- 1Hubei Key Laboratory of Ecological Restoration for River-Lakes and Algal Utilization, School of Civil Engineering, Architecture and Environment, Hubei University of Technology, Wuhan, China
- 2Key Laboratory of Intelligent Health Perception and Ecological Restoration of Rivers and Lakes, Ministry of Education, Hubei University of Technology, Wuhan, China
- 3Hubei Key Laboratory of Regional Development and Environmental Response, Faculty of Resources and Environmental Sciences, Hubei University, Wuhan, China
Microplastics (MPs) are colonized by biofilm-forming microbes. Biodegradable plastics, popular replacements for traditional plastics, still have unknown biofilm formation characteristics. We conducted a 60-day indoor experiment, where sediment was exposed to traditional MPs (polypropylene, PP), biodegradable MPs (polylactic acid, PLA), and glass beads (GLASS). The microbial communities in the MPs-biofilm were analyzed using high-throughput sequencing. Results indicated that Proteobacteria was the dominant phylum on all substrates, followed by Actinobacteria, and Firmicutes. At the genus level, the majority of microorganisms colonizing PP possessed nitrification and denitrification capabilities, while the dominant bacteria on PLA were capable of degrading lignin, cellulose and carbon metabolism. The genus Sphingomonas, a promising bacteria capable of degrading biodegradable microplastics, was particularly discovered on the PLA biofilm, meanwhile, bacterial colonization of PLA indirectly increased the potential for human transmission of pathogens. Redundancy analysis revealed that the pH and moisture significantly affected the bacterial communities. Pearson correlation heatmap indicated that the abundance of the majority of dominant bacterial genera of two MPs biofilms is negatively correlated with the physicochemical parameters of sediment (pH, moisture, TN, TP), except for salinity. The microbial communities associated with PP and PLA exhibited distinct differences caused by the combined effects of changes in physicochemical properties of sediment and different material substrates. This study provides further evidence of the significant selective features exhibited by microbial colonization on these two MPs when exposed to the same source community, offering insights into the exploration of promising bacteria for MPs degradation.
1 Introduction
Plastic products are widely used in daily necessities packaging, transportation, cutting-edge industries, fishing gear applications and other fields (Ribic et al., 2010; Wagner et al., 2014; Karkanorachaki et al., 2021). Plastic fragments or particles with diameter < 5 mm are defined as microplastics (MPs), which are widely distributed in oceans and freshwater (Law and Thompson, 2014; Auta et al., 2017; Gigault et al., 2018; Yang et al., 2022; Kurniawan et al., 2023). Zhang et al. summarized relevant research of MPs on surface water in China in the past decade: the MPs abundances of rivers and lakes were 3.9–7900 and 340–8900 n/m3, respectively (Zhang et al., 2020). Due to the influence of hydrological conditions, biological enrichment, and microplastic sedimentation, microplastics in the water column would settle into the sediment, resulting in the abundance of microplastics in the sediment often higher than that in the water column (Kukulka et al., 2012; Long et al., 2015; Scherer et al., 2020; Chauhan et al., 2021; Zhao et al., 2023).
Microplastics remain in sediment as an emerging pollutant, becoming a carrier for microbial colonization. As early as 1972, Carpenter et al. discovered that floating small pieces of plastic with attached microorganisms, such as diatoms, in the Sargasso Sea area of the North Atlantic Ocean (Carpenter et al., 1972). Microplastic biofilm enrichment special microorganisms, such as antibiotic-resistant bacteria, pathogenic microorganisms and plastic-degrading microorganisms that will have an impact on environmental microecological processes (Liu et al., 2022; Fajardo et al., 2023; Perveen et al., 2023; Zheng et al., 2023; Vercauteren et al., 2024); they may also increase the surface biology of microplastics, making them easier to be swallowed by organisms and enter the food chain (Rios et al., 2007). The community composition of plastic-associated biofilms was affected by microplastic properties, water types and environmental factors (Arias-Andres et al., 2018; Frère et al., 2018; Nguyen et al., 2022; Saygin et al., 2024). The polymer type of microplastics directly affects the carbon source of microorganisms colonizing its surface, and therefore is considered to be the most important factor affecting the microbial community on its surface (Qiang et al., 2021). It was reported that PS microplastic showed different bacterial communities than PE and PP in the Bay of Brest (Frère et al., 2018). Biofilms were more often detected on plastics (PS, PET, PA, PP) than on natural materials (stone, wood), furthermore, the biofilms detected on plastics were more complex, with higher microbial diversity (Raposo et al., 2022). Studying the interactions between MPs and biofilms is crucial for understanding the degradation of MPs in aquatic environments. As synthetic polymers are not water soluble, biofilm-forming bacteria degrade such materials more efficiently than planktonic strains.
Biodegradable plastics as an alternative of conventional plastics have been considered as a sustainable solution to plastic pollution, therefore, there will be an increasing presence of biodegradable MPs in water. It is worth noting that the accumulation of biodegradable MPs may cause more potential risk in aquatic environment than conventional MPs (Bandopadhyay et al., 2018). Among currently available bioplastics (biodegradable plastics), PLA is one of the bio-based and biodegradable plastics of great commercial value (Karan et al., 2019). It was reported that the interaction between PLA-MPs and M. aeruginosa could promote the degrading of PLA-MPs meanwhile favoring M. aeruginosa growth after 63 days, causing increasing harmful cyanobacteria biomass (Tang et al., 2024). To date, most studies have focused on the effects of nondegradable plastic types commonly found in aquatic environments on biofilm formation (Akdogan and Guven, 2019; Chen et al., 2020; Shabbir et al., 2020; Sun et al., 2023). Furthermore, metabolic pathway analysis showed that microorganisms embedded in microplastic biofilms have greater potential for biodegradation and metabolism (Jiang et al., 2018). Biofilms formed on wood and the biodegradable polymer (polycaprolactone, PCL) exhibited a higher abundance of Alphaproteobacteria, whereas biofilms on conventional synthetic polymers like polypropylene (PP), polystyrene (PS), and polyvinyl chloride (PVC) were predominantly comprised of Gammaproteobacteria (Bhagwat et al., 2021). Therefore, studying the characteristics of biodegradable MPs biofilms is more beneficial for a comprehensive understanding of the interactions between such MPs and microbial communities in water, as well as understanding the behavior and potential toxicity of biodegradable plastic polymers in aquatic environment.
Up to now, our understanding of biofilm formation on microplastics mainly relies on in situ observations. Field studies have shown that various factors such as seasonality, geography, and hydrology can affect the community composition of plastic-associated biofilms (Oberbeckmann et al., 2014). Furthermore, field sampling in situ is subject to inherent variability and uncertainty, while replicable studies with controlled exposure conditions are lacking. In this study, we aimed to 1) examine the effects of substrate type on microbial community assembly, using traditional MPs (polypropylene, PP), biodegradable plastics (polylactic acid, PLA), and a non-plastic substrate (glass beads). The glass beads acted as an inert substrate, providing a surface for colonization but not serving as an energy source for microorganisms; 2) Notably, sediment samples collected from Nan Lake in Wuhan city were subjected to a 60-day exposure to these substrates under precisely controlled conditions. The correlation analysis between changes in soil physicochemical properties caused by two different types of MPs and microbial communities on the biofilms also needs to be explored. This study contributes to a comprehensive understanding of the relationship between MPs and microbial communities, providing a reference method for screening promising microorganisms capable of degrading MPs. It will be beneficial for the prevention and control of MPs pollution in water. Additionally, it allows for a more comprehensive understanding of the behavior and potential toxicity of MPs in aquatic environments.
2 Materials and methods
2.1 Sediment and water sampling
The test sediment and water were collected from Nan Lake, a typical urban lake (30°28′59.2″N, 114°23′3.6″E) in Wuhan, China. Water samples were pre-filtered with a 6-µm sieve to remove potential grazers that may feed on bacteria during the incubation. Sediment samples were sealed in aluminum foil bags and transported back to the lab. After removing impurities (such as plant residues and shells), the sediment was thoroughly mixed and then naturally dried before biofilm experiment.
Microplastics were extracted from sediment samples using a density separation method (Rodrigues et al., 2018; He et al., 2020), three replicates were carried out for each sample. Under a stereo microscope (XTD-7045A, China Coissm) with magnifications ranging from 40 to 100 x, observations and documentation of the color and shapes of plastic particles on the filter paper are documented. Subsequently, particles within the size range of 0.63–5.00 mm are analyzed through a Fourier Transform Infrared Spectrometer (FTIR, Vertex 70, Germany Bruker) within a scan frequency spectrum of 4000 to 400 cm−1. This analysis aims to identify the types of polymers present in the sediment of Nan Lake.
