
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Environ. Sci. , 26 April 2022
Sec. Water and Wastewater Management
Volume 10 - 2022 | https://doi.org/10.3389/fenvs.2022.875157
This article is part of the Research Topic Emerging Approaches for Sustainable Management for Wastewater View all 8 articles
Microbial manganese peroxidases (MnPs) are ligninolytic enzymes primarily responsible for degrading lignin, but they are also capable of degrading dyes, organic pollutants, and emerging contaminants in wastewater. They have been isolated and characterized from many white-rot fungi and few bacteria. For microbial MnPs, the optimum pH range is between 3.5 and 9.0; the optimum temperature range is between 25°C and 70°C. Their molecular weights range between 25 kDa and 68 kDa. By oxidizing Mn2+ to Mn3+, they can oxidize a wide range of phenolic and non-phenolic substrates. MnPs can be used in a wide range of industrial applications, including delignification of pulp, wastewater treatment, biofuel production, dye removal, biopulping, biobleaching, and juice extract clarification. The purpose of this review is to summarize biotechnological applications of manganese peroxidases.
Manganese peroxidase (MnP, EC 1.11.1.13) is a H2O2-dependent glycoprotein peroxidase that comprises heme as a cofactor and belongs to the oxidoreductase family. The MnP enzyme is found in most white-rot fungi and some bacteria (Qin et al., 2017). This enzyme was first discovered in the fungus Phanerochaete chrysosporium (Paszczynski et al., 1985). The range of its molecular mass is from 25kDa to 68 kDa. Different MnPs exist in nature with different properties. Based on the lengths of their C-terminal tail, MnPs can be classified as short, long, or extra-long types in white-rot fungi (Fernández-Fueyo et al., 2014; Chang et al., 2021). In Agaricales, three different subfamilies of the fungal MnP were described based on residues found in their Mn2+-binding sites: MnP-ESD (Glu/Ser/Asp Mn2+-oxidation site), MnP-DGD (Asp/Gly/Asp Mn2+-oxidation site), and MnP-DED (Asp/Glu/Asp Mn2+-oxidation site) (Chang et al., 2021; Ruiz-Dueñas et al., 2021). Mn2+-binding sites are not unique in MnPs. Those peroxidases, which were originated from MnPs, also contain structures identical to the Mn2+-binding site and can oxidize Mn2+ to Mn3+ (Ayuso-Fernández et al., 2018; Chang et al., 2021).
MnPs need hydrogen peroxidase and Mn2+ for oxidation of various aromatic and phenolic compounds (Bermek et al., 2004; Asgher et al., 2014). MnPs have the greatest capacity for enzymatic actions, and they are extraordinarily flexible in nature with a wide variety of industrial uses (Nayanashree and Thippeswamy, 2015a). MnP has biotechnological applications in the treatment of textile waste effluents (Bilal and Asgher, 2015). Various kinds of environmental pollutants can be found in textile effluents, such as synthetic dye residues that are toxic and can cause cancer. Based on their eco-toxicological properties, textile effluents containing dyes have been extensively studied for their potential ecological hazards (Iqbal and Bhatti, 2014; Bilal and Asgher, 2015). MnP can be used to oxidize aromatic amines and phenolic compounds in wastewater through the use of one-electron oxidation and produce more reactive free radicals (Heinzkill et al., 1998). In nature, the MnP enzyme catalyzes the depolymerization of plant lignin and is a constituent of the lignin-degrading enzyme complex. It can degrade lignin most commonly, as well as hemicellulose, cellulose, and lignin in agriculture (Torres and Ayala, 2010). In addition, it can degrade recalcitrant aromatic contaminants, including industrial dyes, polycyclic aromatic hydrocarbons, chlorophenols, and antibiotics, which may be hazardous to human health (Qin et al., 2014). Recently, this enzyme has been widely studied in terms of its potential to perform the wastewater treatment. It can be applied to dye decolorization, biopulping, biobleaching, clarifying juices, bioethanol production, and degradation of various xenobiotic compounds (Asgher and Iqbal, 2013; Nousiainen et al., 2014; Nayanashree and Thippeswamy, 2015b).
MnP is a versatile enzyme with numerous applications in many industries, such as pulp and paper, textile, food, and biofuel. Only a few review articles have been published on MnP-catalyzed wastewater treatment. Chowdhary et al. (2019) presented an overview of the current scenario on MnPs. Chang et al. (2021) discussed decolorization of textile dyes by MnPs. Currently, their wide range of applications is rarely reviewed. This review discusses the industrial and biotechnological applications of MnPs.
MnPs are produced by fungi and bacteria (Champagne and Ramsay, 2005; Chowdhary et al., 2019). In this section, we described various fungal and bacterial sources of MnPs.
MnPs are produced and characterized from a number of fungi that belong to the class Basidiomycetes (Tello et al., 2000; Dong et al., 2014). Examples are Agaricus bisporus (Hilden et al., 2013), Irpex lacteus F17 (Yang et al., 2016), Trametes sp. 48424 (Zhang et al., 2016), Echinodontium taxodii 2538 (Kong et al., 2016), Ganoderma lucidum IBL-05 (Bilal et al., 2015), Fusarium sp. strain ZH-H2 (Muhammad et al., 2011), Agrocybe praecox (Hilden et al., 2014), Penicillium sp. (Oliveira et al., 2009), Irpex lacteus CD2 (Qin et al., 2014), Trametes versicolor (Shi et al., 2021), Phlebia radiata (Hakala et al., 2006), Ceriporiopsis subvermispora (Tanaka et al., 2009), Pleurotus ostreatus (Kamitsuji et al., 2004), and Pseudolagarobasidium sp. PP17-33 (Thamvithayakorn et al., 2019). A list of MnP-producing fungi is presented in Table 1.
Fungal MnPs exhibited the highest activity between pH 3.0 and 5.5. The optimum temperature for fungal MnPs is 30–70°C. Immobilization may increase their thermostability. For example, the chitosan-immobilized MnP from Trichoderma harzianum exhibited stability at 50°C (El-Shora et al., 2017). A fungal MnP has a molecular weight ranging from 36 kDa to 55.2 kDa. Penicillium chrysogenum produces MnP with a molecular weight of 45 kDa that is used in the degradation of natural rubber (Nayanashree and Thippeswamy, 2015a). Galactomyces reessii a subterranean termite-associated yeast species was used for phenol removal from palm oil mill effluents due to its MnP and laccase activities (Chaijak et al., 2018). The MnP from Bjerkandera adusta strain CX-9 named MnP BA30 showed high solvent stability and dye decolorization efficiency (Bouacem et al., 2018). Similarly, Trametes pubescens strain i8 produced MnP TP55 with a molecular weight of 55.2 kDa, which could be used to produce cleaners, decolorize textile dyes, and degrade lignin (Rekik et al., 2019). Trametes polyzona KU-RNW027 produced two MnPs which play essential roles in biodegradation of dyes and pharmaceutical products (Lueangjaroenkit et al., 2019). The main disadvantages of fungal MnPs are their low yields and poor stability.
A number of advantages of bacterial MnPs over fungal MnPs have emerged in recent years from the industrial point of view, including their ability to work in wide ranges of temperatures and pH with a high stability toward inhibitory agents, easy production in a short period of time, broad substrate specificity, and ease of cloning and expression in host cells with appropriate manipulation (Chauhan et al., 2017; Xu et al., 2018). Examples of MnP-producing bacteria include Bacillus subtilis (Nayanashree and Thippeswamy, 2015b), B. amyloliquefaciens SL-7 (Mei et al., 2020), B. aryabhattai (Zainith et al., 2019), B. cereus 11778 (Khelil et al., 2015), and Lactobacillus kefiri (Omar et al., 2021). For bacteria, MnPs show their optimum activity between pH 5.0 and 9.0. For bacterial MnPs, the optimum temperature ranges from 25 to 70°C. The molecular weight of bacterial MnPs ranges from 25 kDa to 68 kDa. Recently, Bacillus velezensis AI-Dhabi 140 produces MnPs of 46 kDa that could degrade tetracycline antibiotic using the fibrous bed reactor (Al-Dhabi et al., 2021). The MnPs from Klebsiella pneumoniae strains have been utilized in the manufacturing of ethanol using rice and wheat bran biomass as substrates (Gaur et al., 2018). A list of MnP-producing bacteria is summarized in Table 2.
