- 1Department of Environment and Resource Sciences, Interdisciplinary Graduate School of Agriculture and Engineering, University of Miyazaki, Miyazaki, Japan
- 2Division of Environmental Chemistry, IDEA Consultants, Inc., Tokyo, Japan
- 3Department of Food, Life and Environmental Science, Faculty of Agriculture, Yamagata University, Yamagata, Japan
- 4Department of Civil and Environmental Engineering, Faculty of Engineering, University of Miyazaki, Miyazaki, Japan
- 5Department of Health Science, School of Allied Health Sciences, Kitasato University, Kanagawa, Japan
- 6Division of Microbiology, Department of Infectious Medicine, Kurume University School of Medicine, Fukuoka, Japan
Studies on the conditions and pollution routes of antibiotic-resistant bacteria (ARB) in rivers can help provide countermeasures against the spread of ARB. This study focused on the pristine uppermost stream of a river, where Escherichia coli (E. coli) and enterococci were detected, although the stream flows through a pristine forest catchment. Antibiotic resistance of E. coli and enterococci isolated from the river water, riverbed sediment, and feces of waterside animals, such as birds and Mustelidae, were investigated throughout the year in the pristine uppermost sites. Antibiotic resistance was present in 1.4% (7/494) of the E. coli strains and 3.0% (24/812) of the enterococcal strains, and was low throughout the year. Although antibiotic resistance of bacteria isolated from feces was not detected in this watershed, the prevalence of multidrug-resistant E. coli was 0.4% (1/246) and 0.6% (1/172) in river water and riverbed sediment samples, respectively were observed. The presence of extended-spectrum β-lactamase (ESBL)-producing E. coli was confirmed in river water samples, and genomic analysis revealed that the samples possessed the CTX-M-15 group. Multidrug-resistant strains and ESBL-producing strains were classified as phylogroups B1 and A, respectively, which are E. coli phenotypes isolated from wild animals. Pulsed-field gel electrophoresis revealed analysis targeting enterococci that strains isolated from river water and bird feces were in the same cluster with 100% similarity. Therefore, bird feces are a source of enterococci in the uppermost stream of the river. Because multidrug-resistant bacteria and ESBL-producing bacteria were present in the pristine uppermost stream of the pristine river, urgent elucidation of the spreading routes of ARB is important.
1 Introduction
The emergence of antibiotic-resistant bacteria (ARB) is one of the most serious problems globally (O’Neill, 2014; Willyard, 2017; Antimicrobial Resistance Collaborators, 2022). The Group of Seven (G7) is promoting global efforts to address this important issue (G7 Health Ministers’ Communiqué, 2022; World Health Organization, 2022b) and Centers for Disease Control and Prevention (CDC) annually update and publish information on ARB (World Health Organization, 2022a; Centers for Disease, 2019), warning of the seriousness of the problem. Currently, a wide variety of ARB have been detected, and the main sources of ARB include clinical institutions, livestock farms, and fish farms, where antibacterial drugs are frequently used. ARB are ubiquitous in other environments, such as the water environment (Zhang et al., 2015; Daniel et al., 2017; Osińska et al., 2020; Grenni, 2022). Strains resistant to clinically important antibiotics, such as cephalosporins, carbapenems, and vancomycin, have been detected in the water environments (Diwan et al., 2018; Givens et al., 2023). In recent years, ARB, and antibiotic resistance genes (ARGs) have been detected in natural environments and wild animals that are not directly affected by humans. Of the six Enterobacterales isolated from pristine freshwater (rivers) in Brazil, 52%–77% were antibiotic resistance (Lima-Bittencourt et al., 2007). ARGs (1.7 ± 1.0 × 106 copies/mL), predominantly those resistant to β-lactam and tetracycline, were detected in the ocean near Antarctica (Jang et al., 2022). In Uganda, 17% of the Escherichia coli (E. coli) strains isolated from wild gorillas were antibiotic resistant. Of these, 4.2% were resistant to antibiotics used for treating lung inflammation in livestock, such as ceftiofur (Rwego et al., 2008). According to a wildlife review, many Enterobacterales isolated from wild animals possessed plasmids encoding ARGs related to β-lactam antibiotics and colistin, causing concerns about the spread of antibiotic resistance in wild animals and the risk to public health (Dolejska and Papagiannitsis, 2018). Therefore, the global spread of ARB cannot be denied.
Information on rivers that transport and spread ARB over a wide area is crucial, but information on ARB in pristine rivers that are not directly affected by human areas remains scarce. Furthermore, it is difficult to identify the appearance of ARB in pristine environments and the specific source and route of contamination. Although limited information is available, 18% of E. coli detected in a pristine upstream site of the Kaeda River in Miyazaki, Japan, where the catchment area was forest, were ARB, including multidrug-resistant strains (Nishimura et al., 2021). In addition, headwaters are surrounded by forests, which have a rich ecosystem and are home to many wild animals. Studies from other countries suggest that the source of ARB load in headwater areas is likely to be wild animals (birds, mammals, etc.) that use waterside areas (Bonnedahl and Järhult, 2014; Rogers et al., 2018; García et al., 2020; Yuan et al., 2021). However, there is a lack of information on whether the development of antibiotic resistance in headwaters is human-introduced or naturally occurring. When focusing on wild animals as a source, direct contact with wild animals to perform study is difficult and risky. Thus, monitoring ARB in headwaters surrounded by forests and identification of contamination routes using scientific methods can be important countermeasures against the spread of ARB. In addition, pristine environments are ideal environments for understanding the mechanisms and interactions of the early stages of evolution, acquisition, and transmission of drug resistance, and can accumulate new information related to drug resistance (Hwengwere et al., 2022).
Elucidating the actual presence and diffusion routes of ARB in pristine rivers is crucial for controlling the transport and diffusion of ARB. The purpose of this study is to determine the prevalence of ARB in pristine river. Then, the presence of clinically important ARB will be confirmed, and the pollution source will be estimated by genomic analysis. The survey focused on the pristine uppermost stream of the Kaeda River (Figure 1) to determine the prevalence of antibiotic-resistant E. coli (AR-E. coli) and antibiotic-resistant enterococci (AR-ENT) in the river water, riverbed sediment, and feces of waterside animals every 2 months for 1 year. Then, information on the detected ARB was organized, and their characteristics and presence of antibiotic-resistant strains, which are important in clinical institutions, were ascertained. Strains from river water, riverbed sediment, and waterside bird feces were analyzed by genomic analysis and pulsed-field gel electrophoresis (PFGE) to trace the source of ARB.
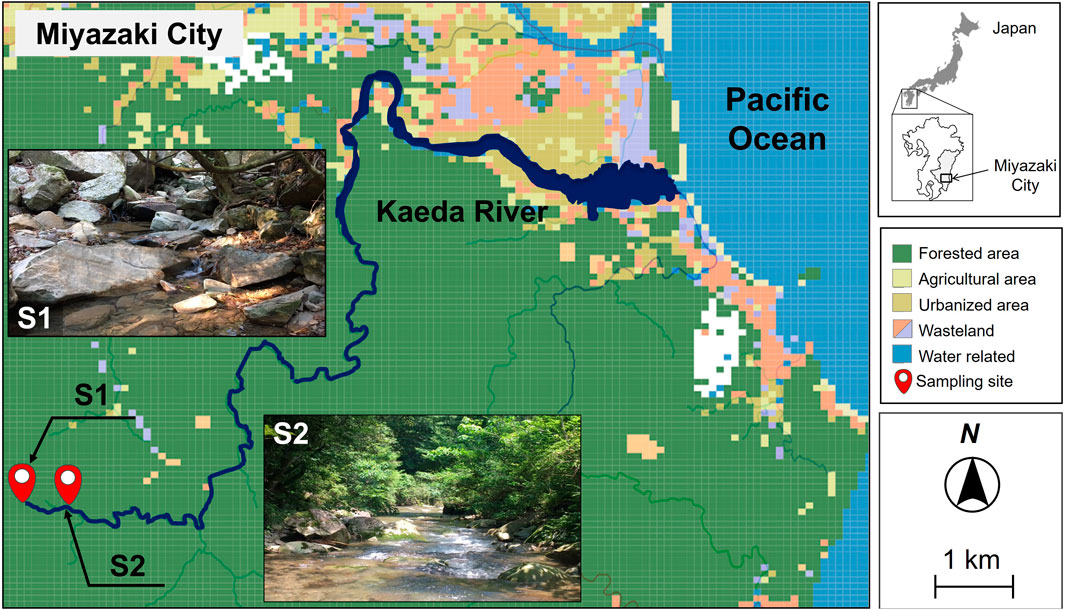
Figure 1. Sampling sites and major land use categories of pristine uppermost stream at the Kaeda River in southern Japan. S1, site1, river source; S2, site2, upstream.
