- Department of Plant Biology, Waksman Institute of Microbiology, Rutgers, The State University of New Jersey, Piscataway, NJ, United States
Plants are exquisitely responsive to their local light and temperature environment utilizing these environmental cues to modulate their developmental pathways and adjust growth patterns. This responsiveness is primarily achieved by the intricate interplay between the photoreceptor phyB (phytochrome B) and PIF (PHYTOCHROME INTERACTING FACTORs) transcription factors (TFs), forming a pivotal signaling nexus. phyB and PIFs co-associate in photobodies (PBs) and depending on environmental conditions, PIFs can dissociate from PBs to orchestrate gene expression. Until recently, the mechanisms governing epigenome modifications subsequent to PIF binding to target genes remained elusive. This mini review sheds light on the emerging role of PIFs in mediating epigenome reprogramming by recruiting chromatin regulators (CRs). The formation of numerous different PIF-CR complexes enables precise temporal and spatial control over the gene regulatory networks (GRNs) governing plant-environment interactions. We refer to PIFs as epigenome landscapers, as while they do not directly reprogram the epigenome, they act as critical sequence-specific recruitment platforms for CRs. Intriguingly, in the absence of PIFs, the efficacy of epigenome reprogramming is largely compromised in light and temperature-controlled processes. We have thoroughly examined the composition and function of known PIF-CR complexes and will explore also unanswered questions regarding the precise of locations PIF-mediated epigenome reprogramming within genes, nuclei, and plants.
Introduction
The major sensor of red (R), far-red (FR) light as well temperature in Arabidopsis is the photoreceptor phyB (Quail et al., 1995; Rockwell et al., 2006; Jung et al., 2016; Legris et al., 2016). After photoactivation, phyB translocates from the cytosol to the nucleus where it compartmentalizes into PBs through liquid-liquid phase separation (LLPS) (Sakamoto and Nagatani, 1996; Kircher et al., 1999; Chen et al., 2003; Chen et al., 2022). PBs function as regulatory light and temperature modules, housing a variety of signaling components that interact directly or indirectly with phyB (Van Buskirk et al., 2012; Jung et al., 2016; Legris et al., 2016; Hahm et al., 2020; Pardi and Nusinow, 2021; Kim et al., 2023; Kwon et al., 2024; Willige et al., 2024). One of the most critical PB component are the PIFs, a family of basic helix-loop-helix (bHLH) TFs which acts a transcriptional activators or repressors (Ni et al., 1998; Huq and Quail, 2002; Huq et al., 2004; Oh et al., 2004; Castillon et al., 2007; Leivar et al., 2008; Shen et al., 2008; Leivar and Quail, 2011; Leivar and Monte, 2014). The Arabidopsis genome encodes eight PIFs (PIF1-PIF8), which integrate phyB’s environmental input into GRNs underlying light and temperature responses (Leivar and Quail, 2011; Jeong and Choi, 2013; Leivar and Monte, 2014; Pham et al., 2018; Bian et al., 2022; Han et al., 2023). PIF1/3/4/5 control the transition from skotomorphogenesis (SM) to photomorphogenesis (PM) by repressing light-responsive gene expression in the dark until light exposure initiates their phytochrome-mediated phosphorylation, ubiquitination and subsequent proteasomal degradation (Leivar and Quail, 2011; Leivar and Monte, 2014; Pham et al., 2018). Furthermore, PIF7 functions as the major regulator alongside PIF1/3/4/5 in orchestrating the shade avoidance syndrome (SAS) (Willige et al., 2021). This syndrome is induced by low R:FR light ratios (referred to as shade) resulting from nearby vegetation, prompting elongation of hypocotyls and petioles, early flowering, and upward leaf positioning (Franklin and Whitelam, 2005; Franklin, 2008; Casal, 2012; Sessa et al., 2018; Casal and Fankhauser, 2023). A phenotypically similar response to SAS is thermomorphogenesis (TM) which refers to the profound effect of elevated ambient temperature (EAT) (up to 28°C, below the heat stress range) on plant growth, development, and immunity (Casal and Balasubramanian, 2019; Burko et al., 2022). In Arabidopsis, TM regulation primarily involves PIF4 and PIF7 (Koini et al., 2009; Kumar et al., 2012; Gangappa et al., 2017; Chung et al., 2020; Fiorucci et al., 2020) whereas TM under shade (low R:FR light) conditions is mainly regulated by PIF7 alone (Burko et al., 2022).
The detailed complex mechanisms governing PIFs at both the transcriptional and protein levels along the crosstalk with other signaling pathways have been extensively discussed in several excellent reviews (Leivar and Quail, 2011; Jeong and Choi, 2013; Shin et al., 2013; Leivar and Monte, 2014; Paik et al., 2017; Pham et al., 2018; Favero, 2020). Additionally, readers are encouraged to explore superb reviews that delve into the exciting connection between light/temperature signaling and chromatin dynamics (Perrella and Kaiserli, 2016; Bourbousse et al., 2019; Jing and Lin, 2020; Perrella et al., 2020). We aim to provide a brief introduction to PIF7, which serves as a partial representation of other PIFs and highlights crucial regulatory aspects such as posttranslational modifications (PTMs), condensate formation, and DNA binding among PIF proteins. In white light (WL) PIF7 is phosphorylated and unlike other PIFs relatively light-stable (Leivar et al., 2008; Huang et al., 2018; Willige et al., 2021; Zhou et al., 2021). PIF7 undergoes LLPS to form biocondensates under WL conditions which subsequently associate with phyB condensates in photobodies (PBs) (Leivar et al., 2008; Willige et al., 2021; Chen et al., 2022; Xie et al., 2023). Upon exposure to shade, PIF7 gets rapidly dephosphorylated and dissociates from PBs to bind to G-boxes (CACGTG) within cis-regulatory elements (CREs) of its targets (Chung et al., 2020; Willige et al., 2021; Xie et al., 2023). PIF7 regulates an extensive GRN encompassing multiple biosynthesis genes for the growth-promoting plant hormone auxin, along with numerous transcription factors (TFs) such as ATHB2 (ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2) (Chung et al., 2020; Willige et al., 2021). A similar signaling mechanism operates during TM (Chung et al., 2020; Fiorucci et al., 2020; Burko et al., 2022), where PIF7 mRNA also serves as a direct thermosensor (Chung et al., 2020).
Here, we will discuss the emerging role of PIFs as epigenome landscapers by recruiting various chromatin regulators (CRs) to shape the epigenome at their target genes (Figure 1). The plant epigenome plays a crucial role as a regulatory framework, integrating both developmental signals and environmental cues into spatiotemporal-specific GRNs (Lloyd and Lister, 2022). In a broad sense, the epigenome encompasses not only all chemical modifications of DNA and histone proteins but also other features that control gene expression, including DNA binding of TFs and CRs, chromatin accessibility, 3D chromatin conformation, nucleosome positioning, and long non-coding RNAs (Rivera and Ren, 2013). We utilize the term CR to encompass all regulatory factors capable of modifying the epigenome, chromatin remodeling complexes (CRCs), histone acetyltransferases/deacetylases (HATs/HDACs), histone methyltransferases/demethylases (KMTs/KDMs), and many others. To enhance comprehension, we discuss various epigenome feature dynamics separately, although it is crucial to recognize their intricate interconnections. Finally, we outline some unresolved questions for further exploration.
