
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 30 June 2023
Sec. Soil Processes
Volume 11 - 2023 | https://doi.org/10.3389/fenvs.2023.1210349
 Wenjuan Zhang*
Wenjuan Zhang*Introduction: Although soil microbial populations are a good predictor of soil texture, little is known about how they react to alpine meadow deterioration.
Methods: This study utilized Illumina HiSeq sequencing to investigate the effects of alpine meadow degradation on soil microbial communities in the Yangtze River source basin at five different degradation levels [i.e., non–degraded (ND), slightly degraded (LD), moderately degraded (MD), severely degraded (SD), and very severely degraded (VD)].
Results: The results indicated that bacterial and fungal α-diversity were not substantially different (p > 0.05) across the damaged alpine meadows, while β-diversity significantly differed (p < 0.01), indicating a higher variation in the microbial community due to alpine meadow degradation. Proteobacteria reduced considerably (p < 0.05) by 8.75%, 22.99%, and 24.65%, while Acidobacteria increased significantly (p < 0.05) by 41.67%, 85.20%, and 108.67%, in MD, SD, and VD compared with ND, respectively. Ascomycota declined significantly (p < 0.05) in the MD, whereas unclassified phyla rose significantly (p > 0.05) in the VD compared to the ND. The heatmaps of bacterial and fungal communities revealed two clusters: a ND, LD, and MD group and a SD and VD group, suggesting significant changes in soil microorganisms of alpine meadow in the SD and VD. Redundancy analysis (RDA) revealed that soil moisture, soil bulk density, soil organic carbon, total nitrogen, and plant biomass could explain 73.8% and 39.4% of the variance in bacterial and fungal community structure, respectively.
Discussion: These findings imply that degradation of the alpine meadow impacts both plant and soil qualities, ultimately leading to changes in soil microbial populations in the Yangtze River’s source region.
The Qinghai-Tibet Plateau (QTP) is known as the “Roof of the World” and has an average altitude of more than 4,000 m (Qiu, 2008). Alpine meadows, accounting for about 1/3 of all grasslands, are crucial to Tibetans and their livestock as a primary ecosystem and food source (Xue et al., 2017). These meadows play a significant role in the QTP by storing water (Zhang et al., 2013), conserving biodiversity (López-Pujol et al., 2011), and sequestering carbon (Ding et al., 2016). Unfortunately, climate change, overgrazing, and rodents have damaged approximately 90% of the alpine meadow on the Qinghai–Tibet Plateau (Xue et al., 2009; Harris, 2010; Peng et al., 2018), leading to alterations in plant communities’ total cover, average height, and above-ground biomass, as well as accelerated soil erosion (Niu et al., 2021; Liu et al., 2022a; Liu et al., 2022b). This degradation also endangers herds’ survival and impedes animal husbandry’s sustainable development (Wu et al., 2014).
Soil microorganisms play an important role in nutrient effectiveness and soil quality, and their vulnerability to disturbances has been shown (Allison and Martiny, 2008; Xue et al., 2009; Zhang et al., 2016). Ecosystem processes such as, litter decomposition, CO2 flux, nitrification, and denitrification are directly impacted by the composition of microbial decomposers since microbes use soil organic matter differently (Liu et al., 2021). Soil microbial community is influenced by soil properties and plant characteristics (Chen et al., 2012; Kumar et al., 2016), including a plant’s type that affects the physical environment and nutrient availability (Chu et al., 2011). Soil substrate availability and heterogeneity also affect microbial communities (Qi et al., 2017). Degradation of grasslands can alter soil nutrients, moisture, plant composition, and biomass, leading to changes in soil microbial communities (Dong et al., 2012; Zhang XF. et al., 2014; Wu et al., 2014). In previous studies on alpine meadow degradation, plant and soil characteristics have been mainly examined, while soil microbial biomass has only been assessed in a few instances (Li et al., 2016; Che et al., 2019; Yu et al., 2019). Studies on soil bacteria under different degradation levels in alpine meadows are rare (Zhou et al., 2019), highlighting the need for a better understanding of how meadow degradation affects soil microbial communities and their diversity. It is essential to understand how alpine meadow declination affects soil microorganisms in the Yangtze River source region and consider the key influencing factors acting on them. The study of soil microbial features in different degeneration levels can shed light on the process and mechanism of alpine meadow degeneration.
We propose to investigate the possibility that changes in bacterial and fungal communities in alpine meadow soils are induced by both vegetation and soil deterioration. As a result, we used a “space-for-time” replacement strategy to examine the characteristics of vegetation biomass, soil physicochemical parameters, and microorganisms in alpine meadows at various stages of deterioration. The goal of our study was to analyze changes in bacterial and fungal communities in degraded meadows and to find factors that have a substantial influence on the organization of microbial communities in degraded alpine meadows to illustrate this.
This study area, Beiluhe, is situated at the Yangtze River’s source (34°49.747′N, 92°56.109′E; 4,628 m) as shown in Figure 1. The climate is distinct seasons, and the cold season lasts for 8 months (October to May) (Jiang et al., 2018), showing an annual average temperature of −4.2∼ −2.8°C from 2005 to 2014, with an average value of −3.5°C, an annual average precipitation of 232–467 mm, accounting for over 90% of annual rainfall (Lin et al., 2020). The vegetation cover is sparse, and the species diversity is poor. Purple-flowered needlegrass is the dominant population in the grassland area, while alpine artemisia and Tibetan artemisia are the dominant populations in the meadow area (Jiang et al., 2018).
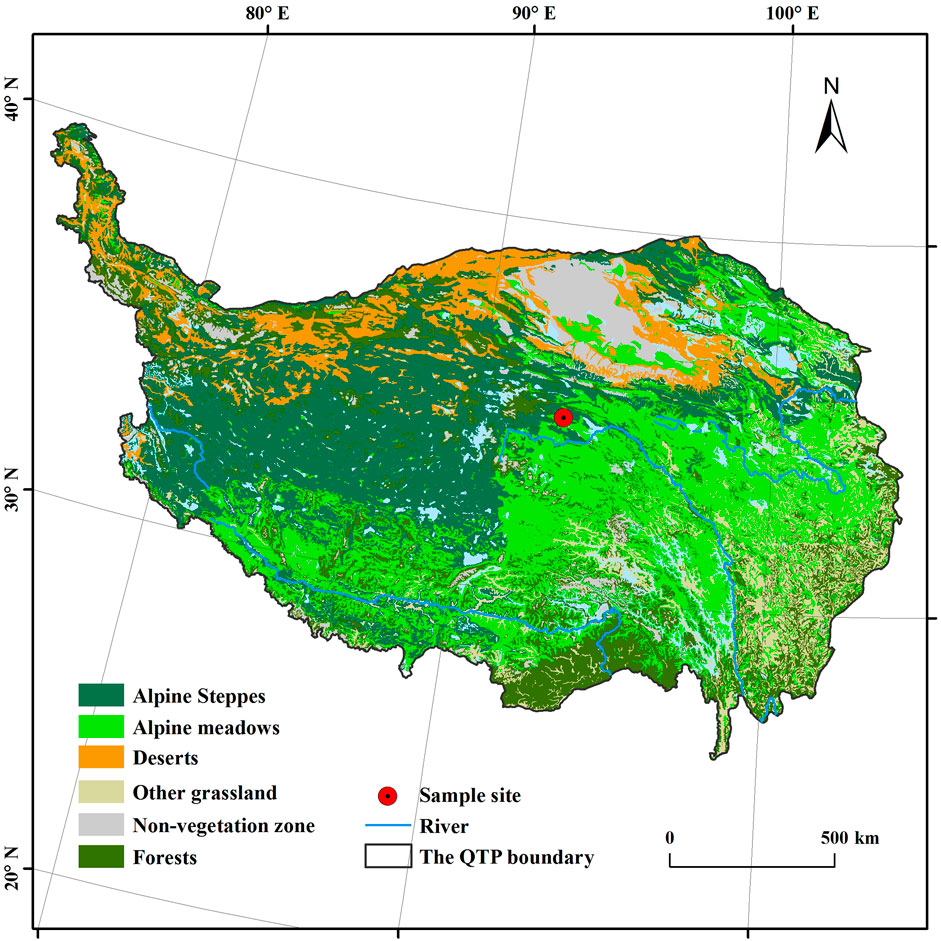
FIGURE 1. Sample site location (data provided by West China Environmental and Ecological Science Data Center, National Natural Science Foundation of China, http://westdc.westgis.ac.cn, based on the vegetation atlas of China 1:1,000,000).
