
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci., 31 March 2023
Sec. Water and Wastewater Management
Volume 11 - 2023 | https://doi.org/10.3389/fenvs.2023.1148872
This article is part of the Research TopicWastewater Treatment: Removal of Recalcitrant CompoundsView all 7 articles
 Karabelo M. Moloantoa1,2*
Karabelo M. Moloantoa1,2* Zenzile P. Khetsha3
Zenzile P. Khetsha3 Gueguim E. B. Kana2
Gueguim E. B. Kana2 Maleke M. Maleke4
Maleke M. Maleke4 Esta Van Heerden5
Esta Van Heerden5 Julio C. Castillo1
Julio C. Castillo1 Errol D. Cason1,6*
Errol D. Cason1,6*Nitrate contamination in water remains to be on the rise globally due to continuous anthropogenic activities, such as mining and farming, which utilize high amounts of ammonium nitrate explosives and chemical-NPK-fertilizers, respectively. This study presents insights into the development of a bioremediation strategy to remove nitrates (NO3−) using consortia enriched from wastewater collected from a diamond mine in Lesotho and a platinum mine in South Africa. A biogeochemical analysis was conducted on the water samples which aided in comparing and elucidating their unique physicochemical parameters. The chemical analysis uncovered that both wastewater samples contained over 120 mg/L of NO3− and over 250 mg/L of sulfates (SO42-), which were both beyond the acceptable limit of the environmental surface water standards of South Africa. The samples were atypical of mine wastewaters as they had low concentrations of dissolved heavy metals and a pH of over 5. A metagenomic analysis applied to study microbial diversities revealed that both samples were dominated by the phyla Proteobacteria and Bacteroidetes, which accounted for over 40% and 15%, respectively. Three consortia were enriched to target denitrifying bacteria using selective media and then subjected to complete denitrification experiments. Denitrification dynamics and denitrifying capacities of the consortia were determined by monitoring dissolved and gaseous nitrogen species over time. Denitrification optimization was carried out by changing environmental conditions, including supplementing the cultures with metal enzyme co-factors (iron and copper) that were observed to promote different stages of denitrification. Copper supplemented at 50 mg/L was observed to be promoting complete denitrification of over 500 mg/L of NO3−, evidenced by the emission of nitrogen gas (N2) that was more than nitrous oxide gas (N2O) emitted as the terminal by-product. Modification and manipulation of growth conditions based on the microbial diversity enriched proved that it is possible to optimize a bioremediation system that can reduce high concentrations of NO3−, while emitting an environmentally-friendly N2 instead of N2O, that is, a greenhouse gas. Data collected and discussed in this research study can be used to model an upscale NO3− bioremediation system aimed to remove nitrogenous and other contaminants without secondary contamination.
Anthropogenic activities associated with urbanization and industrialization can be considered the primary source of water pollution (Xue et al., 2016). With the establishment of multiple mega companies under different industries comes great chemical usage and waste disposal into the environment, which adds excessive pressure to the already stressed water resources worldwide (Mohapatra and Kirpalani, 2017). The intensive mining and agricultural activities in most developing African countries introduce various contaminants into the environment mostly due to the utilization of commercial fertilizers and the use of nitrogen-based explosives during blasting operations (Bijay-Singh and Craswell, 2021; Moloantoa et al., 2022). Owing to other uses of these explosives in most activities that involve rock detonation, such as mining, construction of roads, and mega infrastructure, surface and groundwater bodies around these sites are easily contaminated with high NO3− concentrations from unexploded NH4NO3, which is oxidized to NO3− (Degnan et al., 2016; Feng et al., 2020). Heaps of rock tailings, containing some of the undetonated explosives and processing chemicals, such as nitric acid, also serve as sources of nitrogenous chemicals that leach into the environment, hence exuberating nitrogenous waste in the environment if their disposals are not managed (Ochieng et al., 2010; Sharma and Bhattachyarya, 2017). Owing to these anthropogenic activities, NO3− concentrations can accumulate in water up to 400 mg/L, which is 40 times the acceptable maximum limit of 10 mg/L suggested by the World Health Organization (WHO) and 11 mg/L according to the South Africa National Standards (SANS) (Lawniczak et al., 2016; Xue et al., 2016; Verlicchi and Grillini, 2020). Other localized industrial wastewaters, such as those from dairy waste, can have a NO3− concentration as high as 250 mg/L when discharged from factories (Foglar et al., 2005), while mining wastewater from one mine discharge point can accumulate to over 74,000 mg/L over three decades if the water is not treated, diluted, or released (Thorgersen et al., 2019).
The presence of NO3− in water at concentrations above 10 mg/L poses a serious health threat as it can cause diseases such as methemoglobinemia, blue baby syndrome in infants, automatic abortions (mostly in animals), and colon cancer in humans and animals (Dhania and Rani, 2014; Oladeji, 2017; Rajta et al., 2019). Other impacts of NO3− in water include eutrophication, which results in excessive growth of algae (algal bloom) and other fast-growing plants over the rivers and streams, causing dead zones in the stationary water bodies by depleting and misbalancing nutrients in water (Withers et al., 2014; Feng et al., 2020). Owing to the NO3− contamination challenge being a threat to fresh water availability and a source of health hazards, various treatment options have been applied involving physical, chemical, and biological approaches. In the past three decades, methods applied in NO3− remediation in water, including chemical precipitation, distillation, reverse osmosis, electrodialysis, ion-exchange, and bio-denitrification, have been applied extensively in an attempt to curb the continuation and worsening of NO3− contamination in water (Dahab, 1991; Matei and Racoviteaun, 2021). Recently, because of the fastidious nature of intermediate N species (NO2−, NO, and N2O) from partial denitrification, approaches of combining both chemical and biological methods have been explored (Puigserver et al., 2022). Nevertheless, prospects of optimizing microbial complete denitrification that would produce mainly N2 gas as the terminal by-product has drawn attention as a possible, sustainable, and efficient NO3− remediation method from water (Moloantoa et al., 2022). Nitrogen gas is a desired denitrification by-product as it is an environmentally friendly gas released into the atmosphere to further be cycled spontaneously in the ecosystem, and this contrasts with NO and N2O, which are greenhouse gases (Levy-Booth et al., 2014). Microbial denitrification is a complex yet well-studied biochemical process that is controlled by many factors (Huang et al., 2011; Levy-Booth et al., 2014), including metals such as Cu and Fe, which act as co-factors in the metabolic activities of NO3− reduction (Bowles et al., 2012; Marietou, 2016; Martínez-Espinosa, 2020).
Oxidized nitrogen (N) species (NO3−, NO2−, NO, and N2O) occurring naturally can be used as an electron acceptor by many bacteria, archaea, and fungi that use the nitrogenous compounds in their protein and nucleic acid synthesis; hence the involvement of microorganisms facilitating the N-cycle in the terrestrial and aquatic environments (Levy-Booth et al., 2014; Kamp et al., 2015). Denitrifying bacteria capable of reducing NO3− to N2 gas are generally found in NO3− contaminated wastewater and have been isolated, enriched, and applied in NO3− bioremediation systems to mitigate NO3− contamination (Correa-Galeote et al., 2013; Huang et al., 2015). Denitrifying bacteria are members of various phyla, such as Proteobacteria (Pseudomonas sp., Citrobacter sp., and Paracoccus sp.), Firmicutes (Bacillus sp.), and Actinobacteria (Intrasporagium sp.), and are characterized by containing functional denitrifying genes: narG/napA, nirK/S, norB, and nosZ (Nogales et al., 2002). These genes transcribe proteins that facilitate the stepwise reduction of NO3− to nitrite (NO2−), NO2− to nitric oxide (NO), NO to nitrous oxide (N2O), and eventually N2O to dinitrogen gas (N2) (Green et al., 2010; Huang et al., 2011).
Microbial N metabolisms, particularly denitrification as an enzymatic system, depend on environmental conditions that dictate the rates and the efficiencies of the enzymes facilitating the process (Torres et al., 2016; Rajta et al., 2019; Martínez-Espinosa, 2020). Denitrifying proteins are metallo-enzymes with selected heavy metals, such as copper (Cu), iron (Fe), and molybdenum (Mo), playing vital roles as co-factors to induce and catalyze enzymatic activation, rates, and efficiency during denitrification (Glass and Orphan, 2012). The production of the terminal denitrification product, nitrogen (N2), is because of Cu-dependent nitrous oxide reductase (nosZ) enzyme, and its activity can be greatly regulated by pH and Cu concentrations in the environment (Shen et al., 2020). As Cu promotes the transcription of nitrous oxide reductases (nosZ) and its functionality as a co-factor, its deficiency in the environment might result in the accumulation of N2O rendering the denitrification system ineffective in increasing the production of greenhouse gases, such as NO and N2O (Black et al., 2016; Gao et al., 2017).
To date, the majority of research conducted on optimizing complete denitrification has been conducted on pure cultures of bacteria with the aim of understanding the biochemical pathways involved in bio-denitrification. In recent biotechnological applications of bacteria for the remediation of NO3−-contaminated water, the focus remains primarily on known and benchmarked bacterial isolates used as consortia. In studies conducted by Jiang et al. (2023), an enriched consortium from a sewage treatment plant contained model denitrifiers: P. stutzeri and Paracoccus sp., which were reported to have participated in the removal of NO3−, NO2−, N2O, and total nitrogen (TN). Batch bioreactors are common in experimental bioremediation studies, and sewage as a source of wide microbial communities has become a source of various other denitrifying bacteria (Wang et al., 2022). In most denitrification studies in which enriched consortia are used, dynamics are monitored with the removal of dissolved nitrogen species: NO3−, NO2−, NH4+, and TN, without validation if N2O (potent greenhouse gas) has also been reduced to N2, which is an environmentally friendly gas and desired by-product to be emitted by a denitrification system (Adouani et al., 2015; Wang et al., 2022; Jiang et al., 2023).
With different bacterial groups beside model isolates (P. stutzeri and Paracoccus sp.) that are capable of facilitating different steps of denitrification, this study focuses on optimizing their synergistic approach as a consortium to facilitate complete denitrification. Insights into the manipulation of metabolic pathways through supplementing enzyme co-factors, switching of carbon sources, and creating conducive environmental conditions as optimization approaches are collectively explored. This study presents stepwise development and optimization of a bioremediation system that promotes the complete reduction of NO3− and intermediate N-species (NO2−, NO, and N2O) with the aim of directing the system to yield N2 gas as a final product in conditions mimicking the natural environment.
Two mining companies, 1) a platinum mine situated in South Africa (Pt-SA) and 2) a diamond mine in Lesotho (D-Lst) with contamination alerts of high nitrate concentrations (over 100 mg/L-NO3-) in their wastewaters, were used as study sites for the project. The names and precise locations of the companies cannot be revealed due to a non-disclosure agreement (NDA) that is in place. Site 1: Pt-SA is an underground platinum (Pt) mine situated in the Northwest Province of South Africa with water around the mine reported to contain elevated NO3− concentrations from the nearby mine wastewater catchment and sewage drainages. The mine has a wastewater catchment for potential purification and reuse in the mining processes; however, the water has not been successfully treated, hence its storage. Site 2: D-Lst is a diamond mine situated on the north-eastern side of the Kingdom of Lesotho, a landlocked country within South Africa. The mine is an open cast mine with water catchments near the river for which if it overflows during heavy rains, the water will easily overflow into the river causing a potential threat to the community using water from the river. Both mines utilize tons of ammonium-based explosives, which are most likely to be the source of elevated NO3− concentration in their wastewater catchments.
Water samples were collected using sterile 25 L polyethylene drums. The drums were rinsed three times with site water before collection. A generator-powered water pump connected to a two-way hose pipe was used to transfer water from the catchments into the drums. The collecting end of the hose pipe was immersed 50 cm vertically into the water source before pumping into the sampling drum. The following chemical parameters were measured on site: pH and total dissolved solids (TDS) using an ExStik®II probe; Model EC500 (EXTECH Instruments). On arrival at the laboratory, the working samples were harvested from the drums and hydrogeochemical analysis was performed within 24 h of sample storage for further water sample characterization.
The working samples were subjected to complete chemical analysis to determine their physicochemical characteristics. The concentration of dissolved anions, such as NO3−, SO42-, and Cl−, was determined using ion chromatography (IC), while total organic carbon (TOC) and CaCo3 were determined using a TOC analyzer. Concentrations of elemental cations in the wastewater samples (i.e., Ca, Mg, K, P, Na, Fe, Al, Mn, Ni, and Zn) were determined by inductively coupled plasma-optical emission spectroscopy (Prodigy 7 High Dispersion ICP-OES). For the ICP-OES sample preparation, 200 mL of each wastewater sample was acidified to a pH of less than 2 with HNO3 (2%) and filtered through 0.22 µm pore-sized filters. The samples were then stored at 4°C until they were sent to the Institute of Groundwater Studies (IGS, RSA) at the University of the Free State (UFS, RSA) for the abovementioned chemical analysis.
Molecular analysis of the bacterial content in the water was conducted by concentrating bacterial cells through centrifugation (Eppendorf-Centrifuge 5810R) of 400 mL of the two wastewater samples (6000 RCF, 10 min) in 50 mL portions. The pellets were combined for each sample and used for genomic DNA (gDNA) extraction using the NucleoSpin® Soil kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions. The quality (pure dsDNA concentration and ds/ssRNA contamination concentration) and integrity (DNA sheering) of the DNA were determined using a NanoDrop 3300 fluorospectrometer (Thermo Scientific, Waltham, MA, United States) and 1% agarose gel stained with ethidium bromide (EtBr), respectively. Agarose gels were visualized under UV illumination with a Chem-Doc XRS (Bio-Rad Laboratories) gel documentation system as a quality control measure.
To explore microbial communities in the wastewater samples, the extracted gDNA samples were subjected to Illumina MiSeq sequencing analysis at the Centre for Proteomic Genomic Research (CPGR), Cape Town, South Africa. The sequence libraries were prepared by amplifying approximately the 460 bp region located in the hyper-variable V3/4 region of the 16S rRNA gene with overhang Illumina adapter overhang nucleotide sequences (Klindworth et al., 2013). A polymerase chain reaction (PCR) was performed with a broad-spectrum 16S rRNA primer set: forward primer: 5′-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG-3′ and reverse primer: 5′-GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C-3’ (Fadeev et al., 2021). The 16S V3/4 amplicons were purified using the Agencourt-AMPure XP bead clean-up kit (Beckman Coulter Genomics, Danvers, MA, United States), followed by a second amplification to attach dual indices and Illumina sequencing adapters using the Nextera XT Index kit (Illumina, San Diego, CA, United States). Final purification using the AMPure XP bead clean-up kit (Beckman Coulter Genomics) was carried out, followed by library quantification, normalization, pooling, and denaturing before being subjected to a 2 × 301 cycle sequencing on the Illumina MiSeq using the MiSeq v3 reagent kit (Illumina). The obtained 16S rRNA gene sequence data were analyzed using QIIME v1.9.1 as described by Caparoso et al. (2010) with adopted modifications by Cason et al. (2020). Operational taxonomic unit (OTU) tables were rarefied to 39,787 reads per sample. Metabolic activities of OTUs detected in the wastewater samples were predicted using Functional Annotation of Prokaryotic Taxa (FAPROTAX) v1.2.2. Analyses of the abundance tables were carried out using R v3.6.1 (www.r-project) (R Development Core Team, 2011) and the phyloseq package (McMurdie and Holmes, 2013).
