
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Environ. Sci. , 01 June 2023
Sec. Conservation and Restoration Ecology
Volume 11 - 2023 | https://doi.org/10.3389/fenvs.2023.1187545
 M. Mousavi-Derazmahalleh1,2*
M. Mousavi-Derazmahalleh1,2* R. J. Ellis3
R. J. Ellis3 B. L. D’Rozario3
B. L. D’Rozario3 T. E. Berry2
T. E. Berry2 G. Peverley2
G. Peverley2 K. L. Dawkins2
K. L. Dawkins2 M. Campbell1
M. Campbell1 N. E. White1
N. E. White1 M. E. Allentoft1,4*
M. E. Allentoft1,4*Environmental DNA (eDNA) research is transforming biomonitoring at a global scale, but applicability to reptiles has been restricted because of their presumed low shedding rate. Consequently, eDNA may have considerable limitations as a biomonitoring tool in Australia where 40% of the terrestrial vertebrates are reptiles. However, there is a need to evaluate if method improvements, such as targeting certain substrates, improve the ability to detect reptile eDNA. The Pilbara olive python (Liasis olivaceus barroni) is an uncommon and elusive Australian top predator with a high conservation priority. Like many other snake species, Pilbara olive pythons are challenging to monitor with traditional survey methods; therefore, exploring an eDNA-based approach is highly relevant. The pythons are known to occasionally reside in rock pools. Thus, the development of a reliable eDNA-based approach to detect the pythons in water would provide a needed alternative method. Here, we use a previously developed metabarcoding assay targeting reptiles, to sequence a total of 228 water samples collected from 40 rock pools and drainage pools from six broad locations across the Pilbara region of Western Australia, and we confirm the presence of Pilbara olive python eDNA in 37 samples from 12 of those pools at three of the six broad sampling locations. Other vertebrate taxa, including other reptiles, amphibians, mammals, and birds, were also detected. Our documented ability to detect Pilbara olive python eDNA from rock pool water samples represents an important step toward eDNA-based precision monitoring of this species.
The olive python (Liasis olivaceus) is a member of the terrestrial snake family Pythonidae, and one of the largest snake species in Australia, where its distribution occurs across the northern parts of the country from northwest Western Australia (WA) through the Kimberley and Top End to Queensland. The species is often observed within or in close proximity to natural and anthropogenic water features, primarily within major drainage or rocky gorge, gully, and/or breakaway habitats. Individuals are regularly found partially or entirely submerged, often at the edge of water features, in ambush for prey approaching the water (Pearson, 2003; Ellis, 2013; Ellis and Johnstone, 2016).
As confirmed by the application of eight microsatellite and three mitochondrial markers, two distinct subspecies of olive pythons are recognized in Australia, namely, the northern subspecies (L. o. olivaceus) across northern Australia and the Pilbara subspecies (L. o. barroni), which occurs throughout the Pilbara and south into the Gascoyne bioregions. Morphologically, the Pilbara olive python differs from the northern subspecies by having fewer midbody scale rows and a greater number of ventral scales (Storr et al., 2002; Pearson et al., 2013). The subspecies is listed as vulnerable under the Western Australian Biodiversity Conservation Act 2016 and Commonwealth Environment Protection and Biodiversity Conservation Act 1999 (Department of Climate Change, Energy, the Environment and Water, 2022). The Pilbara olive python population is believed to be in decline due to a number of threats, including a low reproduction rate and predation of juveniles by foxes and cats (Pearson, 2003). Additionally, the ongoing development of the mining infrastructure within its distribution range may adversely affect the species by the reduction or loss of suitable habitat, alteration of prey availability, and an increase in the risk of road fatalities (Pearson, 1993; Pearson, 2003). Therefore, biological surveys of the occurrence, habitat use, trophic interactions, and distribution of the Pilbara olive python are key factors for impact assessments and the development of informed management plans for the sustainable development and use of natural resources. Nevertheless, the implementation of conventional visual biological surveys of snakes are challenging and inefficient, as pythons are often elusive, experts at camouflage, occurring in low abundance, and often nocturnal in warmer regions (Orzechowski et al., 2019).
