- 1Biosciences Department, KEMRI-Wellcome Trust Research Programme, Kilifi, Kenya
- 2ONA Systems Inc, Burlington, VT, United States
- 3Nuffield Department of Medicine, Centre for Clinical Vaccinology and Tropical Medicine, Churchill Hospital, University of Oxford, Oxford, United Kingdom
There is a growing concern for malaria control in the Horn of Africa region due to the spread and rise in the frequency of Plasmodium falciparum Histidine-rich Protein (hrp) 2 and 3 deletions. Parasites containing these gene deletions escape detection by the major PfHRP2-based rapid diagnostic test. In this study, the presence of Pfhrp2/3 deletions was examined in uncomplicated malaria patients in Kilifi County, from a region of moderate-high malaria transmission. 345 samples were collected from the Pingilikani dispensary in 2019/2020 during routine malaria care for patients attending this primary health care facility. The Carestart™ RDT and microscopy were used to test for malaria. In addition, qPCR was used to confirm the presence of parasites. In total, 249 individuals tested positive for malaria by RDT, 242 by qPCR, and 170 by microscopy. 11 samples that were RDT-negative and microscopy positive and 25 samples that were qPCR-positive and RDT-negative were considered false negative tests and were examined further for Pfhrp2/3 deletions. Pfhrp2/3-negative PCR samples were further genotyped at the dihydrofolate reductase (Pfdhfr) gene which served to further confirm that parasite DNA was present in the samples. The 242 qPCR-positive samples (confirmed the presence of DNA) were also selected for Pfhrp2/3 genotyping. To determine the frequency of false negative results in low parasitemia samples, the RDT- and qPCR-negative samples were genotyped for Pfdhfr before testing for Pfhrp2/3. There were no Pfhrp2 and Pfhrp3 negative but positive for dhfr parasites in the 11 (RDT negative and microscopy positive) and 25 samples (qPCR-positive and RDT-negative). In the larger qPCR-positive sample set, only 5 samples (2.1%) were negative for both hrp2 and hrp3, but positive for dhfr. Of the 5 samples, there were 4 with more than 100 parasites/µl, suggesting true hrp2/3 deletions. These findings revealed that there is currently a low prevalence of Pfhrp2 and Pfhrp3 deletions in the health facility in Kilifi. However, routine monitoring in other primary health care facilities across the different malaria endemicities in Kenya is urgently required to ensure appropriate use of malaria RDTs.
Introduction
The 2021 Malaria indicator survey highlights that the disease still presents a significant burden in Kenya, with about 70% of the population being at risk of infection. Malaria accounts for up to 15% of in-patient consultations in the country and individuals in endemic areas are at the greatest risk of disease (1). This is the case for most countries in sub-Saharan Africa, which carries the greatest burden of malaria globally (2). In Kenya, malaria transmission is heterogenous and varies based on factors such as altitude, temperature, and rainfall patterns. Malaria control mainly depends on effective diagnosis and treatment. The Kenyan National Malaria Control Program recommends the use of Microscopy and rapid diagnostic tests (RDTs) as first line modalities for malaria diagnosis (3). Microscopy plays a pivotal role in providing parasite density data for accurate diagnosis of disease. However, the process requires well trained personnel, adequate space, availability of quality microscopes, consistent power supply and laboratory resources and it has a relatively long turnaround time, a luxury that remote and resource limited settings might not afford. On the other hand, malaria RDTs have proven to be an effective and time-saving tool for diagnosis as they are used as a point of care test, with prompt delivery of results (results can be obtained in 15–20 min) (4). The World Health Organization also recommends the use of malaria RDTs as convenient alternatives for diagnosis in malaria endemic regions, particularly in Africa (5).
Malaria RDTs contain antibodies against Plasmodium antigens, including lactose dehydrogenase, aldolase, and the histidine rich protein 2 (HRP2). However, the HRP2-based RDTs are the most preferred for their ability to uniquely detect infection with Plasmodium falciparum, the Plasmodium species that causes the greatest burden of malaria (6). They have also been shown to be more sensitive and heat stable, compared to the other commercially available RDTs (7, 8). While HRP2-based RDTs are primarily designed to detect the HRP2 antigen, they are also able to detect its isoform, HRP3, which has high sequence similarity to HRP2, jointly contributing to RDT test results (9). These RDTs have been instrumental in most malaria endemic areas, where accurate diagnosis is vital for effective control of the disease. The HRP2 protein is 60–105 kDa in size, water-soluble and abundantly expressed by the asexual stages of Plasmodium falciparum (10). Nonetheless, its main function in the parasite remains largely unclear.
Deletions in Plasmodium falciparum hrp2/3 (Pfhrp2/3) genes, compromise the quality of patient care as they result in false negative tests. In addition, they present a significant setback to malaria control because the parasite escapes detection, limiting effective diagnosis and treatment. Deletions in Pfhrp2/3 were originally reported in Peru (4) and have since been documented at relatively high frequencies in other South American countries such as Suriname (11), Brazil (12, 13) and Colombia (14). In Africa, Pfhrp2/3 deletions have been reported at fairly low frequencies of <5%, except in Ethiopia and Eritrea where proportions of up to 80% have been documented, leading to policy changes in the use RDTs based on these proteins (9, 15–21). Nonetheless, the lack of a uniform denominator for calculating these frequencies, limits the accurate interpretation of deletion data and comparison of results across regions (22, 23). The World Health Organization (WHO) recommends that use of HRP2/3 based RDTs be discontinued in regions where these deletions exceed 5% (24). While RDTs remain effective in most African countries, including Kenya, routine surveillance for possible mutations is important for policy makers as it guides appropriate use of these first-line diagnostic tools.
In Kenya, limited data exist on the status of Pfhrp2/3 deletions, although the RDTs in current use are still effective. Routine data on the status of these deletions would however benefit national malaria control programs by guiding policies on appropriate RDT use. The prevalence of Pfhrp2/3 deletions in the south of Kilifi County, a moderate to high malaria transmission setting in Kenya was first determined based on the WHO recommendations of a RDT result and secondly from a criterion set in this study of a real-time PCR test result, which also included a cycle threshold (Ct) cut-off to increase the sensitivity of detecting parasite DNA. Although the Pfhrp2/3 deletions observed occurred at relatively low frequencies, these data provide the baseline for regular surveillance of malaria RDTs used in the country.