2.2 Biofilm development experiment
According to the results of MPs investigation, traditional microplastic beads (PP), biodegradable microplastics (PLA), as well as non-plastic beads (GLASS) within a diameter of 2 mm were used to evaluate biofilm formation. To prepare for the indoor experiment, the above-mentioned substrates were first soaked in a 2% SDS solution. Afterward, they were rinsed several times with sterile water to remove any leftover reagents from the surface. Following this, the substrates were soaked in 75% ethanol for 24 h and rinsed three more times with sterile water. They were then dried in a drying oven. To minimize the presence of microorganisms, the substrates were exposed to ultraviolet light for 1 hour for sterilization. Lastly, they were stored in sterile bags at a temperature of 4°C until ready for use. The ratio of substrate (M) to sediment (M) was 0.70 g:1 kg. Each substrate weighing 0.7 g was added separately to 1 kg of sediment. This process was repeated three times, ensuring thorough mixing for a duration of 60 days. The incubation was performed under controlled light (12:12 h of light: dark) and temperature (25°C). The whole process of microplastic dosing was carried out on the aseptic operation table. At the beginning of the experiment, a sterile sealing film was used to seal the beaker mouth to avoid contamination by external microorganisms. Pure water was added daily to keep the water level at 1 cm.
2.3 DNA extraction and sequencing
On the day 60, the substrate material was vacuum filtered on a hydrophilic nylon membrane (0.2 µm Millipore), rinsed with sterile water and separated from the filter using sterile stainless-steel pincers or scraper into Eppendorf tubes, after which half of each sample was used for DNA extraction. The TGuide S96 Magnetic soil DNA kit (Tiangen Biotech Beijing Co., Ltd.) was used to extract genomic DNA from substrate samples according to the manufacturer’s protocol. Primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) were used to amplify the 16S rRNA hypervariable region V3-V4. PCR merchandise was examined with agarose gel and purified by omega DNA ablution kit (Omega Inc., Norcross, GA, United States). Illumina Novaseq 6000 platform was used for the collection of pure PCR products.
2.4 Data analysis
The quality of the original DNA sequence was optimized through the QIIME program, and the optimized sequences were clustered into operational taxonomic units (OTUs) for species classification based on 97% similarity. The diversity index (Shannon, Simpson), richness index (Chao1) and coverage index of bacterial communities were calculated using the QIIME2 software (Bolyen et al., 2019). Weighted principal coordinate analysis (PCoA) based on Bray-Curtis distance was used to determine the changes in microbial community structure. Differentially abundant taxa were identified by linear discriminated analysis (LDA) with effect size (LEfSe) (Segata et al., 2011). Redundancy analysis (RDA) and correlation heatmap analyses of physicochemical factors and microbial community were carried out with Majorbio Cloud Platform (www.majorbio.com).
3 Results
3.1 Link between bacterial community on biofilms and physicochemical factors of sediments
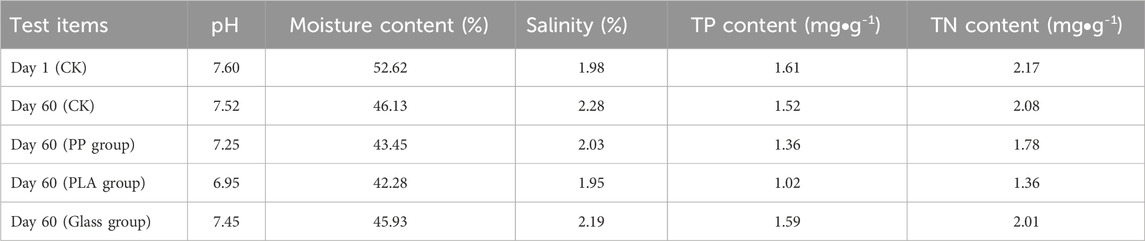
The physicochemical properties of test sediment, as well as the characteristics of MPs were determined. The results showed that the abundance of MPs in sediment was relatively low (68 ± 1.98 n/kg), thus, the influence of background value on subsequent experiment can be ignored. Besides, as shown in Table 1, MPs identified in the sediment of Nan Lake are commonly found and include PP, with all particles having a diameter less than 4 mm. Although PLA was not detected in this sampling, it is not excluded that other sampling sites or freshwater lakes may contain such biodegradable microplastics. Therefore, we selected PP and PLA for comparative analysis of the dissimilarity in microbial communities growing on biofilms. The effects of adding three matrix materials on the physicochemical of sediment were also analyzed as shown in Table 2, the addition of both PP and PLA decreased the physicochemical indicators, including total phosphorus (TP), total nitrogen (TN), and pH value (p < 0.05). However, the addition of GLASS had no effect on the physicochemical properties of sediment (p > 0.05).
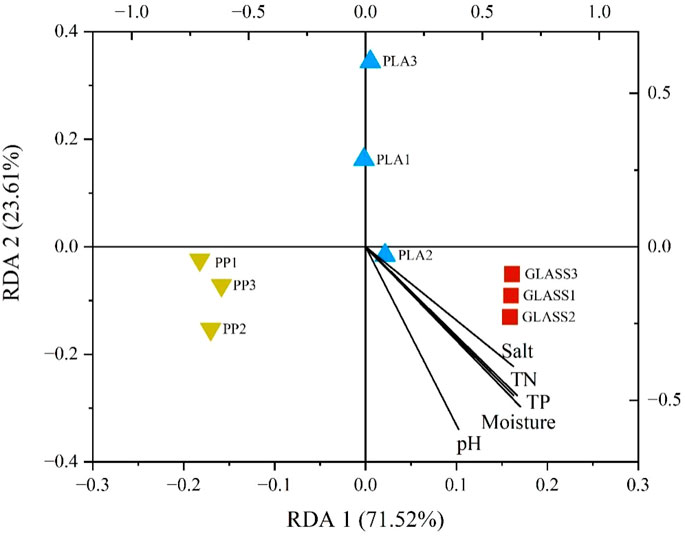
Based on redundancy analysis (RDA), as shown in Figure 1, results revealed that TN, TP and moisture have a strong correlation with each other. Furthermore, the effects of pH and moisture on the overall diversity distribution of the sample communities are the most significant. Compared to GLASS, the bacterial communities of MPs are most influenced by the physicochemical factors. The spearman correlation heatmap analysis was used to investigate the relationship between the sediment physicochemical parameters and relatively microbial abundance of dominated bacterial genera (top 20) during 60-days biofilms formation (Figure 2). Results showed that four physicochemical parameters (pH, moisture, TP and TN) showed a significant negative correlation with the dominant bacterial genera of PLA, such as Sphingomonas and Arthrobacter, while these four parameters showed no correlation with genus Acinetobacter and Staphylococcus, some species of these bacteria being common pathogens. Salinity did not correlate with the above bacteria on the PLA biofilm; conversely, it exhibited a positive correlation with Rhizobacter, Staphylococcus, and Arthrobacter on the PP biofilm. The three parameters (moisture, TP and TN) indicated a negative correlation with the predominant bacterial genera in PP, specifically Nitrospira and Hydrogenophaga, both of which were involved in nitrification and denitrification processes. TP and TN are not correlated with the dominant genera on GLASS, but salinity showed a significant positive correlation with them.
