- Key Laboratory of Soil Resource Sustainable Utilization for Jilin Province Commodity Grain Bases, College of Resource and Environmental Science, Jilin Agricultural University, Changchun, China
The excessive application of phosphorus (P) fertilizer is becoming a major agricultural problem, which reduces the utilization rate of the P fertilizer and degrades soil quality. The following five P fertilizer treatments were investigated to know how they affect soil properties, enzyme activity, bacterial and fungal community structure. 1) no P fertilizer (P0); 2) farmers’ traditional P fertilization scheme (FP); 3) 30% reduction in P fertilizer application (P1, microbial blended fertilizer as base fertilizer); 4) 30% reduction in P fertilizer application (P2, diammonium phosphate as starting fertilizer); 5) 30% reduction in P fertilizer application (P3, microbial inoculum seed dressing). The P fertilizer reduction combined with microbial fertilizer significantly increased soil organic matter (SOM), total phosphorus (TP), available phosphorus (AP) available potassium (AK) contents, and acid phosphatase activity (ACP), however, soil urease activity was significantly reduced. Moreover, the P fertilizer reduction combined with microbial fertilizer significantly increased the relative abundance of a potential beneficial genus (i.e., Bacillus, Pseudomonas, Penicillium, and Acremonium) and potentially pathogenic genus (i.e., Fusarium, Gibberella, and Drechslera). The structural equation model (SEM) revealed that different P fertilizer reduction systems had significant indirect effects on bacterial and fungal community structures. The results suggested that the P fertilizer reduction combined with microbial fertilizer systems regulated the pathogenic and beneficial genus which created a microbial community that is favorable for maize growth. Moreover, the findings highlighted the importance of soil properties in determining the soil bacterial and fungal community structure.
Introduction
Phosphorus (P) is the most abundant essential nutrient in plants after nitrogen and potassium. According to reports, about 40% of the world’s arable land is limited by P deficiency, which affects crop yield. As such the application of P fertilizer has become one of the important agronomic measures to improve crop yield and soil quality (Elser, 2012). However, P fertilizers are easily adsorbed and fixed after being applied to the soil, and their utilization rate in the planting season is only 10%–25%, and the remaining 85% accumulates in the soil in an ineffective state (Zhu et al., 2018). This does not only reduce the application efficiency of P fertilizers but also accelerates the depletion of non-renewable P resources, increasing the risk of environmental pollution (Meinikmann et al., 2015). Therefore, finding ways to improve the utilization efficiency of P fertilizers and slow down the depletion of P rock resources is of great significance for the realization of sustainable agricultural development. This will optimize the application of P fertilizers in order to fully benefit the biological potential of crops and soil microorganisms.
Microorganisms are the key drivers in the cyclic transformation of fertilizer nutrients into the soil and are directly related to nutrient use efficiency and its agronomic and environmental effects (Wang et al., 2022). Soil microorganisms play a very important role in the transformation of soil P and about 40% of soil microorganisms can activate insoluble P and convert it into microbial P to form a soil microbial biomass P pool (Jorquera et al., 2008). Application of P fertilizer can increase the content of soil available P, promote the release of P in soil microorganisms, and provide available P for crop absorption and utilization. Studies have shown that different fertilization methods can directly or indirectly affect the structure and composition of soil microbial communities and their functions by changing the soil microenvironment. This process is driven by factors such as soil pH, texture, moisture, organic carbon content, and nutrient availability (Zhang et al., 2017; Ding et al., 2018; Lin et al., 2019). However, long-term excessive application of P fertilizers not only leads to the decline of soil organic matter content and quality (Zhang and Zhang, 2010), but also leads to the depletion of soil resources and energy, water, air pollution, and other environmental problems (Liu et al., 2007). Therefore, the partial replacement of chemical fertilizers with microbial fertilizers has gradually become an important way to improve soil fertility and realize the reduction and efficiency of chemical fertilizers in this era of intensified agricultural activities. Microbiome studies have proved that the microbial flora in the ecological environment is complex and diverse, and it is difficult to achieve artificial directional control. The most effective solution to promote the growth and development of plants is to inoculate crops with beneficial microbial fertilizers. This will promote the formation of new microbial flora at the root of crops and improve the soil environment (Pineda et al., 2017). Microbial fertilizers mainly include microbial inoculants, compound microbial fertilizers, and bio-organic fertilizers. Studies have reported that microbial fertilizers can promote corn growth and increase yield and can replace 10%–30% of compound chemical fertilizers. At present, there is a lack of targeted research in the case of reducing P fertilizers and replacing them with microbial fertilizers (Atieno et al., 2020). Although there have been much research on the effects of different fertilization measures on soil nutrient transformation and the changes in microbial community structure etc, however, the microbiological mechanism and regulation principle of the changes in soil nutrients and enzyme activities in the case of reducing P fertilizers and replacing it with microbial fertilizers remains unknown. Therefore, this study used Illumina Miseq sequencing technology to analyze the effect of different P reduction systems on soil bacterial and fungal community structure, to elucidate the changes in soil microbial community composition, diversity, and functional bacteria caused by P reduction. The results can provide a theoretical reference to understand the microbiology related to P fertilizer reduction and synergy in the black soil region of Northeast China.
Materials and Methods
Experimental Design
This experiment was set up in an experimental field in the southern area of Jilin Agricultural University. The average annual temperature in the area is 4.9°C and the average annual precipitation is 593.8 mm. The soil tested was typical black soil and the experimental site was a large-scale test site with a test area of 312 m2. The study included five treatments: 1) no P fertilizer (P0); 2) farmers’ traditional P fertilization scheme (FP, 120 kg/hm2 P2O5 of diammonium phosphate as base fertilizer); 3) 30% reduction in P fertilizer application (P1, 84.0 kg P2O5/hm2 and microbial blended fertilizer as base fertilizer); 4) 30% reduction in P fertilizer application (P2, 67.2 kg P2O5/hm2 and microbial blend fertilizer as base fertilizer +16.8 kg P2O5/hm2 and diammonium phosphate as starting fertilizer); 5) 30% reduction in P fertilizer application (P3, microbial inoculum seed dressing +84.0 kg P2O5/hm2 and diammonium phosphate as base fertilizer). The fertilizer application rate of each treatment is shown in Supplementary Table S1, and the field management followed the conventional management system. The planting density was 65,000 plants/hm2 and the tested corn variety was Liangyu 99. The microbial blended fertilizer and microbial inoculum were purchased from Jilin Province Jiabo Biotechnology Co., Ltd. (the number of viable bacteria ≥200 million/g).