The crystal structure of MnP refined at 0.93 Å resolution is presented in Figure 1. The active site of MnP is composed of one heme propionate and the side chains of Glu35, Glu39, and Asp179 (Sundaramoorthy et al., 2010). Two calcium ions are essential components of MnP that provide thermal stability. In MnPs, five disulfide bridges are present to stabilize them, which are formed by 10 cysteine amino acid residues (Chang et al., 2021). Heme propionate is found in the internal hydrophobic cavity of the enzyme, and Mn2+ binds to the center of the active site. As a resting-state structure, MnP contains a penta-coordinated Fe3+ structure integrated with the porphyrin ring of the heme cofactor and a proximal histidine (Chang et al., 2021). At the distal end, a catalytic His and an Arg residue form a peroxide-binding pocket (Sundaramoorthy et al., 2010). This structure is highly conserved among heme peroxidases. Minor variations are due to the Phe residues that are Trp in intercellular peroxidases, cytochrome c, and ascorbate peroxidase (Chang et al., 2021). The Asp242–His173 pair is also conserved (Sundaramoorthy et al., 2010).
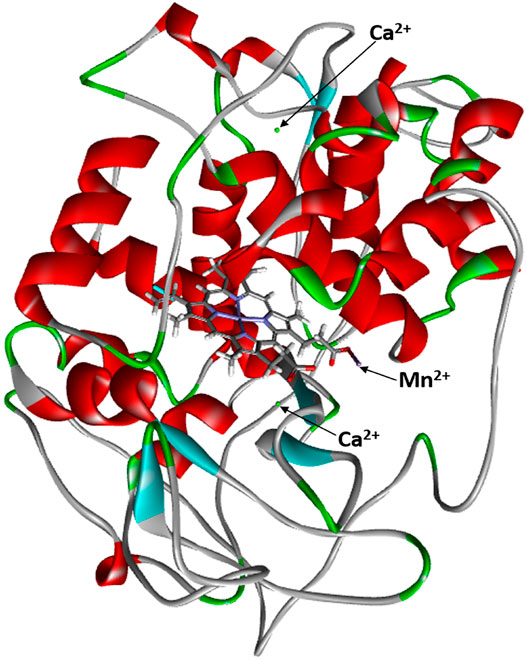
FIGURE 1. Crystal structure of MnP complexed with Mn2+ (Sundaramoorthy et al., 2010).
The catalytic cycle of MnP begins when the heme group of MnP transfers two electrons to hydrogen peroxidase in its resting state, resulting in the production of water and compound I (Hofrichter, 2002). In the next step, compound I oxidizes Mn2+ and produces compound II, a free radical, and Mn3+. Compound II then catalyzes the oxidation of Mn2+ to form Mn3+, allowing MnP to revert to its original form (Figure 2). These catalytic biochemical reactions of MnP are summarized as follows:
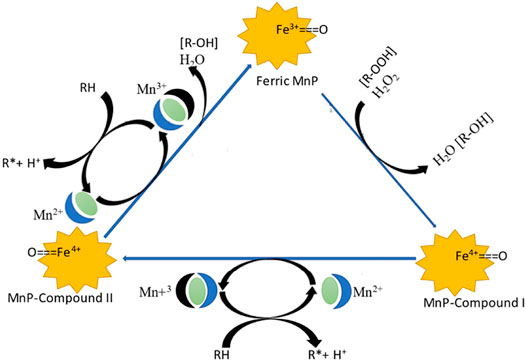
FIGURE 2. Catalytic cycle of MnP (Hofrichter, 2002).
The growth conditions and culture media composition influence the gene expression and total enzyme activity of microbial MnPs (Zhang et al., 2021). It varies from species to species and strain to strain. In this section, we described the effect of various factors on the MnP’s activity and production.
MnP is likely to be produced more efficiently with appropriate carbon sources. Glycerol and walnut pericarp, for example, provided the highest MnP activity compared to other carbohydrates (such as glucose, xylose, and sucrose) in Cerrena unicolor (Zhang et al., 2021). In contrast, in Phanerochaete chrysosporium, glucose is the preferred carbon source (Urek and Pazarlioglu, 2007; Zhou et al., 2013). The most effective carbon source for the MnP enzyme activity in Trichoderma harzianum is sucrose rather than any other carbon source.
High nitrogen concentration in the medium suppresses the MnP activity of several fungal species, including P. chrysosporium (Leatham and Kirk, 1983; Janusz et al., 2013). In contrast, higher amounts of MnP are produced in P. ostreatus and Trametes trogii in the presence of high concentration of nutrient nitrogen (Levin et al., 2002; Levin et al., 2010). Meanwhile, MnP from Dichomitus squalens cultures is secreted under both high- and low-nitrogen conditions (Perie and Gold, 1991). The nitrogen source also effects the production and activities of MnPs. The optimum production of MnP by four Pleurotus species occurred using (NH4)2SO4 or NH4NO3 as the inorganic nitrogen source (Stajić et al., 2006). The enzyme activity of Trichoderma harzianum is optimum when yeast extract is used as a nitrogen source (El-Shora et al., 2017). As a nitrogen source, peptone has been shown to be extremely effective in MnP production under solid state fermentation by many fungi including Cerrena unicolor strain IBB 62, Bjerkandera adusta, Bjerkandera sp. strain BOS55, Lentinus edodes, P ostreatus, and P. chrysosporium (Kaal et al., 1995; Elisashvili et al., 2001; Janusz et al., 2013).
Increases in Mn2+ ions in the culture medium enhance the production of the MnP enzyme by activating the transcription of the mnp gene (Urek and Pazarlioglu, 2007; El-Shora et al., 2017). MnP of Trametes sp. 48424 is strongly resistant to metal ions such as Ni2+, Li+, Ca2+, K+, and Mn2+ (Zhang et al., 2016), whereas Mn2+, Ca2+, Co+, Cu2+, Li+, and K+ enhance the production of MnP BA 30 of the Bjerkandera adusta strain CX–9 (Bouacem et al., 2018). The activity of MnP TP55 of Trametes pubescens strain i8 is induced by Mn2+, Fe2+, Cu2+, and Mg2+ whereas it was completely inhibited by Cd2+, Ni2+, and Hg2+ (Rekik et al., 2019).
Synthetic inducers such as Tween 80 when added in low quantities into the growth medium stimulate the enzyme activity in comparison to control by increasing the surface area for the microbial growth (Urek and Pazarlioglu, 2007; Muhammad et al., 2011). The initial concentration of hydrogen peroxide increases the MnP activity whereas the latter exerts the inhibitory effect on the MnP enzyme activity (Vrsanska et al., 2015; Rekik et al., 2019).
Phenylmethylsulphonyl fluoride (PMSF), an inhibitor of serine protease, when supplemented to a growth medium suppresses the protease production that participates in the regulation of the MnP activity, resulting in inhibiting the MnP activity (Urek and Pazarlioglu, 2007).
Sodium azide, l-cysteine, sodium sulfate, β-mercaptoethanol, potassium cyanide, ethylenediaminetetraacetic acid (EDTA), sodium dodecyl sulfate (SDS), and hydroxylamine have inhibitory effects on enzyme activity (Kong et al., 2016; Bouacem et al., 2018; Gaur et al., 2018; Rekik et al., 2019). Sodium azide can break the disulfide bonds that maintain the active site of the enzyme. SDS removes the hydrophobic interactions that are present in protein, resulting in the degradation of tertiary and quaternary structures. EDTA, a chelating agent of metals, has the property to form complexes with complex or metal ions, resulting in the decreased activity of the enzyme.
Enzyme activity decreases with the increase in the concentration of organic solvents such as methanol, ethanol, DMSO, ethylene glycol, iso-propyl alcohol, butanediol, and glycerine (Kong et al., 2016; Liu et al., 2020).
Chelators such as oxalate, malate, malonate, and lactate are added to the growing media; these chelators stabilized the Mn2+ ions by chelating them, hence increasing the MnP enzyme activity (El-Shora et al., 2017).
The MnP enzyme is affected by temperature and pH. MnPs from different species and strains of microorganisms responded differently to temperature and pH. Each MnP has its own optimal temperature and pH range.
MnPs may degrade several toxic dyes, antibiotics, phenolic compounds, and polycyclic aromatic hydrocarbons under peroxide-dependent conditions, making them a better biocatalyst for the biodegradation of harmful environmental pollutants in wastewater (Singh et al., 2021). In addition, MnPs may be used for delignification of lignin, clarifying juices, and bioethanol production. In this section, various applications of MnPs are summarized.
