- Department of Biology, Boston University, Boston, MA, United States
Projections for the northeastern United States indicate that mean air temperatures will rise and snowfall will become less frequent, causing more frequent soil freezing. To test fungal responses to these combined chronic and extreme soil temperature changes, we conducted a laboratory-based common garden experiment with soil fungi that had been subjected to different combinations of growing season soil warming, winter soil freeze/thaw cycles, and ambient conditions for 4 years in the field. We found that fungi originating from field plots experiencing a combination of growing season warming and winter freeze/thaw cycles had inherently lower activity of acid phosphatase, but higher cellulase activity, that could not be reversed in the lab. In addition, fungi quickly adjusted their physiology to freeze/thaw cycles in the laboratory, reducing growth rate, and potentially reducing their carbon use efficiency. Our findings suggest that less than 4 years of new soil temperature conditions in the field can lead to physiological shifts by some soil fungi, as well as irreversible loss or acquisition of extracellular enzyme activity traits by other fungi. These findings could explain field observations of shifting soil carbon and nutrient cycling under simulated climate change.
Introduction
Warming in high-latitude biomes is projected to decrease snowpack in winter, causing soils to freeze more frequently (Groffman et al., 2001). These dual soil temperature shifts – warming during the growing season and increased frequency of freeze/thaw cycles in winter – can shift soil microbial communities, as well as their growth rates and C-cycling enzyme activities, either separately (Lipson and Schmidt, 2004; Allison and Treseder, 2008; Aanderud et al., 2013; Buckeridge et al., 2013; Luo et al., 2014; DeAngelis et al., 2015; Pold et al., 2015; Fernandez et al., 2016), or in combination (Sorensen et al., 2018; Garcia et al., 2020). However, it is still unclear why such shifts occur. Is it due to plasticity in traits of some taxa, in which rates of respiration or other physiological activities adjust to compensate for changes in temperature (Malcolm et al., 2008; Allison et al., 2010; Yuste-Lisbona et al., 2010; Crowther and Bradford, 2013; Tucker et al., 2013); evolution of certain taxa in response to new environmental conditions (Romero-Olivares et al., 2015); or selection for taxa that can survive extreme environmental changes, leading to the establishment or loss of species with narrow climate envelopes (Rousk et al., 2012; Luo et al., 2014; Treseder et al., 2016)? Answering this question for soil fungal communities is particularly important, as fungi are major decomposers in northern terrestrial systems (Schneider et al., 2012), responsible for the release of at least 36 Gt of carbon (C) to the atmosphere annually as carbon dioxide (CO2) through respiration (Talbot, 2017). Because specific fungal taxa can have unique roles in cycling C, nitrogen (N), and other elements through the biosphere (Hanson et al., 2008; Koide and Malcolm, 2009; McGuire et al., 2010), understanding how fungal species respond to projected climate change (i.e., their response traits) and how these responses influence ecosystem biogeochemistry (i.e., their effect traits) will help us predict impacts of climate change on biogeochemistry of ecosystems (Koide et al., 2014).
Soil freeze/thaw cycles may exert particularly strong selection for taxa that can survive extreme fluctuations in temperature, moisture, and physical structure (Ostroumov and Siegert, 1996), as well as changes in plant C availability belowground due to root damage (Sanders-DeMott et al., 2018). In field soils, combined warming and soil freeze/thaw cycles that have been applied experimentally reduced microbial biomass and nutrient immobilization in soil (Sorensen et al., 2018). These biogeochemical changes indicate an overall reduction in soil microbial activity that could be driven by selection for taxa that can survive extreme environmental changes, but have low intrinsic capacity to cycle C and nutrients (Garcia et al., 2020). Fungi originating from polar climates, for example, have developed mechanisms to survive extreme conditions, including higher concentrations of sugars, alcohols, lipids and fatty acids, or antifreeze proteins in their cells compared to their mesophilic relatives (Robinson, 2001). Indeed, combined warming and increased soil freeze/thaw cycles in winter cause greater shifts in soil microbial community composition than increased warming alone (Garcia et al., 2020). Increases in the frequency of freeze/thaw cycles in temperate ecosystems may select for fungal species that intrinsically allocate more of their resources to stress-tolerance than to growth and decomposition traits, relative to communities experiencing growing season warming alone.
In addition, evidence is accumulating that many fungi can acclimate or adjust their behavior to compensate for changes in climate. In the laboratory, some taxa display thermal plasticity, increasing respiration rates with increasing temperature (Luo et al., 2001; Malcolm et al., 2008; Crowther and Bradford, 2013). This activity change may be due to decreased efficiency of substrate use (i.e., carbon use efficiency) (Frey et al., 2013), which represents the amount of fungal biomass produced (vs. C respired) per unit C uptake. Short-term warming can increase microbial production of exoenzymes that target labile plant biopolymers (i.e., cellulose) (Pold et al., 2016; Anthony et al., 2021), indicating rapid uptake of labile C, while other studies note initial positive effects of experimentally warmed soils on rates of microbial respiration (Romero-Olivares et al., 2017). By contrast, selection for taxa that generate high oxidase activity (which targets more recalcitrant C molecules, e.g., lignin) – can occur after longer-term (>20 years) warming conditions deplete labile C pools in soil (Pold et al., 2015; Chen et al., 2018, 2020). Because fungal species differ in their intrinsic ability to adjust morphology, behavior, or physiological states based on environmental conditions (Westeberhard, 1989), the ability of some species (and not others) to acclimate to new temperatures—especially in terms of their labile C (e.g., cellulose) use or their carbon use efficiency—may account for shifts in fungal community composition observed with experimental warming (Allison and Treseder, 2008; Luo et al., 2014; DeAngelis et al., 2015; Pold et al., 2015; Fernandez et al., 2016). Alternatively, some fungal taxa may evolve in response to elevated temperatures on relatively short timescales—as short as a few weeks or months in some cases—aided by their relatively short generation times (Leu and Murray, 2006; Bradford et al., 2008; Dettman et al., 2008; Romero-Olivares et al., 2015). For example, Neurospora discreta grown at elevated temperatures produces more spores per unit biomass and higher rates of respiration after 1,500 generations relative to the parental strains, but appears to trade off mycelial growth and biomass accumulation for increases in spore production (Romero-Olivares et al., 2015). Resilience of fungal communities to other environmental changes, like N deposition, also appears to be related to the evolution of fundamental biological activities (e.g., growth rates and extracellular enzyme activities) in fungi after long-term exposure to N-enriched field conditions (van Diepen et al., 2017).
The goal of this study was to determine how warmer growing season temperatures and more frequent winter freeze/thaw cycles affect soil fungi—whether through plasticity, irreversible acquisition of traits (potentially through evolutionary responses), or selection for unique taxa—and how fungal responses to year-round climate change are linked to their potential to affect soil biogeochemistry (specifically, growth and extracellular enzyme activity). If extreme soil temperature conditions (e.g., warming and soil freeze/thaw cycles) cause fungal species to acclimate or evolve, we would expect to find similar suites of species as in ambient conditions; individuals exposed to extreme conditions would likely have higher capacities to withstand stress, but lower abilities to degrade soil organic matter and build biomass compared to fungi that have been exposed to ambient conditions. If fungi evolved, rather than acclimated, to climate change conditions, their activity (characterized by growth and enzyme activity) would not be readily reversed when grown under reference conditions. Alternatively, extreme climate conditions could simply select for species—those already adapted to extreme climate conditions (warming and freeze/thaw cycles)—that have unique physiologies (e.g., high capacity to produce cellulase activity, rather than oxidase activity) compared to ambient conditions.
To explore these potential fungal responses to changing soil temperatures, we isolated fungi from the Climate Change Across Seasons Experiment (CCASE) at the Hubbard Brook Experimental Forest, where soils experience one of three experimentally applied climate treatments: +5°C warming during the growing season, +5°C warming in the growing season plus up to four soil freeze/thaw cycles in winter, or ambient soil temperatures (Templer et al., 2017). We then measured the growth and enzyme activity of fungal isolates in their respective “home” (i.e., origin) and “away” soil temperature environments, as a part of a lab-based common garden experiment, similar to previous tests of fungal response to N deposition (van Diepen et al., 2017).
Materials and Methods
Field Site
We cultured fungi from soils of three CCASE treatments. CCASE is a field-based climate change experiment established in 2013 at the Hubbard Brook Experimental Forest in New Hampshire, United States (43°56′N, 71°45′W). The forest at CCASE is dominated by red maple (Acer rubrum; 64% basal area) and American beech (Fagus grandifolia; 21% basal area), mixed with other hardwood and conifer tree species. The climate is continental: mean air temperatures ranges from an average of 19°C in July to −9°C in January, and precipitation averages 1,400 mm annually, 25–33% of which accumulates as snow (Bailey et al., 2003). Summers are short and cool, and winters are long and cold, with periods of low temperatures ranging between −12 and −18°C (Campbell et al., 2010). Historically, snowpack has accumulated at the Hubbard Brook Experimental Forest between mid-December and mid-April.
