
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 05 August 2022
Sec. Conservation and Restoration Ecology
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.952414
This article is part of the Research Topic Water and Ecological System: Response, Management, and Restoration View all 15 articles
Environmental DNA (eDNA) technology has become an alternative tool for monitoring aquatic communities due to its sensitive, economical, and non-invasive properties. However, the application of this technique is often limited by the complexity of environmental conditions, which often poses a barrier to the transmission of biological information. Here, we conducted a series of experiments with grass carp as the target species to evaluate the effects of total dissolved gas (TDG) supersaturation and sediment on the persistence of eDNA under different flow conditions. The results showed TDG supersaturation promoted eDNA decay in still water but with no significant effect in flowing water for rapid dissipation of TDG. For sediment, its presence accelerated the decay of eDNA no matter the flow conditions. The grass carp eDNA showed an exponential decay pattern in water and the decay rate constant decreased gradually with time. Our study highlights the importance of integrating experimental results with the natural environment and provides an important reference for species monitoring using eDNA technology in aquatic ecosystems with high dams built.
The widespread environmental DNA (eDNA) technology utilizes DNA molecules that organisms continuously release into the environment (Li et al., 2018), which consist predominately of genetic materials in the form of cell-free DNA (Lydolph et al., 2005), mucus, feces (Merkes et al., 2014) and cells (Turner et al., 2014). As an emerging method, eDNA technology combines molecular biology with species monitoring and improves the ability of species detection by eliminating the effects of morphological characteristics (Sansom and Sassoubre, 2017). Additionally, it has higher temporal and spatial sampling resolution and could improve sampling accuracy compared to traditional biomonitoring methods (Seymour et al., 2018). Thus, eDNA technology is gradually complementing or even replacing the application of traditional methods (Czeglédi et al., 2021). Nowadays, eDNA technology is used frequently for invasive species detection (Crane et al., 2021; Whitaker et al., 2021), endangered species monitoring (Budd et al., 2021), species biomass estimation (Tillotson et al., 2018), and biodiversity evaluation (Port et al., 2016; Vašutová et al., 2021). Among many ecosystems, aquatic ecosystems are especially appropriate for eDNA analysis because water is a highly efficient medium for transport and deposition, and eDNA can be easily sampled and detected in all water bodies (Bylemans et al., 2019; Lawson Handley et al., 2019; Djurhuus et al., 2020; Broadhurst et al., 2021). Since eDNA technology was first applied to investigate invasive species in French (Ficetola et al., 2008), it has been common in research work on aquatic organisms from a range of different ecological communities, such as fish (Dully et al., 2021; Sales et al., 2021), crustaceans (Johnsen et al., 2020; Crane et al., 2021), amphibians (Valentini et al., 2016), and mollusks (Sansom and Sassoubre, 2017).
Although eDNA technology is currently being increasingly used in aquatic ecosystems, the biological information conveyed by eDNA is often ambiguous at a given sampling time and site (Jo and Minamoto, 2021). Available study shows that the removal of eDNA in aquatic ecosystems mainly includes three processes: degradation, sedimentation, and transport (Harrison et al., 2019). The degradation of eDNA in water is affected by various factors, such as temperature (Eichmiller et al., 2016), pH (Seymour et al., 2018), and UV(Machler et al., 2018), and bacteria (Wei et al., 2018). These biotic and abiotic elements increase eDNA degradation by disrupting DNA structure directly and altering microbial metabolism and enzyme kinetics indirectly (Barnes et al., 2014; Strickler et al., 2015). For sedimentation and transport, different flow conditions may cause their contribution to eDNA persistence differently. First in the lentic environment (lakes and reservoirs), when eDNA molecules are released into the water column, they will spread around in a short time and some larger eDNA molecules will settle down over time, which leads to a decrease in eDNA concentration. However, additional removal processes occur in a lotic environment (rivers and streams) because the eDNA molecules might be transported downstream with the flow before settling down. eDNA of organisms might decay more rapidly in flowing water and could not even be detectable for a short time (Stoeckle et al., 2017).
Although different flow conditions determine the difference in the removal process of eDNA, sediment might be an important influencing factor in two different aquatic ecosystems. Because both in the lentic and lotic environment, eDNA could be removed from the water by falling into sediment interstices and being adsorbed on the surface of the sediment. Additionally, after a period of preservation in sediment, eDNA molecules could be resuspended into the water, which always leads to a problem of false-positive eDNA detection (Stoeckle et al., 2017). Therefore, the presence of sediment may increase the complexity of eDNA technology application and its impact on eDNA persistence in aquatic ecosystems still needs further validation. For the complex, open and dynamic ecosystem of rivers, in addition to their flow characteristics, artificial structures such as dams can also have an impact on aquatic species by altering river structures and creating a range of environmental problems (Consuegra et al., 2021). When high dams release water during flood season, a large amount of air below the dams could be swept into rivers and dissolves in water under very high pressure, leading to the supersaturation of total dissolved gas (TDG) in water and gas bubble disease (GBD) in aquatic organisms (Weitkamp and Katz, 1980; Wang et al., 2017). These problems significantly affect the survival of aquatic species (especially farmed fish) and are harmful to the maintenance of biodiversity in rivers and reservoirs. Existing studies have focused questions on the effects of river fragmentation and GBD on aquatic organisms caused by high dam construction (Weitkamp and Katz, 1980; Consuegra et al., 2021), while the effect of TDG supersaturation on eDNA persistence is still unknown, which leads to uncertainty in the application of eDNA technology in affected water bodies and thus hinders the transfer of biological information.
Here, we conducted a series of experiments to investigate the effects of total dissolved gas supersaturation and sediment on the persistence of eDNA in water. The grass carp (Ctenopharyngodon Idella), an important freshwater farmed fish in China, is used as the target species. The results are intended to provide theoretical support for applications of eDNA technology in rivers and reservoirs downstream of high dams.
A total of six grass carp used in this study were kept in 100 L tanks after being purchased from the aquaculture section of Baijia Agricultural Products Wholesale Market in Chengdu, China. To ensure the stability of eDNA concentration in water, the grass carp were not fed during the experiment. We used the TDG supersaturation generation device invented by Sichuan University to prepare supersaturated total dissolved gas water (Li et al., 2010). The principle of the device is to mix the high-speed circulating water flow (experimental water, without grass carp eDNA) with the high-pressure gas generated by air compressor to create the conditions for supersaturated TDG water. TDG saturation was controlled in the experiment by adjusting the amount of water pumped into the reactor vessel. Previous study has shown that the TDG saturations in rivers downstream of high dams range from 122 to 140% (Feng et al., 2014). To keep the initial TDG saturation in water at a high level and to maintain TDG supersaturation over a long time, the TDG saturation of water used in the experiment was set to about 145%. Homogeneous river sand with a particle size of 1 mm was selected as sediment in this study, which was purchased from a river sand manufacturer in Chengdu, China. Before the experiment, the sediment was sifted, cleaned and dried to remove impurities.
