
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci., 07 October 2022
Sec. Water and Wastewater Management
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.990855
Wastewater discharges by textile industries constitute a major environmental concern as they exert grave human health dangers. Innovative, cost-effective and economical strategy aimed at ameliorating the effect of textile effluent in the environment is highly desirable. Hence, this study investigated the decolorization efficiency of selective reactive azo dyes by textile effluent non-adapted Lysinibacillus sphaericus MTCC 9523 in optimized operational conditions. Visual color changes with 96.30% and 92.71% decolorization for Reactive Yellow F3R (RYF3R) and Joyfix Red RB (JRRB) elucidated by UV-visible spectrophotometric analysis affirmed decolorization of the dyes. Meanwhile, Fourier Transform Infrared (FTIR) Spectroscopy of the metabolites obtained after degradation further revealed the functional groups and loss of peak for azo group indicated the breaking of azo bond. On High-Performance Liquid Chromatography (HPLC) analysis, difference in control dye’s retention time compared to treated sample with peaks at 2.413 and 2.895 min for RYF3R and 2.466 min and 1.640 min for Joyfix Red RB supported biodegradation by this organism. The chemical identities of the metabolites revealed after Gas Chromatography-Mass Spectrometry (GC-MS) analysis was used in the prediction of each dye’s metabolic pathway of degradation based on their molecular weight and mass-to-charge ratio. They were found to be (2Z)-but-2-ene, 1,3,5-triazine, (3E)-penta-1,3-diene for RYF3R and (methylsulfonyl)benzene, phenol, buta-1,3-diene, 1,3,5-triazine-2,4(1H,3H)-dione, nitrobenzene for JRRB. Overall, the use of adapted and non-adapted microbes is highly promising as an efficient, cost-effective, time-saving and universal green strategy for biodegradation of textile effluent containing azo dyes.
The textile dyeing industry is one of the oldest and greatest employment providers, contributing significantly to the economic development of most developing nations. These industries use a wide variety of chemicals in their processing. Effluents and wastewater released into the environment indiscriminately by the textile industries is one of the most potent contributor to high chemical oxygen demand (COD) (Li et al., 2015). They generate waste effluents containing toxic dyes that pollute water and soil posing a serious threat to humans and the environment in many countries. They also serve as repositories for compounds with potential carcinogenic and mutagenic characteristics (Alves de Lima et al., 2007). Synthetic azo dyes are largely used by the textile manufacturing industries due to their broad application, color and light fastness properties and versatility (Bankole et al., 2019). Nevertheless, the depressing reality is that the need for azo dyes is associated with a substantial quantity of outflow, largely as recalcitrant compounds, that pose a significant hazard to the marine environments and significantly contaminate drinkable water sources (Carmen and Daniela 2012). Accumulation of sludge causes difficulties in proper disposal resulting in environmental pollution. They change the pH of rivers, lower oxygen levels, and decrease penetration of light, causing photosynthesis to be disrupted and the aquatic ecosystem to be harmed (Molina-Guijarro et al., 2009). Color also diminishes the water’s aesthetic appeal for reuse. The decline in availability of water in developing nations have made farmers resort to using textile wastewater which most times contain traces of dye contaminants (Kumar et al., 2016). Taken together, the aggravated levels of contaminant in wastewater which poses a major threat to the environment have made it expedient to fashion out an efficient treatment of textile industry effluents. Constant attention is being given to the development of innovative, eco-friendly and cost-effective technologies for the treatment of textile dye-containing wastewater. Various physico-chemical methods deployed in the treatment of azo dyes have been proven to be effective and reasonably promising albeit uneconomical, energy-demanding and time-consuming and in most cases results in the accumulation of sludge which further aggravates environmental degradation (Forgacs et al., 2004; Zhang et al., 2004). Hence, an eco-friendly and efficient alternative for the degradation of azo dyes is required.
Microorganisms being natural recyclers, have been studied for decades by scientists worldwide for application at the field level. Pollutants are physiologically or genetically predisposed to be broken down by bioremediation with the natural ability of microorganisms that use them as food or energy sources in their metabolism (Vidali 2001). Basically, the mechanism involved in decolorization process relies largely on the active induction of oxido-reductase enzymes (Wu et al., 2010). The large scale application of this eco-friendly method depends on the optimization of operational condition for enhanced dye treatment processes. Bacterial degradation of azo dyes has attracted the attention of several workers over the years due to its relative efficiency, economical and vast applicability (Saratale et al., 2011). Microbes are selected for decolorization activities because of their immense capacity to adapt favorably to varying environmental conditions. Moreover, in a bid to thrive in such environment, they typical secrete enzymes which are capable of mineralizing diverse contaminants. Depending on their nature and growth, bacterial systems may be aerobic, anaerobic or facultatively anaerobic. Bacteria proliferate fast and easily with relatively high fecundity in both aerobic and anaerobic conditions which in turn aids in the induction of selective and non-selective enzymes (Solís et al., 2012). They are regarded as an environmentally safe solution for the breakdown and full disintegration of a diverse variety of dyes employing anaerobic, aerobic, microaerophilic, or combined anaerobic-aerobic processes that result in non-toxic final products (Forgacs et al., 2004). These enzymes produced by bacteria are well-suited to treating dye-contaminated textile wastewaters because of their tolerance to heat, different substrate sensitivity, short production period, and capacity to function in neutral to alkaline pH levels (Lee et al., 2003; Wells et al., 2006; Hilden et al., 2009; Reiss et al., 2011).
Aeromonas, Bacillus as well as Pseudomonas microbes obtained from diverse wastewater sites were observed to degrade a variety of dyes, despite the time and effort required to isolate such organisms (Banat et al., 1997). Two Stenotrophomonas strains isolated from textile effluent in Mexico were found to decolorize different textile dyes at various concentrations as confirmed by Fourier-transform infrared spectroscopy and ultra-performance liquid chromatography-mass spectrometry (Vilchis-Carmona et al., 2021). Optimization of operational environmental conditions brings about enhanced microbial growth and faster degradation activity for efficient dye treatment processes thus reducing them to less harmful metabolites through enzymatic digestion. As a result, given optimum process conditions of required nutrients, ideal pH, proper aeration-agitation, presence of the donors as well as receptors of electrons) these microorganisms can eliminate pollutants in the environment. The bacteria could be native to the contaminated area or isolated elsewhere and transported here, where they transform the toxicants through their metabolic processes.
Any organic substance can be degraded naturally or through evolution and adaptation by using the right strains in the right environment (Gayle 1952). Textile effluent-adapted soil microorganisms found in contaminated regions are effective at decomposing azo dyes, according to empirical evidence. However, the screening, identification, and optimization processes are time-consuming and do not always result in the isolation of viable strains suitable for industrial use. Discovering an aptly characterized non-adapted microbe to evaluate one’s potential and specific activity for pollutant degradation using the literature and exhaustive carefully selected data sources currently available in culture collections and bioinformatics-based predictive databases, on the other hand, is important to recognize to decrease the initial screening period. Non-native strains are also capable of reducing any dye with an aromatic ring.
