- Department of Plant Biology and Genome Center, University of California, Davis, CA, United States
Plant epigenetic studies have revealed that developmental or environmental events can trigger both local and global epigenetic remodeling. In multiple cases, transposable elements (TE) respond to the trigger and act as mediators. Epigenetic remodeling results in mitotically and even meiotically persistent states that impact phenotype and could contribute to its plasticity. The challenge is to understand the mechanisms that trigger and mediate remodeling, their evolutionary role, and their potential in breeding.
Introduction
Plants life cycles display great diversity, from the short sexual cycles of annuals such as arabidopsis and cultivated rice, to the clonal persistence of invasive weeds such as knotweed (Hollingsworth and Bailey, 2000) and cordgrass (Ainouche et al, 2009), and to the long lives of forest species, such as juniper, fir, and aspen (de Witte and Stöcklin, 2010). Plants are sessile and must adapt to their varying environment. Mitotically stable changes alone have great importance for large and long-lived clones that encounter great spatial and temporal environmental variation. For plants with shorter cycles, both mitotic and meiotically persistent epigenetic changes could play a role in fitness. The existence of this potential is a critical question at a time of accelerated climate change.
Developmental epigenetics in plants
We have made progress in understanding programmed epigenetic switches occurring during reproductive development. These are recurring changes associated with the development of flowers, fruits and seeds. Flowering in arabidopsis is one of the best characterized epigenetic switches. FLOWERING LOCUS C (FLC), a repressor of flowering, integrates signals from three pathways, including one involving antisense RNAs, to undergo persistent silencing by the Polycomb Repressive Complex (PRC) and generational resetting by histone demethylases (Whittaker and Dean, 2017). Certain specialized tissues essential for reproduction undergo large-scale epigenetic changes: fleshy tissues in fruits, the central cell and endosperm, and the vegetative cell of pollen. These tissues are distinct, non-proliferating branches of plant development. Fleshy fruits exhibit two physiological ripening responses: climacteric ones, such as tomato, exhibit a postharvest, ethylene-triggered respiratory burst (Chalmers and Rowan, 1971; Gallusci et al, 2016). Non-climacteric, such as oranges, do not. In both types, ripening can be regulated by large-scale changes in DNA methylation (Gallusci et al, 2016). In tomato fruit, DNA methylation at many loci opposes the action of ethylene, preventing early ripening. Demethylating enzymes (Liu et al, 2015) remove methyl-C marks. Demethylation plays a role in strawberry, a non-climacteric fruit, and is mediated by downregulation of RNA-dependent DNA methylation (RdDM) (Cheng et al, 2018). The opposite happens in oranges, where non-climacteric ripening correlates with hypermethylation (Huang et al, 2019). In developing seed endosperm, a nutritional organ resulting from independent fertilization, parental imprinting of key seed factors depends on the maternal action of DNA demethylases and on the maternal establishment of PRC-dependent repressive chromatin (Gehring, 2013; Moreno-Romero et al, 2019). In the pollen vegetative cell, a non-dividing cell supporting male reproduction, epigenetic marks in transposable elements (TE) undergo programmed relaxation (Slotkin et al, 2009) and produce small RNA capable of interacting with sperm targets (Calarco and Martienssen, 2011). Small RNAs are thought to orchestrate epigenetic genomic changes at various stages of reproduction, including the haploid gametophytic stage although the specific mechanisms are still being elucidated (Martinez and Köhler, 2017). TEs are not the only genomic feature to lose a genomic mark persistent in the Soma: centromeres undergo programmed loss of their characteristic chromatin via proteasomal processing of sumoylated CENH3 (Mérai et al, 2014). Taken together, the epigenetic changes documented at the different developmental stages of flowering, fruiting, and seed production demonstrate the flexible and varied regulation of developmental chromatin marks and indicate a key role in reproductive fitness.
Epigenetic responses to stress
Many studies have documented the effect of stress, biotic and abiotic, on epigenetic regulation: see Gallusci (2023) for a recent review. How does stress affect epigenetic regulation? A take-home lesson from FLOWERING LOCUS C regulation is that cold and day length signaling are integrated into the developmental regulation leading to epigenetic imprinting (Whittaker and Dean, 2017). Because many plant species are exposed to cold winters, they have evolved sophisticated chromatin-based mechanisms to ensure that they flower only in favorable times. Other environmental stresses recur. In most environments, even mesic ones, drought and heat periods are expected on a yearly basis. Multiple papers have documented environmentally triggered epigenetic changes affecting DNA and histone marks of key genes (Chang et al, 2020; Gallusci et al, 2023). These findings suggest that stress signals engender fitness-promoting epigenetic memories. It remains unclear what molecular mechanisms transduce environmental and physiological clues into epigenetic remodeling, what regulates their persistence, and to what degree these states endure through generations.
