
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. For. Glob. Change , 08 December 2022
Sec. Pests, Pathogens and Invasions
Volume 5 - 2022 | https://doi.org/10.3389/ffgc.2022.971916
This article is part of the Research Topic Invasive Pathogens and Arthropods: Biogeography, Drivers of Success, Impacts, and Emerging Management Strategies View all 5 articles
 Julien Roy1,2*
Julien Roy1,2* Ioanna Kyritsi1
Ioanna Kyritsi1 Nadja Reinwarth1
Nadja Reinwarth1 Julien B. Bachelier1
Julien B. Bachelier1 Matthias C. Rillig1,2
Matthias C. Rillig1,2 Robert Lücking3
Robert Lücking3Plant fungal pathogens are an increasing emerging threat as climate change progresses. Sphaeropsis sapinea (syn. Diplodia sapinea), the causal fungal agent of Diplodia tip blight, is a major pathogen of pines of forestry and ornamental relevance in Europe and worldwide. Here, we combined molecular-based field surveys in a common-garden setting and across an elevation gradient with historical records, cultivation-based growth experiments and microscopy to report on host and abiotic constraints on the distribution of S. sapinea. Using the arboretum at the Botanical Garden Berlin, Germany, to control for environmental variability, S. sapinea was detected on all seven host Pinus species we studied. However, P. sylvestris is the only species in which the fungus was detected in symptomless needles at the arboretum, and the most frequently recorded host for over a century, suggesting that it is the main, and perhaps, potential original host. In addition, sampling symptomatic needles in four out of the seven same species across a gradient from 200 to 2,100 m of elevation in the French Alps, S. sapinea was not detected at elevation higher than 800 m. Abiotic constraints were also supported by reduced growth of isolates of S. sapinea at low temperature under controlled conditions, but a 35°C prior stress exposure increased the subsequent growth of S. sapinea within its optimal temperature range (20-30°C). Altogether, our study thus not only suggests that S. sapinea is more likely to cause tip blight in P. sylvestris compared to the other species we studied, but also that in the current context of global climate change with predicted temperature increases, the fungus could infect a wider range of pine hosts and locations worldwide.
Plant fungal pathogens are an increasing emerging threat as climate change progresses (Anderson et al., 2004; Seidl et al., 2017). Among these, Sphaeropsis sapinea (Fr.) Dyko & Sutton [Botryosphaeriaceae, Botryosphaeriales; selected synonyms: Diplodia sapinea (Fr.) Fuckel, D. pinea (Desm.) J. Kickx f.] is responsible for Diplodia tip blight disease (or Diplodia shoot blight, Figure 1). The fungus is one of the major and most widespread pathogens of pines or other conifers of forestry or ornamental relevance in Europe and worldwide (Bußkamp, 2018; CABI Invasive Species Compendium1).
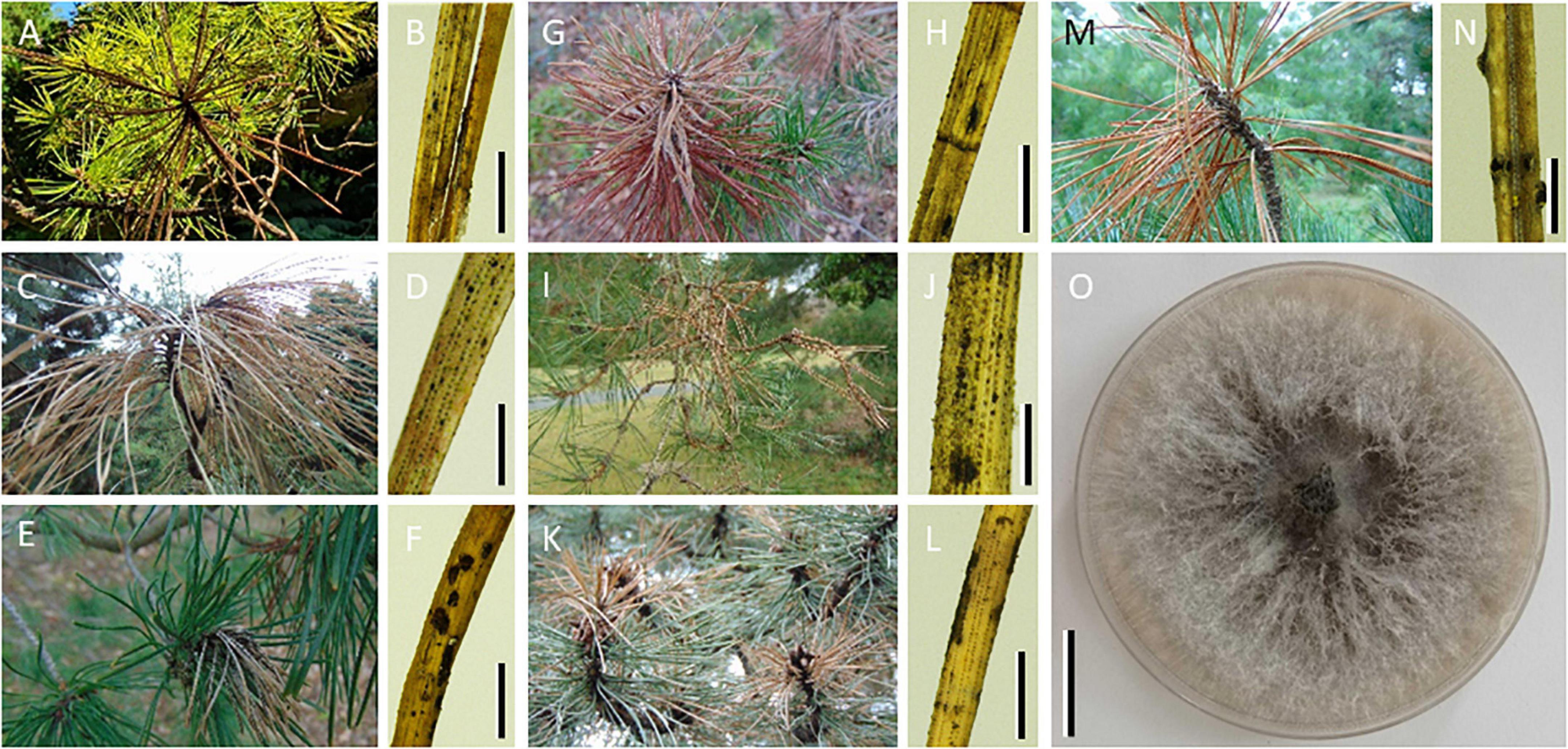
Figure 1. Photographs of tip blight symptoms across seven pine species where Sphaeropsis sapinea was detected. Macroscopic photographs of shoots with tip blight symptoms (A–G) and of needles from those shoots under stereomicroscope (H–N) collected on Pinus rigida [(A,B); scale bar = 4 mm] Pinus ponderosa [(C,D); scale bar = 2 mm] Pinus mugo [(E,F); scale bar = 4 mm] Pinus uncinata subsp. rotundata [(G,H); scale bar = 4 mm] Pinus sylvestris [(I,J); scale bar = 2 mm] Pinus nigra [(K,L); scale bar = 4 mm] Pinus cembra [(M,N); scale bar = 750 μm] trees tested positive for S. sapinea. Note the multiple symptoms on a single needle putatively caused by different fungal pathogens. (O) Colony of S. sapinea growing on 1.5% malt extract agar at 21°C (scale bar = 2 cm).
Sphaeropsis sapinea was first described in the 19th century and occurs mainly as saprotroph on dead needles, cone bracts, bark or shoots of pine or fir, and coarse woody debris (Piou et al., 1991; Smith et al., 1996; Santini et al., 2008; Fabre et al., 2011). The fungus has more recently been reported as an endophyte in symptomless trees (Smith et al., 1996; Stanosz et al., 1997, 2005, 2007; Flowers et al., 2001; Maresi et al., 2007; Bihon et al., 2011; Bußkamp et al., 2020; Terhonen et al., 2021). The fungus is of interest as it also is an opportunistic pathogen, causing disease in trees subjected to stress, especially drought, extreme temperature, excess nitrogen, and wounding caused by hail, insects or mechanical (Piou et al., 1991; Swart and Wingfield, 1991; Zwolinski et al., 1995; Blodgett et al., 1997; Stanosz et al., 2001; Brodde et al., 2019; Blumenstein et al., 2021b). As a latent saprotroph and pathogen that can live as a symptomless endophyte in its host tree, the fungus goes unnoticed before the host becomes stressed or damaged and starts to show symptoms, and possibly dies.
Sphaeropsis sapinea has been reported from most pine species worldwide2. In Europe, the fungus can infect P. nigra (including the subspecies P. nigra subsp. laricio), P. sylvestris, P. radiata, P. pinaster, P. halepensis, P. pinea, P. mugo, and P. leucodermis with various degrees of host susceptibility (Fabre et al., 2011; Oliva et al., 2013; Luchi et al., 2014; Adamson et al., 2015; Aragonés et al., 2021). As a symptomless endophyte, the fungus has been reported on P. sylvestris in Germany (Bußkamp et al., 2020; Blumenstein et al., 2021a), Finland (Terhonen et al., 2021), and in the USA (Flowers et al., 2001), on P. nigra in Italy (Maresi et al., 2007), and in the USA (Flowers et al., 2001), on P. patula (Bihon et al., 2011), and on P. banksiana (Stanosz et al., 2007) and P. resinosa (Stanosz et al., 1997, 2005; Munck et al., 2009; Santamaria et al., 2012) in the USA, and on P. elliottii, P. patula, P. radiata, and P. taeda in South Africa (Smith et al., 1996). However, studies of its host spectrum are not exhaustive at local scale, and whether S. sapinea lives as an endophyte in other pine species in Germany is unknown. A better knowledge of its host distribution, especially as a symptomless endophyte, is necessary to better assess the risk this fungus poses to European pine forests.
Host identity, environmental conditions and geography together shape the spatial distribution of forest fungal pathogens (Drenkhan et al., 2016; Woods et al., 2016). Therefore, certain pine species can appear uninfected not because of non-susceptibility but because of their occurrence outside the climatic or biogeographic range of the fungus. This can be analyzed by sampling a diversity of potential pine species co-occurring within the climatic niche and geographical range of the fungus, such as in botanical gardens or arboretums.