Various researchers have provided evidence that diverse endogenous bacteria isolates adapt easily to the dye wastewater rich environment and have shown promising potentials to degrade azo dyes. Due to the prevalence of a range of chemical substances, it has been demonstrated that a horizontally movable microbe genetic pool plays a key role in the acclimation to stressful conditions (Frost et al., 2005; Thomas and Nielsen, 2005). The transfer of some genes horizontally aids in the rapid microbial transformation of xenobiotic substances, and the genes for degradative enzymes are transported by some movable genetic elements like transposons as well as plasmids or could be located inside the chromosome themselves (Sinha et al., 2009). However, some authors have reported azo dyes decolorization by non-adapted bacterial strains isolated from sources other than the textile effluents, different geographic regions and environments (Chen et al., 2003; Leena and Selva Raj 2008; Hsueh et al., 2009; Bouraie and Din 2016; Srinivasan and Sadasivam 2021). As a result, genes encoding for the bacterial enzymes that can break the dyes do not always need to be encoded using plasmids. Inherent bacteria genes are naturally over-expressed in high stressed condition such as exposure to toxic azo dyes resulting in their degradation. Thus, well-characterized non-adapted strains possessing azo dye degrading abilities naturally or through genetic manipulation can be regarded as potential candidates for application in textile effluent bioremediation. Thus, with this understanding, this study intended to appraise the prospect to decolorize and degrade textile azo dyes, i.e., Reactive Yellow F3R (RYF3R) and Joyfix Red RB (JRRB) utilizing a textile effluent non-autochthonous bacteria Lysinibacillus sphaericus MTCC 9523. There is an abundance of material on dye degradation, but there still does not seem to be a workable answer. Since dye degrading enzymes are naturally present but perform best under specific inductive conditions, we need to shift our attention, time, and resources away from textile effluent-polluted soil or water microbes and toward a futuristic and viable approach that uses non-native bacterial strains for bioremediation due to their capacity to degrade a wide range of dye toxicants without specificity. Thus, this innovative study reports effective decolorization as well as degradation of two extensively used textile azo dyes, Reactive Yellow F3R and Joyfix Red RB, by the L. sphaericus MTCC 9523 textile effluent non-adapted strain. It validated our novel goal of using it as an easy, less expensive, and more effective alternative for changing wastewater treatment because it showed good decolorization percentage and degraded the metabolites.
The Reactive Yellow F3R (RYF3R) and Joyfix Red RB (JRRB) dyes used in this study were obtained through a generous gift by a textile factory in Tamil Nadu, India. A spectrophotometer was used to find dye the maximum absorption wavelength for dyes by UV-Vis spectroscopy (UV–Vis, 1800; Shimadzu, Japan). The composition of chemicals in the nutrient medium was purchased from Hi Media Laboratory, Mumbai, India. Other chemicals used were of high purity and analytical grade.
IMT (Institute of Microbial Technology) in Chandigarh housing the Microbial Type Culture Collection and Gene Bank (MTCC), India provided L. sphaericus MTCC 9523, validated by biochemical tests and pure culture is retained at 4°C in nutrient agar slants having Beef and Yeast extract 1.5 g L−1 each, peptone and NaCl 5.0 g L−1 each, along with 15.0 g L−1 agar. Bergey’s Manual of Determinative Bacteriology was used to characterize the organisms visually and biochemically.
Bacteria were cultivated till the log phase in the nutrient medium at 37 ± 2°C in a shaking setting of 120 rpm for 16 h at pH 7. They were used for the decolorization studies with 5% of bacterial inoculum, 100 ml broth of nutrient media, 100 mg L−1 of azo dye, at a pH of 8 temperature of 37°C for 72 h in an aerobic (shaking at 120 rpm, 24 h)-microaerophilic (static) environment. The optimization of operations condition was carried out in our earlier studies (Kumar et al., 2016; Srinivasan and Sadasivam, 2018). An aerobic (shaking) environment provided oxygen for the efficient growth of bacterial biomass on dye-containing medium, and microaerophilic (static) conditions were effective for the decolorization of azo dyes (Ogugbue et al., 2012). Due to the fact that oxygen hinders the activity of azoreduction, oxygen deprivation in microaerophilic cultures enables facultative anaerobic bacteria to dissimulate the reduction of azo dyes. It was found that maintaining the flasks in shaking (120 rpm for about 18–24 h) till Optical Density 1.0 is reached was optimal for obtaining bacterial culture in lag phase so that all the enzymes may have been produced. Once maximum bacterial biomass culture is reached, it is followed by keeping the flask at static conditions and observing it for maximum decolorization till 72 h. Keeping the flask further did not increase the decolorization % further. Hence, the strategy to keep the flask in shaking (aerobic) for 24 h and further in static (microaerophilic) conditions was adopted. Samples were withdrawn periodically from the flasks for centrifugation. The clear supernatant obtained were used in the measuring of the absorbance. Decolorization percentages and indeed decolorization efficiency were evaluated using absorbance measurements taken at respective λmax of dyes. Control flasks (devoid of microorganisms) were included in the study.
UV–Vis spectrophotometry was used to undertake qualitative characterization of dyes and biologically damaged dye samples (UV-1800, Shimadzu, Japan). After the decolorization was completed, the supernatant was obtained from the culture broth by centrifugation in an Eppendorf centrifuge (5804 R, Germany) at 10,000 rpm, 10 min with the same amount of ethyl acetate for extracting dried metabolites and suspended in methanol to store till investigation. Shimadzu LC40102010 instrument with a built-in detector for dual absorbance and C18 column was used to perform HPLC at room temperature. The mobile phase consisted of HPLC grade methanol flowing at 1.0 ml min−1 for a while of 10 min at 470 nm. The functional groups were identified in biodegraded samples through FTIR scanned in the range of 400–4,000 cm−1 at a speed of 16 with the instrument PerkinElmer (spectrum RX I). The results of the treated sample were compared with the control dye. Pure KBr (potassium bromide) was mixed with samples in 5:95 ratios to make pellets to secure in the holder. Gas chromatography (45XGC-44 GC)-mass spectroscopy had been used to determine the mass spectrum in degradation compounds and correlate them to fragmented patterns in the NIST library (MS-40 Scion, Bruker). It was performed at 70 eV ionization voltage made of a column of DB-WAX with dimensions of 0.25–30 mm in programming mode for temperature with initially the column’s temperature set at 80°C, 2 min, in addition, amplified to a temperature of 250°C increasing at a rate of 10°C min−1. In this process, Helium was utilized as the carrier gas flowing at 1.0 ml min−1 for 26 min.
L. sphaericus MTCC 9523 was incubated in the nutrient broth at the temperature of 37°C, 24 h, pH 7 and supernatant was extracted on centrifugation for 10 min at 10,000 rpm. Cells were sonicated at 4°C on 40 amp with 8 strokes, each one for 30 s, with an intermission of 2 min and were combined in potassium phosphate buffer of pH 7.4 (50 mM). The supernatant obtained after centrifugation of the crude extract was utilized as an enzyme source. The fully decolorized samples were used to obtain products similarly. The extract was used to assess the presence of laccase and azoreductase enzymes as per the protocols described by Hatvani and Mecs (2001) and Olukanni et al. (2010). The experiments were performed in triplicates to measure the average value of enzyme activity. Blank comprised all components except the enzyme in the tests.
The compounds obtained after L. sphaericus MTCC 9523 decolorized and degraded the azo dyes, namely Reactive Yellow F3R and Joyfix Red RB were compared using UV–Vis, FTIR, HPLC, and GC-MS analysis for identifying and confirming the outcomes of degradation for the chosen dyes by the bacteria. The dyes used in this work are commonly used azo group of dyes and were procured directly from the textile industry in Tamil Nadu. In several of our prior experiments, these dyes also demonstrated good decolorization as well as degrading capabilities (Kumar et al., 2016; Srinivasan et al., 2019). As a result, these have been chosen for this work.
UV-vis analysis of Reactive Yellow F3R control dye showed maximum absorbance at 450 nm and for Joyfix Red RB at 520 nm and disappearance of these peaks in treated samples indicated decolorization of these dyes (Figure 1). At 72 h, there was a change in the maximum peak of Reactive Yellow F3R, as well as 96.30% decolorization by L. sphaericus MTCC 9523. The disappearance of λmax peak of Joyfix Red RB at 520 nm along with 92.71% decolorization by L. sphaericus MTCC 9523 was observed at 72 h (Supplementary Figure S1; Table 1). At 72 h, the maximum peak of Drimaren Red CL-B at 520 nm and Remazol Yellow RR at 450 nm had vanished, accompanied by 88.35% and 90.66% decolorization by L. sphaericus MTCC 9523 respectively (Srinivasan, 2018; Srinivasan and Sadasivam 2018). Visual color change and comparison of peaks obtained by UV-vis analysis of control and dye samples treated by L. sphaericus MTCC 9523 indicated decolorization of these azo dyes. Keeping the flask further did not increase the decolorization percentage additionally. Hence, further decolorization till 96 h was not included in the study.
