
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Epigenet. Epigenom. , 19 July 2024
Sec. Chromatin Epigenomics
Volume 2 - 2024 | https://doi.org/10.3389/freae.2024.1430294
This review provides an overview of common assays used to screen for gene methylation and early biomarkers of methylation in various cancers. DNA methylation, one of the most well-studied epigenetic modifications, plays a crucial role in normal cell and tissue development. It is increasingly utilized as a biomarker for early cancer and precancerous lesion detection. In this review, we describe common methods associated with gene methylation, including bisulfite sequencing PCR (BSP), pyrosequencing technology (PYR), methylation-specific polymerase chain reaction (MS-PCR/MSP), methylation-sensitive high-resolution melting (MS-HRM), methylation sensitive single nucleotide primer extension (MS-SnuPE), Epityper, Droplet digital PCR (ddPCR), methylation-sensitive restriction enzyme (MSRE) analysis, COBRA and PacBio SMRT sequencing. Additionally, we summarize methylation markers and their sample types for early cancer screening, focusing on colorectal cancer, hepatocellular carcinoma, gastric cancer, pancreatic cancer, esophageal cancer (digestive system), lung cancer (respiratory system), breast cancer, ovarian cancer, cervical cancer (female reproductive system), bladder cancer, and prostate cancer (urinary system). Furthermore, we discuss the recent detection of methylation biomarkers in clinical samples such as blood, urine, sputum, feces, and tissues. The aim of this review is to summarize early methylation biomarkers that are expected or have already been clinically applied. For future large-scale studies or the integration of available methylome level data, the discovery of sufficiently sensitive clinical biomarkers is essential.
DNA methylation is a crucial epigenetic regulatory mechanism that plays a pivotal role in tumor development. The human genome contains approximately 2,80,000 CpG sites (cytosine-phosphate-guanine sites). These CpG sites exhibit different distributions across the genome, and CpG dinucleotide clusters are found at expected frequencies, forming regions known as “CpG islands” (CGI) (Gardiner-Garden and Frommer, 1987). In mammalian cells, DNA is methylated at approximately 70%–80% of CpG cytosine C5 sites, but also at non-CpG sites (He and Ecker, 2015). Specific DNA methylation at CpG and non-CpG sites is essential for chromatin regulation. The DNA methyltransferase DNMT3A interacts with its TRD and RD loops with target sites surrounded by variable DNA sequences. DNA methylation typically occurs at CpG sites, where cytosine in the CpG dinucleotide environment is methylated at the fifth carbon position by DNA methyltransferases (DNMTs), resulting in 5-methylcytosine (5 mC) (Bestor, 1988) (Figure 1). During tumor development, normal epigenetic processes are disrupted, leading to global changes in DNA methylation patterns (Jones and Baylin, 2007). Cancer cells are characterized by genome-wide hypomethylation, but tumor suppressor initiators may contain associated CpG islands and therefore can become hypermethylated before or during tumorigenesis (Baylin and Jones, 2011). Hypermethylation of promoter CpG islands is a common phenomenon and represents an alternative mechanism for inactivating tumor suppressor genes, DNA repair genes, cell cycle regulators, and transcription factors.
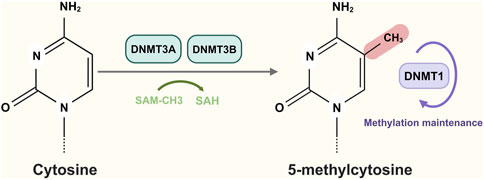
Figure 1. DNA methylation occurs at CpG sites and cytosine is methylated by DNA methyltransferases (DNMT3A and DNMT3B) to generate 5-methylcytosine. Figure created with BioRender.com.
Accurate early diagnosis and prediction of cancer cell metastasis are crucial for effective treatment. Biomarkers play a significant role in aiding clinicians in determining tumor classification, degree of progression, and molecular dysregulation mechanisms. MLH1 promoter CpG sites have been found to exhibit high specificity in detecting MLH1 methylation, making them suitable for screening Lynch syndrome (LS) (Tan et al., 2024). Recent studies (Butler et al., 2020; Della Monica et al., 2022; Leske et al., 2023) have demonstrated that methylation of the O6-methylguanine-DNA methyltransferase (MGMT) promoter can help evaluate prognosis and predict drug response in glioblastoma patients. Moreover, nanopore sequencing has been identified as an effective method for detecting the methylation status of the MGMT promoter (Halldorsson et al., 2024). Hypermethylated BCAT1, IKZF1, and SEPT9 are found to be differentially methylated in both colorectal and gastroesophageal adenocarcinomas. The detection of methylated circulating tumor DNA (ctDNA) of SEPT9 in blood is used as a screening method for colorectal cancer (CRC) (Laven-Law et al., 2024). Early screening of methylation markers and cancer type diagnosis through specific methods can greatly improve the effectiveness of existing cancer treatments and enhance patient recovery rates. Methylation biomarkers are not only useful for diagnosing and staging patients but also for subsequent monitoring and assessing patient prognosis. Therefore, the examination of DNA methylation profiles is increasingly recognized as an important biomarker for tumor diagnosis, prognosis, treatment response, and disease surveillance (Heyn and Esteller, 2012). DNA methylation biomarkers are also considered valuable noninvasive diagnostic tools for cancer detection.
Liquid biopsy (LB) is a less invasive method that reduces the need for tissue biopsies. However, LB faces challenges in clinical validation, sensitivity, specificity, standardization, and scalability. The most common form of LB is cfDNA testing, but it can be influenced by improper specimen collection, anticoagulant use, and storage. To accurately interpret and make clinical decisions, it is crucial to understand the advantages and limitations of cfDNA analysis. LB shows promise as a tool for new precision medicine strategies in cancer treatment. In recent years, LB has been widely used in various studies, such as the development of a Multiple Genome Liquid Biopsy for Prostate Cancer in Whole Blood (PROSTest) (Modlin et al., 2024) and the rapid detection of ctDNA (circulating tumor DNA) in metastatic breast cancer using ultrasensitive and rapid circulating tumor DNA liquid biopsies with surface-restricted gene amplification on dispersible magnetic nanoelectrodes (Park et al., 2024). CtDNA has also emerged as a non-invasive tool to detect minimal residual disease and early detection of recurrence in early stage non-small cell lung cancer (NSCLC) (Tan et al., 2024). Free DNA has shown potential for preoperative malignancy risk assessment in ovarian masses, but further validation is needed through larger clinical studies (Gaillard et al., 2024). Li et al. (2024) developed a cfDNA-based multi-modal epigenetic sequencing analysis (MESA) method for the detection of CRC that captures and integrates cfDNA methylation and nucleosome occupancy. Overall, the detection of cfDNA is a growing trend in cancer research and provides new opportunities for non-invasive diagnosis and prognosis of cancer.
Methylation detection methods have evolved significantly, offering a range of techniques to study DNA methylation patterns. Methylation assays are summarized in Figure 2. The advantages and disadvantages of methylation methods are summarized in Table 1.
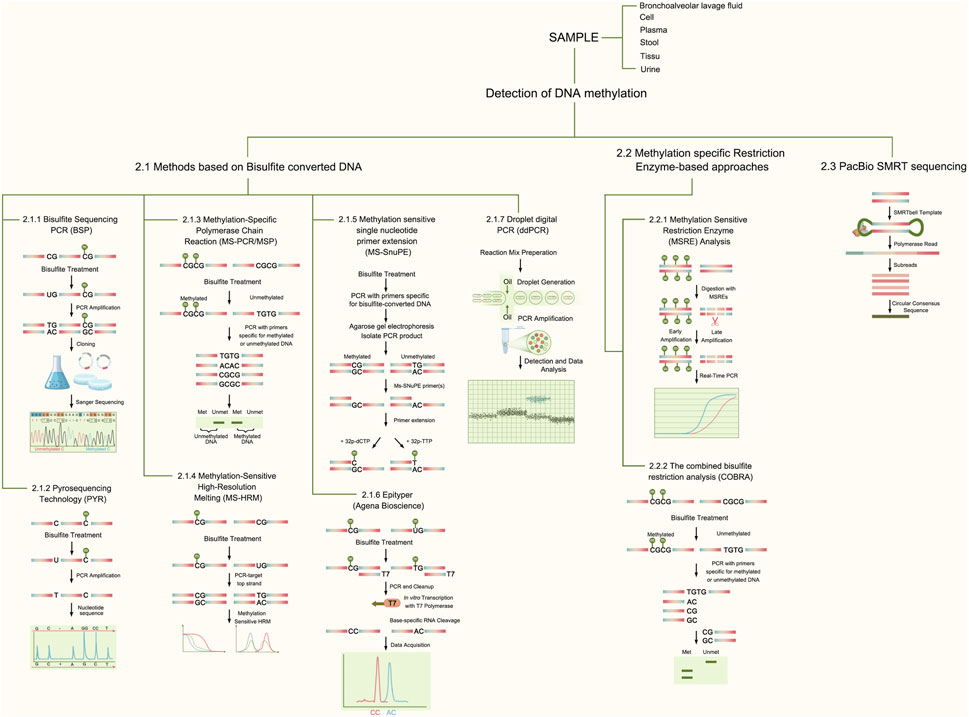
Figure 2. Methods Schematic for Methylation Detection. This figure describes the principles of common methods associated with gene methylation, including bisulfite sequencing PCR (BSP), pyrosequencing technology (PYR), methylation-specific polymerase chain reaction (MS-PCR/MSP), methylation-sensitive high-resolution melting (MS-HRM), methylation-sensitive single-nucleotide primer extension (MS-SnuPE), Epityper, Droplet digital PCR (ddPCR), methylation-sensitive restriction enzyme (MSRE) analysis, COBRA, and PacBio SMRT sequencing.
Genomic sequencing of bisulfite converted DNA is a popular technique for studying DNA methylation. Bisulfite treatment converts unmethylated cytosine to uracil, while leaving 5 mC unaffected. However, one major drawback of bisulfite treatment is DNA degradation, which limits its use for sequencing long DNA molecules. To overcome this, single-molecule sequencing methods like PacBio SMRT sequencing can be employed. We will discuss this method in more detail later. Commonly used tools for detecting DNA methylation in bisulfite-converted DNA include Bismark, BS-Seeker2, and BWA-Meth. Recently novel BSBolt methylation callers have emerged offering more accurate and faster methylation calls without the need for additional formatting (Farrell et al., 2021). High-throughput sequencing combined with bisulfite conversion is a widely used approach for analyzing genome-wide DNA methylation. In one method, biotinylated RNA probes are used to capture a small portion of the genome (approximately 4 Mb) before bisulfite conversion and sequencing (Morselli et al., 2021). This method efficiently handles a large amount of targeted bisulfite sequencing data, allowing for quantitative determination of known methylation statuses.
The PYR method enables the accurate quantification of methylation frequencies at individual contiguous CpG sites, making it possible to detect subtle alterations in methylation levels. This is achieved by determining the proportion of C-T at a single site through the incorporation of C and T during sequence extension. Therefore, the PYR method can accurately detect methylation variants at different loci. It is a relatively simple and rapid method that provides comparable results. Additionally, PYR is suitable for analyzing CpG-poor or CpG-rich regions. The PYR assay consists of three steps (Harrington et al., 2013):① Biotinylated primers are used for PCR amplification and labeling.② Streptavidin separates the PCR products and hybridizes to sequencing primers.③ Sequencing is performed, with nucleotides added one by one according to the template sequence. This releases pyrophosphate (PPi) under the action of polymerase. Sulfatase is used to produce ATP together with adenosine phosphate (APS), and luciferase converts ATP to oxyfluorescein. The light intensity in the thermogram is presented as a peak. The PYR method can be used in combination with MSP to assess tumor tissue methylation in glioblastoma patients (Estival et al., 2019). It can also be employed to assess DNA methylation levels of microRNA-137 as a differential diagnosis between benign and malignant ascites (Bose et al., 2023). Furthermore, the PYR methods has been used to detect methylation of CADM1 and MAL promoter regions in cervical cancer tissue samples as well as some basal methylation in healthy women (Mersakova et al., 2014).
MSP is a methylation detection method specifically designed for CGI. It utilizes two sets of primers for methylated and unmethylated alleles in selected regions. While gel electrophoresis is commonly used to assay PCR products, real-time PCR (qPCR) is now frequently employed for more sensitive results. The detect on of TWIST1 and NID2 gene methylation in urine sediment using MSP offers a highly sensitive and specific non-invasive method for detecting BCa (Renard et al., 2010). Currently, there are two methods for determining methylation products depending on the probes used for qPCR. The method using TaqMan probes is called MethyLight (Eads et al., 2000), and the method using SYBR Green I is called MethylQuant (Thomassin et al., 2004). MethyLight is a highly sensitive assay that can detect methylated alleles even in the presence of a 10,000-fold excess of unmethylated alleles. The development of this technology significantly enhances our ability to rapidly and accurately generate epigenetic profiles of tumor samples. Methylation levels of SFRP1, SFRP2, SDC2, and PRIMA1 were determined using a MethyLight assay after cfDNA isolation and bisulfite conversion of samples (Barták et al., 2019). MethyLight can also quantitatively detect SOX21 gene promoter methylation, serving as a fecal-based noninvasive biomarker for early diagnosis of colorectal cancer (Moradi et al., 2021). Furthermore, the MethyLight assay accurately detects B3GAT2 and ZNF793 methylation levels in endoscopic barrett’s esophagus (BE) samples (Yu et al., 2015). In validation experiments, the MS-HRM assay showed better sensitivity than the MethyLight assay (Wojdacz and Dobrovic, 2007). On the other hand, MethylQuant is an economically efficient and relatively simple technique that enables quantitative analysis of the methylation status of individual cytosines at specific locations in DNA (Dugast-Darzacq and Grange, 2009).
MS-HRM are utilized for comparing the melting curves of DNA sequences with varying base compositions. By analyzing these melting curves, we can identify differences between methylated and unmethylated DNA after bisulfite treatment. This method is highly efficient and can accurately distinguish between methylated and unmethylated sites, making it ideal for cases where only a small portion of the sample sites are methylated. To overcome PCR bias, specific primer design is crucial, especially after bisulfite treatment. Therefore, specific primer design requirements must be met for DNA sequences. Before conducting MS-HRM analysis, DNA must undergo bisulfite modification to differentiate the melting temperature between methylated and unmethylated templates. MS-HRM analysis relies on different melting temperatures (Tm) for C-G and A-T pairs. Tm refers to the temperature at which two DNA strands separate, resulting in a sudden decrease in fluorescence signal as the dye (e.g., SYBR Green) is released. The simplicity and high reproducibility of MS-HRM protocols have made it the preferred method for methylation assessment in various diagnostic and research applications (Wojdacz and Dobrovic, 2007). In a recent study (Liu et al., 2020), MS-HRM was employed to evaluate the methylation status of CASK (calmodulin-dependent serine protein kinase) in 296 tumor tissues. The combined use of MS-HRM and dMS-HRM allowed for the assessment of the extent and complexity of heterogeneous methylation revealing visible patterns.
MS-SnuPE is a commonly used method for analyzing cytosine methylation and quantification. It is performed after conversion and PCR amplification. In this method, the PCR product is used in a second reaction, that specifically targets the 3’ end before CpG after primer annealing. The resulting product is then separated by electrophoresis and analyzed using a phosphorimager (Gonzalgo and Jones, 1997). MS-SnuPE is particularly suitable for examining abnormal methylation patterns during tumorigenesis and monitoring changes induced by DNA methylation inhibitors (Gonzalgo and Liang, 2007). Previous studies have demonstrated the effectiveness of MS-SnuPE in investigating methylation markers such as AGTR1, GALR1, SLC5A8, ZMYND10, and NTSR1 in non-small cell lung cancer (NSCLC) (Guo et al., 2015).
Epityper is an analytical method that utilizes base-specific cleavage and MALDI-TOF-MS (matrix-assisted laser desorption/ionization time-of-flight mass spectrometry). Although it requires expensive instruments, its high-throughput nature makes it ideal for testing a large number of samples. Epityper technology enables accurate, rapid, and reproducible quantification of DNA methylation status at single or multiple CpG positions in numerous samples. This technique can analyze DNA from various sample types, including liquid biopsies, fresh frozen tissues, and formalin-fixed paraffin-embedded tissues. Epityper has also been used to the detect XXYLT1 methylation in lung adenocarcinoma (Zeng et al., 2021).
Droplet digital PCR (ddPCR) is a sequencing technique that uses water-oil emulsion droplet technology. PCR amplification occurs in each droplet, and the proportion of PCR-positive droplets in the original sample is then determined through analysis. Methylation targets are quantified using a standard mutation/SNP singleplex assay strategy in ddPCR analysis of bisulfite converted DNA samples. In the analysis of cell-free circulating DNA (cfDNA), the samples are bisulfite treated and the SEPT9 and BMP3 methylation status is analyzed using a ddPCR system (Lima et al., 2023). Droplet digital methylation-specific PCR (ddMSP) is a combined method of ddPCR and MSP for the detection of methylation markers (RASGRF1, CPXM1, HOXA10, and DACH1) in breast cancer (Uehiro et al., 2016).
