
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 03 June 2022
Sec. Toxicology, Pollution and the Environment
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.902321
This article is part of the Research Topic Fate and toxicity of emerging pollutants in the environment View all 6 articles
Strawberry fruits are easily contaminated with antibiotic resistance genes (ARGs) in the soil fertilized with livestock manure, which is widely used by farmers in order to increased yield and fruit quality. Composting can reduce the ARGs contamination in livestock manure. However, there is a knowledge gap in understanding and comparing the ARGs contamination of strawberries with the long-term use of raw, aerobic composting, and anaerobic composting livestock manure. In this study, our results suggested that compared with the long-term use of raw livestock manure, both aerobic and anaerobic composting livestock manure reduced the ARGs diversity and abundance of strawberries. Mobile genetic elements (MGEs) closely determined the ARGs dissemination. The correlation analysis between ARGs and MGEs revealed the ARGs dissemination from fruits to human was attenuated when strawberries were fertilized with long-term use of composting livestock manure. In addition, this study illustrated the correlations between the antibiotic resistome and microbiome via network co-occurrence analysis, finding that the phylum Proteobacteria, Firmicutes, and Myxococcota were identified as the potential ARGs hosts which exerted the vital role in the environmental behavior of ARGs. The most important highlight was that the contamination of ARGs of strawberries was alleviated by the long-term use of composting livestock manure, especially the anaerobiotic composting manure.
At present, the contamination with antibiotic resistance bacteria (ARB) and antibiotic resistance genes (ARGs) is a global concern (Brown and Wright, 2016). Many studies have focused on the environmental behavior and risk of ARGs in urban sewage, oceans, rivers, lakes, farmland, and other environments (Joy et al., 2013; Nnadozie and Odume, 2019; Cuadrat et al., 2020; Zhang M.-S. et al., 2021; Nguyen et al., 2021). ARGs can be transferred from soil to vegetables and fruits, posing a direct threat to human health (Marti et al., 2013; Zhu et al., 2016; Mei et al., 2021).
The fate of ARGs in livestock manure is significantly influenced by composting method (Shi et al., 2020; Shen et al., 2021). Although the results of some studies on the transformation of ARGs are partly contracted with each other (Wainaina et al., 2020), numerous studies have shown that both aerobic and anaerobic composting can reduce ARG diversity, abundance, and the capacity of dissemination (Selvam et al., 2012; Qian et al., 2017; Gou et al., 2018; Awasthi et al., 2019; Huang et al., 2021). Many studies have concentrated on the behavior of ARGs in soil amended with kinds of livestock manure, but little information of demonstrating and comparing the ARGs contamination of vegetables and fruits fertilized with different organic manures. The strawberry is an important fruit worldwide, its flavor is enjoyed by people across cultures, and it is indispensable to the kitchen table. ARG-contaminated edible parts of plants can enter the human digestive tract along the food chain, threatening human health (Abriouel et al., 2008; Jung and Rubin, 2020; Mei et al., 2021). Therefore, ARG-contaminated agricultural products are of global concern. Some vegetables and fruits can be peeled or excessively rinsed to avoid this problem (Mei et al., 2021); however, these methods cannot be used for strawberries, which are soft, juicy fruits. The use of livestock manure to farmlands is an important method of ARG dissemination from the human world into nature (Pu et al., 2020; Zhang M.-S. et al., 2021). Strawberry plants crawl on the ground, and the fruits are more easily contaminated by soil ARGs than fruits off the ground (Zhang H. et al., 2020). To increase the yield and flavor of strawberries, the use of livestock manure has become a common fertilizing method for farmers (Song et al., 2020; Kilic et al., 2021). The fertilization with composting livestock manure is very common in developed countries, however, for developing and underdeveloped countries, the use of raw livestock manure is also very usual. Therefore, it is very essential to understand and compare the ARGs contamination of strawberries with the long-term use of raw, aerobic composting, and anaerobic composting livestock manure.
In this study, we hypothesized that the contamination of ARGs of strawberries was alleviated by the long-term use of composting livestock manure by comparison of raw livestock manure, and verified it via field trials. High-throughput quantitative PCR (HT-qPCR) was used to detect ARGs and mobile genetic elements (MGEs). Additionally, the bacterial community was determined by performing 16S rRNA gene high-throughput sequencing. By combining the antibiotic resistome and microbiome data, the potential ARGs microbial hosts were identified via the non-random co-occurrence network analysis.
