- Department of Biomolecular Engineering, Graduate School of Engineering, Nagoya University, Nagoya, Japan
The widespread use of carbon nanotubes (CNTs) has raised concerns about the human health and ecological effects of CNTs released into the environment. Bacteria play an important role in bioremediation and waste treatment, and their enzymes are mostly responsible for the degradation of contaminants. However, there are still only a few reports about the bacterial degradation of CNTs, and evidence showing the involvement of bacterial enzymes in CNT degradation with their mechanisms has never been reported. The purpose of this study is to clarify whether CNTs can be degraded by bacterial enzymes. In this study, the degradation of oxidized (carboxylated) single-walled CNTs (O-SWCNTs) by mt2DyP, a dye-decolorizing peroxidase of Pseudomonas putida mt-2, a common soil bacterium, was investigated. After incubation of O-SWCNTs with recombinant mt2DyP and its substrate H2O2 for 30 d, the optical absorbance and Raman spectra revealed the degradation of O-SWCNTs. However, inactivation of the enzyme was observed within 60 min of the start of incubation, suggesting that the degradation of O-SWCNTs occurred nonenzymatically. The inactivation of mt2DyP was accompanied by the release of iron, the active center metal, and degradation of O-SWCNTs was significantly inhibited in the presence of diethylenetriamine pentaacetic acid, a chelating agent, indicating that O-SWCNTs were degraded by the Fenton reaction with iron released from mt2DyP and H2O2. The same phenomenon was observed with P450, which is also a heme enzyme. Furthermore, we investigated the contribution of the Fenton reaction to the O-SWCNT degradation by horseradish peroxidase (HRP), which was reported to enzymatically and rapidly degrade O-SWCNTs. Our results revealed that the degradation of O-SWCNTs in the presence of HRP is also mainly due to the Fenton reaction, with negligible enzymatic degradation. This contradicts the report showing enzymatic degradation of O-SWCNTs by HRP but supports the subsequent report quantitatively showing very slow transformation of O-SWCNTs by HRP. The current results emphasize that the Fenton reaction, which has received little attention in CNT degradation by heme enzymes, must be taken into consideration and will contribute to the development of a simple disposal method for CNTs, utilizing the Fenton reaction with bacteria/bacterial enzymes and H2O2.
1 Introduction
Carbon nanotubes (CNTs) have been applied in a wide range of fields, such as electronics (Xiang et al., 2018), energy storage (Yang et al., 2019b; Chen et al., 2021), drug delivery (Ho et al., 2021), biosensor devices (Sireesha et al., 2018; Anzar et al., 2020; Zhao et al., 2021), and water treatment (Dong et al., 2021), owing to their excellent mechanical strength, optical properties, and electrical and thermal conductivity (Dresselhaus et al., 2004; Byrne et al., 2018). Among CNTs, single-walled CNTs (SWCNTs) have particularly excellent physical properties for application. The technology for highly efficient production of SWCNTs has been established in recent years, and the problem of high production costs has been solved (Hata et al., 2004; Almarasy et al., 2021). This will lead to the expansion of the CNT market and the development of a wider range of applications. With the use of CNTs becoming more widespread, the amount of CNTs released into the environment is expected to increase, either accidentally or as waste, raising concerns about their impact on human health and ecosystems. Recent studies have reported that some CNTs exhibit toxicity to plants, animals, and microorganisms (Chen et al., 2015b; Kumarathasan et al., 2015; Hatami, 2017; Mendonca et al., 2017; Chen et al., 2018).
To date, knowledge of the biodegradation and remediation of CNTs is lacking, although many efforts have been made to understand these processes. Since Allen et al. reported the degradation of oxidized (carboxylated) SWCNTs (O-SWCNTs), which are functionalized SWCNTs, by horseradish peroxidase (HRP) (Allen et al., 2008; Allen et al., 2009), the biodegradation of CNTs by incubation with heme enzymes from several organisms, such as human myeloperoxidase (Kagan et al., 2010), human eosinophil peroxidase (Andon et al., 2013), bovine lactoperoxidase (Bhattacharya et al., 2015), fungal manganese peroxidase (Zhang et al., 2014) and lignin peroxidase (Chandrasekaran et al., 2014) in the presence of their oxidizing substrate H2O2, has been reported. However, Flores-Carvantes et al. were the first to quantitatively investigate the enzymatic degradation of CNTs and estimated a half-life of 80 years for CNT degradation by HRP (Flores-Cervantes et al., 2014), which is significantly different from a previous report (Allen et al., 2009). Quantitative studies and reproducibility have not yet been conducted for enzymes other than HRP. There is still much, that is, not well understood about the enzymatic degradation of CNTs.
Bacteria have an immense and diverse metabolic system and play an important role in environmental bioremediation and waste treatment. However, there are still only a few reports of bacterial degradation of CNTs. The degradation of CNTs by bacterial cometabolism with additional carbon sources has been reported, but the mechanism remains to be elucidated (Zhang et al., 2013; You et al., 2017). Labrys sp. WJW was shown to utilize carbon nanomaterials, including CNTs, as the sole carbon source for growth via an extracellular biogenic Fenton-like reaction using siderophores that are secreted only under iron-deficient conditions (Wang et al., 2020). Though bacterial enzymes significantly contribute to the bacterial treatment of contaminants, their involvement in CNT degradation has not yet been confirmed. Elucidation of the function of bacterial enzymes in the degradation of CNTs may lead to more rapid remediation and waste treatment of CNTs under a wide range of conditions using bacteria.
Fungal enzymes involved in the degradation of lignin, a huge persistent organic compound, have attracted attention for their potential to degrade other persistent organic compounds (Kadri et al., 2017; Singh et al., 2021), including CNTs (Chandrasekaran et al., 2014; Zhang et al., 2014). However, white-rot fungi that secrete lignin-degrading enzymes have a limited habitat and are more difficult to apply to waste treatment than bacteria. The purpose of this study is to clarify the degradation behavior of CNTs by bacterial heme enzymes. We focused on dye-decolorizing peroxidases (DyPs), which are widely found mainly in bacteria and have been studied for their ability to degrade lignin (Singh et al., 2013; Colpa et al., 2014; Rahmanpour and Bugg, 2015; Yang et al., 2018). Thus, we investigated the degradation of O-SWCNTs by DyP from Pseudomonas putida mt-2 (mt2DyP), a common soil bacterium reported to have high lignin degradation capacity (Bugg et al., 2011). O-SWCNTs were degraded by incubation with mt2DyP. However, the degradation of O-SWCNTs proceeded nonenzymatically via the Fenton reaction with iron released from the enzyme. The same phenomenon was observed with other heme enzymes. The results of this study provide insight that will lead to the accurate evaluation of CNT biodegradation and the development of new remediation and/or waste treatment methods.
2 Materials and methods
2.1 Materials
Dispersion of O-SWCNTs (Product No. ZEONANOR-SG101) was produced and provided by Zeon Nanotechnology Co., Ltd. (Japan). 2SYN medium consisted of 2% glucose, 4% soytone, 0.5% yeast extract, 0.015% CaCl2・2H2O, and 50 μg/ml neomycin. The pH of the 2SYN medium was adjusted to 7.2 with NaOH and H2SO4. Hemin chloride, 30% H2O2, diethylenetriaminepentaacetic acid (DTPA), 2,2′-azino-bis(3-ethylthiazoline-6-sulfonate) (ABTS), ferrozine, methanol, and acetonitrile were purchased from FUJIFILM Wako Pure Chemical Co., Ltd. (Japan). 3′-(p-hydroxyphenyl) fluorescein (HPF) was purchased from Goryo Chemical, Inc. (Japan). Lyophilized HRP type VI, the same enzyme used by Allen et al. (2009), was purchased from Sigma-Aldrich, Ltd. (United States).
2.2 Recombinant protein production
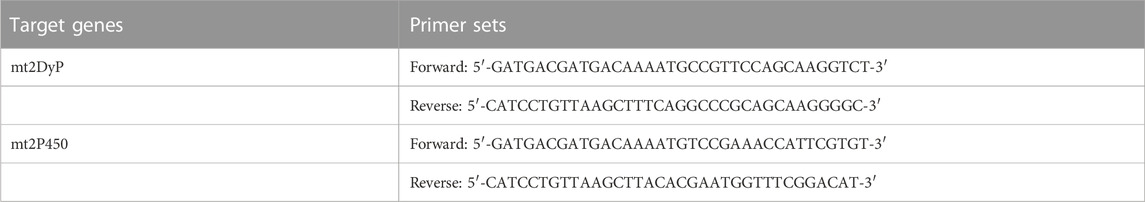
To produce recombinant enzymes, the Brevibacillus secretory expression system (TaKaRa Bio Inc., Japan) was used according to the manufacturer’s instructions. First, for the cloning of DyP and cytochrome P450 homologs from Pseudomonas putida mt-2 (mt2DyP and mt2P450, respectively), polymerase chain reaction (PCR) primers were designed on the basis of the DNA sequence of P. putida KT2440. They consisted of a sequence (20 bases) amplifying a target gene and a 15-base 5′ overhang that was identical to the pBIC3 expression vector (Table 1). Chromosomal DNA from P. putida mt-2 was extracted and purified using a commercial kit (Cica Geneus DNA extraction reagent, Kanto Chemical Co. Inc., Japan) for PCR as a template. The resulting PCR fragments were mixed with the plasmid vector and competent cells of Brevibacillus choshinensis to obtain transformants. Transformed cells were grown in 2SYN medium supplemented with 30 μM hemin chloride for 48 h at 30°C, and recombinant protein with an N-terminal His6-tag was extracellularly produced in the culture supernatant. After removing the cells by centrifugation, the culture supernatant was applied onto a Ni Sepharose column (Ni-Sepharose 6 Fast Flow column, GE Healthcare, Sweden), which was then washed with binding buffer (20 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole, pH 7.4). The recombinant protein was eluted with elution buffer (20 mM sodium phosphate, 500 mM NaCl, 500 mM imidazole, pH 7.4). The obtained eluate was subjected to ultrafiltration using a centrifugal concentrator (Vivaspin Turbo 15, 10,000 MWCO, Sartorius, Germany) to remove imidazole and exchange buffer into ultrapure water (resistivity at 25°C, >18 MΩ cm; TOC, <5 ppb). The recombinant protein was further purified by gel filtration using an AKTA purifier system (GE Healthcare Bio-Sciences AB, Sweden) equipped with a column (Superdex G-75, Pharmacia, Sweden), as needed. The protein sample was loaded onto the gel filtration column pre-equilibrated with 20 mM phosphate buffer (pH 7.0) and eluted with the same buffer at a flow rate of 2.0 ml/min. The fractions containing the enzyme were collected, and the buffer was exchanged by ultrafiltration. All abovementioned purification steps were performed at 4°C, and the effluent from the column was monitored by absorbance at 280 nm. The protein purity was assessed by SDS-PAGE on a 12.5% polyacrylamide gel. The concentration of protein was measured with a bicinchoninic acid protein assay kit (Pierce, Thermo Fisher Scientific, United States). Purified enzymes were stored at −30°C until further use.
