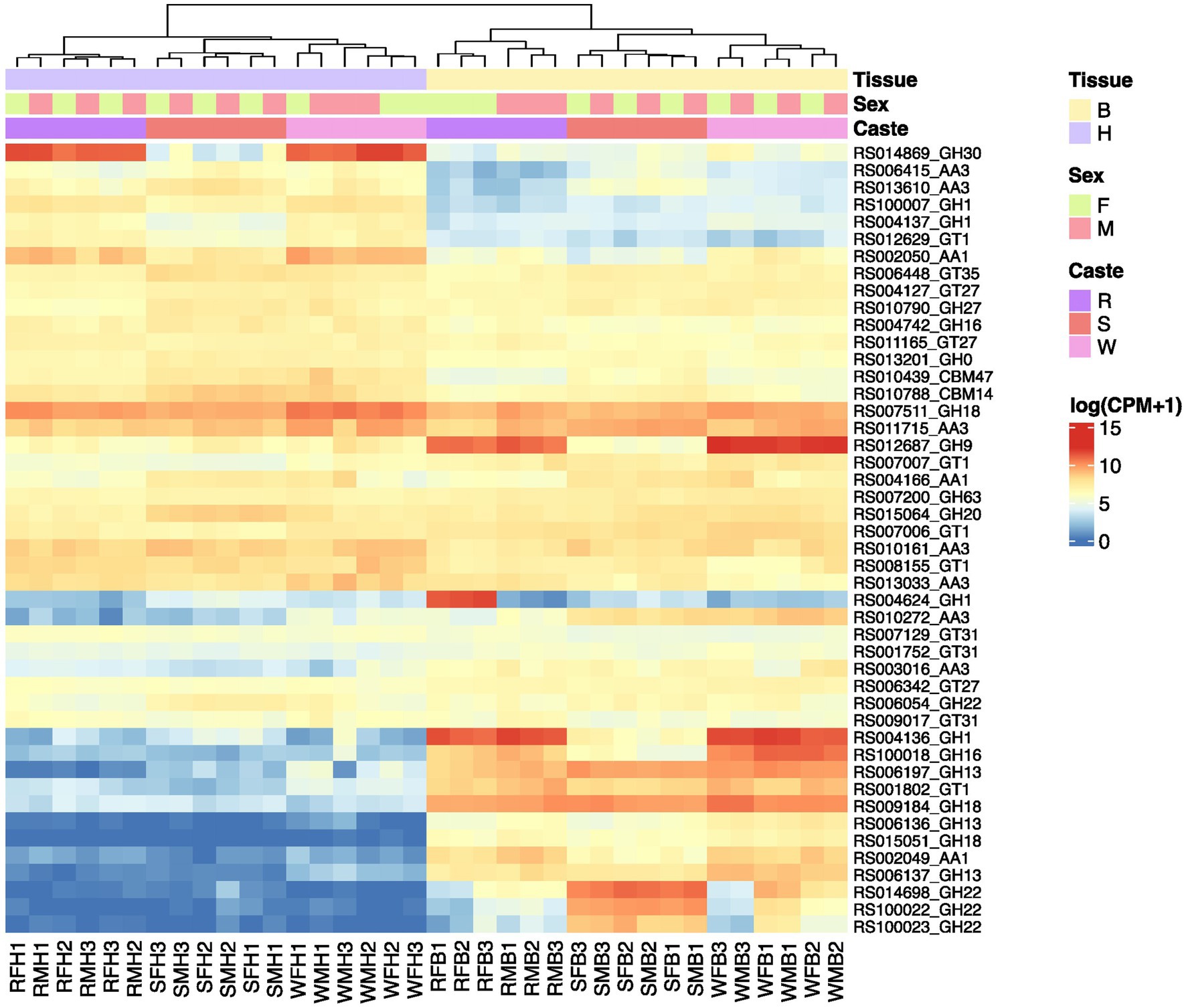
Figure 6. The expression of a number of identified CAZymes in R. speratus mentioned in the manuscript. R, reproductives, S, soldiers, W, workers, M, males, F, females, H, heads, B, body (throax+abdomen). The original data is from a previously published study (Shigenobu et al., 2022) and the expression data are presented as log10(cpm + 1).
