
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 13 November 2023
Sec. Toxicology, Pollution and the Environment
Volume 11 - 2023 | https://doi.org/10.3389/fenvs.2023.1270599
 Wenjie Chen1,2†
Wenjie Chen1,2† Yilin Zuo1,2†
Yilin Zuo1,2† Zhuonan Hou1†
Zhuonan Hou1† Bo Wang3
Bo Wang3 Shangao Xiong4
Shangao Xiong4 Xiaoyan Ding2
Xiaoyan Ding2 Bihui Peng1,2
Bihui Peng1,2 Kaiyun Zhou1,2
Kaiyun Zhou1,2 Ji Li1,2
Ji Li1,2 Rui Liu1,5*
Rui Liu1,5* Guochun Ding1,2
Guochun Ding1,2 Yuquan Wei1,2*
Yuquan Wei1,2* Ting Xu1,2
Ting Xu1,2Bioremediation can be effective method for achieving polycyclic aromatic hydrocarbons (PAHs) degradation in soil contaminated with petroleum. The aim of this study was to compare the effect of Rhodococcus bioaugmentation (BIOE) and biostimulation (BIOS) on dibenzothiophene biodegradation and bacterial community interaction in petroleum-contaminated soils. The findings revealed that compared to natural degradation treatment (NAT) and BIOS, BIOE had the highest dibenzothiophene (DBT) and the majority of DBT degradation occurred within the first 30 days. BIOS had a positive impact in the early stage but an opposite effect in the later stages for degrading DBT. Beta diversity analysis revealed significant differences of bacterial composition among NAT, BIOS, and BIOE. Sequencing results indicated that Bacillus and Paenibacillus were dominant genera involved in DBT degradation. Network analysis revealed co-occurrence patterns and connectivity, with BIOE exhibiting higher connectivity and the highest number of links in BIOS. In summary, Rhodococcus bioaugmentation was the simplest and effective method to enhance the clustering degree of bacterial network and DBT degradation in petroleum-contaminated soil, compared to NAT and BIOS.
Soil contamination caused by petroleum hydrocarbons is a critical environmental issue that has severe consequences for ecosystems and human health (Adipah, 2019). It has been reported that there was extensive occurrence of polycyclic aromatic hydrocarbons (PAHs) in soil contaminated with petroleum, which are hard to be biodegraded than aromatic compounds with single rings or branched hydrocarbons (Rodrigo et al., 2014). Bioremediation is widely used for removal of these pollutants in the environment; however, it is always limited by the existence of indigenous degrading species and abiotic factors such as nutrient availability (Lu et al., 2019). Consequently, it has gained great attention in recent decades for improving the PAHs biodegradation method and efficiency (Ossai et al., 2020).
Soil microbial communities play a crucial role in the degradation of organic contaminants, including PAHs (Duarte et al., 2001). These microbial communities consist of diverse bacterial taxa that possess the metabolic capabilities to break down complex hydrocarbons (Patel et al., 2021). Previous studies have demonstrated the involvement of specific bacterial groups, such as Actinobacteria and Firmicutes, in soil with PAHs pollution (Yuan et al., 2015). However, the dynamics of bacterial communities and their responses to different pollution conditions and bioremediation strategies remain poorly understood. Biostimulation is a kind of bioremediation method by adding specific nutrients, mainly nitrogen (N) and phosphorus (P) to increase the microbial population and metabolic activities, and bioaugmentation is a feasible strategy to enhance the existing microbial community by adding indigenous or commercial microorganisms for soil bioremediation (Hamdi et al., 2007). Thus, it becomes imperative to comprehend the dynamics of soil bacterial communities and their roles in PAHs biodegradation in the process of bioaugmentation and biostimulation.
Microbial interactions and community dynamics within soil ecosystems are essential factors influencing the degradation of organic pollutants (Semple et al., 2007). Microorganisms can form intricate networks of cooperative or competitive relationships, which affect pollutant degradation rates and ecosystem functioning (Liang et al., 2016). Network analysis provides a powerful tool for deciphering the complex interactions and co-occurrence patterns among soil bacterial taxa (Barberán et al., 2012). Therefore, identifying keystones or functional groups within these networks can offer insights into the microbial consortia driving PAHs degradation. Furthermore, investigating the structural properties of these networks can reveal the resilience and stability of microbial communities during PAHs degradation, which provides insights into the functional potential and metabolic versatility of microbial communities and helps to the development of strategies to manipulate microbial interactions.
