- 1Plant Genetic Engineering Laboratory, National Center for Biotechnology (NCB), Nur-Sultan, Kazakhstan
- 2L.N.Gumilyov Eurasian National University, Nur-Sultan, Kazakhstan
Soil contamination caused by industrial and agricultural activities is an environmental problem that poses a serious risk to human health and the ecosystem. Persistent organic pollutants (POPs) are organic chemicals that persist in the environment for long periods because of their high resistance to photolytic, chemical, and biological degradation. Besides POPs, high concentrations of non-essential heavy metals and metalloids, such as arsenic, cadmium, and lead, are increasingly becoming a problem worldwide. Remediation strategies for organic and inorganic pollutants in the environment have received global attention. For organic or inorganic contaminants, phytoremediation is the strategy of choice because of a green technology that uses plants and solar energy to clean hyper-accumulated toxic pollutants from the environment. Some plant species have a high capacity to grow and survive in elevated levels of contaminants. With a long cultivation history and adaptability to a wide range of territories, alfalfa has not only widely been used for animal feed and a medicinal herb but is also an ideal natural resource and model plant for remediation of contaminated soils, offering a variety of elite characteristics. This review provides, firstly, abundant genomic information on the genetic diversity and population structure of alfalfa. Secondly, we focused on the transgenic alfalfa plants for enhanced phytoremediation of POPs, such as atrazine, polychlorinated biphenyl (PCB), and trichloroethylene (TCE), as well as phytoremediation of petroleum and heavy metals. Thirdly, the future perspective of enhancement of phytoremediation efficiency was discussed in depth. This review is intended to provide a comprehensive overview of the phytoremediation capabilities of transgenic alfalfa plants, presenting fundamental information for future research studies for enhancing phytoremediation efficiency.
Introduction
Alfalfa and Core Collections Worldwide
Alfalfa (Medicago sativa L.) belongs to the Fabaceae family, Trifolieae tribe (Rhouma et al., 2017), and Medicago genus and includes several diploid alfalfa (2n = 2x = 16) with a small genome size of 550 Mbp and autotetraploid cultivated alfalfa (2n = 4x = 32) with a genome size of 800–1,000 Mbp (Blondon et al., 1994; Havananda et al., 2011). This plant grows and develops well at 22°C–30°C. Under sufficient humidity, it can also survive at 37°C–42°C. Moreover, it is characterized by high cold tolerance, ranging from −15°C to −30°C (Stepanova et al., 2016). Alfalfa has been suggested to have two feasible origins based on its abundant diversity. One is in the Asian Minor, including Northwestern Iran, highlands of Armenia, Georgia, and Eastern Turkey, and the other is in Central Asia, including Kazakhstan, Uzbekistan, and Afghanistan (Wang and Şakiroğlu, 2021). Unfortunately, it is difficult to determine its precise origins.
Nowadays, alfalfa has been grown in approximately 30 million hectares of hay, silage, and pasture areas globally (Hawkins and Yu 2018). More than 21 thousand Medicago sativa L. accessions are reported from 37 country collections worldwide, and the major germplasm banks of alfalfa accessions are in the United States (4,771 accessions), followed by Australia (3,843 accessions), the Russian Federation (3,647 accessions), the United Kingdom (1,085 accessions), and Italy (989 accessions) (reviewed in (Irish and Greene 2021). Detailed accession-associated passport information on 1966 accessions is given in the online platform of the Genesys database (https://www.genesys-pgr.org/) It is important to study the genotypic diversity of alfalfa due to the large ecological distribution and demographic expansion of the species and the abundant germplasm collection.
Genotypic Diversity of Alfalfa
Genetic Diversity and Population Structure of Alfalfa
Many studies over the last decade have investigated the genetic diversity of alfalfa assessed by genetic markers [reviewed in (Li and Brummer, 2012; Hawkins and Yu 2018)]. Here we concentrate on research from 2018 onwards. The genetic diversity and population structure of six alfalfa cultivars from the United States were evaluated by amplifying SSR markers with 312 discernible alleles from the whole genome (Yin et al., 2018). Twenty wild alfalfa germplasms from the Xinjiang province of China were analyzed for genetic diversity by six pairs of random amplified polymorphic DNA (RAPD) and six pairs of inter-simple sequence repeat (ISSR) markers (Ma et al., 2018). The genetic diversity among 242 worldwide alfalfa accessions was studied using 102 simple sequence repeat (SSR) primers; it was found to consist of 471 alleles distributed on eight chromosomes (He H et al., 2019). Seventy-seven alfalfa genotypes from Brazil were explored using 10 SSR markers with 54 alleles (dos Santos et al., 2020). Inter-retrotransposon-amplified polymorphism (IRAP) markers have been used to assess alfalfa’s genetic diversity (Badr et al., 2020). Twelve Iranian genotypes were studied based on seed storage proteins patterns (Kakaei and Ahmadian 2021), while the genetic diversity of 25 Russian alfalfa cultivars was examined by sequence-related amplified polymorphism (SRAP) markers (Shamustakimova et al., 2021). In 2020, the phylogenetic and evolutionary analyses of diploid alfalfa had been done by 793.2 Mb genome sequencing, comprising 47,202 annotated protein-coding genes (Li et al., 2020). Worldwide, 162 autotetraploid alfalfa germplasms comprising 3.92 Mb of contig length covered 50% of the genome, and 49,165 annotated genes (Shen et al., 2020) have undergone phylogenetic and evolutionary analyses. The comparative and evolutionary analyses in these two research studies revealed that Medicago sativa L. diverged from Medicago truncatula approximately 5.2–8 million years ago (Li et al., 2020; Shen et al., 2020). Altogether, 220 global alfalfa core germplasms from China, Turkey, other Asian regions, America, and a mixed group from Europe and America were sequenced and characterized for their population structure, kinship, and population differentiation. In addition to these analyses, 875,023 high-quality SNPs, 350 common candidate genetic regions, and 686 genes were identified (Chen et al., 2021). Based on the results of these studies, alfalfa has a very high genetic diversity arising from a long history of cultivation and domestication.
Alfalfa for Phytoremediation
With its long cultivation history and adaptability to a wide range of territories, alfalfa has been commonly used for animal feed (Biazzi et al., 2020). It has also been utilized as a medicinal herb since it is a good source of vitamins (A, C, E, and K), protein, and minerals such as calcium, potassium, phosphorus, and iron (for overview see https://www.webmd.com/vitamins/ai/ingredientmono-19/alfalfa). Apart from these uses, alfalfa is an ideal natural resource and model plant for remediation of contaminated soils, offering a variety of elite characteristics, including a highly productive biomass, drought tolerance, a fast-growing and prosperous root system, and availability in large amounts over several months of the year (Ren et al., 2018). Wild-type alfalfa has been used for phytoremediation of soils contaminated with heavy metals (Ju et al., 2020; He et al., 2021; Wang X et al., 2021) and organic pollutants (Chekol and Vough 2001).
