- 1University of Belgrade, Faculty of Biology, Department of Morphology, Systematics and Phylogeny of Animals, Belgrade, Serbia
- 2University of Zagreb, Faculty of Agriculture, Department of Fisheries, Apiculture, Wildlife Management and Special Zoology, Zagreb, Croatia
- 3Department of Ecology and Vertebrate Zoology, Faculty of Biology and Environmental Protection, University of Lodz, Lodz, Poland
- 4University of Zagreb, Faculty of Science, Department of Biology, Zagreb, Croatia
Introduction: The genetic diversity of brown trout in the Western Balkans has been disrupted by the introduction of non-native Atlantic phylogenetic lineages and non-native haplotypes of the Danubian phylogenetic lineage. The Western Balkans is characterized by the greatest phenotypic and genotypic diversity of trout populations, and a large part of the internal territory belongs to the Black Sea basin, where the Danubian Da1 haplotype is native. Artificial propagation of non-native lineages in the Western Balkans has a long history, and these populations are often the only available material for stocking rivers attractive for fishing.
Material and Methods: Fifteen populations in the Danube basin of the continental Croatia were analysed. The analysis of eight microsatellite loci was performed to determine the structure of brown trout populations, as well as the degree of introgression of non-native genetic material into the native.
Results and Disscusion: The results of this study showed significant genetic similarity among brown trout populations, confirming a long history of introduction with non-native genetic material. The main reason was uncontrolled stocking with inadequate material, which is available in fish farms and consists mainly of brown trout of the Atlantic phylogenetic lineage. The results of this study also indicated stocking with brown trout of the non-native haplotypes of the Danubian phylogenetic lineage. The potential breeding origin of brown trout carrying the Danubian Da2 mtDNA haplotype and ways of its introduction into rivers have yet to be investigated. For the survival of the unique gene pool of brown trout in Croatian rivers, it is of fundamental importance to know the structure of wild and farmed populations with the aim of proposing and implementing conservation measures.
Introduction
The commercial value of brown trout Salmo trutta, along with its significant importance to humans, is reflected in various aspects, particularly its widespread artificial propagation in fish farms globally. Farming is carried out for various purposes, of which stocking notably impacts the survival of native, genetically pure, and geographically unique brown trout populations.
Numerous studies on this species, still considered the S. trutta species complex, have shown that the life history, phenotypic plasticity, and high adaptability to diverse environmental conditions have not yet been clarified, more than likely as a consequence of their exceptionally high morphological and genetic variability (Behnke, 1972; Weiss et al., 2001; Sanz, 2018; Škraba Jurlina et al., 2018). The Western Balkans exhibit a high degree of endemism in salmonid genera and species (Behnke, 1986; Oikonomou et al., 2014). Together with the Iberian Peninsula and the Apennines, the Western Balkans is considered the area with the greatest phenotypic and genetic diversity among trout populations (Bernatchez, 2001) with complex evolutionary mechanisms, including the occurrence of secondary contacts between ancestral lineages, as well as local adaptation (Sanz et al., 2002; Snoj et al., 2008; Vera et al., 2010). The Western Balkans is the home range for populations of three phylogenetic lineages of Salmo cf. trutta which are defined based on the control region (CR) of mitochondrial DNA (mtDNA): Danubian (DA), Adriatic (AD) and marmoratus (MA). The DA lineage inhabits the northern, eastern, and central parts of the Balkan Peninsula, i.e., the Black Sea basin, and the AD lineage inhabits the northwestern and southwestern regions, including numerous lotic and some lentic habitats in the continental part of the Adriatic and Ionian Seas’ basins (Georgiev, 2003). The MA lineage is found close to the dispersal range of brown trout of the Danubian and Adriatic lineages, in the narrowest area in the Western Balkans, inhabiting the Adriatic drainages in Bosnia and Herzegovina and Montenegro (Snoj et al., 2010; Mrdak eta al., 2012; Škraba Jurlina et al., 2020; Righi et al., 2023). The status of the MA lineage, which is still often identified as marble trout, has not yet been resolved in Croatia (Ćaleta et al., 2019).
The rivers of the Western Balkans within the Black Sea basin drain a considerable territory and are very important in the context of studying the diversity and life history of different phylogenetic lineages (Škraba Jurlina et al., 2018). The DA lineage, native to the Danube basin, consists of numerous haplotypes, with the Da1 haplotype being the most widespread. Still, not all of the DA haplotypes are considered native to the Western Bakans area/region. The presence of non-native DA haplotypes, as well as other phylogenetic lineages, in most of the studied rivers presents a significant threat to the native population’s genetic structure (Snoj, 2004; Marić et al., 2006; Jadan et al., 2007; Tošić et al., 2016; Simonović et al., 2017a; Buj et al., 2020; Ivić et al., 2021).
Breeding brown trout in the Balkans is a profitable enterprise which makes it highly popular across all countries in the region. With the aim of intensive artificial propagation, and thus, a greater commercial profit (Simonović et al., 2017b), the initially domesticated native DA lineage (Gridelli, 1936; Kohout et al., 2012) proved to be insufficient. Consequently, breeders introduced breeding technology from Western Europe and the Atlantic lineage of brown trout suitable for this purpose (Taler, 1949; Pofuk et al., 2017; Piria et al., 2020).
Human-induced translocations of organisms and anthropogenic alterations of natural habitats are the most common causes of hybridization and introgression of non-native genotypes into native populations (Allendorf et al., 2001). Salmo trutta is characterized by genetic diversity at the intrapopulation level, which is the reason for the existence of genetically differentiated local populations. The loss of these unique populations could lead to a significant reduction in genetic variability within the species (Laikre et al., 1999). Stocking became a common practice to increase and support wild stocks of many fish species, including brown trout (Righi et al., 2023). In the beginning of the 20th century, it was pointed out that supportive breeding programs could also pose a threat to gene-level biodiversity, by releasing cultured individuals into the wild, with the aim of increasing the size of natural populations (Laikre et al., 2008). These activities could lead to manipulation of reproductive rates, causing reduced effective population size, increased rate of genetic drift and loss of genetic variability (Ryman et al., 1995; Wang and Ryman, 2001; Laikre et al., 2008), which is observed in many populations of brown trout in the Balkans (Jadan et al., 2007; Buj et al., 2020; Piria et al., 2020; Škraba Jurlina et al., 2020).
Although the research on genetic variability of brown trout has been extensively conducted to assess the introgression of farmed lineages, gene-level monitoring programs are still notably absent, despite repeated calls for their implementation (Laikre et al., 2008; Tošić et al., 2016; Simonović et al., 2017a; Škraba-Jurlina et al., 2020). To ensure the survival of a population or species, it is necessary to preserve its genetic diversity, and thus its evolutionary potential (Ryman et al., 1995). Unfortunately, the genotyping of brown trout individuals in brood stocks has been limited to a small number of fish farms in the Western Balkans.
In Croatian fish farms, brown trout of the AT lineage are mainly bred, so consequently the rivers are most likely stocked with these individuals due to their availability (Marić et al., 2022). In this way, non-native genetic material enters natural watercourses (Piria et al., 2020). Brown trout of the AT lineage are already recognized as the main cause of the loss of the original genetic diversity of brown trout (Weiss et al., 2001; Simonović et al., 2017b) and show an invasive character (Simonović et al., 2013; 2015; Piria et al., 2020), but their stocking is not regulated by any law in Croatia.
The goal of this research was to determine the genetic structure of brown trout populations in the Croatian part of the Danube basin, and to assess the degree of introgression of non-native genetic material within studied populations, as well as its possible origin.
Material and methods
A total of 141 samples were collected and analyzed from 15 locations: in the Gorski Kotar area (six locations), in Žumberak (three locations) and on the slopes of Mount Papuk (six locations) (Figure 1). The sampling was carried out in the period of April/May during 2017 and 2018.