China has the largest strawberry planting acreage in the world. Jurong, located in Jiangsu Province, China, has an intensive strawberry planting area (16,000 ha). Baitu (31°98 N, 119°28 E), the core planting area in Jurong, was selected for the long-term fertilization experiment. The fruits that were collected from five replicate fields (5 × 1333 m2) with the use of raw poultry manure were defined as PM (2000 kg ha−1); the fruits collected from five replicate fields (5 × 1333 m2) with the use of aerobic composting poultry manure were defined as APM (2000 kg ha−1); the fruits that were collected from five replicate fields (5 × 1333 m2) with the use of anaerobic composting poultry manure were defined as AAPM (2000 kg ha−1). The manure was obtained from poultry farms. Additionally, urea was with 600 kg ha−1a. All manure was used before transplantation, while urea was used during the berry expansion period. The fields were fertilized with the raw or composting livestock manure for 9 years in succession. Five sampling sites were randomly selected across each field. Ten fruits were plucked at each site within an area of 4 m2 (2 m × 2 m). The sampling sites were separated by a distance of 50 m. Fruits from the same field were mixed gently and immediately stored in an ice box. Microbes attached to strawberries were collected within 2 h. The method of collecting microbes was modified according to the description by Zhu et al. (2016). Five fruits (∼20 g per fruit) from the same field were picked out and put into a 500 ml conical flask containing 200 ml of sterilized phosphate-buffered saline (0.01 M, pH 7.4). The ultrasonic probe was put into PBS and sonicated for 10 min. Flasks were shaken for 0.5 h at 150 rpm and 30°C. The fruits were then filtered using sterilized gauze, and the phosphate buffer was filtered through a 0.22 μm cellulose membrane.
DNA was extracted and stored as described by Zhu et al. (2016). The primers (Supplementary Table S1) and HT-qPCR protocol used in this study were described by Zhang W.-G. et al. (2021). Analysis of amplification results and calculation of ARG abundance were the same as those described by Guo et al. (2018). HT-qPCR reactions were performed using SmartChip Real-time PCR Systems (WaferGen Inc., United States); there were altogether 295 primer sets targeting 285 ARGs, 8 transposases, 1 class1 intergron, and 16S ribosomal RNA (rRNA) gene. The 285 ARG assays in this research conferred resistance to almost all major antibiotics and covered three resistance mechanisms. Amplification was conducted in 100 nL reaction containing (final concentration) 1×LightCycler 480 SYBR® Green I Master Mix (Roche Inc., United States), nuclease-free PCR-grade water, 1 ng μL−1 BSA, 3 ng μL−1 DNA template, and 1 μM of each forward and reverse primers. The thermal cycle was: Initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, finally with a melting curve analysis auto-generated by the program. For each primer set, amplification was conducted in triplicate and a non-template control was included.
The results of the high-throughput qPCR were analyzed using SmartChip qPCR software (V2.7.0.1), wells with multiple melting peak as well as wells with amplification efficiency beyond the range (1.8–2.2) were discarded. A threshold cycle (Ct) 31 was used as the detection limit, only samples with three replicates which had amplification were regarded as positive. Relative copy number was calculated referring to a previous study (Looft et al., 2012):
Relative gene copy number = 10 ((31-Ct)/(10/3))
Where Ct is the threshold cycle. The absolute copy number of 16S rRNA gene was quantified by Roche 480 using a SYBR® Green approach separately. The qPCR reaction (20 μL) consisted of 10 μL 2× LightCycler 480 SYBR® Green I Master Mix (Roche Inc., United States), 1 μM of each primer (same primer used in high-throughput qPCR), 1 μL DNA template, 7 μL nuclease-free water. Amplification was conducted with Roche 480 (Roche Inc., United States) as follows: Initial pre-incubation at 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 60°C for 1 min, and 72°C for 15 s. A standard plasmid containing a cloned and sequenced 16S rRNA gene fragment (1.39×1010 copies L−1) was used as eight-point calibration curves from 10-fold dilutions of it to apply external standard calculation, and the PCR reaction was run in triplicate with negative and positive controls. A significantly high correlation between the statistics from SmartChip Real Time System and Roche 480 was observed (r = 0.96, p < 0.01); thus, the relative copy number of ARGs generated by the high-throughput qPCR could be transformed to absolute copy number by normalized to absolute 16S rRNA gene copy number.
The V4 fragment of the 16S rRNA gene was the target region for characterizing the bacterial communities (Bates et al., 2011). Before obtaining the effective sequences, low-quality reads were trimmed, separated, and identified (Edgar et al., 2011). All 16S rRNA gene sequences in this study were uploaded to the National Center for Biotechnology Information (NCBI) database under the accession number PRJNA825758.
The bipartite network was constructed using ggClusterNet in R. All possible pairwise Spearman’s rank correlations between ARGs and MGEs were calculated to construct a correlation matrix. Correlations meeting the standard of Spearman’s correlation coefficient (ρ) > 0.8 and p < 0.05 were regarded as statistically significant. A similar method was used to determine the correlation between the microbiome and the antibiotic resistome. Network co-occurrence analysis was visualized on the Gephi interactive platform. The Mantel test was used to build a correlation matrix between the pathogens and ARGs. A correlation was considered statistically significant when the p-value was <0.05.