2.3 Holoenzyme reconstitution
Holoenzymes were reconstituted by incubating purified enzyme in an aqueous solution of hemin chloride at twice the molar concentration of the enzyme for 2 h at room temperature (25°C). The excess hemin was removed by passing the mixture through a Ni Sepharose 6 Fast Flow column, and imidazole in the eluate was removed in the same manner as described above. The spectral characteristics of holoenzymes were obtained using a UV-Vis spectrophotometer (Cary 60 UV-Vis, Agilent Technologies, United States). Purified and reconstituted enzymes were stored at −30°C until further use.
2.4 Incubation of O-SWCNTs with enzymes
The enzyme was added to the O-SWCNT dispersion diluted with ultrapure water and incubated for 24 h at 4°C in the dark; thereafter, phosphate-buffered saline (PBS) and H2O2 were added to initiate the reaction. For incubation with mt2DyP or mt2P450, the 5 ml reaction mixture contained 30 μg/ml O-SWCNTs, 4 μM enzyme, and 1, 10, or 100 mM H2O2 in PBS (adjusted to pH 7.0) and was incubated at 37°C in the dark for 30 d, with a daily supply of each concentration of H2O2 chaser using a pipette. DTPA was added to the reaction mixture at a final concentration of 200 μM when its inhibition for CNT degradation was investigated. For incubation with HRP, the 5 ml reaction mixture contained the same concentrations of O-SWCNTs (30 μg/ml) and the enzyme (4 μM = 160 μg/ml) as above in PBS (adjusted to pH 6.0), and the reaction was initiated with 40 μM H2O2 and incubated for 30 d at 25°C in the dark with a daily supply of 1 μM H2O2 chaser using a pipette (Allen et al., 2009; Flores-Cervantes et al., 2014). HRP stock solution was prepared by dissolving lyophilized powder of HRP into ultrapure water at 5 mg/ml. During the incubation, samples were taken periodically, an equal volume of 5% sodium dodecyl sulfate (SDS) was added to disperse the O-SWCNT aggregate, sonicated for 10 min, and the absorbance at 750 nm (A750) was measured to calculate the concentration of O-SWCNTs from a calibration curve prepared using stocks of O-SWCNT dispersions. It was confirmed that the 10 min sonication itself did not alter the A750 and physical property (Raman spectrum) of O-SWCNTs (Supplementary Figure S1). Incubation of O-SWCNTs with free iron was performed under the same conditions as for incubation with enzymes, except that the enzyme was replaced by the same molar concentration of FeCl3.
The release of free iron from holoenzymes during incubation with O-SWCNTs was measured colorimetrically using ferrozine, following a previous method with slight modifications (Paumann-Page et al., 2013). Briefly, a 1 ml sample taken from the incubated mixture was lyophilized and resuspended in 60 µl of ultrapure water. An equal volume (60 μl) of 1.13 mM ascorbic acid solution in 0.2 M HCl was added and left for 5 min at room temperature. The protein was then precipitated by adding 60 μl of 11.3% trichloroacetic acid, and the samples were left on ice for 5 min followed by a short fast spin at 4°C. To the supernatant, 72 μl of 10% ammonium acetate was added, followed by 18 μl of 6.1 mM ferrozine, and the absorbance at 563 nm (A563) was measured after 5 min at room temperature (ε = 28,000 M−1cm−1). Hydroxyl radical (·OH) concentration of O-SWCNT suspension was measured using a fluorescent probe (HPF, 5 μM) (Hessler et al., 2012; Wang et al., 2020).
2.5 Measurement of enzyme activity
The enzyme activity during incubation with O-SWCNTs was determined by the initial velocity assay using ABTS as a substrate. The reaction was initiated by adding 10 μl of 100 mM H2O2 to 990 μl of the reaction buffer containing 15 μl of 30 mM ABTS and the sample solution (25 μl for mt2DyP/mt2P450 or 5 μl for HRP). As the buffer solution, acetate buffer of the optimum pH for each enzyme was used for mt2DyP or mt2P450, and PBS (adjusted to pH 6.0) was used for HRP. The absorbance of oxidized ABTS at 413 nm (A413) was continuously monitored for 30 s after the start of the reaction at room temperature (25 °C), and the enzyme activity was calculated from the slope of the linear approximation of the absorbance change. When the optimum pH of enzymes was determined, the enzyme activity for ABTS oxidation was measured in a Britton-Robinson buffer (50 mM phosphoric acid, 50 mM boric acid, and 50 mM acetic acid mixed with NaOH to the desired pH in the range of 3–9) containing 0.1 μM of purified enzyme.
To validate enzyme activity assays, the activity of HRP was measured using Amplex Red or ABTS as substrates, following the method employed in previous studies by Allen et al. (2009) and by Flores-Cervantes et al. (2014) or in this study. To follow the previous studies, we performed the assay under conditions described in their papers, including the concentrations of H2O2 and Amplex Red, the buffer used, and temperature. HRP solutions of various concentrations (0.6, 1.2, 2.4, and 4 μM) were prepared by dissolving lyophilized HRP into PBS (pH 6.0). Instead of sample solution, 500 μl of the HRP solution of various concentrations (or PBS for 0 μM) was added to a mixture of 468 μl of ultrapure water, 30 μl of 800 μM H2O2, and 2 μl of 10 mM Amplex Red (dissolved in dimethyl sulfoxide). Therefore, the final concentrations of HRP in this assay ranged from 0 to 2 μM. The mixture was gently mixed for 5 min at room temperature, and the UV-vis spectrum (300–700 nm) of oxidized Amplex Red was acquired. The relative activity of each HRP solution was determined from the absorbance at 570 nm (A570) of the UV-vis spectrum obtained. To validate our assay method in this study, the HRP activity was assayed using ABTS as a substrate under the conditions described above. Instead of sample solution, 5 μL of the HRP solution prepared above (0.6, 1.2, 2.4, or 4 μM) or PBS (for 0 μM) was added to 995 μl of the reaction mixture so that its final concentration ranged from 0 to 20 nM. The enzyme activity was determined by the initial velocity assay described above and the UV-vis spectrum (350–700 nm) of oxidized ABTS was also acquired 30 s after the start of the reaction. The activity of HRP was also assayed using Amplex Red instead of ABTS as a substrate, although basically following the method in this study. The reaction was initiated by adding 10 μl of 100 mM H2O2 to a mixture of 987 μl of PBS, 2 μl of 10 mM Amplex Red, and 1 μl of HRP solution of various concentrations (0.6, 1.2, 2.4, or 4 μM), and A570 was monitored at room temperature. HRP activity was determined from the initial velocity assay at various final concentrations of HRP (from 0 to 4 nM). The UV-vis spectra of oxidized ABTS at different HRP concentrations were also acquired 30 s after the start of the reaction.
2.6 Instrumental analyses
For Raman spectroscopy, samples were subjected to ultracentrifugation at 28000 rpm for 30 min, the supernatant was discarded by decantation, and the pellet was resuspended in methanol through sonication. Specimens were prepared by drop-casting approximately 100 μl of the samples on carbon tape or glass microscope slide and drying. All spectra were collected on a micro-Raman spectrometer (inVia Reflex, Renishaw, UK) using an excitation wavelength of 532 nm. The samples were scanned from 1000 to 1900 cm−1 to visualize the D-band and G-band of O-SWCNTs. Spectra were collected with a 10 s exposure time and averaged across 5 scans per sample. Samples analyzed by Raman spectroscopy were also used for X-ray photoelectron spectroscopy (XPS) (ESCALAB250, VG Scientific, UK). XPS was performed using monochromatized Al Kα radiation (hv= 1486.6 eV) as an X-ray source.
To analyze the degradation products of O-SWCNTs using high-performance liquid chromatography (HPLC), 3-ml samples taken from the incubated O-SWCNT suspension were lyophilized, resuspended in 200 μL methanol by vortexing for 1 min, and centrifuged again at 10000 rpm for 1 min. The supernatant was filtered through a 0.22 μm membrane filter (Millex-GV, Millipore, Ireland), and 20 μl of the filtrate was injected into an HPLC system (LC-20AT, Shimadzu, Japan) equipped with a reverse-phase C18 column (InertSustain C18, 4.6 × 150 mm, 5 μm, GL sciences, Japan) and a UV detector (SPD-10A VP, Shimadzu, Japan). The mobile phase, acetonitrile solution with a linear gradient from 0% to 70% for the initial 5 min followed by 70% isocratic elution, flowed at 1 mL/min. The detection wavelength was set to 254 nm based on the compounds identified as O-SWCNT degradation products in a previous study (Allen et al., 2009). In liquid chromatography-mass spectrometry (LC-MS), the column and separation conditions followed HPLC analysis. Approximately 10 μl of the sample was injected into the HPLC (1200 Series, Agilent, United States) tandem mass spectrometer (Compact, Bruker Daltonics, United States) and analyzed for positive ions using electrospray mass spectrometry.
