- 1State Key Laboratory of Environmental Criteria and Risk Assessment, Chinese Research Academy of Environmental Sciences, Beijing, China
- 2College of Oceanography and Ecological Science, Shanghai Ocean University, Shanghai, China
- 3Basin Research Center for Water Pollution Control, Chinese Research Academy of Environmental Sciences, Beijing, China
- 4Shanghai Universities Key Laboratory of Marine Animal Taxonomy and Evolution, Shanghai Ocean University, Shanghai, China
- 5Engineering Research Center of Environmental DNA and Ecological Water Health Assessment, Shanghai Ocean University, Shanghai, China
- 6Department of Plant Protection, College of Horticulture and Landscape, Tianjin Agricultural University, Tianjin, China
Fish are considered objective indicators of environmental health and ecosystem stability. Establishing regional reference databases of mitochondrial genome sequences from local fish communities can significantly enhance fish monitoring using environmental DNA (eDNA) analysis. For non-native species, the eDNA technique provides early detection and rapid monitoring. It is also crucial to include fundamental genetic information for both native and non-native species in genetic databases. This study presents the complete mitochondrial genomes of 17 fish species inhabiting the Baiyangdian Basin, a national key wetland in China. The mitochondrial DNA of these fish was analyzed to investigate their characteristics, and their phylogeny was determined using maximum likelihood (ML) methods. Various analyses were performed, including the examination of nucleotide composition, evaluation of AT-skew and GC-skew, analysis of codon frequency, and determination of relative synonymous codon usage (RSCU) values, and assessment of selection pressure on protein-coding genes (PCGs). The analysis showed that all PCGs in all fish underwent purifying selection. Using Xenocyprididae as a representative, this study investigated the genetic selection tendencies of native and non-native fish species in the Baiyangdian Basin. Significant differences were found in five of the 13 PCGs: COI, COII, COIII, Cytb, and ATP8. Except for ATP8, the findings indicated that the genes of non-native species underwent stronger purifying selection during evolution compared to native species. Additionally, comparing the population ω values of non-native species to those of native species showed that the Cytb and COIII genes exhibited greater differential purification selection than COI and COII. These differences may be the result of the evolution of non-native species to migrate and adapt to the Baiyangdian Basin, thereby affecting the evolution of related genes.
1 Introduction
With advancements in molecular theory, mitochondrial techniques have become increasingly mature. The mitochondrial genome, which is inherited maternally, is characterized by a high mutation rate (Chinnery and Hudson, 2013; Q. L; Wu et al., 2014). Due to this characteristic, the mitochondrial genome is commonly employed in evolutionary analysis and species identification in fish (Ceruso et al., 2019; Sharma et al., 2020). A phylogenetic tree utilizes tree-like branching patterns to represent the evolutionary relationships and ancestral connections among species (Hayati et al., 2019). Although phylogenetic trees do not directly trace the origin of species, this method offers valuable insights into the evolutionary history of organisms and their interrelationships (Levin et al., 2012). For example, the complete mitochondrial genome of an unknown fish has been utilized to classify its species using a phylogenetic tree (J. Hu et al., 2023). Mitochondria also play important roles in the context of invasive species. Invasive species are costly to manage, with increasing economic impacts worldwide, and are recognized as one of the main drivers of biodiversity loss (Diagne et al., 2021; Hoffmann et al., 2010). However, accurately assessing the invasion history and negative impacts of non-native species can be challenging due to the lack of systematic surveys and historical data on non-native species (Chen et al., 2017). Examining the patterns of genetic variation in mitochondrial DNA is one approach to understanding the geographic history of invasive species (Du et al., 2018).
The mitochondrial DNA of all fish species includes 37 genes, consisting of two ribosomal RNAs (12S rRNA and 16S rRNA), 13 protein-coding genes (PCGs) (Cytb, ND6, ND5, ND4, ND4L, ND3, COIII, ATP6, ATP8, COII, COI, ND2 and ND1) and 22 transfer RNAs (tRNAs) (Brown, 2008; Montag et al., 2023). Among these genes, genes such as Cytb, 12S rRNA, and COI are particularly valued for their utility in environmental DNA (eDNA) metabarcoding (Feng et al., 2023; Leray et al., 2013; Levy et al., 2023). Published mitochondrial genomes have become a valuable resource for primer design and are considered to be among the most crucial components of eDNA metabarcoding (Collins et al., 2019; Ficetola et al., 2008). eDNA metabarcoding is widely recognized as a highly effective technique for monitoring the diversity of organisms and community mechanisms (Qian et al., 2023; Sahu et al., 2023; Yan et al., 2023). This method offers advantages such as low time consumption and minimal cost compared with traditional detection (Cristescu and Hebert, 2018; M; Shen et al., 2022). The mitochondrial genome is often preferred over other genes as a molecular marker for eDNA metabarcoding (Jensen et al., 2021). Research suggests that establishing a local database of mitochondrial genomes for aquatic organisms can promote the application of the eDNA technique (S. Wang et al., 2021). Early monitoring of invasive species is crucial, as effective and low-cost methods are essential during the initial stages of invasion. Once invasive species are established, eradication becomes exceedingly challenging (Larson et al., 2020; Vander Zanden et al., 2010). The use of eDNA had emerged as an important monitoring tool for both native and non-native species (Sepulveda et al., 2020). eDNA monitoring can facilitate the early detection of invasive species (Banerjee et al., 2021).
The Baiyangdian Basin is the largest freshwater basin in the North China Plain, with an area of 366 km2 (S. Hu et al., 2012), as well as a national key wetland (Li et al., 2023). As apex predators within aquatic ecosystems, fish play a crucial role in maintaining the ecological balance and contribute significantly to economic activities (Harris, 1995; Konar et al., 2019). The biodiversity of fish is a critical indicator for assessing the ecosystem health of the Baiyangdian Basin. However, fish communities in the Baiyangdian Basin have not been investigated systematically. Therefore, establishing a local mitochondrial genome reference database for the Baiyangdian Basin is through systematic investigations is necessary.
This paper investigated the mitochondrial genomes of 17 fish species, aiming to provide a comprehensive overview of the mitochondrial genomes in these species. The primary goal of this study was to document this genomic data to contribute to a genetic database that will aid in the recording and management of both native and non-native fish species by the relevant authorities. Furthermore, this study aimed to reveal selection pressures on PCGs in native and non-native fish species of the Xenocyprididae family within the Baiyangdian Basin.
