
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 15 September 2022
Sec. Toxicology, Pollution and the Environment
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.979922
This article is part of the Research Topic Soil and Sediment Pollution, Processes and Remediation, Volume II View all 17 articles
Soil microorganisms play an important role in maintaining soil quality and function, although the response of soil microbial biodiversity to heavy metals has been extensively investigated, the microbe-microbe associations under the influence of both native plant species and extremely high heavy metal contamination are not well documented. We examined the diversity and composition of microbial communities and the physicochemical properties in the rhizosphere of three native plant species, Carex breviculmis, Buddleja davidii, and Artemisia annua growing on and around a Pb-Zn waste heap with a nearly 100-year history of natural recovery. Both plant species and heavy metals influence soil microbial diversity and composition. C. breviculmis and A. annua showed a prominent advantage in increasing rhizosphere microbial diversity and richness as well as network complexity compared with plant Buddleja davidii at severely contaminated soil, which was mainly related to the accumulation of soil nutrients such as soil organic carbon (SOC), total nitrogen ammonium nitrogen and nitrate nitrogen rather than a reduction in heavy metal concentrations. Moreover, the heavy metal concentration and soil nutrient levels significantly affected the microbial groups affiliated with Proteobacteria, Chloroflexi, Ascomycota, and Basidiomycota, in which those affiliated with Chloroflexi and Ascomycota were positively associated with heavy metals. Soil microbial network on the Pb-Zn waste heap exhibited higher average degree and a higher proportion of positive links than those around the waste heap, and thus soil microbial structure became more complexity and unstable with increasing heavy metal pollution.
Over the past several decades, prosperous mining activities have promoted the development of the Chinese economy. However, they have generated vast amounts of solid waste, which results in the degradation of the surrounding ecosystem (Becerra-Castro et al., 2012; Li et al., 2021). Li (2006) and Liu et al. (2020) reported that mining activities alone have generated approximately 3.2 Mha of wasteland and that this value is increasing by 46,700 ha/year in China. Waste materials contain large amounts of heavy metals and metalloids that often reach toxic levels and are susceptible to dispersing into the adjacent ecosystem via wind dispersion and water erosion (Luo et al., 2019), imposing a threat to life, including humans, plants, and microorganisms (Epelde et al., 2014). Some heavy metals and metalloids, such as Pb, Cd, Cr, and As, are nonessential for living organisms. However, because of their reactivity with thiols or other groups, these heavy metals can interfere with physiological processes even at low concentrations by disturbing enzymatic reactions, triggering cell membrane damage, and modifying deoxyribose nucleic acid (DNA) structure (Dubey et al., 2018; DalCorso et al., 2019). In China, soil heavy metal contamination resulting from mining activities has become a severe environmental problem, especially in areas rich in nonferrous metal resources (Li Q. et al., 2020).
Heavy metals impact ecosystems through a variety of effects on plants, soils, and microbial communities (Azarbad et al., 2013; Barra Caracciolo and Terenzi, 2021). Metalliferous soils, such as waste heaps, are often sandy and dry, with low organic matter and nutrient availability, as well as elevated heavy metal contents (Borymski et al., 2018). This leads to strong selective pressure driving the evolution and adaptation of plants and microorganisms (Epelde et al., 2014). In this context, some metal-tolerant plants have been the focus of previous studies (Becerra-Castro et al., 2012; Elbehiry et al., 2020) because of their biological mechanisms that resist and tolerate heavy metals. Pseudo-metallophyte populations of Carex breviculmis (Lei and Duan, 2008; Xu et al., 2020), Buddleja davidii (Margui et al., 2006), and Artemisia annua (Petriccione et al., 2013; Inelova et al., 2021) are metal-tolerant plant species that have exhibited good abilities in colonizing Pb, Zn, and Cd contaminated soils. Soil microorganisms play an important role in maintaining soil quality and function by decomposing organic matter, participating in nutrient cycling, and reducing heavy metal toxicity (Banning et al., 2011). Heavy metal/metalloids alter various metabolic processes of microorganisms, including blocking essential functional groups, displacing other metal ions, and inactivating biological molecules (Chen et al., 2020). The effect of heavy metal contamination on soil microbial communities is usually adverse, resulting in the reduction of microbial biomass and activity, the disturbance of specific taxa, and shifts in the composition of the microbiome (Azarbad et al., 2013; Epelde et al., 2014). Although previous studies have revealed that soil microbial structure, composition, and diversity, can change under heavy metal contamination, it is unclear how microbe-microbe associations change with increasing heavy metal pollution, especially when considering the complex interactions between heavy metal/metalloid-contaminated soils and plants (Borymski et al., 2018; Chun et al., 2021). Therefore, identifying inter-taxa correlations is crucial for comprehensively understanding microbial community structures and predicting the response of soil ecosystems to future degenerative and pollution processes (Cheng et al., 2020).
In addition to the phytoextraction and phytostabilization of soil heavy metals, metal-tolerant plants strongly affect soil microbial biomass and activity through the rhizosphere effect, which is thought to arise from the deposition of various organic compounds, including amino acids, carbohydrates, and flavonoids, from plants into soils (Haichar et al., 2008; Montiel-Rozas et al., 2016). The nutrient-rich microenvironment provided by rhizosphere deposition allows certain microbial communities to thrive, as different plant species produce different exudates with specific compositions (Haichar et al., 2008; Borymski et al., 2018). Ali et al. (2013) reported that the metabolic activities of rhizosphere microorganisms are 10–100 times higher than those in bulk soil because of the exudates secreted by plants. Metal-tolerant plants select specialized metal-tolerant microorganisms to inhabit their rhizosphere, with a preference for plant growth-promoting bacteria that enhance the phytoremediation of heavy metal-contaminated soils (Hamidpour et al., 2019; Li Z. et al., 2020). Therefore, the interactions between metal-tolerant plants and accompanying microorganisms, as well as the surrounding environment, play key roles in maintaining nutrient cycling and element turnover (Epelde et al., 2010). On this basis, it is important to study the biodiversity of rhizosphere microbial communities linked with plant-soil-microbe interactions to better understand soil functions impacted by both heavy metal pollution and plant species to improve phytoremediation of mine sites in the future.
Northwestern Guizhou, China, is an area rich in nonferrous metal resources, especially lead zinc ore. This region has an approximately 300-year-old history of zinc mining and smelting using indigenous methods, and large volumes of Pb-Zn waste (more than 20 million tons by 2000) have been left and piled onto the surrounding farmland with almost no safety treatment (Yang et al., 2016). In this study, we examined a Pb-Zn waste heap with a nearly 100-year history, which has been naturally colonized by some native wild plant species (C. breviculmis, B. davidii, and A. annua) with limited human disturbance. We hypothesized that plant species will significantly affect rhizosphere microbial richness, diversity and interaction, and that soil microbe-microbe association will be different on and around the Pb-Zn waste heap. To test this hypothesis, we collected the rhizosphere of the aforementioned plant species both on and around the Pb-Zn waste heap. Heavy metals/metalloids and soil properties were quantified, and microbial diversity was measured using high-throughput sequencing to: 1) assess the response of soil properties to plant species and sampling sites, including soil nutrients and heavy metal contents, as well as the diversity and composition of microbial communities; 2) explore the major environmental factors contributing to soil microbial community variations; and 3) profile the microbial association in the rhizosphere soil of three metal-tolerant plants on and around the Pb-Zn waste heap. The results of this study will provide crucial insights into the restoration of heavy metal-contaminated soils.