Environmental DNA (eDNA) (i.e., all the genetic material obtained from a given substrate, such as water and soil) metabarcoding (Taberlet et al., 2012) has been used as a successful alternative or complementary approach for biodiversity assessment and species detection in various ecosystems (Taberlet et al., 2018). However, the application of eDNA methods to reptiles has been limited compared with other major vertebrate groups. To date, studies targeting snakes are only represented 14 times in the eDNA literature (Nordstrom et al., 2022). This low number may reflect the hypothesis that animals with keratinized skin (such as reptiles) shed less DNA into the environment and as a result it may be challenging to detect them using an eDNA approach (Adams et al., 2019; Andruszkiewicz Allan et al., 2021). To better understand the potential and limitations of the use of eDNA for reptile biomonitoring, more studies targeting reptiles are required and various factors including species microhabitat use, behavior, lifespan, and substrate sampling strategies must be considered on a case-by-case basis. In the case of the Pilbara olive python, we propose to test the utility of eDNA monitoring on rock pool samples because in an arid and harsh environment like the Pilbara, biota congregate around these natural water sources, where pythons have also been reported. Thus, instead of sampling “random” soil samples across the Pilbara (a vast and remote area), we focus on rock pools as an obvious and feasible focal point for eDNA sampling.
Here, we collate and analyze eDNA metabarcoding data generated as part of ongoing biomonitoring efforts in the Pilbara region. The aim is two-fold: first, to test if water samples collected from natural rock pools in the Pilbara can be used for python eDNA detection; second, to confirm that a 16S metabarcoding assay that was developed for the detection of aquatic reptiles (West et al., 2021) is applicable to olive python DNA. As part of this effort, we use the 16S metabarcoding assay to also generate nucleotide reference sequences for eight different taxa in the family Pythonidae that may assist in future biomonitoring programs.
The result of this eDNA survey, regardless of successful python detection, represents an important stepping-stone toward improved monitoring of the Pilbara olive python, and it increases our knowledge about reptile, particularly snake, eDNA biomonitoring in general. As such, this study serves as a proof of concept for eDNA biomonitoring of a terrestrial/semi-aquatic reptile.
The 16S reptile assay was tested with the aim to generate an in-house custom database from eight python taxa (n = 39 samples): L. o. barroni (Pilbara olive python), L. o. olivaceus (northern olive python), Antaresia perthensis (pygmy python), Antaresia childreni (children’s python, also containing samples previously assigned to A. s. stimsoni and A.s. orientalis), Morelia spilota variegata (diamond python), M.s. imbricata (southwestern carpet python), Aspidites melanocephalus (black-headed python), and Aspidites ramsayi (woma python). DNA extracted from a variety of sources (skin, scale clips, mouth swabs, muscle, and liver tissue) was amplified following the PCR conditions described in the following section. The PCR products were sent to the Australian Genome Research Facility (AGRF, Perth, Australia) for Sanger sequencing (GenBank accession OQ595374-OQ595412). Additionally, a DNA extract from L. o. barroni (positive control) was MID-tagged (described in the following section) and sequenced on a MiSeq instrument. We chose this method because this reference sequence was longer in length, due to the sequencing binding primer site being before the gene-specific primer region (i.e., reptile 16S) and did not produce the unreadable areas typically located adjacent to the primer-binding sites for Sanger sequencing.
A total of 228 water samples, including biological replicates (54 unique samples), were collected from six broad locations across the Pilbara region (Figure 1), during different times of the year from 2020 to 2022 (Supplementary Figure S1). The water samples were collected from natural water features, including intermittent rock pools (following rainfall), spring-fed pools (continuous/near continuous naturally occurring feed of water into a pool), and drainage pools (pooling water along drainage lines, primarily following rainfall) as well as one artificial water source (turkey nest dam which was used to hold groundwater welled to the surface). Additionally, 33 field, extraction, positive, and negative template controls (NTCs) were included in the analysis, making the total number of processed samples n = 261.
Water filtering was performed in the field by filtering up to 1 L of water through 0.45 µm pore size MCE filters using Sentino peristaltic pumps (Pall Life Sciences, New York, United States of America). Individual filter membranes were folded in half, stored in separate sample bags, and frozen (−20°C) for transport to the eDNA frontiers laboratory for further processing. DNA was extracted from half of each filter paper with a modified Qiagen DNeasy blood and tissue kit protocol (Qiagen, Germany) using an automated QIAcube extraction platform (Qiagen). All extractions were undertaken in a dedicated PCR-free laboratory, and extraction controls processed alongside samples. Extractions were eluted in a final volume of 100 µL AE buffer.