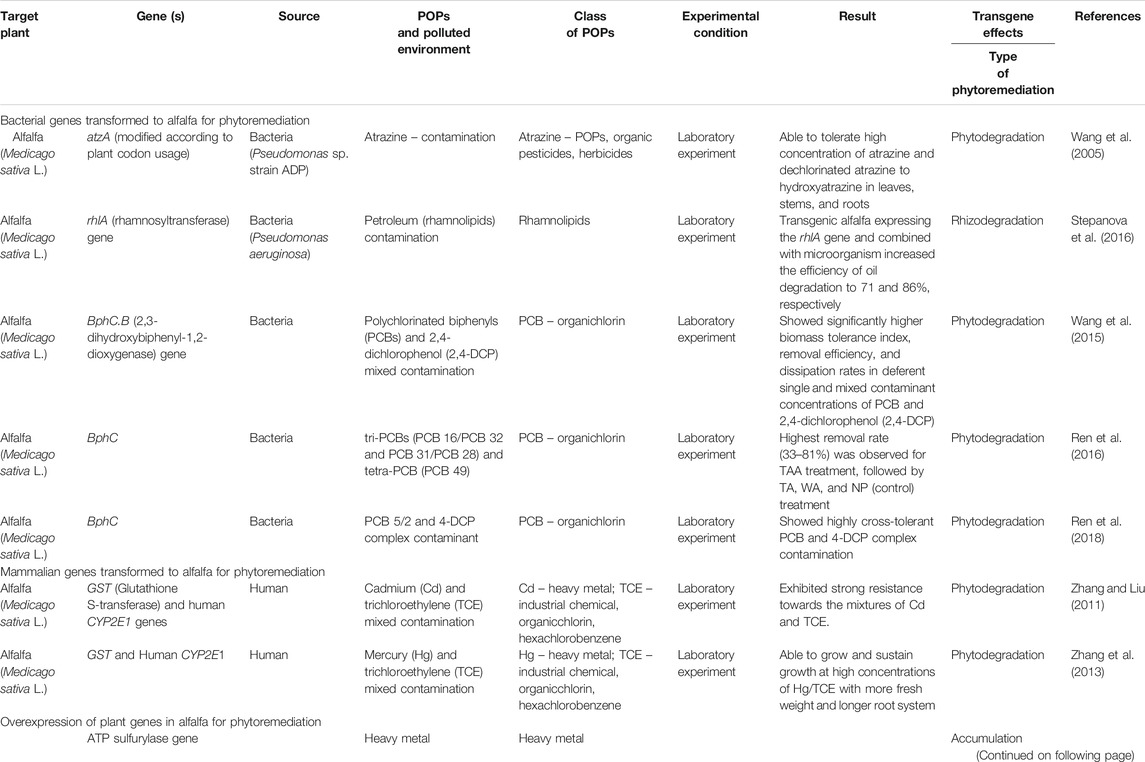
The genetic engineering of plants is one of the advances in the field of phytoremediation, overcoming many of the obstacles of traditional phytoremediation (Aken et al., 2010; Rylott et al., 2015). The primary reason for the need to use transgenic plants for phytoremediation is that plants lack the necessary enzymatic machinery, which is involved in bacteria or mammals. Some of the genes or traits manipulated in transgenic plants for improved tolerance and phytoremediation potential are summarized by (Cherian and Oliveira 2005). Alternatively, the transgenic approach involves the incorporation of specific and useful genes into alfalfa to improve the traits of interest (Kumar 2011). In this review, we focus on the transgenic alfalfa plants for enhanced phytoremediation of POPs, such as atrazine, PCB and TCE, and petroleum and heavy metals (Table 1).
Phytoremediation for Removing Persistent Organic Pollutants
Persistent organic pollutants (POPs) are toxic organic chemicals that possess a combination of physical and chemical properties. They can persist in the environment for long periods because of high resistance to photolytic, chemical, and biological degradation (Mamirova et al., 2019). The 12 initial POPs, including pesticides, industrial chemicals, and by-products, were listed in the Stockholm Convention (http://chm.pops.int/TheConvention/ThePOPs/ListingofPOPs/tabid/2509/Default.aspx). POPs are distributed in soil, sediments, air, and water. The five countries that use the most pesticides are China, the United States, Argentina, Thailand, and Brazil (Tarla et al., 2020). According to the official data of the Ministry of Agriculture of the Republic of Kazakhstan, 2,054 and 1,396 L of pesticides were used for non-agricultural purposes (weed control on roadsides) in 2018 and 2019, respectively (https://ipen.org/sites/default/files/documents/final_en_kazakhstan_hhp_report_summary_18_may_2020_0.pdf.). However, Nurzhanova et al. (2011) reported the total amount of obsolete pesticides in two regions (Almaty and Akmola) of Kazakhstan in 2011 to be 388.6 tons (Nurzhanova et al., 2011). A thorough analysis of the foci of pollution with POP pesticides and the development of methods for the remediation of contaminated areas is one of the key areas of the Kazakhstan Republic strategic development plan (Mamirova et al., 2019).
Climate change has the potential of affecting the behavior and distribution of organic pollutants, including POPs. Direct effects of climate change, like temperature increase, modification of wind and precipitation patterns, sea-level rise, snow, and ice cover, may alter the partitioning of POPs among the environmental compartments (Dalla Valle et al., 2007). The regular remediation strategies of POPs, comprising physical, chemical, and biological methods, have drawn considerable international attention over the past several decades because of such limitations as the cost-intensive, lower efficiency, and negative effects on natural habitats (Cherian and Oliveira 2005). Among these strategies, bioremediation and phytoremediation can be considered to be cost-effective, environmentally suitable, and appropriate for public acceptance (Ren et al., 2018). Bioremediation is a process used to clean up the contaminated environment, including water, soil, and subsurface materials, by microorganisms to detoxify and degrade the target pollutants. In this review, phytoremediation is examined in detail.
The term “phytoremediation” emerged in scientific research in the 1990s (Raskin et al., 1994; Salt et al., 1995) and refers to a green biotechnology using plants, associated microorganisms, and solar energy to clean hyper-accumulated toxic pollutants from the environment (Cobbett and Meagher 2002; Wu et al., 2015) and metabolize or transform them into less toxic metabolites (Cherian and Oliveira 2005). For example, the uptake and translocated processing of organic pollutants involve three detoxification stages. The first stage comprises conversion or transformation of the chemical compounds (oxidations by cytochrome P450 monooxygenases; reduction by dehydrogenases; and hydrolysis by esterases). The second stage is the conjugation to glutathione or sugars by glutathione-S-transferases or glycosyltransferases, leading to conjugated and frequently fewer toxic metabolites. The final stage is the compartmentation of these compounds in plant vacuoles or bound to the cell wall and lignin by the mechanism of ATP- or proton-dependent transporters or exocytosis (Schäffner et al., 2002; Cherian and Oliveira 2005). Phytoremediation is a better alternative approach that is appropriate for in situ degradation of pollutants over a large area and is cost-effective compared with the other methods. Besides these advantages, phytoremediation could support the creation of habitats for animals, promote biodiversity, reduce soil erosion by wind or water, help speed the restoration of damaged ecosystems, and supply nutrients for rhizosphere bacteria, allowing the growth and maintenance of a microbial community (Kumar et al., 2019).
Five special types of phytoremediation have been defined for removing POPs from the environment: 1) Phytodegradation, degrading organic pollutants in the soil or within the plant tissue through specific enzymes that the plant roots secrete; 2) Phytovolatilization, taking up pollutants from the soil or water and releasing them into the atmosphere through plant transpiration; 3) Phytostabilization, focusing mainly on soil pollutants near the roots, resulting in a reduction of soil erosion; 4) Phytoextraction, using the ability of plants or algae to remove contaminants from soil or water and concentrating them above ground in the plant biomass; 5) Phytostimulation, removing dangerous organic contaminants from soil using organisms within the rhizosphere (Nwoko 2010; Mishra et al., 2020) (Figure 1).