2 Materials and methods
2.1 Sample collection and sequencing
Fish samples were collected from 30 sites in the Baiyangdian Basin using traditional fishing methods from April 12 to 20 April 2019. The collected samples were initially identified based on morphological features. DNA extraction was performed using the QIAquick Gel Extraction kit (Qiagen, GER). Following extraction, the DNA underwent quality control, genomic library construction, and screening. Pre-sequencing quality assessment was also conducted. The sequenced samples were processed using the Illumina NovaSeq platform, and the resulting data were assembled with the Fast-Plast software.
2.2 Sequence annotation
The mitochondrial DNA was annotated using the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/) and the MITOS (http://mitos2.bioinf.uni-leipzig.de/). This process involved retrieving the reference sequence from NCBI (Wolfsberg et al., 2001), and annotating the assembled sequence using MITOS (Bernt et al., 2013; D; Wu et al., 2022). Geneious Prime Software and CGView (https://cgview.ca/) were employed for data visualization.
2.3 Sequence analyses
The data analyzed consisted of the mitochondrial genomes of 17 fish species. The AT-skew and GC-skew information was obtained using MEGA 11 based on the following formulas: AT-skew = (A−T)/(A + T) and GC-skew = (G−C)/(G + C) (Perna and Kocher, 1995). The codon usage data of all 17 fish species were collected using MEGA 11 (Tamura et al., 2021), and Python 3.11.2 was employed to design heatmaps of the related data. The non-synonymous substitution rate (Ka) and synonymous substitution rate (Ks) for 13 PCGs were calculated in DnaSP6. To avoid phylogenetic inertia, five genera of native species and three genera of non-native species were selected from the Xenocyprididae family. The mitochondrial DNA of the selected species was analyzed to determine the Ka/Ks ratio. The ω value referred to the ratio of the non-synonymous substitution rate to the synonymous substitution rate. In this study, the ω values from native and non-native species served as quantitative data. If the quantitative data were normally distributed, intergroup comparisons were conducted based on one-way analysis of variance (ANOVA); if the quantitative data did not conform to a normal distribution, the non-parametric Wilcoxon signed-rank test was employed. A p-value <0.05 was considered statistically significant, and the p-value was calculated using the SPSS27 software.
2.4 Phylogenetic analyses
The mitochondrial genomes of 17 fish species were analyzed to investigate their phylogenetic evolution. A phylogenetic tree was constructed using the whole mitochondrial DNA sequences of the fish, to illustrate the kinship between native and non-native species. The 13 PCGs were concatenated into a single dataset using PhyloSuitev1. Subsequently, these 13 PCGs were processed to construct a maximum likelihood (ML) tree in MEGA 11 software using the general time-reversible model. One hundred bootstrap repeat tests were conducted to verify branches. The phylogenetic tree was visualized using FigTree software.
3 Results and discussion
3.1 Basic information
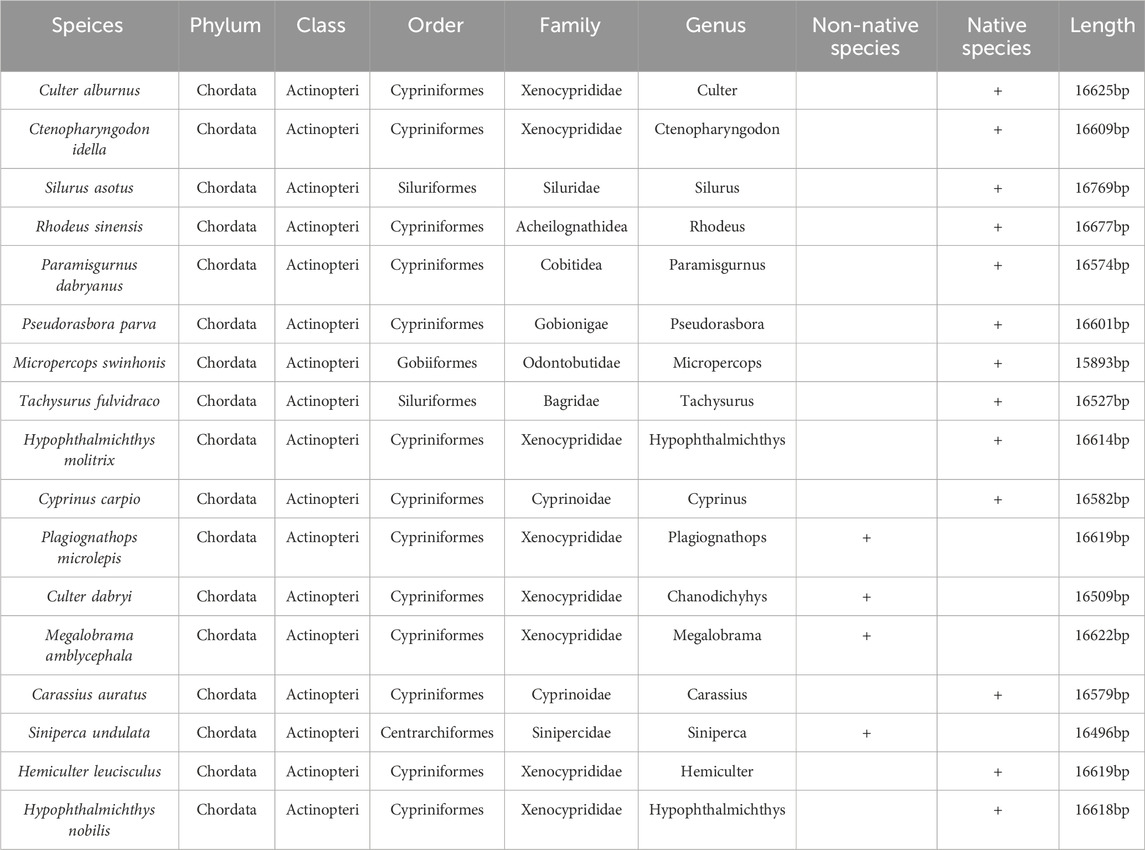
Out of 17 fish species investigated in this study, examining the historical records of fish in the Baiyangdian Basin revealed that 13 were native species, while four were non-native species. The native species belonged to eight families and 13 genera, while the non-native species belonged to two families and four genera. All 17 fish species were in the Xenocyprididae family (Table 1). The four non-native species were S. undulata, Megalobrama amblycephala, Culter dabryi, and Plagiognathops microlepis. Figure 1 illustrates the entire mitochondrial DNA of R. sinensis, a representative fish species. The mitochondrial genomes exhibited similar nucleotide composition. A small difference was observed in the A + T content, which ranged from 54.60% (S. undulata) to 58.76% (Pseudorasbora parva). The AT-skews were positive in all species except Micropercops swinhonis (−0.02261), and the GC-skews were negative across all species. In all 17 fish species, the A + T content of PCGs was exhibited a narrow range, from 54.20% to 59.40%. All GC-skews of the PCGs in the 17 fish species were negative, while only five fish species exhibited negative AT-skews for PCGs (M. swinhonis, P. parva, Paramisgurnus dabryanus, R. sinensis, and S. undulata). The most common PCG start codon was ATG, although some start codons of PCGs, such as COI, were in the form of GTG. The termination codons of PCGs predominantly consisted of variants of T- (T, TA, TAA, and TAG), with TAA being the most common at 48.42%, followed by T at 30.77%. The occurrences of TA and TAG were similar, accounting for 10.41% and 9.95%, respectively. In all 17 species, the termination codon was T or TA, possibly due to polyadenylation after translation (Ojala et al., 1981; T; Wang et al., 2019).