In our previous study (Nishimura et al., 2021), we confirmed the presence of ARB in the upstream stream of the Kaeda River, and its prevalence was comparable between the upstream and downstream streams. However, we have not been able to elucidate the source of ARB in upstream streams, where deep in the forest and unaffected by humans. Therefore, this study was designed to focus on the uppermost reaches of the Kaeda River in the forest and the animals that use their banks. As the target bacteria, we focused on E. coli and enterococci. These bacteria, whose hosts are warm-blooded animals, including humans, livestock, and wild animals, are ubiquitous in aquatic environments (Farnleitner et al., 2010). They are also classified as fecal indicator bacteria and are important for the protection of public health (World Health Organization, 2022a; Ishii and Sadowsky, 2008). In addition, some important bacteria are resistant to antibiotics, and information is accumulated in the aquatic environment (Alonso et al., 2001; Baquero et al., 2008).
2 Materials and methods
2.1 Sampling sites and sample collection
Figure 1; Supplementary Table S1 show the sampling site map as land use data around the Kaeda River and the number of samples, respectively. The Kaeda River (channel length, 17.5 km; basin area, 53.8 km2) originates from headwaters dominated by forests in their catchment areas, flows through a deep valley with land with various uses, and drains into the Hyuganada (Pacific Ocean). In this study, two sites were selected as pristine uppermost streams not directly affected by human activities: 1) a headwaters site (S1: width, 0.3 m; depth, 0.2 m; flow rate, 2 m3/s) and 2) approximately 400 m below the headwaters site (S2: width, 4 m; depth, 0.4 m; flow rate, 13 m3/s). The catchments of S1 and S2 were exclusively forested and consisted of natural and planted forests, respectively. There is no anthropogenic impact from human activity in this study area. Sampling was conducted every 2 months for 1 year (31 August 2017–30 August 2018).
River water, riverbed sediment, and feces from wild animals that use the waterside were collected. River water samples were stored in sterile 5-L polyethylene bottles for each site. Riverbed sediments were collected by shoveling 2–5 cm of the surface layer from 3 m radius of the water sample points. The feces of wild animals (birds and Mustelidae) on waterside rocks situated within a 10 m radius of the water sample points were collected with a sterile cotton swab and placed into 15 mL sterilized polyethylene tubes.
Water temperature and dissolved oxygen (DO) were determined using a fluorescent-type dissolved oxygen meter (HQ40d, Hach Company, COL., Tokyo, Japan) at the sampling site. A benchtop pH/water quality analyzer (LAQUA, HORIBA, Ltd., Kyoto, Japan) was used to measure pH and electrical conductivity. Turbidity was determined using a turbidity meter (SEP-PT-706D, Mitsubishi Chemical Co., Tokyo, Japan). All samples were transported back to the laboratory and used for experiments within 4 h of collection (Japanese Standard Association, 2016).
2.2 Counting and isolation of fecal indicator bacteria
The number of E. coli and enterococci, as well as other coliforms, was counted in each sample by the membrane filter method. Five liters of river water was collected from each sampling site. Bacterial counts varied greatly depending on the survey period, so the amount of passing water sample was set at three levels (10, 100, and 1,000 mL) for each survey and filtered through a 0.45 μm pore membrane filter (47 mm in diameter, mixed cellulose ester; Advantec, Tokyo, Japan). Membranes were placed on CHROMagar ECC agar plates (CHROMagar, Paris, France), and incubated at 37°C for 24 h. Subsequently, filters with appropriate flow volume for bacterial counting and isolation were selected and analyzed. On the filter, blue colonies were considered presumptive E. coli and mauve colonies were considered other presumptive coliform. Enterococci were counted using membrane-enterococci indoxyl-β-D-glucoside (mEI) agar plate (United States Environmental Protection Agency, 2014). The samples (10, 100, and 500 mL) were filtered through a membrane filter. Membranes were incubated on mEI agar plates at 41°C for 24 h. On the filter, blue colonies were considered presumptive enterococci. The number of bacteria in each sample was counted in three replicates and the mean was calculated as colony forming unit (CFU) per 100 mL. The detection limit of this method was 0.3 CFU/100 mL. Riverbed sediment samples (5 g) were mixed with 40 mL of sterilized physiological saline solution or phosphate-buffered saline (Boehm et al., 2009) and allowed to settle for 1 min. Supernatant liquid was filtered through a membrane filter. Then bacteria were isolated in a manner identical to that of water sample analysis. The detection limit of riverbed sediment samples was 7 CFU/100 g. Samples of wild animal feces (1 g) were mixed with 9 mL of sterilized physiological saline solution or phosphate-buffered saline and allowed to settle for 1 min. Then, 0.1–0.001 mL of the supernatant was filtered through a membrane filter. Bacterial counts of feces were not measured, only strains were isolated. Thirty single colonies were randomly isolated from CHROMagar ECC and mEI agar plates for each sample at S1 and S2. The plates were streaked twice on Brain Heart Infusion agar (BD, New Jersey, United States) or Todd–Hewitt agar (Becton, Dickinson, NJ, United States) to isolate E. coli and enterococci, respectively. When less than 30 isolates were available, all single colonies were isolated. The plates were incubated for 24 h at 37°C. For E. coli-positive strains and enterococci-positive strains, max 30 strains were isolated from each sample at S1 and S2. Supplementary Tables S2, S3 show the number of E. coli-positive strains or enterococci-positive strains isolated from each sample at S1 and S2.
2.3 Identification of E. coli and enterococci by MALDI-TOF MS
For E. coli-positive (n = 567) and enterococci-positive (n = 839) strains isolated from all the samples, the identification of bacterial species was performed using MALDI-TOF MS (Fenselau and Demirev, 2001; Christner et al., 2014; Suzuki et al., 2018). All samples were analyzed using an Autoflex III TOF/TOF mass spectrometer (Bruker Daltonics, Billerica, MA, United States). Measurements were performed using flexControl 3.0 software (Bruker Daltonics, Billerica, MA, United States) for database construction and validation. The software settings were based on Suzuki et al. (2018). The instrument was calibrated using a Bruker bacterial test standard (part no. 8255343, Bruker Daltonics, Billerica, MA, United States). Recorded mass spectra were processed with the MALDI Biotyper Compass microbial identification system (Bruker Daltonics, Billerica, MA, United States) using standard settings. The MALDI Biotyper output score had a range of 0.00–3.00, and E. coli and enterococci identification scores were ≥ 2.00.
2.4 Antibiotic susceptibility testing
Antibiotic susceptibility testing was performed on strains identified as E. coli (n = 494) and enterococci (n = 813). The minimum inhibitory concentration (MIC) of each antibiotic was determined using the agar dilution method, according to the guidelines of Clinical Laboratory Standards Institute (CLSI) (Clinical Laboratory Standards Institute, 2017). Twelve antibiotics that are important in the resistance of E. coli and enterococci were used. The preculture, dissolution and dilution of antibiotics, and test procedure were based on Nishimura et al. (2021) for E. coli and Nishiyama et al. (2017) for enterococci. According to the recommendations of CLSI, 12 antibiotics were tested using plates containing 2-fold dilutions of antibiotics with five graded concentrations. MIC breakpoints for resistance were based on CLSI (CLSI, 2012) criteria. Quality control was used for E. coli ATCC 25922 (E. coli) and enterococci ATCC 29212 (Enterococcus faecalis).
2.5 Genomic analysis
Genomic analysis was performed on strains determined as ESBL-producing E. coli (n = 1) and multidrug-resistant E. coli (n = 1). In advance, the double-disk synergy test was performed for the ESBL-producing E. coli strain (Clinical Laboratory Standards Institute M100-S26). Genomic DNA was purified from 1 mL overnight culture of E. coli strains using DNeasy Blood and Tissue Kit (Qiagen). Libraries were prepared using Lotus DNA Library Prep Kit (Integrated DNA Technologies, Coralville, IA, United States) and NEBNext Multiplex Oligos for Illumina (96 Unique Dual Index Primer Pairs) (New England BioLabs Japan, Tokyo, Japan) and sequenced on an Illumina HiseqX Ten platform (Illumina, San Diego, CA, United States) to generate 151-bp paired-end reads. Genome assembly was performed using Platanus_b v1. 3.2 (Kajitani et al., 2020) with default parameters. Assembly quality was assessed using CheckM v1. 2.0 (Parks et al., 2015). Phylogroup and sequence type were determined by ClermonTyping v20.06 (Beghain et al., 2018) and srst2 v2.0 (Inouye et al., 2014), respectively. ARGs were identified by ABRicate v0.9.8 (https://github.com/tseemann/abricate) using the ARG-ANNOT database (Gupta et al., 2014) with default parameters. Mutations for quinolone resistance and colistin resistance were analyzed using AMRFinderPlus 3.11.14 (Feldgarden et al., 2021) with default settings.