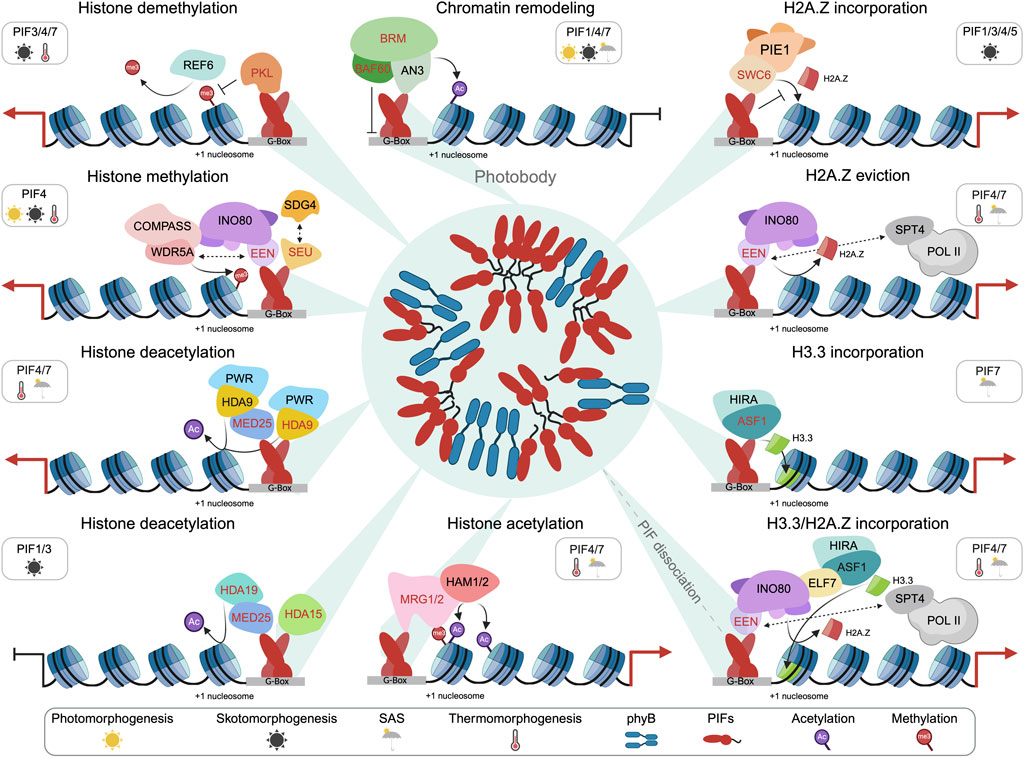
Figure 1. The illustration provides an overview of the current understanding of different PIF-CR complexes. The PBs containing phyB-PIF act as a reservoir for PIFs, enabling their dissociation based on environmental cues. Various PIF-controlled features are depicted. While the complexes are presented separately for clarity, they likely function concurrently. The respective PIFs implicated in specific epigenomic features are highlighted, along with symbols denoting the four major processes (SM, PM, SAS, and TM) where the PIF-CR complexes act. EEN-SPT4 and EEN-WDR5A interactions are shown as dashed double arrows for better visualization. Abbreviations: AN3 (ANGUSTIFOLIA3), ASF1 (ANTI-SILENCING FUNCTION 1A/B), BAF60 (BRG1/BRM associated factor 60), BRM (BRAHMA), COMPASS (Complex Proteins Associated with Set1), EEN (EIN6 ENHANCER), ELF7 (EARLY FLOWERING 7), HAM1/2 (HISTONE ACETYLTRANSFERASE OF THE MYST FAMILY 1 and 2), HDA9 (HISTONE DEACETYLASE 9), HDA15 (HISTONE DEACETYLASE 15), HDA19 (HISTONE DEACETYLASE 19), HDA9 (HISTONE DEACETYLASE 9), HIRA (HISTONE REGULATORY HOMOLOG A), INO80 (INOSITOL REQUIRING 80), MED25 (MEDIATOR 25), MRG1/2 (MORF-RELATED GENE 1/2), PIE1 (PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1), PKL (PICKLE), POL II (RNA Polymerase II), PWR (POWERDRESS), REF6 (RELATIVE OF EARLY FLOWERING 6), SDG4 (SET DOMAIN GROUP 4), SEU (SEUSS), SPT4 (Suppressor of Ty 4), SWC6 (SWR1 complex subunit 6), and WDR5A (WD40 REPEAT 5).
H2A.Z and H3.3 dynamics
Histone variants are histone proteins that differ from canonical histones in their amino acid sequence, structure, and functions (Yi et al., 2006; Borg et al., 2021). One of the best studied histone variants in Arabidopsis is H2A.Z that confers gene responsiveness to environmentally-responsive genes (Coleman-Derr and Zilberman, 2012). The Arabidopsis genome encodes three functionally redundant H2A.Z genes (HTA8, HTA9, HTA11) whose mutations leads to pleiotropic effects such as early flowering, enhanced disease resistance and DNA hypermethylation (Choi et al., 2007; March-Diaz et al., 2008; Coleman-Derr and Zilberman, 2012; Nie et al., 2019). H2A.Z exerts a repressive influence on transcription, and its eviction is a common characteristic of transcriptional activation in plant-environment interactions (Kumar and Wigge, 2010; Kumar et al., 2012; Boden et al., 2013; Cortijo et al., 2017; Sura et al., 2017; Zander et al., 2019; Willige et al., 2021; Xue et al., 2021).
The discovery of H2A.Z’s involvement in PIF-regulated processes occurred with a forward genetic screen aiming to identify temperature sensors in Arabidopsis (Kumar and Wigge, 2010). This screen revealed ARP6 (ACTIN-RELATED PROTEIN 6), a subunit of the SWR1 (SWI2/SNF2 (SWITCH/SUCROSE NONFERMENTABLE)-related 1) chromatin remodeler (Choi et al., 2005; Martin-Trillo et al., 2006; Choi et al., 2007), as a negative TM regulator (Kumar and Wigge, 2010). Other key components of the multi-subunit plant SWR1 complex (SWR1-C) are the ATPase PIE1 (PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1) and accessory units such as SWC6 (SWR1 COMPLEX 6) and SEF (SERRATED LEAVES AND EARLY FLOWERING) (Noh and Amasino, 2003; Choi et al., 2007; March-Di et al., 2007; Luo et al., 2020). Consistent with SWR1-C’s role in incorporating H2A.Z into chromatin (Deal et al., 2007), arp6 mutants have reduced H2A.Z levels resulting in the elevation of thermo-responsive gene expression and longer hypocotyls and petioles even without an EAT stimulus (Kumar and Wigge, 2010). As PIF4 governs the expression of EAT-induced genes (Koini et al., 2009), it was suggested that H2A.Z-containing nucleosomes occlude PIF4 from binding to its target genes. This hindrance is lifted in arp6 mutants, thereby facilitating higher thermo-responsive gene expression (Kumar and Wigge, 2010).
The first direct connection between H2A.Z and PIFs was shown for PIF4-dependent EAT-induced flowering in Arabidopsis (Kumar et al., 2012). EAT-induced expression of FT (FLOWERING LOCUS T) requires direct PIF4 binding at FT which is facilitated by EAT-induced eviction of H2A.Z nucleosomes (Kumar et al., 2012). To elucidate the PIF-H2A.Z interplay in more detail, PIF7 DNA binding as well as H2A.Z occupancy in wildtype and pif457 mutants were tracked simultaneously during SAS over time using ChIP-seq (Chromatin immunoprecipitation followed by sequencing) (Willige et al., 2021). PIF7 initiates shade-induced H2A.Z eviction at its target genes through the association with the INO80 (INOSITOL REQUIRING 80) chromatin remodeling complex (INO80-C) via direct interaction with its essential subunit EEN (EIN6 ENHANCER) (Zander et al., 2019; Willige et al., 2021). Strikingly, this PIF-INO80-C regulatory module is also operational with PIF4 during TM (Figure 1) (Sureshkumar and Balasubramanian, 2021; Xue et al., 2021).
INO80-C belongs to the INO80-type subfamily of SWI2/SNF2 chromatin remodeler (Clapier and Cairns, 2009; Han et al., 2015) and it was shown in various species that INO80-C can facilitate H2A.Z eviction to regulate gene expression (Papamichos-Chronakis et al., 2011; Alatwi and Downs, 2015; Brahma et al., 2017; Zhao et al., 2022). In plants, INO80-C’s role in H2A.Z eviction was shown for ethylene-, low R:FR light-, and EAT-induced genes (Zander et al., 2019; Willige et al., 2021; Xue et al., 2021). INO80-C additionally acts as a platform for recruiting other CRs, thereby potentially expanding the functional capabilities of the PIF-INO80-C regulatory module (Shang et al., 2021; Xue et al., 2021; Zhao et al., 2023). Its subunit EEN interacts with WDR5a (WD40 REPEAT 5), an integral component of the COMPASS (Complex Proteins Associated with Set1) histone H3K4 methyltransferase complex, to facilitate trimethylation of histone 3 lysine 4 (H3K4me3) at PIF4 target genes (Figure 1) (Xue et al., 2021). The association of INO80-C with other major COMPASS histone H3K4 methyltransferase complex components was independently shown (Shang et al., 2021). In addition, EEN interacts with the transcription elongation factors (TEFs) SPT4 (Suppressor of Ty 4)-1 and SPT4-2 to mediate RNA Polymerase II (RNAPII) elongation during TM (Figure 1) (Xue et al., 2021). Interestingly, PIF1/3/4/5 can also associate with SWR1-C via SWC6 during PM to inhibit H2A.Z deposition (Figure 1) (Chen H. et al., 2023). Under this scenario, PIFs inhibit SWR1-C activity at auxin-responsive genes in the dark through an unknown mechanism (Chen H. et al., 2023). All these findings indicate that PIFs can use multiple strategies to alter the H2A.Z landscape at their target genes thereby fine-tuning their expression in an environmental stimulus-dependent manner.
The histone variant H3.3 forms together with the replicative H3.1/H3.2, and the centromeric CenH3 the H3 (Histone 3) family (Henikoff and Ahmad, 2005; Borg et al., 2021). H3.3 is incorporated during transcription and rapid upregulation of environmentally-responsive genes is compromised in h3.3 knockdown mutants (Wollmann et al., 2017). The deposition of H3.3 is in part mediated by the histone chaperones ASF1A (ANTI-SILENCING FUNCTION 1A) and ASF1B in conjunction with the histone chaperone HIRA (HISTONE REGULATORY HOMOLOG A) (Tagami et al., 2004; Zhu et al., 2011; Nie et al., 2014; Duc et al., 2015; Zhong et al., 2022). During SAS, PIF7 recruits ASF1A/B and HIRA to facilitate H3.3 deposition at shade-responsive genes (Figure 1) (Yang et al., 2023). In addition, asf1ab and hira mutants show also reduced hypocotyl elongation under EAT (Yang et al., 2023; Zhao et al., 2023) which suggest that the PIF-ASF1A/B-HIRA module is also operational during TM. Although a PIF4-ASF1A/B interaction has not been confirmed, ASF1A/B could also be indirectly recruited by PIF4 through the INO80 ATPase that associates with ASF1A/B through the TEF PAF1c (Polymerase-Associated Factor 1 complex) subunit ELF7 (EARLY FLOWERING 7) (Figure 1) (Zhao et al., 2023). These findings highlight again the prominent role of PIFs in initiating epigenomic reprogramming through the recruitment of functionally diverse CRs.