Figure 1. Mitogenome map of Rhodeus sinensis. Fluctuation in numerical values of GC-skew was visualized. Locations of PCGs and tRNAs were indicated. It also showcased the length of all portions.
Half of the species exhibited high-frequency codons including GCC(A), GAA(E), UUU(F), UUC(F), CAU(H), CAC(H), AUU(I), AUC(I), AAA(K), AAG(K), UUA(L), CUU(L), CUA(L), AUA(M), AAU(N), AAC(N), CCU(P), CCC(P), CCA(P), CAA(Q), UCU(S), UCC(S), UCA(S), AGC(S), ACU(T), ACC(T), ACA(T), UAU(Y), UAC(Y), UAA (*), and UAG(*). In this study, high-frequency codons were defined as codons with frequency values above the median. The high-frequency codons for all the fish species were AUU(I), AAA(K), CUU(L), CUA(L), and UAU(Y). Relative synonymous codon usage (RSCU) values greater than 1 indicate positive codon usage biases. Conversely, RSCU values lower than 1 suggest negative codon usage biases, while a value of 1 signifies no bias (Sheikh et al., 2020). The RSCU values for AUU(I), AAA(K), AUA(M), ACC(T), and UAA (*) were positive, while the RSCU values of GAG(E), AUC(I), AAG(K), CUG(L), AUG(M), CCG(P), UCG(S), ACG(T), and AGA (*) were negative in all species. According to the standard established by some researchers, an RSCU value exceeding 1.6 indicates the over-representation of synonymous codons, while an RSCU value below 0.6 reflects the under-representation of synonymous codons (Wong et al., 2010). In this study, GCG(A) (except for in H. nobilis) CCG(P) and UCG(S) (except for in S.undulata and C. dabryi); and ACG(T) (except for in H. nobilis) were under-represented with relatively high-frequency. Conversely, GCC(A) (in four species), GGA(G) (in three species), CUA(L) (in three species), and UAA (*) (in seven species) were over-represented with relatively high-frequency. The study of codon usage is of great significance for understanding the theory of gene expression during the process of evolution (Figures 2, 3) (Chakraborty et al., 2017).
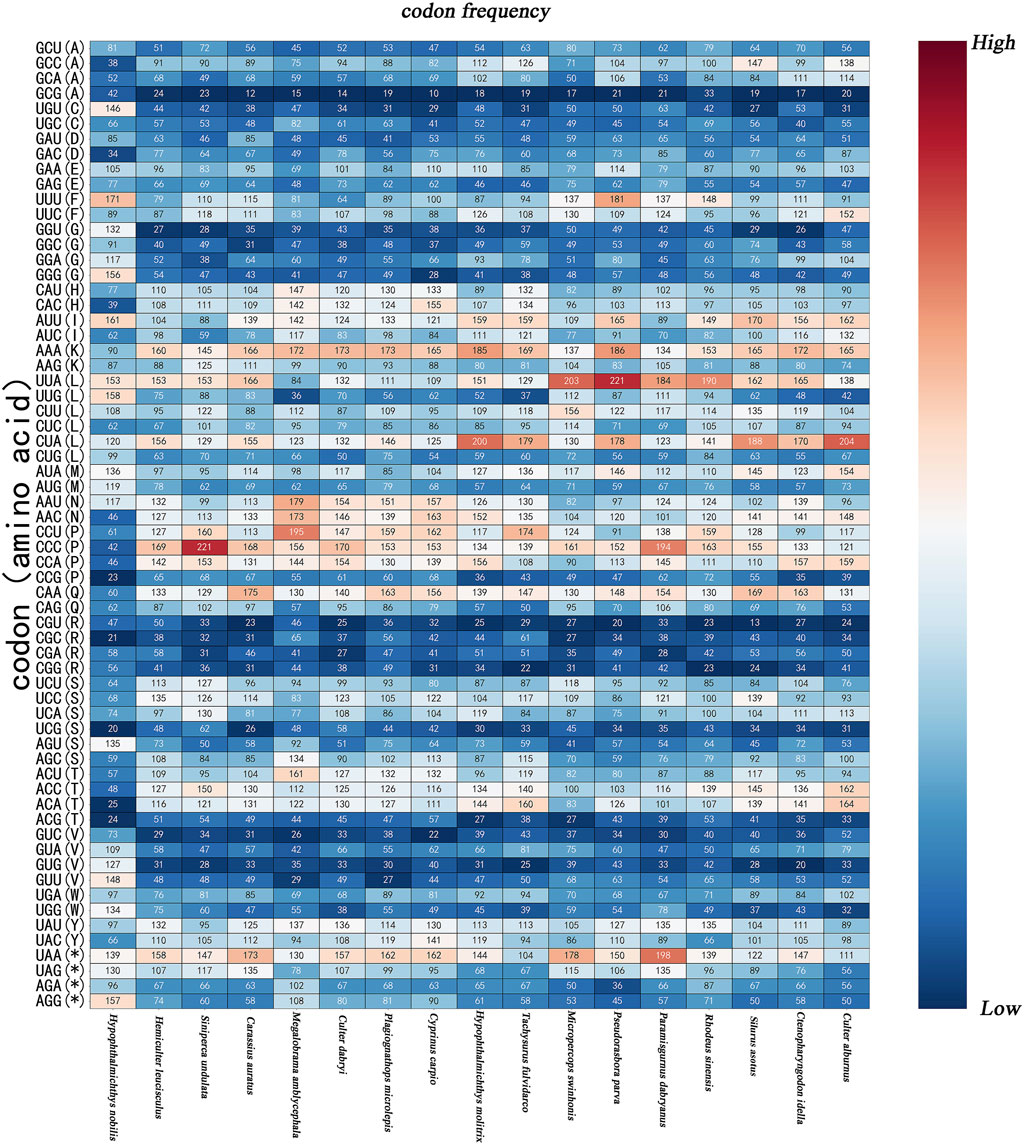
Figure 2. The graph is codon frequency of the 17 species species. The X-axis represents the species, The Y-axis records the types of amino acids and codons; * = the termination codon, A = Ala, F=Phe, C=Cys, D = Asp, N=Asn, E = Glu, Q = Gln, G = Gly, H=His, L = Leu, I=Ile, K = Lys, M = Met, P=Pro, R = Arg, S=Ser, T = Thr, V=Val, W = Trp, Y = Tyr.