Dibenzothiophene (DBT) as one of persistent and toxic aromatic compounds in the family of condensed thiophenes containing sulphur, is commonly used as a model compound in the PAHs biodegradation studies (Davoodi-Dehaghani et al., 2010; Rodrigo et al., 2014). Therefore, the aim of this study was to (1) investigate the influence of exogenous microbial agents and nutrients on the degradation of DBT in oil contaminated soil; (2) assess the response of soil bacterial communities in bioaugmentation and biostimulation; (3) identify key DBT-degrading bacteria and examine their interactions within the bacterial networks in soil. This research provides the potential bacterial resources and sustainable approaches for petroleum-contaminated soil bioremediation.
A sandy loam soil sample was collected from the 0–10 cm soil layer with a previous history of oil contamination in an oil field of Shanxi, stored at 4°C and transferred to laboratory. The main characteristics of soil sample was provided in Supplementary Table S1.
The DBT degrader was isolated from sandy loam soil samples contaminated with oil and purified using liquid and solid mineral salt medium with increasing DBT concentration (100, 200, 300, and 400 mg/L) according to the method of Jiang et al. (2022). The isolated cultivable DBT degrader was identified as Rhodococcus sp. CGMCC 24903 and preserved in the ecological engineering laboratory of China Agricultural University and China General Microbiological Culture Collection Center. Prior to experimentation, the strain was cultured in liquid LB medium at 180 rpm for 3 days to 109 cells/mL determined by the microplate method (Estrada-Bonilla et al., 2017), and subsequently centrifuged bacterial cell was used as inoculum for further study on DBT degradation microcosms.
Three treatments with 1,000 g soil were carried out to study the effect of Rhodococcus bioaugmentation and biostimulation on DBT biodegradation performance in soils and experiment lasted 40 days (Liu et al., 2023). The natural degradation treatment (NAT) consisted of original soil. The biological augmentation treatment (BIOE) involved soil combined with Rhodococcus sp. CGMCC 24903 inoculation at the total populations of 109 cells/g soil. The biostimulation treatment (BIOS) included soil with optimizing nutrient levels by adding nutrient solution, e.g., CH4N2O and NaH2PO3 to a final C:N:P ratio of 100:10:1, with concentrations of C at 24.36 mg/g, N at 2.427 mg/g, and P at 0.24 mg/g. (Xu and Lu, 2010). The soil used in Microcosms was air-dried, and particles larger than 2 mm were removed to achieve soil homogeneity. DBT in the three treatments was set at a final concentration of 100 mg/kg. The microcosms were incubated at 25°C ± 1°C for 40 days in the dark in triplicates, with soil moisture maintained at 20% through periodic addition of distilled water. 50 g of soils were sampled on day 0, 10, 20, 30, and 40. All samples were stored at −20°C until further physicochemical and DNA analysis.
All solvents used were laboratory grade chemicals obtained from Beijing Chemical Company (China). Dibenzothiophene (CAS 132-65-0, purity >99%) were obtained from Sigma Aldrich. Deionized water was used to prepare all media and solutions.
DBT analysis was performed by extracting 5 g of soil using ethyl acetate at a ratio of 1 mL per Gram of soil. Each soil-solvent mixture was sonicated for 1 h, followed by nitrogen drying and resuspension of the residue in 4 mL of the same solvent. Prior to GC-MS analyses (Agilent 7890A-5975C) using a TG-5SILMS column (30 × 0.25 mm, 0.25 μm), samples were filtered (0.22 μm filter, Pall). The GC oven temperature was programmed to increase from 50°C to 250°C at a rate of 15°C per minute, and held at 250°C for 2 min. The injector temperature was set to 250°C. The DBT determination were conducted in triplicate, and the mean values were taken as the results.
Total DNA of soil samples was extracted using the FastDNA spin Kit for soil (MP Biomedicals, Santa Ana, Carlsbad, CA, United States). PCR amplification of 16S rRNA gene fragments was performed using the primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). The PCR products were sequenced in a 2 × 300-bp paired-end format on the Illumina MiSeq platform, which was conducted by Majorbio BioPharm Technology Co. Ltd. (Shanghai, China). The sequences were deposited in the NCBI database under the project accession number PRJNA914518.
The raw 16S rRNA gene sequencing reads were demultiplexed and quality-filtered using fastp version 0.20.0 (Chen et al., 2018), and then merged using FLASH version 1.2.7 (Magoč and Salzberg, 2011). Operational taxonomic units (OTUs) with a 97% similarity cutoff were clustered using UPARSE version 7.1 (Stackebrandt and Goebel, 1994; Edgar, 2013), and chimeric sequences were identified and removed. The taxonomic classification of each OTU representative sequence was determined using RDP Classifier version 2.2 (Wang et al., 2007) with a confidence threshold of 0.7, based on the silva reference database (Quast et al., 2012).