We examined the expression levels for each gene family to gain further insights into the expression patterns of the gene families whose gene trees were constructed. High gene expression were found across all samples for three AA3s (RS011715 and RS010161, and RS013033), one GH18 (RS007511), one GH20 (RS015064), one GH22 (RS006054), three GT1s (RS008155, RS007006, RS007007), three GT27s (RS006342, RS004127, RS011165), and three GT31s (RS007129, RS009017, RS001752) (Supplementary Figure S6). Furthermore, we observed an increased expression of one AA1 (RS002049), two AA3 (RS003016 and RS010272), three GH13s (RS006197, RS006136, RS006137), two GH18s (RS009184 and RS015051), one GT1 (RS001802) in the bodies of all castes. A slightly overall higher expression of one AA1 (RS004166) was also found in both bodies and heads of soldiers than in the other two castes. In the GH1 gene family, we found a high expression of RS004136 in the bodies of workers and reproductives. Moreover, we found increased expression levels of one GH1 (RS012436) in the heads of workers, two GH1s (RS004137 and RS100007) and one AA1 (RS002050) in the heads of all castes.
Termites have a diverse array of CAZyme genes belonging to four major classes, with a notable abundance of genes from the GH and GT classes, as previously reported in C. formosanus (Zhang et al., 2012). Most gene families are conserved with minor changes during termite evolution, suggesting their conserved roles in termite biology. The GT is the most diverse and abundant CAZyme class among the identified gene families. GT enzymes catalyze the formation of glycoside bonds by using activated nucleotide sugar and are involved in multiple physiological activities in insects, such as the detoxification of plant compounds, participation in various developmental processes, chemosensation, and stress response (Nagare et al., 2021). GT1, the largest family in the GT class due to their excellent glycosylation capacities (Zhang et al., 2020) and playing a pivotal role in insect detoxification of xenobiotics (Nagare et al., 2021). It is the most abundant family within GT gene families in all termites. The GT1 gene family is also commonly known as UDP-glycosyltransferase (UGT) in insects with various numbers in different species. A previous study on the nine insect genomes showed that the UGT numbers range from 12 in Apis mellifera to 58 in Acyrthosiphone pisum (Ahn et al., 2012); moreover, a recent report showed different numbers of UGTs ranging from 29 to 50 in various Drosophila species (Ahn and Marygold, 2021). As termites primarily feed on wood and consume a wide range of plant metabolites, high GT1 gene numbers would provide sufficient repertoire genes for detoxification. The GT1 gene family could also use a wide range of natural products, including glycolipids, flavonoids and macrolides (Zhang et al., 2020), which might be related to the various gene groups of the gene family in termites.
Interestingly, we found several losses but a few duplications in different termites, while a large gene number in the termite ancestor. Along with the collinearity blocks, these losses might be the consequence of the functional redundancy of gene duplicates during termite evolution. The retained GT1 genes during termite evolution might gain additional functions other than detoxification, such as olfaction in Bombyx mori (Huang et al., 2008), which could be supported by higher expression of certain GT1 genes observed in the head than the body of R. speratus. Intriguingly, we found that groups 10, 11 and 12 of GT1 gene tree contain no M. natalensis GT1 gene; however, why the higher termite lost these genes is yet to be investigated.
Another two GT gene families with large gene numbers in termites, GT27 and GT31, are related to insect development (Ji et al., 2018; Nagare et al., 2021). These genes have a stable number in termites, but the domain analysis revealed that a few genes encoded additional or truncated domains. This suggests that the genes had undergone evolutionary changes during termite evolution, supported by the inferred duplications and losses. Both gene families had likely been through duplications and strong selection due to the physiological significance of maintaining their functions. This might be corroborated by the constitutive expression of the corresponding genes in the different body parts of different castes of R. speratus. However, as some of these genes are related to embryo development (Brückner et al., 2000), it would be necessary to investigate their expression patterns through different developmental stages to provide insights into their roles in termite development.
The second most abundant gene class in termites is GH, which breaks down glycol bounds in carbohydrates, suggesting their significant contribution to termite wood feeding. Among them, GH1 is the most variable in termites, which might be related to their diversified functions in insects (He et al., 2022). The primary function of GH1 in termites is wood digestion (Tokuda et al., 2002, 2009), reflecting the ancestral function of GH1 in insects (He et al., 2022). As the ancestors of termites were wood feeders, it is not surprising that many GH1 genes were inferred from their ancestor. However, we found a large number of gene losses during termite evolution, indicating that the presence of termite gut symbionts might have compensated for some of the functions related to wood digestion. Furthermore, a GH1 gene specifically expressed in the accessory glands of the ovary (Shigenobu et al., 2022) was associated with termite caste formation (Korb et al., 2009; Matsuura et al., 2009). This is likely an ancestral function of GH1 in termites (He et al., 2022; Shigenobu et al., 2022).