The goal of CCASE is to determine the combined effects of growing season warming and increased frequency of soil freeze/thaw cycles on northern hardwood forests, following projected climate changes over the next century for the region (Campbell et al., 2010). The experiment consists of three climate treatments applied across six plots: two plots are experimentally warmed 5°C above ambient air temperature during the growing season using buried heating cables (hereafter referred to as the “warmed” treatment), two plots are warmed 5°C above ambient air temperature during the growing season and subjected to two to four soil freeze/thaw events in winter (hereafter referred to as “warmed + FTC” treatment), and two plots are unmanipulated reference plots (Templer et al., 2017). Plots are each 11 m2 × 13.5 m2 and spaced approximately 5–60 m apart. Each treatment type is separated by 30-cm-deep rubber membranes to inhibit root and mycelial growth between plots, such that the presence of common fungal species across treatment plots is likely due primarily to dispersal, rather than colonization by the same organism. To warm plots, 4 km (2.5 mi) of heating cable is buried across the four climate treatment plots and turned on continuously during the growing season (snowmelt in spring to the first snow in the following winter). To induce winter soil freeze/thaw events, snow is removed from the forest floor in winter by shoveling and then heating cables are turned on after freezing events to induce soil thaw. A shallower layer of snow (3–5 cm) is left on the snow-removal plots to maintain albedo and reduce disturbance to the forest floor. Heating cables were installed at CCASE in summer 2012 and experimental treatments (starting with winter soil freeze/thaw cycles) began in December 2013. See Templer et al. (2017) for more details about the CCASE experiment.
Study Organisms
We cultured fungi from soil collected from all six plots at CCASE in July 2017, 3.5 years after soil temperature treatments began. Each plot consists of four sampling quadrants; we collected a single soil sample to 10 cm depth from each quadrant. Soils were sampled using a knife and a 10-cm × 10-cm frame that was ethanol-sterilized between samples, following Garcia et al. (2020). Soils were stored on ice, transported back to the laboratory, and sieved through a 2 mm mesh sieved within 24 h of sampling. Soil for fungal isolation was kept at 4°C. To isolate fungi from soil samples, 5 g of soil was diluted in 50 mL of a 0.9% saline solution, serially diluted to 10–3, and plated on solid Modified Melan-Norkins (MMN) media, a medium commonly used to isolate soil fungi (Burke et al., 2014; Talbot et al., 2015). 100 μl of solution was plated on each 9 cm plate. Three replicate aliquots of diluted soil suspension were plated per quadrant (3 CCASE treatments × 2 plots × 4 quadrants × 3 dilution replicates = 72 total plates). After 1 week of incubation, we then picked isolates from plates based on morphology. We cultured and identified approximately 120 isolates across the six plots. We identified fungal isolates taxonomically via Sanger sequencing of the intergenic transcribed spacer region (ITS) of rRNA of each isolate. We extracted DNA from culture tissue using the Extract-N-Amp Tissue PCR kit (Sigma-Aldrich, Inc., St. Louis, MO, United States) and PCR amplified the ITS region of DNA using ITS1F and ITS4 primers (Manter and Vivanco, 2007). PCR products were sequenced using single pass Sanger sequencing (MGH CCIB DNA Core, Boston, MA, United States). Unidentified bases at the ends of sequences were trimmed using Geneious software (Biomatters Ltd., Auckland, New Zealand). Sequences were assigned taxonomy by searching against GenBank using BLASTn. Based on this approach, we collapsed isolates into 74 unique strains (i.e., isolates that were found in different plots or different species within a plot) across 43 unique fungal species.
We determined the relative abundance of each fungal strain in CCASE soils by comparing culture sequences to a database of representative ITS rDNA sequences of fungal stands for operational taxonomic units (OTUs) identified in CCASE soils using high-throughput sequencing (Garcia et al., 2020). We used strains in our laboratory experiment only if they had a close match (>97% similarity) to an existing field OTU. Of the 74 unique strains we identified, 46 strains had an unequivocal match to a field OTU. These strains represented 29 species, which we used in our laboratory experiment (Supplementary Table 1). Species were categorized as “common” or “unique” depending on if they had strains across treatments (“common” species) or within a single treatment (“unique” species). Of the fungal species we identified, 20 were represented by a single strain that originated from only one CCASE treatment. Nine species had two strains that each originated from different CCASE treatments: reference and warmed (n = 4), reference and warmed + FTC (n = 3), and warmed and warmed + FTC (n = 2). No species were found in all three CCASE treatments.
Phylogenetic trees of fungal taxa were constructed to determine if (1) CCASE field treatment (origin) of isolates was confounded with phylogenetic relatedness among species, or if (2) there was a significant phylogenetic signal to response variables (growth rate, enzyme activities), such that relatedness among species would need to be accounted for in the statistical models relating field treatment to response variables. We constructed two phylogenetic trees; one for the fungal species that occurred in two CCASE treatments (“common species”) and one for fungal species that were unique to a CCASE treatment (or found in soils of only one CCASE treatment). To construct trees, one high-quality 28S gene sequence for each species was downloaded from the SILVA rRNA database v. 115 (Quast et al., 2013). In cases where we could not assign taxonomy of a culture to the species level with confidence, the 28S was pulled from the full ITS1-5.8S-ITS2-28S DNA sequence from the best BLASTn match for our strain’s ITS sequence in Genbank. All sequences were aligned to the SILVA SEED LSU nucleotide alignment and used to construct a tree in RAxML using the GTRGAMMA model of nucleotide substitution rates, with 100 bootstrap replicates followed by ML optimization (Stamatakis, 2014). The final phylogenies were visually checked against the currently accepted fungal phylogeny according to James et al. (2006). Branch lengths were calculated by the Grafen (1989) method with the compute.brlen command in the ape package in R (Paradis, 2012).
Laboratory Experiment
To test the responses of fungi to different climate conditions, we conducted a common garden experiment in which we incubated all fungal strains under laboratory conditions that simulated CCASE temperature treatments (Supplementary Table 2). We use the term isolate “origin” to indicate the CCASE field treatment (warmed, warmed + FTC, or reference) from which we cultured each fungal isolate and “laboratory environment” to indicate the climate treatment applied in our common garden experiment (van Diepen et al., 2017). Laboratory environment conditions replicated the average soil temperature of each season (fall, winter, spring, and summer) at CCASE, with each field season represented by a 1-week incubation in the lab (4 weeks total incubation time). We calculated the average soil temperature of each season as the soil temperature observed in each CCASE field plot over a 3-month period, averaged across the two plots per treatment (Sorensen et al., 2018). We simulated the warmed CCASE treatment in the laboratory environment by increasing reference soil temperatures by +5°C in the spring, summer, and fall months (Supplementary Table 2). We induced winter soil freeze/thaw cycles in the laboratory environment by freezing the winter plates to −9°C for approximately 6 h and defrosting for 18 h at 4°C. We repeated this process four times throughout our simulated “winter” week to simulate the warmed + FTC CCASE treatment. While the minimum winter soil temperature in the warmed + FTC CCASE plots from 2015 to 2016 was −2.84°C, the average minimum soil temperature from this CCASE treatment since 2013 falls below this value (Sorensen et al., 2018). Past work has shown that compared to mild or no frost, severe soil frost can reduce fungal growth, dissolved organic C, and cumulative gaseous fluxes compared to soil experiencing mild frost or no frost at the Hubbard Brook Experimental Forest and elsewhere (Haei et al., 2011; Reinmann et al., 2012). However, temperature variations within the severe frost range (−6 versus −12°C) have not been found to have significant impact on fungal growth in soils (Haei et al., 2011).
The common garden laboratory experiment lasted a total of 28 days (we simulated each season for 1 week). We grew isolates on a soil extract media prepared with a composite soil created from soil collected across CCASE plots as the sole carbon and nutrient source. We soaked 400 g of soil in 1 L of water for 2 days, then strained the solution through paper towels twice to ensure no large particulate debris or roots remained in the solution. We combined the solution with 15 g of agar, autoclaved it, and plated 10 ml onto 50 mm Sterilin petri dishes. We used five replicate plates for each of the three simulated CCASE treatments, resulting in 15 plates per isolate (three climate treatments × 46 isolates × 5 replicates = 690 total samples). After 28 days of incubation, we harvested four agar plugs from each replicate experimental petri dish, flash-froze them in liquid N, and stored them frozen at −80°C until analysis.
Enzyme and Growth Rate Measurements
To measure the response of individual fungal isolates to changes in climate conditions, we measured growth rate and extracellular enzyme activity of each isolate in the laboratory experiment. We recorded growth as biomass area on each petri dish at four time points prior to harvesting; we scanned each replicate plate of each isolate at the end of each week during the growth period (28 days) on an Epson Perfection V600 photo scanner. We quantified total growth of each sample as area in mm2 by highlighting all pixels containing fungal biomass in each scanned image using Adobe Photoshop and dividing this number by the number of pixels in a 100 mm2 area. We also calculated lag time and maximum growth rate for each sample using the fit_easylinear function in the growthrates package in R (Hall et al., 2013). None of the isolates reached the plate edge by the end of the incubation experiment (Supplementary Tables 4, 5).