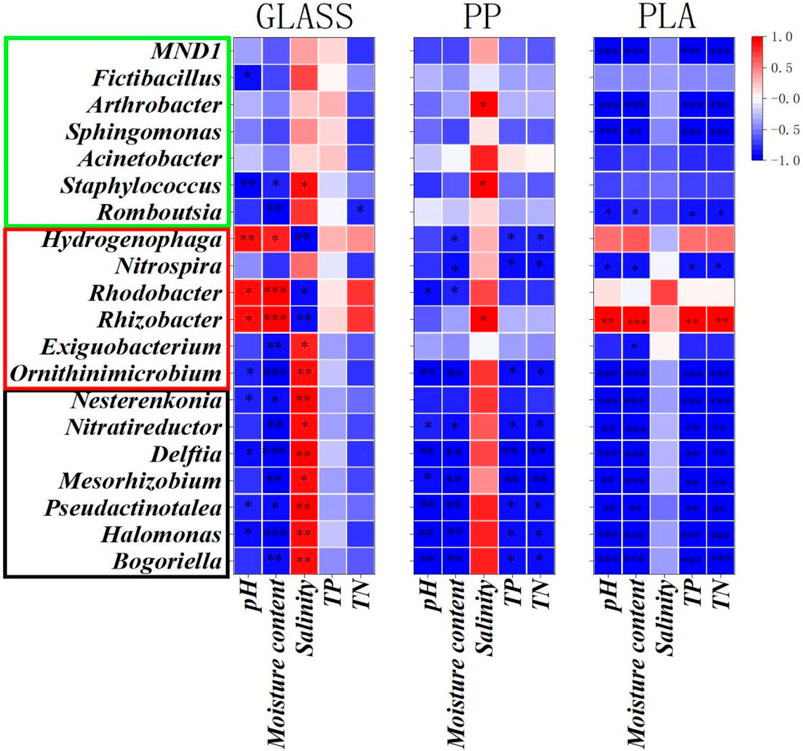
Figure 2. Correlation heatmap of the top 20 genera and physicochemical factors during the 60-day biofilms formation in GLASS, PP, and PLA. The bacteria within the red box, green box, and black box represent the dominant genera in PLA, PP, and GLASS biofilms, respectively. (P < 0.05, P<0.01 and P < 0.001 are demoted as *, **, and ***, respectively.).
3.2 Alpha diversity analysis of bacterial community composition on different substrate surfaces
After 60 days of incubation, a total of 12,149 OTUs were acquired. The numbers of bacterial OTUs on the surfaces of GLASS, PP and PLA were 3125, 3095, and 2944 respectively. The results of Shannon, Simpson, Chao1 and Goods_coverage indexes of each substrate surface as shown in Figure 3. In each treatment group, there was no significant difference in Goods_coverage index, and all were greater than 0.98, indicating that the 16S sequences in the samples were completely detected. The Shannon and Simpson index of the bacterial community on the PP surface were both higher than those of other substrates, especially the Simpson index of the bacterial community on the PP surface was significantly higher than that of other substrates (p < 0.05), indicating that the bacterial community diversity on the PP biofilm was higher.
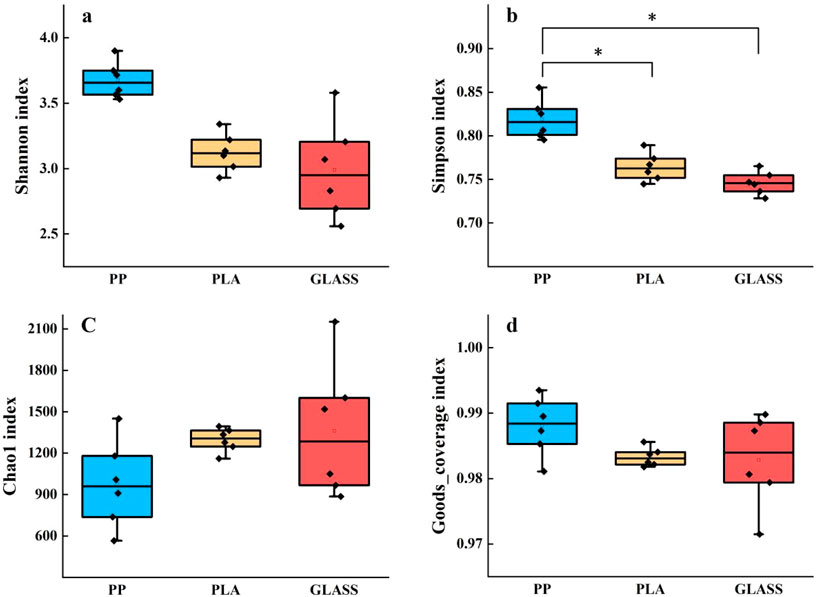
Figure 3. α diversity of bacterial communities on different substrates. (A) Shannon index; (B) Simpson index; (C) Chao1 index; (D) Goods_coverage index (* represented a significant difference between groups (p < 0.05)).
3.3 Beta diversity analysis of bacterial community composition on different substrate surfaces
In Figure 4, the PCoA analysis showed alterations in microbial communities (OTUs based) across three treatments. A significant disparity (ANOISM test, p < 0.01) was observed among the different treatments, with PC1 and PC2 explaining 74% and 10% of the total variation, respectively. The PCoA analysis reveals that the microbial community structures on the PLA and GLASS surfaces are remarkably similar, whereas those on the PLA and PP surfaces show much less similarity. As a non-plastic material, GLASS itself does not contain carbon sources and cannot sustain the growth and proliferation of microorganisms; it only provides a colonization surface for microorganisms. Therefore, the microbial community on the glass surface can indicate the native microorganisms in the sediment.
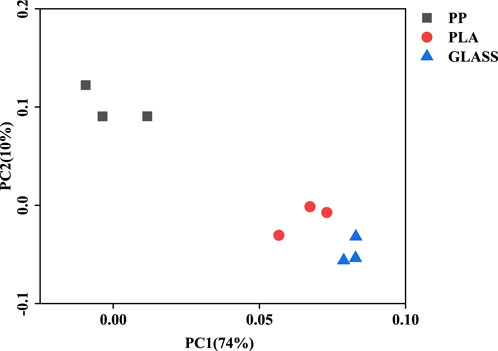
Figure 4. Principal coordinate analysis (PCoA) analysis of bacterial communities based on Bray-Curtis distance across three treatments at the OTUs level.
3.4 Distinct bacterial community structures in different substrates
The distribution of relative abundance of different bacterial community structures at phylum levels of different substrates was shown in Figure 5. The top 10 bacterial categories on each substrate surface were listed. It can be seen that Proteobacteria was the dominant phylum in each substrate (66.43%–71.83%), followed by Actinobacteria (20.52%–23.23%) and Firmicutes (1.59%–6.13%). The relative abundance of Firmicutes on the surface of PLA (6.13%) was higher than that of PP (1.59%) and GLASS (3.82%) (p < 0.05). It was worth noting that Cyanobacteria phylum occupied a higher abundance on the PP surface compared to PLA and GLASS (p < 0.001).
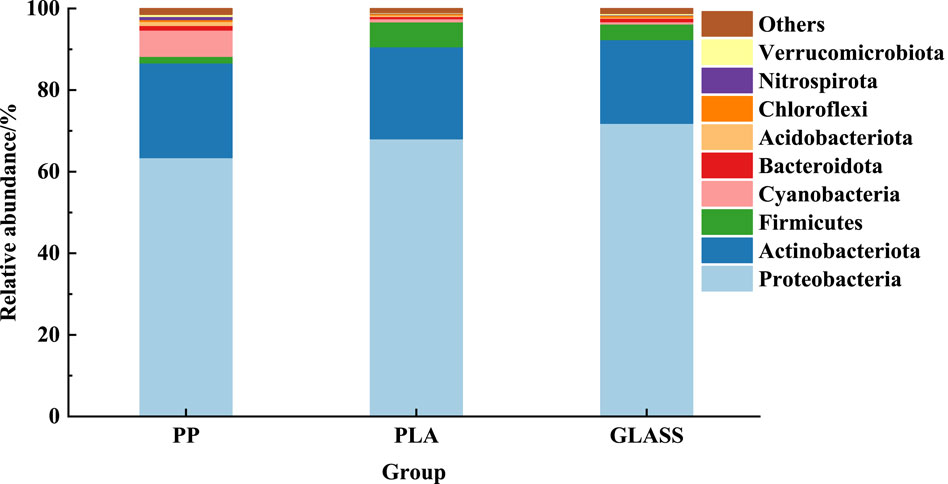
Figure 5. Distribution of relative abundance of different bacterial community structures at phylum levels of different substrates.