MnPs act as biocatalysts for decolorizing dyes in wastewater. Several MnPs have been purified and characterized for their ability to decolorize dyes. Two MnPs isolated from Trametes polyzona KU-RNW027 degraded Remazol brilliant blue (25 mg/L) in 10–30 min and showed high degradation activities toward Remazol navy blue and Remazol brilliant yellow (Lueangjaroenkit et al., 2019). Both of these MnPs were found to be highly stable and were activated by various organic solvents and metal ions. Rekik et al. (2019) purified the MnP (named MnP TP55) from the fungus Trametes pubescens strain i8. Several types of dyes were degraded using MnP TP55, including anthraquinone dyes (Remazol Brilliant Blue Reactif and Cibacet Brilliant Blue BG), triphenylmethane dye (methyl green), indigo dye (indigo carmine), azo dyes (Direct Red 5B and Remazol Brilliant Violet 5R), polymeric dye (Poly R-478), and acid dye (Acid Blue 158). The degradation efficiency of MnP TP55 varied from dye to dye and ranged from 42 to 95% when the dye concentration was 50 µM. Emami et al. (2020) studied the effects of various stabilizers including manganese (IV) dioxide (MnO2), veratryl alcohol (VA), dimethyl sulfoxide (DMSO), polyethylene glycol (PEG), and iron (III) oxide (Fe2O3) on production, stability, and decolorization efficiency of MnP produced by Phanerochaete chrysosporium. The Fe3O4-containing calcium alginate beads increased MnP production by 67%. Furthermore, PEG and DMSO boosted the decolorization efficiency of MnP toward Acid Orange 7 and crystal violet. Gomathy et al. (2021) reported that MnP produced by an alkaliphilic bacterial strain Pseudomonas mendocina was able to degrade mixed azo dyes at alkaline pH. Furthermore, immobilization of this alkaline MnP with the Luffa fibers increased the dye decolorization efficiency. Bouacem et al. (2018) reported that MnP from Bjerkandera adusta strain CX-9 (called MnP BA30) decolorized various dyes including indigo carmine, Remazol Brilliant Violet 5R, Remazol Brilliant Blue Reactif, Poly R-478, Cibacet Brilliant Blue BG, and Acid Blue 158 dyes with decolorization percentages of 42, 70, 38, 80, 77, and 91 within 12 h. In Mn2+-independent processes, MnPs from Bjerkandera adusta strain DSM 11310 and Pleurotus eryngii strain ATCC 90787 decolorized four azo and two phthalocyanine dyes by their direct interaction with the dye (Heinfling et al., 1998).
MnPs may reduce the amount of lignin components and chemicals in the paper industry wastewater through the process of biopulping and pulp biobleaching.
Biopulping is the process of treating wood chips with lignin-degrading fungus or MnPs before pulping. By pre-treating the composted wood with fungi before mechanical pulping, we can reduce electrical energy consumption during the refining process, improve paper strength, and reduce pitch (extractives) content, thereby reducing the environmental impact of the pulping process (Viikari et al., 2009). In biopulping, MnPs from white-rot fungi can be used to degrade lignin partially and loosen lignin structures. Jha and Patil (2011) demonstrated that MnP obtained from Penicillium oxalicum isolates-1 was effective in de-lignifying paper pulp and kraft liquor.
Biobleaching is the process of bleaching pulps with enzymes or lignolytic fungi, thereby reducing the quantity of the chemical bleach required (Kumar, 2021). Chemicals such as H2SO4, Cl2, ClO2, NaOH, and H2O2 are used during bleaching (Kumar, 2021). By reacting with lignin and carbohydrates, these chemicals generate a substantial amount of pollutants in bleach effluents. The pulp and paper industry generates high-strength, dark brown-colored wastewater during various stages of the paper-making process which is discharged into the environment.
In addition to reducing the level of lignin within the pulp, MnPs are generally used in pulp and paper industries as biobleaching agents. Kaneko et al. (1995) reported biobleaching of oxygen-alkaline–treated hardwood kraft pulp using a fungal MnP. In the presence of Tween 20, ISO brightness was improved by five points, while without Tween 20, ISO brightness did not improve significantly. Tween 20 could prevent mechanical inactivation of MnP caused by agitation or reduce the absorption of enzymes that would result in enzyme inactivation. Kondo et al. (1994) reported bleaching of hardwood kraft pulp using MnP from fungus Phanerochaete sordida YK-624 in the presence of MnSO4, Tween 80, and sodium malonate. In comparison to the untreated pulp, the brightness of the pulp increased by approximately 10 points, and the Kappa number decreased by approximately six points. In addition, multiple MnP treatments increased pulp brightness by 43 points to 75.5%. Harazono et al. (1996) investigated effects of various manganese chelators (oxalate, malonate, or gluconate) on MnP-mediated bleaching of hardwood kraft pulp, and they concluded that the brightness of the pulp was increased by about 10 points when oxalate was present in 2 mM. To enhance the bleaching effect of MnP on pulp, Bermek et al. (2002) investigated the effects of a large number of organic compounds such as organic acids, thiol-containing compounds, and their combinations on pulp bleaching. The pulp bleaching effect of linoleic acid, linolenic acid, and some unsaturated fatty acids was better than that of Tween 80. The bleaching power of MnP pulp was higher when Tween 80 and carboxylic acid were combined. Masarin et al. (2016) investigated the degradation of lignin in milled Eucalyptus grandis wood and kraft pulp by MnP from the fungus Ceriporiopsis subvermispora. The MnP-mediated elemental chlorine-free bleaching of eucalyptus kraft pulp resulted in a reduction of 3.7 kg/ton of ClO2, with an increase in ISO brightness from 88 to 90 points. MnP produced by Trametes versicolor during pulp bleaching, demethylates and delignifies kraft pulp and oxidizes mainly the phenolic lignin structures (Paice et al., 1993). Sasaki et al. (2001) developed a chlorine-free pulp-biobleaching system using MnP immobilized with mesoporous silica. There was 88% improvement in brightness when MnP treatment and pulp bleaching were performed in a two-stage reactor at temperatures of 39°C and 70°C, respectively.
By biobleaching of pulp, pulp can be of higher quality without using chlorine-based chemicals in large amounts (Kumar, 2021). When pulp is pretreated with MnPs, the kappa number of the pulp is reduced, and brightness is improved. Biobleaching of pulp has also resulted in improved physical strength properties and pulp viscosity (Kumar, 2021).
Lignin and lignin-like compounds are either discharged from agricultural waste or from the pulping and paper-making industries into water (De los Santos Ramos et al., 2009). Lignin is the second most abundant organic material after cellulose. It occurs in combination with other carbohydrates such as hemicellulose and cellulose at the cell wall level. It is a polymer with a high molecular mass, which makes it resistant to degradation by diverse types of microorganisms in the environment (Wang et al., 2011). Chemically, lignin is a heterogeneous polymer with phenylpropanoid units that are connected via a variety of linkages, e.g., carbon–carbon, aryl–aryl, and aryl–ether (Mohammad et al., 2020). In addition, lignin may be used as a feedstock for the production of chemical compounds such as substituted aromatics (Ragauskas et al., 2006). This type of valorization would require controlled depolymerization of lignin, which is hampered by its high level of resistance to biodegradation (Martinez et al., 2005). There are a variety of lignin degradation methods, such as pyrolysis, hydrogenation, and selective oxidation, as well as degradation under hydrothermal and supercritical conditions (Ma et al., 2015; Dai et al., 2016). Nevertheless, many of these processes are hazardous, environmentally unfriendly, and require large amounts of energy (Ward and Singh, 2002). Most of the processes use expensive chemical catalysts containing metals such as molybdenum or palladium (Chauhan, 2020). Therefore, it is essential to find a cheap and eco-friendly method for bioremediation or degradation of lignin. Hence, microbial enzymes appear to be a better and more suitable alternative to be used in these methods to solve the problems mentioned earlier. By using enzymes specific to their substrate, the selectivity of reactions can be increased and byproducts can therefore be minimized (Janusz et al., 2017; Chauhan, 2020). White-rot fungi have received extensive attention in recent years for their ability to degrade lignin through MnPs.