We measured extracellular enzyme activities on each replicate of each isolate using a standard fluorometric/colorimetric protocol adapted from German et al. (2011). We chose to measure five hydrolytic enzymes; cellobiohydrolase (CBH), β-Glucosidase (BG), and α-Glucosidase (AG)—each involved in the breakdown of more labile plant carbohydrates in soil—as well as N-acetyl-D-glucosaminidase (NAG), which is involved in the breakdown of chitin found in fungal biomass, and acid phosphatase (AP), an organic phosphorus-acquiring enzyme (Sinsabaugh et al., 2002). We also measured polyphenol oxidase (PPO) and peroxidase (PER) activities as an estimate of more recalcitrant C (e.g., lignin and aromatic soil organic matter) breakdown (Talbot et al., 2015). To measure enzyme activity, we blended frozen plugs in 15 ml sodium acetate buffer and mixed an aliquot with either fluorometric enzyme substrates (for measurement of hydrolase activities), 3,4-Dihydroxy-L-phenylalanine (L-DOPA) reagent (for measurement of oxidative enzyme activity), or methylumbelliferone standard (to create a standard curve). We calculated enzyme activity on a sample mass basis (nmol mg agar–1 h–1) and on an area basis (nmol mm tissue–1 h–1) by using an isolate-specific sample mass-to-area (mg/mm) conversion factor. Our data are publicly available through the Hubbard Brook Experimental Forest data archives (Finestone et al., 2022).
Statistical Analyses
All statistical analyses were done in R version 4.0.3 and figures were made using the “ggplot2” package. We analyzed “common” fungal species occurring in two CCASE treatments separately from fungal species that were “unique” to a CCASE treatment. To determine if CCASE field treatment (origin) of isolates was confounded with phylogenetic relatedness among species, we fit a likelihood model for field treatment to phylogenetic trees of fungal taxa using the fitDiscrete function in R’s geiger package (Harmon et al., 2007) and compared them to a null model using a chi squared test. CCASE field treatment (origin) of isolates was not confounded with relatedness among taxa for the common species (P = 0.923) or for the uncommon species (P = 0.999). We also checked for a phylogenetic signal in fungal growth and extracellular enzyme activity by calculating the K statistic (Blomberg et al., 2003) for each metric using the multiPhylosignal command in the Picante package in R (Kembel et al., 2010). There was no significant phylogenetic signal to growth or the activity of most enzymes for species common to multiple CCASE treatments, or for species unique to a single CCASE treatment (Supplementary Table 3). Because common species did not vary across field treatments (origin), we did not control for phylogenetic relatedness in subsequent statistical analyses of treatment effects on growth or enzyme activities.
To test the influence of field climate conditions (origin) and fungal responses to laboratory climate (environment), we conducted a factorial analysis of variance (ANOVA) with CCASE field treatment (origin) of isolates, laboratory treatment (environment), and their interaction as fixed independent variables and plot as a random variable. Both common species and unique species were unevenly distributed across field treatments. However, because relatedness of species did not covary with field treatment, we included species as a fixed independent variable in both ANOVA models, following recent suggestions for factorial experiments conducted on multiple species with traits that do not have strong phylogenetic signals (Freckleton and Rees, 2019). For common species, the “species” variable was crossed with origin and environment treatments, whereas for the unique species, the “species” variable was nested within field treatment (origin). Growth metrics (total growth area in mm2, maximum growth rate, and lag time) or extracellular enzyme activities of each biological replicate per strain (for common species) or per species (for unique species) were used as the dependent variable. Enzyme activities normalized to mm hyphae produced the same quantitative results as activities normalized to sample weight (data not shown), so we report the sample weight-normalized data only.
To explore carbon use of fungal taxa under different climate conditions, we calculated fungal growth relative to carbon-acquiring enzyme activity. The enzyme assays we employed are not typically used in calculations of carbon use efficiency; they measure potential substrate degradation rate under saturating substrate concentrations (German et al., 2011), rather than C uptake per se (Hill et al., 2010). Instead, enzyme activity generated by individual isolates under these conditions provide an indication of the inherent ability of fungi to use C to make more substrates available. Our previous work has established a strong positive linear relationship between growth rate and biomass production (based on qPCR) in these fungi (data not shown). When compared to total growth over the course of the experiment, differences among fungi in enzyme activity may reflect fundamental differences in C allocation to growth vs. enzyme (or other) activities per unit substrate available across isolates. We ran a linear regression with total growth as the dependent variable and CBH and BG enzyme activities as the independent variable. We also included climate “origin,” laboratory “environment,” and their interaction as covariates in the regression model. For all statistical models, we ensured normality of residuals and homogeneity of variance by log-transforming variables prior to fitting each model. Normality and homogeneity of variance was checked visually (Garamszegi, 2014).
Results
We found that fungi isolated from different CCASE temperature treatments (origin) differed in their potential to generate hydrolytic enzyme activity that targeted relatively labile C, N, and phosphorus (P)-rich molecules in soil. For the nine common species, isolates originating from the warmed and warmed + FTC CCASE treatment plots had significantly lower activity of AP relative to isolates from reference plots, but higher activity of BG, a cellulose-degrading enzyme (Figure 1). Activity of AP was also significantly lower in the warmed + FTC laboratory environment than in reference environment, while the activity of polysaccharidases BG, AG, and NAG was higher under the warmed + FTC laboratory environment than under the other environments (Supplementary Figure 1). There was also a significant interaction between CCASE field treatment and lab environment for AP activity. Isolates originating from the warmed + FTC CCASE treatment had significantly lower AP activity than isolates from the reference treatment under the warmed laboratory environment, but not under the warmed + FTC laboratory environment. Fungal isolates originating from warmed plots had significantly lower CBH activity than isolates from warmed + FTC CCASE plots and lower NAG activity than isolates from reference plots (Figure 1). Oxidative enzyme activities did not vary significantly by CCASE field treatment across the unique species [polyphenol oxidase: F(2/60) = 1.76, P = 0.193; peroxidase: F(2/60) = 2.36, P = 0.110].
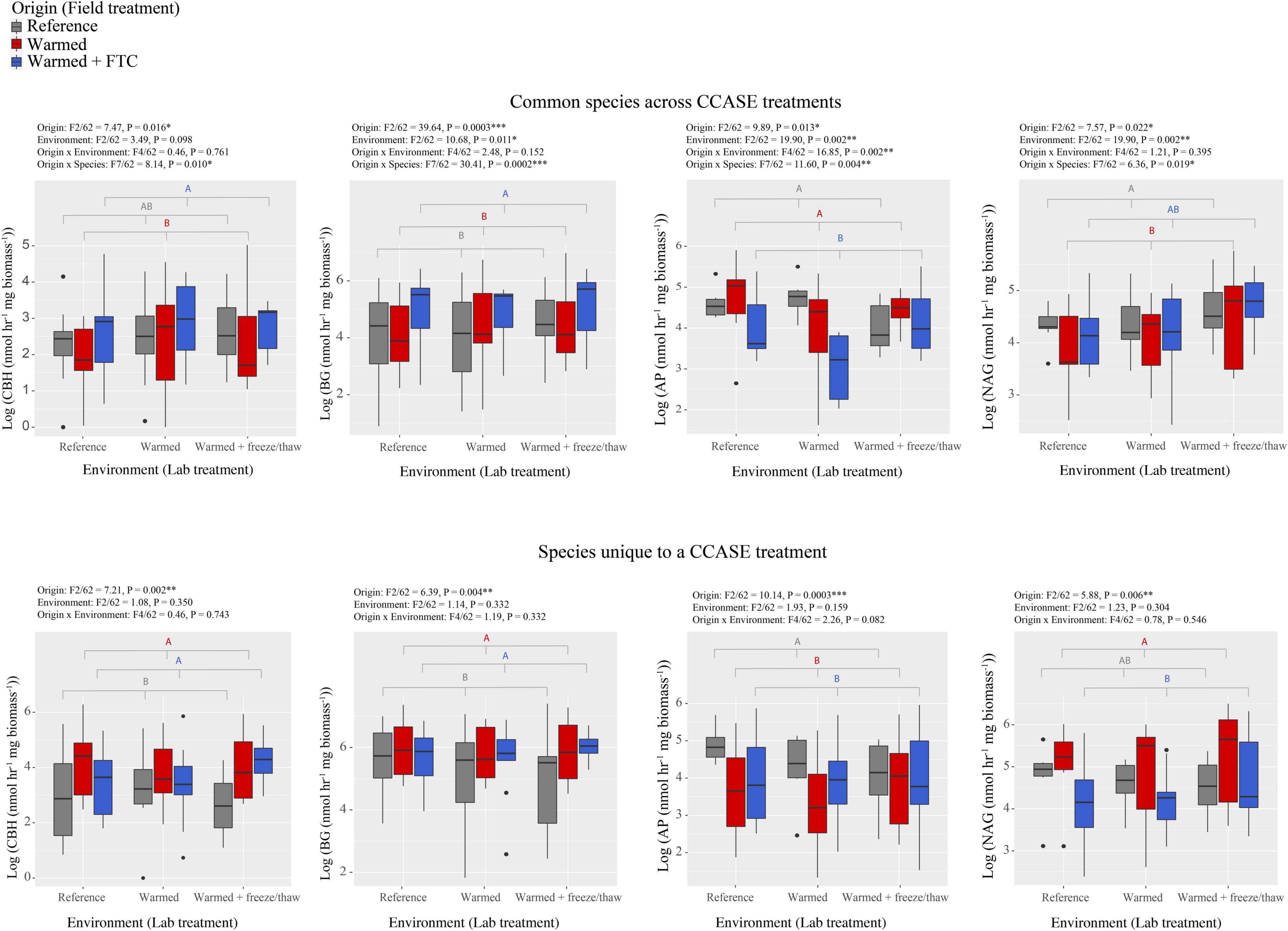
Figure 1. Enzyme activities generated by fungi isolated from climate change across seasons experiment (CCASE) plots and incubated in the laboratory common garden experiment. Common species consisted of nine species, each isolated from two CCASE treatments: reference and warmed plots (n = 4), reference and warmed + FTC plots (n = 3), or warmed and warmed + FTC plots (n = 2). Unique species consisted of 20 species, each of which were found only in one CCASE treatment. Enzymes showing significant activity differences among fungal isolates from different CCASE treatments include cellobiohydrolase (CBH), β-glucosidase (BG), acid phosphatase (AP), and N-acetyl glucosaminidase (NAG). Statistics were generated from factorial analysis of variance (ANOVA) with CCASE field treatment (origin) of isolates, laboratory treatment (environment), their interaction, and species as fixed independent variables and plot as a random variable.