2.7 Statistical analysis
The standard errors (SE) were calculated from three independent experiments. The quantitative data are expressed as the mean with SE (mean ± SE). Details about each figure are provided in the legend. Significance was calculated by Student’s t-test, and 95% confidence intervals (Cls) and p values of 0.05 or less were considered to indicate a statistically significant correlation or difference.
3 Results
3.1 O-SWCNT degradation during incubation in the presence of DyP from Pseudomonas putida mt-2
First, we attempted to clone the gene encoding DyP of P. putida mt-2. Although the whole genome of this strain was not revealed, we found a gene homologous to E-type DyPs (PP_3248 gene; accession number, NC_002947.4) in the genome of P. putida KT2440, the strain that lost the plasmid from the strain mt-2 (Regenhardt et al., 2002; Singh and Eltis, 2015). Utilizing this information for PCR, we successfully obtained the amplicon of mt2DyP, whose DNA sequence was completely identical to that of the DyP gene of strain KT2440. The cloned DNA was used for the production of the recombinant protein by B. choshinensis. The secreted enzyme was purified from the culture supernatant and subjected to SDS-PAGE to confirm its isolation and purity in the sample. As a result, a band was detected on the gel at ∼32 kDa, corresponding to the molecular mass of mt2DyP with a 6 × His tag (Figure 1A). The reconstituted mt2DyP exhibited an absorbance maximum at 410 nm, a Soret band derived from heme, with a Reinheit Zahl (RZ) value (A410/A280 ratio), an indicator for heme insertion, of 1.5 (Figure 1B), which was comparable to those of DyP from other bacteria (Chen et al., 2015a; Rahmanpour et al., 2016; Habib et al., 2019). The oxidation activity of mt2DyP for ABTS, a standard substrate of peroxidase, was examined at various pH values. This revealed that the enzyme activity was the highest at pH 4 (Figure 1C), which was similar to the optimum pH values of DyPs previously reported (Roberts et al., 2011; Li et al., 2012; Santos et al., 2014; Chen et al., 2015a). Then, CNTs were incubated in the presence of mt2DyP at pH 7, which is close to normal environmental conditions, to obtain useful information for predicting the environmental fate of CNTs, even though it is different from the optimum conditions.
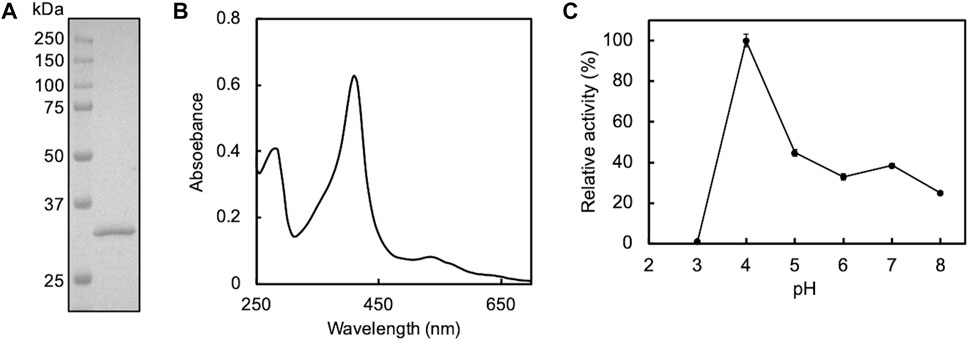
FIGURE 1. SDS-PAGE (A) and UV-Vis spectrum (B) of purified mt2DyP and the pH dependence of the enzyme activity on the ABTS substrate (C). Error bars indicate the SE from three independent assays.
A photograph of the appearance of the O-SWCNT suspensions during incubation in the presence of mt2DyP under different H2O2 concentrations (1, 10, 100 mM) is shown in Figure 2A. The black color of the O-SWCNTs decreased with the passage of time in the suspensions with 10 and 100 mM H2O2. Because biologically derived materials have no absorbance near 750 nm, the concentration of O-SWCNTs can be obtained by measuring A750 (Yang et al., 2019a) after adding SDS to remove proteins adsorbed on O-SWCNTs and dispersing the CNT aggregate. The O-SWCNT concentrations calculated from the A750 values of these samples are shown in Figure 2B The degradation of O-SWCNTs was more pronounced at higher H2O2 concentrations. The measurement of A750 also revealed that the O-SWCNTs degraded at 1 mM H2O2, although there was little change in appearance observed over time.
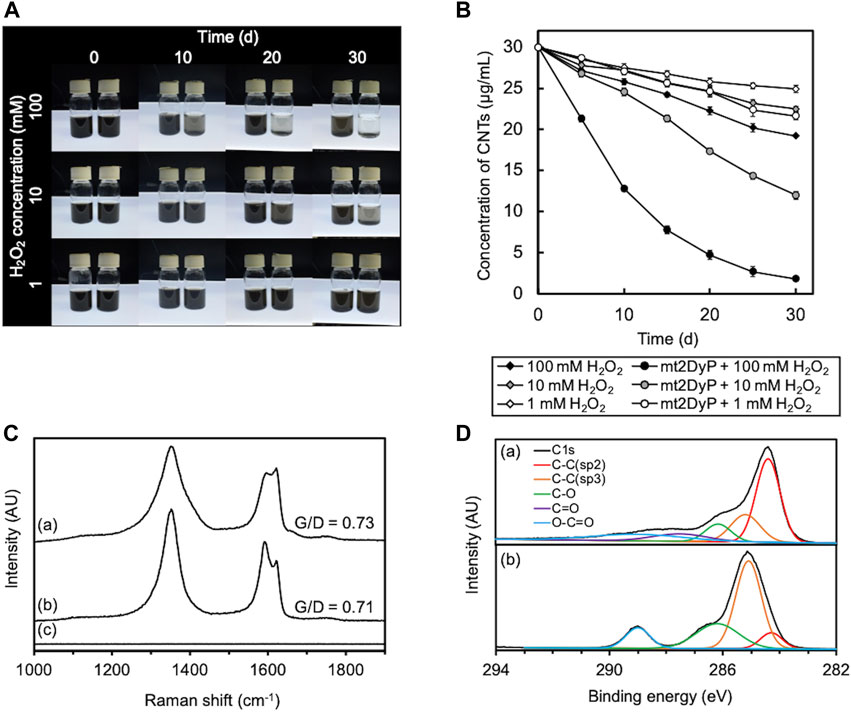
FIGURE 2. Degradation of O-SWCNTs during incubation at 37°C in the presence of mt2DyP under different H2O2 concentration conditions (1, 10, and 100 mM). (A) Photographs of the appearance of the O-SWCNT suspensions during incubation with (right) and without (left) mt2DyP at different time points (0, 10, 20, and 30 d). (B) Time courses of the O-SWCNT concentrations in suspensions incubated with and without mt2DyP. Error bars indicate the SE from three independent incubations. (C) Raman spectra (excitation wavelength, 532 nm) of untreated O-SWCNTs (A) and O-SWCNTs incubated with mt2DyP at an H2O2 concentration of 100 mM for 25 d (B) and 30 d (C). AU, arbitrary units. (D) XPS spectra of untreated O-SWCNTs (A) and O-SWCNTs incubated with mt2DyP at an H2O2 concentration of 100 mM for 25 d (B). AU, arbitrary units.
The degradation of O-SWCNTs was also confirmed by Raman spectroscopy. The G-band (graphite) peak at 1591 cm−1 and the D-band (disordered) peak at ∼1350 cm−1 that were present in the Raman spectra of pristine O-SWCNTs are characteristic Raman bands of graphitic carbon materials (Figure 2C) (Allen et al., 2008; Flores-Cervantes et al., 2014; You et al., 2017). The ratio of G-band to D-band (G/D) decreased with incubation with mt2DyP, which was consistent with the trend observed during CNT degradation in previous studies, and suggested an increase in O-SWCNT surface defects and amorphous carbon (Andon et al., 2013; Chandrasekaran et al., 2014; You et al., 2017; Wang et al., 2020). After incubation with mt2DyP for 30 d, both peaks disappeared, indicating the disappearance of O-SWCNTs. XPS analysis was performed to investigate in more detail the changes in the chemical composition of O-SWCNTs during incubation with mt2DyP. The main peak of C1s spectrum shifted to the lower binding energy because the peak attributed to sp2 C-C identified in pristine O-SWCNTs decreased and that attributed to sp3 C-C increased during incubation with mt2DyP (Figure 2D). In addition, oxygen functional groups (C-O and O-C=O) increased after the incubation. These results suggest that the graphite frame in O-SWCNTs was oxidized, supporting the above interpretation obtained from the Raman spectral change.
Subsequently, the degradation products of O-SWCNTs that were produced by incubation with mt2DyP were analyzed by HPLC. After 7 d of incubation, a peak unobserved at the beginning of incubation appeared at a retention time of 3.3 min, which was expected to be the peak of an O-SWCNT degradation product (Figures 3A, B). To identify this degradation product, the samples were subjected to LC-MS. A mass to charge (m/z) value of 132.1 was observed only in the sample after incubation with mt2DyP (Figures 3C, D), which corresponds to cinnamaldehyde, one of the previously reported degradation products of O-SWCNTs and pristine SWCNTs by HRP or Fenton reaction (Allen et al., 2009).