Pretreatment of lignin by biological means (microorganisms or MnPs) is an emerging method for biodelignification. In these methods, either fungi or bacteria are directly applied to the lignin treatment, or MnPs are used in enzymatic treatments. The white-rot fungi depolymerize lignin more efficiently than bacteria. Examples of lignin-depolymerizing fungi are Trametes pubescens strain i8 (Rekik et al., 2019), Pleurotus ostreatus (Kamitsuji et al., 2004), Echinodontium taxodii 2538 (Kong et al., 2016), Phanerochaete chrysosporium IBL-3 (Muhammad et al., 2011), Trametes versicolor (Shi et al., 2021), and Schizophyllum commune (Shi et al., 2021). Delignification is highly variable, depending on the species of fungi and the carbohydrates in the lignocellulose raw material (Suryadi et al., 2022). Although microorganism-mediated pretreatment boasts numerous advantages, it is still plagued with problems such as lengthy processing times, large space requirements, and regular monitoring of microbial growth (Suryadi et al., 2022). .
In contrast to using fungi directly, MnP-mediated degradation of lignin has several advantages (Suryadi et al., 2022). These include the ability to overcome challenges related to large-scale growth, long incubation processes, penetration of the mycelium into the substrate, excessive polysaccharide consumption which causes carbohydrate degradation, and contamination issues. MnPs can be used for direct lignocellulose treatment, which minimizes the problems encountered in fungal pretreatment. Due to its ability to operate in a short reaction time, it only requires a few hours, and it requires fewer nutrients for enzymatic reactions; this method is well-suited for further exploration (Suryadi et al., 2022).
MnP is crucial in fruit juice clarification to decompose phenolic compounds. The use of MnP in the clarification of fruit juices showed its future utility in food and beverage processing, Manavalan et al. (2015) reported clarification of various fruit juices using a solvent-tolerant MnP isolated from Ganoderma lucidum. They reported that pomegranate, lemon, and apple juices pretreated with MnP exhibited significant reductions in phenolic compounds up to 89, 85, and 78%, respectively. Immmobilization of MnP may increase its stability and effectiveness in clarifying juices. For example, Bilal et al. (2016) used gelatin-immobilized MnP to de-bitter apple juice by reducing the color by 42.7%, as well as reducing turbidity by 36.3%. Moreover, Bilal et al. (2017a) found that treating apple, grape, orange, and pomegranate juices with an immobilized ligninolytic enzyme reduced turbidity by 84.02%, 57.84%, and 82.13%, respectively.
Agricultural wastes (e.g., wood, straw, alfalfa, switch grass, and sugarcane bagasse) are raw materials for production of bioethanol due to presence of carbohydrates. The carbohydrates must be converted into simple sugars (glucose) before they can be fermented into ethanol. Since lignin and cellulose are present in these agri-wastes, pretreatment with lignolytic enzymes such as laccase and MnPs is a pre-requisite for ethanol production. Asgher et al. (2014) pre-treated biomass of wheat straw with ligninolytic enzymes including MnPs, laccase, and lignin peroxidase and produced 22.6 g/L of ethanol through sequential saccharification and fermentation (SSF) using Saccharomyces cerevisiae. Bilal et al. (2017b) observed that S. cerevisiae was able to synthesize bioethanol from de-lignified newspaper waste using ligninolytic enzymes from Ganoderma lucidum IBL-05 followed by saccharification using cellulose extract from Trichoderma harzianum. Gaur et al. (2018) produced bioethanol from wheat and rice bran biomass using the ligninolytic enzymes of Klebsiella pneumoniae strains.
MnPs may degrade phenolic and non-phenolic compounds in wastewater. Kong et al. (2016) isolated MnP from Echinodontium taxodii and tested its ability to oxidize three phenols and three non-phenols present in lignin. It was observed that MnP degrades three lignin phenolic compounds more effectively than non-phenolic compounds. MnP degraded phenolic compounds 4-hydroxy-3-methoxy cinnamic acid, 4-hydroxy cinnamic acid, and 4-hydroxy-3,5-dimethoxy cinnamic acid by 97.79, 28.35, and 99.92%, respectively. The degradative rate of n-cinnamic acid, 3-methoxy cinnamic acid, and 3,5-dimethoxycinnamic acid were 26.19, 30.03, and 44.84% for MnP, respectively.
MnP catalyzed the degradation of mycotoxins in foods. Wang et al. (2019) studied degradation of four major mycotoxins (aflatoxin B1, zearalenone, deoxynivalenol, and fumonisin B1) by the eight MnPs, which belong to four lignocellulose-degrading fungi (five from Irpex lacteus, one from Phanerochaete chrysosporium, one from Ceriporiopsis subvermispora, and another from Nematoloma frowardii). MnP-mediated mycotoxin degradation is only possible in the presence of a dicarboxylic acid malonate, where free radicals play a major role. MnPs from C. subvermispora and I. lacteus transformed mycotoxins similarly, outperforming those from P. chrysosporium and N. frowardii. The ability to degrade multiple mycotoxins is not exclusive to any one MnP but is shared by many MnPs with different sequences.
Recombinant MnPs are constructed to improve their stability, efficiency, and yields using both homologous and heterologous expression systems. The MnP-coding genes of various white-rot fungi were isolated, and then, they were cloned into the appropriate vectors and expressed in the appropriate host strains. An example of this is the MnP gene cloned from Ganoderma lucidum 00679 (GluMnP1), which was expressed in Pichia pastoris SMD1168H. This recombinant MnP (r-GluMnP1) was capable of degrading four dyes and phenol (Xu et al., 2017). Similarly, Zhang et al. (2020) expressed the MnP gene from Cerrena unicolor BBP6 in Pichia pastoris X33 to produce recombinant MnP (rMnP3-BBP6) that plays a significant role in denim bleaching and dye decolorization. Cortés-Espinosa et al. (2011) expressed the MnP gene (mnp1) from Phanerochaete chrysosporium in Aspergillus niger SBC2-T3. This recombinant fungus degraded phenanthrene. Lee et al. (2016) expressed MnP genes from Peniophora incarnata KUC8836 in Saccharomyces cerevisiae and confirmed that the recombinant yeast had improved anthracene degradation. Liu et al. (2020) cloned a MnP gene from Aspergillus sp. TS-A and expressed it in Pichia pastoris GS115. Recombinant MnP (GS115-MnP) was more efficient at degrading dyes. Chen et al. (2015) expressed the MnP gene (imnp) from Irpex lacteus F17 in Escherichia coli Rosetta (DE3). Recombinant MnP exhibited improved pH stability and dye degradation efficiency.
Pech-Canul et al. (2020) used a new approach to obtain a functional recombinant MnP1 (rMnP1) from P. chrysosporium (Chang et al., 2021). A chemically synthesized mnp1 gene was cloned into a pTXB1 vector using NdeI/SapI restriction sites and then transferred into Escherichia coli DH5α cells. The rMnP1 was expressed as a fusion protein, which was recovered from inclusion bodies and was then purified by intein-based protein purification techniques followed by affinity chromatography.
MnPs were isolated and characterized from a variety of white-rot fungi and bacteria and play a significant role in decolorizing dyes, bio-pulping, pulp biobleaching, removing mycotoxins, making ethanol, de-lignifying lignin, and clarifying fruit juices. It was possible to produce recombinant MnPs in large quantities with improved stability, production, and degradation efficiency. The immobilization of MnPs also improved their stability and capability.
Delignification of various lignocellulose materials using purified MnPs should be further investigated. The application of MnPs in dye decolorization and wastewater treatment should be carried out on a large scale. Genetic engineering should be used to develop more efficient MnPs in order to increase biodegradability. The CRISPR/Cas9-mediated genome editing in microorganisms may be used in order to improve industrial applications of MnPs.
AK wrote the manuscript, and PKA checked and edited it.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
PKA acknowledges the Department of Biotechnology, India, for awarding him DBT-Ramalingaswami Re-entry Fellowship.