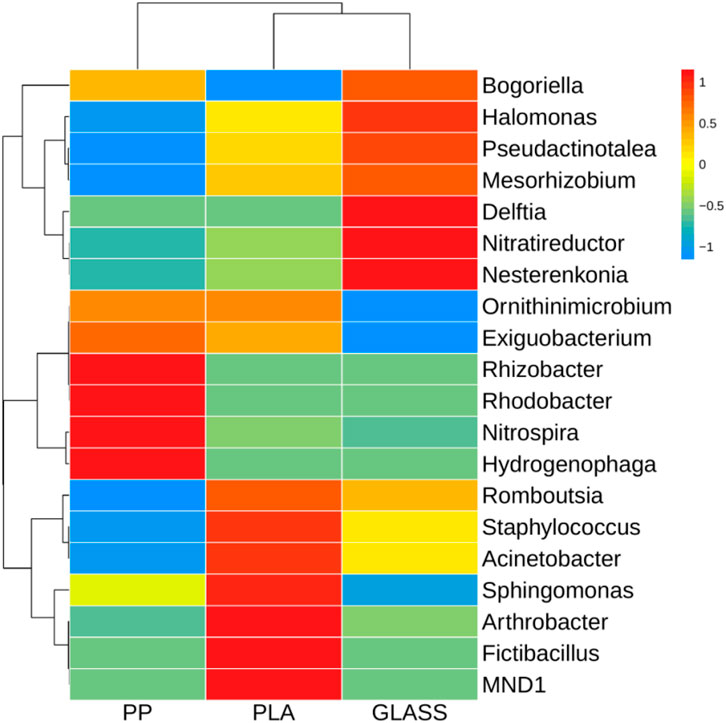
As shown in Figure 6, the heatmap indicated that the dominant genera (top 20) on the surface of different substrates were different from each other. In terms of the microbial communities formed on the PP, the prominent genera were Hydrogenophaga, Nitrospira, Rhodobacter and Rhizobacter. While the bacterial communities of PLA are dominated by MND1, Fictibacillus, Arthrobacter, Sphingomonas, Acinetobacter and Staphylococcus. The bacteria in GLASS were mainly Nesterenkonia, Nitratireductor, Delftia and Halomonas, which were significantly higher than those two MPs substrates (p < 0.01).
In order to determine the differences in the abundance of bacterial species on the surface of three substrates, LEfSe was used to search for metagenomic biomarkers. Differential information for all bacteria at the phylum, class, order, family, and genus level was represented in Figure 7. At the phylum level, there were significant differences in bacteria belonging to the Verrucomicrobiota in the PP samples. At the genus level, bacteria of the genus Halomonas was significantly different in GLASS, bacteria of the genus Aeromicrobium, Ruminococcus_gnavus_group and Acidibacter were significantly different in GLASS, respectively. In the PP sample, there were significant differences in bacteria of the genus Nitrospira from the phylum Nitrospirota, as well as bacteria of the genus Rhodobacter, Tabrizicola, Rhizobacter and Methyloversatilis from the phylum Proteobacteria.
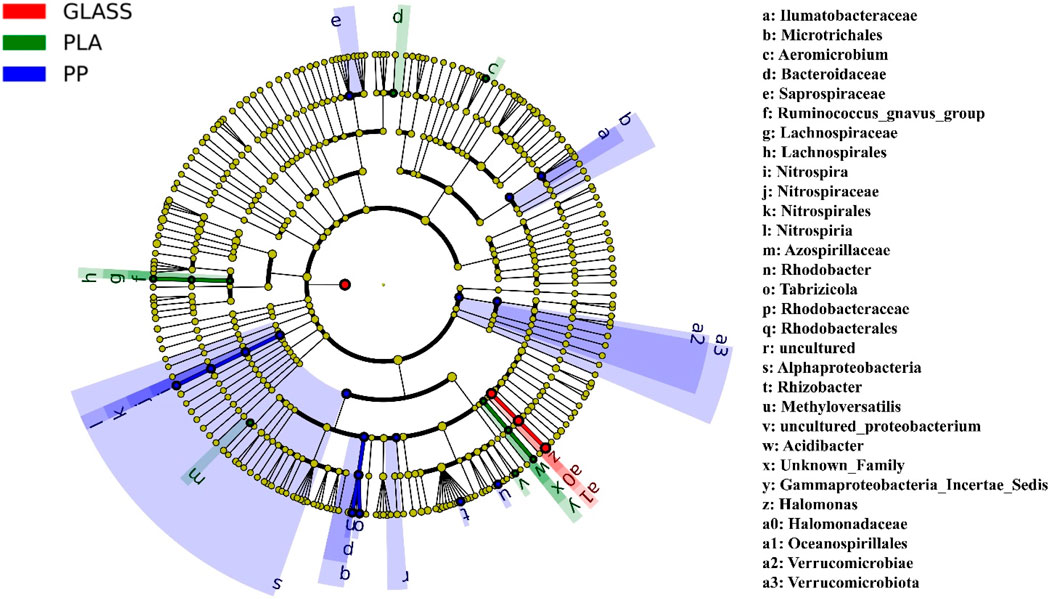
Figure 7. Cladogram indicating the phylogenetic distribution of microbial lineages associated with three substrates; lineages with LDA values of 3.0 or higher determined by LEfSe are displayed. Differences are represented in the color of the most abundant genus (red indicating GLASS, green PLA, blue PP). Each circle’s diameter is proportional to the taxon’s abundance. Circles represent phylogenetic levels from domain to genus inside out. Labels from the inside to the outside, the order is phylum, class, order, family and genus.
4 Discussion
In this indoor study, Proteobacteria was the dominant phylum in each substrate, which is in agreement with previous findings (Roager and Sonnenschein, 2019; Bae and Yoo, 2022), followed by Actinobacteria and Firmicutes. Most of studies have shown that Proteobacteria can adhere to large amounts of the substrate at the early stage of biofilm formation, showing non-specific selection for plastics (Roager and Sonnenschein, 2019). It was also reported that Actinobacteria are able to degrade bioplastics in either laboratory conditions or field trials (Butbunchu and Pathom-Aree, 2019). Notably, we found that the relative abundance of Firmicutes on the surface of PLA was higher than other two matrix. It has also been reported that biodegradable MPs can increase the abundance of Firmicutes (Fan et al., 2023). Microorganisms from the Firmicutes secrete enzymes such as cellulase, lipase, and protease, which may be one of the reasons why these bacteria can stably adsorb on microplastics (Blasco et al., 2011; Ma et al., 2015; Yadav et al., 2016; Dong et al., 2021). Most research studies currently report that the phylum Firmicutes can be isolated from animal bodies and possess the ability to degrade MPs (Lwanga et al., 2018; Janakiev et al., 2023). Besides, in our study, it was worth noting that Cyanobacteria occupied a higher abundance on the PP surface, which was significantly different from the other matrix. Studies have demonstrated that under the stimulation of microplastics (PS, PP), cyanobacteria will secrete more extracellular polymeric substances (EPS) to form hetero-aggregates with microplastics (Jiao et al., 2022; Parsai et al., 2022; Wu et al., 2023). However, the interaction between biodegradable microplastics (such as PLA) and cyanobacteria is still unclear. This phenomenon deserves further study to provide more data for exploration of the interactions between different polymer types and Cyanobacteria.
In particular, the genus Sphingomonas, recognized as an eco-friendly bacterium, was dominated on the PLA biofilm. A recent study indicated that the genera Sphingomonas with higher abundance in the biofilm of Mater-Bi (innovative biodegradable materials) during in situ incubation (Sabatino et al., 2024), this finding is somewhat in-line with our current results. However, Nguyen et al. conducted in situ experiments on HDPE, PLA and PHBV biofilms in freshwater reservoirs, and found that the genus Gemmatimonas and Crenothrix were increased on PLA (Nguyen et al., 2023). Di Pippo et al. investigated LDPE, PET, PLA, and starch-based MaterBi (Mb) biofilms exposed to lakes and found that LDPE, PET, and PLA surface biofilms were predominantly Actinobacteria during early development. The main Actinobacteria genera are Cutibacterium, Propionicella and Olsenella (Di Pippo et al., 2023). Given the variation in habitats, whether analyzing MPs biofilms through laboratory experiments or in situ studies, there exist significant background differences in the formation of microbial communities on these materials. Sphingomonas, has a very rich set of degradation genes, which have application value in degrading aromatic compounds such as pesticides, dioxins, polycyclic aromatic hydrocarbons, and heterocyclic aromatic hydrocarbons, and can coordinate multiple degradation pathways (Zhang et al., 2021). Other studies indicated that PLA-degraded bacteria included Bacillus, Sphingomonas, Pseudomonas (Satti et al., 2018; Zhang et al., 2019). It has been reported that Pseudomonas and Sphingomonas were found to positively related to the degradation of PLA/PBAT film (Zhang et al., 2022). Therefore, we believe that Sphingomonas is a promising friendly bacteria capable of degrading biodegradable microplastics such as polylactic acid (PLA).