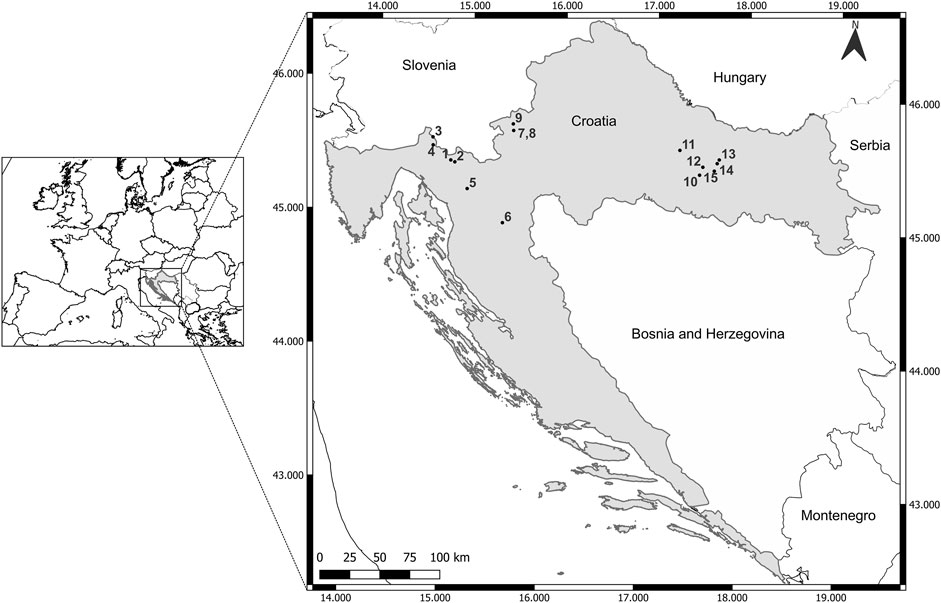
Figure 1. Map of sampling sites of brown trout in the Danube River basin of Croatia. Gorski Kotar region: 1—Mala Lešnica, 2—Curak, 3—Čabranka, 4—Bresni Potok, 5—Jasenak, 6—Lička Jesenica; Žumberak region: 7—Kupčina, 8—“Vrabac” fish farm, 9—Slapnica; Mt. Papuk region: 10—Orljava, 11—Toplica, 12—Brzaja, 13—Jankovac Stream, 15—Jankovac Lake, 15—Veličanka.
For the purposes of genetic analyses, the anal fin clips of each individual were taken and stored in 96% ethanol. The extraction of DNA was conducted according to the protocol of manufacturer of the Quick-gDNA™ MiniPrep (Zymo Research Corporation, United States) extraction kits.
The analysis of the control region of mitochondrial DNA (CR mtDNA) was used to identify the phylogenetic lineages of brown trout and their haplotypes, while restriction analysis of the L-lactate dehydrogenase nuclear locus (LDH-C*) was used to detect if hybrids between AT and DA lineages exist. For a detailed description of the procedures and software analyses refer to Kanjuh et al. (2020).
Population diversity was accessed by analyses of eight microsatellite loci: Str73INRA (Estoup et al., 1993) and Ssa410Uos (Cairney et al., 2000), SsaD190 and SsaD71 (King et al., 2005), Ssa85 (O’Reilly et al., 1996) and SSsp2216 (Paterson et al., 2004), and OMM1064 (Rexroad et al., 2002) and SsoSL438 (Slettan et al., 1995), amplified in four duplex reactions. The total volume of the mixture for all four duplex reactions was 10 μL, and its composition was as follows: forward and reverse primers for a specific locus (with final concentrations: 0.2 µM for SsaD190, SsaD71, SSsp2216 and OMM1064, 0.4 µM for Ssa410Uos and SsoSL438, 0.1 µM for Ssa85 and 0.15 µM for Str73INRA), dH2O, 1X PCR buffer, 0.2 mM dNTP, 1.5 mM MgCl2, 0.5 U Taq polymerase, and 2 μL DNA sample. The amplification conditions for the first three duplex reactions (Str73INRA and Ssa410Uos, SsaD190 and SsaD71, Ssa85 and SSsp2216) were identical: initial denaturation (94°C, 3 min), 30 cycles of denaturation (94°C, 45 s), primer binding (60°C, 1 min), elongation (72°C, 30 s) and final elongation (72°C, 1 h). An exception in the protocol was the fourth duplex reaction (OMM1064 and SsoSL438), in which the primer binding was set to 57°C, and the number of cycles was 35. Fragment analysis of microsatellite loci was performed in the laboratories of MACROGEN® Europe. For several samples, fragment analysis was performed at the Center for Human Molecular Genetics, Faculty of Biology, University of Belgrade, where the fragment analysis was performed using GeneScan™ 500 LIZ® Size Standard (Applied Biosystems, United States) on an ABI-3130 Genetic Analyzer (Applied Biosystems, United States). Analysis was done using GeneMapper® ID v3.2.1 software (Applied Biosystems, United States).
Statistical analyses of microsatellite loci
Hardy-Weinberg equilibrium (HWE) and p-values for FIS within samples, based on 120,000 randomizations, were observed in Fstat 2.9.3.2 software (Goudet, 2002). Observed and expected heterozygosity values, the average number of alleles per locus, allele frequencies, fixation indices, Nei’s distances between populations per 1,000 permutations and the Factorial Correspondent Analysis (FCA) were calculated using GENETIX 4.05 software (Belkhir et al., 1996). The software POPULATIONS 1.2.31 (Langella, 2002) was used to calculate shared allele distances (DAS) and to generate a Neighbor-Joining (NJ) tree. The results of Nei’s distances were used for CLUSTER analysis using the method of Unweighted Pair-group Average (UPGMA) in STATISTICA 8.0 software (StatSoft, Inc, 2001). Population structure analysis was done using STRUCTURE 2.3.4 software (Pritchard et al., 2000). The structure analysis for all 141 individuals was conducted with assumed number of groups K = 15 and the test period was set at 100,000 to 200,000, which was repeated seven times for each group K. The STRUCTURE Harvester tool (Earl and vonHoldt, 2012) was used to estimate the most likely value of K according to Evanno et al. (2005). Additional structure analysis was done only for DA individuals, who according to the LDH-C* molecular marker, were homozygous for the LDH-C*100 allele. The assumed number of groups was K = 15 and the test period was set at 100,000 to 200,000, which was again repeated seven times for each value K. The bottleneck effect was evaluated using the BOTTLENECK 1.2.02 software (Cornuet and Luikart, 1996; Piry et al., 1999), within Sign test and Wilcoxon test (Sign-rank test) and two mutation models, two-phase mutation model (TPM) and stepwise mutation model (SMM). As parameters for the assessment of significance for these tests, the proportion of a mutation step in the TPM was set at 95% and the mutation variance for the TPM at 12,000 and 100,000 iterations.
Results
Control region of mitochondrial DNA and L-lactate dehydrogenase nuclear locus
The results of CR mtDNA and LDH-C* locus analyses are presented in detail in Kanjuh et al. (2020). In short, DA and AT phylogenetic lineages were identified in analyzed samples. The presence of non-native brown trout haplotypes was detected in all sampling areas: At1 (At-H3) in eight localities (including the fish farm), Da2 in seven localities and Da22 in three localities (Table 1). The only river where all individuals (five in total) carried the native Da1 haplotype was the Jankovac Stream. Analysis of the LDH-C* locus revealed interbreeding between DA and AT individuals, indicating the presence of hybrids. Even in the Jankovac Stream the crossbreeding between Da1 and At1 individuals was recorded, with one exception that was homozygous for the LDH-C*100 allele. The only genetically pure population was the one from the fish farm, where all the analysed individuals were non-native AT lineage by maternal (CR mtDNA) and paternal (LDH-C*) inheritance. The results obtained with these two molecular markers indicate a strong introgression of non-native genetic material into native, which is most likely a consequence of crosses in the F1 generation, resulting from uncontrolled, long-term stocking with farmed trout. This is supported by the results showing that 21 individuals of the DA lineage were homozygous for the LDH-C*90 allele, which is characteristic trait of the AT lineage, while six individuals of AT haplogroups were homozygous for the LDH-C*100 allele typically found in the DA haplogroup.