The basic indicators were visualized using Origin Pro 2022. Heatmaps were generated using the pheatmap package in R 4.1.3. Bacterial diversity metrics, including Shannon diversity, Pielou’s evenness, Chao1 index, and Faith’s phylogenetic diversity, were estimated using the Picante and vegan packages. Principal coordinates analysis (PCoA) was conducted based on Bray-Curtis dissimilarity index matrices (Sheng et al., 2023).
Correlation network analysis was conducted to ascertain the role of core microorganisms in DBT degradation. Based on Pearson correlation coefficients, bacterial taxonomic groups at the OTU level that exhibited significant correlations with DBT degradation (p < 0.05, r > 0.97) were selected as core microorganisms, and the visualization was carried out using Cytoscape 3.9.1. Co-occurrence networks at the OTU level were conducted using Molecular Ecology Network Analysis with a cutoff value of 0.85. The networks were visualized using Gephi 0.9.2. Keystones, including connectors, network hubs, and module hubs, were categorized based on within-module connectivity (Zi) and among-module connectivity (Pi) in the network, following the classification proposed by (Deng et al., 2012).
Gas chromatographic analysis revealed that the degradation efficiency of DBT in BIOE was significantly higher than that in NAT and BIOS over a period of 40 days (Figure 1). Within the first 20 days of soil incubation, the DBT degradation efficiency in BIOE and BIOS was significantly higher compared to natural degradation, suggesting that both Rhodococcus bioaugmentation and biostimulation could enhance the initial biodegradation ability of soil microbes. In the subsequent 20 days of incubation, though bioaugmentation group still had the highest degradation rate and increased to ∼60% at day 40, the DBT degradation performance in BIOS was lower than that in NAT. After day 30, the degradation level of the three treatments was gradually flatten, suggesting that the majority of DBT degradation occurred within the first 30 days. Biostimulation had only a positive impact at the early stage of soil incubation, which could be attributed to the proliferation of microorganisms capable of degrading DBT that were promoted by the additional nutrients. However, during the later stage of soil incubation, the nutrients were depleted and the dominant species might be changed to unfavorable conditions for DBT-degrading microorganisms, which reflected the complex interaction between microbial populations and nutrient availability (Fuhrman et al., 2015). The above results suggested that the addition of inoculum (Rhodococcus) for bioaugmentation significantly stimulated the biodegradation of DBT and improved the bioremediation performance in petroleum-contaminated soils.
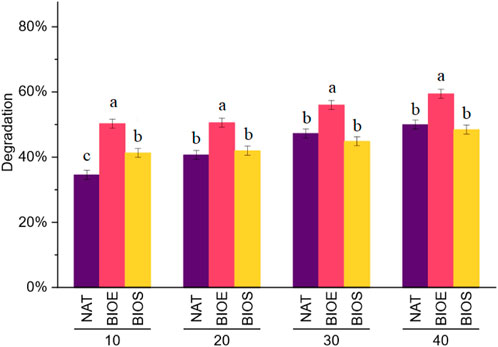
FIGURE 1. Degradation profile of dibenzothiophene in the soil. NAT: natural degradation treatment, BIOE: biological augmentation treatment, BIOS: biostimulation treatment. Different letters indicate significant difference at p < 0.05.
After the optimization of sequencing, a total of 1,125,468 bacterial reads of the 16S rRNA gene were obtained, which were subsequently clustered into 1,362 bacterial operational taxonomic units (OTUs). Among these OTUs, the NAT, BIOE, and BIOS treatments had 119, 6 and 42 unique OTUs, respectively, and had 344 OTUs shared among all treatments (Figure 2A). Firmicutes and Actinobacteria were identified as the dominant phyla in all treatments (Figure 2B). Notably, the addition of nutrients increased the relative abundance of Firmicutes, while reduced the levels of Actinobacteria. To gain deeper insights into the dynamic changes within the bacterial community, a heatmap analysis was performed on the 20 most abundant genera (Figure 2C). Based on the relative abundance, the genera were classified into two clusters. Bacillus, Micromonospora, Streptomyces, Bhargavaea, and Chungangia formed one cluster, while Sporosarcina, Sphingobacterium, Paracoccus, and others clustered together, indicating potential functional similarities or ecological interactions among these taxa (Bao et al., 2023). Bacillus and Streptomyces were dominant genera during soil incubation, which had the ability to degrade dibenzothiophene through their participation in the 4S pathway (Buzanello et al., 2014). This finding highlights their potential as key DBT-degrading bacteria in the remediation of petroleum-contaminated soil. Surprisingly, Rhodococcus genus was not shown in the heatmap even though it was inoculated in the BIOE, which might be due to its low relative abundance in soil and its colonization related to the concentration of PAHs (Cunliffe and Kertesz, 2006).