To determine the required dilution for optimal amplification, PCR reactions were performed in duplicate on each extraction by adding DNA template directly to the PCR master mix (neat) and then performing a 1/10 serial dilution. PCRs were performed at a final volume of 25 µL, where each reaction comprised the following: 1 × PCR gold buffer (Applied Biosystems), 0.25 mM dNTP mix (Astral Scientific, Australia), 2 mM MgCl2 (Applied Biosystems), 1 U AmpliTaq Gold DNA polymerase (Applied Biosystems), 0.4 mg/mL bovine serum albumin (Fisher Biotec), 0.4 µM forward and reverse primers (16S Reptile; Wests et al., 2021), 0.6 μL of a 1:10,000 solution of SYBR Green dye (Life Technologies), and 2 µL template DNA. PCRs were performed on StepOnePlus instruments (Applied Biosystems) with the following cycling conditions: 95°C for 5 min, followed by 50 cycles of 95°C for 30 s, 52°C for 30 s, and 72°C for 45 s, and then a melt curve analysis of 95°C for 15 s, 60°C for 1 min, and 95°C for 15 s, finishing with a final extension stage at 72°C for 10 min.
After selection of the optimal dilution, PCRs were repeated in duplicate using unique, single use combinations of 8 bp multiplex identifier-tagged (MID-tag) primers as described in Koziol et al. (2019) and van der Heyde et al. (2020) and under the conditions described previously. Tags were aliquoted to the correct positions on each plate using a QIAgility instrument (Qiagen) in an ultra-clean lab facility, with negative and positive PCR controls included on every plate. A sequencing library was created by combining samples into mini-pools based on the PCR amplification results, with these mini-pools combined in equimolar amounts based on their concentration (QIAxcel–QIAgen). The library was then size-selected using a Pippin Prep instrument (Sage Science), cleaned using a QIAquick PCR purification kit, quantified on a Qubit (Thermo Fisher), and diluted to 2 nM. The library was sequenced on an Illumina MiSeq instrument using a 500 V2 Cycle Kit with custom sequencing primers.
Raw sequences were analyzed using the eDNAFlow pipeline (Mousavi-Derazmahalleh et al., 2021) as follows: first, data from each library were quality-filtered and demultiplexed separately using OBITools (Boyer et al., 2016) (implemented in eDNAFlow by --onlyDemux, --minAlignLeng ‘12’, --minLen ‘70’, and --minQuality ‘20’). Then, the quality-filtered demultiplexed files were concatenated into one and used as an input for eDNAFlow (--skipDemux and --demuxedInput) to perform denoising and creation of zero-radius operational taxonomic units (ZOTUs) and their abundance table with Unoise3 algorithm of USEARCH64 (--minsize ‘2’; Edgar, 2010), BLASTN to the nt database of GenBank (--perc_identity ‘85’ and --qcov ‘100’; Altschul et al., 1990), and post-clustering curation with LULU (--minMatch_lulu ‘89’; Frøslev et al., 2017).
Sequences generated with Sanger sequencing were manually trimmed to remove primer sequences and low-quality bases in Geneious Prime 2022.1.1. The custom python database consisted of a total of 39 nucleotide sequences (from eight taxa), of which 38 generated with Sanger sequencing and one using Illumina MiSeq. Because the custom database sequences obtained with Sanger technology resulted in sequences shorter (189–195 bp) than our metabarcoding amplicon length (< 250 bp), a query coverage threshold was relaxed to 90% when blasting against the custom database.
After taxonomy assignment using the eDNAFlow LCA module (--taxonomyAssignment, --lca_qcov ‘100’, --lca_pid ‘97’, and --lca_diff ‘0.5’), the results were further curated as follows: data from the extraction and NTC controls were checked for sequence contamination. ZOTUs assigned to Homo sapiens and bacteria were removed. The results from the custom database BLAST were manually checked and assigned manually with respect to the GenBank BLAST results. After removal of ZOTUs with a total count of below 10 in all samples, biodiversity analyses were performed using the R package phyloseq (McMurdie and Holmes, 2013). Principal coordinate analysis (PCA) was performed using the R package mixOmics (Rohart et al., 2017).