To investigate the effects of total dissolved gas supersaturation and sediment, eight treatments were designed based on flow conditions, whether the total dissolved gas was supersaturated, and whether the sediment was present (Table 1; Figure 1). The effect of a single factor and the combined effect of multiple factors on the persistence of eDNA was investigated through a controlled variables approach.
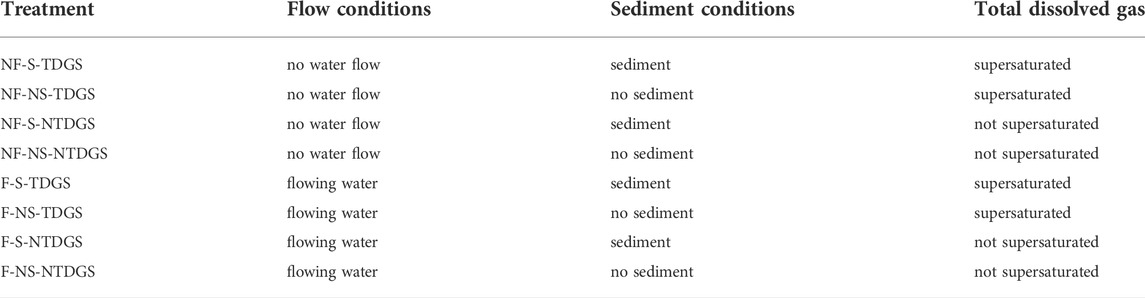
TABLE 1. Abbreviations of the treatments and the corresponding influencing factor settings. NF/F means no water flow/flowing water, NS/S means no sediment/sediment, and NTDGS/TDGS means total dissolved gas not supersaturated/total dissolved gas supersaturated.
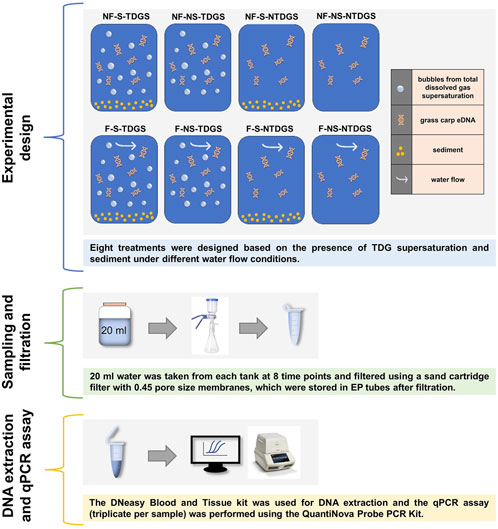
FIGURE 1. Overall flowchart for the tank experiments. Eight treatments were carried out based on flow conditions, whether the total dissolved gas was supersaturated, and whether the sediment was present. 20 ml water was collected at 8 time points and filtered with 0.45 pore size membranes. After extracting eDNA, the copy numbers of DNA were estimated by qPCR.
The eight treatments were carried out in water tanks of the same size, which were 50 cm in length, 10 cm in width, and 20 cm in height and made of polyethylene plastic. Four tanks of the flowing water condition were equipped with external short pipes (2 cm in length and 25 cm in diameter) on both sides, and two soft PVC rubber pipes (1 m in length and 25 cm in diameter) were externally connected. The other end of the PVC rubber pipes was connected with water pumps with the same power of 55W, and water in tanks was driven to circulate by water pumps to achieve the same flow conditions. We set the sediment thickness to 2 cm and the sediment gravel was evenly distributed at the bottom of the tanks. We added the water uniformly and slowly to achieve a homogeneous mixture of grass carp eDNA water and supersaturated TDG water or pure water in tanks. The volume of the total water in one tank was 4 L and the ratio of two different water is controlled at 1:1. To detect changes in eDNA concentration over time, we collected 20 ml water from each tank at the initial time and then at 1, 3, 6, 11, 23, 35, and 48 h elapsed time.
An additional experiment was conducted to investigate the changes in TDG saturation for those TDGS treatments to eliminate the interference of TDG saturation measurement on eDNA concentration. The supersaturated TDG water was added with the same dissolved gas saturation (TDG saturation changed to about 120% after mixing the two kinds of water). Other experimental settings were consistent with the eDNA experiment mentioned above. The TDG saturation level was recorded with a portable TGP meter (OxyGuard, Denmark) at eight time points: Pre, 0, 0.5, 1, 3, 6, 18, and 24 h.
The collected water samples were filtered using a sand cartridge filter with 0.45 pore size membranes. After filtration, filter membranes were stored in numbered 1.5 ml centrifugal tubes and immediately followed by DNA extraction.
The DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) was used for DNA extraction. We added 180 µl of DNA lysate Buffer ATL and 20 µl of Proteinase K to the centrifugal tube and incubated it in a thermostatic metal bath at 56°C for 12 min. We then added 200 µl of Buffer AL to the tube and vortexed it for 15s and incubated it again at 56°C for 12 min. After short centrifugation at 8,000 rpm, we transferred 400 µl supernatant to a new centrifugal column and centrifuged for 1 min (8,000 rpm) to ensure all liquid entered the collection tube of the centrifuge column. We then added 500 µl of Buffer AW1 to the column and centrifuged it at 8,000 rpm for 1 min. After replacing the collection tube with a new column, we added 500 µl of Buffer AW2 to the column and centrifuged it for 1 min (8,000 rpm). Finally, we added 100 µl of Buffer AE to the column and centrifuged it again for 1 min (8,000 rpm), the solution retained in the tube was the desired eDNA sample solution. All eDNA samples obtained were stored at −20°C for subsequent qPCR assay.
To detect and quantify the DNA of grass carp using qPCR, primers and probe were designed with Primer3 version 4.0.0 based on the full-length sequence of grass carp mtDNA, which is available from the National Center of Biotechnology Information (NCBI) (GenBank, https://www.ncbi.nlm.nih.gov). To improve the amplification efficiency of the designed primer-probe, we selected two primer-probes for the specificity test (Table 2).