During the maize tasseling period, soil samples of each treatment were taken from the 0–20 cm layer, using a random sampling method with three replicates. After collection, the samples were placed in an icebox and brought back to the laboratory. Part of each sample was stored in the refrigerator at −80°C for total soil DNA extraction, and the other part was naturally air-dried in a cool place from which a small sample was taken, ground, and sieved according to the quartering method, for the determination of soil physicochemical properties.
Soil Physicochemical Properties and Enzyme Activity Determination
Soil pH was measured by pH meter (the water-soil ratio was 2.5:1), soil organic matter (SOM) was measured by potassium dichromate oxidation-external heating method, and soil total nitrogen (TN) was measured by Kjeldahl method. The alkaline hydrolysis diffusion method was used to determine the available nitrogen (AN) content of the soil. The total phosphorus (TP) content of the soil was digested with H2SO4-HClO4 and antimony resistance was measured by the colorimetric method. Soil total potassium (TK) and soil available potassium (AK) were measured by a flame spectrophotometer. The phenol-sodium hypochlorite colorimetric method was used for the determination of soil urease by using urea as the matrix and the soil phosphatase was determined by the phenyl disodium phosphate colorimetric method.
Soil DNA Extraction
DNA was extracted using the FastDNA® SPIN Kit for Soil (MP Biomedicals, United States), and the extracted DNA was detected and analyzed using gel electrophoresis.
Illumina MiSeq Sequencing
The corresponding primers for the bacterial 16S rRNA V3-V4 region were: 338F (5′-GTG CCA GCM GCC GCG G-3′) and 806R (5′-CCG TCA ATT CMT TTR AGT TT-3′) (Xu et al., 2016). The fungal ITS (ITS1-ITS2) primers used were ITS1F (5′-CTT GGT CAT TTA GAG GAA GTA A-3′) and ITS2 (5′-GCT GCG TTC ATC GAT GC-3′) (Adams et al., 2013). 16S rRNA and ITS gene were performed using a 25 μl reaction system: forward Primer (5 μM) 1 μl, reverse Primer (5 μM) 1 μl, BSA (2 ng/μl) 3 μl, template DNA 30 ng, 2xTaq plus master mix 12.5 μl, ddH2O 7.5 μl. 16S rRNA gene PCR amplification procedure was 94°C 5 min, 94°C 30 s, 55°C 30 s, 72°C 60 s, 72°C 7 min, for 28cycles. ITS gene PCR amplification procedure was as follows: 94°C 5 min, 94°C 30 s, 55°C 30 s, 72°C 60 s, 72°C 7 min, for 34 cycles. All samples were replicated three times, and PCR products from the same sample were pooled. The axyprep DNA recovery kit was used to recover the resulting product. Samples were eluted with Tris-HCl and detected by electrophoresis.
Illumina Miseq Analytical Methods for High Throughput Data
The off-machine data was split by the QIIME software (V1.8.0) according to the barcode sequence, and the Pear software (V0.9.6) was used to filter and splice the data. The minimum overlap during splicing was set to 10 bp, and the mismatch rate was set to 0.1. Short and chimeric sequences were removed using the Vsearch software (V2.7.1) and denovo method. The high-quality sequences obtained were clustered by OTU (Operational Taxonomic Units) using the Vsearch software (V2.7.1) and UPARSE algorithm (version7.1 http://drive5.com/uparse/), and the similarity threshold was set to 97% (Edgar, 2013). The OTUs obtained from the 16S rRNA gene were aligned on the Silva database (Release128 http://www.arb-silva.de) and the minimum similarity was 0.8. The OTUs obtained from the ITS gene were aligned on the UNITE database (version7.2 https://unite.ut.ee/) and the lowest similarity was set to 0.8 (Koljalg et al., 2014). In order to analyze the microbial communities at the same sequencing depth, the lowest sequencing number was randomly selected per sample of which 24,477 sequences were selected for bacterial 16S rRNA gene and 27,192 sequences for fungal ITS gene. The raw sequences were deposited into NCBI under the accession number SRP365800.
Statistical Analysis
The significant difference between treatments was analyzed by the F test in one-way ANOVA and multiple comparisons of means (p < 0.05) were conducted with a Fisher’s protected least significant difference (LSD) using SPSS 20. Pearson analysis was used to analyze the correlation between soil enzyme activity and soil physicochemical properties. Heatmaps were drawn using the “pheatmap” package in R (V3.4.2) (R-project, 2010). Partial least squares discriminative analysis (PLS-DA) was completed in the muma package of R software (version 3.5.0). Bacterial function prediction analysis was done by using FAPROTAX (http://www.zoology.ubc.ca/louca/FAPROTAX). The fungal function was predicted by FUNGuild (http://www.stbates.org/guilds/app.php) (Nguyen et al., 2016). Canonical correspondence analysis (CCA) was done with Canoco software (version 5.0) and the structural equation model (SEM) was established using AMOS 24 (Malaeb et al., 2000). This was used to establish the direct and indirect effects of different P reduction systems on soil bacterial and fungal community diversity and composition. A priori model of predictors were constructed and the soil bacterial and fungal community structures were represented by PC1. Model accuracy was tested with χ2 (p > 0.05), goodness-of-fit index (GFI>0.9), and Akaike information criterion (AIC) (Markland, 2007).
Results
Soil Physicochemical Properties
The application of P significantly increased soil nutrient content compared with P0. Compared with P0, treatments FP, P1, P2, and P3 significantly increased the content of SOM, TP, AN, and AK, while P2 and P3 significantly increased the content of TN (Table 1). Moreover, P1, P2, and P3 significantly increased the content of AP. Compared with high P application (FP), the treatments with phosphorus reduction systems (P1, P2, and P3) significantly improved the contents of SOM, TN, TP, AN, AP, and AK (Table 1). Compared with FP, the SOM and TP contents of P1, P2, and P3 were significantly increased by 10.81%, 16.93%, 13.67% and 13.73%, 22.61%, 23.69%, respectively. In addition, the TN and AN contents of P2 significantly increased by 7.50% and 8.20%, respectively. The AP contents of P2 and P3 were significantly increased by 24.50% and 36.51%, respectively.
