Abstract
Dumpsites are reservoirs of persistent organic pollutants (POPs) and heavy metals (HMs), constituting environmental hazards to humanity. Autochthonous microorganisms in dumpsites exhibit various degrees of responses to contaminants. Unfortunately, there is a dearth of information on the types and concentration of pollutants and the array of microorganisms in these dumpsites which may play important roles in the metabolism of such pollutants or other community processes. Therefore, determining the microbial community structure in such contaminated sites across a municipality is essential for profiling the taxa that would serve as consensus degraders of the pollutants. In this study, soil samples from three dumpsites (Cele, CS; Solous, SS; and Computer Village, CVS) were characterized for geochemical properties using GC-MS, MP-AES, and other analytical protocols, while the dynamics of bacterial communities were evaluated based on their 16S rRNA gene barcodes. A significant difference in the bacterial communities was observed among the dumpsites in relation to the extent of pollution caused by POPs and HMs. CVS, with the highest HM contamination, was rich in Actinobacteria (41.7%) and Acidobacteria (10.2%), in contrast to CS and SS. Proteobacteria (34.1%) and Firmicutes (20%) were the dominant phyla in CS (highest POP contamination), while Bacteroidetes (45.5%) and Proteobacteria (39.9%) were dominant in SS soil. Bacillus was the dominant genus in the most polluted dumpsite. Canonical correspondence analysis revealed that polycyclic aromatic hydrocarbons (PAHs) and HMs shaped the structure of the bacterial operational taxonomic units (OTUs) in the most polluted dumpsite. Out of a total of 706 OTUs, 628 OTUs exhibited a significant correlation (>50%) with benzo(b)fluoranthene, azobenzene, dibenzofurans, pyrene, dibenzo(a,l)pyrene, Cu, and Zn. In particular, Proteobacteria (Achromobacter sp. and Serratia sp.), Bacteroidetes (Zhouia sp.), and Firmicutes (Bacillus sp.) were suggested to be pivotal to the ecophysiology of dumpsite soils contaminated with POPs and HMs. The results generally underscored the importance of metagenomic and physicochemical analyses of polluted systems in enabling correlations for useful prediction of drivers of such ecosystems. This will further improve our understanding of the metabolic potential and adaptation of organisms in such systems.
1 Introduction
Soil is a highly complex environment that provides a habitat for a diversity of plants, animals, and microorganisms. Soil has a mean prokaryotic density of roughly 108 organisms per gram, with Proteobacteria, Acidobacteria, and Actinobacteria making up the majority of the biomass within the bacteria domain (Janssen, 2006; Delmont et al., 2011; Xavier and Naoise, 2014; Xu et al., 2014). The type and number of microorganisms in the soil are influenced by soil characteristics and wastes that flow into the soil, which either inhibits or stimulates their growth (Zhang et al., 2010; Tang et al., 2013). Microorganisms are an essential part of terrestrial ecosystems, playing important roles in soil biogeochemical cycles as well as in the fate of organic pollutants (Basu et al., 2021). In addition, there is enough evidence confirming the important role played by soil microorganisms in several ecosystem services such as erosion control, soil formation, nutrient cycling, and plant health (Nannipieri et al., 2003; Gardi et al., 2009; Ravi et al., 2019). However, microbes are potentially one of the most sensitive organisms to anthropogenic activities. Soil impacted by different pollutants would consequently have a significant detrimental effect on its quality and will also have a deleterious effect on the flora, fauna, and microorganisms. Soil microorganisms are generally considered the best indicators of soil pollution, as they are very responsive and provide important information about the changes occurring in soil (Sumampouw and Risjani, 2014). Studies have shown the alteration of soil microbial composition and diversity of dumpsites, revealing the dominance and loss of different microorganisms (Wang et al., 2017; Salam and Varma, 2019). Therefore, gaining in-depth knowledge of the impacts of pollution on the microbial communities and potential degraders in these environments is important.
Globally, waste generation rates are increasing because of urbanization and rapid population growth. Approximately 4.3 billion urban residents are projected to generate approximately 1.42 kg of solid waste per person, totaling 6.1 million metric tons per day by 2025, resulting in faster generation of waste than of other pollutants such as greenhouse gases (Hoornweg et al., 2013). In developing countries, waste management strategies include recycling, landfill dumping, and incineration (Igbinomwanhia, 2011). In recent years, attention has been drawn to open dumping sites where large amounts of municipal solid wastes have been dumped, and improper disposal of e-waste leads to high concentrations of heavy metals (HMs) leaching into the surrounding environments and bioaccumulation in humans (Longe and Balogun, 2010). HMs are non-biodegradable and bioaccumulate in the environment, and they can severely affect the microbial diversity and abundance because of the toxicity induced (Zhao et al., 2019; Hu et al., 2021). However, some microorganisms are tolerant to HMs and may convert them to less-toxic forms or remove them from the polluted soil (Qiao et al., 2019). Unfortunately, most of such open dump areas, incineration sites, and landfills are located near residential areas and as such, exposure to various toxic chemicals originating from the sites is of serious concern because of the effects on human health, wildlife, and environmental quality. Indiscriminate burning of solid waste, generation of methane gas, lack of advanced waste incineration technology, and natural low-temperature burning are major problems in dumpsites. These are favorable factors for the formation of polycyclic aromatic hydrocarbons (PAHs), dioxins, furans, and their chlorinated analogs which can persist in the environment undegraded for years (Duan et al., 2012). These dioxins can permeate the soil and contaminate groundwater and nearby vegetation; not only does ecological contamination negatively affect overall ecosystem functions, but it also affects the health and functioning of living organisms.
Microbial activities in dumpsite soil play a crucial role in greenhouse gas emissions (Bajar et al., 2021). However, only a few studies have examined the metagenomic microbial profiles in landfills, e-waste, and incinerators simultaneously, especially in African contaminated systems and particularly in Nigeria, where no such investigations have ever been applied to dumpsites or contaminated soils. Nevertheless, quantification of pollutants alone does not provide cogent information about the potential biological impact; a combination of pollutant speciation with an in-depth evaluation of the microbial community structure will give a more informative and definitive assessment. Therefore, this study aims to gain a better understanding of the structure of microbial communities in three dumpsites that are exposed to different types of pollutants and at varying concentrations, with a view to identify the microorganisms that are important in natural biogeochemical cycles that may be adversely affected by pollution. The study also intends to determine whether they may serve as degraders of environmental contaminants. The objective was to establish the specific pollutants or abiotic factors that are the main drivers of the community structure and function. To this end, the correlation between soil physiochemistry and microbial community structure was deduced, as was the level of similarity among the dumpsites. The information obtained would be key to revealing the possible impact of the pollutants and their effects on microbial succession and metabolic mechanism, which overall would be valuable in designing effective bioremediation schemes for the polluted sites.
2 Materials and methods
2.1 Study sites and soil sample collection
Three major dumpsites were mapped out in the Lagos metropolitan area of Lagos State, Nigeria. Lagos metropolis is Africa’s model megacity, which contends with waste generated by its residents. The study areas were ‘Cele’ dumpsite (CS) at Itire, where open burning occurs consistently; ‘Computer Village’ dumpsite (CVS) at Ikeja, which is an electronic recycling site that largely contained electronic wastes (e-wastes); and ‘Solous’ dumpsite (SS) located at Igando, which is an open landfill comprising varieties of domestic and industrial wastes. The GPS coordinates of the dumpsites are delineated in the map presented in Figure 1. Soil samples (10 g apiece) were collected randomly from a depth of 0–20 cm at six different points at each site, pooled to form a composite of each dumpsite in sterile ziplock bags, and transported on ice to the laboratory for further analysis. One part of each soil sample was stored at −80°C for microbial community analysis, while another part was stored at 4°C for physicochemical analysis.
FIGURE 1

Geographical map of Lagos state showing the sampling dumpsites. Sampling sites are given in red dots: Cele, Computer village, and Solous.
2.2 Physicochemical analysis
The physicochemical parameters of the samples were determined using standard protocols. Soil particle size was determined using the hydrometer method according to the protocol described by Day (1965). The pH was determined in situ using a pH meter (British Standard, 1995), and the moisture content was determined by using the drying method (Chopra and Kanwar, 1998). The total organic carbon and total organic matter were determined by using the Walkley–Black chromic acid wet oxidation method. Other physicochemical parameters were determined as follows: total nitrogen using the Kjeldahl method, phosphate using the spectrophotometric method, and oil and grease using the solvent extraction method. All these methods were applied according to protocols previously described by Nelson and Sommers (1983), Storer (1984), AOAC (1990), and Chopra and Kanwar (1998).
For the HM analysis, a sieved air-dried soil sample (1 g) was first digested with HCl/HNO3/H2SO4 (1:2:2 v/v) for 15–20 min on a heated platform. The sample was cooled, after which 20 mL of distilled water was added and again heated to boiling point to bring the metals into solution. The sample was filtered (Whatman 42 filter paper and <0.45 µm Millipore filter paper) and stored in plastic bottles. The contents of metals in the soil samples were evaluated through the microplasma atomic emission spectrophotometer (MP-AES 4200) coupled to a computing system and operated on the MP-AES software interface which enabled elemental analysis at mg/kg for a wide range of metals. Standard solutions used in the calibration of the instrument prior to analysis were prepared by diluting multi-element standard solutions to a concentration of 100 mg/L (AOAC, 1990). For quality control measures, all standards, replicates, and blanks were prepared at the same time and used immediately. The resulting calibrated curve was verified by analyzing initial calibrated curve verification (ICV) and initial calibrated blank samples. The ICV standard was ±20% of its true value.
Persistent organic pollutants (POPs) present in the soil samples were evaluated based on the USEPA method 8270D (USEPA, 2007). Ten gram of homogenized soil was sonically extracted with 20 mL of a 1:1 acetone:dichloromethane solvent complex at 70°C for 30 min. The extracts were dried with sodium sulfate, concentrated, exchanged into cyclohexane, and finally cleaned up via passage over silica gel. The purified extracts were subsequently converted back to acetone:dichloromethane and analyzed via gas chromatography mass spectrometry (GC-MS). The extract composition was examined by total ion scans, and the USEPA 16 priority pollutant PAHs were identified and quantified by using primary and secondary ions. Polychlorinated biphenyls (PCBs), nitroaromatic compounds, dioxins, and furans in the soil were also determined using GC-MS equipped with a HP-5 capillary column coated with 5% phenylmethylsiloxane (30 m length × 0.32 mm diameter × 0.25 µm film thickness) (Agilent Technologies). The injector and detector temperatures were kept at 250°C and 300°C, respectively. The carrier gas (helium) was maintained at a flow rate of 1 mL/min (initial pressure, 1.715 psi; average velocity, 37.758 cm/s). The chromatograph was programmed at 130°C for 2 min initially, then ramped to 200°C at 10°C min-1 for 16 min, then to 265°C at 5°C min-1 for 7 min, and finally to 300°C at 5°C min-1 and held for 3 min. The MS was operated in electron-impact ionization mode at 70 eV with an ion source temperature of 230°C, a quadrupole temperature of 150°C, and a transfer line temperature of 300°C. Acquisition of ions was carried out via scan mode (scanning from m/z 50–500 amu at 2.0 s/scan rate) and selective ion mode (SIM). In all preparations, octachloronaphthalene solution was added to the soil solutions as an internal standard prior to extraction. This was necessary to assess the recovery of the organic pollutants from the soil. Prior to analysis, the instrument was properly calibrated by injecting a series of calibration standards. The typical coefficient of correlation r for the calibrated curve was ≥0.995. The efficiency and accuracy of GC were constantly ascertained by injecting the standards prior to sample analysis.