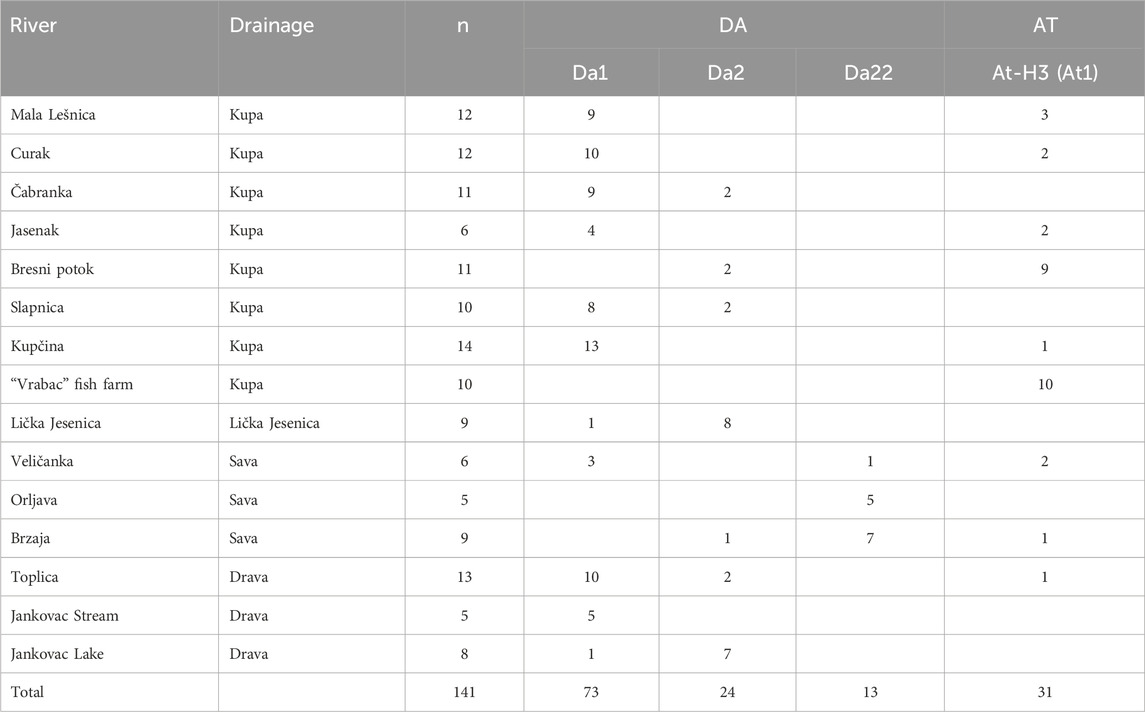
Table 1. Results of control region of mitochondrial DNA showing distribution of the brown trout haplotypes detected in the studied rives in Croatia. n—number of individuals, DA—Danubian lineage, AT—Atlantic lineage (from Kanjuh et al., 2020).
Microsatellite loci
The highest expected (Hexp.) and observed (Hobs.) heterozygosities were recorded in the Jasenak River population (Table 2), in which the highest number of alleles, i.e., the highest allelic richness at the loci OMM1064 (16 alleles) and SsoSL438 (14 alleles) was observed (Supplementary Table S1). The lowest values of Hexp. were observed for populations from Jankovac Stream (0.537) and Orljava River (0.543). The lowest values of Hobs. were recorded for populations from the Orljava River (0.475) and the Curak River (0.458). The lowest average number of alleles was recorded in the Orljava River population (Table 2).
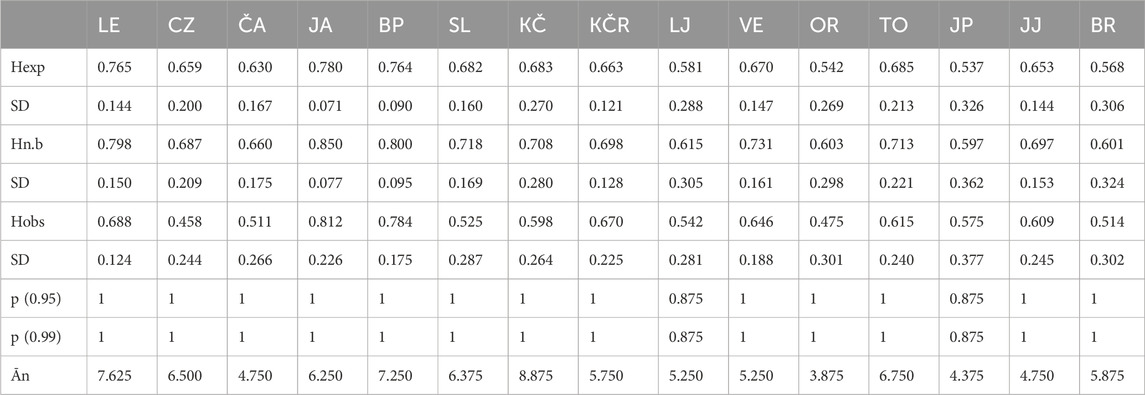
Table 2. Average heterozygosity of microsatellite loci in the studied populations. Hexp.—expected heterozygosity, Hn.b.—objective heterozygosity, Hobs.—observed heterozygosity, SD—standard deviation, p—probability, Ān—average number of alleles per locus, LE—Mala Lešnica, CZ—Curak, ČA—Čabranka, JA—Jasenak, BP—Bresni potok, SL—Slapnica, KČ—Kupčina, KČR—“Vrabac” fish farm, LJ—Lička Jesenica, VE—Veličanka, OR—Orljava, TO—Toplica, JP—Jankovac Stream, JJ—Jankovac Lake, BR—Brzaja
The GENETIX software analysis (Table 3) showed the highest genetic distances (FST) between “Vrabac” fish farm and the Jankovac Stream (0.27), as well as the smallest gene flow (Nm) between these two populations (0.69). Similarly, Nei’s distances (Table 4) showed the highest values between populations from the “Vrabac” fish farm and those from the Veličanka (1.458) and Čabranka rivers (1.408) and Jankovac Stream (1.318). The significant genetic distance between the populations from the “Vrabac” fish farm and the Jankovac Stream coincides with the results of the CR mtDNA analysis which showed that all individuals from the fish farm belong to At1 haplotype, while all individuals from the Jankovac Stream belong to Da1 haplotype (Table 1).
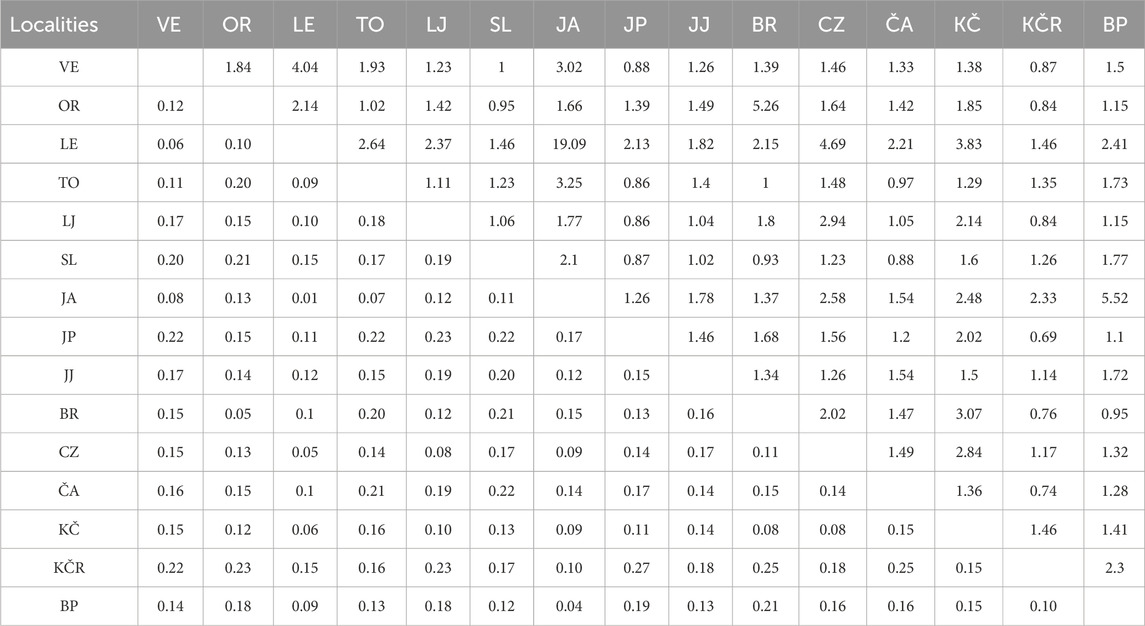
Table 3. Genetic distances for populations pairs, based on allele frequencies across the eight microsatellite loci, are shown by fixation index values (FST) are shown under diagonal, and estimated gene flow values (Nm) are shown above diagonal. VE—Veličanka, OR—Orljava, LE—Mala Lešnica, TO—Toplica, LJ—Lička Jesenica, SL—Slapnica, JA—Jasenak, JP—Jankovac Stream, JJ—Jankovac Lake, BR—Bresni potok, CZ—Curak, ČA—Čabranka, KČ—Kupčina, KČR—“Vrabac” fish farm, BP—Brzaja
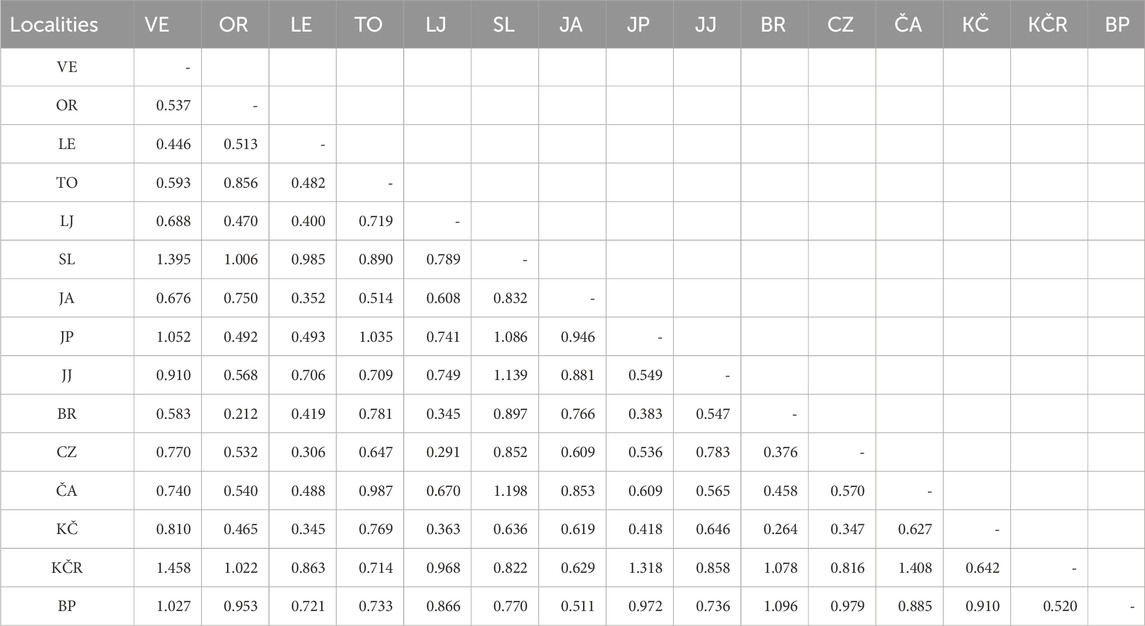
Table 4. Nei’s distances values based on the eight microsatellite loci between pairs of populations. VE—Veličanka, OR—Orljava, LE—Mala Lešnica, TO—Toplica, LJ—Lička Jesenica, SL—Slapnica, JA—Jasenak, JP—Jankovac Stream, JJ—Jankovac Lake, BR—Bresni potok, CZ—Curak, ČA—Čabranka, KČ—Kupčina, KČR—“Vrabac” fish farm, BP—Brzaja