In this study, Acinetobacter and Staphylococcus were also particularly identified in the PLA-attached biofilm, with some species of these bacteria being common pathogens associated with high resistance rates and causing nosocomial infections (Antunes et al., 2014; Oliveira et al., 2018; Iruegas et al., 2023; Yao et al., 2023). Microplastics, as an attachment matrix for microorganisms, may also serve as carriers of pathogens, resulting in ecological risks that may harm human health (Bhatt et al., 2021 Lai et al., 2022; Bydalek et al., 2023; Kim and Yoo, 2024). Kesy et al. (2019) reported that salinity, as well as the “life history” of the particles, can greatly influence the composition of biofilm assemblages on microplastics. Our results also showed that the PLA-added group had the smallest salinity at the end of the experiment, and the biofilm was in a mature and stable phase after 60 days cultivation. It was found that microplastic biofilm (PVC) selectively enriched opportunistic human pathogens, but no enrichment was observed in two natural biofilms (rock and leaf) (Wu et al., 2019). Moreover, our findings suggested that there was no correlation between the abundances of the two pathogenic bacteria detected on the PLA biofilm and the variations in the physicochemical properties of the sediment. However, there is currently a lack of relevant research on whether the biodegradable microplastic-biofilms can carry more pathogens than traditional microplastic-biofilms, which requires further validation by indoor and outdoor experimental studies.
In this experiment, we found that the predominant bacterial genera on the surface of PP and GLASS biofilms exhibit the capability for both nitrification and denitrification (Xie et al., 2018; Yang et al., 2020; Ye et al., 2020; Wang et al., 2023). In contrast, the dominant bacterial genera on the PLA surface possess the ability to degrade lignin, cellulose, and engage in carbon metabolism processes (Zhang et al., 2021). This highlights the distinct micro-biological activities and functionalities that are supported by different materials under study. It is noteworthy that there were distinct differences in the relative abundance of genera between PLA and PP. The relative abundance of Fictibacillus on PLA was 37.6 times that of GLASS, but this genus was not found on PP. It has been reported that Fictibacillus phosphorivorans as a common bacterial specie isolated from the gut of Chironomus riparius, one of the sediment-dwelling invertebrate in freshwater ecosystems, exhibited the most effective proteolytic activity to degrade PVC and PA, but not PE, after 1 week of cultivation (Janakiev et al., 2023). In particular, the relative abundance of the genus Arthrobacter on the surface of PLA was 136 times and 15.3 times that of PP and GLASS respectively. A study has shown that Arthrobacter and Sphingomonas were enriched on biodegradable plastic mulch films compared to non-biodegradable PE, meanwhile, these microbial consortia were able to degrade the plastics in laboratory enrichment cultures (Bandopadhyay et al., 2018).
Most studies have demonstrated that the addition of MPs alters the physicochemical properties of soil as well as its microbial community structure (Seeley et al., 2020; Yin et al., 2023; Li et al., 2024). Li et al. reported that PBAT, one of biodegradable microplastic, is beneficial to soil nitrogen fixation, but it will significantly reduce the soil P content (Li et al., 2023). It has been reported that age-PE MPs can significantly reduce soil TN, TP (Zhang et al., 2023). Our results also revealed that compared to the GLASS group, MPs reduced P content in sediments after 60 days, particularly observed in the PLA group. In addition, some studies have reported that during the process of microbial degradation of MPs, extracellular enzymes (hydrolases) are secreted, causing the pH of the culture medium decreased (Wallace et al., 2017; Joo et al., 2018). Therefore, we hypothesize that bacteria with potential degradation capabilities also exist in the biofilms on the surface of MPs in this experiment, leading to a decrease in the pH value of the sediment, especially noticeable in the PLA group.
Furthermore, this study reveals a correlation between changes in the physicochemical properties of sediments and the abundance of specific bacterial taxa on the MPs. In our study, salinity did not correlate with most bacteria in the biofilms of MPs; conversely, it exhibited a significant positive correlation with the dominant bacterial genera in the GLASS biofilm. It has been reported that salinity was the primary factor affecting bacterial diversity of the colonies on plastic debris in Haihe Estuary (Li et al., 2019). In this study, on one hand, we focus on the environmentally friendly bacterium Sphingomonas that appears on PLA, which has a significant negative correlation with pH, moisture, TN, and TP; on the other hand, the dominant bacterial genera (Nitrospira and Hydrogenophaga) appearing on PP also show a negative correlation with moisture, TN, and TP. A study revealed that Pearson correlation analysis between soil properties and bacterial communities showed that both Sphingomonas and Nitrospira were negatively correlated with pH, TN, and TP (Sun et al., 2020).
Whether obtained from field in situ experiments or indoor experiments, the bacterial communities on MP biofilms show distinct differences at the genus and species levels due to geographical factors, physicochemical factors of water bodies, and different seasons, even with the same microplastic polymers. It has been reported that the structure and composition of plastisphere communities showed notable differences among the locations, and the taxonomic composition on MP samples was associated with their origins in sedimentary and aquatic environments (Jiang et al., 2018). According to the report: the organic content in the water emerged as the primary determinant of biofilm communities on MPs (Nguyen et al., 2022). MP biofilm colonization will also be affected by factors such as polymer type, particle size, surface charge, aging, and hydrophobicity, etc. (Amaral-Zettler et al., 2015; Didier et al., 2017; Mercier et al., 2017; Liu et al., 2020). In our study, the addition of MPs to sediment leads to alterations in the micro-environment, which in turn influences the abundance of specific bacterial genera on various materials. The disparity in bacterial communities between MPs and GLASS is predominantly attributed to the intrinsic properties of the materials themselves along with changes in the physical chemical attributes of the sediments. The impact on biofilm formation by combining the properties of MPs themselves and sedimental environmental factors, which deserves further study in the future.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YJ: Funding acquisition, Writing–original draft. AZ: Conceptualization, Formal Analysis, Writing–review and editing. DZ: Project administration, Writing–review and editing. MC: Supervision, Visualization, Writing–review and editing. LW: Funding acquisition, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (No. 42307514), the National Natural Science Foundation of China (No. 41807452), the National Natural Science Foundation of China (No. 42107305), and the Innovation Demonstration Base of Ecological Environment Geotechnical and Ecological Restoration of Rivers and Lakes (2020EJB004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
MPs, microplastics; PP, polypropylene; PLA, polylactic acid; Glass beads, GLASS; PS, polystyrene; PET, polyethylene terephthalate; PA, polyamide; PE, polyethylene; PCL, polycaprolactone; PVC, polyvinyl chloride; PUF, polyurethane; PBAT, polyadipate/butylene terephthalate; LDPE, low density polyethylene; HDPE, high-density polyethylene; PHBV, 3-hydroxybutyrate-co-3-hydroxyvalerate; M. aeruginosa, Microcystis aeruginosa; TP, total phosphorus; TN, total nitrogen; EPS, extracellular polymeric substances; Mb, MaterBi.