The most abundant gene family of GH in termites is GH18, which is widely spread in all groups of organisms (Karlsson and Stenlid, 2009) and contains hydrolytic chitinases and β-N-acetylglucosaminidases as well as non-hydrolytic proteins such as lectins or xylanase inhibitors (Chen C. et al., 2020). In insects the GH18s were classed into 11 groups and contribute to diverse functions, including molting, nutrition, cell proliferation, and immune defense (Chen W. et al., 2020); the functional GH18s, including groups 1, 2, 3, and 4, previously found in other insects are present in termites. The presence of nine groups in termites, including five groups having a single copy and three groups having duplicates, suggests conserved functions of GH18 genes. However, the continual loss of GH18s in termites indicates the reshaping of the gene families during evolution, as also supported by the presence of fragmented domains in GH18 proteins. In addition, we found one group of termite GH18s does not cluster together with any classified groups, which might be due to the misplacement of the group in the gene phylogeny as shown with a low support value.
Another GH family, GH13, is the largest family of glycoside hydrolases and encodes several enzymes acting on several substrates (Kuriki and Imanaka, 1999; da Costa-Latge et al., 2021). In termites most GH13 genes belong to GH13_17 subfamilies, which encode α-glucosidase, with a few copies of GH13_15, encoding α-Amylase (Stam et al., 2006). The GH20 gene family, involved in insect cuticle formation or degradation (Intra et al., 2008; Yang and Chen, 2019), is another conserved large gene family in termites, suggesting their conserved role in termites. The other two gene families, GH22 and GH9, contain lysozymes and cellulases and are involved in digestion (Davison and Blaxter, 2005; Moraes et al., 2014). A large gene number of GH22 with high expression in the body of R. speratus soldiers suggest their roles other than digestion, possibly related to the division of labor or defense in R. speratus (Shigenobu et al., 2022). However, GH9, a well-studied cellulase in termites (Xu et al., 2010; Bujang et al., 2014), shows a large gene number in Z. nevadensis. The GH9 genes had an ancient origin but experienced diversification in termite species (Davison and Blaxter, 2005; Xu et al., 2010), which might explain the large number of cellulase in termites. A previous study on the evolution of GH9 in higher termites showed duplications of GH9 in the highly diversified feeding group (Bujang et al., 2014). Therefore, a comprehensive understanding of its evolution would require further analysis with a larger size of termite genomes.
Among the other CAZyme gene classes, AA is related to lignocellulose digestion. A large number of AA3, which are closely related FAD-dependent enzymes required for class II peroxidases to oxidize lignin (Levasseur et al., 2013), suggest their involvement in wood-feeding in termites. The observed losses during termite evolution suggest potential compensation by the termite symbionts in lignin degradation. However, we found relatively constant gene numbers of AA1, which contains laccase, ferroxidase and laccase-like multicopper oxidase in all termites, suggesting their conserved role in termite digestion.
The CAZyme class with the least number of members is CE, with only one CE9 in each species, which is the second largest group in CAZyme families and catalyzes the de-O or de-N-acylation by removing the ester decorations from carbohydrates (Nakamura et al., 2017). In addition, we found no PL in termites, which cleaves uronic acid-containing polysaccharides using an elimination instead of a hydrolytic mechanism (Lombard et al., 2010). In the CAZyme database, the CE and PL classes currently have 20 and 42 classified families, respectively; the studies on the function or characterization of both classes are rather limited in insects. Among these families, CE4, the largest family of CE, chitin deacetylases (CDA) in insects, plays roles in molting, pupation, and chitin modification (Li et al., 2021). However, the limited presence of both classes suggests that termites may not rely heavily on them for digestion or other physiological activities, which might be compensated by other mechanisms or gene families. For instance, the role of PL gene families in termites is likely to be compensated by their gut microbiota, which possesses a suite of PL gene families (Arora et al., 2022). This kind of complementary relationship has been proposed in M. natalensis (Poulsen et al., 2014), and we assume it should be common in most termite species because of their close relation to gut symbionts (Bourguignon et al., 2018). This has been also observed in tortoise leaf beetles and their pectinolytic Stammera symbionts that encode rhamnogalacturonan lyase, a PL4, for the adaptation of beetles to herbivory (Salem et al., 2020).