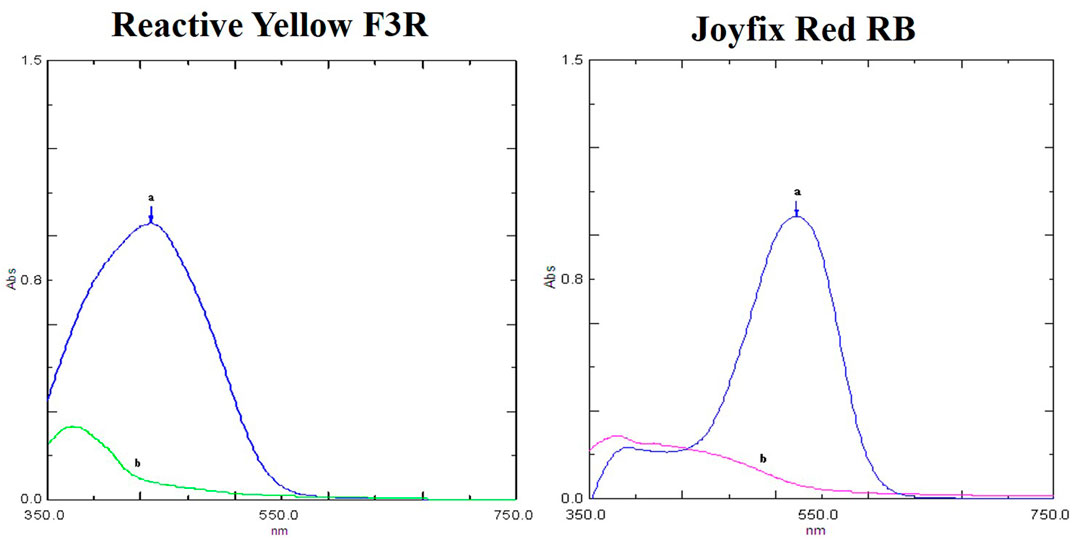
FIGURE 1. UV-vis spectra of dyes (A) before and (B) after decolorization by L. sphaericus MTCC 9523.
For understanding decolorization mechanism, activities of oxidoreductive enzymes namely laccase and azoreductase were tested. L. sphaericus MTCC 9523 non-autochthonous bacteria displayed higher activities for laccase and azoreductase enzymes while degrading the dyes. Laccase activity was highest for Joyfix Red RB degradation while azoreductase activity was highest during degradation of Reactive Yellow F3R. An enhancement in enzyme activities of both enzymes in the samples after decolorization when related to control indicate a combinatorial action of the oxidoreductive enzymes for decolorization as well as biodegradation of azo dyes to simpler products by the non-adapted bacteria (Table 2). Changes in the concentration of important oxidoreductive enzymes were studied widely for their dye decolorizing capabilities (Saratale et al., 2013). In our earlier studies, the strains namely Aeromonas hydrophila MTCC 1739 and L. sphaericus MTCC 9523 exhibited higher activities for laccase, veratryl alcohol oxidase and azoreductase enzymes after decolorization of Drimaren Red CL-5B dye compared to pure culture (Srinivasan and Sadasivam 2018). When exposed to Remazol Yellow RR dye also, these enzyme concentrations amplified while using L. sphaericus MTCC 9523 for degradation (Srinivasan 2018). Non-adapted Bacillus bacteria presented greater dye degradation in comparison to adapted strain (Olukanni et al., 2006). A. hydrophila MTCC 1739 non-adapted bacteria revealed better activity for all three oxidoreductive enzymes when degrading Remazol Yellow RR, Reactive Yellow F3R, Joyfix Red RB (Srinivasan et al., 2021). Hence, well-described, easily available non-autochthonous bacterial cultures may be utilized for their capability to degrade toxic azo dyes due to the presence of suitable enzymes (Srinivasan et al., 2020).
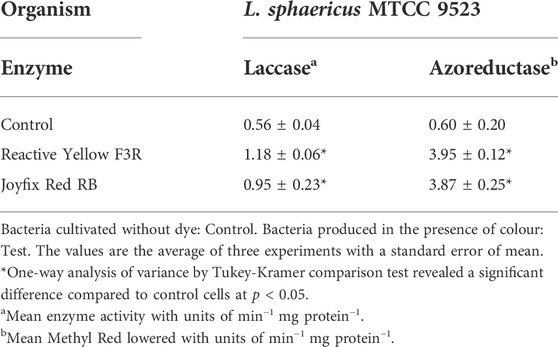
TABLE 2. Laccase and azoreductase enzyme activities of L. sphaericus MTCC 9523 before and after dye decolorization.
Figure 2 showed the HPLC study of control as well as the biodegraded dyes through chromatograms. The RYF3R control dye’s retention time dye sample was 2.495 min, which corresponded to the retention time of pure reactive dye. When this dye was decolorized by the L. sphaericus MTCC 9523, it led to the formation of new peaks at 2.413 and 2.895 min which show that the dye has broken down into smaller molecules. As a result of the change in peak pattern as well as absorbance, HPLC examination supported the biodegradation of Reactive Yellow F3R by this organism.
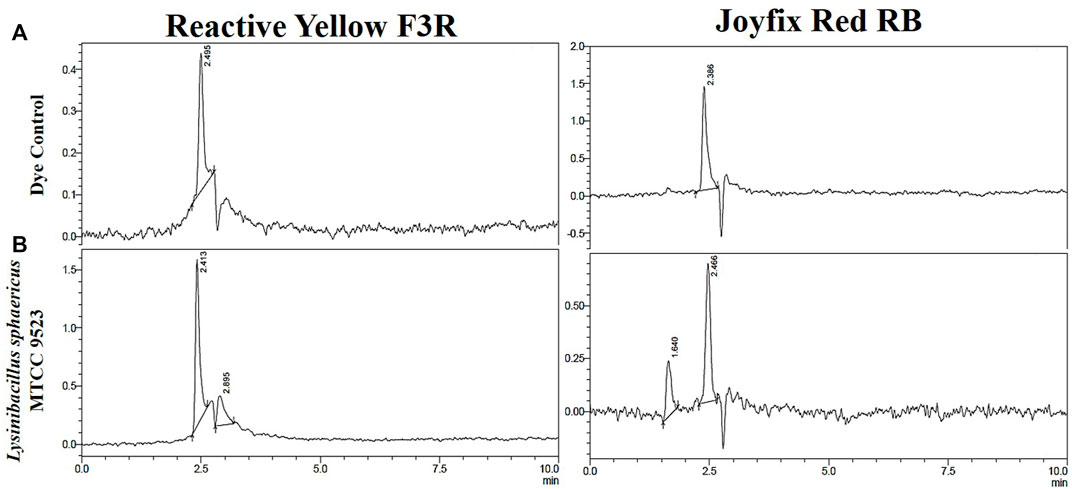
FIGURE 2. HPLC profile of (A) Reactive Yellow F3R, Joyfix Red RB dyes and (B) degradation products by L. sphaericus MTCC 9523.
The HPLC examination of the control Joyfix Red RB dye sample revealed a strong peak at 2.386 min retention duration. Decolorization by L. sphaericus MTCC 9523 resulted in a drop in peak intensity at 2.466 min and the appearance of a new peak at 1.640 min, indicating that the dye had been reduced into smaller molecules (Figure 2).