We also found that CCASE treatments selected for fungi with different capacity to produce enzyme activity. When assessing only the 20 fungal species unique to a single CCASE treatment, fungal species originating from either the warmed or warmed + FTC CCASE treatments had significantly lower activity of AP and higher activity of CBH and BG than fungal species originating from reference plots (Figure 1). In addition, fungal species unique to the warmed + FTC CCASE treatments had significantly lower NAG activity than species unique to the warmed plots. Oxidative enzyme activities did not vary significantly by CCASE field treatment across the common species [polyphenol oxidase: F(2/60) = 3.03, P = 0.123; peroxidase: F(2/60) = 1.65, P = 0.27].
Among the unique species, enzyme activities were generally not significantly affected by laboratory environment with the exception of AG, which was significantly higher under the warmed + FTC laboratory environment than under the warmed and reference environment (Supplementary Figure 2). By contrast, enzyme activity traits varied across species, regardless of their origin. Hydrolytic enzyme activities varied strongly among different fungi species, for both “common” species across CCASE treatments and species that were “unique” to a particular CCASE treatment (Supplementary Figures 3, 4).
Growth rates of fungi changed in response to extreme laboratory temperature conditions, regardless of where the fungal isolates originated. We found a significant effect of laboratory environment on growth rate, such that fungi incubated in warmed + FTC laboratory environments had significantly lower total growth (Figure 2) and lower maximum growth rate (Supplementary Figure 5) than those incubated in reference or warmed laboratory environments. This response to the warmed + FTC laboratory environment occurred across species originating from a single CCASE treatment (20 species total). For the nine common species with strains originating from multiple CCASE treatments, fungi incubated in warmed + FTC laboratory environments had significantly lower growth rates than those incubated in the warmed laboratory environments (Figure 2).
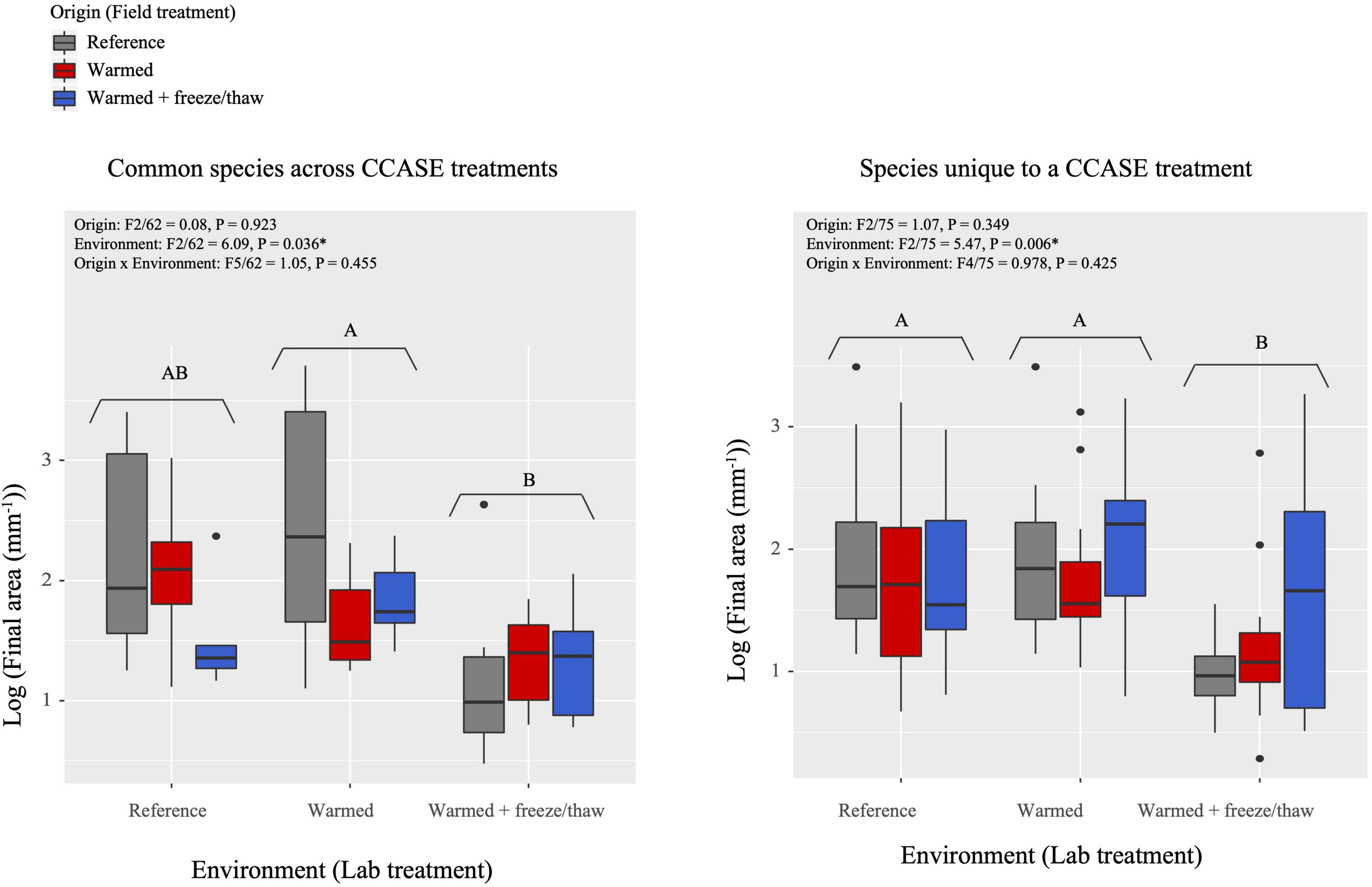
Figure 2. Fungal growth rates, as measured by final area of fungi isolated from CCASE plots after 4 weeks’ incubation in the laboratory common garden experiment. Common species (A) consist of nine species, each isolated from two CCASE treatments: reference and warmed plots (n = 4), reference and warmed + FTC plots (n = 3), and warmed and warmed + FTC plots (n = 2). Unique species include 20 species, each found in only one CCASE treatment.
We found a positive correlation between growth rate and enzyme activity across all fungal species (Figure 3), where growth rate of fungi increased with the activity of enzymes targeting plant cellulose (CBH and BG). The relationship was significantly improved by including laboratory temperature environment on the model for both enzymes CBH (Figure 3) and BG [common species: F(2/14) = 6.81, P = 0.009; species unique to a CCASE treatment: F(2/77) = 5.83, P = 0.004]. Specifically, cultures incubated in the reference and warmed laboratory environments grew faster per unit enzyme activity than cultures incubated in the warmed + FTC lab environment.