References
Akdogan, Z., and Guven, B. (2019). Microplastics in the environment: a critical review of current understanding and identification of future research needs. Environ. Pollut. 254 (Pt A), 113011. doi:10.1016/j.envpol.2019.113011
Amaral-Zettler, L. A., Zettler, E. R., Slikas, B., Boyd, G. D., Melvin, D. W., Morrall, C. E., et al. (2015). The biogeography of the Plastisphere: implications for policy. Front. Ecol. Environ. 13 (10), 541–546. doi:10.1890/150017
Antunes, L. C. S., Visca, P., and Towner, K. J. (2014). Acinetobacter baumannii: evolution of a global pathogen. Pathogens Dis. 71 (3), 292–301. doi:10.1111/2049-632x.12125
Arias-Andres, M., Kettner, M. T., Miki, T., and Grossart, H. P. (2018). Microplastics: new substrates for heterotrophic activity contribute to altering organic matter cycles in aquatic ecosystems. Sci. Total Environ. 635, 1152–1159. doi:10.1016/j.scitotenv.2018.04.199
Auta, H. S., Emenike, C. U., and Fauziah, S. H. (2017). Distribution and importance of microplastics in the marine environment: a review of the sources, fate, effects, and potential solutions. Environ. Int. 102, 165–176. doi:10.1016/j.envint.2017.02.013
Bae, S., and Yoo, K. (2022). Microplastic contamination and microbial colonization in coastal area of Busan City, Korea. Front. Mar. Sci. 9, 1030476. doi:10.3389/fmars.2022.1030476
Bandopadhyay, S., Martin-Closas, L., Pelacho, A. M., and DeBruyn, J. M. (2018). Biodegradable plastic mulch films: impacts on soil microbial communities and ecosystem functions. Front. Microbiol. 9, 819. doi:10.3389/fmicb.2018.00819
Bhagwat, G., Zhu, Q. Y., O’Connor, W., Subashchandrabose, S., Grainge, I., Knight, R., et al. (2021). Exploring the composition and functions of plastic microbiome using whole-genome sequencing. Environ. Sci. Technol. 55 (8), 4899–4913. doi:10.1021/acs.est.0c07952
Bhatt, P., Pathak, V. M., Bagheri, A. R., and Bilal, M. (2021). Microplastic contaminants in the aqueous environment, fate, toxicity consequences, and remediation strategies. Environ. Res. 200, 111762. doi:10.1016/j.envres.2021.111762
Blasco, L., Kahala, M., Tupasela, T., and Joutsjoki, V. (2011). Determination of aspartase activity in dairy Propionibacterium strains. FEMS Microbiol. Lett. 321 (1), 10–13. doi:10.1111/j.1574-6968.2011.02299.x
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37 (8), 852–857. doi:10.1038/s41587-019-0209-9
Butbunchu, N., and Pathom-Aree, W. (2019). Actinobacteria as promising candidate for polylactic acid type bioplastic degradation. Front. Microbiol. 10, 2834. doi:10.3389/fmicb.2019.02834
Bydalek, F., Webster, G., Barden, R., Weightman, A. J., Kasprzyk-Hordern, B., and Wenk, J. (2023). Microplastic biofilm, associated pathogen and antimicrobial resistance dynamics through a wastewater treatment process incorporating a constructed wetland. Water Res. 235, 119936. doi:10.1016/j.watres.2023.119936
Carpenter, E. J., Anderson, S. J., Harvey, G. R., Miklas, H. P., and Peck, B. B. (1972). Polystyrene spherules in coastal waters. Science 178 (4062), 749–750. doi:10.1126/science.178.4062.749
Chauhan, J. S., Semwal, D., Nainwal, M., Badola, N., and Thapliyal, P. (2021). Investigation of microplastic pollution in river Alaknanda stretch of Uttarakhand. Environ. Dev. Sustain. 23 (11), 16819–16833. doi:10.1007/s10668-021-01388-y
Chen, X. C., Chen, X. F., Zhao, Y. H., Zhou, H., Xiong, X., and Wu, C. (2020). Effects of microplastic biofilms on nutrient cycling in simulated freshwater systems. Sci. Total Environ. 719, 137276. doi:10.1016/j.scitotenv.2020.137276
Didier, D., Anne, M., and Alexandra, T. H. (2017). Plastics in the North Atlantic garbage patch: a boat-microbe for hitchhikers and plastic degraders. Sci. Total Environ. 599-600, 1222–1232. doi:10.1016/j.scitotenv.2017.05.059
Di Pippo, F., Bocci, V., Amalfitano, S., Crognale, S., Levantesi, C., Pietrelli, L., et al. (2023). Microbial colonization patterns and biodegradation of petrochemical and biodegradable plastics in lake waters: insights from a field experiment. Front. Microbiol. 14, 1290441. doi:10.3389/fmicb.2023.1290441
Dong, L., Wu, Y., Bian, Y. Z., Zheng, X., Chen, L., Chen, Y., et al. (2021). Carbon nanotubes mitigate copper-oxide nanoparticles-induced inhibition to acidogenic metabolism of Propionibacterium acidipropionici by regulating carbon source utilization. Bioresour. Technol. 330, 125003. doi:10.1016/j.biortech.2021.125003
Fajardo, C., Fortún, S. S., Quintela, D. V., Martin, C., Nande, M., D´ors, A., et al. (2023). Biofilm formation on polyethylene microplastics and their role as transfer vector of emerging organic pollutants. Environ. Sci. Pollut. Res. 30, 84462–84473. doi:10.1007/s11356-023-28278-2
Fan, Z. Q., Jiang, C. L., Muhammad, T., Ali, I., Feng, Y., Sun, L., et al. (2023). Impacts and mechanism of biodegradable microplastics on lake sediment properties, bacterial dynamics, and greenhouse gasses emissions. Sci. Total Environ. 900, 165727. doi:10.1016/j.scitotenv.2023.165727
Frère, L., Maignien, L., Chalopin, M., Huvet, A., Rinnert, E., Morrison, H., et al. (2018). Microplastic bacterial communities in the Bay of Brest: influence of polymer type and size. Environ. Pollut. 242 (Pt A), 614–625. doi:10.1016/j.envpol.2018.07.023
Gigault, J., Halle, A. T., Baudrimont, M., Pascal, P. Y., Gauffre, F., Phi, T. L., et al. (2018). Current opinion: what is a nanoplastic? Environ. Pollut. 235, 1030–1034. doi:10.1016/j.envpol.2018.01.024
He, B. B., Goonetilleke, A., Ayoko, G. A., and Rintoul, L. (2020). Abundance, distribution patterns, and identification of microplastics in Brisbane River sediments, Australia. Sci. Total Environ. 700, 134467. doi:10.1016/j.scitotenv.2019.134467
Iruegas, R., Pfefferle, K., Göttig, S., Averhoff, B., and Ebersberger, I. (2023). Feature architecture aware phylogenetic profiling indicates a functional diversification of type IVa pili in the nosocomial pathogen Acinetobacter baumannii. PLoS Genetics. PLoS Genet. 19 (7), e1010646. doi:10.1371/journal.pgen.1010646
Janakiev, T., Milošević, Đ., Petrović, M., Miljković, J., Stanković, N., Zdravković, D. S., et al. (2023). Chironomus riparius larval gut bacteriobiota and its potential in microplastic degradation. Microb. Ecol. 86 (3), 1909–1922. doi:10.1007/s00248-023-02199-6
Jiang, P. L., Zhao, S. Y., Zhu, L. X., and Li, D. (2018). Microplastic-associated bacterial assemblages in the intertidal zone of the Yangtze Estuary. Sci. Total Environ. 624, 48–54. doi:10.1016/j.scitotenv.2017.12.105
Jiao, Y. Y., Zhu, Y. J., Chen, M., Wan, L., Zhao, Y., Gao, J., et al. (2022). The humic acid-like substances released from Microcystis aeruginosa contribute to defending against smaller-sized microplastics. Chemosphere 303 (Pt 1), 135034. doi:10.1016/j.chemosphere.2022.135034
Joo, S., Cho, I. J., Seo, H., Son, H. F., Sagong, H. Y., Shin, T. J., et al. (2018). Structural insight into molecular mechanism of poly (ethylene terephthalate) degradation. Nat. Commun. 9 (1), 382. doi:10.1038/s41467-018-02881-1
Karan, H., Funk, C., Grabert, M., Oey, M., and Hankamer, B. (2019). Green bioplastics as part of a circular bioeconomy. Trends Plant Sci. 24 (3), 237–249. doi:10.1016/j.tplants.2018.11.010
Karkanorachaki, K., Syranidou, E., and Kalogerakis, N. (2021). Sinking characteristics of microplastics in the marine environment. Sci. Total Environ. 793, 148526. doi:10.1016/j.scitotenv.2021.148526