Acevedo, F., Pizzul, L., Castillo, M. d., González, M. E., Cea, M., Gianfreda, L., et al. (2010). Degradation of Polycyclic Aromatic Hydrocarbons by Free and Nanoclay-Immobilized Manganese Peroxidase from Anthracophyllum Discolor. Chemosphere 80, 271–278. doi:10.1016/j.chemosphere.2010.04.022
Al-Dhabi, N. A., Esmail, G. A., and Valan Arasu, M. (2021). Effective Degradation of Tetracycline by Manganese Peroxidase Producing Bacillus Velezensis Strain Al-Dhabi 140 from Saudi Arabia Using Fibrous-Bed Reactor. Chemosphere 268, 128726. doi:10.1016/j.chemosphere.2020.128726
Asgher, M., Bashir, F., and Iqbal, H. M. N. (2014). A Comprehensive Ligninolytic Pre-treatment Approach from Lignocellulose green Biotechnology to Produce Bio-Ethanol. Chem. Eng. Res. Des. 92, 1571–1578. doi:10.1016/j.cherd.2013.09.003
Asgher, M., and Iqbal, H. M. N. (2013). Enhanced Catalytic Features of Sol-Gel Immobilized MnP Isolated from Solid State Culture of Pleurotus Ostreatus IBL-02. Chin. Chem. Lett. 24, 344–346. doi:10.1016/j.cclet.2013.02.019
Ayeronfe, F., Kassim, A., Hung, P., Ishak, N., Syarifah, S., and Aripin, A. (2019). Production of Ligninolytic Enzymes by Coptotermes Curvignathus Gut Bacteria. Rīgas Teh. Univ. Zināt. Raksti 23, 111–121. doi:10.2478/rtuect-2019-0008
Ayuso-Fernández, I., Ruiz-Dueñas, F. J., and Martínez, A. T. (2018). Evolutionary Convergence in Lignin-Degrading Enzymes. Proc. Natl. Acad. Sci. 115, 6428–6433. doi:10.1073/pnas.1802555115
Bermek, H., Li, K., and Eriksson, K.-E. L. (2002). Studies on Mediators of Manganese Peroxidase for Bleaching of wood Pulps. Bioresour. Tech. 85, 249–252. doi:10.1016/s0960-8524(02)00132-3
Bermek, H., Yazıcı, H., Öztürk, H., Tamerler, C., Jung, H., Li, K., et al. (2004). Purification and Characterization of Manganese Peroxidase from wood-degrading Fungus Trichophyton Rubrum LSK-27. Enzyme Microb. Tech. 35, 87–92. doi:10.1016/j.enzmictec.2004.04.004
Bilal, M., and Asgher, M. (2015). Dye Decolorization and Detoxification Potential of Ca-Alginate Beads Immobilized Manganese Peroxidase. BMC Biotechnol. 15, 111–114. doi:10.1186/s12896-015-0227-8
Bilal, M., Asgher, M., and Ramzan, M. (2015). Purification and Biochemical Characterization of Extracellular Manganese Peroxidase from Ganoderma Lucidum IBL-05 and its Application. Sci. Res. Essays 10, 456–464. doi:10.5897/sre2015.6268
Bilal, M., Asgher, M., Iqbal, H. M. N., Hu, H., and Zhang, X. (2016). Gelatin-immobilized Manganese Peroxidase with Novel Catalytic Characteristics and its Industrial Exploitation for Fruit Juice Clarification Purposes. Catal. Lett. 146, 2221–2228. doi:10.1007/s10562-016-1848-9
Bilal, M., Asgher, M., Iqbal, H. M. N., Hu, H., and Zhang, X. (2017a). Delignification and Fruit Juice Clarification Properties of Alginate-Chitosan-Immobilized Ligninolytic Cocktail. LWT 80, 348–354. doi:10.1016/j.lwt.2017.02.040
Bilal, M., Asgher, M., Iqbal, H. M. N., and Ramzan, M. (2017b). Enhanced Bio-Ethanol Production from Old Newspapers Waste through Alkali and Enzymatic Delignification. Waste Biomass Valor. 8, 2271–2281. doi:10.1007/s12649-017-9871-7
Bouacem, K., Rekik, H., Jaouadi, N. Z., Zenati, B., Kourdali, S., El Hattab, M., et al. (2018). Purification and Characterization of Two Novel Peroxidases from the Dye-Decolorizing Fungus Bjerkandera Adusta Strain CX-9. Int. J. Biol. Macromolecules 106, 636–646. doi:10.1016/j.ijbiomac.2017.08.061
Chaijak, P., Lertworapreecha, M., and Sukkasem, C. (2018). Phenol Removal from palm Oil Mill Effluent Using Galactomyces Reessii Termite-Associated Yeast. Pol. J. Environ. Stud. 27, 39–44. doi:10.15244/pjoes/75205
Champagne, P.-P., and Ramsay, J. A. (2005). Contribution of Manganese Peroxidase and Laccase to Dye Decoloration by Trametes versicolor. Appl. Microbiol. Biotechnol. 69, 276–285. doi:10.1007/s00253-005-1964-8
Chang, Y., Yang, D., Li, R., Wang, T., and Zhu, Y. (2021). Textile Dye Biodecolorization by Manganese Peroxidase: A Review. Molecules 26, 4403. doi:10.3390/molecules26154403
Chauhan, P. S., Goradia, B., and Saxena, A. (2017). Bacterial Laccase: Recent Update on Production, Properties and Industrial Applications. 3 Biotech. 7, 323. doi:10.1007/s13205-017-0955-7
Chauhan, P. S. (2020). Role of Various Bacterial Enzymes in Complete Depolymerization of Lignin: A Review. Biocatal. Agric. Biotechnol. 23, 101498. doi:10.1016/j.bcab.2020.101498
Chen, W., Zheng, L., Jia, R., and Wang, N. (2015). Cloning and Expression of a New Manganese Peroxidase from Irpex Lacteus F17 and its Application in Decolorization of Reactive Black 5. Process Biochem. 50, 1748–1759. doi:10.1016/j.procbio.2015.07.009
Chowdhary, P., Shukla, G., Raj, G., Ferreira, L. F. R., and Bharagava, R. N. (2019). Microbial Manganese Peroxidase: a Ligninolytic Enzyme and its Ample Opportunities in Research. SN Appl. Sci. 1, 1–12. doi:10.1007/s42452-018-0046-3
Cortés-Espinosa, D. V., Absalón, Á. E., Sánchez, N., Loera, O., Rodríguez-Vázquez, R., and Fernández, F. J. (2011). Heterologous Expression of Manganese Peroxidase in Aspergillus niger and its Effect on Phenanthrene Removal from Soil. J. Mol. Microbiol. Biotechnol. 21, 120–129. doi:10.1159/000331563
Dai, J., Patti, A. F., and Saito, K. (2016). Recent Developments in Chemical Degradation of Lignin: Catalytic Oxidation and Ionic Liquids. Tetrahedron Lett. 57, 4945–4951. doi:10.1016/j.tetlet.2016.09.084
De los Santos Ramos, W., Poznyak, T., Chairez, I., and Córdova R., I. (2009). Remediation of Lignin and its Derivatives from Pulp and Paper Industry Wastewater by the Combination of Chemical Precipitation and Ozonation. J. Hazard. Mater. 169, 428–434. doi:10.1016/j.jhazmat.2009.03.152
Dong, Y.-C., Dai, Y.-N., Xu, T.-Y., Cai, J., and Chen, Q.-H. (2014). Biodegradation of Chestnut Shell and Lignin-Modifying Enzymes Production by the white-rot Fungi Dichomitus Squalens, Phlebia Radiata. Bioproc. Biosyst. Eng. 37, 755–764. doi:10.1007/s00449-013-1045-9
El-Shora, H., Ibrahim, M., and Elmekabaty, M. (2017). Immobilization and Thermostability of Manganese Peroxidase from Trichoderma harzianum. Microbiol. Res. J. Int. 21, 1–11. doi:10.9734/mrji/2017/35754
Elisashvili, V., Parlar, H., Kachlishvili, E., Chichua, D., and Kvesitadze, G. (2001). Ligninolytic Activity of Basidiomycetes Grown under Submerged and Solid-State Fermentation on Plant Plant Raw Material (Sawdust of grapevine Cuttings). Adv. Food Sci. 23, 117–123.
Emami, E., Zolfaghari, P., Golizadeh, M., Karimi, A., Lau, A., Ghiasi, B., et al. (2020). Effects of Stabilizers on Sustainability, Activity and Decolorization Performance of Manganese Peroxidase Enzyme Produced by Phanerochaete Chrysosporium. J. Environ. Chem. Eng. 8, 104459. doi:10.1016/j.jece.2020.104459
E. Torres, and M. Ayala (Editors) (2010). Biocatalysis Based on Heme Peroxidases: Peroxidases as Potential Industrial Biocatalysts (Springer Science and Business Media).