2.3 Soil DNA extraction, 16S rRNA gene amplification, and sequencing
Total DNA of the soil samples (0.25 g) was extracted using a MOBIO PowerSoil DNA Extraction Kit (MO BIO, Carlsbad, CA, USA) following the manufacturer’s protocol. DNA was quantified using a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, United States), the quality was visualized through 1% agarose gel electrophoresis, and the DNA was stored at −20°C until further use. The V4–V5 hypervariable regions of the bacterial 16S rRNA gene were amplified (98°C, 30 s; 12 cycles: 98°C, 45 s; 62°C , 30 s, and 72°C, 30 s; the final primer extension at 72°C, 5 min) using primers Hyb515F (5′-GTGYCAGCMGCCGCGGTA -3′) and Hyb909R (3′-TGARTTTMCTTAACYGCCCC-5′) as previously described (Nguyen and Rodrigues, 2018). PCR products were purified using the AMPure XP bead purification system and subsequently barcoded with Hyb primers. Sample libraries were generated from the purified amplicons. The quantity and quality of the libraries were assessed using a Bioanalyzer DNA chip (Agilent). Sequencing was performed using an Illumina MiSeq system (Illumina, San Diego, CA, USA) using primers Hyb515F and Hyb909R with Illumina adapters and barcodes. Pair-end sequencing was employed using 600-cycle MiSeq® Reagent Kit V3 (Illumina, U.S.A.) to generate 2 bp × 300 bp reads at the Genomic Sequencing and Analysis Facility, University of Texas, Austin, United States.
2.4 Processing of sequencing data and taxonomic classification
Sequences were obtained in FASTQ format and analyzed using the Quantitative Insights into Microbial Ecology (QIIME) V. 1.9.1 software pipeline to demultiplex and filter out low-quality and chimera sequences (Caporaso et al., 2010). High-quality sequences were assigned using the ribosomal database project (RDP) classifier at a 97% similarity, and α-diversity indices (Shannon and Chao1) were calculated. The clustering and classification of OTUs at several taxonomic levels (kingdom, phylum, class, order, family, genus, and species) were deduced from the processed dataset. The Illumina sequence data reported here have been deposited in the NCBI Sequence Read Archive database under accession number: PRJNA540227.
2.5 Statistical data analysis
The relative abundances of the phylum and class levels for the three sites were plotted as a bar graph using the Prism 9.0 software program (GraphPad Software, San Diego, CA, USA). The R language platform (XLSTAT) was used to generate a heatmap based on the data summary for the three sites. Principal component analysis (PCA), a multivariate data dimensionality reduction method, was used to further analyze the environmental factors affecting the microbial community. The biological dataset was square root-transformed in order to downweight the effects of the highly abundant OTUs, and the Bray–Curtis similarity index was used to construct the resemblance matrix. Transformation and normalization of environmental data were used in calculating the resemblance matrix using Euclidean distance. Beta diversity patterns of the soil communities were examined by PCA ordination developed on Cluster routine using PRIMER v 7 software (PRIMER-E Ltd., Lutton, UK) (Clarke and Gorley, 2015). Canonical correspondence analysis (CCA) was performed using paleontological statistics software (PAST), which was used to test the relationship between physicochemical parameters and bacterial diversity. Pearson’s correlation was used to test the strength of the association between OTU abundance and soil physicochemistry. Statistical significance was determined at the 95% level (p < 0.05).
3 Results
3.1 Environmental conditions across dumpsites
The dumpsite soils lacked clay particles and were mostly made up of sand (84%–95%), with trace amounts of gravel and silt particles (Table 1). Further investigation of the soil physicochemistry revealed that the pH in all samples was slightly basic and did not differ significantly. The moisture content recorded in CS and CVS samples was more than twice the value obtained for SS. The highest proportion of oil and grease was recorded in CS, followed by SS and CVS in that order. A similar trend was also reported for the TOC and TOM (Table 1). The geochemistry with respect to HM composition in the dumpsites is presented in Table 2. The dumpsites exhibited high concentrations of HMs, with Fe (246.85 mg/L) and Zn (38.01 mg/L) having the highest concentrations, especially in CS soil, which, respectively, correspond to 1.7–3.2 and 25 times more than the levels encountered in other sites. In contrast to Fe and Zn, the highest concentration of Al was found in the CVS site at a threshold that was over ten times the level recorded for CS. Relative concentrations of the HMs in the three sites were of the order Fe > Al > Zn > Cd > Cu > Ni > Pb > Cr. It is noteworthy that the concentrations of Ba, Co, and V were all below the detection limit in the three study sites (Table 2). With the exception of Cu, the concentration of which was within the Federal Ministry of Environment (FMEnv) regulatory limit (except for CS soil), the concentrations of other HMs were above the recommended limit.
TABLE 1
| Soil description | CS | CVS | SS |
|---|---|---|---|
| Gravel (%) | 3 | 8 | 1 |
| Sand (%) | 84 | 86 | 95 |
| Silt (%) | 13 | 6 | 4 |
| pH | 8.4 | 8.7 | 8.5 |
| Moisture content | 8.7 | 8.8 | 3.5 |
| TOC (%) | 20.2 | 5.3 | 11.2 |
| TOM (%) | 34.8 | 9.2 | 19.2 |
| Total nitrogen (mg/kg) | 0.18 | 1.27 | 0.49 |
| Phosphorus (mg/kg) | 1.07 | 1.35 | 1.22 |
| Oil and grease (mg/kg) | 17,550 | 12,042 | 14,688 |
Dumpsite physicochemical properties.
CS, cele dumpsite; CVS, computer village dumpsite; SS, solous dumpsite.
TABLE 2
| Heavy metal (mg/kg) | CS | CVS | SS | FMEnv |
|---|---|---|---|---|
| Al | 25.33 | 255.67 | 102.00 | 0.20 |
| Ba | <0.0001 | <0.0001 | <0.0001 | NA |
| Cd | 4.8594 | 3.1688 | 1.4360 | NA |
| Co | <0.0001 | <0.0001 | <0.0001 | NA |
| Cr | 0.2283 | 0.2219 | 0.1795 | 0.0500 |
| Cu | 8.1288 | 0.4591 | 0.2057 | 2.0000 |
| Fe | 246.85 | 142.45 | 76.14 | 3.00 |
| Mn | 4.5254 | 1.3974 | 1.0232 | 0.1000 |
| Ni | 2.4682 | 2.4624 | 2.6496 | 0.0700 |
| Pb | 4.5235 | 1.5696 | 0.6428 | 0.0500 |
| V | <0.0001 | <0.0001 | <0.0001 | NA |
| Zn | 38.0098 | 1.5604 | 1.4781 | 0.3000 |
Heavy metal composition of the dumpsite soils.
CS, cele dumpsite; CVS, computer village dumpsite; SS, solous dumpsite; FMEnv, Federal Ministry of Environment; NA, not applicable; <0.0001 implies that the composition is less than the detection limit.
Among the 23 PAHs determined in the soil samples collected, dibenz(a,h)anthracene, benzo(b)fluoranthene, 3-methylcholanthrene, benzo(g,h,i)perylene, and acenaphthene constituted the major POPs associated with all the dumpsites (Table 3). Total PAHs in the dumpsites were in the order: CS (8.04 mg/kg) > SS (6.46 mg/kg) > CVS (5.29 mg/kg), which were not significantly different among the sites (p < 0.05). Dibenzofuran, a model compound of dioxins, was detected only in CS (7.61 mg/kg), unlike in the other two dumpsites, where it was below the detection limit. As presented in Table 3, halogenated organic compounds (PCBs and chlorinated dioxin congeners) were not detected in any of the three soils. However, concentrations of other compounds including aniline, nitrobenzene, diphenylamine, carbazole, benzidine, and pentachlorophenol were all below detection limits across the dumpsites. The highest concentration of the total POPs detected was in CS (23.76 mg/kg), with azobenzene (7.95 mg/kg) predominating other POPs. In addition, there was a significant difference in the total POPs detected in the three dumpsites (p > 0.5).
TABLE 3
| Organic compound (mg/kg) | CS | CVS | SS |
|---|---|---|---|
| Naphthalene | 0.137 | 0.114 | <0.0001 |
| Acenaphthylene | <0.0001 | <0.0001 | 0.299 |
| Acenaphthene | 0.446 | 0.353 | 0.472 |
| Fluorene | 0.160 | 0.276 | 0.243 |
| Anthracene | 0.297 | 0.216 | 0.296 |
| Phenanthrene | 0.099 | 0.239 | 0.296 |
| Fluoranthene | 0.084 | 0.085 | 0.091 |
| Pyrene | 0.091 | <0.0001 | <0.0001 |
| Benzo(c)phenanthrene | 0.062 | 0.022 | 0.013 |
| Benz(a)anthracene | 0.327 | 0.196 | 0.206 |
| Chrysene | 0.285 | 0.505 | 0.272 |
| Benzo(e)pyrene | 0.315 | 0.066 | 0.283 |
| Benzo(b)fluoranthene | 0.690 | 0.448 | 0.426 |
| Benzo(k)fluoranthene | 0.305 | 0.199 | 0.202 |
| Benzo(a)pyrene | 0.385 | 0.352 | 0.356 |
| 7,12-Dimethyl benz(a)anthracene | 0.254 | 0.203 | 0.365 |
| 3-Methylcholanthrene | 0.844 | 0.361 | 0.311 |
| Indo(1.2,3-cd)pyrene | 0.332 | 0.247 | 0.341 |
| Dibenz(a,h)anthracene | 0.624 | 0.567 | 0.835 |
| Dibenzo(a,l)pyrene | 0.594 | 0.352 | 0.351 |
| Benzo(g,h,i)perylene | 0.586 | 0.486 | 0.393 |
| Dibenzo(a,i)pyrene | 0.604 | <0.0001 | 0.073 |
| Dibenzo(a,h)pyrene | 0.516 | <0.0001 | 0.333 |
| Total PAHs | 8.039 | 5.285 | 6.458 |
| 2-Nitro-phenol | ND | 1.863 | ND |
| 2,4-Dinitro phenol | 2.482 | ND | ND |
| Dibenzofuran | 7.609 | BDL | BDL |
| 4-Nitro-phenol | 0.023 | 0.014 | 0.020 |
| Azobenzene | 7.952 | 0.288 | BDL |
| 2-Methyl-4,6-dinitro phenol | 3.373 | ND | 10.666 |
| p-Nitroaniline | 0.702 | ND | 0.706 |
| [1,1′-Biphenyl]-4-amine | 1.614 | 1.148 | 3.773 |
| Total organic compound (excluding PAHs) | 23.289 | 3.313 | 15.165 |
Concentration of organic compound pollutants in the dumpsite soils.
CS, cele dumpsite; CVS, computer village dumpsite; SS, solous dumpsite; BDL, below detection limit; ND, not detected.
3.2 Alpha-diversity characteristics of the three dumpsites
A total of 19,567, 17,173, and 22,641 high-quality sequence reads were recovered, respectively, from CS, CVS, and SS following quality filtering and subsequently assigned to corresponding 1,008, 3,293, and 2,653 OTUs. The metagenomics analysis and characteristics of alpha-diversity indices are summarized in Table 4. These alpha diversity parameters (richness and evenness of OTUs) differed greatly among the dumpsites. The lowest OTU richness and evenness were observed in the CS, while CVS exhibited the highest OTU richness and evenness. The Choa1 and Shannon indices showed that the bacterial community in the latter region (CVS) displayed higher abundance and was more diverse than the other two dumpsites (SS and CS). It is noteworthy that the level of richness observed in CVS was more than twice the level observed in the other sites. Whittaker plots, otherwise called rank abundance plots, display species OTU evenness as line slopes and OTU richness as line lengths (Figure 2). The figure revealed relatively evenly distributed OTUs in CVS, unlike a single dominant OTU that accounted for 18% of reads in the libraries as evidence of a less uniform distribution in SS, where the curve was initially rather steep. OTU abundance was highest in SS, while dominant OTU was absent in CVS, where OTU abundance was comparatively evenly distributed. The CVS site also recorded the greatest OTU richness, as evidenced by the longest sample line, while the least was found in CS (Figure 2).
TABLE 4
| Soil | |||
|---|---|---|---|
| CS | CVS | SS | |
| Number of reads | 21,891 | 18,640 | 24,307 |
| Number of quality filtered amplicons | 19,567 | 17,173 | 22,641 |
| Median amplicon length (bp) | 413 | 415 | 413 |
| Number of OTUs | 1,008 | 3,293 | 2,603 |
| Chao1 | 1855.21 | 5,745.66 | 2,653 |
| Shannon | 6.16 | 9.85 | 6.60 |
Characteristics of Illumina libraries and alpha diversity metrics of soil communities.