A total of 261 samples were analyzed resulting in 19, 555, 660 sequences in total, yielding 2,088 ZOTUs, of which 1,204 remained after post-clustering curation with LULU (Frøslev et al., 2017) and 346 of those had at least one BLAST match in the GenBank database. The LCA taxonomy assignment yielded 147 nominations. There was some contamination noted in the field, extraction, and negative controls, which were assigned to H. sapiens and Flavobacterium; the ZOTUs responsible were removed from the dataset. Additionally, being mindful of the aim of this study and to reduce background “noise,” all ZOTUs assigned to bacteria were removed from downstream analyses. Following this data curation, the abundance of six samples (highlighted in Supplementary Figure S1) was zero and removed from downstream analyses. Additionally, four ZOTUs assigned to Insecta and Clitellata classes had a total abundance of below 10 and were hence removed. Two curated ZOTUs (ZOTU3 and ZOTU114) had BLAST matches to the custom database generated in this study. ZOTU3 had a 100% match at both query coverage and percent identity (herein qCov and pid, respectively) to a Pilbara olive python sequence generated from the Illumina MiSeq. ZOTU114 had three matches (at qCov between 90% and 91% and pid 98.96%–99.48%) to A. perthensis sequences generated via Sanger sequencing. This information was used to rectify taxonomy assignment of these ZOTUs with respect to the GenBank BLAST results. As a result, ZOTU3, which was initially assigned to A. melanocephalus (GenBank; qCov 100% and pid 98.60%), was confidently re-assigned to L. o. barroni (custom database; qCOV and pid 100%). Additionally, ZOTU114, which was assigned to Python brongersmai (GenBank; query coverage 100% and pid of 99.53%), was dropped to LCA at Pythonidae family to take into account the result of the custom BLAST database and account for the fact that Python brongersmai has not been reported in Australia (Fowler, 2022). This result is likely attributed to either the presence of a small fragment of DNA that could not be attributed to a more specific taxon (i.e., limited data that were broadly attributed to the species) or that this was the closest taxon available in the BLAST database used (i.e., of the taxa available, it was most similar to Python brongersmai).
After curation, the data revealed a variety of taxa, comprising numerous mammals, birds, fish, insects, flatworms, amphibians, and reptiles, including the Pilbara olive python (Figure 2; Supplementary Figure S2).
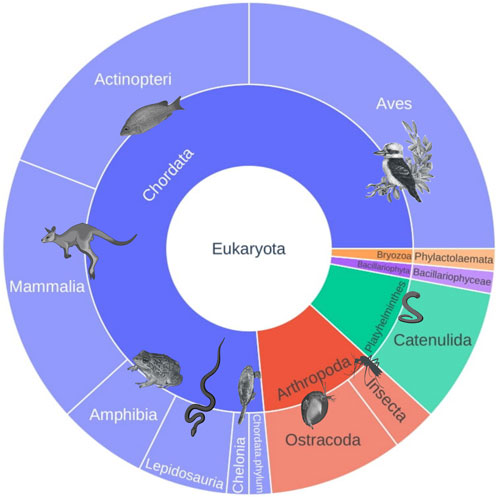
FIGURE 2. Pie chart displaying overall diversity found in the samples. The size of each piece is proportional to the number of taxonomic assignments after data curation.
Using the 16S reptile primer (West et al., 2021), we detected seven taxa from six Mammalia (mammal) families, including assignments made to three species of chiropterans (bats; Taphozous georgianus, Chalinolobus gouldii, and Saccolaimus flaviventris) and one species of macropod (wallaroo; Osphranter robustus), and fourteen taxa from 11 families in class Aves (birds), including the laughing kookaburra (Dacelo novaeguineae) and the brahminy kite (Haliastur indus). Eight taxa from seven families of class Actinopteri (fish) were detected, which included one species of conservation significance endemic to Western Australia, Fortescue grunter (Leiopotherapon aheneus) (The IUCN Red List of Threatened Species 2019). We also detected two taxa from two Amphibia (frog) families, including Main’s frog (Cyclorana maini), desert tree frog (Litoria rubella), and one that could not be resolved at the species level, assigned to the genus Uperoleia. A total of five taxa from four Reptilia (reptile) families were also recorded, which importantly included the Pilbara olive python. Pilbara olive python DNA was detected in samples from 12 individual pools at three of the six broad sampling locations across the Pilbara (Figure 3). One species of a Western Australian freshwater turtle (Chelodina steindachneri), one species of varanid (Varanus panoptes), and one skink species (Ctenotus pantherinus) were also detected. Reptiles and amphibians detected across various regions and seasons are shown in Figure 4. It should be noted that the detection of taxa other than reptiles (i.e., non-specific amplification) using the 16S reptile primer was expected because during primer development, this assay was shown to detect other taxa as well (West et al., 2021). However, no clear separation between locations and seasons was noted using PCA (Supplementary Figure S1).
