- Institute of Land and Urban-Rural Development, Zhejiang University of Finance and Economics, Hangzhou, China
The increasing “non-grainization” of paddy fields affecting large-scale grain cultivation and impacting the sustainable eco-functions protection of soil bacterial in China. The same problem of “non-grainization” has threatened food security and farmland soil ecological functions in other areas of the world. Although previous research has demonstrated that soil microorganisms are strongly affected by land use change, little is known about the effects of paddy field non-grainization consolidation (NGC) on soil microorganisms. This study examined soil samples before and after paddy field NGC in Zhejiang province, China, measured soil environmental factors and performed 16S rDNA amplicon sequencing to analyze the changes in soilbacterial communities and ecosystem functions before and after NGC. Results show that NGC increased the relative abundances of Proteobacteria (27.89%) and Actinobacteria (25.25%) in the project zones. Total soil bacteria increased in all samples after NGC implementation in terms of absolute soil bacterial community content, but there were large differences. NGC improved the alpha diversity indices, including Ace, Chao1, Coverage, and Shannon indices (p < 0.01) in terms of soil bacterial community diversity. The data analysis of RDA and the significance test showed that the environmental factors which were close-knitted with the soil bacterial diversity and structure of the sample sites were TN, AP, pH, SOM, FIQ, and AK. Wilcoxon rank sum test results showed that NGC also significantly enhanced the amino acid transport and metabolic functions of soil bacterial. Our findings suggest that NGC is beneficial to improve the paddy field soil bacterial diversity, enhance the versatility of soil ecosystems, and promote sustainable arable land soil ecosystem protection. The quantitative results would be useful for better studying the use of land remediation engineering measures to the remediation and maintenance of bacterial diversity and sustainable eco-functions protection in paddy fields.
Introduction
The term “non-grain production of arable land” refers to the conversion of arable land originally used for grain crop production into cash crops or even agricultural production such as pond farming and livestock breeding (Zhang et al., 2023). As arable land serves as an important basic resource to ensure national food security, the excessive non-grain production will seriously affect food production and thereby national food security to certain extent (Lu et al., 2021). High-quality arable land resources equipped with food production and ecological functions, paddy fields are valued by the Chinese central government. However, factors such as low farmer income from grain production and the structural adjustment of the agricultural sector have resulted in the increasing “non-grainization” of paddy fields, affecting large-scale grain cultivation and impacting the sustainable eco-functions protection of soil bacterial (Su et al., 2020; Guo and Wang, 2021). In 2020, the General Office of the State Council of China successively issued the “Notice on Resolutely Stopping the “Non-Agriculturalization” of Arable Land” and “Opinions on Preventing the Non-Grainization of Arable Land and Stabilizing Grain Production.” In addition, the local governments of southern China have been actively carrying out land consolidation against the non-grainization of paddy fields. Engineering measures, such as consolidating non-grain paddy fields, restoring grain production functions, and improving the quality of arable land, have been implemented in project zones, seeking to improve the parcel shape, parcel size, and ecological landscape indices of paddy fields, thereby significantly increasing the scale of grain cultivation and effectively resolving issues with the non-grainization and fragmentation of paddy fields (Shi et al., 2018).
Soil bacteria are an indispensable component of the soil ecosystem and are involved in various soil physical and biochemical processes, including the decomposition of soil organic matter (SOM), the formation of humus, and the transformation and circulation of nutrients. Thus, the quality and health of soil ecosystems are closely linked with the changes in the community structure of soil bacteria (Bitas et al., 2013; Lozano et al., 2014; He et al., 2020; Guo et al., 2021). According to existing studies, the different cultivation and utilization methods of arable land can alter their soil physicochemical properties, including pH, organic carbon, available potassium (AK), and moisture content, significantly impacting soil bacteria (Angel et al., 2010; Legrand et al., 2018). From the perspective of relative abundance (Lu et al., 2020), found that the implementation of land consolidation engineering led to improvements in soil heavy metal contamination while also enhancing the diversity and functionality of soil bacteria. Based on the indices of relative abundance obtained through high-throughput sequencing (Lin et al., 2019), demonstrated that land consolidation could improve SOM, pH, moisture content, and other physicochemical properties, thereby enhancing soil bacterial diversity. Conversely, other researchers have found empirical evidence indicating that land consolidation had a substantial impact on the community composition of soil bacteria, and the intensity of engineering measures disrupted and reduced soil ecosystem stability (Li et al., 2019). Thus, researchers have mainly focused on the crucial role of soil bacteria in arable land ecology and food production, arguing that the implementation of land consolidation engineering may have an impact on soil bacteria. However, little is known about the effects of large-scale non-grainization consolidation (NGC) on eco-functions of soil bacterial in the double- and single-cropping rice sub-regions in the middle and lower reaches of the Yangtze River (Jin and Zhong, 2022), key rice production regions with an immense impact on the trends of national grain output. High-throughput sequencing for the analysis of community structure allows the in-depth investigation of information on bacterial community structure. It greatly expands the information obtained on bacteria at the molecular level, further enhancing the acquisition of bacterial community structure information. Previous studies involving high-throughput sequencing experiments have concluded that land consolidation could optimize the soil bacterial communities in farmland (Lin et al., 2019). Nevertheless, employing high-throughput sequencing alone to study the community structure of soil bacteria only provides the species information and relative abundances of the community structure but not the absolute content index, which is one of the three key elements in the evaluation of ecosystem community structure (Lou et al., 2018; Yang et al., 2018; Jiang et al., 2019). Absolute content is a critical indicator for the evaluation of bacterial community structure, and its absence implies that the true quantities of bacterial communities cannot be accurately reflected (Chen et al., 2020). Therefore, we explored the influence mechanism of the effects of paddy field NGC on the soil bacterial diversity and its ecological function by innovatively combining of the relative abundance and absolute content indicators of soil bacteria. Which will have important theoretical and practical implications for sustainable eco-functions protection of soil bacterial in the study area (Yang et al., 2018).
In this study, soil samples were collected before and after paddy field NGC. Soil bacterial DNA was extracted from these samples, and 16S rDNA amplicon sequencing was performed to explore the effects of paddy field NGC on soil bacterial communities. It is hoped that our findings will provide basic data and supporting evidence for the sustainable eco-functions protection of soil bacterial for implementing the NGC of arable land.