In all dynamic and optimization experiments, monitoring of parameters was conducted by regular sampling from batch cultivations with defined media, incubation periods, and conditions. Cultivations were carried out in 100 mL volumes in 150 mL serum vials for anaerobic cultivations and 250 mL Erlenmeyer flasks for aerobic cultivations unless otherwise reported. Samples were collected using sterile syringes into Eppendorf tubes, and the analysis was conducted within 1 h after collection using standardized methods described as follows based on the parameters monitored. All batch experiments were incubated at a constant temperature of 30°C in a shacking incubator rotating at 150 rpm over a defined period according to each experiment. Microbial growth in each culture was monitored by measuring the optical density at 600 nm (OD600nm) using a Spectronic® GENESYSTM 5 spectrophotometer. Fresh medium was used to blank the spectrophotometer prior to analyzing samples. The concentrations of dissolved N-species, NO2− and NO3−, were determined using the Griess assay adopted from Bhusal and Muriana (2021). Fresh reagents used in the assay were prepared every 14 days during the analysis with the validation of their efficacy tested using stand solutions. The amount of biogenic gaseous N-species, N2O and N2, produced from denitrification experiments was determined by analyzing collected gas samples from vials using gas chromatography (GC) (SHIMADZU Scientific Instruments). Two different columns were used to analyze and verify N2 and N2O percentages due to the interference of Ar(g) used as an inert gas in the vials. Helium was used as a carrier gas in both analyses in which N2 gas was analyzed using the Molsieve GC capillary column (Agilent J&W CP-Molsieve 5Å) and N2O gas was analyzed using the Shincarbon-ST column (Okada et al., 2005). The TCD injector, oven, and detector were preheated to 200°C prior to the manual gas injection. Owing to the instability of NO that spontaneously and rapidly oxidizes to NH4+ or NO2− and eventually NO3−, it was not analyzed but accounted for when analyzing NO3− in the medium. Concentrations of carbon sources used in the experiments: glucose and glycerol, were determined by high-performance liquid chromatography (HPLC) with a Shimadzu Prominence Liquid Chromatograph fitted with a refractive index detector (RID) and a BioRad Aminex HPX 87H (300 mm × 7.8 mm) analytical column. The RID cell temperature was set at 40 °C, and the column was held at 65 °C. Sulfuric acid (0.5 mM) was employed as an eluent at 0.6 mL/min. Analytical-grade glucose in a series of concentrations from 0.1 to 5 g/L was employed to construct a calibration curve. Shimadzu LabSolutions were employed for instrument control, and data analysis was conducted with Microsoft Excel. The final selected consortia were subjected to molecular analysis to identify and characterize the denitrifying bacterial groups in the culture. The culture was centrifuged to pellet the cells from a 100 mL culture, and genomic DNA was extracted from the cells as previously described. The extracted DNA was sent to Illumina MiSeq sequencing analysis similar to that carried out for wastewater samples.
Enrichment of denitrifying bacteria was conducted in a nutrient-rich nitrate-reducing medium (NRM) with medium contents indicated in Supplementary Table S1. The medium was prepared with key ingredients of 1 g of nitrate (NO3−) as the main electron acceptor and 1 g of glucose as the sole electron donor with the pH adjusted to 6.5 (Ogden et al., 2002; Romano et al., 2019; Böllmann and Martienssen, 2020). Medium (90 mL) was aliquoted into 150 mL serum vials, deoxygenated with Ar(g), autoclaved, and then, inoculated with 10% (v/v) of the wastewater samples to make up 100 mL cultures (Calderer et al., 2010). After inoculation, the cultures were immediately sampled and then incubated at 30°C in an automated shaker with a rotating speed of 150 rpm for 120 h (5 days). From each culture, 2 mL of samples were collected every 24 h for analyzing the following parameters: microbial growth analysis by measuring OD at 600 nm, denitrification dynamics by quantifying reduced NO3−, and NO2− produced and reduced using a Griess assay as described previously. For all cultivations, each batch was conducted in triplicates for all modified conditions during the enrichment and optimization experiments. The natural selection of denitrifying bacteria from the enriched consortia was induced by sub-culturing the primary enrichment to subsequent generations in the NRM with conditions that promoted the proliferation and dominance of nitrate-reducing bacteria in anaerobic conditions. The 10 mL cultures were passaged from one generation as inoculum into 90 mL of fresh NRM until the third (tertiary) generation (Brűck et al., 2002). Among the cultures that were attained, those that attained less than 80% NO3− reduction were discontinued from the enrichment cycle, while cultures with over 95% NO3− and NO2− reduction were subjected to further denitrification optimization experiments.
The selected consortia from the enrichments were subjected to complete denitrification tests with a focus on consortia that could reduce NO3−, NO2−, and N2O to produce N2 gas as a terminal byproduct. The consortia were cultivated and maintained in the NRM for growth optimization and to promote denitrification over other metabolic processes (Green et al., 2010; Fan et al., 2018). All cultures were inoculated at 10% (v/v) inoculum to medium ratio with an inoculum of not more than 7 days old for maintenance and all optimization batch experiments below unless otherwise stated (Brűck et al., 2002). Glycerol was used as the sole carbon source in all batch experiments applied at 1:1 ratio with nitrates (C/N) to benchmark feasible denitrification with an economical carbon source used in wastewater treatment plants as opposed to glucose or lactate (Fountoulakis et al., 2010; Schroeder et al., 2020; PlÖhn et al., 2022).
Three selected consortia: C1, C2, and C3, which produced more N2 gas with the compete removal of dissolved N-species, were subjected to higher NO3− and NO2− removal optimization tests. The three consortia were cultured in the NRM with 1,000 mg/L of NO3− and further subjected to a high-NO3− concentration removal test and tolerance to biogenic intermediate NO2− with the aim of determining the minimum amount of NO2− that hinders bacterial growth and the continuation of complete denitrification. The cultures were inoculated in the NRM with increasing NO3− concentrations of up to 10,000 mg/L, ranging as follows: 1,000, 2,000, 5,000, and 10,000 mg/L (Pinar et al., 1997; Ogden et al., 2002; Böllmann and Martienssen, 2020). The tests were conducted in triplicates for each of the three cultures under anaerobic conditions. The experiment was conducted for 7 days at 30°C with daily analytical monitoring of bacterial growth (OD600nm) and NO3−, and NO2− concentrations, as previously discussed.
A metal-induced denitrification test was conducted with 2,000 mg/L of NO3− in the NRM supplemented with 5 mg/L each of Cu(II) and Fe(II) supplied separately in the form of CuSO4 (1.99 mg/L of Cu(II)) and FeSo4 (1.84 mg/L of Fe(II)), respectively. The three consortia were assessed individually for Cu2+ effects in promoting the reduction of NO3− and lowering of biogenic N2O emission to promote terminal N2 gas production (Zou et al., 2013; He et al., 2019). Bacterial growth was monitored by the reading of OD600nm to assess the impact of the metals on bacterial growth, as well as other complete denitrification dynamics, as monitored previously with the included analysis of biogenic gas production of N2O and N2 quantified using gas chromatography (GC), as described above. The best consortium was selected for downstream experiments based on the complete denitrification detected by 100% removal of NO3− and NO2− and reduction of N2O detected by increasing amounts of N2 gas.
To mimic environmental settings like the ones where the samples were collected, a synthetic wastewater (SWW) medium adopted from the studies conducted by Foglar et al. (2005) was used in denitrification optimization. The medium was prepared with modifications to match the chemical composition of the wastewater samples collected from the mining sites. The medium was prepared using pure grade chemicals as listed in Supplementary Table S1. Glycerol was used as the sole carbon source with 2,000 mg/L of NO3− at a ratio of 1:1 C/N. One consortium was used for this section of the study and subsequent optimization studies.
Nitrite (NO2−) accumulation from the reduction of NO3− in SWW was observed and a change of carbon source among other preliminary troubleshooting approaches revealed a positive response evident with the reduction of NO2−. Carbon source was then changed from glycerol to glucose to assess and evaluate NO3− reduction capacities in SWW. Individual reduction dynamics of NO3− and NO2− added separately as sole N-sources were evaluated in SWW with glucose as the sole carbon source. The cultures were incubated in anaerobic conditions at 30°C while monitoring OD600nm, NO3−, and NO2− reduction for the effectivity of glucose in the reduction of dissolved N-sources.
The consortium was subjected to complete denitrification optimization by supplementing the media with increasing concentrations of Cu2+ from 0, 10, 25, 50, and 75 mg/L. Glucose was used as the sole carbon source in a 1:1 and 2:1 (C/N) ratio with 1,000 and 2,000 mg/L of NO3− in SWW. Denitrification dynamics were monitored as described previously with daily measurements of OD600nm, NO3−, and NO2− concentrations, and finally, determination of the percentages of N2O and N2 gases produced as terminal denitrification products was carried out.
The selected consortium was finally subjected to molecular analysis to identify and characterize the denitrifying bacterial groups responsible for complete denitrification. The cultures (triplicate from one consortium) were centrifuged to pellet the cells from a 100 mL culture, and the genomic DNA was extracted and sent to CPGR for Illumina MiSeq sequencing analysis as described previously.
Physicochemical parameters of the wastewater samples (Pt-SA and D-Lst) were determined, and the obtained results are recoded in Table 1. Owing to potential sewage spillages in Pt-SA, the introduction of fecal microorganisms in water can alter the water chemistry, including the elevation of NO3− concentrations (Sharma and Bhattacharya, 2017; Olds et al., 2018). The pH of both samples was higher than 5 with the evidence that it could have resulted in low metal leaching from tailings, thus limiting their concentrations in water (Król et al., 2020; Matodzi et al., 2020; Park et al., 2020). Chemical compositions of industrial wastewaters, such as mining, differ in dissolved metal concentrations due to the type of rocks associated with the mineral being mined (McCarthy, 2011; Guo et al., 2013). The Pt-SA sample is collected from an active platinum mine where the geological landscape is composed of mainly mafic and ultra-mafic igneous rocks from the crystallization of magma, which is less soluble and resistant to erosion predominantly forming reefs. In contrast, the D-LSt sample was collected from an active diamond mine where the geology of the environment is predominantly Kimberlite. The Kimberlite rock is composed of various chemicals and harbors not only diamonds as minerals but also Olivine [(Mg2+, Fe2+)2SiO4], which is mainly magnesium and iron (Robles-Cruz et al., 2012). Overall, water chemistry revealed close similarities of the physicochemical parameters of the wastewaters with low dissolved metal concentrations. Amongst parameters that exceeded the SANS limits, NO3− was found to be over 100 mg/L in both samples.
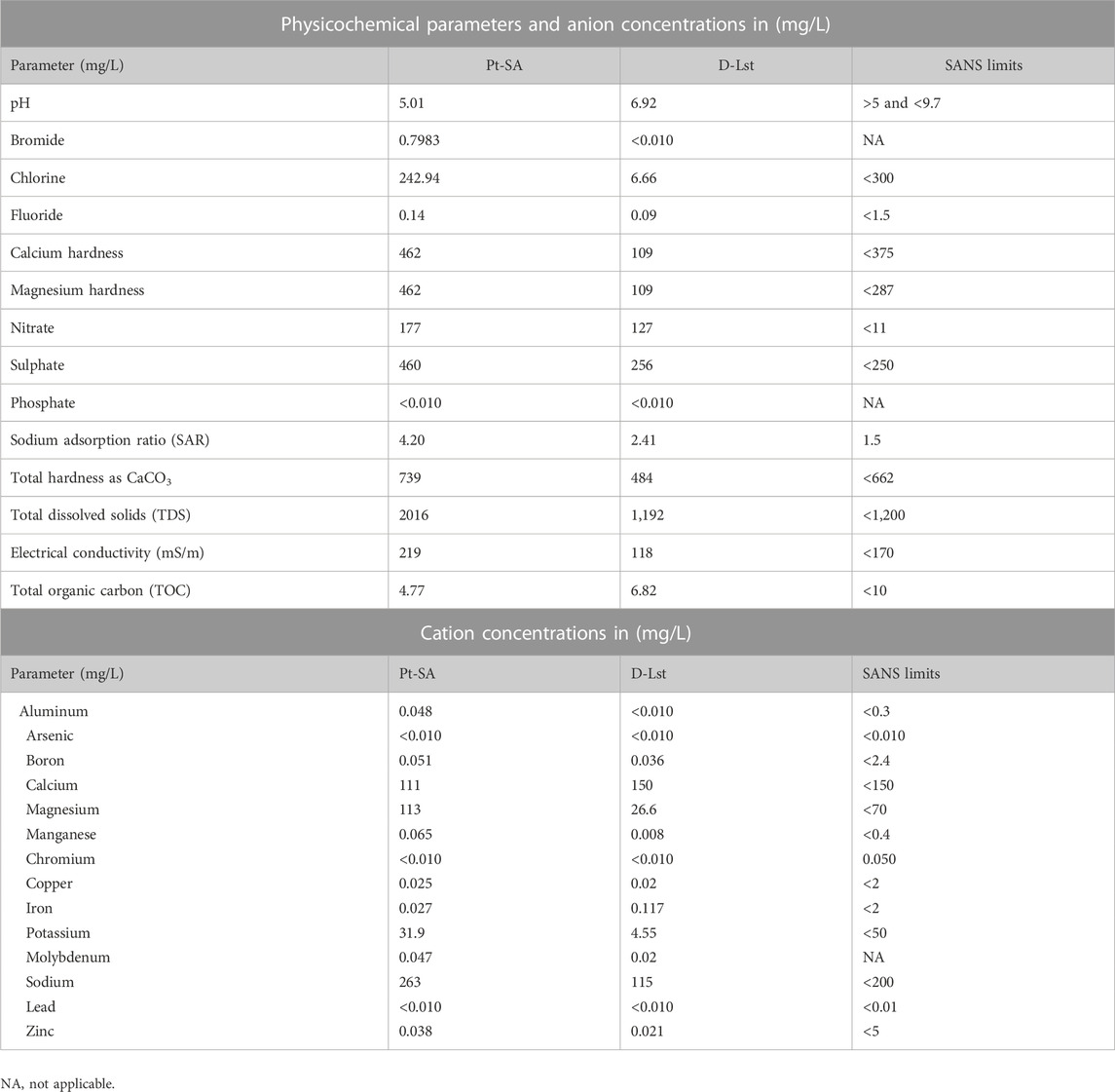
TABLE 1. Physicochemical parameters and anion and cation concentrations measured in the collected mine wastewater samples, with values above the SANS limits highlighted in grey.
TDS of 2,016 mg/L determined in the Pt-SA sample can be attributed to the high dissolved inorganic salts and organic particles (Table 1), most likely due to the influx of sewage into the water source (Butler and Ford, 2018). Comparatively, the D-Lst wastewater sample had a lower TDS of 1,192 mg/L with the lowest metal concentrations. Although below the SANS, Fe concentrations were higher in sample D-Lst at 0.117 mg/L compared to 0.027 mg/L of sample Pt-SA. In both samples, three heavy metal cations, arsenic (As), chromium (Cr), and lead (Pb), had concentrations below the detection limit of the ICP, which further corroborates that the wastewater samples do not have typical acid mine wastewater characteristics (McCarthy, 2011; Egashira et al., 2012; Modoi et al., 2014). It can further be suggested that based on these physicochemical parameters, the wastewater samples present an oligotrophic environment rich in nutrients, specifically nitrogen with a slightly acidic to neutral pH, and reducing micro-toxic conditions suitable for facultative anaerobes, including most denitrifying bacteria (Ponce-Soto et al., 2015). The majority of the detected anions (Table 1) were within the SANS limits except for NO3−, SO42-, and PO43-, specifically in Pt-SA. The wastewaters had high Mg2+ and CaCO3 concentrations, which is indicative of water hardness (Ahn et al., 2018). Ordinarily, sulfates (SO42-) and carbonates (CaCO3) occur in higher concentrations in mine wastewaters due to their abundance in the earth’s crust and are hence associated with most mineral-bearing rocks that get mined for mineral extraction (Fang et al., 2018). Although not alarmingly high, SO42+ appeared to be the second dominant contaminant also above the SANS limits in both samples. Pt-SA, with sewage contamination, had higher anion concentrations as compared to the D-Lst sample with more water hardness containing elevated Mg2+, Ca2+, and CaCO3 concentrations (Grochowska, 2020). According to the Department of Water Affairs and Forestry (DWAF) regulations in South Africa, the value of sodium adsorption ratio (SAR) above 1.5 in irrigation water is considered toxic for the environment and both samples’ higher SAR values of 4.2 in Pt-SA and 2.41 in D-Lst indicate their toxicity, even for farming activities (DWAF, 1996; Kgopa et al., 2018).