2.6 Analysis of PFGE typing for E. coli and enterococci
In total, E. coli (n = 30) and enterococci (n = 29) strains were randomly selected from river water, riverbed sediment, and bird feces from the October 2017 (autumn) survey for genotyping using PFGE. Traces of wild animals at the study site were considered important for examining the bacterial load in the surface waters. Therefore, the analysis focused on October 2017, when the highest number of bird fecal samples were collected at the study site. For E. coli genotyping, PFGE was performed according to the standardized PulseNet protocol for PFGE provided by CDC (Centers for Disease Control, 2017). For enterococci genotyping, PFGE was performed using CHEF Bacterial Genomic DNA Plug Kit (Bio-Rad, Hercules, California, United States), according to the manufacturer’s protocol, with slight modifications. A lambda DNA ladder (range 48.5–873 kb; Lonza, Rockland, ME, United States) was used as a size marker. The details of analysis are shown in Supplementary Text S1.
Band-based PFGE patterns were clustered using Gene Profiler software (Scanalytics, Buckinghamshire, United Kingdom). Levels of similarity between fingerprints were expressed as Dice coefficient. PFGE patterns were clustered using the unweighted pair group method with arithmetic mean. PFGE patterns with 100% similarity were considered identical genotypes.
3 Results and discussion
3.1 Water quality of the pristine uppermost stream in the Kaeda River
Supplementary Table S4 shows the water quality of the pristine uppermost stream in the Kaeda River. Water temperatures at S1 (22.8°C) and S2 (24.7°C) were highest in August (summer) and ranged from 6.6°C to 20.4°C in other surveys (spring, autumn, and winter). The difference in water temperature between S1 and S2 was 1°C–2°C. DO was the highest in winter (10.9 mg/L, near the saturation concentration) and decreased in summer. pH ranged from 6.0 to 6.9, slightly lower than neutral. Electrical conductivity differed between S1 and S2 (47.9 and 58.9 μS/cm, respectively), presumably due to increased mineral components due to the flow processes. Turbidity was extremely low across all surveys, with mean values of 0.3 turbidity units at both sites, and the sample water was extremely clear. Analysis of DO, pH, and turbidity, the water quality parameters of river water, showed no significant differences in water quality between S1 and S2 (p > 0.05), resulting in the water quality in both stations being similar. It is unlikely that the process of flow downstream in these two points would directly affect bacterial inactivation.
3.2 Bacteria counts of in river water and riverbed sediment
Figure 2 shows changes in the numbers of E. coli, enterococci, and other coliforms at S1 and S2 in 1 year. In river water at S1 and S2, E. coli count was low throughout the year (range: 1.7 ± 0.3–20.0 ± 3.3 CFU/100 mL). In summer (August 2017 and 2018), enterococci count was higher than E. coli count, ranging from 47.3 ± 2.6 to 101.3 ± 14.6 CFU/100 mL. In winter (February 2018), enterococci count decreased to <0.3 CFU/100 mL and increased again in summer. Thus, seasonal variation in E. coli and enterococci count differed. Other coliform count showed seasonal variation similar to enterococci count, ranging from 9.2 × 101 ± 20.8 to 1.9 × 104 ± 24.9 CFU/100 mL. By contrast, the seasonal variation of E. coli and enterococci counts in the riverbed sediment from both S1 and S2 were similar. The numbers of E. coli and enterococci were high in summer (6.5 × 102 ± 4.7 × 102–7.9 × 103 ± 4.0 × 103 CFU/100 g) and decreased in winter (7.7 ± 10.9 to 7.9 × 103 ± 4.0 × 103 CFU/100 g). However, bacterial counts were not significantly different between summer and winter (E. coli: p > 0.05, enterococci: p > 0.05).
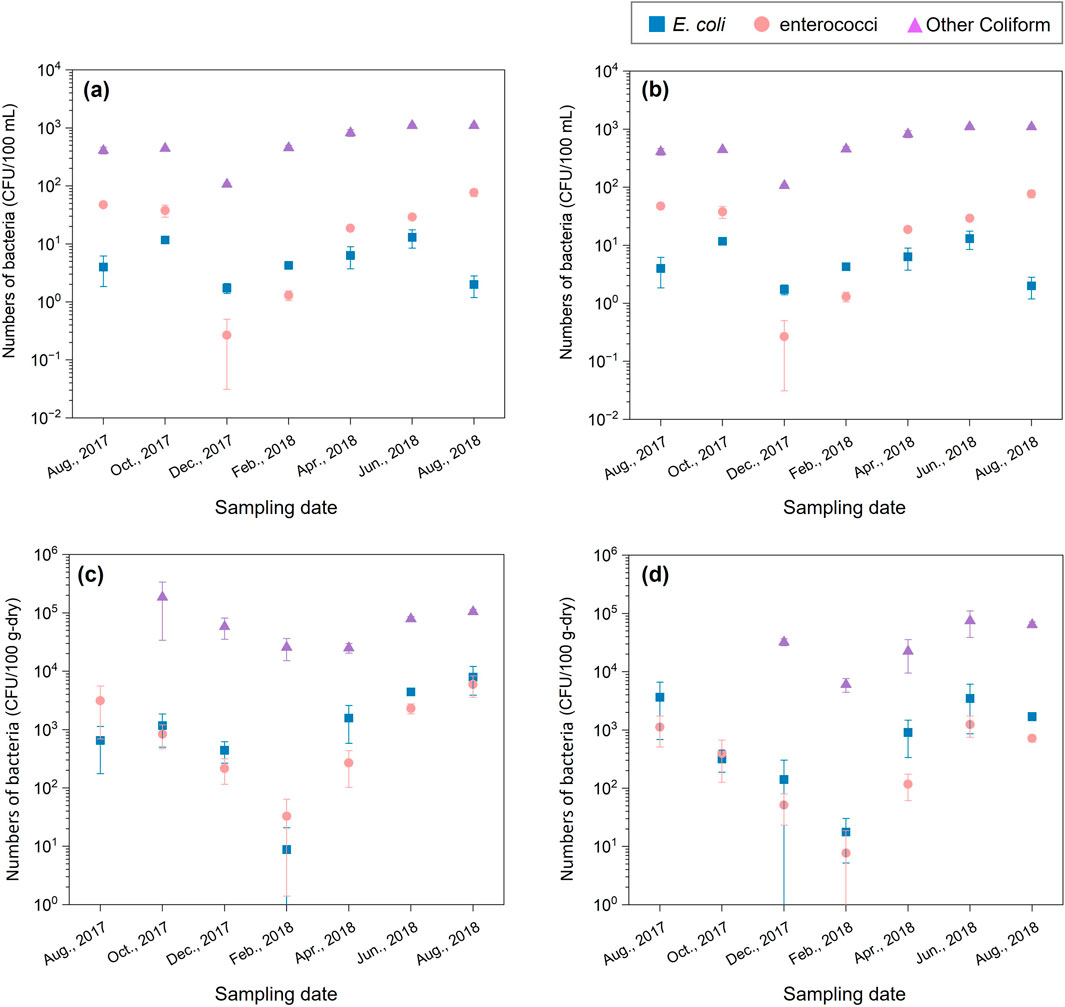
Figure 2. Changes in the numbers of Escherichia coli (E. coli) and enterococci at the S1 and S2 over a 1-year period. Bacteria counts of river water at S1 (A), at S2 (B). Bacteria counts of riverbed sediment at S1 (C), at S2 (D).
Escherichia coli and enterococci were detected throughout the year, although the stream flows through a pristine forest catchment. The sampling site are not a human living area. Seasonal variations in bacterial counts in river water and riverbed sediment are associated with the activity of wild animals. Some species of wild animals are more active in summer, leading to a potentially higher impact of fecal coliform bacteria in the water due to waste deposition. Therefore, bacteria in the river may have been loaded by wild animals (Hansen et al., 2020; Afolabi et al., 2023). In addition to wild animals, other environmental factors, such as rainfall, water temperature, and decomposition of organic matter also significantly influence bacterial counts.
3.3 Identification rate of each positive strain
In total, 567 strains were isolated as E. coli-positive strains from all samples, and 494 (87.1%) were identified as E. coli after MALDI-TOF MS. The identification rate of E. coli was 88.4% (252/285) in river water, 88.8% (175/197) in riverbed sediment, and 78.8% (67/85) in wild animal feces. The major bacterial species of the pseudo-positive strains detected in this study were Escherichia marmotae, Citrobacter freundii, and Serratia fonticola.