We used the DNeasy blood tissue kit (Qiagen, Hilden, Germany) to extract DNA from grass carp tissues and made the extracted solution as a positive DNA sample. We then PCR-amplified the DNA sample using two designed primer-probes and subjected the products to electrophoresis and sequencing. Analysis of the electrophoresis results showed the length and sequence of the product obtained from the No.2 primer-probe matched the expected product better. Accordingly, the No. 2 primer-probe was selected for subsequent experiments in this study.
qPCR assay was performed using the QuantiNova Probe PCR Kit (Qiagen, Hilden, Germany). Individual qPCR reactions (in triplicate) consisted of 20 µl of Probe PCR Master Mix, 6 µl of RNase-Free Water, 1.6 µl of forward and reverse primers (0.8 µl each), 0.4 µl of probe, and 2 µl of eDNA sample. The reactions were performed with the following steps: a PCR initial activation of 95°C for 2 min, 39 cycles with 95°C for 5 s (denaturation), and 60°C for 5 s (combined annealing/extension). The standard curve of this experiment was determined by a series of standard samples of the target gene with known concentration gradients. The regression equation was y = −3.4706x+42.582 with correlation coefficient (r2) of 0.9997 and amplification efficiency of 94% (Supplementary Material). The eDNA concentration at each time point was calculated by the Ct value and sampled water volume (i.e.,20 ml) combined with the standard curve. Samples were considered as not detected if there was no exponential growth during the 39cycles, or if only 1 amplification was detected in triplicate qPCR assays.
The decay of aquatic organisms’ eDNA mostly showed an exponential pattern in water. Accordingly, we evaluated the decay of grass carp eDNA by a Single First-Order rate model (SFO) in this study. The equation of the SFO model is as follows:
where
The decay of aquatic organisms’ eDNA is also a multiphase process, and different forms of DNA may decay at different rates in water. When DNA molecules that decay more easily are removed from water, eDNA concentration might decrease and the decay rate constant might change in the absence of new eDNA sources, which could make the application of the SFO model inappropriate. Therefore, a Weibull decay model was fitted to the data to verify whether the grass carp eDNA decay rate constant would change with time in this study. The Weibull decay model equation is as follows:
Where β is the Weibull parameter, allowing the decay rate constant to vary with time, β < 1 means the decay rate constant decreases with time; β > 1 means the decay rate constant increases with time, when β = 1, the model is simplified to the SFO model. The goodness of fit of the two models was compared by calculating the determinate coefficient R2.
Multi-factor analysis of variance was used to test whether TDG supersaturation and sediment under different flow conditions had a significant effect on decay rate constant. A significant effect was determined by comparing the p-value to the significance level (α = 0.05), with p < 0.05 indicating a significant effect and p > 0.05 indicating a non-significant effect.
We quantified the changes in grass carp eDNA concentration over time in still and flowing water and found that eDNA concentration decreased with time in all eight treatments (Figure 2). In still water, grass carp eDNA concentration of NF-S-TDGS and NF-S-NTDGS decreased more rapidly within the first 11 h and tended to be 0 copies/ml at 48 h. For NF-NS-TDGS, although the rate of decline in eDNA concentration was slower than NF-S-TDGS and NF-S-NTDGS in the first 11 h, the eDNA concentration was similarly close to 0 copies/ml in the last detection. As expected, NF-NS-NTDGS had higher eDNA concentration than the other three treatments at all time points and the slowest rate of decline. Thus, it can be easily found that TDG supersaturation and sediment contributed to grass carp eDNA decay and eDNA decayed fastest when both were present in still water.
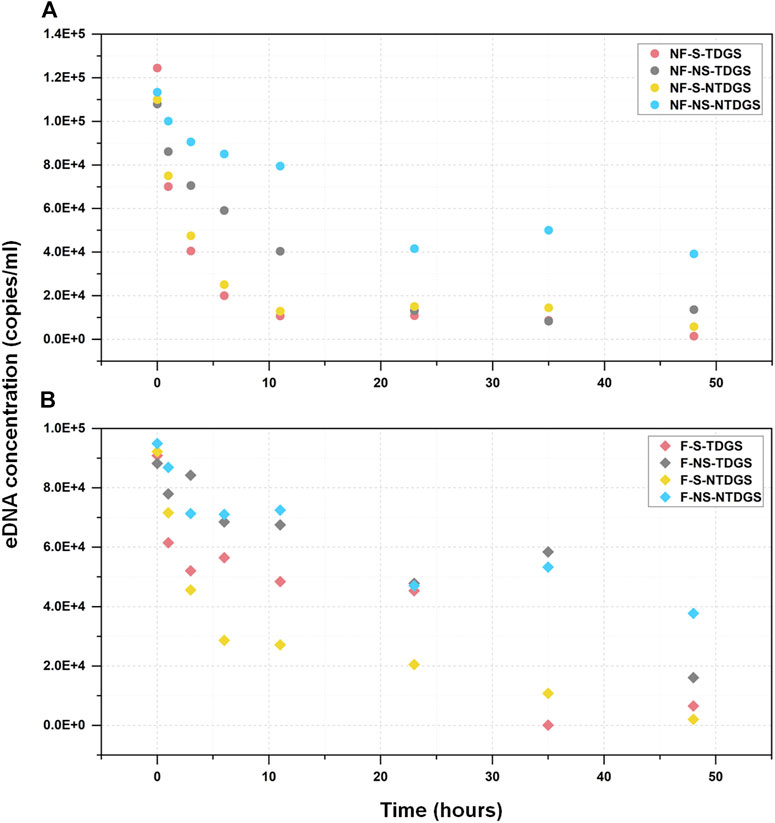
FIGURE 2. The relationship between the grass carp eDNA concentration (mean value) and sampling time. The circular and prismatic dots represent the eDNA concentration at each time point in still water (A) and flowing water (B) respectively. The colors represent different treatments: red, NF-S-TDGS/F-S-TDGS; gray, NF-NS-TDGS/F-NS-TDGS; yellow, NF-S-NTDGS/F-S-NTDGS; blue, NF-NS-NTDGS/F-NS-NTDGS.
The variation pattern of grass carp eDNA concentration in flowing water was more chaotic. But in the same way, the decline rate was faster for F-S-TDGS and F-S-NTDGS where sediment was present in flowing water, which indicated the presence of sediment accelerated the decline of eDNA concentration no matter what the flow conditions were. However, for F-NS-TDGS and F-NS-NTDGS, the eDNA concentration of both treatments was basically at the same level during the experimental period, and TDG supersaturation did not promote the decrease of eDNA concentration, which showed the effects of TDG supersaturation varied with flow conditions in this study.