Materials and methods
Profile of study area
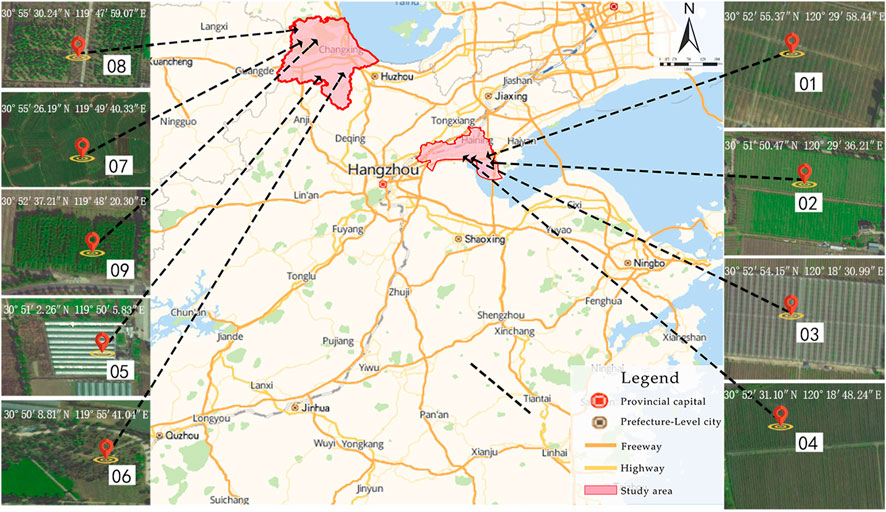
Non-grain production of arable land, as a severe threat to food security, has attracted great attention in China and other countries of the world (Zhuang et al., 2022). China has been implementing the world’s most strict cultivated land protection system to ensure staple food security, and has given priority to cultivated land for grain crops (Lu et al., 2021), but the problem of non-grain cultivation in China has become increasingly prominent, with the rate of “non-grain cultivation” of about 27% (Kong, 2020), which has threatened China’s food security and sustainable agricultural development. Arable land is, by definition, a “living” organism whose health depends on the beneficial interactions between soil organisms, crops, and the environment in which soil organisms are an important part of the arable land ecosystem (Zhuang et al., 2022). As the research on arable land conservation progresses, more studies have paid closer heed to the possibility of repairing arable land biodiversity. Therefore, to tease out the characteristics of the impact of non-grainization consolidation on arable land soil bacterial communities and their ecological functions with the data from the typical high rice production area in China can provide a useful reference for the research on the conservation and repair of arable land biodiversity and the function as per sustainable utility of arable land. The northern Zhejiang plain, located in the northern part of Zhejiang Province, is a typical sub-region for double- and single-cropping rice in the middle and lower reaches of the Yangtze River. This region has accessible waterways and predominantly consists of loam paddy fields, with a history of rice cultivation that spans several millennia (Li et al., 2016). However, due to the large gap in the comparative income of rice cultivation, there has been a continuous increase in non-grain cultivation within the region, leading to the cross-distribution of grain-producting paddy fields with non-grain-producing arable land. This pattern of distribution is unsuitable for large-scale and mechanized rice production and has affected the utilization of paddy fields and the improvement of grain production capacity (Li et al. 2021b). Since 2020, under the leadership of the Zhejiang provincial government, the counties and cities in the northern Zhejiang plain have successively undergone the NGC arable land. The engineering measures include the removal of dryland crops, removal of root systems, plough pan remediation, soil fertility remediation, and restoration of rice cultivation, achieving some success. Therefore, project zones undergoing the NGC of arable land in Jiaxing City and Huzhou City, Zhejiang Province were selected as the study area.
Soil sample collection and sequencing
Soil samples were collected before (August 2019) and after (August 2020) the NGC of arable land was implemented (Figure 1). On the basis of the land-use survey data of the project zones, the crop cultivation status of the sampled plots is shown in Table 1. All samples were collected at a temperature of 27–30°C in the absence of rainfall. The S-shape multi-point sampling method was employed to collect 0–10 cm of the topsoil, which was evenly mixed to form a single soil sample weighing 500 g each. The nine project zones were numbered 01 to 09, while samples collected before NGC implementation were labelled A; those collected after NGC implementation were labelled B, yielding a total of 18 samples. A total of 20 g was weighed out from each sample and stored in a −80°C refrigerator. The samples were later sent to Shanghai Genesky Biotech Co., Ltd. for soil bacterial DNA extraction and 16S rDNA amplicon sequencing (Beckers et al., 2016; Wan et al., 2021). DNA extraction was performed in triplicate for each sample (Dimitrov et al., 2017; Wan et al., 2021). The remaining 480 g of the sample was used for soil physicochemical testing.
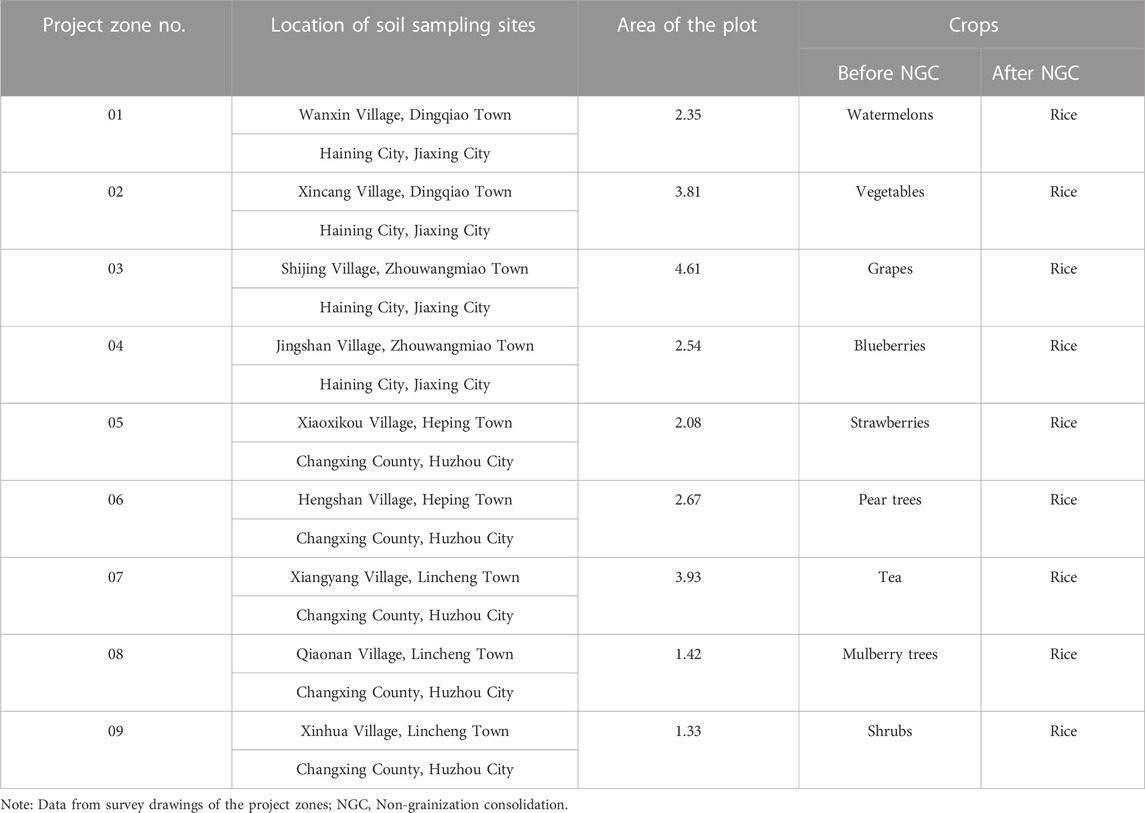
TABLE 1. Changes in arable land-use patterns before and after non-grainization consolidation in project zones Unit: ha.