In total, 839 strains were isolated as enterococci-positive strains from all samples, and 813 (96.9%) were identified as enterococci (Figure 3). Of the 813 strains identified as enterococci, 83.9% (704/839) were E. faecalis, 9.1% (76/839) were E. cassleflavus, 1.7% (14/839) were E. faecium, 1.3% (11/839) were E. hirae, 0.8% (7/839) were E. gallinarum, and 0.1% (1/839) were E. saccharolyticus. The identification rate of enterococci for river water, riverbed sediment, and wild animal feces was 97.1% (371/382), 95.2% (300/315), and 100.0% (142/142) strains, respectively. E. faecalis was predominant in all samples (80.4%–88.0%). E. faecium was slightly detected (0.3%–3.4%). E. faecalis and E. faecium were frequently isolated in the clinical institutions as the major caUnited Statestive species of nosocomial infection (E. faecalis: 80%–90%, E. faecium: 5%–15%) (Ruoff et al., 1990; Gordon et al., 1992). The detection of E. faecalis and E. faecium in the pristine uppermost sites of the Kaeda River is a serious threat to public health, and it is important to investigate their antibiotic resistance.
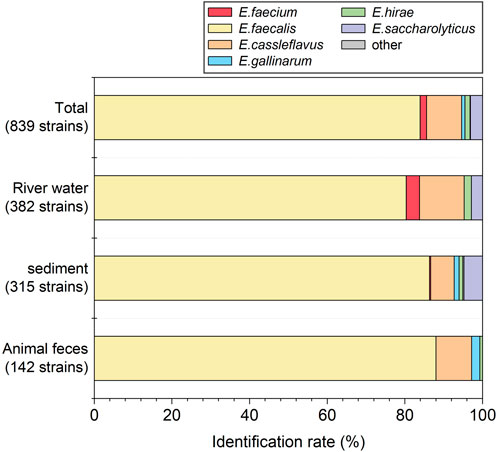
Figure 3. Identification rate of enterococci isolates collected from each sample by MALDI-TOF MS based on the MALDI biotyper software database.
3.4 Detection and seasonal variation of ARB in the pristine uppermost stream
The MIC values of the identified strains of E. coli (n = 494) and enterococci (n = 813) were tested for 8 and 9 antibiotics, respectively. One strain of enterococci was excluded because it did not grow. The antibiotic resistance rates of E. coli and enterococci strains isolated from all samples were 1.4% (7/494) and 3.0% (24/812), which were very low throughout the year (Figure 4). In river water, E. coli was highest in spring 4.8% (1/21) and enterococci was highest in winter 4.8%–9.6% (December 2017–February 2018). The riverbed sediment samples had a similar trend of antibiotic resistance as river water samples. Figure 4 shows the prevalence of ARB was very low throughout the year. The resistance rate of river water and riverbed sediment was slightly higher in spring than in summer and did not significantly correlate with the results of bacterial count (p > 0.05). In addition, ARB were not detected in the feces of wild animals.
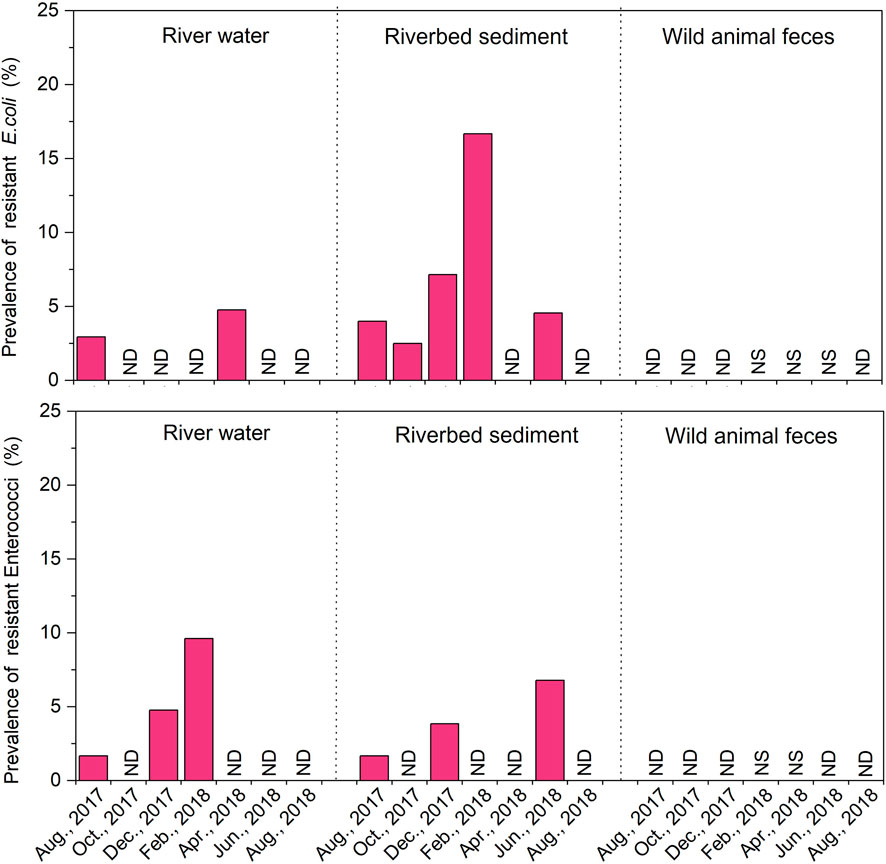
Figure 4. Changes in the prevalence of antibiotic resistant to one or more agents for Escherichia coli (E. coli) and enterococci strains isolated from the river water, riverbed sediment and wild animal feces samples over a 1-year period. No Date (ND), No Sample (NS).
In our previous survey, the antibiotic resistance rate of E. coli in the Kaeda River was 46% in the summer (July 2016) at S2 (Nishimura et al., 2021). By contrast, this value was significantly lower in this study. Although abrupt changes in the value can only be discussed by assumption, comparing this study with our previously reported study (Nishimura et al., 2021), the surrounding environment at the same site was altered. Parts of the artificial forest and riparian forest were cleared, and the number of trees covering the river was reduced. Although quantitative data are not available, the amount of light incident on the river increased. Therefore, the possibility that ecological changes or other factors affected the water cannot be excluded. By contrast, the resistance rate of AR-E. coli was 0.0%–2.5% upstream of the Tama River, which flows through Tokyo, Japan (Terada et al., 2012). We found that ARB were dispersed at a low rate in the pristine uppermost stream in Japan.
ARB were not detected in wild animal feces. This result contrasts with other study reporting detection of ARB in feces of wildlife (Ardiles-Villegas et al., 2011; Khare et al., 2020; Agga et al., 2021). Moreover, there are a few studies that indicate wildlife can also have an impact on ARB by spreading antibiotic resistance (Dolejska and Literak, 2019; Ramey and ahistrom, 2020; Laborda et al., 2022). In addition, several patterns of ARB occurrence in pristine environments have been considered in addition to the effects of wild animals. For example, plants are primary producers in pristine environments, and plants possess a diverse resistome (Berendonk et al., 2015; Chen et al., 2019; Obermeier et al., 2021). These resistomes may contribute to the antibiotic resistance of the bacteria in pristine environments. Thus, the occurrence of ARB in pristine rivers may need to consider both warm-blooded animal loading and natural occurrence (Allen et al., 2010; Hwengwere et al., 2022). Notably, ARB were detected in the uppermost pristine stream. When conducting surveys in such niche fields, it is important and prospects of this research to consider the occurrence of antibiotic resistance from multiple perspectives.
3.5 Characteristics of ARB
3.5.1 Resistance to various classes of antibiotics
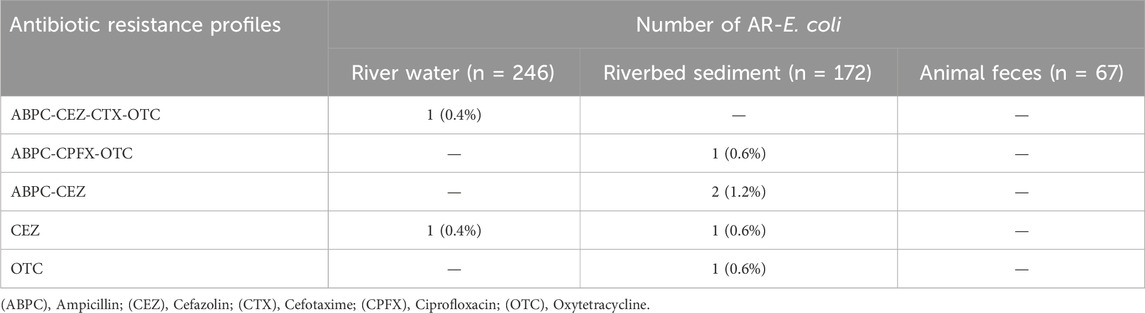
Supplementary Table S5 shows the antibiotic resistance rates of E. coli isolated from all samples. Ampicillin (ABPC)-resistant strains (0.4%–1.7%), cefazolin (CEZ)-resistant strains (0.8%–1.7%), and oxytetracycline (OTC)-resistant strains (0.4%–1.2%) were detected in river water and riverbed sediment. The antibiotic resistance rates were similar between the two sample types. Additionally, ciprofloxacin (CPFX)-resistant strains were detected in riverbed sediment.