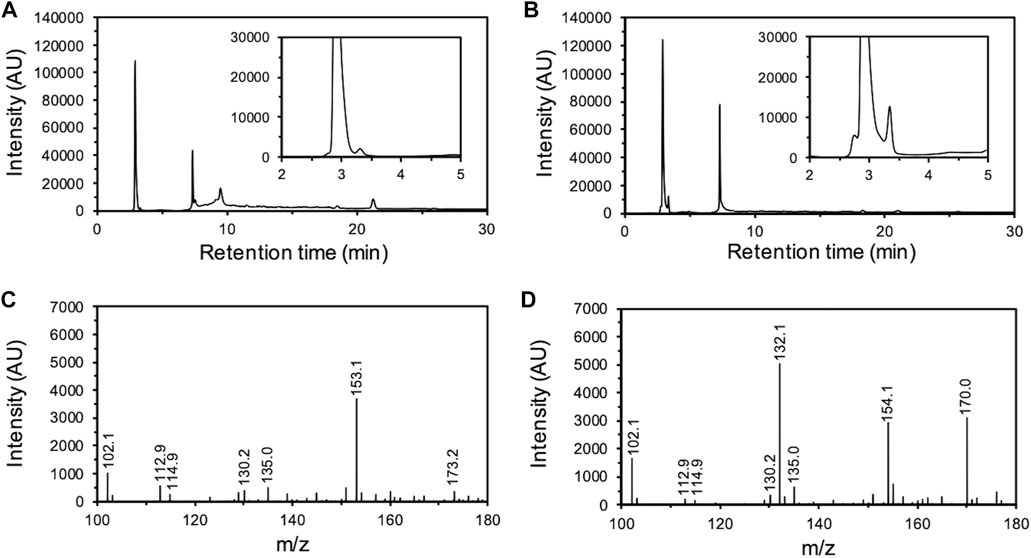
FIGURE 3. Analysis of O-SWCNT degradation products formed by incubation with mt2DyP. HPLC chromatograms of samples after 0 d (A) and 7 d (B) of incubation. LC-MS spectrum of samples after 0 d (C) and 7 d (D) of incubation. AU, arbitrary units.
3.2 Degradation mechanism of O-SWCNTs by incubation in the presence of mt2DyP
To determine whether the enzyme truly remained active during the degradation of the CNTs for as long as a month, the enzymatic activity to oxidize ABTS of mt2DyP during incubation with O-SWCNTs was measured over time. Surprisingly, at every H2O2 concentration, mt2DyP was inactivated within 1 h after the start of the incubation (Figure 4A). This implies that degradation of O-SWCNTs proceeded in the presence of mt2DyP, even after the enzyme was inactivated. In the absence of H2O2, the activity of mt2DyP was largely maintained, indicating that H2O2 caused inactivation of the enzyme. In fact, heme enzymes are known to be inactivated by heme degradation by H2O2, releasing free iron (Villegas et al., 2000; Valderrama et al., 2002; Nagababu and Rifkind, 2004; Albertolle and Guengerich, 2018).
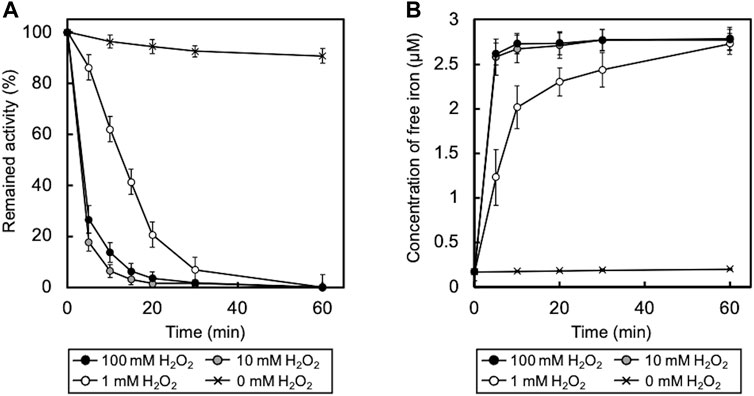
FIGURE 4. (A) Time course of enzyme activity of mt2DyP during incubation with O-SWCNTs. Error bars indicate the SE from three independent incubations. (B) Time course of free iron concentration in samples during incubation. Error bars indicate the SE from three independent incubations.
We supposed that the degradation of O-SWCNTs may have been caused by the Fenton reaction with iron released by the inactivation of mt2DyP. Measurement of the concentration of free iron in the samples revealed its increase during the incubation along with the inactivation of mt2DyP (Figure 4B). To confirm that free iron released from the enzyme mainly contributed to the degradation of O-SWCNTs via the Fenton reaction, incubation with mt2DyP was performed in the presence of DTPA, a chelating agent. The percentage of O-SWCNTs remaining after 30 d of incubation in the presence or absence of DTPA is shown in Figure 5. The degradation of O-SWCNTs was significantly inhibited by the addition of DTPA. The formation of ·OH, the main product of the Fenton reaction, was also detected in the samples (Supplementary Figure S2). In addition, when the enzyme was replaced by free iron at the same stoichiometric concentration as that of mt2DyP in the incubation, the rate of O-SWCNT degradation was similar to that in the incubation with the enzyme (Supplementary Figure S1). These results confirmed our hypothesis that O-SWCNTs were degraded by the Fenton reaction caused by iron released from mt2DyP’s heme. This suggested that the same phenomenon may occur in the presence of other heme enzymes.
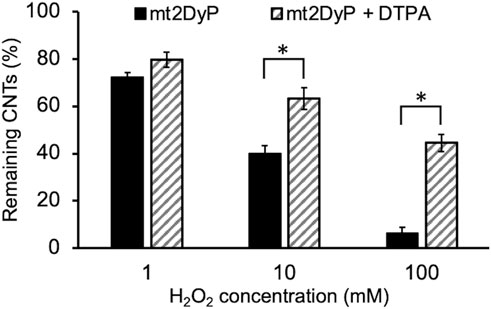
FIGURE 5. Concentration of O-SWCNTs after 30 d of incubation at 37°C with mt2DyP in the presence of DTPA (the initial O-SWCNT concentration is considered to be 100%). Error bars indicate the SE from three independent incubations. *p < 0.05 means significantly different (Student’s t-test).
3.3 Degradation of O-SWCNTs by incubation in the presence of another heme enzyme of P. putida
Cytochrome P450, a heme enzyme found in many organisms, including bacteria, was examined as another heme enzyme for O-SWCNT degradation. We designed PCR primers on the basis of the DNA sequence of the cytochrome P450 gene (PP_1955 gene; accession number, NC_002947.4) on the genome of P. putida KT2440 for the cloning of a cytochrome P450 homolog from P. putida mt-2, mt2P450, and successfully obtained the amplicon by PCR using chromosomal DNA of mt-2 as a template. The DNA sequence of this amplicon was completely identical to that of the cytochrome P450 gene of strain KT2440, and this cloned DNA was used for recombinant protein production by B. choshinensis. On the SDS-PAGE gel of purified mt2P450, a band was detected at ∼45 kDa corresponding to the molecular mass of mt2P450 with a 6 × His tag (Supplementary Figure S4A). The reconstituted mt2P450 exhibited an absorbance maximum at 410 nm, similar to mt2DyP, with an RZ value (A410/A280 ratio) of 0.4 (Supplementary Figure S4B). Because there are no reports of the RZ value for cytochrome P450, it is impossible to evaluate the RZ value of mt2P450 in comparison with others. The ABTS oxidation activity of mt2P450 was highest at pH 5 (Supplementary Figure S4C). As expected, the incubation of O-SWCNTs in the presence of mt2P450 resulted in its degradation. As in the case with mt2DyP, when incubated with mt2P450 in the presence of 10 or 100 mM H2O2, the black color of the samples became distinctly transparent over time (Figure 6A). The concentration of O-SWCNTs measured with A750 decreased with incubation time, and the decrease was faster with higher H2O2 concentrations (Figure 6B). The change in Raman and XPS spectra during incubation in the presence of mt2P450 also showed the same tendency as that in the presence of mt2DyP. The G/D ratio decreased at 25 d of incubation with mt2P450 at 100 mM H2O2 in the Raman spectrum, and both the G- and D-band peaks in O-SWCNTs disappeared after 30 d of incubation (Figure 6C). The XPS spectrum showed a decrease in the sp2 C-C peak and an increase in the sp3 C-C peak and oxygen functional group peaks during the incubation with mt2P450 (Figure 6D). HPLC and LC-MS analyses of degradation products of O-SWCNTs by incubation with mt2P450 showed similar peaks to those observed in the case of incubation with mt2DyP (Supplementary Figure S5). The release of free iron accompanied by the inactivation of mt2P450 enzymatic activity and the formation of ⋅OH during incubation with O-SWCNTs was also observed (Figure 6E; Supplementary Figure S6). In the absence of H2O2, the enzyme activity was largely maintained, indicating that H2O2 also caused inactivation of mt2P450 (Figure 6E). It was described above that the degradation rates of O-SWCNT were almost identical when mt2P450 was replaced by free iron of the same stoichiometric concentration (Supplementary Figure S3); note that the molar concentrations (4 μM) of mt2P450 and mt2DyP used for incubation with O-SWCNTs were same. These results confirmed that O-SWCNTs were degraded by the Fenton reaction caused by iron released from mt2P450, a heme enzyme other than mt2DyP, inactivated by H2O2 during incubation in the presence of H2O2.