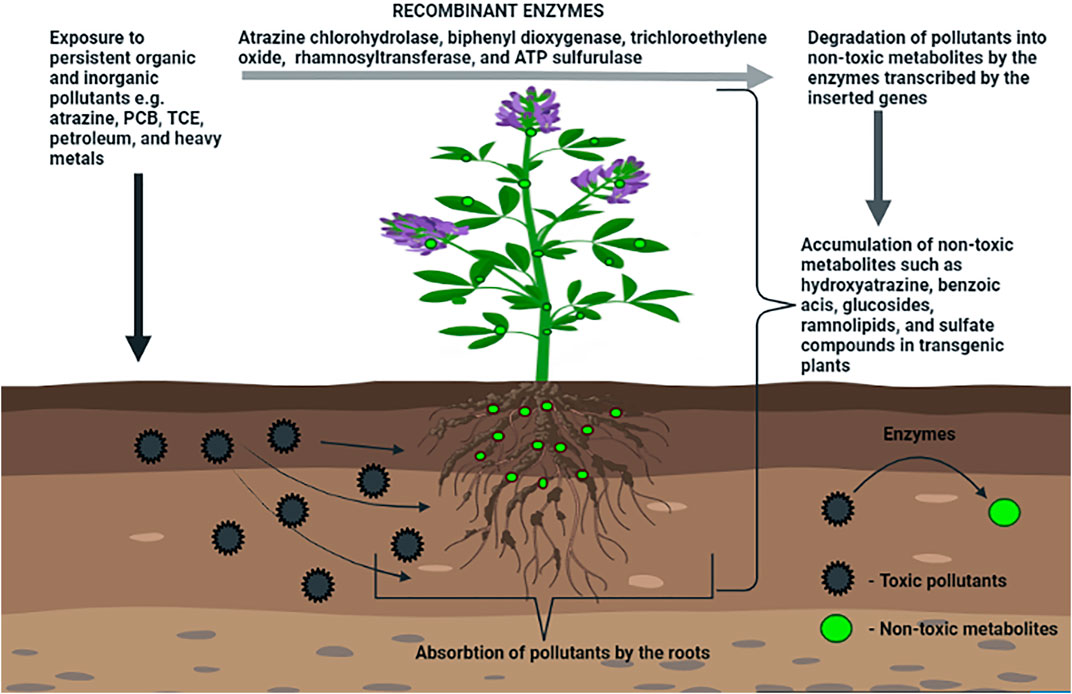
FIGURE 1. Schematic representation of phytoremediation strategies by transgenic alfalfa plants. Phytoremediation processes by transgenic plants include phytoaccumulation (plants take up persistent organic and inorganic pollutants with soil water and store the contaminants in their tissues) and phytodegradation (metabolism of contaminants within plants due to expression of transgenes). The phytotoxic effects of atrazine, polychlorinated biphenyl (PCB), and petroleum are overcome by expressing the bacterial genes AtzA, bphC, B, and rhlA. Expression of mammalian genes CYP2E1 and GST enhance degradation of trichloroethylene (TCE). Overexpression of plant gene ATPS improves heavy metal tolerance. Degradation of pollutants into non-toxic metabolites occurs by the enzymes transcribed by the inserted genes.
Bacterial Genes Transformed to Alfalfa for Phytoremediation
In the past 20 years, many pollutant-degrading bacteria have been isolated and characterized from polluted soils and reported to be capable of bioremediation or phytoremediation in various environments affected by different pollutants. The bacteria-associated plants exhibit contaminant degradation, growth-promoting activities, and carbon sequestration (Deng and Cao 2017). The role of bacteria in phytoremediation and some research examples on use of degradative bacteria for phytoremediation of heavy metals and organic compounds has been disclosed in several reviews (Kong and Glick 2017a; Kong and Glick 2017b; Mamirova et al., 2019; He et al., 2020; Tarla et al., 2020; Alkorta and Garbisu 2021; Basit et al., 2021; Girolkar et al., 2021; González Henao and Ghneim-Herrera 2021; Nedjimi 2021; Sharma 2021; Song et al., 2021; Gajić et al., 2022). González Henao and Ghneim-Herrera (2021), for instance, analyzed 85 research articles published over the past 15 years, which included a total of 62 bacterial genera, comprising 424 bacterial strains associated with 50 plant species in their systematic review article, which was published in Frontiers in Environmental Science in 2021. In this systematic review, Agrobacterium, Bacillus, Klebsiella, Enterobacter, Microbacterium, Pseudomonas, Rhodococcus, and Mesorhizobium were identified as the most commonly evaluated genera having crucial molecular mechanisms associated with plant growth promotion. In particular, Klebsiella and Enterobacter were considered as the best potential candidates for bioremediation and bacteria-assisted phytoremediation strategies in contaminated soil (González Henao and Ghneim-Herrera 2021). Rhodococcus is another genus famous for its ability to catabolize a wide range of organic compounds, including alkanes, cycloalkanes, and various aromatic hydrocarbons (Shevtsov et al., 2013).
Here, we summarize some studies not included in these reviews. Mello et al. (2020) noted that the plants inoculated with endophytic bacteria, including Acinetobacter baumannii BacI43, Enterobacter sp. BacI14, Klebsiella pneumoniae BacI20, Pseudomonas sp. BacI7, Pseudomonas sp. BacI38, and Serratia marcescens BacI56, had increased mercury volatilization and bioaccumulation in plant tissues. Anees et al. (2020) reported increased efficiency of Na phytoremediation by Pseudomonas, Sanguibacter, Bacillus, and Bacillus species. In the study of Yang et al. (2020), Spartina alterniflora-associated rhizo- and endobacterial communities showed resistance against at least one of the tested heavy metal ions. Mesa-Marín et al. (2020) reported that S. ramosissima was able to accumulate high concentrations of heavy metals, including arsenic, cadmium, copper, nickel, lead, and zinc. Silambarasan et al. (2020) revealed the feasibility of using the Pseudomonas citronellolis strain SLP6 to enhance phytostabilization of Cu-contaminated saline soils. The application of the B. altitudinis strain KP-14 remarkably increased the bio-parameters of M. xgiganteus cultivated on V, Cr, Mn, Ni, Cu, Zn, Sr, and Pb-contaminated soils and impacted phytoremediation parameters such as bioconcentration and translocation factors (Pidlisnyuk et al., 2020).
Bacteria-associated phytoremediation is used for heavy metals, as mentioned above, and is also used for organic compounds. In the study of Nurzhanova et al. (2021), Bacillus vallismortis and Bacillus aryabhattai were reported to have a high capability for utilization of dichlorodiphenyldichloroethylene (DDE). The biochemical and genomic characterization of 16 glyphosate-tolerant bacterial strains were described by Massot et al. (2021). As a result, they noted that L. corniculatus in combination with O. haematophilum can be adopted for phytoremediation of glyphosate on agricultural soils. These studies indicated that many microorganisms have the potential to degrade toxic compounds and are ideal candidates for bioremediation of various pollutants. The next goal of scientific research was the transgenic expression of bacterial genes on different plant species to improve phytoremediation efficiency.
Transgenic Alfalfa Plants Expressing Bacterial Atrazine Chlorohydrolase (AtzA) Gene for Phytoremediation
Atrazine (2-chloro-4-ethylamino-6-isopropylamino-1, 3, 5-triazine, molecular formula as C8H14 ClN 5), a herbicide belonging to the triazine class, has been used worldwide for over 50 years as a chemical herbicide (Ye et al., 2016) for weed control due to its good weeding effect and low price (He Q et al., 2019). The annual use of atrazine globally is 70,000–90,000 tons. This herbicide induces oxidative stress, inflammation, mitochondrial dysfunction, and apoptosis in exposed cells. It also passes through biological barriers such as the blood–brain barrier and the blood–testis barrier (Sadeghnia et al., 2021). Studies aimed at removal of atrazine from the environment are important due to its stable structure, difficult degradation, long residence time in the environment, and toxicity to organisms and human beings. The advantages and disadvantages of various methods of atrazine degradation, including physicochemical, chemical, biological, and material-microbial-integrated aspects, were systematically reviewed recently by (He H et al., 2019).
Atrazine is degraded and mineralized to produce 15 metabolites or to remain chemically intact in soil. Among these identified metabolites, the four major ones are dese-thylatrazine, deisopropylatrazine, didealkylatrazine, and hydroxyatrazine (Houjayfa et al., 2020).
Atrazine catabolism is initiated by a plasmid-borne atzA gene encoding atrazine chlorohydrolase. It is the first enzyme that was discovered and purified from the soil organism Pseudomonas sp. strain ADP in the early 1990s (Souza et al., 1996). It catalyzes atrazine to hydroxyatrazine, which is a non-herbicidal, non-toxic, more strongly sorbed form (Wang et al., 2005). The open reading frame for chlorohydrolase was determined by sequencing to 1,419 nucleotides and encoding a 473-amino acid protein with a predicted subunit molecular weight of 52,421 (Souza et al., 1996).
The first transgenic alfalfa plants for phytoremediation were described together with tobacco and Arabidopsis plants expressing a codon-optimized atzA gene from Pseudomonas sp. strain ADP by Wang et al. (2005). The results showed that the transgenic alfalfa, tobacco, and Arabidopsis plants were not only able to survive and grow in the presence of atrazine up to 15 μg/ml in MS medium but could also dechlorinate atrazine to hydroxyatrazine up to 98.6% in leaves, stems, and roots (Wang et al., 2005).