Histone acetylation dynamics
One of the most extensively studied PTM of histones is acetylation, which plays a crucial role in numerous gene regulatory processes (Jiang et al., 2020; Shvedunova and Akhtar, 2022; Chen Y. et al., 2023). Acetylation occurring at various lysine residues of histones H3 and H4, such as H3K9ac or H3K27ac, typically corresponds to gene activation, whereas deacetylation is associated with gene repression (Pandey et al., 2002). The balance of histone acetylation is regulated by the interplay between HATs and HDACs (Eberharter and Becker, 2002). The Arabidopsis genome encodes for 12 HATs and 18 HDACs (Pandey et al., 2002; Hollender and Liu, 2008), and although no direct interactions between PIFs and HATs have been documented, an active role of HATs in PIF-mediated chromatin reprogramming can be inferred (Martínez-García and Moreno-Romero, 2020). During SAS, H3K9ac levels rapidly increase at gene bodies of shade-responsive genes in a PIF4/5/7-dependent manner (Willige et al., 2021). Furthermore, levels of H4K5ac, H3K9ac, and H3K27ac increase in response to shade and EAT at YUC8 in a PIF4/7-dependent manner (Peng et al., 2018; Zhou et al., 2024).
PIF4/7 directly associate with MRG1/2 (MORF-RELATED GENE 1/2) which are histone methylation readers that bind to H3K4/H3K36 trimethylation (H3K4me3/H3K36me3) and can interact with the HATs HAM1/2 (HISTONE ACETYLTRANSFERASE OF THE MYST FAMILY 1 and 2) (Xu et al., 2014). The recruitment of HAM1/2 through the PIF4/7-MRG1/2 module is the current model of PIF4/7-driven histone acetylation dynamics during SAS and TM (Figure 1) (Peng et al., 2018; Zhou et al., 2024). However, additional experimental support is needed because no direct association of HAM1/2 with PIF target genes has been shown so far (Latrasse et al., 2008).
HDA9 (HISTONE DEACETYLASE 9) was found to positively regulate hypocotyl elongation during TM and SAS (Tasset et al., 2018; van der Woude et al., 2019; Nguyen et al., 2023). HDA9’s positive role is still puzzling since expression of PIF4 and YUC8, both essential regulators of EAT-induced hypocotyl elongation (Koini et al., 2009; Franklin et al., 2011; Sun et al., 2012; Proveniers and van Zanten, 2013; Bellstaedt et al., 2019), requires HDA9-mediated H3K9ac/14ac deacetylation at its +1 nucleosome (van der Woude et al., 2019). Interestingly, mutation of the HDA9-interacting SANT-domain containing protein PWR (POWERDRESS), phenocopies hda9 mutants regarding reduced PIF4/YUC8 expression due to higher H3K9ac levels (Tasset et al., 2018). EAT-induced H2A.Z eviction was also compromised in hda9 mutants (van der Woude et al., 2019), however, the mechanism of how HDA9 regulates H2A.Z eviction is still unclear. Although PIF4 was not found to interact with HDA9 (van der Woude et al., 2019), PIF7 was recently identified as an interactor of HDA9 (Nguyen et al., 2023). Given PIF7’s critical role in TM and its capacity to heterodimerize with PIF4 (Kidokoro et al., 2009; Fiorucci et al., 2020), it’s possible that a PIF7/PIF4 heterodimer recruits the HDA9-PWR module to its target genes (Figure 1). The hypothesis of HDA9 recruitment to shade-induced genes via PIF7 has also been proposed for SAS (Nguyen et al., 2023).
During SM, HDA15 (HISTONE DEACETYLASE 15) is recruited by PIF3 to repress the expression of photosynthesis genes through H4 deacetylation (Liu et al., 2013). In addition, PIF1 also interacts with HDA15 to repress genes via histone deacetylation (H3ac) during seed germination in the dark (Figure 1) (Gu et al., 2017). In contrast to HDA9’s positive role, HDA15 is a negative TM regulator directly associating at thermo-responsive genes potentially through HFR1 (LONG HYPOCOTYL IN FAR-RED) (Shen et al., 2019). A current hypothesis that elucidates the contradictory functions of HDA9 and HDA15 proposes that at higher temperatures, the HFR1-HDA15 complex is displaced by PIF4, possibly forming a complex with HDA9 and PWR (Shen et al., 2019). HDA19 is an additional PIF1/3-interacting HDAC that represses PM via H3 deacetylation at the PM genes BBX21 (B-BOX CONTAINING PROTEIN 21) and GLK1 (GOLDEN2-LIKE1) (Guo et al., 2023). Another possible route of connecting PIFs with HDACs is HOS1 (HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 1), a RING E3 ligase which can directly interact with PIF4 but also with HDA6 (HISTONE DEACETYLASE 6) and HDA15 (Jung et al., 2013; Kim et al., 2017).
An additional functional link between PIFs and HDACs are the subunits MED25 (MEDIATOR 25) and MED14 (MEDIATOR 14) of the Mediator complex (Bajracharya et al., 2022; Guo et al., 2023; Shapulatov et al., 2023), which is an evolutionary conserved large multi-subunit protein complex regulating RNAPII function on various levels (Allen and Taatjes, 2015; Soutourina, 2018). MED25 or PFT1 (PHYTOCHROME AND FLOWERING TIME 1) was first discovered as a SAS regulator and can interact with PIF4 in Arabidopsis and tomato to recruit RNAPII (Sun et al., 2020; Shapulatov et al., 2023). During TM, MED25 can associate with HDA9, thereby potentially facilitating the PIF4-mediated recruitment of HDA9 for histone deacetylation at the PIF4 and YUC8 gene (Figure 1) (Shapulatov et al., 2023). Moreover, PIF1 and PIF3 also interact with MED25 and HDA19 to down-regulate the expression of positive the positive PM regulators BBX21 and GLK1 via histone deacetylation and reducing chromatin accessibility (Figure 1) (Guo et al., 2023). Intriguingly, MED25 also undergoes LLPS to form biomolecular condensates with PIF1/3 and HDA19 (Guo et al., 2023). Moreover, PIF4 as well as its coactivator HMR (HEMERA) interact with MED14 to positively regulate thermo-responsive genes (Bajracharya et al., 2022).
Also noteworthy is the recently resolved protein composition of PBs, which has identified the Groucho/Tup1-type co-repressors TPL (TOPLESS) and TPR1 (TOPLESS-RELATED 1) as PB components (Kim et al., 2023). TPL and TPRs putatively exert their repressive function through the direct association with various HDACs such HDA6 and HDA19 (Long et al., 2006; Krogan et al., 2012; Wang et al., 2013; Plant et al., 2021). The confirmation of whether PIFs indeed interact with TPL/TPRs at their target genes remains uncertain at this point. However, the spatial proximity of PIFs and TPL/TPRs in PBs implies a potential functional connection (Kim et al., 2023). In addition, an interaction of PIF1 with LUH (LEUNIG_HOMOLOG), another member of the Groucho/Tup1-type co-repressor family, was shown to regulate expression of PIF1 target genes during seed germination through an unknown mechanism (Lee et al., 2015; Kim et al., 2023).
Histone methylation dynamics
Methylation of various lysine (K) residues of histones H3 and H4 can occur in plants and depending on the modified lysine residue and degree of methylation (mono-, di-, and/or tri), gene expression is instructed differently (Liu et al., 2010; Xiao et al., 2016; Cheng et al., 2020). H3K4me3 on the +1 nucleosome is a critical epigenome feature indicating active genes, and assessing H3K4me3 occupancy is commonly employed as an indicator of an active chromatin state (Bernstein et al., 2002). Mutants in PIF-interacting CRs frequently exhibit altered levels of H3K4me3 at PIF target genes (Huai et al., 2018) However, whether this alteration is regulatory in nature or merely a consequence of transcriptional changes remains unclear. Until very recently, the exact function of H3K4me3 remained elusive. Using an elegant acute depletion method, all SET1/COMPASS complexes were eliminated from mouse embryonic stem cells, unveiling the critical involvement of H3K4me3 in regulating RNAPII pausing, elongation, and eviction (Wang et al., 2023).