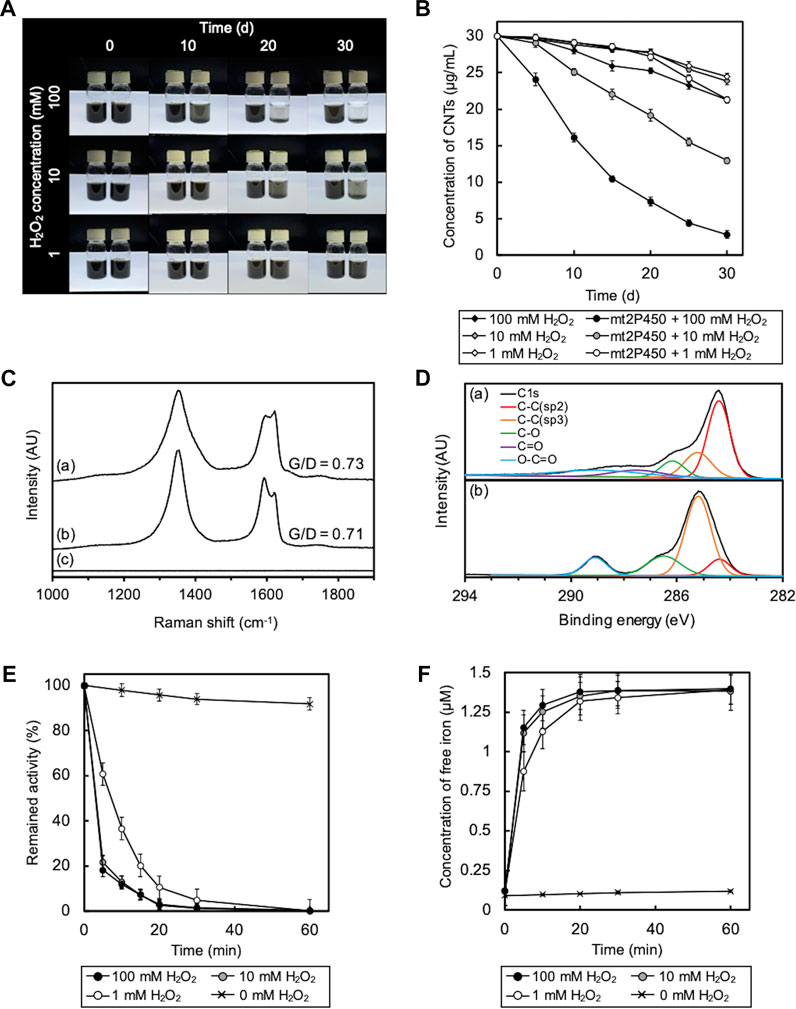
FIGURE 6. Degradation of O-SWCNTs during incubation in the presence of mt2P450 under different H2O2 concentration conditions (1, 10, and 100 mM) at 37 °C. (A) Photographs of the degradation of O-SWCNTs by incubation with mt2P450 (right) and without mt2P450 (left) at different time points (0, 10, 20, and 30 d). (B) Time course of the O-SWCNT concentrations in samples incubated with and without mt2P450. Error bars indicate the SE from three independent incubations. (C) Raman spectra (excitation wavelength, 532 nm) of untreated O-SWCNTs (A) and O-SWCNTs incubated with mt2P450 at an H2O2 concentration of 100 mM for 25 d (B) and 30 d (C). AU, arbitrary units. (D) XPS spectra of untreated O-SWCNTs (A) and O-SWCNTs incubated with mt2P450 at an H2O2 concentration of 100 mM for 25 d (B). AU, arbitrary units. (E) Time course of enzyme activity of mt2P450 during incubation with O-SWCNTs. Error bars indicate the SE from three independent incubations. (F) Time course of free iron concentration in samples during incubation. The color of the marker indicates the concentration of H2O2: 1 mM (white), 10 mM (gray), and 100 mM (black). Error bars indicate the SE from three independent incubations.
3.4 Degradation of O-SWCNTs by incubation in the presence of HRP
Most previous studies on the enzymatic degradation of CNTs have been performed without consideration of the Fenton reaction, despite the use of heme enzymes (Allen et al., 2008; Andon et al., 2013; Chandrasekaran et al., 2014; Zhang et al., 2014; Bhattacharya et al., 2015). Therefore, it is unclear whether O-SWCNT degradation in these studies was solely due to enzymatic reactions. To investigate the contribution of the Fenton reaction to the degradation, O-SWCNTs were incubated with HRP under the same experimental conditions as described by Allen et al. (2009), including H2O2 concentration (40 μM at the initial and 1 μM/d for the chaser), temperature, and pH; however, the O-SWCNTs used, their concentration, and the enzyme concentration differed (enzyme concentration was higher in this study). No visible reduction in the black color of the O-SWCNT suspension was observed even after 30 d of incubation with HRP (Figure 7A), unlike the results shown in the article by Allen et al. (2009). The measurement of A750 revealed that the concentration of O-SWCNTs decreased by 4.6% after 30 d of incubation (Figure 7B). However, the addition of DTPA inhibited the decrease in the O-SWCNT concentration to 1.3%. In addition, more than 80% inactivation of HRP was observed after 10 d of incubation with O-SWCNTs (Figure 7C), accompanied by the release of free iron into the solution (Figure 7D). Even when HRP was replaced by free iron at the same stoichiometric concentration, the degradation rate of O-SWCNTs was almost identical (Supplementary Figure S7). HRP was also not inactivated in the absence of H2O2 (Supplementary Figure S8). These results imply that the Fenton reaction by iron released from HRP inactivated by H2O2 greatly contributed to the degradation of O-SWCNTs and that enzymatic degradation was negligible.
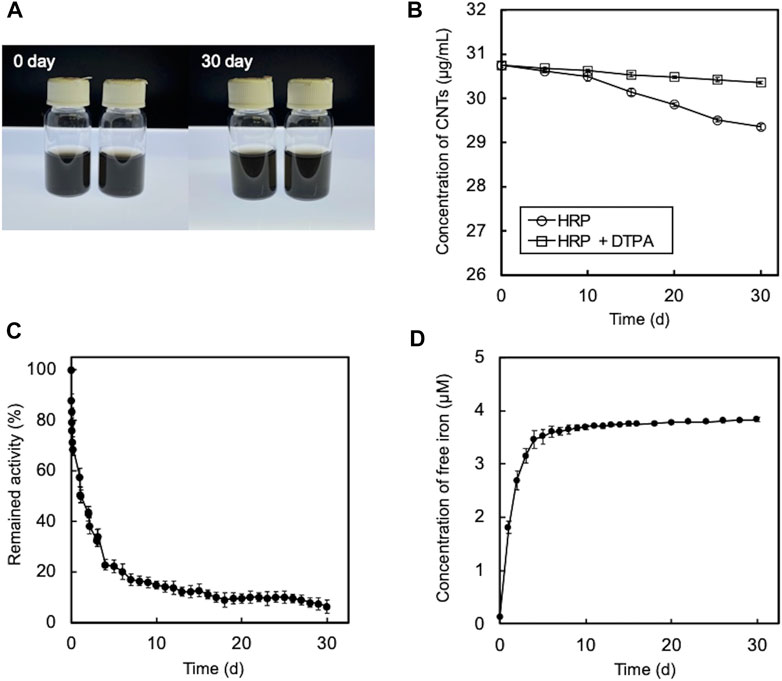
FIGURE 7. Degradation of O-SWCNTs during incubation at 25°C in the presence of HRP and 40 μM H2O2. (A) Photographs of the degradation of O-SWCNTs by incubation with (right) and without HRP (left). (B) Time course of the O-SWCNT concentrations in samples incubated with HRP. The color of the marker indicates with (white) and without (black) DTPA. Error bars indicate the SE from three independent incubations. (C) Time course of the enzyme activity of HRP during incubation with O-SWCNTs. Error bars indicate the SE from three independent incubations. (D) Time course of free iron concentration in samples during incubation. Error bars indicate the SE from three independent incubations.
The rapid inactivation of HRP observed in this study differed from the results of the previous studies by Allen et al. (2009) and by Flores-Cervantes et al. (2014), in which the activity of HRP was maintained for long periods of time. The methods used to measure HRP activity differed between this study and the previous studies. Therefore, we assessed the validity of both methods. To validate the previous assay methods, we measured the oxidation of Amplex Red by HRP at different concentrations, according to the assay conditions described in the previous studies. As a result, the UV-vis spectrum of oxidized Amplex Red was unchanged even when the HRP concentration was changed (Figure 8A). In accordance with the previous studies, the relative activity for Amplex Red oxidation by HRP at different concentrations was determined from A570 of the UV-vis spectra. The relative activity was independent of the HRP concentration (Figure 8B), suggesting inaccuracy in the enzyme activity measurements in the previous studies. To validate the assay method used in this study, we also measured the oxidation of ABTS by HRP at different concentrations. As shown in Figures 8C, D, the UV-vis spectrum of oxidized ABTS was changed by the HRP concentration, and the relative activity determined from the slope of the linear approximation of A413 change (Fig. S9) depended on the HRP concentration. Thus, it was demonstrated that the HRP activity was accurately measured by the method used in this study.