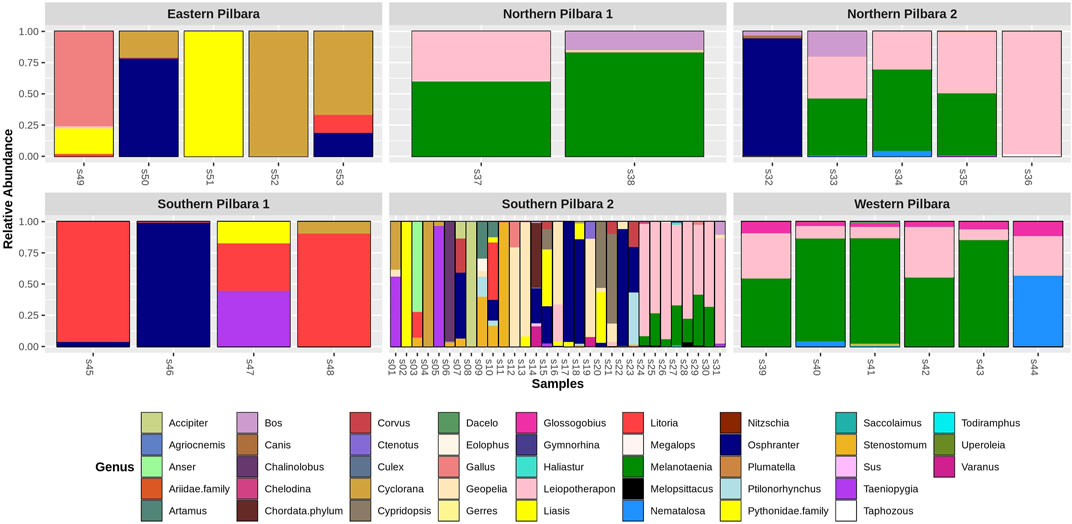
FIGURE 3. Relative abundance of taxa detected using 16 S rRNA assay at the six study sites in the Pilbara region. Genus Liasis represents detection of Pilbara olive python.
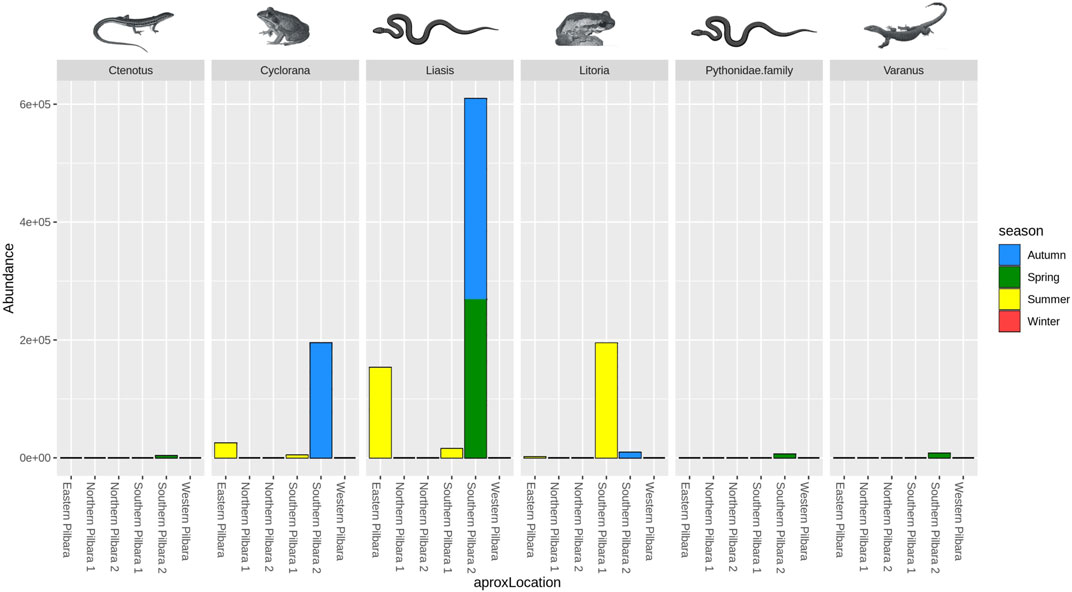
FIGURE 4. Abundance of reads assigned to reptiles and amphibians genera across the study locations and seasons.
Reptiles are disappearing at an alarming rate across the world (Cox et al., 2022), highlighting the urgency for conservation strategies and improved monitoring of this group. eDNA metabarcoding has great potential as a biomonitoring tool; however, due to the limited application of this technique in reptile monitoring to date, it is important that any result–including negative results–are made available publicly.