Methylation-sensitive restriction enzyme (MSRE) is initially used to selectively digest unmethylated DNA while leaving methylated targets intact. The first method for assessing DNA methylation levels is MSRE analysis using methylation-sensitive restriction enzymes such as HpaII, AatII, and ClaI. These enzymes do not require bisulfite conversion of DNA (Cedar et al., 1979). Research has demonstrated that methylation-sensitive restriction enzyme bisulfite sequencing (MREBS) provides sufficient sequencing coverage and reduces measurable cytosine damage compared to whole-genome bisulfite sequencing (WGBS) and reduced representation bisulfite sequencing (RRBS) (Bonora et al., 2019). With the specificity of MSRE, the high amplification efficiency of Lambda-assisted RPA, and the self-amplification effect of CRISPR/Cas12a, as little as 0.05% of methylated DNA can be distinguished (Zhou et al., 2022). In a recent study, the researchers combined the high specificity of the methylation-sensitive restriction enzyme GlaI for methylase digestion with the high amplification efficiency of rolling circle amplification (RCA) to detect GlaI digestion products. The GlaI-RCA technology confirmed that methylated Septin9 in colorectal cancer tissue samples correlated well with standard bisulfite sequencing (Dong et al., 2022).
COBRA is a technique that combines bisulfite conversion with restriction enzymes order to determine DNA methylation levels at specific gene loci in small amounts of genomic DNA. This technology offers ease of use, quantitative accuracy, and compatibility with paraffin sections (Xiong and Laird, 1997). In the COBRA assay, DNA is modified with sulfite esters and then amplified by PCR. The amplified products are then digested with endonucleases that can distinguish between methylated and unmethylated sequences. The COBRA assay allows for the analysis of methylation status at specific sites (Bilichak and Kovalchuk, 2017). Additionally, COBRA can be combined with bisulfite genome sequencing (BGS) to analyze two critical CpG sites in the ROR2 promoter of endometrial cancer (EC) (Liu D. et al., 2021). Methylation levels at two positions in the promoter region of the MGMT gene were quantitatively analyzed using a combination of COBRA and native capillary gel electrophoresis (Goedecke et al., 2015).
PacBio SMRT sequencing is a third-generation methylation assay. Currently, BSP is the mainstream method, but it has certain disadvantages, such as potential DNA degradation after bisulfite treatment. Additionally, BSP is unsuitable for sequencing long DNA molecules, especially with lower methylation levels that are more susceptible to degradation. Therefore, it is not well suited for thrid-generation sequencing. In contrast, PacBio SMRT sequencing can sequence a single DNA molecule and produce an average read length of 13.5 kb with 99.8% sequencing accuracy (Wenger et al., 2019). Choy et al., (2022) demonstrated the presence of long cfDNA in the plasma of hepatocellular carcinoma (HCC) using PacBio SMRT sequencing. They analyzed fragment size and methylation to determine the tissue origin of individual plasma DNA molecules. PacBio sequencing kinetics allows for direct detection of base modifications, such as N6-methyladenine (m6A) and N4-methylcytosine (m4C). Tse et al. (2021) developed a method for single-molecule real-time sequencing to examine 5 mC directly. This method achieves over 90% accuracy and sensitivity in detecting 5 mC at CpG sites in humans and mice based on polymerase signal characteristics. While PacBio SMRT sequencing technology is primarily used for methylation analysis of microbial genomes (Coy et al., 2020), further exploration is needed for sequencing analysis of methylation markers.
According to the 2020 Global Cancer Data Statistics, colorectal cancer ranks third among common cancers worldwide and second among cancer-related mortality worldwide (Sung et al., 2021). CRC is a heterogeneous disease, with patients exhibiting different phenotypes associated with varying prognoses and treatment regimens. Approximately 20% of CRC patients will develop metastases, resulting in a less than 5-year survival rate of only 14% with conventional treatment (Rumpold et al., 2020). The tumor stage at diagnosis is crucial for prognosis, highlighting the significance of considering CRC as an important condition. Carcinoembryonic antigen (CEA) is a well-known serum biomarker for CRC. However, it can also be elevated in various benign conditions, rendering it ineffective for early detection or screening. Many studies on methylation biomarkers have focused on plasma or fecal samples from CRC patients, such as blood samples from blood KCNJ12 and ZNF132 (Zhang et al., 2021), and stool samples from BMP3, GATA4/GATA5, HLTF and others (Liu et al., 2019). These studies have described DNA methylation biomarkers for CRC metastasis, which show promise for early clinical diagnosis. This article primarily outlines 11 methylation biomarkers (ADHFE1, BMP3, CDX2, CYP24A1, DKK3, NSD2, SDC2, SEPT9, SFRP1, VIM) that can be used for early screening of CRC. Additional biomarkers for CRC are listed in Table 2; Figure 3.
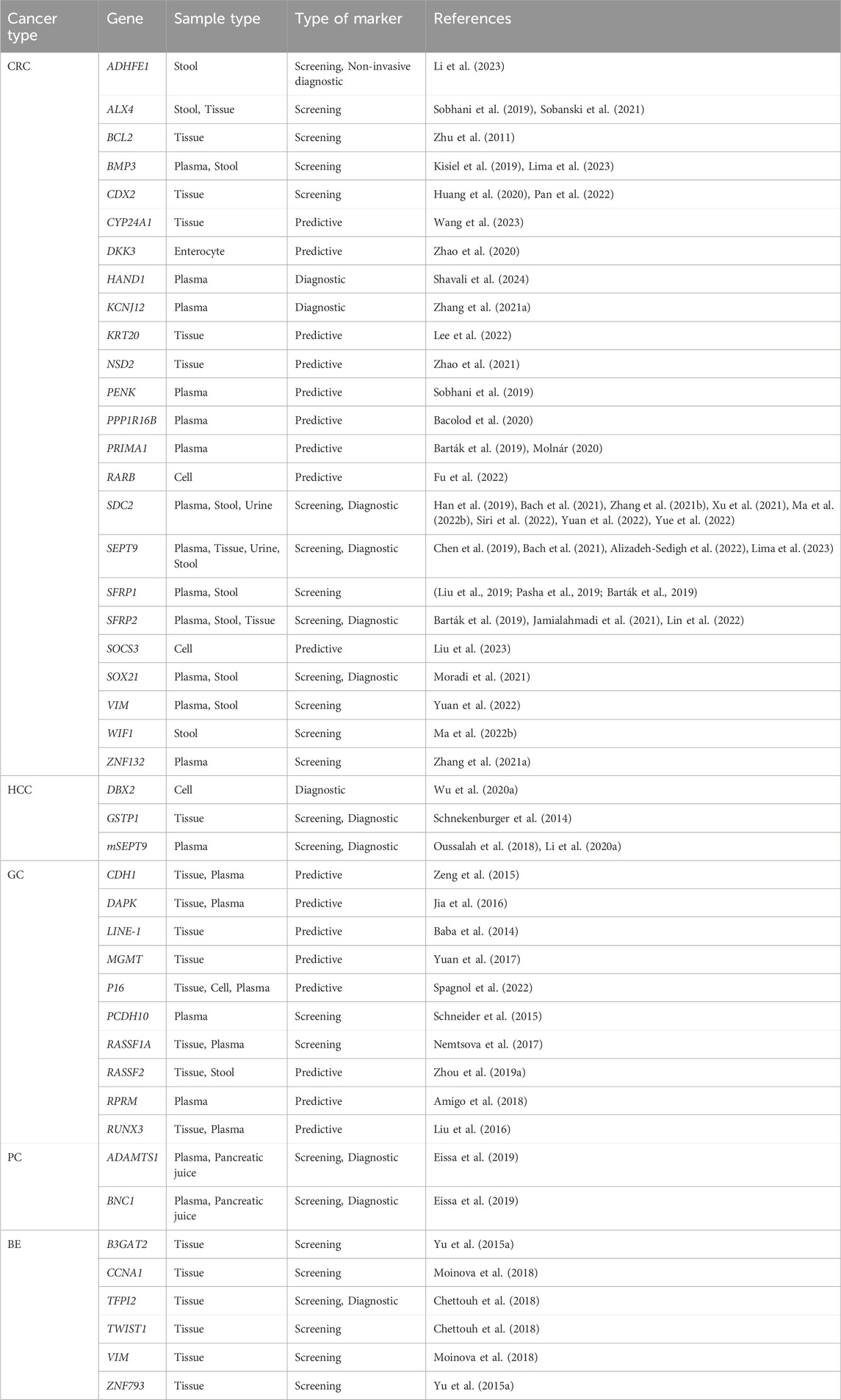
Table 2. Candidate biomarkers of methylation in early diagnosis of colorectal, hepatocellular, gastric, pancreatic and esophageal cancers.
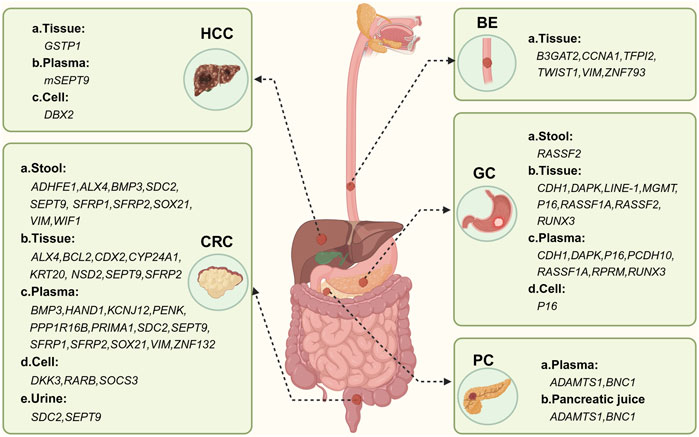
Figure 3. Methylation-based biomarker candidates in different samples for digestive system cancer diagnosis. Abbreviations: HCC: a. Tissue:GSTP1 (glutathione S-transferase pi 1); b. Plasma: mSEPT9; c. cell: DBX2 (developing brain homeobox 2). CRC: a. Stool: ADHFE1 (alcoholdehydrogenase, ironcontaining, 1), ALX4 (ALX homeobox 4), BMP3 (bone morphogenetic protein 3), SDC2 (syndecan 2), SEPT9 (septin 9), SFRP1 (secreted frizzled-related protein 1), SFRP2 (secreted frizzled-related protein 2), SOX21 (sex determining region Y box 21), VIM (vimentin), WIF1 (WNT inhibitory factor 1); b. Tissue: ALX4 (ALX homeobox 4), BCL2 (B-cell CLL/lymphoma 2), CDX2 (caudal type homeobox 2), CYP24A1 (cytochrome P450, family 24, subfamily A, polypeptide 1), KRT20, NSD2 (Wolf-Hirschhorn syndrome candidate 1), SEPT9, SFRP2; c. Plasma: BMP3, HAND1 (heart and neural crest derivatives expressed 1), KCNJ12 (potassium channel, inwardly rectifying subfamily J, member 12), PENK (proenkephalin), PPP1R16B (protein phosphatase 1, regulatory subunit 16B), PRIMA1 (proline rich membrane anchor 1), SDC2 (syndecan 2), SEPT9, SFRP1, SFRP2, SOX21, VIM; d. Cell: DKK3 (dickkopf WNT signaling pathway inhibitor 3), RARB (retinoic acid receptor, beta), SOCS3 (suppressor of cytokine signaling 3); e. Urine: SDC2, SEPT9. BE: a. Tissue: B3GAT2 (beta-1,3-glucuronyltransferase 2), CCNA1 (cyclin A1), TFPI2 (tissue factor pathway inhibitor 2), TWIST1 (twist family bHLH transcription factor 1), VIM, ZNF793 (zinc finger protein 793). GC: a. Stool: RASSF2 (Ras association domain family member 2);b. Tissue: CDH1 (cadherin 1), DAPK (death-associated protein kinase 1), LINE-1, MGMT (O-6-methylguanine-DNA methyltransferase), P16 (cyclin-dependent kinase inhibitor 2A), RASSF1A (Ras association domain family member 1), RASSF2, RUNX3 (runt-related transcription factor 3); c. Plasma: CDH1, DAPK (death-associated protein kinase 1), P16, PCDH10 (protocadherin 10), RASSF1A, RPRM(reprimo), RUNX3 (runt-related transcription factor 3); d. Cell: P16. PDA: a. Plasma: ADAMTS1 (ADAM metallopeptidase with thrombospondin type 1 motif, 1), BNC1 (basonuclin 1); b. Pancreatic juice: ADAMTS1, BNC1. Figure created with BioRender.com.
Hypermethylation of ADHFE1 (Alcohol dehydrogenase iron containing 1) promotes the proliferation of CRC cells by regulating cell cycle progression (Hu et al., 2019). This hypermethylation can serve as an early screening indicator, and restoring ADHFE1 may offer a new treatment option for CRC patients. A recent study has demonstrated that the combined detection of ADHFE1/SDC2/PPP2R5C methylation could potentially be used as a non-invasive diagnostic approach for CRC and screening for precancerous lesions (Li et al., 2023). BMP3 (bone morphogenetic protein 3) belongs to the transforming growth factor-β (TGFB) superfamily. It is used for multi-target fecal DNA detection (Tepus and Yau, 2020). The promoters of BMP3 and SEPT9 (septin 9) in plasma samples can serve as indicators for early detection and screening of CRC in the population (Lima et al., 2023). Methylation levels of BMP3 and VAV3 in stool samples are highly specific for CRC surveillance (Kisiel et al., 2019). BRAF transcription factor CDX2 (caudal type homeobox 2) undergoes DNA methylation silencing in proximal colon cancer (Yang et al., 2024). JARID1B regulates CDX2 expression by demethylation of H3K4me3, which inhibits upstream methylation and promotes cell proliferation in CRC through the Wnt/beta-catenin pathway (Huang et al., 2020). CDX2 is also inhibited by SMYD2-mediated MEX3A to enhance CRC progression (Pan et al., 2022), Decreased expression of CDX2 is more dependent on the CpG island hypermethylation phenotype (CIMP-H) (Lee et al., 2022). The methylation status of CYP2R1 (cytochrome P450) can influence contributors to vitamin D serum levels and predict response to vitamin D supplementation. It has been shown that elevated serum levels of 25-hydroxyvitamin D (25 (OH) D) result in impaired methylation of CYP24A1 (Forouhari et al., 2023). Recent studies have shown that cumulative methylation levels at CpG sites in CYP24A1 are inversely associated with CRC risk (Wang et al., 2023). In most human cancer cells, the expression of DKK3 (Dickkopf-related protein 3) is epigenetically silenced by methylation of CpG islands in promoters (Nozaki et al., 2001). The mRNA expression of DKK3 is inversely correlated with its promoter methylation and may inhibit tumorigenesis and serve as a molecular diagnostic target (Zhao et al., 2020). NSD2 (Histone methyl transferase nuclear receptor-binding SET domain 2) is a crucial enzyme that catalyzes histone H3 lysine 36 dimethylation (H3K36me2). Elevated expression of NSD2 mRNA has been observed in colon cancer. Furthermore, silencing or knocking down NSD2 leads to a significant reduction in the activation of multiple oncogenes (ADAM2, EGFR, Sox2, Bcl-2, SYK, and MET). This suggests that NSD2 may serve as a novel target for detection and therapeutic intervention (Zhao et al., 2021). SDC2 (syndecan 2) is a transmembrane heparan sulfate proteoglycan. It encodes the SDC2 protein, which plays a role in the interaction regulation of cell-environment interactions. SDC2 affects cell proliferation, migration, invasion, and tumor development (Hua et al., 2020). The methylation status of the SDC2 promoter in blood samples can be detected using the methylation quantitative endonuclease resistance DNA assay (MethyQESD). This assay has shown high accuracy in differentiating CRC patients at an early stage of the disease (Siri et al., 2022). SDC2 can be combined with many biomarkers for methylation detection. For example, SDC2-SEPTIN9-BCAT1 combined with ctDNA methylation demonstrates high sensitivity and specificity (Xu et al., 2021). Similarly, the combined detection of SDC2-SEPTIN9-VIM has the potential to serve as a biomarker for early diagnosis of CRC (Yuan et al., 2022). The detection of aberrantly methylated DNA in feces has proven to be an effective early screening method for CRC. The combined detection of methylated SDC2-TFPI2 in feces holds promise as one of the effective and noninvasive tools for early CRC screening (Zhang et al., 2021). Additionally, the combined detection of SDC2-ADHFE1-PPP2R5C methylation could serve as a potentially non-invasive diagnostic method for CRC and precancerous lesion screening (Li et al., 2023). In clinical applications, the combined SDC2-NDRG4 methylation detection assay has shown excellent performance in the detection of CRC and advanced adenomas (Zhang et al., 2024). A study found that the sensitivity of CRC detection increased to 87.25% when methylated SDC2-A and methylated SDC2-B were combined. When using methylated SDC2-A alone, the sensitivity was 85.29%, and when using methylated SDC2-B alone, the sensitivity was 83.33% (Ma et al., 2022a). SDC2-SFRP2 methylation was detected using quantitative PCR (qPCR) or methylation-specific quantitative PCR (MSP) (Lin et al., 2022). There are also new methods for detecting SDC2, such as linear target enrichment quantitative methylation-specific real-time PCR (LTE-qMSP), which is highly specific for early detection of SDC2 methylation in fecal DNA in CRC patients (Han et al., 2019). Droplet digital PCR (ddPCR) indicated that the sensitivity and specificity of SDC2 gene methylation detection could be reduced to 0.1% methylation level and 5 ng methylated DNA input (Yue et al., 2022). The ColoCaller test allows for simultaneous detection of the methylation status of SDC2, TFPI2, WIF1, and NDRG4 genes in fecal DNA, optimizing and screening for colorectal cancer (Ma et al., 2022b). Marker groups consisting of SDC2 and SEPT9 were detected in urine sediment of CRC cases with a specificity of 70%, showing promise for non-invasive CRC detection through urine liquid biopsy (Bach et al., 2021). SEPT9 plays a crucial role in cell division, and its hypermethylation process is linked to carcinogenesis and can alter the conformation of gene sequences. Methylated CpG sites bind to methylated CpG-binding proteins and interact with other proteins to form abnormal chromatin, which inhibits gene expression (Wasserkort et al., 2013). Additionally, the combination of plasma biomarkers SEPT9-BMP3 has been used for early screening of CRC in a Brazilian population (Lima et al., 2023). The methylation panel consisting of SEPT9-SPG20-FBN1 has higher diagnostic value compared to individual biomarkers or other combinations. This panel serves as a simple noninvasive methylation marker for early detection of CRC (Alizadeh-Sedigh et al., 2022). Studies have shown that 90% of CRC patients (27 out of 30) exhibit hypermethylation in the SEPT9 gene, while 83% (25 out of 30) show hypermethylation in the HAND1 gene. This emphasizes the potential of SEPT9 and HAND1 methylation as promising biomarkers for the diagnosis of CRC, leading to early detection and improved patient outcomes (Shavali et al., 2024). Moreover, SEPT9 in urine (Bach et al., 2021) and stool (Chen et al., 2019) samples can also serve as methylation markers for early CRC screening. In recent years, a noninvasive test called methylated CpG tandem amplification and sequencing (MCTA-Seq) has been developed for CRC, specifically for detecting SEPT9. SFRP1 (secreted frizzled-related protein 1) promoter DNA hypermethylation reduces the mRNA expression levels of SFRP1, and it is regulated by H3K27me3 and DNA methylation (Wu et al., 2023). The combined detection of the SFRP1-RUNX3-CEA panel may serve as noninvasive biomarkers for early detection and diagnosis of CRC (Pasha et al., 2019). MethyLight assay can be used to determine the methylation status of SFRP1 (Barták et al., 2019), SFRP2 (Li et al., 2019), and SOX21 (Moradi et al., 2021). VIM (vimentin) and SEPT9 are epigenetic markers in CRC (Bacolod et al., 2020). The use of methylation markers for three genes, VIM-SEPTIN9-SDC2, enhances signal detection and can improve the accuracy of early CRC diagnosis. These markers show great potential for clinical application (Yuan et al., 2022).