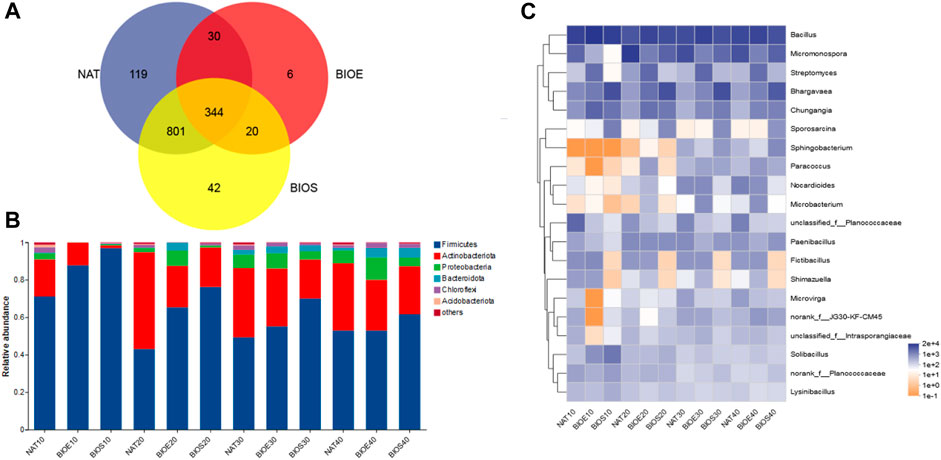
FIGURE 2. Variation of bacterial communities in soil during incubation with different treatments. (A) The quantity difference of common and unique OTUs. (B) Relative abundance of bacterial phylum. (C) Relative abundance of genera (top 20). NAT: natural degradation treatment, BIOE: biological augmentation treatment, BIOS: biostimulation treatment.
The alpha diversity of soil bacterial community during the incubation periods was shown in Figure 3A. The Shannon diversity of the NAT treatment was significantly higher than that of the BIOE and BIOS treatments on the 10th day of incubation. However, after the 40 days of incubation, there were no significant differences in Shannon diversity among the three treatments. During the incubation period, the BIOE treatment exhibited significantly lower Chao1 richness and phylogenetic diversity compared to the other treatments. However, the Simpson evenness of the BIOE treatment was significantly higher than that of the other treatments. The BIOS treatment showed higher Simpson evenness but lower Chao1 richness and phylogenetic diversity compared to the NAT treatment during the incubation period. The above results indicated that Rhodococcus bioaugmentation increased the dominance of species in petroleum-contaminated soil while reducing the richness of total bacterial community. principal coordinates analysis (PCoA) based on Bray-Curtis distance matrix was conducted to visualize the succession patterns of soil bacterial communities in diverse groups (Figure 3B). The first two principal components accounted for 74.7% of the community variance. Notably, the bacterial communities during incubation in all samples were divided into three classes, indicating dissimilar bacterial composition in the microcosms of NAT, BIOE and BIOS. The results highlighted the significant impact of different treatments on the beta diversity of bacterial communities, especially the critical role of nutrient addition and inoculation on bacterial community composition in the degradation process of DBT during soil incubation (Strickland et al., 2009; Wang et al., 2018).