Soil sample DNA was extracted using the FastDNA SPIN Kit for Soil. Polymerase chain reaction (PCR) amplification was performed using the ABI GeneAmp® 9,700 thermal cycler and TransStart FastPfu DNA polymerase. PCR amplification products were measured using the QuantiFluor™-ST Fluorometer. The MiSeq library was prepared using the TruSeqTM DNA Sample Prep Kit based on the Illumina HiSeq gene sequencing system. Quality control and filtering were then performed on the sequencing data using the Trimmomatic software, followed by clustering to form operational taxonomic units (OTUs) with 97% identity using the Ribosomal Database Project (RDP) Bayesian classifier on the Usearch platform (version 7.0) and OTU annotation using the Greengenes database (http://greengenes.secondgenome.com).
Measurement of soil environmental factors
Measurements were performed on 7 environmental factors before and after the NGC of arable land in the project zones, which included the soil moisture index (SMI), pH, Soil organic matter content (SOM), available phosphorous (AP), available potassium (AK), total nitrogen (TN), and parcel fragmentation (FIQ). SMI was determined using the oven drying method, in which the samples were dried at 105°C for 6 h in an electric constant-temperature oven. Then, pH was determined using the potentiometric method. SOM was determined using the potassium dichromate volumetric method. AP was measured using an ultraviolet spectrophotometer. AK was measured using a flame photometer (F-100). TN was measured using an automatic Kjeldahl nitrogen analyser (K9860).
FIQ was calculated by the equation
NT is the total number of arable patches in the project zone, ATmin is the minimum area of a single arable patch in the project zone, and LAT is the total area of arable patches in the project zone. A smaller FIQ value indicates a larger scale of arable land.
Analysis of soil bacterial communities
Raw DNA sequencing data were processed on the Usearch platform (version 7.0). Then, the RDP classifier module was used to calculate the number of OTUs at a threshold of 97% identity, and the community composition was determined at the phylum level (Tipayno et al., 2018). Based on the results of OTU analysis, the R tool was used to plot and analyse the community composition of soil bacteria. The alpha diversity of the soil bacterial communities was characterized by calculating the Shannon, ACE, Chao1, Coverage and Simpson indices using mothur 1.30.1, which were non-dimensionalised for comparative analysis. Larger Shannon, ACE, Chao1 and Coverage values indicate greater diversity, whereas a higher Simpson value indicates lower diversity (Pitta et al., 2014; Wan et al., 2021). Statistics from Student’s test and Wilcoxon rank-sum test were used for the analysis of significant differences indices in diversity between groups. The OTU abundance tables were normalized using the PICRUS software, and the Greengenes OTU IDs were mapped to the COG and KEGG databases to obtain the COG family information and KO information of the corresponding OTUs. Finally, the function abundance profile and the abundances of each functional category were obtained to analyse the changes in soil bacterial function (Langille et al., 2013). The Wilcoxon rank-sum test was performed to test the significance of functional differences in soil bacteria.
Analysis of the effects of environmental factors on soil bacterial diversity and structure
Using the Canoco software, canonical correlation analysis (CCA) was carried out based on the bacterial OTU composition and soil environmental factors to analyse the relationship between soil bacterial community diversity and soil environmental factors before and after the NGC of arable land (Ter Braak, 1986). In addition, to further analyse the correlation between soil bacterial phyla and soil environmental factors, the vegan software package in R was used to perform Spearman’s correlation analysis between the two.
Results
Effects of NGC on soil environmental factors
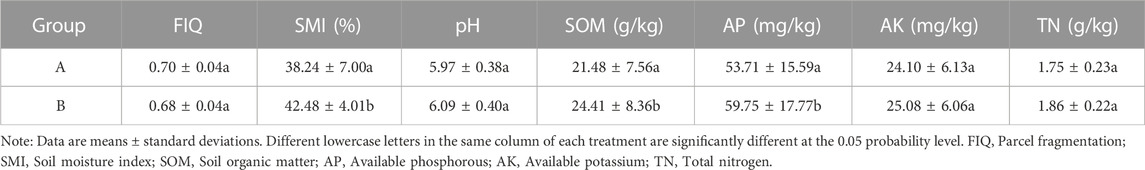
The change characteristics of environmental factor in 18 soil samples showed in Table 2. The comparison of the different items (01–09) in a single group (group A) suggests that there are significant differences in the physicochemical properties of the soil samples at different spatial locations, which indicates the existence of spatial heterogeneity in the soil with the exclusion of the deviations in the sampling and measurement process. This study focused on the changes of soil and its microorganisms before and after the implementation of paddy field NGC, thus the subsequent analysis mainly narrowed to analyze the effects of paddy field NGC on soil and its microorganisms by comparing the data of group A and group B.
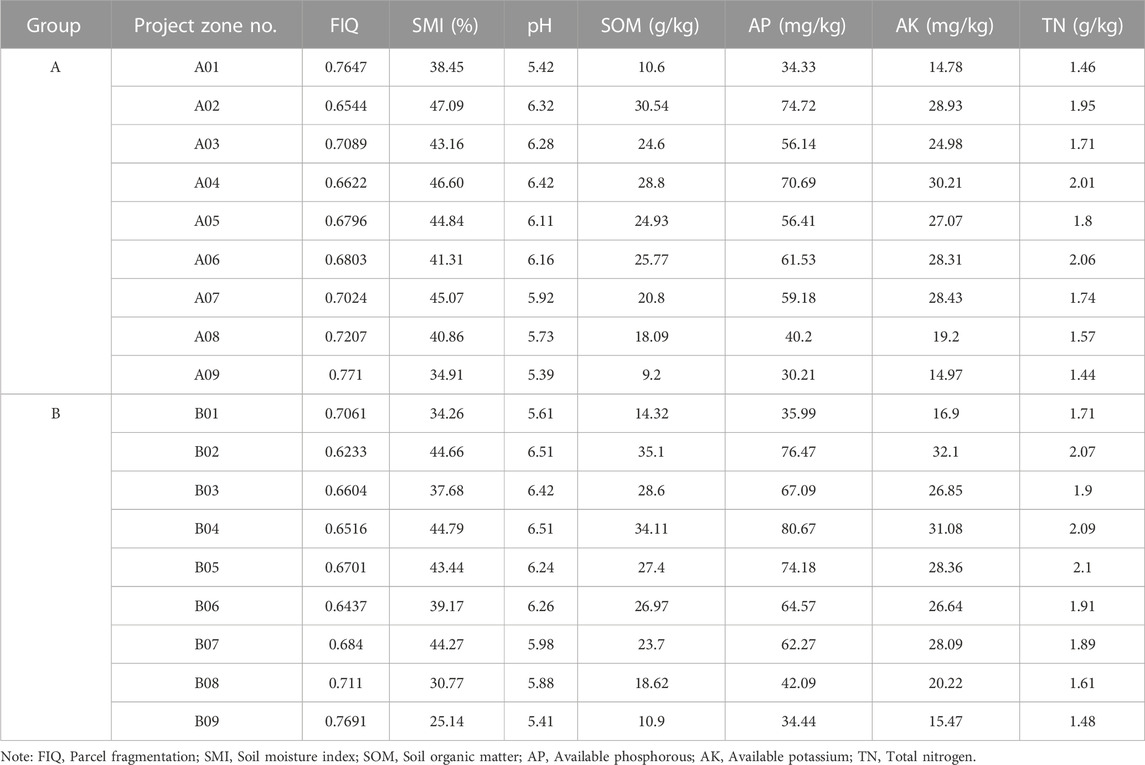
TABLE 2. Changes in soil environmental factors before (group A) and after (group B) paddy field non-grainization consolidation.