The dark-to-light transition at the beginning of PM leads to a PIF-dependent rapid upregulation of gene expression and H3K4me3 levels (Calderon et al., 2022). How this achieved in a PIF-dependent manner is not clear but the PIF4 interacting transcriptional co-regulator SEU (SEUSS) might provide a link between PIFs and active H3K4me3 regulation (Figure 1) (Huai et al., 2018). SEU is a negative PM regulator under red, far-red, and blue light conditions, but interestingly a positive TM regulator (Huai et al., 2018). It has been demonstrated for SEUSS that it controls H3K4me3 deposition at WOX5 (WUSCHEL-RELATED HOMEOBOX 5) targets genes by associating with the H3K4 methyltransferase SDG4 (SET DOMAIN GROUP 4) (Zhai et al., 2020). Therefore, it is plausible to hypothesize the existence of an operational PIF-SEU-SDG4 complex. Additionally, PIFs regulate the local H3K4me3 environment at their target genes by associating with INO80-C/COMPASS complexes during TM (Figure 1) (Shang et al., 2021; Xue et al., 2021).
Removal of histone methylation marks is facilitated by jumonji domain-containing histone demethylases (Lu et al., 2008; Mosammaparast and Shi, 2010). The primary demethylase in Arabidopsis is REF6 (RELATIVE OF EARLY FLOWERING 6) whose mutation causes ectopic gain of H3K27me3 at thousands of genes (Lu et al., 2011; Zander et al., 2019). REF6 was found to cooperatively regulate EAT-induced gene expression with PIF4 via H3K27me3 demethylation (Figure 1) (He et al., 2022). Whether REF6 can interact with PIFs is unknown but it can interact with INO80-C (Smaczniak et al., 2012), suggesting a potential regulatory pathway through which PIFs could influence the H3K27me3 landscape. During SM, PIF3 interacts with the SWI/SWF chromatin-remodeler (PKL/EPP1) (PICKLE/ENHANCED PHOTOMORPHOGENIC1) to repress H3K27me3 deposition at PIF3 target sites (Figure 1) (Zhang et al., 2014). How PKL inhibits the H3K27me3 deposition is not understood since PKL acts cooperatively with the SWR1-C ATPase PIE1 and the H3K27 methyltransferase CLF (CURLY LEAF) to establish and maintain the H3K27me3 landscape in Arabidopsis (Carter et al., 2018).
Chromatin remodeling
Chromatin remodeling is a key process in genome organization, transcriptional regulation, DNA repair and replication (Clapier and Cairns, 2009). Of particular importance for gene expression is the accessibility of cis-regulatory elements and incorporation/eviction of histone variants which is regulated by multi-subunit ATP-dependent SWI2/SNF2 CRCs (Clapier and Cairns, 2009). Besides the interaction of PIFs with the INO80-type remodelers SWR1-C and INO80-C (Willige et al., 2021; Xue et al., 2021; Chen H. et al., 2023), PIFs also interact with various SWI/SNF-type remodelers (Zhang et al., 2017; Hussain et al., 2022). The SWI/SNF-type family consists of the BRM (BRAHMA)-, SYD (SPLAYED)-, and MINU1/2 (MINUSCULE1/2)-associated SWI/SNF (BAS, SASc (to avoid confusion with SAS), and MAS) complexes (Guo et al., 2022). PIF1 recruits BRM to photosynthesis genes in the dark to repress their expression though mechanism that remains unidentified (Zhang et al., 2017). Moreover, PIF7 interacts with AN3 (ANGUSTIFOLIA3), a subunit of either the BAS or SASc complex (Guo et al., 2022) to regulate leaf cell proliferation during shade (Hussain et al., 2022). Shade-stabilized PIF7 outcompetes AN3 at it target genes and represses their expression (Hussain et al., 2022). An additional antagonism between PIFs and a SWI/SNF subunit was shown for PIF4 and BAF60 (BRG1/BRM associated factor 60) (Jegu et al., 2017). BAF60 can be a subunit of the BAS, SASc, and MAS complex but because of a reported direct interaction between PIF4 and BRM, an association within the BAS complex is likely (Zhang et al., 2017; Guo et al., 2022). BAF60 target sites overlap with PIF4 DNA binding sites and strikingly BAF60 antagonizes PIF4 binding through the diurnal regulation of DNA accessibility at PIF4 binding sites (Figure 1) (Jegu et al., 2017).
Discussion
Where in the gene, where in the nucleus, and where in the plant do the PIF-CR complexes act? For PIFs and their respective CRs to interact, they must be brought into proximity. Unlike CRs, which typically occupy gene bodies, PIFs span a wide spectrum, ranging from proximal to distal, intronic, and 3′ CRE binding (Chung et al., 2020; Willige et al., 2021). Most CRE-promoter communications are established by chromatin loops where TF-bound distal CREs or enhancers make direct physical with the proximal promoter/gene body region (Panigrahi and O'Malley, 2021). Indeed, a COP1 (CONSTITUTIVELY PHOTOMORPHOGENIC 1)-controlled phyB-PRC2 (POLYCOMB REPRESSIVE COMPLEX 2)-mediated chromatin loop has been identified at the positive SAS regulator ATHB2 (Steindler et al., 1999; Kim et al., 2021; Wang et al., 2024), which is one of the most prominent shade and EAT-induced PIF7 targets, possessing an unusually large CRE with five PIF7 binding peaks 3–9 kb (kilobase) upstream of its transcription start site (Chung et al., 2020; Willige et al., 2021; Burko et al., 2022). Notably, the PIF interactor MED25 facilitates chromatin looping during active jasmonic acid (JA) signaling through interaction with the JA master TF MYC2 (Wang et al., 2019). Like MYC2 which can form tetramers to support loop formation (Lian et al., 2017), PIF4 can also form tetramers suggesting the potential of PIF4 to form loops (Gao et al., 2022).
This leads us to the nuclear 3D space and our second question: where precisely within the nucleus do the PIF-CR complexes act? We pose this question due to the rapid nature of shade-induced PIF7 DNA binding, which occurs at a few hundred genes within 5 min of shade exposure (Willige et al., 2021), potentially even earlier. Since we lack information regarding the DNA scanning speed of PIF7, we cannot ascertain whether the swift DNA targeting of PIF7 is attributable to PIF7’s search throughout the entire nucleus, or as previously speculated, solely in proximity to PBs (Van Buskirk et al., 2012). Various nuclear bodies in mammals are known to directly regulate chromatin activities (Shan et al., 2023), and intriguingly, a temperature-dependent chromatin association has been demonstrated for phyB (Jung et al., 2016). Moreover, recent findings have unveiled an active role of phyB in chromatin loop formation at the PIF target ATHB2 (Kim et al., 2021; Wang et al., 2024). To further explore this exciting scenario of PB-associated chromatin structures, future research necessitates 3D chromatin conformation analyses as well as single locus imaging technologies.
All findings presented here stem from bulk-level analyses, which may obscure crucial spatial information (Cole et al., 2021). It has been demonstrated that for EAT-induced hypocotyl elongation, it is crucial for PIF4 to function within the epidermis (Kim et al., 2020), in conjunction with the DOF TF CDF2 (CYCLING DOF FACTOR 2) (Gao et al., 2022). Similarly, epidermal phyB plays a pivotal role in regulating most light responses, including SAS (Kim et al., 2016). Recent advancements in single-cell (sc) technologies, such as single-cell RNA sequencing (scRNA-seq), now facilitate the capturing of gene expression profiles at the single-cell level (Han et al., 2023). Thus far, only one scRNA-seq study has been reported for a pif mutant revealing that expression levels of PIF1/3/4/5 remain relatively uniformly across cells in wildtype aerial tissues, but interestingly, the expression of PIF target genes varies among different cell types in pifq mutants (Han et al., 2023). This suggests that the cell-type specificity of PIF signaling may stem from cell type-specific epigenome disparities at PIF target genes (Han et al., 2023). Considering that PIFs initiate epigenome reprogramming at their target genes by recruiting various CR complexes, we hypothesize the existence of cell type-specific PIF-CR complexes to establish gene expression patterns unique to each cell type.
Highlighting these discoveries underscores the pivotal role of PIFs as epigenome landscapers. From our viewpoint, three key elements of PIFs’ epigenome landscaping abilities are particularly noteworthy. Firstly, binding of PIFs to CREs at their target genes is the starting point of stimulus-induced epigenome reprogramming. Secondly, most epigenome features can be directly and simultaneously governed by functionally diverse PIF-CR complexes. Thirdly, several PIF-CR modules, such as PIF-INO80-C, PIF-MRG1/2, and PIF-ASF1-HIRA are operational in multiple response pathways like SAS and TM (Shang et al., 2021; Willige et al., 2021; Yang et al., 2023; Zhao et al., 2023; Zhou et al., 2024), indicating that these complexes belong to the general repertoire of PIF-mediated transcriptional regulation.