Conversely, sampling susceptible hosts potentially outside of the climatic range of the fungus would confirm that the fungus is not associated with tip blight in such environmental conditions and would reveal the abiotic constraints on its distribution. This can be analyzed by sampling needles with tip blight symptoms of susceptible hosts across a climatic gradient, such as provided by difference in elevation. Sphaeropsis sapinea occurs under mediterranean and temperate climate and declines at higher elevations and latitudes, probably driven by the mean daily minimum temperature in winter (Fabre et al., 2011). Yet, records of occurrences of the fungus are primarily based on visual observation of symptoms, which could limit the ability to detect the fungus, in particular in the absence of reproductive structures and owing to the difficulties in correctly identifying species based on visual observation of symptoms or reproductive structures (de Wet et al., 2003). The continental distribution and high clonality of this fungus suggest that it is not limited by dispersal (Burgess et al., 2004). Thus, if the fungus is confirmed to have low host constraints but being rather limited by (low) temperature (Brookhouser and Peterson, 1970; Chou, 1982; Zlatkovic et al., 2016), these results would suggest that changes in climate could lead to infection of a wider range of pine species and range expansion.
Here, we combined molecular-based field surveys in a common-garden setting and across an elevation gradient with historical records and cultivation-based growth experiments to report on host and abiotic constraints on the distribution of S. sapinea. We conducted an initial survey across seven pine species planted in close proximity at a single locality, the Botanical Garden of Berlin, Germany, to complement previous observations on the host spectrum of S. sapinea while controlling for environmental variability and biogeographic differentiation. This survey was complemented with S. sapinea host incidence data from the Global Biodiversity Information Facility (GBIF) and from fungarium specimens from the Botanical Museum of Berlin. We conducted a second survey across a gradient from 200 to 2,100 m of elevation in the French Alps, to complement observations on the elevational range and limits of S. sapinea occurrence. Symptomatic and asymptomatic needles from the host trees were collected to investigate the host and elevational range of the presence of S. sapinea. We used a polymerase chain reaction (PCR)-based assay with previously proposed mtSSU rRNA primers for the specific detection of S. sapinea, and the closely related, but less aggressive Diplodia tip blight agent, S. scrobiculata (formerly Diplodia pinea morphotype group B; de Wet et al., 2003). We further conducted growth experiments for a more mechanistic understanding of the temperature constraints on S. sapinea occurrence. We specifically addressed how prior exposure to temperature stress affects subsequent growth at other temperatures. Finally, we also generated a series of stereomicroscope, fluorescence, and scanning electron microscopy images to document natural infection of needles of Pinus sylvestris by S. sapinea and on other pine species, and better understand the biology of the fungus on different pine hosts. Documenting mycelium distribution within needles and recurrent patterns of infection across hosts would help inform on the host-fungus interactions.
To assess the host range of S. sapinea as a putative pathogen and as symptomless endophyte, we sampled pine trees in the pine arboretum at Botanical Garden and Botanical Museum (BGBM) Berlin, Berlin, Germany (52.4544°, 13.3052°) during June 2021. We carried out a survey of seven pine species, namely P. sylvestris (n = 5 trees), P. mugo (n = 5), P. uncinata subsp. rotundata (n = 3), P. nigra (n = 3), P. rigida (n = 2), P. ponderosa (n = 1) and P. cembra (n = 5) and one individual of a distantly-related gymnosperm, Juniperus communis subsp. communis which is host to a closely related species of S. sapinea, S. sapinea f. sp. cupressi (Solel et al., 1987). For each tree, twigs with asymptomatic or with symptomatic needles were randomly collected and pooled according to the presence of tip blight symptoms. The sampling thus covered a total of seven pine species and one Juniperus communis across 25 trees, totaling 58 samples (25 samples of asymptomatic needles and 33 samples of symptomatic needles, Supplementary Table 1).
To complement the field-based sampling survey 1 to assess the host range of S. sapinea, we collected data of S. sapinea records using the Global Biodiversity Information Facility (GBIF). Occurrences of S. sapinea were searched for, using a search filter based on binomial names (Sphaeropsis sapinea, Diplodia sapinea and Diplodia pinea; taxon key: 2591884; accessed on July 27, 2022; dataset available3). Out of 1,057 records, we could obtain host data for 351 records. We further focused on historical records made before 1910 to support results of survey 1 on which pine host is the potential main and perhaps, original host. In addition, 20 fungarium specimens at BGBM identified as S. sapinea were included (Supplementary Table 1). Duplicates between the GBIF records and the fungarium specimens that are preserved at BGBM were removed for the analysis. The fungarium specimens were mostly bark material, but also cones where pycnidia supposedly of S. sapinea were observed. We suspect some entries in GBIF did not correspond to S. sapinea (especially those made on non-coniferous trees) but it was marginal. Host names in Latin were updated to current accepted names. The Latin host name in GBIF or fungarium specimens at BGBM was not always possible to update to current accepted names due to conflicts (e.g., Pini montanae corresponding to both P. sylvestris, P. mugo and P. cembra, or also Pini pinastri corresponding to unknown pine species).
To assess the elevational range of S. sapinea as a putative pathogen associated with tip blight, we sampled trees from four out of the seven pine species surveyed in survey 1 (namely, P. sylvestris, P. nigra, P. mugo and P. cembra) at elevations from 200 to 2,100 m in four mountain ranges across Grenoble, France, in September 2021, January 2022 and March 2022. On each tree, we collected a single twig with tip blight symptomatic needles of the pine species present at the particular elevation and locality. At 200 m of elevation, trees (n = 2, one P. nigra, one P. sylvestris) come from a single location and are 500 m distant from each other. At 400 m of elevation, trees (n = 3, two P. sylvestris, one P. mugo) come from a single location, but are up to 1 km apart from each other. From 800 to 1,000 m of elevation, trees (n = 3, three P. sylvestris) were sampled in two distinct mountain massifs, distant from ∼50 km. Above 1,000 m of elevation (1,600-2,100 m), trees (n = 20, ten P. mugo, nine P. sylvestris and one P. cembra) were sampled in four distinct mountain massifs. Asymptomatic needles from a single twig were also collected on a subset of the trees to ensure the absence of S. sapinea as endophyte when it was absent in symptomatic needles (one P. nigra and one P. sylvestris at 200 m; two P. sylvestris at 400 m; two P. sylvestris from 800 to 1,000 m; and two P. sylvestris at 1,600 m). The sampling thus covered a total of 4 pine species across 28 trees for 37 samples (9 samples of asymptomatic needles and 28 samples of symptomatic needles, Supplementary Table 1).
For surveys 1 and 2, needles were cut into ∼1 cm pieces and ∼10 pieces were ground using either beads for beating or liquid nitrogen for crushing the material. For beads, needle pieces were placed in 1.5 ml Eppendorf tubes with two to three tungsten beads of 3 mm diameter. Beating was performed using a Restch MM40 instrument, at a frequency of 25 mH for 1 to 2 min, to obtain tissue powder. Beads were cleaned using 70% ethanol followed by bleaching. Crushing with liquid nitrogen was performed using mortars and pestles, previously sterilized using 70% ethanol followed by bleaching. For survey 1, DNA extractions were performed using two methods, a DNA extraction kit allowing relatively fast processing of numerous samples, and a traditional, cost-effective method yielding higher quantities of DNA. For kit-based extraction we used the PowerSoil DNA isolation kit (MoBio Laboratories Inc., Carlsbad, CA, USA), following the manufacturer’s instructions. As alternative cost-effective extraction, a cetyltrimethyl ammonium bromide (CTAB)-based protocol was employed, as successfully used for pine needles in other studies (Smith and Stanosz, 2006). For survey 2, only the CTAB-based protocol was employed.
For the fungarium samples from the BGBM, pycnidia emerging from plant tissues were collected using a scalpel by cutting and scratching. Material (10-50 mg) of all specimens was grinded with liquid nitrogen (and additionally with bead-beating for a subset of the specimens). Given the difficulties in extracting high quality DNA, we used a CTAB-based protocol developed for plant herbarium material, following instructions (Borsch et al., 2003).
We used a PCR-based assay on the small subunit of the mitochondrial RNA marker (mtSSU), for the specific detection of the fungal pathogens S. sapinea and S. scrobiculata, using the primers Dpf-BotR and Dsf-BotR (Zhou and Stanosz, 2001; Smith and Stanosz, 2006). PCR reactions were performed in 25 μl containing 2 μl of DNA template, 0.2 mM of each dNTP, 0.3 μM of forward primers, 0.3 μM of reverse primers, 1X of KAPA-hifi buffer and 0.5 U of KAPA-hifi polymerase. PCR thermo-cycle conditions were as follows: initial denaturation at 95°C for 3 min, 30 cycles (40 cycles for fungarium specimens) of denaturation at 98°C for 20 s, annealing at 53°C for 30 s and elongation at 72°C for 45 s, and a final elongation at 72°C for 5 min. DNA amplifications were recorded by electrophoresis on 1.2% TBE agarose gels stained with RedGel before photographing. Failed PCRs were repeated, diluting DNA template to 1:10 and 1:100. Negative amplifications were considered when none of the dilutions gave positive amplification. To ensure that negative amplifications were not a result of failed DNA extractions, successful fungal DNA extractions were checked for each sample through PCR, using universal fungal primers ITS1-ITS4 (White et al., 1990; Gardes and Bruns, 1993). PCR reactions were performed in 25 μl containing 2 μl of DNA template, 0.2 mM of each dNTP, 0.3 μM of forward primers, 0.3 μM of reverse primers, 1X of KAPA-hifi buffer and 0.5 U of KAPA-hifi polymerase. PCR thermo-cycle conditions were as follows: initial denaturation at 95°C for 3 min, 30 cycles of denaturation at 98°C for 20 s, annealing at 55°C for 30 s and elongation at 72°C for 1 min, and a final elongation at 72°C for 5 min. The results of PCR amplification are summarized in Supplementary Table 1.