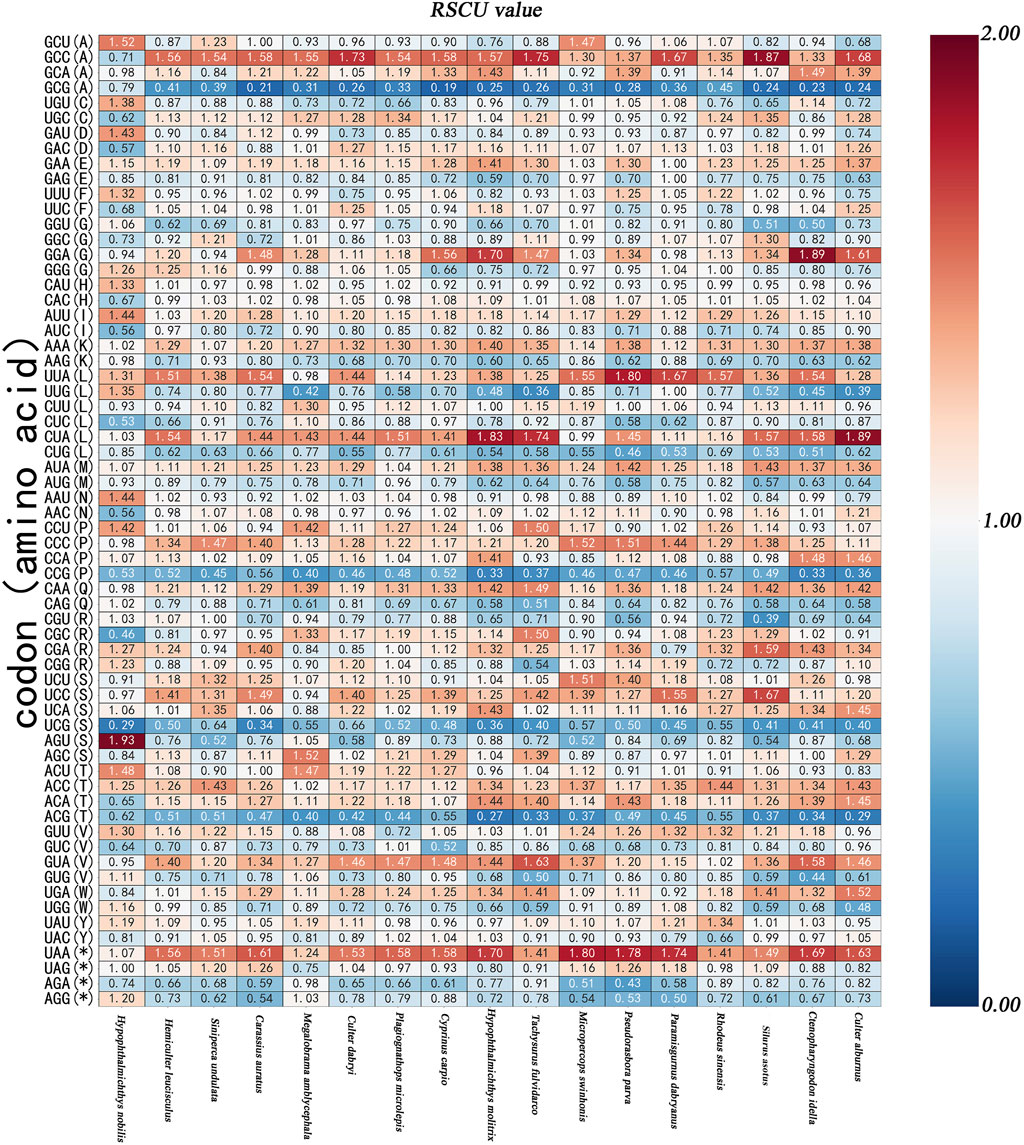
Figure 3. The graph shows relative synonymous codon usage (RSCU) of the 17 species species. The X-axis represents the species, The Y-axis records the types of the amino acids and codons.
The ability of a molecular sequence to adapt through evolution can be determined based on the ω value (Dos Reis, 2015). Purifying selection is indicated when the Ka/Ks value is smaller than 1 (especially less than 0.5) (Ivanova et al., 2017). The present study found that the ω values of 13 PCGs in all 17 fish species met the requirements for purifying selection during the evolutionary process (Figure 4). ATP8 exhibited the maximum
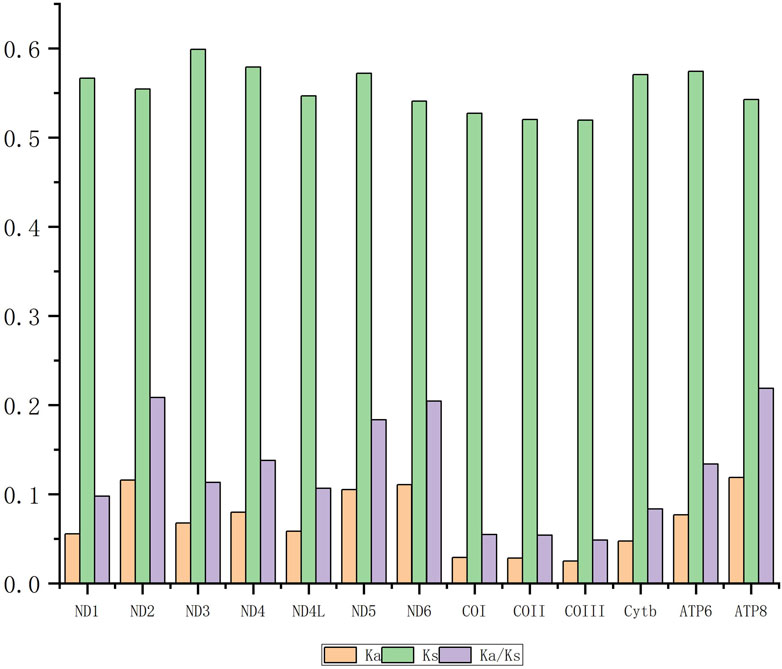
Figure 4. The Ka/Ks values of 13 PCGs from the 17 species species. Ka refers to nonsynonymous nucleotide substitutions. Ks refers to synonymous nucleotide substitutions.