Author contributions
MA: Conceptualization, Data curation, Visualization, Writing–original draft. KM: Conceptualization, Data curation, Writing–original draft. MZ: Conceptualization, Data curation, Funding acquisition, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. MZ is supported by a National Science Foundation (NSF) CAREER grant IOS-2339927. KM is supported by a Fulbright Foreign Student Program (PS00316055).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alatwi, H. E., and Downs, J. A. (2015). Removal of H2A.Z by INO80 promotes homologous recombination. EMBO Rep. 16 (8), 986–994. doi:10.15252/embr.201540330
Allen, B. L., and Taatjes, D. J. (2015). The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 16 (3), 155–166. doi:10.1038/nrm3951
Bajracharya, A., Xi, J., Grace, K. F., Bayer, E. E., Grant, C. A., Clutton, C. H., et al. (2022). PHYTOCHROME-INTERACTING FACTOR 4/HEMERA-mediated thermosensory growth requires the Mediator subunit MED14. Plant Physiol. 190 (4), 2706–2721. doi:10.1093/plphys/kiac412
Bellstaedt, J., Trenner, J., Lippmann, R., Poeschl, Y., Zhang, X. X., Friml, J., et al. (2019). A mobile auxin signal connects temperature sensing in cotyledons with growth responses in hypocotyls. Plant Physiol. 180 (2), 757–766. doi:10.1104/pp.18.01377
Bernstein, B. E., Humphrey, E. L., Erlich, R. L., Schneider, R., Bouman, P., Liu, J. S., et al. (2002). Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. U. S. A. 99 (13), 8695–8700. doi:10.1073/pnas.082249499
Bian, Y., Chu, L., Lin, H., Qi, Y., Fang, Z., and Xu, D. (2022). PIFs- and COP1-HY5-mediated temperature signaling in higher plants. Stress Biol. 2 (1), 35. doi:10.1007/s44154-022-00059-w
Boden, S. A., Kavanova, M., Finnegan, E. J., and Wigge, P. A. (2013). Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biol. 14 (6), R65. doi:10.1186/gb-2013-14-6-r65
Borg, M., Jiang, D., and Berger, F. (2021). Histone variants take center stage in shaping the epigenome. Curr. Opin. Plant Biol. 61, 101991. doi:10.1016/j.pbi.2020.101991
Bourbousse, C., Barneche, F., and Laloi, C. (2019). Plant chromatin catches the Sun. Front. Plant Sci. 10, 1728. doi:10.3389/fpls.2019.01728
Brahma, S., Udugama, M. I., Kim, J., Hada, A., Bhardwaj, S. K., Hailu, S. G., et al. (2017). INO80 exchanges H2A.Z for H2A by translocating on DNA proximal to histone dimers. Nat. Commun. 8, 15616. doi:10.1038/ncomms15616
Burko, Y., Willige, B. C., Seluzicki, A., Novak, O., Ljung, K., and Chory, J. (2022). PIF7 is a master regulator of thermomorphogenesis in shade. Nat. Commun. 13 (1), 4942. doi:10.1038/s41467-022-32585-6
Calderon, R. H., Dalton, J., Zhang, Y., and Quail, P. H. (2022). Shade triggers posttranscriptional PHYTOCHROME-INTERACTING FACTOR-dependent increases in H3K4 trimethylation. Plant Physiol. 190 (3), 1915–1926. doi:10.1093/plphys/kiac282
Carter, B., Bishop, B., Ho, K. K., Huang, R., Jia, W., Zhang, H., et al. (2018). The chromatin remodelers PKL and PIE1 act in an epigenetic pathway that determines H3K27me3 homeostasis in Arabidopsis. Plant Cell 30 (6), 1337–1352. doi:10.1105/tpc.17.00867
Casal, J. J., and Balasubramanian, S. (2019). Thermomorphogenesis. Annu. Rev. Plant Biol. 70, 321–346. doi:10.1146/annurev-arplant-050718-095919
Casal, J. J., and Fankhauser, C. (2023). Shade avoidance in the context of climate change. Plant Physiol. 191 (3), 1475–1491. doi:10.1093/plphys/kiad004
Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12 (11), 514–521. doi:10.1016/j.tplants.2007.10.001
Chen, D., Lyu, M., Kou, X., Li, J., Yang, Z., Gao, L., et al. (2022). Integration of light and temperature sensing by liquid-liquid phase separation of phytochrome B. Mol. Cell 82 (16), 3015–3029.e6. doi:10.1016/j.molcel.2022.05.026
Chen, H., Wang, W., Chen, X., Niu, Y., Qi, Y., Yu, Z., et al. (2023a). PIFs interact with SWI2/SNF2-related 1 complex subunit 6 to regulate H2A.Z deposition and photomorphogenesis in Arabidopsis. J. Genet. Genomics 50 (12), 983–992. doi:10.1016/j.jgg.2023.04.008
Chen, M., Schwab, R., and Chory, J. (2003). Characterization of the requirements for localization of phytochrome B to nuclear bodies. Proc. Natl. Acad. Sci. U. S. A. 100 (24), 14493–14498. doi:10.1073/pnas.1935989100
Chen, Y., Guo, P. G., and Dong, Z. C. (2023b). The role of histone acetylation in transcriptional regulation and seed development. Plant Physiol. 194, 1962–1979. doi:10.1093/plphys/kiad614
Cheng, K., Xu, Y., Yang, C., Ouellette, L., Niu, L., Zhou, X., et al. (2020). Histone tales: lysine methylation, a protagonist in Arabidopsis development. J. Exp. Bot. 71 (3), 793–807. doi:10.1093/jxb/erz435
Choi, K., Kim, S., Kim, S. Y., Kim, M., Hyun, Y., Lee, H., et al. (2005). SUPPRESSOR of FRIGIDA3Encodes a nuclear ACTIN-RELATED PROTEIN6 required for floral repression inArabidopsis w⃞. Plant Cell 17 (10), 2647–2660. doi:10.1105/tpc.105.035485
Choi, K., Park, C., Lee, J., Oh, M., Noh, B., and Lee, I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134 (10), 1931–1941. doi:10.1242/dev.001891
Chung, B. Y. W., Balcerowicz, M., Di Antonio, M., Jaeger, K. E., Geng, F., Franaszek, K., et al. (2020). An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat. Plants 6 (5), 522–532. doi:10.1038/s41477-020-0633-3
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi:10.1146/annurev.biochem.77.062706.153223
Cole, B., Bergmann, D., Blaby-Haas, C. E., Blaby, I. K., Bouchard, K. E., Brady, S. M., et al. (2021). Plant single-cell solutions for energy and the environment. Commun. Biol. 4 (1), 962. doi:10.1038/s42003-021-02477-4
Coleman-Derr, D., and Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8 (10), e1002988. doi:10.1371/journal.pgen.1002988
Cortijo, S., Charoensawan, V., Brestovitsky, A., Buning, R., Ravarani, C., Rhodes, D., et al. (2017). Transcriptional regulation of the ambient temperature response by H2A.Z nucleosomes and HSF1 transcription factors in Arabidopsis. Mol. Plant 10 (10), 1258–1273. doi:10.1016/j.molp.2017.08.014
Deal, R. B., Topp, C. N., McKinney, E. C., and Meagher, R. B. (2007). Repression of flowering inArabidopsisRequires activation ofFLOWERING LOCUS CExpression by the histone variant H2A.Z. Plant Cell 19 (1), 74–83. doi:10.1105/tpc.106.048447
Duc, C., Benoit, M., Le Goff, S., Simon, L., Poulet, A., Cotterell, S., et al. (2015). The histone chaperone complex HIR maintains nucleosome occupancy and counterbalances impaired histone deposition in CAF-1 complex mutants. Plant J. 81 (5), 707–722. doi:10.1111/tpj.12758
Eberharter, A., and Becker, P. B. (2002). Histone acetylation: a switch between repressive and permissive chromatin - second in review series on chromatin dynamics. Embo Rep. 3 (3), 224–229. doi:10.1093/embo-reports/kvf053
Favero, D. S. (2020). Mechanisms regulating PIF transcription factor activity at the protein level. Physiol. Plant. 169 (3), 325–335. doi:10.1111/ppl.13075
Fiorucci, A. S., Galvao, V. C., Ince, Y. C., Boccaccini, A., Goyal, A., Allenbach Petrolati, L., et al. (2020). PHYTOCHROME INTERACTING FACTOR 7 is important for early responses to elevated temperature in Arabidopsis seedlings. New Phytol. 226 (1), 50–58. doi:10.1111/nph.16316
Franklin, K. A. (2008). Shade avoidance. New Phytol. 179 (4), 930–944. doi:10.1111/j.1469-8137.2008.02507.x
Franklin, K. A., Lee, S. H., Patel, D., Kumar, S. V., Spartz, A. K., Gu, C., et al. (2011). PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc. Natl. Acad. Sci. 108 (50), 20231–20235. doi:10.1073/pnas.1110682108
Franklin, K. A., and Whitelam, G. C. (2005). Phytochromes and shade-avoidance responses in plants. Ann. Bot. 96 (2), 169–175. doi:10.1093/aob/mci165
Gangappa, S. N., Berriri, S., and Kumar, S. V. (2017). PIF4 coordinates thermosensory growth and immunity in Arabidopsis. Curr. Biol. 27 (2), 243–249. doi:10.1016/j.cub.2016.11.012