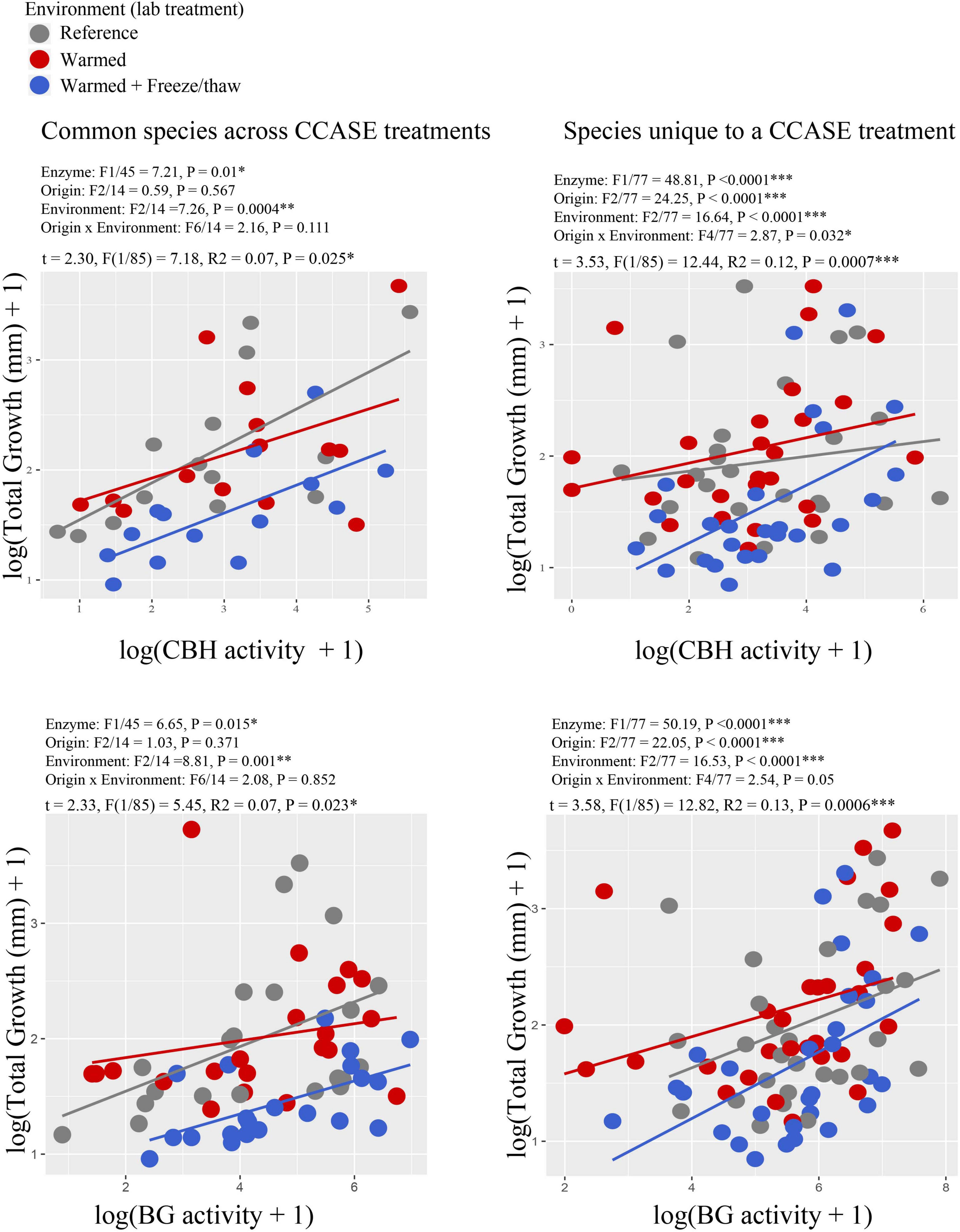
Figure 3. Correlation between enzyme activity (cellobiohydrolase, CBH) and growth rate for strains of the same species originating from different CCASE plots. Points are color-coded by laboratory environment in the common garden experiment. Other enzymes also correlated with growth rate, including β-Glucosidase (BG) (common species: R2 = 0.21, P = 0.001, uncommon species: R2 = 0.08, P = 0.003). Enzyme activities are reported in units of nmol substrate h–1 mg biomass–1.
Discussion
We found evidence that all three responses we investigated—irreversible acquisition of traits (potentially through evolutionary responses), plasticity, and shifts in fungal communities—may have occurred under simulated soil temperature changes at CCASE. Specifically, fungi from warmed and warmed + FTC CCASE treatment (origins) showed both increased and decreased potential enzyme activities, depending on the enzyme, that could not be reversed in the lab (Figure 1). Fungal species showed the ability to acclimate to warmed + FTC laboratory environments by reducing their growth (Figure 2) and growth per unit C decomposed (Figure 3), a potential indicator of carbon use efficiency. In addition, enzyme activity profiles varied among fungal species that were unique to a CCASE treatment (Figure 1), possibly reflecting selection for species in the field that can tolerate specific climate conditions.
Evidence for Evolutionary Responses to Field Climate Manipulations
We found evidence that fungal species have evolved in response to changes in historical climate where they are living. For the nine common fungal species, strains originating from the warmed CCASE treatment exhibited reduced activity of cellulose-degrading enzymes (cellobiohydrolase, CBH) relative to the warmed + FTC CCASE treatments, while isolates originating from warmed + FTC CCASE treatments had reduced activity of phosphorus-acquiring enzymes (acid phosphatase, AP) relative to strains from other treatments: in all cases, these activities could not be reversed when grown under reference conditions (Figure 1). This observation suggests that soil warming – in combination with soil freeze/thaw cycles – may drive irreversible shifts in fungal traits, possibly through evolutionary responses. These data are consistent with studies that have shown adaptive responses of fungi to either warming (Bradford et al., 2008; Romero-Olivares et al., 2015) or freeze/thaw events (Robinson, 2001; Feng et al., 2007). Previous work has also shown that warming decreases the proportion of carbohydrate-degrading genes from eukaryotes in soil (Pold et al., 2016). Warming can increase activity of fungi (Bradford, 2013), yet this may be due to increases in metabolism of low-molecular weight C, rather than decay of plant carbohydrates. If a decay-stress tolerance tradeoff exists in fungi, species may have evolved to allocate C toward either certain enzyme activities or toward mechanisms that allow fungi to tolerate the stress of elevated temperatures (Anthony et al., 2020). Our results are also consistent with observations that freeze/thaw impacts P availability to soil microbes. Freeze/thaw cycles can increase mineral P availability in soil (Zhao et al., 2008) by increasing fine root mortality that releases mineral P and prevents plant P uptake (Fitzhugh et al., 2001). Under these conditions, increased mineral P availability would reduce the need for fungi to invest in C- and N-expensive phosphatases to acquire P from organic sources in soil. Our results parallel those in bulk soils at CCASE, where acid phosphatase activity tends to decline in the warmed + FTC treatment relative to reference plots (Sorensen et al., 2018).
We also found that the direction of fungal response to field temperature treatments varied by species (Supplementary Figures 3–5). Among common species of fungi found across two CCASE field treatments, we found strong differences in enzyme activities and a consistent origin-by-species interaction (Figure 1), indicating that not all fungal species responded the same way to climate conditions in the field. These results suggest that species may have intrinsic differences in their abilities to tolerate stress and shift activity with new climatic conditions. Some fungal species have been shown to utilize a common genetic environmental stress response (ESR), which is a set of approximately 900 diverse genes thought to maintain homeostasis during periods of stress (Gasch et al., 2000; Crowther et al., 2014). Differences in the adaptive abilities of species may eventually lead to long-term shifts in fungal community structure with climate change due to the survival of some taxa and the mortality of others. The dramatic differences in enzyme activities among species (Supplementary Figures 3, 4), may also have masked the effect of climate “origin” (CCASE treatment) on most enzyme activities across these taxa.
Evidence for Plasticity of Growth Rate in Response to Laboratory Climate Conditions
Growth rates of fungi varied consistently by laboratory environment, indicating a highly plastic response of fungal biomass accumulation to temperature changes. Total growth of fungi over the course of the experiment was consistently lower when fungi were incubated under warmed + FTC laboratory environments (Figure 2), primarily because maximum growth rates were lowest under this treatment, perhaps because total growth was arrested at the very low temperatures (−9°C) experienced during that period. In the common species of fungi found in two CCASE field treatments, growth hindrance was particularly dramatic for strains originating from the reference or warmed CCASE treatments, whereas isolates originating from the warmed + FTC treatment showed less response, although there was no significant interaction between laboratory environment and climate origin of isolates (Figure 2).
