
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 13 September 2022
Sec. Drylands
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.993973
This article is part of the Research Topic Advances in Soil and Water Management for Dryland Areas View all 8 articles
Long-term irrational fertilizer inputs affect soil nutrients conditions in the agro-pastoral ecotone of North China. However, the mechanisms by which biotic and abiotic factors are affected by different fertilizer types remain unclear. A 16-year, long-term fertilization experiment was conducted to explore how soil physicochemical properties and microbial communities respond to different fertilizer types at an experimental site in North China. The key environmental factors that drove changes in soil microbial communities were also determined. In September 2019, soils were collected from plots of four fertilizer treatments: 1) non-fertilization control (CK), 2) chemical fertilization only (CF), 3) organic manure fertilization only (M), and 4) chemical fertilization plus organic manure (CFM). Compared with CK, soil organic matter, total nitrogen, available nitrogen, available phosphorus, and available potassium contents were higher in M and CFM, whereas soil pH was significantly lower in CF. Abundances of dominant soil bacterial phyla Proteobacteria, Bacteroidetes, and Gemmatimonadetes were higher in M and CFM than CK. Abundances of dominant soil fungal phyla Ascomycota was lower in CFM than in other treatments. The pathogenic fungi Fusarium, Paramyrothecium, Cladosporium, and Alternaria had the highest abundances in CK and CF, whereas abundances of the beneficial fungi Mortierella were significantly higher in M and CFM than in CF and CK. According to partial least squares path modeling, differences in fertilizer types had direct positive effects on fungal communities but little effect on bacterial communities. Overall, CFM maintained higher soil fertility and a healthy ecosystem because it increased beneficial microorganisms and inhibited pathogenic microorganisms, whereas CF increased the risk of crop infection with soil-borne diseases. The study provided a better understanding of how long-term fertilization affects microbial community composition and their associated ecosystem functions.
Fertilizer application is an important practice in agricultural production (Lenssen et al., 2020), and proper fertilizer application is a key factor to increase yields and improve soil quality (Bouwman, 1998; Bhattacharyya et al., 2007). However, irrational fertilization practices, including long-term over-application, indiscriminate use of chemical fertilizers, and decreased organic fertilizer use, can lead to ecological problems, including reductions in soil organic matter, soil slumping, groundwater contamination, and reduction of soil productivity (Afreh et al., 2018; Bansal et al., 2020; Shinoto et al., 2020). Microorganisms are important in the conversion and cycling of nutrients and organic matter in soil ecosystems, and they also have crucial roles in preserving soil ecological processes (Saha et al., 2019). Moreover, soil fungi are particularly sensitive to environmental changes (Yang et al., 2021). Therefore, research has increasingly focused on effects of fertilization on soil bacterial and fungal communities.
Different fertilization practices can affect soil microbial activities (Hicks et al., 2020; Zhang et al., 2022). Fertilization alters soil fertility by affecting nutrient levels, such as those of soil organic carbon (C) and total nitrogen (N), which directly composition of soil microbial communities (Sradnick et al., 2013; Chen et al., 2015; Zhang et al., 2015). Fertilization can also indirectly affect soil microorganisms by altering soil properties (Xun et al., 2016). For example, a decrease in soil pH following N application was the primary cause of changes of bacterial communities (Zeng et al., 2016), and Yu et al. (2013) observed that changes in C/N ratios caused by fertilization significantly affected the distribution of soil microbial communities. However, Ren et al. (2020) concluded that Proteobacteria and Gemmatimonadetes were not affected by fertilization and soil environments, whereas abundances of Acidobacterial and Actinobacterial were negatively correlated with fertilization treatments. Fertilization may also influence below-ground microorganisms by affecting above-ground plant processes (Zeng et al., 2016). Therefore, fertilization can affect soil microbial communities by a variety of mechanisms, with associated specific effects and critical factors related to cropping systems, tillage practices, and fertilizer application methods. Thus, targeted studies are needed to investigate effects of fertilizer application on microbial communities in order to improve scientific assessment of such effects as well timely and effective alteration of fertilizer application guidelines.
The agro-pastoral ecotone of Inner Mongolia is a typical dry farming region that is also a vital ecological barrier for agricultural and pastoral production bases in North China (Tang et al., 2018; Tang et al., 2020). Farmyard livestock and poultry manure has long been the primary fertilizer source in the region (Li et al., 2021). However, accelerated urbanization and continuous food demand have led to the gradual replacement of organic fertilizers as the primary fertilizers with chemical fertilizers, leading to the emergence of problems such as soil acidification and nutrient imbalances due to improper fertilization practices (Yan et al., 2015; Tang et al., 2021). Thus, reductions in agricultural production inputs are critical, in addition to stabilization of crop production and improvement in soil ecology using scientifically guided and rational fertilization measures. These steps are particularly important because the current rapid economic and social development coincides with several outstanding problems including human population increases, agricultural land area reduction, resource scarcity, and degradation of farmland ecology.
In this study, a 16-year long-term application of organic manure and chemical fertilizer experiment was used, our objectives were to 1) assess the optimum optimal fertilization system combined with soil properties and microbial communities, 2) investigate the responses of soil microbial community composition to different fertilizer systems, and 3) find the main soil chemical parameters that drive the change in soil microbial communities composition in three fertilization systems. The results will provide a foundation for further studies on how long-term fertilization practices affect soil properties and microbial communities in dryland farmland.
The experimental site was established in 2004 at the National Field Scientific Observation and Research Station for Dry Crop Farming Systems in the Inner Mongolia Autonomous Region, China (41° 08′ 22.8″ N and 111° 17′ 43.6″ E). The area features a mid-temperate continental monsoon climate at an altitude of 1,570 m, annual rainfall of 250–400 mm, potential evaporation of 1,848.3 mm, and more than 80% of the rain falls between June and September. The area exhibits an average annual temperature of 1.5–3.7°C and a frost-free period of 90–120 d. The ecosystem represents a typical semi-arid agro-pastoral ecotone. The cropping system is a one-season crop and the soil type is chestnut soil. The initial soil physical-chemical properties in 2004 (in the 0–20 cm layer) were shown in Table 1. The precipitation (195.5–414.9 mm, average 300.08 mm) and the average temperature change (14.6–17.9°C, average 16.2°C) during the crop growth period (from May to September) during the 2004–2019 experiment.
Different fertilization treatments were established within the long-term fertilization experimental site including chemical fertilization alone (CF), organic manure alone (M), organic manure with chemical fertilization (CFM), and a non-fertilization control (CK). All of the treatments were established in a randomized complete block design with three replicates and plot sizes of 50 m2 (5 m × 10 m). Fertilizer dosages and ratios were designed and recommended by the International Plant Nutrition Institute (IPNI) based on soil testing results. Urea was used for nitrogen nutrients, calcium superphosphate for phosphate, potassium chloride for potash, and sheep dung for organic manure. Its water content is 60–65%, organic matter content is 36–45%, and nutrient content (%) is N-P2O5-K2O = 0.57-0.23-1.04. Sheep dung was added at 7,500 kg hm−2 in 2004–2005 and 2007-2012, and 15,000 kg hm−2 in 2006 and 2013-2019, the specific fertilization amounts are shown in Table 2, including the N, P2O5, and K2O components within sheep dung. Potato, rape, and oat were used as rotation crops, with one crop planted every year. In 2019, the potato was planted. Crops were generally planted in early May and harvested in late September. During the 16 years (2004–2019), organic manure was evenly spread on the soil surface each year before sowing and artificially poured into the soil. Chemical fertilization was applied when sowing and all fertilizers were only applied once. The experiment was conducted under rainfed conditions, while other field management measures including pest control followed standard tillage measures.