When selecting ND, LD, MD, SD, and VD alpine meadows (Table 1, see abbreviations Supplementary Table S1), vegetation cover and pasture quality were taken into consideration. Plant species in different degradation levels are presented in Supplementary Table S2.
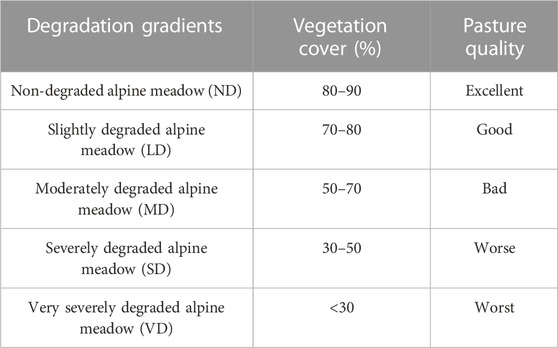
TABLE 1. Classification of alpine meadow degradation (Yushou et al., 2002; Liu et al., 2018).
In July 2019, a random plot (5 m × 5 m) was selected for each degraded level of alpine meadow near the Beiluhe observation station with over 50 m intervals. Three replicates were sampled from each plot. Plant coverage of each plot was determined by point-intercept sampling, using a 30 cm × 30 cm square frame (100 equidistantly spaced sampling points) (Wang et al., 2012). Aboveground biomass was collected by trimming it at the surface of the ground and putting it in paper bags, while belowground plant biomass was taken using soil steel augers to a depth of 10 cm (inner diameter = 50 mm). Soil samples were collected from each data plots (three replicates), stored in a cooler box, then transported to the lab. Soil samples were screeened through a 2 mm mesh (Xiong et al., 2014). Subsamples of these fresh soil samples were archived at −80°C for total DNA extraction. Fifteen individual samples were obtained from each plot (three replicates) and labeled ND1, ND2, ND3, LD1, LD2, LD3, MD1, MD2, MD3, SD1, SD2, SD3, VD1, VD2, and VD3 before being sent to Biomarker Bioinformatics Technology Co., Ltd., Beijing, China (http://www. biomarker. bioon.com.cn) for analysis. Subsamples of the fresh soil samples were archived at 4°C to measure soil pH, while other subsamples were air-dried to measure soil organic carbon (SOC), total nitrogen (TN), phosphorus (TP), total potassium (TK), ammonium-nitrogen (NH4+-N), nitrate-nitrogen (NO3−-N), available phosphorus (AP), and available potassium (AK) (Wang et al., 2007; He et al., 2016). Bulk density (BD) samples were taken using a 5.0 cm cylindrical sampler, and soil moisture (SM) samples were collected in aluminum boxes (v = 100 cm3).
Plant biomass was measured after 48 h drying at 65°C. Soil pH measurements were taken with glass electrodes using a soil/water ratio of 1/2.5 (weight/volume). Soil moisture (SM) was estimated through drying soil samples, at 105°C, 48 h. Soil organic carbon (SOC) was determined by the K2Cr2O7-H2SO4 (Zeng et al., 2013), while soil TN determined by the semimicro Kjeldahl digestion procedure (You et al., 2014). The NH4+-N, NO3−-N, TP, AP, TK and AK content of typic alpine meadow soil were determined using fire atomic absorption spectrophotometry (Purkinje General Instrument Co., Ltd., Beijing, China).
The PowerSoil DNA Isolation Kit was used to extract DNA from 0.5 g of soil samples (MO BIO Laboratories, Carlsbad, CA, United States). A Nanodrop spectrophotometer (ND-1000, NanoDrop Technologies, United States) was used to measure the quantity and quality of DNA.
The 338F primers (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Dennis et al., 2013) were used to amplify the 16S rRNA gene, while ITS1F primer (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2 primer (5′-A-TCCTCCGCTTATTGATATGC-3′) were employed to amplify the ITS1 region of fungi (White et al., 1990). Polymerase chain reaction (PCR) amplification was conducted in a 25 μL reaction system using TransGen AP221-02, namely, TransStart Fastpfu DNA Polymerase (TransGen Biotech, Beijing, China) and carried out in an ABI GeneAmp 9700 (Applied Biosystems, Inc., Carlsbad, United States of America) under the following conditions: 95°C for 5 min, 28 (bacteria)/32 (fungi) cycles at 95°C for 45 s, 55°C for 50 s and 72°C for 45 s, and a final extension at 72°C for 10 min. PCR amplification was carried out in an ABI GeneAmp 9700 (Applied Biosystems, Inc., Carlsbad, United States) using TransGen AP221-02, specifically TransStart Fastpfu DNA Polymerase (TransGen Biotech, Beijing, China). PCR amplification products were purified and recovered using a 1.8% agarose gel electrophoresis method. Following PCR product purification and recovery, the 16S rRNA and ITS1 genes were sequenced using an Illumina Hiseq 2500 PE 100 (San Diego, CA, United States).
This study’s 16S and ITS1 rRNA gene sequences were deposited in the NCBI as Accession No. PRJNA563013.
Initially, raw data with sequence lengths less than 200 bp, low quality scores (≤20), ambiguous bases, or non-matching primer sequences and barcode tags were excluded. Then, Illumina Analysis Pipeline Version 2.6 was utilized to separate qualified reads based on sample-specific barcode sequences. Subsequently, the Quantitative Insights into Microbial Ecology (QIIME) software version 1.8.0 was used to analyze sequences (Quast et al., 2012). OTUs with up to 97% similarity were grouped using UCLUST (Edgar, 2013). Diversity indexes such as ACE, Chao1, Shannon’s diversity, and Simpson’s diversity were calculated for OTUs at a 97% similarity threshold. The ß-diversity of microbial communities was evaluated by performing Principal coordinates analysis (PCoA) and analysis of similarities (Anosim) (Anderson, 2006; Che et al., 2019). Taxonomical assignments of OTUs were also conducted at the 97% similarity level. Additionally, linear discriminant analysis (LDA) effect size (LEfSe) method was applied to identify groups or species responsible for significant differences among degradation levels (Segata et al., 2011).
One-way ANOVA (analysis of variance) was performed to evaluate significance levels for all factors analyzed. Ordination analysis was conducted to assess relationships between environmental and soil bacteria and fungi variables, and Monte Carlo Permutation Tests were used to determine the significance of each environmental variable in the RDA using CANOCO for Windows version 4.5 (Fay et al., 2011). In order to determine the appropriate model for direct gradient analysis, detrended correspondence analysis (DCA) was performed, which indicated that a linear model with gradient lengths less than 3 standard units would be suitable for the data (Jumpponen and Jones, 2014). Redundancy analysis (RDA) was then conducted using this model. The significance of each environmental variable in the RDA was evaluated by using Monte Carlo Permutation Tests (Number of permutations = 999).