Besides transgenic alfalfa plants, transgenic tobacco expressing Pseudomonas sp. atzA (Wang et al., 2010), transgenic rice (Oryza sativa) (Inui and Ohkawa 2005; Kawahigashi et al., 2005; Kawahigashi et al., 2006; Kawahigashi et al., 2008), and potato (Inui and Ohkawa 2005) expressing human CYP1A1, CYP2B6, or CYP2C19, potato expressing rat CYP1A1 (Yamada et al., 2002), Brassica juncea overexpressing Brassica juncea γ-Glutamycysteine synthetase (γ-ECS, GS) gene, and Nicotiana tabaccum and Medicago sativa expressing p-atzAa gene (Wang et al., 2005) were used for the phytoremediation of atrazine and other herbicides, insecticides, and industrial chemicals.
Transgenic Alfalfa Plants Expressing Biphenyl Dioxygenase (bphC, B) Gene for Phytoremediation of Polychlorinated Biphenyl Contamination
PCBs are organic chemicals of chlorinated hydrocarbons. PCB has been applied for industrial and agricultural purposes since 1930 because of its unique physical and chemical properties. The production of PCB was stopped around the late 1970s to the early 1980s worldwide because of its high toxicity and negative impact on fauna, flora, and human health. The efficiency of bioremediation of PCB-polluted objects depends strongly on degree of pollution (Vasil’eva and Strizhakova 2007).
Biphenyl dioxygenase is the first enzyme of the bacterial catabolic pathway involved in the degradation of polychlorinated biphenyls; it catalyzes the ring cleavage reaction of 2,3-dihydroxybiphenyl into 2-hydroxy-6-oxo-6-phenyl-hexa-2,4-dienoic acid. In 2015, Wang et al. generated transgenic alfalfa plants expressing 2, 3-dihydroxybiphenyl-1, 2-dioxygenase gene (bphC, B) from bacteria to degrade PCB, 2,4-dichlorophenol (2,4-DCP), and mixed contamination of PCB 5/2,4-DCP. Their results showed that, firstly, the transgenic lines expressing bphC and bphB genes had a higher fresh weight with 34% higher tolerance index and higher dissipation rates of PCBs in root and shoot in a hydroponic medium containing 0.075 mg/L PCBs at the seeding stage. Secondly, the bphC, bphB transgenic lines had significantly higher biomass and tolerance index and showed 98.8% removal efficiency in 25 mg/L 2,4-DCP treatment after 144 h. Thirdly, the bphC, bphB transgenic lines were able to sustain growth in PCB 5/2,4-DCP mixed contaminants at a concentration up to 120/50 mg/L, whereas wild-type alfalfa plants could scarcely survive in a 40/10 mg/L concentration (Wang et al., 2015).
Apart from PCBs, biphenyl dioxygenase can catalyze the oxygenation of many persistent xenobiotics such as polyaromatic hydrocarbons (PAHs), chlorodibenzofurans, and chlorodioxins (Wackett et al., 2002). Various biphenyl dioxygenase genes have been characterized and cloned in different bacterium strains (Erickson and Mondello 1992; Taira et al., 1992; Sylvestre et al., 1996). The possibility of cloning of the bphC gene encoding 2,3-dihydroxybiphenyl-1,2-dioxygenase, from the Comamonas testosteroni B-356 and Pandoraea pnomenusa B-356, was demonstrated by Novakova et al. (2009), Novakova et al. (2010) for the production of transgenic tobacco plants to remediate of PCBs. However, only the transgenic expression of the targeted gene was reported; the remediation efficiency for PCB was not tested (Novakova et al., 2009; Novakova et al., 2010).
Low water solubility is one of the major limitations in bioremediation processes of PCBs (Wang et al., 2014). Therefore, it is known that the solubilization of high-chlorinated PCBs using biosurfactants is an effective and eco-friendly approach. For example, biosurfactant AlnA protein from Acinetobacter radioresistens was first used to enhance the solubility and bioavailability of PCB in soil (Wang et al., 2014). Thereafter, in 2016, Ren et al. described some crucial physiological traits and PCB remediation efficiency under four treatments, i.e. transgenic alfalfa plants expressing bphC gene with the combination of biosurfactant AlnA protein (TAA treatment), transgenic alfalfa (TA), wild-type alfalfa (WT), and unplanted soil (NP) for PCB-contaminated soil. The highest removal rate (33%–81%) was observed for TAA treatment, followed by TA, WA, and NP (control) treatments (Ren et al., 2016). In 2018, Ren et al. described the transgenic alfalfa plant tolerance to the combination of PCB 5 and 2,4-DCP. The tested transgenic alfalfa plant exhibited strong resistance up to 120/50 mg/L of PCB 5/2, 4-DCP complex contaminants without any visual evidence of damage. Higher remediation efficiency was observed in the use of transgenic alfalfa plants with the combination of recombinant AlnA protein than in the transgenic alfalfa mutants (Ren et al., 2018). In both studies of Ren et al, the microbial community composition of contaminated soil was analyzed, and it showed a different distribution of the phylum and genus of microorganisms. These results suggest that the detected abundance of indigenous microorganisms by transgenic alfalfa plants with the combination of biosurfactant AlnA protein increases PCB degradation (Ren et al., 2016; Ren et al., 2018).
Transgenic Alfalfa Plants Expressing Rhamnosyltransferase (rhlA) Gene for Phytoremediation of Petroleum Contamination
Petroleum hydrocarbons are highly toxic to the environment because of their inherent properties like solubility, volatility, and biodegradability (Hatami et al., 2019). However, the low water solubility and dissolution rate of oil products negatively affect the biodegradation efficiency of oil. The main method for phytoremediation of oil-polluted soil is rhizodegradation. In this context, oil is known to be effectively utilized not only by bacteria but also by fungi. Therefore, most studies focus on assessing the influence of petroleum remediation with the most frequent bacterial species of Pseudomonas fluorescens (Gutiérrez et al., 2020), Pseudomonas aeruginosa (Wang et al., 2017; Dobler et al., 2020; Varjani et al., 2020; Varjani and Upasani 2021), and Bacillus subtilis (Montagnolli et al., 2015; Chen et al., 2017; Wang et al., 2019), and some fungi, including Candida and Yarrowia, have a great potential for phytoremediation (Salam et al., 2017; Cecchi et al., 2021). Some review studies recommend the use of fungi for petroleum phytoremediation since fungi are capable of utilizing a wider spectrum of hydrocarbons, element cycling, bioweathering, and mycogenic mineral formation (Deng and Cao 2017).
Rhamnolipids, glycolipid biosurfactants produced by various bacterial species such as Pseudomonas sp. and Burkholderia sp. can enhance the bioavailability of oil and are therefore widely applied for crude oil biodegradation (Chong and Li 2017). The chemical structure of different identified rhamnolipids and their microbial origins and roles have been thoroughly discussed by Abdel-Mawgoud et al. (Abdel-Mawgoud et al., 2010). Due to the antimicrobial properties of rhamnolipids, they have potential applications in crop protection to confer immunity against fungal and bacterial pathogens [for an extensive review, see (Crouzet et al., 2020)]. Rhamnolipids are well known to also protect plants from harmful effects of metals (Singh and Cameotra 2013; Ayangbenro and Babalola 2018).
Rhamnolipid biosynthesis is regulated exclusively by key enzymes such as RhlA, RhlB, and RhlC, which are found in Pseudomonas sp. and Burkholderia sp (Zheng et al., 2015; Chong and Li 2017). Early in 1994, the necessity of the RhiR gene, which encodes rhamnosyltransferase, for rhamnolipid biosynthesis, was confirmed clearly by Ochsner et al. (1994). In their study, a single open reading frame of the RhiR gene, which regulated rhamnolipid biosynthesis from the strain PG201 of P. aeruginosa, was cloned into a rhamnolipid-deficient mutant strain (65E12) of P. aeruginosa (Ochsner et al., 1994). The results of this research provided a platform for use of genes encoding rhamnosyltransferase to enhance phytoremediation of petroleum-contaminated soil. Therefore, the transgenic expression of rhamnosyltransferase genes in different plant species for increased phytoremediation capacity of the oil has been investigated extensively. For example, Stepanova et al. (2016) evaluated the rationale for using transgenic alfalfa plants expressing the rhlA gene encoding rhamnosyltransferase from Pseudomonas aeruginosa and combined with Candida maltosa for remediation of oil-polluted soil. They identified increasing efficiency of oil degradation of up to 86%, whereas 71% oil degradation was observed by using transgenic alfalfa expressing the rhlA gene without the combination (Stepanova et al., 2016). Similarly, transgenic Arabidopsis thaliana and Nicotiana tabacum expressing RhlA and RhlB genes from Pseudomonas sp. showed tolerance to crude oil and several metals, and higher degradation efficiency was obtained with oil hydrocarbons (27%) than with the wild type (2%) (Brychkova et al., 2004).