Table 3 lists the soil environmental factors changes before (group A) and after (group B) the NGC of paddy fields. The results indicate that paddy field NGC increased SMI and reduced FIQ by consolidating plots and re-cultivation of non-grain-producing parcels (p < 0.05). pH, SOM, AP, and AK are also increased (p < 0.05) after the paddy field NGC, and the soil physical and chemical properties improved obviously.
Effects of NGC on soil bacterial community structure
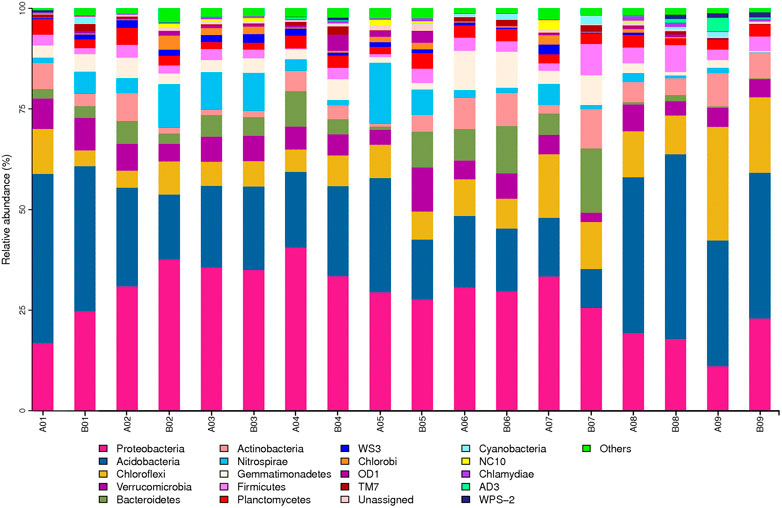
According to the results for the relative abundances of soil bacteria at the phylum level (Figure 2), the bacteria found in the 18 soil samples obtained from the 9 project zones before and after the paddy field NGC were mainly from the following 11 phyla: Proteobacteria (27.89%), Actinobacteria (25.25%), Chloroflexi (10.02%), Verrucomicrobia (5.69%), Bacteroidetes (4.99%), Acidobacteria (4.75%), Nitrospirae (4.41%), Gemmatimonadetes (3.76%), Firmicutes (3.00%), and Planctomycetes (2.71%), which accounted for 92.47% of the OTUs. In terms of the changes in bacterial relative abundances before and after implementation, Proteobacteria (which has the greatest abundance) increased significantly after implementation in project zones 01, 02, and 09 but decreased in the other project zones. Actinobacteria increased markedly in project zones 03, 04, 08, and 09 but decreased in the other project zones.
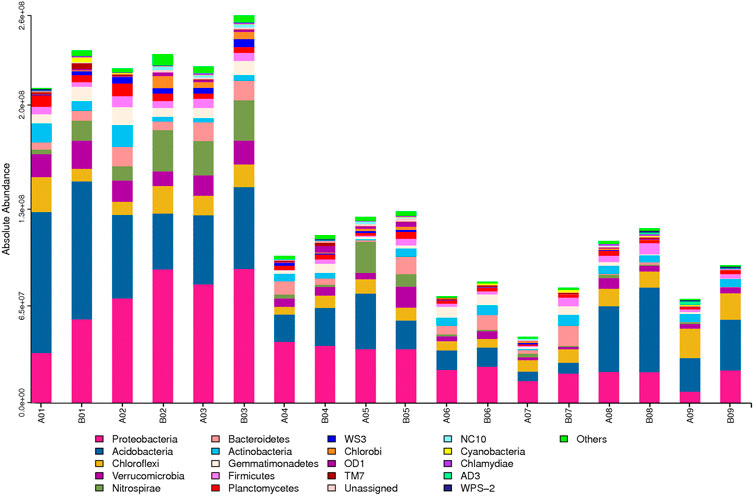
The absolute contents of soil bacteria at the phylum level were calculated (Figure 3), and marked differences were observed in the total amounts of soil bacteria across the different samples, ranging from a minimum of 4.42 × 107 copies/g (A07) to a maximum of 2.61 × 108 copies/g (B03). By comparing the absolute amounts of soil bacteria in soil samples A01 to A09 before the NGC project, it can be seen that the higher absolute amounts of soil bacteria in A01 (planted watermelons), A02 (planted vegetables), and A03 (planted grapes), which were all above 2.11 × 108 copies/g, and the lower will be the absolute amounts of soil bacteria in A05 (planted strawberries), A08 (planted mulberries), A4 (planted blueberries), A06 (planted pear trees), A09 (planted shrubs), and A07 (planted tea) which were all below 1.25 × 108 copies/g, and declined sequentially in absolute soil bacterial amount. This indicates that the planting of different crops on the paddy fields had an impact on the soil bacterial community before the implementation of NGC.
On the other hand, there were marked differences in the trends of change between the absolute content and relative abundance of soil bacteria before and after the NGC of paddy fields. The total amount of soil bacteria was higher after implementation (group B) than that before implementation (group A), with project zone 05 showing the smallest increase (3.05%) and project zone 07 showing the largest increase (74.18%). The absolute content of the dominant phylum, Proteobacteria, increased markedly after implementation in project zones 01, 02, 03, 05, 06, 07, and 09, and decreased slightly in project zones 08 and 04. The relative abundance of soil bacteria is used to indicate the changes in the proportions of the different bacteria within the community, whereas the absolute content is used to indicate the changes in the quantities of the different bacteria. Thus, by integrating these two indicators, we can obtain a more comprehensive and accurate understanding of the effects of arable land NGC on soil bacterial communities (DeAngelis et al., 2015).
Effects of NGC on soil bacterial alpha diversity
The alpha diversity indices of soil bacteria was calculated based on absolute content. Analysis of variance was analyzed using SPSS (version 20) software, Levene’s test for chi-squaredness was used, and Tukey’s test for significance if the variance was chi-squared, Tamhane T2 test for significance if the variance was not chi-squared.
The results show that NGC is the factor that significantly affects the diversity of bacterial communities (p < 0.01) (Table 4). The alpha diversity indices (ACE, Chao1, Coverage, Shannon) of soil bacteria in group B (after NGC implementation) were clearly superior to those in group A (before NGC implementation), implying that engineering measures for paddy field NGC (e.g., re-cultivation of non-grain land, parcel merging, land levelling, ditch construction, and application of organic fertilisers) contributed to improving the alpha diversity of soil bacteria.