The HPLC-based investigation of Remazol Yellow RR control dye and the decolorized product showed a noticeable difference in the peak patterns supporting the biodegradation of Remazol Yellow RR by L. sphaericus MTCC 9523 (Srinivasan, 2018) (Figure 2). The comparable difference in the retention time and peaks of pure DRCL-B and this dye’s decolorization through L. sphaericus MTCC 9523 indicated that the dye has broken down into smaller molecules (Srinivasan and Sadasivam, 2018) (Figure 2). HPLC elution profile of Navy blue with a peak at retention time (RT) 2.77 and the appearance of smaller but prominent peaks with 2.28–2.50 min RT confirming the breakdown of dye into simpler metabolites (Dhanve et al., 2008). According to Sahasrabudhe et al. (2014), the peaks with retention times of 2.56 and 2.78 min were seen in the HPLC chromatogram of the RR 195 dye which was decolorized using M. glutamicus NCIM 2168, whereas the reference dye peaks had been at 1.95 along with 2.25 min. HPLC of RY-84A dye showed a major peak at RT 3.0 min and four minor peaks at different RT, while RY-84A degraded by Galactomyces geotrichum showed three peaks at RT 2.3, 2.7, 2.9 min indicating degradation of RY-84A (Govindwar et al., 2014). According to Telke et al. (2008), the HPLC analysis of Reactive Red 141 revealed a medium peak at 1.89 min and a sharp peak at 2.243 min, whereas the metabolites produced by Rhizobium radiobacter MTCC 8161 had two sharp peaks at 2.241 and 2.498 min and minor peaks at 4.80 and 6.84 min.
Loss of peak for an azo group indicates the breaking of azo bond by azoreductase (Figure 3). The biodegradation of the dye molecule into distinct metabolites by L. sphaericus MTCC 9523 was shown by differences in FTIR spectra of control dyes and its derivatives. Different peaks signifying distinct functional groups were visible in the FTIR spectrum of the control Reactive Yellow F3R (RYF3R), Remazol Yellow RR (RYRR), Joyfix Red RB (JRRB) dyes (Srinivasan and Sadasivam 2021).
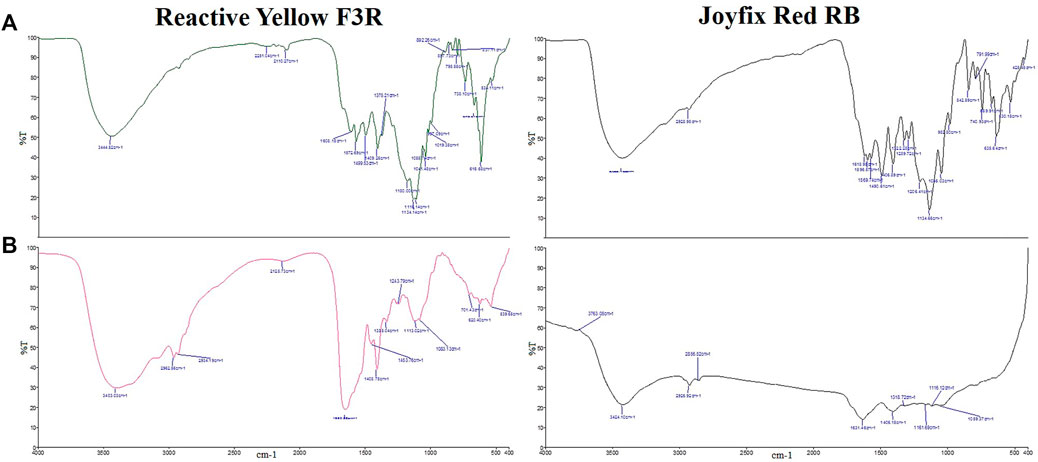
FIGURE 3. FTIR spectra of (A) Reactive Yellow F3R, Joyfix Red RB control dyes and (B) degradation products by L. sphaericus MTCC 9523.
The FTIR spectrum of the products formed after decolorization of Reactive Yellow F3R by L. sphaericus MTCC 9523 showed different peaks at 3,403.03 cm−1 for N-H stretching of primary and secondary amines, 2,962.56 cm−1 for C-H stretching of alkanes, 2,934.19 cm−1 for O-H stretching of chelate compounds, 2,128.73 cm−1 for NH+ vibrations of charged amine derivatives, 1,650.88 cm−1 for C=C stretching of alkenes, 1,453.76 cm−1 for C= S stretching, 1,408.75 cm−1 for S = O stretching of sulfonic acids, 1,335.04 cm−1 for C-N vibrations in aromatic amines, 1,243.79 cm−1 for N-H deformation plus C-N stretching of amides, 1,113.02 cm−1 for C-O stretching of secondary and tertiary alcohols, 1,083.13 cm−1 for C-O stretching of acyclic anhydrides, 701.43 cm−1 for C-N stretching of acyclic compounds, 620.40 cm−1 for C-S stretching and 539.65 cm−1 for N-H deformation plus C-N stretching of acyclic compounds. Disappearance of peak at 1,608 cm−1 for an azo group clearly indicates the breaking of azo bond by azoreductase (Figure 3). Difference in FTIR spectra of Reactive Yellow F3R and its products indicated the biodegradation of dye molecule into different metabolites by L. sphaericus MTCC 9523.
FTIR spectra of metabolites collected after decolorization revealed the loss of several peaks and the emergence of new peaks, indicating successful biotransformation of Joyfix Red RB by L. sphaericus MTCC 9523. Peaks were observed at 3,424.10 cm−1 for N–H stretching vibrations of a secondary amide and primary amines, 2,926.92 and 2,856.52 cm−1 for C-H stretching in alkanes, 1,631.46 cm−1 for C=N stretching vibrations in cyclic α, β unsaturated compounds and oximes, 1,406.15 cm−1 for O-H deformation in phenols and C=S stretching, 1,318.72 cm−1 for C–N vibrations in aromatic amines, 1,161.69 cm−1 for S=O asymmetric stretching in sylfates and sylfonamides, 1,116.12 cm−1 for C-O stretching, 1,059.37 cm−1 for C-O stretching of alcohols and ring C-H deformations (Figure 3).
FTIR of Remazol Yellow RR degraded by L. sphaericus MTCC 9523 showed several peaks for N-H stretching of amides and amines, C-H stretching of alkanes, C-H stretching of aldehydes, stretching of C=O or C=C in alkenes or amides, C=S stretching, O-H deformation of alcohols, S=O stretching of sulfites, C-O stretching of acyclic anhydrides, N-H deformation plus C-N stretching of acyclic compounds (Srinivasan, 2018).
The Drimaren Red CL-B’s FTIR spectra after treatment with L. sphaericus MTCC 9523 showed peaks for –N=O stretching in aromatic intermediates supporting the formation of naphthalene, N–H stretching of secondary amides, S=O stretching, O-H bond vibration, C–O stretching in phenols, and C-H stretching with a lack of an amine group within 1,630–1,575 cm−1 range suggesting an outright removal of the azo group (Srinivasan and Sadasivam, 2018).
Similarly, Dhanve et al. (2009) and Govindwar et al. (2014) found differences in the FTIR spectrum of original dye and bacteria-degraded reactive yellow 84A. Priyadharshini et al. (2016) observed significant variation in functional groups for Reactive Red 195 and Reactive Orange 105 and real textile effluent after the combined treatment process by FT-IR analysis. Telke et al. (2008) also reported substantial biodegradation indicated by a difference in the FTIR spectrum of Reactive Red 141 and the degradation product. Loss of bands between 900 and 675 cm−1 confirmed the mineralization of aromatic rings of azo dyes (Balapure et al., 2016).