Global cancer statistics in 2020 revealed that liver cancer ranks as the sixth most prevalent cancer, with 905,677 new cases per year (Sung et al., 2021). HCC is a growing health concern and the second leading cause of cancer-related deaths worldwide, and it is the ninth most common cancer in women and the fifth most common cancer in men (Cao et al., 2021). Risk factors for HCC include cirrhosis, hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, metabolic syndrome, hemochromatosis, α1-antitrypsin deficiency, and alcohol consumption. The development of HCC typically follows the path of hepatitis → cirrhosis → precancerous lesions → hepatocellular carcinoma, with 80%–90% of patients progressing through this sequence. Patients with early stage (I) liver cancer infected with hepatitis B virus have a significantly higher 5-year survival rate compared to those with advanced stage (III) liver cancer. However, HCC is often asymptomatic in the early stage, and it is frequently detected in the late stage. In recent years, alterations in DNA methylation have been proposed as “liquid biopsies”, which ctDNA, circulating tumor cells (CTC), circulating miRNAs, and exosomes (Li et al., 2018). Among these, ctDNA shows potential as an early diagnostic tool for HCC and can be found in a various early stage tumers such as colorectal cancer, ovarian cancer, lung cancer and breast cancer (Phallen et al., 2017). Currently, about 50% of HCC biomarkers are being investigated at the gene methylation level, with ctDNA methylation profiles in plasma being one of the most extensively studied areas in HCC liquid biopsies. Epigenetically altered genes are often associated with specific etiologic factors. For example, alpha-fetoprotein (AFP), miR-122, and circulating TERT promoter mutations are biomarkers used for diagnostic screening of HBV-related HCC (Trung et al., 2020). The following provides an overview of two well-studied methylation biomarkers that can be used for early HCC screening in recent years. Additional biomarkers are presented in Table 2; Figure 3.
GSTP1 (glutathione S-transferase pi 1) is a key metabolic enzyme gene involved in phase II metabolism of various carcinogens. It plays an important role in maintaining cellular metabolism by decomposing carcinogens, protecting against oxidative damage, and the development of cancer cells. GSTP1 has a large CpG-rich region that is frequently affected by methylation. It is frequently silenced by CpG island promoter hypermethylation in liver cancer, prostate cancer, breast cancer, and others. Some studies have shown that GSTP1 methylation can serve as a diagnostic tool for HCC with a sensitivity of approximately 50%–75% and a specificity of 70%–91% (Gurioli et al., 2018). When combined with other biomarkers such as APC or SOCS (Liu et al., 2015), it can be used for early cancer detection in individuals who are genetically predisposed or exposed to carcinogens (Schnekenburger et al., 2014). Plasma mSEPT9 is a novel circulating free DNA epigenetic biomarker for the diagnosis of HCC (Oussalah et al., 2018). In a study by Li et al. (2020), the diagnostic performance of plasma SEPT9 for HCC was assessed using methylation-specific fluorescent quantitative PCR. The sensitivity of SEPT9 for detecting early HCC increased to 91.3% and 87.7% when combined with serum AFP. The combination of mSEPT9-AFP-PIVKA-II is optimal compared to any single marker and may be useful for opportunistic screening of HCC in the cirrhotic population (Zheng et al., 2023).
According to World Health Organization (WHO) statistics in 2020, gastric cancer is the fifth most common cancer worldwide and the fourth leading cause of cancer death (Sung et al., 2021). The development of GC involves multiple factors, including Helicobacter pylori and Epstein-Barr virus infections, exposure to carcinogens, suppression of tumor suppression genes, abnormal cell apoptosis, and disruption of cell cycle regulation. The progression of GC can be observed through various stages, starting from superficial gastritis, then progressing to atrophic gastritis, intestinal metaplasia, gastric epithelial dysplasia, and finally gastric cancer. Early detection of GC, either in its pre-malignant or early stages, can greatly improve the 5-year survival rate. Some countries with high GC incidence, such as Japan and Korea (Xia and Aadam, 2022), have established screening programs to enhance early detection and overall survival rates. DNA methylation, a process that involves specific genetic and epigenetic changes in the early stages of GC, can be detected in various bodily fluids, such as blood, gastric juice, and feces. Therefore, DNA methylation biomarkers can serve as indicators for the pathogenesis and early screening of GC. This article will outline five potential methylation biomarkers for the early screening of GC, with additional details presented in Table 2; Figure 3.
DNMT3A interacts with YAP/TAZ and recognizes CpG islands within the CDH1 promoter. This interaction leads to hypermethylation of the CDH1 (cadherin 1) promoter, resulting in the transcriptional silencing of CDH1 and the acceleration of epithelial mesenchymal transition (Xu et al., 2024). CDH1 hypermethylation levels are higher in the gastric mucosa adjacent to cancer compared to normal gastric mucosa, and this hypermethylation is closely associated with H. pylori (HP) status (Zeng et al., 2015) Pyrosequencing is commonly used to quantify the methylation of the CDH1 gene. In gastric mucosa, LINE-1 hypomethylation is linked to H. pylori infection, and LINE-1 methylation levels hold promise as a surrogate marker for epigenetic defects in gastric carcinogenesis (Kosumi et al., 2015). MGMT is a DNA repair enzyme that plays a role in maintaining gene stability directly reversing guanine O6 alkylation through methyl groups, which helps prevent tumorigenesis. However, MGMT gene methylation is involved in GC development. A meta-analysis of MGMT promoter methylation and GC suggests that MGMT may be an important biomarker for GC progression (Yuan et al., 2017). The P16 gene (cyclin-dependent kinase inhibitor 2A) located in the chromosome 9P21 region and serve as a key regulator in the cell cycle. The positive rate of P16 gene methylation is significantly higher in lymph node metastasis of GC Considered the second largest tumor suppressor gene after TP53, the P16 gene belongs to the INK4 gene family. Methylation of the P16 gene promoter has relatively low sensitivity but high specificity for the diagnosis of GC (Peng et al., 2014). The Reprimo gene family consists of single exonic genes found only in the vertebrate lineage, including RPRM and RPRM-like (RPRML). Loss of RPRM expression is associated with increased proliferation and growth of GC cells, and this poorly understood gene family has the potential for new translational applications as biomarkers for early screening in the future (Amigo et al., 2018).
Pancreatic cancer is a highly aggressive malignancy that is often not detected until it has reached an advanced stage. According to changes in cancer survival rates in China from 2003 to 2015, the 5-year standardized survival rate for PC is only 7.2% (Zeng et al., 2018). Unfortunately, there are currently no widely used early diagnostic tests for PC, and only a few prognostic tests are available. This is largely due to the lack of reliable biomarkers that can accurately detect the early progression of the disease. However, there is promising research being conducted on the use of circulating tumor DNA (ctDNA) for the early diagnosis of PC. At present, many studies are currently analyzing the methylation of various genes and exploring the possibility of combining multiple candidate genes to create combined biomarkers. These biomarkers can be detected in body fluids such as pancreatic juice and blood. One such promising biomarker is BNC1 (basonuclin 1), which has shown potential for the early diagnosis of PC. Combining the cfDNA methylation of BNC1 with ADAMTS1 (ADAM metallopeptidase with thrombospondin type 1 motif 1) has proven to be a powerful tool for the early detection of PC (Eissa et al., 2019).
DNA methylation can be used as an early detection marker for esophageal cancer. For example, methylation of HIN1, TFPI2, DACH1, and SOX17 (Ma et al., 2016) has shown promising results. Barrett’s esophagus (BE) is a precancerous lesion that occurs due to gastroesophageal reflux. In BE, the normal esophageal squamous epithelium is replaced by intestinal metaplasia, which is believed to be a precursor of esophageal adenocarcinoma (EAC). DNA methylation is a common epigenetic change in BE. Methylation of B3GAT2 (beta-1,3-glucuronyltransferase 2), CCNA1, TFPI2, TWIST1, VIM, and ZNF793 (Zinc finger protein 793) can be confirmed using MethyLight PCR and Cytosponge, a non-endoscopic tool (Chettouh et al., 2018). Among these, TFPI2 has shown promise as a diagnostic biomarker for BE. Combined detection of B3GAT2 and ZNF793 may serve as an early biomarker for noninvasive detection of BE (Yu M. et al., 2015). Additionally, a combination of CCNA1 (cyclin A1) and VIM biomarkers has demonstrated a sensitivity of 90.3% and a specificity of 91.7% in detecting BE metaplasia following DNA methylation (Moinova et al., 2018). Methylation of TFPI2 (Tissue factor pathway inhibitor 2) and TWIST1 (Twist family bHLH transcription factor 1) can also be detected early using Cytosponge non-endoscopic cell collection devices (Chettouh et al., 2018).
Lung cancer (LC) is the most common cancer and the leading cause of cancer death in men. However, there are currently limited effective and practical methods available for its early detection. According to the latest World Health Organization statistics in 2020, lung cancer is the second leading cause of cancer-related death worldwide and ranks first in cancer-related mortality worldwide (Sung et al., 2021). The early symptoms of LC are not typical, and by the time symptoms appear, the cancer has often already progressed significantly. Over the last 2 decades, low-dose computed tomography (LDCT) has been the most commonly used method for LC screening (Liang et al., 2019). However, the high sensitivity of LDCT can result in frequent false positives, and further investigation is needed to address its limitations. Therefore, there is a pressing need to explore biomarkers that can improve the early detection rate of LC. Abnormal DNA methylation typically occurs in the early stages of tumorigenesis and can provide new insights for the early detection of LC. There are two main types of LC: non-small cell lung cancer (NSCLC) and small cell lung cancer (SCLC). NSCLC is more closely associated with methylation. Recent studies have shown that liquid biopsy can be performed to detect lung cancer-related biomarkers in various bodily fluids, including plasma, sputum, bronchial fluid (such as bronchoalveolar lavage fluid, bronchial aspirate, and bronchial washings), and urine (Li et al., 2022). In patients with malignant tumors, ctDNA mainly consists of DNA from primary or metastatic tumor cells. The DNA is released into the blood from both healthy cells and tumor cells due to necrosis or apoptosis. CtDNA methylation in the blood may serve as an effective biomarker for LC. Sputum cytology is now known to have very low sensitivity for detecting LC in the early stages (Su et al., 2016). However, DNA hypermethylation status in sputum has been detected in LC patients in recent years (Hubers et al., 2017). Similarly, bronchoalveolar lavage fluid (BALF), bronchial aspirates, and bronchial washings, which are obtained by fiberoptic bronchoscopy, have characteristics similar to those of the LC tumor microenvironment and are less invasive. For example, a combination of SHOX2 and RASSF1A in bronchoalveolar lavage fluid has been shown to perform methylation analysis, with higher sensitivity and specificity compared to conventional cytology and serum CEA in the diagnosis of LC (Zhang et al., 2017). Additionally, DNA methylation of SHOX2 in bronchial aspirates can distinguish LC patients from healthy individuals with high sensitivity and specificity (Schmidt et al., 2010). DNA methylation biomarkers in bronchial washings can be used for early diagnosis of NSCLC (Um et al., 2018). Recent studies have also found that DNA methylated promoters of CDO1, TAC1, HOXA9 and SOX17 can be detected in the urine of patients with NSCLC (Liu et al., 2020; Bach et al., 2022). The following mainly outlines six methylation biomarkers that may be used as early screening biomarkers for NSCLC, with additional information provided in Table 3; Figure 4.
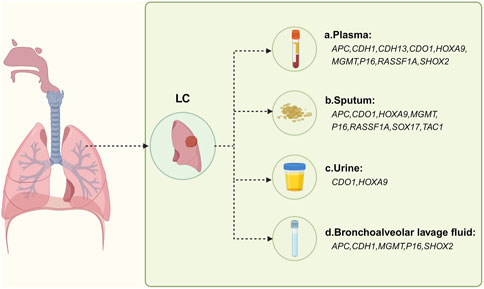
Figure 4. Methylation-based biomarker candidates in different samples for respiratory cancer diagnosis. Abbreviations: LC: a. Plasma: APC (adenomatous polyposis coli), CDH1 (cadherin 1), CDH13 (cadherin 13), CDO1 (cysteine dioxygenase type 1), HOXA9 (homeobox A9), MGMT (O-6-methylguanine-DNA methyltransferase), P16 (cyclin-dependent kinase inhibitor 2A), RASSF1A (Ras association domain family member 1), SHOX2 (short stature homeobox 2); b. Sputum: APC, CDO1, HOXA9, MGMT, P16, RASSF1A, SOX17 (sex determining region Y-box 17), TAC1 (tachykinin precursor 1); c. Urine: CDO1, HOXA9; d. Bronchoalveolar lavage fluid: APC, CDH1, MGMT, P16, SHOX2. Figure created with BioRender.com.
APC (adenomatous polyposis coli) encodes a tumor suppressor protein that acts as an antagonist of the Wnt signaling pathway. Defects in this gene result in familial adenomatous polyposis (FAP), an autosomal dominant precancerous condition that often progresses to malignancy. The methylation of the APC gene promoter is a potential biomarker for the diagnosis of LC. It is particularly useful when detected in serum, sputum, or bronchial lavage fluid of patients with NSCLC, as it exhibits high specificity. However, its low sensitivity makes it unsuitable for lung cancer screening in the general population (Liu et al., 2021). CDH1 (cadherin 1) is a calcium-dependent cell-cell adhesion glycoprotein that is believed to promote cancer progression through proliferation, invasion, and/or metastasis. A meta-analysis (Yu et al., 2015) revealed higher methylation rates of CDH1 in stages III and IV compared to stages I and II, suggesting its potential as an early screening biomarker for LC. The combination of three genes, CDO1 (cysteine dioxygenase type 1), HOXA9 (homeobox A9), and TAC1 (tachykinin, precursor 1) has shown great promise in both the early diagnosis and staging of NSCLC (Wrangle et al., 2014). Hypermethylation of CpG island-rich gene promoters is one of the epigenetic mechanisms involved in carcinogenesis. It is considered a potential biomarker for LC. HOXA9 is one of the genes most associated with LC through this mechanism. Hypermethylation of the HOXA9 promoter leads to its transcriptional inactivation in LC (Rhee et al., 2015). The low expression of HOXA9 is associated with epithelial-mesenchymal transition (EMT), which may be a feature of tumor invasion in NSCLC patients. Abou-Zeid et al., (2023) studied a potential diagnostic for NSCLC that combines HOXA9 gene promoter methylation with SOX2 and HV2 genes in cfDNA. They used PYR and real-time PCR to detect the methylation of the three promoters in blood samples, aiming to determine whether they could serve as non-invasive diagnostic markers for NSCLC. Paschidis et al. (2022) observed MGMT promoter methylation in 30% of patients using methylation-sensitive high-resolution melting analysis (MS-HRM) and bisulfite pyrosequencing or Sanger sequencing analysis. Methylation of the MGMT gene promoter can be used as a biomarker for the early diagnosis of LC. A recent meta-analysis (Chen et al., 2021) showed that MGMT is hypermethylated in the plasma and bronchoalveolar lavage fluid of LC patients. RASSF1A (Ras association domain-containing protein 1A) is a well-studied tumor suppressor gene that plays an important role in cell proliferation, tumorigenesis, and migration. Deficiency in RASSF1A leads to YAP activation and is a key factor in acquiring of malignant phenotypes, invasiveness, anti-apoptotic properties, and eventually transforming from bronchial epithelial cells into cancer cells (Dubois et al., 2016). Frequent promoter methylation of APC and RASSF1A found in 49% and 56% of patients, respectively. The detection of P16 gene promoter methylation in serum or bronchoalveolar fluid/sputum has high specificity but low sensitivity. It may serve as a potential biomarker for NSCLC diagnosis but is not suitable for mass screening due to its low sensitivity (Tuo et al., 2018). However, the combined detection of P16 and RASSF1A may be an effective biomarker for the early diagnosis of LC (Han et al., 2016). In a study by Feng et al. (2020) involving 89 Chinese NSCLC patients and 9 non-tumor patients, methylation of SHOX2 (short stature homeobox 2) was significantly associated with NSCLC (p = 0.003 < 0.05) when using methylation-specific PCR (MSP). MSP has the advantages of rapidity, simplicity, specificity, and low cost, making it suitable for large-scale early screening of LC. The combined detection of SHOX2 and RASSF1A gene methylation is also considered an excellent method for the early screening and monitoring of LC due to its high sensitivity and specificity (Li et al., 2020).