Gao, H., Song, W., Severing, E., Vayssieres, A., Huettel, B., Franzen, R., et al. (2022). PIF4 enhances DNA binding of CDF2 to co-regulate target gene expression and promote Arabidopsis hypocotyl cell elongation. Nat. Plants 8 (9), 1082–1093. doi:10.1038/s41477-022-01213-y
Gu, D. C., Chen, C. Y., Zhao, M. L., Zhao, L. M., Duan, X. W., Duan, J., et al. (2017). Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 45 (12), 7137–7150. doi:10.1093/nar/gkx283
Guo, J., Cai, G., Li, Y. Q., Zhang, Y. X., Su, Y. N., Yuan, D. Y., et al. (2022). Comprehensive characterization of three classes of Arabidopsis SWI/SNF chromatin remodelling complexes. Nat. Plants 8 (12), 1423–1439. doi:10.1038/s41477-022-01282-z
Guo, Q., Jing, Y. J., Gao, Y., Liu, Y. T., Fang, X. F., and Lin, R. C. (2023). The PIF1/PIF3-MED25-HDA19 transcriptional repression complex regulates phytochrome signaling in Arabidopsis. New Phytol. 240 (3), 1097–1115. doi:10.1111/nph.19205
Hahm, J., Kim, K., Qiu, Y., and Chen, M. (2020). Increasing ambient temperature progressively disassembles Arabidopsis phytochrome B from individual photobodies with distinct thermostabilities. Nat. Commun. 11 (1), 1660. doi:10.1038/s41467-020-15526-z
Han, S. K., Wu, M. F., Cui, S., and Wagner, D. (2015). Roles and activities of chromatin remodeling ATPases in plants. Plant J. 83 (1), 62–77. doi:10.1111/tpj.12877
Han, X., Zhang, Y., Lou, Z., Li, J., Wang, Z., Gao, C., et al. (2023). Time series single-cell transcriptional atlases reveal cell fate differentiation driven by light in Arabidopsis seedlings. Nat. Plants 9 (12), 2095–2109. doi:10.1038/s41477-023-01544-4
He, K. X., Mei, H. L., Zhu, J. P., Qiu, Q., Cao, X. F., and Deng, X. (2022). The histone H3K27 demethylase REF6/JMJ12 promotes thermomorphogenesis in Arabidopsis. Natl. Sci. Rev. 9 (5), nwab213. doi:10.1093/nsr/nwab213
Henikoff, S., and Ahmad, K. (2005). Assembly of variant histones into chromatin. Annu. Rev. Cell Dev. Biol. 21, 133–153. doi:10.1146/annurev.cellbio.21.012704.133518
Hollender, C., and Liu, Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50 (7), 875–885. doi:10.1111/j.1744-7909.2008.00704.x
Huai, J., Zhang, X., Li, J., Ma, T., Zha, P., Jing, Y., et al. (2018). SEUSS and PIF4 coordinately regulate light and temperature signaling pathways to control plant growth. Mol. Plant 11 (7), 928–942. doi:10.1016/j.molp.2018.04.005
Huang, X., Zhang, Q., Jiang, Y. P., Yang, C. W., Wang, Q. Y., and Li, L. (2018). Shade-induced nuclear localization of PIF7 is regulated by phosphorylation and 14-3-3 proteins in Arabidopsis. Elife 7, e31636. doi:10.7554/elife.31636
Huq, E., Al-Sady, B., Hudson, M., Kim, C. H., Apel, K., and Quail, P. H. (2004). PHYTOCHROME-INTERACTING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 (5692), 1937–1941. doi:10.1126/science.1099728
Huq, E., and Quail, P. H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21 (10), 2441–2450. doi:10.1093/emboj/21.10.2441
Hussain, E., Romanowski, A., and Halliday, K. J. (2022). PIF7 controls leaf cell proliferation through an AN3 substitution repression mechanism. Proc. Natl. Acad. Sci. U. S. A. 119 (5), e2115682119. doi:10.1073/pnas.2115682119
Jegu, T., Veluchamy, A., Ramirez-Prado, J. S., Rizzi-Paillet, C., Perez, M., Lhomme, A., et al. (2017). The Arabidopsis SWI/SNF protein BAF60 mediates seedling growth control by modulating DNA accessibility. Genome Biol. 18 (1), 114. doi:10.1186/s13059-017-1246-7
Jeong, J., and Choi, G. (2013). Phytochrome-interacting factors have both shared and distinct biological roles. Mol. Cells 35 (5), 371–380. doi:10.1007/s10059-013-0135-5
Jiang, J. J., Ding, A. B., Liu, F. Q., and Zhong, X. H. (2020). Linking signaling pathways to histone acetylation dynamics in plants. J. Exp. Bot. 71 (17), 5179–5190. doi:10.1093/jxb/eraa202
Jing, Y., and Lin, R. (2020). Transcriptional regulatory network of the light signaling pathways. New Phytol. 227 (3), 683–697. doi:10.1111/nph.16602
Jung, J. H., Domijan, M., Klose, C., Biswas, S., Ezer, D., Gao, M., et al. (2016). Phytochromes function as thermosensors in Arabidopsis. Science 354 (6314), 886–889. doi:10.1126/science.aaf6005
Jung, J. H., Park, J. H., Lee, S., To, T. K., Kim, J. M., Seki, M., et al. (2013). The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. Plant Cell 25 (11), 4378–4390. doi:10.1105/tpc.113.118364
Kidokoro, S., Maruyama, K., Nakashima, K., Imura, Y., Narusaka, Y., Shinwari, Z. K., et al. (2009). The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151 (4), 2046–2057. doi:10.1104/pp.109.147033
Kim, C., Kwon, Y., Jeong, J., Kang, M., Lee, G. S., Moon, J. H., et al. (2023). Phytochrome B photobodies are comprised of phytochrome B and its primary and secondary interacting proteins. Nat. Commun. 14 (1), 1708. doi:10.1038/s41467-023-37421-z
Kim, J., Bordiya, Y., Kathare, P. K., Zhao, B., Zong, W., Huq, E., et al. (2021). Phytochrome B triggers light-dependent chromatin remodelling through the PRC2-associated PHD finger protein VIL1. Nat. Plants 7 (9), 1213–1219. doi:10.1038/s41477-021-00986-y
Kim, J., Song, K., Park, E., Kim, K., Bae, G., and Choi, G. (2016). Epidermal phytochrome B inhibits hypocotyl negative gravitropism non-cell-autonomously. Plant Cell 28 (11), 2770–2785. doi:10.1105/tpc.16.00487
Kim, J. H., Lee, H. J., Jung, J. H., Lee, S., and Park, C. M. (2017). HOS1 facilitates the phytochrome B-mediated inhibition of PIF4 function during hypocotyl growth in Arabidopsis. Mol. Plant 10 (2), 274–284. doi:10.1016/j.molp.2016.11.009
Kim, S., Hwang, G., Kim, S., Thi, T. N., Kim, H., Jeong, J., et al. (2020). The epidermis coordinates thermoresponsive growth through the phyB-PIF4-auxin pathway. Nat. Commun. 11 (1), 1053. doi:10.1038/s41467-020-14905-w
Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., et al. (1999). Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11 (8), 1445–1456. doi:10.2307/3870974
Koini, M. A., Alvey, L., Allen, T., Tilley, C. A., Harberd, N. P., Whitelam, G. C., et al. (2009). High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr. Biol. 19 (5), 408–413. doi:10.1016/j.cub.2009.01.046
Krogan, N. T., Hogan, K., and Long, J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139 (22), 4180–4190. doi:10.1242/dev.085407
Kumar, S. V., Lucyshyn, D., Jaeger, K. E., Alos, E., Alvey, E., Harberd, N. P., et al. (2012). Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484 (7393), 242–245. doi:10.1038/nature10928
Kumar, S. V., and Wigge, P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140 (1), 136–147. doi:10.1016/j.cell.2009.11.006
Kwon, Y., Kim, C., and Choi, G. (2024). Phytochrome B photobody components. New Phytol. 242, 909–915. doi:10.1111/nph.19675
Latrasse, D., Benhamed, M., Henry, Y., Domenichini, S., Kim, W., Zhou, D. X., et al. (2008). The MYST histone acetyltransferases are essential for gametophyte development in Arabidopsis. BMC Plant Biol. 8, 121. doi:10.1186/1471-2229-8-121
Lee, N., Park, J., Kim, K., and Choi, G. (2015). The transcriptional coregulator LEUNIG_HOMOLOG inhibits light-dependent seed germination in Arabidopsis. Plant Cell 27 (8), 2301–2313. doi:10.1105/tpc.15.00444
Legris, M., Klose, C., Burgie, E. S., Rojas, C. C., Neme, M., Hiltbrunner, A., et al. (2016). Phytochrome B integrates light and temperature signals in Arabidopsis. Science 354 (6314), 897–900. doi:10.1126/science.aaf5656
Leivar, P., and Monte, E. (2014). PIFs: systems integrators in plant development. Plant Cell 26 (1), 56–78. doi:10.1105/tpc.113.120857
Leivar, P., Monte, E., Al-Sady, B., Carle, C., Storer, A., Alonso, J. M., et al. (2008). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20 (2), 337–352. doi:10.1105/tpc.107.052142
Leivar, P., and Quail, P. H. (2011). PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 16 (1), 19–28. doi:10.1016/j.tplants.2010.08.003