An abandoned Pb-Zn waste heap located in the northwest of the Guizhou Province, China (26°54′N, 104°54′W) was used as the study site. The local climate is subtropical humid, with a mean annual precipitation of 800–1000 mm and an average temperature of 10 °C. Rainfall is irregularly distributed throughout the year, with 55% occurring from June to August. This area is famous for its long history of Zn smelting using indigenous methods, which generated large amounts of waste slag (more than 20 million tons by 2000) and formed numerous Pb-Zn waste heaps (Yang et al., 2016). Waste slag from Zn smelting is a mixture of ore smelting slag and small amounts of unburned cinders and contains high levels of toxic heavy metals, including Cu, Pb, Zn, and Cd (Luo et al., 2019).
At the time of this study, the selected Pb-Zn waste heap is nearly 100 years old, during which period it has been restored naturally, almost without human interference. C. breviculmis, B. davidii, and A. annua are common native wild plant species on and around the Pb-Zn waste heap. C. breviculmis is the dominant perennial herb. A. annua is a late-successional plant in the herbaceous stage, and B. davidii is a pioneer plant in the shrub stage.
Four replicate rhizosphere samples of C. breviculmis, B. davidii, and A. annua were collected on and around the abandoned Pb-Zn waste heap. Relatively isolated plants were selected to reduce the impact of nearby plants and consistent environmental conditions were ensured to reduce the impact of environmental heterogeneity, when collecting the rhizosphere of each plant. The rhizosphere soil samples were collected by shaking off the loosely attached bulk soil adhered to the root systems. In addition, unvegetated soil was collected from the Pb-Zn waste heap as a control (CK). Each sample was divided into two subsamples after all roots, stones, and litter were removed. One subsample was immediately stored at -80 °C for microbiological analysis, and a second subsample was air-dried for physicochemical analysis. Sampling sites on and around the Pb-Zn waste heap were defined as S1 and S2, respectively, and the rhizosphere samples of C. breviculmis, B. davidii, and A. annua were defined as CB, BD, and AA, respectively.
Soil organic carbon (SOC), total nitrogen (TN), total phosphorus (TP), available phosphorus (AP) ammonium nitrogen (AN), nitrate nitrogen (NN), soil pH, and heavy metals were quantified using standard methods and detailed descriptions are provided in the Supplementary Materials.
DNA was extracted from 0.5 g of each soil sample using a FastDNA Spin Kit (MP Biomedicals, Cleveland, United States) following the manufacturer’s specifications. DNA extracts were diluted ten-fold and assessed for quality and quantity using a spectrophotometer (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, United States). The integrity of the DNA extracts was confirmed by 1% agarose gel electrophoresis. The bacterial 16S rRNA and fungal 18S rRNA genes were amplified using the primer pairs 338F/806R (Deng et al., 2018) and ITS1F/ITS2R (Bellemain et al., 2010), respectively. The primers were tagged with unique barcodes for each sample. A PCR was performed in a 30 µL solution containing 2 µL of sterile ultrapure water, 15 µL of Phusion Master Mix (2X), 3 µL of 6 µM primer, and 10 µL of template DNA (5–10 ng). A PCR was conducted at 98°C for 60 s, followed by 30 cycles of 98°C for 10 s, 50°C for 30 s, 72°C for 30 s, and 72°C for 5 min. The amplicons were purified and high-quality libraries were generated using the TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, United States) following the manufacturer’s recommendations. All samples were pooled in equimolar amounts and sequenced on an Illumina HiSeq PE300 platform.
Bacterial and fungal sequences were analyzed using the UPARSE pipeline (Edgar 2013). Briefly, paired-end reads were merged into single sequences, after which the chimeras were removed using the UCHIME algorithm (Caporaso et al., 2010). Sequences with 97% similarity were clustered into operational taxonomic units (OTUs). Low-abundance OTUs were eliminated from the OTU table if they did not present at least two counts across all samples in the experiment. Bacteria were identified using the Silva reference database (http://www.arb-silva.de) with the RDP classifier (Wang et al., 2007) and fungi were identified using the Unite database (https://unite.ut.ee/) with the BLAST tool in QIIME (Abarenkov et al., 2010).
Soil properties, microbial diversity, and abundance at each sampling site were evaluated among the three plant species, using a one-way analysis of variance, followed by an LSD post hoc test at p < 0.05. Soil properties, microbial diversity, and abundances between the two sampling sites were compared using a t-test (p < 0.05). The Shannon and Richness indices were used to represent plant diversity, and Chao1 and Shannon indices were used to indicate the richness and diversity of the soil microbial communities. A two-way ANOVA was conducted to determine the individual and interacting effects of plant species and sampling sites on each soil property, bacterial and fungal composition and α-diversity. Principal Co-ordinates Analysis (PCoA) and an ANOSIM test based on the Bray–Curtis distances of the sequencing data was used to assess differences in the structures of the microbial communities. Mantel tests based on Bray-Curtis distance similarities calculated at the OTU (operational taxonomic unit) level were used to identify the plant and soil factors correlated with microbial community composition. A hierarchical partitioning was used to estimate the importance of individual and group of environmental variables (edaphic variables and heavy metals) in predicting soil microbial communities. Statistical tests and graphic plotting were performed using the packages vegan, reshape2, sem, packfor, picante, and ggplot2 in R v.4.0.3 (R Development Core Team, 2019).
Networks were constructed for the microbial communities for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2. All pairwise Sparse Correlations for Compositional (SparCC) data among microbial nodes using the Fastspar algorithm with 1000 bootstraps and 1000 permutations were calculated to control the false discovery rate. A robust correlation coefficient of |r| > 0.9 and p < 0.01 were retained to construct networks. The networks were visualized using Gephi (version 0.9.2; https://gephi.org/). The nodes in the networks represent OTUs, in which the node size is proportional to the number of connections (degrees), and the node color represents microbial taxonomy. The edges between each pair of nodes (i.e., links) correspond to positive or negative correlations between the nodes.
In general, plant species significantly affected the soil nutrient contents, especially SOC, TN, and pH, while heavy metals significantly differed between the two sampling sites (Table 1). The rhizosphere of AA exhibited the highest contents of SOC (11.669 and 15.656 g kg−1) and TN (4.367 and 4.534 g kg−1) compared with the other two plant species at both S1 and S2 and that in the non-vegetated soil (CK) at S1. The pH of the CB rhizosphere was the highest at S1 and S2, and was significantly higher than that of CK at S1. Overall, the heavy metal content and pH values in the rhizosphere of all three plant species had higher values at S1 than at S2.
A total of 1,558,675 high-quality sequences of bacteria and 1,840,010 high-quality sequences of fungi were obtained. The richness (Richness index) and diversity (Shannon index) of the soil bacterial communities in the rhizosphere of plant BD were significantly lower than in plants CB and AA but were not significantly different from that of the CK at S1; similarly, the richness (Richness index) and diversity (Shannon index) of the soil fungal communities in the rhizosphere of plant BD were significantly lower than in AA but were not significantly different from that of the plant CB and CK at S1. The diversity of the bacterial community was significantly higher in the rhizosphere of plants CB and BD than in AA, and the richness of the fungal community was significantly higher in the rhizosphere of plant CB than in plant BD at S2. Moreover, the richness and diversity of the bacterial and fungal communities in the rhizosphere of plant BD were significantly lower at S1 than at S2 (Table 2).