By comparing the fitting curves and determination coefficient (R2) of the two models (Figure 3; Table 3), it can be found that the Weibull decay model had better goodness of fit and can better describe the removal process of grass carp eDNA in water. Additionally, the parameter β for each treatment was less than 1, which indicated the decay rate constant of grass carp eDNA gradually decreased with time.
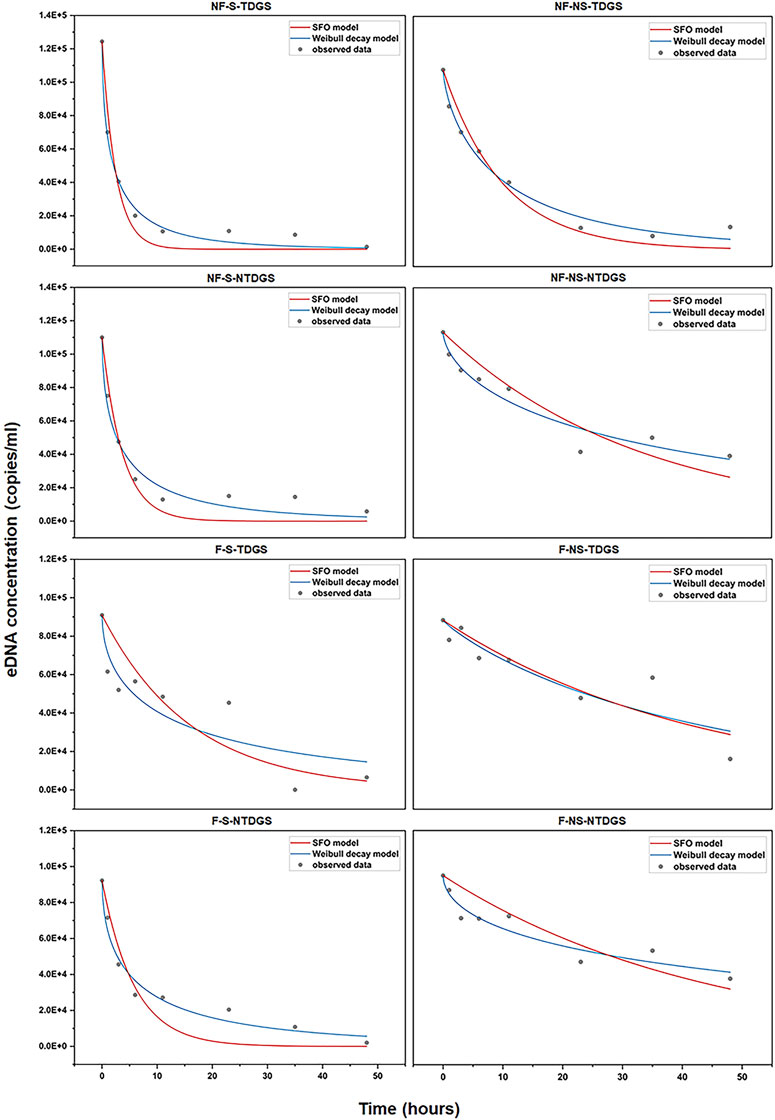
FIGURE 3. Fit of the SFO model (red curve) and the Weibull decay model (blue curve) for each treatment, the grey dots represent the grass carp eDNA concentration detected at each time point.
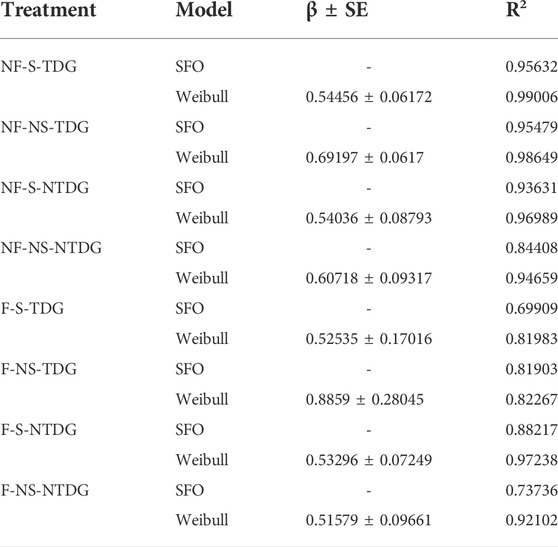
TABLE 3. The Weibull parameter (β ± SE) and determination coefficient (R2) of the Weibull decay model and the SFO model.
Although the Weibull decay model indicated the decay rate constant varied with time, we chose to further analyze the decay rate constant (mean value) calculated by the Weibull decay model as it was more applicable than the SFO model. The results (Figure 4) showed the k value increased due to the existence of TDG supersaturation or sediment and reached a maximum when both factors were present (NF-S-TDGS, k ± SE = 0.61229 ± 0.05838) in still water. But in flowing water, only the presence of sediment increased the decay rate constant, and TDG supersaturation did not accelerate the decay of grass carp eDNA. It can be seen that the effects of sediment in still and flowing water were consistent, but TDG supersaturation showed different effects with different flow conditions.
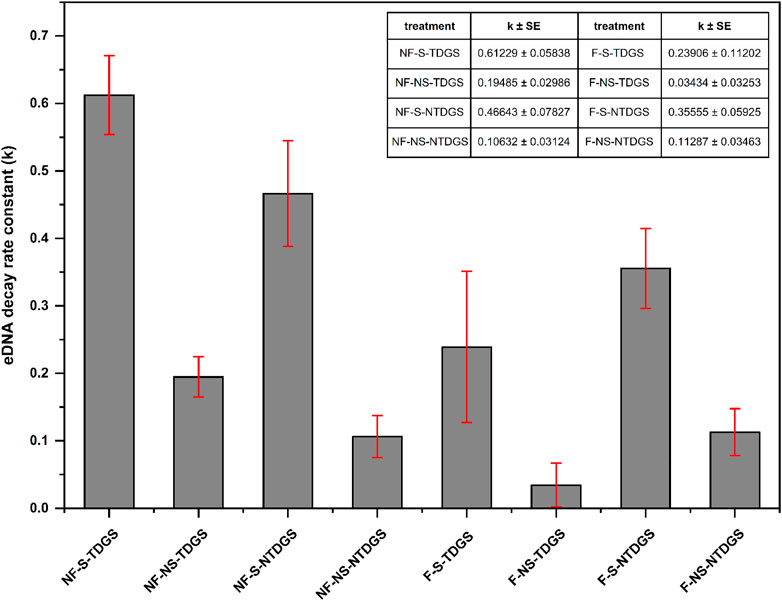
FIGURE 4. The decay rate constant (k) estimated from the Weibull decay model for each treatment, the error bars represent standard error (SE).