In addition to enzyme classes, CAZymes include carbohydrate-binding modules that lack catalytic activities but play an essential role in carbohydrate digestion. Among the CBM families, CBM14 is the largest in termites and is present in all domains of life. CBM14 is known to bind to chitin (Chang and Stergiopoulos, 2015a,b), suggesting its involvement in termite chitin metabolism. In insects, CBM14, also known as the peritrophin-A domain, contributes to the formation of insect peritrophic matrix (Shen and Jacobs-Lorena, 1999; Tellam et al., 1999). Half of them contain multiple repeats within a single protein, indicating their potential interactions with other catalytic ligninolytic enzymes. In termites the different CBM14 gene groups might have different functions as the CBM14s in Tribolium castaneum, which were classified into three subfamilies with different functions, including PM and cuticle formation (Jasrapuria et al., 2010).
Overall, this study compared the CAZymes in different termite species, providing preliminary insights into their evolution. The prevalence of GH and GT enzyme classes in termite hosts and their gut symbionts (Arora et al., 2022) indicates their importance in termite wood digestion. A large number of losses, along with the relatively stable number of genes across species, might be attributed to the abundance of these gene families in their ancestors and the subsequent redundancy during termite evolution. However, it is worth noting that this study analyzed the genomes of only five termite species from two of five feeding termite groups, including Group I (all wood-feeding lower termites), Group II (wood-feeding higher termites), Group IIF (fungus-cultivating higher termites), Group III (soil feeders with a large amount of plant materials), and Group IV (true soil feeders) (Inward et al., 2007). Among the relatively stable CAZyme repertoire, we found fewer gene numbers of GT1 and AA3 in fungus-cultivating higher termites (M. natalensis) than wood-feeding lower termites (the other four termite species). The effect of feeding preferences on the CAZyme repertoire would require multiple genomes of termite species with other feeding preferences. In addition, these genomes were constructed by different projects and pipelines, which may affect the prediction accuracy of gene numbers, domain structures and duplication modes investigated in this study. To gain a more comprehensive understanding of their evolution, further investigations involving a larger number of high-quality termite genomes and the genomes of closely related species would be necessary. Nevertheless, the present study will aid in formulating some testable hypotheses, such as GT1 duplicates functioning in termite olfaction, evolution of CAZyme gene expression being related to the division of labor, and higher termites largely relying on symbionts for lignin degradation, that can be functionally validated in the future and facilitate identifying suitable targets for future species-specific targets for RNAi-mediated termite management practices (Mogilicherla et al., 2022).
The NCBI datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/-, GCF_000696155.1, GCF_002891405.2 and GCA_013340265.1.
SH conceived this study and analyzed the data. All authors contributed to the writing and revision of the manuscript.
SH is supported by the Foundation of Chongqing Normal University (no. 22XLB028) and Natural Science Foundation of Chongqing (no. 2022NSCQ-MSX2875). AC is financed by “EVA 4.0” (no. CZ.02.1.01/0.0/0.0/16 019/0000803) by the OP RDE and “Excellent Team Grants” (2023–2024) from the Faculty of Forestry and Wood Sciences, Czech University of Life Sciences, Prague, Czechia. BJ is supported by Anhui Provincial Key Laboratory of Molecular Enzymology and Mechanism of Major Diseases in Anhui Normal University (no. fzmx202007) and Special Funds for Supporting Innovation and Entrepreneurship for Returned Oversea-students in Anhui Province (no. 2020LCX035).
We acknowledge Amit Roy, Forest Molecular Entomology Lab, Faculty of Forestry and Wood Sciences, Czech University of Life Sciences for his constructive comments in the earlier version of the manuscript. We appreciate the helpful feedback provided by the reviewers and especially the handling editor.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2023.1240804/full#supplementary-material