The deployment of Brevibacillus laterosporus and G. geotrichum as a novel bacteria-yeast consortium accounted for the loss of major peaks and occurrence of new peaks in FTIR spectrum of dye decolorized metabolites implying efficient biotransformation and mineralization of RR 198 into simple metabolites (Kurade et al., 2011). FTIR spectrum of 6 h extracted metabolites of Reactive Red 2 degraded by Pseudomonas sp. SUK1 compared to the control dye, revealed a substantial difference in peak locations indicating the breaking of azo linkage and formation of phenyl radical intermediates (Kalyani et al., 2009). Franciscon et al. (2009) reported a sequential treatment of azo dyes by a facultative Klebsiella sp. strain VN-31 in a microaerophilic-aerobic environment isolated from the activated sludge process of textile industry where during microaerophilic decolorization bands in 1,610–1,630 cm−1 and 1,402 cm−1 region disappeared and two new bands appeared attributing to amine formation which disappeared during aerobic stage with a new peak at 1,680 cm−1 denoting oxidative biodegradation of the amines. The differences in the FTIR spectra of Congo red and the metabolites obtained after decolorization showed no peak for N=N extending at 1,584 cm−1 for degradation products denoting cleavage of azo bond in Congo red by S. Xiamenensis BC01 (Ng et al., 2014). Biotransformation of reactive textile dye Red BLI into the nitrosocompound, oximes and imines by Pseudomonas sp. SUK1 was confirmed by FTIR analysis (Kalyani et al., 2008). Biodegradation of Reactive Red 141 to the metabolites namely naphthalene diazonium, p-dinitrobenzene, as well as naphthol using R. radiobacter MTCC 8161 was indicated by a significant difference in FTIR spectra of Reactive Red 141 and degraded metabolites (Telke et al., 2008). FTIR analyses of control and decolorized products confirmed degradation of Reactive Red 198 by Tagetes patula L. (Marigold) hairy roots (Patil et al., 2009). Similarly, a combination of dyes was degraded by a consortium of bacteria called ASD, giving small molecular weight products like naphthalene, aniline along with benzene (Chattaraj et al., 2015).
The breakdown of Reactive Yellow F3R by L. sphaericus MTCC 9523 was investigated via GCMS for probable detection of different metabolites relying and a probable degradation mechanism was identified based on mass/charge readings, molecular weight, and chemical structure (Figure 4). The azoreductase cleaved the azo bond of standard dye, resulting in intermediates I and II. Subsequently, non-symmetric fragmentation of intermediate I through laccase resulted in the emergence of intermediate III, 6-chloro-1,3,5-triazine-2,4-diamine (Molecular Weight: 143.55; mass/charge ratio: 142.9; Retention Time 15.201), and 1-(2-aminophenyl)urea (Molecular Weight: 151.165; mass/charge ratio: 152.8; Retention Time 16.935), which contributed to the formation of benzene (Molecular Weight: 78.11; mass/charge ratio: 77; Retention Time 16.209) and (2Z)-but-2-ene (Molecular Weight: 56.11; mass/charge ratio: 57; Retention Time 17.548) from intermediate III, 2-chloro-1,3,5-triazine (Molecular Weight: 115.52; mass/charge ratio: 116; Retention Time 16.553) and 1,3,5-triazine (Molecular Weight: 81.08; mass/charge ratio: 83; Retention Time 17.728), and aniline (Molecular Weight: 93.13; mass/charge ratio: 90.9; Retention Time 17.299). Intermediate II was transformed to naphthalen-1-amine (Molecular Weight: 143.19; mass/charge ratio: 142.9; Retention Time 15.201), generating naphthalene (Molecular Weight: 128.17; mass/charge ratio: 129.8; Retention Time 16.209) and (3E)-penta-1,3-diene (Molecular Weight: 68.11; mass/charge ratio: 69.9; Retention Time 16.935) as products (Table 3).
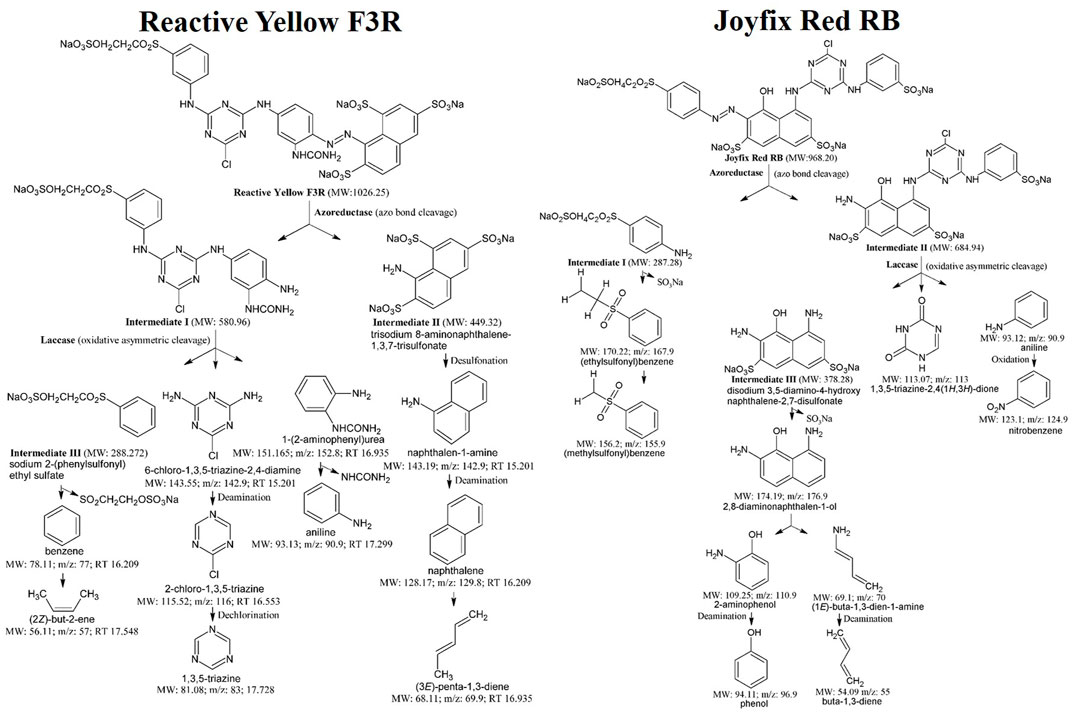
FIGURE 4. Proposed pathways for degradation of Reactive Yellow F3R, Joyfix Red RB dyes by L. sphaericus MTCC 9523.
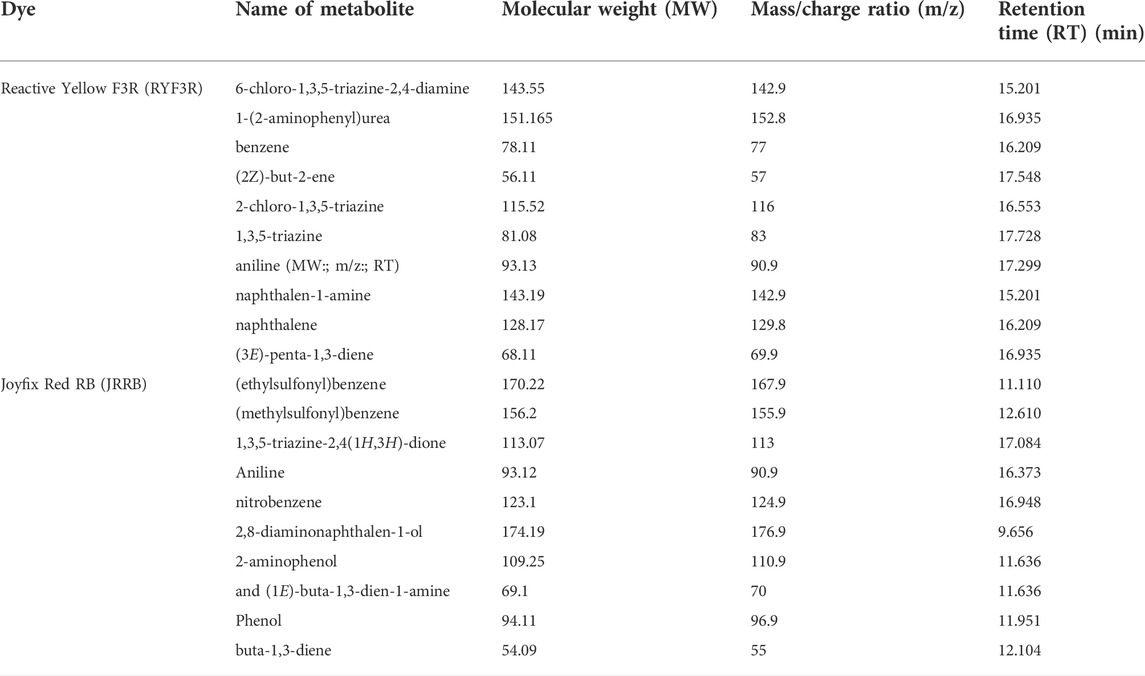
TABLE 3. GCMS spectral data of biodegradation products of RYF3R and JRRB, degraded by L. sphaericus MTCC 9523.