The association between host pine species and the presence or absence of S. sapinea in symptomatic or asymptomatic needles (Survey 1) was tested using Fisher’s exact test for count data and positive or negative associations were considered when Chi-squared test residuals were < −1.96 or > 1.96. To test for the effect of elevation on the probability of presence of S. sapinea (Survey 2), a logistic regression was performed using a generalized linear model (logit link function). This analysis was conducted for samples of symptomatic needles due to insufficient number of samples of asymptomatic needles.
Needles of P. sylvestris presenting shoot blight symptoms were cut into pieces of 1 cm and placed on Petri plates containing 1.5% malt extract agar. The plates were incubated at room temperature until colonies presenting S. sapinea characters were observed. The plates were subcultured until pure colonies presenting S. sapinea characters were obtained (Figure 1). Colonies presenting S. sapinea characters were selected for molecular specific detection of S. sapinea using a PCR-based assay as described above for the molecular detection of S. sapinea on pine needles and used for the growth experiments.
The experiments were conducted with the original isolate of Sphaeropsis sapinea (Figure 1O) that we isolated from symptomatic needles from a Pinus sylvestris tree from the Botanical Garden of Berlin. Plugs of fresh mycelium were extracted from the original isolate and cultivated in the dark in 8 cm diameter Petri dishes on 1.5% MAE, in incubators set at six different temperatures (4°C, 12°C, 20°C, 25°C, 30°C, and 35°C). Prior to incubation, we exposed one set of replicates to two temperature stresses induced by growing the fresh mycelium plug at 4°C or at 35°C overnight. Note that with this design, growth data at 4°C or 35°C after a stress at 4°C or 35°C, respectively, are exactly equivalent to continuously growing at 4°C or 35°C, respectively. Colony size was measured every day for one week. We calculated the mean colony extension over the week to obtain the mean growth rate per day. The experiment was conducted with three replicates per growth temperature.
We fit thermal reaction norms to growth rate measurements across the temperature range to estimate temperature niche parameters, namely optimal temperature, temperature minimal and maximal of growth and temperature niche width (the range of temperature where growth rate >0). We used a modified left-skewed model that describes the left-skewed shape of a thermal reaction norm following Thomas et al. (2012) and fitted using mle2 in the bbmle R package (Bolker and R Development Core Team, 2022). We further analyzed how prior exposure to a (low or high) temperature stress affected growth rate at (higher and lower, respectively) other temperatures. We calculated unpaired mean differences (effect size) between growth rates at a given temperature with and without a prior temperature stress exposure and estimated bootstrap 95% confidence interval using the dabestr R package (Ho et al., 2019).
Macroscopic photographs of shoot blight symptoms across the seven pine species were taken directly on individuals sampled for this study using a Sony DSC-H300 camera (Sony, Japan). Photographs of fresh symptomatic needles were taken using a Leica MDG41 stereomicroscope mounted with a Leica M165C camera with the software LASX 3.7.23463 (Leica Microsystems, Germany).
We performed scanning electron and fluorescence microscopy imaging of symptomatic needles collected from a Pinus sylvestris tree with clear S. sapinea emerging pycnidia and tested positive for S. sapinea with molecular methods. For scanning electron microscopy, fixed material was dehydrated using an increasing ethanol dilution series up to 100% and placed in acetone overnight before being critical-point dried using liquid CO2 and an automated Leica EM CPD 300 (Leica, Germany). Samples were then mounted on aluminum stub using nail polish and coated with gold using an automated Leica EM ACE 200 sputter coater (Leica, Germany), and observed and imaged using a scanning electron microscope Zeiss EVO10 at 20 kV (Zeiss, Germany). Any changes to the brightness, contrast or color balance were uniformly applied to every pixel.
For fluorescent microscopy, needles freshly collected or dried were placed in a mix of 1% calcofluor and 0.01% Auramine-O, observed under fluorescence illumination and a combination of BP and Alexa430 filters (blue and yellow in plates, respectively), and imaged using a microscope Zeiss Axio Imager M2 mounted with a Axiocam 506 color camera (Zeiss, Germany). Fluorescence microscopy was also performed on four other pine species to document recurrent patterns of infection similar to those observed on P. sylvestris.
Colonies with the characteristic morphology of S. sapinea were isolated from symptomatic P. sylvestris needles and were confirmed by PCR using the species-specific primers (Figure 1 and Supplementary Table 1). Tip blight symptoms were observed on all seven pine species but the presence of characteristic emerging pycnidia of S. sapinea on symptomatic needles, clearly observed on P. sylvestris, was not systematically observed on the other pine species (Figure 1).
Fluorescence and scanning electron microscopy imaging showed fungal hyphae supposedly of S. sapinea, emerging and entering through stomata, and that tended to agglomerate around the stoma while single running hyphae connected stomata (Figure 2). When present, pycnidia emerged from stomata and from which single hyphae were sometimes colonizing yet-uncolonized stomata. Fluorescence microscopy revealed diverse structures and patterns of fungal colonization on symptomatic needles where S. sapinea was detected by PCR sampled from the other pine species (Supplementary Figure 1), which agrees with the presence of multiple symptoms caused by different fungal pathogens on single needles (Figure 1). Different hyphal morphology and mycelium colonization patterns occurred within single needles, which could sometimes be observed as differently stained structures (blue or yellow, Supplementary Figure 1). Only in P. rigida could we observe colonization patterns similar to as observed on needles of P. sylvestris (Supplementary Figure 1).
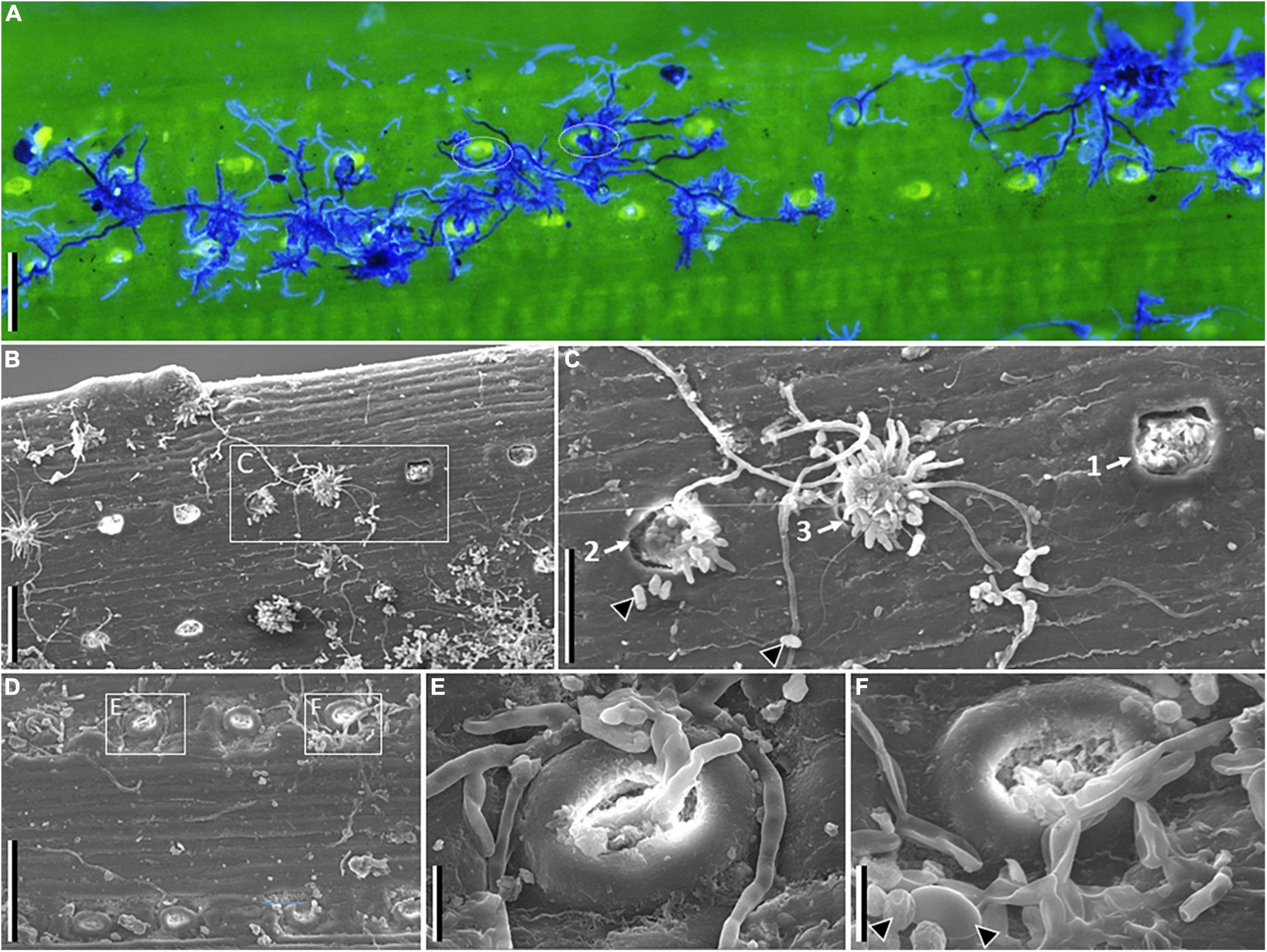
Figure 2. Microscopy images of symptomatic needles of Pinus sylvestris tested positive for Sphaeropsis sapinea. Fluorescence (A) and scanning electron (B–F) images. (A) Shows fungal hyphae in dark and light blue agglomerating around or growing between stomata. (B) Shows stomata at various stages of infection, and (C) developmental sequence of pycnidia emergence and hyphal growth (from 1 to 3). (D) Shows infected stomata, with details showing fungal hyphae emerging (E) or entering (F) stomata (arrowheads point to conidia in F). Scale bars: A, B, D = 100 μm; C = 50 μm; E, F = 10 μm.
In survey 1, S. sapinea was more often detected in symptomatic needles than asymptomatic needles (Fisher’s exact test for count data p-value = 9.546e-06; Figure 3A and Supplementary Table 2). In general, despite successful fungal DNA extraction for all samples (Supplementary Table 1), the CTAB protocol rendered a slightly higher probability of detection of S. sapinea than the soil kit protocol (Supplementary Figure 2). Among the three samples (3 trees) of asymptomatic needles (in P. sylvestris) in which S. sapinea was detected, two detections were obtained with CTAB only, and one with the soil kit only. Among the 23 samples (17 trees) of symptomatic needles in which S. sapinea was detected, detections were obtained with both methods for 12 samples (12 trees), only with CTAB for 6 samples (3 trees) and only with the soil kit for 5 samples (2 trees).