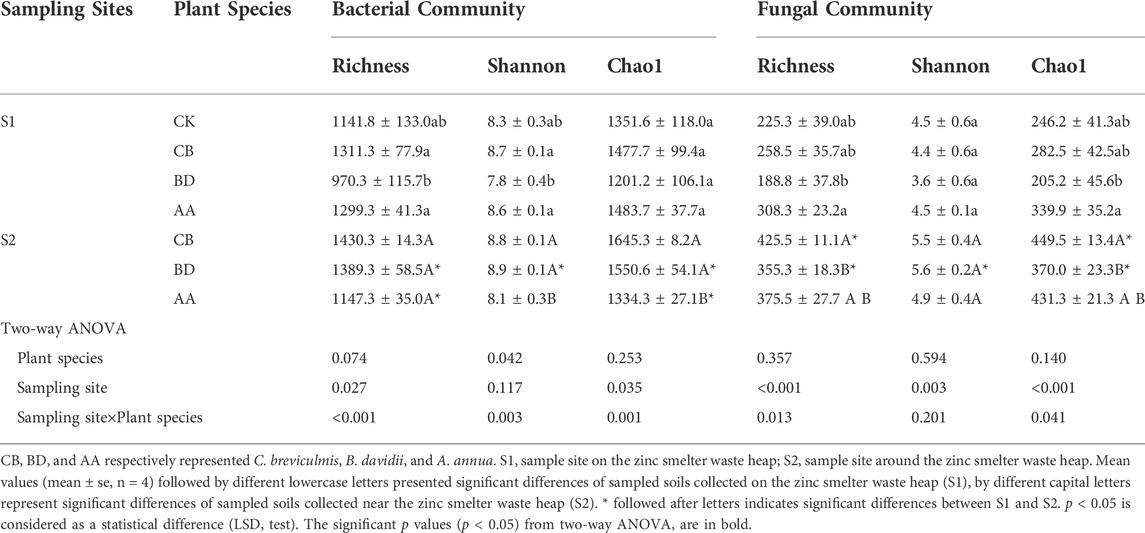
TABLE 2. Soil bacterial and fungal diversities for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2.
A total of eight soil bacterial phyla (relative abundances >1%) were obtained, decreasing in relative abundance as follows: Actinobacteria (25.7%), Proteobacteria (21.2%), Chloroflexi (19.4%), Acidobacteria (15.6%), Gemmatimonadota (3.6%), Myxococcota (2.5%), Planctomycetota (2.1%), and Firmicutes (1.9%) (Figure 1 and Supplementary Table S1). Plant species had a significant effect on Proteobacteria and Chloroflexi, of which Proteobacteria showed a significantly higher relative abundance in the rhizosphere of AA than in CB at both S1 and S2, while Chloroflexi exhibited the opposite trend. Between sampling sites, the relative abundance of Proteobacteria in the rhizosphere of AA was significantly lower at S1 than at S2. Regarding the fungal community, four fungal phyla with a relative abundance >1% were obtained, including Ascomycota (61.2%), Mortierellomycota (13.8%), Basidiomycota (13.7%), and Rozellomycota (8.6%). The relative abundance of Ascomycota was significantly higher and that of Rozellomycota was significantly lower in the rhizosphere of CB than in the rhizospheres of BD and AA at both S1 and S2. For the rhizospheres of BD and AA, the relative abundances of Basidiomycota and Rozellomycota were significantly lower and higher, respectively, at S1 than at S2.
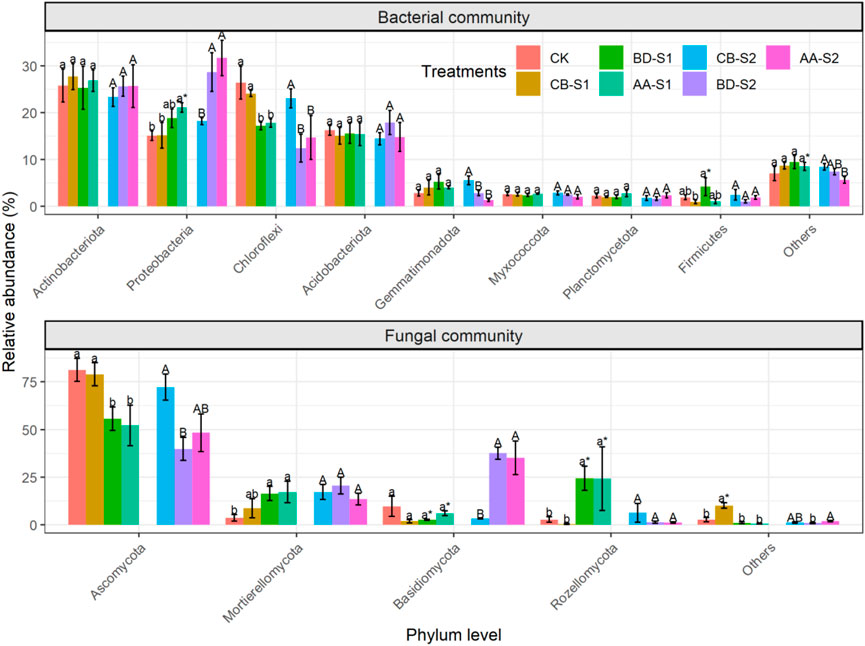
FIGURE 1. Soil microbial composition at the phylum level for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2. CB, BD and AA respectively represented C. breviculmis, B. davidii, and A. annua. S1, sample site on the zinc smelter waste heap; S2, sample site around the zinc smelter waste heap. Mean values (mean ± se, n = 4) followed by different lowercase letters presented significant differences of sampled soils collected on the zinc smelter waste heap (S1), by different capital letters represent significant differences of sampled soils collected near the zinc works (S2). * followed after letters indicates significant differences between S1 and S2. p < 0.05 is considered as a statistical difference (LSD test).
PCoA accompanied with ANOSIM test showed results that rhizosphere bacterial and fungal community composition was significantly different among plant species as well as between sampling sites (Figure 2). Furthermore, pairwise ANOSIM test showed that both bacterial and fungal community from S1 were different from those of S2. Rhizosphere bacterial and fungal community of plants CB and BD were not significantly different but they were significantly distinct from those of plant AA at S1. Moreover, rhizosphere bacterial community of plant BD and rhizosphere fungal community of plants CB and BD were significantly different from those of CK. At sampling site of S2, rhizosphere bacterial and fungal community of plant CB were significantly different from those of plants BD and AA, which showed no significant difference (Supplementary Table S2). Additionally, some soil microbial functional profiles also differed significantly between S1 and S2 (Supplementary Figure S1). For example, the proportion of bacterial functional profiles related to glycine, serine, and threonine metabolism and the fungal functional profiles related to endophyte-plant pathogens were remarkably higher at S1 than at S2. Conversely, samples from S2 had significantly higher fungal functional profiles related to endophyte-soil saprotrophs, animal pathogen-clavicipitaceous endophyte-fungal parasites, wood saprotrophs and plant saprotroph-wood saprotrophs, as compared with those from S1.
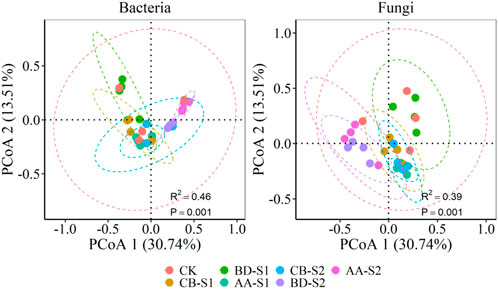
FIGURE 2. The NMDS ordinations of bacterial and fungal communities (OTU level) for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2. Individual data points with different colors represent individual soil samples. Analysis of similarity (ANOSIM) test showing the difference of bacterial and fungal community structure among bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2. CB, BD and AA respectively represented C. breviculmis, B. davidii, and A. annua. S1, sample site on the zinc smelter waste heap; S2, sample site around the zinc smelter waste heap.