Mammalian Genes Transformed to Alfalfa for Phytoremediation
Cytochrome P450s are a family of monooxygenases that catalyze the oxidation and metabolism of a diverse range of xenobiotics and endogenous compounds (Shankar and Mehendale 2014; Cook et al., 2016). CYP450s enzymes are found in all kingdoms of life: animals (over 13,000), plants (over 16,000), fungi (more than 85,000), and bacteria (more than 62,000) (Lamb et al., 2009; Nelson 2018). In 2009, David Nelson created a platform named “The Cytochrome P450 Homepage”, which provides information on evolutionary schemes of 11,512 CYPs, confirming that members of CYP families and subfamilies were from common ancestors (Nelson 2009) (http://drnelson.utmem.edu/CytochromeP450.html). Therefore, in 2018, more than 300,000 CYP450s sequences in plants, vertebrates, fungi, bacteria, and prokaryotic have been mined and collected, and more than 41,000 CYP sequences have been sorted into clans, families, and subfamilies in preparation for naming by BLAST searches (Nelson 2018). In plants, CYP450s can catalyze more than 20 types of reactions in primary and secondary metabolism (Mizutani 2012; Shang and Huang 2020; Wang H et al., 2021). The database of 181 plant P450s was constructed based on sequences, structures, and functions (PCPD: http://p450.biodesign.ac.cn/) (Wang et al., 2015). The role of mammalian cytochrome P450s in the metabolism of xenobiotics has been investigated in much more detail than plant CYPs. Overexpression of mammalian genes in appropriate plant species results in effective phytoremediation by enhancing the metabolic activities of the plant.
Transgenic Alfalfa Plant Co-expressing Human CYP2E1 and GST Genes for Phytoremediation of Trichloroethylene (TCE) Contamination
Trichloroethylene (TCE) is a non-flammable, sweet-smelling, colorless, and chlorinated organic compound that has been used extensively as an industrial solvent and degreaser (Dumas et al., 2018). It is the most common halogenated organic pollutant in the environment. As shown by Li et al. (2019), the high concentration of TCE (greater than 10 mg/kg) negatively affects the carbon and nitrogen cycling and metabolism of soil microorganisms. As a result, soil quality declines and soil decomposition of organic matter and cycling of mineral nutrients are inhibited (Li et al., 2019).
Phytoremediation of TCE can be accomplished using transgenic plants expressing mammalian genes. For instance, one of the key enzymes in mammalian metabolism CYP2E1 co-expressed with glutathione S-transferase (GST) gene in transgenic alfalfa plants showed healthy growth in the coexistence of TCE (0.2 mmol/L) and CdSO 4 (0.15 mmol/L) compared with the transgenic alfalfa plants expressing a single gene (GST or CYP2E1) and the non-transgenic control plants (Zhang and Liu 2011). In 2013, the same study was done on enhancing remediation of mixed mercury (Hg) and TCE by the transgenic alfalfa plants co-expressing GST and human P450 2E1. The result showed that the transgenic alfalfa plants were able to grow in up to 50/500 µM Hg/TCE and sustain growth at concentrations of 20/200 µM of Hg/TCE mixed-contaminated MS medium, accumulating Hg/TCE from 2.6- to 4.2-fold more than the non-transgenic plants. Additionally, the transgenic alfalfa plants had 3.0-fold more fresh weight at 40/400 μM Hg/TCE soil treatment and longer roots at 20/200 μM Hg/TCE soil treatment than non-transgenic plants in mixed-contaminated soil (Zhang et al., 2013).
In the past decade, the CYP2E1 gene has been introduced into various plants, including tobacco, poplar, Sesbania grandiflora, and Arabidopsis thaliana plants, for phytoremediation of different contaminations, including TCE (Doty et al., 2000; Doty et al., 2007; James et al., 2008; Legault et al., 2017), and DDT (Mouhamad et al., 2012).
As discussed earlier, GSTs are a group of small proteins (200–250 amino acids) (Gao et al., 2020) that are critical for certain life processes, as well as for detoxication and toxification mechanisms of pharmaceuticals and environmental pollutants (Nebert and Vasiliou 2004). The GST gene has been introduced in various plants, including tobacco to enhance polyaromatic hydrocarbon degradation (Dixit et al., 2011) and Arabidopsis to enhance trichlorophenol degradation (Su et al., 2012). Besides phytoremediation purposes, some studies have indicated that silencing or overexpression of the GST gene in various plants significantly increased resistance to different stressors (reviewed in (Kumar and Trivedi 2018).
Overexpression of Plant Genes in Alfalfa for Phytoremediation
Transgenic Alfalfa Plants Overexpressing Arabidopsis ATP Sulfurylase Gene for Phytoremediation of Heavy Metal Contamination
High concentrations of non-essential heavy metals and metalloids (arsenic, cadmium, and lead) are a threat to fauna and flora, even to human health (González Henao and Ghneim-Herrera 2021). ATP sulfurylase is the first and key enzyme in the sulfate assimilation pathway in plants. Four ATPS genes (ATPS1, -2, -3, and -4) are present in both chloroplasts and the cytosol of Arabidopsis leaves (Bohrer et al., 2015). The study of Kumar et al. (2019) demonstrated that transgenic alfalfa plants expressing ATP sulfurylase gene from Arabidopsis improved heavy metal (cadmium, vanadium, nickel, copper, and lead) tolerance and metal accumulation ability in the root and shoot tissues compared with non-transgenic plants. Some studies have shown that overexpression of the ATP sulfurylase gene in Indian mustard plants (Brassica juncea L) led to a higher tolerance and accumulation of Se (Pilon-Smits et al., 1999; LeDuc et al., 2006), As(III), As(V), Cd, Cu, Hg, and Zn (Wangeline et al., 2004).
Discussion and Future Prospects
Improvement of the Use of Local Plant Species for Phytoremediation
Numerous plant species have been identified for the purpose of phytoremediation [reviewed in (Cherian and Oliveira 2005)]. However, the phytoremediation process is limited by seasonal and local climatic conditions. There are some studies on the use of local plant species for phytoremediation of crude oil (Ho and Juan 2018; Abbaspour et al., 2020), industrial wastewater (Al-Thani and Yasseen 2020), and metal contamination (Hu et al., 2020). Nurzhanova et al. (2013) have provided a list of 17 pesticide-tolerant plants, including biannual plant sp E. canadensis, B. vulgaris, O. acanthium, A. absinthium, A. cylindrica, and R. confertus, and A. annua, X. strumarium, K. scoparia, K. sieversiana, A. tricolor, S. dulcamara, and A. artemisifoli from the Karasajsk district of Almaty oblast in Kazakhstan (Nurzhanova et al., 2013). It is important to identify readily available local plant species for effective phytoremediation of different organic and inorganic pollutants in the local conditions.
Phytoremediation Process Needs to Establish a Healthy Soil Ecosystem
The purpose of phytoremediation is not only to clean the polluted environment but also to establish a healthy soil ecosystem based on restoring the aboveground vegetation and stability of the underground soil microbial community composition. On this topic, Ren and his team have published significant studies, as outlined above, and provide a good protocol for future researchers. Alfalfa has been proposed as an ideal candidate for effective remediation approaches (Ren et al., 2016; Ren et al., 2018).
Metabolism of Pollutants in Plant Cells to Inert Forms Is Most Desirable
There are several requirements for plants used for successful phytoremediation. First, a well-developed root system is necessary. Second, the plants must produce sufficient biomass to remediate widespread high concentrations of pollutants in the field. Third, the plants must have the ability to metabolize and immobilize the pollutants. Fourth, the plants used in phytoremediation should be widely used in agricultural practice that allows repeated planting and harvesting (Paz-Alberto 2013; Eapen and D'Souza 2005). In this case, one limitation with some forms of phytoremediation is that plants can move pollutants from the ground and transpire them unaltered into the air.