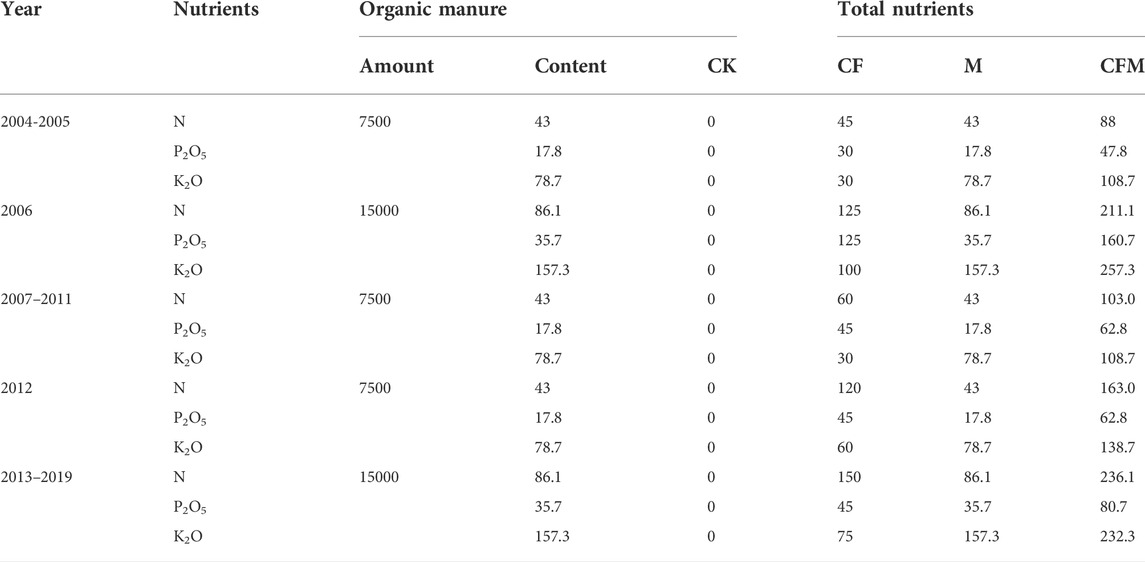
TABLE 2. Fertilizer application rates (equivalent to N, P2O5 and K2O) in the treated plots from 2004 to 2019.
A total of 60 soil samples (four treatments × five random points × three replicates) (2 cm diameter and 0–20 cm depth) were collected from each plot after potato harvest in 2019 in this experiment, and then totally mixed the soils from five random points in each plot. Finally, and the 12 soil samples were obtained, the 12 soil samples were divided into two portions after removing impurities, with one taken to the laboratory in a self-sealing bag, followed by natural airy-drying and filtering with a 2 mm sieve to determine soil nutrient contents. The other portion was sealed in sterile bags and immediately taken back to the laboratory for storage at −80°C for microbial community analyses.
A total of 36 soil samples (four treatments × three random points × three replicates) for the surface soil (0–20 cm) bulk density calculation were taken using cutting rings (volume 100 cm3, inner diameter 5.05 cm, 200–300 g) on three randomly selected points in each plot.
Soil pH was measured with a compound electrode (PE-10, Sartorius, Germany) (water: soil = 2.5:1). Other parameters were measured as previously described including soil bulk density (BD) (O’connell, 1975), organic matter (OM) (Nelson and Sommers, 1982), total nitrogen (TN) (Sparks et al., 1996), available nitrogen (AN), available phosphorus (AP), and available potassium (AK) (Lu 2000).
Total genomic DNA was extracted from 0.5 g of composite soil samples using the CTAB extraction method following the manufacturer’s protocol (MP Biomedicals, Illkirch, France). DNA concentration and purity were assessed with 1% agarose gel electrophoresis. Fungal ITS sequences were amplified with the primer ITS5F (5′GGAAGTAAAAGTCGTAACAAGG3′) and ITS2R (5′TCCTCCGCTTATTGATATGC-3′) (White et al., 1990) while the V4 hypervariable region of bacterial 16S rRNA genes were amplified using the primer pair 515F (5′-GTGCCAGCMGCCGCCGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Berthrong et al., 2013). Illumina MiSeq sequencing of the amplicons was then conducted at Beijing Novogene Technology Co., Ltd. (Novogene, Beijing, China).
PCR reactions were conducted in 30 μL reactions including 15 μl of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM of forward and reverse primers, and about 10 ng of template DNA. The PCR conditions included an initial denaturation at 98°C for 1 min, followed by 30 cycles with denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, all followed by a final extension at 72°C for 5 min. PCR amplification success was evaluated with gel electrophoresis using 1x loading buffer (containing SYBR green) and PCR products, with a 2% agarose gel. PCR products were mixed in equimolar ratios and the product pools were purified with the GeneJETTM Gel Extraction Kit (Thermo Scientific). Sequencing libraries were generated using the Ion Plus Fragment Library Kit (Thermo Scientific) following manufacturer’s recommendations. Sequencing library quality was assessed with the Qubit@ 2.0 Fluorometer (Thermo Scientific). The library was then sequenced on an Ion S5TM XL platform by generating 400 bp/600 bp single-end reads (Sheng et al., 2019; Wang et al., 2021).
Single-end reads were assigned to samples based on their unique barcodes, followed by truncation via removing barcode and primer sequences. Quality filtering of raw reads was performed to obtain high-quality clean reads using the Cutadapt (Martin, 2011) (V1.9.1, http://cutadapt.readthedocs.io/en/stable/) quality control pipeline. The reads were compared against the SILVA reference database (https://www.arb-silva.de/) (Quast et al., 2013) using the UCHIME algorithm (http://www.drive5.com/usearch/manual/uchime_algo.html) (Edgar et al., 2011) to detect chimeric sequences, followed by chimera removal (Haas et al., 2011). Sequence clustering was conducted with the UPARSE software package (v.7.0.1001, http://drive5.com/uparse/) (Edgar, 2013), wherein sequences with ≥97% nucleotide similarity was assigned to the same operational taxonomic units (OTUs). Representative sequences for each OTU were compared against the SILVA Database (https://www.arb-silva.de/) (Quast et al., 2013) using the Mothur classification algorithm to taxonomically annotate the OTUs. To investigate the phylogenetic relationships among OTUs and differences in dominant species among different sample groups, multiple sequence alignments were conducted using the MUSCLE software program (v.3.8.31, http://www.drive5.com/muscle/) (Edgar, 2004).
The raw sequences were submitted to the Sequence Read Archive (SRA) database under the BioProject database of the National Center for Biotechnology Information (NCBI) platform (project identification numbers PRJNA785382 (https://submit.ncbi.nlm.nih.gov/subs/bioproject/SUB10748118/overview) and PRJNA787050 (https://submit.ncbi.nlm.nih.gov/subs/bioproject/SUB10779674/overview) for bacterial and fungal sequences, respectively).
Soil physical and chemical properties, relative abundances of dominant bacterial and fungal taxa (average relative abundances ≥1%), and microbial diversity were analyzed using the least significant difference (LSD) at 5% level by SPSS program (v.24.0, IBM, United States). Permutational multivariate analysis of variance based on Bray–Curtis dissimilarity matrices using the “adonis” function in R and Principal coordinates analysis (PCoA) were used to test and visualize cultivar on the microbial communities using the R packages “vegan”, “pairwise”, “ade4” and “ggplot2”. Taxa with differential abundances among treatments were identified using linear discriminant analysis (LDA) effect size (LEFSe) analyses. Differentially enriched taxa (e.g., biomarkers) were defined based on discriminative characteristics of LDA score thresholds (log10 value) > 3.0 and p values <0.05 for between-group factorial Kruskal-Wallis tests. The relationships among soil properties and dominant soil microbe were examined using Pearson correlation analysis by SPSS program. Redundancy analysis (RDA) was used to evaluate changes in soil microbial community composition in association with physico-chemical properties and to determine the primary factors that drove these changes. RDA was performed using the CANOCO software program (v. 5.0, Ithaca, NY). Partial least squares path models (PLS-PMs) were used to model the relationships among soil physico-chemical properties in association with bacterial and fungal diversity and community composition. The path coefficients and the coefficients of determination (R2) in the path models were estimated in R (4.0.3) using the plspm package (1,000 bootstraps) (Sheik et al., 2012; McMurdie et al., 2013).