Transposon regulation
Studies of epigenetic changes caused by tissue culture suggest that triggers are mediated by transposon regulation, consistent with early insights by Barbara (McClintock, 1984). Tissue culture involving shoot regeneration from organ explants or from single cell protoplasts combines multiple stresses because plant cells are exposed to wounding, to cell wall digestion by fungal enzymes (in the case of protoplasts), to unusual light and hormonal conditions, and are induced to proliferate and organize into shoot meristems. A few hours after exposure to protoplasting treatments, multiple species display strong activation of silent TEs (Pouteau et al, 1991; Hirochika, 1993; Pouteau et al, 1994). Rapid proliferation induced during tissue culture can destabilize DNA methylation (Borges et al, 2021), but the timing of TE induction suggests that at the least the early response is independent of the replication-dependent dilution of methyl-C observed during cell cycle acceleration. While methylation marks can be re-established, transposon activation has persistent effects. A disrupting effect depends on transposon excision and insertion at novel sites, resulting in DNA breaks, genome instability, and relocation of potential regulatory elements (Bennetzen and Wang, 2014). Indeed, both genetic changes from DNA instability and increased epigenetic changes contribute to somaclonal variation (Phillips et al, 1994; Kaeppler et al, 2000; Ong-Abdullah et al, 2015; Fossi et al, 2019; Borges et al, 2021). Another effect involves remodeling of chromatin states, particularly those associated with homologous copies of the activated transposons (Figure 1). Transposons can confer upon associated genes novel regulatory properties that depend on the TE epigenetic state (Bennetzen and Wang, 2014). Tissue culture of oil palm is associated with demethylation of the Karma TE and expression of the connected MANTLED gene resulting in undesirable mantling of the seed (Ong-Abdullah et al, 2015). Remarkably, transposons are often sensitive to stress and can confer stress sensitive regulation to nearby genes (Wessler, 1996; Cavrak et al, 2014). Independent insertion of two retroelements near the Ruby gene of sweet orange conferred cold-dependent activation of anthocyanin expression (Butelli et al, 2012). In rice, novel transposition of TE mPing to new sites resulted in stress-dependent activation of the connected genes (Naito et al, 2009). In summary, when a transposon or a transposon fragment is located near a gene, it can sensitize that gene to the status of cognate TEs. Because these are often sensitive to stress, the gene can become stress responsive. In conclusion, TE can sense multiple stresses and remodel the epigenome.
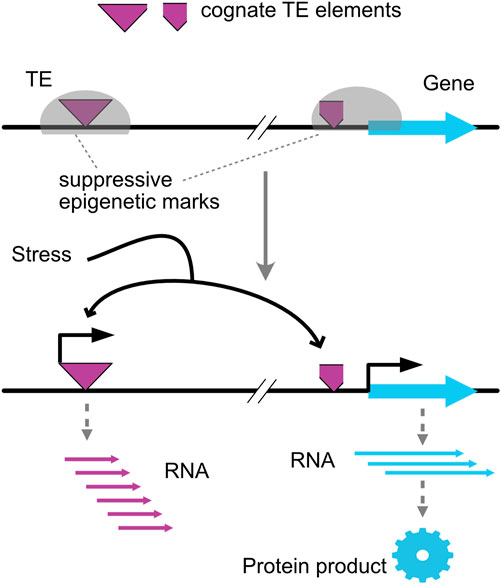
FIGURE 1. TE-mediated regulation of genes. A transposable element (TE) and its cognate element share epigenetic states. Stress can trigger the activation of the TE and induce expression of the proximal gene (Wessler, 1996; Butelli et al, 2012; Cavrak et al, 2014).