We also found a positive relationship between cellulose-degrading enzyme activities (CBH and BG) and growth rate of fungal strains, which varied by laboratory environment. Fungi incubated under warmed + FTC laboratory environments grew less per unit enzyme activity than under reference laboratory conditions (Figure 3). This pattern was driven by reduced growth rate of fungi under the warmed + FTC laboratory environment, rather than a strong difference in enzyme activities. This relationship shift indicates that fungi may adjust their physiology to extreme, immediate temperature changes by re-allocating C to activities other than growth. Fungi have been shown to trade off growth rate for stress-tolerance traits – such as the capacity to dehydrate (Klironomos et al., 2001); produce osmolytes (e.g., trehalose), thick cell walls (Schimel et al., 2007), anti-shock proteins (Robinson, 2001), or high numbers of multi-layer spores; and survive anoxia (Hodkinson and Wookey, 1999) – under the stress of freeze-thaw. Our data does not represent fungal carbon use efficiency directly, as potential cellulase activities may not represent total C taken up from the substrate under all conditions (Lashermes et al., 2016), especially if C uptake rates become more or less rapid with environmental stress. Nevertheless, our results show a strong decrease in growth rate with continued C allocation to enzyme activity under warmed + FTC, in a trade-off that likely reduces the efficiency of C use regardless of actual C uptake rates. Our results parallel those of other studies that have found that when fungi invest in stress-tolerance molecules under freezing and drought stress, this often coincides with reduced fungal growth rates (Crowther et al., 2014). Many microbial taxa are thought to adjust their physiology in response to growing season warming, leading to a change in carbon use efficiency (Frey et al., 2013). Until now, it has not been clear that warming in the winter season (leading to more frequent soil freeze/thaw cycles) could have similar effects.
Evidence for Selection of Fungal Taxa Adapted to Extreme Conditions
We found that species unique to a single CCASE treatment had different enzyme activities (CBH, BG, AP, and NAG) depending on which climate of origin (i.e., CCASE treatment) they grew in, indicating that climate manipulations in the field may select for species with different resource capture abilities. Specifically, fungi from the warmed and warmed + FTC CCASE treatments had significantly higher activities of CBH relative to fungi originating from reference plots (Figure 1). These data are consistent with recent work in New England showing higher relative abundances of fungal hydrolytic enzyme encoding genes in warmed versus control plots using a fungal gene capture technique (Anthony et al., 2021). The growing season is expected to lengthen under climate change conditions, allowing more time for fine root and microbial biomass to accrue (Huntington et al., 2009; Groffman et al., 2012), but also increasing mortality of these fine roots and microorganisms (Tierney et al., 2001; Herrmann and Witter, 2002). These dynamics could increase easily accessible labile C stocks in the soil and potentially select for taxa that specialize in breaking down this type of C. Past studies have shown a boost in microbial activity after freeze/thaw cycles, largely attributed to this influx of labile organic matter (Schimel and Clein, 1996; Herrmann and Witter, 2002; Feng et al., 2007). Microbes exposed to warming alone often preferentially target cellulose and other C-rich plant polysaccharides (Zhou et al., 2012; Luo et al., 2014; Pold et al., 2016), accelerating decomposition of these easily-decomposable (i.e., labile) C sources in soil (Melillo et al., 2002). Other studies have demonstrated that microorganisms isolated from experimentally-warmed temperate forest soil had higher capability to decompose cellulose substrates compared to isolates from ambient soils (Pold et al., 2016), which may account for the often observed increase in CO2 release from soil with warming (Rustad et al., 2001; Romero-Olivares et al., 2017).
Conclusion
Soil microbial communities are shaped by a combination of selection by current environmental conditions and longer-term processes like adaptation to historical climate conditions (Hanson et al., 2012). Our study gives insight into how these processes shape soil fungal communities under rapid and extreme climate change conditions. The treatments simulated at CCASE are based on temperature projections for the region (Templer et al., 2017), such that this study acts as a model for how fungi may potentially acclimate and evolve in response to severe year-round climate changes in northern hardwood ecosystems. Fungal taxa that persist in these new climate conditions have unique decomposition traits, including increased abilities to decompose abundant labile organic matter (i.e., plant cellulose), such that new fungal communities that emerge under these climate conditions may alter the future rates of decomposition and nutrient cycling within forest soil on large geographic scales.
Data Availability Statement
The original data used in this study are available for download from the Environmental Data Initiative Repository at doi: 10.6073/pasta/f4a66de07c2945115956996dc8af980b.
Author Contributions
JF and JB conceived the project idea. PT maintained the CCASE site at Hubbard Brook Experimental Forest. JB cultured fungi from soil. JF conducted assays and wrote the manuscript. JB and PT edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by a Boston University Peter Paul Professorship award and a U.S. Department of Energy BER Environmental System Science Award (DE-SC0022194) to JB. JF acknowledges the UROP program at Boston University for financial support. This research was also supported by a National Science Foundation (NSF) Long Term Ecological Research (LTER) grant to Hubbard Brook (NSF 1114804 and 1637685) and a NSF CAREER grant to PT (NSF DEB1149929). This manuscript is a contribution of the Hubbard Brook Ecosystem Study. Hubbard Brook is part of the LTER network, which is supported by the NSF. The Hubbard Brook Experimental Forest is operated and maintained by the USDA Forest Service, Northern Research Station.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the HBEF staff for granting us access to the CCASE experiment, as well as J. Gewirtzman and the Templer lab for collecting soil cores and subsampling for our culture analysis. We thank M. O. Garcia and S. Cruz-Ramirez for conducting initial culturing of fungi used in this experiment. We also thank E. Macdonald and M. Chen for assistance with extracellular enzyme assays, S. Vivelo and Z. Werbin for aid with statistical analyses, and all members of the Bhatnagar lab for feedback on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/ffgc.2022.800335/full#supplementary-material
References
Aanderud, Z. T., Jones, S. E., Schoolmaster, D. R., Fierer, N., and Lennon, J. T. (2013). Sensitivity of soil respiration and microbial communities to altered snowfall. Soil Biol. Biochem. 57, 217–227. doi: 10.1016/j.soilbio.2012.07.022
Allison, S. D., McGuire, K. L., and Treseder, K. K. (2010). Resistance of microbial and soil properties to warming treatment seven years after boreal fire. Soil Biol. Biochem. 42, 1872–1878. doi: 10.1016/j.soilbio.2010.07.011
Allison, S. D., and Treseder, K. K. (2008). Warming and drying suppress microbial activity and carbon cycling in boreal forest soils. Global Change Biol. 14, 2898–2909.
Anthony, M. A., Crowther, T. W., Maynard, D. S., van den Hoogen, J., and Averill, C. (2020). Distinct Assembly Processes and Microbial Communities Constrain Soil Organic Carbon Formation. One Earth 2, 349–360. doi: 10.1016/j.oneear.2020.03.006
Anthony, M. A., Knorr, M., Moore, J. A. M., Simpson, M., and Frey, S. D. (2021). Fungal community and functional responses to soil warming are greater than for soil nitrogen enrichment. Elementa 9:1 doi: 10.1525/elementa.2021.000059
Bailey, A. S., Hornbeck, J. W., Campbell, J. L., and Eagar, C. (2003). Hydrometeorological database for Hubbard Brook Experimental Forest: 1955-2000. Gen. Tech. Rep. NE-305, (Newtown Square, PA: Northeastern Research Station).
Blomberg, S. P., Garland, T., and Ives, A. R. (2003). Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution 57, 717–745.
Bradford, M. A. (2013). Thermal adaptation of decomposer communities in warming soils. Front. Microbiol. 4:333. doi: 10.3389/fmicb.2013.00333
Bradford, M. A., Davies, C. A., Frey, S. D., Maddox, T. R., Melillo, J. M., Mohan, J. E., et al. (2008). Thermal adaptation of soil microbial respiration to elevated temperature. Ecol. Lett. 11, 1316–1327. doi: 10.1111/j.1461-0248.2008.01251.x
Buckeridge, K. M., Banerjee, S., Siciliano, S. D., and Grogan, P. (2013). The seasonal pattern of soil microbial community structure in mesic low arctic tundra. Soil Biol. Biochem. 65, 338–347. doi: 10.1016/j.soilbio.2013.06.012
Burke, D. J., Smemo, K. A., and Hewins, C. R. (2014). Ectomycorrhizal fungi isolated from old-growth northern hardwood forest display variability in extracellular enzyme activity in the presence of plant litter. Soil Biol. Biochem. 68, 219–222. doi: 10.1016/j.soilbio.2013.10.013
Campbell, J. L., Ollinger, S. V., Flerchinger, G. N., Wicklein, H., Hayhoe, K., and Bailey, A. S. (2010). Past and projected future changes in snowpack and soil frost at the Hubbard Brook Experimental Forest. New Hampshire U.S.A. 24, 2465–2480. doi: 10.1002/hyp.7666
Chen, J., Elsgaard, L., van Groenigen, K. J., Olesen, J. E., Liang, Z., Jiang, Y., et al. (2020). Soil carbon loss with warming: new evidence from carbon-degrading enzymes. Glob. Chang. Biol. 26, 1944–1952. doi: 10.1111/gcb.14986
Chen, J., Luo, Y. Q., Garcia-Palacios, P., Cao, J. J., Dacal, M., Zhou, X. H., et al. (2018). Differential responses of carbon-degrading enzyme activities to warming: implications for soil respiration. Glob. Chang. Biol. 24, 4816–4826. doi: 10.1111/gcb.14394
Crowther, T. W., and Bradford, M. A. (2013). Thermal acclimation in widespread heterotrophic soil microbes. Ecol. Lett. 16, 469–477. doi: 10.1111/ele.12069
Crowther, T. W., Maynard, D. S., Crowther, T. R., Peccia, J., Smith, J. R., and Bradford, M. A. (2014). Untangling the fungal niche: the trait-based approach. Front. Microbiol. 5:579. doi: 10.3389/fmicb.2014.00579
DeAngelis, K. M., Pold, G., Topçuoðlu, B. D., van Diepen, L. T., Varney, R. M., Blanchard, J. L., et al. (2015). Long-term forest soil warming alters microbial communities in temperate forest soils. Front. Microbiol. 6:104. doi: 10.3389/fmicb.2015.00104
Dettman, J. R., Anderson, J. B., and Kohn, L. M. (2008). Divergent adaptation promotes reproductive isolation among experimental populations of the filamentous fungus Neurospora. Bmc Evol. Biol. 8:35. doi: 10.1186/1471-2148-8-35
Feng, X. J., Nielsen, L. L., and Simpson, M. J. (2007). Responses of soil organic matter and microorganisms to freeze-thaw cycles. Soil Biol. Biochem. 39, 2027–2037. doi: 10.1016/j.soilbio.2007.03.003
Fernandez, C. W., Nguyen, N. H., Stefanski, A., Han, Y., Hobbie, S. E., Montgomery, R. A., et al. (2016). Ectomycorrhizal fungal response to warming is linked to poor host performance at the boreal-temperate ecotone. Glob. Chang. Biol. 23, 1598–1609. doi: 10.1111/gcb.13510
Finestone, J., Templer, P. H., and Bhatnagar, J. M. (2022). Climate Change Across Seasons Experiment (CCASE) at the Hubbard Brook Experimental Forest: Growth and Enzyme Activity Traits of Soil Fungi Isolated from CCASE in July 2017, Grown Under a Common Garden Experiment in the Laboratory that Mimicked CCASE Soil Temperature Treatments Ver 1. Environmental Data Initiative. Available online at: https://doi.org/10.6073/pasta/f4a66de07c2945115956996dc8af980b (accessed February 22, 2022).