CS, cele dumpsite; CVS, computer village dumpsite; SS, solous dumpsite.
FIGURE 2
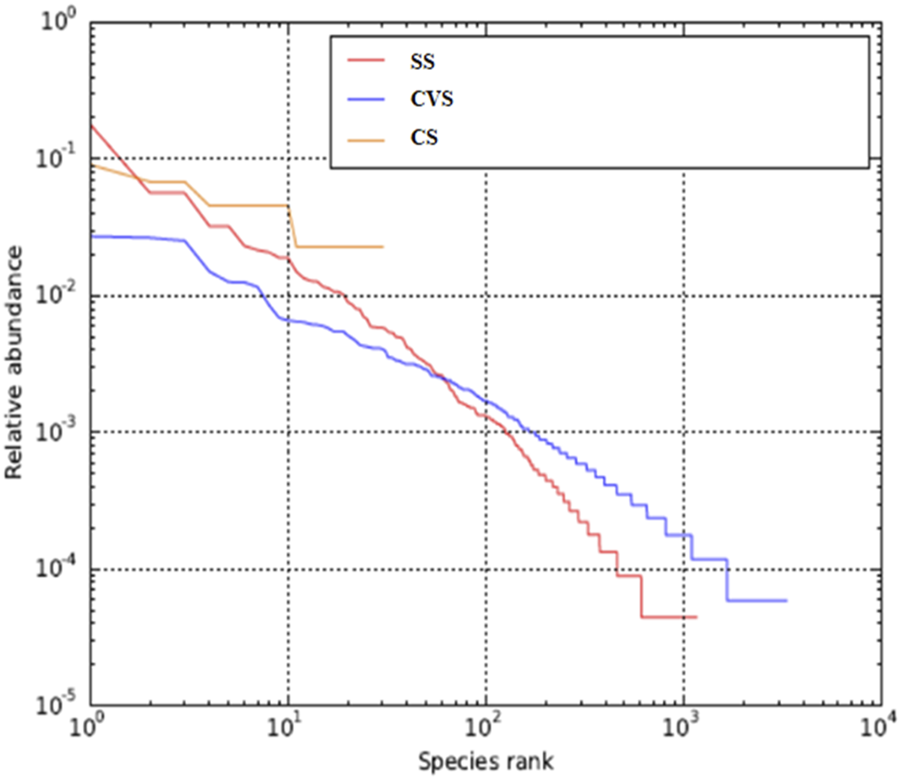
Rank abundance curve of average OTU abundance in CS, CVS, and SS dumpsite soil communities.
3.3 Interactions between the microbial communities and environmental factors
PCA based on Euclidean distance revealed the transformation of environmental data with the OTUs as monitored with principal coordinate ordination analysis (PCO) (Figure 3). As summarized in Figure 3, the first PCO accounts for a large proportion of the data, 63.9% of the variation in the samples, separating CS from the rest of the sites. Most of the environmental variables (POPs) had more effect on the CS bacterial community, and most of the PAHs had a correlation with dibenzofurans and a few of the HMs (Mn, Zn, and Cu). However, organic pollutants such as (1,1′-biphenyl)-4-amine, 7,12-dimethylbenz(a)anthracene, 2-methyl-4,6-dinitrophenol, and 2,4-dinitro phenol exhibited more impacts and were the drivers of the SS ecosystem compared to other sites. Total nitrogen, phosphate, chrysene, 2-nitro-phenol, and Al had more effect in CVS, which, interestingly, was the least polluted site (Figure 3). Furthermore, CCA demonstrated that environmental factors were strongly correlated with the bacterial communities (p < 0.05) (Figure 4). Moreover, it was established that the three different dumpsites ordinated differently, whereby most of the POPs and HMs shaped the bacterial community of the CS.
FIGURE 3

Principal component ordinations of bacterial communities in CS (Cele), CVS (Computer village) and SS (Solous) soils depicting levels of similarity and environmental variables [(A) POPs; (B) physicochemical] associated with the segregation of communities along the PCO axes. 1-5: Ni; [1,1′-biphenyl]-4-amine; 7,12-dimethylbenz(a)anthracene; 2-methyl-4,6-dinitrophenol; 2,4-ninitro phenol; 6–7: p-nitroaniline; Anthracene; 8–22: Mn; Zn; Cu; Azobenzene; Dibenzofuran; Dibenzo(a,i)pyrene; Dibenzo(a,l)pyrene; Dibenzo(a,h)anthracene; 3-methylcholanthrene; Benzo(a)pyrene; Benzo(k)fluoranthene; Benzo(b)fluoranthene; Benzo(e)pyrene; Benz(a)anthracene; Pyrene.
FIGURE 4
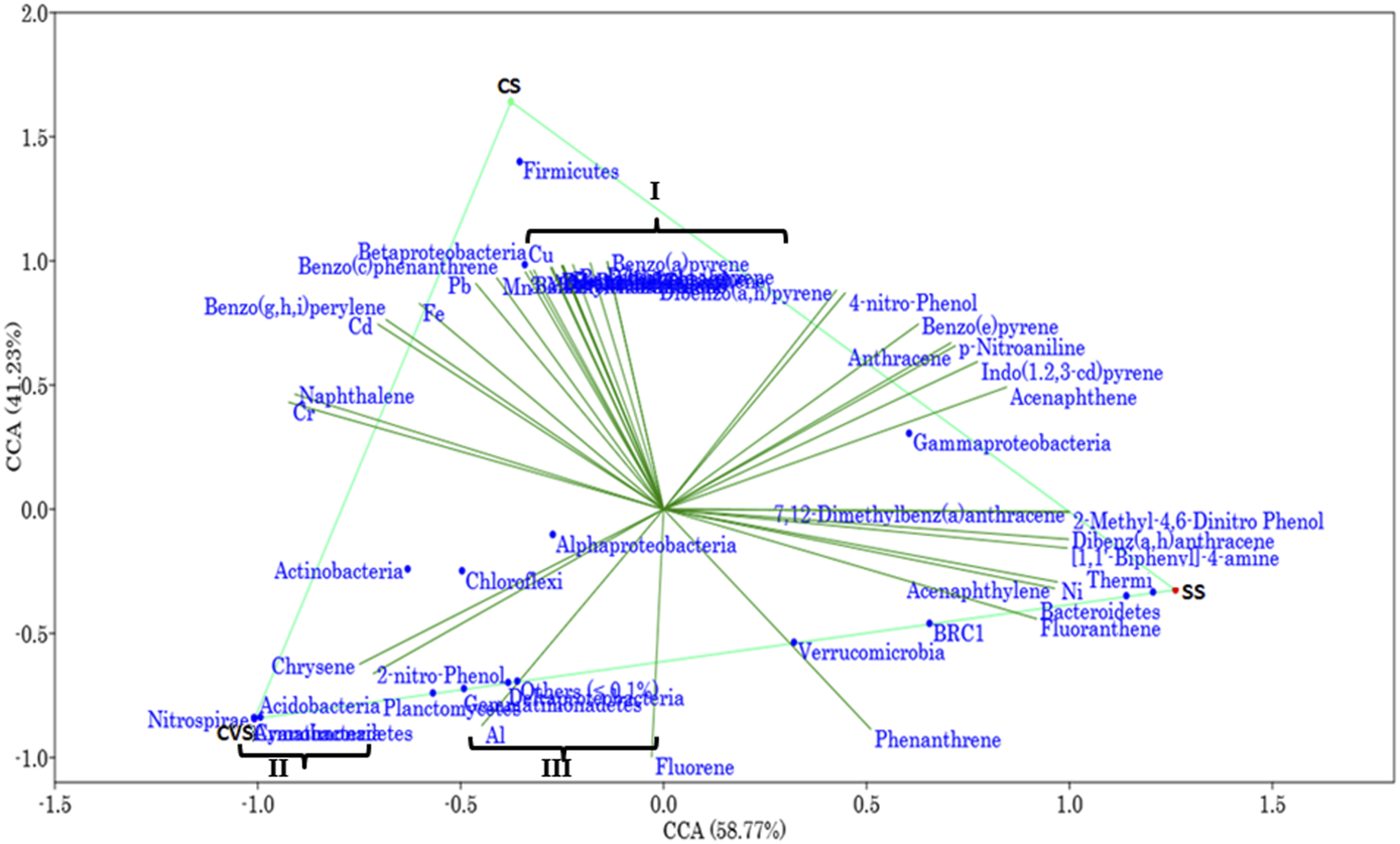
Canonical correspondence analysis, two-axis plot of variation partitioning of bacterial communities and environmental variable in CS, CVS, and SS dumpsites. The environmental variables clustered in I are Zn, azobenzene, dibenzofurans, pyrene, 2,4-dinitrophenol, dibenzo(a,i)pyrene, dibenzo(a,l)pyrene, 3-methylcholanthrene, benzo(a)anthracene, benzo(k)fluoranthrene, and benzo(b)fluoranthene; clustered OTUs in II are Armatimonadetes and Cyanobacteria; and those in III are Deltaproteobacteria and Gemmatimonadetes.
The CCA bi-plot resolved the geographic differences of the CS and CVS by 58.77% variance along the x-axis. The bacterial communities in relation to environmental variations were subdivided across a slope gradient through 41.2% variability. A critical scrutiny of the data summarized in Figure 4 readily suggested a positive effect of 2-nitrophenol and chrysene on the abundance of the dominant OTUs; Acidobacteria, Actinobacteria, Chloroflexi, Planctomycetes, and α-Proteobacteria. On the other hand, γ-Proteobacteria seemed to be negatively correlated with 2-nitrophenol and chrysene and positively correlated with indo(1,2,3-cd)pyrene and acenaphthene. Organic compounds such as benzo(a)pyrene, benzo(c)phenanthrene, and benzo(g,h.i)perylene as well as HMs Cd, Pb, and Fe were strongly associated with the phylum Firmicutes. Similarly, the same environmental variables that were positively associated with the Firmicutes were negatively correlated with the phyla Verrucomicrobia and Bacteroidetes. However, both phyla shared a positive correlation with phenanthrene, fluoranthene, and acenaphthylene.
Out of a total of 706 OTUs, 628 OTUs had a significant correlation with environmental variables such as benz(a)anthracene, benzo(b)fluoranthene, azobenzene, dibenzofurans, pyrene, dibenzo(a,l)pyrene, 2,4-dinitrophenol, Cu, and Zn, with each variable accounting for over 50% of the OTU correlation (Table 5). A dendrogram displaying the groupings of environmental characteristics that frequently co-occurred as variables significantly correlated with OTU abundance had three main branches, as shown in Figure 5. Branches IA and IB contained dibenzofurans, some PAHs, Zn, and Cu, whereas branch II was predominantly represented by PAHs and other organic pollutants. Within branch III, there were three sub-clusters (III A–C) of frequently co-occurring PAHs and/or metals. In addition, 4-nitrophenol and acenaphthylene significantly correlated with the abundance of Proteobacteria and Firmicutes, respectively, in the soils. Anthracene and p-nitroaniline had a significant negative correlation with Acidobacteria, Cyanobacteria, Chloroflexi, and Nitrospirae, while in contrast, there was a positive correlation between some of the HMs (i.e., Cu, Zn, and Ni), dibenzofurans, azobenzene, and pyrene, with an abundance of Bacteroidetes and Firmicutes (Supplementary Table S1). Heatmap analysis of both the sequence and environmental data further reinforces the microbial community composition divergence of the CVS from the rest of the sites. Having revealed the patterns of correlation between environmental variables and OTUs (phyla) and correlations among the three sites vis-à-vis the taxonomic composition of the bacterial community, it was established that CVS and SS had relatively similar responses (Figure 6A). As seen in Figure 6B, the side dendrogram consists of two main clusters, where the lower cluster had a strong correlation with OTU abundance in CS when compared with the other two sites, which exhibited a low correlation with OTU abundance.