Co-occurrence networks of soil microbial community for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2 were constructed (Figure 3), and topological properties were calculated to describe the complex pattern of network and to compare the real network with an identically sized Erdös-Réyni random network (Table 3). Overall, the structure properties of the real networks were greater than the Erdös-Réyni random networks, indicating that the former are more clustered than the latter. Soil microbial network of CK was the largest (more edges) and most complex (higher average degree) compared with rhizosphere microbial networks of three plants at S1. Furthermore, rhizosphere microbial networks of plant CB and AA were larger and more complex than that of plant BD at S1, but rhizosphere microbial networks showed no obvious different among plant species at S2. Compared with site S2, rhizosphere microbial networks at S1 were larger and more complex, and the ratio of positive to negative edges of the microbial network was higher. Moreover, there were fewer nodes affiliated with Ascomycota but more nodes affiliated with Chlofoflexi, Planctomycetota and Myxococcota at S1 compared with S2 (Supplementary Table S3). Ascomycota, Actinobacteriota, Chloroflexi, Actinobacteriota and Rozellomycota were the keystone taxa in networks for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2 (Supplementary Table S4).
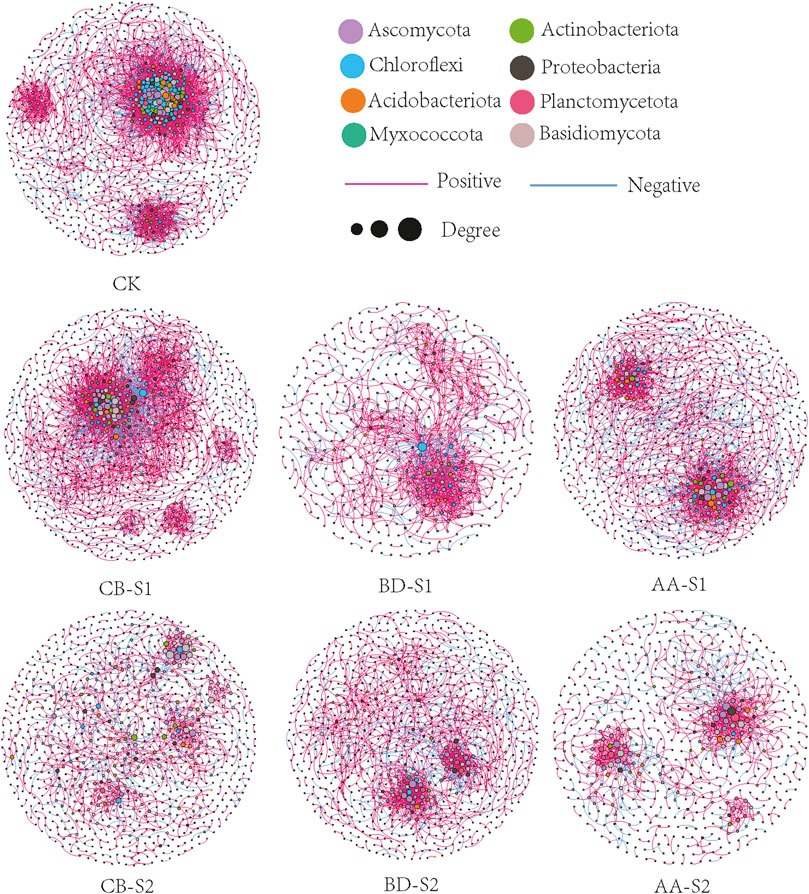
FIGURE 3. Co-occurrence networks of soil microbial community for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2. The nodes are colored by the phylum level, and size of each node is proportional to its degree. The line between each pair of nodes represent positive (red) and negative (blue) correlation with p < 0.01 and |r| > 0.9. CB, BD and AA respectively represented C. breviculmis, B. davidii, and A. annua. S1, sample site on the zinc smelter waste heap; S2, sample site around the zinc smelter waste heap.
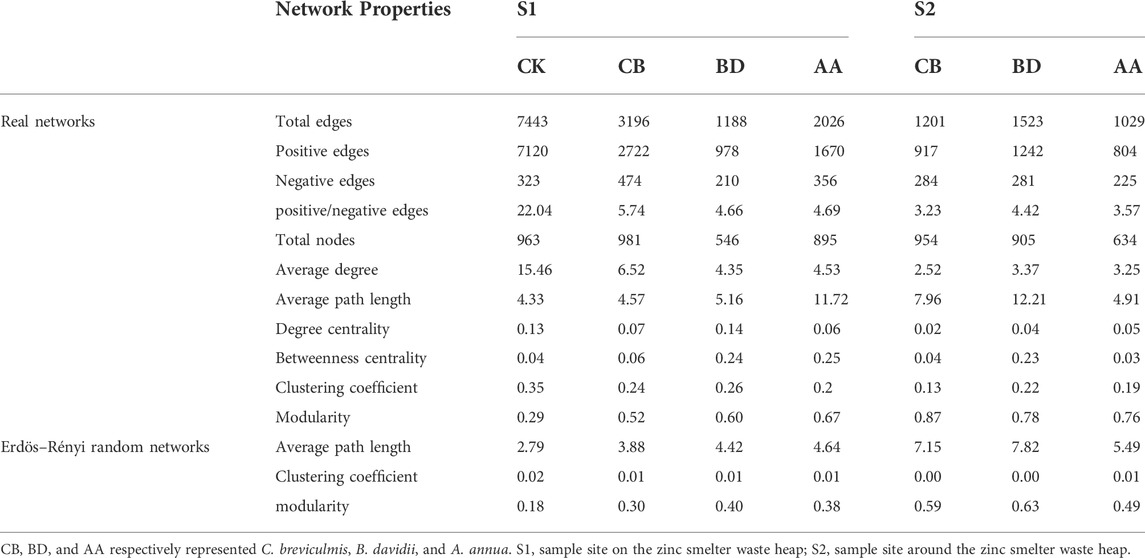
TABLE 3. Topological properties of empirical networks and Erdös–Rényi random networks for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2.
The Mantel test results for the correlation between community composition and environmental variables revealed that both the bacterial and fungal communities were significantly correlated with heavy metals (Cu, Zn, As, Cd and Pb). In addition, the bacterial community had significant correlations with SOC, TN, AN, pH, and moisture, while the fungal community only had significant correlations with SOC and AN. Additionally, there was a significant positive correlation within indices of soil nutrients, which were negatively correlated and heavy metals (Supplementary Figure S2). In general, the edaphic variables showed a higher relative importance in predicting both bacterial and fungal communities compared with heavy metals, and edaphic variables of SOC, AN, NN and pH together with heavy metals of Zn, As and Pb were important predictors of bacterial and fungal communities compared with the other environmental variables (Figure 4). The correlation heatmaps showed that all heavy metals and pH were negatively correlated with Proteobacteria and Basidiomycota, and positively correlated with Ascomycota and Chloroflexi. Conversely, SOC, TN, and AN were significantly positively correlated with Proteobacteria and Basidiomycota and negatively correlated with Ascomycota (Figure 5).