Materials and methods
Study area and sample selection
EDTA blood samples from 345 patients between November 2019 and February 2020 were obtained from a malaria monitoring study that aimed to compare the performance of two malaria RDTs used for diagnosis in Pingilikani dispensary (25). Pingilikani is located within the Kilifi Health and Demographic Surveillance System (KHDSS), approximately 30 km south of Kilifi town (26). The malaria monitoring study (approved by the ethics review committee of the Kenya Medical Research Institute under protocol number SERU 2617) routinely consents patients with malaria for a dried blood spot (DBS) and EDTA blood sample alongside the diagnostic Carestart™ RDT and a microscopy test. The microscopy was conducted in the KEMRI-Wellcome Trust Research Programme GCLP accredited labs by competent expert microscopists, as per the WHO criteria, who routinely support clinical trials. The lab is also registered for quantitative Polymerase Chain Reaction (qPCR), UKNEQAS. The qPCR assay targeted the P. falciparum 18S ribosomal DNA. The 345 samples were categorized as either RDT and microscopy positive, RDT positive and microscopy negative, RDT and microscopy negative, or RDT negative and microscopy positive. The latter category (RDT negative and microscopy positive) was of interest as it highlighted a discordant sample set that were suspected false negatives, hence explored further for Pfhrp2 and hrp3 gene deletions, the primary targets in the Carestart™ RDT. Within the broader sample set, we explored the utility of using clinical surveillance samples for routine molecular monitoring of Pfhrp2 and Pfhrp3 gene deletions. Since the samples had been analyzed by qPCR alongside RDT for clinical diagnosis, all the samples positive by qPCR were genotyped for the Pfhrp2/3 gene deletions along with 30% of the qPCR negative samples. The latter samples were first genotyped for Pfdhfr to confirm parasite DNA quality and those that tested positive were subjected to hrp2/3 PCRs. We also analyzed a subset of 25 samples that were positive by qPCR but negative by RDT and 23 samples that were positive by RDT but negative by both qPCR and microscopy (to determine the rate of detecting false negative results at very low parasitemia). The sample selection criteria based on RDT and qPCR are shown in Figure 1.
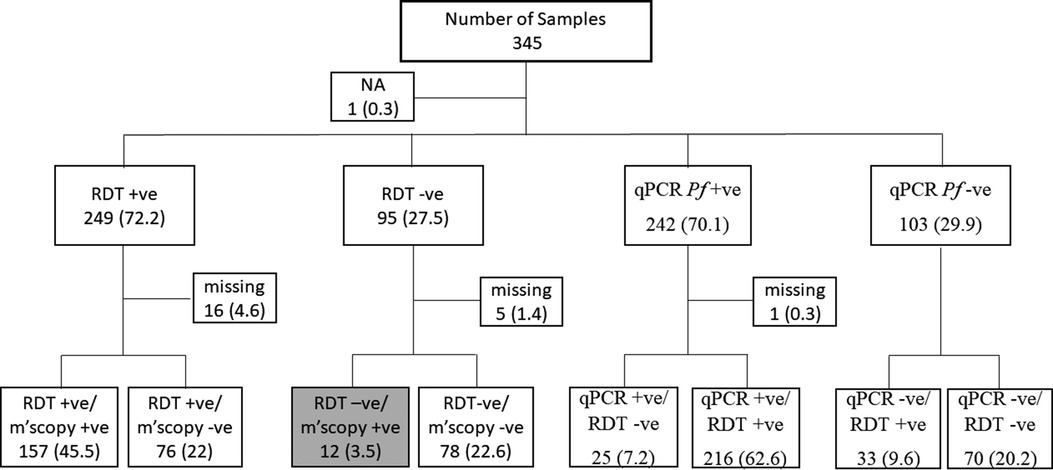
Figure 1. The sample selection criteria for the study. Samples were selected based on RDT the routine malaria diagnostic test conducted at health facilities. Thus, 12 discordant samples were RDT negative but positive by microscopy (grey box). Sample selection was also based on qPCR results following a routine assessment done within this study. NA, sample with no RDT data; Missing, samples with RDT but no microscopy data or qPCR but no RDT data. The values in brackets represent the frequencies of the respective categories relative to the overall sample size of 345.
DNA extraction and Pfhrp2/3 PCR
Following the manufacturer's instructions, parasite genomic DNA was extracted from the EDTA whole blood samples using the QIAamp® DNA Blood Mini kit (QIAGEN). The QIAcube HT (QIAGEN) platform was used for automated extraction, and the Nanodrop, ND 1000 Spectrophotometer was used to assess the concentration and purity (A260/A280 ratios) of the isolated genomic DNA. To create working concentrations of 5 ng/µl that could be used in the following procedures, the extracted DNA samples were diluted in sterile, double-distilled water and aliquots stored at −20°C. The Pfhrp2/3 PCRs were conducted in a semi-nested approach, using previously published primers (targeting Exon 2 of both genes) (17) listed in Table 1. The Plasmodium falciparum dhfr gene was also included in the PCRs, to confirm the presence and quality of parasite DNA in samples with possible Pfhrp2/3 deletions. The Expand High Fidelity PCR system (Roche™) was used to conduct conventional PCR. The annealing temperature for each primer combination (Supplementary Table S1) was determined through gradient PCR, conducted using Pf3D7 DNA as the template. Given that the Pfhrp2 and Pfhrp3 PCRs were semi-nested, the first round of amplification was conducted using the F1&R1 primer combination for each gene. Amplicons from the first amplification round were then used as templates for the second PCR which was performed using the F2&R1 primer combinations. A single reaction volume of 10.5 µl consisted of 1μl of template DNA (5 ng/μl), 6.56 µl of ultrapure DNase/RNase free distilled water, 0.2 µl of 10 mM deoxynucleotides triphosphate (dNTPs), 1 µl of 25 mM MgCl2, 0.3 µl of 10pmol/µl forward primer (F1or F2), 0.3 µl of 10pmol/µl reverse primer, 1 µl of 10X PCR buffer with 15 mM MgCl2 and 0.14 µl of 3.5 U/µl Expand High fidelity PCR Taq polymerase (Roche). The thermal cycling conditions for each round of PCR were 94°C for 2 min, 10 cycles of 94°C for 15 s, 54°C - 30 s and 72°C for 2 min, 25 cycles of 94°C for 15 s, 54- 30 s and 72°C for 2 min then a final extension at 72°C for 2 min. The positive controls were DNA from the P. falciparum 3D7 (with both Pfhrp2 and Pfhrp3), Dd2 (Pfhrp2 deleted) and HB3 (Pfhrp3 deleted) laboratory isolates. The Pfdhfr PCRs were performed using published primers (27) in a 10.5 µl reaction (dhfr had one forward primer, unlike hrp2/3 which had two, each). The thermal cycling conditions included 94°C for 2 min, 10 cycles of 94°C for 15 s, 58°C for 30 s and 68°C for 2 min, 25 cycles of 94°C for 15 s, 58- 30 s and 68°C for 2 min then a final extension at 72°C for 2 min. P. falciparum 3D7 DNA was similarly used as the positive control. Nuclease-free water was used as the template for the negative controls, for all three genes. 2% (w/v) agarose gels stained with 5 µl of 20,000X RedSafe DNA staining solution, were used to visualize amplification products. 5 µl of each PCR product was used for gel electrophoresis. All gels were run at 100 V for 40 min then imaged using the Bio-Rad ChemiDoc XRS + TM imaging system.