The mass spectra, molecular weight, and chemical structure of Joyfix Red RB (MW: 968.20) were used to conduct a GC-MS analysis of biodegraded metabolites. The action of the azo reductase enzyme was responsible for the disintegration of Joyfix Red RB dye’s azo bond leading to the formation of intermediates I and II (Figure 4). The mineralization of azo dye by L. sphaericus MTCC 9523 is caused by the breakdown of aromatic amines to some aliphatic compounds, resulting in the generation of smaller molecular weight molecules (Oturkar et al., 2011). Depending on GCMS evaluation of degraded compounds and previous literature, these species’ degradation pathways for Joyfix Red RB were proposed. The Intermediate I (MW: 287.28) undergoes desulfonation generating (ethylsulfonyl)benzene (Molecular Weight: 170.22; mass/charge ratio: 167.9; Retention Time: 11.110 min), subsequently forming (methylsulfonyl)benzene (Molecular Weight: 156.2; mass/charge ratio: 155.9; Retention Time: 12.610 min). Further, oxidative asymmetric cleavage by laccase on Intermediate II (Molecular Weight: 684.94) lead to formation of Intermediate III disodium 3,5-diamino-4-hydroxy naphthalene-2,7 disulfonate (Molecular Weight: 378.28), 1,3,5-triazine-2,4(1H,3H)-dione (Molecular Weight: 113.07; mass/charge ratio: 113; Retention Time: 17.084 min) as well as aniline (Molecular Weight: 93.12; mass/charge ratio: 90.9; Retention Time: 16.373 min) that is oxidized to nitrobenzene (Molecular Weight: 123.1; mass/charge ratio: 124.9; Retention Time: 16.948 min). Intermediate III is desulfonated to give 2,8-diaminonaphthalen-1-ol (Molecular Weight: 174.19; mass/charge ratio: 176.9; Retention Time: 9.656 min) which is then broken down into 2-aminophenol (Molecular Weight: 109.25; mass/charge ratio: 110.9; Retention Time: 11.636 min) as well as (1E)-buta-1,3-dien-1-amine (Molecular Weight: 69.1; mass/charge ratio: 70; Retention Time: 11.636 min) which is then transformed into phenol (Molecular Weight: 94.11; mass/charge ratio: 96.9; Retention Time: 11.951 min) as well as buta-1,3-diene (Molecular Weight: 54.09 mass/charge ratio: 55; Retention Time: 12.104 min) respectively (Table 3). Hence, the formation of lower molecular weight intermediates suggests the mineralization of the dyes due to the dissipation of enzymes by L. sphaericus MTCC 9523. The GCMS peaks and related data was shown in Supplementary Table S1.
The disintegration of Remazol Yellow RR with L. sphaericus MTCC 9523 by its oxidoreductive enzymes was investigated by GCMS for possible determination of metabolites. It revealed the formation of phenol, propanal and benzene as final degraded products (Srinivasan, 2018). Analysis of degradation products of Drimaren Red CL-B dye by azoreductase as well as laccase enzymes based on GCMS also resulted in the formation of low molecular weight metabolites such as naphthalene, phenol, and 2-aminophenol after treatment with L. sphaericus MTCC 9523 (Srinivasan and Sadasivam, 2018).
Sahasrabudhe et al. (2014) reported the formation of low molecular weight products on the degradation of Reactive Red 195 by Micrococcus glutamicus NCIM 2168. Priyadharshini et al. (2016) successfully deployed naphthalene-degrading bacteria consortia for the decolorization of Reactive Red 195 and Reactive Orange 105 dyes. They reported that the consortia exhibit capacity to cleave and mineralize aromatic intermediates into simpler organic acids and aldehydes were used. Similarly, Telke et al. (2008) reported degradation of Reactive Red 141 into naphthalene diazonium, 1, 3, 5-triazine 2, 4-diol, p-dinitrobenzene and 2-nitroso naphthol by R. radiobacter MTCC 8161. The genotoxic, cytotoxic, as well as phytotoxicity of the azo dye Reactive Orange 16 were studied. It was found that lesser toxic products were formed after its degradation using the bacteria Lysinibacillus sp. RGS (Bedekar et al., 2021).
To the best of knowledge, this is the first report on the efficient decolorization of Reactive Yellow F3R and Joyfix Red RB by the textile effluent non-autochthonous L. sphaericus MTCC 9523 bacteria. The use of non-adapted bacterial cultures with the capacity to decolorize and biodegrade a wide assortment of toxic azo dyes through a non-specific mechanism was explicated as an encouraging method for effluent treatment. Less/non-toxic end product formation substantiated our purpose of utilizing non-adapted bacteria as a simple, inexpensive and proficient substitute in the conventional treatment methodologies usually comprising a strenuous primary isolation, screening and identification process for the greatest dye degrading adapted bacteria. This is possible due to the presence of various oxidoreductive dye degrading enzymes inherently present in these organisms which start working well under a standardized condition. Standard cultures already researched and readily available in laboratories will lessen the time as well as budget of wastewater treatment. In future, genetic modifications may also enhance the enzyme’s activity and degradative ability to exist microorganisms and consortia to consume the dyes as selective carbon as well as nitrogen source. The autochthonous bacterial cultures can also be screened using bioinformatics tools and software for their effectiveness in degradation of textile dyes. Inter-disciplinary studies of dry lab and wet lab technologies can be a positive futuristic and active approach to bioremediation of textile effluent.
The future prospects include toxicity assessment, transforming laboratory research to real time treatment of textile industry wastewater. The in silico studies comprising analysis of azo dye degrading proteins of any organism, modeling and enzyme-dye docking studies could be used to construct a dynamic computational interface for virtually selecting best species available in the vicinity for actual microbial bioremediation. Finally, the nature of degraded metabolites after treatment need to be investigated by microbial toxicity, cytotoxicity and phytotoxicity analysis prior to large scale bioremediation.
This is the first extensive report on efficient degradation of reactive azo dyes Reactive Yellow F3R and Joyfix Red RB by textile effluent non-adapted L. sphaericus MTCC 9523 bacteria. Adapted and non-adapted bacterial strains have proven to exhibit great capacity in the biodegradation of vast majority of textile azo dyes which elucidated their promising potentials in eco-friendly remediation technology. Here, the L. sphaericus showed great potency in reducing the reactive textile azo dyes from high molecular weights to less molecular weight metabolites. Treatment of RYF3R and JRRB dyes led to the formation of simpler metabolites as revealed by GC-MS analysis. These metabolites were used in the prediction of metabolic pathway of degradation of each dye. The enzyme analysis revealed roles played by azo reductase and laccase in the degradation of the dyes. This current study further affirmed the efficiency of non-adapted bacteria strains as an eco-friendly, less-laborious, economical strategy for the treatment of dye wastewater. Going forward, deploying the knowledge of genetic engineering and molecular biology to enhance induction of enzymes and degradation efficiency of the strains will promote bioremediation of organic pollutants as a promising strategy. It is however recommended that there should be a synergy between academia and industries on the effective application of this technology on the bioremediation of dye contaminated sites to ensure safe environment for humans, plants, and animals.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
ShS: Conceptualization, experimental design, methodology, statistical analysis, writing—original draft preparation, investigation, visualization, validation. PB: Experimental design, methodology, statistical analysis, writing—reviewing and editing. SeS: Writing—review and editing of manuscript original draft, supervision.