Nitrate concentrations in the Pt-SA and D-Lst samples were 177 and 127 mg/L, respectively, which were both over 10 fold higher than the maximum acceptable limit of <11 mg/L according to the SANS. Most mining explosives contain NH4NO3, which can be oxidized to NO3−, which completely dissolves in water, resulting in their accumulation over time (Cyplik et al., 2012; Degnan et al., 2016; Feng et al., 2020). Water contamination poses environmental and health threats that need an urgent remediation strategy to mitigate the impacts incurred by high NO3− concentrations (Moloantoa et al., 2022). With such wastewater physicochemical characteristics, the samples provide a suitable growth environment for nitrogen-metabolizing microorganisms that can be isolated, enriched, and applied in a NO3− bioremediation system.
Microbial diversity was assessed using 16S rRNA-targeted metagenomic sequencing, and the data acquired revealed 3,938 and 2,931 OTUs in samples PT-SA and D-Lst, respectively. Sample PT-SA seemed to be the most diverse relative to D-Lst with a total of nine against five dominant phyla detected, respectively (Figure 1A). The most abundant bacterial phyla in both samples were Proteobacteria, Bacteroidetes, Verrucomicrobia, Actinobacteria, and Planctomycetes, which are common in most mine wastewaters (Dadrasnia et al., 2017; Kisková et al., 2018; Osunmakinde et al., 2019). The remaining four phyla, Acidobacteria, Chlamydiae, Cyanobacteria, and Firmicutes, were exclusively found in sample Pt-SA, which had a slightly acidic to neutral pH of 5.01 and sewage flowing into the water promoting the dominance of various microorganisms that use different metabolites in the wastewaters (Cai et al., 2014). Furthermore, Proteobacteria was the most dominant phylum in both mine wastewater samples, accounting for 42.8% and 47.7% in Pt-SA and D-Lst, respectively, which is common in most mine wastewater samples (Ma et al., 2015; Kisková et al., 2018). Likewise, Zhang et al. (2018) reported the dominance of Proteobacteria as compared to other phyla in lakes with high NO3− concentrations. The second dominant bacterial phylum was Bacteroidetes, accounting for 15.8% and 15.7% in Pt-SA and D-Lst, respectively. The phylum Bacteroidetes has been regarded as the human-associated bacterial indicator in wastewater assessments, and its presence in both samples occurring in similar percentages of dominance indicates human-associated contamination in the samples (Cai et al., 2014; Olds et al., 2018). The presence of these two bacterial groups, Proteobacteria and Bacteroidetes, is common in NO3−-contaminated water with a near-neutral pH, high NO3− concentration, and denitrification bioreactors in which their dominance in the wastewater samples can serve as a source of bacteria for bioremediation purposes (Safonov et al., 2018; Jinxiang et al., 2020).
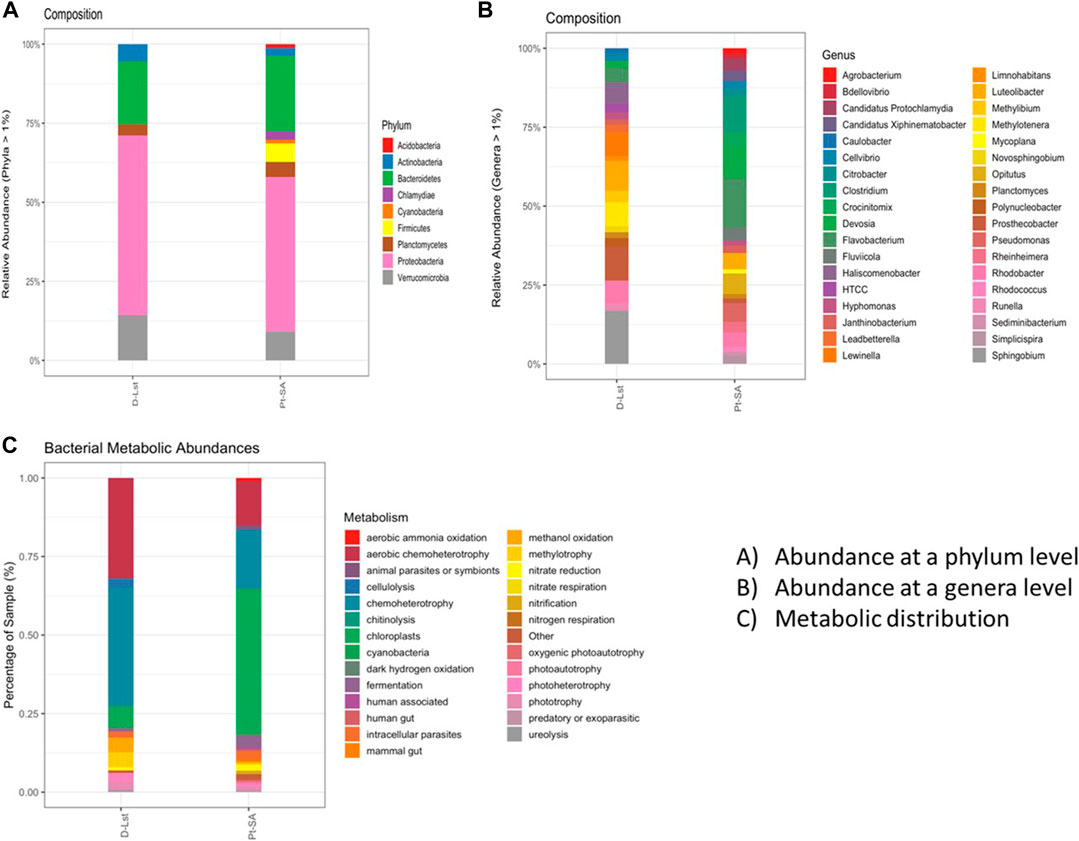
FIGURE 1. Taxonomic distribution and characteristics of bacterial communities in the D-Lst and Pt-SA samples: (A) Microbial relative abundance on a phylum level. (B) Microbial relative abundance on a genera level. (C) Functional annotation of OTUs, using FAPROTAX, of metabolic abundances from the two wastewater samples.
In addition to sewage being the source of various bacterial groups in the Pt-SA sample, the near-neutral pH of 6.92 in sample D-Lst could have also been the reason why Acidobacteria were not dominant in the wastewater but prevalent in the Pt-SA sample with a pH of 5.01. For example, Sait et al. (2006) isolated more colonies of Acidobacteria at pH 5.5 than at pH 7, which further substantiated the relevance of lower pH in promoting the prevalence of Acidobacteria, even in environmental samples. The presence of specific bacterial communities in water systems, such as dams, lakes, and rivers, is strongly linked to pH and chemical composition that become selective to the microbiome of the environment (Sait et al., 2006; Wu et al., 2017; Paruch et al., 2019). The presence of Cyanobacteria in the Pt-SA wastewater sample serves as an indicative bacterial group for an environment that is stable and nutrient-rich, while in this case, sewage could have been the main source of excess nutrients as the phylum was abundant only in Pt-SA (Romanis et al., 2021). Among the phyla that were exclusively dominant in Pt-SA, Firmicutes was present and commonly known to dominate anaerobic digestors degrading polymers such as polysaccharides, proteins, and lipids, which are present in high concentrations in sewage, hence their absence in the D-Lst sample (Jabari et al., 2016; Nascimento et al., 2018). Sewage is mainly composed of human waste that includes pathogenic and gut microbiota associated with humans, which could be the result of Chlamydiae as a known human pathogenic bacteria group becoming prevalent in the Pt-SA sample (Collingro et al., 2020). A taxonomic comparison of the two wastewater samples at the genus level revealed 54 dominant genera and characterized their percentage of abundance in the samples; 39 were beyond 1% in dominance as annotated in Figure 1B. Among the detected genera, well-studied and reported denitrifiers, such as Pseudomonas, Citrobacter, and Clostridium, were among the dominant groups that drive the N-cycle in the wastewater systems, which aids the quest to enrich some known and novel denitrifying bacterial groups in the samples (Huang and Tseng, 2001; Kim et al., 2005; Lv et al., 2017; Rajta et al., 2019).
FAPROTAX analysis in Figure 1C revealed a total of 26 inferred metabolic pathways by the bacterial communities in the wastewater samples. Functional metabolic predictions in the D-Lst sample revealed that over 50% of the metabolic network consisted of chemo-heterotrophy, which is carried out by both the organotrophs and lithotrophs in the environment (Rajta et al., 2019). With high TOC (6.82), excess amounts of NO3− (127 mg/L) and SO42- (256 mg/L) in the D-Lst sample relatively concurred with the presence of oligotrophs and lithotrophs utilizing chemical compounds, such as NH3+, NO3−, and SO42-, as metabolites. The most abundant predicted metabolic pathways in Pt-SA were the ones facilitated by the Cyanobacteria that accounted for over 40% of the total functional groups. In a nutrient- and carbon-rich environment, such as the one presented by Pt-SA, photoautotrophic (Woo, 2018) and chloroplast-aided photosynthetic (Tang et al., 2011) metabolisms facilitated by the Cyanobacteria tend to be more prevalent. Among the common metabolisms predicted in both samples, there was nitrate reduction and nitrogen and nitrate respiration, which play roles in N-cycling in the environment. Genera of bacteria responsible for nitrate metabolisms, such as Bacillus, Clostridium, and Citrobacter, were concurrently detected in the samples, and the metabolic predictions further validate their presence in the mine wastewater samples (Kisková et al., 2018; Tiburcio et al., 2020). Nitrification and ammonium oxidation metabolic pathways both result in the formation of NO3− and were exclusively predicted in the Pt-SA sample that had the highest nitrate concentrations (177 mg/L), which strongly suggests that nitrifying bacteria converts NH4+ and NO2− to NO3− (Spangler and Gilmour, 1966; Kim et al., 2005). In addition, the Pt-SA sample had more human-associated and mammal gut-related metabolisms, as reported previously (Cai et al., 2014; Kho and Lal, 2018; Collingro et al., 2020).
The composition of bacterial species and their functional potential can be influenced by geochemical parameters, such as pH, metal concentrations, and electron acceptors/donors, in the wastewater. For instance, wastewater samples and treatment plants can serve as excellent sources of indigenous bacteria capable of bioremediation of contaminated wastewater (Correa-Galeote et al., 2013; Nascimento et al., 2018). In fact, several authors have successfully enriched NO3−-reducing bacteria from a wastewater treatment plant and used the enrichment cultures as inoculum for bioremediation research studies (Liu et al., 2016; Yao et al., 2020). The readily available NO3− and SO42- in the samples serve as terminal electron acceptors in the metabolic activities of large groups of nitrogen- and sulfur-metabolizing bacteria. The presence of ammonium oxidizing microorganisms, which contain the amoA gene, indicates that the enrichments of all microbial groups without selection could lead to the conversion of NH3+ to NO3− for denitrification to take place when oxygen levels become lower and conducive for denitrification (Rajta et al., 2019). In contrast, the same nitrifying microorganisms could also cause a reversal effect of NO3− production in a system aimed to reduce the contaminant. Indeed, the difference in the microbial diversities of the samples signified the importance of chemical differences and sources of contamination in wastewater samples.
As FAPROTAX only infers metabolic potential from the taxonomy data, further validation was required to identify the presence of nitrate-reducing genes in the samples analyzed. The cultivation and enrichments of bacterial species containing these genes could result in their application for NO3−bioremediation (Huang et al., 2011; Levy-Booth et al., 2014; Rossi et al., 2015). Viable bacteria in the samples were successfully enriched in the NRM with glycerol as an electron donor and other components listed in Supplementary Table S1 (Liu et al., 2011). In general, nitrate-reducing bacteria are expected to be present in high-NO3−-contaminated environments as they use oxidized nitrogen compounds as electron acceptors. However, their presence does not equate to their activity due to metabolic limitations imposed by high metal concentrations (Gang et al., 2013; He et al., 2020) and environmental conditions, such as low temperature, low pH, and high dissolved oxygen and carbon source availability (Saleh-Lakha et al., 2009; Igiri et al., 2018). The utilization of nitrate as the terminal electron acceptor under anoxic conditions served as a force of selection in growing facultative and obligate anaerobic nitrate-reducing bacteria (Melton et al., 2014). A total of 30 consortia were characterized based on their NO3− and NO2− reduction capacities as well as the volumetric measure of biogenic gas produced over time. A cut-off limit of 95% NO3− removal from an initial 1,000 mg/L of NO3− was set for the selection of consortia to characterize and optimize for complete denitrification. Overall, three of the 30 consortia attained 100% NO3− reduction within 48 h.
The three selected consortia (C1, C2, and C3) that attained 100% NO3− reduction were further subjected to denitrification tests in batch experiments to determine their potential and capacity to completely remove dissolved NO3− and NO2− at elevated concentrations. A minimum of 1,000 mg/L was applied during enrichment and a maximum of 10,000 mg/L was applied during tolerance studies by Pinar et al. (1997).
From the obtained results, all consortia attained a 100% reduction of 1,000 mg/L NO3− and all biogenic NO2− that formed subsequently over a 7-day incubation period (168 h). Bacterial tolerance to NO3− and NO2− was evaluated in experiments with up to 10,000 mg/L of initial NO3− and biogenic NO2− forming from nitrate reduction. Remarkably, microbial proliferation for all concentrations was not affected by the NO3− concentrations from 1,000 to 10,000 mg/L (Figures 2A–C). It is worth noting that the lag and exponential phases of the consortia cultures were all within 12 h. The maximum bacterial growth rate (μmax) above 0.36 h in all cultures was attained with their doubling time (Td) of less than 2 h except by C1 at NO3− concentrations of 5,000 and 10,000 mg/L, which had a Td of over 2 h. Although not uniform for all cultures, C1 and C2 had the same trend of lag phases that merged with the exponential phases indicating favorable growth and denitrifying conditions (Miyahara et al., 2010). For C3, there was a dip at the beginning of the lag phase indicating an adaptation period between 0 and 4 h followed by the sharp exponential phase. These results demonstrated a similar trend of dipping lag phases as observed by Ling and Juanjuan, (2018) during their biological NO3− removal studies with a consortium from a sewage plant. The differences in the lag and exponential phases for the three consortia could be due to the different bacterial compositions growing at different rates. Among all three consortia, C2 had the shortest doubling time (Td = 1.3) and the highest growth rate of more than 0.5, indicating favorable growth conditions for the consortium at 2,000 mg/L (Cua and Stein, 2014). Cultures cultivated in the lowest NO3− concentrations (1,000 mg/L) were the first to reach deceleration phases, most likely due to the depleted electron acceptor (NO3−) in the medium. This was indicated by the gradual decline of the growth curves after 40 h of incubation (Pinar et al., 1997; Miyahara et al., 2010). In contrast, cultures in higher NO3− concentrations (2,000, 5,000, and 10,000 mg/L) maintained their stationary phases for up to 168 h. Evidently, the enriched consortia demonstrated great tolerance to high NO3− concentrations as the concentrations tested did not hinder or drastically affect microbial growth.
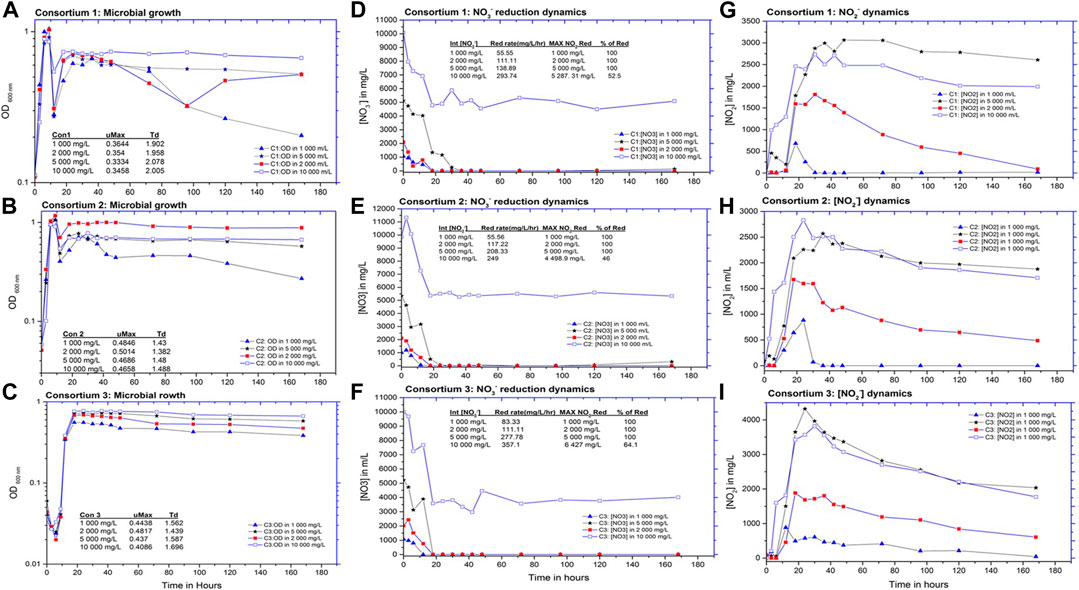
FIGURE 2. Microbial and denitrification dynamics in increasing nitrate concentrations. (A–C) Microbial growth curves of C1, C2, and C3 cultures with 1,000, 2000, 5,000, and 10,000 mg/L of NO3. (D–F) Nitrate reduction profiles by C1, C2, and C3 consortia. (G–I) Nitrite reduction profiles of C1, C2, and C3 consortia.