Plant AGB and BGB decreased considerably (p < 0.05) in the MD, SD, and VD, but not in the LD, while SOC, TN, and NH4+-N levels declined in the MD, SD, and VD by comparison with the ND, with an opposite pattern observed in AP, BD, and pH. Soil AK did not change in the LD or MD, but decreased with SD, while soil TK increased only (p < 0.05) in the SD contrasted to the ND. The degraded alpine meadows had significantly lower soil NO3−-N, TP, and SM levels (p < 0.05) (Table 2).
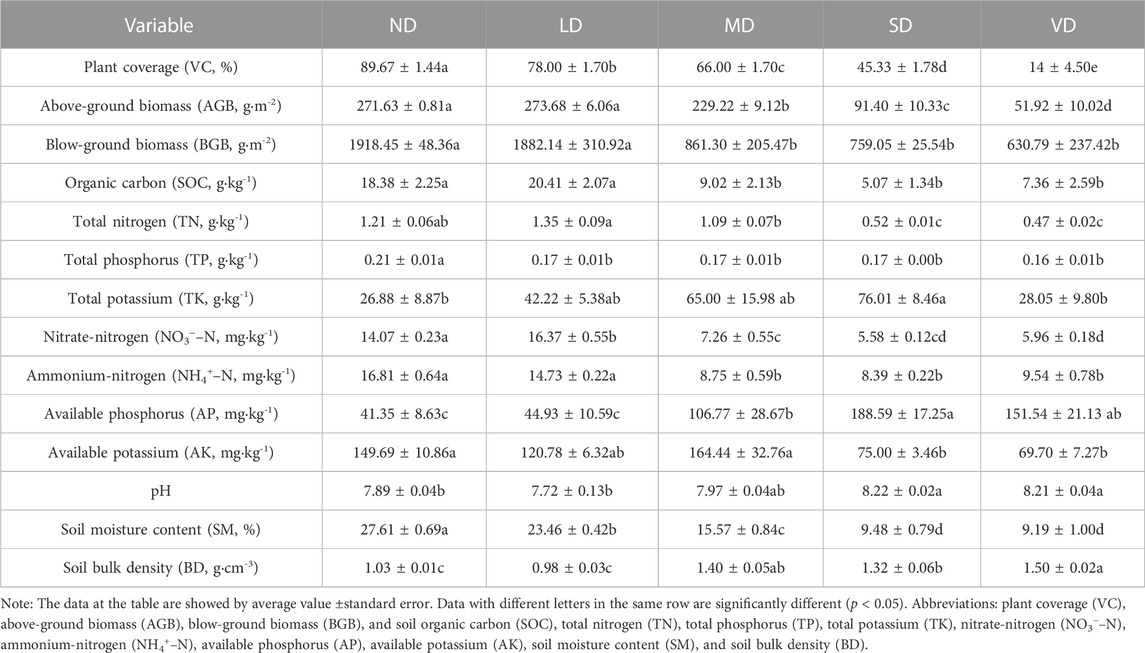
TABLE 2. Summaries of vegetation characteristics and soil properties in the non-degraded (ND), slightly degraded (LD), moderately degraded (MD), severely degraded (SD), and very severely degraded (VD) alpine meadow (n = 3, soil depth = 0–10 cm).
Bacterial OTUs from 19 phyla, 206 families, and 343 genera were identified. Proteobacteria was the most abundant division, contributing to over 99% of total reads in all libraries and significantly decreased by 8.75%, 22.99%, and 24.65% in the MD, SD, and VD compared with ND, respectively. Conversely, Acidobacteria significantly increased by 41.67%, 85.20%, and 108.67% in the MD, SD, and VD compared with ND, respectively. Chloroflexi, Gemmatimonadetes, and Nitrospirae also showed significant increases with increasing degradation stage, while Patescibacteria exhibited an opposite response pattern. Except for Gemmatimonadetes and Patescibacteria, the top 10 phyla showed no change in the ND and LD (Figures 2A, B).
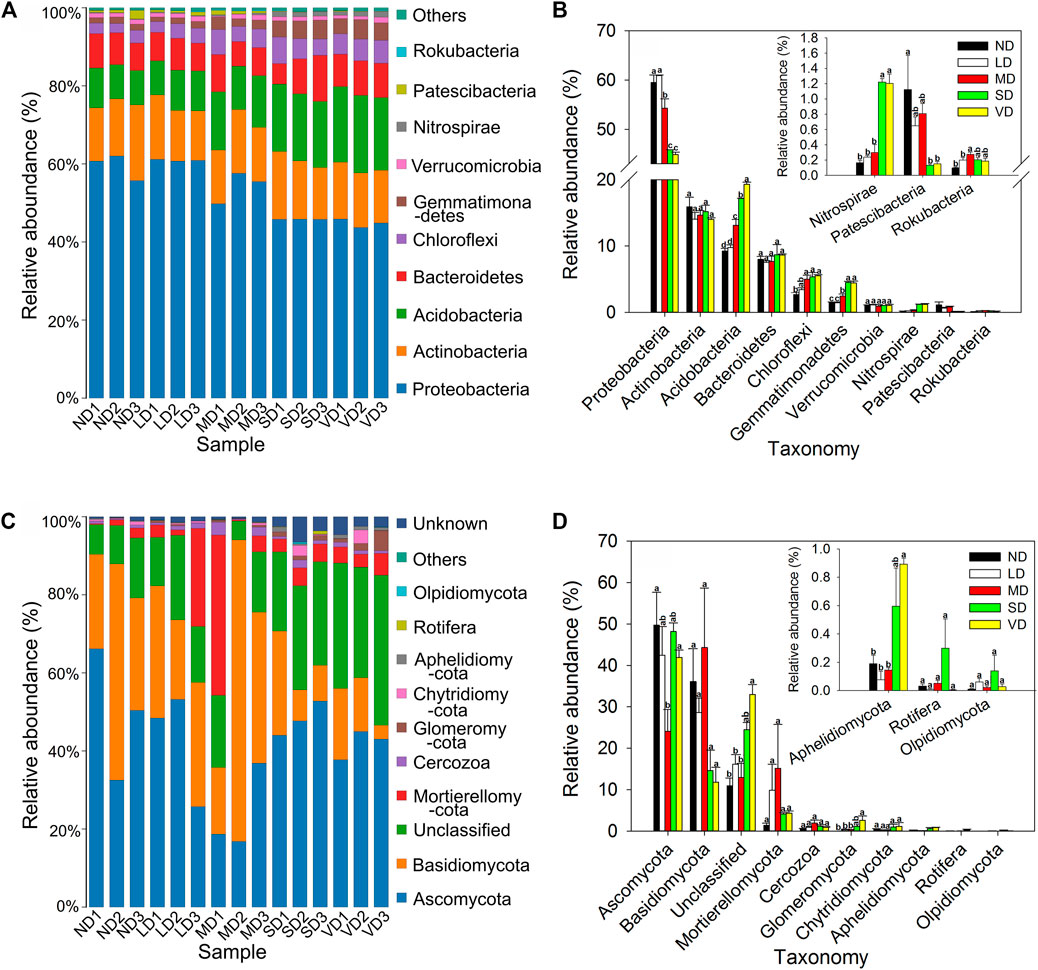
FIGURE 2. The phylum-level abundance of bacteria (A,B) and fungi (C,D) in non-degraded (ND), slightly degraded (LD), moderately degraded (MD), severely degraded (SD), and very severely degraded (VD) alpine meadow. A 97% similarity cluster of OTUs was used to calculate relative abundances, and each letter indicates a significant difference between soil samples according to Duncan’s test (p < 0.05). The insert figure in the top right corner shows an enlargement of Nitrospirae, Patescibacteria, Rokubacteria, Aphelidiomycota, Rotifera, and Olpidiomycota for ND, LD, MD, SD, and VD. Error bars represent standard errors (n = 3).