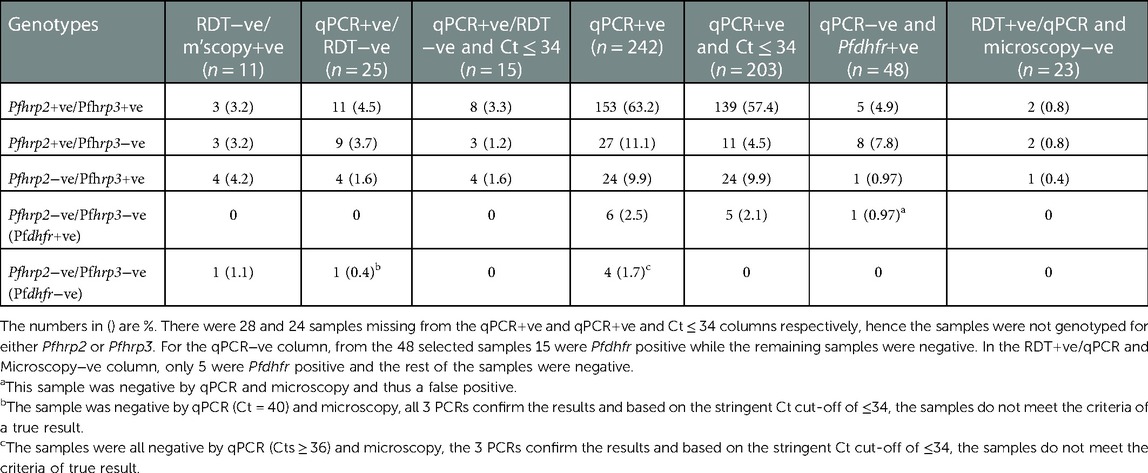
Table 1. A summary of the Pfhrp2, Pfhrp3 and pfdhfr genotyping results from different sample categories.
P. falciparum qPCR analysis
Real-time amplification for detection and absolute detection of P. falciparum were performed on the ABI 7500 Real-Time PCR system (Applied Biosystems) as previously described (28) using TaqMan based P. falciparum specific probes. The reaction was performed in a final volume of 25 μl containing: 2.5 µl each of P. falciparum 18S rDNA forward and reverse primers (10 pmol/µl), 0.625 µl of 18S probe (10 pmol/µl), 12.5 µl TaqMan universal PCR master mix (2X), 6.75 µl of sample or 3D7 control samples, with the remaining volume PCR clean water. The following RT-PCR conditions were used: 50°C for 2 min, 95°C for 10 min, and then 45 cycles of 95°C for 15 s and 60°C for 1 min. All qPCR assays were run with appropriate controls including the Non-Template Control (NTC). For parasite quantification, eight defined standards with varying parasitemias were included, Standard 1: 460,000 parasites/μl, Standard 2: 27,000 parasites/μl, Standard 3: 15,000 parasites/μl, Standard 4: 10,000, Standard 5: 100 parasites/μl, Standard 6: 10 parasites/μl, Standard 7: 1 parasites/μl and Standard 8: 0.1 parasites/μl. Following an extrapolation from the standard curve, samples with a Ct value of above 34 were considered P. falciparum negative (<5 parasites/ul).
Sequencing and sequence analysis
We sequenced samples that were Pfhrp2 and Pfhrp3 positive, from the 12 that were microscopy positive but RDT negative. Following PCR, amplicons were purified using the ExoSap-IT™ (Affymetrix™) reagent, then sequenced using the BigDye® Terminator v3.1 cycle sequencing kit from Applied Biosystems, UK. Sequencing was conducted using the same primers utilized for PCR. Briefly, a single reaction volume of 10 µl consisted of 2 µl, purified template DNA, 0.5 µl BigDye® Terminator enzyme mix, 1.75 µl 5X BigDye® sequencing buffer, 4.75 µl ultrapure DNase/RNase-free distilled water and 1 µl of 5pmol/µl sequencing primer. The cycling conditions for sequencing were as follows; an initial denaturation at 95°C for 30 s, 25 cycles of 96°C for 10 s, 50°C for 5 s and 60°C for 4 min, then a final hold at 15°C for 10 min. The products of sequencing were thereafter subjected to ethanol precipitation then capillary electrophoresis on the 3730xl DNA analyser (ILRI, Kenya), to generate sequence chromatograms. Finally, sequence analysis was conducted using CLC Workbench Version 7.7.1 (QIAGEN).
Statistical analysis
Statistical analyses were conducted using Version 4.2.1 of the R software (29), where frequencies were calculated. A log10 transformation of parasitaemia was applied and used to compare the different Pfhrp2/3 genotypes using a Student's t-test with the statistical significance set at a p value <0.05. Box plots were used to show the 25th percentiles, geometric mean and 75th percentiles of the log transformed parasitemia.