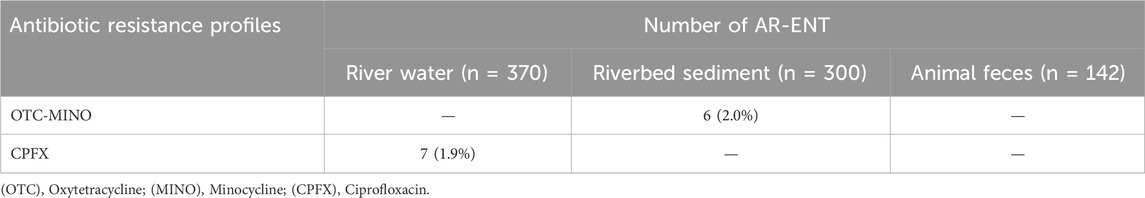
Supplementary Table S6 shows the antibiotic resistance rates of enterococci isolated from all samples. CPFX-resistant strains (1.9%; 7/371) were detected in river water. OTC-resistant (2.0%; 6/300) and minocycline-resistant (2.0%; 6/300) strains were detected in riverbed sediment. Among the AR-ENT detected, antibiotic-resistant E. faecium, an important enterococci species in clinical institutions, was not observed.
Notably, E. coli and enterococci resistant to semisynthetic or synthetic antibiotics were detected in each sample. Bacteria resistant to semisynthetic or synthetic antibiotics have been detected in pristine environments (Lenart-Boroń et al., 2022; Sajjad et al., 2023). This result confirms similar trends to previous studies.
3.5.2 Comparison of antibiotic-resistant profiles
Tables 1, 2 show the antibiotic resistance profiles of AR-E. coli and AR-ENT. In this study, strains resistant to ≥3 antibiotics were defined as multidrug-resistant strains. The resistance combinations of the 494 E. coli strains were profiled and compared between sample types (Table 1), revealing 2 and 4 patterns in river water and riverbed sediment, respectively. The major resistance patterns of river water strains were ABPC-CEZ-CTX-OTC (0.4%, 1/246) and CEZ (0.4%, 1/246), and the presence of multidrug-resistant E. coli was confirmed. In addition, the presence of multidrug-resistant E. coli was confirmed in riverbed sediment (profile: ABPC-CPFX-OTC; 0.6%, 1/172). Multidrug-resistant strains of enterococci were not identified (Table 2). Notably, multidrug-resistant E. coli (profile: ABPC-CEZ-CTX-OTC) detected in river water may be an important ESBL-producing strain in clinical institutions. In recent years, the rate of dissemination of ESBL-producing bacteria has been accelerating worldwide, making the control of their spread a significant challenge (Kawamura et al., 2017; Husna et al., 2023). The presence of ESBL-producing genes encoded on Enterobacteriaceae plasmids deserves is an issue of particular concern (Pana and Zaoutis, 2018). These genes can spread and disseminate across bacterial species, raising concerns about transmission (Stadler et al., 2018). Globally, the presence of ESBL-producing bacteria and associated genes has been confirmed in pristine environments (Hernandez et al., 2012), raising concerns about their dissemination. The presence of ESBL-producing bacteria in the pristine headwaters of rivers is highly significant information, and the spread and transportation of ARB in river water or downstream processes must be avoided. Therefore, our future challenge is to elucidate the mechanisms of antibiotic resistance in bacteria isolated pristine environment and to accumulate information on the horizontal transmission of antibiotic resistance.
3.5.3 Genomic analysis of multidrug-resistant E. coli and ESBL-producing E. coli
Genomic analysis of ESBL-producing E. coli (1 strain) and multidrug-resistant E. coli (1 strain) isolated from the Kaeda River was used to estimate the possession of ARGs and hosts. Table 3 shows the results of genomic analysis of ESBL-producing E. coli (ID:KS_R23, profile: ABPC-CEZ-CTX-OTC) isolated from river water and multidrug-resistant E. coli (ID: KS_S24, profile: ABPC-CPFX-OTC) isolated from riverbed sediment.
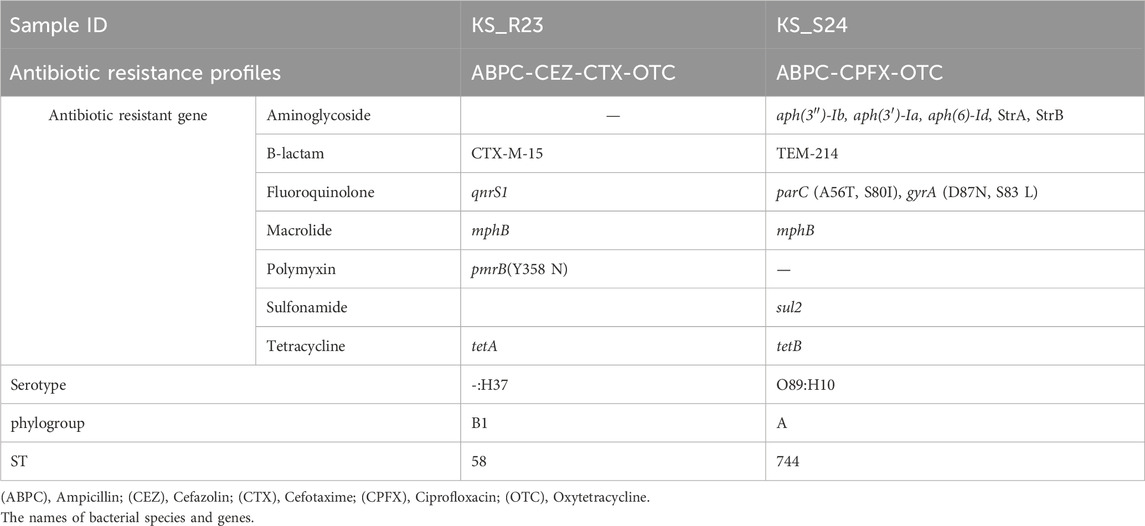
Table 3. The results of genomic analysis of ESBL-producing Escherichia coli (ID:KS_R23) isolated from the river water and multidrug-resistant Escherichia coli (ID:KS_S24) isolated from riverbed sediment.
KS_R23 harbored CTX-M-15 and tetA. KS_S24 harbored TEM-214 and tetB, mutations in perC (A56T, S80I) and gyrA (D87N, S83L). CTX-M can break down a wide range of beta-lactam antibiotics, including penicillins and cephalosporins (Canton et al., 2012). The gene tet provides resistance to tetracycline antibiotics by encoding the efflux pumps that discharges the antibiotics out of the bacterial cell (Li and Nikaido, 2009). These were resistant genes and mutations that could explain the results of susceptibility testing. ARGs associated with antibiotic resistance to macrolide antibiotics (mphB) and polypeptide antibiotics [pmrB (Y358N)], which were not tested in this study, were detected from KS_R23. This strain may exhibit resistance to these two antibiotics. ARGs associated with antibiotic resistance to aminoglycoside antibiotics [aph(3'')-Ib, aph(3')-La, aph(6)-Id, strA, strB]; macrolide antibiotics [pmrB (Y358N)]; and sulfonamide antibiotics (sul2) were detected from KS_S24. This strain may exhibit resistance to these three antibiotics. Among the detected genes, the difference between the resistance genes that could explain the results of susceptibility testing and other genes was considered to be ARGs originally possessed by the bacteria and ARGs acquired through horizontal spread via mobile genetic factors such as plasmids. KS_R23 and KS_S24 were classified into phylogroups B1 and A, respectively. E. coli isolated from wild animals was mainly classified into phylogroups A and B1 (Johnson et al., 2017; Cristovao et al., 2017; Haenni et al., 2020). Therefore, ESBL-producing E. coli and multidrug-resistant E. coli may be derived from wild animals. However, ARB were not detected in the feces of wild animals in this study. The only wild animals targeted in this study were bird and Mustelidae that excrete fecal matter on riverbanks. It is possible that other wild animals may have loaded ESBL-producing bacteria. It is possible that fecal matters from wild boars and deer live in the catchment may have been input into the river as surface water via forest surface soils with rainfall. Therefore, future items to be considered are feces of other major wild animals and surface water flows into the river.