Soil Enzymatic Activity
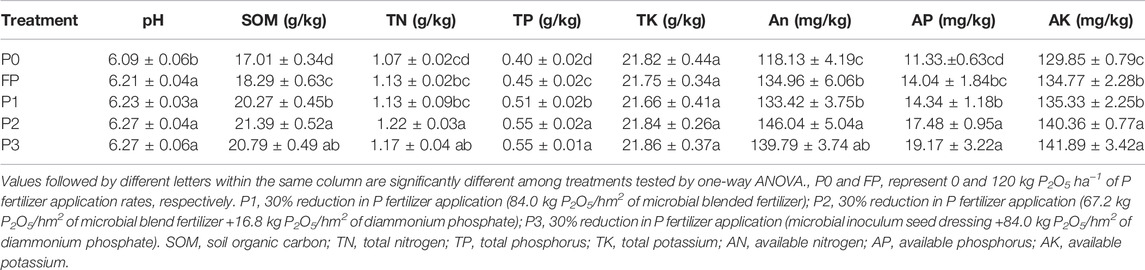
Compared with P0, the application of P fertilizer significantly reduced soil urease activity. Compared with FP, P2 and P3 significantly reduced soil urease activity (Figure 1). Also, P2 and P3 significantly increased acid phosphatase (ACP) activity compared with P0 and FP. There was no significant difference in neutral phosphatase (NEP) activity among the different P application systems. Compared with P0, the application of P fertilizer significantly increased alkaline phosphatase (ALP) activity. Urease activity was significantly negatively correlated with TP and AP, while ACP activity and ALP activity were significantly positively correlated with TP and AP (Supplementary Figure S1). Neutral phosphatase was significantly positively correlated with AP.
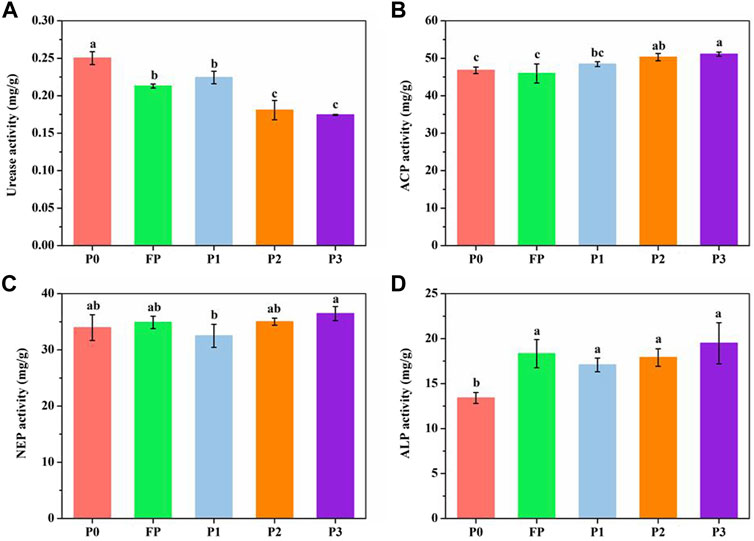
FIGURE 1. Soil enzymatic activity under different P fertilizer application systems. Urease (A); ACP, acid phosphatase (B); NEP, neutral phosphatase (C); ALP, alkaline phosphatase (D). P0 and FP represent 0 and 120 kg P2O5 ha−1 of P fertilizer application rates, respectively. P1, 30% reduction in P fertilizer application (84.0 kg P2O5/hm2 of microbial blended fertilizer); P2, 30% reduction in P fertilizer application (67.2 kg P2O5/hm2 of microbial blend fertilizer +16.8 kg P2O5/hm2 of diammonium phosphate); P3, 30% reduction in P fertilizer application (microbial inoculum seed dressing +84.0 kg P2O5/hm2 of diammonium phosphate).
Soil Bacterial and Fungal Community Composition
In the Illumina-miseq sequencing of the 16S rRNA gene in this experiment, 596,004 optimized sequences were obtained from all samples, with 31,258–48,530 optimized sequences per sample (an average of 39,734 optimized sequences) (Supplementary Table S2). In the Illumina-miseq sequencing of ITS genes, 660,412 optimized sequences were obtained from all samples, with 35,632–49,544 optimized sequences per sample (44,027 optimized sequences on average) (Supplementary Table S3). The dominant bacterial phyla under different phosphorus application systems were Proteobacteria (29.5%–40.3%), Actinobacteria (19.2%–22.8%), Acidobacteria (12.0%–16.0%), Bacteroidetes (5.84%–9.77%), and Saccharibacteria (3.95%–9.53%) (Supplementary Figure S2A). Moreover, P2 and P3 significantly increased the relative abundance of Saccharibacteria compared with P0 and FP. The relative abundance of Ascomycota (57.3%–85.8%) was the highest under the different P application systems, followed by Basidiomycota and Mortierellomycota (Supplementary Figure S2B). Compared with FP, the combination of P reduction and microbial fertilizer significantly increased the relative abundance of Ascomycota.
The relative abundance of some functional genera changed after P reduction and microbial fertilizer application. P3 significantly increased the relative abundance of Pseudarthrobacter (Figure 2A), P1 significantly increased the relative abundance of Mucilaginibacter and Streptomyces, and P1 and P3 significantly increased the relative abundance of Bacillus. The relative abundance of Pseudomonas and Marmoricola significantly increased in P2. The dominant fungal genus under the different P application systems was Penicillium (25.4%–65.7%) (Figure 2B). The relative abundance of Penicillium increased significantly after P reduction and microbial fertilizer application. P2 and P3 significantly increased the abundance of Fusarium and Gibberella. Also, P3 significantly increased the relative abundance of Exophiala, Acremonium, Drechslera, and P2 significantly increased the relative abundance of Bionectria.
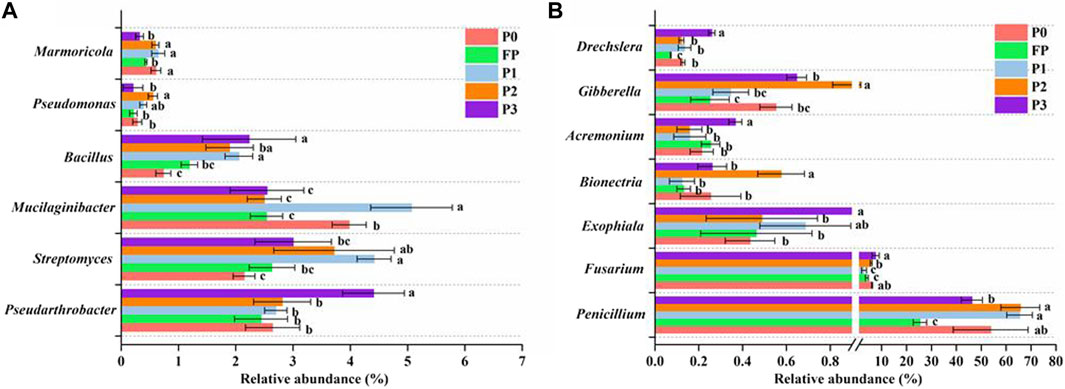
FIGURE 2. Relative abundance of the dominant functional bacterial genera (A) and functional fungal genera (B) under different P fertilizer application systems. P0 and FP represent 0 and 120 kg P2O5 ha−1 of P fertilizer application rates, respectively. P1, 30% reduction in P fertilizer application (84.0 kg P2O5/hm2 of microbial blended fertilizer); P2, 30% reduction in P fertilizer application (67.2 kg P2O5/hm2 of microbial blend fertilizer +16.8 kg P2O5/hm2 of diammonium phosphate); P3, 30% reduction in P fertilizer application (microbial inoculum seed dressing +84.0 kg P2O5/hm2 of diammonium phosphate).