The application of livestock manure can increase the ARGs in vegetables, indicating that ARGs migrate from contaminated environments to vegetables (Duan et al., 2017; Chen et al., 2018; Mei et al., 2021). This migration mechanism might function via 1) environmental bacteria that colonize roots, migrating up and colonizing the interiors of stems, leaves, and fruits (Chi et al., 2005); or 2) the antibiotic-resistant bacteria from water, soil, and air directly attaching to the exterior of vegetables and then colonizing (Marti et al., 2013; Liu et al., 2017; Zhang Y. J. et al., 2020). Studies have shown that both aerobic and anaerobic composting could significantly decrease the diversity and abundance of ARGs in livestock manure (Selvam et al., 2012; Qian et al., 2017; Gou et al., 2018). The high temperature generated during the process of aerobic or anaerobic composting functioned in decreasing the diversity and abundance of ARB. Moreover, the high temperature restrained the process of horizontal gene transfer (HGT) of ARGs (Huang et al., 2021). Theoretically, the ARGs contamination of strawberries fertilized with composting livestock manure was lower than those fertilized with raw livestock manure.
In this study, we detected 243, 159, and 135 ARGs in PM, APM, and AAPM, respectively. The results of Chao1 and Shannon indices showed that the ARGs of PM were more diverse than those of APM and AAPM (Figure 1A). Furthermore, principal coordinates analysis (PCoA) showed that strawberries fertilized with raw livestock manure clustered together, distinguishing them from those fertilized with composting livestock manure (Figure 1B). The detected ARGs were distributed across a variety of antibiotic-resistant classes, including those resistant to aminoglycoside, beta-lactamase, chloramphenicol, MLSB, multidrug, sulfonamide, tetracycline, and vancomycin (Figure 2A). The bipartite network showed that there were many more unique ARGs in PM than in APM or AAPM (Figure 2B). The abundance of ARGs in PM, APM, and AAPM is shown in detail in Figure 3. The absolute ARGs copy numbers of PM, APM, and AAPM were ranged from 2.07×103 copies g−1∼ 4.54×108 copies g−1, 115 copies g−1∼ 5.43×107 copies g−1, and 202 copies g−1∼ 6.31 ×107 copies g−1 respectively. The majority of the ARGs detected in PM were more abundant than those detected in APM and AAPM. The results verified that the ARGs contamination of strawberries is alleviated by the long-term use of composting livestock manure.
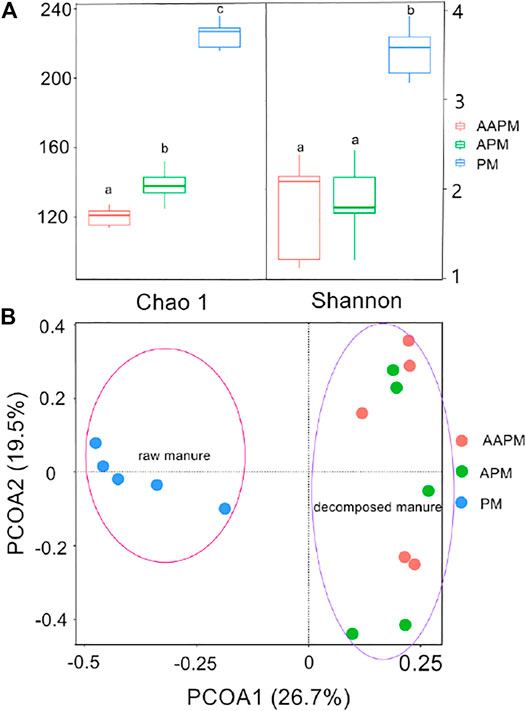
FIGURE 1. α-diversity index of ARGs attached to strawberries collected from fields with long-term use of raw poultry manure (PM), aerobic composting poultry manure (APM), and anaerobic composting poultry manure (AAPM) (A). Principal coordinates analysis (B) (n = 5).
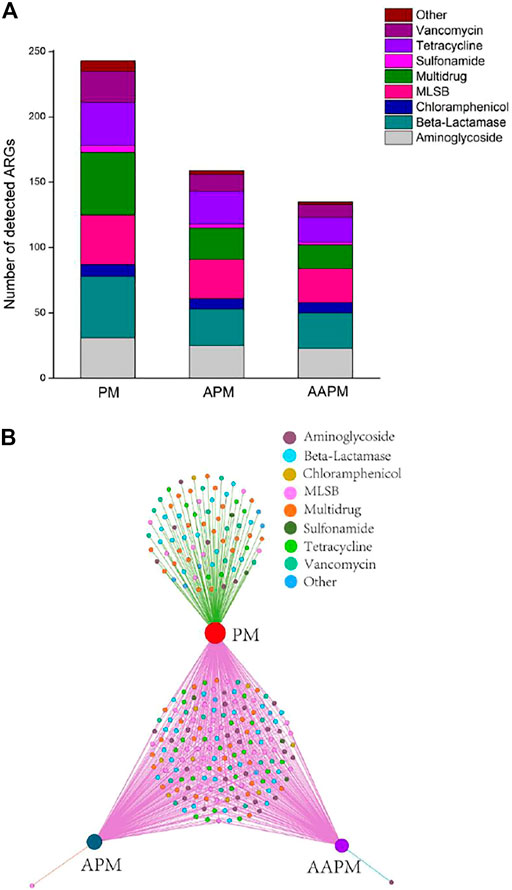
FIGURE 2. Number of detected ARGs attached to strawberries collected from fields with long-term use of raw poultry manure (PM), aerobic composting poultry manure (APM), and anaerobic composting poultry manure (AAPM) (A); Bipartite network analysis showing unique ARGs (B).