Here, we have contributed to this pool of knowledge by exploring the detection of the threatened Pilbara olive python using eDNA metabarcoding. Uncovering reptile DNA from environmental samples using either species-specific or metabarcoding assays has shown varying efficiency (Adams et al., 2019). For instance, while species-specific assays have successfully been applied for the detection of Burmese python (Python bivittatus) using eDNA from both aquatic water (Piaggio et al., 2014) and terrestrial soil samples (Kucherenko et al., 2018), and eastern massasauga rattlesnake (Sistrurus catenatus) was only detected in two of 100 environmental samples despite their known presence, as confirmed by visual observation in 12 samples (Baker et al., 2020). The application of metabarcoding assays has been shown to be effective for the detection of three North American species of snakes: redbelly snake (Storeria occipitomaculata), northern watersnake (Nerodia sipedon), and milksnake (Lampropeltis triangulum) (Lacoursière-Roussel et al., 2016).
Our results support applicability of eDNA metabarcoding for Pilbara olive python biomonitoring, as shown by the successful detection of the species from multiple pools across the Pilbara region. These detections also occurred over a temporal spread including autumn, spring, and summer sampling periods. However, due to uneven sample size, no definitive conclusion can be made regarding the spatial or temporal variation of these results as demonstrated by PCA (Supplementary Figure S1). Continuation of Pilbara olive python biomonitoring programs with inclusion of broader geographic sampling and season variation will provide more reliable insights in this regard. Nevertheless, successful detection of Pilbara olive python DNA using a 16S assay (West et al., 2021) in the rock pools provides a functional framework for biomonitoring of this species, which is a considerable improvement over the existing methods. For instance, the Pilbara olive python was recorded only once during a 4-year survey of the Pilbara region comprising predominantly conventional pitfall trap lines (Doughty et al., 2011).
Further method improvements could be developed following comparison of our proposed sampling approach (i.e., using water from rock pools) with other substrates, such as dirt from other microhabitats including potential den sites, caves, crevices, and scats Moreover, conducting a study to understand the persistence of python DNA in the water is necessary to shed light on the temporal resolution of these results. Clearly, a positive result implies that pythons live in the area around the rock pool and utilize the rock pool present; however, the temporal scale of these detections is unknown, i.e., if individuals detected occurred at the pool within a period of 3 h or 3 weeks prior to water sampling. Extracellular DNA detection probability depends on density and ratio between the released and degraded DNA (Dejean et al., 2011) and can vary depending on the substrate used as well (van der Heyde et al., 2020). Therefore, the knowledge about DNA persistence of a species in an environment and the appropriate choice of substrates for metabarcoding will have a great impact on future planning of the biodiversity survey.
Finally, this study clearly demonstrates the importance of completeness and robustness of underlying reference libraries for accurate taxonomic identification of eDNA. In the absence of the custom database generated in this study, the sequences (i.e., ZOTU3 and ZOTU114), which were able to assign to L. o. barroni and Pythonidae family, would have been assigned to an incorrect species, due to the lack of information in the reference library. This highlights the importance of filling gaps in barcoding reference databases.
Australia is home to approximately 10% of all known species of reptiles, the largest number of any country globally (Tingley et al., 2019). Despite this, many species are poorly resolved taxonomically, and data deficiency hinders appropriate conservation planning (Geyle et al., 2020). While additional studies are required to understand factors affecting the shedding rate and longevity of DNA from different reptiles, our result add to a growing field, illuminating the promise of eDNA for reptile biomonitoring in Australia and elsewhere.
The custom database generated in this study is available from NCBI GenBank under the accession numbers OQ595374 to OQ595412. Data used for analyses have been deposited in DRYAD: https://doi.org/10.5061/dryad.3tx95x6m4.
Ethical review and approval was not required for the animal study because the study was conducted using environmental DNA water samples and as such does not require ethics approval.
MM-D and MA conceived the study and drafted the manuscript. RE and BD’R collected the samples. TB, GP, and KD performed lab analyses. MC and NW provided and prepared the samples for Sanger sequencing. MM-D performed data analyses. All authors contributed to the manuscript and approved the submitted version.