Lian, T. F., Xu, Y. P., Li, L. F., and Su, X. D. (2017). Crystal structure of tetrameric Arabidopsis MYC2 reveals the mechanism of enhanced interaction with DNA. Cell Rep. 19 (7), 1334–1342. doi:10.1016/j.celrep.2017.04.057
Liu, C., Lu, F., Cui, X., and Cao, X. (2010). Histone methylation in higher plants. Annu. Rev. Plant Biol. 61, 395–420. doi:10.1146/annurev.arplant.043008.091939
Liu, X., Chen, C. Y., Wang, K. C., Luo, M., Tai, R., Yuan, L., et al. (2013). PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25 (4), 1258–1273. doi:10.1105/tpc.113.109710
Lloyd, J. P. B., and Lister, R. (2022). Epigenome plasticity in plants. Nat. Rev. Genet. 23 (1), 55–68. doi:10.1038/s41576-021-00407-y
Long, J. A., Ohno, C., Smith, Z. R., and Meyerowitz, E. M. (2006). TOPLESS regulates apical embryonic fate in Arabidopsis. Science 312 (5779), 1520–1523. doi:10.1126/science.1123841
Lu, F., Cui, X., Zhang, S., Jenuwein, T., and Cao, X. (2011). Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat. Genet. 43 (7), 715–719. doi:10.1038/ng.854
Lu, F., Li, G., Cui, X., Liu, C., Wang, X. J., and Cao, X. (2008). Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J. Integr. Plant Biol. 50 (7), 886–896. doi:10.1111/j.1744-7909.2008.00692.x
Luo, Y. X., Hou, X. M., Zhang, C. J., Tan, L. M., Shao, C. R., Lin, R. N., et al. (2020). A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. EMBO J. 39 (7), e102008. doi:10.15252/embj.2019102008
March-Diaz, R., Garcia-Dominguez, M., Florencio, F. J., and Reyes, J. C. (2007). SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 143 (2), 893–901. doi:10.1104/pp.106.092270
March-Diaz, R., Garcia-Dominguez, M., Lozano-Juste, J., Leon, J., Florencio, F. J., and Reyes, J. C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53 (3), 475–487. doi:10.1111/j.1365-313x.2007.03361.x
Martínez-García, J. F., and Moreno-Romero, J. (2020). Shedding light on the chromatin changes that modulate shade responses. Physiol. Plant. 169 (3), 407–417. doi:10.1111/ppl.13101
Martin-Trillo, M., Lazaro, A., Poethig, R. S., Gomez-Mena, C., Pineiro, M. A., Martinez-Zapater, J. M., et al. (2006). EARLY IN SHORT DAYS 1 (ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development 133 (7), 1241–1252. doi:10.1242/dev.02301
Mosammaparast, N., and Shi, Y. (2010). Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179. doi:10.1146/annurev.biochem.78.070907.103946
Nguyen, N. H., Sng, B. J. R., Chin, H. J., Choi, I. K. Y., Yeo, H. C., and Jang, I. C. (2023). HISTONE DEACETYLASE 9 promotes hypocotyl-specific auxin response under shade. Plant J. 116 (3), 804–822. doi:10.1111/tpj.16410
Ni, M., Tepperman, J. M., and Quail, P. H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 (5), 657–667. doi:10.1016/s0092-8674(00)81636-0
Nie, W. F., Lei, M., Zhang, M., Tang, K., Huang, H., Zhang, C., et al. (2019). Histone acetylation recruits the SWR1 complex to regulate active DNA demethylation in Arabidopsis. Proc. Natl. Acad. Sci. U. S. A. 116 (33), 16641–16650. doi:10.1073/pnas.1906023116
Nie, X., Wang, H., Li, J., Holec, S., and Berger, F. (2014). The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol. Open 3 (9), 794–802. doi:10.1242/bio.20148680
Noh, Y. S., and Amasino, R. M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15 (7), 1671–1682. doi:10.1105/tpc.012161
Oh, E., Kim, J., Park, E., Kim, J. I., Kang, C., and Choi, G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 (11), 3045–3058. doi:10.1105/tpc.104.025163
Paik, I., Kathare, P. K., Kim, J. I., and Huq, E. (2017). Expanding roles of PIFs in signal integration from multiple processes. Mol. Plant 10 (8), 1035–1046. doi:10.1016/j.molp.2017.07.002
Pandey, R., Muller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30 (23), 5036–5055. doi:10.1093/nar/gkf660
Panigrahi, A., and O'Malley, B. W. (2021). Mechanisms of enhancer action: the known and the unknown. Genome Biol. 22 (1), 108. doi:10.1186/s13059-021-02322-1
Papamichos-Chronakis, M., Watanabe, S., Rando, O. J., and Peterson, C. L. (2011). Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell 144 (2), 200–213. doi:10.1016/j.cell.2010.12.021
Pardi, S. A., and Nusinow, D. A. (2021). Out of the dark and into the light: a new view of phytochrome photobodies. Front. Plant Sci. 12, 732947. doi:10.3389/fpls.2021.732947
Peng, M., Li, Z., Zhou, N., Ma, M., Jiang, Y., Dong, A., et al. (2018). Linking PHYTOCHROME-INTERACTING FACTOR to histone modification in plant shade avoidance. Plant Physiol. 176 (2), 1341–1351. doi:10.1104/pp.17.01189
Perrella, G., and Kaiserli, E. (2016). Light behind the curtain: photoregulation of nuclear architecture and chromatin dynamics in plants. New Phytol. 212 (4), 908–919. doi:10.1111/nph.14269
Perrella, G., Zioutopoulou, A., Headland, L. R., and Kaiserli, E. (2020). The impact of light and temperature on chromatin organization and plant adaptation. J. Exp. Bot. 71 (17), 5247–5255. doi:10.1093/jxb/eraa154
Pham, V. N., Kathare, P. K., and Huq, E. (2018). Phytochromes and phytochrome interacting factors. Plant Physiol. 176 (2), 1025–1038. doi:10.1104/pp.17.01384
Plant, A. R., Larrieu, A., and Causier, B. (2021). Repressor for hire! The vital roles of TOPLESS-mediated transcriptional repression in plants. New Phytol. 231 (3), 963–973. doi:10.1111/nph.17428
Proveniers, M. C. G., and van Zanten, M. (2013). High temperature acclimation through PIF4 signaling. Trends Plant Sci. 18 (2), 59–64. doi:10.1016/j.tplants.2012.09.002
Quail, P. H., Boylan, M. T., Parks, B. M., Short, T. W., Xu, Y., and Wagner, D. (1995). Phytochromes: photosensory perception and signal transduction. Science. 268 (5211), 675–680. doi:10.1126/science.7732376
Rivera, C. M., and Ren, B. (2013). Mapping human epigenomes. Cell 155 (1), 39–55. doi:10.1016/j.cell.2013.09.011
Rockwell, N. C., Su, Y. S., and Lagarias, J. C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858. doi:10.1146/annurev.arplant.56.032604.144208
Sakamoto, K., and Nagatani, A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10 (5), 859–868. doi:10.1046/j.1365-313x.1996.10050859.x
Sessa, G., Carabelli, M., Possenti, M., Morelli, G., and Ruberti, I. (2018). Multiple pathways in the control of the shade avoidance response. Plants (Basel) 7 (4), 102. doi:10.3390/plants7040102
Shan, L., Li, P., Yu, H., and Chen, L. L. (2023). Emerging roles of nuclear bodies in genome spatial organization. Trends Cell Biol. doi:10.1016/j.tcb.2023.10.012
Shang, J. Y., Lu, Y. J., Cai, X. W., Su, Y. N., Feng, C., Li, L., et al. (2021). COMPASS functions as a module of the INO80 chromatin remodeling complex to mediate histone H3K4 methylation in Arabidopsis. Plant Cell 33 (10), 3250–3271. doi:10.1093/plcell/koab187
Shapulatov, U., van Zanten, M., van Hoogdalem, M., Meisenburg, M., van Hall, A., Kappers, I., et al. (2023). The Mediator complex subunit MED25 interacts with HDA9 and PIF4 to regulate thermomorphogenesis. Plant Physiol. 192 (1), 582–600. doi:10.1093/plphys/kiac581
Shen, H., Zhu, L., Castillon, A., Majee, M., Downie, B., and Huq, E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20 (6), 1586–1602. doi:10.1105/tpc.108.060020
Shen, Y., Lei, T. T., Cui, X. Y., Liu, X. Y., Zhou, S. L., Zheng, Y., et al. (2019). Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. Plant J. 100 (5), 991–1006. doi:10.1111/tpj.14492
Shin, J., Anwer, M. U., and Davis, S. J. (2013). Phytochrome-interacting factors (PIFs) as bridges between environmental signals and the circadian clock: diurnal regulation of growth and development. Mol. Plant 6 (3), 592–595. doi:10.1093/mp/sst060
Shvedunova, M., and Akhtar, A. (2022). Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 23 (5), 329–349. doi:10.1038/s41580-021-00441-y