Fitzhugh, R. D., Driscoll, C. T., Groffman, P. M., Tierney, G. L., Fahey, T. J., and Hardy, J. P. (2001). Effects of soil freezing disturbance on soil solution nitrogen, phosphorus, and carbon chemistry in a northern hardwood ecosystem. Biogeochemistry 56, 215–238. doi: 10.1023/A:1013076609950
Freckleton, R. P., and Rees, M. (2019). Comparative analysis of experimental data. Methods Ecol. Evol. 10, 1308–1321.
Frey, S. D., Lee, J., Melillo, J. M., and Six, J. (2013). The temperature response of soil microbial efficiency and its feedback to climate. Nat. Clim. Chang. 3, 395–398.
Garamszegi, L. Z. (2014). Modern Phylogenetic Comparative Methods and Their Application in Evolutionary Biology: Concepts and Practice. London, UK: Springer-Verlag Berlin Heidelberg.
Garcia, M. O., Templer, P. H., Sorensen, P. O., Sanders-DeMott, R., Groffman, P. M., and Bhatnagar, J. M. (2020). Soil Microbes Trade-Off Biogeochemical Cycling for Stress Tolerance Traits in Response to Year-Round Climate Change. Front. Microbiol. 11:616. doi: 10.3389/fmicb.2020.00616
Gasch, A. P., Spellman, P. T., Kao, C. M., Carmel-Harel, O., Eisen, M. B., Storz, G., et al. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257.
German, D. P., Weintraub, M. N., Grandy, A. S., Lauber, C. L., Rinkes, Z. L., and Allison, S. D. (2011). Optimization of hydrolytic and oxidative enzyme methods for ecosystem studies. Soil Biol. Biochem. 43, 1387–1397. doi: 10.1016/j.soilbio.2011.03.017
Grafen, A. (1989). The phylogenetic Regression. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 326, 119–157.
Groffman, P. M., Driscoll, C. T., Fahey, T. J., Hardy, J. P., Fitzhugh, R. D., and Tierney, G. L. (2001). Colder soils in a warmer world: a snow manipulation study in a northern hardwood forest ecosystem. Biogeochemistry 56, 135–150.
Groffman, P. M., Rustad, L. E., Templer, P. H., Campbell, J. L., Christenson, L. M., Lany, N. K., et al. (2012). Long-Term Integrated Studies Show Complex and Surprising Effects of Climate Change in the Northern Hardwood Forest. Bioscience 62, 1056–1066. doi: 10.1525/bio.2012.62.12.7
Haei, M., Rousk, J., Ilstedt, U., Öquist, M., Bååth, E., and Laudon, H. (2011). Effects of soil frost on growth, composition and respiration of the soil microbial decomposer community. Soil Biol. Biochem. 43, 2069–2077.
Hall, B. G., Acar, H., Nandipati, A., and Barlow, M. (2013). Growth Rates Made Easy. Mol. Biol. Evol. 31, 232–238. doi: 10.1093/molbev/mst187
Hanson, C. A., Allison, S. D., Bradford, M. A., Wallenstein, M. D., and Treseder, K. K. (2008). Fungal Taxa Target Different Carbon Sources in Forest Soil. Ecosystems 11, 1157–1167. doi: 10.1007/s10021-008-9186-4
Hanson, C. A., Fuhrman, J. A., Horner-Devine, M. C., and Martiny, J. B. (2012). Beyond biogeographic patterns: processes shaping the microbial landscape. Nat. Rev. Microbiol. 10, 497–506.
Harmon, L. J., Weir, J. T., Brock, C. D., Glor, R. E., and Challenger, W. (2007). GEIGER: investigating evolutionary radiations. Bioinformatics 24, 129–131.
Herrmann, A., and Witter, E. (2002). Sources of C and N contributing to the flush in mineralization upon freeze-thaw cycles in soils. Soil Biol. Biochem. 34, 1495–1505. doi: 10.1016/S0038-0717(02)00121-9
Hill, B. H., McCORMICK, F. H., Harvey, B. C., Johnson, S. L., Warren, M. L., and Elonen, C. M. (2010). Microbial enzyme activity, nutrient uptake and nutrient limitation in forested streams. Freshw. Biol. 55, 1005–1019.
Hodkinson, I. D., and Wookey, P. (1999). Functional ecology of soil organisms in tundra ecosystems: towards the future. Appl. Soil Ecol. 11, 111–126.
Huntington, T. G., Richardson, A. D., McGuire, K. J., and Hayhoe, K. (2009). Climate and hydrological changes in the northeastern United States: recent trends and implications for forested and aquatic ecosystems. Can. J. For. Res. 39, 199–212. doi: 10.1139/X08-116
James, T. Y., Kauff, F., Schoch, C. L., Matheny, P. B., Hofstetter, V., Cox, C. J., et al. (2006). Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443, 818–822.
Kembel, S. W., Cowan, P. D., Helmus, M. R., Cornwell, W. K., Morlon, H., Ackerly, D. D., et al. (2010). Picante: r tools for integrating phylogenies and ecology. Bioinformatics 26, 1463–1464.
Klironomos, J. N., Hart, M. M., Gurney, J. E., and Moutoglis, P. (2001). Interspecific differences in the tolerance of arbuscular mycorrhizal fungi to freezing and drying. Can. J. Bot. 79, 1161–1166.
Koide, R. T., Fernandez, C., and Malcolm, G. (2014). Determining place and process: functional traits of ectomycorrhizal fungi that affect both community structure and ecosystem function. New Phytol. 201, 433–439. doi: 10.1111/nph.12538
Koide, R. T., and Malcolm, G. M. (2009). N concentration controls decomposition rates of different strains of ectomycorrhizal fungi. Fungal Ecol. 2, 197–202. doi: 10.1016/j.funeco.2009.06.001
Lashermes, G., Gainvors-Claisse, A., Recous, S., and Bertrand, I. (2016). Enzymatic Strategies and Carbon Use Efficiency of a Litter-Decomposing Fungus Grown on Maize Leaves, Stems, and Roots. Front. Microbiol. 7:1315. doi: 10.3389/fmicb.2016.01315
Leu, J. Y., and Murray, A. W. (2006). Experimental evolution of mating discrimination in budding yeast. Curr. Biol. 16, 280–286. doi: 10.1016/j.cub.2005.12.028
Lipson, D. A., and Schmidt, S. K. (2004). Seasonal Changes in an Alpine Soil Bacterial Community in the Colorado Rocky Mountains. Appl. Environ. Microbiol. 70, 2867–2879. doi: 10.1128/aem.70.5.2867-2879.2004
Luo, C., Rodriguez-R, L. M., Johnston, E. R., Wu, L., Cheng, L., Xue, K., et al. (2014). Soil microbial community responses to a decade of warming as revealed by comparative metagenomics. Appl. Environ. Microbiol. 80, 1777–1786.