The CLUSTER analysis of Nei’s distances and shared allele distances values showed populations’ grouping. Four populations from the Gorski Kotar and Žumberak areas were clustered together (SL, JA, BP and KČR) (Figure 2), and the second cluster consisted of the remaining 11 populations. Two populations stood out compared to the others: the Slapnica River and the Jankovac Stream (Figure 2). The shared allele distances values (DAS) showed small deviations from Nei’s distances (Figure 3). Analysis of both DAS and Nei’s distances values showed sister relationships between the populations of Čabranka and Jankovac Lake, which follows the low FST value between these two populations (0.14) (Table 3). In contrast to Nei’s distances values, DAS values showed clustering of the population from the Jasenak River with populations from the Toplica River, but also the separation of Veličanka, Mala Lešnica and Kupčina rivers.
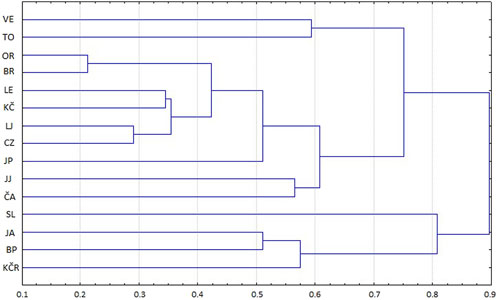
Figure 2. CLUSTER analysis of matrix distances of 15 investigated populations according to Nei’s distances based on eight microsatellite loci. VE—Veličanka, OR—Orljava, LE—Mala Lešnica, TO—Toplica, LJ—Lička Jesenica, SL—Slapnica, JA—Jasenak, JP—Jankovac Stream, JJ—Jankovac Lake, BR—Bresni potok, CZ—Curak, ČA—Čabranka, KČ—Kupčina, KČR—“Vrabac” fish farm, BP—Brzaja
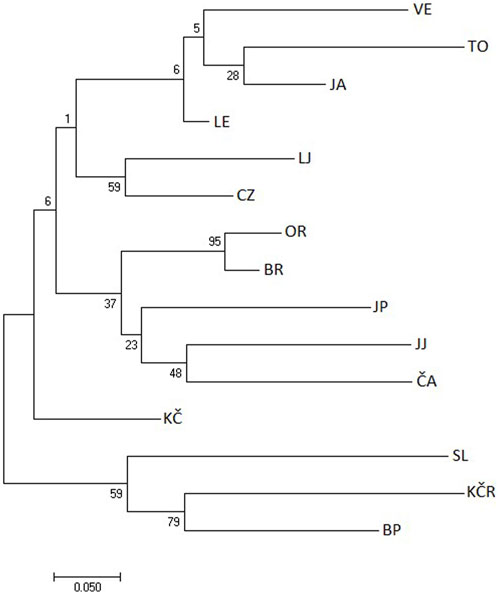
Figure 3. Clustering of populations using the Neighbor-Joining rooted bootstrap tree method according to shared allele distances values based on eight microsatellite loci. VE—Veličanka, OR—Orljava, LE—Mala Lešnica, TO—Toplica, LJ—Lička Jesenica, SL—Slapnica, JA—Jasenak, JP—Jankovac Stream, JJ—Jankovac Lake, BR—Bresni potok, CZ—Curak, ČA—Čabranka, KČ—Kupčina, KČR—“Vrabac” fish farm, BP—Brzaja
The corresponding factorial analysis (CFA) (Figures 4A–C) showed a clear separation of the Slapnica River population (Figure 4A), while a large genetic overlap is shown for the other populations.
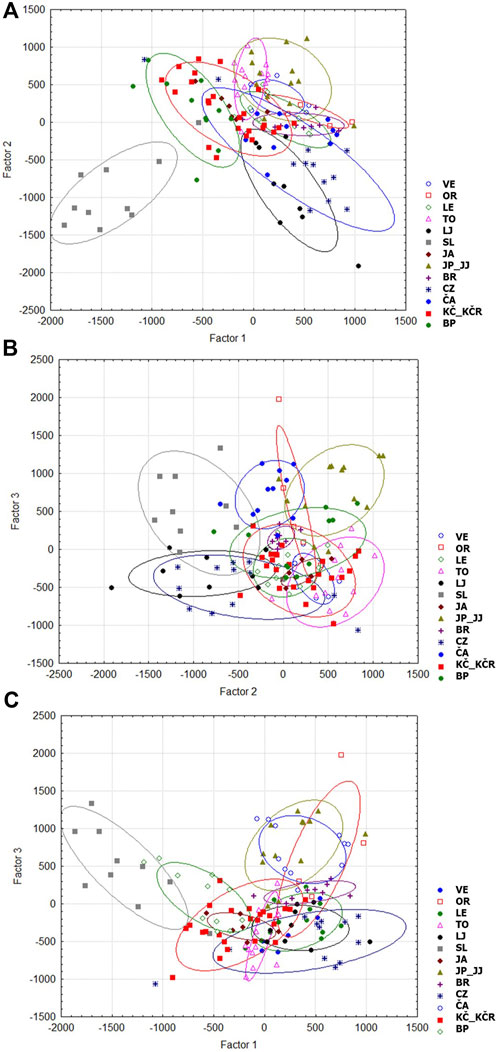
Figure 4. Corresponding factor analysis of 15 analysed populations according to factors: (A) 1 and 2, (B) 2 and 3 and (C) 1 and 3. VE—Veličanka, OR—Orljava, LE—Mala Lešnica, TO—Toplica, LJ—Lička Jesenica, SL—Slapnica, JA—Jasenak, JP—Jankovac Stream, JJ—Jankovac Lake, BR—Bresni potok, CZ—Curak, ČA—Čabranka, KČ—Kupčina, KČR—“Vrabac” fish farm, BP—Brzaja
The evaluation of the bottleneck effect showed that the He value was higher than the Heq value in the populations from the Orljava River (at five loci according to TPM), Jankovac Stream (at five loci according to TMP and SMM), Brzaja River (at six loci according to TPM and SMM), and Čabranka River (at six loci according to TPM, and at five loci according to SMM) (Table 5). However, no locus had a probability lower than 0.05. Based on both mutation models (TMP and SMM), a significant heterozygosity deficit was recorded in the rivers Brzaja, Curak and the “Vrabac” fish farm, while for the population from the Kupčina River, a heterozygosity deficit was recorded only according to SMM. These results indicate a recent expansion of the above mentioned populations (Table 5).