Sixteen years of fertilization led to significant changes in soil physico-chemical properties (Table 3). Soil BD decreased in M and CFM compared with that in CF and CK. Soil TN, AN, AP, and AK contents had the greatest increases in CFM. Soil OM content increased significantly in M and CFM, but decreased in CF compared with CK. The CF treatment significantly reduced soil pH compared with that in CK.
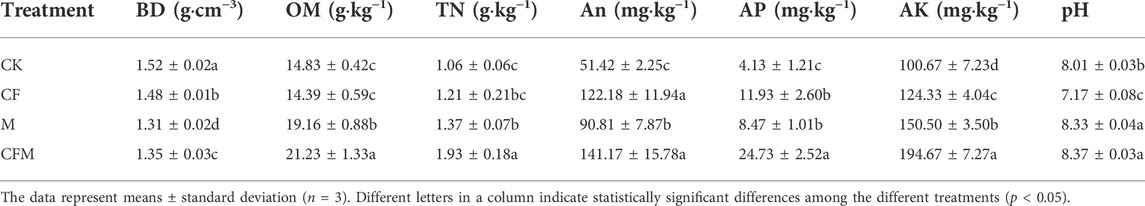
TABLE 3. Soil physical and chemical properties among the different treatments after 16 years (2004–2019).
The number of bacterial operational taxonomic units (OTUs) in M was significantly higher than that in CK, whereas the Chao1 index in CF was lower than that in the other treatments. Fungal OTU numbers were higher in M and CFM than in CK, the Chao1 index was significantly higher in M, whereas it was not significantly different in CF and CFM. The Shannon index of bacterial and fungal communities were not significantly different among treatments (Table 4).
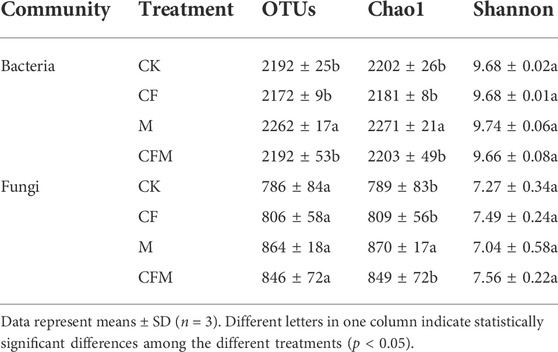
TABLE 4. Soil microbial community richness and diversity indices among the different treatments after 16 years (2004–2019).
Variation in structure of soil bacterial and fungal communities in different treatments was investigated using principal coordinates analysis (PCoA) (Figure 1). Treatments M and CFM were clustered together, indicating that structure of those microbial communities was similar, and they were also significantly separated from CF and CK along the PCo1 axis.
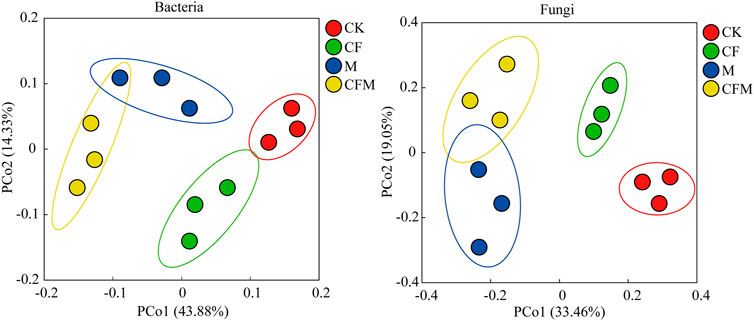
FIGURE 1. Principal coordinates analysis (PCoA) representing variation of soil bacterial and fungal communities across the different treatments after 16 years (2004–2019). Only OTUs with relative abundances >0.01% were included in the analysis.
In bacterial community, the dominant phyla across all samples included Proteobacteria (28.6–35.4%), Actinobacteria (25.4–33.5%), Acidobacteria (11.9–14.8%), Bacteroidetes (7.1–12.1%), with Gemmatimonadetes, Chloroflexi, Verrucomicrobia, Thaumarchaeota, Firmicutes, and Nitrospirae accounting for 13.5–16.8%. At the phylum level, 2.6%–3.3% of the bacterial reads were unclassified (Figure 2A). Abundances of Proteobacteria were significantly higher in M and CFM than in CK and CF, whereas abundances of Actinobacteria and Chloroflexi were significantly lower than those in CK and CF. Abundance of Acidobacterial was highest in CK, whereas abundances of Bacteroidetes and Gemmatimonadetes were significantly higher in CFM than in CK (Supplementary Table A1). Responses of the 20 most abundant genera of bacteria indicated that abundances of seven dominant genera were significantly different among fertilization treatments. Abundances of Sphingomonas, unidentified_Acidobacteria, and Bryobacter were higher in M than in other treatments. In addition, abundances of Lysobacter and Iamia were higher in CFM, whereas abundances of Nocardioides and Pseudarthrobacter were higher in CF communities than in other treatments. Blastococcus and Gaiella abundances were lower in CF, M, and CFM than in CK (Supplementary Table A2). Cluster analysis indicated that dominant genera in CFM were similar to those in M and CF but were significantly different compared with those in CK (Figure 2C).
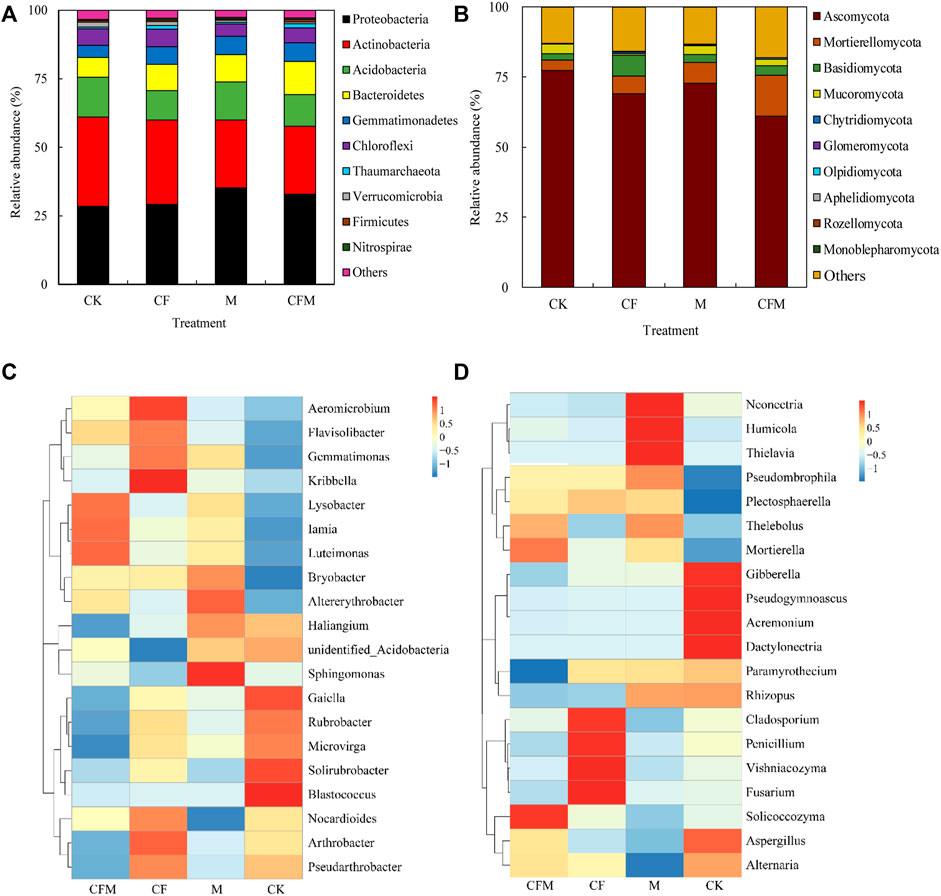
FIGURE 2. Relative abundances of the dominant bacterial and fungal taxa in soils among the different treatments after 16 years (2004–2019). Relative abundances of bacterial (A), phylum level; (C), genus level) and fungal (B), phylum level; (D), genus level) taxa among the treatment soils. The 10 most abundant phyla and the 20 most abundant genera are shown in the panels. The relative abundance of each taxon was calculated using the average relative abundance of that taxa across all of the soils divided by the average total relative abundance of all of the phyla in each treatments.