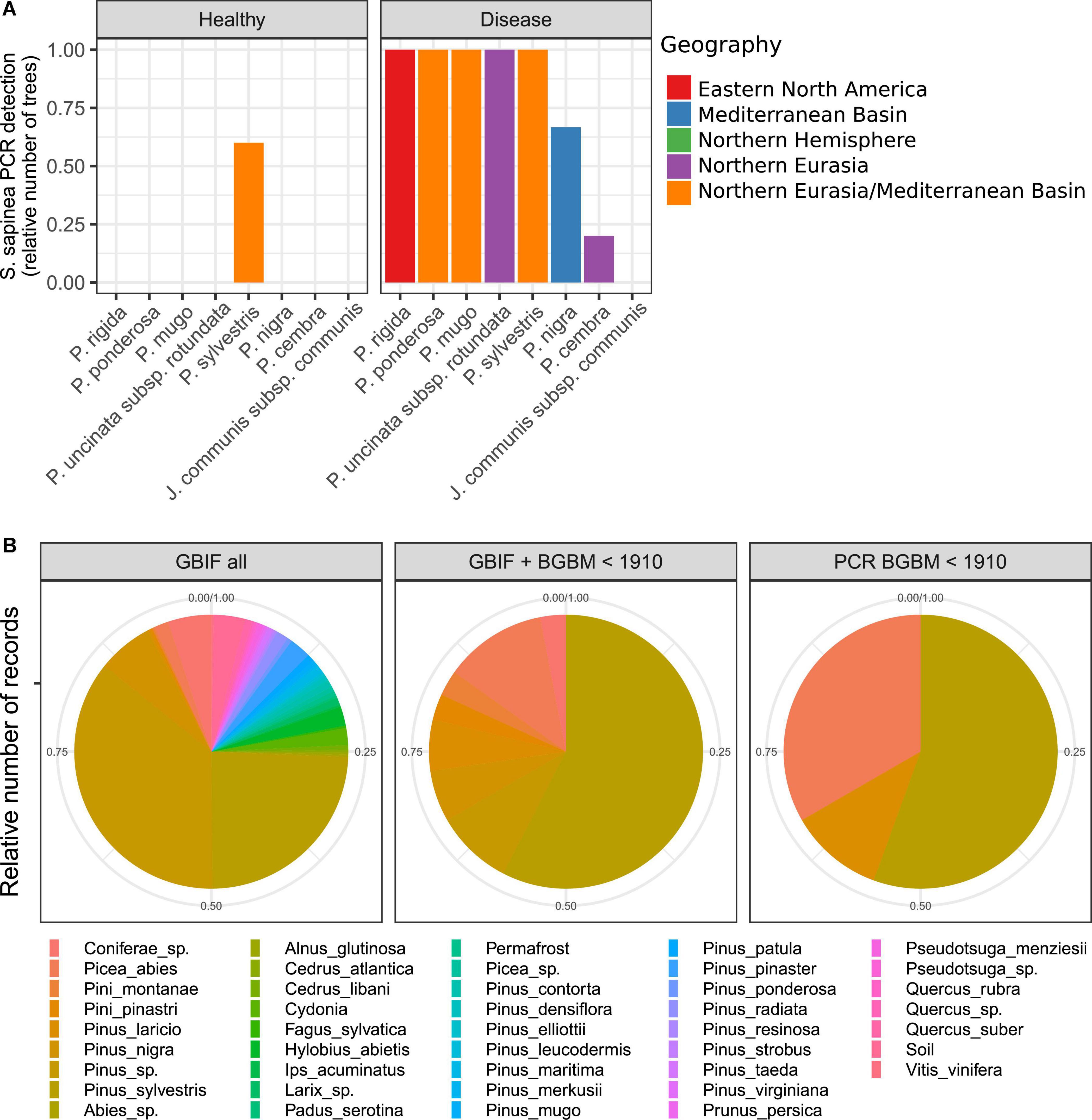
Figure 3. Distribution of Sphaeropsis sapinea across host pine species. PCR detection of Sphaeropsis sapinea across pine species and needle health status using species-specific DpF-BotR primers. (A) Positive PCR detection of S. sapinea across pine species and health status. Pinus rigida (n = 2 trees), P. ponderosa (n = 1), P. mugo (n = 5), P. uncinata subsp. rotundata (n = 3), P. sylvestris (n = 5), P. nigra (n = 3), P. cembra (n = 5), Junniperus communis (n = 1). (B) Host of records of S. sapinea based on all GBIF records (GBIF all; n = 351), on combined data from GBIF and fungarium specimens at Botanical Garden and Botanical Museum (BGBM) of Berlin before 1910 with a recorded host (GBIF + BGBM < 1910; n = 33), and the subset of specimens for which the identity of S. sapinea was confirmed by PCR species-specific assay (PCR BGBM < 1910; n = 9).
Aggregating PCR results across DNA extractions, S. sapinea was detected on all seven pine species (Figure 3A). In symptomatic needles, the fungus was detected in 100% of the trees sampled, with the exception of P. nigra (two detections out of three trees; Chi-squared residuals for the absence of S. sapinea = 0.38), the pine species exclusively of a Mediterranean Basin origin in this survey, and P. cembra (one detection out of five trees; Chi-squared residuals for the absence of S. sapinea = 2.68), the pine species exclusively of the Quinquefolia section in this survey, supporting a slight host effect (Fisher’s exact test for count data p-value = 0.009; Figure 3A and Supplementary Table 2). However, in asymptomatic needles, S. sapinea was observed only in P. sylvestris (non-supported host effect: Fisher’s exact test for count data p-value = 0.1226; but Chi-squared residuals for the presence of S. sapinea in P. sylvestris = 2.90; Figure 3A and Supplementary Table 2). As expected, S. sapinea was not detected in Juniperus communis.
A survey at GBIF records of S. sapinea occurrences and fungarium specimens at BGBM covering a period from 1873 to 2022 revealed 39 different hosts out of 351 entries with a reported host, of a total of 1,057 entries, of which 18 hosts were Pinus spp. (Figure 3B). P. sylvestris was the host for which most records were obtained (85 records), and this was even stronger before 1910 (19 records out of 33 entries with a reported host, of a total of 45 entries) (Figure 3B). Species-specific PCR assays on the subset of records based on material at BGBM, confirming the identity of the fungus, revealed similar results: out of 13 specimens with a recorded host and collected before 1910, a positive PCR amplification of S. sapinea was obtained for 9 specimens, of which 5 were collected from P. sylvestris (Figure 3B).
In survey 2, there was a clear decrease in S. sapinea detection with increasing elevation (Figure 4; symptomatic needles: estimate = −0.006158, p-value = 0.0315). In symptomatic needles, S. sapinea was detected in the two trees sampled at 200 m (one P. sylvestris and one P. nigra), in two out of three trees at 400 m of elevation (present in one P. sylvestris and in one P. mugo), and in one out of three trees (all P. sylvestris) at 800 m to 1,000 m of elevation. This tree was located in the most southern part of the survey and at 800 m of elevation (Supplementary Table 1). However, irrespective of the location of the sampled trees, S. sapinea was never detected between 1,000 m and 2,100 m of elevation despite relatively larger sampling of trees at high elevation (8 trees at or below 1,000 m against 20 trees above 1,000 m of elevation). In asymptomatic needles, the fungus was detected in one tree only (P. nigra) at the lowest elevation. Sphaeropsis scrobiculata was never detected, neither in survey 1 nor in survey 2 (Supplementary Table 1).
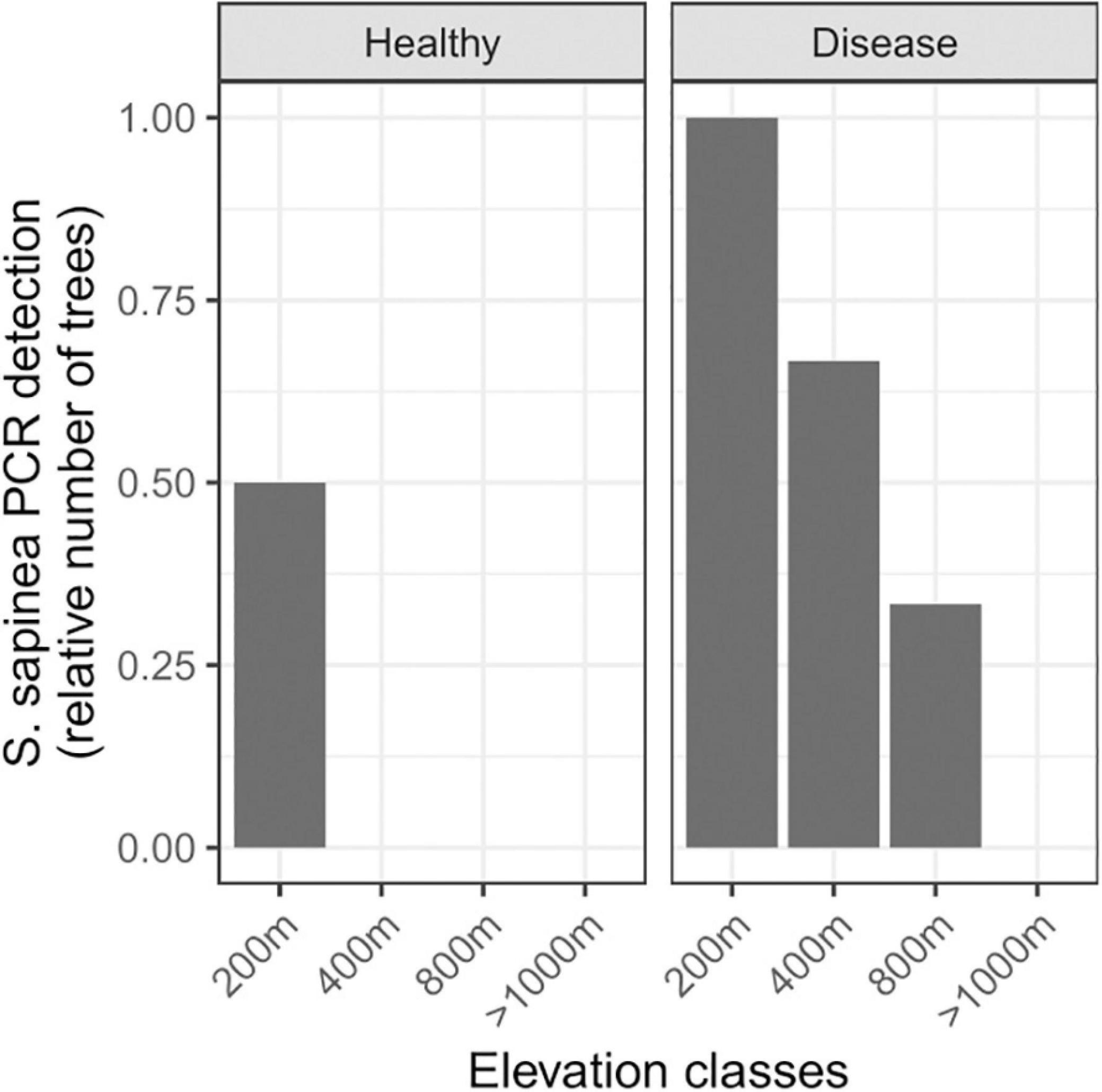
Figure 4. Distribution of Sphaeropsis sapinea across an elevation gradient in the French Alps. PCR detection of S. sapinea across health status and elevation. Symptomatic needle samples: from 200 m above sea level (asl) (n = 2), 400 m asl (n = 3), 800 m asl (n = 3), >1,000 m asl (up to 2,100 m asl, n = 20); asymptomatic needle samples: from 200 m asl (n = 2), 400 m asl (n = 2), 800 m asl (n = 2), >1,000 m asl (up to 1,600 m asl, n = 2).