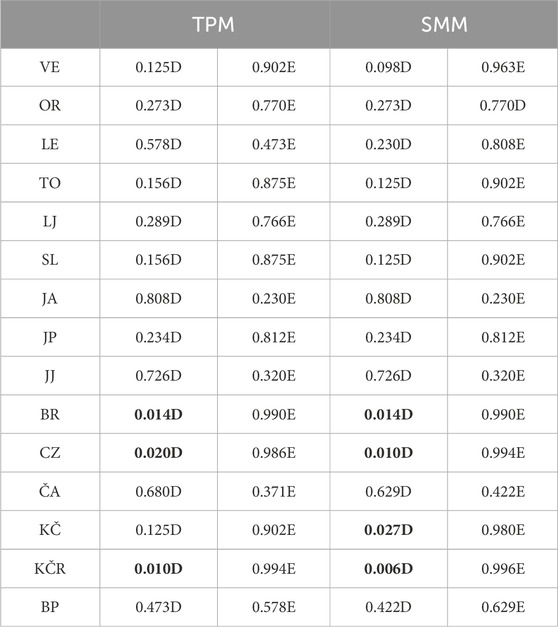
Table 5. Wilcoxon test results based on two-phase mutation model (TPM) and stepwise mutation model (SMM). Bold values are significantly lower than 0.05. D—heterozygosity deficiency, E—heterozygosity excess, VE—Veličanka, OR—Orljava, LE—Mala Lešnica, TO—Toplica, LJ—Lička Jesenica, SL—Slapnica, JA—Jasenak, JP—Jankovac Stream, JJ—Jankovac_Lake, BR—Brzaja, CZ—Curak, ČA—Čabranka, KČ—Kupčina, KČR—“Vrabac” fish farm, BP—Bresni potok
The STRUCTURE analysis revealed two distinct brown trout genetic clusters (when K = 2, Figure 5A). The structuring of the populations was very much congruent with the mtDNA haplogroup to which the individuals belong (Kanjuh et al., 2020). One group of individuals colored green were those of the DA lineage, while the others colored red were of the AT lineage (Figure 5B). This genetic population structure can best be seen in the brown trout from the Curak River. Twelve individuals of this population were separated during grouping. Ten of them of the DA lineage were grouped with other individuals of the DA lineage (i.e., colored green). Remaining two individuals from the Curak River belonging to AT lineage (one in homozygous state for LDH-C*90, and the other in heterozygous state) were grouped with individuals of the AT lineage (i.e., colored red). The green colored group of individuals comprised both DA-AT hybrids and genetically pure DA individuals. Populations from the Čabranka and Lička Jasenica rivers were grouped together (i.e., colored green). All individuals belonged to DA lineage, but among them there were also DA-AT hybrids: two individuals in homozygous state for LDH-C*90 (LJ41 and LJ42), and five individuals in heterozygous state (LJ37, LJ43, ČA5, ČA8 and ČA10) (Figure 5). The grouping with only DA individuals suggested three genetic clusters (K = 3) (Figure 6A). Individuals from the Curak River (CZ) separated into one genetic cluster, while the other two genetic clusters showed overlapping genetic structure (Figure 6B).

Figure 5. (A) Value of ΔK and (B) genetic population structuring. VE—Veličanka, OR—Orljava, LE—Mala Lešnica, TO—Toplica, LJ—Lička Jesenica, SL—Slapnica, JA—Jasenak, JP—Jankovac Stream, JJ—Jankovac Lake, BR—Brzaja, CZ—Curak, ČA—Čabranka, KČ—Kupčina, KČR—“Vrabac” fish farm, BP—Bresni potok
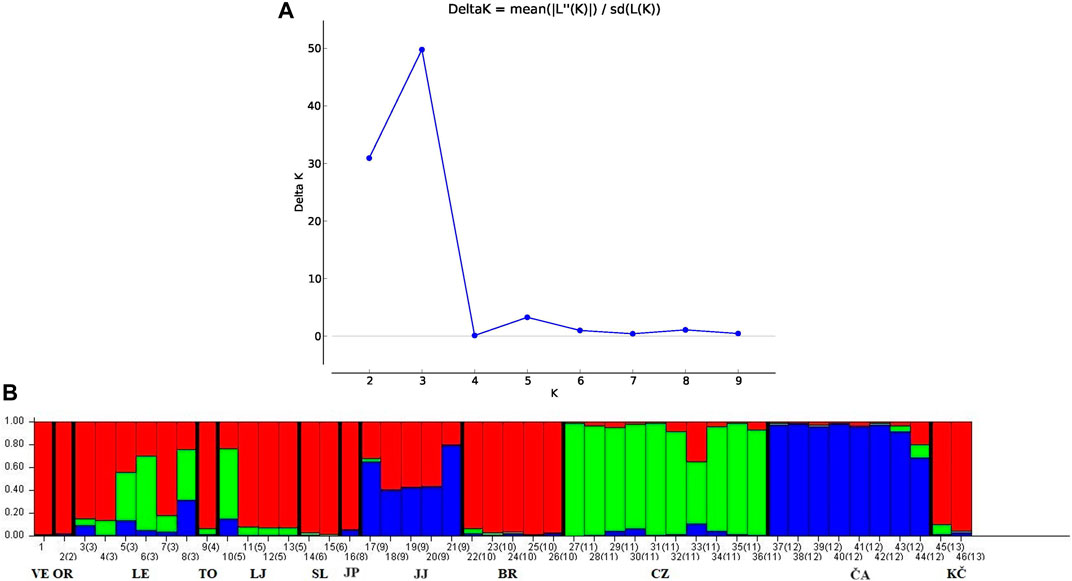
Figure 6. (A) Value of ΔK and (B) genetic population structuring of the Danubian lineage individuals. VE—Veličanka, OR—Orljava, LE—Mala Lešnica, TO—Toplica, LJ—Lička Jesenica, SL—Slapnica, JP—Jankovac Stream, JJ—Jankovac Lake, BR—Brzaja, CZ—Curak, ČA—Čabranka, KČ—Kupčina.
Discussion
The results of this study indicated a decrease in genetic variability in the Orljava River and Jankovac Stream. Analysis of CR mtDNA revealed that only one haplotype is present in both populations, non-native Da22 in Orljava River and Da1 in Jankovac Stream (Table 1). Neither Jankovac Stream nor Orljava River population proved to be genetically “pure”, considering that DA-AT hybrids were identified there (Kanjuh et al., 2020). In this regard, a bottleneck effect was observed for these two populations. Therefore, to continuously follow the status of these two populations, ongoing monitoring and further investigation are necessary. Evaluating the bottleneck effect in other populations revealed a heterozygosity deficit in the Brzaja, Curak and Kupčina rivers, indicating population expansions, most likely due to the introduction of the non-native farmed At1 haplotype. The same was observed in the “Vrabac” fish farm, but that was to be expected knowing that within the fish farm non-native At1 individuals are farmed.