Fernández-Fueyo, E., Acebes, S., Ruiz-Duenas, F. J., Martínez, M. J., Romero, A., Medrano, F. J., et al. (2014). Structural Implications of the C-Terminal Tail in the Catalytic and Stability Properties of Manganese Peroxidases from Ligninolytic Fungi. Acta Crystallogr. D Biol. Crystallogr. 70, 3253–3265. doi:10.1107/s1399004714022755
Gaur, N., Narasimhulu, K., and Y, P. (2018). Biochemical and Kinetic Characterization of Laccase and Manganese Peroxidase from novelKlebsiella Pneumoniaestrains and Their Application in Bioethanol Production. RSC Adv. 8, 15044–15055. doi:10.1039/c8ra01204k
Gomathy, S., Sridharan, R., Kumar, P. S., Gayathri, K. V., Al Farraj, D. A., and Chen, T.-W. (2021). Application of Alkaline MnP Immobilized Luffa Fibers in Mixed Azo Dyes Degradation. Environ. Tech. Innovation 24, 101964. doi:10.1016/j.eti.2021.101964
Govarthanan, M., Fuzisawa, S., Hosogai, T., and Chang, Y.-C. (2017). Biodegradation of Aliphatic and Aromatic Hydrocarbons Using the Filamentous Fungus Penicillium Sp. CHY-2 and Characterization of its Manganese Peroxidase Activity. RSC Adv. 7, 20716–20723. doi:10.1039/c6ra28687a
Hakala, T. K., Hildén, K., Maijala, P., Olsson, C., and Hatakka, A. (2006). Differential Regulation of Manganese Peroxidases and Characterization of Two Variable MnP Encoding Genes in the white-rot Fungus Physisporinus Rivulosus. Appl. Microbiol. Biotechnol. 73, 839–849. doi:10.1007/s00253-006-0541-0
Harazono, K., Kondo, R., and Sakai, K. (1996). Bleaching of Hardwood Kraft Pulp with Manganese Peroxidase from Phanerochaete Sordida YK-624 without Addition of MnSO(inf4). Appl. Environ. Microbiol. 62, 913–917. doi:10.1128/aem.62.3.913-917.1996
Heinfling, A., Martínez, M. J., Martínez, A. T., Bergbauer, M., and Szewzyk, U. (1998). Transformation of Industrial Dyes by Manganese Peroxidases from Bjerkandera Adusta and Pleurotus Eryngii in a Manganese-independent Reaction. Appl. Environ. Microbiol. 64, 2788–2793. doi:10.1128/aem.64.8.2788-2793.1998
Heinzkill, M., Bech, L., Halkier, T., Schneider, P., and Anke, T. (1998). Characterization of Laccases and Peroxidases from Wood-Rotting Fungi (Family Coprinaceae ). Appl. Environ. Microbiol. 64, 1601–1606. doi:10.1128/aem.64.5.1601-1606.1998
Hildén, K., Mäkelä, M. R., Lankinen, P., and Lundell, T. (2013). Agaricus Bisporus and Related Agaricus Species on Lignocellulose: Production of Manganese Peroxidase and Multicopper Oxidases. Fungal Genet. Biol. 55, 32–41. doi:10.1016/j.fgb.2013.02.002
Hildén, K., Mäkelä, M. R., Steffen, K. T., Hofrichter, M., Hatakka, A., Archer, D. B., et al. (2014). Biochemical and Molecular Characterization of an Atypical Manganese Peroxidase of the Litter-Decomposing Fungus Agrocybe Praecox. Fungal Genet. Biol. 72, 131–136. doi:10.1016/j.fgb.2014.03.002
Hofrichter, M. (2002). Review: Lignin Conversion by Manganese Peroxidase (MnP). Enzyme Microb. Tech. 30, 454–466. doi:10.1016/s0141-0229(01)00528-2
Iqbal, M., and Bhatti, I. A. (2014). Re-utilization Option of Industrial Wastewater Treated by Advanced Oxidation Process. Pak. J. Agric. Sci. 51, 1141–1147.
Irshad, M., and Asgher, M. (2011). Production and Optimization of Ligninolytic Enzymes by white Rot Fungus Schizophyllum Commune IBL-06 in Solid State Medium Banana Stalks. Afr. J. Biotechnol. 10, 18234–18242. doi:10.5897/ajb11.2242
Janusz, G., Kucharzyk, K. H., Pawlik, A., Staszczak, M., and Paszczynski, A. J. (2013). Fungal Laccase, Manganese Peroxidase and Lignin Peroxidase: Gene Expression and Regulation. Enzyme Microb. Tech. 52, 1–12. doi:10.1016/j.enzmictec.2012.10.003
Janusz, G., Pawlik, A., Sulej, J., Świderska-Burek, U., Jarosz-Wilkołazka, A., and Paszczyński, A. (2017). Lignin Degradation: Microorganisms, Enzymes Involved, Genomes Analysis and Evolution. FEMS Microbiol. Rev. 41, 941–962. doi:10.1093/femsre/fux049
Jha, H., and Patil, M. (2011). Biopulping of Sugarcane Bagasse Using Manganese Peroxidase from Penicillium oxalicum Isolate-1. Rom. Biotechnol. Lett. 16, 6809–6819.
Kaal, E. E. J., Field, J. A., and Joyce, T. W. (1995). Increasing Ligninolytic Enzyme Activities in Several white-rot Basidiomycetes by Nitrogen-Sufficient media. Bioresour. Tech. 53, 133–139. doi:10.1016/0960-8524(95)00066-n
Kamitsuji, H., Honda, Y., Watanabe, T., and Kuwahara, M. (2004). Production and Induction of Manganese Peroxidase Isozymes in a white-rot Fungus Pleurotus Ostreatus. Appl. Microbiol. Biotechnol. 65, 287–294. doi:10.1007/s00253-003-1543-9
Kaneko, R., Iimori, T., Miyawaki, S., Machida, M., and Murakami, K. (1995). Biobleaching with Manganese Peroxidase Purified from the Pulp Bleaching Fungus SKB-1152. Biosci. Biotechnol. Biochem. 59, 1584–1585. doi:10.1271/bbb.59.1584
Khelil, O., Choubane, S., and Cheba, B. A. (2015). Co-production of Cellulases and Manganese Peroxidases by Bacillus Sp. R2 and Bacillus Cereus 11778 on Waste Newspaper: Application in Dyes Decolourization. Proced. Tech. 19, 980–987. doi:10.1016/j.protcy.2015.02.140
Kondo, R., Harazono, K., and Sakai, K. (1994). Bleaching of Hardwood Kraft Pulp with Manganese Peroxidase Secreted from Phanerochaete Sordida YK-624. Appl. Environ. Microbiol. 60, 4359–4363. doi:10.1128/aem.60.12.4359-4363.1994
Kong, W., Chen, H., Lyu, S., Ma, F., Yu, H., and Zhang, X. (2016). Characterization of a Novel Manganese Peroxidase from white-rot Fungus Echinodontium Taxodii 2538, and its Use for the Degradation of Lignin-Related Compounds. Process Biochem. 51, 1776–1783. doi:10.1016/j.procbio.2016.01.007
Kumar, A. (2021). Biobleaching: an Eco-Friendly Approach to Reduce Chemical Consumption and Pollutants Generation. Phys. Sci. Rev. 6, 20190044. doi:10.1515/psr-2019-0044
Kumar, V., and Chandra, R. (2018). Characterisation of Manganese Peroxidase and Laccase Producing Bacteria Capable for Degradation of Sucrose Glutamic Acid-Maillard Reaction Products at Different Nutritional and Environmental Conditions. World J. Microbiol. Biotechnol. 34, 32–18. doi:10.1007/s11274-018-2416-9
Leatham, G. F., and Kent Kirk, T. (1983). Regulation of Ligninolytic Activity by Nutrient Nitrogen in white-rot Basidiomycetes. FEMS Microbiol. Lett. 16, 65–67. doi:10.1111/j.1574-6968.1983.tb00260.x
Lee, A. H., Kang, C.-M., Lee, Y. M., Lee, H., Yun, C.-W., Kim, G.-H., et al. (2016). Heterologous Expression of a New Manganese-dependent Peroxidase Gene from Peniophora Incarnata KUC8836 and its Ability to Remove Anthracene in Saccharomyces cerevisiae. J. Biosci. Bioeng. 122, 716–721. doi:10.1016/j.jbiosc.2016.06.006
Levin, L., Forchiassin, F., and Ramos, A. M. (2002). Copper Induction of Lignin-Modifying Enzymes in the white-rot Fungus Trametes Trogii. Mycologia 94, 377–383. doi:10.2307/3761771
Levin, L., Melignani, E., and Ramos, A. M. (2010). Effect of Nitrogen Sources and Vitamins on Ligninolytic Enzyme Production by Some white-rot Fungi. Dye Decolorization by Selected Culture Filtrates. Bioresour. Tech. 101, 4554–4563. doi:10.1016/j.biortech.2010.01.102
Liu, S., Xu, X., Kang, Y., Xiao, Y., and Liu, H. (2020). Degradation and Detoxification of Azo Dyes with Recombinant Ligninolytic Enzymes from Aspergillus Sp. With Secretory Overexpression in Pichia pastoris. R. Soc. Open Sci. 7, 200688. doi:10.1098/rsos.200688
Lueangjaroenkit, P., Teerapatsakul, C., Sakka, K., Sakka, M., Kimura, T., Kunitake, E., et al. (2019). Two Manganese Peroxidases and a Laccase ofTrametes polyzonaKU-Rnw027 with Novel Properties for Dye and Pharmaceutical Product Degradation in Redox Mediator-free System. Mycobiology 47, 217–229. doi:10.1080/12298093.2019.1589900
Ma, R., Xu, Y., and Zhang, X. (2015). Catalytic Oxidation of Biorefinery Lignin to Value-Added Chemicals to Support Sustainable Biofuel Production. ChemSusChem 8, 24–51. doi:10.1002/cssc.201402503
Manavalan, T., Manavalan, V., Thangavelu, K. P., Kutzner, A., and Heese, K. (2015). Characterization of a Solvent‐Tolerant Manganese Peroxidase (MnP) from Ganoderma lucidum and Its Application in Fruit Juice Clarification. J. Food Biochem. 39, 754–764.