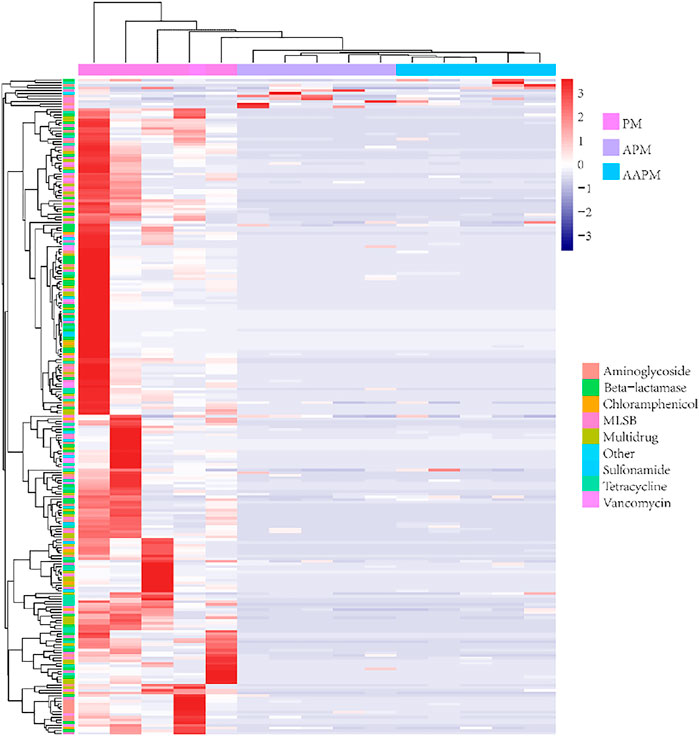
FIGURE 3. Heatmap analysis of ARGs attached to strawberries collected from fields with long-term use of raw poultry manure (PM), aerobic composting poultry manure (APM), and anaerobic composting poultry manure (AAPM). The ARGs detected in this study are listed on the vertical axis. The horizontal axis shows the sampling group (n = 5).
ARGs are disseminated via vertical gene transfer (VGT) and HGT. Compared to VGT, HGT is a pathway that can disseminate ARGs among microorganisms, posing a greater environmental risk (Gogarten and Townsend, 2005; Gillings et al., 2015). Mobile genetic elements (MGEs) act as manipulators in this gene transfer process (Gillings et al., 2008; Partridge et al., 2009; Gaze et al., 2011). Therefore, the correlation between ARGs and MGEs is commonly used to assess the dissemination capacity of ARGs in various environments (Zhang W.G. et al., 2021).
Twelve MGEs were selected for detection in this study (Supplementary Table S1). All 12 MGEs were detected in PM, whereas 8 and 7 MGEs were detected in APM and AAPM, respectively. For strawberries fertilized with raw livestock manure, there were 286 robust connections between ARGs and MGEs (Figure 4A), while there were 237 and 119 connections for the strawberries fertilized with aerobic and anaerobic composting manure, respectively (Figures 4B,C). Our field trial results revealed that compared with the use of raw livestock manure, the long-term use of composting manure, especially anaerobic composting manure, is a better strategy to attenuate the HGT process of ARGs. Although the exact effects and mechanisms of ingesting exogenous ARGs on human health are unclear, many studies have indicated that intestinal microbiota can acquire exogenous ARGs via horizontal gene transfer (Marti et al., 2013; Zhu et al., 2016; Mei et al., 2021). If exogenous ARGs are acquired by pathogenic bacteria, it would be detrimental to curing diseases. Therefore, our results suggested that the use of composting livestock manure especially anaerobic composting manure could mitigate the threat of ARGs to human health.