Breast cancer is the most common cancer in women and the leading cause of cancer death. According to the latest statistics in 2020, female breast cancer ranks first among common cancers worldwide and is the fifth leading cause of cancer-related deaths worldwide (Sung et al., 2021). Traditional diagnostic methods for BC include mammography, MRI, biopsy, and histological analysis of tumor tissue. However, the incidence of BC is increasing year by year, especially in developing countries where screening and awareness are lacking. Therefore, liquid biopsy based on blood samples has emerged as a valuable tool for the early detection of BC. Biomarkers may enhance early detection of BC and provide symptomatic treatment options, thereby reducing the risk of death. Epigenetic features are considered good biomarkers for early detection, prognosis, and targeted therapy of BC, with DNA methylation being a potential biomarker for its early detection. For example, the down-regulation of PAQR3 (progestin and adipoQ receptor family member III) expression in BC tissues is significantly associated with abnormal methylation of gene promoters (Chen et al., 2016). Methylation of APC and RARB genes can be detected in serum samples from patients with early BC (Swellam et al., 2015). Most promoters such as ALDH1A2, ALDH1L1, GSTP1, p16, ESR1 and PITX2 have been studied regarding BC methylation markers focusing on the prognosis of operable patients with BC (Sheng et al., 2017; Zhang et al., 2022). Recent studies (Lønning et al., 2022; Al-Moghrabi et al., 2024) have shown that BRCA1 epigenetic mutations increase the risk of triple-negative breast cancer (TNBC). Monitoring the DNA methylation status of BRCA1 has great potential as a pre-screening tool for these cancer forms. However, there have been few studies on the early diagnosis of BC so far. Methylation markers for early diagnosis of BC in recent years are summarized in Table 4; Figure 5.
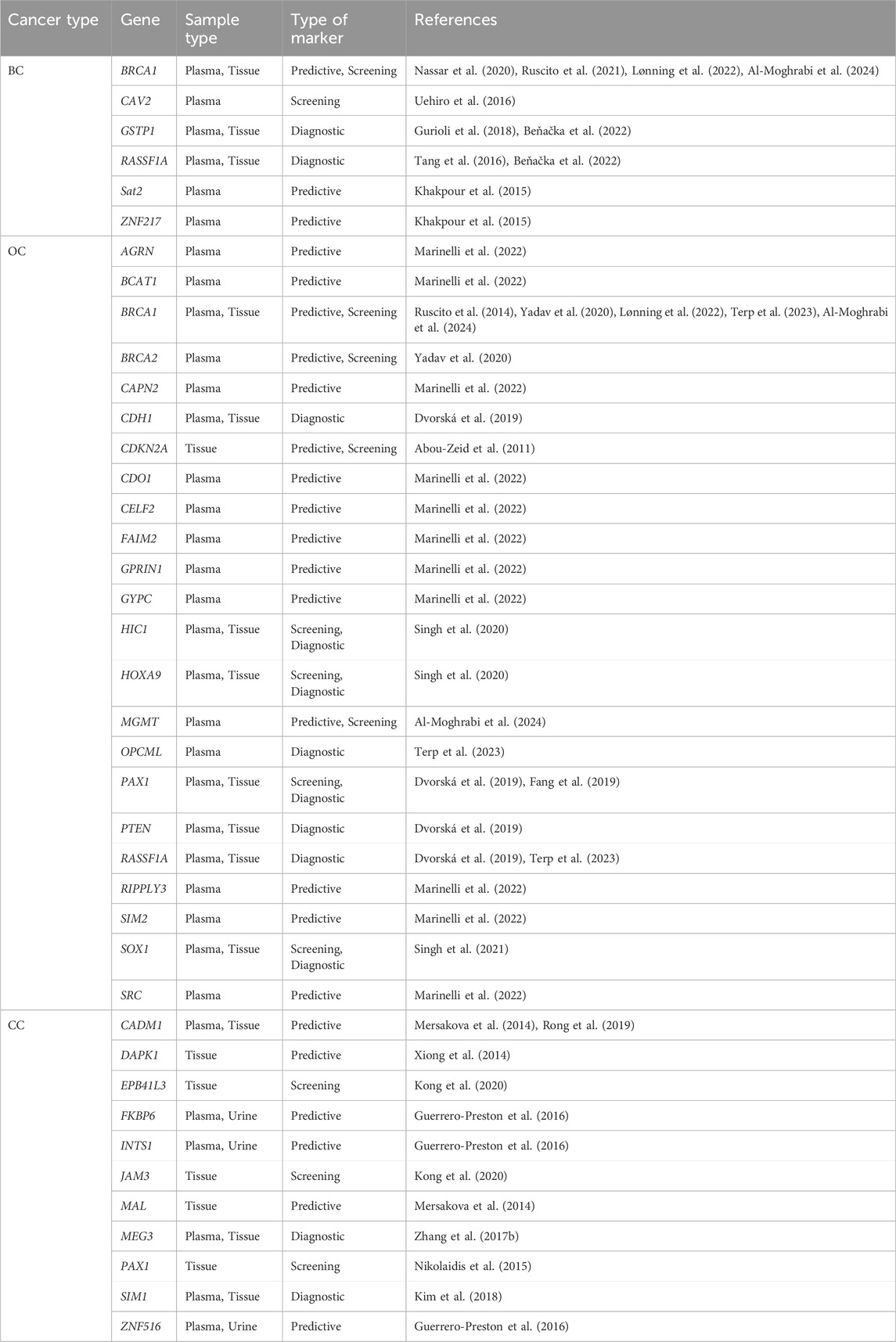
Table 4. Candidate biomarkers of methylation in early diagnosis of breast, ovarian and cervical cancer.
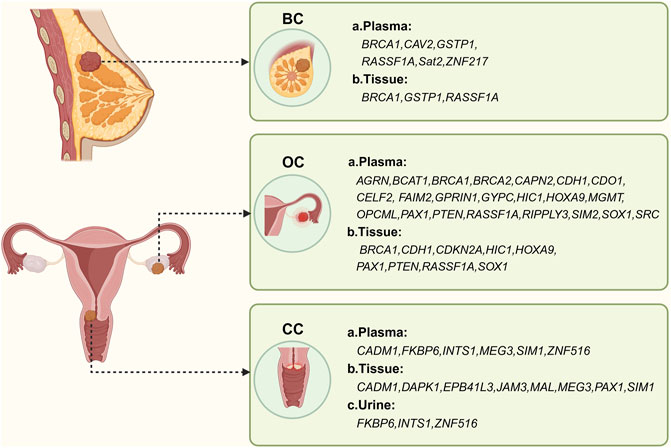
Figure 5. Methylation-based biomarker candidates in different samples for female reproductive system cancer diagnosis. Abbreviations: BC: a. Plasma: BRCA1 (breast cancer 1), CAV2 (caveolin 2), GSTP1 (glutathione S-transferase pi 1), RASSF1A (Ras association domain family member 1), Sat2 (spermine N1-acetyltransferase family member 2), ZNF217 (zinc finger protein 217); b. Tissue: BRCA1, GSTP1, RASSF1A. OC: a. Plasma: AGRN (agrin), BCAT1 (branched chain amino-acid transaminase 1), BRCA1, BRCA2 (breast cancer 2), CAPN2 (calpain 2), CDH1 (cadherin 1), CDO1 (cysteine dioxygenase type 1), CELF2 (CUGBP, Elav-like family member 2), FAIM2 (Fas apoptotic inhibitory molecule 2), GPRIN1 (G protein regulated inducer of neurite outgrowth 1), GYPC (glycophorin C), HIC1 (hypermethylated in cancer 1), HOXA9 (homeobox A9), MGMT (O-6-methylguanine-DNA methyltransferase), OPCML (opioid binding protein/cell adhesion molecule-like), PAX1 (paired box 1), PTEN (phosphatase and tensin homolog), RASSF1A, RIPPLY3 (ripply transcriptional repressor 3), SIM2 (single-minded family bHLH transcription factor 2), SOX1 (sex determining region Y-box 1), SRC (SRC proto-oncogene, non-receptor tyrosine kinase); b. Tissue: BRCA1, CDH1, CDKN2A (cyclin-dependent kinase inhibitor 2A), HIC1, HOXA9, PAX1, PTEN, RASSF1A, SOX1. CC: a. Plasma: CADM1 (cell adhesion molecule 1), FKBP6 (FK506 binding protein 6), INTS1 (integrator complex subunit 1), MEG3 (maternally expressed 3), SIM1 (single-minded family bHLH transcription factor 1), ZNF516 (zinc finger protein 516); b. Tissue: CADM1, DAPK1 (death-associated protein kinase 1), EPB41L3 (erythrocyte membrane protein band 4.1-like 3), JAM3 (junctional adhesion molecule 3), MAL (mal, T-cell differentiation protein), MEG3 (maternally expressed 3), PAX1 (paired box 1), SIM1; c. Urine: FKBP6 (FK506 binding protein 6), INTS1 (integrator complex subunit 1), ZNF516. Figure created with BioRender.com.
According to the latest data from the American Cancer Society (SEER) in 2023, ovarian cancer is the most lethal gynecological malignancy, with a 5-year survival rate of 50.8%. High-grade serous ovarian cancer (HGSOC) accounts for 70% of ovarian cancer cases. Currently, the commonly used screening methods are clinical trials that combine the tumor markers CA-125 and transvaginal ultrasound. However, most patients do not exhibit specific symptoms in the early stages of OC and CA125 has limited sensitivity and specificity for early cancer detection (Gupta et al., 2019). The development of proteomics and genomics has led to the discovery of novel non-invasive methods for cancer detection and screening, such as tumor-associated autoantibodies and serum/plasma protein markers (Fortner et al., 2017). Unfortunately, most of these markers are only useful for advanced disease, so recent attention has focused on DNA methylation biomarkers for early screening of OC. DNA methylation is an early event in tumorigenesis and can be detected in ctDNA, making it a promising biomarker for early detection of OC (Liu et al., 2020). Data suggests that serum/plasma cfDNA methylation testing has a strong diagnostic accuracy for OC with a median accuracy of 85% (range 40–91%) (Guo et al., 2021). Several studies have identified candidate methylation biomarkers in plasma, such as AGRN, BCAT1, CAPN2, CDO1, CELF2, FAIM2, GPRIN1, GYPC, RIPPLY3, SRC, and SIM2, using sulfite sequencing and methylation-specific PCR. These biomarkers have shown high sensitivity and specificity for OC (Marinelli et al., 2022). Approximately 10%–20% of HGSOCs have BRCA1 promoter methylation, which is an important risk factor (Lønning et al., 2022). MGMT methylation is also an important risk factor for HGSOCs (Al-Moghrabi et al., 2024). Dvorská et al. (2019) analyzed the regulation of CDH1, PAX1, PTEN and RASSF1 tumor suppressor genes in benign and malignant ovarian tissues, as well as corresponding plasma samples, and demonstrated that the CDH1 gene is a potential non-invasive diagnostic marker for OC. Methylation of the CDH1 and VIM genes in epithelial ovarian cancer (EOC) can be detected using MS-HRM and is a reliable and sensitive method for detecting promoter methylation and early detection of EOC (Thakur et al., 2019). The MethyLight assay quantitatively analyzed the promoter methylation of HIC1 and HOXA9 in cfDNA from tissue samples, cancerous samples, and non-cancerous samples. This novel marker combination showed a sensitivity of 88.9% in serum cfDNA, making it suitable as a serum biomarker for early screening of OC (Singh et al., 2020). Studies have shown that SOX1 can be used for indirect detection of serum cfDNA, along with HIC1 and HOXA9 multiplex detection (Singh et al., 2021), specifically in the northern Indian population. Research on the early diagnosis of OC is still ongoing, and recent years have seen a summary of OC methylation biomarkers, as shown in Table 4; Figure 5.
Cervical cancer is ranked as the fourth most common cancer and the fourth leading cause of cancer death in women, with an estimated 6,04,000 new cases and 3,42,000 deaths worldwide in 2020 (Sung et al., 2021). Currently, routine screening for CC includes cytological tests, Pap tests, and HPV-DNA methods (Huh et al., 2015). Human papillomavirus (HPV) is a necessary but insufficient cause of CC, and preventive measures such as the HPV vaccine and screening can be taken. During the development of CC, various genetic and epigenetic changes occur in both the HPV and host genomes. After HPV gene methylation, HPV L1, HPVE6, and HPVE7 are present. E7 inactivates the retinoblastoma protein, while E6 inactivates p53. E6 can induce DNMT1 expression by inhibiting p53, and E7 can directly activate DNMT1 (Herbst et al., 2022). Methylation of gene promoters such as CADM1, CCNA1, CDH1, DAPK1, FHIT, MAL, P16, PAX1, RAR-β, and RASSF1 can be considered as non-invasive biomarkers for early cervical lesions (El et al., 2021). The combination of plasma CADM1 methylation and D-dimer testing is a biomarker that can be used for early screening of CC (Rong et al., 2019). Methylation of EPB41L3 and JAM3 is an accurate and feasible screening method for more severe lesions (CIN2) in CC tissues (Kong et al., 2020). A combination of promoter methylation tests for FKBP6, INTS1, and ZNF516 can be detected early in blood cfDNA and urine (Guerrero-Preston et al., 2016). Abnormal methylation of MEG3 (Zhang et al., 2017) and SIM1 (Kim et al., 2018) may serve as potential plasma biomarkers for CC. In summary, eight methylation biomarkers for CC screening and early detection are described, while the others are listed in Table 4; Figure 5.
Bladder cancer is the tenth most common cancer worldwide, with higher incidence in men than in women. It ranks as the sixth most common cancer and the ninth leading cause of cancer death in men (Sung et al., 2021). The most common malignancy of the urinary tract is BCa, and early detection is crucial for bladder preservation and improved survival. Cancer is caused by interactions between the environment and genetic and epigenetic factors, with DNA methylation being a common representation of epigenetics. Fluorescence in situ hybridization (FISH) is widely used for routine clinical detection of BCa, with a high sensitivity of 60%–80%. The utMeMA method, developed by Chen et al. (2020), is a rapid, high-throughput, noninvasive and promising method for early diagnosis of BCa. It can be used for detection of minimal residual tumor and assessment of DNA methylation in urine samples. Some studies have developed combined methylation assays for HOXA9, PCDH17, POU4F2, and ONECUT2, which are effective markers for early detection of BCa in urine samples from patients with hematuria (Wu et al., 2020). Another highly sensitive and specific noninvasive method for detecting primary BCa is the detection of methylated NID2 and TWIST1 genes in urine sediment using MSP (Renard et al., 2010). SOCS1 (suppressor of cytokine signaling 1) protein is a negative regulator of the Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathway and has tumor suppressor activity. In certain cancers, the methylation of CpG islands in the promoter region of the SOCS1 gene inactivates the gene. The aberrant methylation of the CpG island in the promoter region of the SOCS1 gene can be used as a biomarker for early prediction and diagnosis of BCa (Guan et al., 2020). Overall, there is limited research on methylation biomarkers for early screening and detection of BCa, as shown in Table 5; Figure 6.
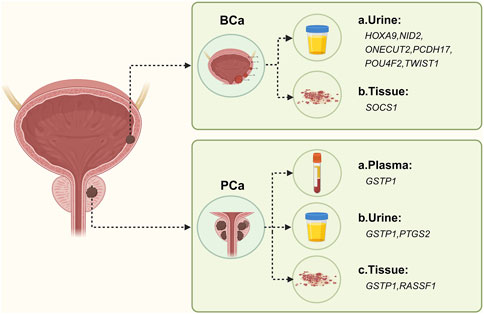
Figure 6. Methylation-based biomarker candidates in different samples for urologic cancer diagnosis. Abbreviations: BCa: a. Urine: HOXA9 (homeobox A9), NID2 (nidogen 2), ONECUT2 (one cut homeobox 2), PCDH17 (protocadherin 17), POU4F2 (POU class 4 homeobox 2), TWIST1 (twist family bHLH transcription factor 1); b. Tissue: SOCS1 (suppressor of cytokine signaling 1). PCa: a. Plasma: GSTP1 (glutathione S-transferase pi 1); b. Urine: GSTP1, PTGS2 (prostaglandin-endoperoxide synthase 2); c. Tissue: GSTP1, RASSF1 (Ras association domain family member 1). Figure created with BioRender.com.