The fungal communities were identified to have 9 phyla and 100 families. Ascomycota, Unclassified, Basidiomycota, and Mortierellomycota were the dominant phyla, accounting for over 90% of the total reads in all degradation levels. Ascomycota was the most abundant division and significantly (p < 0.05) decreased by 51.48% in the MD compared with ND. Conversely, Unclassified, Glomeromycota, and Aphelidiomycota significantly (p < 0.05) increased in the VD compared with ND, while no significant changes were observed among ND, LD, MD, and SD (Figures 2C, D).
The heatmap for bacteria was created using the top 50 most abundant bacterial genera (Figure 3A), using a two-sample cluster partition. The first cluster had ND, LD, and MD, whereas the second contained SD and VD. The heatmap for fungi was based on the top 50 fungal families (Figure 3B), demonstrating a two-cluster sample division: ND, LD, and MD in one group, and SD and VD in another.
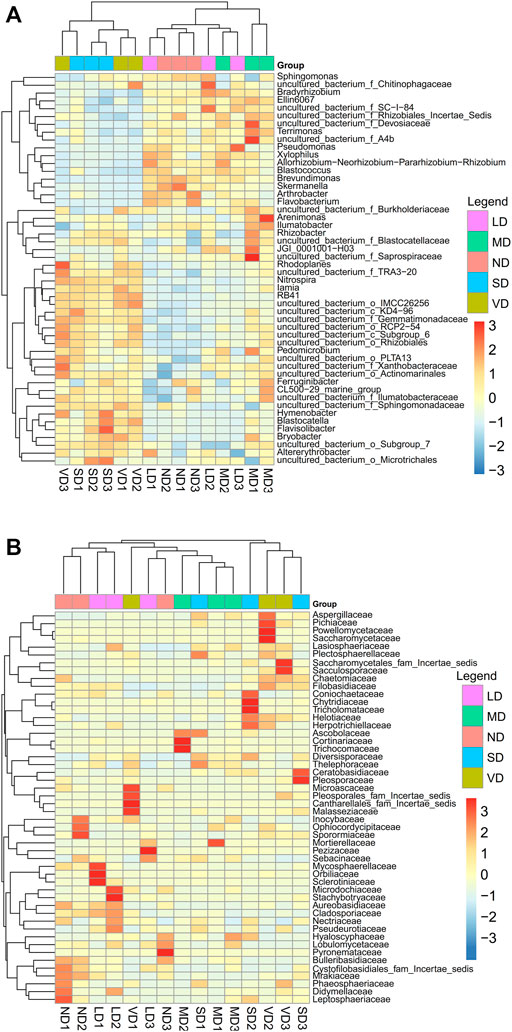
FIGURE 3. Heatmaps representation and cluster analysis of bacterial (A) and fungal (B) communities for the fifteen samples.
In terms of microbial diversity, the fungal and bacterial a-diversity indexes, including ACE, Shannon’s, Chao1, and Simpson’s, showed no significant (p > 0.05) variation between alpine meadow deterioration stages (Supplementary Table S5). The examination of similarities (anosim) indicated, however, that the samples from distinct damaged alpine meadows differed considerably (p < 0.01) from one other (Figures 4A, C).
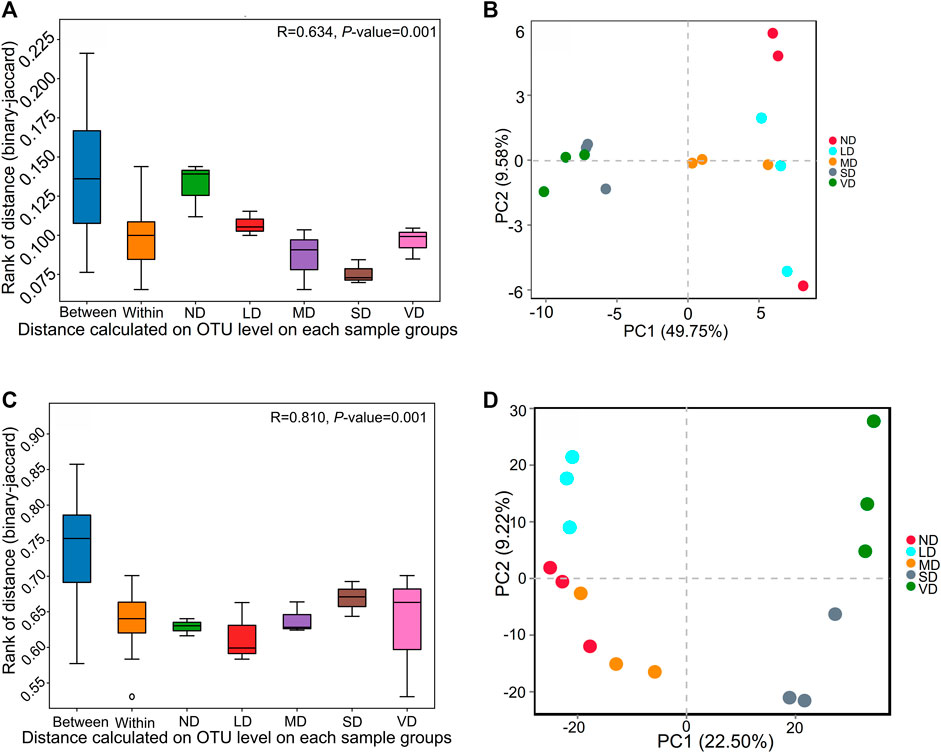
FIGURE 4. The analysis of similarities (ANOSIM) (A,C) and Principal coordinates analysis (PCoA) (B,D) of all bacteria (A,B) and fungi (C,D) community for ND, LD, MD, SD, and VD alpine meadow. PCoA is based on total OTU level information.
Bacterial (Figure 4B) and fungal (Figure 4D) communities in ND, LD, and MD clustered together and were distinct from those in SD and VD. For bacteria, PCoA analysis revealed that the overall explanatory powers of measure variables interpreted 49.75% and 9.58% of the total variation on the coordinate axes 1 and 2, respectively, whereas for fungus, first and second axes interpreted 22.50% and 9.22% of the variation (Figures 4B, D).