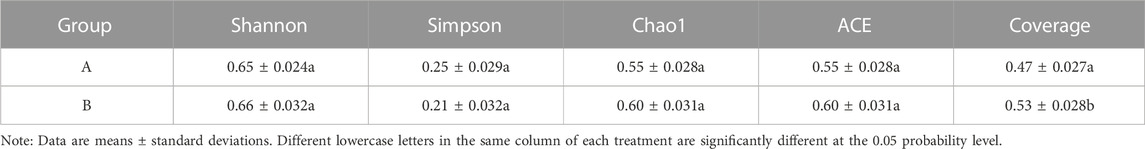
TABLE 4. Alpha diversity indices of soil bacteria before (groupA) and after (group B). Paddy fields non-grainization consolidation.
The effects of NGC, environmental factors, and geographical location on soil bacterial
Geographical location and environmental factors may play key roles in the mechanism of NGC effects on soil bacterial changes in arable land. Therefore, VPA was employed to determine the effects of NGC, Environmental factors Geographical location (PCNM matrix). The results showed that NGC, Environmental factors and geographical location separately had significant effects on the bacterial (NGC, F = 2.6248, p < 0.01; Environmental factors, F = 3.1907, p < 0.05; Geographical location, F = 1.0337, p < 0.05) (Figure 4). As a result, NGC, Environmental factors and Geographical location interacted and systematically influenced the process of soil bacterial community construction.
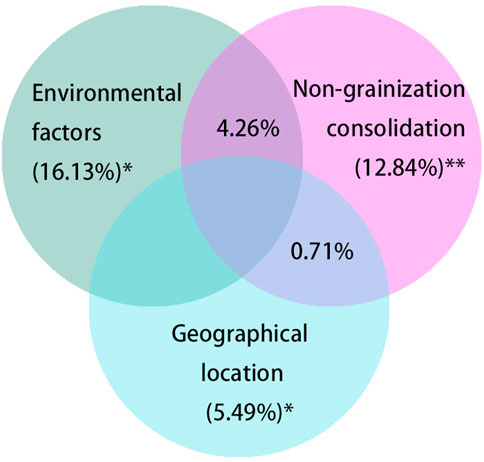
FIGURE 4. VPA Venn diagram of the effects of non-grainization consolidation, environmental factors and geographical location on soil bacteria. Note: Asterisks represent significance level, * and ** corresponding to 0.05 and 0.01 level, respectively. VPA, Variation partitioning analysis.
Effects of environmental factors on soil bacterial diversity and structure
Redundancy Analysis (RDA) refers to an expanded method of multi-response regression analysis and testing is often used in microbial community analysis to decompose changes in species abundance into variance associated with environmental variables and to explore the significance of community species composition constrained by environmental variables through data testing. The data analysis of RDA and the significance test showed that the environmental factors which were close-knitted with the soil bacterial community structure of the sample sites were TN (r2 = 0.99797, p = 0.037), AP (r2 = 0.99560, p = 0.017), pH (r2 = 0.99536, p = 0.006), SOM (r2 = 0.98950, p = 0.004), FIQ (r2 = −0.98068, p = 0.007), SMI (r2 = 0.90261, p = 0.018), and AK (r2 = 0.85988, p = 0.024) (Figure 5) were highly correlated with RDA axis 1.
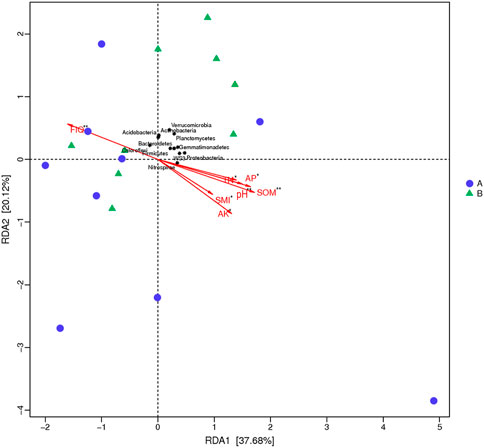
FIGURE 5. RDA ranking of soil bacterial composition and environmental factors. Note: asterisks represent significance level, * indicates 0.01 < p ≤ 0.05, ** indicates 0.001 < p ≤ 0.01; RDA, Redundancy Analysis.
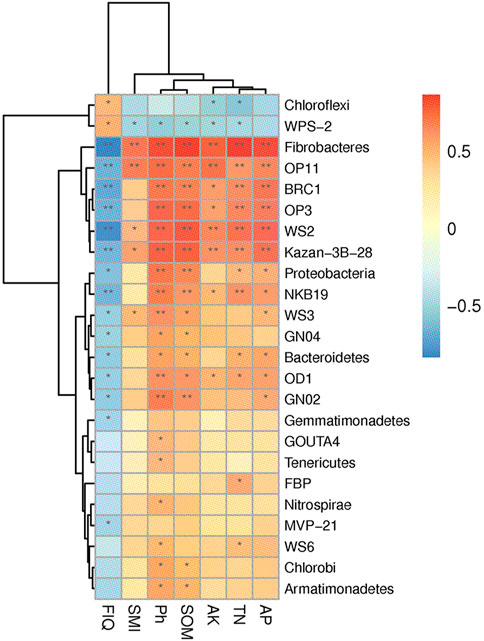
Spearman correlation analysis and test showed that Fibrobacteres, OP11, GN02, WS2, and Kazan-3b-28 etc. in soil samples were significantly correlated with TN, AP, pH, SOM, FIQ, SMI, and AK (p < 0.01) (Figure 6). These findings suggest that paddy field NGC altered the TN, AP, pH, SOM, FIQ, SMI, and AK, which in turn affected the diversity and structure of the soil bacterial communities.
(The figure shows the correlation between environmental factors and soil fungi, with warm tones indicating positive correlation and cool tones indicating negative correlation. * indicates 0.01 < p ≤ 0.05, ** indicates 0.001 < p ≤ 0.01).
Effects of NGC on soil bacterial functions
The 16s rRNA gene is a key tool to researching the function of biological communities. Using PICRUSt software to calculate the corresponding functional abundance of bacterial communities. Wilcoxon rank sum test results showed that soil bacterial functions improved significantly (p < 0.01) after the implementation of paddy field NGC (group B) compared to before the implementation (group A), and there were differences in the magnitude of improvement of each bacterial community function, E amino acid transport and metabolism, C energy production and conversion, T signal transduction mechanism and M Cell membrane biogenesis were the soil bacterial functions that changed significantly before and after project implementation, which was mainly the result of the combined effect of the change in bacterial species dominance and change in quantity (Figure 7).