According to statistics from the World Health Organization in 2020, prostate cancer is the fourth most common cancer worldwide and the second most common cancer and the fifth leading cause of cancer death in men in 2020 (Sung et al., 2021). The identified risk factors are limited to increasing age, family history of malignancy, certain genetic mutations (e.g., BRCA1 and BRCA2) and disease (Lynch syndrome). Epigenetic mechanisms play an important role in the initiation and development of PCa, with gene-specific hypermethylation and hypomethylation being associated with PCa (Willard and Koochekpour, 2012). Currently, the standard diagnostic tools for PCa are prostate-specific antigen (PSA) serum tests and confirmation through histopathological examination. However, various biomarkers investigating epigenetic genetic alterations and their variants may be more predictive than PSA. The etiology of most cases of PCa is not fully understood, so there is a need to identify the molecular basis and early diagnostic markers for PCa. GSTP1 (Glutathione S-transferase pi 1) expression is low in normal prostate luminal cells. However, in prostate intraepithelial neoplasia (PIN) and prostate adenocarcinoma (CaP), GSTP1 is silenced due to cytosine-guanine phosphate (CpG) island promoter hypermethylation. Several studies have been conducted on PCa tissue samples, and they consistently demonstrate that GSTP1 methylation is an early event in the carcinogenic process (Zelic et al., 2016). The most commonly used method for analyzing GSTP1 methylation status in PCa research is methylation-specific polymerase chain reaction (MS-PCR). Other methods include bisulfite genome sequencing, pyrosequencing, methylation-specific dot blot assay, among others (Gurioli et al., 2018). GSTP1 exhibits high specificity but low sensitivity as a biomarker for PCa. Combining GSTP1 with traditional prostate-specific antigen (PSA) and other methylated genes can enhance early detection of PCa (Ye et al., 2023). In fact, a combination of three genes, GSTP1, GAS6, and HAPLN3, has been found to accurately classify and screen early PCa (Patel et al., 2019). Some meta-analyses suggest that P16 methylation may serve as a potential biomarker for PCa (Feng et al., 2015). Although there have been few studies on methylation markers in PCa, GSTP1 methylation remains the most extensively studied epigenetic marker in PCa. Other markers are listed in Table 5; Figure 6.
In 2020, the global incidence of thyroid cancer was 586,000, ranking ninth in terms of incidence, with a rate of 10.1/100,000 in women worldwide, which is three times higher than that in men (Sung et al., 2021). Thyroid cancer remains the most common endocrine malignancy, and its incidence has been gradually increasing in recent years. Sheikholeslami et al. (2021) noted that the methylation status of DAPK1, P16, RASSF1, RUNX3, SLC5A8, TIMP3, and TSHR promoters is a common and likely early event in lymph node metastasis and additional thyroid invasion. This information can be used for early diagnosis and differentiation of malignant and benign thyroid diseases.
Glioblastoma (GBM) accounts for the majority of gliomas (56.6%), making it the most common malignant primary brain tumor in adults with the worst prognosis (Ostrom et al., 2018). MGMT is potentially important for guiding the use of the chemotherapeutic agent temozolomide (TMZ) in GBM, and MGMT promoter methylation is significantly associated with improved overall survival in GBM patients (Sareen et al., 2022). TP53 mutations can be detected in the early stages of 65% of secondary GBM multiforme (Rumpold et al., 2020), which provides a potential biomarker for early screening for extraneural metastases.
In conclusion, cancer epigenetics is prevalent in tumors of the digestive, respiratory, female reproductive, and urinary systems, and significant advancements have been made in this field. Epigenetic markers offer promising prospects for cancer screening and treatment. Tissue and free DNA-based epigenomics methods have been proposed for early detection, diagnosis, and treatment monitoring of various cancers, including colorectal cancer, lung cancer, breast cancer, bladder cancer, prostate cancer, and glioblastoma (Leal et al., 2020).
However, from a biological standpoint, despite substantial progress in gene methylation over the past decade, many unresolved questions persist. There is a need to develop optimized combinations of methylation markers for early tumor screening, and promising candidate methylation markers should be validated in prospective study cohorts and tested in large screening populations using high-quality studies.
Emerging liquid biopsy methods (free circulating nucleic acids, CTCs, and exosomes) have been utilized in recent years to discover, detect, and validate biomarkers. Data from studies conducted over the last 5 years demonstrate the feasibility of using epigenetic-based markers in body fluids and their potential for clinical translation. Therefore, there is an urgent need for large-scale, methodologically rigorous epidemiological studies to elucidate the potential role of methylation alterations in the development of various cancers and their impact on early detection in clinical samples such as blood, urine, sputum, feces, and tissues. With the rapid development of cfDNA analysis technology, liquid biopsy has a broader clinical application and prospect in oncology.
XX: Conceptualization, Data curation, Formal Analysis, Writing–original draft, Writing–review and editing. YG: Investigation, Supervision, Writing–original draft, Writing–review and editing. ML: Investigation, Supervision, Writing–original draft, Writing–review and editing. YH: Conceptualization, Methodology, Writing–original draft, Writing–review and editing. SL: Supervision, Writing–original draft, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
We thank BioRender (www.biorender.com) providing mapping assistance during the preparation of this manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abou-Zeid, A., Hashad, D., Baess, A., Mosaad, M., and Tayae, E. (2023). HOXA9 gene promotor methylation and copy number variation of SOX2 and HV2 genes in cell free DNA: a potential diagnostic panel for non-small cell lung cancer. BMC Cancer 23 (1), 329. doi:10.1186/s12885-023-10793-7
Abou-Zeid, A. A., Azzam, A. Z., and Kamel, N. A. (2011). Methylation status of the gene promoter of cyclin-dependent kinase inhibitor 2A (CDKN2A) in ovarian cancer. Scand. J. Clin. Laboratory Investigation 71 (7), 542–547. doi:10.3109/00365513.2011.590224
Ahmed, H. (2010). Promoter methylation in prostate cancer and its application for the early detection of prostate cancer using serum and urine samples. Biomarkers Cancer 2, BIC.S3187–33. doi:10.4137/bic.s3187
Alizadeh-Sedigh, M., Fazeli, M. S., Mahmoodzadeh, H., Sharif, S. B., and Teimoori-Toolabi, L. (2022). Methylation of FBN1, SPG20, ITF2, RUNX3, SNCA, MLH1, and SEPT9 genes in circulating cell-free DNA as biomarkers of colorectal cancer. Cancer Biomarkers 34 (2), 221–250. doi:10.3233/cbm-210315
Al-Moghrabi, N., Al-Showimi, M., Alqahtani, A., Almalik, O., Alhusaini, H., Almalki, G., et al. (2024). Constitutional BRCA1 and MGMT methylation are significant risk factors for triple-negative breast cancer and high-grade serous ovarian cancer in Saudi women. Int. J. Mol. Sci. 25 (6), 3108. doi:10.3390/ijms25063108
Amigo, J. D., Opazo, J. C., Jorquera, R., Wichmann, I. A., Garcia-Bloj, B. A., Alarcon, M. A., et al. (2018). The reprimo gene family: a novel gene lineage in gastric cancer with tumor suppressive properties. Int. J. Mol. Sci. 19 (7), 1862. doi:10.3390/ijms19071862
Baba, Y., Murata, A., Watanabe, M., and Baba, H. (2014). Clinical implications of the LINE-1 methylation levels in patients with gastrointestinal cancer. Surg. Today 44 (10), 1807–1816. doi:10.1007/s00595-013-0763-6
Bach, S., Paulis, I., Sluiter, N. R., Tibbesma, M., Martin, I., van de Wiel, M. A., et al. (2021). Detection of colorectal cancer in urine using DNA methylation analysis. Sci. Rep. 11 (1), 2363. doi:10.1038/s41598-021-81900-6
Bach, S., Wever, B. M. M., van de Wiel, M. A., Veltman, J. D., Hashemi, S. M. S., Kazemier, G., et al. (2022). Dynamics of methylated cell-free DNA in the urine of non-small cell lung cancer patients. Epigenetics 17 (10), 1057–1069. doi:10.1080/15592294.2021.1982511
Bacolod, M. D., Mirza, A. H., Huang, J., Giardina, S. F., Feinberg, P. B., Soper, S. A., et al. (2020). Application of multiplex bisulfite PCR-ligase detection reaction-real-time quantitative PCR assay in interrogating bioinformatically identified, blood-based methylation markers for colorectal cancer. J. Mol. Diagnostics 22 (7), 885–900. doi:10.1016/j.jmoldx.2020.03.009
Barták, B. K., Kalmár, A., Galamb, O., Wichmann, B., Nagy, Z. B., Tulassay, Z., et al. (2019). Blood collection and cell-free DNA isolation methods influence the sensitivity of liquid biopsy analysis for colorectal cancer detection. Pathol. Oncol. Res. 25 (3), 915–923. doi:10.1007/s12253-018-0382-z
Baylin, S. B., and Jones, P. A. (2011). A decade of exploring the cancer epigenome - biological and translational implications. Nat. Rev. Cancer 11 (10), 726–734. doi:10.1038/nrc3130
Beňačka, R., Szabóová, D., Guľašová, Z., Hertelyová, Z., and Radoňák, J. (2022). Classic and new markers in diagnostics and classification of breast cancer. Cancers (Basel) 14 (21), 5444. doi:10.3390/cancers14215444
Bestor, T. H. (1988). Cloning of a mammalian DNA methyltransferase. Gene 74 (1), 9–12. doi:10.1016/0378-1119(88)90238-7
Bilichak, A., and Kovalchuk, I. (2017). The combined bisulfite restriction analysis (COBRA) assay for the analysis of locus-specific changes in methylation patterns. Methods Mol. Biol. Clift. N.J. 1456, 63–71. doi:10.1007/978-1-4899-7708-3_5
Bonora, G., Rubbi, L., Morselli, M., Ma, F., Chronis, C., Plath, K., et al. (2019). DNA methylation estimation using methylation-sensitive restriction enzyme bisulfite sequencing (MREBS). PLoS One 14 (4), e0214368. doi:10.1371/journal.pone.0214368
Borg, M., Wen, S. W. C., Andersen, R. F., Timm, S., Hansen, T. F., and Hilberg, O. (2023). Methylated circulating tumor DNA in blood as a tool for diagnosing lung cancer: a systematic review and meta-analysis. Cancers (Basel) 15 (15), 3959. doi:10.3390/cancers15153959
Bose, K., Scurt, F. G., Thon, C., Franke, S., Schulz, C., Malfertheiner, P., et al. (2023). Factors affecting performance of DNA methylation as a potential biomarker in ascites for peritonitis and peritoneal carcinomatosis. J. Gastrointest. Liver Dis. 32 (2), 206–215. doi:10.15403/jgld-4710
Butler, M., Pongor, L., Su, Y. T., Xi, L., Raffeld, M., Quezado, M., et al. (2020). MGMT status as a clinical biomarker in glioblastoma. Trends Cancer 6 (5), 380–391. doi:10.1016/j.trecan.2020.02.010
Cao, W., Chen, H. D., Yu, Y. W., Li, N., and Chen, W. Q. (2021). Changing profiles of cancer burden worldwide and in China: a secondary analysis of the global cancer statistics 2020. Chin. Med. J. 134 (7), 783–791. doi:10.1097/cm9.0000000000001474
Cedar, H., Solage, A., Glaser, G., and Razin, A. (1979). Direct detection of methylated cytosine in DNA by use of the restriction enzyme Mspl. Nucleic Acids Res. 6 (6), 2125–2132. doi:10.1093/nar/6.6.2125
Chen, B., Ying, X., and Bao, L. (2021). MGMT gene promoter methylation in humoral tissue as biomarker for lung cancer diagnosis: an update meta-analysis. Thorac. Cancer 12 (23), 3194–3200. doi:10.1111/1759-7714.14186
Chen, J., Sun, H., Tang, W., Zhou, L., Xie, X., Qu, Z., et al. (2019). DNA methylation biomarkers in stool for early screening of colorectal cancer. J. Cancer 10 (21), 5264–5271. doi:10.7150/jca.34944
Chen, J., Wang, F., Xu, J., He, Z., Lu, Y., and Wang, Z. (2016). The role of PAQR3 gene promoter hypermethylation in breast cancer and prognosis. Oncol. Rep. 36 (3), 1612–1618. doi:10.3892/or.2016.4951
Chen, X., Zhang, J., Ruan, W., Huang, M., Wang, C., Wang, H., et al. (2020). Urine DNA methylation assay enables early detection and recurrence monitoring for bladder cancer. J. Clin. Investigation 130 (12), 6278–6289. doi:10.1172/jci139597
Chettouh, H., Mowforth, O., Galeano-Dalmau, N., Bezawada, N., Ross-Innes, C., MacRae, S., et al. (2018). Methylation panel is a diagnostic biomarker for Barrett's oesophagus in endoscopic biopsies and non-endoscopic cytology specimens. Gut 67 (11), 1942–1949. doi:10.1136/gutjnl-2017-314026
Choy, L. Y. L., Peng, W., Jiang, P., Cheng, S. H., Yu, S. C. Y., Shang, H., et al. (2022). Single-molecule sequencing enables long cell-free DNA detection and direct methylation analysis for cancer patients. Clin. Chem. 68 (9), 1151–1163. doi:10.1093/clinchem/hvac086
Coy, S. R., Gann, E. R., Papoulis, S. E., Holder, M. E., Ajami, N. J., Petrosino, J. F., et al. (2020). SMRT sequencing of paramecium bursaria chlorella virus-1 reveals diverse methylation stability in adenines targeted by restriction modification systems. Front. Microbiol. 11, 887. doi:10.3389/fmicb.2020.00887
Della Monica, R., Cuomo, M., Buonaiuto, M., Costabile, D., Franca, R. A., Del Basso De Caro, M., et al. (2022). MGMT and whole-genome DNA methylation impacts on diagnosis, prognosis and therapy of glioblastoma multiforme. Int. J. Mol. Sci. 23 (13), 7148. doi:10.3390/ijms23137148
Dong, N., Wang, W., Qin, Y., Wang, Y., and Shan, H. (2022). Sensitive lateral flow assay for bisulfite-free DNA methylation detection based on the restriction endonuclease GlaI and rolling circle amplification. Anal. Chim. Acta 1227, 340307. doi:10.1016/j.aca.2022.340307
Dubois, F., Keller, M., Calvayrac, O., Soncin, F., Hoa, L., Hergovich, A., et al. (2016). RASSF1A suppresses the invasion and metastatic potential of human non-small cell lung cancer cells by inhibiting YAP activation through the GEF-H1/RhoB pathway. Cancer Res. 76 (6), 1627–1640. doi:10.1158/0008-5472.can-15-1008
Dugast-Darzacq, C., and Grange, T. (2009). MethylQuant: a real-time PCR-based method to quantify DNA methylation at single specific cytosines. Methods Mol. Biol. 507, 281–303. doi:10.1007/978-1-59745-522-0_21
Dvorská, D., Braný, D., Nagy, B., Grendár, M., Poka, R., Soltész, B., et al. (2019). Aberrant methylation status of tumour suppressor genes in ovarian cancer tissue and paired plasma samples. Int. J. Mol. Sci. 20 (17), 4119. doi:10.3390/ijms20174119
Eads, C. A., Danenberg, K. D., Kawakami, K., Saltz, L. B., Blake, C., Shibata, D., et al. (2000). MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 28 (8), 322–E40. doi:10.1093/nar/28.8.e32
Eissa, M. A. L., Lerner, L., Abdelfatah, E., Shankar, N., Canner, J. K., Hasan, N. M., et al. (2019). Promoter methylation of ADAMTS1 and BNC1 as potential biomarkers for early detection of pancreatic cancer in blood. Clin. Epigenetics 11 (1), 59. doi:10.1186/s13148-019-0650-0
El, A. A., El-Abid, H., El Mallali, Y., Attaleb, M., Ennaji, M. M., and El Mzibri, M. (2021). Association between gene promoter methylation and cervical cancer development: global distribution and A meta-analysis. Cancer Epidemiol. Biomarkers Prev. 30 (3), 450–459. doi:10.1158/1055-9965.epi-20-0833
Estival, A., Sanz, C., Ramirez, J. L., Velarde, J. M., Domenech, M., Carrato, C., et al. (2019). Pyrosequencing versus methylation-specific PCR for assessment of MGMT methylation in tumor and blood samples of glioblastoma patients. Sci. Rep. 9 (1), 11125. doi:10.1038/s41598-019-47642-2
Fang, C., Wang, S. Y., Liou, Y. L., Chen, M. H., Ouyang, W., and Duan, K. M. (2019). The promising role of PAX1 (aliases: HUP48, OFC2) gene methylation in cancer screening. Mol. Genet. Genomic Med. 7 (3), e506. doi:10.1002/mgg3.506
Farrell, C., Thompson, M., Tosevska, A., Oyetunde, A., and Pellegrini, M. (2021). BiSulfite Bolt: a bisulfite sequencing analysis platform. Gigascience 10 (5), giab033. doi:10.1093/gigascience/giab033
Feng, H., Shao, W., Du, L., Qing, X., Zhang, Z., Liang, C., et al. (2020). Detection of SHOX2 DNA methylation by methylation-specific PCR in non-small cell lung cancer. Transl. Cancer Res. 9 (10), 6070–6077. doi:10.21037/tcr-20-887
Feng, W., Han, Z., Zhu, R., Liu, P., and Liu, S. (2015). Association of p16 gene methylation with prostate cancer risk: a meta-analysis. J. buon 20 (4), 1074–1080.