In comparison with all the analysed populations, it was shown that the Slapnica River population is genetically different. The presence of AT lineage individuals was not recorded there, so this population consists only of individuals of the DA lineage. Nevertheless, the presence of non-native Da2 and DA-AT hybrids in the Slapnica River indicates that its native character was disturbed (Kanjuh et al., 2020). Population from the Slapnica River most likely separates due to the presence of private alleles in hybrid individuals, at microsatellite loci Str73INRA, Ssa410Uos, SsaD190 and OMM1064 (Supplementary Table S1). In all other populations, a large overlap was observed (Figure 2; Figure 3), both within geographically close and among geographically distant ones, as clearly evidenced in populations from the Čabranka River (Gorski Kotar) and Jankovac Lake (Mt. Papuk). This level of overlap indicates a high genetic similarity of the populations, which is a consequence of stocking from hatcheries that import and breed, most likely, the same (AT) phylogenetic lineage of brown trout (Piria et al., 2020). In fish farms throughout Croatia, brown trout carrying the At1 mtDNA haplotype is probably the most farmed and used for stocking of streams attractive for fishing. Minor genetic differences between populations observed in this study, most likely caused by stocking with the same genetic material, were additionally confirmed by the results of the population structure analysis. Separation of only two genetic clusters indicates strong introgression of the non-native genotypes, which is consistent with molecular variance results for CR mtDNA haplotypes (Kanjuh et al., 2020), which show little variation among populations. Structuring of only DA individuals also showed genetic similarity between populations. This result indicated the stocking with not only Atlantic (AT) lineage, but also non-native Danubian (DA) haplotypes of brown trout. Two non-native DA haplotypes were recorded in the analysed rivers of the Danube basin in Croatia (Kanjuh et al., 2020). The Da22 haplotype was limited to three localities (Veličanka River, Orljava River and Brzaja River) at the northern slopes of the Mt. Papuk (Kanjuh et al., 2020). This haplotype is native to the Lohnbach and Daglesbach rivers in Austria (Duftner et al., 2003) and in the Una River drainage in Bosnia and Herzegovina (Škraba et al., 2017). The non-native Da2 haplotype was much more frequent than Da22 haplotype (recorded in seven out of fifteen analysed populations) (Kanjuh et al., 2020). The Da2 haplotype is native to the streams and rivers in southern Germany (Bernatchez, 2001) and streams belonging to the Austrian part of the Danube drainage (Weiss et al., 2001). The stocking of various freshwaters of Europe during the 19th century was carried out with fish material from southern Germany (Kohout et al., 2012). It is most likely that these stockings included the territory of today’s Croatia. Precise data on fish farm breeding of non-native Danubian haplotypes of brown trout do not exist, but first data on the presence of the Da2 haplotype in freshwater of Croatia was recorded in the Gacka River, together with the non-native At1 haplotype (Jadan et al., 2007). The separation of DA individuals from the Curak River into a single genetic cluster (Figure 6B) is interesting information from the conservation point-of-view. All DA individuals from the Curak River belong to the native Da haplotype and are homozygous to the native LDH-C*100 allele (Kanjuh et al., 2020). It is possible that the influence of the non-native At1 haplotype was suppressed in this population, considering that out of a total of twelve individuals, only one was genetically pure AT and only one was AT-DA hybrid (Kanjuh et al., 2020).
Genetic research of brown trout and their susceptibility to the presence of the introduced AT lineage in Croatia was until now mainly analysed using cytb and CR mtDNA as molecular markers. Buj et al. (2020) and Ivić et al. (2021) emphasized the negative impact of non-native AT lineage on the native populations of the DA lineage (nominal Salmo labrax) and the need for conservation of its genetic character in the Plitvice Lakes National Park and Žumberak-Samobor Hills Nature Park, respectively. The presence of the At1 haplotype has been reported for many streams attractive for fishing throughout the Western Balkans (Snoj, 2004; Marić et al., 2006; 2012; Jadan et al., 2007; Simonović et al., 2015; Tošić et al., 2016; Škraba Jurlina et al., 2018; 2020). In this regard, it is significant that apart from the At1 haplotype, the presence of the rare At17 haplotype was recorded in the upper reaches of the Mrtvica River (Adriatic Basin), undoubtedly indicating a unique event of stocking of this river (Škraba Jurlina et al., 2018). As its presence was not detected in the lower part of the river, the authors assume that the trout spawned only in the upper part of the river, producing hybrids with brown trout of the resident Adriatic lineage. They also pointed out that gene flow between the populations in the upper and lower parts of the river was low (despite the absence of physical and/or reproductive barriers), indicating the impossibility of individuals from the upper reaches of the Mrtvica River to migrate downstream and hybridize with the brown trout of the native Adriatic lineage consisting of both resident (predominantly males) and lake-dwelling, migratory (predominantly females) fish (Škraba Jurlina et al., 2018). The negative influence of the At1 haplotype was also recorded in non-fishing rivers, which are characterized by the presence of specific and narrowly distributed haplotypes. One such example is the population in the Vratna River, a tributary of the Danube River at the downstream border of the Iron Gate Gorge, where the rare haplotype Da23c occurs, which is currently threatened by the appearance of the stocked brown trout of the At1 haplotype (Tošić et al., 2016; Škraba Jurlina et al., 2020).
In addition to the genetic analyses, the invasive character of the AT lineage has been highlighted in recent research, which showed the rapid adaptation of this lineage to the different conditions in natural watercourses, the consumption of a wide range of available food and competition for food and space, as well as diet overlap with native DA lineage individuals (Piria et al., 2020). Also, in assessment of invasiveness risk in the Danube and Adriatic basins across four Balkan countries, out of 13 existing and four horizon non-native salmonid species, S. trutta of the AT lineage showed a very high risk of invasiveness in the current climate conditions (Marić et al., 2022). In addition to At1, the non-native Da2 haplotype has also a significant impact on disrupting the genetic structure of the native Croatian populations of brown trout. Therefore, it would be important for future research to focus on obtaining data on the potential breeding origin and ways of introduction of the Da2 haplotype, but also on assessing the risk of invasiveness to the native genetic pool of the brown trout complex. Apart from the presence of the Da2 haplotype in the rivers of Croatia (Kanjuh et al., 2020), the presence of Da2 haplotype was recorded in the rivers of Serbia (Marić et al., 2006; Simonović et al., 2015; Tošić et al., 2016), Bosnia and Herzegovina (Mrdak et al., 2012; Škraba et al., 2017), and Montenegro (Mrdak et al., 2012).
Everything said above points out to a long history and the current management practice of stocking with the hatchery-reared brown trout. There are clear warnings of the negative impact of the AT hyplotypes on native and wild brown trout populations. Although the detrimental effects of strong introgression in wild brown trout populations are visible, management procedures in many countries are apparently not strict enough and properly applied. Despite proposals for potential management plans, the native gene pool of brown trout remains under serious threat. Studying the wild, native, and locally specific populations of brown trout on the territory of Croatia, determining of their status and genetic structure should be a priority to maintain and preserve their original genetic stocks. Genotyping the remaining brown trout stock from inland waters is essential. It is to be implemented into the national legislation, in order to set a mandatory genotyping of hatchery brood stocks and to identify and register native brood fish. This practice is currently weakly enforced in Croatia, and in many countries of the Balkan Peninsula. It is important to examine the genetic structure of brown trout grown in fish farms, as well as the stocks used for stocking, which, so far, has not been legally regulated or implemented as a necessary practice in majority of countries in the Western Balkans. Although some authors point out that stocking as a measure of fisheries management should be re-examined (Škraba Jurlina et al., 2018), for the recovery of the degraded status of the brown trout populations, stocking would be desirable. Stocking should imply a controlled, strictly defined, and planned approach, and the stocked material should be bred as a “foundation brood stock”. In addition, all of the populations that would be studied and marked as genetically “pure” and/or unique to a certain locality should be protected from the negative influence of fishing pressure. Implementing a mandatory unconditional “catch-and-release” angling regime (Simonović et al., 2018) as a compulsory legal protection measure is proposed as a possible solution that would ensure the self-sustainability of the trout stock without the need for restocking, but also the sustainability of the fishery. Many authors additionally pointed out that long-term stabilization of the density of trout populations is often not possible without environmental improvement strategies, such as habitat restoration, removal of migration barriers, improvement of hydrological regimes, which are generally more effective than stocking (Cowx, 1994; Fjellheim et al., 2003; Oosterhout et al., 2005; Ferguson, 2007; Škraba Jurlina et al., 2018), and the brown trout habitats are certainly special in their characteristics, being very sensitive to all kinds of anthropogenic pressures.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by Ministry of Agriculture and Ministry of Environmental Protection and Energy. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
TK: Conceptualization, Formal Analysis, Methodology, Resources, Writing–original draft. AM: Formal Analysis, Methodology, Resources, Writing–review and editing. DŠ: Writing–review and editing. PS: Resources, Writing–review and editing. IS: Formal Analysis, Resources, Writing–review and editing. MP: Resources, Writing–review and editing. IM: Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was conducted as part of the scientific project “Climate change and invasive species—determining the impact on the biodiversity of native freshwater crayfish and trout and their conservation (CLINEinBIOta)” (project no. IP-06-2016) funded by the Croatian Science Foundation.