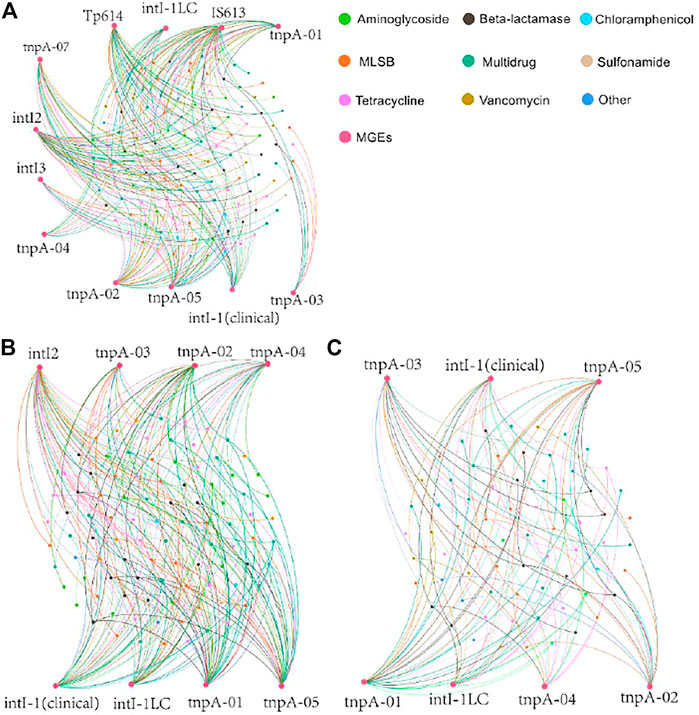
FIGURE 4. Network analysis depicting the correlations among ARGs and MGEs of strawberries collected from the fields with long-term use of raw poultry manure (PM) (A), aerobic composting poultry manure (APM) (B), and anaerobic composting poultry manure (AAPM) (C). Each significant Spearman’s correlation (p < 0.05) is presented by a connection line. Co-occurrence patterns among antibiotic resistome and microbiome.
As the host, the microbial community determines the pattern and environmental behavior of ARGs (Hu et al., 2017; Guo et al., 2018; Du et al., 2020; Zhang M.-S. et al., 2021). The non-random co-occurrence network correlation between ARGs and bacterial taxa was regarded as an effective way to track the potential ARGs hosts in various environments (Hu et al., 2017; Guo et al., 2018). In our study, the microbial taxa at the phylum level exhibited no significant differences among the PM, APM, and AAPM, except for the phylum Firmicutes. Proteobacteria was the predominant taxa in all samples, accounting for 85.6–95.1%, followed by Firmicutes (Supplementary Figure S1). Our results showed that the predominant phyla Proteobacteria and Firmicutes built intensive connections with ARGs (Figure 5). This indicated that the dominant bacterial taxa Proteobacteria and Firmicutes were the potential ARGs hosts. Our finding was consistent with the previous study where the pattern of endophytic ARGs of strawberries was investigated. The study found that the endophytic bacteria hosts of ARGs were preferably distributed in the domination phylum Proteobacteria (Zhang et al., 2020). Additionally, it should be noted that the phylum Myxococcota, a non-dominant bacterial taxon, also built intensive correlations with ARGs, indicating the important role of Myxococcota in influencing the pattern of antibiotic resistome of strawberries. The similar results were shown in other environments where the non-dominant microbial taxa were the potential ARGs hosts (Guo et al., 2018; Zheng et al., 2018; Zhang M.-S. et al., 2021).
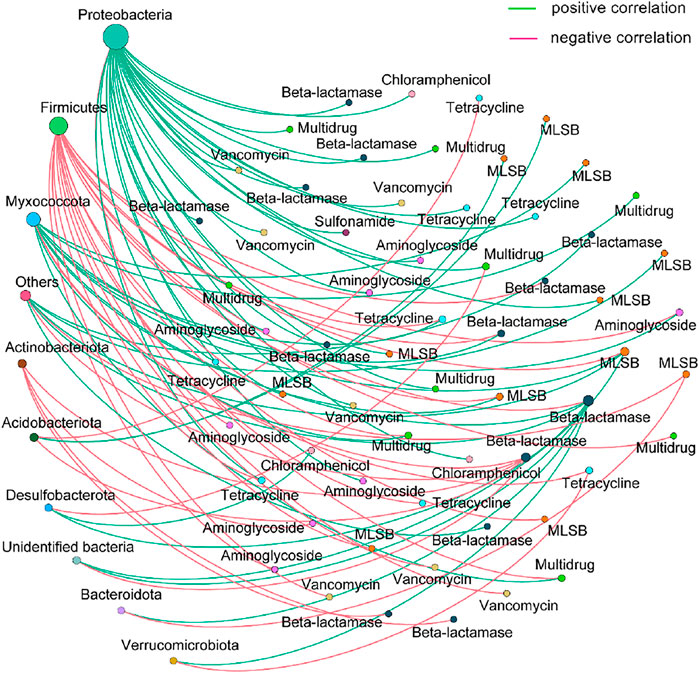
FIGURE 5. Network co-occurrence analysis between the microbiome and antibiotic resistome. The size of each node is proportional to the number of connections.
A comparable study was conducted to understand whether the long-term use of aerobic and anaerobic composting manure can mitigate the ARGs contamination of strawberries by comparison with raw livestock manure. The results demonstrated that 1) the ARGs diversity and abundance of strawberries decreased when fertilized with the aerobic and anaerobic composting manure; 2) ARGs of strawberries with the long-term use of raw livestock manure built more intensive and robust correlations with MGEs than those with the use of composting livestock manure.; 3) the use of composting livestock manure especially anaerobic composting manure can the threat of ARGs to human health; 4) ARGs were mostly harbored by the phyla Proteobacteria, Firmicutes, and Myxococcota.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA825758.