Soil Bacterial and Fungal Community Diversity
Compared with FP, P3 significantly increased the Shannon index of bacteria (Figure 3A), as well as the Shannon index and Chao1 index of fungi (Figure 3B). PLS-DA analysis showed that the soil bacterial and fungal communities on PC1 both formed their cluster distributions between high and low P, and the bacterial and fungal communities of P3 were significantly different on PC2 (Figure 4).
FIGURE 3. Soil bacterial (A) and fungal (B) community Shannon and Chao1 index under different P fertilizer application systems. P0 and FP represent 0 and 120 kg P2O5 ha−1 of P fertilizer application rates, respectively. P1, 30% reduction in P fertilizer application (84.0 kg P2O5/hm2 of microbial blended fertilizer); P2, 30% reduction in P fertilizer application (67.2 kg P2O5/hm2 of microbial blend fertilizer +16.8 kg P2O5/hm2 of diammonium phosphate); P3, 30% reduction in P fertilizer application (microbial inoculum seed dressing +84.0 kg P2O5/hm2 of diammonium phosphate).
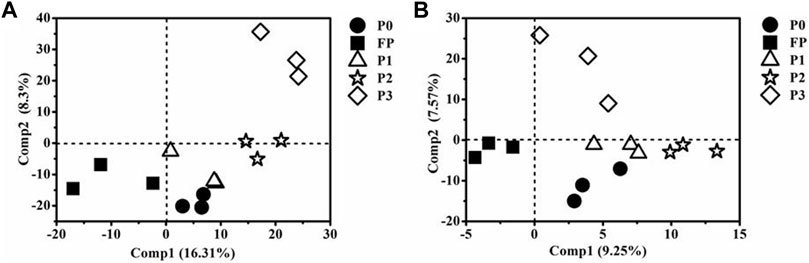
FIGURE 4. Partial least squares discriminant analysis (PLS-DA) of soil bacterial (A) and fungal (B) community under different P fertilizer application systems. P0 and FP represent 0 and 120 kg P2O5 ha−1 of P fertilizer application rates, respectively. P1, 30% reduction in P fertilizer application (84.0 kg P2O5/hm2 of microbial blended fertilizer); P2, 30% reduction in P fertilizer application (67.2 kg P2O5/hm2 of microbial blend fertilizer +16.8 kg P2O5/hm2 of diammonium phosphate); P3, 30% reduction in P fertilizer application (microbial inoculum seed dressing +84.0 kg P2O5/hm2 of diammonium phosphate).
Correlation Analysis Between Microbial Community and Environmental Factors
The CCA analysis showed that AP was the main driver of changes in bacterial and fungal community structure (p < 0.05) (Figure 5, Supplementary Tables S4, S5). SEM was used to predict the direct and indirect effects of different P reduction systems on soil microbial diversity and composition. The final fitted model complies with the established criteria of this experiment and gives a better model effect GFI = 0.997, AIC = 54.161, df = 1, χ2 = 0.161 (Figure 6B, Figure 6). Different P reduction systems indirectly affected the bacterial community through their effects on soil pH, SOM, AP, AN, TP, ACP, NEP, and urease. Again, the P reduction systems indirectly affected the fungal community structure through their effects on soil pH, SOM, AN, TN, ACP, and urease (Figures 6A, B).
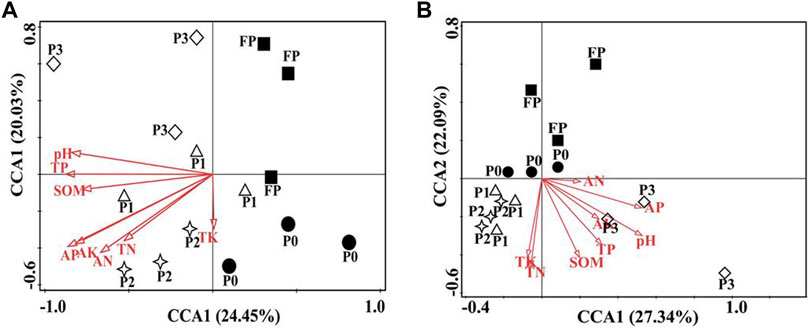
FIGURE 5. Canonical correspondence analysis (CCA) of soil bacterial (A) and fungal (B) community structure and soil properties under different P fertilizer application systems. P0 and FP represent 0 and 120 kg P2O5 ha−1 of P fertilizer application rates, respectively. P1, 30% reduction in P fertilizer application (84.0 kg P2O5/hm2 of microbial blended fertilizer); P2, 30% reduction in P fertilizer application (67.2 kg P2O5/hm2 of microbial blend fertilizer +16.8 kg P2O5/hm2 of diammonium phosphate); P3, 30% reduction in P fertilizer application (microbial inoculum seed dressing +84.0 kg P2O5/hm2 of diammonium phosphate).