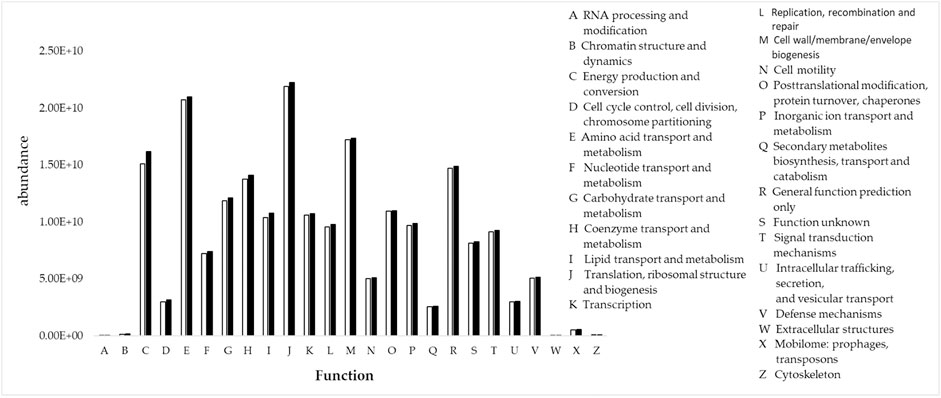
FIGURE 7. PICRUSt inferred functional abundance of soil samples before (white) and after (black) the non-grainization consolidation of paddy fields.
Discussion
Uncovering the patterns of change in the bacterial communities of arable land is a key topic in the field of sustainable eco-functions protection that has been extensively explored in microbial ecology (Tecon and Or, 2017). However, very few studies have comprehensively examined the patterns of change in the bacterial communities and ecological functions of arable land by combining the three dimensions of species diversity, relative abundance, and absolute abundance. In view of the above, we employed 16S amplicon sequencing to explore the effects of paddy field NGC on soil bacterial communities and ecological functions which through combining the relative abundance and total bacterial quantities. By performing experiments at the parcel scale, we have found powerful evidence supporting the crucial role of NGC engineering in improving the soil bacterial communities and ecological functions of arable land.
The need for economic benefits has driven a gradual increase in the non-grainization of paddy fields in China, which has led to a significant impact on the sustainable ecosystem structure and function of arable land (Su et al., 2020; Li et al., 2022). Changes in land use types and land remediation can significantly affect soil properties and microbial community composition in many areas. Studies, with high-throughput sequencing analyzing the relative abundance of soil bacteria at disposal, reveal the changes in soil bacterial communities (Bai et al., 2022; Li et al., 2016). In this study, we observed that implementing the NGC of paddy fields led to a decrease in the relative abundances of Proteobacteria and Actinobacteria in project zones 05, 06, and 07. Based solely on the decreases found in these two phyla, we can only conclude that the NGC of paddy fields was unfavourable for their growth. However, by calculating the absolute content, we found that the quantities of the two bacterial phyla had increased to some extent after the NGC of paddy fields, with project zone 07 showing increases of up to 33.78% and 17.55% in the absolute contents of Proteobacteria and Actinobacteria, respectively. Previous studies have demonstrated that combined evaluation with relative abundance and absolute content was more suitable for describing the changes in soil bacterial communities (Aitchison, 1982; Ter Braak, 1986; Gloor et al., 2017; Knight et al., 2018; Morton et al., 2019). In ecology, relative abundance is mainly used to describe whether a given species is more or less abundant relative to the rest of the community, whereas absolute content is an indicator that truly describes the actual quantity of a given species (Hubbell and Luis, 2004). Existing studies have reported similar trends involving the improvement of soil bacterial communities by rational land cnsolidation engineering (Lin et al., 2019; Li et al., 2021a). Soil bacteria in paddy fields are significantly affected by the non-grain dryland farming, implying that the non-grainization of paddy fields in the economically developed areas of southern China has exacerbated the loss of soil microbial diversity (Qi et al., 2022). Thus, engineering measures for land consolidation, such as the re-cultivation of rice in non-grain parcels, and the application of organic fertilisers, can effectively prevent the degradation of arable land, the loss of biodiversity, and the decline of paddy field ecosystem functions. Our study also revealed that when evaluating the bacterial community of a single sample, the NGC of paddy fields, the increase in the absolute contents of bacteria at the phylum level was not significant in project zone 02 at 4.27%. Absolute content is better suited to describe and evaluate the actual quantitative changes in soil microbial community structure and the relationships between microorganisms across multiple samples (Lou et al., 2018). Relative abundance is more suitable for describing and evaluating the relationship between microorganisms within a single soil sample (Yang et al., 2018). The relative abundances indicate that the proportions of Nitrospirae, Chloroflexi, and Chlorobi increased by an order of magnitude. Thus, although paddy field NGC did not significantly increase the quantity of bacteria, it led to increases in Nitrospirae, Chloroflexi, and Chlorobi relative to other bacterial phyla, which were able to improve the capacity for ammonia-nitrite-nitrate conversion and energy production and conversion to a greater extent. According to previous studies, the long-term dryland farming of paddy fields can lead to systematic and abrupt changes in multiple ecosystem attributes (Breidenbach et al., 2017). In particular, our findings suggest that to reduce the negative effects caused by the non-grainization of arable land on the diversity and ecological functions of soil bacterial communities, the changes in soil bacterial diversity and ecological functions should not be overlooked during the formulation of arable land NGC and policies for the management of agricultural production. In short, the two indicators of bacterial community structure (i.e., relative abundance and absolute content) each have their own advantages and disadvantages when used to perform evaluations. Relative abundance is more suitable for describing and evaluating the relationships among different bacteria within a single sample with respect to bacterial community structure. In contrast, absolute content is more suitable for describing and evaluating the actual quantitative variations in bacteria and their community structure, as well as the relationship between bacteria across different samples (Yang et al., 2018; Chen et al., 2020).
Elucidating the mechanisms underlying the effects of paddy field NGC on soil bacterial community and ecological functions has significant implications for better understanding the remediation and maintenance of bacterial diversity and sustainable eco-functions protection in paddy fields. In this study, the comparison of bacterial community structure, relative abundance, and absolute abundance before and after the NGC of paddy fields showed that engineering measures involving the restoration of rice cultivation in non-grain dryland farming were closely associated with the remediation and balance of soil bacterial communities. The parcel-scale experiments conducted in this study further confirmed the association between land consolidation engineering and soil bacterial communities. In particular, the implementation of various engineering measures, such as the removal of dryland crops, removal of root systems, plough pan remediation, remediation of soil fertility, and restoration of rice cultivation, led to the optimisation and improvement of soil bacterial eco-functions. This may be due to the complex interactions between soil bacteria and soil physicochemical properties (Hermans et al., 2020). For example, certain soil bacteria can regulate amino acid transport and metabolic functions, which can provide substrates for crop growth (Wang et al., 2019). The implementation of NGC engineering altered the soil physicochemical properties, optimised the relative abundance and absolute content of soil bacteria, as well as strengthened the amino acid transport and metabolic functions of soil bacteria. In this study, we first established the mechanisms underlying the effects of paddy field NGC on soil bacterial communities and then emphasised the potential role of changes in soil bacterial community structure, relative abundance, and absolute content in achieving the sustainable eco-functions protection of paddy field.
There are certain potential limitations that should be considered within the scope of this study. First, many sequences were obtained from the 16S rDNA high-throughput sequencing of the 18 soil samples, which can provide theoretical support for future studies to some extent. However, since NGC engineering may exert continuous effects on soil microbes, meanwhile, differences in geographic location may also lead to different effects of NGC on soil microbes. Samples from different years and representative areas should be added in future studies to comprehensively reflect the dynamic effects of the different stages and spaces of NGC engineering on soil microbes. In addition, the effects of NGC engineering on soil microbes should be evaluated using a multi-disciplinary approach involving multiple methods to obtain more effective and holistic research conclusions.