Results
Semi-nested PCR amplification of false RDT negative samples based on microscopy
In this study, DNA from 345 samples tested for malaria by RDT, microscopy and qPCR were used and 249 (72.2%), 170 (49.3%) and 242 (70.1%) samples were positive for each test, respectively. There were 12 (3.47%) discordant samples that were P. falciparum negative by RDT but positive by microscopy (Figure 1), considered as false negative RDTs, were genotyped for Pfhrp2 and Pfhrp3. Pfdhfr was used as a secondary gene to confirm the presence of parasite DNA in samples that tested negative for both Pfhrp2 and Pfhrp3. One of the 12 discordant samples was missing and was therefore excluded from subsequent analysis. Among the remaining 11 samples, 3 (0.87%) tested positive for both Pfhrp2 and Pfhrp3, 3 (0.87%) were positive for Pfhrp2 but negative for Pfhrp3, while 4 (1.16%) were negative for Pfhrp2 but positive for Pfhrp3 (Table 1). Six of these samples had low parasitemias of <1,000 parasites/µl and the remaining sample was negative for all the genes tested (Pfhrp2, Pfhrp3 and Pfdhfr). Notably, this sample was negative by qPCR but contained 3,520 parasites/µl.
Semi-nested PCR amplification of false negative RDT samples based on qPCR
There were 25 (7.25%) samples that were positive by qPCR but negative by RDT, considered as discordant based on the qPCR test, and these were assessed for Pfhrp2 and Pfhrp3 deletions. Within this sample set, 6 samples were also microscopy positive. The results obtained, identified 11 (3.19%) samples that were positive for both Pfhrp2 and Pfhrp3, 9 (2.61%) were positive for pfhrp2 but negative for Pfhrp3, while 4 (1.16%) were positive for pfhrp3 but negative for Pfhrp2. None of the samples were negative for both Pfhrp2 and Pfhrp3 but positive for dhfr (Table 1). We extended the analysis further and grouped the samples based on a Cycle threshold (Ct) cut-off of 34, which represented at least 5 parasites/µl (an extrapolation from our standard curve). This stringent cut-off yielded 17 discordant samples, from which the Pfhrp3 negative only samples reduced to 4 (1.16%) with no change to the number of to the Pfhrp2 negative only samples. There were no samples that tested negative for both hrp2 and hrp3 but positive for dhfr. In addition, the lone sample that was Pfhrp2−ve/Pfhrp3−ve (Pfdhfr−ve) but qPCR positive when Ct values were not considered as a criteria was further confirmed as lacking parasite DNA and dropped off with the stringent Ct cut-off.
Pfhrp2/3 genotyping results from qPCR positive samples
In addition to the discordant samples above, the analysis included a larger sample set that were P. falciparum positive by qPCR. A total of 242 samples were qPCR positive, the large majority, 44.4% were positive for both hrp2 and hrp3, while only 6 (1.74%) were negative for both genes but positive for Pfdhfr. The remaining samples were either Pfhrp2 negative only, Pfhrp3 negative only, negative for all three genes or missing with no genotyping data. The stringent cut-off of Ct ≤ 34 resulted in 218 samples that met the criteria of a parasite positive sample. Similar to the qPCR discordant samples, there was reduction by about half [13, (4.1%)] in the numbers of Pfhrp2 negative only samples. Only 1 sample was not accurately determined as Pfhrp2 and Pfhrp3 negative plus Pfdhfr positive, this sample was also microscopy negative. The four samples that were Pfhrp2−ve/Pfhrp3−ve (Pfdhfr−ve) were further confirmed as parasite negative samples and dropped off with the stringent Ct cut-off.
Pfhrp2/3 genotyping results from qPCR negative samples
As a final confirmation of the variation in qPCR and PCR specificity, 48 (∼47%) samples that were negative for P. falciparum by qPCR were selected and tested for the Pfhrp2 and Pfhrp3 deletions. All the 48 samples were genotyped for Pfdhfr to confirm that parasite DNA was present, yielding 15 positive samples, 69% of which were microscopy negative. Only 1 (0.29%) sample was negative for both genes but positive for Pfdhfr and further examination of the sample revealed that it was negative by microscopy and thus a false positive. Most, 8 (2.3%), of the remaining samples were negative for Pfhrp3 only and only 2 of these samples were microscopy positive; and within the 5 Pfhrp2 and Pfhrp3 positive samples, 3 were microscopy positive.
Pfhrp2/3 genotyping results from RDT positive but microscopy and qPCR-negative samples
We had an additional category of samples (23) that were positive for P. falciparum by RDT, but negative by both microscopy and qPCR. Like the qPCR negative group, the samples were genotyped at the dhfr locus to select positive DNA samples for Pfhrp2 and Pfhrp3 testing. Of the 23, only 5 (1.45%) were positive for dhfr and of the 5, Pfhrp2/3 data was obtained from 4 samples. Two (0.6%) of the four samples was positive for both hrp2 and hrp3, 2 (0.6%) were Pfhrp3 negative only, while 1 (0.3%) was Pfhrp2 negative only. None of the samples was negative for both Pfhrp2 and Pfhrp3.
Effect of parasitemia on Pfhrp2/3 genotyping
We further assessed if the level of parasitemia influenced whether a sample tested positive or negative for Pfhrp2, Pfhrp3 or both. There were 231 samples with Pfhrp2/3 genotypes out of the total 345 samples. The geometric mean parasitemia of the respective Pfhrp2/3 genotypes was 61,965 parasites/μl (Pfhrp2/3 + ve), 6,088 parasites/μl (Pfhrp2 + ve, Pfhrp3−ve), 44,247 parasites/μl (Pfhrp2−ve, Pfhrp3 + ve), and 32,212 parasites/μl (Pfhrp2/3−ve). Overall, the analysis revealed that the Pfhrp2 and Pfhrp3 negative and Pfhrp2−ve only samples had similar levels of parasitemia as those that tested positive for both genes (Figure 2). However, there was a significant difference in parasitemia between the Pfhrp3−ve only and Pfhrp2 + ve/hrp3 + ve samples (p = 0.033, Figure 2). Parasitemia levels were generally lower between samples that were discordant by either microscopy or qPCR and those that were non-discordant. Samples that were microscopy positive, but RDT negative had significantly (p = 0.0087) lower levels of parasitemia compared to those that were both microscopy and RDT-positive (Figure 3A), though cognizance is taken of the expected low numbers of discordant samples. There were 25 qPCR + ve/RDT−ve samples in total, however only 6 of these had microscopy data, and only 150 of the 216 qPCR + ve/RDT + ve samples had microscopy data. The low numbers of qPCR + ve/RDT−ve samples are likely to have attributed to the lack of significance observed in the comparison with qPCR + ve/RDT + ve samples (p = 0.065, Figure 3B).