In fungal communities, Ascomycota was the dominant phylum (69.1–72.6%), followed by Mortierellomycota (3.7–14.4%), Basidiomycota (2.3–7.5%), and Mucoromycota (0.5–3.4%). In addition, Chytridiomycota, Glomeromycota, Olpidiomycota, Aphelidiomycota, Rozellomycota and Monoblepharomycota accounted for 0.4–0.9% of all fungal communities. At the phylum level, 12.9%–18.0% of all fungal reads could not be classified (Figure 2B). Abundances of Mortierellomycota and Basidiomycota were significantly higher in CF than in the other treatments, whereas abundances of Ascomycota were significantly lower in CFM than in CK, CF, and M (Supplementary Table A3). Only six of the dominant genera had abundances that were significantly different among fertilization treatments. Compared with CK, Mortierella was significantly more abundant in CF, M, and CFM, whereas Acremonium, Alternaria, and Dactylonectria were significantly less abundant in CF. In addition, Fusarium was significantly more abundant in CF, whereas Rhizopus was less abundant in CF and CFM compared with CK (Supplementary Table A4). Cluster analysis indicated that dominant genera in CFM and CF were similar, whereas genera in CK were significantly different from those in the other treatments (Figure 2D).
In a LEfSe analysis, LDA values >3.5 indicated significant enrichment of microbial taxa, and those responsive taxa were considered “biomarkers” for different treatments. Forty-five taxa, inclusive of all taxonomic levels, were identified that best separated soil bacterial communities among the four fertilization treatments. Twenty-two enriched taxa were identified in CFM, fourteen in CK, seven in M, and only two in CF (Supplementary Figure A1). The class Gammaproteobacteria was one of the most predominant biomarkers in CFM, whereas relatively high abundances identified the families Sphingobacteriaceae and Pyrinomonadaceae as biomarkers in M and CK. The CF treatment included only a single biomarker, the class Chloroflexia. Fifty significantly enriched fungal taxa (LDA score >3.5) distinguished soil fungal communities among different treatments, including 12 taxa in M, 18 in CK, 10 in CFM, and 10 in CF (Supplementary Figure A2). The genus Mortierella, order Hypocreales, and the class Sordariomycetes were identified as biomarkers in CFM, M, and CK, respectively. In addition, abundances of the family Cladosporlaceae were significantly higher in CF than in other treatments.
Correlation analysis revealed that soil chemical properties affected microbial community composition (Table 5). In bacterial communities, Proteobacteria was significantly positively correlated with soil OM and TN, whereas Actinobacteria was significantly negatively correlated with soil TN, AN, and pH. In fungal communities, Ascomycota was significantly negatively correlated with soil AN, AP, AK, and pH. Redundancy analysis (RDA) revealed that soil chemical properties were associated with phylum-level composition of bacterial and fungal communities in CFM. Composition of the bacterial communities in CF and composition of fungal communities at the phylum-level in M and at the genus-level in CK were greatly affected by soil pH and BD. Consequently, changes in soil pH induced by fertilization were reflected in responses in microbial abundances (Figure 3).
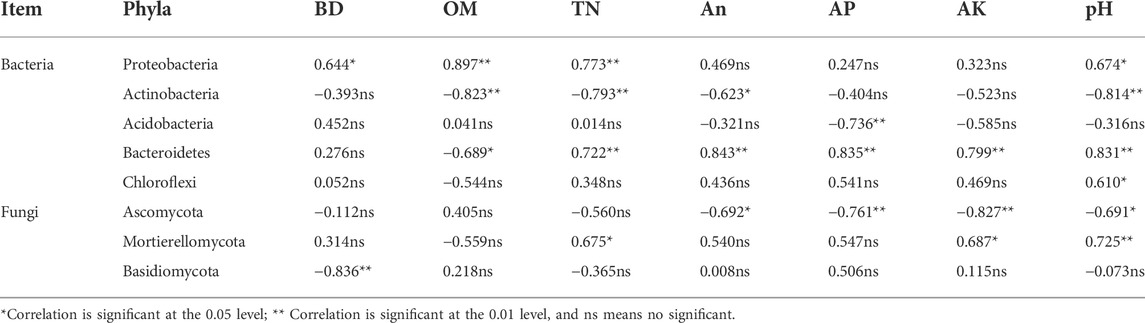
TABLE 5. Correlation analysis of the dominant soil microbial phyla and soil environmental factors after 16 years (2004–2019).
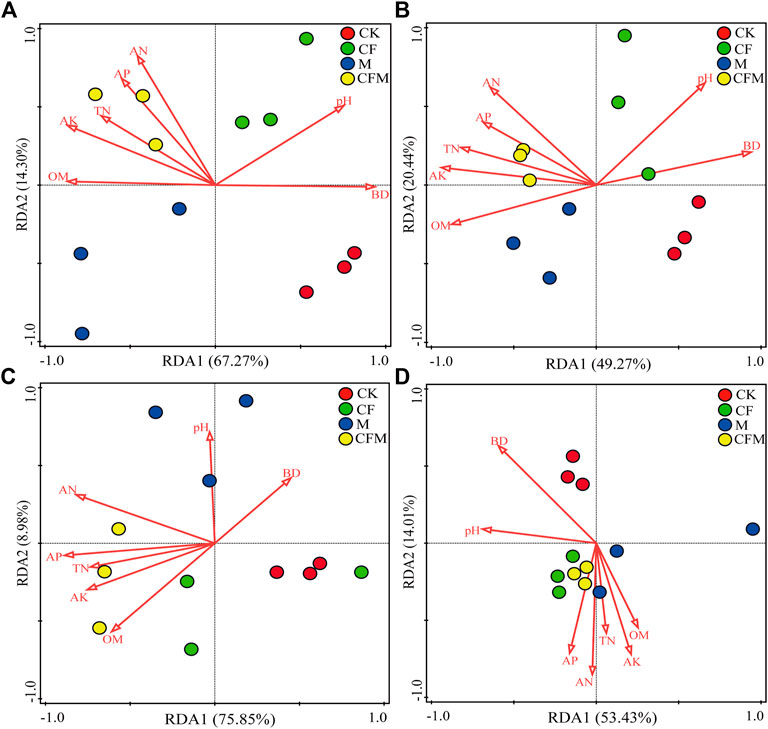
FIGURE 3. Redundancy analysis (RDA) explaining the influence of soil physical and chemical parameters on the composition of soil bacterial and fungal communities across the different treatments after 16 years (2004–2019). Only the 10 most abundant phyla and 20 most abundant genera were used in the analyses.