Conclusion
In summary, the application of 16S rDNA sequencing technology, together with testification from the two aspects of relative abundance and absolute content, verifies that the non-grain paddy field rearrangement has changed the soil bacterial community structure and improved the soil bacterial diversity, which will profoundly affect the sustainability of paddy field ecological function. Therefore, it is urgent to incorporate the diversity of soil bacterial diversity and its ecological significance of non-grain paddy field rearrangement into the research framework and theoretical system of ecological protection of cultivated land. Meanwhile, it should be noted that since this observative study only focuses on the changes before and after the implementation of non-grain paddy field rearrangement within one year span, it is still uncertain for the author to reach qualitative outcomes, that is, whether the results of this study are the temporary response or long-term impact on soil bacterial diversity to non-grain remediation in paddy fields. In addition, the results also justify the positive response between soil environmental factors and soil bacterial function and non-grain paddy field remediation. Therefore, it needs further effort to ponder over the coupling research between soil bacteria and environmental factors and the evolution process of various ecological functions in cultivated land to further reveal the nutrient differences of paddy ecosystem caused by the change of soil bacterial community under non-grain paddy field rearrangement and its feedback effect on the sustainability of ecological functions, and most importantly to provide a scientific basis for optimizing the diversity of soil bacterial communities and the sustainable protection of paddy ecological functions under non-grain remediation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA839191.
Author contributions
BG: Conceptualization, methodology, data curation, validation, and supervision. YL: Investigation, software, writing- original draft preparation, writing- reviewing, and editing.
Funding
This study was supported by the National Natural Science Foundation of China (NO. 42201288).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aitchison, J. (1982). The statistical analysis of compositional data. J. R. Stat. Soc. Ser. B Methodol. 44 (2), 139–160. doi:10.1111/j.2517-6161.1982.tb01195.x
Angel, R., Soares, M. I. M., Ungar, E. D., and Gillor, O. (2010). Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 4, 553–563. doi:10.1038/ismej.2009.136
Bai, Z. Y., Zheng, L. B., Bai, Z. J., Jia, A. M., and Wang, M. J. (2022). Long-term cultivation alter soil bacterial community in a forest-grassland transition zone. Front. Microbiol. 13, 1001781. doi:10.3389/FMICB.2022.1001781
Beckers, B., De Beeck, M. O., Thijs, S., Truyens, S., Weyens, N., Boerjan, W., et al. (2016). Performance of 16s rDNA primer pairs in the study of rhizosphere and endosphere bacterial microbiomes in metabarcoding studies. Front. Microbiol. 7, 650–715. doi:10.3389/fmicb.2016.00650
Bitas, V., Kim, H. S., Bennett, J. W., and Kang, S. (2013). Sniffing on microbes: Diverse roles of microbial volatile organic compounds in plant health. Mol. Plant-Microbe Interact. 26 (8), 835–843. doi:10.1094/mpmi-10-12-0249-cr
Breidenbach, B., Brenzinger, K., Brandt, F. B., Blaser, M. B., and Conrad, R. (2017). The effect of crop rotation between wetland rice and upland maize on the microbial communities associated with roots. Plant Soil 419, 435–445. doi:10.1007/s11104-017-3351-5
Chen, H., Zhu, T., Li, B., Fang, C., and Nie, M. (2020). The thermal response of soil microbial methanogenesis decreases in magnitude with changing temperature. Nat. Commun. 11, 5733–5737. doi:10.1038/s41467-020-19549-4
DeAngelis, K. M., Pold, G., Topçuoğlu, B. D., van Diepen, L. T. A., Varney, R. M., Blanchard, J. L., et al. (2015). Long-term forest soil warming alters microbial communities in temperate forest soils. Front. Microbiol. 6, 104–113. doi:10.3389/fmicb.2015.00104
Dimitrov, M. R., Veraart, A. J., de Hollander, M., Smidt, H., van Veen, J. A., and Kuramae, E. E. (2017). Successive DNA extractions improve characterization of soil microbial communities. Peerj 5, e2915–e2929. doi:10.7717/peerj.2915
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V., and Egozcue, J. J. (2017). Microbiome datasets are compositional: And this is not optional. Front. Microbiol. 8, 2224–2316. doi:10.3389/fmicb.2017.02224
Guo, J., Wu, Y. Q., Wu, X. H., Ren, Z., and Wang, G. B. (2021). Soil bacterial community composition and diversity response to land conversion is depth-dependent. Glob. Ecol. Conservation 32, 019233–e2012. doi:10.1016/j.gecco.2021.e01923
Guo, Y. Z., and Wang, J. Y. (2021). Identifying the determinants of nongrain farming in China and its implications for agricultural development. Land 10 (9), 902–916. doi:10.3390/land10090902
He, H., Miao, Y. J., Gan, Y. D., Wei, S. D., Tan, S. J., Rask, K. A., et al. (2020). Soil bacterial community response to long-term land use conversion in Yellow River Delta. Appl. Soil Ecol. 156, 103709–103714. doi:10.1016/j.apsoil.2020.103709
Hermans, S. M., Buckley, H. L., Case, B. S., Curran-Cournane, F., Taylor, M., and Lear, G. (2020). Using soil bacterial communities to predict physico-chemical variables and soil quality. Microbiome 8 (1), 79–13. doi:10.1186/s40168-020-00858-1
Hubbell, S. P., and Luis, B-D-Á. (2004). The unified neutral theory of biodiversity and biogeography: Reply. Ecology 85 (11), 3175–3178. doi:10.1890/04-0808
Jiang, S. Q., Yu, Y. N., Gao, R. W., Wang, H., Zhang, J., Li, R., et al. (2019). High-throughput absolute quantification sequencing reveals the effect of different fertilizer applications on bacterial community in a tomato cultivated coastal saline soil. Sci. Total Environ. 687, 601–609. doi:10.1016/j.scitotenv.2019.06.105
Jin, T., and Zhong, T. Y. (2022). Changing rice cropping patterns and their impact on food security in southern China. Food Secur. 14 (4), 907–917. doi:10.1007/s12571-022-01254-3
Knight, R., Vrbanac, A., Taylor, B. C., Aksenov, A., Callewaert, C., Debelius, J., et al. (2018). Best practices for analysing microbiomes. Nat. Rev. Microbiol. 16 (7), 410–422. doi:10.1038/s41579-018-0029-9
Kong, X. B. (2020). The problem of "non-grain" of arable land, causes and countermeasures. China Land 11, 17–19. doi:10.13816/j.cnki.ISSN1002-9729.2020.11.05
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31 (9), 814–821. doi:10.1038/nbt.2676