Notwithstanding the tolerance toward elevated concentrations of NO3−, the consortia could not attain 100% NO3− and NO2− removal at higher NO3− concentrations (from 5,000 to 10,000 mg/L), probably due to NO2− accumulation and toxicity (Figures 2D–I). Regardless of the incomplete NO3− removal in the cultures with the highest NO3− concentrations (10,000 mg/L), over 4,000 mg/L of NO3− was removed, which highlights the tolerance and great capacity of the consortia to reduce elevated amounts of NO3−. Ideally, the consortia had to be continuously fed with carbon source to combat NO2− accumulation as carried out in continuous cultivations (Sun et al., 2009), while in this study, no additional carbon source was supplied so as to notice the effect of NO2− on the consortia. The reduction of NO3− by the enriched consortium and pure cultures at such high concentrations is rarely obtained (Vijaya-Chitra and Lakshmanaperumalsamy, 2006). In C1 and C2, 100% NO3− reduction was attained at 18 h for both 1,000 and 2,000 mg/L NO3− attaining cultures at 55.55 and 111.11 mg/L per hour, respectively. The fastest NO3− reduction of 1,000 mg/L among the three consortia was 83.33 mg/L-1 attained by C3 achieving 100% NO3− reduction within 12 h. For the consortia incubated in higher NO3− concentrations of 5,000 mg/L, the complete reduction was attained in all cultures at 36, 24, and 18 h by C1, C2, and C3, respectively, with C3 exhibiting the fastest reduction rate, removing 277.78 mg/L-1. From all the reduction capacities displayed by the three consortia, it was evident that the enriched bacteria could reduce high NO3− concentrations beyond the 3,000 mg/L of NO3− reported by Cyplik et al. (2012). The biological denitrification rates and effectiveness in most remediation systems are determined solely by the removal of NO3−. However, other intermediate nitrogen species, such NO2−, NO, NH4+, and N2O besides N2 gas, serve as an indication of incomplete denitrification (Krishna Reddy et al., 2017; Kong et al., 2018). In cultures with higher NO3− concentrations, the accumulation of more than 2,000 mg/L of NO2− was observed mostly due to the formation of reactive oxygen species (ROS) from molecular oxygen and nitrogen species (RNS), such as NO from the accumulation of NO2− and NO as NO3− (Durzan and Pedroso, 2002; Poole, 2005; Damiani et al., 2016; Yao et al., 2020). For consortia grown on 2,000 mg/L, C1 and C2 reduced over 90% and 50% of the formed NO2−, respectively, while C3 had low NO2− production and a lower reduction capacity of NO2−. The ability of consortium C3 to rapidly reduce high NO3− concentration and its lack of ability to reduce the NO2− could be attributed to the presence of nitrate-respiring bacteria that used NO3− for their metabolic activities but lack nitrite reductase genes that transcribe genes that facilitate NO2− reduction, hence its accumulation (Nogales et al., 2002; Philips et al., 2002). Owing to the overall performances of the consortia for NO3− reduction and denitrification under all concentrations, 2,000 mg/L of NO3− was deemed as the benchmark target for the downstream experiments to optimize and evaluate complete denitrification capacities of the selected consortia.
Heavy metals such as copper (Cu), iron (Fe), and molybdenum (Mo) have been reported as co-factors in protein activity, including those involved in denitrification (Glass and Orphan, 2012; Wang et al., 2013; Black et al., 2016). Both iron (Fe2+) and copper (Cu2+) have the capacity to promote enzymatic activities of various denitrification steps and have been successfully explored in studies to enhance NO3− reduction by nitrate reductases (narG) (Schaedler et al., 2018), NO2− reduction by nitrite reductases (NirK) (Green et al., 2010), and N2O reduction by nitrous oxide reductases (nosZ) (Sanford et al., 2012). In this study, both Cu2+ and Fe2+ added at 5 mg/L (Lee et al., 2009) were used as standardized test concentrations to explore their dynamic effects on the reduction rates of dissolved NO3− with the initial concentration of 2,000 mg/L, while monitoring concentrations of biogenic NO2− and the production of N2 as the terminal product for complete denitrification.
The obtained results recorded in Table 2 revealed that cultures behaved differently when exposed to different conditions; for instance, four of the nine (three consortia: C1, C2, and C3 conducted in triplicates) metal-supplemented consortia, C3:Cu-NO3; C3:Cu and Fe-NO3; C2:Cu and Fe-NO3; and C2:Fe-NO3, attained 100% NO3− reduction within 6 h followed by the other three consortia, C1:Cu-NO3; C2:Cu-NO3, and C1:Cu and Fe-NO3, attaining 100% NO3− reduction at 12 h. All these cultures besides C3:Fe-NO3 contained Cu2+ in the medium, which indicates the promoting effect Cu2+ (5 mg/L) had on NO3− reduction. In consortia supplemented with only Fe2+, the cultures demonstrated slow NO3− removal, with C2:Fe-NO3 attaining 100% NO3− reduction after 24 h, while C1:Fe-NO3 attained less than 50% NO3− reduction after 36 h. There have been reports showing that Fe promotes NO3− reduction, but in this study, it seemed not to be the case (Glass and Orphan, 2012). Herein, our findings with the selected consortia supported the use of Cu2+ instead of Fe2+ to optimize NO3− reduction through metal co-factor supplementing.
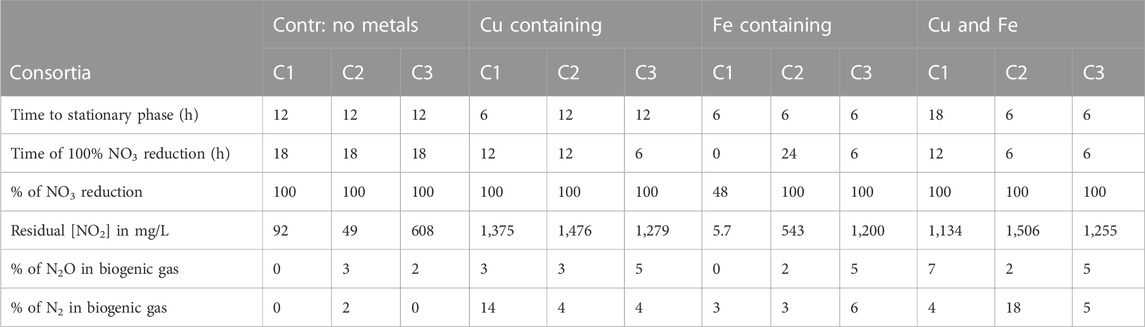
TABLE 2. Summary of the biogeochemical effects of metals (Cu and Fe) on denitrification dynamics on the three selected consortia.
Biogenic NO2− concentrations in the Fe2+-supplemented cultures C1:Fe-NO3 and C2:Fe-NO3 were the lowest, which correlated with the low amount of NO3− reduced. In other cultures, NO2− was observed to start accumulating after the third hour of incubation and became stagnant until all the NO3− was reduced. The same effects of poor NO2− formation and its removal was observed in cultures with Fe2+ as a sole metal cofactor, and a similar observation was noticed by Giannopoulos et al. (2020) when assessing the effects of different heavy metals on denitrification. The sole Fe2+-containing cultures performing poorly with respect to reducing NO3− could be due to NO2− toxicity that effects the growing bacteria needing Cu2+ to promote the catalytic effect on nitrite reductases to reduce NO2− as nirK genes are induced by Cu2+ ions to transcribe the nitrate reductase enzyme and promote its functionality (Smith et al., 2007). The biogenic gas analysis by GC revealed that most N2 gas formed in the cultures containing Cu (C1:Cu-NO3 and C2:Cu and Fe-NO3), which further supported the findings reported above on Cu2+ inducing NO3− and NO2− reduction. High N2 percentages in Cu2+ containing cultures, as opposed to the non-metal supplemented cultures, indicate the potential of the enriched consortia to have the capacity to completely reduce NO3− to N2 when supplemented with Cu2+ (Sullivan et al., 2013; Zhang et al., 2019; Giannopoulos et al., 2020; He et al., 2020). Copper toxicity in N metabolism is known for forming lipid peroxidation and protein damage in most Cu2+ intolerant bacteria, including denitrifiers (Dupont et al., 2011; Ochoa-Herrera et al., 2011). The accumulation of NO2− beyond 1,000 mg/L has been reported to form nitrous acid (HNO2), which is toxic to bacteria and has the potential to halt denitrification (Glass et al., 1997). To uncover and validate the possibility of NO2− accumulation being the reason for the truncation of complete denitrification, more optimization tests to bypass the hurdle were conducted, with the three consortia subjected to complete denitrification optimization tests to choose one consortium for downstream optimization experiments.
The inducing effects of Cu2+ on complete denitrification as a fundamental metal cofactor of denitrifying enzymes was further assessed on the three consortia in the NRM with 2,000 mg/L of NO3− with a two-fold increase (10 mg/L from 5 mg/L, as previously tested). Bacterial growth curves (Figure 3A) showed that C1 and C2 had a similar growth trend as opposed to C3, which had a longer lag phase. In this set of experiments, C1 and C2 attained stationary phases much faster (within 6 h) than C3 (within 12 h). Bacterial growth in all three cultures was not affected by the presence of 10 mg/L of Cu2+ as it was expected to negatively affect the bacteria given the metal toxicity at higher concentrations (Ochoa-Herrera et al., 2011). However, the Td and μmax of the cultures differed as C1 was observed to have the shortest Td (0.88 h) and the fastest μmax (0.788 h) when compared to C2 and C3 (Td of 0.934 and 1.193 h, respectively). Denitrification dynamics revealed unique effects of Cu2+ on all three consortia in which C2 and C3 had the fastest NO3− and NO2− removal rates, attaining 100% NO3− reduction simultaneously within 5 h as compared to C1, which attained complete NO3− removal after 72 h of incubation (Figure 3B). The presence of Cu2+ seemed to have boosted the metabolic activity of bacteria in C2 and C3 cultures to reduce 100% of the produced NO2− at 18 h and 30 h, respectively, which corroborates with the results obtained by Wagner et al. (2018), who observed an increase in the microbial activity through Cu2+ dosing that promoted bacterial growth and denitrification. The inability to reduce NO3− by C1 was observed and this could be due to the different denitrifying bacteria that do not positively respond to either 5 or 10 mg/L of the added Cu2+ as reported by Ochoa-Herrera et al. (2011). The rapid reduction of NO3−, formation of NO2−, and its subsequent reduction in C2 and C3 (Figure 3C) indicated a positive response of the microorganisms to the two-fold increase of the added Cu2+. Theoretically, copper addition would also assist in the reduction of N2O to N2 as Cu2+ is known to determine the transcription and activity rate of nitrous oxide reductases (Giannopoulos et al., 2020; Shen et al., 2020). After 7 days (168 h) of monitoring the denitrification dynamics, only C1 still failed to reduce 100% of NO2−, indicating the consortium’s poor ability to thrive in the presence of the dosed metal cofactor.
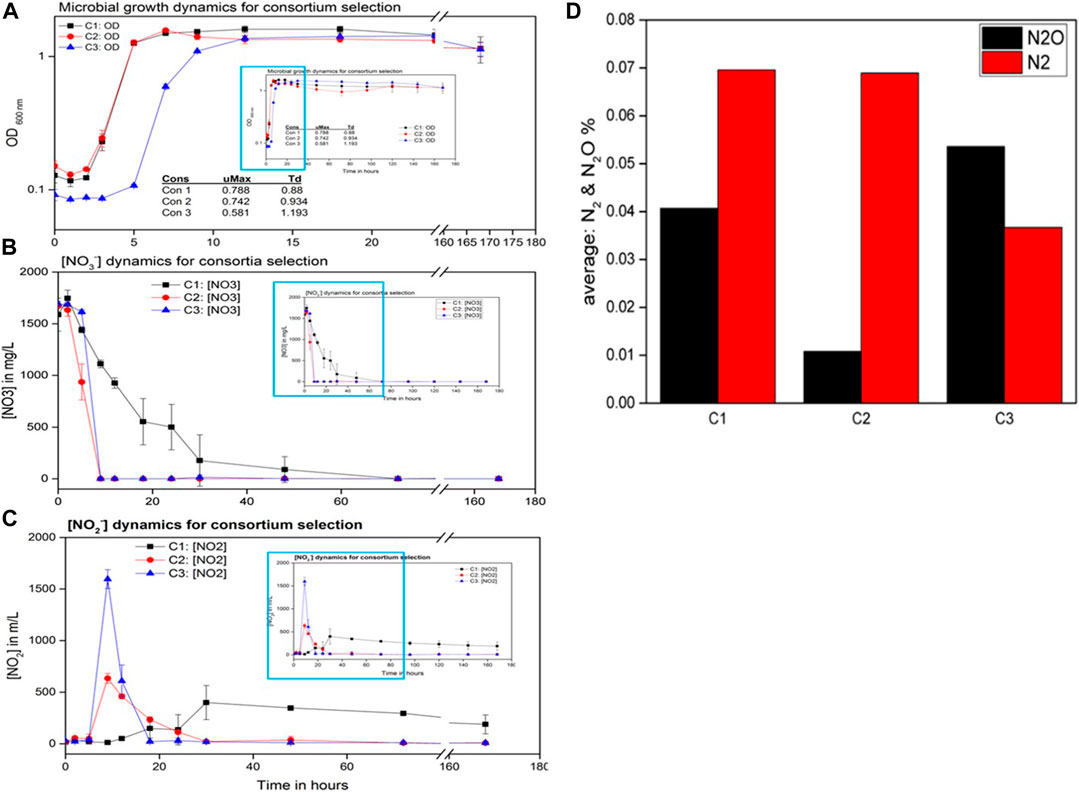
FIGURE 3. Complete denitrification dynamics for consortium selection studies. (A) Microbial growth curves, (B) NO3− reduction dynamics, (C) NO2− reduction dynamics, and (D) copper effect on the reduction of nitrous oxide (N2O) and production of nitrogen (N2) percentages in the three consortia (C1, C2, and C3).
Gas chromatography revealed that the lowest N2O gas (Figure 3D) was detected in C2 cultures with an average of approximately 0.01% followed by C1 with N2O levels as low as 0.04% of the biogenic gas with both consortia producing similar amounts of N2 gas. C1, which was unable to reduce most of the biogenic NO2−, also accumulated N2O. C3 had the highest N2O quantities produced by the bacteria with the lowest N2 quantities, indicating the hindrance of N2O reduction to N2, which further indicates poor utilization of Cu2+. Another possible explanation could be the inherent toxic effect of Cu2+ lowering N2O reduction capacities where the bacteria are unable to tolerate the toxicity of the metal, hence their inability to perform complete denitrification (Granger and Ward, 2003). This inability to reduce N2O to N2 could also be due to the absence of complete denitrifiers that expresses nitrous oxide reductases (nosZ gene) (Nogales et al., 2002). With low N2O quantities in C2 cultures, it was expected that the N2 quantities would have also increased beyond the N2 quantities of C1, but this was not the case probably due to the incomplete reduction of N2O that accounted for over 200 mg/L of residual NO2− that is not converted to gaseous nitrogen species.