The growth rate showed a left-skewed unimodal thermal reaction norm, irrespective of prior temperature stress, with an exponential increase from 4°C to 30°C and abrupt decrease after 30°C, and minimal growth at 35°C and 4°C (R2 = 0.94-0.99 for the three stress exposures; Figure 5A and Table 1). When the fungus had not been exposed to a previous temperature stress, the optimal growth temperature was estimated at 26.95°C (measure: 25°C), with a maximal growth rate of 5.986 mm.day–1, but remained high within 20°C to 30°C. Minimal and maximal estimated growth temperatures were 3.6°C and 35.35°C, respectively, resulting in a temperature niche width of 31.7°C.
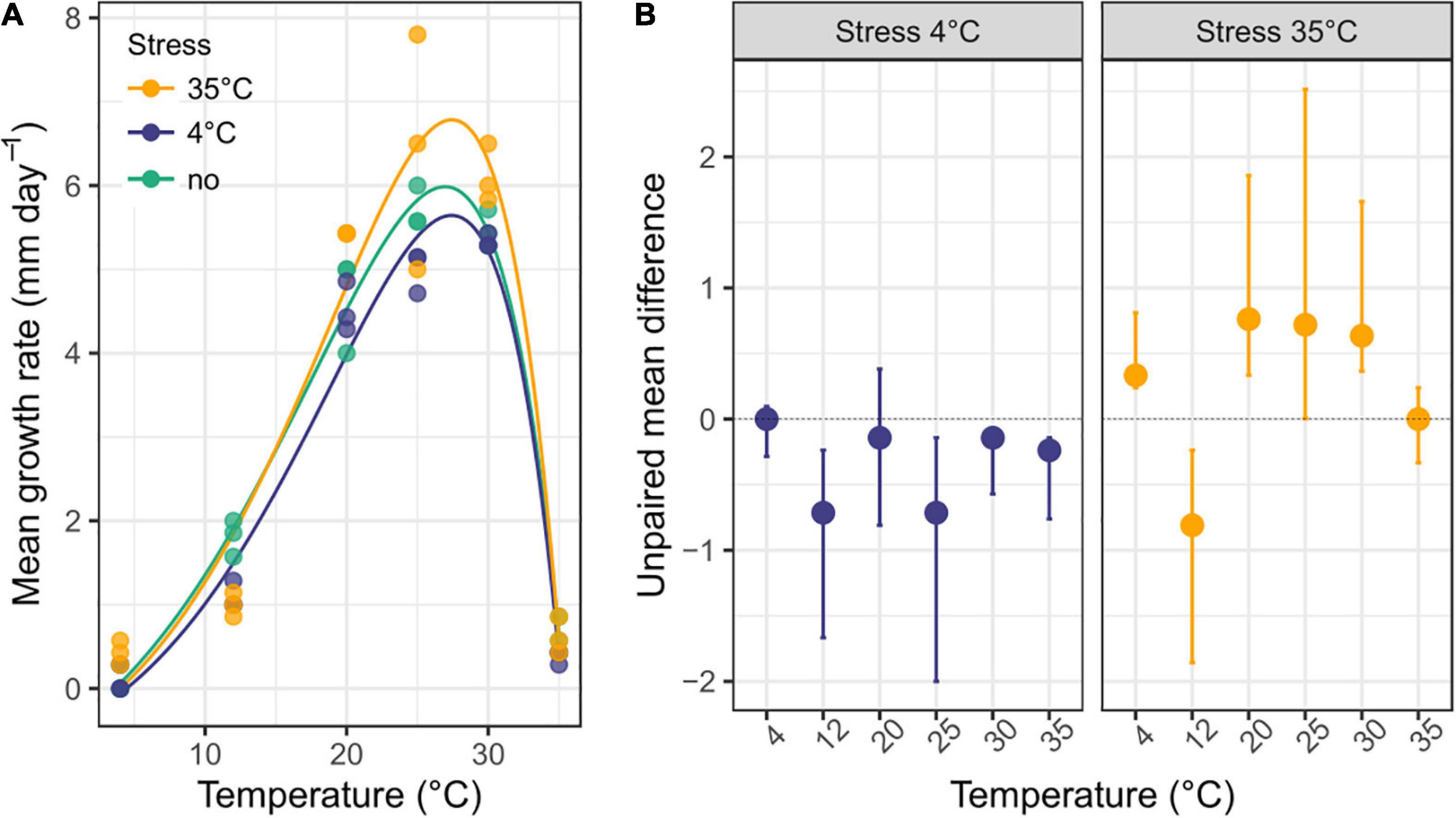
Figure 5. Thermal reaction norm of Sphaeropsis sapinea previously exposed or not to a temperature stress. (A) Mean growth rate (n = 3) of S. sapinea on 1.5% MAE across temperature without prior temperature stress or with a prior 4°C and 35°C temperature stress. The curve depicts the thermal reaction norm model fitted to the data. Parameter estimates of the thermal reaction norm are reported in Table 1. (B) Effect size (unpaired mean difference) and bootstrapping 95% confidence interval of prior temperature stress exposure on growth rate compared to when the fungus was not exposed to prior temperature stress.
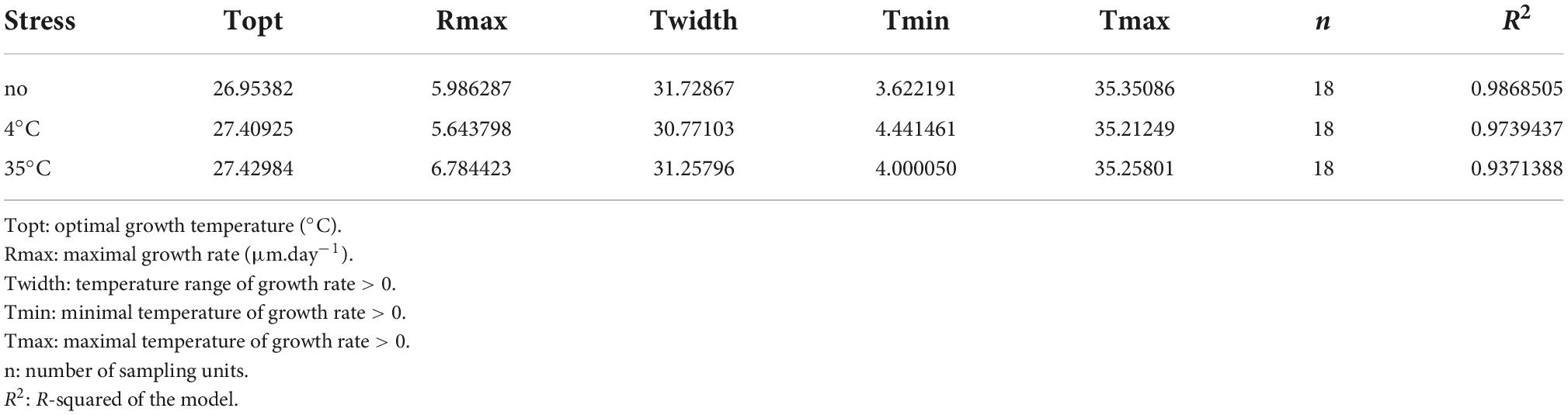
Table 1. Parameter estimates of the thermal reaction norm when Sphaeropsis sapinea has been exposed to a previous temperature stress or having not been exposed.
In general, S. sapinea grew respectively less and better after a previous 4°C and 35°C stress exposure (Table 1 and Figure 5B) compared to when the fungus was not exposed (that is, grown continuously at the same temperature). Overall, the maximum growth rate was lower after a previous 4°C stress exposure (5.64 mm.day–1), especially between 25 and 30°C, but higher after exposure at 35°C (6.78 mm.day–1), especially at 4°C, 20°C and 30°C (Table 1; Figure 5B). After both stresses, the maximal growth rate was achieved at optimal growth temperature 0.5°C higher (27.4°C; measured: 25°C) compared to when the fungus was not exposed to previous stress. However, the minimal and maximal growth temperatures were, respectively higher (4°C stress: 4.4°C; 35°C stress: 4.0°C) and lower (4°C stress: 35.2°C; 35°C stress: 35.3°C), resulting in a slightly smaller temperature niche width (4°C stress: 30.8°C; 35°C stress: 31.3°C) (Table 1).