The A + T content of 22 tRNAs from all 17 fish species in this investigation varied from 57.00% (S. undulata) to 54.45% (P. dabryanus). Additionally, all values exhibited positive AT-skew and GC-skew. The length of the base pairs varied, with a maximum of 946 bp and a minimum of 962 bp in 12S rRNA sequences, and the A + T content of ranged from 49.60% to 54.60%. Concurrently, the length of the 16S rRNA ranged between 1581 and 1692 bp, with its A + T content falling within the range of 54.20%–58.30%. Both 12S and 16S rRNA showed positive AT-skew but negative GC-skew. The highest A + T content was found in the D-loop, ranging from 61.40% to 67.50%. Additionally, all GC-skew values were less than zero, and most AT-skew values were greater than zero. Regarding the AT-skew from the D-loop, Carassius auratus had a value of zero, while Tachysurus fulvidraco (−0.0195) and R. sinensis (−0.0475) had negative values.
3.2 Phylogenetic analyses
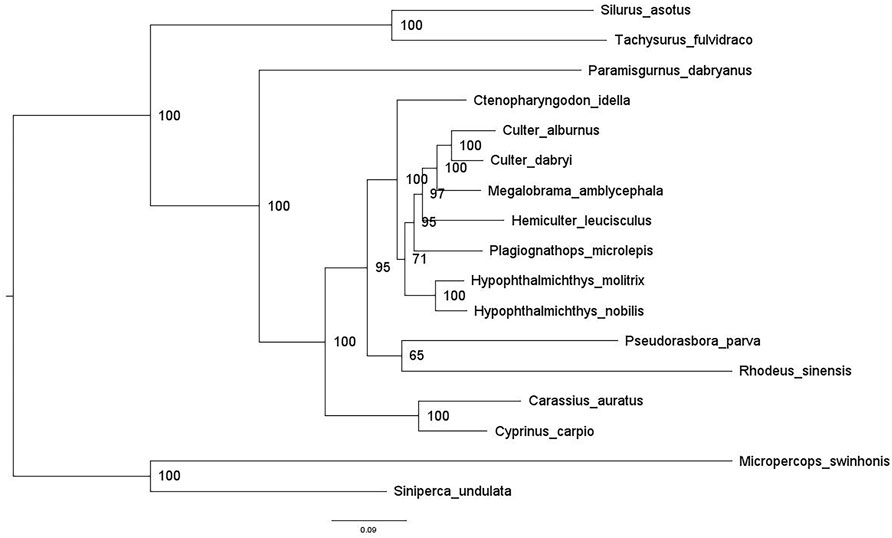
A phylogenetic tree (Figure 5) for the 17 fish species was constructed using ML methods. For this survey, there were four orders and nine families. Among these four orders, Cypriniformes dominated with a majority share of 76.47%, followed by Siluriformes (11.76%), Gobiiformes (5.88%), and Centrarchiformes (5.88%). Xenocyprididae, Cyprinoidae, Gobionigae, Cobitidea, and Acheilognathidea were included in Cypriniformes. Siluriformes includes two families, Bagridae and Siluridae. Gobiiformes and Centrarchiformes contain Odontobutidae and Sinipercidae, respectively. Most bootstrap values exceeded 70, with strong node support. The relationship of this branch for all orders was ((Gobiiformes + Centrarchiformes) + (Siluriformes + Cypriniformes)). The relationship of this branch of Cypriniformes was (Cobitidea + ((Xenocyprididae + (Gobionigae + Acheilognathidea)) + Cyprinoidae)).
3.3 Analysis of ω values between native and non-native fishes
In the Xenocyprididae family, the average ω value for the 13 PCGs was less than 1 for both the five native species and the three non-native species analyzed to determine the Ka/Ks ratio (Figure 6). This indicated that the genes were subject to purifying selection during the evolutionary process. Among both native and non-native species, there were significant differences in the ω values for five genes among the 13 PCGs. These five genes were ATP8 (p = 0.026, ANOVA), COI (p = 0.007, Wilcoxon), COII (p = 0.001, Wilcoxon), COIII (p = 0.023, Wilcoxon), and Cytb (p = 0.023, ANOVA). Among the five genes, except for ATP8, the ω values of all other genes demonstrated that the genes of non-native species experienced stronger purifying selection during evolution compared to the genes of native species. Studies have shown that COX and Cytb have a higher number of conserved amino acid sites (Satoh et al., 2016). ATP8 is a component of Complex V in the mitochondrial respiratory chain that participates in the process of oxidative phosphorylation within the cell (Vercellino and Sazanov, 2022). In some fish species, ATP8 is the most adaptive and fastest-evolving gene site within the mitochondrial genome (Ragauskas et al., 2023). In the non-native fish species in the Xenocyprididae family, the results showed that ATP8 gene underwent significantly less purifying selection during the evolutionary process compared to native species. Studies have indicated that within the Gobiidae family, positively selected genes may be linked to energy metabolism adaptations across various environments. ATP8 has positively selected gene sites, and its ω value is higher (but less than 1) than in environments that do not require adaptation (Shang et al., 2022). Therefore, we speculate that non-native species may exhibit a higher ω value than native species due to their adaptations to new environments. The ω values of COX and Cytb in non-native species in the Baiyangdian Basin were significantly smaller than those found in native species. Studies have indicated that the ω values of genes in birds with different flight capabilities vary, with birds that have strong migratory abilities exhibiting smaller ω values compared to birds with weaker flight capabilities (Y. Y. Shen et al., 2009). Research also suggests that higher energy requirements in fish are accompanied by stronger purifying selection on genes (Sun et al., 2011). COX is a component of Complex IV in the mitochondrial respiratory chain and is involved in important processes such as energy supply, apoptosis, and metabolism (Uddin et al., 2019; Vercellino and Sazanov, 2022). Cytb, the only cytochrome encoded by mitochondrial DNA, is a component of Complex III2 in the mitochondrial respiratory chain and participates in electron transport within the mitochondrial respiratory chain (Vercellino and Sazanov, 2022). Mitochondrial protein genes produce not only the energy required for exercise and other purposes, but also the heat that aids in the regulation of body temperature, especially for organisms living in cold climates (Ruiz-Pesini et al., 2004). The Baiyangdian Basin is in the warm temperate, semi-humid continental monsoon climate zone with cold winters. Different genes in non-native species exhibit a variety of functions that play roles of differing importance, with varying effects attributed to energy production (such as Cytb and COIII) and heat generation (such as COI and COII). Depending on which factor plays a more significant role, different selective constraints are observed (Sun et al., 2011). In this study, in terms of the ratios of the ω values between non-native species gene populations and native species gene populations, the ratio of Cytb was 53.4494%, that of COIII was 49.0398%, that of COI was 84.2803%, and that of COII was 68.0333%. Here, it was found that the Cytb and COIII genes, relative to the COI and COII genes, exhibited greater differential purifying selection in non-native species compared to native species gene populations. Therefore, it is reasonable to speculate that there is a greater demand for energy under the cold climate conditions of the Baiyangdian Basin and for non-native species that need to migrate. This leads to a stronger purifying effect on genes such as COX and Cytb in non-native species compared to native species. Thus, it is likely that energy production plays a more important role than heat generation in the invasion and adaptation of non-native species to the environment of Baiyangdian Basin.