3.6 Comparison of genotypic similarities between strains isolated from river water and bird feces by PFGE analysis
Based on genomic analysis, there was a high commonality of antibiotic resistance between strains obtained from river sample and bird feces. Therefore, PFGE analysis was used to compare genotypic similarities between strains isolated from river samples and bird feces. In a survey conducted on 27 October 2017, E. coli strains (n = 30) were randomly selected from river water, riverbed sediment, and bird feces collected at S2. A dendrogram based on the similarity of PFGE types of each E. coli strain is shown in Supplementary Figure S1. When the clusters were classified into groups with 100% similarity, PFGE types of E. coli were highly diverse, and no information on the source of the E. coli pollution. The 29 randomly selected enterococci strains from each sample were analyzed for the similarity of PFGE types for each strain (Figure 5). Three strains isolated from bird feces were excluded because PFGE band patterns could not be identified. When the clusters were classified into groups with 100% similarity, 4 clusters (A, B, C and D types) were identified, consisting of 2–4 strains each. Type A consisted of one strain isolated from river water and three strains isolated from bird feces strains. Thus, an identical enterococci clone was present in both the river water and bird feces. The results of enterococcal PFGE analysis suggest that bird feces is one of the sources of enterococci contamination of the pristine uppermost stream of the Kaeda River. In addition, the number of target strains needs to be increased to increase the significance of the PFGE analysis. This is an issue to be addressed in the future.
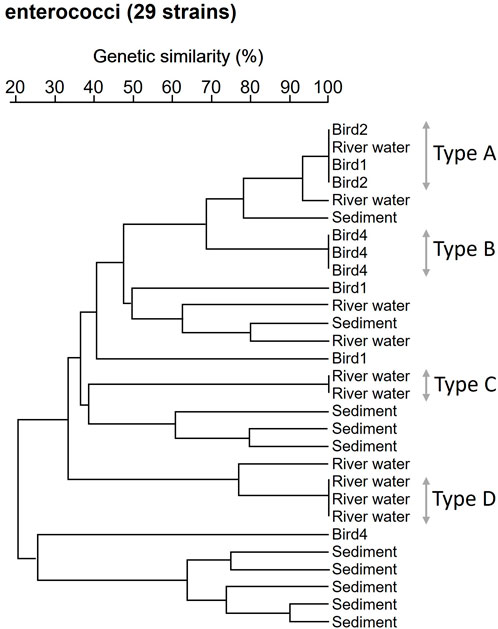
Figure 5. Dendrogram analysis based on the similarity of enterococci PFGE types. Enterococci strains isolated from the river water, riverbed sediment and of bird feces at S2 site. Bird 1, 2, 4: Enterococci strains isolated from bird 1, 2, 4.
4 Conclusion
AR-E. coli and AR-ENT were investigated in river water, riverbed sediment, and feces of waterside animals, such as birds and Mustelidae, at pristine uppermost sites throughout the year. The resistance rates of E. coli and enterococci fluctuated at low levels (E. coli: 2.9%–17%, enterococci: 1.7%–9.6%). ESBL-producing E. coli (0.4%) with multidrug resistance (0.6%) were detected among the AR-E. coli detected in the river. Notably, multidrug-resistant bacteria and clinically important ESBL-producing bacteria were found in the pristine environment. This shows that rapid elucidation of the routes of ARB dissemination is important. Although we could not determine the sources of the detected ARB in this study, birds were one of the sources of bacterial contamination of the pristine uppermost stream of the Kaeda River. To elucidate the sources of ARB in pristine natural environments, conducting investigations from a multifaceted perspective (e.g., large wild animals, production by plants, climate change) and not limited to the influence of birds is necessary (Torres et al., 2020; Caliz et al., 2022).
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author contributions
EN: Data curation, Investigation, Writing–original draft, Writing–review and editing. HX: Validation, Writing–original draft. ST: Validation, Writing–original draft. MN: Investigation, Writing–original draft. KN: Validation, Writing–original draft. YH: Investigation, Writing–original draft. YO: Investigation, Writing–original draft. YS: Conceptualization, Investigation, Methodology, Project administration, Supervision, Validation, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank laboratory member for technical cooperate with the experiments. Finally, we are grateful to the referees for useful comments.
Conflict of interest
Author EN was employed by IDEA Consultants, Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2024.1439174/full#supplementary-material
References
Afolabi, E. O., Quilliam, R. S., and Oliver, D. M. (2023). Persistence of E. coli in streambed sediment contaminated with faeces from dairy cows, geese, and deer: legacy risks to environment and health. Int. J. Environ. Res. Public Health 20 (7), 5375. doi:10.3390/ijerph20075375
Agga, G. E., Silva, P. J., and Martin, R. S. (2021). Third-generation cephalosporin- and tetracycline-resistant Escherichia coli and antimicrobial resistance genes from metagenomes of mink feces and feed. Foodborne Pathog. Dis. 18 (3), 169–178. doi:10.1089/fpd.2020.2851
Allen, H. K., Donato, J., Wang, H. H., Cloud-Hansen, K. A., Davies, J., and Handelsman, J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8 (4), 251–259. doi:10.1038/nrmicro2312
Alonso, A., Sánchez, P., and Martínez, J. L. (2001). Environmental selection of antibiotic resistance genes. Environ. Microbiol. 3 (1), 1–9. doi:10.1046/j.1462-2920.2001.00161.x00161.x
Antimicrobial Resistance Collaborators, Ikuta, K. S., Sharara, F., Swetschinski, L., Robles Aguilar, G., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lance 399 (10325), 629–655. doi:10.1016/S0140-6736(21)02724-0
Ardiles-Villegas, K., González-Acuña, D., Waldenström, J., Olsen, B., and Hernández, J. (2011). Antibiotic resistance patterns in fecal bacteria isolated from Christmas shearwater (Puffinus nativitatis) and masked booby (Sula dactylatra) at remote Easter Island. Avian Dis. 55 (3), 486–489. doi:10.1637/9619-122010-ResNote.1
Baquero, F., Martínez, J. L., and Cantón, R. (2008). Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 19 (3), 260–265. doi:10.1016/j.copbio.2008.05.006
Beghain, J., Bridier-Nahmias, A., Le Nagard, H., Denamur, E., and Clermont, O. (2018). ClermonTyping: an easy-to-use and accurate in silico method for Escherichia genus strain phylotyping. Microb. Genom. 4 (7), e000192. doi:10.1099/mgen.0.000192
Berendonk, T. U., Manaia, C. M., Merlin, C., Fatta-Kassinos, D., Cytryn, E., Walsh, F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13 (5), 310–317. doi:10.1038/nrmicro3439
Boehm, A. B., Griffith, J., McGee, C., Edge, T. A., Solo-Gabriele, H. M., Whitman, R., et al. (2009). Faecal indicator bacteria enumeration in beach sand: a comparison study of extraction methods in medium to coarse sands. J. Appl. Microbiol. 107 (5), 1740–1750. doi:10.1111/j.13652672.2009.0444-0.x
Bonnedahl, J., and Järhult, J. D. (2014). Antibiotic resistance in wild birds. Ups. J. Med. Sci. 119 (2), 113–116. doi:10.3109/03009734.2014.905663
Caliz, J., Subirats, J., Triadó-Margarit, X., Borrego, C. M., and Casamayor, E. O. (2022). Global dispersal and potential sources of antibiotic resistance genes in atmospheric remote depositions. Environ. Int. 160, 107077. doi:10.1016/j.envint.2022.107077
Canton, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3, 110. doi:10.3389/fmicb.2012.00110
Centers for Disease (2019). Antibiotic resistance threats in the United States 2019, 1–148. Atlanta, GA, UNITED STATES: U.S. Department of Health and Human Services. Available at: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on May 7, 2024).
Centers for Disease Control (2017). Standard operating procedure for PulseNet PFGE of Escherichia coli O157: H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. PNL05 last updated. Available at: https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf (accessed on May 7, 2024).