We wish to express our profound gratitude to the authorities of National College (Autonomous), Tiruchirappalli, India and Kalasalingam University, Krishnankoil, India for logistics and experimental support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.990855/full#supplementary-material
SUPPLEMENTARY FIGURE S1Analysis of decolorization of chosen textile dyes by Lysinibacillus sphaericus MTCC 9523 at various time intervals.
Alves de Lima, R. O., Bazo, A. P., Salvadori, D. M., Rech, C. M., de Palma Oliveira, D., and de Aragão Umbuzeiro, G. (2007). Mutagenic and carcinogenic potential of a textile azo dye processing plant effluent that impacts a drinking water source. Mutat. Research/Genetic Toxicol. Environ. Mutagen. 626, 53–60. doi:10.1016/j.mrgentox.2006.08.002
Balapure, K., Jain, K., Bhatt, N., and Madamwar, D. (2016). Exploring bioremediation strategies to enhance the mineralization of textile industrial wastewater through sequential anaerobic-microaerophilic process. Int. Biodeterior. Biodegrad. 106, 97–105. doi:10.1016/j.ibiod.2015.10.008
Banat, I. M., Nigam, P., McMullan, G., Marchant, R., and Singh, D. (1997). The isolation of thermophilic bacterial cultures capable of textile dyes decolorization. Environ. Int. 23, 547–551. doi:10.1016/s0160-4120(97)00061-5
Bankole, P. O., Adekunle, A. A., and Govindwar, S. P. (2019). Demethylation and desulfonation of textile industry dye, Thiazole Yellow G by Aspergillus niger LAG. Biotechnol. Rep. (Amst). 20, e00327. doi:10.1016/j.btre.2019.e00327
Bedekar, P. A., Saratale, R. G., Saratale, G. D., and Govindwar, S. P. (2014). Oxidative stress response in dye degrading bacterium Lysinibacillus sp. RGS exposed to reactive orange 16, degradation of RO16 and evaluation of toxicity. Environ. Sci. Pollut. Res. 21, 11075–11085. doi:10.1007/s11356-014-3041-2
Bouraie, M. E., and Din, W. S. E. (2016). Biodegradation of Reactive Black 5 by Aeromonas hydrophila strain isolated from dye-contaminated textile wastewater. Sustain. Environ. Res. 26, 209–216. doi:10.1016/j.serj.2016.04.014
Carmen, Z., and Daniela, S. (2012). “Textile organic dyes-characteristics, polluting effects and separation/elimination procedures from industrial effluents- a critical overview,” in Organic pollutants ten years after the Stockholm convention-environmental and analytical update (Croatia: InTech), 55–81.
Chattaraj, S., Johnson, J., and Madamwar, D. (2015). Biotransformation of mixture of dyes by enriched bacterial consortium ASD. Desalin Water Treat., 1–13. doi:10.1080/19443994.2015.1124345
Chen, K. C., Wu, J. Y., Liou, D. J., and Hwang, S. C. J. (2003). Decolorization of the textile dyes by newly isolated bacterial strains. J. Biotechnol. 101, 57–68. doi:10.1016/s0168-1656(02)00303-6
Dhanve, R. S., Kalyani, D. C., Phugare, S. S., and Jadhav, J. P. (2009). Coordinate action of exiguobacterial oxidoreductive enzymes in biodegradation of reactive yellow 84A dye. Biodegradation 20, 245–255. doi:10.1007/s10532-008-9217-z
Dhanve, R. S., Shedbalkar, U. U., and Jadhav, J. P. (2008). Biodegradation of diazo reactive dye Navy blue HE2R (Reactive blue 172) by an isolated Exiguobacterium sp. RD3. J. Bioprocess Biotechnol. Eng. 13, 53–60. doi:10.1007/s12257-007-0165-y
Forgacs, E., Cserhati, T., and Oros, G. (2004). Removal of synthetic dyes from wastewaters: A review. Environ. Int. 30, 953–971. doi:10.1016/j.envint.2004.02.001
Franciscon, E., Zille, A., Dias, G. F., de Ragagnin, M. C., Durrant, L. R., and Cavaco-Paulo, A. (2009). Biodegradation of textile azo dyes by a facultative Staphylococcus arlettae strainVN-11 using a sequential microaerophilic/aerobic process. Int. Biodeterior. Biodegrad. 63, 280–288. doi:10.1016/j.ibiod.2008.10.003
Frost, L. S., Leplae, R., Summers, A. O., and Toussaint, A. (2005). Mobile genetic elements: The agents of open source evolution. Nat. Rev. Microbiol. 3, 722–732. doi:10.1038/nrmicro1235
Govindwar, S. P., Kurade, M. B., Tamboli, D. P., Kabra, A. N., Kim, P. J., and Waghmode, T. R. (2014). Decolorization and degradation of xenobiotic azo dye Reactive Yellow-84A and textile effluent by Galactomyces geotrichum. Chemosphere 109, 234–238. doi:10.1016/j.chemosphere.2014.02.009
Hatvani, N., and Mecs, I. (2001). Production of laccase and manganese peroxidase by Lentinus edodes on malt containing by product of the brewing process. Process Biochem. 37, 491–496. doi:10.1016/s0032-9592(01)00236-9
Hilden, K., Hakala, T. K., and Lundell, T. (2009). Thermotolerant and thermostable laccases. Biotechnol. Lett. 31, 1117–1128. doi:10.1007/s10529-009-9998-0
Hsueh, C. C., Chen, B. Y., and Yen, C. Y. (2009). Understanding effects of chemical structure on azo dye decolorization characteristics by Aeromonas hydrophila. J. Hazard. Mat. 167, 995–1001. doi:10.1016/j.jhazmat.2009.01.077
Kalyani, D. C., Patil, P. S., Jadhav, J. P., and Govindwar, S. P. (2008). Biodegradation of reactive textile dye Red BLI by an isolated bacterium Pseudomonas sp. Bioresour. Technol. 99, 4635–4641. doi:10.1016/j.biortech.2007.06.058
Kalyani, D. C., Telke, A. A., Dhanve, R. S., and Jadhav, J. P. (2009). Eco-friendly biodegradation and detoxification of Reactive Red-2 textile dye by newly isolated Pseudomonas sp. SUK1. J. Haz Mat. 163, 735–742. doi:10.1016/j.jhazmat.2008.07.020
Kumar, S. S., Shantkriti, S., Muruganandham, T., Murugesh, E., Rane, N., and Govindwar, S. P. (2016). Bioinformatics aided microbial approach for bioremediation of wastewater containing textile dyes. Ecol. Inf. 31, 112–121. doi:10.1016/j.ecoinf.2015.12.001
Kurade, M. B., Waghmode, T. R., and Govindwar, S. P. (2011). Preferential biodegradation of structurally dissimilar dyes from a mixture by Brevibacillus laterosporus. J. Hazard. Mat. 192, 1746–1755. doi:10.1016/j.jhazmat.2011.07.004
Lee, S. Y., Choi, J. H., and Xu, Z. (2003). Microbial cell-surface display. Trends Biotechnol. 21, 45–52. doi:10.1016/s0167-7799(02)00006-9
Leena, R., and Selva Raj, D. (2008). Bio-decolourization of textile effluent containing Reactive Black-B by effluent-adapted and non-adapted bacteria. Afr. J. Biotechnol. 7, 3309–3313. doi:10.5897/AJB08.045
Li, C., Zhang, Z., Li, Y., and Cao, J. (2015). Study on dyeing wastewater treatment at high temperature by MBBR and the thermotolerant mechanism based on its microbial analysis. Process Biochem. 50, 1934–1941. doi:10.1016/j.procbio.2015.08.007
Molina-Guijarro, J. M., Perez, J., Munoz-Dorado, J., Guill´en, F., Moya, R., Hern´andez, M., et al. (2009). Detoxification of azo dyes by a novel pH-versatile salt-resistant laccase from Streptomyces ipomoea. Int. Microbiol. 12, 13–21. doi:10.2436/20.1501.01.77
Ng, I. S., Chen, T., Lin, R., Zhang, X., Ni, C., and Sun, D. (2014). Decolorization of textile azo dye and Congo red by an isolated strain of the dissimilatory manganese-reducing bacterium Shewanella xiamenensis BC01. Appl. Microbiol. Biotechnol. 98, 2297–2308. doi:10.1007/s00253-013-5151-z
Ogugbue, C. J., Sawidis, T., and Oranusi, N. A. (2012). Bioremoval of chemically different synthetic dyes by Aeromonas hydrophila in simulated wastewater containing dyeing auxiliaries. Ann. Microbiol. 62, 1141–1153. doi:10.1007/s13213-011-0354-y
Olukanni, O. D., Osuntoki, A. A., and Gbenle, G. O. (2006). Textile effluent biodegradation potentials of textile effluent-adapted and non-adapted bacteria. Afr. J. Biotechnol. 5, 1980–1984.