MM-D and NW are supported by the BHP Social Investment Fund, eDNA for Global Biodiversity (eDGES) programme in projects managed by MA. This work was supported by resources provided by the Pawsey Supercomputing Research Centre with funding from the Australian Government and the Government of Western Australia. We thank Shane Herbert for facilitating collation and use of the data. We also thank those who assisted in sample collection: Chris Knuckey, Andrew Hide, and Ashleigh Kimpton of Biologic Environmental Survey. Field sampling and sample collection were supported by Tanya Carroll and Alice Taysom of BHP Western Australia Iron Ore.
Authors MM-D, TB, GP, and KD are wholly or part-employed by the fee-for-service entity “eDNA Frontiers” that operates in a commercial manner out of Curtin University. The samples included in this publication were originally processed as a paid independent biomonitoring study. The authors declare that relevant client permissions for inclusion in this publication have been sought and granted.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1187545/full#supplementary-material
Adams, C. I., Hoekstra, L. A., Muell, M. R., and Janzen, F. J. (2019). A brief review of non-avian reptile environmental DNA (eDNA), with a case study of painted turtle (Chrysemys picta) eDNA under field conditions. Diversity 11, 50. doi:10.3390/d11040050
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi:10.1016/s0022-2836(05)80360-2
Andruszkiewicz Allan, E., Zhang, W. G., C Lavery, A., and F Govindarajan, A. (2021). Environmental DNA shedding and decay rates from diverse animal forms and thermal regimes. Environ. DNA 3, 492–514. doi:10.1002/edn3.141
Baker, S. J., Niemiller, M. L., Stites, A. J., Ash, K. T., Davis, M., Dreslik, M. J., et al. (2020). Evaluation of environmental DNA to detect Sistrurus catenatus and Ophidiomyces ophiodiicola in crayfish burrows. Conserv. Genet. Resour. 12, 13–15. doi:10.1007/s12686-018-1053-9
Boyer, F., Mercier, C., Bonin, A., Le Bras, Y., Taberlet, P., and Coissac, E. (2016). obitools: A unix-inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16, 176–182. doi:10.1111/1755-0998.12428
Cox, N., Young, B. E., Bowles, P., Fernandez, M., Marin, J., Rapacciuolo, G., et al. (2022). A global reptile assessment highlights shared conservation needs of tetrapods. Nature 605, 285–290. doi:10.1038/s41586-022-04664-7
Dejean, T., Valentini, A., Duparc, A., Pellier-Cuit, S., Pompanon, F., Taberlet, P., et al. (2011). Persistence of environmental DNA in freshwater ecosystems. PloS One 6, e23398. doi:10.1371/journal.pone.0023398
Doughty, P., Rolfe, J. K., Burbidge, A. H., Pearson, D. J., and Kendrick, P. G. (2011). Herpetological assemblages of the Pilbara biogeographic region, western Australia: Ecological associations, biogeographic patterns and conservation. Rec. West. Aust. Mus. 78, 315–341. doi:10.18195/issn.0313-122x.78(2).2011.315-341
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi:10.1093/bioinformatics/btq461
Ellis, R., and Johnstone, (2016). Liasis olivaceus barroni (Pilbara olive Python) diet. Herpetol. Rev. 47, 685.
Fowler, J. (2022). The reptiles of Australia - pythons. http://reptilesofaustralia.com/snakes/pythons/pythons.htm#.Y1-ZEtJBzmE (Accessed 10 31, 2022).