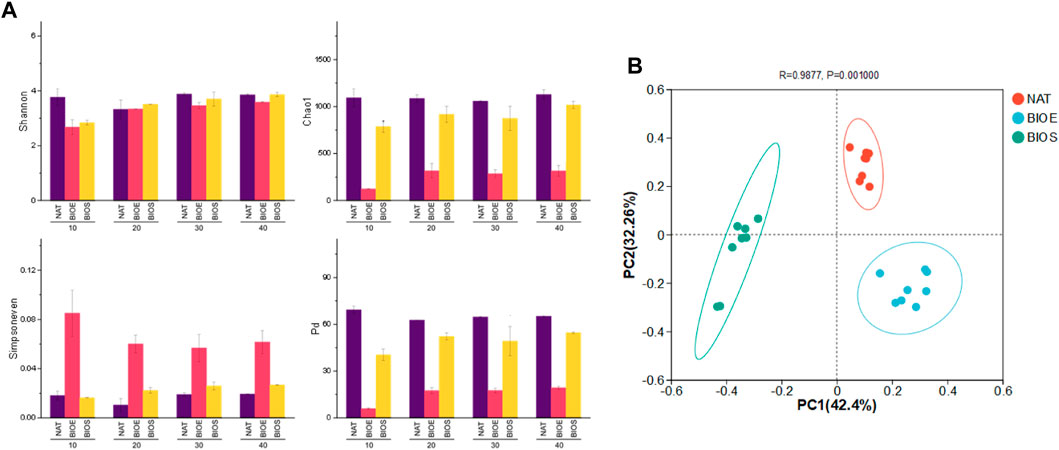
FIGURE 3. (A) Alpha diversity indices including Shannon diversity, Chao1 richness, Simpson evenness and Faith’s phylogenetic diversity (PD) of bacterial communities during incubation in different treatments. (B) The principal coordinates analysis (PCoA) of bacterial communities in different treatments. NAT: natural degradation treatment, BIOE: biological augmentation treatment, BIOS: biostimulation treatment.
The degradation of DBT was inevitably influenced by the dynamics of the bacterial community and their interaction during soil incubation process. Here, network analysis was employed to identify the key microbial taxa at the OTU level that were associated with DBT degradation, which were screened based on significant correlations with DBT degradation (p < 0.05). In NAT, BIOE, and BIOS groups, there were 15, 1, and 7 nodes, respectively, significantly participating in the DBT degradation process (Figure 4). The OTU1291 (Shimazuella), OTU1532 (Paenibacillus) in NAT, OTU 1851 (Bacillus) in BIOE and OTU1292 (Bacillus), OTU1322 (Bacillus) in BIOS were not only the core OTUs involved in DBT degradation but also belonged to the top 20 bacterial genera during soil incubation (Figure 2C; Supplementary Table S2). Previous studies also reported that Shimazuella and Bacillus had strong capabilities in DBT degradation (Buzanello et al., 2014).
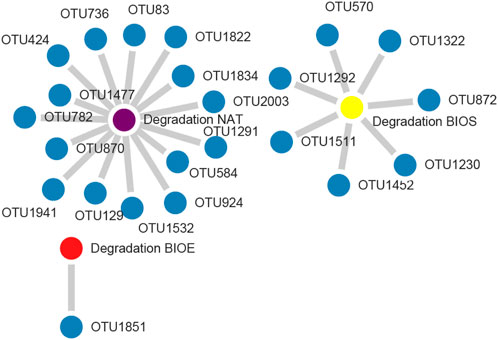
FIGURE 4. Network analysis of dibenzothiophene degradation and related bacterial OTUs according to Pearson correlation analysis (p < 0.05, r > 0.97) in incubation. NAT: natural degradation treatment, BIOE: biological augmentation treatment, BIOS: biostimulation treatment.
The bacterial interactions under different treatments were investigated by constructing co-occurrence networks based on Pearson correlations among OTUs (Figure 5A). All co-occurrence networks demonstrated general network properties of scale-free, small-world, and modularity (Supplementary Table S3), with the network topology following a power-law distribution (R2 = 0.886, 0.756, 0.804 in NAT, BIOE and BIOS, respectively). Compared to other treatments, BIOE network showed fewer nodes (163) and links (606), but displayed higher average degree (avgK) and average clustering coefficient clustering coefficient (avgCC), as well as lower average path distance (GD) and average clustering coefficient (HD). These findings suggested that the BIOE network was more tightly connected, and external disturbances could quickly propagate throughout the entire network. Compared to NAT, the BIOS network also exhibits tight connectivity based on its topological characteristics, with the highest number of links (2467). The correlations between bacterial species in the network varied, with NAT showing a higher percentage of negative correlations (45.85%) compared to BIOE (16.17%) and BIOS (35.63%). These findings collectively indicated significant differences in the networks among the three treatments, and more bacteria in BIOE and BIOS groups showed strongly cooperative symbiosis to the environmental pressure (DBT pollution) (Zhang et al., 2020).