The effects of TDG supersaturation, sediment and flow conditions on the decay rate constant (k, the Weibull decay model) were variable (Table 4). The main effect of TDG supersaturation on the decay rate constant was not significant (p = 0.569), but the main effect of sediment and flow conditions was significant (p < 0.001). The interaction effect of TDG supersaturation with sediment was not significant (p = 0.275), but the interaction effect of TDG supersaturation with flow conditions was significant (p = 0.001). There was also a significant interaction effect between sediment and flow conditions (p = 0.011), but the interaction effect between TDG supersaturation, sediment, and flow conditions was not significant (p = 0.364).

TABLE 4. Results of multi-factor analysis of variance. Analysis of the main effect and interaction effect of TDG supersaturation, sediment and flow conditions on the decay rate constant (k). Significant p-values are indicated in bold.
On the premise of significant interaction, we conducted a simple effect test to analyze the different effects of TDG supersaturation and sediment on decay rate constant under different flow conditions. The results (Table 5) showed that the decay rate constant of eDNA in still water was significantly higher than that in flowing water when dissolved gas was supersaturated and sediment existed in water (mean difference (I-J) = −0.263, p < 0.001; mean difference (I-J) = −0.233, p < 0.001).

TABLE 5. Results of simple effect test. Analysis of the different effects of TDG supersaturation and sediment on decay rate constant under different flow conditions. Significant p-values are indicated in bold.
Taken together, TDG supersaturation had a significant contribution to eDNA decay in still water but not in flowing water. The effect of sediment, although slightly different depending on the flow conditions, showed a significant effect on grass carp eDNA decay in general.
TDG is a gas mixture with the same composition as the atmosphere (Weitkamp and Katz, 1980), in which the partial pressure of oxygen is about 21%. As a biologically active gas, a change in oxygen content affects microbial metabolism in water (Barnes et al., 2014). Existing studies have shown DNA can be rapidly degraded in the aquatic environment due to oxidation and microbial activity (Lindahl, 1993; Torti et al., 2015). However, when oxygen content and microbial action are reduced or absent, these genetic materials can last for a long time (Seymour et al., 2018). Accordingly, for this study, the effect of TDG supersaturation which accelerated the decay of eDNA in still water was likely to alter the oxygen content of water and thus ultimately enhanced the microbial metabolism. However, in flowing water, TDG supersaturation did not have the same effect. Combined with the results of dissolved gas saturation monitoring (Figure 5), it can be found that the rate of TDG dissipation in flowing water was faster than in still water, and the gas dissolved saturation was already below the standard limit of TDG supersaturation (110%) (Engineers, 2005) within half an hour. This suggested that water flow affected the rate of total dissolved supersaturated gas dissipation, which was consistent with the previous study (Li et al., 2013). If the turbulence intensity of the flow increases, the TDG dissipation rate will increase. Consequently, it can be hypothesized that TDG supersaturation in flowing water did not significantly affect the decay of eDNA can be attributed to the rapid dissipation of TDG caused by flow turbulence, eventually failing to significantly affect microbial metabolic activities as in still water.
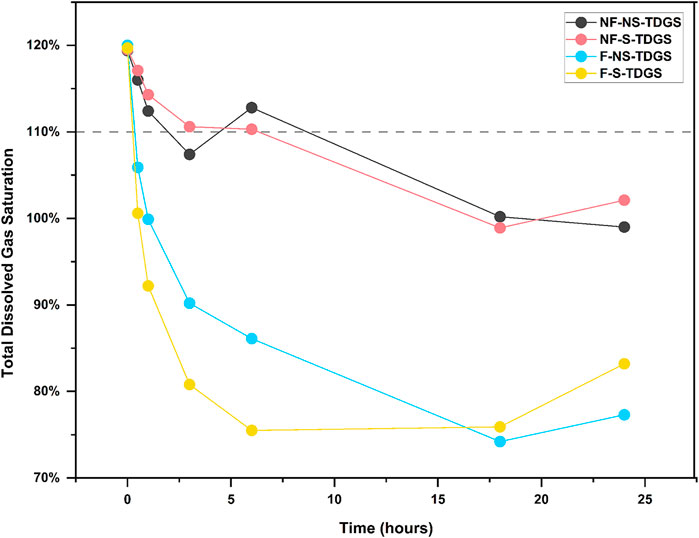
FIGURE 5. Variation of TDG saturation in TDGS treatments during the experiment period. The dots indicate the TDG saturation at each time point for different treatments: black, NF-NS-TDGS; red, NF-S-TDGS; blue, F-NS-TDGS, yellow, F-S-TDGS. The gray dashed line represents the minimum standard limit of 110% for TDG supersaturation.
Unlike the effect of TDG supersaturation, sediment played a crucial role in the persistence of grass carp eDNA both in still and flowing water, which is in line with previous research (Stoeckle et al., 2017). When biological DNA molecules and cells are released into the water column, in addition to their gradual decay, they will settle under the force of gravity and eventually attach to the sediment surface or fall into the sediment interstices. When these eDNA molecules come into contact with sediment particles, they will bind to gravel particles due to electrostatic interactions and other forces (Hou et al., 2014) and adhere to biofilms on the sediment surface. eDNA from organisms will be degraded by components such as heterotrophic microorganisms in biofilm as a carbon source (Jerde et al., 2016), eventually reducing the detectable eDNA concentration in the water column. Therefore, even though a small fraction of eDNA molecules in this study will be resuspended into water under the disturbance of water flow (Turner et al., 2015), the presence of sediment still significantly reduced the concentration of eDNA in water.