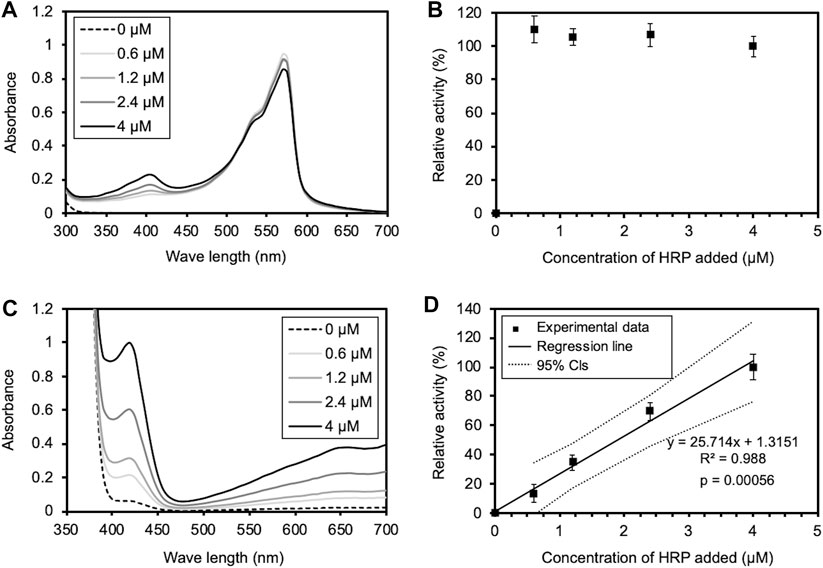
FIGURE 8. Validation of enzyme activity assays used in the previous and this studies. (A) The UV-Vis spectrum of oxidized Amplex Red under the assay conditions in the previous studies. 500 μl of a HRP solution of various concentrations (0, 0.6, 1.2, 2.4, and 4 μM) was added to the equal volume of the reaction mixture. The final concentrations of Amplex Red, H2O2, and HRP in PBS (pH 6.0) were 20, 24 μM, and 0, 0.3, 0.6, 1.2, and 2 μM. (B) Relative activity of Amplex Red oxidation at different HRP concentrations, determined from A570 of the UV-Vis spectrum in (A), according to the previous studies. The activity at the highest HRP concentration is shown as 100%. Error bars indicate the SE from three independent assays. (C) The UV-Vis spectrum of oxidized ABTS under the assay conditions in this study. 5 μl of a HRP solution of various concentrations (0, 0.6, 1.2, 2.4, and 4 μM) was added to 995 μl of the reaction mixture. The final concentrations of ABTS, H2O2, and HRP in PBS (pH 6.0) were 0.45, 1 mM, and 3, 6, 12, and 20 nM. (D) Relative activity of ABTS oxidation at different HRP concentrations, determined from A413 of the UV-Vis spectrum in (C). The activity at the highest HRP concentration is shown as 100%. Error bars indicate the SE from three independent assays. The solid and dotted lines indicate a regression line and 95% confidence intervals, respectively. The statistical p-value is also provided.
4 Discussion
At least under the experimental conditions used in this study, the degradation of O-SWCNTs in the presence of a heme enzyme was found to be primarily due to the Fenton reaction because the degradation proceeded during and after inactivation of the enzyme but was significantly inhibited by DTPA. This would have been the case even in O-SWCNT degradation in the presence of HRP, which was previously shown to enzymatically degrade O-SWCNTs (Allen et al., 2009). Flores-Cervantes et al. first quantitatively investigated CNT degradation by heme enzymes and showed that O-SWCNTs were biotransformed by HRP by 0.51% in 30 d (Flores-Cervantes et al., 2014), which was largely different from the rapid degradation (became colorless within 10 d) in the article by Allen et al. (2009). Of course, not all experimental conditions in these two studies were the same, but the difference in results between the two studies was too great. Therefore, to date, the degradation of CNTs by HRP is still not well-understood. Our results support the results of the study by Flores-Cervantes et al. (2014).
In addition, this study revealed that the assay methods to determine the activity of HRP during incubation with CNTs in the previous studies (Allen et al., 2009; Flores-Cervantes et al., 2014) were not accurate. This should be because the concentration of H2O2 used in the previous studies was too low relative to that of HRP. The assays were performed at the molar concentration ratio of HRP to H2O2 of 1:20 in the previous studies, whereas it was 1:50000 in this study. Since a lack of substrate leads to underestimation of the enzyme activity, an excess of substrates relative to the enzyme concentration is usually employed to accurately measure peroxidase activity (Zhao et al., 1992; Monier et al., 2010; Yu et al., 2019). In addition, in the previous studies, the enzyme activity was determined by measuring the absorbance at one time point, 5 min after the start of the reaction; this method has rarely been used for measuring peroxidase activity. By contrast, we determined the enzyme activity by continuously monitoring the change in the absorbance for 30 s from the start of the reaction to get the slope of the linear approximation; this initial velocity assay is common for measuring peroxidase activity. Under the assay conditions used in this study, the absorbance of oxidized ABTS increased gradually and linearly for 30 s from the start of the reaction, and the slope of the approximate line depended on the HRP concentration (Fig. S9). We also attempted to monitor the change in the absorbance of oxidized Amplex Red under the conditions used in the previous studies. As a result, Amplex Red was instantly oxidized at the start of the reaction, and the change in absorbance could not be measured accurately; the reaction ended before the start of absorbance measurement at time 0, and absorbance did not change or conversely decreased thereafter (Supplementary Figure S10), suggesting that the assay conditions used in the previous studies were inappropriate. Thus, the initial velocity assay is also useful to validate the assay conditions. We examined HRP activity using Amplex Red as a substrate, following our assay conditions with a slight modification; the molar concentration ratio of HRP to H2O2 was set at 1:250000 because Amplex Red is more sensitive than ABTS as a substrate for HRP. Under these conditions, the absorbance was saturated at around 2 min and therefore, measurement of the absorbance at one time point of 5 min cannot determine the correct enzyme activity (Supplementary Figure S11). As shown in Supplementary Figure S12, it was revealed that HRP activity can be accurately measured under appropriate conditions even using Amplex Red. The precise activity assay in this study showed that the activity of HRP is not maintained for very long during incubation with CNTs in the presence of H2O2. In the presence of an oxidizing substrate, H2O2, and the absence of a reducing substrate, the inactivation of peroxidase is accelerated by heme degradation, called suicide inactivation (Valderrama et al., 2002). The fast inactivation of HRP observed in this study implies that CNTs were not utilized as a reducing substrate, i.e., they were not enzymatically oxidized. However, on one hand, since there are various types of CNTs with different numbers of walls, morphologies, and attached functional groups, further investigation is needed to rule out the possibility that there are CNTs that are susceptible to enzymatic degradation by HRP. On the other hand, the results of this study emphasize that the Fenton reaction, which has received little attention in the degradation of CNTs by heme enzymes, must be taken into consideration as well as quantitative analyses for accurate assessment of CNT degradation, and that accurate measurement of enzyme activity is essential.
At each H2O2 concentration, even in the control sample without enzyme, O-SWCNTs were slightly degraded (Figures 2B, 6B). This occurred probably due to trace amounts of residual iron used as a catalyst for O-SWCNT synthesis (Hata et al., 2004). In addition, the degradation of O-SWCNTs proceeded to some extent even in the presence of DTPA (Figure 5). This occurred probably because chelating agents reduce the rate of the Fenton reaction but do not prevent it completely (Li et al., 2005). Thus, the Fenton reaction, which is highly dependent on the H2O2 concentration, is more effective and stable than the enzymatic reaction in the degradation of CNTs, suggesting that its contribution to degradation should be carefully considered. The O-SWCNT degradation rates were much higher for mt2DyP and mt2P450 than for HRP, which is likely due to the much higher H2O2 concentrations used for the mt2 enzymes than for HRP. Conversely, the inactivation of HRP was slower than that of the mt2 enzymes, probably for the same reason. In this study, degradation experiments were performed at pH 7, which is not optimal for mt2DyP and mt2P450, to match the natural environmental conditions. Moreover, H2O2 concentrations were high enough to inactivate enzymes. Therefore, degradation experiments should be conducted at the optimum pH and H2O2 concentrations for enzymes to confirm whether enzymatic degradation of O-SWCNTs truly occurs.
There is an estimation that 1 g of soil contains 1010–1011 bacteria (Horner-Devine et al., 2003). Enzymes externally secreted by bacteria are also abundant in soil, and the activity and role of enzymes after secretion into the soil have been extensively studied (Burns et al., 2013). Soil enzymes also include heme enzymes such as peroxidase (Sinsabaugh, 2010). Iron is widely distributed in the environment, mostly as iron oxide minerals that are less efficient than free iron in the Fenton reaction (Huang et al., 2013; Lai et al., 2021). Therefore, the external addition of H2O2 to release free iron from reproducible heme enzymes into the environment and induce the Fenton reaction is expected to be an effective method of CNT degradation for environmental remediation and waste treatment. In fact, Fenton or Fenton-like reactions using H2O2 have been studied as a relatively inexpensive method for the remediation of environments contaminated with various toxic substances (Vicente et al., 2011; Yap et al., 2011; Sun et al., 2014; Yu et al., 2019b; Li et al., 2023). These studies applied H2O2 concentrations similar to or higher than those used in this study but much lower than those used for disinfection purposes (Zeng and Wu, 2021). Therefore, our results will contribute to the development of a simple disposal method for CNTs without the risk of secondary contamination.