PLS-PM analysis was used to evaluate relations among fertilization, soil physico-chemical properties, and microbial community composition and diversity (Figure 4). The model constructed indicated that differences in fertilization types (CK→CF→M→CFM) had significant and direct positive effects on OM (1.052), nutrient contents (a combination of N, P, and K; 0.895), and especially fungal communities (1.527), in addition to significant and direct negative effects on soil BD (−0.610) (Supplementary Table A5). However, differences in fertilization did not directly affect on bacterial communities. In addition, soil pH directly and strongly affected composition of bacterial and fungal communities, whereas soil OM and nutrient contents did not significantly affect composition of microbial communities.
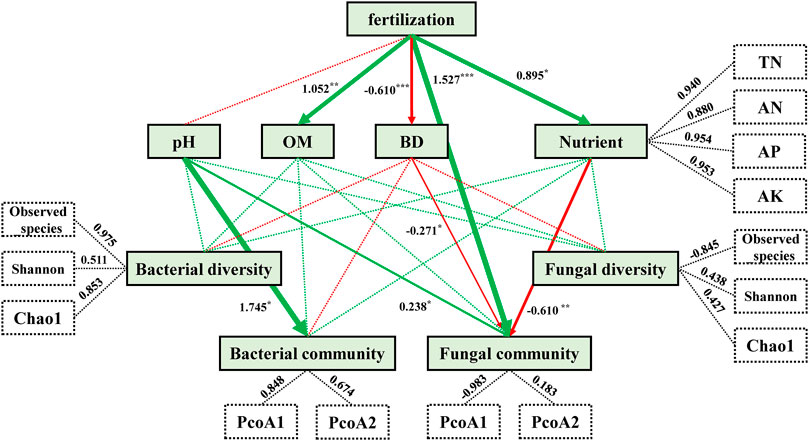
FIGURE 4. Directed graph demonstrating the partial least squares path model (PLS-PM) of the different treatments on soil microbial communities after 16 years (2004–2019). Each box represents an observed (i.e., measured) or latent variable (i.e., constructs). Dashed rectangles indicate nutrient loads and three soil microbial diversity indices that introduce potential variability. Green arrows indicate positive effects, while red arrows indicate negative effects, and arrow width is proportional to effect sizes. Dashed lines indicate the lack of significant effects (p > 0.05). Path coefficients were calculated after 1,000 bootstraps and are reflected in arrow widths. Asterisks indicate the degree of influence: *p < 0.05, **p < 0.01, ***p < 0.001. The model was assessed using the Goodness of Fit (GoF) statistic, with a corresponding GoF value of 0.702.
Proper fertilization is a critical practice to stabilize soil productivity, improve soil quality, and promote the sustainable development of farmland ecosystems. Soil BD is a direct physicochemical indicator of soil quality conditions and is one of the most sensitive soil properties to long-term fertilization (Lin et al., 2018). In CK in this study, soil BD increased by 5.56% compared with that in 2004. By contrast, soil BD significantly decreased in CFM and M, which could be because organic manure supplements provided a cementing agent for soils, thereby resulting in changes to soil structure and promoting crop root growth. The combination of root secretions and mineral colloids can help improve soil BD (Qiao et al., 2020). Soil OM and nutrient (TN, AN, AP, and AK) contents were higher in M and CFM than CK. Humic acids in soil organic C pools can increase plant root vigor and increase root respiration along with nutrient uptake, thereby facilitating accumulation of soil nutrients (Furukawa et al., 2014). The CF treatment significantly decreased soil pH, with a clear trend toward soil acidification. Low pH due to proton deficiency caused by chemical fertilizer application also reduced soil nutrients (Tian and Niu, 2015). In M and CFM, the application of organic manure increased levels of alkaline cations in soils and helped to neutralize soil acidity. Thus, M and CFM treatments maintained a high soil pH, and protonation of organic anions to form central molecules was the main mechanism to resist soil acidification (Shi et al., 2019). Alternatively, the experimental area had arid chestnut soils with high CaCO2 contents, which could have reduced the risk of soil acidification from chemical fertilizer application (Sarfraz et al., 2017). In addition, the pH in both M and CFM soils decreased slightly compared with the initial soil pH (Table 1), and it was inferred that the mechanism of the slight decrease in soil pH might be related to the high initial pH of the alkaline soils. Most studies conclude that chemical fertilizer application does not directly increase soil nutrient contents, but primarily increases accumulation of soil nutrients by increasing plant root stubble, root systems, and root secretions while stimulating the decomposition of pre-existing soil nutrients. Thus, although the CF treatment also increased soil nutrients, overall increases were limited (Knoblauch et al., 2017). The CF treatment reduced soil OM content because of long-term deficiencies in soil organic matter inputs. In this study, CFM was the treatment that most effectively improved soil properties, while also providing a supplementary input of in-season nutrient sources to soils. However, the risk of further soil acidification associated with the long-term application of chemical fertilizers is also a concern and reinforces the importance of using organic manure–chemical fertilizer to improve soil chemical properties.
Microorganisms have significant roles in forming soil fertility, improving ecological characteristics, and preventing soil borne diseases. As a consequence, abundances and diversity of microorganisms affect the conversion of soil nutrients and the modulation of biological effectiveness, which are critical signs of soil health (Ji et al., 2014). According to the PCoA changes in fertilization had relatively large effects on composition of microbial communities. Proteobacteria, Bacteroidetes, and Gemmatimonadetes are copiotrophic taxa that can effectively deplete labile C pools in soils, and they also have high nutrient requirements (Ghosh et al., 2016). In this study, Proteobacteria, Bacteroidetes, and Gemmatimonadetes were significantly more abundant in CFM and M, which also had higher OM and nutrient contents than those in CK. Organic manure application favors the growth of copiotrophs, whereas high abundances of copiotrophic taxa also indicate high soil fertility. The Proteobacteria comprises many taxa involved in soil N cycling, including Sphingomonas and Lysobacter, which varied in abundance similarly to that of Proteobacteria overall. Thus, organic manure application can improve soil N fixation capacity. Acidobacteria and Chloroflexi are oligotrophic taxa that consume recalcitrant organic C pools and have high nutrient affinities, despite relatively slow growth rates (Delgado-Baquerizo et al., 2017). In this study, Acidobacteria and Chloroflexi were most abundant in CK with low OM contents and soil nutrients and therefore overall low fertility because of prolonged lack of fertilization. Such conditions can drive bacterial communities toward dominance by oligotrophs. Unidentified_Acidobacteria and Bryobacter are members of Acidobacteria, and abundances of those taxa varied similarly to that of Acidobacteria overall. Actinobacteria are mostly pathogenic bacteria, and in this study, abundances were highest in CK and CF. Thus, long-term chemical fertilization and the lack of fertilization can lead to soil nutrient imbalances and increased abundances of pathogenic bacteria, thereby increasing the risk of soil bacterial diseases that endanger healthy soil environments. Blastococcus, Nocardioides, Gaiella, Pseudarthrobacter, and Iamia are members of Actinobacteria and abundances of those taxa varied similarly to that of Actinobacteria overall, except for Iamia. Lysobacter and Sphingomonas are associated with antagonistic activities toward plant pathogens (Khan et al., 2014; Liu et al., 2020), and in this study, those taxa had high abundances in CFM, suggesting that application of organic manure with chemical fertilizer improved soil microenvironments.