The SFO model suggests the decay processes for intracellular DNA and extracellular free DNA fragments occur on similar time scales, and the rate of cell decomposition and DNA decay in water is the same (Saito and Doi, 2021). Therefore, researchers can use different combinations of experiments to quantify the decay rate constant under different conditions and compare their difference (Bylemans et al., 2018). However, in this study, the Weibull decay model was proved to have a better description of the decay pattern of grass carp eDNA, with a rapid decay period at the initial time and then the decay rate constant decreasing with time. Previous studies have also shown that the decay rate decreases with time in a model that better reflects the decay process of biological DNA in water (Bylemans et al., 2018; Eichmiller et al., 2016; Shogren et al., 2018). Thus, it can be inferred that eDNA does exist in water with different decay rates (e.g., nuclear and mitochondrial DNA, long fragments and short fragments of DNA). Some eDNA fragments decay at a faster rate, and the DNA concentration in water decreases gradually with time, ultimately resulting in a decrease in the total decay rate constant. Therefore, when considering the persistence of eDNA in natural environments, the use of the Weibull decay model can more accurately predict the residence time of biological eDNA in water or more accurately predict the initial eDNA concentration in this area based on the concentration of eDNA detected at a certain moment. This allows researchers to better apply eDNA analysis methods and more accurately understand the basic information of eDNA in the natural environment.
Experimental variables in this study, including total dissolved gas supersaturation, sediment, and flow conditions, had measurable effects on the eDNA persistence of grass carp. However, all the results were obtained under controlled experimental conditions, which did not fully reflect the patterns in the natural environment. Because in aquatic ecosystems, these factors may play a more complex role in biological eDNA. For TDG supersaturation, the experiment was conducted to explore its effect on eDNA persistence by adding dissolved gas supersaturated water. Rapid dissipation of TDG resulted in almost no effect on eDNA decay in flowing water. But in the natural environment, TDG supersaturation continues to occur in rivers downstream of high dams, and it is difficult to release quickly into the atmosphere and could be transported over long distances (Yuan et al., 2020). Accordingly, in natural rivers, the effect of TDG supersaturation on biological eDNA might be similar to that in still water of this study. Secondly, this study demonstrated the presence of sediment can improve the removal of eDNA from water through adsorption and biofilms. But at the same time, sediment can also protect eDNA molecules from nuclease degradation and chemical damage (Romanowski et al., 1991; Hou et al., 2014) and protect cell and tissue fragments from shear forces and microbial degradation (Bradford et al., 2013). These effects tend to make the persistence of eDNA much higher in sediment (Troth et al., 2021), thus creating an opportunity for resuspension of eDNA and blurring the inference of spatial and temporal scales of eDNA in water columns (Turner et al., 2015). Finally, the flow conditions in this study are fundamentally designed to simulate the turbulence of natural rivers, where the dynamic processes of DNA molecules in water are dominated by dilution, degradation, sedimentation, retention, and resuspension. However, eDNA in rivers could also be affected by long-distance transportation along with water flow (Shogren et al., 2017), which tends to reduce the detectable concentration of eDNA at the original location and increases the uncertainty of eDNA technology application. Given the above, results based on controlled experiments must be carefully interpreted and analyzed in close relation to factors in the natural environment, only in this way can eDNA technology be applied to species monitoring properly.
This study investigated the effects of TDG supersaturation and sediment on the persistence of grass carp eDNA in still and flowing water, and the results showed:
1) the effect of TDG supersaturation varied with flow conditions, with TDG supersaturation promoting eDNA decay in still water, but with no significant effect in flowing water due to rapid dissipation of TDG.
2) The effect of sediment on eDNA persistence is consistent in different flow conditions. The presence of sediment accelerated the decay of grass carp eDNA in water.
3) The Weibull decay model with decay rate constant varies with time can better describe the decay process of eDNA than the Single First-Order rate model (SFO model), which suggested grass carp eDNA decayed rapidly during the initial period, and then the decay rate constant decreased with time.
In conclusion, TDG supersaturation and sediment can affect the persistence of aquatic eDNA in different ways under still and flowing water conditions, and all factors interact with each other and act together. Meanwhile, it is worth noting that results based on controlled experiments might deviate from the true patterns in the natural environment. Therefore, in practical applications, the interpretation of sampling results should always be integrated with the prevailing environmental conditions to get the most accurate species information.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Ethics Committee, College of Life Sciences, Sichuan University.
All authors conceived and planned the experiments. LH performed the experiment, analyzed the data, interpreted the results, wrote the manuscript, and designed the figures. JZ helped perform the analysis and edited the manuscript with constructive discussions. YW acquired funding, participated in data analysis, and revised it critically for important intellectual content. All authors approved of the final draft.