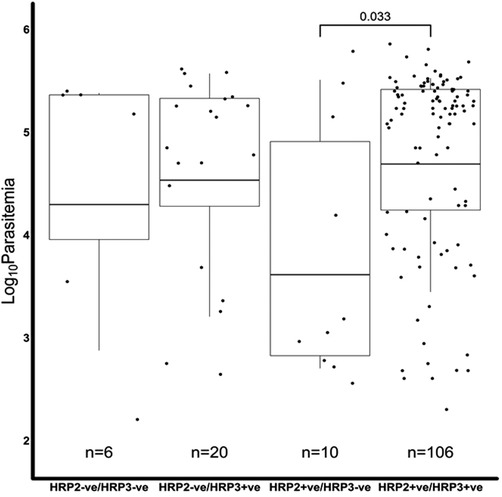
Figure 2. A comparison of the microscopy log base 10 parasitemia of the Pfhrp2/3 genotypes. The log10 geometric mean of the Pfhrp2/3 genotypes was, 4.30 (32,212 parasites/µl), 4.53 (44,247 parasites/µl), 3.61 (6,088 parasites/µl) and 4.69 (61,965 parasites/µl) for hrp2−ve/hrp3−ve, hrp2−ve/hrp3 + ve, hrp2 + ve/hrp3−ve and hrp2 + ve/hrp3 + ve, respectively. The hrp2 + ve/hrp3−ve group had significantly lower levels of parasitemia than the hrp2 + ve/hrp3 + ve category (t-test p < 0.05), in the overall sample set. There was no difference (p > 0.05) in parasitemia among the other hrp2/3 genotypes.
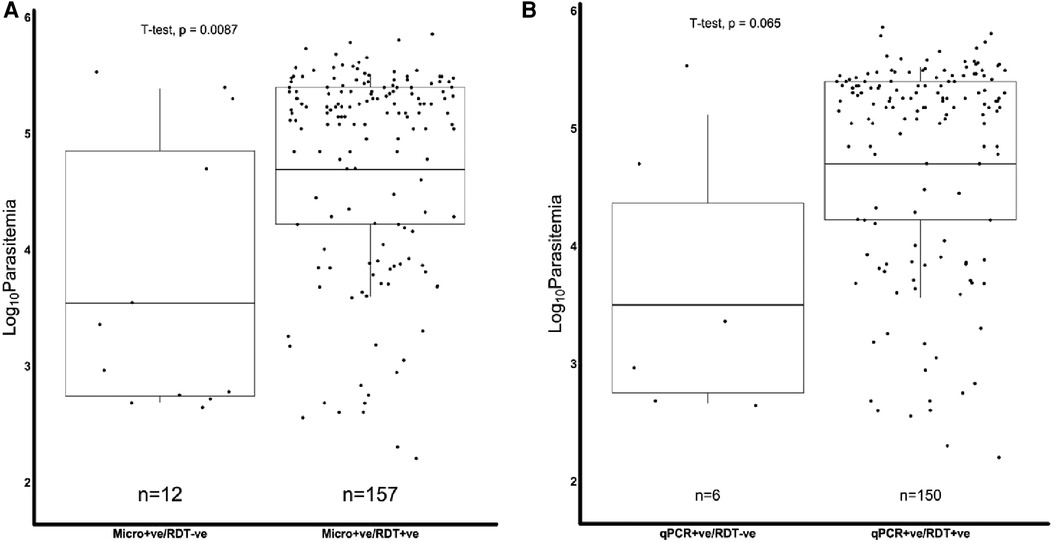
Figure 3. Combined Box and scatter diagrams comparing the microscopy log10 parasite densities of discordant and non-discordant samples. Horizontal lines represent the geometric means of log10 parasitemia. (A) Microscopy positives where the geometric mean parasitemia (log 10 parasitemia) was 3.5 (4978.0 parasites/μl) for m'scopy + ve/RDT−ve and 4.7 (61309.7 parasites/μl) for m'scopy + ve/RDT + ve. (B) qPCR positives, where the geometric mean parasitemia (log 10 parasitemia) was 3.5 (4427.3 parasites/μl) for qPCR + ve/RDT−ve and 4.7 (62589.9 parasites/μl) for qPCR + ve/RDT + ve.
Pfhrp2/3 sequencing
We sequenced 12 and 13 samples for Pfhrp3 and Pfhrp2, respectively. The 7 and 9 good quality sequences obtained for Pfhrp2 and Pfhrp3 respectively, were subsequently analyzed for the presence of 3-, 6- and 9-amino acid repeats. A total of 20 repeat types were identified. Type 4 (AHH) and type 7 (AHHAAD) repeats were common in Pfhrp2 and Pfhrp3 and an additional 8 repeat types were described for Pfhrp2, while 5 more repeats were defined for Pfhrp3 (Supplementary Table S2). Thus, demonstrating the similarity between both proteins and with the global population (30, 31).
Discussion
This study was conducted in a primary healthcare facility that routinely conducts RDT tests to diagnose malaria. It had the advantage of examining microscopy and qPCR that were also routinely conducted for all the malaria positive samples presenting to the clinic. This provided a unique setting to interrogate Pfhrp2 and Pfhrp3 gene deletion frequencies, given the current expected low prevalence of the mutations in Kenya (21, 32). Pfhrp2 and Pfhrp3 deletions were therefore examined within this context of routine malaria monitoring in a clinical set up to detect uncomplicated malaria cases. Thus, malaria RDTs was the baseline selection criteria to initially identify discordant malaria test results when compared to microscopy and qPCR data.
The first observation was the lack of Pfhrp2−ve and Pfhrp3−ve parasites and the low (<5%) presence of Pfhrp2−ve only and Pfhrp3−ve only parasites within the RDT + ve and microscopy−ve discordant sample set. These samples were also significantly lower in parasitemia compared to the RDT + ve and microscopy + ve category, highlighting the difficulty in obtaining a true negative PCR result for either Pfhrp2 or Pfhrp3 and a true positive for the single copy confirmatory gene, Pfdhfr. This was also demonstrated by the lone sample that was qPCR negative, Pfhrp2 and Pfhrp3 semi-nested-PCR negative and Pfdhfr negative, albeit with a parasitemia of about 3,000 parasites/µl by microscopy. Thus, raising concerns on the specificity of the semi-nested-PCR approach used to determine Pfhrp2 and Pfhrp3 gene deletion frequencies.