Kesy, K., Oberbeckmann, S., Kreikemeyer, B., and Labrenz, M. (2019). Spatial environmental heterogeneity determines young biofilm assemblages on microplastics in Baltic Sea mesocosms. Front. Microbiol. 10, 1665. doi:10.3389/fmicb.2019.01665
Kim, H., and Yoo, K. (2024). Marine plastisphere selectively enriches microbial assemblages and antibiotic resistance genes during long-term cultivation periods. Environ. Pollut. 344, 123450. doi:10.1016/j.envpol.2024.123450
Kukulka, T., Proskurowski, G., Morét-Ferguson, S., Meyer, D. W., and Law, K. L. (2012). The effect of wind mixing on the vertical distribution of buoyant plastic debris. Geophys. Res. Lett. 39 (7). doi:10.1029/2012gl051116
Kurniawan, T. A., Haider, A., Ahmad, H. M., Mohyuddin, A., Umer Aslam, H. M., Nadeem, S., et al. (2023). Source, occurrence, distribution, fate, and implications of microplastic pollutants in freshwater on environment: a critical review and way forward. Chemosphere 325, 138367. doi:10.1016/j.chemosphere.2023.138367
Lai, K. P., Tsang, C. F., Li, L., Yu, R. M. K., and Kong, R. Y. C. (2022). Microplastics act as a carrier for wastewater-borne pathogenic bacteria in sewage. Chemosphere 301, 134692. doi:10.1016/j.chemosphere.2022.134692
Law, K. L., and Thompson, R. C. (2014). Microplastics in the seas. Science 345 (6193), 144–145. doi:10.1126/science.1254065
Li, C. T., Li, Z. H., Cui, Q., Hassan, A., Zhang, K., Lu, X., et al. (2023). Effect of different additions of low-density polyethylene and microplastics polyadipate/butylene terephthalate on soil bacterial community structure. Environ. Sci. Pollut. Res. 30, 55649–55661. doi:10.1007/s11356-023-26159-2
Li, W. J., Zhang, Y., Wu, N., Zhao, Z., Xu, W., Ma, Y., et al. (2019). Colonization characteristics of bacterial communities on plastic debris influenced by environmental factors and polymer types in the Haihe Estuary of Bohai Bay, China. Environ. Sci. Technol. 53 (18), 10763–10773. doi:10.1021/acs.est.9b03659
Li, X. W., Qin, H. J., Tang, N., and Xing, W. (2024). Microplastics enhance the invasion of exotic submerged macrophytes by mediating plant functional traits, sediment properties, and microbial communities. J. Hazard. Mater. 469, 134032. doi:10.1016/j.jhazmat.2024.134032
Liu, P., Dai, J. M., Bie, C. Y., Li, H., Zhang, Z. X., and Guo, X. T. (2022). Bioaccessibility of microplastic-associated antibiotics in freshwater organisms: highlighting the impacts of biofilm colonization via an in vitro protocol. Environ. Sci. Technol. 56 (17), 12267–12277. doi:10.1021/acs.est.2c02782
Liu, P., Zhan, X., Wu, X. W., Li, J., Wang, H., and Gao, S. (2020). Effect of weathering on environmental behavior of microplastics: properties, sorption and potential risks. Chemosphere 242, 125193. doi:10.1016/j.chemosphere.2019.125193
Long, M., Moriceau, B., Gallinari, M., Lambert, C., Huvet, A., Raffray, J., et al. (2015). Interactions between microplastics and phytoplankton aggregates: impact on their respective fates. Mar. Chem. 175, 39–46. doi:10.1016/j.marchem.2015.04.003
Lwanga, E. H., Thapa, B., Yang, X. M., Gertsen, H., Salánki, T., Geissen, V., et al. (2018). Decay of low-density polyethylene by bacteria extracted from earthworm's guts: a potential for soil restoration. Sci. Total Environ. 624, 753–757. doi:10.1016/j.scitotenv.2017.12.144
Ma, J., Ma, Y. S., Wei, W., and Wei, D. z. (2015). In vivo functional expression of an extracellular Ca2+-independent Bacillus pumilus lipase in Bacillus subtilis WB800N. Ann. Microbiol. 65 (4), 1973–1983. doi:10.1007/s13213-015-1035-z
Mercier, A., Gravouil, K., Aucher, W., Brosset-Vincent, S., Kadri, L., Colas, J., et al. (2017). Fate of eight different polymers under uncontrolled composting conditions: relationships between deterioration, biofilm formation, and the material surface properties. Environ. Sci. Technol. 51 (4), 1988–1997. doi:10.1021/acs.est.6b03530
Nguyen, H. T., Choi, W., Kim, E. J., and Cho, K. (2022). Microbial community niches on microplastics and prioritized environmental factors under various urban riverine conditions. Sci. Total Environ. 849, 157781. doi:10.1016/j.scitotenv.2022.157781
Nguyen, N. H. A., Marlita, M., El-Temsah, Y. S., Hrabak, P., Riha, J., and Sevcu, A. (2023). Early stage biofilm formation on bio-based microplastics in a freshwater reservoir. Sci. Total Environ. 858, 159569. doi:10.1016/j.scitotenv.2022.159569
Oberbeckmann, S., Loeder, M. G. J., Gerdts, G., and Osborn, A. M. (2014). Spatial and seasonal variation in diversity and structure of microbial biofilms on marine plastics in Northern European waters. Fems Microbiol. Ecol. 90, 478–492. doi:10.1111/1574-6941.12409
Oliveira, D., Borges, A., and Simoes, M. (2018). Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins 10 (6), 252. doi:10.3390/toxins10060252
Parsai, T., Figueiredo, N., Dalvi, V., Martins, M., Malik, A., and Kumar, A. (2022). Implication of microplastic toxicity on functioning of microalgae in aquatic system. Environ. Pollut. 308, 119626. doi:10.1016/j.envpol.2022.119626
Perveen, S., Pablos, C., Reynolds, K., Stanley, S., and Marugán, J. (2023). Growth and prevalence of antibiotic-resistant bacteria in microplastic biofilm from wastewater treatment plant effluents. Sci. Total Environ. 856 (Pt 2), 159024. doi:10.1016/j.scitotenv.2022.159024
Qiang, L. Y., Cheng, J. P., Mirzoyan, S., Kerkhof, L. J., and Häggblom, M. M. (2021). Characterization of microplastic-associated biofilm development along a freshwater-estuarine gradient. Environ. Sci. Technol. 55 (24), 16402–16412. doi:10.1021/acs.est.1c04108
Raposo, A., Mansilha, C., Veber, A., Melo, A., Rodrigues, J., Matias, R., et al. (2022). Occurrence of polycyclic aromatic hydrocarbons, microplastics and biofilms in Alqueva surface water at touristic spots. Sci. Total Environ. 850, 157983. doi:10.1016/j.scitotenv.2022.157983
Ribic, C. A., Sheavly, S. B., Rugg, D. J., and Erdmann, E. S. (2010). Trends and drivers of marine debris on the Atlantic coast of the United States 1997-2007. Mar. Pollut. Bull. 60 (8), 1231–1242. doi:10.1016/j.marpolbul.2010.03.021
Rios, L. M., Moore, C., and Jones, P. R. (2007). Persistent organic pollutants carried by synthetic polymers in the ocean environment. Mar. Pollut. Bull. 54 (8), 1230–1237. doi:10.1016/j.marpolbul.2007.03.022
Roager, L., and Sonnenschein, E. C. (2019). Bacterial candidates for colonization and degradation of marine plastic debris. Environ. Sci. Technol. 53 (20), 11636–11643. doi:10.1021/acs.est.9b02212
Rodrigues, M. O., Abrantes, N., Goncalves, F. J. M., Nogueira, H., Marques, J., and Gonçalves, A. (2018). Spatial and temporal distribution of microplastics in water and sediments of a freshwater system (Antuã River, Portugal). Sci. Total Environ. 633, 1549–1559. doi:10.1016/j.scitotenv.2018.03.233
Sabatino, R., Zullo, R., Di Cesare, A. D., Piscia, R., Musazzi, S., Corno, G., et al. (2024). Traditional and biodegradable plastics host distinct and potentially more hazardous microbes when compared to both natural materials and planktonic community. J. Hazard. Mater. 465, 133166. doi:10.1016/j.jhazmat.2023.133166
Satti, S. M., Shah, A. A., Marsh, T. L., and Auras, R. (2018). Biodegradation of Poly (lactic acid) in soil microcosms at ambient temperature: evaluation of natural attenuation, bioaugmentation and bio-stimulation. J. Polym. Environ. 26, 3848–3857. doi:10.1007/s10924-018-1264-x
Saygin, H., Tilkili, B., Kayisoglu, P., and Baysal, A. (2024). Oxidative stress, biofilm-formation and activity responses of P. aeruginosa to microplastic-treated sediments: effect of temperature and sediment type. Environ. Res. 248, 118349. doi:10.1016/j.envres.2024.118349