Sphaeropsis sapinea is a worldwide pathogen observed on multiple hosts in their native and exotic distribution, especially Pinus species, but has also been reported from other Gymnosperms and Angiosperms to a lesser extent (see text footnote 2). Here, we provide novel data on host and abiotic constraints on the distribution of S. sapinea combining molecular-based field survey with historical records, cultivation-based growth experiments and microscopy. We report for the first time S. sapinea infecting P. mugo and P. uncinata (subsp. rotundata) in Germany, and P. cembra, P. rigida, and P. ponderosa in Europe. Our study is the second report of infection of P. cembra by S. sapinea, replicating the findings of Oblinger et al. (2009). Our results also confirm that S. sapinea is an endophyte in P. sylvestris (Flowers et al., 2001; Bußkamp et al., 2020; Blumenstein et al., 2021a; Terhonen et al., 2021) but not in the other species tested here, as we detected S. sapinea only in healthy needles of P. sylvestris and not in those of the other pine species screened at the single location of the Botanical Garden of Berlin.
In general, the available data suggest that at least in central Europe, P. sylvestris may be the main, and perhaps original, host of S. sapinea, one region suggested to be the centre of origin of the fungus (Burgess et al., 2004). In support of this hypothesis, most records of S. sapinea were obtained from P. sylvestris, and this was clear before 1910. Recent population genetic studies revealed high intraspecific diversity within S. sapinea in Baltic countries where P. sylvestris is native, suggesting that the fungus is long established in this region (Adamson et al., 2021). On the other hand, the higher intraspecific diversity in populations of North America suggests that S. sapinea may originate from North America, or in Asia or central America (Adamson et al., 2021). However, S. sapinea was not found here to be endophytic in P. rigida, the host species of North American origin. Nonetheless, P. sylvestris has been widely introduced in North America and certainly there is a good population size for some time to perhaps trigger diversification of the fungus. One scenario could be an introduction in Europe through a species of Pinus and then a switch to P. sylvestris, with rapid expansion. Another could be a switch to P. sylvestris from Europe in another pine species in North America and then back-introduction of infected P. sylvestris into Europe. Sphaeropsis sapinea has also been reported in healthy shoots of other pine species (this study: one P. nigra tree in France, Smith et al., 1996; Stanosz et al., 1997, 2005, 2007; Flowers et al., 2001; Maresi et al., 2007; Bihon et al., 2011), indicating that its presence as an endophyte in different pine species may also depend on additional factors. Shoots are probably the primary habitat for S. sapinea, at least in P. sylvestris (Flowers et al., 2001; Bußkamp et al., 2020), suggesting that by screening needles only, we possibly underestimated the susceptibility to infection for different hosts. On the other hand, infection in artificial settings, such as botanical gardens, may not be a reliable indicator whether the same species are similarly susceptible to infections in natural populations.
The geographical location of our first survey, the Botanical Garden in Berlin, Germany, near sea level, and the very nature of the location, a botanical garden, have important implications. First, botanical gardens and arboretums condense multiple potential host plants for fungal and other pathogens, many of them outside their native distribution, that would otherwise not naturally co-occur. Thus, while infection of novel hosts by fungal pathogens may not easily happen in the wild, it may be facilitated in such artificial settings. Infections as recorded here might thus not represent the true risk of pine species to be infected, but on the other hand, they could be a source for host switches that subsequently extend the novel association to natural populations of the new host. For example, the very first and only previous report of S. sapinea from P. cembra was observed in the University of Wisconsin-Madison campus, on an ornamental individual tree positioned in-between two infected P. nigra trees (Oblinger et al., 2009). Pine trees growing in arboretums are more susceptible to foliar fungal pathogens than wild trees under natural regeneration (Drenkhan et al., 2016). Thus, despite their invaluable scientific, educational and recreational contributions, botanical gardens, arboretums or ornamental gardens may be potential incubators for host-switching that could spread into the wild, a notion that deserves further attention (Newcombe and Dugan, 2010; Gougherty and Davies, 2021; Wondafrash et al., 2021).
Focusing on the Botanical Garden in Berlin also allowed sampling in the sense of a common garden experiment, controlling for broad variations in environmental conditions. It allowed us to screen for the infection by S. sapinea on hosts that naturally occur outside of the climatic niche of the fungus. Pinus cembra and P. mugo are typical alpine species, found above 1,400 m of elevation in alpine regions of Europe. Our second survey across an elevation gradient confirmed the absence of the fungus at higher elevation (Fabre et al., 2011), in particular above 800 m elevation, where mostly P. cembra and P. mugo were sampled. It was not found either in P. sylvestris at similar elevations. Our results based on molecular data are consistent with climatic niche modeling of S. sapinea based on visual assessment that has shown that the occurrence of the fungus, and severity of its infection, decreases with increasing elevation, a pattern probably related to temperature (especially mean daily minimum temperature in winter; Fabre et al., 2011). Accordingly, our experiment in controlled conditions showed that temperature strongly affects the growth of S. sapinea. Consistent with previous studies (Brookhouser and Peterson, 1970; Chou, 1982; Zlatkovic et al., 2016), the optimal growth was at 25-30°C, and it grew very slowly at lower temperature, especially at 4°C and 12°C. Moreover, a prior 4°C temperature stress decreased the ability of the fungus to grow at its optimal temperature. Thus, shoot blight at high elevation was not caused by S. sapinea. Shoot blight was also unlikely to be caused by S. scrobiculata since we did not detect this species. Which fungal species cause shoot blight at high elevation deserves further screening and experiments to assess risks fungal diseases pose to alpine pine forests.
Given these findings, Pinus cembra and P. mugo are therefore probably at low risk of infection in their natural distribution ranges, both because the species occur in a climatic niche barely overlapping with that of the fungus, and because they usually do not grow in close proximity to potentially infected individuals of the main host, P. sylvestris. However, as temperatures rise, larger areas of P. cembra and P. mugo forests will be within the climatic niche of S. sapinea (Bosso et al., 2017), most likely first at their distribution trailing edge. A rise in temperature may not only affect growth and distribution of the fungus, but also its life-history life-cycle, potentially resulting in higher germination, infection and sporulation rates and in an increased probability of overwintering (Brookhouser and Peterson, 1970). In addition, we show that the fungus may benefit from prior (temperature) stress exposure, a priming effect that could result in a positive feedback on its infection capability. Our growth experiments revealed that a prior exposure at 35°C of young mycelium increased the growth rate of the fungus at optimal temperature, and are supported by similar results of a previous study (Chou, 1982). This suggests that a positive effect through stress priming is possible (Hilker et al., 2016), which would then be an additional factor influencing the fungus-host dynamics under climate change scenarios when potentially new hosts are encountered. High intraspecific diversity has been revealed within S. sapinea (Adamson et al., 2021) with different virulences between strains (Bußkamp et al., 2021). Conducting similar growth experiments assessing temperature niche parameters and maximum growth rate coupled with niche modeling with isolates of different haplotypes would help predict the future distribution of S. sapinea strains and their potential risk.
Another issue is that P. sylvestris is being widely planted in alpine regions in close proximity to P. cembra or P. mugo forests, in ecosystems where it is not native, thus replicating the “botanical garden effect” in situ. Given the high dispersal capacity of S. sapinea (Burgess et al., 2004), it can be envisioned that S. sapinea will follow this artificial expansion of its main host, particularly under climate change scenarios. Our findings that P. cembra trees were little infected by S. sapinea support a low susceptibility of this species for this fungus (Oblinger et al., 2009) and suggest a low risk of outbreak. However, the susceptibility of P. cembra to S. sapinea could change as a function of increasing temperature or decreasing precipitation (increasing the water stress incurred by the trees) under climate change scenarios, as already observed for P. sylvestris (Blumenstein et al., 2021b) and in other tree-fungal pathogen interactions (Desprez-Loustau et al., 2006). In addition, host-switching in artificial landscapes, such as botanical gardens, and through planting of trees in situ outside their natural ecosystems may foster the evolution of new pathogen strains better adapted to novel hosts, possibly enabling them to extend their ranges across those of the new hosts. In contrast to P. cembra, P. mugo appears highly susceptible for the fungal pathogen, which raises additional concerns. Our results showed that the closely related species, P. uncinata (subsp. rotundata) (Jin et al., 2021) was also susceptible to S. sapinea and would therefore be unlikely to replace P. mugo in forestry programs if the goal was higher resistance against this fungal pathogen.
There are few microscopic photographs of shoot blight symptomatic needles and/or naturally-infected needles by Sphaeropsis sapinea (but see Li et al., 2019). Diverse macroscopic symptoms and microscopic patterns of colonization were observed among and within individual needles of the pine species investigated, suggesting the simultaneous infection by multiple fungi as observed by community-scale screening (Bußkamp et al., 2020, Oliva et al., 2021). Koch postulates have been fulfilled confirming S. sapinea alone being the causal agent of shoot tip blight (Oliva et al., 2013; Blumenstein et al., 2021b). However, this does not exclude the possibility of the disease being caused by species complexes (Drenkhan et al., 2016), especially for particular hosts, and determined by fungus-fungus interactions (Bußkamp et al., 2020; Blumenstein et al., 2021b; Oliva et al., 2021). Sphaeropsis sapinea is often associated with other pine fungal pathogens (Drenkhan et al., 2016; Markovskaja et al., 2016), and so it could be a secondary saprotrophic colonizer following infection by other fungal pathogens. The molecular data presented here suggest that S. sapinea might be more often the cause of shoot blight in Pinus sylvestris than in other pine species, in which it could be more often a secondary colonizer. A recent study shows that infected P. sylvestris trees are more likely to die from S. sapinea infection than trees from P. nigra, P. pinea or P. halepensis (Caballol et al., 2022).