Acknowledgments
Prof. Dušanka Savić-Pavićević, Ph.D., Jelena Karanović, Ph.D., Miloš Brkušanin, Ph.D., and Jovan Pešović, Ph.D., from the Center for Human Molecular Genetics, Faculty of Biology, University of Belgrade, helped in performing the fragment analysis of several samples and contributed to the understanding of the obtained results.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2024.1379878/full#supplementary-material
References
Allendorf, F., Leary, R., Spruell, P., and Wenburg, J. (2001). The problems with hybrids: setting conservation guidelines. Trends Ecol. Evol. 16 (11), 613–622. doi:10.1016/s0169-5347(01)02290-x
Behnke, R. J. (1972). The systematics of salmonid fishes of recently glaciated lakes. J. Fish. Res. Board Can. 29, 639–671. doi:10.1139/f72-112
Belkhir, L. C., Raufaste, N., and Bonhomme, F. (1996). “GENETIX 4.05 Logiciel Sous Windows TM Pour la Génétique des Populations,” in Laboratoire génome P, interactions, CNRS UMR 5171 (Montpellier: Université de Montpellier II).
Bernatchez, L. (2001). The evolutionary history of brown trout (Salmo trutta L.) interred from phylogenetic, nested clade, and mismatch analyses of mitochondrial DNA variation. Evolution 55 (2), 351–379. doi:10.1554/0014-3820(2001)055[0351:tehobt]2.0.co;2
Buj, I., Raguž, L., Marčić, Z., Ćaleta, M., Duplić, A., Zanella, D., et al. (2020). Plitvice Lakes National Park harbors ancient, yet endangered diversity of trout (genus Salmo). J. Appl. Ichthyol. 00, 20–37. doi:10.1111/jai.14120
Cairney, M., Taggart, J. B., and Hoyheim, B. (2000). Characterization of microsatellite and minisatellite loci in Atlantic salmon (Salmo salar L.) and cross-species amplification in other salmonids. Mol. Ecol. 9, 2175–2178. doi:10.1046/j.1365-294x.2000.01053-12.x
Ćaleta, M., Marčić, Z., Buj, I., Zanella, D., Mustafić, P., Duplić, A., et al. (2019). A review of extant Croatian freshwater fish and lampreys – annotated list and distribution. Croat. J. Fish. 77, 137–234. doi:10.2478/cjf-2019-0016
Cornuet, J. M., and Luikart, G. (1996). Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144 (4), 2001–2014. doi:10.1093/genetics/144.4.2001
Cowx, I. G. (1994). Stocking strategies. Fish. Manag. Ecol. 1 (1), 15–30. doi:10.1111/j.1365-2400.1970.tb00003.x
Earl, D. A., and vonHoldt, B. M. (2012). STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conser. Genet. Resour. 4 (2), 359–361. doi:10.1007/s12686-011-9548-7
Estoup, A., Presa, P., Krieg, F., Vaiman, D., and Guyomard, R. (1993). (CT)n and (GT)n microsatellite: a new class of genetic markers for Salmo trutta L. (brown trout). Heredity 71, 488–496. doi:10.1038/hdy.1993.167
Evanno, G., Regnaut, S., and Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Ferguson, A. (2007). Genetic impacts of stocking on indigenous brown trout populations. Environment Agency Science Report No. SC040071/SR. Accessed https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/291703/scho0707bmzi-e-e.pdf.
Fjellheim, A., Barlaup, B. T., Gabrielsen, S. E., and Raddum, G. G. (2003). Restoring fish habitat as an alternative to stocking in a river with strongly reduced flow. Ecohydrol. Hydrobiol. 3, 17–26.
Goudet, J. (2002). FSTAT version 2.9.3.2, a program to estimate and test gene diversities and fixation indices. Lausanne, Switzerland: Institute of Ecology.
Gridelli, E. (1936). I pesci d’aquaDolce della Venezia Giulia. Bolletino della Soc. Adriatica di Sci. Nat. Trieste 35, 7–40.
Ivić, L., Buj, I., Raguž, L., Marčić, Z., Ćaleta, M., Zanella, D., et al. (2021). Diversity and structure of trout populations (Salmo sp., Salmonidae, Actinopteri) in the Žumberak-Samoborsko Gorje Nature Park in Croatia. Fundam. Appl. Limnol. 194, 215–225. doi:10.1127/fal/2020/1283
Jadan, M., Čož-Rakovac, R., Tupić-Popović, N., and Strunjak-Perović, I. (2007). Presence of unexpected phylogenetic lineages of brown trout Salmo trutta L. in Gacka River, Croatia. Aquac. Res. 38 (15), 1682–1685. doi:10.1111/j.1365-2109.2007.01832.x
Kanjuh, T., Marić, A., Piria, M., Špelić, I., Maguire, I., and Simonović, P. (2020). Diversity of brown trout, Salmo trutta (actinopterygii: salmoniformes: salmonidae), in the Danube River basin of Croatia revealed by mitochondrial DNA. Acta Ichthyol. Piscat. 50 (3), 291–300. doi:10.3750/aiep/02939
King, T. L., Eackles, M. S., and Letcher, B. H. (2005). Microsatellite DNA markers for the study of Atlantic salmon (Salmo salar) kinship, population structure and mixed-fishery analyses. Mol. Ecol. Notes. 5 (1), 130–132. doi:10.1111/j.1471-8286.2005.00860.x
Kohout, J., Jašková, I., Papoušek, I., Šedivá, A., and Šlechta, V. (2012). Effects of stocking on the genetic structure of brown trout, Salmo trutta, in Central Europe inferred from mitochondrial and nuclear DNA markers. Fish. Manag. Ecol. 19 (3), 252–263. doi:10.1111/j.1365-2400.2011.00828.x
Laikre, L., Antunes, A., Apostolidis, A., Berrebi, P., Duguid, A., Ferguson, A., et al. (1999). Conservation genetic management of brown trout (Salmo trutta) in Europe. Report by the Concerted action on identification, management, and exploitation of genetic resources in the brown trout (Salmo trutta). Troutconcert, EU Fair CT97-3882.
Laikre, L., Larsson, L. C., Palmé, A., Charlier, J., Josefsson, M., and Ryman, N. (2008). Potentials for monitoring gene level biodiversity: using Sweden as an example. Biodivrs. Conserv. 17, 893–910. doi:10.1007/s10531-008-9335-2
Langella, O. (2002). POPULATIONS 1.2. 28. Population genetic software (individuals or populations distances, phylogenetic trees). France: CNRS.