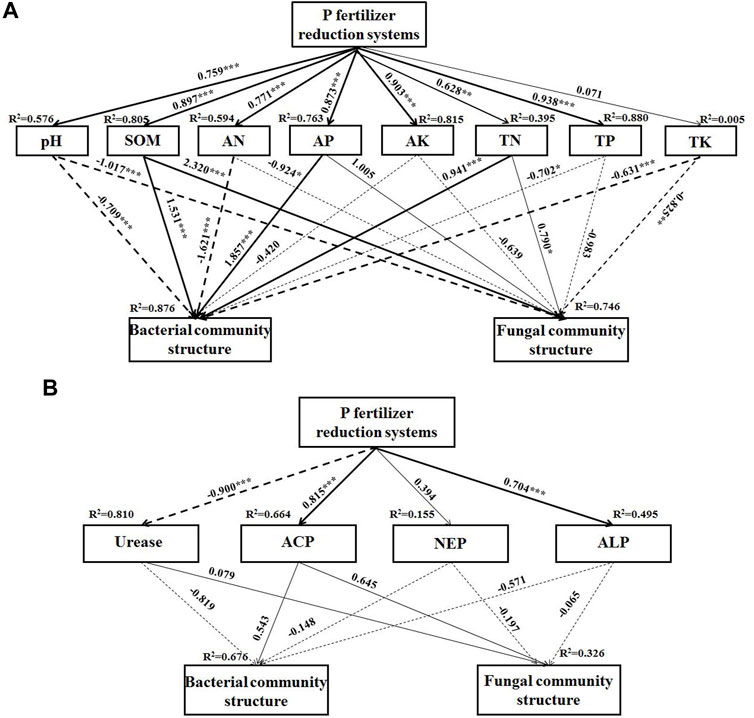
FIGURE 6. The structural equation model (SEM) for the relationship between P fertilizer reduction systems and soil properties (A), soil enzymatic activities and their relationship with soil bacterial and fungal community structure (B). The thickness of the arrow was proportional to the strength of the path coefficient. A solid arrow represents a positive path coefficient, a dotted arrow represents a negative path coefficient. *, **, *** respectively indicated p < 0.05, p < 0.01, p < 0.001. SOM, soil organic carbon; TN, total nitrogen; TP, total phosphorus; TK, total potassium; AN, available nitrogen; AP, available phosphorus; AK, available potassium. ACP, acid phosphatase; NEP, neutral phosphatase; ALP, alkaline phosphatase. P0 and FP represent 0 and 120 kg P2O5 ha−1 of P fertilizer application rates, respectively. P1, 30% reduction in P fertilizer application (84.0 kg P2O5/hm2 of microbial blended fertilizer); P2, 30% reduction in P fertilizer application (67.2 kg P2O5/hm2 of microbial blend fertilizer +16.8 kg P2O5/hm2 of diammonium phosphate); P3, 30% reduction in P fertilizer application (microbial inoculum seed dressing +84.0 kg P2O5/hm2 of diammonium phosphate).
Discussion
Shifts in Soil Physicochemical Properties and Enzymatic Activity in Response to Different P Fertilizer Reduction Systems
The results of this study showed that compared with FP, treatments P1, P2, and P3 significantly increased the contents of SOM and TP, indicating that soil organic matter conditions were improved after P reduction and microbial fertilizer application, and P fixation was reduced, thereby improving soil P availability. This may be due to the fact that the microbial fertilizer contains a variety of microorganisms, which can increase the content of SOM, thereby improving soil fertility (Juwarkar and Jambhulkar, 2008; Tosic et al., 2016). P2 and P3 significantly increased ACP activity compared with FP, as similarly indicated by Clarholm (1993) that chronic P application decreased ACP activity. This indicated that there was a positive interaction between microbial fertilizers with reduced P fertilizers and host microorganisms in the soil. This may be because the rich active substances in microbial fertilizer can have an impact on the activity of soil ACP to promote the absorption and utilization of P nutrients by crops and also enhance the availability of soil available P, which leads to an increased ACP activity (Prasanna et al., 2012).
Effects of Different P Fertilizer Reduction Systems on Soil Microbial Community Composition
In this study, Proteobacteria, Actinobacteria, and Acidobacteria were the most abundant bacterial phyla at the phylum level (Supplementary Figure S2A), which is consistent with previous studies which have shown that Proteobacteria, Actinobacteria, and Acidobacteria are the most common bacterial phyla in different agricultural systems (Shen et al., 2014). Similarly as has been indicated in previous studies almost all fungi isolated from farmland are Ascomycota (Viaud et al., 2000), in this study the dominant fungal phylum with the highest relative abundance was Ascomycota (Supplementary Figure S2B). This study showed the relative abundance of most potential beneficial genera (i.e., Pseudarthrobacter, Mucilaginibacter, Streptomyces, Bacillus, Pseudomonas, Penicillium, and Acremonium) and pathogenic genera (i.e., Fusarium, Gibberella, Exophiala, and Drechslera) increased in P reduction and microbial fertilizer application. Some studies have reported that the application of microbial bacterial fertilizer can significantly increase the diversity of microbial communities and promote the proliferation of beneficial bacteria (Araujo et al., 2020; Wang et al., 2022). In Chen et al. (2021) Pseudarthrobacter defluvii E5 was isolated from agricultural soils and showed efficient phthalic acid esters degradation and mineralization abilities. Mucilaginibacter is known for the degradation of cellulose and hemicellulose (López-mondéjar et al., 2016) while Streptomyces from a disease-suppressive soil produced volatile organic compounds (Cordovez et al., 2015). Bacillus with P solubilizing function may be a potential inoculant to regulate the biotic process of P transformation (Zhang, et al., 2021). Pseudomonas resistance against pathogen growth improves P cycling and also serves as inorganic P solubilizing bacteria that can be cultured (Jones and Oburger, 2011; Panpatte et al., 2016). Penicillium is an antagonistic phytopathogenic fungus and plays a critical role in the control of plant diseases. Penicillium oxalicum controls tomato fusarium wilt (Sabuquillo et al., 2006) while Penicillium bilaii has been shown to increase P solubility (Karamanos et al., 2010). Large and complex fungal genera containing many plant pathogens such as F. oxysporum and F. equiseti are important pathogenic bacteria that cause root rot (Zhou et al., 2018). Additionally, Gibberella zeae causes head blight in wheat, and can produce estrogenic toxins such as zearalenone, trichothecene, and deoxynivalenol (Khan et al., 2001). Exophiala are common saprobes in soil, opportunistic pathogens of animals, or endophytes in plant roots (Cheikh-Ali et al., 2015). Acremonium is an antagonistic plant pathogenic fungus and plays a certain role in the control of plant diseases. Acremonium obclavatum controls peanut rust (Sathiyabama and Balasubramanian, 2018). Drechslera tritici-repentis was noted in a previous study as a potentially pathogenic bacteria genus that caused wheat tan spots (Carmona et al., 2006). After the P reduction and microbial fertilizer application, crops can stimulate microorganisms to increase the number of beneficial microorganisms. This is done by increasing the number of certain secretions and crop root exudates that can act as signal molecules to regulate microbial populations and increase the number of antagonistic pathogenic bacteria in the soil. This promotes the healthy development of the soil, which is ultimately beneficial to the growth of crops (Henkes et al., 2011). Microbial function is closely related to soil function. Functional microorganisms can fix nitrogen and dissolve phosphorus, and also produce polysaccharides and other substances through reproduction and metabolism. This improves soil physical and chemical properties, alleviate soil hardening, improve soil fertilizer and water retention, enhance root absorption capacity and improve nutrient utilization. Functional microorganisms donot only ensure proper soil function, but also serve as the basis for restoring soil function (Yang et al., 2020).