Scherer, C., Weber, A., Stock, F., Vurusic, S., Egerci, H., Kochleus, C., et al. (2020). Comparative assessment of microplastics in water and sediment of a large European river. Sci. Total Environ. 738, 139866. doi:10.1016/j.scitotenv.2020.139866
Seeley, M. E., Song, B., Passie, R., and Hale, R. C. (2020). Microplastics affect sedimentary microbial communities and nitrogen cycling. Nat. Commun. 11 (1), 2372. doi:10.1038/s41467-020-16235-3
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12 (6), R60. doi:10.1186/gb-2011-12-6-r60
Shabbir, S., Faheem, M., Ali, N., Kerr, P. G., Wang, L. F., Kuppusamy, S., et al. (2020). Periphytic biofilm: an innovative approach for biodegradation of microplastics. Sci. Total Environ. 717, 137064. doi:10.1016/j.scitotenv.2020.137064
Sun, J. H., Yang, L., Wei, J., Quan, J., and Yang, X. (2020). The responses of soil bacterial communities and enzyme activities to the edaphic properties of coal mining areas in Central China. PLoS One 15 (4), e0231198. doi:10.1371/journal.pone.0231198
Sun, X. L., Xiang, H., Xiong, H. Q., Fang, Y. C., and Wang, Y. (2023). Bioremediation of microplastics in freshwater environments: a systematic review of biofilm culture, degradation mechanisms, and analytical methods. Sci. Total Environ. 863, 160953. doi:10.1016/j.scitotenv.2022.160953
Tang, B. R., Zhang, L. X., Salam, M., Yang, B., He, Q., Yang, Y., et al. (2024). Revealing the environmental hazard posed by biodegradable microplastics in aquatic ecosystems: an investigation of polylactic acid's effects on Microcystis aeruginosa. Environ. Pollut. 344, 123347. doi:10.1016/j.envpol.2024.123347
Vercauteren, M., Lambert, S., Hoogerwerf, E., Janssen, C. R., and Asselman, J. (2024). Microplastic-specific biofilm growth determines the vertical transport of plastics in freshwater. Sci. Total Environ. 910, 168399. doi:10.1016/j.scitotenv.2023.168399
Wagner, M., Scherer, C., Alvarez-Muñoz, D., Brennholt, N., Bourrain, X., Buchinger, S., et al. (2014). Microplastics in freshwater ecosystems: what we know and what we need to know. Environ. Sci. Eur. 26 (1), 12. doi:10.1186/s12302-014-0012-7
Wallace, P. W., Haernvall, K., Ribitsch, D., Zitzenbacher, S., Schittmayer, M., Steinkellner, G., et al. (2017). PpEst is a novel PBAT degrading polyesterase identified by proteomic screening of Pseudomonas pseudoalcaligenes. Appl. Microbiol. Biotechnol. 101 (6), 2291–2303. doi:10.1007/s00253-016-7992-8
Wang, C. J., Hu, H. Y., Shi, J. J., Chen, L., Wang, L., and Bao, Z. (2023). Copper-ion-mediated removal of nitrous oxide by a salt-tolerant aerobic denitrifier Halomonas sp. 3H. Environ. Technol. Innovation 30, 103045. doi:10.1016/j.eti.2023.103045
Wu, Q. R., Li, S. X., Wang, H. Z., Wang, W., Gao, X., Guan, X., et al. (2023). Construction of an efficient microalgal-fungal co-cultivation system for swine wastewater treatment: nutrients removal and extracellular polymeric substances (EPS)-mediated aggregated structure formation. Chem. Eng. J. 476, 146690. doi:10.1016/j.cej.2023.146690
Wu, X. J., Pan, J., Li, M., Li, Y., Bartlam, M., and Wang, Y. (2019). Selective enrichment of bacterial pathogens by microplastic biofilm. Water Res. 165, 114979. doi:10.1016/j.watres.2019.114979
Xie, C. L., Xia, J. M., Wang, J. S., Lin, D. H., and Yang, X. W. (2018). Metabolomic investigations on Nesterenkonia flava revealed significant differences between marine and terrestrial actinomycetes. Mar. Drugs 16 (10), 356. doi:10.3390/md16100356
Yadav, A. N., Sachan, S. G., Verma, P., Kaushik, R., and Saxena, A. K. (2016). Cold active hydrolytic enzymes production by psychrotrophic Bacilli isolated from three sub-glacial lakes of NW Indian Himalayas. J. Basic Microbiol. 56 (3), 294–307. doi:10.1002/jobm.201500230
Yang, R. L., Li, j, Wei-Xie, L. Y., and Shao, L. (2020). Oligotrophic nitrification and denitrification bacterial communities in a constructed sewage treatment ecosystem and nitrogen removal of Delftia tsuruhatensis NF4. Pol. J. Microbiol. 69 (1), 99–108. doi:10.33073/pjm-2020-013
Yang, S. Y., Zhou, M., Chen, X., Hu, L., Xu, Y., Fu, W., et al. (2022). A comparative review of microplastics in lake systems from different countries and regions. Chemosphere 286 (Pt 2), 131806. doi:10.1016/j.chemosphere.2021.131806
Yao, Y. K., Chen, Q., and Zhou, H. (2023). Virulence factors and pathogenicity mechanisms of Acinetobacter baumannii in respiratory infectious diseases. Antibiotics 12 (12), 1749. doi:10.3390/antibiotics12121749
Ye, Y. H., Yan, C., Nie, Y. F., Zhang, J., Zhao, Z., Zhang, R., et al. (2020). Nitratireductor mangrovi sp. nov., a nitrate-reducing bacterium isolated from mangrove soil. Curr. Microbiol. 77 (7), 1334–1340. doi:10.1007/s00284-020-01885-9
Yin, M. Y., Yan, B., Wang, H., Wu, Y., Wang, X., Wang, J., et al. (2023). Effects of microplastics on nitrogen and phosphorus cycles and microbial communities in sediments. Environ. Pollut. 318, 120852. doi:10.1016/j.envpol.2022.120852
Zhang, M., Jia, H., Weng, Y. X., and Li, C. (2019). Biodegradable PLA/PBAT mulch on microbial community structure in different soils. Int. Biodeterior. and Biodegrad. 145, 104817. doi:10.1016/j.ibiod.2019.104817
Zhang, X. Y., Xia, X. J., Dai, M., Cen, J., Zhou, L., and Xie, J. (2021). Microplastic pollution and its relationship with the bacterial community in coastal sediments near Guangdong Province, South China. Sci. Total Environ. 760, 144091. doi:10.1016/j.scitotenv.2020.144091
Zhang, Y., Yang, S., Zeng, Y. P., Chen, Y., Liu, H., Yan, X., et al. (2023). A new quantitative insight: interaction of polyethylene microplastics with soil-microbiome-crop. J. Hazard. Mater. 460, 132302. doi:10.1016/j.jhazmat.2023.132302
Zhang, Y. L., Gao, W., Mo, A. Y., and Jiang, J. (2022). Degradation of polylactic acid/polybutylene adipate films in different ratios and the response of bacterial community in soil environments. Environ. Pollut. 313, 120167. doi:10.1016/j.envpol.2022.120167
Zhang, Z. Y., Mamat, Z., and Chen, Y. G. (2020). Current research and perspective of microplastics (MPs) in soils (dusts), rivers (lakes), and marine environments in China. Ecotoxicol. Environ. Saf. 202, 110976. doi:10.1016/j.ecoenv.2020.110976
Zhao, X. L., Liu, Z. H., Lu, C., and Han, J. (2023). Occurrence and distribution of microplastics in surface sediments of a typical river with a highly eroded catchment, a case of the Yan River, a tributary of the Yellow River. Sci. Total Environ. 863, 160932. doi:10.1016/j.scitotenv.2022.160932
Keywords: microplastic, PP, PLA, sediment, biofilm, bacterial community structure
Citation: Jiao Y, Zhou A, Zhang D, Chen M and Wan L (2024) Distinct microbial community structures formed on the biofilms of PLA and PP, influenced by physicochemical factors of sediment and polymer types in a 60-day indoor study. Front. Environ. Sci. 12:1452523. doi: 10.3389/fenvs.2024.1452523
Received: 21 June 2024; Accepted: 23 August 2024;
Published: 02 September 2024.
Edited by:
Weitao Liu, Nankai University, ChinaReviewed by:
Shangwei Zhang, Beijing Normal University, ChinaHuacheng Xu, Chinese Academy of Sciences (CAS), China
Liandong Zhu, Wuhan University, China
Copyright © 2024 Jiao, Zhou, Zhang, Chen and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiying Jiao, amlhb3lpeWluZzEyMjNoZ2RAMTYzLmNvbQ==
 Yiying Jiao
Yiying Jiao Anqi Zhou1
Anqi Zhou1