Significance analysis of differences between degradation levels revealed that 40 bacterial clades and 20 fungal clades exhibited statistically significant differences with an LDA threshold of 4.0 (Figures 5A, B). Alphaproteobacteria, Actinobacteria, Micrococcales, Caulobacterales, Micrococcaceae, Arthrobacter, and Brevundimonas were enriched in the ND, while Proteobacteria, Gammaproteobacteria, Pseudomonadales, Pseudomonadaceae, Rhizobiaceae, and Pseudomonas were enriched in the LD. Betaproteobacteriales and Rhizobiales were enriched in the MD. Gemmatimonadales, Gemmatimonadetes, and Gemmatimonadaceae were enriched in both SD and VD, whereas uncultured_bacterium_g_RB41 was enriched only in VD (Figure 5A). Tremellales, Bulleribasidiaceae, Pyronemataceae, Vishniacozyma, Acicuseptoria, Acicuseptoria_rumicis, Dioszegia_hungarica, and Inocybe_substraminipes were enriched in the ND, while Mycosphaerellaceae was abundant in the LD. Glomeromycota, Diversisporales, Pleosporales_fam_Incertae_sedis, Sacculosporaceae, Paraphoma, Sacculospora, Thyrostroma, Sacculospora_baltica, and Thyrostroma_cornicola were enriched in the VD. Gemmatimonadales, Gemmatimonadetes, and Gemmatimonadaceae were enriched in SD, while no significant difference was found for most differential fungi among ND, LD, and VD (Figure 5B).
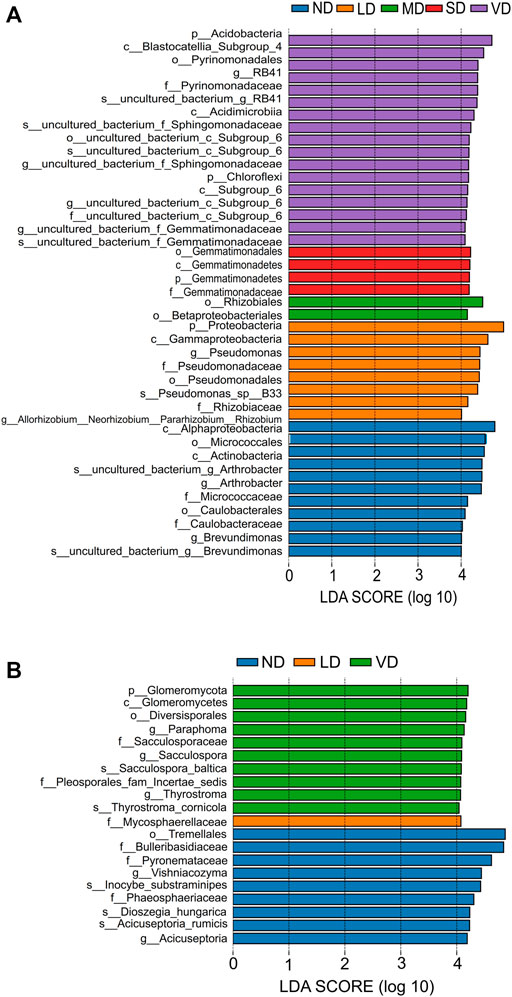
FIGURE 5. The Linear Discriminant Analysis (LDA) Effect Size (LEfSe) of bacterial (A) and fungal (B) abundance in ND, LD, MD, SD, and VD alpine meadow. The histograms represent LDA scores computed for differentially abundant microbes among degraded alpine meadow identified with a threshold value of 4.0. The length of the histogram represents the effect of different species, and different colors represent different levels of species.
RDA analysis revealed that the first and second principal components explained 53.9% and 19.9% of soil bacterial community variation, respectively (Figure 6A). Diverse degradation levels resulted in diverse bacterial populations in soil, which were substantially related to soil pH, SM, BD, SOC, TN, NO3−-N, TP, AP, AK, VC, AGB, and BGB (Supplementary Table S6), although soil TK and NO4+-N had no influence on bacteria (p > 0.05).
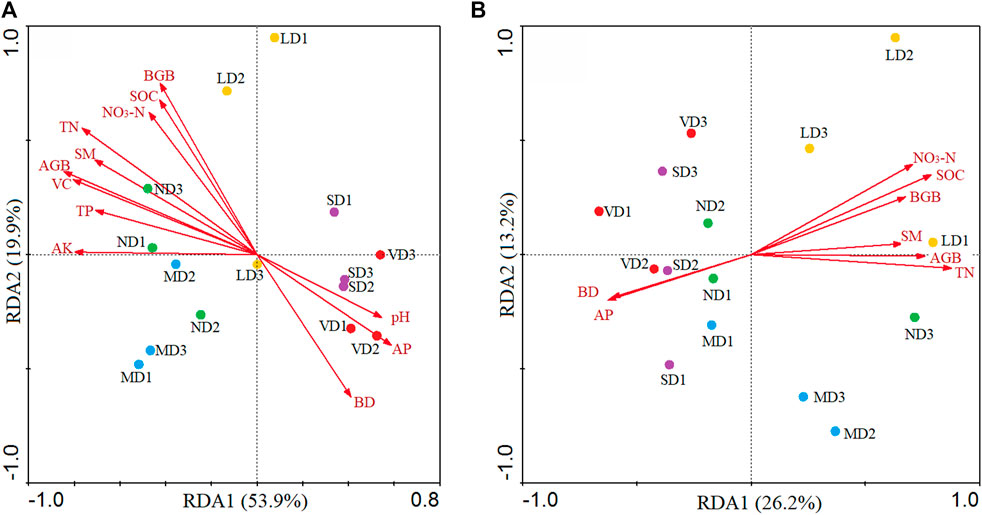
FIGURE 6. Relationships between soil samples (circles) and soil and plant properties (arrows) for bacterial (A) and fungal (B) community structure based on the redundancy analysis. Abbreviations: plant coverage (VC), above-ground biomass (AGB), blow-ground biomass (BGB), and soil organic carbon (SOC), total nitrogen (TN), total phosphorus (TP), total potassium (TK), nitrate-nitrogen (NO3−–N), ammonium-nitrogen (NH4+–N), available phosphorus (AP), available potassium (AK), soil moisture content (SM), and soil bulk density (BD).
The RDA analysis demonstrated that the first and second principal components explained 26.2% and 13.2% of the variation, respectively, in the case of fungus (Figure 6B). Soil SM, BD, SOC, TN, NO3−-N, AP, AGB, and BGB were all substantially connected to fungal community structure (p > 0.05), however TK had no influence on fungus (Supplementary Table S6).
In accordance with earlier studies (Li et al., 2016; Zhou et al., 2019), this study discovered that alpine meadow degradation dramatically changed the ß-diversity of soil microbial communities but had no significant impact on the a-diversity (Figures 4A–D; Supplementary Table S5). A freshly generated population of bacteria could be replacing lost ones, which would explain why there have not been any noticeable changes in biodiversity (Figures 2A–D). The structure of the soil microbial community can be altered by changes in the relative abundance of bacteria, which can have a considerable impact on -diversity. Based on these observations, it was discovered that alpine meadows’ soil microbial community structure drastically changed throughout the deterioration process in the study area. This has ramifications for biological functions including carbon mineralization, which are closely linked to the organization of microbial communities (Che et al., 2019). According to the findings, degraded alpine meadows may change the organization of microbial communities more than the number of species.
According to prior research (Ho et al., 2017), SD and VD increased oligotrophic lineages such as Acidobacteria, Gemmatimonadetes, Chloroflexi, and Nitrogenaspirae while decreasing copiotrophic lineages such as Proteobacteria (Figures 2A–D). According to the LDA analysis, protozoa and Acidobacteria were the most prevalent biomarker bacteria, and their relative abundance rose or decreased as degradation severity increased (Figure 5A). Two mechanisms can explain the increase and reduction in the fraction of oligotrophic and copiotrophic microorganisms. First, a decline in soil nutrients in deteriorated alpine meadows (Che et al., 2017; Zhou et al., 2019; Ali et al., 2021), including SOC, TN, TP, NO3−-N, and NH4+-N (Table 2), may have influenced microbial community composition, as supported by a significant correlation between soil microbial community composition and soil nutrient factors (Supplementary Table S6). Second, pH, a crucial element controlling soil microbial community structure (Zhang YJ. et al., 2014; Ali et al., 2019; Yu et al., 2019), rose considerably with alpine meadow degradation intensity (Table 2), thereby affecting the proportions of Chloroflexi, Gemmatimonadetes, and Proteobacteria.