Effects of Different P Fertilizer Reduction Systems on Soil Microbial Diversity
In the present study, P3 significantly increased bacterial diversity (Figure 3A), fungal diversity, and richness (Figure 3B) compared with FP. This may be due to the increase in soil nutrient content, especially the increase in organic matter content, which is the main reason for the increase in microbial diversity. Yue et al. (2022) also found that the combined application of microbial fertilizers and chemical fertilizers significantly increased the Shannon index of soil bacteria. The results of Chhabra et al. (2013) also showed that the addition of P reduced the microbial Shannon diversity index in the soil. Li et al. (2021) found that adding bio-organic fertilizer increased bacterial diversity but decreased fungal diversity. The results of Liu et al. (2018) showed that different P application systems had no significant effect on bacterial diversity, while fungal richness and diversity indices decreased significantly with the increase of P application rates. The reason for the different responses maybe that fungi are more sensitive to soil fertility and P addition than bacteria. Typically, fungal hyphae can extend into the soil, increasing the surface area in the soil for water and nutrient uptake. This gives fungi easy access to AP in soil culminating in its increased sensitivity to P.
Driving Factors Affecting Changes in Soil Microbial Community Structure
The results of this study showed that AP was the main driver of bacterial and fungal community structural changes (p < 0.05) (Figure 5, Supplementary Tables S4, 5). Liu et al. (2018) also found that AP was the dominant factor affecting fungal community changes in different P application systems. Similarly, Siciliano et al. (2014) showed that soil P content is an important driver of soil bacterial and fungal diversity. The effect of P availability on microbial diversity may depend on increasing the relative abundance of some genera and altering competition patterns between different populations (He et al., 2016). These results suggest that P input is a determinant of changes in soil microbial community structure in agricultural systems. Increased AP content may affect microbial community structure, possibly by exerting selective pressure on microbial communities and altering competitive interactions (Crowther et al., 2011).
Conclusion
Reduced application of P fertilizer combined with the application of microbial fertilizer significantly increased the content of SOM, TP, AP, AK, and ACP activity and also improved bacterial and fungal diversity. Moreover, the relative abundance of potentially beneficial genera (i.e., Bacillus, Pseudarthrobacter, Penicillium, and Acremonium) and potentially pathogenic genera (i.e., Fusarium, Gibberella, and Drechslera) increased significantly. Different environmental factors also have different degrees of influence on the changes in soil bacterial and fungal communities, among which AP had the greatest impact. The results suggested that the P fertilizer reduction combined with microbial fertilizer systems regulated the pathogenic and beneficial fungi populations which created a microbial community that is favorable for maize growth. Therefore we recommend the reduced application of P fertilizer combined with the application of microbial fertilizer as the most effective agronomic practice for improving soil fertility and crop yields. This will help to rationalize the application of P fertilizer and also ensure proper nutrient use efficiency and sustainable agriculture.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI (accession: SRP365800).
Author Contributions
CL and QG designed the study; HL performed the work; SL, RQ, and EL analysed the data; and ZZ revised the manuscript. All authors read and approved the final manuscript.
Funding
This study was funded by the National Key Research and Development Program of China (Grant numbers 2017YFD0200205), and the Special Funds Project for Central Government Guides Local Science and Technology Development in Jilin Province (Grant number 202002013JC).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.899727/full#supplementary-material
References
Adams, R. I., Miletto, M., Taylor, J. W., and Bruns, T. D. (2013). Dispersal in Microbes: Fungi in Indoor Air Are Dominated by Outdoor Air and Show Dispersal Limitation at Short Distances. ISME J. 7, 1262–1273. doi:10.1038/ismej.2013.28
Araujo, R., Dunlap, C., and Franco, C. M. M. (2020). Analogous Wheat Root Rhizosphere Microbial Successions in Field and Greenhouse Trials in the Presence of Biocontrol Agents Paenibacillus Peoriae SP9 and Streptomyces Fulvissimus FU14. Mol. Plant Pathol. 21, 622–635. doi:10.1111/mpp.12918
Atieno, M., Herrmann, L., Nguyen, H. T., Phan, H. T., Nguyen, N. K., Srean, P., et al. (2020). Assessment of Biofertilizer Use for Sustainable Agriculture in the Great Mekong Region. J. Environ. Manag. 275, 111300–111324. doi:10.1016/j.jenvman.2020.111300
Carmona, M., Ferrazini, M., and Barreto, D. (2006). Tan Spot of Wheat Caused by Drechslera Tritici-Repentis: Detection, Transmission, and Control in Wheat Seed. Cereal Res. Commun. 34, 1043–1049. doi:10.1556/CRC.34.2006.2-3.236
Cheikh-Ali, Z., Glynou, K., Ali, T., Ploch, S., Kaiser, M., Thines, M., et al. (2015). Diversity of Exophillic Acid Derivatives in Strains of an Endophytic Exophiala Sp. Phytochemistry 118, 83–93. doi:10.1016/j.phytochem.2015.08.006
Chen, F., Chen, Y., Chen, C., Feng, L., Dong, Y., Chen, J., et al. (2021). High-efficiency Degradation of Phthalic Acid Esters (PAEs) by Pseudarthrobacter Defluvii E5: Performance, Degradative Pathway, and Key Genes. Sci. Total Environ. 794, 148719. doi:10.1016/j.scitotenv.2021.148719
Chhabra, S., Brazil, D., Morrissey, J., Burke, J., O’Gara, F., Dowling, D. N., et al. (2013). Fertilization Management Affects the Alkaline Phosphatase Bacterial Community in Barley Rhizosphere Soil. Biol. Fertil. Soils 49, 31–39. doi:10.1007/s00374-012-0693-2
Clarholm, M. (1993). Microbial Biomass P, Labile P, and Acid Phosphatase Activity in the Humus Layer of a Spruce Forest, after Repeated Additions of Fertilizers. Biol. Fert. Soils 16, 287–292. doi:10.1007/BF00369306