With C1 being unable to reduce NO2− completely and C3 responding negatively to the Cu2+ supplement noticeable by high N2O and low N2 quantities, C2 was selected for downstream experiments for further denitrification optimization. The results further prove and support that Cu2+ in conjunction with the copper-tolerant denitrifiers in the consortium can promote N2O reduction to N2, rendering the consortium capable of complete denitrification.
To mimic the environmental conditions where the wastewater samples were collected, an SWW medium was prepared (Table 1) with the modifications reported by Foglar et al. (2005). The selected consortium (C2) was tested with the aim of reducing 2,000 mg/L of NO3− within 24 h under varied conditions monitored for a maximum of 120 h. Microbial growth curves under six different test conditions (Figure 4A) revealed that the variations imposed had different effects on bacterial growth. Compared to the rapid bacterial proliferation observed in NRM, it was evident that the bacterial growth rate decreased with the increasing Td when cultivated on the SWW. This was also observed by Delgadillo-Mirquezand et al. (2016) when switching media compositions during the optimization studies to remove nitrogen and phosphate from wastewater. Nevertheless, C2 was still able to achieve 100% removal of the 2,000 mg/L. Bacterial growth dynamics were not noticeably different when C2-cultivated glycerol was used as a sole carbon source with no metals, with Cu, Fe, and Cu and Fe combined. When glycerol was used as the sole C-source, an average Td of 7.789 h, with the slowest growth rate of 0.089, was observed, which was in contrast to when glucose was used in which Td decreased to 3.081 h. Similar effects were observed in the literature when glucose produced more bacterial biomass than glycerol in studies conducted by Liu et al. (2011), comparing bacterial fermentation processes in which glycerol and glucose were used independently and as co-substrates. In addition to glucose-shortening microbial Td in SWW, the biomass yields also appeared to be greater when glucose was used, resulting in a growth peak above all other experimental culture sets, validating the poor energy supply from glycerol as compared to glucose (Yao et al., 2016; Zhang et al., 2020). When using glucose as the C-source and different nitrogen sources, NO3− and NO2−, microbial growth rates differed by 0.225 and 0.127 with Td of 3.081 and 5.458 h. respectively. Low growth rate and high Td served as an indication of NO3− preference over NO2− as the sole nitrogen source. Sang et al. (2020) observed similar results when they characterized the use of NO3−, NO2−, and NH4+, whereby NO2− yielded the lowest microbial biomass. From the abovementioned bacterial growth dynamics, glucose was observed to promote higher bacterial proliferation, which may suggest modifications need to be adopted when treating wastewater with NO3− concentrations beyond 2,000 mg/L.
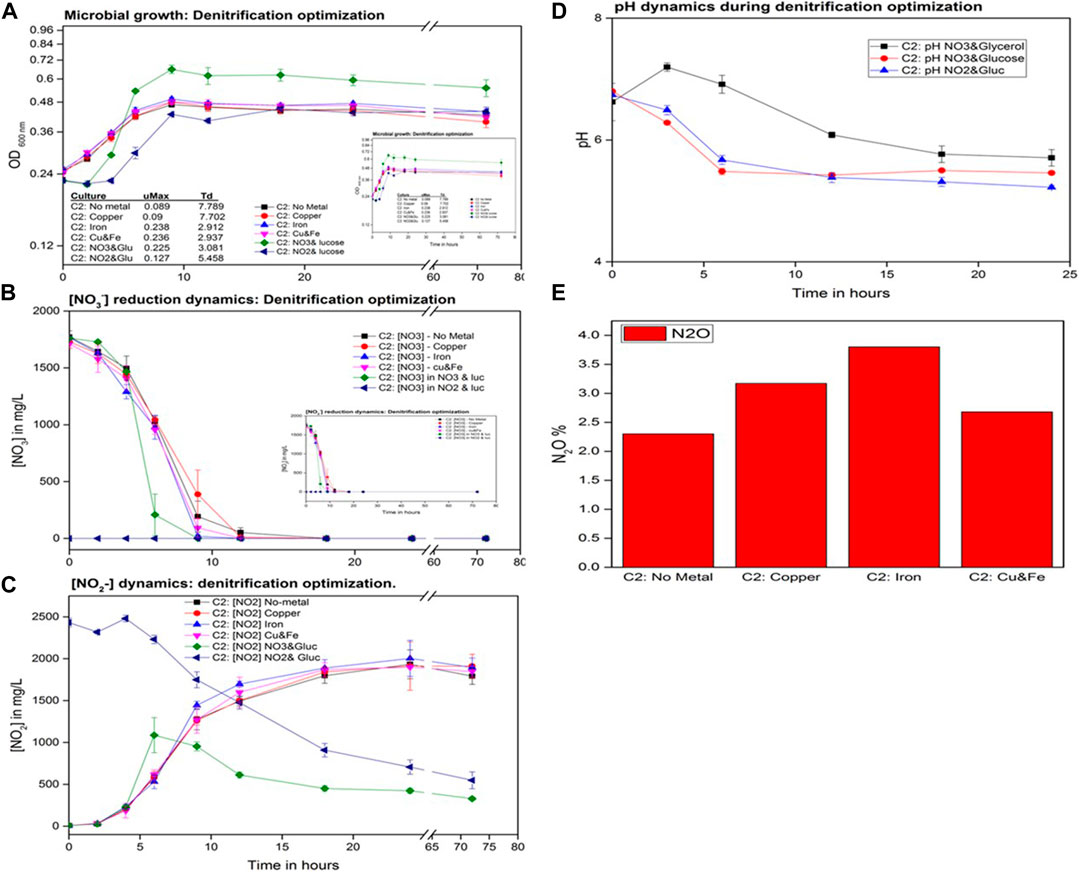
FIGURE 4. Metal kinetics data. (A) Microbial growth curves, (B) nitrate reduction progress curves, (C) nitrite production curves, (D) pH changes in the cultures, and (E) nitrous oxide percentages in biogenic gases.
Fluctuations of pH were monitored for 24 h in three no metal-containing cultures with varying C and N sources (Figure 4D): 1. C2 in SWW with glycerol and NO3−, 2. C2 in SWW with glucose and NO3−, and 3. C2 in SWW with glucose and NO2−. In glucose-containing cultures with both NO3− and NO2− as sole N-sources, pH lowered to below 5 from 6.5 within 6 h, while in the glycerol- and NO2−-containing cultures, pH increased to over neutral lowered below pH 7 after 6 h. The noticeable pH fluctuations occurred between time 0 and 12 h where microbial growth was at its exponential phase, indicating high metabolic activities, hence the change in pH (Sánchez-Clemente et al., 2018). Biochemical oxidation of glycerol results in more proton release during the reduction of an electron acceptor, hence the increase in pH from 6.5 to beyond neutral pH between 0 and 6 h, which remained higher than the pH in cultures with glucose (Baeseman et al., 2006). The oxidation of glucose continuously releases hydrogen ions, hence lowering the pH and also contributing to the pH changes (Liu et al., 2011).
Nitrate and nitrite removal dynamics in all six experimental conditions were observed to be different for each optimization test (Figures 4B, C). Complete NO3− removal was achieved in all five tests but at different rates excluding the one supplied with NO2− as a sole N-source. The first culture to attain 100% NO3− removal with 9 h was the one supplied with glucose without metals, while the other supplied with glycerol without metals attained 100% NO3− removal at 18 h. As previously observed, Fe2+ promoted faster NO3− when supplemented solely, 100% NO3− removal was achieved at 12 h compared to Cu2+, and when Fe and Cu were combined, both tests achieved 100% NO3− removal at 18 h.
Biogenic NO2− accumulated (beyond 1500 mg/L) with a same trend for all culture with Fe2+ containing consortia producing the highest concentrations among all of them. The no metal-containing culture cultivated on glycerol also followed the same trend as the metal tests. The glucose-containing cultures without metals were also observed to accumulate NO2− but only up to a maximum of over 1,000 mg/L at the sixth hour and interestingly started getting reduced. Interestingly, up to 70% of the biogenic NO2− was removed, which was good for the ongoing denitrification process with glucose supplied as a sole C-source (Srinandan et al., 2012). The cultures containing NO2− and glucose as sole N- and C-sources, respectively, also demonstrated the gradual removal of NO2− from 2,500 mg/L to 548 mg/L (78% removal), which concurs with reports made by Yao et al. (2020) that denitrifiers can reduce NO2− directly and not as a biogenic product of NO3− reduction. In denitrification studies in which glycerol was used as a cheaper carbon source from the biodiesel production plant, NO2− accumulated as a terminal byproduct of the denitrification process (Baideme et al., 2017), and for further removal of NO2− by denitrification, C/N ration was re-adjusted to circumvent NO2− accumulation from 1.8 to 3.5, as was performed in studies conducted by Carneiro and Foresti (2017) when running up-flow bioreactors with glycerol as a sole C-source. Biochemical utilization of the three carbon (glycerol) and six carbon (glucose) molecules as energy sources depends on the bacteria using them for denitrification and their energy demands as the two molecules get biodegraded at different rates. Bacteria such as P. stuzeri can degrade glucose much faster than glycerol, promoting the rapid removal of NO3− and NO2− during denitrification (Akunna et al., 1993; Srinandan et al., 2012). It was reported in studies conducted by Bowles et al. (2012) that sulfates and sulfides have denitrification hindrance capacities, which could have been one of the contributing factors resulting in incomplete denitrification in our study as SO4 formed part of the metal compound (CuSO4 and FeSO4) supplements (Yu and Bishop, 2001; Marietou, 2016).
The NO2− reduction dynamics could not be completely relied upon to determine the efficiency of the metals in boosting denitrification; hence, biogenic N2O quantities in each culture were analyzed (Figure 4E) as it is a sensitive indicator of progressive denitrification (de Catanzaro and Beauchamp, 1985). The highest N2O percentages were detected in the culture with Fe2+ as the sole metal co-factor, which indicates the positive boost to nitrite reductases that proceeded with trace quantities of N-metabolisms beyond NO2− reduction. A higher N2O percentage (3.8%) in the Fe2+-containing cultures could be due to the absence of a metal co-factor (Cu2+) that boosts N2O removal to N2 (Baeseman et al., 2006). The culture with the lowest N2O percentages was the non-metal-containing culture, which produced 2.3% of N2O, which could be due to the non-boosting effects of nitrite reductases (Granger and Ward, 2003). The metal co-factor effects were noticeable with varied N2O quantities but with less to no significant NO2− reduction by the consortium growing on glycerol as a sole C-source. The results obtained corroborate those attained by Sun et al. (2009) in which NO2− accumulated from the reduction of NO3− in cultures with four different carbon sources, methanol, ethanol, sodium acetate, and sodium propionate, when glucose was used and no NO3− or NO2− accumulation was observed. With the results obtained in these experiments, it was evident that the removal of dissolved N-species is possible with a change of growth parameters to optimize complete denitrification. For downstream experiments in the optimization of NO2− and N2O reduction to N2, the SWW medium with glucose as the sole C-source and copper as a metal co-factor for N2O removal were conducted.
The effect of supplementing Cu2+ to assess complete denitrification through its catalytic effect to the nosZ gene was explored by adding increasing Cu2+ concentrations (0, 10, 25, 50, and 75 mg/L) to the consortium with 1:1 and 1:2 ratios of C:N (glucose:nitrate). Obtained GC results (Table 3) revealed that biogenic N2 and N2O percentages were noticeably lower than when there was no added Cu2+. Similar findings were noticed in denitrification studies by Sullivan et al. (2013), highlighting how Cu2+ promotes N2O reduction to N2 gas. According to the residual glucose concentrations determined by HPLC (Table 3), cultures that contained two-fold of glucose yielded more CO2 than the cultures that had a 1:1 ratio with NO3−. It is also noticeable that the cultures without Cu2+ and the one with the lowest Cu2+ concentrations used up all the glucose as the residual concentrations were found to be below the detection limit, indicating the complete utilization of the C-source without any hindrance of metal toxicity (He et al., 2020).
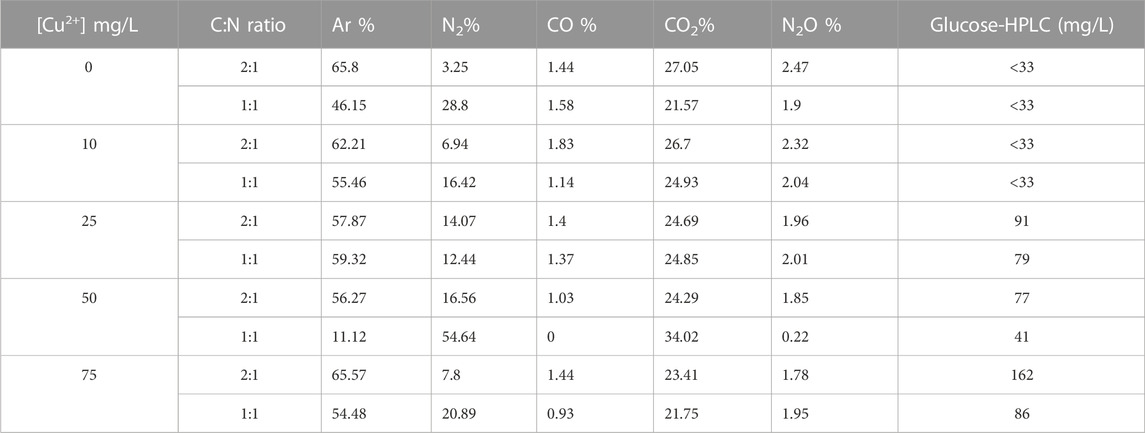
TABLE 3. GC and HPLC analysis results with biogenic gas composition and residual glucose concentrations from the cultures with copper and glucose.
The most biogenic N2 produced in these experiments accounted for over 54% of the total gas observed in cultures containing 50 mg/L of Cu2+ with a C:N ratio of 1:1. The highest Cu2+ concentration (75 mg/L) yielded 7.8% and 20.9% of the total gas in cultures with C/N ratios of 2:1 and 1:1, respectively, which is a sign of metal toxicity in all cultures with 75 mg/L of Cu2+. The bioavailability of Cu2+ in the denitrification system is dependent of the metal dissolution state, which is highly dependent on the pH of the medium (Król et al., 2020). Copper (II) has been reported to dissociate easily in acidic environments and precipitate in neutral to alkaline pH, thus reducing its bioavailability (Cuppett et al., 2006). In this study, the pH of the SWW medium was adjusted to 6.5, like the pH used during bacterial enrichments that yielded the best denitrification results. Owing to the precipitation factor of Cu2+ at a higher pH close to 7, the concentrations used in these experiments could have precipitated reducing the bioavailability of the required Cu2+ ions needed for the biochemical process in denitrification (Cuppett et al., 2006; Król et al., 2020). Alternatively, Cu2+ concentrations below 50 mg/L could have precipitated, thus showing no significant impacts on denitrification, as opposed to concentrations above 50 mg/L in which most of it could have dissolved beyond the limit that the consortium can tolerate.
Copper toxicity at 75 mg/L was noticed with low biogenic nitrogenous gas production in which only 7.8% of N2 and 1.78% of N2O were produced even when the C:N ratio was 2:1. The residual glucose concentrations were also the highest in the culture with the highest Cu2+ concentrations (75 mg/L), which again indicates poor bacterial proliferation with low utilization of glucose. The summed-up data for the cultures cultivated with Cu2+ revealed that 50 mg/L of Cu2+ promoted the best denitrification results with biogenic N2 gas of over 55% and less than 1% of N2O in the cultures after 48 h. Furthermore, the results further validate that supplementing denitrification cultures with Cu2+ does promote N2O reduction to N2 and revealed the toxicity levels of inorganic Cu2+ to be evident at concentrations over 75 mg/L. Owing to the abovementioned details, Cu2+ supplemented at 50 mg/L benchmarks the maximum concentrations that promotes the complete denitrification of over 2,000 mg/L of NO3−. Finally, the abovementioned results provide successful establishment of optimization conditions that can be applied in a bioremediation system to obtain the best complete denitrification results using an indigenous bacterial consortium.