Epialleles
An epiallele affects a trait without underlying DNA changes. Plant epialleles can confer qualitative and quantitative phenotypes and can persist through generations (Cubas et al, 1999; Mittelsten Scheid et al, 2003; Manning et al, 2006; Weigel and Colot, 2012; Quadrana et al, 2014; Zeng and Cheng, 2014; Baduel and Colot, 2021). Most persistent epialleles result from DNA methylation marks that reside inside a TE or spread from a TE. If a novel gene-TE association that results in altered regulation increases fitness, the polymorphism should experience positive selection. When associated with a persistent or potential epigenetic mark, sensitized loci can be identified when DNA methylation homeostasis is perturbed. For example, mutation in MET1 or DDM1, respectively the major CpG methyltransferase and a chromatin factor needed for methylation maintenance, generates epialleles either by loss of methylation (Baduel and Colot, 2021), or by hypermethylation (Saze and Kakutani, 2007; Yi and Richards, 2008). Genome wide removal of methylation marks is largely reversible by complementation of the methylation defect, but some loci remain unmethylated providing allelic variation that can underlie QTL. Reversibility is affected by RdDM and the strength of the silencing signal provided by the cognate TE family (Baduel and Colot, 2021). Thus, biotic and abiotic stresses activate TEs, which in turn create and modulate epialleles. Expressed or silenced epialleles can flip state as a result of extreme life history events.
Discussion
The effects of epigenetic marks on genes raises the questions of what initiates, destabilizes, and resets them. At least in part, the answers may lay in the co-evolution of parasitic genomic elements (and related viruses) with the host. As exemplified by the sophisticated sensing of the host state by lysogenic λ bacteriophage, selfish elements co-wire their regulation with host stress pathways (Rokney et al, 2008). In the case of plants, hormonal networks influence many regulatory decisions, both developmental and stress-dependent (Waadt et al, 2022): for example, imprinting-dependent phenotypes in developing seeds are exquisitely sensitive to regulation by flavonoids and auxins (Dilkes et al, 2008; Köhler et al, 2021). Epigenetic remodelers may thus directly or indirectly respond to hormonal regulation (Liu et al, 2015). Elucidating the changes of epigenetic states during development is a grand challenge. To what degree do different types of marks, such as those determined by the PRC, depend on TEs? Why do many TEs respond to stress? How do hormones participate in their regulation? Activation when the host is in danger makes good sense for viruses, but what do cell-bound elements gain from this strategy? How is cellular and developmental homeostasis coupled to epigenetic states?
Answering these questions will contribute to our basic understanding of epigenetic phenomena. Further, it will help elucidate the contribution of epigenetic state to evolution and the potential exploitation of epigenetic regulation by plant breeders. Over a quarter century ago, Rasmusson and Phillips proposed that unexpected phenotypic variation observed in relatively narrowly based breeding programs could, at least in part, be based on epigenetic changes (Rasmusson and Phillips, 1997). Methylation-dependent epialleles could thus contribute to variation useful for breeding (Tsaftaris and Polidoros, 1999). Experimental evidence supporting the influence of epialleles on quantitative traits relevant to agriculture has multiplied (Baduel and Colot, 2021). Mutation of mitochondrial MutS Homolog 1 affects DNA methylation homeostasis in the nuclear genome, influencing agronomic traits (Yang et al, 2015). It is possible, therefore, that natural and induced epigenetic manipulation could increase breeding efficiency (Springer and Schmitz, 2017; Gahlaut et al, 2020). To play an economic role, however, agronomic epialleles must be sufficiently stable, emphasizing the importance of understanding triggering and resetting mechanisms.
Notwithstanding the considerable interest in their potential, there is no conclusive evidence that epialleles have contributed to evolution and that they can be exploited for plant breeding. Skeptical views are useful: selection on homozygous lines that show phenotypic variation has failed (Charlesworth et al, 2017). Virtually all significant traits characterized have a genetic origin (Doebley et al, 2006; Weigel and Colot, 2012). Furthermore, most epialleles are genes sensitized by a genetic change, such as a TE insertion nearby or the formation of a transactive inverted repeat (Jablonka and Lamb, 1998; Melquist et al, 1999). These considerations should stimulate careful reflections on their role and on the type of evidence necessary to attribute an evolutionary role and to deploy them for breeding. For example, epialleles could serve as crutches conferring short term fitness in dire circumstances and facilitating the development of more persistent genetic solutions (Jablonka and Lamb, 1998; Kalisz and Purugganan, 2004). Plants are useful experimental systems to test these hypotheses.
In conclusion, scientific studies that describe the causes, mechanisms, and impact of changes in epigenetic states will broadly benefit the field of epigenetics and genome regulation.
Author contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Funding
Financial support for this research was provided by National Science Foundation Plant Genome Integrative Organismal Systems (IOS) Grant 1956429 (Variants and Recombinants without Meiosis) to LC.