Ascomycota was dominant in fungal communities in this study. Soil fungi use OM as energy sources and nutrients, participate in OM mineralization, decompose OM from crop residues or organic manure that enter soils, and supply plants with nutrients (Detheridge et al., 2016). However, most fungi are also plant pathogens. In this study, 16 of the 20 most abundant fungal genera were members of Ascomycota, and abundances of those genera were significantly lower in CFM than in the other treatments. To some extent, those results reflected a reduced incidence of fungal soilborne diseases. Fusarium, Gibberella, and Dactylonectria can cause plant stem rot or spikelet rot (Larkin and Griffin, 2007; Cannon et al., 2012), whereas Paramyrothecium and Plectosphaerella can cause plant root rot (Carlucci et al., 2012). Paramyrothecium abundances were significantly lower in CFM than in other treatments. Furthermore, Alternaria, which can cause potato brown spot and potato early blight, was most abundant in CK and CF (Xu et al., 2022). Cladosporium is a saprophytic taxon but is also an important plant pathogen that mainly infects plant leaves, branches, and fruits, resulting in poor quality and reduced yields (Bensch et al., 2012). Cladosporium abundance was significantly higher in CF than in the other treatments. The genus Mortierella in Mortierellomycota is important in improving nutrient uptake efficiency and protecting crops from adverse conditions by improving the availability of soil P, K, and ferric iron (Ozimek and Hanaka, 2021). Vishniacozyma is a member of Basidiomycota, which had high abundances in CF, although the ecological functions of the genus remain unknown. Long-term fertilization increased relative abundances of fungi in communities, which might lead to soil fungalization. Overall, the results of this study suggest that long-term fertilization affects the structure of microbial communities, with chemical fertilization increasing pathogenic fungal loads and thereby increasing the risk of disease infestation. However, combining organic manure and chemical fertilization can reduce fungal pathogen loads and increase abundances of beneficial fungal genera.
Soil physical and chemical properties greatly affect the composition of soil microbial communities. In this study, dominant microbial taxa were consistently positively correlated with soil OM and nutrient contents in CFM. Those results were likely because the combined manure–chemical fertilization application increased soil OM and nutrient contents, which are necessary nutrients for microbial growth. Thus, high OM content contributes to improving soil quality and maintaining soil health (Mishra et al., 2018). High soil fertility results in increased abundances of copiotrophic populations. In this study, Proteobacteria was significantly positively correlated with soil OM and TN, as were microbial taxa that could inhibit some pathogens (Raoul des Essarts et al., 2016), including the fungus Mortierella, which was highly abundant in CFM. Soil fungal diversity and abundances of dominant fungi were significantly higher in CF than in other treatments, although soil OM content was lowest in that treatment. Fungi primarily use C sources and nutrients for growth and reproduction, which are obtained from the decomposition of organic matter. This is particularly evident for large fungi that can consume large molecular-weight organic matter. Such interactions might explain the decrease in OM resulting from chemical fertilization. The dominant microbial taxa in CK were negatively correlated with soil nutrient contents, suggesting that poor fertility induces explosive growth of fungal pathogens, as indicated by significant negative correlations of Ascomycota with soil AN, AP, and AK. The PLS-PM analysis also indicated that fertilization treatments directly affected fungal communities, in contrast to bacterial communities, indicating that fungi were more sensitive to fertilization treatments than bacteria (Cassman et al., 2016; Ai et al., 2018). In addition, the PLS-PM analysis indicated that the soil pH was primary factor that significantly affected soil microbial community composition. Those results are in contrast to those of many studies suggesting that nutrient changes due to fertilization directly drive shifts in microbial communities. Xun et al. (2015) proposed the“rugby ball model”to explain this phenomenon. In that model, near-neutral soils (characterized by the central part of the rugby ball) are strongly influenced by soil nutrients that also affect microbial community transformations. The radius of the central part of the rugby ball is larger and represents the strength and direction of the driving effects of nutrient indicators. When soil acidification or alkalization increases (characterized by the two ends of the rugby ball), soil pH has an increasing effect on microbial community transformations, whereas the effects of soil nutrients are gradually minimized (as represented by movement towards the ends of the ball, with the cross-sectional radius of the rugby ball). In this study, different fertilization treatments resulted in significant changes in soil pH in dryland soils. Changes in pH were especially evident with long-term chemical fertilization alone, with increases in soil pH in that treatment increasing the risk of soil acidification. In this case, the negative effects of soil acidification outweighed improvement in soil fertility, as indicated by the decrease in soil OM content. By contrast, the combined use of organic manure and chemical fertilization significantly improved soil pH while also supplementing soil nutrients, thereby promoting and realizing the full positive effects on soil nutrients.
In this study, different fertilization types affected soil physico-chemical properties and microbial community characteristics. However, only the overall changes in soil microbial communities were evaluated, and microorganisms in unique functional groups were not investigated. Thus, differences in soil metabolic pathways and functions associated with unique functional groups of microorganisms necessitate further research in order to determine whether differences in biomass, diversity, and community structure under different fertilization practices lead to changes in soil fertility, health status, and ecosystem function.
In this study, the CFM maintained high soil fertility, suitable pH, and stable microbial community composition, suggesting it was the best measure among the three amendment types. In contrast, CF decreased soil OM contents and increased the risk of soil acidification, while pathogenic microbial taxa abundances significantly increased, leading to an increased risk of crop infection by soil-borne fungal diseases. Therefore, a fertilization regime comprising combined organic manure and chemical fertilizer application should be used to improve soil fertility and soil microenvironment. These results provide a better understanding of how long-term fertilization affects microbial community composition and their associated ecosystem functions in agro-pastoral ecotone agroecosystems.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra, PRJNA785382, PRJNA787050.
YD and JZ conceived the study. HL, YJ, and PZ designed the research and performed the experiments. RG analyzed the data and wrote the main manuscript text. YR and XL prepared Figures 1–4. All authors reviewed the manuscript.
This study was financially supported by Natural Science Foundation of Inner Mongolia, China (2019MS03032; 2018MS03007) and Central Fund for Guiding Local Science and Technology Development, China (2020ZY0016).