W-GZ: conceptualization, method, data analysis, sampling, writing-original draft, visualization, project administration, funding acquisition. M-SZ and WL: sampling, analysis.
This study was financially supported by the National Natural Science Foundation of China (31600419).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We appreciate the workers for their assistance during sample collection.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.902321/full#supplementary-material
Abriouel, H., Omar, N. B., Molinos, A. C., López, R. L., Grande, M. J., Martínez-Viedma, P., et al. (2008). Comparative Analysis of Genetic Diversity and Incidence of Virulence Factors and Antibiotic Resistance Among Enterococcal Populations from Raw Fruit and Vegetable Foods, Water and Soil, and Clinical Samples. Int. J. Food Microbiol. 123 (1-2), 38–49. doi:10.1016/j.ijfoodmicro.2007.11.067
Awasthi, M. K., Liu, T., Chen, H., Verma, S., Duan, Y., Awasthi, S. K., et al. (2019). The Behavior of Antibiotic Resistance Genes and Their Associations with Bacterial Community during Poultry Manure Composting. Bioresour. Technol. 280, 70–78. doi:10.1016/j.biortech.2019.02.030
Bates, S. T., Berg-Lyons, D., Caporaso, J. G., Walters, W. A., Knight, R., and Fierer, N. (2011). Examining the Global Distribution of Dominant Archaeal Populations in Soil. ISME J. 5 (5), 908–917. doi:10.1038/ismej.2010.171
Brown, E. D., and Wright, G. D. (2016). Antibacterial Drug Discovery in the Resistance Era. Nature 529 (7586), 336–343. doi:10.1038/nature17042
Chen, Q.-L., Fan, X.-T., Zhu, D., An, X.-L., Su, J.-Q., and Cui, L. (2018). Effect of Biochar Amendment on the Alleviation of Antibiotic Resistance in Soil and Phyllosphere of Brassica Chinensis L. Soil Biol. Biochem. 119, 74–82. doi:10.1016/j.soilbio.2018.01.015
Chi, F., Shen, S.-H., Cheng, H.-P., Jing, Y.-X., Yanni, Y. G., and Dazzo, F. B. (2005). Ascending Migration of Endophytic Rhizobia, from Roots to Leaves, Inside Rice Plants and Assessment of Benefits to Rice Growth Physiology. Appl. Environ. Microbiol. 71, 7271–7278. doi:10.1128/aem.71.11.7271-7278.2005
Cuadrat, R., Sorokina, M., Andrade, B. G., Goris, T., and Davila, A. (2020). Global Ocean Resistome Revealed: Exploring Antibiotic Resistance Genes (ARGs) Abundance and Distribution on TARA Oceans Samples Through Machine Learning Tools. GigaScience 9 (5), giaa046. doi:10.1093/gigascience/giaa046
Du, S., Shen, J.-P., Hu, H.-W., Wang, J.-T., Han, L.-L., Sheng, R., et al. (2020). Large-Scale Patterns of Soil Antibiotic Resistome in Chinese Croplands. Sci. Total Environ. 712, 136418. doi:10.1016/j.scitotenv.2019.136418
Duan, M., Li, H., Gu, J., Tuo, X., Sun, W., Qian, X., et al. (2017). Effects of Biochar on Reducing the Abundance of Oxytetracycline, Antibiotic Resistance Genes, and Human Pathogenic Bacteria in Soil and Lettuce. Environ. Pollut. 224, 787–795. doi:10.1016/j.envpol.2017.01.021
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 27 (16), 2194–2200. doi:10.1093/bioinformatics/btr381
Gaze, W. H., Zhang, L., Abdouslam, N. A., Hawkey, P. M., Calvo-Bado, L., Royle, J., et al. (2011). Impacts of Anthropogenic Activity on the Ecology of Class 1 Integrons and Integron-Associated Genes in the Environment. ISME J. 5, 1253–1261. doi:10.1038/ismej.2011.15
Gillings, M., Boucher, Y., Labbate, M., Holmes, A., Krishnan, S., Holley, M., et al. (2008). The Evolution of Class 1 Integrons and the Rise of Antibiotic Resistance. J. Bacteriol. 190, 5095–5100. doi:10.1128/jb.00152-08
Gillings, M. R., Gaze, W. H., Pruden, A., Smalla, K., Tiedje, J. M., and Zhu, Y.-G. (2015). Using the Class 1 Integron-Integrase Gene as a Proxy for Anthropogenic Pollution. ISME J. 9 (6), 1269–1279. doi:10.1038/ismej.2014.226
Gogarten, J. P., and Townsend, J. P. (2005). Horizontal Gene Transfer, Genome Innovation and Evolution. Nat. Rev. Microbiol. 3 (9), 679–687. doi:10.1038/nrmicro1204