TABLE 5
| Environmental variable | OTU correlation | %OTU correlation |
|---|---|---|
| Naphthalene | 5 | 0.70922 |
| Acenaphthylene | 22 | 3.120567 |
| Acenaphthene | 14 | 1.985816 |
| Fluorene | 1 | 0.141844 |
| Anthracene | 134 | 19.00709 |
| Phenanthrene | 9 | 1.276596 |
| Fluoranthene | 4 | 0.567376 |
| Pyrene | 389 | 55.1773 |
| Benzo(c)phenanthrene | 18 | 2.553191 |
| Benz(a)anthracene | 365 | 51.77305 |
| Chrysene | 142 | 20.14184 |
| Benzo(e)pyrene | 34 | 4.822695 |
| Benzo(b)fluoranthene | 397 | 56.31206 |
| Benzo(k)fluoranthene | 352 | 49.92908 |
| Benzo(a)pyrene | 1 | 0.141844 |
| 7,12-Dimethylbenz(a)anthracene | 5 | 0.70922 |
| 3-Methylcholanthrene | 40 | 5.673759 |
| Indo(1.2,3-cd)pyrene | 30 | 4.255319 |
| Dibenz(a,h)anthracene | 4 | 0.567376 |
| Dibenzo(a,l)pyrene | 390 | 55.31915 |
| Benzo(g,h,i)perylene | 11 | 1.560284 |
| 2-Nitro-phenol | 137 | 19.43262 |
| 2,4-Dinitro phenol | 389 | 55.1773 |
| Dibenzofuran | 389 | 55.1773 |
| Azobenzene | 396 | 56.17021 |
| 2-Methyl-4,6-dinitro phenol | 17 | 2.411348 |
| p-Nitroaniline | 125 | 17.7305 |
| [1,1′-Biphenyl]-4-amine | 4 | 0.567376 |
| Cd | 13 | 1.843972 |
| Cr | 3 | 0.425532 |
| Cu | 394 | 55.88652 |
| Fe | 6 | 0.851064 |
| Mn | 36 | 5.106383 |
| Ni | 17 | 2.411348 |
| Pb | 36 | 5.106383 |
| Zn | 372 | 52.76596 |
| Moisture content | 22 | 3.120567 |
Environmental variables that have correlations with different OTUs (genus level).
FIGURE 5
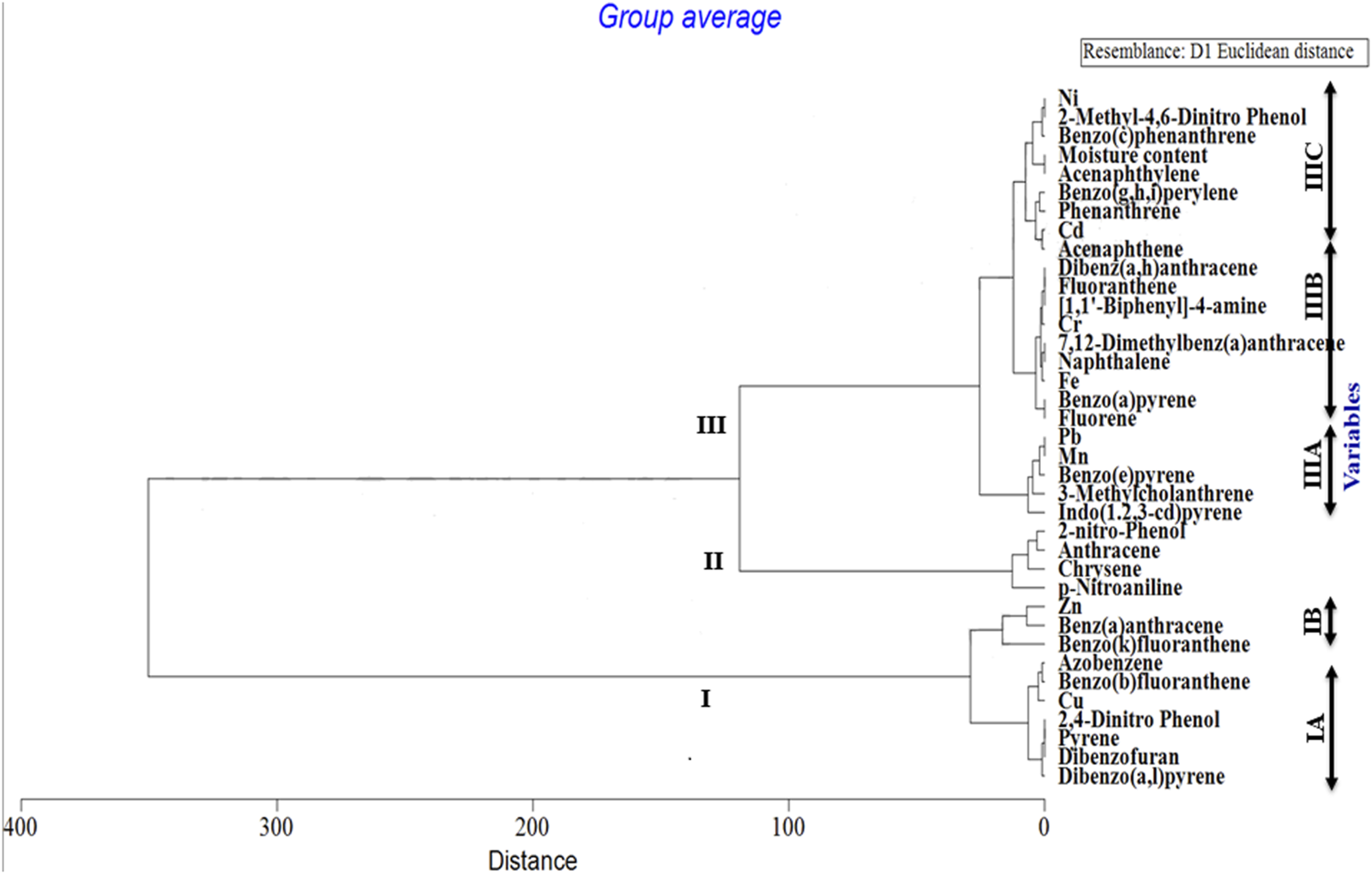
Dendrogram illustrating groupings of environmental properties that frequently co-occurred as variables that significantly correlated with OTU abundance. Three major clusters are represented with sub-clusters within groups I and III.
FIGURE 6
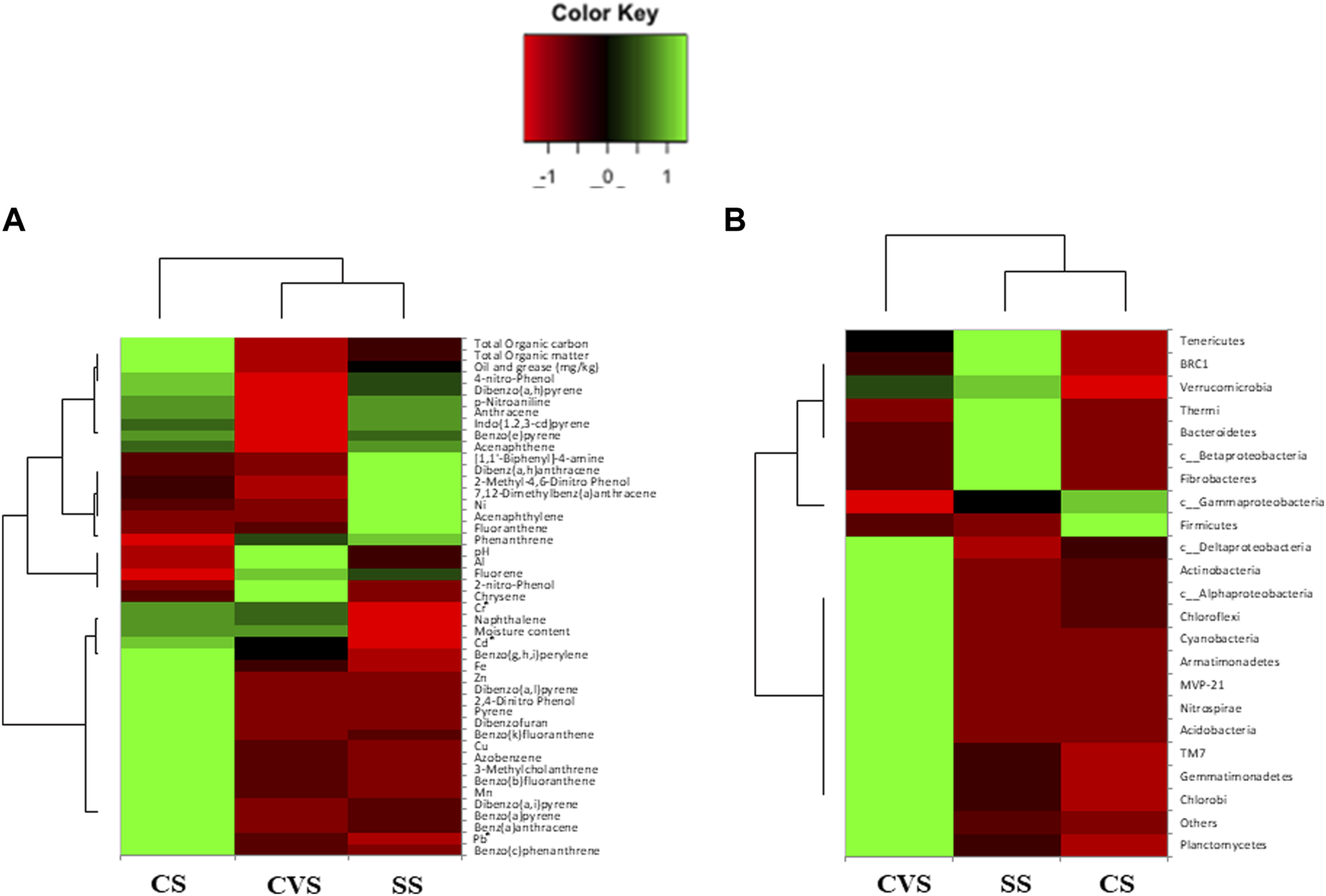
Heatmap comparison of three dumpsite soil environmental variables (A) and OTU abundance at the phylum level (B). Red and green denote negative and positive correlation, respectively, while black represents no correlation. Euclidean distance is computed to evaluate the similarity, and clustering is conducted using the complete linkage method. CS, CVS, and SS dumpsites.
3.4 Analysis of the microbial community composition and structure of the polluted sites
The bacterial community structures of the soil samples as determined by Illumina sequencing of the 16S rRNA gene generated across all libraries were a total of 706 bacterial and five archaeal OTUs (97% of sequence reads). Despite presenting the least diversity among the three sites, the relative abundance of unclassified archaea at the CVS site was greater than that of either SS or CVS soil. The archaeal community profiles in CS comprised 95% of total archaeal OTUs in all three sites, whereas CVS and SS accounted for 4% and 1%, respectively (Figure 7). Irrespective of the extremely low profile in CVS, this site was more diverse in the archaeal community than the other two. Whereas three phyla, namely, Euryarchaeota, Crenarchaeota, and Parvarchaeota, were encountered in the CVS, the archaeal community, in both CS and SS, primarily contained only unclassified archaea (Supplementary Table S2).