Different pathogens have different infection approaches, as observed with the multiple patterns of hyphal colonization on needles: for example, the fungus Rhizoctonia solani, infecting and causing maize sheath blight, can penetrate through the leaf epidermis and stomata (Jie et al., 2000). S. sapinea can colonize new shoots either through stomata or tender shoots (Li et al., 2019). Wounds are also considered as important entry points (Swart and Wingfield, 1991; Blodgett et al., 2021). Detailed images on symptomatic needles of P. sylvestris with clear S. sapinea emerging pycnidia and which tested positive for S. sapinea with molecular methods revealed agglomeration of hyphae around stomata while single running hyphae connected various stomata. Images clearly revealed the emergence of pycnidia from stomata. Hypothesized germ tubes (short outgrowths, non-septate germinating hyphae produced by spores) and mature hyphae emerging from stomata were pictured, probably germinating from conidia produced by hyphae already present within needle tissues. Hyphae entering were also pictured, as observed by other authors (Li et al., 2019), but the original conidia could not be confirmed. No intrusion of germ tubes or mature hyphae extending directly into the needles were observed. Considering the endophytic habit of the fungus (Bußkamp et al., 2020), it is unknown what the entry or exit point in organs such as needles would be. Irrespective of stoma being an entry or exit point for S. sapinea, or likely both, our findings support other findings that leaf wounds are not necessary for infection (Li et al., 2019). Similar patterns of infection were observed on P. rigida. It is possible that the infection process of S. sapinea is similar between P. sylvestris, the potential main host of S. sapinea, and P. rigida, the host of North American origin where the fungus may originate. The absence of recurrent patterns of infections on other hosts suggest a different colonization process and localization. Certain physical and chemical features of the host needle surface can influence conidia germination, germ tube growth, and appressoria formation (Wynn, 1981; Howard and Valent, 1996; Dean, 1997), which would determine the function of the fungus and the host-fungus interaction.
To conclude, the distribution of S. sapinea is more limited by temperature than constrained by pine host species identity. Yet, a slight host effect is also observed, P. sylvestris being more likely exposed to Diplodia tip blight due to the endophytic infection of S. sapinea of its needles. Our results support the notion that botanical gardens and arboretums may act as potential incubators for host-switching of fungal pathogens and that in the current context of global climate change, S. sapinea could infect a wider range of pine species and locations if temperature keeps increasing.
The original contributions presented in this study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
JR designed the research with inputs from RL. JR performed the sampling. JR, IK, and NR performed the molecular analyses. JR and IK performed the growth experiments. JR and JBB performed the microscopy analyses. MCR and JBB contributed to the lab facilities and reagents. JR analyzed the data and wrote the manuscript with inputs from RL, JBB, and MCR. All authors read and approved the final version of the manuscript.
We gratefully thank Gerald Parolly for sampling permission at Botanical Garden Berlin. We thank Nana Silakadze, François Roy, Françoise Roy, Max Fussan, and Eva Troner for help during sampling. We thank the administration at Parc Naturel Régional du Vercors for sampling-related information. We also thank Isabel A. Munck and two reviewers for their valuable comments that improve the manuscript. Open Access funding provided by the Freie Universität Berlin.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2022.971916/full#supplementary-material
Supplementary Table 1 | Sample metadata and results of PCR-detection of Sphaeropsis sapinea.
Adamson, K., Klavina, D., Drenkhan, R., Gaitnieks, T., and Hanso, M. (2015). Diplodia sapinea is colonizing the native Scots pine (Pinus sylvestris) in the northern Baltics. Eur. J. Plant Pathol. 143, 343–350. doi: 10.1007/s10658-015-0686-8
Adamson, K., Laas, M., Blumenstein, K., Busskamp, J., Langer, G. J., Klavina, D., et al. (2021). Highly clonal structure and abundance of one haplotype characterise the Diplodia sapinea populations in Europe and Western Asia. J. Fungi 7:634. doi: 10.3390/jof7080634
Anderson, P. K., Cunningham, A. A., Patel, N. G., Morales, F. J., Epstein, P. R., and Daszak, P. (2004). Emerging infectious diseases of plants: Pathogen pollution, climate change and agrotechnology drivers. Trends Ecol. Evol. 19, 535–544. doi: 10.1016/j.tree.2004.07.021
Aragonés, A., Manzanos, T., Stanosz, G., Munck, I. A., Raposo, R., Elvira-Recuenco, M., et al. (2021). Comparison of Diplodia tip blight pathogens in Spanish and North American pine ecosystems. Microorganisms 9:2565. doi: 10.3390/microorganisms9122565
Bihon, W., Burgess, T., Slippers, B., Wingfield, M. J., and Wingfield, B. D. (2011). Distribution of Diplodia pinea and its genotypic diversity within asymptomatic Pinus patula trees. Australas. Plant Pathol. 40, 540–548. doi: 10.1007/s13313-011-0060-z
Blodgett, J. T., Kruger, E. L., and Stanosz, G. R. (1997). Sphaeropsis sapinea and water stress in a red pine plantation in Central Wisconsin. Phytopathology 87, 429–434. doi: 10.1094/PHYTO.1997.87.4.429
Blodgett, J. T., Smith, D., and Stanosz, G. (2021). First report of Diplodia shoot blight and canker disease caused by Diplodia sapinea on Ponderosa pine in Wyoming, USA. Plant Dis. [Epub ahead of print]. doi: 10.1094/PDIS-03-21-0671-PDN
Blumenstein, K., Bußkamp, J., Langer, G. J., Langer, E. J., and Terhonen, E. (2021a). The Diplodia tip blight pathogen Sphaeropsis sapinea is the most common fungus in Scots pines’ mycobiome irrespective of health status — A case study from Germany. J. Fungi 7:607. doi: 10.3390/jof7080607
Blumenstein, K., Bußkamp, J., Langer, G. J., Schlößer, R., Rojas Parra, N. M., and Terhonen, E. (2021b). Sphaeropsis sapinea and associated endophytes in Scots pine: Interactions and effect on the host under variable water content. Front. For. Glob. Change 4:655769. doi: 10.3389/ffgc.2021.655769
Bolker, B., and R Development Core Team (2022). bbmle: Tools for general maximum likelihood estimation. R package version 1.0.25. Available Online at: https://CRAN.R-project.org/package=bbmle
Borsch, T., Hilu, K. W., Quandt, D., Wilde, V., Neinhuis, C., and Barthlott, W. (2003). Noncoding plastid trnT-trnF sequences reveal a well resolved phylogeny of basal angiosperms. J. Evol. Biol. 16, 558–576. doi: 10.1046/j.1420-9101.2003.00577.x
Bosso, L., Luchi, N., Maresi, G., Cristinzio, G., Smeraldo, S., and Russo, D. (2017). Predicting current and future disease outbreaks of Diplodia sapinea shoot blight in Italy: Species distribution models as a tool for forest management planning. For. Ecol. Manag. 400, 655–664. doi: 10.1016/j.foreco.2017.06.044
Brodde, L., Adamson, K., Camarero, J. J., Castaño, C., Drenkhan, R., Lehtijärvi, A., et al. (2019). Diplodia tip blight on its way to the north: Drivers of disease emergence in Northern Europe. Front. Plant Sci. 9:1818. doi: 10.3389/fpls.2018.01818
Brookhouser, L., and Peterson, G. W. (1970). Infection of Austrian, Scots, and ponderosa pines by Diplodia pinea. Phytopathology 61, 409–414. doi: 10.1094/Phyto-61-409
Burgess, T. I. A., Wingfield, M. J., and Wingfield, B. D. (2004). Global distribution of Diplodia pinea genotypes revealed using simple sequence repeat (SSR) markers. Australas. Plant Pathol. 33, 513–519. doi: 10.1071/AP04067
Bußkamp, J. (2018). Schadenserhebung, kartierung und charakterisierung des Diplodia-triebsterbens“ der kiefer, insbesondere des endophytischen vorkommens in den klimasensiblen räumen und identifikation von den in kiefer (Pinus sylvestris) vorkommenden endophyten. Kassel: Universität Kassel.
Bußkamp, J., Blumenstein, K., Terhonen, E., and Jutta, G. (2021). Differences in the virulence of Sphaeropsis sapinea strains originating from Scots pine and non-pine hosts. For. Pathol. 51:e12712. doi: 10.1111/efp.12712
Bußkamp, J., Langer, G. J., and Langer, E. J. (2020). Sphaeropsis sapinea and fungal endophyte diversity in twigs of Scots pine (Pinus sylvestris) in Germany. Mycol. Prog. 19, 985–999. doi: 10.1007/s11557-020-01617-0
Caballol, M., Ridley, M., Colangelo, M., Valeriano, C., Julio Camarero, J., and Oliva, J. (2022). Tree mortality caused by Diplodia shoot blight on Pinus sylvestris and other Mediterranean pines. For. Ecol. Manag. 505:119935. doi: 10.1016/j.foreco.2021.119935
Chou, C. K. S. (1982). Diplodia pinea infection of Pinus radiata seedlings: Effect of temperature and shoot wetness duration. N. Z. J. For. Sci. 12:425–437.
de Wet, J., Burgess, T., Slippers, B., Preisig, O., Wingfield, B. D., and Wingfield, M. J. (2003). Multiple gene genealogies and microsatellite markers reflect relationships between morphotypes of Sphaeropsis sapinea and distinguish a new species of Diplodia. Mycol. Res. 107, 557–566. doi: 10.1017/S0953756203007706
Dean, R. A. (1997). Signal pathways and appressorium morphogenesis. Annu. Rev. Phytopathol. 35, 211–234. doi: 10.1146/annurev.phyto.35.1.211
Desprez-Loustau, M.-L., Marcais, B., Nageleisen, L.-M., Piou, D., and Vannini, A. (2006). Interactive effects of drought and pathogens in forest trees. Annu. For. Sci. 63, 597–612. doi: 10.1051/forest:2006040
Drenkhan, R., Tomešová-Haataja, V., Fraser, S., Bradshaw, R. E., Vahalík, P., Mullett, M. S., et al. (2016). Global geographic distribution and host range of Dothistroma species: A comprehensive review. For. Pathol. 46, 408–442. doi: 10.1111/efp.12290
Fabre, B., Piou, D., Desprez-Loustau, M.-L., and Marçais, B. (2011). Can the emergence of pine Diplodia shoot blight in France be explained by changes in pathogen pressure linked to climate change? Glob. Change Biol. 17, 3218–3227. doi: 10.1111/j.1365-2486.2011.02428.x
Flowers, J., Nuckles, E., Hartman, J., and Vaillancourt, L. (2001). Latent infection of Austrian and Scots pine tissues by Sphaeropsis sapinea. Plant Dis. 85, 1107–1112.