In fungi, changes in biogeochemical properties induced by degradation may alter the behavior of specific species if their preferences for substrate utilization processes differ (Allison and Treseder, 2008; McGuire et al., 2010). In the VD compared to ND, Glomeromycota and unclassified phyla were significantly more abundant (Figure 2C). Glomeromycota and unclassified phyla were much more common in the VD than in the ND (Figure 2C). Furthermore, significantly deteriorated alpine meadow soils contained more plant pathogenic fungus than non-degraded alpine meadow soils (Li et al., 2016). The LDA analysis indicated that Ascomycota, Basidiomycota, and Glomeromycota were the most abundant biomarker fungi, leading to significant differences in fungal community structure among degradation levels (Figure 3A). Therefore, this study demonstrates that soil fungal communities are extremely vulnerable to alpine meadow deterioration (Hawkes et al., 2008; Dooley and Treseder, 2012), with interactions between plants, soil, and microorganisms potentially influencing fungal community sensitivity to degradation severity (Miki et al., 2010; Cao et al., 2017).
RDA analysis revealed substantial (p < 0.01) variations in the architectures of soil bacterial and fungal communities with degradation (Figures 6A, B), which is consistent with earlier research on the Qinghai-Tibetan Plateau (Li et al., 2016; Zhou et al., 2019). Plant and soil properties changes in alpine meadows may explain observed differences in microbial community structure (Wang et al., 2009; Wu et al., 2014), including a significant decrease (p < 0.05) in soil nutrient levels as meadow degradation increased (Table 2) (Dong et al., 2012; Wu et al., 2014). This might be related to decreases in plant covering and biomass, soil SM, and increases in soil BD and pH as degradation severity rises (Table 2), potentially resulting in nutrient leaching and loss (Mchunu and Chaplot, 2012).
According to the study, soil nutrients (N, P, K), soil moisture level, plant cover, and biomass may be key factors defining changes in microbial communities during meadow degradation, accounting for approximately 73.8% and 39.4% of variations in bacterial and fungal communities, respectively (Figures 6A, B). Previous research has shown that these parameters have an influence on soil bacteria (He et al., 2012; Jumpponen and Jones, 2014; Ali et al., 2020). Changes in soil moisture availability have been linked to decreased C use efficiency and altered biomass or ratios of bacteria and fungi in the soil (Fay et al., 2011; Cregger et al., 2012; Zeglin et al., 2013), whereas nitrogen gradients have been linked to changes in the composition of the entire bacterial community (Ramirez et al., 2012; Paungfoo-Lonhienne et al., 2015). Limitations in TP may hamper the growth of microbial (Liu et al., 2012; He et al., 2016) and affect bacterial community significantly (Supplementary Table S6). Plants regulate microbial communities through substrate supplies and changes to the physical environment in the active soil layer, which may impact soil microbial community diversity and structure (Miki et al., 2010). This study discovered a substantial relationship between plant cover, AGB, and/or BGB and bacterial and fungal communities (Supplementary Table S6), implying that evolution of the composition and biomass of plant species caused by degradation have a significant impact on microbial communities.
The purpose of this study was to examine the variations in microbial communities in alpine meadows with various degrees of deterioration. We used the high-throughput Illumina HiSeq sequencing method to do this. Our research found a significant variation in the microbial communities as well as the degradation of alpine meadows. Instead of factors that are only biological or abiotic in nature, we found that the most likely cause of this difference was a combination of both soil and plant properties. These properties were most closely associated with soil organic carbon, total nitrogen, soil moisture, soil bulk density, nitrate-nitrogen, aboveground biomass, and belowground biomass. This study underscores the need for more investigation into the mechanisms of degradation even if it offers insightful information about the modifications to soil bacterial and fungal communities during alpine meadow degradation. We will be able to better manage and protect these significant biological systems if we understand the causes underlying alpine meadow decline.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/, PRJNA563013.
WZ performed the working concept or design of the study, data collection, drafting the paper, making significant revisions to the paper, and approving the final version of the paper to be published.
The Gansu Provincial Department of Education: Youth Doctoral Fund Project (2022QB195) and Doctoral Start-up Foundation of Lanzhou University of Arts and Sciences (2021BSQD011) supported this research.
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1210349/full#supplementary-material
Ali, A., Ghani, M. I., Elrys, A. S., Ding, H., Iqbal, M., Cheng, Z., et al. (2021). Different cropping systems regulate the metabolic capabilities and potential ecological functions altered by soil microbiome structure in the plastic shed mono-cropped cucumber rhizosphere. Agric. Ecosyst. Environ. 318, 107486. doi:10.1016/j.agee.2021.107486
Ali, A., Ghani, M. I., Haiyan, D., Iqbal, M., Cheng, Z., and Cai, Z. (2020). Garlic substrate induces cucumber growth development and decreases Fusarium wilt through regulation of soil microbial community structure and diversity in replanted disturbed soil. Int. J. Mol. Sci. 21 (17), 6008. doi:10.3390/ijms21176008
Ali, A., Ghani, M. I., Li, Y. H., Ding, H., Meng, H., and Cheng, Z. (2019). Hiseq base molecular characterization of soil microbial community, diversity structure, and predictive functional profiling in continuous cucumber planted soil affected by diverse cropping systems in an intensive greenhouse region of northern China. Int. J. Mol. Sci. 20 (11), 2619. doi:10.3390/ijms20112619
Allison, S. D., and Martiny, J. B. H. (2008). Resistance, resilience, and redundancy in microbial communities. Proc. Natl. Acad. Sci. 105, 11512–11519. doi:10.1073/pnas.0801925105
Allison, S. D., and Treseder, K. K. (2008). Warming and drying suppress microbial activity and carbon cycling in boreal forest soils. Glob. Change Biol. 14, 2898–2909. doi:10.1111/j.1365-2486.2008.01716.x
Anderson, M. J. (2006). Distance-based tests for homogeneity of multivariate dispersions. Biometrics 62, 245–253. doi:10.1111/j.1541-0420.2005.00440.x