Forouhari, A., Heidari-Beni, M., Veisi, S., Poursafa, P., and Kelishadi, R. (2023). Effect of epigenetics on vitamin D levels: a systematic review until December 2020. Arch. Public Health 81 (1), 106. doi:10.1186/s13690-023-01122-2
Fortner, R. T., Damms-Machado, A., and Kaaks, R. (2017). Systematic review: tumor-associated antigen autoantibodies and ovarian cancer early detection. Gynecol. Oncol. 147 (2), 465–480. doi:10.1016/j.ygyno.2017.07.138
Fu, L., Shi, Z., and Chen, B. (2022). Deleted in lymphocytic leukemia 2 induces retinoic acid receptor beta promoter methylation and mitogen activated kinase-like protein activation to enhance viability and mobility of colorectal cancer cells. Bioengineered 13 (5), 12847–12862. doi:10.1080/21655979.2022.2076482
Gaillard, D. H. K., Lof, P., Sistermans, E. A., Mokveld, T., Horlings, H. M., Mom, C. H., et al. (2024). Evaluating the effectiveness of pre-operative diagnosis of ovarian cancer using minimally invasive liquid biopsies by combining serum human epididymis protein 4 and cell-free DNA in patients with an ovarian mass. Int. J. Gynecol. Cancer 34 (5), 713–721. doi:10.1136/ijgc-2023-005073
Gardiner-Garden, M., and Frommer, M. (1987). CpG islands in vertebrate genomes. J. Mol. Biol. 196 (2), 261–282. doi:10.1016/0022-2836(87)90689-9
Ge, Y. Z., Xu, L. W., Jia, R. P., Xu, Z., Feng, Y. M., Wu, R., et al. (2014). The association between RASSF1A promoter methylation and prostate cancer: evidence from 19 published studies. Tumor Biol. 35 (4), 3881–3890. doi:10.1007/s13277-013-1515-3
Goedecke, S., Mühlisch, J., Hempel, G., Frühwald, M. C., and Wünsch, B. (2015). Quantitative analysis of DNA methylation in the promoter region of the methylguanine-O(6) -DNA-methyltransferase gene by COBRA and subsequent native capillary gel electrophoresis. Electrophoresis 36 (23), 2939–2950. doi:10.1002/elps.201500242
Gonzalgo, M. L., and Jones, P. A. (1997). Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE). Nucleic Acids Res. 25 (12), 2529–2531. doi:10.1093/nar/25.12.2529
Gonzalgo, M. L., and Liang, G. (2007). Methylation-sensitive single-nucleotide primer extension (Ms-SNuPE) for quantitative measurement of DNA methylation. Nat. Protoc. 2 (8), 1931–1936. doi:10.1038/nprot.2007.271
Guan, H., Li, X., Lu, J., Meng, L., Guo, Y., Dong, J., et al. (2020). Aberrant methylation of suppressor of cytokine signaling 1 (SOCS1) gene as a biomarker for early prediction and diagnosis of bladder cancer. Clin. Lab. 66 (5). doi:10.7754/clin.lab.2019.190926
Guerrero-Preston, R., Valle, B. L., Jedlicka, A., Turaga, N., Folawiyo, O., Pirini, F., et al. (2016). Molecular triage of premalignant lesions in liquid-based cervical cytology and circulating cell-free DNA from urine, using a panel of methylated human papilloma virus and host genes. Cancer Prev. Res. 9 (12), 915–924. doi:10.1158/1940-6207.capr-16-0138
Guo, S., Yan, F., Xu, J., Bao, Y., Zhu, J., Wang, X., et al. (2015). Identification and validation of the methylation biomarkers of non-small cell lung cancer (NSCLC). Clin. Epigenetics 7 (1), 3. doi:10.1186/s13148-014-0035-3
Guo, X. M., Miller, H., Matsuo, K., Roman, L. D., and Salhia, B. (2021). Circulating cell-free DNA methylation profiles in the early detection of ovarian cancer: a scoping review of the literature. Cancers (Basel) 13 (4), 838. doi:10.3390/cancers13040838
Gupta, K. K., Gupta, V. K., and Naumann, R. W. (2019). Ovarian cancer: screening and future directions. Int. J. Gynecol. Cancer 29 (1), 195–200. doi:10.1136/ijgc-2018-000016
Gurioli, G., Martignano, F., Salvi, S., Costantini, M., Gunelli, R., and Casadio, V. (2018). GSTP1 methylation in cancer: a liquid biopsy biomarker? Clin. Chem. Laboratory Med. (CCLM) 56 (5), 702–717. doi:10.1515/cclm-2017-0703
Halldorsson, S., Nagymihaly, R. M., Patel, A., Brandal, P., Panagopoulos, I., Leske, H., et al. (2024). Accurate and comprehensive evaluation of O6-methylguanine-DNA methyltransferase promoter methylation by nanopore sequencing. Neuropathology Appl. Neurobiol. 50 (3), e12984. doi:10.1111/nan.12984
Han, J. C., Xu, F., Chen, N., Qi, G. B., Wei, Y. J., Li, H. B., et al. (2016). Promoter methylations of RASSF1A and p16 is associated with clinicopathological features in lung cancers. J. Cancer Res. Ther. 12 (1), 340–349. doi:10.4103/0973-1482.154926
Han, Y. D., Oh, T. J., Chung, T. H., Jang, H. W., Kim, Y. N., An, S., et al. (2019). Early detection of colorectal cancer based on presence of methylated syndecan-2 (SDC2) in stool DNA. Clin. Epigenetics 11 (1), 51. doi:10.1186/s13148-019-0642-0
Harrington, C. T., Lin, E. I., Olson, M. T., and Eshleman, J. R. (2013). Fundamentals of pyrosequencing. Archives Pathology Laboratory Med. 137 (9), 1296–1303. doi:10.5858/arpa.2012-0463-ra
He, Y., and Ecker, J. R. (2015). Non-CG methylation in the human genome. Annu. Rev. Genomics Hum. Genet. 16, 55–77. doi:10.1146/annurev-genom-090413-025437
Herbst, J., Pantel, K., Effenberger, K., and Wikman, H. (2022). Clinical applications and utility of cell-free DNA-based liquid biopsy analyses in cervical cancer and its precursor lesions. Br. J. Cancer 127 (8), 1403–1410. doi:10.1038/s41416-022-01868-6
Heyn, H., and Esteller, M. (2012). DNA methylation profiling in the clinic: applications and challenges. Nat. Rev. Genet. 13 (10), 679–692. doi:10.1038/nrg3270
Hindson, B. J., Ness, K. D., Masquelier, D. A., Belgrader, P., Heredia, N. J., Makarewicz, A. J., et al. (2011). High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 83 (22), 8604–8610. doi:10.1021/ac202028g
Hu, Y. H., Ma, S., Zhang, X. N., Zhang, Z. Y., Zhu, H. F., Ji, Y. H., et al. (2019). Hypermethylation of ADHFE1 promotes the proliferation of colorectal cancer cell via modulating cell cycle progression. OncoTargets Ther. 12, 8105–8115. doi:10.2147/ott.s223423
Hua, R., Yu, J., Yan, X., Ni, Q., Zhi, X., Li, X., et al. (2020). Syndecan-2 in colorectal cancer plays oncogenic role via epithelial-mesenchymal transition and MAPK pathway. Biomed. Pharmacother. 121, 109630. doi:10.1016/j.biopha.2019.109630
Huang, D., Xiao, F., Hao, H., Hua, F., Luo, Z., Huang, Z., et al. (2020). JARID1B promotes colorectal cancer proliferation and Wnt/β-catenin signaling via decreasing CDX2 level. Cell Commun. Signal 18 (1), 169. doi:10.1186/s12964-020-00660-4
Hubers, A. J., Heideman, D. A., Duin, S., Witte, B. I., de Koning, H. J., Groen, H. J., et al. (2017). DNA hypermethylation analysis in sputum of asymptomatic subjects at risk for lung cancer participating in the NELSON trial: argument for maximum screening interval of 2 years. J. Clin. Pathol. 70 (3), 250–254. doi:10.1136/jclinpath-2016-203734
Huh, W. K., Ault, K. A., Chelmow, D., Davey, D. D., Goulart, R. A., Garcia, F. A., et al. (2015). Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Gynecol. Oncol. 136 (2), 178–182. doi:10.1016/j.ygyno.2014.12.022
Hwang, S. H., Kim, K. U., Kim, J. E., Kim, H. H., Lee, M. K., Lee, C. H., et al. (2011). Detection of HOXA9 gene methylation in tumor tissues and induced sputum samples from primary lung cancer patients. cclm 49 (4), 699–704. doi:10.1515/cclm.2011.108
Jamialahmadi, K., Azghandi, M., Javadmanesh, A., Zardadi, M., Shams Davodly, E., and Kerachian, M. A. (2021). A DNA methylation panel for high performance detection of colorectal cancer. Cancer Genet. 252-253, 64–72. doi:10.1016/j.cancergen.2020.12.007
Jia, W., Yu, T., Cao, X., An, Q., and Yang, H. (2016). Clinical effect of DAPK promoter methylation in gastric cancer: a systematic meta-analysis. Med. Baltim. 95 (43), e5040. doi:10.1097/md.0000000000005040
Jones, P. A., and Baylin, S. B. (2007). The epigenomics of cancer. Cell 128 (4), 683–692. doi:10.1016/j.cell.2007.01.029
Khakpour, G., Pooladi, A., Izadi, P., Noruzinia, M., and Tavakkoly Bazzaz, J. (2015). DNA methylation as a promising landscape: a simple blood test for breast cancer prediction. Tumor Biol. 36 (7), 4905–4912. doi:10.1007/s13277-015-3567-z
Kim, H. J., Kim, C. Y., Jin, J., Bae, M. K., Kim, Y. H., Ju, W., et al. (2018). Aberrant single-minded homolog 1 methylation as a potential biomarker for cervical cancer. Diagn. Cytopathol. 46 (1), 15–21. doi:10.1002/dc.23838
Kisiel, J. B., Klepp, P., Allawi, H. T., Taylor, W. R., Giakoumopoulos, M., Sander, T., et al. (2019). Analysis of DNA methylation at specific loci in stool samples detects colorectal cancer and high-grade dysplasia in patients with inflammatory bowel disease. Clin. Gastroenterology Hepatology 17 (5), 914–921.e5. doi:10.1016/j.cgh.2018.05.004
Kong, L., Wang, L., Wang, Z., Xiao, X., You, Y., Wu, H., et al. (2020). DNA methylation for cervical cancer screening: a training set in China. Clin. Epigenetics 12 (1), 91. doi:10.1186/s13148-020-00885-7
Kosumi, K., Baba, Y., Ishimoto, T., Harada, K., Miyake, K., Izumi, D., et al. (2015). Relationship between LINE-1 hypomethylation and Helicobacter pylori infection in gastric mucosae. Med. Oncol. 32 (4), 117. doi:10.1007/s12032-015-0571-5
Krishnamurthy, K., Mishra, T. K., Saxena, A., Daga, M. K., Khurana, N., Masroor, M., et al. (2019). Evaluating NISCH and CDH1 promoter hypermethylation in nonsmokers, cancer free smokers and lung cancer patients: a case control study. Indian J. Clin. Biochem. 34 (4), 458–464. doi:10.1007/s12291-018-0767-5
Laven-Law, G., Kichenadasse, G., Young, G. P., Symonds, E. L., and Winter, J. M. (2024). BCAT1, IKZF1 and SEPT9: methylated DNA biomarkers for detection of pan-gastrointestinal adenocarcinomas. Biomarkers 29 (4), 194–204. doi:10.1080/1354750x.2024.2340663
Leal, A., Sidransky, D., and Brait, M. (2020). Tissue and cell-free DNA-based epigenomic approaches for cancer detection. Clin. Chem. 66 (1), 105–116. doi:10.1373/clinchem.2019.303594
Lee, J. A., Seo, M. K., Yoo, S. Y., Cho, N. Y., Kwak, Y., Lee, K., et al. (2022). Comprehensive clinicopathologic, molecular, and immunologic characterization of colorectal carcinomas with loss of three intestinal markers, CDX2, SATB2, and KRT20. Virchows Arch. 480 (3), 543–555. doi:10.1007/s00428-021-03260-w
Leske, H., Camenisch Gross, U., Hofer, S., Neidert, M. C., Leske, S., Weller, M., et al. (2023). MGMT methylation pattern of long-term and short-term survivors of glioblastoma reveals CpGs of the enhancer region to be of high prognostic value. Acta Neuropathol. Commun. 11 (1), 139. doi:10.1186/s40478-023-01622-w
Li, B., Huang, H., Huang, R., Zhang, W., Zhou, G., Wu, Z., et al. (2020a). SEPT9 gene methylation as a noninvasive marker for hepatocellular carcinoma. Dis. Markers 2020, 1–9. doi:10.1155/2020/6289063
Li, B., Liu, S., Gao, Y., Zheng, L., and Lu, Y. (2023). Combined detection of SDC2/ADHFE1/PPP2R5C methylation in stool DNA for colorectal cancer screening. J. Cancer Res. Clin. Oncol. 149 (12), 10241–10253. doi:10.1007/s00432-023-04943-4
Li, H., Wang, Z., Zhao, G., Ma, Y., Chen, Y., Xue, Q., et al. (2019). Performance of a MethyLight assay for methylated SFRP2 DNA detection in colorectal cancer tissue and serum. Int. J. Biol. Markers 34 (1), 54–59. doi:10.1177/1724600818820536
Li, J., Han, X., Yu, X., Xu, Z., Yang, G., Liu, B., et al. (2018). Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J. Exp. Clin. Cancer Res. 37 (1), 213. doi:10.1186/s13046-018-0893-1
Li, N., Zeng, Y., and Huang, J. (2020b). Signaling pathways and clinical application of RASSF1A and SHOX2 in lung cancer. J. Cancer Res. Clin. Oncol. 146 (6), 1379–1393. doi:10.1007/s00432-020-03188-9
Li, P., Liu, S., Du, L., Mohseni, G., Zhang, Y., and Wang, C. (2022). Liquid biopsies based on DNA methylation as biomarkers for the detection and prognosis of lung cancer. Clin. Epigenetics 14 (1), 118. doi:10.1186/s13148-022-01337-0
Li, Y., Xu, J., Chen, C., Lu, Z., Wan, D., Li, D., et al. (2024). Multimodal epigenetic sequencing analysis (MESA) of cell-free DNA for non-invasive colorectal cancer detection. Genome Med. 16 (1), 9. doi:10.1186/s13073-023-01280-6
Liang, W., Zhao, Y., Huang, W., Gao, Y., Xu, W., Tao, J., et al. (2019). Non-invasive diagnosis of early-stage lung cancer using high-throughput targeted DNA methylation sequencing of circulating tumor DNA (ctDNA). Theranostics 9 (7), 2056–2070. doi:10.7150/thno.28119
Lima, A. B., Dos Reis, M. B., Matsushita, M., Dos Reis, M. T., de Oliveira, M. A., Reis, R. M., et al. (2023). Combined <i>SEPT9</i> and <i>BMP3</i> methylation in plasma for colorectal cancer early detection and screening in a Brazilian population. Cancer Med. 12 (15), 15854–15867. doi:10.1002/cam4.6224
Lin, J., Zhang, L., Chen, M., Chen, J., Wu, Y., Wang, T., et al. (2022). Evaluation of combined detection of multigene mutation and SDC2/SFRP2 methylation in stool specimens for colorectal cancer early diagnosis. Int. J. Colorectal Dis. 37 (6), 1231–1238. doi:10.1007/s00384-022-04170-2
Liu, B., Ricarte Filho, J., Mallisetty, A., Villani, C., Kottorou, A., Rodgers, K., et al. (2020b). Detection of promoter DNA methylation in urine and plasma aids the detection of non-small cell lung cancer. Clin. Cancer Res. 26 (16), 4339–4348. doi:10.1158/1078-0432.ccr-19-2896
Liu, D., Enriquez, L., and Ford, C. E. (2021a). ROR2 is epigenetically regulated in endometrial cancer. Cancers (Basel) 13 (3), 383. doi:10.3390/cancers13030383
Liu, D., Peng, H., Sun, Q., Zhao, Z., Yu, X., Ge, S., et al. (2017). The indirect efficacy comparison of DNA methylation in sputum for early screening and auxiliary detection of lung cancer: a meta-analysis. Int. J. Environ. Res. Public Health 14 (7), 679. doi:10.3390/ijerph14070679
Liu, F., Lu, X., Zhou, X., and Huang, H. (2021b). APC gene promoter methylation as a potential biomarker for lung cancer diagnosis: a meta-analysis. Thorac. Cancer 12 (21), 2907–2913. doi:10.1111/1759-7714.14151
Liu, F., Wang, Y., Cao, Y., Wu, Z., Ma, D., Cai, J., et al. (2023). Transcription factor B-MYB activates lncRNA CCAT1 and upregulates SOCS3 to promote chemoresistance in colorectal cancer. Chemico-Biological Interact. 374, 110412. doi:10.1016/j.cbi.2023.110412
Liu, M., Cui, L. H., Li, C. C., and Zhang, L. (2015). Association of APC, GSTP1 and SOCS1 promoter methylation with the risk of hepatocellular carcinoma: a meta-analysis. Eur. J. Cancer Prev. 24 (6), 470–483. doi:10.1097/cej.0000000000000121
Liu, M. C., Oxnard, G. R., Klein, E. A., Swanton, C., Seiden, M. V., Liu, M. C., et al. (2020c). Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 31 (6), 745–759. doi:10.1016/j.annonc.2020.02.011
Liu, Q., Wang, S., Pei, G., Yang, Y., and Huang, Y. (2021c). Diagnostic efficacy of SHOX2 gene hypermethylation for lung cancer: a meta-analysis. Zhongguo Fei Ai Za Zhi 24 (7), 490–496. doi:10.3779/j.issn.1009-3419.2021.101.27
Liu, R., Su, X., Long, Y., Zhou, D., Zhang, X., Ye, Z., et al. (2019). A systematic review and quantitative assessment of methylation biomarkers in fecal DNA and colorectal cancer and its precursor, colorectal adenoma. Mutat. Research/Reviews Mutat. Res. 779, 45–57. doi:10.1016/j.mrrev.2019.01.003
Liu, X., Wang, L., and Guo, Y. (2016). The association between runt-related transcription factor 3 gene promoter methylation and gastric cancer: a meta-analysis. J. Cancer Res. Ther. 12 (Suppl. ment), 50–53. doi:10.4103/0973-1482.191630
Liu, Y., Huang, H., Fu, J., Zhang, Y., Xu, J., Zhang, L., et al. (2020a). Colorectal cancer patients with CASK promotor heterogeneous and homogeneous methylation display different prognosis. Aging (Albany NY) 12 (20), 20561–20586. doi:10.18632/aging.103928
Lønning, P. E., Nikolaienko, O., Pan, K., Kurian, A. W., Eikesdal, H. P., Pettinger, M., et al. (2022). Constitutional BRCA1 methylation and risk of incident triple-negative breast cancer and high-grade serous ovarian cancer. JAMA Oncol. 8 (11), 1579–1587. doi:10.1001/jamaoncol.2022.3846
Ma, K., Cao, B., and Guo, M. (2016). The detective, prognostic, and predictive value of DNA methylation in human esophageal squamous cell carcinoma. Clin. Epigenetics 8, 43. doi:10.1186/s13148-016-0210-9
Ma, L., Gong, J., Zhao, M., Kong, X., Gao, P., Jiang, Y., et al. (2022b). A novel stool methylation test for the non-invasive screening of gastric and colorectal cancer. Front. Oncol. 12, 860701. doi:10.3389/fonc.2022.860701
Ma, L., Qin, G., Gai, F., Jiang, Y., Huang, Z., Yang, H., et al. (2022a). A novel method for early detection of colorectal cancer based on detection of methylation of two fragments of syndecan-2 (SDC2) in stool DNA. BMC Gastroenterol. 22 (1), 191. doi:10.1186/s12876-022-02264-3
Marinelli, L. M., Kisiel, J. B., Slettedahl, S. W., Mahoney, D. W., Lemens, M. A., Shridhar, V., et al. (2022). Methylated DNA markers for plasma detection of ovarian cancer: discovery, validation, and clinical feasibility. Gynecol. Oncol. 165 (3), 568–576. doi:10.1016/j.ygyno.2022.03.018
Martisova, A., Holcakova, J., Izadi, N., Sebuyoya, R., Hrstka, R., and Bartosik, M. (2021). DNA methylation in solid tumors: functions and methods of detection. Int. J. Mol. Sci. 22 (8), 4247. doi:10.3390/ijms22084247
Mersakova, S., Visnovsky, J., Holubekova, V., Nachajova, M., Kudela, E., Danko, J., et al. (2014). Detection of methylation of the promoter region of the MAL and CADM1 genes by pyrosequencing in cervical carcinoma. Neuro Endocrinol. Lett. 35 (7), 619–623.