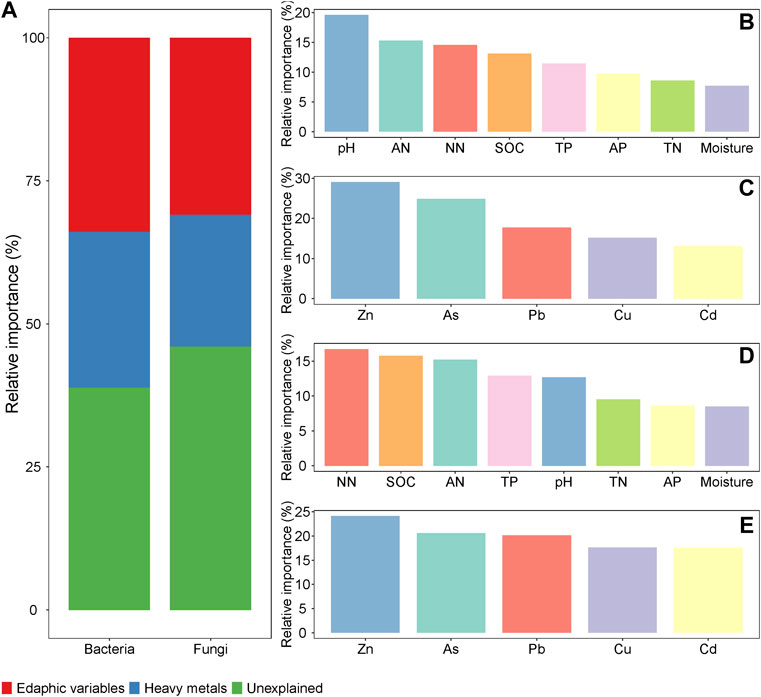
FIGURE 4. The relative importance of individual and group of environmental variables in predicting soil microbial communities. (A) group of edaphic variables and heavy metals variables in predicting soil microbial communities; (B) individual edaphic variables in predicting bacterial communities; (C) individual heavy metal variables in predicting bacterial communities; (D) individual edaphic variables in predicting fungal communities; (E) individual heavy metal variables in predicting fungal communities. SOC, soil organic carbon; TN, total nitrogen; TP, total phosphorus; AP, available phosphorus; AN, ammonium nitrogen; NN, nitrate nitrogen.
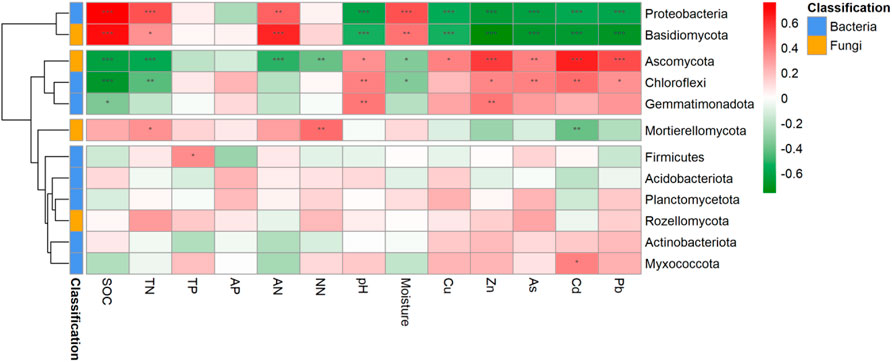
FIGURE 5. Correlation heat maps for the correlation between community composition and environmental variables at the phylum level. The left dendogram linkages represent clustering results of these microbial groups. SOC, soil organic carbon; TN, total nitrogen; TP, total phosphorus; AP, available phosphorus; AN, ammonium nitrogen; NN, nitrate nitrogen. *, <0.05; **, <0.01; ***, <0.001.
In this study, the heavy metal contamination levels at S1 and S2 exceeded the allowable limits set by Chinese standards for agricultural soils (Ministry of Ecological Environment of the People’s Republic of China, 2018), in which the concentrations of multiple heavy metals at S1 were at least two times higher than those at S2. Thus, S1 was defined as “severely contaminated soil,” while S2 was defined as “moderately contaminated soil.” Our results show that AA had a better ability to increase the SOC and TN contents in the rhizosphere, but most heavy metal contents were not significantly affected by plant species at S1. The differences in rhizosphere SOC and TN contents among the three studied plant species could be attributed to differences in the quantity and quality of root exudates and the organic litter inputs of each plant species (Bardgett et al., 2014). However, although there was no obvious difference in the heavy metal contents among the three plant species, this does not indicate that the plants had no effect on the total metal concentrations. This is mainly because the background heavy metal content value in the Pb-Zn waste heap is extremely high, and any changes in heavy metal content caused by plant colonization would be comparatively insignificant, rendering them statistically undetectable. Moreover, the phytoremediation of heavy metals generally involves one or more mechanisms such as phytoaccumulation, phytostabilization, phytodegradation, phytovolatilization, and hydraulic control, depending on the plant species (Muthusaravanan et al., 2018). Thus, phytoremediation is more relevant to the bioavailability of heavy metals (Fu et al., 2019) than their absolute abundance. However, in this study, only the heavy metal contents of the soil samples were measured. Therefore, the response of the heavy metal speciation distributions to plant species must be further studied to confirm these findings. Nevertheless, we found that the contents of Zn and Cd in the rhizosphere of AA was significantly lower than CK at S1, which is consistent with the findings of Inelova et al. (2021) and Petriccione et al. (2013), who found that A. annua has a large accumulation capacity for heavy metals, especially Cd and Zn. Note that the difference among the heavy metal content among the three plant species at S2 was mainly attributable to differences in sampling location rather than the effect of plant species because C. breviculmis was growing closer to S1 than the other plants.
Certain plant species have the ability to take up or immobilize heavy metal contaminants in soils while enriching rhizosphere nutrients by transporting various organic compounds (such as amino acids, organic acids, and phenolic compounds) from plants into the soil (Borymski et al., 2018; Muthusaravanan et al., 2018). Plant characteristics and rhizosphere effects greatly contribute to changes in the rhizosphere microbial community (Borymski et al., 2018; Barra Caracciolo and Terenzi, 2021). Our results showed that, at S1, the richness and diversity of the bacterial communities of BD were lower than those of CB and AA but were not significantly different from those of the CK, and the fungal community showed a similar trend. The difference in soil microbial community among plant species was mainly the result of an accumulation of soil nutrients rather than a reduction in heavy metal concentrations, which was supported by our findings that the edaphic variables showed a higher relative importance in predicting both bacterial and fungal communities compared with the heavy metal variables, especially the SOC, AN, NN and pH (Figure 4). The different effects of plant species on the rhizosphere microbial community were also verified by the NMDS results, which showed that the rhizosphere bacterial and fungal community were significantly different among species both at S1. Borymski et al. (2018) suggested that a nutrient-deficit microenvironment was not effective in attracting and stimulating microbial groups around their roots. Several other previous studies also demonstrated that the rhizosphere effect usually increases microbial diversity (Azarbad et al., 2013; Zhao et al., 2020). Notably, we found the soil bacterial and fungal α-diversity in CK was not significantly different from that in the rhizosphere of the three plant species at S1. This discrepancy may be because the S1 sampling site is on an artificial Pb-Zn waste heap that has undergone natural restoration for nearly a hundred years, and C. breviculmis and A. annua are dominant pioneer species with a large degree of coverage, which may cause its rhizosphere effect to influence the soils uncovered by plants. Moreover, as heavy metal contamination has persisted for nearly a hundred years, the soil microbes have adapted to heavy metal contamination and increased their diversity and richness (Bourceret et al., 2016).
The rhizosphere bacterial composition also responded to plant species, in which Proteobacteria showed a significantly higher relative abundance in the rhizosphere of AA, as compared with CB at both sites, while Chloroflexi showed the opposite trend. As Proteobacteria are usually considered copiotrophic r-strategists and prefer to utilize fresh photosynthates (Bastida et al., 2016), the observed increase in its relative abundance resulted from the higher amount of photosynthates in the rhizosphere of AA than that in CB. Chloroflexi are slow-growing oligotrophic bacteria that are particularly resistant to heavy metals (Rastogi et al., 2011). Herein, the significantly higher relative abundance of Chloroflexi in the rhizosphere of CB was related to its high content of heavy metals, especially Zn and Cd. Regarding fungal composition, the dominant phylum of Ascomycota was significantly higher and Rozellomycota had a significantly lower relative abundance in the rhizosphere of CB than in that of BD and AA at both S1 and S2. The dominance of Ascomycota in the rhizosphere of all three plant species reveals its strong tolerance and profound impact on certain ecological and biogeochemical conditions under severe heavy metal contamination (Lin et al., 2020). Both Ascomycota and Rozellomycota are heavy metal-tolerant fungi (Iram et al., 2009; Lin et al., 2019), and improvements in soil quality usually weaken the dominance of Ascomycota (Yuan et al., 2020). Meanwhile, Rozellomycota has been found to be positively affected by soil organic matter (Lin et al., 2019). In this study, nutrients, such as SOC and TN, were relatively higher for BD and AA (Table 1), which contributed to the differences in the Ascomycota and Rozellomycota abundances in the rhizosphere of each plant species.