5 Conclusion
O-SWCNTs can be degraded in the presence of bacterial heme enzymes via the Fenton reaction with externally added H2O2 and iron released from the inactivated enzymes. Under the experimental conditions in this study, even in the presence of HRP, the degradation of O-SWCNTs was primarily due to the Fenton reaction, and enzymatic degradation was negligible. The obtained results suggest that the Fenton reaction, which has received little attention in CNT degradation by heme enzymes, must be taken into consideration and will contribute to the development of a simple disposal method for CNTs, utilizing the Fenton reaction with bacteria/their enzymes and H2O2.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Author contributions
ST: Investigation, methodology, formal analysis, writing—Original draft. FT: Investigation, methodology. KH: Conceptualization, planning, writing—Review and editing. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Zeon Nanotechnology Co., Ltd. for providing the O-SWCNT sample. We also thank Kanie of Friend Microbe Inc. (Japan) for the helpful discussion about this study. The preprint of the paper has been submitted to ChemRxiv (https://doi.org/10.26434/chemrxiv-2021-t6n20-v2). The authors declare that this study received a contribution from Zeon Nanotechnology Co. Ltd. The company was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1184257/full#supplementary-material
References
Albertolle, M. E., and Guengerich, F. P. (2018). The relationships between cytochromes P450 and H2O2: Production, reaction, and inhibition. J. Inorg. Biochem. 186, 228–234. doi:10.1016/j.jinorgbio.2018.05.014
Allen, B. L., Kichambare, P. D., Gou, P., Vlasova, I. I., Kapralov, A. A., Konduru, N., et al. (2008). Biodegradation of single-walled carbon nanotubes through enzymatic catalysis. Nano Lett. 8 (11), 3899–3903. doi:10.1021/nl802315h
Allen, B. L., Kotchey, G. P., Chen, Y. N., Yanamala, N. V. K., Klein-Seetharaman, J., Kagan, V. E., et al. (2009). Mechanistic investigations of horseradish peroxidase-catalyzed degradation of single-walled carbon nanotubes. J. Am. Chem. Soc. 131 (47), 17194–17205. doi:10.1021/ja9083623
Almarasy, A. A., Hayasaki, T., Abiko, Y., Kawabata, Y., Akasaka, S., and Fujimori, A. (2021). Comparison of characteristics of single-walled carbon nanotubes obtained by super-growth CVD and improved-arc discharge methods pertaining to interfacial film formation and nanohybridization with polymers. Colloids Surfaces a-Physicochemical Eng. Aspects 615, 126221. doi:10.1016/j.colsurfa.2021.126221
Andon, F. T., Kapralov, A. A., Yanamala, N., Feng, W. H., Baygan, A., Chambers, B. J., et al. (2013). Biodegradation of single-walled carbon nanotubes by eosinophil peroxidase. Small 9 (16), 2721–2729. doi:10.1002/smll.201202508
Anzar, N., Hasan, R., Tyagi, M., Yadav, N., and Narang, J. (2020). Carbon nanotube - a review on synthesis, properties and plethora of applications in the field of biomedical science. Sensors Int. 1, 100003. doi:10.1016/j.sintl.2020.100003
Bhattacharya, K., El-Sayed, R., Andon, F. T., Mukherjee, S. P., Gregory, J., Li, H., et al. (2015). Lactoperoxidase-mediated degradation of single-walled carbon nanotubes in the presence of pulmonary surfactant. Carbon 91, 506–517. doi:10.1016/j.carbon.2015.05.022
Bugg, T. D. H., Ahmad, M., Hardiman, E. M., and Singh, R. (2011). The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 22 (3), 394–400. doi:10.1016/j.copbio.2010.10.009
Burns, R. G., DeForest, J. L., Marxsen, J., Sinsabaugh, R. L., Stromberger, M. E., Wallenstein, M. D., et al. (2013). Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 58, 216–234. doi:10.1016/j.soilbio.2012.11.009
Byrne, C., Subramanian, G., and Pillai, S. C. (2018). Recent advances in photocatalysis for environmental applications. J. Environ. Chem. Eng. 6 (3), 3531–3555. doi:10.1016/j.jece.2017.07.080
Chandrasekaran, G., Choi, S. K., Lee, Y. C., Kim, G. J., and Shin, H. J. (2014). Oxidative biodegradation of single-walled carbon nanotubes by partially purified lignin peroxidase from Sparassis latifolia mushroom. J. Industrial Eng. Chem. 20 (5), 3367–3374. doi:10.1016/j.jiec.2013.12.022
Chen, C., Shrestha, R., Jia, K., Gao, P. F., Geisbrecht, B. V., Bossmann, S. H., et al. (2015a). Characterization of dye-decolorizing peroxidase (DyP) from Thermomonospora curvata reveals unique catalytic properties of A-type DyPs. J. Biol. Chem. 290 (38), 23447–23463. doi:10.1074/jbc.M115.658807
Chen, D. R., Adusei, P. K., Chitranshi, M., Fang, Y. B., Johnson, K., Schulz, M., et al. (2021). Electrochemical activation to enhance the volumetric performance of carbon nanotube electrodes. Appl. Surf. Sci. 541, 148448. doi:10.1016/j.apsusc.2020.148448
Chen, M., Zhou, S., Zhu, Y., Sun, Y. Z., Zeng, G. M., Yang, C. P., et al. (2018). Toxicity of carbon nanomaterials to plants, animals and microbes: Recent progress from 2015-present. Chemosphere 206, 255–264. doi:10.1016/j.chemosphere.2018.05.020
Chen, Q. L., Wang, H., Yang, B. S., He, F., Han, X. M., and Song, Z. H. (2015b). Responses of soil ammonia-oxidizing microorganisms to repeated exposure of single-walled and multi-walled carbon nanotubes. Sci. Total Environ. 505, 649–657. doi:10.1016/j.scitotenv.2014.10.044
Colpa, D. I., Fraaije, M. W., and van Bloois, E. (2014). DyP-Type peroxidases: A promising and versatile class of enzymes. J. Industrial Microbiol. Biotechnol. 41 (1), 1–7. doi:10.1007/s10295-013-1371-6
Dong, Y. D., Zhang, H., Zhong, G. J., Yao, G., and Lai, B. (2021). Cellulose/carbon composites and their applications in water treatment - a review. Chem. Eng. J. 405, 126980. doi:10.1016/j.cej.2020.126980
Dresselhaus, M. S., Dresselhaus, G., Charlier, J. C., and Hernandez, E. (2004). Electronic, thermal and mechanical properties of carbon nanotubes. Philosophical Trans. R. Soc. a-Mathematical Phys. Eng. Sci. 362 (1823), 2065–2098. doi:10.1098/rsta.2004.1430
Flores-Cervantes, D. X., Maes, H. M., Schaffer, A., Hollender, J., and Kohler, H. P. E. (2014). Slow biotransformation of carbon nanotubes by horseradish peroxidase. Environ. Sci. Technol. 48 (9), 4826–4834. doi:10.1021/es4053279
Habib, M. H., Rozeboom, H. J., and Fraaije, M. W. (2019). Characterization of a new DyP-peroxidase from the alkaliphilic cellulomonad, cellulomonas bogoriensis. Molecules 24 (7), 1208. doi:10.3390/molecules24071208
Hata, K., Futaba, D. N., Mizuno, K., Namai, T., Yumura, M., and Iijima, S. (2004). Water-assisted highly efficient synthesis of impurity-free single-walled carbon nanotubes. Science 306 (5700), 1362–1364. doi:10.1126/science.1104962
Hatami, M. (2017). Toxicity assessment of multi-walled carbon nanotubes on Cucurbita pepo L. under well-watered and water-stressed conditions. Ecotoxicol. Environ. Saf. 142, 274–283. doi:10.1016/j.ecoenv.2017.04.018
Hessler, C. M., Wu, M. Y., Xue, Z., Choi, H., and Seo, Y. (2012). The influence of capsular extracellular polymeric substances on the interaction between TiO2 nanoparticles and planktonic bacteria. Water Res. 46 (15), 4687–4696. doi:10.1016/j.watres.2012.06.009
Ho, N. T., Siggel, M., Camacho, K. V., Bhaskara, R. M., Hicks, J. M., Yao, Y. C., et al. (2021). Membrane fusion and drug delivery with carbon nanotube porins. Proc. Natl. Acad. Sci. U. S. A. 118 (19), 4118. doi:10.1073/pnas.2016974118
Horner-Devine, M. C., Leibold, M. A., Smith, V. H., and Bohannan, B. J. M. (2003). Bacterial diversity patterns along a gradient of primary productivity. Ecol. Lett. 6 (7), 613–622. doi:10.1046/j.1461-0248.2003.00472.x
Huang, W. Y., Brigante, M., Wu, F., Hanna, K., and Mailhot, G. (2013). Effect of ethylenediamine-N,N'-disuccinic acid on Fenton and photo-fenton processes using goethite as an iron source: Optimization of parameters for bisphenol A degradation. Environ. Sci. Pollut. Res. 20 (1), 39–50. doi:10.1007/s11356-012-1042-6
Kadri, T., Rouissi, T., Brar, S. K., Cledon, M., Sarma, S., and Verma, M. (2017). Biodegradation of polycyclic aromatic hydrocarbons (PAHs) by fungal enzymes: A review. J. Environ. Sci. 51, 52–74. doi:10.1016/j.jes.2016.08.023
Kagan, V. E., Konduru, N. V., Feng, W. H., Allen, B. L., Conroy, J., Volkov, Y., et al. (2010). Carbon nanotubes degraded by neutrophil myeloperoxidase induce less pulmonary inflammation. Nat. Nanotechnol. 5 (5), 354–359. doi:10.1038/Nnano.2010.44
Kumarathasan, P., Breznan, D., Das, D., Salam, M. A., Siddiqui, Y., MacKinnon-Roy, C., et al. (2015). Cytotoxicity of carbon nanotube variants: A comparative in vitro exposure study with A549 epithelial and J774 macrophage cells. Nanotoxicology 9 (2), 148–161. doi:10.3109/17435390.2014.902519