Based on the Illumina MiSeq data analysis (Figure 5) of the C2 consortium, it is evident that bacterial groups detected in the wastewater samples were successfully enriched. The selection in the NRM with high concentrations of NO3− as selective pressure resulted in a consortium comprising two main phyla, Proteobacteria and Firmicutes, which accounted for 84% and 16% of the bacterial community, respectively (Figure 5A). The phylum Proteobacteria has been widely reported as the primary bacterial group in remediation systems comprising most bacteria used in wastewater treatments (Cydzik-Kwiatkowska and Zielińska, 2016). The most abundant detected genera were Asaccharospora, Clostridium_sensu_stricto 18, Paraclostridium, Tissierella, Clostridioides, Escherichia-Shigella, and Tepidimicrobium (Figure 5B). It is worth noting that Paraclostridium accounted for over 50% of the whole microbial population in the selected consortium, whereas in the original water sample, it constituted less than 1% of the reads. It has been reported that Paraclostridium, an obligate anaerobic Firmicute, has the capacity to carry out complete denitrification, reducing NO3− to N2 gas (Kutsuna et al., 2019). Among the dominant bacterial genera, Clostridium and Clostridiales were present and are known to facilitate dissimilatory nitrate reduction to ammonium (DNRA) and fermentative denitrification, respectively. In closed systems, the two bacterial genera can synergistically metabolize N by reducing NO3− to NH4+ and then NH4+ to N2 for Clostridium and Clostridiales, respectively, which makes them key bacteria in the denitrifying consortium used in this study (Ahmad et al., 2021; van den Berg et al., 2017). Interestingly, the consortium attained the best denitrification dynamics without the dominance of any benchmarked known complete denitrifiers, such as Paracoccus and Pseudomonas, commonly used in denitrification kinetics and dynamics studies (Yao et al., 2020). Therefore, it can be concluded that the bacterial groups detected in the enriched consortium possess the synergistic metabolic relationship permitting them to perform complete denitrification, as most are known to be incomplete denitrifiers. Thus, the results suggest that bacteria capable of metabolizing different N-species were co-cultured and acted synergistically to perform complete denitrification as a complete denitrifier would when cultured in a suitable environment with all growth factors.
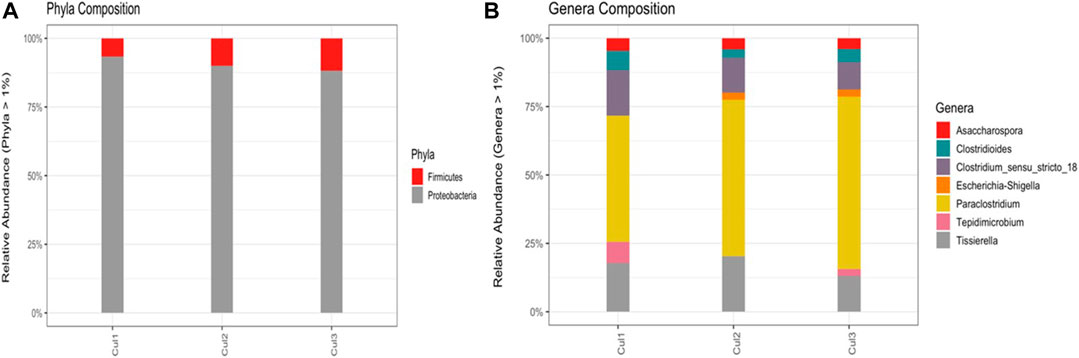
FIGURE 5. Taxonomic distribution of the bacterial groups in the selected consortium at the (A) phylum and (B) genus levels.
Nitrate-contaminated mine wastewater used in the current study served as a source of bacteria that showed the potential for being used in the remediation of the same wastewater when provided with a suitable carbon source, metabolic-based metal co-factors, and a regulated environment. Two mine wastewater samples (D-Lst and Pt-SA) were collected, analyzed both chemically and biologically. Analytical data revealed that explosives used during mining exuberate wastewater contamination by introducing high concentrations of NO3−. Both samples were similar in their chemical characteristics when comparing dissolved cation and anion concentrations but differed in microbial community composition. Sample Pt-SA was found to be relatively more diverse than sample D-Lst, probably due to the sewage spillage in the wastewater catchment, which could have resulted in the bioaugmentation of various bacterial groups widening the microbial community in the wastewater. FAPROTAX data further presented a wide range of functional metabolic pathways associated with the Pt-SA sample, highlighting its diversity and complexity. Enrichment studies successfully yielded consortia that could reduce high NO3− concentrations. In both samples, the most detected taxa belonged to phylum Proteobacteria, commonly known to be applicable in wastewater bioremediation. Anaerobic enrichment studies targeting denitrifying bacteria were successful, of which three consortia that portrayed excellent denitrification dynamics were selected and subjected to downstream experiments. The optimization of complete denitrification by the selected consortia revealed the limits of tolerance of denitrifiers to nitrate concentrations over 10,000 mg/L. Furthermore, it was proved that supplementing growth media with metal cofactors (Fe2+ and Cu2+) stimulated bacterial metabolisms to perform complete denitrification. In-depth evaluation of metal co-factors as stimulants for complete denitrification resulted in the evidence that supplementing inorganic copper to a defined denitrifying consortium can promote the reduction of the fastidious N2O, which is an undesirable by-product as it is a potent greenhouse gas. The investigation of complete denitrification studies revealed that 50 mg/L of inorganic Cu2+ can induce maximum N2O reduction to N2, while over 75 mg/L exerted toxic effects on the bacteria. With further optimization experiments in a quest to rapidly reduce dissolved N-species, optimum conditions were established with the selection of glucose as a suitable C-source to aid complete denitrification in a synthetic medium mimicking collected mine wastewater samples. The obtained results set a new benchmark in the conditions required for the complete denitrification by a consortium enriched from industrial wastewater samples and that can be applied in bench top bioreactors to continuously treat NO3− from industrial wastewater samples. This is a breakthrough in a biotechnological setting to use indigenous bacteria with experimental modifications to optimally reduce NO3− and its intermediate byproducts to yield an environmentally friendly N2 gas as a terminal biological byproduct.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
KM, EV, and EC were responsible for conceptualizing the study. KM, MM, JC, and EC collected wastewater samples. KM conducted the experimental work. KM, EC, JC, and EV conducted the data analysis. KM drafted the first manuscript, KM, MM, ZK, GK, and EC wrote and reviewed the final manuscript. All authors read and approved the final manuscript.
This research was funded by the National Research Foundation (NRF-South Africa. Grant number AEMD170707250199) and the Technology Innovation Agency (TIA. Grant number SABDI16/1070). Granted Ethics Clearance number UFS-ESD2020/0165/1011.
The authors would like to acknowledge NRF and TIA for financial support and the involved departments at the University of the Free State (UFS), Central University of Technology (CUT), and University of Kwa-Zulu Natal (UKZN) where resources, infrastructure, and equipment were used in conducting the experiments and preparing the manuscript. They finally acknowledge the two mining companies in South Africa and Lesotho for granting access to collect samples and the Institute of Groundwater Studies (IGS) at the UFS for assistance with chemical analysis of water samples.
Author EV was employed by iWater Pvt., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1148872/full#supplementary-material
Adouani, N., Limousy, L., Lendornmi, T., and Sire, O. (2015). N2O and NO emissions during wastewater denitrification step: Influence of temperature on the biological process. Comptes Rendus Chime 18, 15–22.
Ahmad, H. A., Guo, B., Zhuang, X., ZhaiAhmad, Y. S., Lee, T., Zhu, J., et al. (2021). A twilight for the complete nitrogen removal via synergistic partial-denitrification, anammox, and DNRA process. Clean. Water 31, 1–11.
Ahn, M. K., Chilakala, R., Han, C., and Thenepalli, T. (2018). Removal of hardness from water samples by a carbonation process with a closed pressure reactor. Water 10 (54), 54–10. doi:10.3390/w10010054
Akunna, J. C., Bizeau, C., and Moletta, R. (1993). Nitrate and nitrite reductions with anaerobic sludge using various carbon sources: Glucose, glycerol, acetic acid, lactic acid and methanol. Water Res. 27 (8), 1303–1312. doi:10.1016/0043-1354(93)90217-6
Baeseman, J. L., Smith, R. L., and Silverstein, J. (2006). Denitrification potential in stream sediments impacted by acid mine drainage: Effects of pH, various electron donors, and iron. Microb. Ecol. 51 (2), 232–241. doi:10.1007/s00248-005-5155-z
Baideme, M., Long, C., Plante, L., Starke, J., Butkus, M., and Chandran, K. (2017). Optimization of partial denitrification to maximize nitrite production using glycerol as an external carbon source – impact of influent COD:N ratio. Proc. Water Environ. Fed. 13, 1356–1360. doi:10.2175/193864717822153869
Bhusal, A., and Muriana, P. M. (2021). Isolation and characterization of nitrate reducing bacteria for conversion of vegetable-derived nitrate to ‘natural nitrite. Appl. Microbiol. 1, 11–23. doi:10.3390/applmicrobiol1010002
Black, A., Hsu, P. L., Hamonts, K. E., and Condron, L. M. (2016). Influence of copper on expression of nirS, norB and nosZ and the transcription and activity of NIR, NOR and N2OR in the denitrifying soil bacteria Pseudomonas stutzeri. Microb. Biotechnol. 9, 381–388. doi:10.1111/1751-7915.12352
Böllmann, J., and Martienssen, M. (2020) Comparison of different media for the detection of denitrifying and nitrate reducing bacteria in mesotrophic aquatic environments by the most probable number method. J. Microbiol. Methods 168(105808), 105808–105810. doi:10.1016/j.mimet.2019.105808
Bowles, M. W., Nigro, L. M., Teske, A. P., and Joye, S. B. (2012). Denitrification and environmental factors influencing nitrate removal in Guaymas Basin hydrothermally altered sediments. Front. Microbiol. 3 (377), 1–11. doi:10.3389/fmicb.2012.00377
Bruck, W. M., Graverholt, G., and Gibson, G. R. (2002). Use of batch culture and a two-stage continuous culture system to study the effect of supplemental ᾂ-lactalbumin and glycomacro peptide on mixed populations of human gut bacteria. Fed. Eur. Microbiol. Soc. 41, 231–237. doi:10.1016/s0168-6496(02)00296-9
Butler, B. A., and Ford, R. G. (2018). Evaluating relationships between total dissolved solids (TDS) and total suspended solids (TSS) in a mining-influenced watershed. Mine Water Environ. 37 (1), 18–30. doi:10.1007/s10230-017-0484-y
Cai, L., Ju, F., and Zhang, T. (2014). Tracking human sewage microbiome in a municipal wastewater treatment plant. Appl. Microbiol. Biotechnol. 98, 3317–3326. doi:10.1007/s00253-013-5402-z
Calderer, M., Gilbert, O., Marti, V., Rovira, M., de Pablo, J., Jordana, S., et al. (2010). Denitrification in presence of acetate and glucose for bioremediation of nitrate-contaminated groundwater. Environ. Technol. 31 (7), 799–814. doi:10.1080/09593331003667741
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 7, 335–336. doi:10.1038/nmeth.f.303
Carneiro, R. B., and Foresti, E. (2017). Nutrient biological removal in an up-flow sludge bed reactor under intermittent aeration using glycerol as the sole carbon source. Braz. J. Chem. Eng. 34 (4), 961–969. doi:10.1590/0104-6632.20170344s2016050
Carrea-Galeote, D., Tortosa, G., and Bedmar, E. J. (2013). Determination of denitrification genes abundance in environmental samples. Metagenomics 2, 1–14. doi:10.4303/mg/235702
Cason, E. D., Mahlomaholo, B. J., Taole, M. M., Abong, G. O., Vermeulen, J., de Smidt, O., et al. (2020). Bacterial and fungal dynamics during the fermentation process of Sesotho, a traditional beer of Southern Africa. Front. Microbiol. 11 (1451), 1–14. doi:10.3389/fmicb.2020.01451
Collingro, A., Köstlbacher, S., and Horn, M. (2020). Chlamydiae in the environment. Trends Microbiol. 28 (11), 877–888. doi:10.1016/j.tim.2020.05.020
Correa-Galeote, D., Marco, D. E., Tortosa, G., Bru, D., Philippot, L., and Bedmar, E. J. (2013). Spatial distribution of N-cycling microbial communities showed complex patterns in constructed wetland sediments. FEMS Microbiol. Ecol. 83 (2), 340–351. doi:10.1111/j.1574-6941.2012.01479.x
Cua, L. S., and Stein, L. Y. (2014). Characterization of denitrifying activity by the alphaproteobacterium, sphingomonaswittichiiRW1. Front. Microbiol. 5 (404), 1–7.
Cuppett, J. D., Duncan, S. E., and Dietrich, A. M. (2006). Evaluation of copper speciation and water quality factors that affect aqueous copper tasting response. Chem. Senses 31, 689–697. doi:10.1093/chemse/bjl010
Cydzik-Kwiatkowska, A., and Zielińska, M. (2016). Bacterial communities in full-scale wastewater treatment system. World J. Microbiol. Biotechnol. 32 (66), 1–8.
Cyplik, P., Marecik, R., Piotrowska-Cyplik, A., Olejnik, A., DroŻdŻyńska, A., and Chrzanowski, L. (2012). Biological denitrification of high nitrate processing wastewaters from explosives production plant. Water, Air Soil Pollut. 223, 1791–1800. doi:10.1007/s11270-011-0984-5
DadrasniaUsmanLim, A. M. M. K. T., Velappan, R. D., Shahsavari, N., Vejan, P., Mahmud, A. F., and Ismail, S. (2017). Microbial aspects in wastewater treatment – a technical review. Environ. Pollut. Prot. 2 (2), 75–86.
Dahab, M. F. (1991). “Nitrate treatment methods: An overview,” in Nitrate contamination. NATO ASI series. Editors I. Bogardi, R. D. Kuzelka, and W. G. Ennenga (Berlin, Heidelberg: Springer), 30.
Damiani, I., Pauly, N., Puppo, A., Brouquisse, R., and Boscari, A. (2016). Reactive oxygen species and nitric oxide control early steps of the legume – rhizobium symbiotic interaction. Front. Plant Sci. 7 (454), 454–458. doi:10.3389/fpls.2016.00454
de Catanzaro, J. B., and Beauchamp, E. G. (1985). The effect of some carbon substrate on denitrification rates and carbon utilization in soil. Biol. Fertil. Soils 1, 183–187.
Degnan, J. R., Bohlke, J. K., Pelham, K., Langlais, D. M., and Walsh, G. J. (2016). Identification of groundwater nitrate contamination from explosives used in road construction: Isotopic, chemical, and hydrologic evidence. Environ. Sci. Technol. 50, 593–603. doi:10.1021/acs.est.5b03671
Delgadillo-Mirquez, L., Lopes, F., Taidi, B., and Pareau, D. (2016). Nitrogen and phosphate removal from wastewater with a mixed microalgae and bacteria culture. Biotechnol. Rep. 11, 18–26. doi:10.1016/j.btre.2016.04.003
Dhania, G., and Rani, K. (2014). Impact of urbanization on ground water pollution-an emerging problem. J. Int. Acad. Res. Multidiscip. 2 (9), 123–133.
Dupont, C. L., Grass, G., and Rensing, C. (2011). Copper toxicity and the origin of bacterial resistance—New insights and applications. Metallomics 3 (11), 1109–1118. doi:10.1039/c1mt00107h
Durzan, D. J., and Pedroso, M. C. (2002). Nitric oxide and reactive nitrogen oxide species in plants. Biotechnol. Genet. Eng. Rev. 19 (1), 293–338. doi:10.1080/02648725.2002.10648032
DWAF (Department of water affairs and forestry, South Africa) (1996). “South African water quality guidelines,”. Domestic water use. 1Second edition.