Ahn, S. J., and Marygold, S. J. (2021). The Udp-glycosyltransferase family in Drosophila melanogaster: nomenclature update, gene expression and phylogenetic analysis. Front. Physiol. 12:648481. doi: 10.3389/fphys.2021.648481
Ahn, S. J., Vogel, H., and Heckel, D. G. (2012). Comparative analysis of the Udp-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 42, 133–147. doi: 10.1016/j.ibmb.2011.11.006
Arora, J., Kinjo, Y., Šobotník, J., Buček, A., Clitheroe, C., Stiblik, P., et al. (2022). The functional evolution of termite gut microbiota. Microbiome 10:78. doi: 10.1186/s40168-022-01258-3
Ashton, L. A., Griffiths, H. M., Parr, C. L., Evans, T. A., Didham, R. K., Hasan, F., et al. (2019). Termites mitigate the effects of drought in tropical rainforest. Science 363, 174–177. doi: 10.1126/science.aau9565
Bourguignon, T., Lo, N., Dietrich, C., Šobotník, J., Sidek, S., Roisin, Y., et al. (2018). Rampant host switching shaped the termite gut microbiome. Curr. Biol. 28, 649–654.e2. doi: 10.1016/j.cub.2018.01.035
Brückner, K., Perez, L., Clausen, H., and Cohen, S. (2000). Glycosyltransferase activity of fringe modulates Notch-Delta interactions. Nature 406, 411–415. doi: 10.1038/35019075
Brune, A. (2014). Symbiotic digestion of lignocellulose in termite guts. Nat. Rev. Microbiol. 12, 168–180. doi: 10.1038/nrmicro3182
Brune, A., and Dietrich, C. (2015). The gut microbiota of termites: digesting the diversity in the light of ecology and evolution. Annu. Rev. Microbiol. 69, 145–166. doi: 10.1146/annurev-micro-092412-155715
Bucek, A., Šobotník, J., He, S., Shi, M., Mcmahon, D. P., Holmes, E. C., et al. (2019). Evolution of termite symbiosis informed by transcriptome-based phylogenies. Curr. Biol. 29, 3728–3734.e4. doi: 10.1016/j.cub.2019.08.076
Bujang, N. S., Harrison, N. A., and Su, N.-Y. (2014). A phylogenetic study of endo-beta-1,4-glucanase in higher termites. Insect. Soc. 61, 29–40. doi: 10.1007/s00040-013-0321-7
Cairo, J. P. L., Cannella, D., Oliveira, L. C., Gonçalves, T. A., Rubio, M. V., Terrasan, C. R. F., et al. (2020). On the roles of Aa15 lytic polysaccharide monooxygenases derived from the termite Coptotermes gestroi. J. Inorg. Biochem. 216:111316. doi: 10.1016/j.jinorgbio.2020.111316
Chang, T.-C., and Stergiopoulos, I. (2015a). Evolutionary analysis of the global landscape of protein domain types and domain architectures associated with family 14 carbohydrate-binding modules. FEBS Lett. 589, 1813–1818. doi: 10.1016/j.febslet.2015.05.048
Chang, T.-C., and Stergiopoulos, I. (2015b). Inter- and intra-domain horizontal gene transfer, gain–loss asymmetry and positive selection mark the evolutionary history of the Cbm14 family. FEBS J. 282, 2014–2028. doi: 10.1111/febs.13256
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). Tbtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, W., Jiang, X., and Yang, Q. (2020). Glycoside hydrolase family 18 chitinases: the known and the unknown. Biotechnol. Adv. 43:107553. doi: 10.1016/j.biotechadv.2020.107553
Da Costa-Latge, S. G., Bates, P., Dillon, R., and Genta, F. A. (2021). Characterization of glycoside hydrolase families 13 and 31 reveals expansion and diversification of α-amylase genes in the phlebotomine Lutzomyia longipalpis and modulation of sandfly glycosidase activities by leishmania infection. Front. Physiol. 12:635633. doi: 10.3389/fphys.2021.635633
Davison, A., and Blaxter, M. (2005). Ancient origin of Glycosyl hydrolase family 9 Cellulase genes. Mol. Biol. Evol. 22, 1273–1284. doi: 10.1093/molbev/msi107
Drost, H.-G., Gabel, A., Grosse, I., and Quint, M. (2015). Evidence for active maintenance of Phylotranscriptomic hourglass patterns in animal and plant embryogenesis. Mol. Biol. Evol. 32, 1221–1231. doi: 10.1093/molbev/msv012
Durand, D., Halldórsson, B. V., and Vernot, B. (2005). A Hybrid Micro-Macroevolutionary Approach to Gene Tree Reconstruction. Research in Computational Molecular Biology: 9th Annual International Conference, Recomb 2005, Cambridge, MA, USA, May 14–18, 2005. Springer, 250–264.