To meet the abovementioned requirements: first, a better understanding is needed of the mechanisms underlying organic and inorganic pollutant accumulation and tolerance of plants. Second, utilization of plant-associated bacteria must be enhanced. Third, transgenic plants expressing bacterial and mammalian genes capable of degrading different pollutants must be generated. Fourth, transgenic plants together with microorganisms and biosurfactant to increase phytoremediation efficiency should be used.
The Main Problem Is Mixed Contamination of the Environment
One of the main problems with remediation of the contaminated environment is that there is generally contamination with multiple chemicals, often including both heavy metals and organic compounds (Olaniran et al., 2013). Most soil remediation strategies are adjusted to “focus on” single contaminant systems. It is essential to use an effective remediation method to clean up mixed contaminated areas.
Solutions to these problems: first, the combination of plants and microorganisms is nowadays often recommended for mixed contaminated soils. Second, the plants that contain genes enabling them to remediate a wider range of pollutants are desirable. The use of transgenic plants co-expressing desirable genes can be used in a variety of ways to increase the effectiveness of phytoremediation. For example, the utilization of transgenic alfalfa plants co-expressing human CYP2E1 and GST genes for phytoremediation of Hg/TCE contaminations yielded positive results compared with non-transgenic plants or transgenic alfalfa plants expressing a single gene (GST or CYP2E1) (Zhang and Liu 2011; Zhang et al., 2013).
The Field Experiment of Transgenic Plants for Phytoremediation Must Be Extended
To our knowledge, most studies on phytoremediation, especially on transgenic plants, have been done only in the laboratory, not in field conditions. There are many challenges to using phytoremediation successfully under natural conditions. The process of creating transgenic plants and adaptation in field conditions requires much time and labor. For example, Legault et al. (2017) spent 6 years on the field study of transgenic poplar expressing cytochrome P450 2E1 to enhance degradation of TCE. Phytoremediation in field conditions requires large areas, and the transgenic plants are limited in number. Another challenge is that while transgenic plants bring favorable advantages to phytoremediation, there are many concerns regarding human health and environmental sustainability (Gunarathne et al., 2019). Thus, appropriate political management and investigation of the status of transgenic plants for wide growth in polluted areas are needed.
Identification of Unique Genes From Hyperaccumulators and Introduction of These Key Genes Into Suitable Plant Species
About 400 plant species considered hyperaccumulators that can accumulate high concentrations of pollutants through their root system are being applied in phytoremediation. However, traditional hyperaccumulators require an extensive period, even years, for remediation of polluted areas. Besides this, there are other potential limitations of phytoremediation such as incomplete metabolism, toxic metabolites, and lower detoxification capacity (Gunarathne et al., 2019). However, the identification and proof of unique genes from the hyperaccumulators and transferring them to fast-growing species have been shown to be highly beneficial by the advanced transgenic technologies. In this case, alfalfa is a model legume for creating transgenic lines.
Genome Editing Plants for Phytoremediation
Genome editing systems, such as CRISPR/Cas, have been used for trait improvement of tubers, fibers, and cereal crops (Gao et al., 2017; Liu et al., 2021; Tussipkan and Manabayeva 2021). To our knowledge, there are few studies on using the CRISPR/Cas9 system for the enhancement of phytoremediation efficiency. Despite the genome sequence of alfalfa (Medicago sativa L.) being known, no genome-edited alfalfa plants used for phytoremediation have been obtained. Thus, it is an important research task to find and knock out sensitive genes by the CRISPR/Cas system to enhance the phytoremediation efficiency of alfalfa. Knocking out sensitive genes has a great significance in trait improvement for crops. Tang et al. (2017) and Yang et al. (2019) reported that knocking out the metal transporter gene OsNramp5 using the CRISPR/Cas9 system showed low Cd accumulation in rice. (Tang et al., 2017; Yang et al., 2019).
Author Contributions
TD and MS wrote the manuscript together. All authors contributed to the article and approved the submitted version.
Funding
This work is supported by a grant from the Ministry of Education and Science of the Republic of Kazakhstan (grant number: AP09260362).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abbaspour, A., Zohrabi, F., Dorostkar, V., Faz, A., and Acosta, J. A., (2020). Remediation of an Oil-Contaminated Soil by Two Native Plants Treated with Biochar and Mycorrhizae. J. Environ. Manage. 254, 109755. doi:10.1016/j.jenvman.2019.109755
Abdel-Mawgoud, A. M., Lépine, F., and Déziel, E. (2010). Rhamnolipids: Diversity of Structures, Microbial Origins and Roles. Appl. Microbiol. Biotechnol. 86 (5), 1323–1336. doi:10.1007/s00253-010-2498-2
Aken, B. V., Correa, P. A., and Schnoor, J. L. (2010). Phytoremediation of Polychlorinated Biphenyls: New Trends and Promises. Environ. Sci. Technol. 44 (8), 2767–2776. doi:10.1021/es902514d
Al-Thani, R. F., and Yasseen, B. T. (2020). Phytoremediation of Polluted Soils and Waters by Native Qatari Plants: Future Perspectives. Environ. Pollut. 259, 113694. doi:10.1016/j.envpol.2019.113694
Alkorta, I., and Garbisu, C. (2021). Reflections and Insights on the Evolution of the Biological Remediation of Contaminated Soils. Front. Environ. Sci. 9, 734628. doi:10.3389/fenvs.2021.734628
Anees, M., Qayyum, A., Jamil, M., Rehman, F. u., Abid, M., Malik, M. S., et al. (2020). Role of Halotolerant and Chitinolytic Bacteria in Phytoremediation of saline Soil Using Spinach Plant. Int. J. phytoremediation. 22 (6), 653–661. doi:10.1080/15226514.2019.1707160
Ayangbenro, A., and Babalola, O. (2018). Metal(loid) Bioremediation: Strategies Employed by Microbial Polymers. Sustainability. 10, 3028. doi:10.3390/su10093028
Badr, A., El-Sherif, N., Aly, S., Ibrahim, S. D., and Ibrahim, M. (2020). Genetic Diversity Among Selected Medicago Sativa Cultivars Using Inter-retrotransposon-amplified Polymorphism, Chloroplast DNA Barcodes and Morpho-Agronomic Trait Analyses. Plants. 9 (8), 995. doi:10.3390/plants9080995
Basit, A., Shah, S. T., Ullah, I., Muntha, S. T., and Mohamed, H. I. (2021). Microbe-assisted Phytoremediation of Environmental Pollutants and Energy Recycling in Sustainable Agriculture. Arch. Microbiol. 203 (10), 5859–5885. doi:10.1007/s00203-021-02576-0
Biazzi, E., Nelson, N., Pecetti, L., and Annicchiarico, P. (2020). “GBS-based Genome-wide Association and Genomic Selection for Alfalfa (Medicago Sativa) Forage Quality Improvement,” in The Model Legume Medicago Truncatula, 923–927. doi:10.1002/9781119409144.ch118
Blondon, F., Marie, D., Brown, S., and Adam, K. (1994). Genome Size and Base Composition in Medicago Sativa and M. Truncatula Species. Genome. 37 (2), 264. doi:10.1139/g94-037
Bohrer, A. S., Yoshimoto, N., Ai, S., Rykulski, N., Saito, K., and Takahashi, H. (2015). Alternative Translational Initiation of ATP Sulfurylase Underlying Dual Localization of Sulfate Assimilation Pathways in Plastids and Cytosol in Arabidopsis thaliana. Front. Plant Sci. 5, 750. doi:10.3389/fpls.2014.00750
Brychkova, G. G., Sorokin, A. P., and Kartel, N. A. (2004). Bioremediation with Ecologically Safe Plants. NATO Science Series. Ser. 1, 147–158.