Marić, A., Špelić, I., Radočaj, T., Vidović, Z., Kanjuh, T., Vilizzi, L., et al. (2022). Changing climate may mitigate the invasiveness risk of non-native salmonids in the Danube and Adriatic basins of the Balkan Peninsula (south-eastern Europe). NeoBiota 76, 135–161. doi:10.3897/neobiota.76.82964
Marić, S., Nikolić, V., Tošić, A., and Simonović, P. (2012). Record of the brown trout Salmo trutta L., 1758 in the main riverbed of the Serbian part of the Danube River. J. Appl. Ichthyol. 28, 135–137. doi:10.1111/j.1439-0426.2011.01881.x
Marić, S., Sušnik, S., Simonović, P., and Snoj, A. (2006). Phylogeographic study of brown trout from Serbia, based on mitochondrial DNA control region analysis. Genet. Sel. Evol. 38 (4), 411–430. doi:10.1186/1297-9686-38-4-411
Mrdak, D., Nikolić, V., Tošić, A., and Simonović, P. (2012). Molecular and ecological features of the soft-muzzled trout Salmo obtusirostris (Heckel, 1852) in the Zeta River, Montenegro. Biologia 67 (1), 222–233. doi:10.2478/s11756-011-0150-y
Oikonomou, A., Leprieur, F., and Leonardos, I. D. (2014). Biogeography of freshwater fishes of the balkan Peninsula. Hydrobiologia 738 (1), 205–220. doi:10.1007/s10750-014-1930-5
Oosterhout, G. R., Huntingdon, C. W., Nickelson, T. E., and Lawson, P. W. (2005). Potential benefits of a conservation hatchery program for supplementing Oregon coast coho salmon (Oncorhynchus kisutch) populations: a stochastic model investigation. Can. J. Fish. Aquat. Sci. 62, 1920–1935. doi:10.1139/f05-080
O'Reilly, P. T., Hamilton, L. C., McConnell, S. K., and Wright, J. M. (1996). Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can. J. Fish. Aquat. Sci. 53 (10), 2292–2298. doi:10.1139/f96-192
Paterson, S., Piertney, S. B., Knox, D., Gilbey, J., and Verspoor, E. (2004). Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Mol. Ecol. Notes. 4 (2), 160–162. doi:10.1111/j.1471-8286.2004.00598.x
Piria, M., Špelić, I., Rezić, A., and Šprem, N. (2020). Morfološke značajke i kondicija potočne pastrve Salmo trutta Žumberačkih i Samoborskih potoka. J. Cent. Eur. Agric. 21 (2), 231–245. doi:10.5513/JCEA01/21.2.2460
Piry, S., Luikart, G., and Cornuet, J. M. (1999). Computer note. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. J. Hered. 90 (4), 502–503. doi:10.1093/jhered/90.4.502
Pofuk, M., Zanella, D., and Piria, M. (2017). An overview of the translocated native and non-native fish species in Croatia: pathways, impacts and management. Manag. Biol. Invasions. 8 (3), 425–435. doi:10.3391/mbi.2017.8.3.16
Pritchard, J. K., Stephens, M., and Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics 155 (2), 945–959. doi:10.1093/genetics/155.2.945
Rexroad, C. E., Coleman, R. L., Hershberger, W. K., and Killefer, J. (2002). Rapid communication: thirty-eight polymorphic microsatellite markers for mapping in rainbow trout. J. Anim. Sci. 80 (2), 541–542. doi:10.2527/2002.802541x
Righi, T., Fasola, E., Iaia, M., Stefani, F., and Volta, P. (2023). Limited contribution of hatchery-produced individuals to the sustainment of wild marble trout (Salmo marmoratus Cuvier, 1829) in an Alpine basin. Sci. Total Environ. 164555. doi:10.1016/j.scitotenv.2023.164555
Ryman, N., Jorde, P. E., and Laikre, L. (1995). Supportive breeding and variance effective population size. Conserv. Biol. 9 (6), 1619–1628. doi:10.1046/j.1523-1739.1995.09061619.x
Sanz, N. (2018). “Phylogeographic structure of brown trout: a review,” in Brown trout: Biology, ecology and management. Editors H. Lobón-Cerviá, and N. Sanz (London: JohnWiley and Sons Ltd), 17–63. doi:10.1002/9781119268352.ch2
Sanz, N., García-Marín, J. L., and Pla, C. (2002). Managing fish populations under mosaic relationships. The case of brown trout (Salmo trutta) in peripheral Mediterranean populations. Conserv. Genet. 3, 385–400. doi:10.1023/A:1020527420654
Simonović, P., Grubić, G., Tošić, A., Čanak Atlagić, J., Škraba Jurlina, D., and Nikolić, V. (2018). Justification for retention of the Catch-and-Release in the wild brown trout Salmo cf. trutta fishery, Fish. Res. 200, 17–24. doi:10.1016/j.fishres.2017.12.010
Simonović, P., Tošić, A., Škraba Jurlina, D., Čanak Atlagić, J., and Nikolić, V. (2017a). Stakeholders’ participation in conservation of brown trout stocks in Serbia. Wild Trout XII Symposium "Science, politics, and wild trout management: who’s driving and where are we going? MT, USA: West Yellowstone, 55–62.
Simonović, P., Tošić, A., Škraba Jurlina, D., Nikolić, V., Piria, M., Tomljanović, T., et al. (2017b). Diversity of brown trout Salmo cf. trutta in the River Danube Basin of Western Balkans as assessed from the structure of their mitochondrial control region haplotypes, J. Ichthyol. 57 (4), 603–616. doi:10.1134/s0032945217040154
Simonović, P., Tošić, A., Vassilev, M., Apostolou, A., Mrdak, D., Ristovska, M., et al. (2013). Risk assessment of non-native fishes in the Balkans region using FISK, the invasiveness screening tool for non-native freshwater fishes. Mediterr. Mar. Sci. 14 (2), 369–376. doi:10.12681/mms.337
Simonović, P., Vidović, Z., Tošić, A., Škraba, D., Čanak Atlagić, J., and Nikolić, V. (2015). Risks to stocks of native trout of the genus Salmo (Actinopterygii: Salmoniformes: Salmonidae) of Serbia and management for their recovery. Acta Ichthyol. Piscat. 45 (2), 161–173. doi:10.3750/aip2014.45.2.06
Škraba, D., Bećiraj, A., Šarić, I., Ićanović, I., Džaferović, A., Piria, M., et al. (2017). Haplotype Diversity of brown trout (Salmo trutta L.) populations from Una River drainage area in Bosnia and Herzegovina: implications for conservation and fishery management. Acta Zool. Bulg. 69 (1), 25–30.
Škraba Jurlina, D., Marić, A., Karanović, J., Nikolić, V., Brkušanin, M., Kanjuh, T., et al. (2018). Effect of the introgression of Atlantic brown trout, Salmo trutta, into Adriatic trout, Salmo farioides in a stream at the drainage area of the Adriatic Sea basin of Montenegro. Acta Ichthyol. pisc. 48, 363–372. doi:10.3750/AIEP/02491
Škraba Jurlina, D., Marić, A., Mrdak, D., Kanjuh, T., Špelić, I., Nikolić, V., et al. (2020). Alternative life-history in native trout (Salmo spp.) suppresses the invasive effect of alien trout strains introduced into streams in the western part of the Balkans. Front. Ecol. Evol. 8, 188. doi:10.3389/fevo.2020.00188
Slettan, A., Olsaker, I., and Lie, Ø. (1995). Atlantic salmon, Salmo salar, microsatellites at the SSOSL25, SSOSL85, SSOSL311, SSOSL417 loci. Anim. Genet. 26, 281–282. doi:10.1111/j.1365-2052.1995.tb03262.x
Snoj, A. (2004). Filogenetska struktura postrvi (Salmo trutta L.) v Sloveniji. Ribič, Glasilo Slovenskog ribištva. Ribiška sveza Slov. 10, 239–243.
Snoj, A., Bogut, I., and Sušnik, S. (2008). Evidence of a genetically distinct population of Vrljika softmouth trout Salmo obtusirostris Heckel evolved by vicariance. J. Fish. Biol. 72, 1945–1959. doi:10.1111/j.1095-8649.2008.01816.x
Snoj, A., Glamuzina, B., Razpet, A., Zablocki, J., Bogut, I., Lerceteau-Köhler, E., et al. (2010). Resolving taxonomic uncertainties using molecular systematics: Salmo dentex and the Balkan trout community. Hydrobiologia 651, 199–212. doi:10.1007/s10750-010-0297-5
StatSoft INC (2001). STATISTICA (data analysis software system), version 6, 150. Tulsa, USA: StatSoft INC.
Tošić, A., Škraba, D., Nikolić, V., Čanak Atlagić, J., Mrdak, D., and Simonović, P. (2016). Haplotype diversity of brown trout Salmo trutta (L.) in the broader Iron Gate area. Turk. J. Zool. 40, 655–662. doi:10.3906/zoo-1510-54
Vera, M., Cortey, M., Sanz, N., and Garcia-Marin, J. L. (2010). Maintenance of an endemic lineage of brown trout (Salmo trutta) within the Duero River basin. J. Zool. Syst. Evol. Res. 48, 181–187. doi:10.1111/j.1439-0469.2009.00547.x
Wang, J., and Ryman, N. (2001). Genetic effects of multiple generations of supportive breeding. Conserv. Biol. 15 (6), 1619–1631. doi:10.1046/j.1523-1739.2001.00173.x
Keywords: brown trout, microsatellites, Croatia, stocking, conservation
Citation: Kanjuh T, Marić A, Škraba Jurlina D, Simonović P, Špelić I, Piria M and Maguire I (2024) Loss of native brown trout diversity in streams of the continental Croatia. Front. Environ. Sci. 12:1379878. doi: 10.3389/fenvs.2024.1379878
Received: 31 January 2024; Accepted: 12 March 2024;
Published: 26 March 2024.
Edited by:
Carla Simone Pavanelli, State University of Maringá, BrazilReviewed by:
Steven Weiss Joseph Weiss, University of Graz, AustriaVladimir Pavan Margarido, Western Parana State University, Brazil
Copyright © 2024 Kanjuh, Marić, Škraba Jurlina, Simonović, Špelić, Piria and Maguire. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tamara Kanjuh, dGFtYXJhLmthbmp1aEBiaW8uYmcuYWMucnM=
 Tamara Kanjuh
Tamara Kanjuh Ana Marić1
Ana Marić1 Predrag Simonović
Predrag Simonović Marina Piria
Marina Piria Ivana Maguire
Ivana Maguire