Egashira, A, R., Tanabe, S., and Habaki, H. (2012). Adsorption of heavy metals in mine wastewater by Mongolian natural zeolite. Procedia Eng. 42, 49–57. doi:10.1016/j.proeng.2012.07.394
Fadeev, E., Cardozo-Mino, M. G., Rapp, J. Z., Beinhold, C., Salter, I., Salman-Carvalho, V., et al. (2021). Comparison of two 16S rRNA primers (V3-V4 ad V4-V5) for studies of arctic microbial communities. Front. Microbiol. 12 (637526), 1–11.
Fan, F., Zhang, B., Morrill, P. L., and Husain, T. (2018). Isolation of nitrate-reducing bacteria from an offshore reservoir and the associated biosurfactant production, 8. United Kingdom: Royal Society of Chemistry, 26596–26609.
Fang, P., Tang, Z., Chen, X., Huang, J., Tang, Z., and Cen, C. (2018). Removal of high-concentration sulfate ions from the sodium alkali FGD wastewater using ettringite precipitation method: Factor assessment, feasibility, and prospect. Hindawi J. Chem. 2018, 1–8. doi:10.1155/2018/1265168
Feng, W., Wang, C., Lei, X., Wang, H., and Zhang, X. (2020). Distribution of nitrate content in groundwater and evaluation of potential health risks: A case study of rural areas in northern China. Environ. Res. Public Health 17 (9390), 1–14. doi:10.3390/ijerph17249390
Foglar, L., Briški, F., Sipos, L., and Vuković, M. (2005). High nitrate removal from synthetic wastewater with mixed bacterial culture. Bioresour. Technol. 96, 897–888.
Fountoulaskis, M. S., Petousi, I., and Manios, T. (2010). Co-digestion of sewage sludge with glycerol to boost biogas production. Waste Manag. 30, 1849–1853. doi:10.1016/j.wasman.2010.04.011
Gang, Z., Ylinen, A., Di Capua, F., Papirio, S., Lakaniemi, A., and Puhakka, J. (2013). Impact of heavy metals on denitrification of simulated mining wastewater. Adv. Mater. Res. 825, 500–508.
Gao, N., Shen, W. S., Camargo, E., Shiratori, Y., Nishizawa, T., Isobe, K., et al. (2017). Nitrous oxide (N2O)-reducing denitrifer-innoculated granular organic fertilizer mitigates N2O emission from agricultural soils. Biol. Fertil. Soils 53, 885–898. doi:10.1007/s00374-017-1231-z
Giannopoulos, G., Hartop, K. R., Brown, B. L., Song, B., Elsgaard, L., and Franklin, R. B. (2020). Trace metal availability affecrs greenhouse gas emissions ad microbial functional group abundance in freshwater wetland sediments. Front. Microbiol. 11 (560861), 1–12.
Glass, C., Silverstein, J., and Oh, J. (1997). Inhibition of denitrification in activated sludge by nitrite. Water Environ. Res. 69 (6), 1086–1093. doi:10.2175/106143097x125803
Glass, J. B., and Orphan, V. J. (2012). Trace metal requirements for microbial enzymes involved in the production and consumption of methane and nitrous oxide. Front. Microbiol. 3 (61), 61–20. doi:10.3389/fmicb.2012.00061
Granger, J., and Ward, B. B. (2003). Accumulation of nitrogen oxides in copper-limited cultures of denitrifying bacteria. Am. Soc. Limnol. Oceanogr. 48 (1), 313–318. doi:10.4319/lo.2003.48.1.0313
Green, S. J., Prakash, O., Gihring, T. M., Akob, D. M., Jasrotia, P., Jardine, P. M., et al. (2010). Denitrifying bacteria isolated from terrestrial subsurface sediments exposed to mixed-waste contamination. Appl. Environ. Microbiol. 76 (10), 3244–3254. doi:10.1128/aem.03069-09
Grochowska, J. (2020). Assessment of water buffer capacity of two morphometrically different, degraded, urban lakes. Water 12 (1512), 1512–1518. doi:10.3390/w12051512
Guo, Y., Huang, P., Zhang, W., Yuan, X., Fan, F., Wang, H., et al. (2013). Leaching of heavy metals from Dexing copper mine tailings pond. Trans. Nonferrous Metals Soc. China 23 (10), 3068–3075. doi:10.1016/s1003-6326(13)62835-6
He, T., Xie, D., Ni, J., Cai, X., and Li, Z. (2019). Investigating the effect of copper and magnesium ions on nitrogen removal capacity of pure cultures by modified non-competitive inhibition model. Ecotoxicol. Environ. Saf. 170, 479–487. doi:10.1016/j.ecoenv.2018.12.019
He, T., Xie, D., Ni, J., Li, Z., and Li, Z. (2020). Effects of cobalt, Cadmium and Manganese on nitrogen removal capacity of AnthrobacterarilaitensisY-10. Water 12 (1701), 1–12.
Huang, H. K., and Tseng, S. K. (2001). Nitrate reduction by Citrobacter diversus under aerobic environment. Appl. Microbiol. Biotechnol. 55, 90–94. doi:10.1007/s002530000363
Huang, S., Chen, C., Yang, X., Wu, Q., and Zhang, R. (2011). Distribution of typical denitrifying functional genes and diversity of the nirS- encoding bacterial community related to environmental characteristics of river sediments. Biogeosciences 8, 3041–3051. doi:10.5194/bg-8-3041-2011
Huang, T-L., Zhou, S-L., Zhang, H-H., Zhou, N., Guo, L., Di, S. Y., et al. (2015). Nitrogen removal from micro-polluted reservoir water by indigenous aerobic denitrifiers. Int. J. Mol. Sci. 16, 8008–8026. doi:10.3390/ijms16048008
Igiri, B. E., Okoduwa, S. I. R., Idoko, G. O., Akabuogu, E. P., Adeyi, A. O., and Ejiogu, I. K. (2018). Toxicity and bioremediation of heavy metals contaminated ecosystem from tannery wastewater: A review. Hindawi J. Toxicol. 2018, 1–16. doi:10.1155/2018/2568038
Jabari, L., Gannoun, H., Khelifi, E., Cayol, J. L., Godon, J. L., Hamdi, M., et al. (2016). Bacterial ecology of abattoir wastewater treated by an anaerobic digestor. Braz. J. Microbiol. 47, 73–84. doi:10.1016/j.bjm.2015.11.029
Jiang, M., Wu, Y., He, P., Hu, S., Li, Q., and Chem, S. (2023). Assembled denitrifiying consortia for efficient nitrate removal under low-COD/N conditions. Chem. Eng. J. 460 (141655), 1–10.
Jinxiang, F., Zhang, Z., and Zhu, J. (2020). Study on the diversity of denitrification bacteria treating with wastewater by using PPGC filler on SBMBBR at low temperature. Int. Conf. Environ. Pollut. Prev. 158, 04002. doi:10.1051.e3sconf/202015804002
Kamp, A., Hogslund, S., Risgaard-Petersen, N., and Stief, P. (2015). Nitrate storage and dissimilatory nitrate reduction by eukaryotic microbes. Front. Microbiol. 6 (1492), 1–15. doi:10.3389/fmicb.2015.01492
Kgopa, P. M., Mashela, P. W., and Manyevere, A. (2018). Suitability of treated wastewater with respect to pH, electrical conductivity, selected cations and sodium adsorption ratio for irrigation in semi-arid region. Water S. Afr. 44 (4), 551–556.
Kho, Z. Y., and Lal, S. (2018). The human gut microbiome - a potential controller of wellness and diseases. Front. Microbiol. 8 (1835), 1–5.
Kim, J. K., Park, K. J., Cho, K. S., Nam, S. W., Park, T. J., and Bajpa, R. (2005). Aerobic nitrification-denitrification by heterotrophic Bacillus strains. Bioresour. Technol. 96 (17), 1897–1906. doi:10.1016/j.biortech.2005.01.040
Kisková, J., Perhácová, Z., Vlcko, L., Sedláková, J., Kvasnová, S., and Pristaš, P. (2018). The bacterial population of neutral mine drainage water of elizabeth’s shaft (slovinky, Slovakia). Curr. Microbiol. 75, 988–996. doi:10.1007/s00284-018-1472-6
Klindworth, A., Pruesse, E., Schweer, T., Peplies, J., Quast, C., Horn, M., et al. (2013). Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing based diversity studies. Nucleic Acids Res. 41, e1. doi:10.1093/nar/gks808
Kong, D., Li, W., Deng, Y., Ruan, Y., Chen, G., Yu, J., et al. (2018). Denitrification-potential evaluation and nitrate-removal-pathway analysis of aerobic denitrification strain Marinobacterhydro carbon oclasticusRAD-2. Water 10 (1298), 1–12.
Krishna Reddy, Y. V., Adamala, S., Levlin, E. K., and Reddy, K. S. (2017). Enhancing nitrogen removal efficiency of domestic wastewater through increased total efficiency in sewage treatment (ITEST) pilot plant in cold climatic regions of Baltic Sea. Int. J. Sustain. Built Environ. 6, 351–358. doi:10.1016/j.ijsbe.2017.05.002
Król, A., izerna, K., and Bozym, M. (2020). An assessment of pH-dependent release and mobility of heavy metals from metallurgical slag. J. Hazard. Mater. 384 (121502), 121502–121509. doi:10.1016/j.jhazmat.2019.121502
Kutsuna, R., Miyoshi-Akiyama, T., Mori, K., Hayashi, M., Tomida, J., Morita, Y., et al. (2019). Description of Paraclostridiumbifermentanssub sp. Muricolitidissub sp. nov., emended description of Paraclostridiumbifermentans(SasiJyothsna et al., 2016), and creation of Paraclostridiumbifermentassubsp. Bifermentanssub sp. nov. Microbiol. Immunol. 63, 1–10. doi:10.1111/1348-0421.12663
Lawniczak, A. E., Zbierska, J., Nowak, B., Achtenberg, K., Grzeskowiak, A., and Kanas, K. (2016). Impact of agriculture and land use on nitrate contamination in groundwater and running waters in central-west Poland. Environ. Monit. Assess. 188 (172), 1–17.
Lee, Y. W., Tian, Q., Ong, S. K., Sato, C., and Chung, J. (2009). Inhibitory effects of copper on nitrifying bacteria in suspended and attached growth reactors. Water, Air Soil Pollut. 203, 17–27. doi:10.1007/s11270-009-9988-9
Levy-Booth, D. J., Prescott, C. E., and Grayston, S. J. (2014). Microbial functional genes involved in nitrogen fixation, nitrification and denitrification in forest ecosystems. Soil Biol. Biochem. 75, 11–25. doi:10.1016/j.soilbio.2014.03.021
Ling, S., and Juanjuan, S. (2018). Optimal growth conditions and nutrient degradation characteristics of an aerobic denitrifying bacterium. BioRxiv. doi:10.1101/353847
Liu, Y., Feng, C., Chen, N., Sheng, Y., Dong, S., Hao, C., et al. (2016). Bioremediation of nitrate and Fe(II) combined contamination in groundwater by heterotrophic denitrifying bacteria and microbial community analysis. R. Soc. Chem. Adv. 6, 108375–108383. doi:10.1039/c6ra22687f
Liu, Y., Zhang, Y., Zhang, R., Zhang, F., and Zhu, J. (2011). Glycerol/glucose Co-fermentation: One more proficient process to produce propionic acid by propionibacterium acidipropionici. Propionibacterium Acidipropionici. Curr. Microbiol. 62, 152–158. doi:10.1007/s00284-010-9683-5
Lv, P., Luo, J., Zhuang, X., Zhang, D., Huang, Z., and Bai, Z. (2017). Diversity of culturable aerobic denitrifying bacteria in the sediment, water and biofilms in Liangshui river of Beijing, China. Nat. Sci. Rep. 7 (10032). doi:10.1038/s41598-017-09556-9
Ma, Q., Qu, Y-Y., Zhang, X-W., Shen, W-L., Liu, Z-Y., Wang, J-W., et al. (2015). Identification of the microbial community composition and structure of coal-mine wastewater treatment plants. Microbiol. Res. 175, 1–5. doi:10.1016/j.micres.2014.12.013
Marietou, A. (2016). Nitrate reduction in sulfate-reducing bacteria. Fed. Eur. Microbiol. Soc. Microbiol. Lett. 363 (15), fnw155–4. doi:10.1093/femsle/fnw155
Martínez-Espinosa, R. M. (2020). Heterologous and homologous expression of proteins from haloarchaea: Denitrification as a case study. Mol. Sci. 21 (82), 1–14.
Matei, A., and Racoviteanu, G. (2021). Review of the technologies for nitrates removal from water intended for human consumption. IOP Conf Earth Environ. Sci. 664 (012024), 1–11. doi:10.1088/1755-1315/664/1/012024
Matodzi, V., Legodi, M. A., and Tavengwa, N. T. (2020). Determination of platinum group metal in dust, water, soil and sediments in the vicinity of a cement manufacturing plant. Springer Nat. Appl. Sci. 2 (1090), 1–10.
McCarthy, T. S. (2011). The impact of acid mine drainage in South Africa. South Afr. J. Sci. 107 (5), 1–7. doi:10.4102/sajs.v107i5/6.712
McMurdie, P. J., and Holmes, S. (2013). phyloseq: An R Package for reproducible interactive analysis and graphics of microbiome census data. Plos One 8 (4), 1–11. doi:10.1371/journal.pone.0061217
Melton, E. D., Rudolph, A., Behrens, S., Schmidt, C., and Kappler, A. (2014). Influence of nutrient concentrations on MPN quantification and enrichment of nitrate-reducing Fe(II)-Oxidizing and Fe(III)-Reducing bacteria from littoral freshwater lake sediments. Geomicrobiol. J. 31, 788–801. doi:10.1080/01490451.2014.892765
Miyahara, M., Kim, S. W., Fushinobu, S., Takaki, K., Yamada, T., Watanabe, A., et al. (2010). Potential of aerobic denitrification by Pseudomonas stutzeri TR2 to reduce nitrous oxide emissions from wastewater treatment plant. Appl. Environ. Microbiol. 76 (14), 4619–4625. doi:10.1128/aem.01983-09
Modoi, O-C., Roba, C., Török, Z., and Ozunu, A. (2014). Environmental risks due to heavy metal pollution of water resulted from mining wastes in NW Romania. Environ. Eng. Manag. J. 13 (9), 2325–2336. doi:10.30638/eemj.2014.260
Mohapatra, D. P., and Kirpalani, D. M. (2017). Process effluents and mine tailings: Sources, effects and management and role of nanotechnology. Nanotechnol. Environ. Eng. 2 (1), 1–12. doi:10.1007/s41204-016-0011-6
Moloantoa, K,M., Khetsha, Z. P., van Heerden, E., Castillo, J. C., and Cason, E. D. (2022). Nitrate water contamination from industrial activities and complete denitrification as a remediation option. Water 14 (799), 1–31. doi:10.3390/w14050799
Nascimento, A. L., Souza, A. J., Andrade, P. A. M., Andreote, F. D., Coscione, A. R., Oliveira, F. C., et al. (2018). Sewage sludge microbial structures and relations to their sources, treatments, and chemical attributes. Front. Microbiol. 9 (1462), 1–11. doi:10.3389/fmicb.2018.01462
Nogales, B., Timmis, K. N., Nedwell, D. B., and Osborn, A. M. (2002). Detection and diversity of expressed denitrification genes in estuarine sediments after reverse transcription-PCR amplification from mRNA. Appl. Environ. Microbiol. 68 (10), 5017–5025. doi:10.1128/aem.68.10.5017-5025.2002
Ochieng, G. M., Seanego, E. S., and Nkwonta, O. I. (2010). Impacts of mining on water resources in SouthSouth Africa: A review. Sci. Res. Essays 5 (22), 3351–3357.