Cecchi, G., Cutroneo, L., Di Piazza, S., Besio, G., Capello, M., and Zotti, M. (2021). Port Sediments: Problem or Resource? A Review Concerning the Treatment and Decontamination of Port Sediments by Fungi and Bacteria. Microorganisms. 9 (6), 1279. doi:10.3390/microorganisms9061279
Chekol, T., and Vough, L. (2001). A Study of the Use of Alfalfa (Medicago Sativa L.) for the Phytoremediation of Organic Contaminants in Soil. Remediation J. 11, 89–101. doi:10.1002/rem.1017
Chen, L., He, F., Long, R., Zhang, F., Li, M., Wang, Z., et al. (2021). A Global Alfalfa Diversity Panel Reveals Genomic Selection Signatures in Chinese Varieties and Genomic Associations with Root Development. J. Integr. Plant Biol. 63 (11), 1937–1951. doi:10.1111/jipb.13172
Chen, Q., Li, J., Liu, M., Sun, H., and Bao, M. (2017). Study on the Biodegradation of Crude Oil by Free and Immobilized Bacterial Consortium in marine Environment. PLoS One. 12 (3), e0174445. doi:10.1371/journal.pone.0174445
Cherian, S., and Oliveira, M. M. (2005). Transgenic Plants in Phytoremediation: Recent Advances and New Possibilities. Environ. Sci. Techn. 39 (24), 9377–9390. doi:10.1021/es051134l
Chong, H., and Li, Q. (2017). Microbial Production of Rhamnolipids: Opportunities, Challenges and Strategies. Microb. Cel Factories. 16 (1), 137. doi:10.1186/s12934-017-0753-2
Cobbett, C. S., and Meagher, R. B. (2002). Arabidopsis and the Genetic Potential for the Phytoremediation of Toxic Elemental and Organic Pollutants. Arabidopsis Book. 1, e0032. doi:10.1199/tab.0032
Cook, D. J., Finnigan, J. D., Cook, K., Black, G. W., and Charnock, S. J. (2016). “Chapter Five - Cytochromes P450: History, Classes, Catalytic Mechanism, and Industrial Application,” in Advances in Protein Chemistry and Structural Biology. Editor C. Z. Christov (Academic Press), 105–126. doi:10.1016/bs.apcsb.2016.07.003
Crouzet, J., Arguelles-Arias, A., Dhondt-Cordelier, S., Cordelier, S., Pršić, J., Hoff, G., et al. (2020). Biosurfactants in Plant Protection against Diseases: Rhamnolipids and Lipopeptides Case Study. Front. Bioeng. Biotechnol. 8, 1014. doi:10.3389/fbioe.2020.01014
Dalla Valle, M., Codato, E., and Marcomini, A. (2007). Climate Change Influence on POPs Distribution and Fate: A Case Study. Chemospher 67 (7), 1287–1295. doi:10.1016/j.chemosphere.2006.12.028
Deng, Z., and Cao, L. (2017). Fungal Endophytes and Their Interactions with Plants in Phytoremediation: A Review. Chemosphere. 168, 1100–1106. doi:10.1016/j.chemosphere.2016.10.097
Dixit, P., Mukherjee, P. K., Sherkhane, P. D., Kale, S. P., and Eapen, S. (2011). Enhanced Tolerance and Remediation of Anthracene by Transgenic Tobacco Plants Expressing a Fungal Glutathione Transferase Gene. J. Hazard. Mater. 192 (1), 270–276. doi:10.1016/j.jhazmat.2011.05.018
Dobler, L., Ferraz, H. C., Araujo de Castilho, L. V., Sangenito, L. S., Pasqualino, I. P., Souza Dos Santos, A. L., et al. (2020). Environmentally Friendly Rhamnolipid Production for Petroleum Remediation. Chemosphere. 252, 126349. doi:10.1016/j.chemosphere.2020.126349
dos Santos, I. G., Rocha, J. d., Vigna, B. B., Damião Cruz, C., Ferreira, R., Basigalup, D. H., et al. (2020). Exploring the Diversity of Alfalfa within Brazil for Tropical Production. Euphytica. 216 (5), 72. doi:10.1007/s10681-020-02606-w
Doty, S., James, C., Moore, A., Vajzovic, A., Singleton, G., Ma, C., et al. (2007). Enhanced Phytoremediation of Volatile Environmental Pollutants with Transgenic Trees. Proc. Natl. Acad. Sci. U. S. A. 104, 16816–16821. doi:10.1073/pnas.0703276104
Doty, S. L., Shang, T. Q., Wilson, A. M., Tangen, J., Westergreen, A. D., Newman, L. A., et al. (2000). Enhanced Metabolism of Halogenated Hydrocarbons in Transgenic Plants Containing Mammalian Cytochrome P450 2E1. Proc. Natl. Acad. Sci. U. S. A. 97 (12), 6287–6291. doi:10.1073/pnas.97.12.6287
Dumas, O., Despreaux, T., Perros, F., Lau, E., Pascal, A., Humbert, M., et al. (2018). Respiratory Effects of Trichloroethylene. Respir. Med. 134, 47–53. doi:10.1016/j.rmed.2017.11.021
Eapen, S., and D'Souza, S. F. (2005). Prospects of Genetic Engineering of Plants for Phytoremediation of Toxic Metals. Biotechnol. Adv. 23 (2), 97–114. doi:10.1016/j.biotechadv.2004.10.001
Erickson, B. D., and Mondello, F. J. (1992). Nucleotide Sequencing and Transcriptional Mapping of the Genes Encoding Biphenyl Dioxygenase, a Multicomponent Polychlorinated-Biphenyl-Degrading Enzyme in Pseudomonas Strain LB400. J. Bacteriol. 174 (9), 2903–2912. doi:10.1128/jb.174.9.2903-2912.1992
Gajić, G., Mitrović, M., and Pavlović, Pa. (2022). “Chapter 12 - Phytobial Remediation by Bacteria and Fungi,” in Assisted Phytoremediation. Editor V. Pandey (Elsevier), 285–344.
Gao, J. J., Zhang, L., He Peng, R., Wang, B., and QuanYao, H. (2020). Recombinant Expression of Thermosynechococcus Elongatus BP-1 Glutathione S-Transferase in Arabidopsis thaliana : an Efficient Tool for Phytoremediation of Thiocyanate. Biotechnol. Biotechnological Equipment. 34 (1), 494–505. doi:10.1080/13102818.2020.1779127
Gao, W., Lu, L., Tian, X., Xu, F., Liu, J., Singh, P. K., et al. (2017). Genome Editing in Cotton with the CRISPR/Cas9 System. Front. Plant Sci. 8, 1364. doi:10.3389/fpls.2017.01364
Girolkar, S., Thawale, P., and Juwarkar, A. (2021). “Chapter 12 - Bacteria-Assisted Phytoremediation of Heavy Metals and Organic Pollutants: Challenges and Future Prospects,” in Bioremediation for Environmental Sustainability. Editors V. Kumar, G. Saxena, and M. P. Shah (Elsevier), 247–267. doi:10.1016/b978-0-12-820318-7.00012-5
González Henao, S., and Ghneim-Herrera, T. (2021). Heavy Metals in Soils and the Remediation Potential of Bacteria Associated with the Plant Microbiome. Front. Environ. Sci. 9 (15), 604216. doi:10.3389/fenvs.2021.604216
Gunarathne, V., Mayakaduwa, S., Ashiq, A., Ranjani Weerakoon, S., Biswas, J. K., and Vithanage, M. (2019). “Chapter 5 - Transgenic Plants: Benefits, Applications, and Potential Risks in Phytoremediation,” in Transgenic Plant Technology for Remediation of Toxic Metals and Metalloids. Editor M. Vara Prasad (Academic Press), 89–102. doi:10.1016/b978-0-12-814389-6.00005-5
Gutiérrez, E. J., Abraham, M. D. R., Baltazar, J. C., Vázquez, G., Delgadillo, E., and Tirado, D. (2020). Pseudomonas Fluorescens: A Bioaugmentation Strategy for Oil-Contaminated and Nutrient-Poor Soil. Int. J. Environ. Res. Public Health. 17 (19), 6959. doi:10.3390/ijerph17196959
Hatami, E., Ali, A., and Dorostkar, V. (2019). Phytoremediation of a Petroleum-Polluted Soil by Native Plant Species in Lorestan Province, Iran. Environ. Sci. Pollut. Res. 26 (24), 24323–24330. doi:10.1007/s11356-018-1297-7
Havananda, T., Brummer, C., and Doyle, J. (2011). Complex Patterns of Autopolyploid Evolution in Alfalfa and allies(Medicago Sativa; Leguminosae). Am. J. Bot. 98, 1633–1646. doi:10.3732/ajb.1000318
Hawkins, C., and Yu, L-X. (2018). Recent Progress in Alfalfa (Medicago Sativa L.) Genomics and Genomic Selection. Crop J. 6 (6), 565–575. doi:10.1016/j.cj.2018.01.006
He, H., Wu, M., Su, R., Zhang, Z., Chang, C., Qi, P., et al. (2021). Strong Phosphorus (P)-zinc (Zn) Interactions in a Calcareous Soil-Alfalfa System Suggest that Rational P Fertilization Should Be Considered for Zn Biofortification on Zn-Deficient Soils and Phytoremediation of Zn-Contaminated Soils. Plant and Soil. 461 (1), 119–134. doi:10.1007/s11104-020-04793-w
He, H., Liu, Y., You, S., Liu, J., Xiao, H., and Tu, Z. (2019). A Review on Recent Treatment Technology for Herbicide Atrazine in Contaminated Environment. Int. J. Environ. Res. Public Health. 16 (24), 5129. doi:10.3390/ijerph16245129
He, Q., Li, Y. Y., Liu, Y. L., Li, Z. P., Sun, S. S., and Zhan, Q. W. (2019). Genetic Diversity in the Worldwide Alfalfa Germplasm Assessed through SSR Markers. Int. J. Agric. Biol. 22 (5), 1205–1210. doi:10.17957/IJAB/15.1188
He, W., Megharaj, M., Wu, C-Y., Subashchandrabose, S. R., and Dai, C-C. (2020). Endophyte-assisted Phytoremediation: Mechanisms and Current Application Strategies for Soil Mixed Pollutants. Crit. Rev. Biotechnol. 40 (1), 31–45. doi:10.1080/07388551.2019.1675582
Ho, D., and Juan, A. (2018). “Phytoremediation of Crude Oil-Contaminated Soil with Local Plant Species,” in 11th Curtin University Technology, Science and Engineering (CUTSE) International Conference.