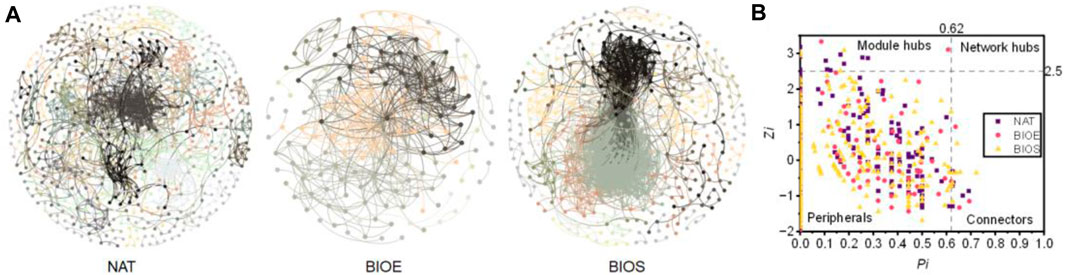
FIGURE 5. (A) The co-occurrence networks of different treatments. The different colors represent the network modules. (B) The Zi-Pi plot of network nodes. The topological role of each node was assessed based on the scatter plot of within-module connectivity (Zi) and among-module connectivity (Pi). NAT: natural degradation treatment, BIOE: biological augmentation treatment, BIOS: biostimulation treatment.
Based on the analysis of within-module connectivity (Zi) and among-module connectivity (Pi) (Olesen et al., 2007), a total of 14, 2, and 4 module hubs were observed in the NAT, BIOE, and BIOS networks, respectively, along with 6, 4, and 8 connectors (Figure 5B; Supplementary Table S4). However, no network hubs were identified in these groups. More key nodes in the NAT network indicated its higher complexity. All these nodes represented keystones that played crucial roles in the network. The 20 keystones in the NAT network mainly belonged to Firmicutes, Proteobacteria, and Actinobacteriota, the 6 keystones in the BIOE network belonged to Firmicutes, Actinobacteriota, and Proteobacteria, and the 12 keystones in the BIOS network were predominantly from Myxococcota, Firmicutes, and Actinobacteriota. Members of Firmicutes and Actinobacteriota were the predominant keystone taxa during the incubation process of petroleum-contaminated soil, which accounted for approximately 70% of all module hubs and connectors. Furthermore, key microorganisms belonging to the genera Paenibacillus (OTU1532, OTU861, OTU 2001, OTU1317, OTU1325, OTU206, OTU1390) and Bacillus (OTU1020, OTU1696) related to DBT degradation were also identified as keystones in bacterial interaction network, suggesting that these microorganisms might be indigenous bacteria with a potential function of DBT degradation by regulating core bacterial collaborative symbiosis in the co-occurrence network. The above results indicated that Rhodococcus bioaugmentation (BIOE) was the simplest and effective method to enhance the clustering degree of bacterial community network compared to natural degradation for enhancing DBT degradation in petroleum-contaminated soil. Additionally, nutrient addition (BIOS) reduces the complexity of bacterial community in petroleum-contaminated soil without obviously enhancing DBT degradation capability.
The study compared the degradation of DBT and the dynamics of bacterial communities in the petroleum-contaminated soil incubation of diverse treatments (NAT, BIOE and BIOS). Rhodococcus bioaugmentation exhibited higher DBT degradation efficiency (∼60%) compared to other groups after 40 days. The addition of nutrients exhibited an initial increase followed by a decrease in the degradation efficiency of DBT with no obvious advantage compared to NAT. The dominant phyla in petroleum-contaminated soil were Firmicutes and Actinobacteria, and biostimulation significantly influenced their relative abundances. Bacillus and Paenibacillus played crucial roles in DBT degradation. There were distinct co-occurrence patterns and connectivity among bacterial species in NAT, BIOE and BIOS. Rhodococcus bioaugmentation was the simplest and effective method to enhance the clustering degree of bacterial community network compared to NAT for enhancing DBT degradation in petroleum-contaminated soil.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
WC: Data curation, Formal Analysis, Writing–original draft. YZ: Data curation, Writing–original draft. ZH: Data curation, Methodology, Writing–original draft. BW: Resources, Validation, Writing–original draft. SX: Formal Analysis, Resources, Writing–original draft. XD: Data curation, Methodology, Writing–original draft. BP: Data curation, Formal Analysis, Writing–original draft. KZ: Data curation, Formal Analysis, Methodology, Writing–original draft. JL: Funding acquisition, Supervision, Writing–review and editing. RL: Funding acquisition, Project administration, Resources, Supervision, Visualization, Writing–review and editing. GD: Methodology, Resources, Software, Writing–review and editing. YW: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing–original