Olukanni, O. D., Osuntoki, A. A., Kalyani, D. C., Gbenle, G. O., and Govindwar, S. P. (2010). Decolorization and biodegradation of reactive blue 13 by Proteus mirabilis LAG. J. Hazard. Mat. 184, 290–298. doi:10.1016/j.jhazmat.2010.08.035
Oturkar, C. C., Nemade, H. N., Mulik, P. M., Patole, M. S., Hawaldar, R. R., and Gawai, K. R. (2011). Mechanistic investigation of decolorization and degradation of Reactive Red 120 by Bacillus lentus BI377. Bioresour. Technol. 102, 758–764. doi:10.1016/j.biortech.2010.08.094
Patil, P., Desai, N., Govindwar, S., Jadhav, J. P., and Bapat, V. ( (2009). Degradation analysis of reactive red 198 by hairy roots of Tagetes patula L. (Marigold). Planta 230, 725–735. doi:10.1007/s00425-009-0980-9
Priyadharshini, A., Hosimin, S., Sergio, F., and Maruthamuthu, S. (2016). An integrated (electro- and bio-oxidation) approach for remediation of industrial wastewater containing azo-dyes: Understanding the degradation mechanism and toxicity assessment. J. Hazard. Mat. 318, 203–215. doi:10.1016/j.jhazmat.2016.07.028
Reiss, R., Ihssen, J., and Thony-Meyer, L. (2011). Bacillus pumilus laccase: A heat stable enzyme with a wide substrate spectrum. BMC Biotechnol. 11, 9. doi:10.1186/1472-6750-11-9
Sahasrabudhe, M., Bhattacharya, A., Pathade, A., and Pathade, G. (2014). Detoxification of reactive red 195 by Micrococcus glutamicus NCIM 2168. J. Environ. Res. Dev. 9, 120–128.
Saratale, R. G., Gandhi, S. S., Purankar, M. V., Kurade, M. B., Govindwar, S. P., Oh, S. E., et al. (2013). Decolorization and detoxification of sulfonated azo dye C.I. Remazol Red and textile effluent by isolated Lysinibacillus sp. RGS. J. Biosci. Bioeng. 115, 658–667. doi:10.1016/j.jbiosc.2012.12.009
Saratale, R. G., Saratale, G. D., Chang, J. S., and Govindwar, S. P. (2011). Bacterial decolorization and degradation of azo dyes: A review. J. Taiwan Inst. Chem. Eng. 42, 138–157. doi:10.1016/j.jtice.2010.06.006
Sinha, S., Chattopadhyay, P., Pan, I., Chatterjee, S., Chanda, P., Bandyopadhyay, D., et al. (2009). Microbial transformation of xenobiotics for environmental bioremediation. Afr. J. Biotechnol. 8, 6016–6027. doi:10.5897/ajb09.740
Solís, M., Solís, A., Perez, H. I., Manjarrez, N., and Floresa, M. (2012). Microbial decolouration of azo dyes: A review. Process Biochem. 47, 1723–1748. doi:10.1016/j.procbio.2012.08.014
Srinivasan, S. (2018). “Biodegradation of textile azo dye, Remazol Yellow RR using non-autochthonous bacteria Lysinibacillus sphaericus MTCC 9523, supported by docking,” in Proceedings of the international conference on biodiversity & sustainable resource management (Chennai: Centre for Environmental Sciences & Centre for Water Resources Management, University of Madras), 242–255.
Srinivasan, S., Kanyaga, P. M., and Nagaraj, S. (2020). “Latest innovations in bacterial degradation of textile azo dyes,” in Emerging technologies in environmental bioremediation. Editors M. P. Shah, S. Rodriguez-Couto, and S. S. Sengor (Elsevier), 285–309.
Srinivasan, S., and Sadasivam, S. K. (2021). Biodegradation of textile azo dyes by textile effluent non-adapted and adapted Aeromonas hydrophila. Environ. Res. 194, 110643. doi:10.1016/j.envres.2020.110643
Srinivasan, S., and Sadasivam, S. K. (2018). Exploring docking and aerobic-microaerophilic biodegradation of textile azo dye by bacterial systems. J. Water Process Eng. 22, 180–191. doi:10.1016/j.jwpe.2018.02.004
Srinivasan, S., Sadasivam, S. K., Gunalan, S., Shanmugam, G., and Kothandan, G. (2019). Application of docking and active site analysis for enzyme linked biodegradation of textile dyes. Environ. Pollut. 248, 599–608. doi:10.1016/j.envpol.2019.02.080
Telke, A., Kalyani, D., Jadhav, J., and Govindwar, S. P. (2008). Kinetics and mechanism of Reactive Red 141 degradation by a bacterial isolate Rhizobium radiobacter MTCC 8161. Acta Chim. Slov. 55, 320–329.
Thomas, C. M., and Nielsen, K. M. (2005). Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 3 (9), 711–721. doi:10.1038/nrmicro1234
Vidali, M. (2001). Bioremediation: An overview. Pure Appl. Chem. 73, 1163–1172. doi:10.1351/pac200173071163
Vilchis-Carmona, J. A., Rodríguez-Luna, I. C., Elufisan, T. O., Sánchez-Varela, A., Bibbins-Martínez, M., Rivera, G., et al. (2021). The decolorization and degradation of azo dyes by two Stenotrophomonas strains isolated from textile effluent (Tepetitla, Mexico). Braz. J. Microbiol. 52 (4), 1755–1767. doi:10.1007/s42770-021-00542-y
Wells, A., Teria, M., and Eve, T. (2006). Green oxidations with laccase mediator systems. Biochem. Soc. Trans. 34, 304–308. doi:10.1042/bst0340304
Wu, J., Kim, K. S., Lee, J. H., and Lee, Y. C. (2010). Cloning, expression in Escherichia coli, and enzymatic properties of laccase from Aeromonas hydrophila WL-11. J. Environ. Sci. 22, 635–640. doi:10.1016/s1001-0742(09)60156-x
Keywords: azo dyes, decolorization, biodegradation, non-autochthonous bacteria, Lysinibacillus sphaericus
Citation: Srinivasan S, Bankole PO and Sadasivam SK (2022) Biodecolorization and degradation of textile azo dyes using Lysinibacillus sphaericus MTCC 9523. Front. Environ. Sci. 10:990855. doi: 10.3389/fenvs.2022.990855
Received: 13 July 2022; Accepted: 26 September 2022;
Published: 07 October 2022.
Edited by:
Ahmed El Nemr, National Institute of Oceanography and Fisheries (NIOF), EgyptReviewed by:
Manikant Tripathi, Avadh University, IndiaCopyright © 2022 Srinivasan, Bankole and Sadasivam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shantkriti Srinivasan, c2hhbnRrcml0aTg5QGdtYWlsLmNvbQ==; Paul Olusegun Bankole, YmFua29sZXBvQGZ1bmFhYi5lZHUubmc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.