We would like to thank the Soil Improvement Team and Dry Farming Team for field and data collection, and Professor Zhihua Pan’s guidance on the papers from China Agricultural University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.993973/full#supplementary-material
Afreh, D., Zhang, J., Guan, D. H., Liu, K. L., Song, Z. W., Zheng, C. Y., et al. (2018). Long-term fertilization on nitrogen use efficiency and greenhouse gas emissions in a double maize cropping system in subtropical China. Soil Tillage Res. 180, 259–267. doi:10.1016/j.still.2018.03.016
Ai, C., Zhang, S. Q., Zhang, X., Guo, D. D., Zhou, W., and Huang, S. M. (2018). Distinct responses of soil bacterial and fungal communities to changes in fertilization regime and crop rotation. Geoderma 319, 156–166. doi:10.1016/j.geoderma.2018.01.010
Bansal, S., Yin, X. H., Savoy, H. J., Jagadamma, S., Lee, J., and Sykes, V. (2020). Long‐term influence of phosphorus fertilization on organic carbon and nitrogen in soil aggregates under no‐till corn–wheat–soybean rotations. Agron. J. 112, 2519–2534. doi:10.1002/agj2.20200
Bensch, K., Braun, U., Groenewald, J. Z., and Crous, P. W. (2012). The genus Cladosporium. Stud. Mycol. 72, 1–401. doi:10.3114/sim0003
Berthrong, S. T., Buckley, D. H., and Drinkwater, L. E. (2013). Agricultural management and labile carbon additions affect soil microbial community structure and interact with carbon and nitrogen cycling. Microb. Ecol. 66, 158–170. doi:10.1007/s00248-013-0225-0
Bhattacharyya, R., Chandra, S., Singh, R. D., Kundu, S., Srivastva, A. K., and Gupta, H. S. (2007). Long-term farmyard manure application effects on properties of a silty clay loam soil under irrigated wheat-soybean rotation. Soil Tillage Res. 94, 386–396. doi:10.1016/j.still.2006.08.014
Bouwman, A. F. (1998). Environmental science: Nitrogen oxides and tropical agriculture. Nature 392, 866–867. doi:10.1038/31809
Cannon, P. F., Buddie, A. G., Bridge, P. D., de Neergaard, E., Lubeck, M., Askar, M. M., et al. (2012). Lectera, a new genus of the Plectosphaerellaceae for the legume pathogen Volutella colletotrichoides. Mycokeys 3, 23–36. doi:10.3897/mycokeys.3.3065
Carlucci, A., Raimondo, M. L., Santos, J., and Phillips, A. J. (2012). Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia 28, 34–48. doi:10.3767/003158512x638251
Cassman, N. A., Leite, M. F., Pan, Y., de Hollander, M., van Veen, J. A., and Kuramae, E. E. (2016). Plant and soil fungal but not soil bacterial communities are linked in long-term fertilized grassland. Sci. Rep. 6, 23680. doi:10.1038/srep23680
Chen, X. F., Li, Z. P., Liu, M., and Che, Y. (2015). Microbial community and functional diversity associated with different aggregate fractions of a paddy soil fertilized with organic manure and/or NPK fertilizer for 20 years. J. Soils Sediments 15, 292–301. doi:10.1007/s11368-014-0981-6
Delgado-Baquerizo, M., Trivedi, P., Trivedi, C., Eldridge, D. J., Reich, P. B., Jeffries, T. C., et al. (2017). Microbial richness and composition independently drive soil multifunctionality. Funct. Ecol. 31, 2330–2343. doi:10.1111/1365-2435.12924
Detheridge, A. P., Brand, G., Fychan, R., Crotty, F. V., Sanderson, R., Griffith, G. W., et al. (2016). The legacy effect of cover crops on soil fungal populations in a cereal rotation. Agric. Ecosyst. Environ. 228, 49–61. doi:10.1016/j.agee.2016.04.022
Edgar, R. C. (2004). Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi:10.1093/nar/gkh340
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi:10.1093/bioinformatics/btr381
Edgar, R. C. (2013). Uparse: Highly accurate OTU sequences from microbial amplicon reads. Nat. methods 10, 996–998. doi:10.1038/nmeth.2604
Furukawa, Y., Reed, A. H., and Zhang, G. P. (2014). Effect of organic matter on estuarine flocculation: A laboratory study using montmorillonite, humic acid, xanthan gum, guar gum and natural estuarine flocs. Geochem. Trans. 15, 1–9. doi:10.1186/1467-4866-15-1
Ghosh, A., Bhattacharyya, R., Dwivedi, B. S., Meena, M. C., Agarwal, B. K., Mahapatra, P., et al. (2016). Temperature sensitivity of soil organic carbon decomposition as affected by long-term fertilization under a soybean based cropping system in a sub-tropical Alfisol. Agric. Ecosyst. Environ. 233, 202–213. doi:10.1016/j.agee.2016.09.010
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454–pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi:10.1101/gr.112730.110
Hicks, L. C., Rousk, K., Rinnan, R., and Rousk, J. (2020). Soil microbial responses to 28 years of nutrient fertilization in a subarctic heath. Ecosystems 23, 1107–1119. doi:10.1007/s10021-019-00458-7
Ji, B. Y., Hu, H., Zhao, Y. L., Mu, X. Y., Liu, K., and Li, C. H. (2014). Effects of deep tillage and straw returning on soil microorganism and enzyme activities. Sci. World J. 2014, 1–12. doi:10.1155/2014/451493
Khan, A. L., Waqas, M., Kang, S. M., Al-Harrasi, A., Hussain, J., Al-Rawahi, A., et al. (2014). Bacterial endophyte Sphingomonas sp. LK11 produces gibberellins and IAA and promotes tomato plant growth. J. Microbiol. 52 (8), 689–695. doi:10.1007/s12275-014-4002-7
Knoblauch, C., Watson, C., Becker, R., Berendonk, C., and Wichern, F. (2017). Change of ergosterol content after inorganic N fertilizer application does not affect short-term C and N mineralization patterns in a grassland soil. Appl. Soil Ecol. 111, 57–64. doi:10.1016/j.apsoil.2016.11.016
Larkin, R. P., and Griffin, T. S. (2007). Control of soilborne potato diseases using Brassica green manures. Crop Prot. 26, 1067–1077. doi:10.1016/j.cropro.2006.10.004
Lenssen, A. W., Sainju, U. M., Jones, C., McVay, K., and Angvick, T. (2020). Nitrogen fertilization rate and method influences water and nitrogen productivity of forage winter wheat. Agron. J. 113, 577–589. doi:10.1002/agj2.20495
Li, Y., Wang, J., Tang, J. Z., Wang, E. L., Pan, Z. H., Pan, X. B., et al. (2021). Optimum planting date and cultivar maturity to optimize potato yield and yield stability in North China. Field Crops Res. 269, 108179. doi:10.1016/j.fcr.2021.108179
Lin, Y., Slessarev, E. W., Yehl, S. T., D’Antonio, C. M., and King, J. Y. (2018). Long-term nutrient fertilization increased soil carbon storage in California grasslands. Ecosystems 22, 754–766. doi:10.1007/s10021-018-0300-y
Liu, Z. X., Liu, J. J., Yu, Z. H., Yao, Q., Li, Y. S., Liang, A. Z., et al. (2020). Long-term continuous cropping of soybean is comparable to crop rotation in mediating microbial abundance, diversity and community composition. Soil Tillage Res. 197, 104503. doi:10.1016/j.still.2019.104503
Lu, R. K. (2000). Soil agricultural chemical analysis method. Beijing: China Agric. Sci. Tech. Press. in Chinese.
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10. doi:10.14806/ej.17.1.200
McMurdie, P., Holmes, S., Kindt, R., Legendre, P., and O’Hara, R. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. doi:10.1371/journal.pone.0061217
Mishra, G., Giri, K., and Pandey, S. (2018). Role of alnus nepalensis in restoring soil fertility: A case study in mokokchung, Nagaland. Natl. Acad. Sci. Lett. 41, 265–268. doi:10.1007/s40009-018-0668-4
Nelson, D., and Sommers, L. E. (1982). “Total carbon, organic carbon, and organic matter,” in Methods of soil analysis Part 3—chemical methods. (methodsofsoilan3). doi:10.2134/agronmonogr9.2
O’connell, D. (1975). The measurement of apparent specific gravity of soils and its relationship to mechanical composition and plant root growth. Tech. Bull. Minist. Agric. Fish. Food 29, 298–313.