draft, Writing–review and editing. TX: Writing–review and editing, Conceptualization, Supervision, Validation, Resources, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work is financially supported by the Key Project of Inter-Governmental International Scientific and Technological Innovation Cooperation (2019YFE0115800), National Key Research and Development Program of China (2020YFC1807901), the Open Research Fund Program of State Environmental Protection Key Laboratory of Food Chain Pollution Control (FC2022YB01), the National Natural Science Foundation of China (32071552), and the Independent Research Project of Science and Technology Innovation Base in Tibet Autonomous Region (XZ2022JR0007G).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1270599/full#supplementary-material
Adipah, S. (2019). Introduction of petroleum hydrocarbons contaminants and its human effects. J. Environ. Sci. Public Health 3 (1), 1–9. doi:10.26502/jesph.96120043
Bao, S., Zhao, L., Liu, Y., Yang, L., Liu, P., Chen, C., et al. (2023). Influencing factors of bioaugmentation treatment of PAH-contaminated soils in slurry bioreactors. J. Environ. Chem. Eng. 11 (3), 109893. doi:10.1016/j.jece.2023.109893
Barberán, A., Bates, S. T., Casamayor, E. O., and Fierer, N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 6 (2), 343–351. doi:10.1038/ismej.2011.119
Buzanello, E. B., Rezende, R. P., Sousa, F. M. O., de Lima Silva Marques, E., and Loguercio, L. L. (2014). A novel Bacillus pumilus-related strain from tropical landfarm soil is capable of rapid dibenzothiophene degradation and biodesulfurization. Bmc Microbiol. 14 (1), 257–310. doi:10.1186/preaccept-1987262613115981
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi:10.1093/bioinformatics/bty560
Cunliffe, M., and Kertesz, M. (2006). Effect of Sphingobium yanoikuyae B1 inoculation on bacterial community dynamics and polycyclic aromatic hydrocarbon degradation in aged and freshly PAH-contaminated soils. Environ. Pollut. 144 (1), 228–237. doi:10.1016/j.envpol.2005.12.026
Davoodi-Dehaghani, F., Vosoughi, M., and Ziaee, A. (2010). Biodesulfurization of dibenzothiophene by a newly isolated Rhodococcus erythropolis strain. Bioresour. Technol. 101 (3), 1102–1105. doi:10.1016/j.biortech.2009.08.058
Deng, Y., Jiang, Y.-H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinforma. 13, 113–120. doi:10.1186/1471-2105-13-113
Duarte, G. F., Rosado, A. S., Seldin, L., de Araujo, W., and van Elsas, J. D. (2001). Analysis of bacterial community structure in sulfurous-oil-containing soils and detection of species carrying dibenzothiophene desulfurization (dsz) genes. Appl. Environ. Microbiol. 67 (3), 1052–1062. doi:10.1128/aem.67.3.1052-1062.2001
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 (10), 996–998. doi:10.1038/nmeth.2604
Estrada-Bonilla, G. A., Lopes, C. M., Durrer, A., Alves, P. R., Passaglia, N., and Cardoso, E. J. (2017). Effect of phosphate-solubilizing bacteria on phosphorus dynamics and the bacterial community during composting of sugarcane industry waste. Syst. Appl. Microbiol. 40 (5), 308–313. doi:10.1016/j.syapm.2017.05.003
Fuhrman, J. A., Cram, J. A., and Needham, D. M. (2015). Marine microbial community dynamics and their ecological interpretation. Nat. Rev. Microbiol. 13 (3), 133–146. doi:10.1038/nrmicro3417
Hamdi, H., Benzarti, S., Manusadžianas, L., Aoyama, I., and Jedidi, N. (2007). Bioaugmentation and biostimulation effects on PAH dissipation and soil ecotoxicity under controlled conditions. Soil Biol. Biochem. 39 (8), 1926–1935. doi:10.1016/j.soilbio.2007.02.008
Jiang, B., Chen, Y., Xing, Y., Lian, L., Shen, Y., Zhang, B., et al. (2022). Negative correlations between cultivable and active-yet-uncultivable pyrene degraders explain the postponed bioaugmentation. J. Hazard. Mater. 423, 127189. doi:10.1016/j.jhazmat.2021.127189
Liang, Y., Zhao, H., Deng, Y., Zhou, J., Li, G., and Sun, B. (2016). Long-term oil contamination alters the molecular ecological networks of soil microbial functional genes. Front. Microbiol. 7, 60. doi:10.3389/fmicb.2016.00060