Conflict of interest
The author LC declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ainouche, M. L., Fortune, P. M., Salmon, A., Parisod, C., Grandbastien, M.-A., Fukunaga, K., et al. (2009). Hybridization, polyploidy and invasion: Lessons from spartina (poaceae). Biol. Invasions 11, 1159–1173. doi:10.1007/s10530-008-9383-2
Baduel, P., and Colot, V. (2021). The epiallelic potential of transposable elements and its evolutionary significance in plants. Philosophical Trans. R. Soc. B Biol. Sci. 376, 20200123. doi:10.1098/rstb.2020.0123
Bennetzen, J. L., and Wang, H. (2014). The contributions of transposable elements to the structure, function, and evolution of plant genomes. Annu. Rev. Plant Biol. 65, 505–530. doi:10.1146/annurev-arplant-050213-035811
Borges, F., Donoghue, M. T. A., LeBlanc, C., Wear, E. E., Tanurdžić, M., Berube, B., et al. (2021). Loss of small-RNA-directed DNA methylation in the plant cell cycle promotes germline reprogramming and somaclonal variation. Curr. Biol. 31, 591–600.e4. doi:10.1016/j.cub.2020.10.098
Butelli, E., Licciardello, C., Zhang, Y., Liu, J., Mackay, S., Bailey, P., et al. (2012). Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell 24, 1242–1255. doi:10.1105/tpc.111.095232
Calarco, J. P., and Martienssen, R. A. (2011). Genome reprogramming and small interfering RNA in the Arabidopsis germline. Curr. Opin. Genet. Dev. 21, 134–139. doi:10.1016/j.gde.2011.01.014
Cavrak, V. V., Lettner, N., Jamge, S., Kosarewicz, A., Bayer, L. M., and Mittelsten Scheid, O. (2014). How a retrotransposon exploits the plant’s heat stress response for its activation. PLoS Genet. 10, e1004115. doi:10.1371/journal.pgen.1004115
Chalmers, D. J., and Rowan, K. S. (1971). The climacteric in ripening tomato fruit. Plant Physiol. 48, 235–240. doi:10.1104/pp.48.3.235
Chang, Y.-N., Zhu, C., Jiang, J., Zhang, H., Zhu, J.-K., and Duan, C.-G. (2020). Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 62, 563–580. doi:10.1111/jipb.12901
Charlesworth, D., Barton, N. H., and Charlesworth, B. (2017). The sources of adaptive variation. Proc. R. Soc. B Biol. Sci. 284, 20162864. doi:10.1098/rspb.2016.2864
Cheng, J., Niu, Q., Zhang, B., Chen, K., Yang, R., Zhu, J.-K., et al. (2018). Downregulation of RdDM during strawberry fruit ripening. Genome Biol. 19, 212. doi:10.1186/s13059-018-1587-x
Cubas, P., Vincent, C., and Coen, E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401, 157–161. doi:10.1038/43657
de Witte, L. C., and Stöcklin, J. (2010). Longevity of clonal plants: Why it matters and how to measure it. Ann. Bot. 106, 859–870. doi:10.1093/aob/mcq191
Dilkes, B. P., Spielman, M., Weizbauer, R., Watson, B., Burkart-Waco, D., Scott, R. J., et al. (2008). The maternally expressed WRKY transcription factor TTG2 controls lethality in interploidy crosses of Arabidopsis. PLoS Biol. 6, e308–e2720. doi:10.1371/journal.pbio.0060308
Doebley, J. F., Gaut, B. S., and Smith, B. D. (2006). The molecular genetics of crop domestication. Cell 127, 1309–1321. doi:10.1016/j.cell.2006.12.006
Fossi, M., Amundson, K., Kuppu, S., Britt, A., and Comai, L. (2019). Regeneration of Solanum tuberosum plants from protoplasts induces widespread genome instability. Plant Physiol. 180, 78–86. doi:10.1104/pp.18.00906
Gahlaut, V., Zinta, G., Jaiswal, V., and Kumar, S. (2020). Quantitative epigenetics: A new avenue for crop improvement. Epigenomes 4, 25. doi:10.3390/epigenomes4040025
Gallusci, P., Agius, D. R., Moschou, P. N., Dobránszki, J., Kaiserli, E., and Martinelli, F. (2023). Deep inside the epigenetic memories of stressed plants. Trends Plant Sci. 28, 142–153. doi:10.1016/j.tplants.2022.09.004