Chen, Q. L., Cui, H. L., Su, J. Q., Penuelas, J., and Zhu, Y. G. (2019). Antibiotic resistomes in plant microbiomes. Trends. Plant Sci. 24 (6), 530–541. doi:10.1016/j.tplants.2019.02.010
Christner, M., Trusch, M., Rohde, H., Kwiatkowski, M., Schlüter, H., Wolters, M., et al. (2014). Rapid MALDI-TOF mass spectrometry strain typing during a large outbreak of Shiga-Toxigenic Escherichia coli. PLoS One 9 (7), e101924. doi:10.1371/journal.pone.0101924
CLSI (2012). M100-S22 performance standards for antimicrobial susceptibility testing; twenty-second informational supplement. Wayne, Pennsylvania, America: Clinical and Laboratory Standards Institute, 1–184.
CLSI (2017). M100 performance standards for antimicrobial susceptibility testing. 27th Edition. Wayne, Pennsylvania, America: Clinical and Laboratory Standards Institute, 1–249. Replaces M100-S26.
Cristovao, F., Alonso, C. A., Igrejas, G., SoUnited States, M., Silva, V., Pereira, J. E., et al. (2017). Clonal diversity of extended-spectrum beta-lactamase producing Escherichia coli isolates in fecal samples of wild animals. FEMS Microbiol/Lett., 364(5), doi:10.1093/femsle/fnx039
Daniel, D. S., Lee, S. M., Gan, H. M., Dykes, G. A., and Rahman, S. (2017). Genetic diversity of Enterococcus faecalis isolated from environmental, animal and clinical sources in Malaysia. J. Infect. Public Health 10 (5), 617–623. doi:10.1016/j.jiph.2017.02.006
Diwan, V., Hanna, N., Purohit, M., Chandran, S., Riggi, E., Parashar, V., et al. (2018). Seasonal variations in water-quality, antibiotic residues, resistant bacteria and antibiotic resistance genes of Escherichia coli isolates from water and sediments of the Kshipra river in central India. Int. J. Environ. Res. Public Health 15 (6), 1281. doi:10.3390/ijerph15061281
Dolejska, M., and Literak, I. (2019). Wildlife is overlooked in the epidemiology of medically important antibiotic-resistant bacteria. Antimicrob. Agents Chemother. 63 (8), 011677-19–e1219. doi:10.1128/AAC.01167-19
Dolejska, M., and Papagiannitsis, C. C. (2018). Plasmid-mediated resistance is going wild. Plasmid 99, 99–111. doi:10.1016/j.plasmid.2018.09.010
Farnleitner, A. H., Ryzinska-Paier, G., Reischer, G. H., Burtscher, M. M., Knetsch, S., Kirschner, A., et al. (2010). Escherichia coli and enterococci are sensitive and reliable indicators for human, livestock and wildlife faecal pollution in alpine mountainous water resources. J. Appl. Microbiol. 109 (5), 1599–1608. doi:10.1111/j.1365-2672.2010.04788.x04788.x
Feldgarden, M., Brover, V., Gonzalez-Escalona, N., Frye, J. G., Haendiges, J., Haft, D. H., et al. (2021). AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 11 (1), 12728. doi:10.1038/s41598-021-91456-0
Fenselau, C., and Demirev, P. A. (2001). Characterization of intact microorganisms by MALDI mass spectrometry. Mass Spectrom. Rev. 20 (4), 157–171. doi:10.1002/mas.10004
G7 Health Ministers’ Communiqué (2022). Overview of the G7 documents: G7 health Ministers’ Communiqué 20 may 2022, Berlin. Berlin, Germany, 1–12. Available at: https://www.g7germany.de/g7-en/g7-documents (accessed on May 7, 2024).
García, L. A., Torres, C., López, A. R., Rodríguez, C. O., Espinosa, J. O., and Valencia, C. S. (2020). Staphylococcus Spp. from wild mammals in Aragón (Spain): antibiotic resistance status. J. Vet. Res. 64 (3), 373–379. doi:10.2478/jvetres-2020-0057
Givens, C. E., Kolpin, D. W., Hubbard, L. E., Meppelink, S. M., Cwiertny, D. M., Thompson, D. A., et al. (2023). Simultaneous stream assessment of antibiotics, bacteria, antibiotic resistant bacteria, and antibiotic resistance genes in an agricultural region of the United States. Sci. Total Environ. 904, 166753. doi:10.1016/j.scitotenv.2023.166753
Gordon, S., Swenson, J. M., Hill, B. C., Pigott, N. E., Facklam, R. R., Cooksey, R. C., et al. (1992). Antimicrobial susceptibility patterns of common and unusual species of enterococci causing infections in the United States. Enterococcal Study Group. J. Clin. Microbiol. 30 (9), 2373–2378. doi:10.1128/jcm.30.9.2373-2378.1992
Grenni, P. (2022). Antimicrobial resistance in rivers: a review of the genes detected and new challenges. Environ. Toxicol. Chem. 41 (3), 687–714. doi:10.1002/etc.5289
Gupta, S. K., Padmanabhan, B. R., Diene, S. M., Lopez-Rojas, R., Kempf, M., Landraud, L., et al. (2014). ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 58 (1), 212–220. doi:10.1128/AAC.01310-13
Haenni, M., Métayer, V., Jarry, R., Drapeau, A., Puech, M. P., Madec, J. Y., et al. (2020). Wide spread of blaCTX-M-9/mcr-9 IncHI2/ST1 plasmids and CTX-M-9-producing Escherichia coli and enterobacter cloacae in rescued wild animals. Front. Microbiol. 11, 601317. doi:10.3389/fmicb.2020.601317
Hansen, S., Messer, T., Mittelstet, A., Berry, E. D., Bartelt-Hunt, S., and Abimbola, O. (2020). Escherichia coli concentrations in waters of a reservoir system impacted by cattle and migratory waterfowl. Sci. Total Environ. 705, 135607. doi:10.1016/j.scitotenv.2019.135607
Hernandez, J., Stedt, J., Bonnedahl, J., Molin, Y., Drobni, M., Calisto-Ulloa, N., et al. (2012). Human-associated extended-spectrum β-lactamase in the antarctic. Appl. Environ. Microbiol. 78 (6), 2056–2058. doi:10.1128/AEM.07320-11
Husna, A., Rahman, M. M., Badruzzaman, A. T. M., Sikder, M. H., Islam, M. R., Rahman, M. T., et al. (2023). Extended-spectrum β-lactamases (ESBL): challenges and opportunities. Biomedicines 11 (11), 2937. doi:10.3390/biomedicines11112937
Hwengwere, K., Paramel Nair, H., Hughes, K. A., Peck, L. S., Clark, M. S., and Walker, C. A. (2022). Antimicrobial resistance in Antarctica: is it still a pristine environment? Microbiome 10 (1), 71. doi:10.1186/s40168-022-01250-x
Inouye, M., Dashnow, H., Raven, L. A., Schultz, M. B., Pope, B. J., Tomita, T., et al. (2014). SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 6 (11), 90. doi:10.1186/s13073-014-0090-6
Ishii, S., and Sadowsky, M. J. (2008). Escherichia coli in the environment: implications for water quality and human health. Microbes. Environ. 23 (2), 101–108. doi:10.1264/jsme2.23.101
Jang, J., Park, J., Hwang, C. Y., Choi, J., Shin, J., Kim, Y. M., et al. (2022). Abundance and diversity of antibiotic resistance genes and bacterial communities in the western Pacific and Southern Oceans. Sci. Total Environ. 822, 153360. doi:10.1016/j.scitotenv.2022.153360-2022.153360
Japanese Standard Association (2016). Testing methods for industrial wastewate, K 0102: 2016. Tokyo, Japan: Japanese Standard Association, 1–370. Available at: https://webdesk.jsa.or.jp/preview/pre-_jis_k_00102_000_000_2016_e_ed10_i4.pdf (accessed on May 7, 2024).