Liu, Q., Sun, S., Chen, S., Su, Y., Wang, Y., Tang, F., et al. (2023). A novel dehydrocoenzyme activator combined with a composite microbial agent TY for enhanced bioremediation of crude oil-contaminated soil. J. Environ. Manag. 331, 117246. doi:10.1016/j.jenvman.2023.117246
Lu, C., Hong, Y., Liu, J., Gao, Y., Ma, Z., Yang, B., et al. (2019). A PAH-degrading bacterial community enriched with contaminated agricultural soil and its utility for microbial bioremediation. Environ. Pollut. 251, 773–782. doi:10.1016/j.envpol.2019.05.044
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27 (21), 2957–2963. doi:10.1093/bioinformatics/btr507
Olesen, J. M., Bascompte, J., Dupont, Y. L., and Jordano, P. (2007). The modularity of pollination networks. Proc. Natl. Acad. Sci. 104 (50), 19891–19896. doi:10.1073/pnas.0706375104
Ossai, I. C., Ahmed, A., Hassan, A., and Hamid, F. S. (2020). Remediation of soil and water contaminated with petroleum hydrocarbon: a review. Environ. Technol. Innovation 17, 100526. doi:10.1016/j.eti.2019.100526
Patel, A. B., Manvar, T., Jain, K. R., Desai, C., and Madamwar, D. (2021). Metagenomic insights into bacterial communities’ structures in polycyclic aromatic hydrocarbons degrading consortia. J. Environ. Chem. Eng. 9 (6), 106578. doi:10.1016/j.jece.2021.106578
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41 (D1), D590–D596. doi:10.1093/nar/gks1219
Rodrigo, J., Boltes, K., and Esteve-Nuñez, A. (2014). Microbial-electrochemical bioremediation and detoxification of dibenzothiophene-polluted soil. Chemosphere 101, 61–65. doi:10.1016/j.chemosphere.2013.11.060
Semple, K. T., Doick, K. J., Wick, L. Y., and Harms, H. (2007). Microbial interactions with organic contaminants in soil: definitions, processes and measurement. Environ. Pollut. 150 (1), 166–176. doi:10.1016/j.envpol.2007.07.023
Sheng, Y., Baars, O., Guo, D., Whitham, J., Srivastava, S., and Dong, H. (2023). Mineral-bound trace metals as cofactors for anaerobic biological nitrogen fixation. Environ. Sci. Technol. 57 (18), 7206–7216. doi:10.1021/acs.est.3c01371
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44 (4), 846–849. doi:10.1099/00207713-44-4-846
Strickland, M. S., Lauber, C., Fierer, N., and Bradford, M. A. (2009). Testing the functional significance of microbial community composition. Ecology 90 (2), 441–451. doi:10.1890/08-0296.1
Wang, H., Liu, S., Zhang, X., Mao, Q., Li, X., You, Y., et al. (2018). Nitrogen addition reduces soil bacterial richness, while phosphorus addition alters community composition in an old-growth N-rich tropical forest in southern China. Soil Biol. Biochem. 127, 22–30. doi:10.1016/j.soilbio.2018.08.022
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 (16), 5261–5267. doi:10.1128/aem.00062-07
Xu, Y., and Lu, M. (2010). Bioremediation of crude oil-contaminated soil: comparison of different biostimulation and bioaugmentation treatments. J. Hazard. Mater. 183 (1-3), 395–401. doi:10.1016/j.jhazmat.2010.07.038
Yuan, J., Lai, Q., Sun, F., Zheng, T., and Shao, Z. (2015). The diversity of PAH-degrading bacteria in a deep-sea water column above the Southwest Indian Ridge. Front. Microbiol. 6, 853. doi:10.3389/fmicb.2015.00853
Keywords: mesocosm, soil bioremediation, polycyclic aromatic hydrocarbons (PAHs), microbial community, molecular ecology network
Citation: Chen W, Zuo Y, Hou Z, Wang B, Xiong S, Ding X, Peng B, Zhou K, Li J, Liu R, Ding G, Wei Y and Xu T (2023) Effect of Rhodococcus bioaugmentation and biostimulation on dibenzothiophene biodegradation and bacterial community interaction in petroleum-contaminated soils. Front. Environ. Sci. 11:1270599. doi: 10.3389/fenvs.2023.1270599
Received: 01 August 2023; Accepted: 01 November 2023;
Published: 13 November 2023.
Edited by:
Fayuan Wang, Qingdao University of Science and Technology, ChinaReviewed by:
Robin Brigmon, Savannah River National Laboratory (DOE), United StatesCopyright © 2023 Chen, Zuo, Hou, Wang, Xiong, Ding, Peng, Zhou, Li, Liu, Ding, Wei and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuquan Wei, d2VpeXEyMDE5QGNhdS5lZHUuY24=, d2VpeXEyMDEzQGdtYWlsLmNvbQ==; Rui Liu, cmxpdUBjYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.