The second analysis selected samples based on qPCR positivity and microscopy negativity; this approach doubled the discordant sample size and only 6 of the 25 samples were detectable by microscopy, indicating the increased sensitivity of qPCR in detecting samples with parasites. This superiority of qPCR over RDTs has been described before (33, 34). Despite the increased discordant sample size, no Pfhrp2−ve/Pfhrp3−ve parasites were detected and the frequency of Pfhrp2−ve only and Pfhrp3−ve only parasites decreased to <2%. The qPCR assay had the added advantage of excluding very low parasitemia levels that would be within the limit of detection of a true negative result. The combined Ct stringency cut-off improved the confidence in specificity of the semi-nested-PCR approach that determined the Pfhrp2 and Pfhrp3 gene deletion frequencies.
Thirdly, qPCR positivity as the first analysis served as a confirmatory test for the presence of parasite DNA in the samples, providing a much larger sample size. Thus, Pfhrp2 and Pfhrp3 gene deletion frequencies were examined without the need for Pfdhfr though it was still genotyped in samples that were Pfhrp2−ve and Pfhrp3−ve. The inclusion of the Ct cut-off increased the frequency of the Pfhrp2−ve only parasites to a more reliable frequency of about 5%, 10% for Pfhrp3−ve only parasites and 2% for Pfhrp2−ve and Pfhrp3−ve parasites.
Since qPCR−ve but Pfdhfr + ve samples may be either RDT + ve or microscopy + ve or both, they were also explored for Pfhrp2 and Pfhrp3 gene deletions. The only reliable results were the two Pfhrp3−ve only samples with microscopy data. Therefore, these analyses demonstrate the need for combined data to improve the accuracy of determining a true negative result.
The final analyses examined the potential to detect Pfhrp2 and Pfhrp3 false negatives using RDT + ve, qPCR−ve and microscopy−ve samples. This selection criteria represents samples that potentially had residual HRP2 and HRP3 in the blood from a cleared infection, since HRP2 can be found in blood up to 40 days following treatment (35). Thus, any amplified sample is likely due to DNA from dead parasites, sub-microscopic infections or infections below the qPCR detection limit. The false negative results for Pfhrp2−ve only and Pfhrp3−ve only samples were <1%.
It is apparent that the Pfhrp2−ve and Pfhrp3−ve parasites are detected in samples with low parasitemia and a larger sample is required to confirm this finding. However, the low parasitemia increases the chances of detecting false negative gene deletion results. The addition of qPCR provided confidence in detecting a true negative result and considerations need to be made when considering RDT positive and microscopy negative samples alone as they can contribute to the number of false negative results if they are not also determined to be qPCR negative, which is overall a more sensitive assay. Furthermore, from this sample size of 345, over a 4-month period, which is close to the current WHO hrp2/3 protocol of 370 per health facility, the RDT positive and microscopy negative criteria and the qPCR positive and RDT negative discordant pairs did not detect any Pfhrp2−ve and Pfhrp3−ve parasites. This is expected considering the absence of the dual gene deletions in 192 febrile individuals studied in 2016 from a health facility in Busia county, Western Kenya (32) and in Mbita, Homa Bay County, where the prevalence of Pfhrp2 deleted parasites was 9% from 89 samples analyzed from a sample size of 274 asymptomatic individuals in 2014 (36).
Similar to the earliest Pfhrp2/3 study (36) conducted in Kenya, qPCR works well to define the sample selection criteria for Pfhrp2/3 gene deletion screening in low prevalence settings. From this study, most parasites were Pfhrp3 negative only at frequency of 10%, followed Pfhrp2 negative only parasites at 5%, and Pfhrp2 and Pfhrp3 negative parasites were at a frequency of 2% from sample size of 345 febrile individuals. The higher prevalence of Pfhrp3 negative only infections was similar to the finding in Ethiopia where Pfhrp3 negative infections were 31% and Pfhrp2 negative infections 27% from 610 febrile individuals presenting to health facilities (37). The analysis of whole genome sequencing data suggested that the Pfhrp3 deletions were older and independently arose multiple times, while Pfhrp2 deletions are under more recent strong selective pressure (37).
Currently, in the Horn of Africa, Ethiopia (19, 37) and Eritrea (16, 38), there has been a switch from HRP2-based to LDH-based RDTs, due to the countries being above the recommended WHO Pfhrp2/3 gene deletion prevalence threshold of 5%. The increasing data for Pfhrp2/3 deletions in Africa, though at a low frequency, calls for continued molecular surveillance of these genetic markers in a region that is heavily reliant on RDTs for malaria diagnosis and downstream case management.
Data availability statement
Original datasets are available in a publicly accessible repository: The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.5281/zenodo.7594474
Ethics statement
The studies involving human participants were reviewed and approved by KEMRI Scientific Ethics Review Unit, protocol number SERU 2617. Written informed consent to participate in this study was provided by the participants' legal guardian/next of kin.
Author contributions
DO: conducting the experiments, data collation and drafting manuscript. LN: conducting the experiments, data collation and drafting manuscript. VO: conducting the experiments, data collation and review of manuscript. NA: conducting the experiments and review of manuscript. JW: coordinating field work and review of manuscript. CN: coordinating microscopy work and review of manuscript. PL-D: funding of field work, conceptualisation and review of manuscript. AS: funding of field work, conceptualisation and review of manuscript. PB: funding of field work, conceptualisation and review of manuscript. LO-O: conceptualization, study design, coordination of the whole study and drafting of manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported through a sub-award from ONA Systems Inc, with support from a Wellcome Trust Intermediate Fellowship, Grant Number 107568/Z/15/Z awarded to LO-O and a Calestous Juma Leadership Fellowship Award from the Bill and Melinda Gates Foundation, INV-036442 to LO-O.
Acknowledgments
The team acknowledges the study participants and sincerely appreciates to their parents. We thank the KEMRI Director for permission to publish this manuscript.
Conflict of interest
PL-D was employed by ONA Systems Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fepid.2023.1083114/full#supplementary-material.
References
1. Kenya Malaria Indicator Survey. Kenya malaria indicator survey 2020 final report Ministry of Health Division of National Malaria Programme Nairobi Republic of Kenya Ministry of Health (2021). Available at: www.nmcp.or.ke.