Luo, Y. Q., Wan, S. Q., Hui, D. F., and Wallace, L. L. (2001). Acclimatization of soil respiration to warming in a tall grass prairie. Nature 413, 622–625. doi: 10.1038/35098065
Malcolm, G. M., Lopez-Gutierrez, J. C., Koide, R. T., and Eissenstat, D. M. (2008). Acclimation to temperature and temperature sensitivity of metabolism by ectomycorrhizal fungi. Glob. Chang. Biol. 14, 1169–1180. doi: 10.1111/j.1365-2486.2008.01555.x
Manter, D. K., and Vivanco, J. M. (2007). Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J. Microbiol. Method 71, 7–14. doi: 10.1016/j.mimet.2007.06.016
McGuire, K. L., Bent, E., Borneman, J., Majumder, A., Allison, S. D., and Treseder, K. K. (2010). Functional diversity in resource use by fungi. Ecology 91, 2324–2332. doi: 10.1890/09-0654.1
Melillo, J. M., Steudler, P. A., Aber, J. D., Newkirk, K., Lux, H., Bowles, F. P., et al. (2002). Soil warming and carbon-cycle feedbacks to the climate system. Science 298, 2173–2176.
Ostroumov, V. E., and Siegert, C. (1996). Exobiological aspects of mass transfer in microzones of permafrost deposits. Adv. Space Res. 18, 79–86. doi: 10.1016/0273-1177(96)00002-6
Pold, G., Billings, A. F., Blanchard, J. L., Burkhardt, D. B., Frey, S. D., Melillo, J. M., et al. (2016). Long-term warming alters carbohydrate degradation potential in temperate forest soils. Appl. Environ. Microbiol. 82, 6518–6530.
Pold, G., Melillo, J. M., and DeAngelis, K. M. (2015). Two decades of warming increases diversity of a potentially lignolytic bacterial community. Front. Microbiol. 6:480. doi: 10.3389/fmicb.2015.00480
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucl. Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Reinmann, A. B., Templer, P. H., and Campbell, J. L. (2012). Severe soil frost reduces losses of carbon and nitrogen from the forest floor during simulated snowmelt: a laboratory experiment. Soil Biol. Biochem. 44, 65–74. doi: 10.1016/j.soilbio.2011.08.018
Robinson, C. H. (2001). Cold adaptation in Arctic and Antarctic fungi. New Phytol. 151, 341–353. doi: 10.1046/j.1469-8137.2001.00177.x
Romero-Olivares, A. L., Allison, S. D., and Treseder, K. K. (2017). Soil microbes and their response to experimental warming over time: a meta-analysis of field studies. Soil Biol. Biochem. 107, 32–40. doi: 10.1016/j.soilbio.2016.12.026
Romero-Olivares, A. L., Taylor, J. W., and Treseder, K. K. (2015). Neurospora discreta as a model to assess adaptation of soil fungi to warming. Bmc Evol. Biol. 15, 1–8. doi: 10.1186/s12862-015-0482-2
Rousk, J., Frey, S. D., and Baath, E. (2012). Temperature adaptation of bacterial communities in experimentally warmed forest soils. Glob. Chang. Biol. 18, 3252–3258. doi: 10.1111/j.1365-2486.2012.02764.x
Rustad, L. E., Campbell, J. L., Marion, G. M., Norby, R. J., Mitchell, M. J., Hartley, A. E., et al. (2001). A meta-analysis of the response of soil respiration, net nitrogen mineralization, and aboveground plant growth to experimental ecosystem warming. Oecologia 126, 543–562.
Sanders-DeMott, R., Sorensen, P. O., Reinmann, A. B., and Templer, P. H. (2018). Growing season warming and winter freeze–thaw cycles reduce root nitrogen uptake capacity and increase soil solution nitrogen in a northern forest ecosystem. Biogeochemistry 137, 337–349.
Schimel, J., Balser, T. C., and Wallenstein, M. (2007). Microbial Stress-Response Physiology and Its Implications for Ecosystem Function. Ecology 88, 1386–1394.
Schimel, J. P., and Clein, J. S. (1996). Microbial response to freeze-thaw cycles in tundra and taiga soils. Soil Biol. Biochem. 28, 1061–1066. doi: 10.1016/0038-0717(96)00083-1
Schneider, T., Keiblinger, K. M., Schmid, E., Sterflinger-Gleixner, K., Ellersdorfer, G., Roschitzki, B., et al. (2012). Who is who in litter decomposition? Metaproteomics reveals major microbial players and their biogeochemical functions. ISME J. 6, 1749–1762.
Sinsabaugh, R. L., Carreiro, M. M., and Alvarez, S. (2002). “Enzyme and microbial dynamics of litter decomposition,” in Enzymes in the Environment, Activity, Ecology, and Applications, eds R. G. Burns and R. P. Dick (Basel: Marcel Dekker), 249–265.
Sorensen, P. O., Finzi, A. C., Giasson, M.-A., Reinmann, A. B., Sanders-DeMott, R., and Templer, P. H. (2018). Winter soil freeze-thaw cycles lead to reductions in soil microbial biomass and activity not compensated for by soil warming. Soil Biol. Biochem. 116, 39–47. doi: 10.1016/j.soilbio.2017.09.026
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
Talbot, J. M. (2017). “Fungal communities and climate change,” in The Fungal Community: its Organization and Role in the Ecosystem, Fourth Edition, eds J. Dighton and J. F. White (Boca Raton, FL: Taylor & Francis Group, LLC).
Talbot, J. M., Martin, F., Kohler, A., Henrissat, B., and Peay, K. G. (2015). Functional guild classification predicts the enzymatic role of fungi in litter and soil biogeochemistry. Soil Biol. Biochem. 88, 441–456. doi: 10.1016/j.soilbio.2015.05.006
Templer, P. H., Reinmann, A. B., Sanders-DeMott, R., Sorensen, P. O., Juice, S. M., Bowles, F., et al. (2017). Climate Change Across Seasons Experiment (CCASE): a new method for simulating future climate in seasonally snow-covered ecosystems. PLoS One 12:e0171928. doi: 10.1371/journal.pone.0171928
Tierney, G. L., Fahey, T. J., Groffman, P. M., Hardy, J. P., Fitzhugh, R. D., and Driscoll, C. T. (2001). Soil freezing alters fine root dynamics in a northern hardwood forest. Biogeochemistry 56, 175–190. doi: 10.1023/A:1013072519889
Treseder, K. K., Marusenko, Y., Romero-Olivares, A. L., and Maltz, M. R. (2016). Experimental warming alters potential function of the fungal community in boreal forest. Glob. Chang. Biol. 22, 3395–3404. doi: 10.1111/gcb.13238
Tucker, M. A., Jayasena, K., Ellwood, S. R., and Oliver, R. P. (2013). Pathotype variation of barley powdery mildew in Western Australia. Australas. Plant Pathol. 42, 617–623. doi: 10.1007/s13313-013-0226-y
van Diepen, L. T. A., Frey, S. D., Landis, E. A., Morrison, E. W., and Pringle, A. (2017). Fungi exposed to chronic nitrogen enrichment are less able to decay leaf litter. Ecology 98, 5–11. doi: 10.1002/ecy.1635
Westeberhard, M. J. (1989). Phenotypic Plasticity and the Origins of Diversity. Annu. Rev. Ecol. Syst. 20, 249–278. doi: 10.1146/annurev.es.20.110189.001341
Yuste-Lisbona, F. J., Lopez-Sese, A. I., and Gomez-Guillamon, M. L. (2010). Inheritance of resistance to races 1, 2 and 5 of powdery mildew in the melon TGR-1551. Plant Breed. 129, 72–75. doi: 10.1111/j.1439-0523.2009.01655.x
Zhao, Q., Zeng, D.-H., and Fan, Z.-P. (2008). Effects of freeze-thaw on soil nitrogen and phosphorus availability at the Keerqin Sandy Lands, China. J. Fores. Res. 19, 44–48. doi: 10.1007/s11676-008-0007-6
Keywords: climate change, carbon use efficiency, cellulase, freeze/thaw, Hubbard Brook Experimental Forest, acid phosphatase, fungi
Citation: Finestone J, Templer PH and Bhatnagar JM (2022) Soil Fungi Exposed to Warming Temperatures and Shrinking Snowpack in a Northern Hardwood Forest Have Lower Capacity for Growth and Nutrient Cycling. Front. For. Glob. Change 5:800335. doi: 10.3389/ffgc.2022.800335
Received: 22 October 2021; Accepted: 31 January 2022;
Published: 04 April 2022.
Edited by:
Mark A. Anthony, ETH Zürich, SwitzerlandReviewed by:
Eric Morrison, University of New Hampshire, United StatesGregory Pec, University of Nebraska at Kearney, United States
Copyright © 2022 Finestone, Templer and Bhatnagar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer M. Bhatnagar, am1iaGF0QGJ1LmVkdQ==
 Julia Finestone
Julia Finestone Pamela H. Templer
Pamela H. Templer Jennifer M. Bhatnagar
Jennifer M. Bhatnagar