Cordovez, V., Carrion, V. J., Etalo, D. W., Mumm, R., Zhu, H., van Wezel, G. P., et al. (2015). Diversity and Functions of Volatile Organic Compounds Produced by Streptomyces from a Disease-Suppressive Soil. Front. Microbiol. 6, 1081. doi:10.3389/fmicb.2015.01081
Crowther, T. W., Boddy, L., and Jones, T. H. (2011). Outcomes of Fungal Interactions Are Determined by Soil Invertebrate Grazers. Ecol. Lett. 14, 1134–1142. doi:10.1111/j.1461-0248.2011.01682.x
Ding, L. J., Su, J. Q., Sun, G. X., Wu, J. S., and Wei, W. X. (2018). Increased Microbial Functional Diversity under Long-Term Organic and Integrated Fertilization in a Paddy Soil. Appl. Microbiol. Biotechnol. 102, 1969–1982. doi:10.1007/s00253-017-8704-8
Edgar, R. C. (2013). UPARSE: Highly Accurate OTU Sequences from Microbial Amplicon Reads. Nat. Methods 10, 996–998. doi:10.1038/nmeth.2604
Elser, J. J. (2012). Phosphorus : a Limiting Nutrient for Humanity. Curr. Opin. Biotechnol. 23, 833–838. doi:10.1016/j.copbio.2012.03.001
He, D., Xiang, X., He, J.-S., Wang, C., Cao, G., Adams, J., et al. (2016). Composition of the Soil Fungal Community Is More Sensitive to Phosphorus Than Nitrogen Addition in the Alpine Meadow on the Qinghai-Tibetan Plateau. Biol. Fertil. Soils 52, 1059–1072. doi:10.1007/s00374-016-1142-4
Henkes, G. J., Jousset, A., Bonkowski, M., Thorpe, M. R., Scheu, S., Lanoue, A., et al. (2011). Pseudomonas Fluorescens CHA0 Maintains Carbon Delivery to Fusarium Graminearum-Infected Roots and Prevents Reduction in Biomass of Barley Shoots through Systemic Interactions. J. Exp. Bot. 62, 4337–4344. doi:10.1093/jxb/err149
Jones, D. L., and Oburger, E. (2011). “Phosphorus in Action, Biological Processes in Soil Phosphorus Cycling,” in Chapter 7 Solubilization phosphorus by soil Microorg (Berlin: Springer) 26. doi:10.1007/978-3-642-15271-9
Jorquera, M. A., Hernández, M. T., Rengel, Z., Marschner, P., De, M., and Mora, L. (2008). Isolation of Culturable Phosphobacteria with Both Phytate-Mineralization and Phosphate-Solubilization Activity from the Rhizosphere of Plants Grown in a Volcanic Soil. Biol. Fert. Soils 8, 1025–1034. doi:10.1007/s00374-008-0288-0
Juwarkar, A. A., and Jambhulkar, H. P. (2008). Restoration of Fly Ash Dump through Biological Interventions. Environ. Monit. Assess. 139, 355–365. doi:10.1007/s10661-007-9842-8
Karamanos, R. E., Flore, N. A., and Harapiak, J. T. (2010). Re-visiting Use of Penicillium Bilaii with Phosphorus Fertilization of Hard Red Spring Wheat. Can. J. Plant Sci. 90, 265–277. doi:10.4141/cjps09123
Khan, N. I., Pathologist, P. P., and Pathology, P. (2001). Selection and Evaluation of Microorganisms for Biocontrol of Fusarium Head Blight of Wheat Incited by Gibberella Zeae. Plant Dis. 85, 1253–1258. doi:10.1094/PDIS.2001.85.12.1253
Koljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., and Bahram, M. (2014). Towards a Unified Paradigm for Sequence-Based Identification of Fungi. Mol. Ecol. 22, 5271–5277. doi:10.1111/mec.12481
Li, W., Zhang, F., Cui, G., Wang, Y., Yang, J., Cheng, H., et al. (2021). Effects of Bio-Organic Fertilizer on Soil Fertility, Microbial Community Composition, and Potato Growth. ScienceAsia 47, 347–356. doi:10.2306/SCIENCEASIA1513-1874.2021.039
Lin, Y., Ye, G., Kuzyakov, Y., Liu, D., Fan, J., and Ding, W. (2019). Long-term Manure Application Increases Soil Organic Matter and Aggregation , and Alters Microbial Community Structure and Keystone Taxa. Soil Biol. biochem. 134, 187–196. doi:10.1016/j.soilbio.2019.03.030
Liu, J. l., Liao, W. h., Zhang, Z. x., Zhang, H. t., Wang, X. j., and Meng, N. (2007). Effect of Phopshate Fertilizer and Manure on Crop Yield, Soil P Accumulation, and the Environmental Risk Assessment. Agric. Sci. China 6, 1107–1114. doi:10.1016/S1671-2927(07)60153-9
Liu, M., Liu, J., Chen, X., Jiang, C., Wu, M., and Li, Z. (2018). Shifts in Bacterial and Fungal Diversity in a Paddy Soil Faced with Phosphorus Surplus. Biol. Fertil. Soils 54, 259–267. doi:10.1007/s00374-017-1258-1
López-mondéjar, R., Zühlke, D., Becher, D., Riedel, K., and Baldrian, P. (2016). Cellulose and Hemicellulose Decomposition by Forest Soil Bacteria Proceeds by the Action of Structurally Variable Enzymatic Systems. Sci. Rep. 6, 25279. doi:10.1038/srep25279
Malaeb, Z. A., Summers, J. K., and Pugesek, B. H. (2000). Using Structural Equation Modeling to Investigate Relationships Among Ecological Variables. Environ. Ecol. Stat. 7, 93–111. doi:10.1023/A:1009662930292
Markland, D. (2007). The Golden Rule Is that There Are No Golden Rules: A Commentary on Paul Barrett ’ S Recommendations for Reporting Model Fit in Structural Equation Modelling. Pers. Individ. Dif. 42, 851–858. doi:10.1016/j.paid.2006.09.023
Meinikmann, K., Hupfer, M., and Lewandowski, J. (2015). Phosphorus in Groundwater Discharge – A Potential Source for Lake Eutrophication. J. Hydrol. 524, 214–226. doi:10.1016/j.jhydrol.2015.02.031
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: An Open Annotation Tool for Parsing Fungal Community Datasets by Ecological Guild. Fungal Ecol. 20, 241–248. doi:10.1016/j.funeco.2015.06.006
Panpatte, D., Jhala, Y., Shelat, H., and Vyas, S. (2016). “Pseudomonas fluorescens: A Promising Biocontrol Agent and PGPR for Sustainable Agriculture,” Microbial Inoculants in Sustainable Agricultural Productivity. Editors D. P. Singh, H. B. Singh, and R. Prabha (New Delhi: Springer), 257–270. doi:10.1007/978-81-322-2647-5_15
Pineda, A., Kaplan, I., and Bezemer, T. M. (2017). Steering Soil Microbiomes to Suppress Aboveground Insect Pests. Trends Plant Sci. 22, 770–778. doi:10.1016/j.tplants.2017.07.002
Prasanna, R., Joshi, M., Rana, A., Shivay, Y. S., and Nain, L. (2012). Influence of Co-inoculation of Bacteria-Cyanobacteria on Crop Yield and C-N Sequestration in Soil under Rice Crop. World J. Microbiol. Biotechnol. 28, 1223–1235. doi:10.1007/s11274-011-0926-9
R-project (2010). A Language and Environment for Statistical Computing. R foundation 571 for statistical computing, ISBN 3-900051-07-0. Available at: http://www.r-project.org (Accessed November 22, 2021).