3. National Malaria Control Program. National guidelines for the diagnosis, treatment and prevention of malaria in Kenya Ministry of Health Fifth Edition. 2016. Available at: http://www.nmcp.or.ke.
4. Gamboa D, Ho MF, Bendezu J, Torres K, Chiodini PL, Barnwell JW, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One. (2010) 5(1):e8091. doi: 10.1371/journal.pone.000809120111602
6. Murray CK, Bennett JW. Rapid diagnosis of malaria. Interdiscip Perspect Infect Dis. (2009) 2009:1–7. doi: 10.1155/2009/415953
7. Coldiron ME, Assao B, Langendorf C, Sayinzoga-Makombe N, Ciglenecki I, de La Tour R, et al. Clinical diagnostic evaluation of HRP2 and pLDH-based rapid diagnostic tests for malaria in an area receiving seasonal malaria chemoprevention in Niger. Malar J. (2019) 18(1):443. doi: 10.1186/s12936-019-3079-131878947
8. Chiodini PL, Bowers K, Jorgensen P, Barnwell JW, Grady KK, Luchavez J, et al. The heat stability of Plasmodium lactate dehydrogenase-based and histidine-rich protein 2-based malaria rapid diagnostic tests. Trans R Soc Trop Med Hyg. (2007) 101(4):331–7. doi: 10.1016/j.trstmh.2006.09.00717212967
9. Kaaya RD, Kavishe RA, Tenu FF, Matowo JJ, Mosha FW, Drakeley C, et al. Deletions of the Plasmodium falciparum histidine-rich protein 2/3 genes are common in field isolates from north-eastern Tanzania. Sci Rep. (2022) 12(1):5802. doi: 10.1038/s41598-022-09878-335388127
10. Sepúlveda N, Phelan J, Diez-Benavente E, Campino S, Clark TG, Hopkins H, et al. Global analysis of Plasmodium falciparum histidine-rich protein-2 (pfhrp2) and pfhrp3 gene deletions using whole-genome sequencing data and meta-analysis. Infect Genet Evol. (2018) 62:211–9. doi: 10.1016/j.meegid.2018.04.039
11. Okoth SA, Abdallah JF, Ceron N, Adhin MR, Chandrabose J, Krishnalall K, et al. Variation in Plasmodium falciparum histidine-rich protein 2 (Pfhrp2) and Plasmodium falciparum histidine-rich protein 3 (Pfhrp3) gene deletions in Guyana and Suriname. PLoS One. (2015) 10(5):e0126805. doi: 10.1371/journal.pone.012680525978499
12. Góes L, Chamma-Siqueira N, Mário Peres J, Nascimento JM, Valle S, Arcanjo AR, et al. Evaluation of histidine-rich proteins 2 and 3 gene deletions in Plasmodium falciparum in endemic areas of the Brazilian Amazon. Public Health. (2021) 18:123. doi: 10.3390/ijerph18010123
13. Viana GMR, Okoth SA, Silva-Flannery L, Barbosa DRL, de Oliveira AM, Goldman IF, et al. Histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in Plasmodium falciparum isolates from select sites in Brazil and Bolivia. PLoS One. (2017) 12(3):e0171150. doi: 10.1371/journal.pone.017115028301474
14. Murillo Solano C, Akinyi Okoth S, Abdallah JF, Pava Z, Dorado E, Incardona S, et al. Deletion of Plasmodium falciparum histidine-rich protein 2 (pfhrp2) and histidine-rich protein 3 (pfhrp3) genes in Colombian parasites. PLoS One. (2015) 10(7):e0131576. doi: 10.1371/journal.pone.013157626151448
15. Bosco AB, Anderson K, Gresty K, Prosser C, Smith D, Nankabirwa JI, et al. Molecular surveillance reveals the presence of pfhrp2 and pfhrp3 gene deletions in Plasmodium falciparum parasite populations in Uganda, 2017-2019. Malar J. (2020) 19(1):300. doi: 10.1186/s12936-020-03362-x32843041
16. Berhane A, Russom M, Bahta I, Hagos F, Ghirmai M, Uqubay S. Rapid diagnostic tests failing to detect Plasmodium falciparum infections in Eritrea: an investigation of reported false negative RDT results. Malar J. (2017) 16(1):105. doi: 10.1186/s12936-017-1752-928264689
17. Berzosa P, González V, Taravillo L, Mayor A, Romay-Barja M, Garciá L, et al. First evidence of the deletion in the pfhrp2 and pfhrp3 genes in Plasmodium falciparum from Equatorial Guinea. Malar J. (2020) 19(1):99. doi: 10.1186/s12936-020-03178-932122352
18. Eyong EM, Etutu SJM, Jerome FC, Nyasa RB, Kwenti TE, Moyeh MN. Plasmodium falciparum histidine-rich protein 2 and 3 gene deletion in the mount Cameroon region. IJID Regions. (2022) 3:300–7. doi: 10.1016/j.ijregi.2022.05.00635755467
19. Golassa L, Swedberg G. Extensive deletion and sequence variation of Plasmodium falciparum histidine rich protein 2/3 (pfhrp2/3) genes in Ethiopia: implication for RDT-based malaria diagnosis and control (2020). Available at: https://doi.org/10.21203/rs.3.rs-22606/v1.
20. McCaffery JN, Nace D, Herman C, Singh B, Sompwe EM, Nkoli PM, et al. Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions among patients in the DRC enrolled from 2017 to 2018. Sci Rep. (2021) 11(1):22979. doi: 10.1038/s41598-021-02452-334837020
21. Rogier E, McCaffery JN, Nace D, Svigel SS, Assefa A, Hwang J, et al. Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions from persons with symptomatic malaria infection in Ethiopia, Kenya, Madagascar, and Rwanda. Emerg Infect Dis. (2022) 28(3):608–16. doi: 10.3201/eid2803.21149935201739
22. Agaba BB, Yeka A, Nsobya S, Arinaitwe E, Nankabirwa J, Opigo J, et al. Systematic review of the status of pfhrp2 and pfhrp3 gene deletion, approaches and methods used for its estimation and reporting in Plasmodium falciparum populations in Africa: review of published studies 2010-2019. Malar J. (2019) 18:355. doi: 10.1186/s12936-019-2987-431694718
23. Cheng Q, Gatton ML, Barnwell J, Chiodini P, McCarthy J, Bell D, et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J. (2014) 13(1):283. doi: 10.1186/1475-2875-13-28325052298