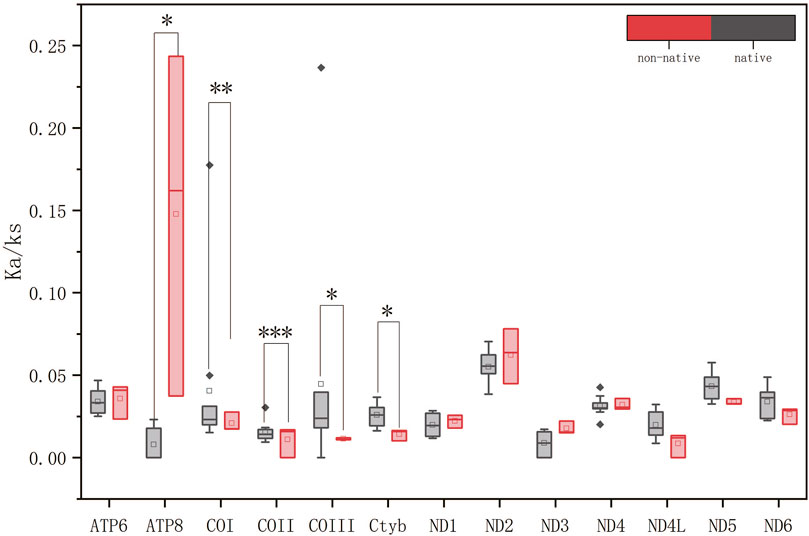
Figure 6. Comparisons of the Ka/Ks values for the 13 PCGs between the native species and non-native species groups. Note: *0.01 < p ≤ 0.05, **0.001 < p ≤ 0.01, ***p ≤ 0.001.
4 Conclusion
This paper analyzed the mitochondrial genomes of 17 fish species in the Baiyangdian Basin for the first time. The basic characteristics of the complete mitochondrial genes of each species, including their basic composition, PCG selection pressure, base preference, and RSCU, were fully elaborated. Phylogenetic relationships were established using ML trees to reveal the evolutionary relationships among species. This study also provided fundamental information on native and non-native species in the Baiyangdian Basin. Using Xenocyprididae as a representative family, this study investigated the selective pressures on PCGs of both native and non-native fish species. Among the 13 PCGs, the ω value of the ATP8 gene was significantly lower in native species than that in non-native species, while the ω values of the COX gene and Cytb gene were significantly lower in non-native species compared to native species. These differences may have arisen because the survival of non-native species during migration and in cold receiving environments depends on the adaptation of genes that encode proteins involved in energy production and heat generation. These results suggest that energy production plays a more important role than heat generation in the invasion and adaptation of non-native species to the environment of the Baiyangdian Basin.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by the Chinese Research Academy of Environmental Sciences Institution Review Board (CRAES IRB) and Chinese Research Academy of Environmental Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
R-QZ: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing. Y-WB: Investigation, Writing–review and editing. J-TH: Methodology, Writing–review and editing. P-YW: Investigation, Writing–review and editing. YQ: Writing–review and editing. T-XZ: Investigation, Writing–review and editing. H-YJ: Writing–review and editing. X-LL: Methodology, Writing–original draft, Writing–review and editing. Z-GY: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Key Research and Development Program (Grant no. 2021YFC3201005 and 2022YFC2601301).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2024.1415150/full#supplementary-material
References
Banerjee, P., Dey, G., Antognazza, C. M., Sharma, R. K., Maity, J. P., Chan, M. W. Y., et al. (2021). Reinforcement of environmental DNA based methods (sensu stricto) in biodiversity monitoring and conservation: a review. Biol. (Basel) 10 (12), 1223. doi:10.3390/biology10121223
Bernt, M., Donath, A., Juhling, F., Externbrink, F., Florentz, C., Fritzsch, G., et al. (2013). MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet Evol. 69 (2), 313–319. doi:10.1016/j.ympev.2012.08.023
Brown, K. H. (2008). Fish mitochondrial genomics: sequence, inheritance and functional variation. J. Fish Biol. 72 (2), 355–374. doi:10.1111/j.1095-8649.2007.01690.x
Ceruso, M., Mascolo, C., Anastasio, A., Pepe, T., and Sordino, P. (2019). Frauds and fish species authentication: study of the complete mitochondrial genome of some Sparidae to provide specific barcode markers. Food control. 103, 36–47. doi:10.1016/j.foodcont.2019.03.028
Chakraborty, S., Nag, D., Mazumder, T. H., and Uddin, A. (2017). Codon usage pattern and prediction of gene expression level in Bungarus species. Gene 604, 48–60. doi:10.1016/j.gene.2016.11.023
Chen, Y., Sun, C., and Zhan, A. (2017). Biological invasions in aquatic ecosystems in China. Aquatic Ecosyst. Health and Manag. 20 (4), 402–412. doi:10.1080/14634988.2017.1403268
Chinnery, P. F., and Hudson, G. (2013). Mitochondrial genetics. Br. Med. Bull. 106 (1), 135–159. doi:10.1093/bmb/ldt017
Collins, R. A., Bakker, J., Wangensteen, O. S., Soto, A. Z., Corrigan, L., Sims, D. W., et al. (2019). Non-specific amplification compromises environmental DNA metabarcoding with COI. Methods Ecol. Evol. 10 (11), 1985–2001. doi:10.1111/2041-210x.13276
Cristescu, M. E., and Hebert, P. D. N. (2018). Uses and misuses of environmental DNA in biodiversity science and conservation. Annu. Rev. Ecol. Evol. Syst. 49 (1), 209–230. doi:10.1146/annurev-ecolsys-110617-062306
Diagne, C., Leroy, B., Vaissiere, A. C., Gozlan, R. E., Roiz, D., Jarić, I., et al. (2021). High and rising economic costs of biological invasions worldwide. Nature 592 (7855), 571–576. doi:10.1038/s41586-021-03405-6
Dos Reis, M. (2015). How to calculate the non-synonymous to synonymous rate ratio of protein-coding genes under the Fisher-Wright mutation-selection framework. Biol. Lett. 11 (4), 20141031. doi:10.1098/rsbl.2014.1031
Du, Z., Liu, H., Li, H., Ishikawa, T., Su, Z. H., Cai, W., et al. (2018). Invasion of the assassin bug Agriosphodrus dohrni (Hemiptera: reduviidae) to Japan: source estimation inferred from mitochondrial and nuclear gene sequences. Int. J. Biol. Macromol. 118 (Pt B), 1565–1573. doi:10.1016/j.ijbiomac.2018.06.191