Ozimek, E., and Hanaka, A. (2021). Mortierella species as the plant growth-promoting fungi present in the agricultural soils. Agriculture 11 (1), 7. doi:10.3390/agriculture11010007
Qiao, Y. X., Zhu, H. Z., Zhong, H. P., and Li, Y. Z. (2020). Stratified data reconstruction and spatial pattern analyses of soil bulk density in the northern grasslands of China. ISPRS Int. J. Geoinf. 9 (11), 682. doi:10.3390/ijgi9110682
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi:10.1093/nar/gks1219
Raoul des Essarts, Y., Cigna, J., Quetu-Laurent, A., Caron, A., Munier, E., Beury-Cirou, A., et al. (2016). Biocontrol of the potato blackleg and soft rot diseases caused by Dickeya dianthicola. Appl. Environ. Microbiol. 82, 268–278. doi:10.1128/aem.02525-15
Ren, N., Wang, Y., Ye, Y. L., Zhao, Y. A., Huang, Y. F., Fu, W., et al. (2020). Effects of continuous nitrogen fertilizer application on the diversity and composition of rhizosphere soil bacteria. Front. Microbiol. 11, 1948. doi:10.3389/fmicb.2020.01948
Saha, A., Basaka, B. B., Gajbhiyea, N. A., Kalariyaa, K. A., and Manivela, P. (2019). Sustainable fertilization through co-application of biochar and chemical fertilizers improves yield, quality of Andrographis paniculata and soil health. Ind. Crops Prod. 140, 111607. doi:10.1016/j.indcrop.2019.111607
Sarfraz, R., Shakoor, A., Abdullah, M., Arooj, A., Hussain, A., and Xing, S. H. (2017). Impact of integrated application of biochar and nitrogen fertilizers on maize growth and nitrogen recovery in alkaline calcareous soil. Soil Sci. Plant Nutr. 63, 488–498. doi:10.1080/00380768.2017.1376225
Sheik, C. S., Mitchell, T. W., Rizvi, F. Z., Rehman, Y., Faisal, M., Hasnain, S., et al. (2012). Exposure of soil microbial communities to chromium and arsenic alters their diversity and structure. PLoS ONE 7, e40059. doi:10.1371/journal.pone.0040059
Sheng, R., Li, K., Zhang, W. Z., Wang, H., Liu, H. L., Zhu, Y., X., et al. (2019). Differentiations of determinants for the community compositions of bacteria, fungi, and nitrogen fixers in various steppes. Ecol. Evol. 9, 3239–3250. doi:10.1002/ece3.4940
Shi, R. Y., Liu, Z. D., Li, Y., Jiang, T., Xu, M., Li, J. Y., et al. (2019). Mechanisms for increasing soil resistance to acidification by long-term manure application. Soil Tillage Res. 185, 77–84. doi:10.1016/j.still.2018.09.004
Shinoto, Y., Otani, R., Matsunami, T., and Maruyama, S. (2020). Analysis of the shallow root system of maize grown by plowing upland fields converted from paddy fields: Effects of soil hardness and fertilization. Plant Prod. Sci. 24, 297–305. doi:10.1080/1343943X.2020.1863823
Sparks, D. L., Page, A. L., Helmke, P. A., and Loeppert, R. H. (1996). Methods of soil analysis Part 3—chemical methods. Soil sci. Soci. Am.. Madison, WI: Am. Soci. Agron..
Sradnick, A., Murugan, R., Oltmanns, M., Raupp, J., and Joergensen, R. G. (2013). Changes in functional diversity of the soil microbial community in a heterogeneous sandy soil after long-term fertilization with cattle manure and mineral fertilizer. Appl. Soil Ecol. 63, 23–28. doi:10.1016/j.apsoil.2012.09.011
Tang, J. Z., Wang, J., Wang, E. L., Yu, Q., Yin, H., He, D., et al. (2018). Identifying key meteorological factors to yield variation of potato and the optimal planting date in the agro-pastoral ecotone in North China. Agric. For. Meteorol. 256, 283–291. doi:10.1016/j.agrformet.2018.03.022
Tang, J. Z., Xiao, D. P., Bai, H. Z., Wang, B., Liu, D. L., Feng, P. Y., et al. (2020). Potential benefits of potato yield at two sites of agro-pastoral ecotone in North China under future climate change. Int. J. Plant Prod. 14, 401–414. doi:10.1007/s42106-020-00092-7
Tang, J. Z., Xiao, D. P., Wang, J., Fang, Q. X., Zhang, J., and Bai, H. Z. (2021). Optimizing water and nitrogen managements for potato production in the agro-pastoral ecotone in North China. Agric. Water Manag. 253, 106945. doi:10.1016/j.agwat.2021.106945
Tian, D. S., and Niu, S. L. (2015). A global analysis of soil acidification caused by nitrogen addition. Environ. Res. Lett. 10, 024019. doi:10.1088/1748-9326/10/2/024019
Wang, X. Y., Bian, Q., Jiang, Y. J., Zhu, L. G., Chen, Y., Liang, Y. T., et al. (2021). Organic amendments drive shifts in microbial community structure and keystone taxa which increase C mineralization across aggregate size classes. Soil Biol. Biochem. 153, 108062. doi:10.1016/j.soilbio.2020.108062
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). in Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Editors M. A. Innis, and D. H. Gelfand.
Xu, X., Zhang, L., Yang, X. L., Cao, H. S., Li, J. J., Cao, P., et al. (2022). Alternaria spp. associated with leaf blight of maize in heilongjiang province, China. Plant Dis. 106 (2), 572–584. doi:10.1094/PDIS-06-21-1151-RE
Xun, W. B., Huang, T., Zhao, J., Ran, W., Wang, B. R., Shen, Q. R., et al. (2015). Environmental conditions rather than microbial inoculum composition determine the bacterial composition, microbial biomass and enzymatic activity of reconstructed soil microbial communities. Soil Biol. Biochem. 90, 10–18. doi:10.1016/j.soilbio.2015.07.018
Xun, W. B., Zhao, J., Xue, C., Zhang, G. S., Ran, W., Wang, B. R., et al. (2016). Significant alteration of soil bacterial communities and organic carbon decomposition by different long-term fertilization management conditions of extremely low-productivity arable soil in South China. Environ. Microbiol. 18, 1907–1917. doi:10.1111/1462-2920.13098
Yan, Q. L., Zhu, J. J., Zheng, X., and Jin, C. J. (2015). Causal effects of shelter forests and water factors on desertification control during 2000-2010 at the Horqin Sandy Land region, China. J. For. Res. 26, 33–45. doi:10.1007/s11676-014-0012-x
Yang, W. H., Li, C. J., Wang, S. H., Zhou, B. Q., Mao, Y. L., Christopher, R., et al. (2021). Influence of biochar and biochar-based fertilizer on yield, quality of tea and microbial community in an acid tea orchard soil. Appl. Soil Ecol. 166, 104005. doi:10.1016/j.apsoil.2021.104005
Yu, W. T., Bi, M. L., Xu, Y. G., Zhou, H., Ma, Q., and Jiang, C. M. (2013). Microbial biomass and community composition in a luvisol soil as influenced by long-term land use and fertilization. Catena 107, 89–95. doi:10.1016/j.catena.2013.02.010
Zeng, J., Liu, X. J., Song, L., Lin, X. G., Zhang, H. Y., Shen, C. C., et al. (2016). Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 92, 41–49. doi:10.1016/j.soilbio.2015.09.018
Zhang, Q., Zhou, W., Liang, G. Q., Sun, J. W., Wang, X. B., and He, P. (2015). Distribution of soil nutrients, extracellular enzyme activities and microbial communities across particle-size fractions in a long-term fertilizer experiment. Appl. Soil Ecol. 94, 59–71. doi:10.1016/j.apsoil.2015.05.005
Keywords: long-term fertilization, fertilizer types, agro-pastoral ecotone, soil physicochemical property, microbial community composition
Citation: Gao R, Duan Y, Zhang J, Ren Y, Li H, Liu X, Zhao P and Jing Y (2022) Effects of long-term application of organic manure and chemical fertilizer on soil properties and microbial communities in the agro-pastoral ecotone of North China. Front. Environ. Sci. 10:993973. doi: 10.3389/fenvs.2022.993973
Received: 14 July 2022; Accepted: 23 August 2022;
Published: 13 September 2022.
Edited by:
Giulio Castelli, University of Florence, ItalyReviewed by:
Qin Qin, Shanghai Academy of Agricultural Sciences, ChinaCopyright © 2022 Gao, Duan, Zhang, Ren, Li, Liu, Zhao and Jing. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peiyi Zhao, emhweTE5NzJAMTYzLmNvbQ==; Yupeng Jing, anlwMjM2QDE2My5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.