Legrand, F., Picot, A., Cobo-Díaz, J. F., Carof, M., Chen, W., and Le Floch, G. (2018). Effect of tillage and static abiotic soil properties on microbial diversity. Appl. Soil Ecol. 132, 135–145. doi:10.1016/j.apsoil.2018.08.016
Li, G. Y., Zhang, M. X., and Wu, C. F. (2021a). Agricultural land consolidation impacted edaphic microbial community dynamics and assembly in China - a case study from Zhejiang Province. Catena 205, 105424–105429. doi:10.1016/j.catena.2021.105424
Li, H., Su, D., Cao, Y., and Wang, J. Y. (2022). Optimizing the compensation standard of cultivated land protection based on ecosystem services in the hangzhou bay area, China. Sustainability 14 (4), 2372. doi:10.3390/su14042372
Li, X. X., Ma, J., Yang, Y. J., Hou, H. P., Liu, G. J., and Chen, F. (2019). Short-term response of soil microbial community to field conversion from dryland to paddy under the land consolidation process in north China. Agriculture-Basel 9 (10), 216–217. doi:10.3390/agriculture9100216
Li, Y., Wu, H. X., and Shi, Z. (2016). Farmland productivity and its application in spatial zoning of agricultural production: A case study in Zhejiang province, China. Environ. Earth Sci. 75 (2), 159–217. doi:10.1007/s12665-015-4887-4
Li, Y. F., Zhao, B. C., Huang, A., Xiong, B. Y., and Song, C. F. (2021b). Characteristics and driving forces of non-grain production of cultivated land from the perspective of food security. Sustainability 13 (24), 14047–14118. doi:10.3390/su132414047
Lin, Y. B., Ye, Y. M., Wu, C. F., Yang, J. H., Hu, Y. M., and Shi, H. K. (2019). Comprehensive assessment of paddy soil quality under land consolidation: A novel perspective of microbiology. Peerj 7, 73511–e7422. doi:10.7717/peerj.7351
Lou, J., Yang, L., Wang, H. Z., Wu, L. S., and Xu, J. M. (2018). Assessing soil bacterial community and dynamics by integrated high-throughput absolute abundance quantification. Peerj 6, e4514–e4519. doi:10.7717/peerj.4514
Lozano, Y. M., Hortal, S., Armas, C., and Pugnaire, F. I. (2014). Interactions among soil, plants, and microorganisms drive secondary succession in a dry environment. Soil Biol. Biochem. 78, 298–306. doi:10.1016/j.soilbio.2014.08.007
Lu, H. L., Wu, Y. X., Liang, P. X., Song, Q. M., Zhang, H. X., Wu, J. H., et al. (2020). Alkaline amendments improve the health of soils degraded by metal contamination and acidification: Crop performance and soil bacterial community responses. Chemosphere 257, 127309–127310. doi:10.1016/j.chemosphere.2020.127309
Lu, X. L., Zhang, Y. W., and Zou, Y. C. (2021). Evaluation the effect of cultivated land protection policies based on the cloud model: A case study of xingning, China. Ecol. Indic. 2021, 108247. doi:10.1016/J.ECOLIND.2021.108247
Morton, J. T., Marotz, C., Washburne, A., Silverman, J., Zaramela, L. S., Edlund, A., et al. (2019). Establishing microbial composition measurement standards with reference frames. Nat. Commun. 10, 2719–2811. doi:10.1038/s41467-019-10656-5
Pitta, D. W. P. N., Patel, A. K., Indugu, N., Kumar, S., Prajapathi, K. B., Patel, A. D., et al. (2014). Bacterial diversity dynamics associated with different diets and different primer pairs in the rumen of kankrej cattle. PLoS ONE 9 (11), e111710. doi:10.1371/journal.pone.0111710
Qi, J. J., Chen, B. B., Gao, J. M., Peng, Z. H., Jiao, S., Wei, G. H., et al. (2022). Responses of soil bacterial community structure and function to dry-wet cycles more stable in paddy than in dryland agricultural ecosystems. Glob. Ecol. Biogeogr. 31 (2), 362–377. doi:10.1111/geb.13433
Shi, Y. S., Cao, X. Y., Fu, D. M., and Wang, Y. C. (2018). Comprehensive value discovery of land consolidation projects: An empirical analysis of Shanghai, China. Sustainability 10 (6), 2039. doi:10.3390/su10062039
Su, Y., Qian, K., Lin, L., Wang, K., Guan, T., and Gan, M. Y. (2020). Identifying the driving forces of non-grain production expansion in rural China and its implications for policies on cultivated land protection. Land Use Policy 92, 104435–104510. doi:10.1016/j.landusepol.2019.104435
Tecon, R., and Or, D. (2017). Biophysical processes supporting the diversity of microbial life in soil. Fems Microbiol. Rev. 41 (5), 599–623. doi:10.1093/femsre/fux039
Ter Braak, C. J. F. (1986). Canonical correspondence analysis: A new eigenvector technique for multivariate direct gradient analysis. Ecology 67 (5), 1167–1179. doi:10.2307/1938672
Tipayno, S. C., Truu, J., Samaddar, S., Truu, M., Preem, J. K., Oopkaup, K., et al. (2018). The bacterial community structure and functional profile in the heavy metal contaminated paddy soils, surrounding a nonferrous smelter in South Korea. Ecol. Evol. 8 (12), 6157–6168. doi:10.1002/ece3.4170
Wan, Y., Li, W. J., Wang, J., and Shi, X. J. (2021). Bacterial diversity and community in response to long-term nitrogen fertilization gradient in citrus orchard soils. Diversity-Basel 13 (7), 282–315. doi:10.3390/d13070282
Wang, W. H., Luo, X., Chen, Y., Ye, X. F., Wang, H., Cao, Z., et al. (2019). Succession of composition and function of soil bacterial communities during key rice growth stages. Front. Microbiol. 10, 421–511. doi:10.3389/fmicb.2019.00421
Yang, L., Lou, J., Wang, H. Z., Wu, L. S., and Xu, J. M. (2018). Use of an improved high-throughput absolute abundance quantification method to characterize soil bacterial community and dynamics. Sci. Total Environ. 633, 360–371. doi:10.1016/j.scitotenv.2018.03.201
Zhang, D., Yang, W., Kang, D., and Zhang, H. (2023). Spatial-temporal characteristics and policy implication for non-grain production of cultivated land in Guanzhong Region. Land Use Policy 125, 106466. doi:10.1016/J.LANDUSEPOL.2022.106466
Keywords: non-grainization, soil bacterial communities, ecosystem functions, 16S rDNA amplicon sequencing technology, paddy fields
Citation: Liang Y and Geng B (2023) Effects of paddy field non-grainization consolidation on sustainable eco-functions protection of soil bacterial: Empirical evidence from Zhejiang province, China. Front. Environ. Sci. 11:1130234. doi: 10.3389/fenvs.2023.1130234
Received: 23 December 2022; Accepted: 01 March 2023;
Published: 10 March 2023.
Edited by:
Xiangbin Kong, China Agricultural University, ChinaReviewed by:
Bangbang Zhang, Northwest A&F University, ChinaYueyu Sui, Northeast Institute of Geography and Agroecology (CAS), China
Copyright © 2023 Liang and Geng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bin Geng, Z2VuZ2Jpbl96amNqZHhAMTI2LmNvbQ==
 Ying Liang
Ying Liang Bin Geng*
Bin Geng*