Feng, X., Li, B., Chen, Y., Zhu, R., Jia, Y., and Sui, X. (2023). Species-level monitoring of rare and invasive fishes using eDNA metabarcoding in the middle and upper Yarlung Zangbo River, Tibet. Water Biol. Secur. 2 (1), 100089. doi:10.1016/j.watbs.2022.100089
Ficetola, G. F., Miaud, C., Pompanon, F., and Taberlet, P. (2008). Species detection using environmental DNA from water samples. Biol. Lett. 4 (4), 423–425. doi:10.1098/rsbl.2008.0118
Harris, J. H. (1995). The use of fish in ecological assessments. Aust. J. Ecol. 20 (1), 65–80. doi:10.1111/j.1442-9993.1995.tb00523.x
Hayati, M., Shadgar, B., and Chindelevitch, L. (2019). A new resolution function to evaluate tree shape statistics. PLoS One 14 (11), e0224197. doi:10.1371/journal.pone.0224197
Hoffmann, M., Hilton-Taylor, C., Angulo, A., Böhm, M., Brooks, T. M., Butchart, S. H. M., et al. (2010). The impact of conservation on the status of the world's vertebrates. Science 330 (6010), 1503–1509. doi:10.1126/science.1194442
Hu, J., Lan, C., and Li, C. (2023). Microdousamblyrhynchos sp. nov., a new member of the small-toothed sleepers (Teleostei, Gobiiformes, Odontobutidae) from Guangxi, southern China. Zookeys 1153, 1–13. doi:10.3897/zookeys.1153.97139
Hu, S., Liu, C., Zheng, H., Wang, Z., and Yu, J. (2012). Assessing the impacts of climate variability and human activities on streamflow in the water source area of Baiyangdian Lake. J. Geogr. Sci. 22 (5), 895–905. doi:10.1007/s11442-012-0971-9
Ivanova, Z., Sablok, G., Daskalova, E., Zahmanova, G., Apostolova, E., Yahubyan, G., et al. (2017). Chloroplast genome analysis of resurrection tertiary relict haberlea rhodopensis highlights genes important for desiccation stress response. Front. Plant Sci. 8, 204. doi:10.3389/fpls.2017.00204
Jensen, M. R., Sigsgaard, E. E., Liu, S., Manica, A., Bach, S. S., Hansen, M. M., et al. (2021). Genome-scale target capture of mitochondrial and nuclear environmental DNA from water samples. Mol. Ecol. Resour. 21 (3), 690–702. doi:10.1111/1755-0998.13293
Konar, M., Qiu, S., Tougher, B., Vause, J., Tlusty, M., Fitzsimmons, K., et al. (2019). Illustrating the hidden economic, social and ecological values of global forage fish resources. Resour. Conservation Recycl. 151, 104456. doi:10.1016/j.resconrec.2019.104456
Larson, E. R., Graham, B. M., Achury, R., Coon, J. J., Daniels, M. K., Gambrell, D. K., et al. (2020). From eDNA to citizen science: emerging tools for the early detection of invasive species. Front. Ecol. Environ. 18 (4), 194–202. doi:10.1002/fee.2162
Leray, M., Yang, J. Y., Meyer, C. P., Mills, S. C., Agudelo, N., Ranwez, V., et al. (2013). A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front. Zool. 10, 34. doi:10.1186/1742-9994-10-34
Levin, B. A., Freyhof, J., Lajbner, Z., Perea, S., Abdoli, A., Gaffaroğlu, M., et al. (2012). Phylogenetic relationships of the algae scraping cyprinid genus Capoeta (Teleostei: cyprinidae). Mol. Phylogenet Evol. 62 (1), 542–549. doi:10.1016/j.ympev.2011.09.004
Levy, N., Simon-Blecher, N., Ben-Ezra, S., Yuval, M., Doniger, T., Leray, M., et al. (2023). Evaluating biodiversity for coral reef reformation and monitoring on complex 3D structures using environmental DNA (eDNA) metabarcoding. Sci. Total Environ. 856 (Pt 2), 159051. doi:10.1016/j.scitotenv.2022.159051
Li, Y., Lu, C., Liu, G., Chen, Y., Zhang, Y., Wu, C., et al. (2023). Risk assessment of wetland degradation in the Xiong’an New Area based on AHP-EWM-ICT method. Ecol. Indic. 153, 110443. doi:10.1016/j.ecolind.2023.110443
Montag, L. F. d. A., Koroiva, R., Ribeiro-dos-Santos, Â., Magalhães, L., Cavalcante, G. C., Silva, C. S., et al. (2023). The complete mitogenome of amazonian Hyphessobrycon heterorhabdus (characiformes: characidae) as a valuable resource for phylogenetic analyses of characidae. Fishes 8 (5), 233. doi:10.3390/fishes8050233
Ojala, D., Montoya, J., and Attardi, G. (1981). tRNA punctuation model of RNA processing in human mitochondria. Nature 290 (5806), 470–474. doi:10.1038/290470a0
Perna, N. T., and Kocher, T. D. (1995). Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 41 (3), 353–358. doi:10.1007/BF00186547
Qian, M. M., Wang, Z. Y., Zhou, Q., Wang, J., Shao, Y., Qiao, Q., et al. (2023). Environmental DNA unveiling the fish community structure and diversity features in the Yangtze River basin. Environ. Res. 239 (Pt 1), 117198. doi:10.1016/j.envres.2023.117198
Ragauskas, A., Ignataviciene, I., Rakauskas, V., Grauda, D., Prakas, P., and Butkauskas, D. (2023). Trends of eurasian perch (Perca fluviatilis) mtDNA ATP6 region genetic diversity within the hydro-systems of the eastern part of the baltic Sea in the anthropocene. Anim. (Basel) 13 (19), 3057. doi:10.3390/ani13193057
Ruiz-Pesini, E., Mishmar, D., Brandon, M., Procaccio, V., and Wallace, D. C. (2004). Effects of purifying and adaptive selection on regional variation in human mtDNA. Science 303 (5655), 223–226. doi:10.1126/science.1088434
Sahu, A., Kumar, N., Pal Singh, C., and Singh, M. (2023). Environmental DNA (eDNA): powerful technique for biodiversity conservation. J. Nat. Conservation 71, 126325. doi:10.1016/j.jnc.2022.126325