Mikhaĭlenko, D. S., Perepechin, D. V., Apolikhin, O. I., Efremov, G. D., and Sivkov, A. V. (2014). Markers for non-invasive molecular genetic diagnosis of oncourological diseases. Urologiia (5), 116–120.
Modlin, I. M., Kidd, M., Drozdov, I. A., Boegemann, M., Bodei, L., Kunikowska, J., et al. (2024). Development of a multigenomic liquid biopsy (PROSTest) for prostate cancer in whole blood. Prostate 84 (9), 850–865. doi:10.1002/pros.24704
Moinova, H. R., LaFramboise, T., Lutterbaugh, J. D., Chandar, A. K., Dumot, J., Faulx, A., et al. (2018). Identifying DNA methylation biomarkers for non-endoscopic detection of Barrett's esophagus. Sci. Transl. Med. 10 (424), eaao5848. doi:10.1126/scitranslmed.aao5848
Molnár, K. B. (2020). Analysis of DNA methylation alterations in cellfree DNA fraction during colorectal cancer development. Magy. Onkol. 64 (1), 70–72.
Moradi, K., Babaei, E., Hosseinpour Feizi, M. A., Safaralizadeh, R., and Rezvani, N. (2021). Quantitative detection of SRY-Box 21 (SOX21) gene promoter methylation as a stool-based noninvasive biomarker for early diagnosis of colorectal cancer by MethyLight method. Indian J. Cancer 58 (2), 217–224. doi:10.4103/ijc.ijc_37_19
Morselli, M., Farrell, C., Rubbi, L., Fehling, H. L., Henkhaus, R., and Pellegrini, M. (2021). Targeted bisulfite sequencing for biomarker discovery. Methods 187, 13–27. doi:10.1016/j.ymeth.2020.07.006
Nassar, F. J., Chamandi, G., Tfaily, M. A., Zgheib, N. K., and Nasr, R. (2020). Peripheral blood-based biopsy for breast cancer risk prediction and early detection. Front. Med. (Lausanne) 7, 28. doi:10.3389/fmed.2020.00028
Nemtsova, M. V., Strelnikov, V. V., Tanas, A. S., Bykov, I. I., Zaletaev, D. V., Rudenko, V. V., et al. (2017). Implication of gastric cancer molecular genetic markers in surgical practice. Curr. Genomics 18 (5), 408–415. doi:10.2174/1389202918666170329110021
Nikolaidis, C., Nena, E., Panagopoulou, M., Balgkouranidou, I., Karaglani, M., Chatzaki, E., et al. (2015). PAX1 methylation as an auxiliary biomarker for cervical cancer screening: a meta-analysis. Cancer Epidemiol. 39 (5), 682–686. doi:10.1016/j.canep.2015.07.008
Nozaki, I., Tsuji, T., Iijima, O., Ohmura, Y., Andou, A., Miyazaki, M., et al. (2001). Reduced expression of REIC/Dkk-3 gene in non-small cell lung cancer. Int. J. Oncol. 19 (1), 117–121. doi:10.3892/ijo.19.1.117
Ooki, A., Maleki, Z., Tsay, J. J., Goparaju, C., Brait, M., Turaga, N., et al. (2017). A panel of novel detection and prognostic methylated DNA markers in primary non-small cell lung cancer and serum DNA. Clin. Cancer Res. 23 (22), 7141–7152. doi:10.1158/1078-0432.ccr-17-1222
Ostrom, Q. T., Gittleman, H., Truitt, G., Boscia, A., Kruchko, C., and Barnholtz-Sloan, J. S. (2018). CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro-Oncology 20 (Suppl. l_4), iv1–iv86. doi:10.1093/neuonc/noy131
Oussalah, A., Rischer, S., Bensenane, M., Conroy, G., Filhine-Tresarrieu, P., Debard, R., et al. (2018). Plasma mSEPT9: a novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine 30, 138–147. doi:10.1016/j.ebiom.2018.03.029
Palanca-Ballester, C., Rodriguez-Casanova, A., Torres, S., Calabuig-Fariñas, S., Exposito, F., Serrano, D., et al. (2021). Cancer epigenetic biomarkers in liquid biopsy for high incidence malignancies. Cancers (Basel) 13 (12), 3016. doi:10.3390/cancers13123016
Pan, L., Fan, Y., and Zhou, L. (2022). SMYD2 epigenetically activates MEX3A and suppresses CDX2 in colorectal cancer cells to augment cancer growth. Clin. Exp. Pharmacol. Physiology 49 (9), 959–969. doi:10.1111/1440-1681.13679
Park, B. C., Soh, J. O., Choi, H. J., Park, H. S., Lee, S. M., Fu, H. E., et al. (2024). Ultrasensitive and rapid circulating tumor DNA liquid biopsy using surface-confined gene amplification on dispersible magnetic nano-electrodes. ACS Nano 18 (20), 12781–12794. doi:10.1021/acsnano.3c12266
Paschidis, K., Zougros, A., Chatziandreou, I., Tsikalakis, S., Korkolopoulou, P., Kavantzas, N., et al. (2022). Methylation analysis of APC, AXIN2, DACT1, RASSF1A and MGMT gene promoters in non-small cell lung cancer. Pathology - Res. Pract. 234, 153899. doi:10.1016/j.prp.2022.153899
Pasha, H. F., Radwan, M. I., Yehia, A. M., and Toam, M. M. (2019). Circulating methylated RUNX3 and SFRP1 genes as a noninvasive panel for early detection of colorectal cancer. Eur. J. Gastroenterology Hepatology 31 (11), 1342–1349. doi:10.1097/meg.0000000000001532
Patel, P. G., Wessel, T., Kawashima, A., Okello, J. B. A., Jamaspishvili, T., Guérard, K. P., et al. (2019). A three-gene DNA methylation biomarker accurately classifies early stage prostate cancer. Prostate 79 (14), 1705–1714. doi:10.1002/pros.23895
Peng, D., Zhang, H., and Sun, G. (2014). The relationship between P16 gene promoter methylation and gastric cancer: a meta-analysis based on Chinese patients. J. Cancer Res. Ther. 10 (Suppl. l), 292–295. doi:10.4103/0973-1482.151535
Phallen, J., Sausen, M., Adleff, V., Leal, A., Hruban, C., White, J., et al. (2017). Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 9 (403), eaan2415. doi:10.1126/scitranslmed.aan2415
Renard, I., Joniau, S., van Cleynenbreugel, B., Collette, C., Naômé, C., Vlassenbroeck, I., et al. (2010). Identification and validation of the methylated TWIST1 and NID2 genes through real-time methylation-specific polymerase chain reaction assays for the noninvasive detection of primary bladder cancer in urine samples. Eur. Urol. 58 (1), 96–104. doi:10.1016/j.eururo.2009.07.041
Rhee, Y. Y., Lee, T. H., Song, Y. S., Wen, X., Kim, H., Jheon, S., et al. (2015). Prognostic significance of promoter CpG island hypermethylation and repetitive DNA hypomethylation in stage I lung adenocarcinoma. Virchows Arch. 466 (6), 675–683. doi:10.1007/s00428-015-1749-0
Rong, G., Zhang, M., Xia, W., Li, D., Miao, J., and Wang, H. (2019). Plasma CADM1 promoter hypermethylation and D-dimer as novel metastasis predictors of cervical cancer. J. Obstetrics Gynaecol. Res. 45 (7), 1251–1259. doi:10.1111/jog.13966
Rumpold, H., Niedersüß-Beke, D., Heiler, C., Falch, D., Wundsam, H. V., Metz-Gercek, S., et al. (2020). Prediction of mortality in metastatic colorectal cancer in a real-life population: a multicenter explorative analysis. BMC Cancer 20 (1), 1149. doi:10.1186/s12885-020-07656-w
Ruscito, I., Dimitrova, D., Vasconcelos, I., Gellhaus, K., Schwachula, T., Bellati, F., et al. (2014). BRCA1 gene promoter methylation status in high-grade serous ovarian cancer patients--a study of the tumour Bank ovarian cancer (TOC) and ovarian cancer diagnosis consortium (OVCAD). Eur. J. Cancer 50 (12), 2090–2098. doi:10.1016/j.ejca.2014.05.001
Ruscito, I., Gasparri, M. L., De Marco, M. P., Costanzi, F., Besharat, A. R., Papadia, A., et al. (2021). The clinical and pathological profile of BRCA1 gene methylated breast cancer women: a meta-analysis. Cancers (Basel) 13 (6), 1391. doi:10.3390/cancers13061391
Sareen, H., Ma, Y., Becker, T. M., Roberts, T. L., de Souza, P., and Powter, B. (2022). Molecular biomarkers in glioblastoma: a systematic review and meta-analysis. Int. J. Mol. Sci. 23 (16), 8835. doi:10.3390/ijms23168835
Schmidt, B., Liebenberg, V., Dietrich, D., Schlegel, T., Kneip, C., Seegebarth, A., et al. (2010). SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer 10, 600. doi:10.1186/1471-2407-10-600
Schneider, B. G., Mera, R., Piazuelo, M. B., Bravo, J. C., Zabaleta, J., Delgado, A. G., et al. (2015). DNA methylation predicts progression of human gastric lesions. Cancer Epidemiol. Biomarkers Prev. 24 (10), 1607–1613. doi:10.1158/1055-9965.epi-15-0388
Schnekenburger, M., Karius, T., and Diederich, M. (2014). Regulation of epigenetic traits of the glutathione S-transferase P1 gene: from detoxification toward cancer prevention and diagnosis. Front. Pharmacol. 5, 170. doi:10.3389/fphar.2014.00170
Šestáková, Š., Šálek, C., and Remešová, H. (2019). DNA methylation validation methods: a coherent review with practical comparison. Biol. Proced. Online 21, 19. doi:10.1186/s12575-019-0107-z
Shavali, M., Moradi, A., Tahmaseb, M., Mohammadian, K., and Ganji, S. M. (2024). Circulating-tumour DNA methylation of HAND1 gene: a promising biomarker in early detection of colorectal cancer. BMC Med. Genomics 17 (1), 117. doi:10.1186/s12920-024-01893-9
Sheikholeslami, S., Zarif-Yeganeh, M., Farashi, S., Azizi, F., Kia, S. K., Teimoori-Toolabi, L., et al. (2021). Promoter methylation of tumor suppressors in thyroid carcinoma: a systematic review. Iran. J. Public Health 50 (12), 2461–2472. doi:10.18502/ijph.v50i12.7928
Sheng, X., Guo, Y., and Lu, Y. (2017). Prognostic role of methylated GSTP1, p16, ESR1 and PITX2 in patients with breast cancer: a systematic meta-analysis under the guideline of PRISMA. Med. Baltim. 96 (28), e7476. doi:10.1097/md.0000000000007476
Singh, A., Gupta, S., Badarukhiya, J. A., and Sachan, M. (2020). Detection of aberrant methylation of HOXA9 and HIC1 through multiplex MethyLight assay in serum DNA for the early detection of epithelial ovarian cancer. Int. J. Cancer 147 (6), 1740–1752. doi:10.1002/ijc.32984
Singh, A., Gupta, S., and Sachan, M. (2021). Evaluation of the diagnostic potential of candidate hypermethylated genes in epithelial ovarian cancer in north Indian population. Front. Mol. Biosci. 8, 719056. doi:10.3389/fmolb.2021.719056
Siri, G., Alesaeidi, S., Dizghandi, S. E., Alani, B., Mosallaei, M., and Soosanabadi, M. (2022). Analysis of SDC2 gene promoter methylation in whole blood for noninvasive early detection of colorectal cancer. J. Cancer Res. Ther. 18 (Suppl. ment), S354–s8. doi:10.4103/jcrt.jcrt_1072_22
Skorodumova, L. O., Babalyan, K. A., Sultanov, R., Vasiliev, A. O., Govorov, A. V., Pushkar, D. Y., et al. (2016). GSTP1, APC and RASSF1 gene methylation in prostate cancer samples: comparative analysis of MS-HRM method and Infinium HumanMethylation450 BeadChip beadchiparray diagnostic value. Biomed. Khim 62 (6), 708–714. doi:10.18097/pbmc20166206708
Sobanski, T., Arantes, L., Dos Santos, W., Matsushita, M., de Oliveira, M. A., Costa, M., et al. (2021). Methylation profile of colon cancer genes in colorectal precursor lesions and tumor tissue: perspectives for screening. Scand. J. Gastroenterology 56 (8), 920–928. doi:10.1080/00365521.2021.1922744
Sobhani, I., Bergsten, E., Couffin, S., Amiot, A., Nebbad, B., Barau, C., et al. (2019). Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. U. S. A. 116 (48), 24285–24295. doi:10.1073/pnas.1912129116
Spagnol, L. W., Polettini, J., Silveira, D. A., Wegner, G. R. M., and Paiva, D. F. F. (2022). P16 gene promoter methylation is associated with oncogenesis and progression of gastric carcinomas: a systematic review and meta-analysis. Crit. Rev. Oncology/Hematology 180, 103843. doi:10.1016/j.critrevonc.2022.103843
Su, Y., Fang, H., and Jiang, F. (2016). Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin. Epigenetics 8, 109. doi:10.1186/s13148-016-0275-5
Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I., Jemal, A., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 71 (3), 209–249. doi:10.3322/caac.21660
Swellam, M., Abdelmaksoud, M. D., Sayed, M. M., Ramadan, A., Abdel-Moneem, W., and Hefny, M. M. (2015). Aberrant methylation of APC and RARβ<sub>2</sub> genes in breast cancer patients. IUBMB Life 67 (1), 61–68. doi:10.1002/iub.1346
Tan, A. C., Lai, G. G. Y., Saw, S. P. L., Chua, K. L. M., Takano, A., Ong, B. H., et al. (2024b). Detection of circulating tumor DNA with ultradeep sequencing of plasma cell-free DNA for monitoring minimal residual disease and early detection of recurrence in early-stage lung cancer. Cancer 130 (10), 1758–1765. doi:10.1002/cncr.35263
Tan, X., Fang, Y., Fan, X., Deng, W., Huang, J., Cai, Y., et al. (2024a). Testing region selection and prognostic analysis of MLH1 promoter methylation in colorectal cancer in China. Gastroenterol. Rep. 12, goae011. doi:10.1093/gastro/goae011
Tang, Q., Cheng, J., Cao, X., Surowy, H., and Burwinkel, B. (2016). Blood-based DNA methylation as biomarker for breast cancer: a systematic review. Clin. Epigenetics 8, 115. doi:10.1186/s13148-016-0282-6
Tepus, M., and Yau, T. O. (2020). Non-invasive colorectal cancer screening: an overview. Gastrointest. Tumors 7 (3), 62–73. doi:10.1159/000507701
Terp, S. K., Stoico, M. P., Dybkær, K., and Pedersen, I. S. (2023). Early diagnosis of ovarian cancer based on methylation profiles in peripheral blood cell-free DNA: a systematic review. Clin. Epigenetics 15 (1), 24. doi:10.1186/s13148-023-01440-w