Microbes do not thrive in isolation but instead interact and adapt to survive in harsh environments (Faust and Raes, 2012; Chun et al., 2021). Soil microbial networks unravel the associations among network members and reflect the response of the microbial community to environmental changes (Banerjee et al., 2019). Co-occurrence networks of soil microbial community for bare soils and rhizosphere soils of three metal-tolerant plants at S1 and S2 showed that rhizosphere microbial networks of plant CB and AA were larger and more complex than that of plant BD at S1, but rhizosphere microbial networks showed no obvious different among plant species at S2. This indicate that interactions or niche-sharing in rhizosphere microbial community of plant CB and AA are maximal relative to plant BD at the severely contaminated soil, however, influence of plant species on microbial interactions are reduced at the moderately contaminated soil. Soil microbial communities with a large proportion of positively connected members are unstable because members may respond in tandem with environmental changes (Deng et al., 2012). In this sense, soil microbial networks at S1 had a higher numbers of edges and higher ratio of positive to negative edges relative to S2 indicate that soil microbial communities at S1 are less stable with multi-metal contamination increasing. A possible explanation for these findings is that the association in microbial communities were strengthened under harsh environmental conditions, such as heat, drought, and heavy metal pollution (Zaefarian et al., 2021). In this study, the extremely high content of multiple heavy metals at S1 affected the microbial community, forcing some metal-tolerant bacteria to survive via stronger positive cooperation and trophic interaction, in which such cooperation may cause the formation of a microbial biofilm (Li Q. et al., 2020; Chun et al., 2021). However, this assumption must be corroborated by further studies. In addition, soil temperature at S1 are higher than S2 because it is located on a lead-zinc waste heap, and the warm environment has been demonstrated to activate dormant microbial members and strengthen the interactions among microbial communities (Chen et al., 2012). One potential limitation of co-occurrence network analysis is that species associations based on correlation analysis can yield spurious results and cannot be automatically interpreted as interactions, consequently, it may not be possible to comprehensively depict the microbial interactions under real-world conditions (Jiao et al., 2021). Even so, the information about negative/positive correlations between taxa is still essential for estimating potential species interrelationships within complex environments, and they are frequently used to investigate microbial interconnection patterns (Banerjee et al., 2018; Jin et al., 2022).
Microbes interact with plants to adapt to and alter soil environments (Chun et al., 2021). In this study, the rhizosphere microbial community was influenced by heavy metals (Pb, Zn and As), soil nutrients (SOC, TN, NN and AN), moisture, and pH (Supplementary Figure S2 and Figure 4). Moreover, the relationship between heavy metals, pH, and the microbial community was opposite to the relationship between soil physical properties and the microbial community (Figure 5). Similar results have been reported in previous studies (Haichar et al., 2008; Li C. et al., 2020; Lopez-Marcos et al., 2020), and the opposing effects of heavy metals and soil nutrients on soil microbial communities may be one of the driving forces for maintaining soil microbial community stability. Metal-contaminated environments exert strong selective pressure on microbial compositions by affecting microbial metabolism, including the production of oxidative stress or the destruction of microbial proteins and DNA (Mejias Carpio et al., 2017). This results in the evolution of several metal-tolerant microbial groups with better metal resistance mechanisms, such as extracellular and intracellular sequestration, exclusion by permeability barriers, and enzymatic detoxification (Epelde et al., 2014). Bacterial groups affiliated with Chloroflexi and fungal groups affiliated with Ascomycota were positively associated with almost all the heavy metals involved in this study, indicating that they are tolerant to heavy metal pollution and play leading roles in the bioremediation of heavy metals. Additionally, soil nutrients (especially SOC, TN, NN and AN), moisture, and pH play important roles in shaping soil microbial communities (Figure 3). Although microorganisms participate in the decomposition of soil organic matter and the generation of available nitrogen and phosphorus, microbial metabolisms are affected by soil moisture and pH, which alter extracellular enzyme activity and the absorption rate of soluble substrates (Schimel and Schaeffer, 2012; Calicioglu et al., 2018).
Both plant species and heavy metals influence soil microbial diversity and composition. The influence of native plant species on the microbial structure and diversity of soils contaminated with different degrees of heavy metal pollution was mainly the result of an accumulation of soil nutrients rather than a reduction in heavy metal concentrations. Herein, AA and CB showed relatively higher rhizosphere microbial diversity, richness and network complexity than BD at the severely contaminated soil (S1), which was mainly because of a significant accumulation of soil nutrients, including SOC, TN, AN and NN. Moreover, the heavy metal concentration and soil nutrient levels significantly affected the microbial groups affiliated with Proteobacteria, Basidiomycota, Ascomycota, and Chloroflexi. In particular, the microbial groups affiliated with Chloroflexi and Ascomycota were tolerant to heavy metals and may play an important role in the bioremediation of heavy metals, as they were positively associated with almost all the heavy metals involved in this study. Effect of plant species on rhizosphere microbial networks was more obvious under higher heavy metal pollution. Furthermore, microbial networks at S1 exhibited higher average degree and a higher proportion of positive links than those at S2, indicating that soil microbial structure became more complexity and unstable with increasing heavy metal pollution.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
CS, GW and XK initiated and designed the research, collected the materials, and performed the experiments. CS and PW obtained funding for this study. CS analyzed the data and wrote the manuscript. All authors read and approved the manuscript.