Satoh, T. P., Miya, M., Mabuchi, K., and Nishida, M. (2016). Structure and variation of the mitochondrial genome of fishes. BMC Genomics 17 (1), 719. doi:10.1186/s12864-016-3054-y
Sepulveda, A. J., Nelson, N. M., Jerde, C. L., and Luikart, G. (2020). Are environmental DNA methods ready for aquatic invasive species management? Trends Ecol. Evol. 35 (8), 668–678. doi:10.1016/j.tree.2020.03.011
Shang, Y., Wang, X., Liu, G., Wu, X., Wei, Q., Sun, G., et al. (2022). Adaptability and evolution of Gobiidae: a genetic exploration. Anim. (Basel) 12 (14), 1741. doi:10.3390/ani12141741
Sharma, A., Siva, C., Ali, S., Sahoo, P. K., Nath, R., Laskar, M., et al. (2020). The complete mitochondrial genome of the medicinal fish, Cyprinion semiplotum: insight into its structural features and phylogenetic implications. Int. J. Biol. Macromol. 164, 939–948. doi:10.1016/j.ijbiomac.2020.07.142
Shen, M., Xiao, N., Zhao, Z., Guo, N., Luo, Z., Sun, G., et al. (2022). eDNA metabarcoding as a promising conservation tool to monitor fish diversity in Beijing water systems compared with ground cages. Sci. Rep. 12 (1), 11113. doi:10.1038/s41598-022-15488-w
Shen, Y. Y., Shi, P., Sun, Y. B., and Zhang, Y. P. (2009). Relaxation of selective constraints on avian mitochondrial DNA following the degeneration of flight ability. Genome Res. 19 (10), 1760–1765. doi:10.1101/gr.093138.109
Sheikh, A., Al-Taher, A., Al-Nazawi, M., Al-Mubarak, A. I., and Kandeel, M. (2020). Analysis of preferred codon usage in the coronavirus N genes and their implications for genome evolution and vaccine design. J. Virol. Methods 277, 113806. doi:10.1016/j.jviromet.2019.113806
Sun, Y. B., Shen, Y. Y., Irwin, D. M., and Zhang, Y. P. (2011). Evaluating the roles of energetic functional constraints on teleost mitochondrial-encoded protein evolution. Mol. Biol. Evol. 28 (1), 39–44. doi:10.1093/molbev/msq256
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38 (7), 3022–3027. doi:10.1093/molbev/msab120
Uddin, A., Mazumder, T. H., and Chakraborty, S. (2019). Understanding molecular biology of codon usage in mitochondrial complex IV genes of electron transport system: relevance to mitochondrial diseases. J. Cell. Physiol. 234 (5), 6397–6413. doi:10.1002/jcp.27375
Vander Zanden, M. J., Hansen, G. J. A., Higgins, S. N., and Kornis, M. S. (2010). A pound of prevention, plus a pound of cure: early detection and eradication of invasive species in the Laurentia Great Lakes. J. Gt. Lakes. Res. 36 (1), 199–205. doi:10.1016/j.jglr.2009.11.002
Vercellino, I., and Sazanov, L. A. (2022). The assembly, regulation and function of the mitochondrial respiratory chain. Nat. Rev. Mol. Cell. Biol. 23 (2), 141–161. doi:10.1038/s41580-021-00415-0
Wang, S., Yan, Z., Hanfling, B., Zheng, X., Wang, P., Fan, J., et al. (2021). Methodology of fish eDNA and its applications in ecology and environment. Sci. Total Environ. 755 (Pt 2), 142622. doi:10.1016/j.scitotenv.2020.142622
Wang, T., Zhang, S., Pei, T., Yu, Z., and Liu, J. (2019). Tick mitochondrial genomes: structural characteristics and phylogenetic implications. Parasit. Vectors 12 (1), 451. doi:10.1186/s13071-019-3705-3
Wolfsberg, T. G., Schafer, S., Tatusov, R. L., and Tatusov, T. A. (2001). Organelle genome resources at NCBI. Trends Biochem. Sci. 26 (3), 199–203. doi:10.1016/s0968-0004(00)01773-4
Wong, E. H., Smith, D. K., Rabadan, R., Peiris, M., and Poon, L. L. (2010). Codon usage bias and the evolution of influenza A viruses. Codon usage biases of influenza virus. BMC Evol. Biol. 10, 253. doi:10.1186/1471-2148-10-253
Wu, D., Zhou, L., Xue, J., Xia, Q., and Meng, L. (2022). Characterization of two new Apodemus mitogenomes (rodentia: muridae) and mitochondrial phylogeny of muridae. Diversity 14 (12), 1089. doi:10.3390/d14121089
Wu, Q. L., Li, Q., Gu, Y., Shi, B. C., van Achterberg, C., Wei, S. J., et al. (2014). The complete mitochondrial genome of Taeniogonalos taihorina (Bischoff) (Hymenoptera: trigonalyidae) reveals a novel gene rearrangement pattern in the Hymenoptera. Gene 543 (1), 76–84. doi:10.1016/j.gene.2014.04.003
Keywords: mitochondrial genome, fish, Baiyangdian basin, phylogeny, Non-native species
Citation: Zhang R-Q, Bai Y-W, Hu J-T, Wang P-Y, Qi Y, Zhang T-X, Jiao H-Y, Lin X-L and Yan Z-G (2024) Complete mitochondrial genomes and phylogenetic analysis of native and non-native fishes in a national key wetland of China. Front. Environ. Sci. 12:1415150. doi: 10.3389/fenvs.2024.1415150
Received: 10 April 2024; Accepted: 16 September 2024;
Published: 25 September 2024.
Edited by:
Wei Wang, Ocean University of China, ChinaReviewed by:
Yiguo Hong, Guangzhou University, ChinaWeiji Wang, Chinese Academy of Fishery Sciences (CAFS), China
Jinzhe Du, Qingdao Agricultural University, China
Baocheng Guo, Institute of Zoology (CAS), China
Copyright © 2024 Zhang, Bai, Hu, Wang, Qi, Zhang, Jiao, Lin and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhen-Guang Yan, emd5YW5AY3JhZXMub3JnLmNu; Xiao-Long Lin, eGxsaW5Ac2hvdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Rui-Qing Zhang1,2†
Rui-Qing Zhang1,2† Yan Qi
Yan Qi Tian-Xu Zhang
Tian-Xu Zhang Xiao-Long Lin
Xiao-Long Lin Zhen-Guang Yan
Zhen-Guang Yan