Johnson, J. R., Johnston, B. D., Delavari, P., Thuras, P., Clabots, C., and Sadowsky, M. J. (2017). Phylogenetic backgrounds and virulence-associated traits of Escherichia coli isolates from surface waters and diverse animals in Minnesota and Wisconsin. Appl. Environ. Microbiol. 83 (24), 013299-17–e1417. doi:10.1128/AEM.01329-17
Kajitani, R., Yoshimura, D., Ogura, Y., Gotoh, Y., Hayashi, T., and Itoh, T. (2020). Platanus_B: an accurate de novo assembler for bacterial genomes using an iterative error-removal process. DNA Res. 27 (3), dsaa014. doi:10.1093/dnares/dsaa014
Kawamura, K., Nagano, N., Suzuki, M., Wachino, J. I., Kimura, K., and Arakawa, Y. (2017). ESBL-Producing Escherichia coli and its rapid rise among healthy people. Food Saf. . (Tokyo) 5 (4), 122–150. doi:10.14252/foodsafetyfscj.2017011
Khare, N., Kaushik, M., Martin, J. P., Mohanty, A., and Gulati, P. (2020). Genotypic diversity in multi-drug-resistant E. coli isolated from animal feces and Yamuna River water, India, using rep-PCR fingerprinting. Environ. Monit. Assess. 192 (11), 681. doi:10.1007/s10661-020-08635-1
Laborda, P., Sanz-García, F., Ochoa-Sánchez, L. E., Gil-Gil, T., Hernando-Amado, S., and Martínez, J. L. (2022). Wildlife and antibiotic resistance. Front. Cell. Infect. Microbiol. 12, 873989. doi:10.3389/fcimb.2022.873989
Lenart-Boroń, A., Prajsnar, J., Guzik, M., Boroń, P., Grad, B., and Żelazny, M. (2022). Antibiotics in groundwater and river water of białka—a pristine mountain river. Appl. Sci. 12 (24), 12743. doi:10.3390/app122412743
Li, X. Z., and Nikaido, H. (2009). Efflux-mediated drug resistance in bacteria: an update. Drugs 69 (12), 1555–1623. doi:10.2165/11317030-000000000-00000
Lima-Bittencourt, C. I., Cursino, L., Gonçalves-Dornelas, H., Pontes, D. S., Nardi, R. M., Callisto, M., et al. (2007). Multiple antimicrobial resistance in Enterobacteriaceae isolates from pristine freshwater. Genet. Mol. Res. 6 (3), 510–521. Available at: https://pubmed.ncbi.nlm.nih.gov/17985304/.
Nishimura, E., Nishiyama, M., Nukazawa, K., and Suzuki, Y. (2021). Comparison of antibiotic resistance profile of Escherichia coli between pristine and human-impacted sites in a river. Antibiot. (Basel) 10 (5), 575. doi:10.3390/antibiotics10050575
Nishiyama, M., Ogura, Y., Hayashi, T., and Suzuki, Y. (2017). Antibiotic resistance profiling and genotyping of vancomycin-resistant enterococci collected from an urban river basin in the provincial city of Miyazaki, Japan. Water 9 (2), 79. doi:10.3390/w9020079
Obermeier, M. M., Wicaksono, W. A., Taffner, J., Bergna, A., Poehlein, A., Cernava, T., et al. (2021). Plant resistome profiling in evolutionary old bog vegetation provides new clues to understand emergence of multi-resistance. ISME J. 15 (3), 921–937. doi:10.1038/s41396-020-00822-9
O’Neill, J. (2014). Review on antimicrobial resistance: antimicrobial resistance: tackling a crisis for the health and wealth of nations. London, United Kingdom: The UK Department of Health, 1–20. Available at: https://wellcomecollection.org/works/rdpck35v (accessed on May 7, 2024).
Osińska, A., Korzeniewska, E., Harnisz, M., Felis, E., Bajkacz, S., Jachimowicz, P., et al. (2020). Small-scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mat. 381, 121221. doi:10.1016/j.jhazmat.2019.121221
Pana, Z. D., and Zaoutis, T. (2018). Treatment of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBLs) infections: what have we learned until now? F1000Res 7, 1347. F1000 Faculty Rev-1347. doi:10.12688/f1000research.14822.1
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25 (7), 1043–1055. doi:10.1101/gr.186072.114
Ramey, A. M., and Ahlstrom, C. A. (2020). Antibiotic resistant bacteria in wildlife: perspectives on trends, acquisition and dissemination, data gaps, and future directions. J. Wildl. Dis. 56, 1–15. doi:10.7589/2019-04-099
Rogers, S. W., Shaffer, C. E., Langen, T. A., Jahne, M., and Welsh, R. (2018). Antibiotic-resistant genes and pathogens shed by wild deer correlate with land application of residuals. Ecohealth 15 (2), 409–425. doi:10.1007/s10393-018-1316-7
Ruoff, K. L., de la Maza, L., Murtagh, M. J., Spargo, J. D., and Ferraro, M. J. (1990). Species identities of enterococci isolated from clinical specimens. J. Clin. Microbiol. 28 (3), 435–437. doi:10.1128/-jcm.28.3.435-437.1990
Rwego, I. B., Isabirye-Basuta, G., Gillespie, T. R., and Goldberg, T. L. (2008). Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park, Uganda. Conserv. Biol. 22 (6), 1600–1607. doi:10.1111/j.1523-1739.2008.01018.x01018.x
Sajjad, W., Ali, B., Niu, H., Ilahi, N., Rafiq, M., Bahadur, A., et al. (2023). High prevalence of antibiotic-resistant and metal-tolerant cultivable bacteria in remote glacier environment. Environ. Res. 239 (2), 117444. doi:10.1016/j.envres.2023.117444
Stadler, T., Meinel, D., Aguilar-Bultet, L., Huisman, J. S., Schindler, R., Egli, A., et al. (2018). Transmission of ESBL-producing Enterobacteriaceae and their mobile genetic elements-identification of sources by whole genome sequencing: study protocol for an observational study in Switzerland. BMJ open 8 (2), e021823. doi:10.1136/bmjopen-2018-021823
Suzuki, Y., Niina, K., Matsuwaki, T., Nukazawa, K., and Iguchi, A. (2018). Bacterial flora analysis of coliforms in sewage, river water, and ground water using MALDI-TOF mass spectrometry. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 53 (2), 160–173. doi:10.1080/10934529.2017.13831-28
Terada, S., Miyake, H., and Urase, T. (2012). Profile of antibiotic resistance of Escherichia coli isolated from different water environments. JSWE 35, 73–80. doi:10.2965/jswe.35.73
Torres, R. T., Fernandes, J., Carvalho, J., Cunha, M. V., Caetano, T., Mendo, S., et al. (2020). Wild boar as a reservoir of antimicrobial resistance. Sci. Total Environ. 717, 135001. doi:10.1016/j.scitotenv.2019.135001
United States Environmental Protection Agency (2014). Method 1600: enterococci in water by membrane filtration using membrane-Enterococcus indoxyl-β-D-glucoside agar (mEI). Washington, D.C., America: United States Environmental Protection Agency. Available at: https://www.epa.gov/sites/-default/files/2018-06/documents/method_1600_sept-2014.pdf (accessed on May 7, 2024).
Willyard, C. (2017). The drug-resistant bacteria that pose the greatest health threats. Nature 543 (7643), 15. doi:10.1038/nature.2017.21550
World Health Organization (2022b). Guidelines for drinking-water quality: fourth edition incorporating the first and second addenda. China: Licence: CC BY-NC-SA 3.0 IGO.
World Health Organization (2022a). WHO and ECDC report: antimicrobial resistance surveillance in Europe 2022–2020. Sweden: European Centre for Disease Prevention and Control: Stockholm County, 1–164. Available at: https://www.ecdc.europa.eu/sites/default/files/documents/Joint-WHO-ECDC-AMR-report-2022.pdf (accessed on May 7, 2024).
Yuan, Y., Liang, B., Jiang, B. W., Zhu, L. W., Wang, T. C., Li, Y. G., et al. (2021). Migratory wild birds carrying multidrug-resistant Escherichia coli as potential transmitters of antimicrobial resistance in China. PLoS One 16 (12), e0261444. doi:10.1371/journal.pone.0261444
Keywords: Escherichia coli, enterococci, antibiotic resistance, antibiotic-resistant bacteria, multidrug-resistant bacteria, extended-spectrum β-lactamase, genomic analysis, pristine uppermost stream
Citation: Nishimura E, Xie H, Tamai S, Nishiyama M, Nukazawa K, Hoshiko Y, Ogura Y and Suzuki Y (2024) Year-round monitoring of antibiotic-resistant bacteria in pristine uppermost stream and estimation of pollution sources. Front. Environ. Sci. 12:1439174. doi: 10.3389/fenvs.2024.1439174
Received: 27 May 2024; Accepted: 19 August 2024;
Published: 09 September 2024.
Edited by:
Visva Bharati Barua, University of North Carolina at Charlotte, United StatesReviewed by:
Raju Sekar, Xi’an Jiaotong-Liverpool University, ChinaChandan Pal, Zespri International Ltd., New Zealand
Copyright © 2024 Nishimura, Xie, Tamai, Nishiyama, Nukazawa, Hoshiko, Ogura and Suzuki. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yoshihiro Suzuki, eXN1enVraUBjYy5taXlhemFraS11LmFjLmpw
 Emi Nishimura1,2
Emi Nishimura1,2 Soichiro Tamai
Soichiro Tamai Masateru Nishiyama
Masateru Nishiyama Yuki Hoshiko
Yuki Hoshiko Yoshihiro Suzuki
Yoshihiro Suzuki