Thakur, G. K., Sharma, T., Krishna Latha, T., Banerjee, B. D., Shah, H. K., and Guleria, K. (2019). High resolution based quantitative determination of methylation status of CDH1 and VIM gene in epithelial ovarian cancer. Asian Pac J. Cancer Prev. 20 (10), 2923–2928. doi:10.31557/apjcp.2019.20.10.2923
Thomassin, H., Kress, C., and Grange, T. (2004). MethylQuant: a sensitive method for quantifying methylation of specific cytosines within the genome. Nucleic Acids Res. 32 (21), e168. doi:10.1093/nar/gnh166
Topaloglu, O., Hoque, M. O., Tokumaru, Y., Lee, J., Ratovitski, E., Sidransky, D., et al. (2004). Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin. Cancer Res. 10 (7), 2284–2288. doi:10.1158/1078-0432.ccr-1111-3
Trung, N. T., Hoan, N. X., Trung, P. Q., Binh, M. T., Van Tong, H., Toan, N. L., et al. (2020). Clinical significance of combined circulating TERT promoter mutations and miR-122 expression for screening HBV-related hepatocellular carcinoma. Sci. Rep. 10 (1), 8181. doi:10.1038/s41598-020-65213-8
Tse, O. Y. O., Jiang, P., Cheng, S. H., Peng, W., Shang, H., Wong, J., et al. (2021). Genome-wide detection of cytosine methylation by single molecule real-time sequencing. Proc. Natl. Acad. Sci. U. S. A. 118 (5), e2019768118. doi:10.1073/pnas.2019768118
Tuo, L., Sha, S., Huayu, Z., and Du, K. (2018). P16<i>INK4a</i> gene promoter methylation as a biomarker for the diagnosis of non-small cell lung cancer: an updated meta-analysis. Thorac. Cancer 9 (8), 1032–1040. doi:10.1111/1759-7714.12783
Uehiro, N., Sato, F., Pu, F., Tanaka, S., Kawashima, M., Kawaguchi, K., et al. (2016). Circulating cell-free DNA-based epigenetic assay can detect early breast cancer. Breast Cancer Res. 18 (1), 129. doi:10.1186/s13058-016-0788-z
Um, S. W., Kim, Y., Lee, B. B., Kim, D., Lee, K. J., Kim, H. K., et al. (2018). Genome-wide analysis of DNA methylation in bronchial washings. Clin. Epigenetics 10, 65. doi:10.1186/s13148-018-0498-8
Wang, B. H., Li, Y. Y., Han, J. Z., Zhou, L. Y., Lv, Y. Q., Zhang, H. L., et al. (2017). Gene methylation as a powerful biomarker for detection and screening of non-small cell lung cancer in blood. Oncotarget 8 (19), 31692–31704. doi:10.18632/oncotarget.15919
Wang, Y. F., Li, L., Deng, X. Q., Fang, Y. J., and Zhang, C. X. (2023). Association of DNA methylation of vitamin D metabolic pathway related genes with colorectal cancer risk. Clin. Epigenetics 15 (1), 140. doi:10.1186/s13148-023-01555-0
Wasserkort, R., Kalmar, A., Valcz, G., Spisak, S., Krispin, M., Toth, K., et al. (2013). Aberrant septin 9 DNA methylation in colorectal cancer is restricted to a single CpG island. BMC Cancer 13, 398. doi:10.1186/1471-2407-13-398
Weiss, G., Schlegel, A., Kottwitz, D., König, T., and Tetzner, R. (2017). Validation of the SHOX2/PTGER4 DNA methylation marker panel for plasma-based discrimination between patients with malignant and nonmalignant lung disease. J. Thorac. Oncol. 12 (1), 77–84. doi:10.1016/j.jtho.2016.08.123
Wenger, A. M., Peluso, P., Rowell, W. J., Chang, P. C., Hall, R. J., Concepcion, G. T., et al. (2019). Accurate circular consensus long-read sequencing improves variant detection and assembly of a human genome. Nat. Biotechnol. 37 (10), 1155–1162. doi:10.1038/s41587-019-0217-9
Willard, S. S., and Koochekpour, S. (2012). Regulators of gene expression as biomarkers for prostate cancer. Am. J. Cancer Res. 2 (6), 620–657.
Wojdacz, T. K., and Dobrovic, A. (2007). Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 35 (6), e41. doi:10.1093/nar/gkm013
Wrangle, J., Machida, E. O., Danilova, L., Hulbert, A., Franco, N., Zhang, W., et al. (2014). Functional identification of cancer-specific methylation of CDO1, HOXA9, and TAC1 for the diagnosis of lung cancer. Clin. Cancer Res. 20 (7), 1856–1864. doi:10.1158/1078-0432.ccr-13-2109
Wu, G., Wang, Q., Wang, D., Xiong, F., Liu, W., Chen, J., et al. (2023). Targeting polycomb repressor complex 2-mediated bivalent promoter epigenetic silencing of secreted frizzled-related protein 1 inhibits cholangiocarcinoma progression. Clin. Transl. Med. 13 (12), e1502. doi:10.1002/ctm2.1502
Wu, X., Li, J., Gassa, A., Buchner, D., Alakus, H., Dong, Q., et al. (2020a). Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int. J. Biol. Sci. 16 (9), 1551–1562. doi:10.7150/ijbs.44024
Wu, Y., Jiang, G., Zhang, N., Liu, S., Lin, X., Perschon, C., et al. (2020b). HOXA9, PCDH17, POU4F2, and ONECUT2 as a urinary biomarker combination for the detection of bladder cancer in Chinese patients with hematuria. Eur. Urol. Focus 6 (2), 284–291. doi:10.1016/j.euf.2018.09.016
Xia, J. Y., and Aadam, A. A. (2022). Advances in screening and detection of gastric cancer. J. Surg. Oncol. 125 (7), 1104–1109. doi:10.1002/jso.26844
Xiong, J., Li, Y., Huang, K., Lu, M., Shi, H., Ma, L., et al. (2014). Association between DAPK1 promoter methylation and cervical cancer: a meta-analysis. PLoS One 9 (9), e107272. doi:10.1371/journal.pone.0107272
Xiong, Z., and Laird, P. W. (1997). COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25 (12), 2532–2534. doi:10.1093/nar/25.12.2532
Xu, F., Yu, S., Han, J., Zong, M., Tan, Q., Zeng, X., et al. (2021). Detection of circulating tumor DNA methylation in diagnosis of colorectal cancer. Clin. Transl. Gastroenterology 12 (8), e00386. doi:10.14309/ctg.0000000000000386
Xu, S., Yuan, Z., Jiang, C., Chen, W., Li, Q., and Chen, T. (2024). DNMT3A cooperates with YAP/TAZ to drive gallbladder cancer metastasis. Adv. Sci. 11 (16), e2308531. doi:10.1002/advs.202308531
Yadav, G., Vashisht, M., Yadav, V., and Shyam, R. (2020). Molecular biomarkers for early detection and prevention of ovarian cancer-A gateway for good prognosis: a narrative review. Int. J. Prev. Med. 11, 135. doi:10.4103/ijpvm.ijpvm_75_19
Yang, L., Tu, L., Bisht, S., Mao, Y., Petkovich, D., Thursby, S. J., et al. (2024). Tissue-location-specific transcription programs drive tumor dependencies in colon cancer. Nat. Commun. 15 (1), 1384. doi:10.1038/s41467-024-45605-4
Ye, J., Wu, M., He, L., Chen, P., Liu, H., and Yang, H. (2023). Glutathione-S-Transferase p1 gene promoter methylation in cell-free DNA as a diagnostic and prognostic tool for prostate cancer: a systematic review and meta-analysis. Int. J. Endocrinol. 2023, 7279243. doi:10.1155/2023/7279243
Yu, M., O'Leary, R. M., Kaz, A. M., Morris, S. M., Carter, K. T., Chak, A., et al. (2015a). Methylated B3GAT2 and ZNF793 are potential detection biomarkers for barrett's esophagus. Cancer Epidemiol. Biomarkers Prev. 24 (12), 1890–1897. doi:10.1158/1055-9965.epi-15-0370
Yu, Q., Guo, Q., Chen, L., and Liu, S. (2015b). Clinicopathological significance and potential drug targeting of CDH1 in lung cancer: a meta-analysis and literature review. Drug Des. Dev. Ther. 9, 2171–2178. doi:10.2147/dddt.s78537
Yuan, R. Q., Zhao, H., Wang, Y., Song, K., Yang, J., He, W., et al. (2022). SEPTIN9-SDC2-VIM methylation signature as a biomarker for the early diagnosis of colorectal cancer. Am. J. Cancer Res. 12 (7), 3128–3140.
Yuan, X., Xu, J., Fang, W., Zhao, Z., Wang, F., and Tong, Z. (2017). The association between MGMT promoter methylation and patients with gastric cancer: a meta-analysis. Genet. Test. Mol. Biomarkers 21 (4), 213–221. doi:10.1089/gtmb.2016.0284
Yue, C., Zhang, Y., Wang, Y., Zhang, Z., Zhang, M., Wang, H., et al. (2022). The application value of syndecan-2 gene methylation for colorectal cancer diagnosis: a clinical study and meta-analyses. Front. Med. (Lausanne) 9, 753545. doi:10.3389/fmed.2022.753545
Zelic, R., Fiano, V., Zugna, D., Grasso, C., Delsedime, L., Daniele, L., et al. (2016). Global hypomethylation (LINE-1) and gene-specific hypermethylation (GSTP1) on initial negative prostate biopsy as markers of prostate cancer on a rebiopsy. Clin. Cancer Res. 22 (4), 984–992. doi:10.1158/1078-0432.ccr-15-0606
Zeng, H., Chen, W., Zheng, R., Zhang, S., Ji, J. S., Zou, X., et al. (2018). Changing cancer survival in China during 2003-15: a pooled analysis of 17 population-based cancer registries. Lancet Glob. Health 6 (5), e555–e567. doi:10.1016/s2214-109x(18)30127-x
Zeng, H., Wang, Y., Wang, Y., and Zhang, Y. (2021). XXYLT1 methylation contributes to the occurrence of lung adenocarcinoma: methylation and lung adenocarcinoma. Med. Baltim. 100 (1), e24150. doi:10.1097/md.0000000000024150
Zeng, W., Zhu, J., Shan, L., Han, Z., Aerxiding, P., Quhai, A., et al. (2015). The clinicopathological significance of CDH1 in gastric cancer: a meta-analysis and systematic review. Drug Des. Dev. Ther. 9, 2149–2157. doi:10.2147/dddt.s75429
Zhang, C., Yu, W., Wang, L., Zhao, M., Guo, Q., Lv, S., et al. (2017a). DNA methylation analysis of the SHOX2 and RASSF1A panel in bronchoalveolar lavage fluid for lung cancer diagnosis. J. Cancer 8 (17), 3585–3591. doi:10.7150/jca.21368
Zhang, J., Yao, T., Lin, Z., and Gao, Y. (2017b). Aberrant methylation of MEG3 functions as a potential plasma-based biomarker for cervical cancer. Sci. Rep. 7 (1), 6271. doi:10.1038/s41598-017-06502-7
Zhang, K., He, Q., Cao, Q., Chuan, J., Qin, A., Tang, L., et al. (2024). Evaluating the clinical performance of SDC2/NDRG4 methylation for colorectal cancer detection. Epigenomics 16 (2), 93–108. doi:10.2217/epi-2023-0290
Zhang, W., Wang, H., Qi, Y., Li, S., and Geng, C. (2022). Epigenetic study of early breast cancer (EBC) based on DNA methylation and gene integration analysis. Sci. Rep. 12 (1), 1989. doi:10.1038/s41598-022-05486-3
Zhang, W., Yang, C., Wang, S., Xiang, Z., Dou, R., Lin, Z., et al. (2021b). SDC2 and TFPI2 methylation in stool samples as an integrated biomarker for early detection of colorectal cancer. Cancer Manag. Res. 13, 3601–3617. doi:10.2147/cmar.s300861
Zhang, Y., Wu, Q., Xu, L., Wang, H., Liu, X., Li, S., et al. (2021a). Sensitive detection of colorectal cancer in peripheral blood by a novel methylation assay. Clin. Epigenetics 13 (1), 90. doi:10.1186/s13148-021-01076-8
Zhao, L. H., Li, Q., Huang, Z. J., Sun, M. X., Lu, J. J., Zhang, X. H., et al. (2021). Identification of histone methyltransferase NSD2 as an important oncogenic gene in colorectal cancer. Cell Death Dis. 12 (11), 974. doi:10.1038/s41419-021-04267-6
Zhao, S., Hao, C. L., Zhao, E. H., Jiang, H. M., and Zheng, H. C. (2020). The suppressing effects of Dkk3 expression on aggressiveness and tumorigenesis of colorectal cancer. Front. Oncol. 10, 600322. doi:10.3389/fonc.2020.600322
Zheng, K., Dai, L., Zhao, Y., Li, L., Li, W., Zhang, X., et al. (2023). Methylated SEPT9 combined with AFP and PIVKA-II is effective for the detection of HCC in high-risk population. BMC Gastroenterol. 23 (1), 260. doi:10.1186/s12876-023-02900-6
Zhou, K., Cai, C., He, Y., Zhou, C., Zhao, S., Ding, X., et al. (2019a). Association between RASSF2 methylation and gastric cancer: a PRISMA-compliant systematic review and meta-analysis. DNA Cell Biol. 38 (10), 1147–1154. doi:10.1089/dna.2019.4922
Zhou, S., Dong, J., Deng, L., Wang, G., Yang, M., Wang, Y., et al. (2022). Endonuclease-assisted PAM-free recombinase polymerase amplification coupling with CRISPR/Cas12a (E-PfRPA/Cas) for sensitive detection of DNA methylation. ACS Sens. 7 (10), 3032–3040. doi:10.1021/acssensors.2c01330
Zhou, X., Jiao, D., Dou, M., Chen, J., Li, Z., Li, Y., et al. (2019b). Association of glutathione-S-transferase p1 gene promoter methylation and the incidence of prostate cancer: a systematic review and meta-analysis. J. Cancer Res. Clin. Oncol. 145 (8), 1939–1948. doi:10.1007/s00432-019-02962-8
Zhu, Q., Jin, Z., Yuan, Y., Lu, Q., Ge, D., and Zong, M. (2011). Impact of MTHFR gene C677T polymorphism on Bcl-2 gene methylation and protein expression in colorectal cancer. Scand. J. Gastroenterology 46 (4), 436–445. doi:10.3109/00365521.2010.537682
Keywords: cancer, gene methylation, biomarkers, methylation detection methods, epigenetics
Citation: Xu X, Guo Y, Liu M, Hu Y and Li S (2024) Advancements in the clinical application of gene methylation for early cancer detection. Front. Epigenet. Epigenom. 2:1430294. doi: 10.3389/freae.2024.1430294
Received: 09 May 2024; Accepted: 02 July 2024;
Published: 19 July 2024.
Edited by:
Raffaele Frazzi, Azienda Unità Sanitaria Locale - IRCCS di Reggio Emilia, ItalyReviewed by:
Lise Lotte Hansen, Aarhus University, DenmarkCopyright © 2024 Xu, Guo, Liu, Hu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shijun Li, bGlzaGlqdW5AZG11LmVkdS5jbg==
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.