24. World Health Organization. Malaria policy advisory committee - March 2017 meeting report (2017).
25. Park C, Ngo H, Lavitt LR, Karuri V, Bhatt S, Lubell-Doughtie P, et al. The design and evaluation of a mobile system for rapid diagnostic test interpretation. Proc ACM Interact Mob Wearable Ubiquitous Technol. (2021) 5(1):1889. doi: 10.1145/3448106
26. Scott JAG, Bauni E, Moisi JC, Ojal J, Gatakaa H, Nyundo C, et al. Profile: the Kilifi health and demographic surveillance system (KHDSS). Int J Epidemiol. (2012) 41(3):650–7. doi: 10.1093/ije/dys06222544844
27. Osoti V, Akinyi M, Wamae K, Kimenyi KM, de Laurent Z, Ndwiga L, et al. Targeted amplicon deep sequencing for monitoring antimalarial resistance markers in western Kenya. Antimicrob Agents Chemother. (2022) 66(4):e0194521. doi: 10.1128/aac.01945-2135266823
28. Hermsen CC, Telgt DSC, Linders EHP, van de Locht LATF, Eling WMC, Mensink JBM, et al. Detection of Plasmodium falciparum malaria parasites in vivo by real-time quantitative PCR. Mol Biochem Parasitol. (2001) 118:247–51. doi: 10.1016/S0166-6851(01)00379-611738714
29. R Core Team. R: a language and environment for statistical computing. R foundation for statistical computing, Vienna, Austria (2019). Available at: https://www.R-project.org/ (Accessed July 21, 2022).
30. Kumar Bharti P, Singh Chandel H, Krishna S, Nema S, Ahmad A, Udhayakumar V, et al. Sequence variation in Plasmodium falciparum histidine rich proteins 2 and 3 in Indian isolates: implications for malaria rapid diagnostic test performance. Sci Rep. (2017) 7(1):1308. doi: 10.1038/s41598-017-01506-928465622
31. Baker J, Ho MF, Pelecanos A, Gatton M, Chen N, Abdullah S, et al. Open access RESEARCH global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar J. (2010) 9:129. doi: 10.1186/1475-2875-9-12920470441
32. Nderu D, Kimani F, Thiong’o K, Akinyi M, Karanja E, Meyer CG, et al. PfHRP2-PfHRP3 diversity among Kenyan isolates and comparative evaluation of PfHRP2/pLDH malaria RDT with microscopy and nested PCR methodologies. Parasitol Int. (2018) 67(6):793–9. doi: 10.1016/j.parint.2018.08.00730138695
33. Wardhani P, Butarbutar TV, Adiatmaja CO, Betaubun AM, Hamidah N, Aryati . Performance comparison of two malaria rapid diagnostic test with real time polymerase chain reaction and gold standard of microscopy detection method. Infect Dis Rep. (2020) 12:8731. doi: 10.4081/idr.2020.873132874462
34. Oboh MA, Oriero EC, Ndiaye T, Badiane AS, Ndiaye D, Amambua-Ngwa A. Comparative analysis of four malaria diagnostic tools and implications for malaria treatment in southwestern Nigeria. Int J Infect Dis. (2021) 108:377–81. doi: 10.1016/j.ijid.2021.05.04934044139
35. Poti KE, Sullivan DJ, Dondorp AM, Woodrow CJ. HRP2: transforming malaria diagnosis, but with caveats. Trends Parasitol. (2020) 36:112–26. doi: 10.1016/j.pt.2019.12.00431848119
36. Beshir KB, Sepúlveda N, Bharmal J, Robinson A, Mwanguzi J, Busula AO, et al. Plasmodium falciparum parasites with histidine-rich protein 2 (pfhrp2) and pfhrp3 gene deletions in two endemic regions of Kenya. Sci Rep. (2017) 7(1):14718. doi: 10.1038/s41598-017-15031-229116127
37. Feleke SM, Reichert EN, Mohammed H, Brhane BG, Mekete K, Mamo H, et al. Plasmodium falciparum is evolving to escape malaria rapid diagnostic tests in Ethiopia. Nat Microbiol. (2021) 6(10):1289–99. doi: 10.1038/s41564-021-00962-434580442
38. Mihreteab S, Anderson K, Pasay C, Smith D, Gatton ML, Cunningham J, et al. Epidemiology of mutant Plasmodium falciparum parasites lacking histidine-rich protein 2/3 genes in Eritrea 2 years after switching from HRP2-based RDTs. Sci Rep. (2021) 11(1):21082. doi: 10.1038/s41598-021-00714-834702923
Keywords: Pfhrp2, Pfhrp3, gene deletions, malaria RDTs, Plasmodium falciparum
Citation: Okanda D, Ndwiga L, Osoti V, Achieng N, Wambua J, Ngetsa C, Lubell-Doughtie P, Shankar A, Bejon P and Ochola-Oyier LI (2023) Low frequency of Plasmodium falciparum hrp2/3 deletions from symptomatic infections at a primary healthcare facility in Kilifi, Kenya. Front. Epidemiol. 3:1083114. doi: 10.3389/fepid.2023.1083114
Received: 28 October 2022; Accepted: 23 January 2023;
Published: 21 February 2023.
Edited by:
Shannon Takala Harrison, University of Maryland, United StatesReviewed by:
Alfredo Mayor, Instituto Salud Global Barcelona (ISGlobal), SpainBilly Ngasala, Muhimbili University of Health and Allied Sciences, Tanzania
© 2023 Okanda, Ndwiga, Osoti, Achieng, Wambua, Ngetsa, Lubell-Doughtie, Shankar, Bejon and Ochola-Oyier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lynette Isabella Ochola-Oyier bGlvY2hvbGFAa2Vtcmktd2VsbGNvbWUub3Jn
Specialty Section: This article was submitted to Infectious Disease Epidemiology, a section of the journal Frontiers in Epidemiology
 Dorcas Okanda1
Dorcas Okanda1 Leonard Ndwiga
Leonard Ndwiga Nicole Achieng
Nicole Achieng Caroline Ngetsa
Caroline Ngetsa Peter Lubell-Doughtie
Peter Lubell-Doughtie Anuraj Shankar
Anuraj Shankar Philip Bejon
Philip Bejon Lynette Isabella Ochola-Oyier
Lynette Isabella Ochola-Oyier