FIGURE 7
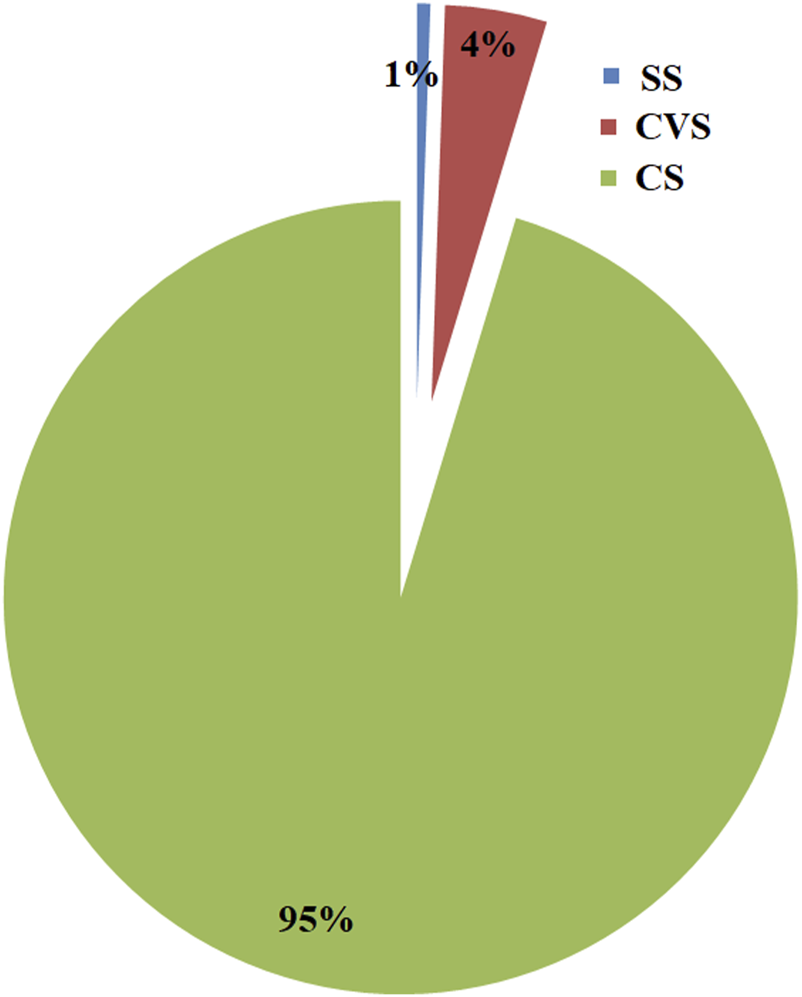
Relative abundance of archaeal taxa in CS, CVS, and SS dumpsites. SS dumpsite soils. Although 95% of the total archaeal OTUs were found in CS soil, CVS had more diversity than other soils.
In terms of the bacterial community structure, soil originating from CVS had the highest unique OTU (34.7%) when compared with others, while the lowest unique OTU was observed in CS (Figure 8). Although the highest similarity index (23.5%) was recorded between SS and CS, the bacterial diversity pattern of the three dumpsites clearly separated CS from the two sites along PCO1, which accounted for 63.9% of the total variation, showing a 40% similarity index for CVS and SS sites (Figure 8). Proteobacteria, Actinobacteria, and Bacteroidetes were the core phyla, accounting for approximately 77.22% of the bacterial taxa in all the sites combined. The phyla Actinobacteria, Chloroflexi, Firmicutes, and Proteobacteria were detected in the three soil communities, though to varying degrees, as depicted in Figure 9. While Proteobacteria (48.32%) was the dominant phylum in CS, Bacteroidetes (45.40%) and Actinobacteria (42.21%) unequivocally dominated the communities in SS and CVS, respectively. Among the Proteobacteria group in the three dumpsites, the class α-Proteobacteria, led by the genus Balneimonas, in the order Rhizobiales and family Bradyrhizobiaceae, predominated in CVS; while in CS, the dominant class was γ-Proteobacteria, led by the family Enterobacteriaceae and genus Serratia (Figure 9). In the case of the SS dumpsite, approximately 75% of the members of Proteobacteria encountered belonged to the class γ-Proteobacteria, which was mainly dominated by the families Alteromonadaceae, Xanthomonadaceae, and Pseudomonadaceae. The community structure of the CS site presented an interesting observation. Unlike the CVS and SS soils, the bacterial community was practically made of four phyla consisting of Proteobacteria in addition to Firmicutes, Actinobacteria, and Chloroflexi. The latter was merely 3.26% of the population. By implication, other phyla, including Acidobacteria, Bacteroidetes, Gemmatimonadetes, and Planctomycetes, which were found in other sites, were absent in CS. It is also noteworthy that Firmicutes, which constituted nearly 30% of the communities in CS, contributed less than 2% to the bacterial flora in both CVS and SS systems (Figure 9). Conversely, the δ-proteobacteria class, which was absent in CS, was found in SS and CVS, but its relative abundance was higher in the latter than in the former. With the exception of Bacteroidetes, Proteobacteria, and Actinobacteria, all other representative phyla in SS constituted generally less than 3% of the soil microflora. Bacteroidetes that was the dominant phylum in the SS dumpsite constituted barely 2.5% of the community in CVS; in contrast, Actinobacteria, which dominated the bacterial community in CVS, constituted less than 7% of the composition in the SS soil.
FIGURE 8
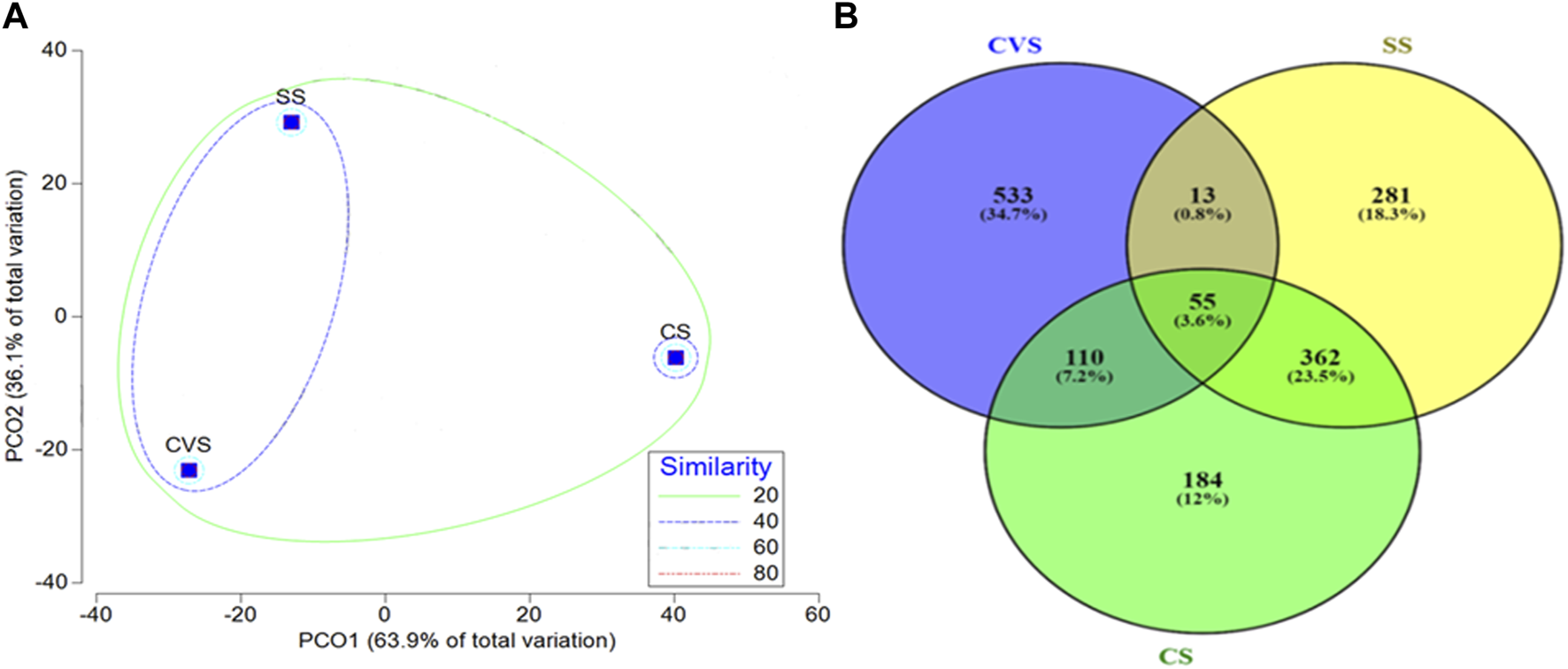
Principal component ordination of bacterial communities in CS, CVS, and SS dumpsite soils, with contours indicating the percent resemblance between samples (A) and Venn diagram displaying distribution of common and unique OTUs (genus) among the dumpsites (B).
FIGURE 9
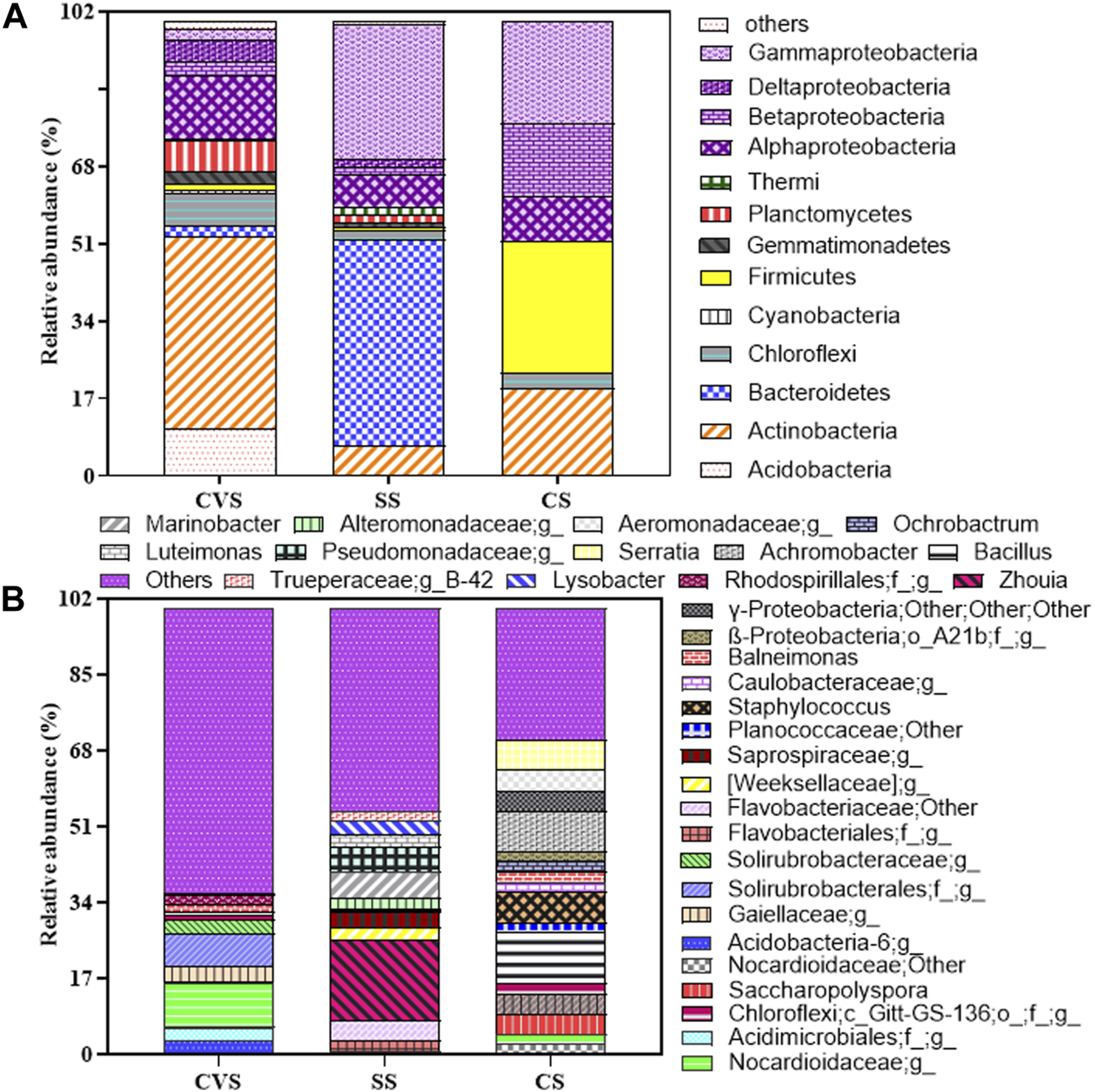
Relative abundance of major bacterial genera (A) and phyla (>0.5% of a given library; (B) in CS, CVS, and SS dumpsite soils. Proteobacteria is subdivided by class unlike other phyla and are all color-coded purple. Taxonomic abundance <0.5% at the phylum level was classified as others. Abundance is the fraction of the total sequences in a given library represented by a single OTU.
Further interrogation of the obtained sequence data revealed Zhouia (18.29%) belonging to the family Flavoacteriaceae as the most abundant genus, and it was only found in the SS soil, thus making it the dominant genus in that site. This genus is more than three orders of magnitude higher than Marinobacter, the next dominant genus in the same dumpsite (Figure 9B). Like Zhouia, the genus Marinobacter was not detected in the other dumpsites. In the case of CS and CVS, Bacillus (11.36%) and an unclassified genus in the family Norcardioidaceae (9.8%) were the respective dominant genera.