Gardes, M., and Bruns, T. (1993). ITS primers with enhanced specificity for Basidiomycetes: Application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118.
Gougherty, A. V., and Davies, T. J. (2021). Towards a phylogenetic ecology of plant pests and pathogens. Philos. Trans. R. Soc. B Biol. Sci. 376:20200359. doi: 10.1098/rstb.2020.0359
Hilker, M., Schwachtje, J., Baier, M., Balazadeh, S., Isabel, B., Geiselhardt, S., et al. (2016). Priming and memory of stress responses in organisms lacking a nervous system. Biol. Rev. 91, 1118–1133. doi: 10.1111/brv.12215
Ho, J., Tumkaya, T., Aryal, S., Choi, H., and Claridge-Chang, A. (2019). Moving beyond P values: Data analysis with estimation graphics. Nat. Methods 16, 565–566. doi: 10.1038/s41592-019-0470-3
Howard, R. J., and Valent, B. (1996). Breaking and entering: Host penetration by the fungal rice blast pathogen Magnaporthe grisea. Annu. Rev. Microbiol. 50, 491–512. doi: 10.1146/annurev.micro.50.1.491
Jie, C., Chaorong, T., Zenggui, G., Chunsheng, X., Xiaofan, N., and Zuoheng, S. (2000). On penetration process of sheath blight pathogen in maize. J. Shenyang Agric. Univ. 31, 503–506.
Jin, W.-T., Gernandt, D. S., Wehenkel, C., Xia, X.-M., and Wei, X.-X. (2021). Phylogenomic and ecological analyses reveal the spatiotemporal evolution of global pines. Proc. Natl. Acad. Sci. U.S.A. 118:e2022302118. doi: 10.1073/pnas.2022302118
Li, X., Liu, B., Ren, X., Cui, J., and Li, H. (2019). Infection approach of Diplodia sapinea, the causal agent of tip blight of Pinus tabulaeformis in China. For. Pathol. 49:e12544. doi: 10.1111/efp.12544
Luchi, B. N., Oliveira Longa, C. M., Danti, R., Capretti, P., and Maresi, G. (2014). Diplodia sapinea: The main fungal species involved in the colonization of pine shoots in Italy. For. Pathol. 44, 372–381. doi: 10.1111/efp.12109
Maresi, B. G., Luchi, N., Pinzani, P., Pazzagli, M., and Capretti, P. (2007). Detection of Diplodia pinea in asymptomatic pine shoots and its relation to the Normalized Insolation index. For. Pathol. 37, 272–280. doi: 10.1111/j.1439-0329.2007.00506.x
Markovskaja, S., Kaèergius, A., Davydenko, K., and Fraser, S. (2016). First record of Neocatenulostroma germanicum on pines in Lithuania and Ukraine and its co-occurrence with Dothistroma spp. and other pathogens. For. Pathol. 46, 522–533. doi: 10.1111/efp.12308
Munck, I. A., Smith, D. R., Sickley, T., and Stanosz, G. R. (2009). Site-related influences on cone-borne inoculum and asymptomatic persistence of Diplodia shoot blight fungi on or in mature red pines. For. Ecol. Manag. 257, 812–819. doi: 10.1016/j.foreco.2008.10.023
Newcombe, G., and Dugan, F. M. (2010). “Fungal pathogens of plants in the homogocene,” in Molecular identification of fungi, eds Y. Gherbawy and K. Voigt (Berlin: Springer), 3–34. doi: 10.1007/978-3-642-05042-8_1
Oblinger, B. W., Smith, D. R., and Stanosz, G. R. (2009). An ornamental Swiss stone pine (Pinus cembra) in Wisconsin is a host of the shoot blight pathogen Diplodia pinea. Plant Dis. 93:845. doi: 10.1094/PDIS-93-8-0845B
Oliva, J., Boberg, J., and Stenlid, J. (2013). First report of Sphaeropsis sapinea on Scots pine (Pinus sylvestris) and Austrian pine (P. nigra) in Sweden. New Dis. Rep. 27:23. doi: 10.5197/j.2044-0588.2013.027.023
Oliva, J., Ridley, M., Redondo, M. A., and Caballol, M. (2021). Competitive exclusion amongst endophytes determines shoot blight severity on pine. Funct. Ecol. 35, 239–254.
Piou, D., Chandelier, P., and Morelet, M. (1991). Un nouveau problème sanitaire des pins en France? Rev. Forestiere Fr. 43:203–213.
Santamaria, O., Smith, D., and Stanosz, G. (2012). Interaction between Diplodia pinea or Diplodia scrobiculata and fungal endophytes isolated from pine shoots. Can. J. For. Res. 42, 1819–1826. doi: 10.1139/x2012-132
Santini, A., Pepori, A., Ghelardini, L., and Capretti, P. (2008). Persistence of some pine pathogens in coarse woody debris and cones in a Pinus pinea forest. For. Ecol. Manag. 256, 502–506. doi: 10.1016/j.foreco.2008.05.010
Seidl, R., Thom, D., Kautz, M., Dario, M. B., Peltoniemi Mikko Vacchiano, G., et al. (2017). Forest disturbances under climate change. Nat. Clim. Change 7, 395–402.
Smith, D. R., and Stanosz, G. R. (2006). A species-specific PCR assay for detection of Diplodia pinea and D. scrobiculata in dead red and jack pines with collar rot symptoms. Plant Dis. 90, 307–313. doi: 10.1094/PD-90-0307
Smith, H., Wingfield, M. J., Crous, P., and Coutinho, T. A. (1996). Sphaeropsis sapinea and Botryosphaeria dothidea endophytic in Pinus spp. and Eucalyptus spp. in South Africa. S. Afr. J. Bot. 62, 86–88. doi: 10.1016/S0254-6299(15)30596-2
Solel, Z., Madar, Z., Kimchi, M., and Golan, Y. (1987). Diplodia canker of cypress. Can. J. Plant Pathol. 9, 115–188.
Stanosz, G. R., Blodgett, J. T., Smith, D. R., and Kruger, E. L. (2001). Water stress and Sphaeropsis sapinea as a latent pathogen of red pine seedlings. New Phytol. 149, 531–538. doi: 10.1046/j.1469-8137.2001.00052.x
Stanosz, B. G. R., Smith, D. R., and Albers, J. S. (2005). Surveys for asymptomatic persistence of Sphaeropsis sapinea on or in stems of red pine seedlings from seven Great Lakes region nurseries. For. Pathol. 35, 233–244. doi: 10.1111/j.1439-0329.2005.00407.x
Stanosz, G. R., Smith, D. R., Guthmiller, M. A., and Stanosz, J. C. (1997). Persistence of Sphaeropsis sapinea on or in asymptomatic shoots of red and jack pines. Mycologia 89, 525–530. doi: 10.1080/00275514.1997.12026813
Stanosz, G. R., Smith, D. R., and Leisso, R. (2007). Diplodia shoot blight and asymptomatic persistence of Diplodia pinea on or in stems of jack pine nursery seedlings. For. Pathol. 37, 145–154. doi: 10.1111/j.1439-0329.2007.00487.x
Swart, W. J., and Wingfield, M. J. (1991). Biology and control of Sphaeropsis sapinea on Pinus species in South Africa. Plant Dis. 75, 761–766. doi: 10.1094/PD-75-0761
Terhonen, E., Babalola, J., Kasanen, R., Jalkanen, R., and Blumenstein, K. (2021). Sphaeropsis sapinea found as symptomless endophyte in Finland. Silva Fenn. 55:10420. doi: 10.14214/sf.10420
Thomas, M. K., Kremer, C. T., Klausmeier, C. A., and Litchman, E. (2012). A global pattern of thermal adaptation in marine phytoplankton. Science 338, 1085–1088. doi: 10.1126/science.1224836
White, T., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics”, in PCR protocols a guide to methods and applications (San Diego CA: Academic Press), 315–322.
Wondafrash, M., Wingfield, M. J., Wilson, J. R., Hurley, B. P., Slippers, B., and Paap, T. (2021). Botanical gardens as key resources and hazards for biosecurity. Biodivers. Conserv. 30, 1929–1946. doi: 10.1007/s10531-021-02180-0
Woods, A. J., Martín-García, J., Bulman, L., Vasconcelos, M. W., Boberg, J., La Porta, N., et al. (2016). Dothistroma needle blight, weather and possible climatic triggers for the disease’s recent emergence. For. Pathol. 46, 443–452. doi: 10.1111/efp.12248
Wynn, W. K. (1981). Tropic and taxic response of pathogens to plants. Annu. Rev. Phytopathol. 19, 237–255. doi: 10.1146/annurev.py.19.090181.001321
Zhou, S., and Stanosz, G. R. (2001). Primers for amplification of mt SSU rDNA, and a phylogenetic study of Botryosphaeria and associated anamorphic fungi. Mycol. Res. 105, 1033–1044. doi: 10.1016/S0953-7562(08)61965-6
Zlatkovic, M., Keca, N. D., Wingfiel, M. J., Jami, F., and Slippers, B. (2016). Botryosphaeriaceae associated with the die-back of ornamental trees in the Western Balkans. Antonie van Leeuwenhoek. 109, 543–564. doi: 10.1007/s10482-016-0659-8
Keywords: biogeography, forest pathogens, global change, host specificity, invasive species, plant disease, stress priming
Citation: Roy J, Kyritsi I, Reinwarth N, Bachelier JB, Rillig MC and Lücking R (2022) Host and abiotic constraints on the distribution of the pine fungal pathogen Sphaeropsis sapinea (= Diplodia sapinea). Front. For. Glob. Change 5:971916. doi: 10.3389/ffgc.2022.971916
Received: 17 June 2022; Accepted: 17 October 2022;
Published: 08 December 2022.
Edited by:
Isabel Alvarez Munck, Forest Health Protection, Forest Service (USDA), United StatesReviewed by:
Angus J. Carnegie, New South Wales Department of Primary Industries, AustraliaCopyright © 2022 Roy, Kyritsi, Reinwarth, Bachelier, Rillig and Lücking. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julien Roy, cm95anVsaWVuQHplZGF0LmZ1LWJlcmxpbi5kZQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.