Gallusci, P., Hodgman, C., Teyssier, E., and Seymour, G. B. (2016). DNA methylation and chromatin regulation during fleshy fruit development and ripening. Front. Plant Sci. 7, 807. doi:10.3389/fpls.2016.00807
Gehring, M. (2013). Genomic imprinting: Insights from plants. Annu. Rev. Genet. 47, 187–208. doi:10.1146/annurev-genet-110711-155527
Hirochika, H. (1993). Activation of tobacco retrotransposons during tissue culture. EMBO J. 12, 2521–2528. doi:10.1002/j.1460-2075.1993.tb05907.x
Hollingsworth, M. L., and Bailey, J. P. (2000). Evidence for massive clonal growth in the invasive weed Fallopia japonica (Japanese Knotweed). Botanical J. Linn. Soc. 133, 463–472. doi:10.1006/bojl.2000.0359
Huang, H., Liu, R., Niu, Q., Tang, K., Zhang, B., Zhang, H., et al. (2019). Global increase in DNA methylation during orange fruit development and ripening. Proc. Natl. Acad. Sci. U. S. A. 116, 1430–1436. doi:10.1073/pnas.1815441116
Jablonka, E., and Lamb, M. J. (1998). Epigenetic inheritance in evolution. J. Evol. Biol. 11, 159–183. doi:10.1046/j.1420-9101.1998.11020159.x
Kaeppler, S. M., Kaeppler, H. F., and Rhee, Y. (2000). Epigenetic aspects of somaclonal variation in plants. Plant Mol. Biol. 43, 179–188. doi:10.1023/a:1006423110134
Kalisz, S., and Purugganan, M. D. (2004). Epialleles via DNA methylation: Consequences for plant evolution. Trends Ecol. Evol. 19, 309–314. doi:10.1016/j.tree.2004.03.034
Köhler, C., Dziasek, K., and Del Toro-De León, G. (2021). Postzygotic reproductive isolation established in the endosperm: Mechanisms, drivers and relevance. Philosophical Trans. R. Soc. B Biol. Sci. 376, 20200118. doi:10.1098/rstb.2020.0118
Liu, R., Halle, S., Liu, M., Kong, J., Wu, C., Degraeve-Guibault, C., et al. (2015). A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. U. S. A. 112, 10804–10809. doi:10.1073/pnas.1503362112
Manning, K., Tör, M., Poole, M., Hong, Y., Thompson, A. J., King, G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38, 948–952. doi:10.1038/ng1841
Martinez, G., and Köhler, C. (2017). Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Curr. Opin. Plant Biol. 36, 22–28. doi:10.1016/j.pbi.2016.12.006
McClintock, B. (1984). The significance of responses of the genome to challenge. Science 226, 792–801. doi:10.1126/science.15739260
Melquist, S., Luff, B., and Bender, J. (1999). Arabidopsis PAI gene arrangements, cytosine methylation and expression. Genetics 153, 401–413. doi:10.1093/genetics/153.1.401
Mérai, Z., Chumak, N., García-Aguilar, M., Hsieh, T.-F., Nishimura, T., Schoft, V. K., et al. (2014). The AAA-ATPase molecular chaperone Cdc48/p97 disassembles sumoylated centromeres, decondenses heterochromatin, and activates ribosomal RNA genes. Proc. Natl. Acad. Sci. U. S. A. 111, 16166–16171. doi:10.1073/pnas.1418564111
Mittelsten Scheid, O., Afsar, K., and Paszkowski, J. (2003). Formation of stable epialleles and their paramutation-like interaction in tetraploid Arabidopsis thaliana. Nat. Genet. 34, 450–454. doi:10.1038/ng1210
Moreno-Romero, J., Del Toro-De León, G., Yadav, V. K., Santos-González, J., and Köhler, C. (2019). Epigenetic signatures associated with imprinted paternally expressed genes in the Arabidopsis endosperm. Genome Biol. 20, 41. doi:10.1186/s13059-019-1652-0
Naito, K., Zhang, F., Tsukiyama, T., Saito, H., Hancock, C. N., Richardson, A. O., et al. (2009). Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature 461, 1130–1134. doi:10.1038/nature08479
Ong-Abdullah, M., Ordway, J. M., Jiang, N., Ooi, S.-E., Kok, S.-Y., Sarpan, N., et al. (2015). Loss of Karma transposon methylation underlies the mantled somaclonal variant of oil palm. Nature 525, 533–537. doi:10.1038/nature15365