Edgar, R. C. (2004). Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Fujita, A., Miura, T., and Matsumoto, T. (2008). Differences in cellulose digestive systems among castes in two termite lineages. Physiol. Entomol. 33, 73–82. doi: 10.1111/j.1365-3032.2007.00606.x
Geng, A., Cheng, Y., Wang, Y., Zhu, D., Le, Y., Wu, J., et al. (2018). Transcriptome analysis of the digestive system of a wood-feeding termite (Coptotermes formosanus) revealed a unique mechanism for effective biomass degradation. Biotechnol. Biofuels 11:24. doi: 10.1186/s13068-018-1015-1
Griffiths, H. M., Ashton, L. A., Evans, T. A., Parr, C. L., and Eggleton, P. (2019). Termites can decompose more than half of deadwood in tropical rainforest. Curr. Biol. 29, 105–119. doi: 10.1016/j.cub.2019.01.012
He, S., Jiang, B., Chakraborty, A., and Yu, G. (2022). The evolution of glycoside hydrolase family 1 in insects related to their adaptation to plant utilization. Insects 13:786. doi: 10.3390/insects13090786
Hervé, V., Liu, P., Dietrich, C., Sillam-Dussès, D., Stiblik, P., Šobotník, J., et al. (2020). Phylogenomic analysis of 589 metagenome-assembled genomes encompassing all major prokaryotic lineages from the gut of higher termites. PeerJ 8:e8614. doi: 10.7717/peerj.8614
Huang, F.-F., Chai, C.-L., Zhang, Z., Liu, Z.-H., Dai, F.-Y., Lu, C., et al. (2008). The Udp-glucosyltransferase multigene family in Bombyx mori. BMC Genomics 9:563. doi: 10.1186/1471-2164-9-563
Huerta-Cepas, J., Serra, F., and Bork, P. (2016). Ete 3: reconstruction, analysis, and visualization of Phylogenomic data. Mol. Biol. Evol. 33, 1635–1638. doi: 10.1093/molbev/msw046
Intra, J., Pavesi, G., and Horner, D. S. (2008). Phylogenetic analyses suggest multiple changes of substrate specificity within the Glycosyl hydrolase 20 family. BMC Evol. Biol. 8:214. doi: 10.1186/1471-2148-8-214
Inward, D. J., Vogler, A. P., and Eggleton, P. (2007). A comprehensive phylogenetic analysis of termites (Isoptera) illuminates key aspects of their evolutionary biology. Mol. Phylogenet. Evol. 44, 953–967. doi: 10.1016/j.ympev.2007.05.014
Itakura, S., Yoshikawa, Y., Togami, Y., and Umezawa, K. (2020). Draft genome sequence of the termite, Coptotermes formosanus: genetic insights into the pyruvate dehydrogenase complex of the termite. J. Asia Pac. Entomol. 23, 666–674. doi: 10.1016/j.aspen.2020.05.004
Jasrapuria, S., Arakane, Y., Osman, G., Kramer, K. J., Beeman, R. W., and Muthukrishnan, S. (2010). Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem. Mol. Biol. 40, 214–227. doi: 10.1016/j.ibmb.2010.01.011
Ji, S., Samara, N. L., Revoredo, L., Zhang, L., Tran, D. T., Muirhead, K., et al. (2018). A molecular switch orchestrates enzyme specificity and secretory granule morphology. Nat. Commun. 9:3508. doi: 10.1038/s41467-018-05978-9
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., Mcanulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinform 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kalleshwaraswamy, C. M., Shanbhag, R. R., and Sundararaj, R. (2022). “Wood degradation by termites: ecology, economics and protection” in Science of Wood Degradation and Its Protection. ed. R. Sundararaj (Springer Singapore: Singapore)
Karlsson, M., and Stenlid, J. (2009). Evolution of family 18 glycoside hydrolases: diversity, domain structures and phylogenetic relationships. J. Mol. Microbiol. Biotechnol. 16, 208–223. doi: 10.1159/000151220
Katoh, K., and Standley, D. M. (2013). Mafft multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Korb, J., Poulsen, M., Hu, H., Li, C., Boomsma, J. J., Zhang, G., et al. (2015). A genomic comparison of two termites with different social complexity. Front. Genet. 6:9. doi: 10.3389/fgene.2015.00009
Korb, J., Weil, T., Hoffmann, K., Foster, K. R., and Rehli, M. (2009). A gene necessary for reproductive suppression in termites. Science 324:758. doi: 10.1126/science.1170660
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645. doi: 10.1101/gr.092759.109
Kuriki, T., and Imanaka, T. (1999). The concept of the α-amylase family: structural similarity and common catalytic mechanism. J. Biosci. 87, 557–565. doi: 10.1016/S1389-1723(99)80114-5