This work was financially supported by the China Postdoctoral Science Foundation (2020M683373), Guizhou Provincial Science and Technology Projects [(2020)1Y192], National Natural Sciences Foundation of China (41967058), United Foundation of the Guizhou Province Government and the Natural Science Foundation of China (No. U1612442), High-Level Talent Training Program in Guizhou [(2016)5664].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.979922/full#supplementary-material
Abarenkov, K., Henrik Nilsson, R., Larsson, K. H., Alexander, I. J., Eberhardt, U., Erland, S., et al. (2010). The UNITE database for molecular identification of fungi - recent updates and future perspectives. New Phytol. 186, 281–285. doi:10.1111/j.1469-8137.2009.03160.x
Ali, H., Khan, E., and Sajad, M. A. (2013). Phytoremediation of heavy metals-Concepts and applications. Chemosphere 91, 869–881. doi:10.1016/j.chemosphere.2013.01.075
Azarbad, H., Niklińska, M., van Gestel, C. A. M., van Straalen, N. M., Röling, W. F. M., and Laskowski, R. (2013). Microbial community structure and functioning along metal pollution gradients. Environ. Toxicol. Chem. 32, 1992–2002. doi:10.1002/etc.2269
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16 (9), 567–576. doi:10.1038/s41579-018-0024-1
Banerjee, S., Walder, F., Büchi, L., Meyer, M., Held, A. Y., Gattinger, A., et al. (2019). Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. Isme J. 13, 1722–1736. doi:10.1038/s41396-019-0383-2
Banning, N. C., Gleeson, D. B., Grigg, A. H., Grant, C. D., Andersen, G. L., Brodie, E. L., et al. (2011). Soil microbial community successional patterns during forest ecosystem restoration. Appl. Environ. Microbiol. 77, 6158–6164. doi:10.1128/AEM.00764-11
Bardgett, R. D., Mommer, L., and De Vries, F. T. (2014). Going underground: Root traits as drivers of ecosystem processes. Trends Ecol. Evol. 29, 692–699. doi:10.1016/j.tree.2014.10.006
Barra Caracciolo, A., and Terenzi, V. (2021). Rhizosphere microbial communities and heavy metals. Microorganisms 9, 1462. doi:10.3390/microorganisms9071462
Bastida, F., Torres, I. F., Moreno, J. L., Baldrian, P., Ondoño, S., Ruiz-Navarro, A., et al. (2016). The active microbial diversity drives ecosystem multifunctionality and is physiologically related to carbon availability in Mediterranean semi-arid soils. Mol. Ecol. 25, 4660–4673. doi:10.1111/mec.13783
Becerra-Castro, C., Monterroso, C., Prieto-Fernández, A., Rodríguez-Lamas, L., Loureiro-Viñas, M., Acea, M. J., et al. (2012). Pseudometallophytes colonising Pb/Zn mine tailings: A description of the plant-microorganism-rhizosphere soil system and isolation of metal-tolerant bacteria. J. Hazard. Mater. 217-218, 350–359. doi:10.1016/j.jhazmat.2012.03.039
Bellemain, E., Carlsen, T., Brochmann, C., Coissac, E., Taberlet, P., and Kauserud, H. (2010). ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 10, 189. doi:10.1186/1471-2180-10-189
Borymski, S., Cycoń, M., Beckmann, M., Mur, L. A. J., and Piotrowska-Seget, Z. (2018). Plant species and heavy metals affect biodiversity of microbial communities associated with metal-tolerant plants in metalliferous soils. Front. Microbiol. 9, 1425. doi:10.3389/fmicb.2018.01425
Bourceret, A., Cébron, A., Tisserant, E., Poupin, P., Bauda, P., Beguiristain, T., et al. (2016). The bacterial and fungal diversity of an aged PAH- and heavy metal-contaminated soil is affected by plant cover and edaphic parameters. Microb. Ecol. 71, 711–724. doi:10.1007/s00248-015-0682-8
Calicioglu, O., Shreve, M. J., Richard, T. L., and Brennan, R. A. (2018). Effect of pH and temperature on microbial community structure and carboxylic acid yield during the acidogenic digestion of duckweed. Biotechnol. Biofuels. 11, 275. doi:10.1186/s13068-018-1278-6
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi:10.1038/nmeth.f.303
Chen, B., Landry, M. R., Huang, B., and Liu, H. (2012). Does warming enhance the effect of microzooplankton grazing on marine phytoplankton in the ocean? Limnol. Oceanogr. 57, 519–526. doi:10.4319/lo.2012.57.2.0519
Chen, X., Zhao, Y., Zhao, X., Wu, J., Zhu, L., Zhang, X., et al. (2020). Selective pressures of heavy metals on microbial community determine microbial functional roles during composting: Sensitive, resistant and actor. J. Hazard. Mater. 398, 122858. doi:10.1016/j.jhazmat.2020.122858
Cheng, C., Li, Y., Long, M., Gao, M., Zhang, Y., Lin, J., et al. (2020). Moss biocrusts buffer the negative effects of karst rocky desertification on soil properties and soil microbial richness. Plant Soil 475, 153–168. doi:10.1007/s11104-020-0460210.1007/s11104-020-04602-4
Chun, S. J., Kim, Y. J., Cui, Y., and Nam, K. H. (2021). Ecological network analysis reveals distinctive microbial modules associated with heavy metal contamination of abandoned mine soils in Korea. Environ. Pollut. 289, 117851. doi:10.1016/j.envpol.2021.117851
DalCorso, G., Fasani, E., Manara, A., Visioli, G., and Furini, A. (2019). Heavy metal pollutions: State of the art and innovation in phytoremediation. Ijms 20 (14), 3412. doi:10.3390/ijms20143412
Deng, J., Yin, Y., Zhu, W. X., and Zhou, Y. (2018). Variations in soil bacterial community diversity and structures among different revegetation types in the Baishilazi nature reserve. Front. Microbiol. 9, 2874. doi:10.3389/fmicb.2018.02874
Deng, Y., Jiang, Y. H., Yang, Y., He, Z., Luo, F., and Zhou, J. (2012). Molecular ecological network analyses. BMC Bioinforma. 13, 113. doi:10.1186/1471-2105-13-113
Dubey, S., Shri, M., Gupta, A., Rani, V., and Chakrabarty, D. (2018). Toxicity and detoxification of heavy metals during plant growth and metabolism. Environ. Chem. Lett. 16, 1169–1192. doi:10.1007/s10311-018-0741-8
Edgar, R. C. (2013). Uparse: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi:10.1038/nmeth.2604
Elbehiry, F., Elbasiouny, H., Ali, R., and Brevik, E. C. (2020). Enhanced immobilization and phytoremediation of heavy metals in landfill contaminated soils. Water Air Soil Pollut. 231, 204. doi:10.1007/s11270-020-04493-2
Epelde, L., Becerril, J. M., Barrutia, O., González-Oreja, J. A., and Garbisu, C. (2010). Interactions between plant and rhizosphere microbial communities in a metalliferous soil. Environ. Pollut. 158, 1576–1583. doi:10.1016/j.envpol.2009.12.013
Epelde, L., Lanzén, A., Blanco, F., Urich, T., and Garbisu, C. (2014). Adaptation of soil microbial community structure and function to chronic metal contamination at an abandoned Pb-Zn mine. FEMS Microbiol. Ecol. 91, 1–11. doi:10.1093/femsec/fiu007
Faust, K., and Raes, J. (2012). Microbial interactions: From networks to models. Nat. Rev. Microbiol. 10, 538–550. doi:10.1038/nrmicro2832
Fu, S., Wei, C., Xiao, Y., Li, L., and Wu, D. (2019). Heavy metals uptake and transport by native wild plants: Implications for phytoremediation and restoration. Environ. Earth. Sci. 78, 103. doi:10.1007/s12665-019-8103-9
Haichar, F., Marol, C., Berge, O., Rangel-Castro, J. I., Prosser, J. I., Balesdent, J., et al. (2008). Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2, 1221–1230. doi:10.1038/ismej.2008.80
Hamidpour, M., Nemati, H., Abbaszadeh Dahaji, P., and Roosta, H. R. (2019). Effects of plant growth-promoting bacteria on EDTA-assisted phytostabilization of heavy metals in a contaminated calcareous soil. Environ. Geochem Health. 42 (8), 2535–2545. doi:10.1007/s10653-019-00422-3
Inelova, Z., Nurzhanova, A., Yerubayeva, G., Aitzhan, M., Djansugurova, L., and Bekmanov, B. (2021). Heavy metal contents in plants of phytocenoses of the point of besqaynar, kyzylkairat and taukaraturyk. Pak. J. Bot. 53, 511–516. doi:10.30848/PJB2021-2(3310.30848/pjb2021-2(33)
Iram, S., Ahmad, I., Javed, B., Yaqoob, S., Akhtar, K., Kazmi, M. R., et al. (2009). Fungal tolerance to heavy metals. Pak J. Bot. 41, 2583–2594.