This work was supported by the National Natural Science Foundation of China (Grant NO. 52079082) and the Open Research Fund of State Key Laboratory of Simulation and Regulation of Water Cycle in River Basin, China Institute of Water Resources and Hydropower Research (Grant NO. IWHR-SKL-KF202107).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.952414/full#supplementary-material
Barnes, M. A., Turner, C. R., Jerde, C. L., Renshaw, M. A., Chadderton, W. L., Lodge, D. M., et al. (2014). Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48, 1819–1827. doi:10.1021/es404734p
Bradford, S. A., Morales, V. L., Zhang, W., Harvey, R. W., Packman, A. I., Mohanram, A., et al. (2013). Transport and fate of microbial pathogens in agricultural settings. Crit. Rev. Environ. Sci. Technol. 43, 775–893. doi:10.1080/10643389.2012.710449
Broadhurst, H. A., Gregory, L. M., Bleakley, E. K., Perkins, J. C., Lavin, J. V., Bolton, P., et al. (2021). Mapping differences in mammalian distributions and diversity using environmental DNA from rivers. Sci. Total Environ. 801, 149724. doi:10.1016/j.scitotenv.2021.149724
Budd, A. M., Cooper, M. K., Port, A. L., Schils, T., Mills, M. S., Deinhart, M. E., et al. (2021). First detection of critically endangered scalloped hammerhead sharks (Sphyrna lewini) in Guam, Micronesia, in five decades using environmental DNA. Ecol. Indic. 127, 107649. doi:10.1016/j.ecolind.2021.107649
Bylemans, J., Furlan, E. M., Gleeson, D. M., Hardy, C. M., and Duncan, R. P. (2018). Does size matter? An experimental evaluation of the relative abundance and decay rates of aquatic environmental DNA. Environ. Sci. Technol. 52, 6408–6416. doi:10.1021/acs.est.8b01071
Bylemans, J., Gleeson, D. M., Duncan, R. P., Hardy, C. M., and Furlan, E. M. (2019). A performance evaluation of targeted eDNA and eDNA metabarcoding analyses for freshwater fishes. Environ. DNA 1, 402–414. doi:10.1002/edn3.41
Consuegra, S., O'Rorke, R., Rodriguez-Barreto, D., Fernandez, S., Jones, J., Garcia de Leaniz, C., et al. (2021). Impacts of large and small barriers on fish assemblage composition assessed using environmental DNA metabarcoding. Sci. Total Environ. 790, 148054. doi:10.1016/j.scitotenv.2021.148054
Crane, L. C., Goldstein, J. S., Thomas, D. W., Rexroth, K. S., and Watts, A. W. (2021). Effects of life stage on eDNA detection of the invasive European green crab (Carcinus maenas) in estuarine systems. Ecol. Indic. 124, 107412. doi:10.1016/j.ecolind.2021.107412
Czeglédi, I., Sály, P., Specziár, A., Preiszner, B., Szalóky, Z., Maroda, Á., et al. (2021). Congruency between two traditional and eDNA-based sampling methods in characterising taxonomic and trait-based structure of fish communities and community-environment relationships in lentic environment. Ecol. Indic. 129, 107952. doi:10.1016/j.ecolind.2021.107952
Djurhuus, A., Closek, C. J., Kelly, R. P., Pitz, K. J., Michisaki, R. P., Starks, H. A., et al. (2020). Environmental DNA reveals seasonal shifts and potential interactions in a marine community. Nat. Commun. 11, 254. doi:10.1038/s41467-019-14105-1
Dully, V., Balliet, H., Frühe, L., Däumer, M., Thielen, A., Gallie, S., et al. (2021). Robustness, sensitivity and reproducibility of eDNA metabarcoding as an environmental biomonitoring tool in coastal salmon aquaculture – an inter-laboratory study. Ecol. Indic. 121, 107049. doi:10.1016/j.ecolind.2020.107049
Eichmiller, J. J., Best, S. E., and Sorensen, P. W. (2016). Effects of temperature and trophic state on degradation of environmental DNA in lake water. Environ. Sci. Technol. 50, 1859–1867. doi:10.1021/acs.est.5b05672
Engineers, U. A. C. o. (2005). Technical analysis of TDG processes. Portland: US Army Corps of Engineers-Northwest Division, Environmental Resources and Fish Planning Offices.
Feng, J., Li, R., Ma, Q., and Wang, L. (2014). Experimental and field study on dissipation coefficient of supersaturated total dissolved gas. J. Central South Univ. 21, 1995–2003. doi:10.1007/s11771-014-2148-4
Ficetola, G. F., Miaud, C., Pompanon, F., and Taberlet, P. (2008). Species detection using environmental DNA from water samples. Biol. Lett. 4, 423–425. doi:10.1098/rsbl.2008.0118
Harrison, J. B., Sunday, J. M., and Rogers, S. M. (2019). Predicting the fate of eDNA in the environment and implications for studying biodiversity. Proc. R. Soc. B 286, 20191409. doi:10.1098/rspb.2019.1409
Hou, Y., Wu, P., and Zhu, N. (2014). The protective effect of clay minerals against damage to adsorbed DNA induced by cadmium and mercury. Chemosphere 95, 206–212. doi:10.1016/j.chemosphere.2013.08.069
Jerde, C. L., Olds, B. P., Shogren, A. J., Andruszkiewicz, E. A., Mahon, A. R., Bolster, D., et al. (2016). Influence of stream bottom substrate on retention and transport of vertebrate environmental DNA. Environ. Sci. Technol. 50, 8770–8779. doi:10.1021/acs.est.6b01761
Jo, T., and Minamoto, T. (2021). Complex interactions between environmental DNA (eDNA) state and water chemistries on eDNA persistence suggested by meta-analyses. Mol. Ecol. Resour. 21, 1490–1503. doi:10.1111/1755-0998.13354
Johnsen, S. I., Strand, D. A., Rusch, J. C., and Vrålstad, T. (2020). Environmental DNA (eDNA) monitoring of noble crayfish Astacus astacus in lentic environments offers reliable presence-absence surveillance – but fails to predict population density. Front. Environ. Sci. 8. doi:10.3389/fenvs.2020.612253
Lawson Handley, L., Read, D. S., Winfield, I. J., Kimbell, H., Johnson, H., Li, J., et al. (2019). Temporal and spatial variation in distribution of fish environmental DNA in England’s largest lake. Environ. DNA 1, 26–39. doi:10.1002/edn3.5
Li, F., Peng, Y., Fang, W., Altermatt, F., Xie, Y., Yang, J., et al. (2018). Application of environmental DNA metabarcoding for predicting anthropogenic pollution in rivers. Environ. Sci. Technol. 52, 11708–11719. doi:10.1021/acs.est.8b03869
Li, R., Hodges, B. R., Feng, J., and Yong, X. (2013). Comparison of supersaturated total dissolved gas dissipation with dissolved oxygen dissipation and reaeration. J. Environ. Eng. New. York. 139, 385–390. doi:10.1061/(ASCE)EE.1943-7870.0000598
Li, R., Huang, X., Li, K., Yi, W., and Li, J. (2010). Device for generation of total dissolved gas supersaturation in water body and research on influence of total dissolved gas supersaturation on fish. CN 201479739 U. Chengdu.