Martínez, Á. T., Speranza, M., Ruiz-Dueñas, F. J., Ferreira, P., Camarero, S., Guillén, F., et al. (2005). Biodegradation of Lignocellulosics: Microbial, Chemical, and Enzymatic Aspects of the Fungal Attack of Lignin. Int. Microbiol. 8, 195–204.
Masarin, F., Norambuena, M., Ramires, H. O., Demuner, B. J., Pavan, P. C., and Ferraz, A. (2016). Manganese Peroxidase and Biomimetic Systems Applied Toin Vitrolignin Degradation inEucalyptus Grandismilled wood and Kraft Pulps. J. Chem. Technol. Biotechnol. 91, 1422–1430. doi:10.1002/jctb.4739
Mei, J., Shen, X., Gang, L., Xu, H., Wu, F., and Sheng, L. (2020). A Novel Lignin Degradation Bacteria-Bacillus Amyloliquefaciens SL-7 Used to Degrade Straw Lignin Efficiently. Bioresour. Tech. 310, 123445. doi:10.1016/j.biortech.2020.123445
Mester, T., and Field, J. A. (1997). Optimization of Manganese Peroxidase Production by the white Rot Fungus Bjerkandera Sp. Strain BOS55. FEMS Microbiol. Lett. 155, 161–168. doi:10.1111/j.1574-6968.1997.tb13873.x
Mohammad, I. N., Ongkudon, C. M., and Misson, M. (2020). Physicochemical Properties and Lignin Degradation of Thermal-Pretreated Oil Palm Empty Fruit Bunch. Energies 13, 5966. doi:10.3390/en13225966
Muhammad, A., Nadeem, A., and Iqbal, H. M. N. (2011). Hyperproductivity of Extracellular Enzymes from Indigenous white Rot Fungi (P. Chrysosporium IBL-03) by Utilizing Agro-Wastes. Bio-Resources 6, 4454–4467.
Nayanashree, G., and Thippeswamy, B. (2015a). Natural Rubber Degradation by Laccase and Manganese Peroxidase Enzymes of Penicillium chrysogenum Penicillium chrysogenum. Int. J. Environ. Sci. Technol. 12, 2665–2672. doi:10.1007/s13762-014-0636-6
Nayanashree, G., and Thippeswamy, B. (2015b). Biodegradation of Natural Rubber by Laccase and Manganese Peroxidase Enzyme of Bacillus Subtilis. Environ. Process. 2, 761–772. doi:10.1007/s40710-015-0118-y
Nousiainen, P., Kontro, J., Manner, H., Hatakka, A., and Sipilä, J. (2014). Phenolic Mediators Enhance the Manganese Peroxidase Catalyzed Oxidation of Recalcitrant Lignin Model Compounds and Synthetic Lignin. Fungal Genet. Biol. 72, 137–149. doi:10.1016/j.fgb.2014.07.008
Oliveira, P. L. D., Duarte, M. C. T., Ponezi, A. N., and Durrant, L. R. (2009). Purification and Partial Characterization of Manganese Peroxidase from Bacillus Pumilus and Paenibacillus Sp. Braz. J. Microbiol. 40, 818–826. doi:10.1590/s1517-83822009000400012
Omar, A. A., Mahgoub, S., Salama, A., Likotrafiti, E., Rhoades, J., Christakis, C., et al. (2021). Evaluation of Lactobacillus Kefiri and Manganese Peroxidase‐producing Bacteria for Decolorization of Melanoidins and Reduction of Chemical Oxygen Demand. Water Environ. J. 35, 704–714. doi:10.1111/wej.12663
Paice, M. G., Reid, I. D., Bourbonnais, R., Archibald, F. S., and Jurasek, L. (1993). Manganese Peroxidase, Produced by Trametes versicolor during Pulp Bleaching, Demethylates and Delignifies Kraft Pulp. Appl. Environ. Microbiol. 59, 260–265. doi:10.1128/aem.59.1.260-265.1993
Paszczynski, A., Huynh, V. B., and Crawford, R. (1985). Enzymatic Activities of an Extracellular, Manganese-Dependent Peroxidase from Phanerochaete chrysosporium. FEMS Microbiol. Lett. 29, 37–41.