Jiao, S., Peng, Z., Qi, J., Gao, J., and Wei, G. (2021). Linking bacterial-fungal relationships to microbial diversity and soil nutrient cycling. mSystems 6 (2), e01052–20. doi:10.1128/mSystems.01052-20
Jin, X., Wang, Z., Wu, F., Li, X., and Zhou, X. (2022). Litter mixing alters microbial decomposer community to accelerate tomato root litter decomposition. Microbiol. Spectr. 10 (3), e0018622. doi:10.1128/spectrum.00186-22
Lei, D., and Duan, C. (2008). Restoration potential of pioneer plants growing on lead-zinc mine tailings in Lanping, southwest China. J. Environ. Sci. 20, 1202–1209. doi:10.1016/S1001-0742(08)62210-X
Li, C., Quan, Q., Gan, Y., Dong, J., Fang, J., Wang, L., et al. (2020a). Effects of heavy metals on microbial communities in sediments and establishment of bioindicators based on microbial taxa and function for environmental monitoring and management. Sci. Total Environ. 749, 141555. doi:10.1016/j.scitotenv.2020.141555
Li, M. S. (2006). Ecological restoration of mineland with particular reference to the metalliferous mine wasteland in China: A review of research and practice. Sci. Total Environ. 357, 38–53. doi:10.1016/j.scitotenv.2005.05.003
Li, Q., You, P., Hu, Q., Leng, B., Wang, J., Chen, J., et al. (2020b). Effects of co-contamination of heavy metals and total petroleum hydrocarbons on soil bacterial community and function network reconstitution. Ecotoxicol. Environ. Saf. 204, 111083. doi:10.1016/j.ecoenv.2020.111083
Li, S., Wu, J., Huo, Y., Zhao, X., and Xue, L. (2021). Profiling multiple heavy metal contamination and bacterial communities surrounding an iron tailing pond in Northwest China. Sci. Total Environ. 752, 141827. doi:10.1016/j.scitotenv.2020.141827
Li, Z., Song, C., Yi, Y., and Kuipers, O. P. (2020c). Characterization of plant growth-promoting rhizobacteria from perennial ryegrass and genome mining of novel antimicrobial gene clusters. Bmc. Genomics 21 (1), 157. doi:10.1186/s12864-020-6563-7
Lin, Y., Ye, Y., Hu, Y., and Shi, H. (2019). The variation in microbial community structure under different heavy metal contamination levels in paddy soils. Ecotoxicol. Environ. Saf. 180, 557–564. doi:10.1016/j.ecoenv.2019.05.057
Lin, Y., Xiao, W., Ye, Y., Wu, C., Hu, Y., and Shi, H. (2020). Adaptation of soil fungi to heavy metal contamination in paddy fields-a case study in eastern China. Environ. Sci. Pollut. Res. 27, 27819–27830. doi:10.1007/s11356-020-09049-9
Liu, K., Zhang, H., Liu, Y., Li, Y., and Yu, F. (2020). Investigation of plant species and their heavy metal accumulation in manganese mine tailings in Pingle Mn mine, China. Environ. Sci. Pollut. Res. 27, 19933–19945. doi:10.1007/s11356-020-08514-9
López-Marcos, D., Turrión, M., and Martínez-Ruiz, C. (2020). Linking soil variability with plant community composition along a mine-slope topographic gradient: Implications for restoration. Ambio 49, 337–349. doi:10.1007/s13280-019-01193-y
Luo, Y., Wu, Y., Qiu, J., Wang, H., and Yang, L. (2019). Suitability of four woody plant species for the phytostabilization of a zinc smelting slag site after 5 years of assisted revegetation. J. Soils Sediments 19, 702–715. doi:10.1007/s11368-018-2082-4
Marguí, E., Iglesias, M., Queralt, I., and Hidalgo, M. (2006). Lead isotope ratio measurements by ICP-QMS to identify metal accumulation in vegetation specimens growing in mining environments. Sci. Total Environ. 367, 988–998. doi:10.1016/j.scitotenv.2006.03.036
Mejias Carpio, I., Ansari, A., and Rodrigues, D. (2017). Relationship of biodiversity with heavy metal tolerance and sorption capacity: A meta-analysis approach. Environ. Sci. Technol. 52 (1), 184–194. doi:10.1021/acs.est.7b04131
Ministry of Ecological Environment of the People’s Republick of China (2018). Soil environment quality-risk control standard for soil contamination of agriculture land (GB 15618-2018). Beijing, China: Environment of the People’s Republick of China.
Montiel-Rozas, M. M., Madejón, E., and Madejón, P. (2016). Effect of heavy metals and organic matter on root exudates (low molecular weight organic acids) of herbaceous species: An assessment in sand and soil conditions under different levels of contamination. Environ. Pollut. 216, 273–281. doi:10.1016/j.envpol.2016.05.080
Muthusaravanan, S., Sivarajasekar, N., Vivek, J. S., Paramasivan, T., Naushad, M., Prakashmaran, J., et al. (2018). Phytoremediation of heavy metals: Mechanisms, methods and enhancements. Environ. Chem. Lett. 16, 1339–1359. doi:10.1007/s10311-018-0762-3
Petriccione, M., Di Patre, D., Ferrante, P., Papa, S., Bartoli, G., Fioretto, A., et al. (2013). Effects of Pseudomonas fluorescens seed bioinoculation on heavy metal accumulation for mirabilis jalapa phytoextraction in smelter-contaminated soil. Water Air Soil Pollut. 224, 1645. doi:10.1007/s11270-013-1645-7
R Development Core Team, (2019). R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. AvaliableAt: http:/www.R-project.org.
Rastogi, G., Barua, S., Sani, R. K., and Peyton, B. M. (2011). Investigation of microbial populations in the extremely metal-contaminated coeur d'Alene river sediments. Microb. Ecol. 62, 1–13. doi:10.1007/s00248-011-9810-2
Schimel, J., and Schaeffer, S. (2012). Microbial control over carbon cycling in soil. Front. Microbio. 3, 348. doi:10.3389/fmicb.2012.00348
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi:10.1128/AEM.00062-07
Xu, J., Zheng, L., Xu, L., and Wang, X. (2020). Uptake and allocation of selected metals by dominant vegetation in Poyang Lake wetland: From rhizosphere to plant tissues. Catena 189, 104477. doi:10.1016/j.catena.2020.104477
Yang, S., Cao, J., Li, F., Peng, X., Peng, Q., Yang, Z., et al. (2016). Field evaluation of the effectiveness of three industrial by-products as organic amendments for phytostabilization of a Pb/Zn mine tailings. Environ. Sci. Process. Impacts 18, 95–103. doi:10.1039/c5em00471c
Yuan, J., Wen, T., Zhang, H., Zhao, M., Penton, C. R., Thomashow, L. S., et al. (2020). Predicting disease occurrence with high accuracy based on soil macroecological patterns of Fusarium wilt. ISME J. 14, 2936–2950. doi:10.1038/s41396-020-0720-5
Zaefarian, F., Rezvani, M., Rejali, F., Ardakani, M. R., and Noormohammadi, G. (2021). Effect of heavy metals and arbuscular mycorrhizal fungal on growth and nutrients (N, P, K, Zn, Cu and Fe) accumulation of alfalfa (medicago sativa L.). American-Eurasian J. Agric. Environ Sci. 11 (3), 346–352. doi:10.1007/s11356-020-08538-1
Keywords: trace element, Pb-Zn waste heap, biodiversity, co-occurrence patterns, plant-soil interaction
Citation: Sun C, Wu P, Wang G and Kong X (2022) Heavy metal pollution decreases the stability of microbial co-occurrence networks in the rhizosphere of native plants. Front. Environ. Sci. 10:979922. doi: 10.3389/fenvs.2022.979922
Received: 28 June 2022; Accepted: 18 August 2022;
Published: 15 September 2022.
Edited by:
Buyun Du, Jiangsu Open University, ChinaReviewed by:
Wenjie Ren, Institute of Soil Science (CAS), ChinaCopyright © 2022 Sun, Wu, Wang and Kong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pan Wu, cHd1QGd6dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.