Cao, C. Y., Zhang, Y., Cui, Z. B., Feng, S., Wang, T., and Ren, Q. (2017). Soil bacterial community responses to revegetation of moving sand dune in semi-arid grassland. Appl. Microbiol. Biotechnol. 101, 6217–6228. doi:10.1007/s00253-017-8336-z
Che, R. X., Wang, F., Wang, W. J., Zhang, J., Zhao, X., Rui, Y., et al. (2017). Increase in ammonia-oxidizing microbe abundance during degradation of alpine meadows may lead to greater soil nitrogen loss. Biogeochemistry 136, 341–352. doi:10.1007/s10533-017-0399-5
Che, R. X., Wang, Y. F., Li, K. X., Xu, Z., Hu, J., Wang, F., et al. (2019). Degraded patch formation significantly changed microbial community composition in alpine meadow soils. Soil Tillage Res. 195, 104426. doi:10.1016/j.still.2019.104426
Chen, S. Y., Liu, W. J., Qin, X., Zhang, T., Chen, K., et al. (2012). Response characteristics of vegetation and soil environment to permafrost degradation in the upstream regions of the Shule River Basin. Environ. Res. Lett. 7, 045406. doi:10.1088/1748-9326/7/4/045406
Chu, H., Neufeld, J. D., Walker, V. K., and Grogan, P. (2011). The influence of vegetation type on the dominant soil bacteria, archaea, and fungi in a low arctic tundra landscape. Soil Sci. Soc. Am. J. 75, 1756–1765. doi:10.2136/sssaj2011.0057
Cregger, M. A., Schadt, C. W., McDowell, N. G., Pockman, W. T., and Classen, A. T. (2012). Response of the soil microbial community to changes in precipitation in a semiarid ecosystem. Appl. Environ. Microbiol. 78, 8587–8594. doi:10.1128/aem.02050-12
Dennis, K. L., Wang, Y., Blatner, N. R., Saadalla, A., Trudeau, E., et al. (2013). Adenomatous polyps are driven by microbe-instigated focal inflammation and are controlled by IL-10-producing T cells. Cancer Res. 73, 5905–5913. doi:10.1158/0008-5472.CAN-13-1511
Ding, J. Z., Li, F., Yang, G. B., Chen, L., Zhang, B., Liu, L., et al. (2016). The permafrost carbon inventory on the Tibetan plateau: A new evaluation using deep sediment cores. Glob. Change Biol. 22, 2688–2701. doi:10.1111/gcb.13257
Dong, S. K., Wen, L., Li, Y. Y., Wang, X. X., Zhu, L., and Li, X. Y. (2012). Soil-quality effects of grassland degradation and restoration on the Qinghai-Tibetan Plateau. Soil Sci. Soc. Am. J. 76, 2256–2264. doi:10.2136/sssaj2012.0092
Dooley, S. R., and Treseder, K. K. (2012). The effect of fire on microbial biomass: A meta-analysis of field studies. Biogeochemistry 109, 49–61. doi:10.1007/s10533-011-9633-8
Edgar, R. C. (2013). Uparse: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi:10.1038/nmeth.2604
Fay, P. A., Blair, J. M., and Smith, M. D. (2011). Relative effects of precipitation variability and warming on grassland ecosystem function. Biogeosciences Discuss. 8, 6859–6900. doi:10.5194/bgd-8-6859-2011
Fierer, N., Bradford, M. A., and Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecol. Soc. Am. 88, 1354–1364. doi:10.1890/05-1839
Harris, R. B. (2010). Rangeland degradation on the Qinghai-Tibetan plateau: A review of the evidence of its magnitude and causes. J. Arid Environ. 74, 1–12. doi:10.1038/nature25753
Hawkes, C. V., Hartley, I. P., Ineson, P., and Fitter, A. H. (2008). Soil temperature affects carbon allocation within arbuscular mycorrhizal networks and carbon transport from plant to fungus. Glob. Change Biol. 14, 1181–1190. doi:10.1111/j.1365-2486.2007.01535.x
He, D., Xiang, X. J., HeWang, J. S. C., Cao, G., Adams, J., et al. (2016). Composition of the soil fungal community is more sensitive to phosphorus than nitrogen addition in the alpine meadow on the Qinghai-Tibetan Plateau. Biol. Fertil. Soils 52, 1059–1072. doi:10.1007/s00374-016-1142-4
He, Z. L., Piceno, Y., DengXu, Y. M., Lu, Z., DeSantis, T., et al. (2012). The phylogenetic composition and structure of soil microbial communities shifts in response to elevated carbon dioxide. ISME J. 6, 259–272. doi:10.1038/ismej.2011.99
Ho, A., Di Lonardo, D. P., and Bodelier, P. L. E. (2017). Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93, fix006. doi:10.1093/femsec/fix006
Jiang, G. L., Wu, Q. B., and Zhang, Z. Q. (2018). Study on the differences of thermal-moisture dynamics in the active layer of permafrost in different alpine ecosystems on the Tibetan Plateau[J]. J. Glaciol. Geocryol. 40 (1), 7–17. doi:10.7522/j.issn.1000-0240.2017.0316
Jumpponen, A., and Jones, K. L. (2014). Tallgrass prairie soil fungal communities are resilient to climate change. Fungal Ecol. 10, 44–57. doi:10.1016/j.funeco.2013.11.003
Kumar, M., Männistö, M. K., van Elsas, J. D., and Nissinen, R. M. (2016). Plants impact structure and function of bacterial communities in Arctic soils. Plant Soil 399, 319–332. doi:10.1007/s11104-015-2702-3
Li, Y. M., Wang, S. P., Jiang, L. L., Zhang, L., Cui, S., Meng, F., et al. (2016). Changes of soil microbial community under different degraded gradients of alpine meadow. Agric. Ecosyst. Environ. 222, 213–222. doi:10.1016/j.agee.2016.02.020
Lin, Z. J., Gao, Z. Y., Fan, X. W., Niu, F., Luo, J., Yin, G., et al. (2020). Factors controlling near surface ground-ice characteristics in a region of warm permafrost,Beiluhe Basin,Qinghai-Tibet Plateau. Geoderma 376, 114540. doi:10.1016/j.geoderma.2020.114540
Liu, L., Gundersen, P., Zhang, T., and Mo, J. (2012). Effects of phosphorus addition on soil microbial biomass and community composition in three forest types in tropical China. Soil Biol. Biochem. 44, 31–38. doi:10.1016/j.soilbio.2011.08.017
Liu, S. B., Zamanian, K., Schleuss, P. M., Zarebanadkouki, M., and Kuzyakov, Y. (2018). Degradation of Tibetan grasslands: Consequences for carbon and nutrient cycles. Agric. Ecosyst. Environ. 252, 93–104. doi:10.1016/j.agee.2017.10.011
Liu, Y., Havrilla, C. A., Jia, C., Liu, X. Z., and Wu, G. L. (2021). Litter crusts enhance soil nutrients through bacteria rather than fungi in sandy ecosystems. Catena 204, 105413. doi:10.1016/j.catena.2021.105413
Liu, Y., Li, S. Y., Shi, J. J., Niu, Y., Cui, Z., Zhang, Z., et al. (2022a). Effectiveness of mixed cultivated grasslands to reduce sediment concentration in runoff on hillslopes in the Qinghai-Tibetan Plateau. Geoderma 422, 115933. doi:10.1016/j.geoderma.2022.115933
Liu, Y., Zhao, L. R., Liu, Y. F., Huang, Z., Shi, J., Wang, Y., et al. (2022b). Restoration of a hillslope grassland with an ecological grass species (Elymus tangutorum) favors rainfall interception and water infiltration and reduces soil loss on the Qinghai-Tibetan Plateau. Catena 219, 106632. doi:10.1016/j.catena.2022.106632
López-Pujol, J., Zhang, F. M., Sun, H. Q., Ying, T. S., and Ge, S. (2011). Centres of plant endemism in China: Places for survival or for speciation? J. Biogeogr. 38, 1267–1280. doi:10.1111/j.1365-2699.2011.02504.x
McGuire, K. L., Bent, E., Borneman, J., Majumder, A., Allison, S. D., and Treseder, K. K. (2010). Functional diversity in resource use by fungi. Ecology 91, 2324–2332. doi:10.1890/09-0654.1