The type and distribution of bacterial genera in the soils were somewhat unique and obviously different from site to site, an inference that further reinforces the data pattern shown in Figure 8. For instance, out of the 32 genera represented in Figure 9B, only 10 were found across the dumpsites, accounting for less than 32% of captured genera, and eight of these had less than 0.1% abundance in either or two of the soil’s microflora. In addition, only two of the ten genera had a relative abundance of >0.1% across the sites. These genera included Bacillus and an uncultured genus in the family Norcardioidaceae.
4 Discussion
Lagos, the commercial hub of Nigeria, is the second fastest-growing city in Africa and seventh in the world and is one of the largest producers of solid waste (if not the largest) in Africa. The latest reports estimated its population to be more than 21 million, making it the largest city in entire Africa. In Lagos, over 12,000 metric tons of waste are generated daily at a generation per capita (GPC) of 0.72 kg/person/day (Olukanni and Oresanya, 2018). Over 4.5 million tons of waste are generated annually in Lagos, and 30% of the waste is recycled to reduce the amount going to landfills (Bakare, 2016). The climate of Lagos State is tropical with alternate dry and wet seasons, with a temperature range of 28°C–33°C.
Dumpsites are the final repository for most of the discarded materials from human society. It is generally known that polluted sites exert environmental pressure on fundamental ecological parameters such as abundance, diversity, and nutrient cycling. Therefore, an in-depth knowledge of the microbial community structure is essential to gain insights into processes that may affect the fate of pollutants specifically and biogeochemical cycling more broadly. The three dumpsites examined in this study were extensively polluted, which may have paved the way for selective pressure and evolution of competent pollutant degraders (Saibu et al., 2020). In Lagos dumpsites, PAHs, oil and grease, HMs, and other organic pollutants were widespread in soils, but the concentrations depended on waste-type input and other activities occurring on such sites (e.g., incineration and open burning). Indiscriminate and extreme incineration activities, which the CS site is constantly subjected to, may have accounted for the highest burden of PAHs, oil, and grease encountered (Ferronato and Torretta, 2019). In the case of dibenzofuran, since its concentration was below the detection limit in both CVS and SS and dioxins are relatively ubiquitous in nature, it is plausible that aerial transport and deposition from air to soil may have occurred, thus reducing concentration at one spot and increasing it at another location (Mahfouz et al., 2020). This remobilization, distribution, and volatilization may also be responsible for the absence of PCBs and polychlorinated dibenzo-p-dioxins/furans (PCDD/Fs) across the sites. Interestingly, this phenomenon has been observed in several environmental matrices by several researchers (Šrédlová and Cajthaml, 2022). Chlorinated organics, particularly PCBs and PCDD/Fs, can travel long distances via air and get deposited in areas remote from where they were released.
Cumulatively, CS may account for the highest proportion of POPs; the highest cumulative concentration of HMs was found in CVS soil, where both Al and Fe occurred at significantly high concentrations. Although Al and Fe may not pose a serious environmental threat compared to other metals, the level of occurrence can, nonetheless, be very dangerous to plants, weakening and eventually killing them (Marzorati et al., 2022). Among the list of priority pollutant metals, Cu, Cr, Ni, Pb, and Zn were detected in all three sites at concentrations above the permissible limit, with the exception of Cu at SS and CVS. The environmental hazards of these metals, particularly on fragile ecosystems, cannot be overemphasized. Sensitive fauna and flora can be wiped out, thus giving rise to the evolution of some HM-resistant microorganisms (Ayangbenro and Babalola, 2017).
Generally, the level of pollutants detected across the sites is not unexpected. What is frightening is the significant threat posed to other ecosystems within their vicinity. In Nigeria, the widely used three main solid waste disposal processes are landfilling, open dumping, and open burning, which are not satisfactory technologies. Landfills, for instance, are poorly designed and poorly managed, creating several adverse environmental impacts such as wind-blown litter, attraction of vermin, and formation of leachate (Ishaq et al., 2022). Leachate could percolate through the waste, contaminating ground and surface water bodies and causing damage to the ecosystem by entering into the food chain (Kamboj et al., 2020; Ishaq et al., 2022). Therefore, the level of pollutants in the dumpsites under investigation is a latent poison to water bodies and soils within their vicinity.
The microbial community structures in the dumpsites were shaped by inherent environmental factors. The impact of organic wastes on CS and SS soils would also be consistent with the much higher levels of PAHs, TOM, and other organic compounds (total) at these two sites compared to CVS. The potential impacts of anthropogenic pollutants on microbial communities in the dumpsites were reflected in variations in alpha diversity. In comparison to CVS, CS and SS microbial communities had significantly low species richness as well as species evenness. For instance, Shannon’s diversity index was highest in CVS, where POP levels were significantly low. Therefore, it follows that the reduction in diversity observed in both CS and SS is a direct function of the amounts of POPs and other organic wastes inputted into the environment. Data obtained from β-diversity suggest that neither HMs nor POPs alone were the major limiting factors for bacterial activities. However, the combination of both variables had a profound impact on bacterial communities. Multiple stressors, such as HMs and POPs, elicit a more complex response from microbial community depending on the type of stressor, soil properties, and other factors (Azarbad et al., 2015). While PCO analysis could not resolve the abiotic factors driving the microbial community, CCA readily did. For example, most of the POPs and HMs were the environmental factors driving the population of Firmicutes and β-Proteobacteria in CS, in which the former was the dominant taxon in that community. Therefore, CCA determined the correlation between the environmental factors and the bacterial community in each of these sites. Such information is necessary for providing baseline knowledge of contaminated sites, which is essential for designing bioremediation strategies in polluted systems (Saibu et al., 2023).
Metagenomics data emanating from several laboratories have often declared Bacteroidetes and Proteobacteria as predominant bacterial taxonomic groups in landfill sites (Wang et al., 2017; Zainun and Samarani, 2018; Sandhu et al., 2022), a trend that is consistent with the results obtained in this study, except for CVS. Generally, the three dumpsites investigated exhibited stark variations in terms of microbial community structure and dominance. However, the configuration of the microbial communities in CS and SS was similar in terms of the relatively high population of α-Proteobacteria, in contrast to CVS, where the α-Proteobacteria was a minor constituent. In the less polluted CVS site, β-Proteobacteria, next to γ-Proteobacteria among the Proteobacteria phylum, predominates, suggesting the vulnerability of this class to POPs and not necessarily HMs. The resilience of α-Proteobacteria in POP-burdened environmental systems noted in this study is consistent with our recent findings (Saibu et al., 2023) and those of Obi et al. (2016). Similar resilience is also observed among the δ-Proteobacteria, a taxon that was not found in the CVS soil. In addition, the organisms in this class are better adapted to POPs and perhaps are a major player in the degradation of these pollutants in both CS and SS since they were notably absent in CVS. It is somewhat amazing that the community conformation of the dumpsites is not only the dominance of Actinobacteria and Bacteroidetes in CVS and SS, respectively, as against the much-expected Proteobacteria taxon, but the decrease in the members of the γ-Proteobacteria class in the SS soil in spite of the huge input of organic pollutants. Organisms in this taxonomic group play fundamental roles in nutrient cycling and are widely known for their metabolic versatilities which are variously exploited for bioremediation of polluted systems (Vinas et al., 2005; Horel et al., 2015; Obi et al., 2016; Vikram et al., 2016). Therefore, it is expected that they would constitute a dominant population in SS at a threshold much higher than that of soil obtained from CVS. Notwithstanding the level of γ-Proteobacteria in SS, the dominance of the genus Marinobacter is a vindication of the existence of organisms with the capacity to metabolize an extensive spectrum of xenobiotic pollutants, cementing their roles in nutrient cycling in that ecosystem (Horel et al., 2015). This is more so because Marinobacter is an organism in the family Pseudomonadaceae which houses Pseudomonas, and both organisms are not represented in either CVS or CS. Just like Pseudomonas, Marinobacter is a key hydrocarbon degrader that has become recognized as obligate hydrocarbonoclastic bacteria and a prolific biosurfactant producer (Raddadi et al., 2017).
These observations further show that there were significant variations in microbial community structure associated with soil contamination that could lead to shifts in pathways of fundamental biogeochemical processes (Bissett et al., 2013). Therefore, speciation and the concentration of (in)organic pollutants notwithstanding, the primary populations in CVS including Acidobacteria, Chloroflexi, Planctomycetes, Gemmatimonadetes, and Cyanobacteria, which were either absent or present at very low abundance in other soils, are phyla that are typically abundant in healthy soils (Xu et al., 2014). Some of these bacteria, like Proteobacteria, also play considerable roles in global carbon and nitrogen cycles. Cyanobacteria, for instance, through their metabolic proficiency, embodied a potentially notable pathway for carbon turnover, aeration of the environment, and direct nitrogen uptake through non-symbiotic nitrogen fixation (Karlson et al., 2015; Adam et al., 2016). This important taxon, though a minority member of the CVS community, is conspicuously absent from the rest of the sites.
In the case of Actinobacteria, its predominance in CVS may be connected with elevated concentrations of Al and Fe and in the sum total of HMs, which was highest at this site. This phylum is dominated by organisms of the order Solirubrobacterales, which, according to Seki et al. (2015), consists of three families, three genera, and eleven species. These organisms showed a propensity for soils with reduced carbon/nutrient availability (Shange et al., 2012) and are also widespread with high abundance in HM-polluted soils (Navarro-Noya et al., 2010; Gołębiewski et al., 2014; Yan et al., 2016; Salam and Varma, 2019). Recently, Goswami et al. (2023) reported a positive correlation between this taxon and Zn and Pb concentration. Interestingly, the group has been identified as an indicative taxon in HM-contaminated mining and agricultural soils (Huang et al., 2021) and similarly demonstrated to associate with metal-accumulating plants and are easily enriched in HM-supplemented agricultural soils (Wu et al., 2021; Kuo et al., 2018). In addition to Actinobacteria, other phyla encountered in the CVS soil including Acidobacteria, Gemmatimonadetes, and Chloroflexi readily suggest their potential importance in metal transformation and sequestration. This finding is supported by those of previous studies, which revealed their strong tolerance to HMs (Macdonald et al., 2011; Li et al., 2015; Girardot et al., 2020). Unfortunately, little is known about the ecology and metabolism of Gemmatimonadetes. However, the presence and increased abundance, together with those taxonomic groups, can suggest their indispensability and essential functions in the contaminated soils.
Another interesting observation in the community structure of the dumpsites is the relative abundance of the Firmicutes in CS at a proportion that was nearly 20 times greater than that of the population found in other sites, a pattern similar to the archaeal domain. This phylum is essentially dominated by the genus Bacillus, and the unabated incineration activity at the site may have accounted for their overriding success. Bacillus are more resilient and better adapted to harsh environmental conditions due to their ability to produce spores that are impervious to heat, radiation, and chemicals, while in the case of archaea, the lipid bilayer cell membrane enables them to function at high temperatures (Koga, 2012; Bruns, 2021). In fact, Bacillus is among the most reported bacteria in soil possessing much higher tolerance to organic pollutants and HMs, which, in this study, exhibited a positive correlation with Ni. Incidentally, this genus emerged as the most dominant when bacterial communities of food waste co-composting degradation of PCDD/F-contaminated soil were investigated (Huang et al., 2019). A connection between dioxin degradation and the genus Bacillus was also established by Mahfouz et al. (2020). These authors reported that bacterial species belonging to Bacillus were randomly distributed in soils highly polluted with PCDD/Fs and unequivocally demonstrated the abilities of several species of Bacillus to degrade 2,3,7,8-tetrachlorodibenzo-p-dioxin.