Phillips, R. L., Kaeppler, S. M., and Olhoft, P. (1994). Genetic instability of plant tissue cultures: Breakdown of normal controls. Proc. Natl. Acad. Sci. U. S. A. 91, 5222–5226. doi:10.1073/pnas.91.12.5222
Pouteau, S., Grandbastien, M.-A., and Boccara, M. (1994). Microbial elicitors of plant defence responses activate transcription of a retrotransposon. Plant J. 5, 535–542. doi:10.1046/j.1365-313x.1994.5040535.x
Pouteau, S., Huttner, E., Grandbastien, M. A., and Caboche, M. (1991). Specific expression of the tobacco Tnt1 retrotransposon in protoplasts. EMBO J. 10, 1911–1918. doi:10.1002/j.1460-2075.1991.tb07717.x
Quadrana, L., Almeida, J., Asís, R., Duffy, T., Dominguez, P. G., Bermúdez, L., et al. (2014). Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 5, 4027. doi:10.1038/ncomms5027
Rasmusson, D. C., and Phillips, R. L. (1997). Plant breeding progress and genetic diversity from de novo variation and elevated epistasis. Crop Sci. 37, 303–310. doi:10.2135/cropsci1997.0011183X003700020001x
Rokney, A., Kobiler, O., Amir, A., Court, D. L., Stavans, J., Adhya, S., et al. (2008). Host responses influence on the induction of lambda prophage. Mol. Microbiol. 68, 29–36. doi:10.1111/j.1365-2958.2008.06119.x
Saze, H., and Kakutani, T. (2007). Heritable epigenetic mutation of a transposon-flanked Arabidopsis gene due to lack of the chromatin-remodeling factor DDM1. EMBO J. 26, 3641–3652. doi:10.1038/sj.emboj.7601788
Slotkin, R. K., Vaughn, M., Borges, F., Tanurdzic, M., Becker, J. D., Feijo, J. A., et al. (2009). Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136, 461–472. doi:10.1016/j.cell.2008.12.038
Springer, N. M., and Schmitz, R. J. (2017). Exploiting induced and natural epigenetic variation for crop improvement. Nat. Rev. Genet. 18, 563–575. doi:10.1038/nrg.2017.45
Tsaftaris, A. S., and Polidoros, A. N. (1999). DNA methylation and plant breeding. Plant Breed. Rev. doi:10.3389/fpls.2021.596236
Waadt, R., Seller, C. A., Hsu, P.-K., Takahashi, Y., Munemasa, S., and Schroeder, J. I. (2022). Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 23, 680–694. doi:10.1038/s41580-022-00479-6
Weigel, D., and Colot, V. (2012). Epialleles in plant evolution. Genome Biol. 13, 249. doi:10.1186/gb-2012-13-10-249
Wessler, S. R. (1996). Plant retrotransposons: Turned on by stress. Curr. Biol. 6, 959–961. doi:10.1016/S0960-9822(02)00638-3
Whittaker, C., and Dean, C. (2017). The FLC locus: A platform for discoveries in epigenetics and adaptation. Annu. Rev. Cell Dev. Biol. 33, 555–575. doi:10.1146/annurev-cellbio-100616-060546
Yang, X., Kundariya, H., Xu, Y.-Z., Sandhu, A., Yu, J., Hutton, S. F., et al. (2015). MutS HOMOLOG1-derived epigenetic breeding potential in tomato. Plant Physiol. 168, 222–232. doi:10.1104/pp.15.00075
Yi, H., and Richards, E. J. (2008). Phenotypic instability of Arabidopsis alleles affecting a disease Resistance gene cluster. BMC Plant Biol. 8, 36. doi:10.1186/1471-2229-8-36
Keywords: chromatin remodeling, stress, epiallele, transposon, methylation, DNA, evolution
Citation: Comai L (2023) Triggers and mediators of epigenetic remodeling in plants. Front. Epigenet. Epigenom. 1:1188733. doi: 10.3389/freae.2023.1188733
Received: 17 March 2023; Accepted: 03 May 2023;
Published: 23 May 2023.
Edited by:
Suresh Kumar, Indian Agricultural Research Institute (ICAR), IndiaReviewed by:
Federica Maria Consiglio, National Research Council (CNR), ItalyElena Caro, Center for Plant Biotechnology and Genomics (UPM-INIA, CSIC), Spain
Copyright © 2023 Comai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luca Comai, bGNvbWFpQHVjZGF2aXMuZWR1
 Luca Comai
Luca Comai