Smaczniak, C., Li, N., Boeren, S., America, T., van Dongen, W., Goerdayal, S. S., et al. (2012). Proteomics-based identification of low-abundance signaling and regulatory protein complexes in native plant tissues. Nat. Protoc. 7 (12), 2144–2158. doi:10.1038/nprot.2012.129
Soutourina, J. (2018). Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 19 (4), 262–274. doi:10.1038/nrm.2017.115
Steindler, C., Matteucci, A., Sessa, G., Weimar, T., Ohgishi, M., Aoyama, T., et al. (1999). Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126 (19), 4235–4245. doi:10.1242/dev.126.19.4235
Sun, J. Q., Qi, L. L., Li, Y. N., Chu, J. F., and Li, C. Y. (2012). PIF4–Mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. Plos Genet. 8 (3), e1002594. doi:10.1371/journal.pgen.1002594
Sun, W., Han, H., Deng, L., Sun, C., Xu, Y., Lin, L., et al. (2020). Mediator subunit MED25 physically interacts with PHYTOCHROME INTERACTING FACTOR4 to regulate shade-induced hypocotyl elongation in tomato. Plant Physiol. 184 (3), 1549–1562. doi:10.1104/pp.20.00587
Sura, W., Kabza, M., Karlowski, W. M., Bieluszewski, T., Kus-Slowinska, M., Paweloszek, L., et al. (2017). Dual role of the histone variant H2A.Z in transcriptional regulation of stress-response genes. Plant Cell 29 (4), 791–807. doi:10.1105/tpc.16.00573
Sureshkumar, S., and Balasubramanian, S. (2021). Complexes and complexities: INO80 takes center stage. Mol. Plant 14 (11), 1776–1778. doi:10.1016/j.molp.2021.08.012
Tagami, H., Ray-Gallet, D., Almouzni, G., and Nakatani, Y. (2004). Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116 (1), 51–61. doi:10.1016/s0092-8674(03)01064-x
Tasset, C., Singh Yadav, A., Sureshkumar, S., Singh, R., van der Woude, L., Nekrasov, M., et al. (2018). POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 14 (3), e1007280. doi:10.1371/journal.pgen.1007280
Van Buskirk, E. K., Decker, P. V., and Chen, M. (2012). Photobodies in light signaling. Plant Physiol. 158 (1), 52–60. doi:10.1104/pp.111.186411
van der Woude, L. C., Perrella, G., Snoek, B. L., van Hoogdalem, M., Novak, O., van Verk, M. C., et al. (2019). HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proc. Natl. Acad. Sci. 116 (50), 25343–25354. doi:10.1073/pnas.1911694116
Wang, H., Fan, Z., Shliaha, P. V., Miele, M., Hendrickson, R. C., Jiang, X. J., et al. (2023). H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature 615 (7951), 339–348. doi:10.1038/s41586-023-05780-8
Wang, H., Li, S., Li, Y., Xu, Y., Wang, Y., Zhang, R., et al. (2019). MED25 connects enhancer-promoter looping and MYC2-dependent activation of jasmonate signalling. Nat. plants 5 (6), 616–625. doi:10.1038/s41477-019-0441-9
Wang, L., Kim, J., and Somers, D. E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. U. S. A. 110 (2), 761–766. doi:10.1073/pnas.1215010110
Wang, W., Kim, J., Martinez, T. S., Huq, E., and Sung, S. (2024). COP1 controls light-dependent chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 121 (8), e2312853121. doi:10.1073/pnas.2312853121
Willige, B. C., Yoo, C. Y., and Saldierna Guzman, J. P. (2024). What is going on inside of phytochrome B photobodies? Plant Cell, koae084. doi:10.1093/plcell/koae084
Willige, B. C., Zander, M., Yoo, C. Y., Phan, A., Garza, R. M., Wanamaker, S. A., et al. (2021). PHYTOCHROME-INTERACTING FACTORs trigger environmentally responsive chromatin dynamics in plants. Nat. Genet. 53 (7), 955–961. doi:10.1038/s41588-021-00882-3
Wollmann, H., Stroud, H., Yelagandula, R., Tarutani, Y., Jiang, D., Jing, L., et al. (2017). The histone H3 variant H3.3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol. 18 (1), 94. doi:10.1186/s13059-017-1221-3
Xiao, J., Lee, U. S., and Wagner, D. (2016). Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 34, 41–53. doi:10.1016/j.pbi.2016.08.002
Xie, Y., Liu, W., Liang, W., Ling, X., Ma, J., Yang, C., et al. (2023). Phytochrome B inhibits the activity of phytochrome-interacting factor 7 involving phase separation. Cell Rep. 42 (12), 113562. doi:10.1016/j.celrep.2023.113562
Xu, Y. F., Gan, E. S., Zhou, J., Wee, W. Y., Zhang, X. Y., and Ito, T. (2014). Arabidopsis MRG domain proteins bridge two histone modifications to elevate expression of flowering genes. Nucleic Acids Res. 42 (17), 10960–10974. doi:10.1093/nar/gku781
Xue, M., Zhang, H., Zhao, F., Zhao, T., Li, H., and Jiang, D. (2021). The INO80 chromatin remodeling complex promotes thermomorphogenesis by connecting H2A.Z eviction and active transcription in Arabidopsis. Mol. Plant 14 (11), 1799–1813. doi:10.1016/j.molp.2021.07.001
Yang, C., Zhu, T., Zhou, N., Huang, S., Zeng, Y., Jiang, W., et al. (2023). PIF7-mediated epigenetic reprogramming promotes the transcriptional response to shade in Arabidopsis. EMBO J. 42 (8), e111472. doi:10.15252/embj.2022111472
Yi, H., Sardesai, N., Fujinuma, T., Chan, C. W., Veena, G. S. B., and Gelvin, S. B. (2006). Constitutive expression exposes functional redundancy between the Arabidopsis histone H2A gene HTA1 and other H2A gene family members. Plant Cell 18 (7), 1575–1589. doi:10.1105/tpc.105.039719
Zander, M., Willige, B. C., He, Y., Nguyen, T. A., Langford, A. E., Nehring, R., et al. (2019). Epigenetic silencing of a multifunctional plant stress regulator. eLife 8, e47835. doi:10.7554/elife.47835
Zhai, H., Zhang, X., You, Y., Lin, L., Zhou, W., and Li, C. (2020). SEUSS integrates transcriptional and epigenetic control of root stem cell organizer specification. EMBO J. 39 (20), e105047. doi:10.15252/embj.2020105047
Zhang, D., Jing, Y., Jiang, Z., and Lin, R. (2014). The chromatin-remodeling factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26 (6), 2472–2485. doi:10.1105/tpc.113.121848
Zhang, D., Li, Y., Zhang, X., Zha, P., and Lin, R. (2017). The SWI2/SNF2 chromatin-remodeling ATPase BRAHMA regulates chlorophyll biosynthesis in Arabidopsis. Mol. Plant 10 (1), 155–167. doi:10.1016/j.molp.2016.11.003
Zhao, F., Xue, M., Zhang, H., Li, H., Zhao, T., and Jiang, D. (2023). Coordinated histone variant H2A.Z eviction and H3.3 deposition control plant thermomorphogenesis. New Phytol. 238 (2), 750–764. doi:10.1111/nph.18738
Zhao, Q., Dai, B. D., Wu, H. Y., Zhu, W. C., and Chen, J. Y. (2022). Ino80 is required for H2A.Z eviction from hypha-specific promoters and hyphal development of Candida albicans. Mol. Microbiol. 118 (1-2), 92–104. doi:10.1111/mmi.14954
Zhong, Z., Wang, Y., Wang, M., Yang, F., Thomas, Q. A., Xue, Y., et al. (2022). Histone chaperone ASF1 mediates H3.3-H4 deposition in Arabidopsis. Nat. Commun. 13 (1), 6970. doi:10.1038/s41467-022-34648-0
Zhou, N., Li, C., Xie, W., Liang, N., Wang, J., Wang, B., et al. (2024). Histone methylation readers MRG1/2 interact with PIF4 to promote thermomorphogenesis in Arabidopsis. Cell Rep. 43 (2), 113726. doi:10.1016/j.celrep.2024.113726
Zhou, Y., Park, S. H., Soh, M. Y., and Chua, N. H. (2021). Ubiquitin-specific proteases UBP12 and UBP13 promote shade avoidance response by enhancing PIF7 stability. Proc. Natl. Acad. Sci. U. S. A. 118 (45), e2103633118. doi:10.1073/pnas.2103633118
Keywords: plant epigenomics, chromatin dynamics, transcription factors, chromatin remodeling complexes, light and temperature signaling
Citation: Ammari M, Maseh K and Zander M (2024) PIF transcription factors-versatile plant epigenome landscapers. Front. Epigenet. Epigenom. 2:1404958. doi: 10.3389/freae.2024.1404958
Received: 22 March 2024; Accepted: 30 April 2024;
Published: 16 May 2024.
Edited by:
Raul Mostoslavsky, Massachusetts General Hospital Cancer Center, United StatesReviewed by:
Jordi Moreno-Romero, Autonomous University of Barcelona, SpainCopyright © 2024 Ammari, Maseh and Zander. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark Zander, bXphbmRlckB3YWtzbWFuLnJ1dGdlcnMuZWR1
†These authors have contributed equally to this work
 Moonia Ammari†
Moonia Ammari† Mark Zander
Mark Zander