Other genera encountered in CS at varying proportions that are either effectively absent or diminutive in other sites were Serratia, Staphylococcus, Achromobacter, and Ochrobactrum. Interestingly, two bacterial species belonging to the genera Serratia and Bacillus which were unambiguously demonstrated to utilize dibenzofuran, 2,8-dichlorodibenzofuran, and 2,7-dibenzo-p-dioxin as the sole sources of carbon and energy were obtained from this site (Saibu et al., 2020). In a related study, we also reported the successful adaptation of both taxa to dioxin-contaminated soil and touted their potential use as bioindicators of dioxin pollution (Saibu et al., 2023). Interestingly, CS was the only site where dioxin was detected, which may likely be responsible for the populations of the encountered genera. Therefore, there is no uncertainty regarding the dioxin metabolic capabilities of these taxa.
Among the genera of organisms identified in this study, the highest percentage was credited to Zhouia. In addition, this genus was the most dominant in the SS site, where Bacteroidetes also dominated. This relatively novel genus is yet to be explored for its environmental benefits. The organism in this taxon was first isolated from marine sediment and mangrove soil (Liu et al., 2006; Hong et al., 2023) and was among the tropical mangrove sediment communities demonstrated to readily degrade Enteromorpha prolifera (Zhao and Ruan, 2011). Similarly, with regard to the sharp increase in population of the genus observed in treated petroleum hydrocarbon-contaminated soil, Rahmeh et al. (2021) attributed this surge to the less complex ring structure derivatives resulting from the degradation of aromatic compounds with complex structures. A critical evaluation of the metagenome data clearly showed that among the population of bacterial phyla reported across the sites, none came close to the Bacteroidetes. The phylum is essentially made up of the family Flavobacteriaceae, of which Zhouia is a representative. The potential of Bacteroidetes in soil ecosystems has been accredited to their ability to degrade complex organic compounds (McBride et al., 2014) and the secretion of carbohydrate-active enzymes that specifically target the highly wide-ranging polysaccharides in soil (Larsbrink and Mckee, 2020). Members of this phylum have also been implicated in the degradation of chemical weapons (Thouin et al., 2019). According to these authors, soil historically contaminated by the burning of chemical ammunition was dominated by Bacteroidetes in addition to Proteobacteria and Acidobacteria. Therefore, the huge number of members of this group of organisms in SS may not be unconnected with these metabolic properties coupled with their capacity to fix nitrogen non-symbiotically. Perhaps, we are beginning to understand the functional roles of the genus Zhouia in polluted systems, where they play important, albeit unknown, roles in pollutant degradation and/or other community processes.
5 Conclusion
We have provided an in-depth insight into the potential impacts that POPs may exert on soil bacterial communities. The MiSeq methodology obtained a high resolution of the microbial community profile in dumpsites. The communities of two of the sites (CS and SS) strongly impacted by POPs were significantly different from the relatively less-impacted site (CVS), which is the obvious deletion of OTUs (Nitrospirae and Cyanobacteria) involved in key biogeochemical processes (nitrogen and carbon fixation). In addition, further understanding of the bacterial communities that may arbitrate PAHs and degradation of other POPs in Lagos dumpsites was given. There were strong indications in the types of potential degraders apparently enriched in the soil compared to those identified in other soil ecosystems impacted by POPs. For instance, γ- and α-Proteobacteria were indicated as key PAH/POP phylotypes responsive to the high concentration of pollutants inherent in the CS site, which, interestingly, also had the highest level of contamination. In contrast, the less PAH/POP-heavily impacted soil (SS), the organisms with the potential metabolism of these pollutants shifted to the α-Proteobacteria taxon. Thus, Proteobacteria were a significant group potentially associated with PAHs/POPs, but the classes varied depending upon the concentration of these pollutants and possibly other factors such as HMs and prevailing environmental parameters. Furthermore, the evident increase in populations of Bacillus and Serratia in CS, which were both absent in SS and CVS sites, highlighted their superior dioxin metabolic functionalities in addition to other POPs, consistent with findings from our previous study (Saibu et al., 2020). Since dioxin was only detected at the CS site, these organisms may have been particularly enriched by the presence of this pollutant.
Overall, our investigation provides a detailed report on the abundance and diversity of bacteria in Lagos municipal dumpsites, revealing the impacts of waste inputs on the soil microflora and the important environmental factors driving the microbial communities in the ecosystem. The bacterial species richness in this study shows the complexity of anthropogenic activities in these ecosystems and calls for further research on archaea and eukarya diversities as well as investigation of the functional dynamics of the microorganisms that have been hypothesized in this study.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI Sequence Read Archive, accession number PRJNA540227.
Author contributions
SS: data curation, formal analysis, investigation, methodology, validation, and writing–original draft. SA: conceptualization, data curation, project administration, supervision, and writing–review and editing. GO: supervision and writing–review and editing. DR: funding acquisition, project administration, supervision, and writing–review and editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
This study was financially supported partially by the Welch foundation award number (E-2011-20190330) and Biological and Environmental Research Science Focus Area grant: U.S. Department of Energy (Grant No. DE-AC52-06NA25396).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1304033/full#supplementary-material
References
1
AdamB.KlawonnI.SvedenJ. B.BergkvistJ.NaharN.WalveJ.et al (2016). N2 fixation, ammonium release and N-transfer to the microbial and classical food web within a plankton community. ISME J.10, 450–459. 10.1038/ismej.2015.126
2
AOAC (Association of Official Analytical Chemists) (1990). Official methods of analysis.
3
AyangbenroA. S.BabalolaO. O. (2017). A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int. J. Environ. Res. Public Health.14, 94. 10.3390/ijerph14010094
4
AzarbadH.NiklińskaM.NikielK.van StraalenN. M.RölingW. F. M. (2015). Functional and compositional responses in soil microbial communities along two metal pollution gradients: does the level of historical pollution affect resistance against secondary stress?Biol. Fertil. Soils.51, 879–890. 10.1007/s00374-015-1033-0
5
BajarS.SinghA.KaushikC. P.KaushikA. (2021). Suitability assessment of dumpsite soil biocover to reduce methane emission from landfills under interactive influence of nutrients. Environ. Sci. Pollut. Res.28, 1519–1532. 10.1007/s11356-020-10441-8
6
BakareW. (2016). Solid waste management in Nigeria. Bio-Energy Newsl.Available at: http://www.bioenergyconsult.solid-waste-Nigeria.
7
BasuS.KumarG.ChhabraS.PrasadR. (2021). Role of soil microbes in biogeochemical cycle for enhancing soil fertility. New Future Dev. Microb. Biotechnol. Bioeng., 149–157. 10.1016/B978-0-444-64325-4.00013-4
8
BissettA.BrownM. V.SicilianoS. D.ThrallP. H. (2013). Microbial community responses to anthropogenically induced environmental change: towards a systems approach. Ecol. Lett.16, 128–139. 10.1111/ele.12109
9
British Standard (1995). Determination of pH. BS 7755-3.2 1995 ISO 10390.
10
BrunsM. A. (2021). “Bacteria and archaea,” in Principles and applications of soil microbiology (Elsevier), 111–148.
11
CaporasoJ. G.KuczynskiJ.StombaughJ.BittingerK.BushmanF. D.CostelloE. K.et al (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods.7, 335–336. 10.1038/nmeth.f.303
12
ChopraS. L.KanwarJ. S. (Editors) (1998). Analytical agricultural Chemistry (London: Macmillian Press).
13
ClarkeK. R.GorleyR. N. (2015). Getting started with PRIMER v7. PRIMER-E Plymouth, Plymouth Mar. Lab.20 (1).
14
DayP. R. (1965). Particle fractionation and particle-size analysis. Methods Soil Analysis Part 1 Phys. Mineralogical Prop. Incl. Statistics Meas. Sampl.9, 545–567.
15
DelmontT. O.PrestatE.KeeganK. P.FaubladierM.RobeP.ClarkI. M.et al (2011). Structure, fluctuation and magnitude of a natural grassland soil metagenome. ISME J.6, 1677–1687. 10.1038/ismej.2011.197
16
DuanH.LiJ.LiuY.YamazakiN.JiangW. (2012). Characterizing the emission of chlorinated/brominated dibenzo-p-dioxins and furans from low temperature thermal processing of waste printed circuit board. Environ. Pollut.161, 185–191. 10.1016/j.envpol.2011.10.033
17
FerronatoN.TorrettaV. (2019). Waste mismanagement in developing countries: a review of global issues. Int. J. Environ. Res. Public Health.16, 1060. 10.3390/ijerph16061060
18
GardiC.MontanarellaL.ArrouaysD.BispoA.LemanceauP.JoviletC.et al (2009). Soil biodiversity monitoring in Europe: ongoing activities and challenges. Eur. J. Soil Sci.60, 807–819. 10.1111/j.1365-2389.2009.01177.x
19
GirardotF.AllégraS.PfendlerS.ConordC.ReyC.GilletB.et al (2020). Bacterial diversity on an abandoned, industrial wasteland contaminated by polychlorinated biphenyls, dioxins, furans and trace metals. Sci. Total Environ.748, 141242. 10.1016/j.scitotenv.2020.141242
20
GołębiewskiM.Deja-SikoraE.CichoszM.TretynA.WróbelB. (2014). 16S rDNA pyrosequencing analysis of bacterial community in heavy metals polluted soils. Microb. Ecol.67, 635–647. 10.1007/s00248-013-0344-7
21
GoswamiA.Adkins-JablonskyS. J.Barreto FilhoM. M.ShillingM. D.DawsonA.HeiserS.et al (2023). Heavy metal pollution impacts soil bacterial community structure and antimicrobial resistance at the Birmingham 35th Avenue Superfund Site. Microbiol. Spectr.11 (2), e0242622–22. 10.1128/spectrum.02426-22
22
HongC. W. L.LiewK. J.LamM. Q.ChongC. S. (2023). Draft genome sequence of Zhouia amylolytica CL16, isolated from mangrove soil. Microbiol. Resour. Announc.12 (5), e0006823–23. 10.1128/mra.00068-23
23
HoornwegD.Bhada-TataP.KennedyC. (2013). Environment: waste production must peak this century. Nature502, 615–617. 10.1038/502615a
24
HorelA.MortazaviB.SobeckyP. A. (2015). Input of organic matter enhances degradation of weathered diesel fuel in sub-tropical sediments. Sci. Total Environ.533, 82–90. 10.1016/j.scitotenv.2015.06.102
25
HouD.ZhangP.ZhangJ.ZhouY.YangY.MaoQ.et al (2019). Spatial variation of sediment bacterial community in an acid mine drainage contaminated area and surrounding river basin. J. Environ. Manag.251, 109542. 10.1016/j.jenvman.2019.109542
26
HuX.WangJ.LvY.LiuX.ZhongJ.CuiX.et al (2021). Effects of heavy metals/metalloids and soil properties on microbial communities in farmland in the vicinity of a metals smelter. Front. Microbiol.12, 707786. 10.3389/fmicb.2021.707786
27
HuangJ.WuY.SunJ.LiX.GengX.ZhaoM.et al (2021). Health risk assessment of heavy metal (loid) s in park soils of the largest megacity in China by using Monte Carlo simulation coupled with Positive matrix factorization model. J. Hazard. Mat.415, 125629. 10.1016/j.jhazmat.2021.125629
28
HuangW.-Y.NgoH.-H.LinC.VuC.-T.KaewlaoyoongA.BoonsongT.et al (2019). Aerobic co-composting degradation of highly PCDD/F-contaminated field soil. A study of bacterial community. Sci. Total Environ.660, 595–602. 10.1016/j.scitotenv.2018.12.312
29
IgbinomwanhiaD. I. (2011). “Status of waste of management,” in Integrated Waste Management. 2nd Edn, Editor KumarS., Vol. 2. Available at: http://www.intechopen.com/books/integrated-waste-management-volume-ii/status-of-waste-management (InTech Open Access Publisher).