Gou, M., Hu, H.-W., Zhang, Y.-J., Wang, J.-T., Hayden, H., Tang, Y.-Q., et al. (2018). Aerobic Composting Reduces Antibiotic Resistance Genes in Cattle Manure and the Resistome Dissemination in Agricultural Soils. Sci. Total Environ. 612, 1300–1310. doi:10.1016/j.scitotenv.2017.09.028
Guo, Y., Liu, M., Liu, L., Liu, X., Chen, H., and Yang, J. (2018). The Antibiotic Resistome of Free-Living and Particle-Attached Bacteria Under a Reservoir Cyanobacterial Bloom. Environ. Int. 117, 107–115. doi:10.1016/j.envint.2018.04.045
Hu, H.-W., Wang, J.-T., Li, J., Shi, X.-Z., Ma, Y.-B., Chen, D., et al. (2017). Long-Term Nickel Contamination Increases the Occurrence of Antibiotic Resistance Genes in Agricultural Soils. Environ. Sci. Technol. 51 (2), 790–800. doi:10.1021/acs.est.6b03383
Huang, C., Tang, Z., Xi, B., Tan, W., Guo, W., Wu, W., et al. (2021). Environmental Effects and Risk Control of Antibiotic Resistance Genes in the Organic Solid Waste Aerobic Composting System: A Review. Front. Environ. Sci. Eng. 15, 127. doi:10.1007/s11783-021-1415-5
Joy, S. R., Bartelt-Hunt, S. L., Snow, D. D., Gilley, J. E., Woodbury, B. L., Parker, D. B., et al. (2013). Fate and Transport of Antimicrobials and Antimicrobial Resistance Genes in Soil and Runoff Following Land Application of Swine Manure Slurry. Environ. Sci. Technol. 47 (21), 12081–12088. doi:10.1021/es4026358
Jung, D., and Rubin, J. E. (2020). Identification of Antimicrobial Resistant Bacteria from Plant-Based Food Products Imported into Canada. Int. J. Food Microbiol. 319, 108509. doi:10.1016/j.ijfoodmicro.2020.108509
Kilic, N., Turemis, N. F., and Dasgan, H. Y. (2021). The Effect of Fertilizers on Crop Yield, Fruit Quality and Plant Nutrition of Organically Grown Strawberry (Fragaria X Ananassa Duch.). Appl. Ecol. Env. Res. 19 (3), 2201–2211. doi:10.15666/aeer/1903_22012211
Liu, H., Carvalhais, L. C., Crawford, M., Singh, E., Dennis, P. G., Pieterse, C. M. J., et al. (2017). Inner Plant Values: Diversity, Colonization and Benefits from Endophytic Bacteria. Front. Microbiol. 8, 2552. doi:10.3389/fmicb.2017.02552
Looft, T., Johnson, T. A., Allen, H. K., Bayles, D. O., Alt, D. P., Stedtfeld, R. D., et al. (2012). In-Feed Antibiotic Effects on the Swine Intestinal Microbiome. Proc. Natl. Acad. Sci. U.S.A. 109, 1691–1696. doi:10.1073/pnas.1120238109
Marti, R., Scott, A., Tien, Y.-C., Murray, R., Sabourin, L., Zhang, Y., et al. (2013). Impact of Manure Fertilization on the Abundance of Antibiotic-Resistant Bacteria and Frequency of Detection of Antibiotic Resistance Genes in Soil and on Vegetables at Harvest. Appl. Environ. Microbiol. 79 (18), 5701–5709. doi:10.1128/aem.01682-13
Mei, Z., Xiang, L., Wang, F., Xu, M., Fu, Y., Wang, Z., et al. (2021). Bioaccumulation of Manure-Borne Antibiotic Resistance Genes in Carrot and its Exposure Assessment. Environ. Int. 157, 106830. doi:10.1016/j.envint.2021.106830
Nguyen, A. Q., Vu, H. P., Nguyen, L. N., Wang, Q., Djordjevic, S. P., Donner, E., et al. (2021). Monitoring Antibiotic Resistance Genes in Wastewater Treatment: Current Strategies and Future Challenges. Sci. Total Environ. 783, 146964. doi:10.1016/j.scitotenv.2021.146964
Nnadozie, C. F., and Odume, O. N. (2019). Freshwater Environments as Reservoirs of Antibiotic Resistant Bacteria and Their Role in the Dissemination of Antibiotic Resistance Genes. Environ. Pollut. 254 (Part B), 113067. doi:10.1016/j.envpol.2019.113067
Partridge, S. R., Tsafnat, G., Coiera, E., and Iredell, J. R. (2009). Gene Cassettes and Cassette Arrays in Mobile Resistance Integrons. FEMS Microbiol. Rev. 33, 757–784. doi:10.1111/j.1574-6976.2009.00175.x
Pu, Q., Zhao, L.-X., Li, Y.-T., and Su, J.-Q. (2020). Manure Fertilization Increase Antibiotic Resistance in Soils from Typical Greenhouse Vegetable Production Bases, China. J. Hazard. Mater. 391, 122267. doi:10.1016/j.jhazmat.2020.122267