Sabuquillo, P., Cal, A. De., and Melgarejo, P. (2006). Biocontrol of Tomato Wilt by Penicillium oxalicum Formulations in Different Crop Conditions. Biol. Control 37, 256–265. doi:10.1016/j.biocontrol.2006.02.009
Sathiyabama, M., and Balasubramanian, R. (2018). Protection of Groundnut Plants from Rust Disease by Application of Glucan Isolated from a Biocontrol Agent Acremonium Obclavatum. Int. J. Biol. Macromol. 116, 316–319. doi:10.1016/j.ijbiomac.2018.04.190
Shen, C., Liang, W., Shi, Y., Lin, X., Zhang, H., Wu, X., et al. (2014). Contrasting Elevational Diversity Patterns between Eukaryotic Soil Microbes and Plants. Ecology 95, 3190–3202. doi:10.1890/14-0310.1
Siciliano, S. D., Palmer, A. S., Winsley, T., Lamb, E., Bissett, A., Brown, M. V., et al. (2014). Soil Fertility Is Associated with Fungal and Bacterial Richness , whereas pH Is Associated with Community Composition in Polar Soil Microbial Communities. Soil Biol. biochem. 78, 10–20. doi:10.1016/j.soilbio.2014.07.005
Tosic, I., Golic, Z., and Radosavac, A. (2016). Effects of the Application of Biofertilizers on the Microflora and Yield of Lettuce (Lactuca sativa L.). Acta Agric. Serbica 21, 91–98. doi:10.5937/aaser1642091t
Viaud, M., Pasquier, A., and Brygoo, Y. (2000). Diversity of Soil Fungi Studied by PCR – RFLP of ITS. Control 104, 1027–1032. doi:10.1017/S0953756200002835
Wang, F., Wang, Q., Adams, C. A., Sun, Y., and Zhang, S. (2022). Effects of Microplastics on Soil Properties: Current Knowledge and Future Perspectives. J. Hazard. Mater 424, 127531. doi:10.1016/j.jhazmat.2021.127531
Xu, N., Tan, G., Wang, H., and Gai, X. (2016). Effect of Biochar Additions to Soil on Nitrogen Leaching, Microbial Biomass and Bacterial Community Structure. Eur. J. Soil Biol. 74, 1–8. doi:10.1016/j.ejsobi.2016.02.004
Yang, T., Siddique, K. H. M., and Liu, K. (2020). Cropping Systems in Agriculture and Their Impact on Soil Health-A Review. Glob. Ecol. Conserv. 23, e01118. doi:10.1016/j.gecco.2020.e01118
Yue, H., Zhang, D., Hou, D., Li, Y., Yao, T., and Huang, G. S. (2022). Effects of Partial Substitution of Chemical Fertilizer by Microbial Fertilizer on Yield of Cucumber and Soil Bacterial Community Structure in Greenhouse. J. Northwest A&F Univ. 7, 2–11. doi:10.13207/j.cnki.jnwafu.2022.07.014
Zhang, M., and Zhang, H. (2010). Co-transport of Dissolved Organic Matter and Heavy Metals in Soils Induced by Excessive Phosphorus Applications. J. Environ. Sci. 22, 598–606. doi:10.1016/S1001-0742(09)60151-0
Zhang, X., Zhan, Y., Zhang, H., Wang, R., Tao, X., and Zhang, L. (2021). Inoculation of Phosphate-Solubilizing Bacteria (Bacillus) Regulates Microbial Interaction to Improve Phosphorus Fractions Mobilization during Kitchen Waste Composting. Bioresour. Technol. 340, 125714. doi:10.1016/j.biortech.2021.125714
Zhang, X., Zhang, R., Gao, J., Wang, X., Fan, F., Ma, X., et al. (2017). Thirty-one Years of Rice-Rice-Green Manure Rotations Shape the Rhizosphere Microbial Community and Enrich Beneficial Bacteria. Soil Biol. biochem. 104, 208–217. doi:10.1016/j.soilbio.2016.10.023
Zhou, Q., Li, N., Chang, K. F., Hwang, S. F., Strelkov, S. E., Conner, R. L., et al. (2018). Genetic Diversity and Aggressiveness of Fusarium Species Isolated from Soybean in Alberta, Canada. Crop Prot. 105, 49–58. doi:10.1016/j.cropro.2017.11.005
Keywords: phosphate fertilizer reduction, microbial fertilizer, bacteria, fungi, high-throughput sequencing
Citation: Liu H, Li S, Qiang R, Lu E, Li C, Zhang J and Gao Q (2022) Response of Soil Microbial Community Structure to Phosphate Fertilizer Reduction and Combinations of Microbial Fertilizer. Front. Environ. Sci. 10:899727. doi: 10.3389/fenvs.2022.899727
Received: 19 March 2022; Accepted: 19 May 2022;
Published: 24 June 2022.
Edited by:
Tim George, The James Hutton Institute, United KingdomReviewed by:
Lin Zhang, China Agricultural University, ChinaXiaohua Long, Nanjing Agricultural University, China
Copyright © 2022 Liu, Li, Qiang, Lu, Li, Zhang and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cuilan Li, Y3VpbGFubGlAMTI2LmNvbQ==; Jinjing Zhang, emhhbmdqaW5qaW5nQDEyNi5jb20=; Qiang Gao, Z3l0MTk5OTYyQDE2My5jb20=
 Hang Liu
Hang Liu Songsong Li
Songsong Li