30
IshaqA.SaidM. I. M.AzmanS.AbdulwahabM. F.AlfaM. I. (2022). Impact, mitigation strategies, and future possibilities of Nigerian municipal solid waste leachate management practices: a review. Niger. J. Technol. Dev.19 (3), 181–194. 10.4314/njtd.v19i3.1
31
JanssenP. H. (2006). Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol.72, 1719–1728. 10.1128/aem.72.3.1719-1728.2006
32
KambojN.BishtA.KambojV.BishtA. (2020). Leachate disposal induced groundwater pollution: a threat to drinking water scarcity and its management. Adv. Environ. Pollut. Manag. Wastewater Impacts Treat. Technol.1, 54–76. 10.26832/aesa-2020-aepm-05
33
KarlsonA. M. L.DubergJ.MotwaniN. H.HogforsH.KlawonnI.PlougH.et al (2015). Nitrogen fixation by cyanobacteria stimulates production in Baltic food webs. Ambio44, S413–S426. 10.1007/s13280-015-0660-x
34
KogaY. (2012). Thermal adaptation of Archaeal and bacterial lipid membranes. Archaea2012, 6. Article ID 789652. 10.1155/2012/789652
35
LarsbrinkJ.McKeeL. S. (2020). Bacteroidetes bacteria in the soil: glycan acquisition, enzyme secretion, and gliding motility. Adv. Appl. Microbiol.110, 63–98. 10.1016/bs.aambs.2019.11.001
36
LiJ.MaY. B.HuH. W.WangJ. T.LiuY. R.HeJ. Z. (2015). Field-based evidence for consistent responses of bacterial communities to copper contamination in two contrasting agricultural soils. Front. Microbiol.2, 31. 10.3389/fmicb.2015.00031
37
LiuZ. P.WangB. J.DaiX.LiuX. Y.LiuS. J. (2006). Zhouia amylolytica gen. nov., sp. nov., a novel member of the family Flavobacteriaceae isolated from sediment of the South China Sea. Int. J. Syst. Evol. Microbiol.56 (12), 2825–2829. 10.1099/ijs.0.64587-0
38
LongeE. O.BalogunM. R. (2010). Groundwater quality assessment near a municipal landfill, Lagos, Nigeria. Res. J. Appl. Sci. Eng. Technol.2, 39–44.
39
MacdonaldC. A.ClarkI. M.ZhaoF. J.HirschP. R.SinghB. K. (2011). Mcgrath, S.P. Long-term impacts of zinc and copper enriched sewage sludge additions on bacterial, archaeal and fungal communities in arable and grassland soils. Soil Biol. biochem.43, 932–941. 10.1016/j.soilbio.2011.01.004
40
MahfouzS.MansourG.MurphyD. J.HananoA. (2020). Dioxin impacts on lipid metabolism of soil microbes: towards effective detection and bioassessment strategies. Bioresour. Bioprocess.7, 59–17. 10.1186/s40643-020-00347-1
41
MarzoratiF.MidaliA.MorandiniP.MurgiaI. (2022). Good or bad? The double face of iron in plants. Front. Young Minds10, 718162. 10.3389/frym.2022.718162
42
McBrideM. J.LiuW.LuX.ZhuY.ZhangW. (2014). The family cytophagaceae. prokaryotes4, 577–593. 10.1007/978-3-642-38954-2_382
43
NannipieriP.AscherJ.CeccheriniM. T.LandiL.PietramellaraG.RenellaG. (2003). Microbial diversity and soil functions. Eur. J. Soil Sci.54, 655–670. 10.1046/j.1351-0754.2003.0556.x
44
Navarro-NoyaY. E.Jan-RobleroJ.González-ChávezM. C.HernándezGamaR.Hernández-RodríguezC. (2010). Bacterial communities associated with the rhizosphere of pioneer plants (Bahia xylopoda and Viguiera linearis) growing on heavy metals-contaminated soils. Ant. Van Leeuwenhoek97, 335–349. 10.1007/s10482-010-9413-9
45
NelsonD. A.SommersL. (1983). Total carbon, organic carbon, and organic matter. Methods soil analysis Part 2 Chem. Microbiol. Prop.9, 539–579.
46
NguyenH. N.RodriguesD. F. (2018). Chronic toxicity of graphene and graphene oxide in sequencing batch bioreactors: a comparative investigation. J. Hazard. Mat.343, 200–207. 10.1016/j.jhazmat.2017.09.032
47
North Central Agricultural Experiment Stations (1998). Recommended chemical soil test procedures for the north central region. Research Publication No, 221.
48
ObiC. C.AdebusoyeS. A.UgojiE. O.IloriM. O.AmundO. O.HickeyW. J. (2016). Microbial communities in sediments of Lagos lagoon, Nigeria: elucidation of community structure and potential impacts of contamination by municipal and industrial wastes. Front. Microbiol.7, 1213. 10.3389/fmicb.2016.01213
49
OlukanniD. O.OresanyaO. O. (2018). Progression in waste management processes in Lagos State, Nigeria. Int. J. Eng. Res. Afri.35, 11–23. 10.4028/www.scientific.net/jera.35.11
50
QiaoZ.CaoM.WangD.LiaoS.WangG. (2019). Sphingosinicella humi sp. nov., isolated from arsenic-contaminated farmland soil and emended description of the genus Sphingosinicella. Int. J. Syst. Evol. Microbiol.69 (2), 498–503. 10.1099/ijsem.0.003186
51
RaddadiN.GiacomucciL.TotaroG.FavaF. (2017). Marinobacter sp. from marine sediments produce highly stable surface-active agents for combatting marine oil spills. Microb. Cell Factories.16, 186–213. 10.1186/s12934-017-0797-3
52
RahmehR.AkbarA.KumarV.Al-MansourH.KishkM.AhmedN.et al (2021). Insights into bacterial community involved in bioremediation of aged oil-contaminated soil in arid environment. Evol. Bioinform.17, 117693432110168–13. 10.1177/11769343211016887
53
RaviR. K.AnusuyaS.BalachandarM.MuthukumarT. (2019). “Microbial interactions in soil formation and nutrient cycling,” in Mycorrhizosphere and pedogenesis (Singapore: Springer), 363–382.
54
SaibuS.AdebusoyeS. A.OyetiboG. O. (2023). Soil microbiome response to 2-chlorodibenzo-p-dioxin during bioremediation of contaminated tropical soil in a microcosm-based study. J. Hazard. Mat.451, 131105. 10.1016/j.jhazmat.2023.131105
55
SaibuS.AdebusoyeS. A.OyetiboG. O.RodriguesD. F. (2020). Aerobic degradation of dichlorinated dibenzo-p-dioxin and dichlorinated dibenzofuran by bacteria strains obtained from tropical contaminated soil. Biodegr31, 123–137. 10.1007/s10532-020-09898-8
56
SalamM.VarmaA. (2019). Bacterial community structure in soils contaminated with electronic waste pollutants from Delhi NCR, India. Electron. J. Biotechnol.41, 72–80. 10.1016/j.ejbt.2019.07.003
57
SandhuM.PaulA. T.JhaP. N. (2022). Metagenomic analysis for taxonomic and functional potential of Polyaromatic hydrocarbons (PAHs) and Polychlorinated biphenyl (PCB) degrading bacterial communities in steel industrial soil. PLoS ONE17 (4), e0266808. 10.1371/journal.pone.0266808
58
SekiT.MatsumotoA.ŌmuraS.TakahashiY. (2015). Distribution and isolation of strains belonging to the order Solirubrobacterales. J. Antibiot.68 (12), 763–766. 10.1038/ja.2015.67
59
ShangeR. S.AnkumahR. O.IbekweA. M.ZabawaR.DowdS. E. (2012). Distinct soil bacterial communities revealed under a diversely managed agroecosystem. PloS one7 (7), e40338. 10.1371/journal.pone.0040338
60
ŠrédlováK.CajthamlT. (2022). Recent advances in PCB removal from historically contaminated environmental matrices. Chemosphere287, 132096. 10.1016/j.chemosphere.2021.132096
61
StorerD. A. (1984). A simple high sample volume ashing procedure for determination of soil organic matter. Comm. Soil Sci. Plant Anal.15, 759–772. 10.1080/00103628409367515
62
SumampouwO. J.RisjaniY. (2014). Bacteria as indicators of environmental pollution: review. Int. J. Ecosyst.4, 251–258. 10.5923/j.ije.20140406.03
63
TangX. J.QiaoJ. N.ChenC.ChenL. T.YuC. N.ShenC. F.et al (2013). Bacterial communities of polychlorinated biphenyls polluted soil around an e-waste recycling workshop. Soil Sediment. Contam.22, 562–573. 10.1080/15320383.2013.750269
64
ThouinH.Battaglia-BrunetF.NoriniM.-P.JoulianC.HellalJ.ForestierL. L.et al (2019). Microbial community response to environmental changes in a technosol historically contaminated by the burning of chemical ammunitions. Sci. Total Environ.697, 134108. 10.1016/j.scitotenv.2019.134108
65
TorsvikV.OvreasL. (2002). Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol.5, 240–245. 10.1016/s1369-5274(02)00324-7
66
US Environmental Protection Agency (2007). Method 8270D Semivolatile organic compounds by gas chromatography/mass spectrometry (GC/MS).
67
VikramA.LipusD.BibbyK. (2016). Metatranscriptome analysis of active microbial communities in produced water samples from the Marcellus Shale. Microb. Ecol.72, 571–581. 10.1007/s00248-016-0811-z
68
VinasM.SabateJ.EspunyM. J.SolanasA. M. (2005). Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl. Environ. Microbiol.71 (11), 7008–7018. 10.1128/aem.71.11.7008-7018.2005
69
WangX.CaoA.ZhaoG.ZhouC.XuR. (2017). Microbial community structure and diversity in a municipal solid waste landfill. Waste Manag.66, 79–87. 10.1016/j.wasman.2017.04.023
70
WuQ.HuW.WangH.LiuP.WangX.HuangB. (2021). Spatial distribution, ecological risk and sources of heavy metals in soils from a typical economic development area, Southeastern China. Sci. Total Environ.780, 146557. 10.1016/j.scitotenv.2021.146557
71
XavierR.NaoiseN. (2014). Spatial ecology of bacteria at the microscale in soil. Plos one9 (1), e87217. 10.1371/journal.pone.0087217
72
XuZ.HansenM. A.HansenL. H.JacquiodS.SorensenS. (2014). Bioinformatic approaches reveal metagenomic characterization of soil microbial community. Plos one9 (4), e93445. 10.1371/journal.pone.0093445
73
YanX.LuoX.ZhaoM. (2016). Metagenomic analysis of microbial community in uranium-contaminated soil. Appl. Microbiol. Biotechnol.100, 299–310. 10.1007/s00253-015-7003-5
74
ZainunM. Y.SimaraniK. (2018). Metagenomics profiling for assessing microbial diversity in both active and closed landfills. Sci. Total Environ.617, 269–278. 10.1016/j.scitotenv.2017.10.266
75
ZhangW.WangH.ZhangR.YuX. Z.QianP. Y.WongM. H. (2010). Bacterial communities in PAH contaminated soils at an electronic waste processing center in China. Ecotoxicol19, 96–104. 10.1007/s10646-009-0393-3
76
ZhaoC.RuanL. (2011). Biodegradation of Enteromorpha prolifera by mangrove degrading micro-community with physical-chemical pretreatment. Appl. Microbiol. Biotechnol.92, 709–716. 10.1007/s00253-011-3384-2
77
ZhaoX.HuangJ.LuJ.SunY. (2019). Study on the influence of soil microbial community on the long-term heavy metal pollution of different land use types and depth layers in mine. Ecotoxicol. Environ. Saf.170, 218–226. 10.1016/j.ecoenv.2018.11.136
Summary
Keywords
soil, dumpsite, bacterial diversity, persistent organic pollutants, heavy metals
Citation
Saibu S, Adebusoye SA, Oyetibo GO and Rodrigues DF (2023) Geochemistry and metagenomics analyses of bacterial community structure in selected waste dumpsites in Lagos Metropolis, Nigeria. Front. Environ. Sci. 11:1304033. doi: 10.3389/fenvs.2023.1304033
Received
28 September 2023
Accepted
13 November 2023
Published
30 November 2023
Volume
11 - 2023
Edited by
Baile Xu, Free University of Berlin, Germany
Reviewed by
Sisay Abebe Debela, Salale University, Ethiopia
Agnieszka Klimkowicz-Pawlas, Institute of Soil Science and Plant Cultivation, Poland
Updates
Copyright
© 2023 Saibu, Adebusoye, Oyetibo and Rodrigues.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sunday A. Adebusoye, sadebusoye@yahoo.com, sadebusoye@unilag.edu.ng
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.