Qian, X., Gu, J., Sun, W., Wang, X. J., Su, J. Q., and Stedfeld, R. (2017). Diversity, Abundance, and Persistence of Antibiotic Resistance Genes in Various Types of Animal Manure Following Industrial Composting. J. Hazard. Mat. 344, 716–722. doi:10.1016/j.jhazmat.2017.11.020
Selvam, A., Xu, D., Zhao, Z., and Wong, J. W. C. (2012). Fate of Tetracycline, Sulfonamide and Fluoroquinolone Resistance Genes and the Changes in Bacterial Diversity during Composting of Swine Manure. Bioresour. Technol. 126, 383–390. doi:10.1016/j.biortech.2012.03.045
Shen, Q., Tang, J., Wang, X., Li, Y., Yao, X., Sun, H., et al. (2021). Fate of Antibiotic Resistance Genes and Metal Resistance Genes During the Thermophilic Fermentation of Solid and Liquid Swine Manures in an Ectopic Fermentation System Composted Chicken Manure for Anaerobic Soil Disinfestation Increased the Strawberry Yield and Shifted the Soil Microbial Communities. Ecotox. Environ. Safe. Ecotoxicology Environ. Saf. 21312 (716), 1119811–1119815. doi:10.1016/j.ecoenv.2021.111981
Shi, Z., Zhao, R., Wan, J., Li, B., Shen, Y., Zhang, S., et al. (2020). Metagenomic Analysis Reveals the Fate of Antibiotic Resistance Genes in Two-Stage and One-Stage Anaerobic Digestion of Waste Activated Sludge. J. Hazard. Mat. 406, 124595. doi:10.1016/j.jhazmat.2020.124595
Song, Z., Massart, S., Yan, D., Cheng, H., Eck, M., Berhal, C., et al. (2020). Composted Chicken Manure for Anaerobic Soil Disinfestation Increased the Strawberry Yield and Shifted the Soil Microbial Communities Sustainability-Basel 12 (16), 1–15. doi:10.1016/j.envint.2016.11.001
Wainaina, S., Awasthi, M. K., Sarsaiya, S., Chen, H., Singh, E., Kumar, A., et al. (2020). Resource Recovery and Circular Economy from Organic Solid Waste Using Aerobic and Anaerobic Digestion Technologies. Bioresour. Technol. 301, 122778. doi:10.1016/j.biortech.2020.122778
Zhang, H., Zhang, Q., Chen, S., Zhang, Z., Song, J., Long, Z., et al. (2020). Enterobacteriaceae Predominate in the Endophytic Microbiome and Contribute to the Resistome of Strawberry. Sci. Total Environ. 727, 138708. doi:10.1016/j.scitotenv.2020.138708
Zhang, M.-S., Li, W., Zhang, W.-G., Li, Y.-T., Li, J.-Y., and Gao, Y. (2021). Agricultural Land-Use Change Exacerbates the Dissemination of Antibiotic Resistance Genes via Surface Runoffs in Lake Tai Basin, China. Ecotoxicol. Environ. Saf. 220 (5), 112328. doi:10.1016/j.ecoenv.2021.112328
Zhang, W.-G., Wen, T., Liu, L.-Z., Li, J.-Y., Gao, Y., Zhu, D., et al. (2021). Agricultural Land-Use Change and Rotation System Exert Considerable Influences on the Soil Antibiotic Resistome in Lake Tai Basin. Sci. Total Environ. 771, 144848. doi:10.1016/j.scitotenv.2020.144848
Zhang, Y. J., Hu, H. W., Chen, Q. L., Yan, H., Wang, J. T., Chen, D., et al. (2020). Manure Application Did Not Enrich Antibiotic Resistance Genes in Root Endophytic Bacterial Microbiota of Cherry Radish Plants. Appl. Environ. Microbiol. 86, 12. doi:10.1128/aem.02106-19
Zheng, J., Zhou, Z., Wei, Y., Chen, T., Feng, W., and Chen, H. (2018). High-Throughput Profiling of Seasonal Variations of Antibiotic Resistance Gene Transport in a Peri-Urban River. Environ. Int. 114, 87–94. doi:10.1016/j.envint.2018.02.039
Keywords: antibiotic resistance genes, strawberry, composting manure, public health, raw manure
Citation: Zhang W-G, Zhang M-S and Li W (2022) The Antibiotic Resistance Genes Contamination of Strawberries With the Long-Term Use of Raw, Aerobic Composting, and Anaerobic Composting Livestock Manure: A Comparative Study. Front. Environ. Sci. 10:902321. doi: 10.3389/fenvs.2022.902321
Received: 23 March 2022; Accepted: 09 May 2022;
Published: 03 June 2022.
Edited by:
Anita Mukherjee, University of Calcutta, IndiaReviewed by:
Naresh Devarajan, University of California, Davis, United StatesCopyright © 2022 Zhang, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Guo Zhang, d2VpZ3VvemhhbmdAamFhcy5hYy5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.