Lindahl, T. (1993). Instability and decay of the primary structure of DNA. Nature 362, 709–715. doi:10.1038/362709a0
Lydolph, M. C., Jacobsen, J., Arctander, P., Gilbert, M. T. P., Gilichinsky, D. A., Hansen, A. J., et al. (2005). Beringian paleoecology inferred from permafrost-preserved fungal DNA. Appl. Environ. Microbiol. 71, 1012–1017. doi:10.1128/AEM.71.2.1012-1017.2005
Machler, E., Osathanunkul, M., and Altermatt, F. (2018). Shedding light on eDNA: Neither natural levels of UV radiation nor the presence of a filter feeder affect eDNA-based detection of aquatic organisms. Plos One 13, e0195529. doi:10.1371/journal.pone.0195529
Merkes, C. M., McCalla, S. G., Jensen, N. R., Gaikowski, M. P., and Amberg, J. J. (2014). Persistence of DNA in carcasses, slime and avian feces may affect interpretation of environmental DNA data. Plos One 9, e113346. doi:10.1371/journal.pone.0113346
Port, J. A., O'Donnell, J. L., Romero-Maraccini, O. C., Leary, P. R., Litvin, S. Y., Nickols, K. J., et al. (2016). Assessing vertebrate biodiversity in a kelp forest ecosystem using environmental DNA. Mol. Ecol. 25, 527–541. doi:10.1111/mec.13481
Romanowski, G., Lorenz, M. G., and Wackernagel, W. (1991). Adsorption of plasmid DNA to mineral surfaces and protection against DNase I. Appl. Environ. Microbiol. 57, 1057–1061. doi:10.1128/aem.57.4.1057-1061.1991
Saito, T., and Doi, H. (2021). Degradation modeling of water environmental DNA: Experiments on multiple DNA sources in pond and seawater. Environ. DNA 3, 850–860. doi:10.1002/edn3.192
Sales, N. G., Wangensteen, O. S., Carvalho, D. C., Deiner, K., Praebel, K., Coscia, I., et al. (2021). Space-time dynamics in monitoring neotropical fish communities using eDNA metabarcoding. Sci. Total Environ. 754, 142096. doi:10.1016/j.scitotenv.2020.142096
Sansom, B. J., and Sassoubre, L. M. (2017). Environmental DNA (eDNA) shedding and decay rates to model freshwater mussel eDNA transport in a river. Environ. Sci. Technol. 51, 14244–14253. doi:10.1021/acs.est.7b05199
Seymour, M., Durance, I., Cosby, B. J., Ransom-Jones, E., Deiner, K., Ormerod, S. J., et al. (2018). Acidity promotes degradation of multi-species environmental DNA in lotic mesocosms. Commun. Biol. 1, 4. doi:10.1038/s42003-017-0005-3
Shogren, A. J., Tank, J. L., Egan, S. P., August, O., Rosi, E. J., Hanrahan, B. R., et al. (2018). Water flow and biofilm cover influence environmental DNA detection in recirculating streams. Environ. Sci. Technol. 52, 8530–8537. doi:10.1021/acs.est.8b01822
Shogren, A. J., Tank, J. L., Andruszkiewicz, E., Olds, B., Mahon, A. R., Jerde, C. L., et al. (2017). Controls on eDNA movement in streams: Transport, retention, and resuspension. Sci. Rep. 7, 5065. doi:10.1038/s41598-017-05223-1
Stoeckle, B. C., Beggel, S., Cerwenka, A. F., Motivans, E., Kuehn, R., Geist, J., et al. (2017). A systematic approach to evaluate the influence of environmental conditions on eDNA detection success in aquatic ecosystems. Plos One 12, e0189119. doi:10.1371/journal.pone.0189119
Strickler, K. M., Fremier, A. K., and Goldberg, C. S. (2015). Quantifying effects of UV-B, temperature, and pH on eDNA degradation in aquatic microcosms. Biol. Conserv. 183, 85–92. doi:10.1016/j.biocon.2014.11.038
Tillotson, M. D., Kelly, R. P., Duda, J. J., Hoy, M., Kralj, J., Quinn, T. P., et al. (2018). Concentrations of environmental DNA (eDNA) reflect spawning salmon abundance at fine spatial and temporal scales. Biol. Conserv. 220, 1–11. doi:10.1016/j.biocon.2018.01.030
Torti, A., Lever, M. A., and Jorgensen, B. B. (2015). Origin, dynamics, and implications of extracellular DNA pools in marine sediments. Mar. Genomics 24 Pt 3, 185–196. doi:10.1016/j.margen.2015.08.007
Troth, C. R., Sweet, M. J., Nightingale, J., and Burian, A. (2021). Seasonality, DNA degradation and spatial heterogeneity as drivers of eDNA detection dynamics. Sci. Total Environ. 768, 144466. doi:10.1016/j.scitotenv.2020.144466
Turner, C. R., Barnes, M. A., Xu, C. C. Y., Jones, S. E., Jerde, C. L., Lodge, D. M., et al. (2014). Particle size distribution and optimal capture of aqueous macrobialeDNA. Methods Ecol. Evol. 5, 676–684. doi:10.1111/2041-210x.12206
Turner, C. R., Uy, K. L., and Everhart, R. C. (2015). Fish environmental DNA is more concentrated in aquatic sediments than surface water. Biol. Conserv. 183, 93–102. doi:10.1016/j.biocon.2014.11.017
Valentini, A., Taberlet, P., Miaud, C., Civade, R., Herder, J., Thomsen, P. F., et al. (2016). Next-generation monitoring of aquatic biodiversity using environmental DNA metabarcoding. Mol. Ecol. 25, 929–942. doi:10.1111/mec.13428
Vašutová, M., Jiroušek, M., and Hájek, M. (2021). High fungal substrate specificity limits the utility of environmental DNA to detect fungal diversity in bogs. Ecol. Indic. 121, 107009. doi:10.1016/j.ecolind.2020.107009
Wang, Y., An, R., Li, Y., and Li, K. (2017). Swimming performance of rock carp Procypris rabaudi and prenant’s schizothoracin Schizothorax prenanti acclimated to total dissolved gas supersaturated water. North Am. J. Fish. Manag. 37, 1183–1190. doi:10.1080/02755947.2017.1353558
Wei, N., Nakajima, F., and Tobino, T. (2018). A microcosm study of surface sediment environmental DNA: Decay observation, abundance estimation, and fragment length comparison. Environ. Sci. Technol. 52, 12428–12435. doi:10.1021/acs.est.8b04956
Weitkamp, D. E., and Katz, M. (1980). A review of dissolved gas supersaturation literature. Trans. Am. Fish. Soc. 109, 659–702. doi:10.1577/1548-8659(1980)109<659:arodgs>2.0.co;2
Whitaker, J. M., Brower, A. L., and Janosik, A. M. (2021). Invasive lionfish detected in estuaries in the northern Gulf of Mexico using environmental DNA. Environ. Biol. Fishes 104, 1475–1485. doi:10.1007/s10641-021-01177-6
Keywords: decay, environmental DNA, sediment, supersaturation, total dissolved gas
Citation: Huang L, Zhang J and Wang Y (2022) Effects of total dissolved gas supersaturation and sediment on environmental DNA persistence of grass carp (Ctenopharyngodon idella) in water. Front. Environ. Sci. 10:952414. doi: 10.3389/fenvs.2022.952414
Received: 25 May 2022; Accepted: 08 July 2022;
Published: 05 August 2022.
Edited by:
Chunhui Li, Beijing Normal University, ChinaReviewed by:
Wei Huang, China Institute of Water Resources and Hydropower Research, ChinaCopyright © 2022 Huang, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yurong Wang, d2FuZ3l1cm9uZ0BzY3UuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.