Pech-Canul, A. D. L. C., Carrillo-Campos, J., Ballinas-Casarrubias, M. D. L., Solis-Oviedo, R. L., Hernández-Rascón, S. K., Hernández-Ochoa, L. R., et al. (2020). Functional Expression and One-step Protein Purification of Manganese Peroxidase 1 (rMnP1) from Phanerochaete Chrysosporium Using the E. Coli-Expression System. Int. J. Mol. Sci. 21, 416. doi:10.3390/ijms21020416
Périé, F. H., and Gold, M. H. (1991). Manganese Regulation of Manganese Peroxidase Expression and Lignin Degradation by the white Rot Fungus Dichomitus Squalens. Appl. Environ. Microbiol. 57, 2240–2245. doi:10.1128/aem.57.8.2240-2245.1991
Qin, X., Zhang, J., Zhang, X., and Yang, Y. (2014). Induction, Purification and Characterization of a Novel Manganese Peroxidase from Irpex Lacteus CD2 and its Application in the Decolorization of Different Types of Dye. PLoS One 9, e113282. doi:10.1371/journal.pone.0113282
Qin, X., Sun, X., Huang, H., Bai, Y., Wang, Y., Luo, H., et al. (2017). Oxidation of a Non-phenolic Lignin Model Compound by Two Irpex Lacteus Manganese Peroxidases: Evidence for Implication of Carboxylate and Radicals. Biotechnol. Biofuels 10, 103–113. doi:10.1186/s13068-017-0787-z
Ragauskas, A. J., Williams, C. K., Davison, B. H., Britovsek, G., Cairney, J., Eckert, C. A., et al. (2006). The Path Forward for Biofuels and Biomaterials. Science 311, 484–489. doi:10.1126/science.1114736
Rekik, H., Zaraî Jaouadi, N., Bouacem, K., Zenati, B., Kourdali, S., Badis, A., et al. (2019). Physical and Enzymatic Properties of a New Manganese Peroxidase from the white-rot Fungus Trametes Pubescens Strain I8 for Lignin Biodegradation and Textile-Dyes Biodecolorization. Int. J. Biol. Macromolecules 125, 514–525. doi:10.1016/j.ijbiomac.2018.12.053
Ruiz-Dueñas, F. J., Barrasa, J. M., Sánchez-García, M., Camarero, S., Miyauchi, S., Serrano, A., et al. (2021). Genomic Analysis Enlightens Agaricales Lifestyle Evolution and Increasing Peroxidase Diversity. Mol. Biol. Evol. 38, 1428–1446. doi:10.1093/molbev/msaa301
Sasaki, T., Kajino, T., Li, B., Sugiyama, H., and Takahashi, H. (2001). New Pulp Biobleaching System Involving Manganese Peroxidase Immobilized in a Silica Support with Controlled Pore Sizes. Appl. Environ. Microbiol. 67, 2208–2212. doi:10.1128/aem.67.5.2208-2212.2001
Shi, K., Liu, Y., Chen, P., and Li, Y. (2021). Contribution of Lignin Peroxidase, Manganese Peroxidase, and Laccase in lignite Degradation by Mixed white-rot Fungi. Waste Biomass Valor. 12, 3753–3763. doi:10.1007/s12649-020-01275-z
Singh, A. K., Bilal, M., Iqbal, H. M. N., Meyer, A. S., and Raj, A. (2021). Bioremediation of Lignin Derivatives and Phenolics in Wastewater with Lignin Modifying Enzymes: Status, Opportunities and Challenges. Sci. Total Environ. 777, 145988. doi:10.1016/j.scitotenv.2021.145988
Stajić, M., Persky, L., Friesem, D., Hadar, Y., Wasser, S. P., Nevo, E., et al. (2006). Effect of Different Carbon and Nitrogen Sources on Laccase and Peroxidases Production by Selected Pleurotus Species. Enzyme Microb. Technol. 38, 65–73. doi:10.1016/j.enzmictec.2005.03.026
Sundaramoorthy, M., Gold, M. H., and Poulos, T. L. (2010). Ultrahigh (0.93Å) Resolution Structure of Manganese Peroxidase from Phanerochaete Chrysosporium: Implications for the Catalytic Mechanism. J. Inorg. Biochem. 104, 683–690. doi:10.1016/j.jinorgbio.2010.02.011
Suryadi, H., Judono, J. J., Putri, M. R., Eclessia, A. D., Ulhaq, J. M., Agustina, D. N., et al. (2022). Biodelignification of Lignocellulose Using Ligninolytic Enzymes from white-rot Fungi. Heliyon 8, e08865. doi:10.1016/j.heliyon.2022.e08865
Tanaka, H., Koike, K., Itakura, S., and Enoki, A. (2009). Degradation of wood and Enzyme Production by Ceriporiopsis Subvermispora. Enzyme Microb. Tech. 45, 384–390. doi:10.1016/j.enzmictec.2009.06.003
Tello, M., Corsini, G., Larrondo, L. F., Salas, L., Lobos, S., and Vicuña, R. (2000). Characterization of Three New Manganese Peroxidase Genes from the Ligninolytic Basidiomycete Ceriporiopsis Subvermispora. Biochim. Biophys. Acta (Bba) - Gene Struct. Expr. 1490, 137–144. doi:10.1016/s0167-4781(99)00227-4
Thamvithayakorn, P., Phosri, C., Pisutpaisal, N., Krajangsang, S., Whalley, A. J. S., and Suwannasai, N. (2019). Utilization of Oil palm Decanter Cake for Valuable Laccase and Manganese Peroxidase Enzyme Production from a Novel white-rot Fungus, Pseudolagarobasidium Sp. PP17-33. 3 Biotech. 9, 417. doi:10.1007/s13205-019-1945-8
Urek, R. O., and Pazarlioglu, N. K. (2007). Enhanced Production of Manganese Peroxidase by Phanerochaete Chrysosporium. Braz. Arch. Biol. Technol. 50, 913–920. doi:10.1590/s1516-89132007000700001
Viikari, L., Suurnäkki, A., Grönqvist, S., Raaska, L., and Ragauskas, A. (2009). “Forest Products: Biotechnology in Pulp and Paper Processing,” in Encyclopedia of Microbiology (Academic Press), 80–94. doi:10.1016/b978-012373944-5.00123-1
Vrsanska, M., Buresova, A., Damborsky, P., and Adam, V. (2015). Influence of Different Inducers on Ligninolytic Enzyme Activities. J. Metallomics Nanotechnol. 3, 64–70.
Wang, W., Yan, L., Cui, Z., Gao, Y., Wang, Y., and Jing, R. (2011). Characterization of a Microbial Consortium Capable of Degrading Lignocellulose. Bioresour. Tech. 102, 9321–9324. doi:10.1016/j.biortech.2011.07.065
Wang, X., Qin, X., Hao, Z., Luo, H., Yao, B., and Su, X. (2019). Degradation of Four Major Mycotoxins by Eight Manganese Peroxidases in Presence of a Dicarboxylic Acid. Toxins 11, 566. doi:10.3390/toxins11100566
Ward, O. P., and Singh, A. (2002). Bioethanol Technology: Developments and Perspectives. Adv. Appl. Microbiol. 51, 53–80. doi:10.1016/s0065-2164(02)51001-7
Xu, H., Guo, M. Y., Gao, Y. H., Bai, X. H., and Zhou, X. W. (2017). Expression and Characteristics of Manganese Peroxidase from Ganoderma Lucidum in Pichia pastoris and its Application in the Degradation of Four Dyes and Phenol. BMC Biotechnol. 17, 19–12. doi:10.1186/s12896-017-0338-5
Xu, R., Zhang, K., Liu, P., Han, H., Zhao, S., Kakade, A., et al. (2018). Lignin Depolymerization and Utilization by Bacteria. Bioresour. Tech. 269, 557–566. doi:10.1016/j.biortech.2018.08.118
Yang, X., Zheng, J., Lu, Y., and Jia, R. (2016). Degradation and Detoxification of the Triphenylmethane Dye Malachite green Catalyzed by Crude Manganese Peroxidase from Irpex Lacteus F17. Environ. Sci. Pollut. Res. 23, 9585–9597. doi:10.1007/s11356-016-6164-9
Zainith, S., Purchase, D., Saratale, G. D., Ferreira, L. F. R., Bilal, M., and Bharagava, R. N. (2019). Isolation and Characterization of Lignin-Degrading Bacterium Bacillus Aryabhattai from Pulp and Paper Mill Wastewater and Evaluation of its Lignin-Degrading Potential. 3 Biotech. 9 (3), 92–11. doi:10.1007/s13205-019-1631-x
Zhang, H., Zhang, S., He, F., Qin, X., Zhang, X., and Yang, Y. (2016). Characterization of a Manganese Peroxidase from white-rot Fungus Trametes sp.48424 with strong Ability of Degrading Different Types of Dyes and Polycyclic Aromatic Hydrocarbons. J. Hazard. Mater. 320, 265–277. doi:10.1016/j.jhazmat.2016.07.065
Zhang, H., Zhang, J., Zhang, X., and Geng, A. (2018). Purification and Characterization of a Novel Manganese Peroxidase from white-rot Fungus Cerrena Unicolor BBP6 and its Application in Dye Decolorization and Denim Bleaching. Process Biochem. 66, 222–229. doi:10.1016/j.procbio.2017.12.011
Zhang, H., Zhang, X., and Geng, A. (2020). Expression of a Novel Manganese Peroxidase from Cerrena Unicolor BBP6 in Pichia pastoris and its Application in Dye Decolorization and PAH Degradation. Biochem. Eng. J. 153, 107402. doi:10.1016/j.bej.2019.107402
Zhang, Y., Dong, Z., Luo, Y., Yang, E., Xu, H., Chagan, I., et al. (2021). The Manganese Peroxidase Gene Family of Trametes Trogii: Gene Identification and Expression Patterns Using Various Metal Ions under Different Culture Conditions. Microorganisms 9, 2595. doi:10.3390/microorganisms9122595
Keywords: bioethanol, textile dye, bioremediation, biopulping, biobleaching
Citation: Kumar A and Arora PK (2022) Biotechnological Applications of Manganese Peroxidases for Sustainable Management. Front. Environ. Sci. 10:875157. doi: 10.3389/fenvs.2022.875157
Received: 13 February 2022; Accepted: 21 March 2022;
Published: 26 April 2022.
Edited by:
Vineet Kumar, National Environmental Engineering Research Institute (CSIR), IndiaReviewed by:
Hanuman Singh Jatav, Sri Karan Narendra Agriculture University, IndiaCopyright © 2022 Kumar and Arora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pankaj Kumar Arora, YXJvcmE0ODRAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.