Frøslev, T. G., Kjøller, R., Bruun, H. H., Ejrnæs, R., Brunbjerg, A. K., Pietroni, C., et al. (2017). Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 8, 1188. doi:10.1038/s41467-017-01312-x
Geyle, H. M., Tingley, R., Amey, A. P., Cogger, H., Couper, P. J., Cowan, M., et al. (2020). Reptiles on the brink: Identifying the Australian terrestrial snake and lizard species most at risk of extinction. Pac. Conserv. Biol. 27, 3–12. doi:10.1071/pc20033
Koziol, A., Stat, M., Simpson, T., Jarman, S., Dibattista, J. D., Harvey, E. S., et al. (2019). Environmental DNA metabarcoding studies are critically affected by substrate selection. Mol. Ecol. Resour. 19, 366–376. doi:10.1111/1755-0998.12971
Kucherenko, A., Herman, J. E. E. M. E., and Urakawa, H. (2018). Terrestrial snake environmental DNA accumulation and degradation dynamics and its environmental application. Herpetologica 74, 38–49. doi:10.1655/herpetologica-d-16-00088
Lacoursière-Roussel, A., Dubois, Y., Normandeau, E., and Bernatchez, L. (2016). Improving herpetological surveys in eastern North America using the environmental DNA method. Genome 59, 991–1007. doi:10.1139/gen-2015-0218
Mcmurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PloS one 8, e61217. doi:10.1371/journal.pone.0061217
Mousavi-Derazmahalleh, M., Stott, A., Lines, R., Peverley, G., Nester, G., Simpson, T., et al. (2021). eDNAFlow, an automated, reproducible and scalable workflow for analysis of environmental DNA sequences exploiting Nextflow and Singularity. Mol. Ecol. Resour. 21, 1697–1704. doi:10.1111/1755-0998.13356
Nordstrom, B., Mitchell, N., Byrne, M., and Jarman, S. (2022). A review of applications of environmental DNA for reptile conservation and management. Ecol. Evol. 12, e8995. doi:10.1002/ece3.8995
Orzechowski, S. C., Frederick, P. C., Dorazio, R. M., and Hunter, M. E. (2019). Environmental DNA sampling reveals high occupancy rates of invasive Burmese pythons at wading bird breeding aggregations in the central Everglades. Plos one 14, e0213943. doi:10.1371/journal.pone.0213943
Pearson, D. J. (1993). “Distribution, status and conservation of pythons in Western Australia” in Herpetology in Australia: A Diverse Discipline, eds. D. Lunney, and D. Ayers (Sydney, Australia: Royal Society of New South Wales), 383–395.
Pearson, D., Spencer, P., Hillyer, M., and How, R. (2013). Genetcis survey of the Pilbara olive Python (Liasis olivaceaus barroni). final report. https://library.dbca.wa.gov.au/static/FullTextFiles/071985.pdf.
Piaggio, A. J., Engeman, R. M., Hopken, M. W., Humphrey, J. S., Keacher, K. L., Bruce, W. E., et al. (2014). Detecting an elusive invasive species: A diagnostic PCR to detect B urmese python in F lorida waters and an assessment of persistence of environmental DNA. Mol. Ecol. Resour. 14, 374–380. doi:10.1111/1755-0998.12180
Rohart, F., Gautier, B., Singh, A., and Lê Cao, K.-A. (2017). mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 13, e1005752. doi:10.1371/journal.pcbi.1005752
Storr, G. M., Smith, L. A., and Johnstone, R. E. (2002). Snakes of western Australia. Perth, Australia: Western Australian Museum.
Taberlet, P., Bonin, A., Zinger, L., and Coissac, E. (2018). Environmental DNA: For biodiversity research and monitoring. Oxford, UK: Oxford University Press.
Taberlet, P., Coissac, E., Pompanon, F., Brochmann, C., and Willerslev, E. (2012). Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. doi:10.1111/j.1365-294x.2012.05470.x
Tingley, R., Macdonald, S. L., Mitchell, N. J., Woinarski, J. C., Meiri, S., Bowles, P., et al. (2019). Geographic and taxonomic patterns of extinction risk in Australian squamates. Biol. Conserv. 238, 108203. doi:10.1016/j.biocon.2019.108203
Van Der Heyde, M., Bunce, M., Wardell-Johnson, G., Fernandes, K., White, N. E., and Nevill, P. (2020). Testing multiple substrates for terrestrial biodiversity monitoring using environmental DNA metabarcoding. Mol. Ecol. Resour. 20, 732–745. doi:10.1111/1755-0998.13148
Keywords: eDNA, biomonitoring, reptiles, python, Liasis olivaceus barroni
Citation: Mousavi-Derazmahalleh M, Ellis RJ, D’Rozario BL, Berry TE, Peverley G, Dawkins KL, Campbell M, White NE and Allentoft ME (2023) Rock pools as a source of environmental DNA for the detection of the threatened Pilbara olive python (Liasis olivaceus barroni). Front. Environ. Sci. 11:1187545. doi: 10.3389/fenvs.2023.1187545
Received: 16 March 2023; Accepted: 15 May 2023;
Published: 01 June 2023.
Edited by:
Lei Xu, Chinese Academy of Fishery Sciences (CAFS), ChinaReviewed by:
Victor Hugo Valiati, University of the Rio dos Sinos Valley, BrazilCopyright © 2023 Mousavi-Derazmahalleh, Ellis, D’Rozario, Berry, Peverley, Dawkins, Campbell, White and Allentoft. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Mousavi-Derazmahalleh, cy5tb3VzYXZpZEBjdXJ0aW4uZWR1LmF1; M. E. Allentoft, bW9ydGVuLmFsbGVudG9mdEBjdXJ0aW4uZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.