Mchunu, C., and Chaplot, V. (2012). Land degradation impact on soil carbon losses through water erosion and CO2 emission. Geoderma 177–178, 72–79. doi:10.1016/j.geoderma.2012.01.038
Miki, T., Ushio, M., Fukui, S., and Kondoh, M. (2010). Functional diversity of microbial decomposers facilitates plant coexistence in a plant-microbe-soil feedback model. Proc. Natl. Acad. Sci. 107, 14251–14256. doi:10.1073/pnas.0914281107
Niu, Y. L., Li, S. Y., Liu, Y., Shi, J., Wang, Y., Ma, Y., et al. (2021). Regulation of alpine meadow patch coverage on runoff and sediment under natural rainfall on the eastern Qinghai-Tibetan Plateau. J. Hydrology 603, 127101. Part C. doi:10.1016/j.jhydrol.2021.127101
Paungfoo-Lonhienne, C., Yeoh, Y. K., Kasinadhuni, N. R. P., Lonhienne, T. G. A., Robinson, N., Hugenholtz, P., et al. (2015). Nitrogen fertilizer dose alters fungal communities in sugarcane soil and rhizosphere. Sci. Rep. 5, 8678. doi:10.1038/srep08678
Peng, F., You, Q. G., Xue, X., Huang, C., Dong, S., Liao, J., et al. (2018). Changes of soil properties regulate the soil organic carbon loss with grassland degradation on the Qinghai-Tibet Plateau. Ecol. Indic. 93, 572–580. doi:10.1016/j.ecolind.2018.05.047
Qi, Q., Zhao, M., Wang, S., Ma, X., Wang, Y., Gao, Y., et al. (2017) The biogeographic pattern of microbial functional genes along an altitudinal gradient of the Tibetan pasture. Front. Microbiol. 8, 976. doi:10.3389/fmicb.2017.00976
Qiu, J. (2008). Climate change is coming fast and furious to the Tibetan plateau. Nature 454, 4. doi:10.1038/454393a
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi:10.1093/nar/gks1219
Ramirez, K. S., Craine, J. M., and Fierer, N. (2012). Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Change Biol. 18, 1918–1927. doi:10.1111/j.1365-2486.2012.02639.x
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60. doi:10.1186/gb-2011-12-6-r60
Wang, C. T., Long, R. J., Wang, Q. L., Jing, Z. C., and Shi, J. J. (2009). Changes in plant diversity, biomass and soil C, in alpine meadows at different degradation stages in the headwater region of three rivers, China. Land Degrad. Dev. 20, 187–198. doi:10.1002/ldr.879
Wang, G. X., Wang, Y. B., Li, Y. S., and Cheng, H. (2007). Influences of alpine ecosystem responses to climatic change on soil properties on the Qinghai–Tibet Plateau, China. CATENA 70 (3), 506–514. doi:10.1016/j.catena.2007.01.001
Wang, S. P., Duan, J. C., Xu, G. P., Wang, Y., Zhang, Z., Rui, Y., et al. (2012). Effects of warming and grazing on soil N availability, species composition, and ANPP in an alpine meadow. Ecology 93, 2365–2376. doi:10.1890/11-1408.1
White, T. J., Bruns, T., and Lee, S. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR protocols (Elsevier), 315–322. ISBN 978-0-12-372180-8.
Wu, G. L., Ren, G. H., Dong, Q. M., Shi, J. J., and Wang, Y. L. (2014). Above- and belowground response along degradation gradient in an alpine grassland of the Qinghai-Tibetan plateau. Clean. - Soil, Air, Water 42, 319–323. doi:10.1002/clen.201200084
Xiong, J. B., Peng, F., Sun, H. B., Xue, X., and Chu, H. (2014). Divergent responses of soil fungi functional groups to short-term warming. Microb. Ecol. 68, 708–715. doi:10.1007/s00248-014-0385-6
Xue, X., Guo, J., Han, B. S., Sun, Q., and Liu, L. (2009). The effect of climate warming and permafrost thaw on desertification in the Qinghai–Tibetan Plateau. Geomorphology 108, 182–190. doi:10.1016/j.geomorph.2009.01.004
Xue, X., You, Q. G., Peng, F., Dong, S., and Duan, H. (2017). Experimental warming aggravates degradation-induced topsoil drought in alpine meadows of the Qinghai-Tibetan Plateau. Land Degrad. Dev. 28, 2343–2353. doi:10.1002/ldr.2763
You, Q. G., Xue, X., Peng, F., Xu, M., Duan, H., and Dong, S. (2014). Comparison of ecosystem characteristics between degraded and intact alpine meadow in the Qinghai-Tibetan Plateau, China. Ecol. Eng. 71, 133–143. doi:10.1016/j.ecoleng.2014.07.022
Yu, C. Q., Han, F. S., and Fu, G. (2019). Effects of 7 years experimental warming on soil bacterial and fungal community structure in the Northern Tibet alpine meadow at three elevations. Sci. Total Environ. 655, 814–822. doi:10.1016/j.scitotenv.2018.11.309
Yushou, M., Beining, L., Qingyun, L., Jianjun, S., and Quanmin, D. (2002). Study on rehabilitating and rebuilding technologies for degenerated alpine meadow in the Changjiang and Yellow river source region. Pratcultural Sci. 19 (9), 1–5. doi:10.3969/j.issn.1001-0629.2002.09.001
Zeglin, L. H., Bottomley, P. J., Jumpponen, A., Rice, C. W., Arango, M., Lindsley, A., et al. (2013). Altered precipitation regime affects the function and composition of soil microbial communities on multiple time scales. Ecology 94, 2334–2345. doi:10.1890/12-2018.1
Zeng, C., Zhang, F., Wang, Q. J., Chen, Y., and Joswiak, D. R. (2013). Impact of alpine meadow degradation on soil hydraulic properties over the Qinghai-Tibetan Plateau. J. Hydrology 478, 148–156. doi:10.1016/j.jhydrol.2012.11.058
Zhang, L. L., Su, F. G., Yang, D. Q., Hao, Z., and Tong, K. (2013) Discharge regime and simulation for the upstream of major rivers over Tibetan Plateau. J. Geophys. Res. Atmos. 118, 8500–8518. doi:10.1002/jgrd.50665
Zhang, X. F., Xu, S. J., Li, C. M., Zhao, L., Feng, H., Yue, G., et al. (2014a). The soil carbon/nitrogen ratio and moisture affect microbial community structures in alkaline permafrost-affected soils with different vegetation types on the Tibetan plateau. Res. Microbiol. 165, 128–139. doi:10.1016/j.resmic.2014.01.002
Zhang, Y., Dong, S. K., Gao, Q. Z., Liu, S., Zhou, H., Ganjurjav, H., et al. (2016). Climate change and human activities altered the diversity and composition of soil microbial community in alpine grasslands of the Qinghai-Tibetan Plateau. Sci. Total Environ. 562, 353–363. doi:10.1016/j.scitotenv.2016.03.221
Zhang, Y. J., Cong, J., Lu, H., Li, G., Qu, Y., Su, X., et al. (2014b). Community structure and elevational diversity patterns of soil Acidobacteria. J. Environ. Sci. 26, 1717–1724. doi:10.1016/j.jes.2014.06.012
Keywords: microbial diversity, microbial community structure, land degradation, alpine meadow, Qinghai-Tibet Plateau
Citation: Zhang W (2023) Influence of alpine meadow deterioration on soil microbial communities in the Yangtze River source region. Front. Environ. Sci. 11:1210349. doi: 10.3389/fenvs.2023.1210349
Received: 25 April 2023; Accepted: 21 June 2023;
Published: 30 June 2023.
Edited by:
Qibing Wang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2023 Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjuan Zhang, emhhbmd3ZW5qdWFuNTkxQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.