Levasseur, A., Drula, E., Lombard, V., Coutinho, P. M., and Henrissat, B. (2013). Expansion of the enzymatic repertoire of the Cazy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 6:41. doi: 10.1186/1754-6834-6-41
Li, H. (2023). Protein-to-genome alignment with miniprot. Bioinform 39:btad014. doi: 10.1093/bioinformatics/btad014
Li, Y., Liu, L., Yang, J., and Yang, Q. (2021). An overall look at insect chitin deacetylases: promising molecular targets for developing green pesticides. J. Pestic. Sci. 46, 43–52. doi: 10.1584/jpestics.D20-085
Lombard, V., Bernard, T., Rancurel, C., Brumer, H., Coutinho, P. M., and Henrissat, B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochemist 432, 437–444. doi: 10.1042/BJ20101185
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., and Zdobnov, E. M. (2021). Busco update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38, 4647–4654. doi: 10.1093/molbev/msab199
Marynowska, M., Goux, X., Sillam-Dussès, D., Rouland-Lefèvre, C., Roisin, Y., Delfosse, P., et al. (2017). Optimization of a metatranscriptomic approach to study the lignocellulolytic potential of the higher termite gut microbiome. BMC Genomics 18:681. doi: 10.1186/s12864-017-4076-9
Matsuura, K., Yashiro, T., Shimizu, K., Tatsumi, S., and Tamura, T. (2009). Cuckoo fungus mimics termite eggs by producing the cellulose-digesting enzyme β-glucosidase. Curr. Biol. 19, 30–36. doi: 10.1016/j.cub.2008.11.030
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., Von Haeseler, A., et al. (2020). Iq-tree 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi: 10.1093/molbev/msaa015
Mogilicherla, K., Chakraborty, A., Taning, C., Smagghe, G., and Roy, A. (2022). Rnai in termites (Isoptera): current status and prospects for pest management. Entomol Gen 43, 55–68. doi: 10.1127/entomologia/2022/1636
Moraes, C. D. S., Diaz-Albiter, H. M., Faria, M. D. V., Sant'anna, M. R. V., Dillon, R. J., and Genta, F. A. (2014). Expression pattern of glycoside hydrolase genes in Lutzomyia longipalpis reveals key enzymes involved in larval digestion. Front. Physiol. 5:276. doi: 10.3389/fphys.2014.00276
Nagare, M., Ayachit, M., Agnihotri, A., Schwab, W., and Joshi, R. (2021). Glycosyltransferases: the multifaceted enzymatic regulator in insects. Insect Mol. Biol. 30, 123–137. doi: 10.1111/imb.12686
Nakamura, A. M., Nascimento, A. S., and Polikarpov, I. (2017). Structural diversity of carbohydrate esterases. Biotechnol Res Innov 1, 35–51. doi: 10.1016/j.biori.2017.02.001
Poulsen, M., Hu, H., Li, C., Chen, Z., Xu, L., Otani, S., et al. (2014). Complementary symbiont contributions to plant decomposition in a fungus-farming termite. Proc. Natl. Acad. Sci. U. S. A. 111, 14500–14505. doi: 10.1073/pnas.1319718111
Rust, M. K., and Su, N. Y. (2012). Managing social insects of urban importance. Annu. Rev. Entomol. 57, 355–375. doi: 10.1146/annurev-ento-120710-100634
Salem, H., Kirsch, R., Pauchet, Y., Berasategui, A., Fukumori, K., Moriyama, M., et al. (2020). Symbiont digestive range reflects host plant breadth in herbivorous beetles. Curr. Biol. 30, 2875–2886.e4. doi: 10.1016/j.cub.2020.05.043
Shelomi, M., Wipfler, B., Zhou, X., and Pauchet, Y. (2020). Multifunctional cellulase enzymes are ancestral in Polyneoptera. Insect Mol. Biol. 29, 124–135. doi: 10.1111/imb.12614
Shen, Z., and Jacobs-Lorena, M. (1999). Evolution of chitin-binding proteins in invertebrates. J. Mol. Evol. 48, 341–347. doi: 10.1007/PL00006478
Shigenobu, S., Hayashi, Y., Watanabe, D., Tokuda, G., Hojo, M. Y., Toga, K., et al. (2022). Genomic and transcriptomic analyses of the subterranean termite Reticulitermes speratus: gene duplication facilitates social evolution. Proc. Natl. Acad. Sci. U. S. A. 119:e2110361119. doi: 10.1073/pnas.2110361119
Stam, M. R., Danchin, E. G., Rancurel, C., Coutinho, P. M., and Henrissat, B. (2006). Dividing the large glycoside hydrolase family 13 into subfamilies: towards improved functional annotations of alpha-amylase-related proteins. Protein Eng. Des. Sel. 19, 555–562. doi: 10.1093/protein/gzl044
Tartar, A., Wheeler, M. M., Zhou, X., Coy, M. R., Boucias, D. G., and Scharf, M. E. (2009). Parallel metatranscriptome analyses of host and symbiont gene expression in the gut of the termite Reticulitermes flavipes. Biotechnol. Biofuels 2:25. doi: 10.1186/1754-6834-2-25