Lai, C., Shi, X. X., Li, L., Cheng, M., Liu, X. G., Liu, S. Y., et al. (2021). Enhancing iron redox cycling for promoting heterogeneous Fenton performance: A review. Sci. Total Environ. 775, 145850. doi:10.1016/j.scitotenv.2021.145850
Li, J., Liu, C., Li, B. Z., Yuan, H. L., Yang, J. S., and Zheng, B. W. (2012). Identification and molecular characterization of a novel DyP-type peroxidase from Pseudomonas aeruginosa PKE117. Appl. Biochem. Biotechnol. 166 (3), 774–785. doi:10.1007/s12010-011-9466-x
Li, X. M., Xu, J. L., and Yang, Z. L. (2023). Insight on efficiently oriented oxidation of petroleum hydrocarbons by redistribution of oxidant through inactivation of soil organic matter coupled with passivation of manganese minerals. J. Hazard. Mater. 443, 130192. doi:10.1016/j.jhazmat.2022.130192
Li, Y. C., Bachas, L. G., and Bhattacharyya, D. (2005). Kinetics studies of trichlorophenol destruction by chelate-based Fenton reaction. Environ. Eng. Sci. 22 (6), 756–771. doi:10.1089/ees.2005.22.756
Mendonca, M. C. P., Rizoli, C., Avila, D. S., Amorim, M. J. B., and de Jesus, M. B. (2017). Nanomaterials in the environment: Perspectives on in vivo terrestrial toxicity testing. Front. Environ. Sci. 5, 71. doi:10.3389/fenvs.2017.00071
Monier, M., Ayad, D. M., Wei, Y., and Sarhan, A. A. (2010). Immobilization of horseradish peroxidase on modified chitosan beads. Int. J. Biol. Macromol. 46 (3), 324–330. doi:10.1016/j.ijbiomac.2009.12.018
Nagababu, E., and Rifkind, J. M. (2004). Heme degradation by reactive oxygen species. Antioxidants Redox Signal. 6 (6), 967–978. doi:10.1089/ars.2004.6.967
Paumann-Page, M., Furtmuller, P. G., Hofbauer, S., Paton, L. N., Obinger, C., and Kettle, A. J. (2013). Inactivation of human myeloperoxidase by hydrogen peroxide. Archives Biochem. Biophysics 539 (1), 51–62. doi:10.1016/j.abb.2013.09.004
Rahmanpour, R., and Bugg, T. D. H. (2015). Characterisation of dyp-type peroxidases from Pseudomonas fluorescens pf-5: Oxidation of Mn(II) and polymeric lignin by Dyp1B. Archives Biochem. Biophysics 574, 93–98. doi:10.1016/j.abb.2014.12.022
Rahmanpour, R., Rea, D., Jamshidi, S., Fulop, V., and Bugg, T. D. H. (2016). Structure of Thermobifida fusca DyP-type peroxidase and activity towards Kraft lignin and lignin model compounds. Archives Biochem. Biophysics 594, 54–60. doi:10.1016/j.abb.2016.02.019
Regenhardt, D., Heuer, H., Heim, S., Fernandez, D. U., Strompl, C., Moore, E. R. B., et al. (2002). Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ. Microbiol. 4 (12), 912–915. doi:10.1046/j.1462-2920.2002.00368.x
Roberts, J. N., Singh, R., Grigg, J. C., Murphy, M. E. P., Bugg, T. D. H., and Eltis, L. D. (2011). Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry 50 (23), 5108–5119. doi:10.1021/bi200427h
Santos, A., Mendes, S., Brissos, V., and Martins, L. O. (2014). New dye-decolorizing peroxidases from Bacillus subtilis and Pseudomonas putida MET94: Towards biotechnological applications. Appl. Microbiol. Biotechnol. 98 (5), 2053–2065. doi:10.1007/s00253-013-5041-4
Singh, A. K., Bilal, M., Iqbal, H. M. N., and Raj, A. (2021). Lignin peroxidase in focus for catalytic elimination of contaminants ? A critical review on recent progress and perspectives. Int. J. Biol. Macromol. 177, 58–82. doi:10.1016/j.ijbiomac.2021.02.032
Singh, R., and Eltis, L. D. (2015). The multihued palette of dye-decolorizing peroxidases. Archives Biochem. Biophysics 574, 56–65. doi:10.1016/j.abb.2015.01.014
Singh, R., Grigg, J. C., Qin, W., Kadla, J. F., Murphy, M. E. P., and Eltis, L. D. (2013). Improved manganese-oxidizing activity of DypB, a peroxidase from a lignolytic bacterium. Acs Chem. Biol. 8 (4), 700–706. doi:10.1021/cb300608x
Sinsabaugh, R. L. (2010). Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biol. Biochem. 42 (3), 391–404. doi:10.1016/j.soilbio.2009.10.014
Sireesha, M., Babu, V. J., Kiran, A. S. K., and Ramakrishna, S. (2018). A review on carbon nanotubes in biosensor devices and their applications in medicine. Nanocomposites 4 (2), 36–57. doi:10.1080/20550324.2018.1478765
Sun, S. P., Zeng, X., Li, C., and Lemley, A. T. (2014). Enhanced heterogeneous and homogeneous Fenton-like degradation of carbamazepine by nano-Fe3O4/H2O2 with nitrilotriacetic acid. Chem. Eng. J. 244, 44–49. doi:10.1016/j.cej.2014.01.039
Valderrama, B., Ayala, M., and Vazquez-Duhalt, R. (2002). Suicide inactivation of peroxidases and the challenge of engineering more robust enzymes. Chem. Biol. 9 (5), 555–565. doi:10.1016/S1074-5521(02)00149-7
Vicente, F., Rosas, J. M., Santos, A., and Romero, A. (2011). Improvement soil remediation by using stabilizers and chelating agents in a Fenton-like process. Chem. Eng. J. 172 (2-3), 689–697. doi:10.1016/j.cej.2011.06.036
Villegas, J. A., Mauk, A. G., and Vazquez-Duhalt, R. (2000). A cytochrome c variant resistant to heme degradation by hydrogen peroxide. Chem. Biol. 7 (4), 237–244. doi:10.1016/S1074-5521(00)00098-3
Wang, J. W., Ma, Q., Zhang, Z. J., Li, S. Z., Diko, C. S., Dai, C. X., et al. (2020). Bacteria mediated Fenton-like reaction drives the biotransformation of carbon nanomaterials. Sci. Total Environ. 746, 141020. doi:10.1016/j.scitotenv.2020.141020
Xiang, L., Zhang, H., Hu, Y. F., and Peng, L. M. (2018). Carbon nanotube-based flexible electronics. J. Mater. Chem. C 6 (29), 7714–7727. doi:10.1039/c8tc02280a
Yang, C. X., Yue, F. F., Cui, Y. L., Xu, Y. M., Shan, Y. Y., Liu, B. F., et al. (2018). Biodegradation of lignin by Pseudomonas sp Q18 and the characterization of a novel bacterial DyP-type peroxidase. J. Industrial Microbiol. Biotechnol. 45 (10), 913–927. doi:10.1007/s10295-018-2064-y
Yang, M., Zhang, M. F., Nakajima, H., Yudasaka, M., Iijima, S., and Okazaki, T. (2019a). <p>Time-dependent degradation of carbon nanotubes correlates with decreased reactive oxygen species generation in macrophages</p>. Int. J. Nanomedicine 14, 2797–2807. doi:10.2147/Ijn.S199187
Yang, Z. F., Tian, J. R., Yin, Z. F., Cui, C. J., Qian, W. Z., and Wei, F. (2019b). Carbon nanotube- and graphene-based nanomaterials and applications in high-voltage supercapacitor: A review. Carbon 141, 467–480. doi:10.1016/j.carbon.2018.10.010
Yap, C. L., Gan, S. Y., and Ng, H. K. (2011). Fenton based remediation of polycyclic aromatic hydrocarbons-contaminated soils. Chemosphere 83 (11), 1414–1430. doi:10.1016/j.chemosphere.2011.01.026
You, Y., Das, K. K., Guo, H., Chang, C. W., Navas-Moreno, M., Chan, J. W., et al. (2017). Microbial transformation of multiwalled carbon nanotubes by Mycobacterium vanbaalenii PYR-1. Environ. Sci. Technol. 51 (4), 2068–2076. doi:10.1021/acs.est.6b04523
Yu, B., Cheng, H., Zhuang, W., Zhu, C. J., Wu, J. L., Niu, H. Q., et al. (2019a). Stability and repeatability improvement of horseradish peroxidase by immobilization on amino-functionalized bacterial cellulose. Process Biochem. 79, 40–48. doi:10.1016/j.procbio.2018.12.024
Yu, Q. Q., Feng, L., Chai, X. N., Qiu, X. H., Ouyang, H., and Deng, G. Y. (2019b). Enhanced surface Fenton degradation of BPA in soil with a high pH. Chemosphere 220, 335–343. doi:10.1016/j.chemosphere.2018.12.141
Zeng, Y. C., and Wu, G. (2021). Electrocatalytic H2O2 generation for disinfection. Chin. J. Catal. 42 (12), 2149–2163. doi:10.1016/S1872-2067(20)63781-0
Zhang, C., Chen, W., and Alvarez, P. J. J. (2014). Manganese peroxidase degrades pristine but not surface-oxidized (carboxylated) single-walled carbon nanotubes. Environ. Sci. Technol. 48 (14), 7918–7923. doi:10.1021/es5011175
Zhang, L. W., Petersen, E. J., Habteselassie, M. Y., Mao, L., and Huang, Q. G. (2013). Degradation of multiwall carbon nanotubes by bacteria. Environ. Pollut. 181, 335–339. doi:10.1016/j.envpol.2013.05.058
Zhao, J. G., Henkens, R. W., Stonehuerner, J., Odaly, J. P., and Crumbliss, A. L. (1992). Direct electron-transfer at horseradish-peroxidase colloidal gold modified electrodes. J. Electroanal. Chem. 327 (1-2), 109–119. doi:10.1016/0022-0728(92)80140-Y
Keywords: carbon nanotubes, degradation, heme enzyme, Fenton reaction, bacteria
Citation: Takahashi S, Taguchi F and Hori K (2023) Contribution of the Fenton reaction to the degradation of carbon nanotubes by enzymes. Front. Environ. Sci. 11:1184257. doi: 10.3389/fenvs.2023.1184257
Received: 11 March 2023; Accepted: 31 May 2023;
Published: 13 June 2023.
Edited by:
Vishal Tripathi, Graphic Era University, IndiaReviewed by:
Jong-Rok Jeon, Gyeongsang National University, Republic of KoreaXiaojia He, Chemical Insights Research Institute, United States
Copyright © 2023 Takahashi, Taguchi and Hori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katsutoshi Hori, a2hvcmlAY2hlbWJpby5uYWdveWEtdS5hYy5qcA==
 Seira Takahashi
Seira Takahashi Katsutoshi Hori
Katsutoshi Hori