Ochoa-Herrera, V., Leon, G., Banihani, Q., Field, J. A., and Sierra-Alvarez, R. (2011). Toxicity of copper (II) ions to microorganisms in biological wastewater treatment systems. Sci. Total Environ. 412-413, 380–385. doi:10.1016/j.scitotenv.2011.09.072
Ogden, K. L., Gadgill, J., and Akin, T. (2002). Biological denitrification of hydrolysates from octahydro-1,3,5,7 tetranitro-1,3,5,7-tetrazocine. Water Environ. Res. 74 (4), 338–345. doi:10.2175/106143002x140099
Okada, N., Nomura, N., Nakajima-Kambe, T., and Uchiyama, H. (2005). Characterization of the aerobic denitrifiication in Mesorhizobium sp. strain NH-14 in comparison with that in related Rhizobia. Microbes Environ. 20 (4), 208–215. doi:10.1264/jsme2.20.208
Oladeji, S. O. (2017). Impact of wastewater on nitrate concentrations in soil and vegetables grown along kubanni river, zaria in kaduna state, Nigeria. Archives Agric. Environ. Sci. 2 (4), 318–324. doi:10.26832/24566632.2017.020413
Olds, H. T., Corsi, S. R., Dila, D. K., Halmo, K. M., Bootsma, M. J., and McLellan, S. L. (2018). High levels of sewage contamination released from urban areas after storm events: A quantitative survey with sewage specific bacterial indicators. PLOS Med. 15 (7), e1002614–e1002623. doi:10.1371/journal.pmed.1002614
Osunmakinde, C. O., Selvarajan, R., Mamba, B. B., and Msgadi, T. A. M. (2019). Profiling bacterial diversity and potential pathogens in wastewater treatment plants using High-throughput sequencing analysis. Microorganisms 7 (506), 1–18.
Park, J., Kwon, E., Chung, E., Kim, H., Battogtokh, B., and Woo, N. C. (2020). Environmental sustainability of open-pit coal mining practices at Baganuur, Mongolia. Sustainability 12 (248), 1–20. doi:10.3390/su12010248
Paruch, L., Paruch, A. M., Eiken, H. G., and Sørheim, R. (2019). Faecal pollution affects abundance and diversity of Pt-SAuatic microbial community in anthropogenically influenced lotic systems. Nat. Sci. Rep. 9, 1–13.
Philips, S., Laanbroek, H. J., and Versraete, W. (2002). Origin, causes and effects of increased nitrite concentrations in aquatic environments. Rev. Environ. Sci. Biotechnol. 1, 115–141. doi:10.1023/a:1020892826575
Pinar, G., Duque, E., Haidour, A., Oliva, J. M., Sanchez-Barbero, L., Calvo, V., et al. (1997). Removal of high concentrations of nitrate from industrial wastewaters by bacteria. Appl. Environ. Microbiol. 63 (5), 2071–2073. doi:10.1128/aem.63.5.2071-2073.1997
PlÖhn, M., Scherer, K., Stagge, S. m. J. Öhnsson, L. J., and Funk, C. (2022). Utilization od different carbon sources by Nordic microalgae grown under mixotrophic conditions. Front. Mar. Sci. 9 (830800), 1–11.
Ponce-Soto, G. Y., Aguirre-von-Wobeser, E., Eguiarte, L. E., Elser, J. J., Lee, Z. M.-P., and Souza, V. (2015). Enrichment experiment changes microbial interactions in an ultra-oligotrophic environment. Front. Microbiol. 6 (246), 1–11. doi:10.3389/fmicb.2015.00246
Poole, R. K. (2005). Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 33 (1), 176–180. doi:10.1042/bst0330176
Puigserver, D., Herrero, J., and Carmona, J. M. (2022). Nitrate removal by combining chemical and biostimulation approaches using micro-zero valent iron and lactic acid. Sci. Total Environ. 843 (156841), 1–12. doi:10.1016/j.scitotenv.2022.156841
R Development Core Team (2011). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Rajta, A., Bhatia, R., Setia, H., and Pathania, P. (2019). Role of heterotrophic aerobic denitrifying bacteria in nitrate removal from wastewater. J. Appl. Microbiol. 128, 1261–1278. doi:10.1111/jam.14476
Robles-Cruz, S. E., Melgarejo, J. C., Gali, S., and AmdEscayol, M. (2012). Major- and trace-element compositions of indicator minerals that occur as macro- and megacrysts, and of xenoliths, from kimberlites in northeastern Angola. Minerals 2, 318–337. doi:10.3390/min2040318
Romanis, C. S., Pearson, L. A., and Neilan, B. A. (2021). Cyanobacterial blooms in wastewater treatment facilities: Significance and emerging monitoring strategies. J. Microbiol. Methods 180, 106123. doi:10.1016/j.mimet.2020.106123
Romano, I., Abbate, M., Poli, A., and D’Orazio, L. (2019). Bio-cleaning of nitrate salt efflorescence on stone samples using extremophilic bacteria. Sci. Rep. 9 (1668), 1–11. doi:10.1038/s41598-018-38187-x
Rossi, F., Motta, O., Matrella, S., Proto, A., and Vigliotta, G. (2015). Nitrate removal from wastewater through biological denitrification with OGA 24 in a batch reactor. Water 7, 51–62. doi:10.3390/w7010051
Safonov, A. V., Babich, T. L., Sokolova, D. S., Grouzdev, D. S., Tourova, T. P., Poltaraus, A. B., et al. (2018). Microbial community and in situ bioremediation of groundwater by nitrate removal in the zone of a radioactive waste surface repository. Front. Microbiol. 9 (1985), 1–17. doi:10.3389/fmicb.2018.01985
Sait, M., Davis, K. E. R., and Janssen, P. H. (2006). Effect of pH on isolation and distribution of members of subdivision 1 of the phylum Acidobacteria occurring in soil. Appl. Environ. Microbiol. 72 (3), 1852–1857. doi:10.1128/aem.72.3.1852-1857.2006
Saleh-Lakha, S., Shannon, K. E., Henderson, S. L., Goyer, C., Trevors, J. T., Zabarth, B. J., et al. (2009). Effect of pH and temperature on denitrification gene expression and activity in Pseudomonas mandelii. Pseudomonas mandelii. Appl. Environ. Microbiol. 75 (12), 3903–3911. doi:10.1128/aem.00080-09
Sánchez-Clemente, R., Ingeńo, M. I., Población, A. G., Guijo, M. I., Merchan, F., and Blasco, R. (2018). Study of pH changes in media during bacterial growth of several environmental strains. Proceedings 2 (1297), 1–5.
Sanford, R. A., Wagner, D. D., Wu, Q., Chee-Sanford, J. C., Thomas, S. H., Cruz-Garcia, C., et al. (2012). Unexpected non-denitrifier nitrous oxide reductase gene diversity and abundance in soils. Proceeding Natl. Acad. Sci. U. S. A. 109 (48), 19709–19714. doi:10.1073/pnas.1211238109
Sang, C., Fu, Y., Guo, S., Luo, J., and Zhang, Q. (2020). Isolation and characterization of an aerobic denitrifiers Bacillus sp. SC16 from an intensive aquaculture pond. Water 12 (3559), 1–14.
Schaedler, F., Lockwood, C., Lueder, U., Glombitza, C., Kappler, A., and Schmidt, C. (2018). Microbially mediated coupling of Fe and Ncycles by nitrate-reducing Fe(II)-oxidizing bacteria un Littoral freshwater sediments. Appl. Environ. Microbiol. 84 (2), 1–14.
Schroeder, A., Souza, D. H., Fernandes, M., Rodrigues, E. B., Trevisan, V., and Skoronski, E. (2020). Application of glycerol as carbon source for continuous drinking water denitrification using microorganisms from natural biomass. J. Environ. Manag. 256, 1–8.
Sharma, S., and Bhattacharya, A. (2017). Drinking water contamination and treatment techniques. Appl. Water Sci. 7, 1043–1067. doi:10.1007/s13201-016-0455-7
Shen, W., Xue, H., Gao, N., Shiratori, Y., Kamiya, T., Fujiwara, T., et al. (2020). Effects of copper on nitrous oxide (N2O) reduction in denitrifiers and N2O emissions from agricultural soils. Biol. Fertil. Soils 56, 39–51. doi:10.1007/s00374-019-01399-y
Singh, B., and Craswell, E. (2021). Fertilizers and nitrate pollution of surface and ground water: An increasingly pervasive global problem. Springer Nat. Appl. Sci. 3 (518), 518–524. doi:10.1007/s42452-021-04521-8
Smith, C. J., Nedwell, D. B., Dong, L. F., and Osborn, A. M. (2007). Diversity and abundance of nitrate reductase gene (narG and napA), nitrite reductase genes (nirS and nirK), and their transcripts in estuarine sediments. Appl. Environ. Microbiol. 73 (11), 3612–3622. doi:10.1128/aem.02894-06
Spangler, W. J., and Gilmour, C. M. (1966). Biochemistry of nitrate respiration in Pseudomonas stutzeri. Aerobic and nitrate respiration routes of carbohydrate. Am. Soc. Microbiol. J. Clin. Microbiol. 91 (1), 245–250. doi:10.1128/jb.91.1.245-250.1966
Srinandan, C. S., D’souza, G., Srivastava, N., Bhusan, B., Nayak, B. B., and Nerurkar, A. S. (2012). Carbon sources influence the nitrate removal activity, community structure and biofilm architecture. Bioresour. Technol. 117, 292–299. doi:10.1016/j.biortech.2012.04.079
Sullivan, M. J., Gates, A. J., Appia-Ayme, C., Rowley, G., and Richardson, D. J. (2013). Copper control of bacterial nitrous oxide emission and its impact on vitamin B12-dependent metabolism. Proceeding Natl. Acad. Sci. United States America110 49, 19926–19931. doi:10.1073/pnas.1314529110
Sun, S., Yang, Q., Peng, Y., Shi, X., Wang, S., and Zhang, S. (2009). Nitrite accumulation during the denitrification process in SBR for the treatment of pre-treated landfill leachate. Biotechnol. Bioeng. 17 (6), 1027–1031. doi:10.1016/s1004-9541(08)60312-2
Tang, K. H., Tang, Y. J., and Blankenship, R. E. (2011). Carbon metabolic pathway in phototrophic bacteria and their broader evolutionary implications. Front. Microbiol. 2 (165), 1–23.
Thorgersen, M. P., Ge, X., Poole, F. L., Price, M. N., Arkin, A. P., and Adams, M. W. W. (2019). Nitrate-utilizing microorganisms resistant to multiple metals from the heavily contaminated Oak Ridge Reservation. Appl. Environ. Microbiol. 85 (17), 1–15. doi:10.1128/aem.00896-19
Tiburcio, S., Macrae, A., Peixoto, R., and Rachid, C. (2020). Desulfovibrio and Pseudomonas dominated enriched produced water reinforcing their importance in oilfields and production process. Res. Square, 1–19. doi:10.21203/re.3
Torres, M. J., Simon, J., Rowley, G., Bedmar, E. J., Richardson, D. J., Gates, A. J., et al. (2016). Nitrous oxide metabolism in nitrate-reducing bacteria: Physiology and regulatory mechanisms. Adv. Microb. Physiology 2 (007), 353–432. doi:10.1016/bs.ampbs.2016.02.007
Van den Berg, E. M., Elisário, M. P., Kuenen, J. G., Kleerebezem, R., and van Loosdrecht, M. C. M. (2017). Fermentative bacteria influence the competition between denitrifiers and DNRA bacteria. Front. Microbiol. 8 (1684), 1–13. doi:10.3389/fmicb.2017.01684
Verlicchi, P., and Grillini, V. (2020). Surface water and groundwater quality in South Africa and Mozambique-Analysis of the most critical pollutants for drinking purposes and challenges in water treatment selection. Water 12 (305), 305–321. doi:10.3390/w12010305
Vijaya-Chitra, A., and Lakshmanaperumalsamy, P. (2006). Biodegradation of nitrate in waste streams from explosives manufacturing plants. Res. J. Microbiol. 1, 142–151. doi:10.3923/jm.2006.142.151
Wagner, F. B., Nielsen, P. B., Boe-Hansen, R., and Albrechtsen, H. (2018). Remediation of incomplete nitrification and capacity increase of biofilters at different drinking water treatment plants through copper dosing. Water Res. 132, 42–51. doi:10.1016/j.watres.2017.12.061
Wang, K., Jiang, J., Ma, L., Zheng, L., Zhang, G., and Wu, D. (2022). Study on advanced nitrogen removal and microbial community structure of traditional Chinese medicine wastewater by a novel system coupling anaerobic sequencing batch reactor and modified sequencing batch biofilm reactor. Front. Environ. Sci. 10, 1–11. doi:10.3389/fenvs.2022.932657
Wang, Q., Burger, M., Doane, T. A., Horwath, W. R., Castillo, A. R., and Mitloehner, F. M. (2013). Effects of inorganic v. organic copper on denitrification in agricultural soil. Adv. Animal Biosci. 4 (1), 42–49. doi:10.1017/s2040470013000307
Withers, P. J. A., Neal, C., Jarvie, H. P., and Doody, D. G. (2014). Agriculture and eutrophication: Where do we go from here? Sustainability 6, 5853–5875. doi:10.3390/su6095853
Woo, H. M. (2018). Metabolic pathway rewiring in engineered Cyanobacteria for solar-to-chemical and solar-to-fuel production from CO2. Bioengineering 9 (1), 2–5. doi:10.1080/21655979.2017.1317572
Wu, Y., Zeng, J., Zhu, Q., Zhang, Z., and Lin, X. (2017). pH is the primary determinant of the bacterial community structure in agricultural soils impacted by polycyclic aromatic hydrocarbon pollution. Sci. Rep. 7, 40093. doi:10.1038/srep40093
Xue, Y., Song, J., Zhang, Y., Kong, F., Wen, M., and Zhang, G. (2016). Nitrate pollution and preliminary source identification of surface water in a semi-arid river basin, using isotopic and hydrochemical approaches. Water 8 (328), 1–12. doi:10.3390/w8080328
Yao, R., Xiong, D., Hu, H., Wakayama, M., Yu, W., Zhang, X., et al. (2016). Elucidation of the co-metabolisms of glycerol and glucose in Escherichia coli by genetic engineering, transcription profiling and 13C metabolic flux analysis. Biotechnol. Biofuels 9 (175), 1–14.
Yao, R., Yuan, Q., and Wang, K. (2020). Enrichment of denitrifying bacterial community using nitrate as an electron acceptor for nitrogen removal from wastewater. Water 12 (48), 1–12.
Yu, T., and Bishop, P. (2001). Stratification and oxidation-reduction potential change in an aerobic and sulfate-reducing biofilm studied using microelectrodes. Water Environ. Res. 73 (3), 368–373. doi:10.2175/106143001x139399
Zhang, H., Wang, Y., Chen, S., Zhao, Z., Feng, J., Zhang, Z., et al. (2018). Water bacterial and fungal community composition associated with irban lakes, Xi’an, China. Int. J. Environ. Res. Public Health 15 (469), 1–18.
Zhang, L., Wűst, A., Prasser, B., Műller, C., and Einsle, O. (2019). Functional assembly of nitrous oxide reductase provides insights into copper site maturation. Proceeding Natl. Acad. Sci. U. S. A. 116 (26), 12822–12827. doi:10.1073/pnas.1903819116
Zhang, Q., Yang, P., Liu, L., and Liu, Z. (2020). Formulation ad characterization of heterotrophic nitrification-aerobic denitrification synthetic microbial community and its application to livestock wastewater treatment. Water 12 (218), 1–15.
Keywords: bioremediation, complete denitrification, copper toxicity, metal cofactors, metagenomics, microbial denitrification, nitrate contamination, optimization
Citation: Moloantoa KM, Khetsha ZP, Kana GEB, Maleke MM, Van Heerden E, Castillo JC and Cason ED (2023) Metagenomic assessment of nitrate-contaminated mine wastewaters and optimization of complete denitrification by indigenous enriched bacteria. Front. Environ. Sci. 11:1148872. doi: 10.3389/fenvs.2023.1148872
Received: 20 January 2023; Accepted: 17 March 2023;
Published: 31 March 2023.
Edited by:
Julie Joseane Murcia Mesa, Universidad Pedagógica y Tecnológica de Colombia, ColombiaReviewed by:
Alejandro Rodriguez-Sanchez, University of Granada, SpainCopyright © 2023 Moloantoa, Khetsha, Kana, Maleke, Van Heerden, Castillo and Cason. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karabelo M. Moloantoa, TW9sb2FudG9hS0B1a3puLmFjLnph; Errol D. Cason, Y2Fzb25lZEB1ZnMuYWMuemE=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.