Tellam, R. L., Wijffels, G., and Willadsen, P. (1999). Peritrophic matrix proteins. Insect Biochem. Mol. Biol. 29, 87–101. doi: 10.1016/S0965-1748(98)00123-4
Thompson, J. D., Plewniak, F., Ripp, R., Thierry, J. C., and Poch, O. (2001). Towards a reliable objective function for multiple sequence alignments. J. Mol. Biol. 314, 937–951. doi: 10.1006/jmbi.2001.5187
Thompson, J. D., Thierry, J. C., and Poch, O. (2003). Rascal: rapid scanning and correction of multiple sequence alignments. Bioinform 19, 1155–1161. doi: 10.1093/bioinformatics/btg133
Tokuda, G. (2019). Plant cell wall degradation in insects: recent progress on endogenous enzymes revealed by multi-omics technologies. Adv in Insect Phys 57, 97–136. doi: 10.1016/bs.aiip.2019.08.001
Tokuda, G., Lo, N., Watanabe, H., Arakawa, G., Matsumoto, T., and Noda, H. (2004). Major alteration of the expression site of endogenous cellulases in members of an apical termite lineage. Mol. Ecol. 13, 3219–3228. doi: 10.1111/j.1365-294X.2004.02276.x
Tokuda, G., Miyagi, M., Makiya, H., Watanabe, H., and Arakawa, G. (2009). Digestive β-glucosidases from the wood-feeding higher termite, Nasutitermes takasagoensis: intestinal distribution, molecular characterization, and alteration in sites of expression. Insect Biochem. Mol. Biol. 39, 931–937. doi: 10.1016/j.ibmb.2009.11.003
Tokuda, G., Saito, H., and Watanabe, H. (2002). A digestive β-glucosidase from the salivary glands of the termite, Neotermes koshunensis (Shiraki): distribution, characterization and isolation of its precursor cdna by 5′- and 3′-race amplifications with degenerate primers. Insect Biochem. Mol. Biol. 32, 1681–1689. doi: 10.1016/S0965-1748(02)00108-X
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). McscanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Watanabe, H., Noda, H., Tokuda, G., and Lo, N. (1998). A cellulase gene of termite origin. Nature 394, 330–331. doi: 10.1038/28527
Wickham, H. (2016). Ggplot2: Elegant Graphics for Data Analysis, Cham, Switzerland, Springer International Publishing.
Wu, C., Ulyshen, M. D., Shu, C., Zhang, Z., Zhang, Y., Liu, Y., et al. (2021). Stronger effects of termites than microbes on wood decomposition in a subtropical forest. For. Ecol. Manag. 493:119263. doi: 10.1016/j.foreco.2021.119263
Xu, Q., Zhou, Y., Li, J., Chen, H., Lu, J., Chen, K.-P., et al. (2010). Diversifying evolution of endo-beta-1,4-glucanase in termites (Isoptera). Sociobiology 56, 623–636.
Yang, Q., and Chen, W. (2019). Structural and biochemical analysis completes the puzzle of chitin hydrolysis in insects. FASEB J. 33:798.10. doi: 10.1096/fasebj.2019.33.1
Yuki, M., Moriya, S., Inoue, T., and Kudo, T. (2008). Transcriptome analysis of the digestive organs of Hodotermopsis sjostedti, a lower termite that hosts mutualistic microorganisms in its hindgut. Zool. Sci. 25, 401–406. doi: 10.2108/zsj.25.401
Zhang, D., Lax, A. R., Henrissat, B., Coutinho, P. M., Katiya, N., Nierman, W. C., et al. (2012). Carbohydrate-active enzymes revealed in Coptotermes formosanus (Isoptera: Rhinotermitidae) transcriptome. Insect Mol. Biol. 21, 235–245. doi: 10.1111/j.1365-2583.2011.01130.x
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). dbcan2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, 95–101. doi: 10.1093/nar/gky418
Keywords: wood digestion, glycosyl transferase, glycoside hydrolase, gene duplications, gene losses
Citation: He S, Chakraborty A, Li F, Zhou C, Zhang B, Chen B and Jiang B (2023) Genome-wide identification reveals conserved carbohydrate-active enzyme repertoire in termites. Front. For. Glob. Change. 6:1240804. doi: 10.3389/ffgc.2023.1240804
Received: 15 June 2023; Accepted: 13 October 2023;
Published: 27 October 2023.
Edited by:
Bernard Slippers, University of Pretoria, South AfricaReviewed by:
Gaku Tokuda, University of the Ryukyus, JapanCopyright © 2023 He, Chakraborty, Li, Zhou, Zhang, Chen and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shulin He, c2h1bGluaGVAaG90bWFpbC5jb20=; Bin Jiang, YmluLmppYW5nQGFobnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.