Houjayfa, O. M., Noubissié, E., and Ngassoum, M. B. (2020). Mobility Studies of Atrazine in the Soil-Plant System in Two Cameroonian Vegetables Amaranthus hybridus and Corchorus Olitorius. Environ. Sustainability Indicators. 6, 100036. doi:10.1016/j.indic.2020.100036
Hu, Z., Wang, Y., Fang, Z., Shi, G., Lou, L., Ren, K., et al. (2020). Italian Ryegrass–rice Rotation System for Biomass Production and Cadmium Removal from Contaminated Paddy fields. J. Soils Sediments. 20 (2), 874–882. doi:10.1007/s11368-019-02470-9
Inui, H., and Ohkawa, H. (2005). Herbicide Resistance in Transgenic Plants with Mammalian P450 Monooxygenase Genes. Pest Manag. Sci. 61 (3), 286–291. doi:10.1002/ps.1012
Irish, B. M., and Greene, S L. (2021). “Germplasm Collection, Genetic Resources, and Gene Pools in Alfalfa,” in The Alfalfa Genome. Editors L-Xi. Yu, and C. Kole (Cham: Springer International Publishing), 43–64. doi:10.1007/978-3-030-74466-3_4
James, C. A., Xin, G., Doty, S. L., and Strand, S. E. (2008). Degradation of Low Molecular Weight Volatile Organic Compounds by Plants Genetically Modified with Mammalian Cytochrome P450 2E1. Environ. Sci. Technol. 42 (1), 289–293. doi:10.1021/es071197z
Ju, W., Liu, L, Jin, X., Duan, C., Cui, Y., Wang, J, et al. (2020). Co-inoculation Effect of Plant-Growth-Promoting Rhizobacteria and Rhizobium on EDDS Assisted Phytoremediation of Cu Contaminated Soils. Chemosphere. 254, 126724. doi:10.1016/j.chemosphere.2020.126724
Kakaei, M., and Ahmadian, S. (2021). Genetic Diversity Study of Some Iranian Alfalfa Genotypes Based on Seed Storage Proteins Patterns. Iranian J. Sci. Technol. Trans. A: Sci. 45 (4), 1223–1228. doi:10.1007/s40995-021-01142-z
Kawahigashi, H., Hirose, S., Ohkawa, H., and Ohkawa, Y. (2006). Phytoremediation of the Herbicides Atrazine and Metolachlor by Transgenic rice Plants Expressing Human CYP1A1, CYP2B6, and CYP2C19. J. Agric. Food Chem. 54 (8), 2985–2991. doi:10.1021/jf052610u
Kawahigashi, H., Hirose, S., Ohkawa, H., and Ohkawa, Y. (2005). Transgenic rice Plants Expressing Human CYP1A1 Remediate the Triazine Herbicides Atrazine and Simazine. J. Agric. Food Chem. 53 (22), 8557–8564. doi:10.1021/jf051370f
Kawahigashi, H., Hirose, S., Ohkawa, H., and Ohkawa, Y. (2008). Transgenic rice Plants Expressing Human P450 Genes Involved in Xenobiotic Metabolism for Phytoremediation. J. Mol. Microbiol. Biotechnol. 15 (2-3), 212–219. doi:10.1159/000121332
Kong, Z., and Glick, B. R. (2017a). “Chapter Two - the Role of Plant Growth-Promoting Bacteria in Metal Phytoremediation,” in Advances in Microbial Physiology. Editor R. K. Poole (Academic Press), 97–132. doi:10.1016/bs.ampbs.2017.04.001
Kong, Z., and Glick, B. R. (2017b). The Role of Bacteria in Phytoremediation. Applied Bioengineering., 327–353. doi:10.1002/9783527800599.ch11
Kumar, S. (2011). Biotechnological Advancements in Alfalfa Improvement. J. Appl. Genet. 52 (2), 111–124. doi:10.1007/s13353-011-0028-2
Kumar, S., and Trivedi, P. K. (2018). Glutathione S-Transferases: Role in Combating Abiotic Stresses Including Arsenic Detoxification in Plants. Front. Plant Sci. 9, 751. doi:10.3389/fpls.2018.00751
Kumar, V., AlMomin, S., Al-Shatti, A., Al-Aqeel, H., Al-Salameen, F., Shajan, A. B., et al. (2019). Enhancement of Heavy Metal Tolerance and Accumulation Efficiency by Expressing Arabidopsis ATP Sulfurylase Gene in Alfalfa. Int. J. Phytoremediation. 21 (11), 1112–1121. doi:10.1080/15226514.2019.1606784
Lamb, D. C., Lei, L., Warrilow, A. G., Lepesheva, G. I., Mullins, J. G., Waterman, M. R., et al. (2009). The First Virally Encoded Cytochrome P450. J. Virol. 83 (16), 8266–8269. doi:10.1128/JVI.00289-09
LeDuc, D., AbdelSamie, M., Montes-Bayón, M., Wu, C., Reisinger, S., and Terry, N. (2006). Overexpressing Both ATP Sulfurylase and Selenocysteine Methyltransferase Enhances Selenium Phytoremediation Traits in Indian Mustard. Environ. Pollut. (Barking, Essex). 144, 70–76. doi:10.1016/j.envpol.2006.01.008
Legault, E. K., James, C. A., Stewart, K., Muiznieks, I., Doty, S. L., and Strand, S. E. (2017). A Field Trial of TCE Phytoremediation by Genetically Modified Poplars Expressing Cytochrome P450 2E1. Environ. Sci. Technol. 51 (11), 6090–6099. doi:10.1021/acs.est.5b04758
Li, A., Liu, A, Du, X., Chen, J-Y., Yin, M., Hu, H-Y., et al. (2020). A Chromosome-Scale Genome Assembly of a Diploid Alfalfa, the Progenitor of Autotetraploid Alfalfa. Hortic. Res. 7 (1), 194. doi:10.1038/s41438-020-00417-7
Li, P., Zhang, Y., Meng, Q., Liu, Y., Tuyiringire, D., Chen, Z., et al. (2019). Effects of Trichloroethylene Stress on the Microbiological Characteristics of Mollisol. Ecotoxicology Environ. Saf. 184, 109595. doi:10.1016/j.ecoenv.2019.109595
Li, X, and Brummer, E. C. (2012). Applied Genetics and Genomics in Alfalfa Breeding. Agronomy. 2 (1), 40–61. doi:10.3390/agronomy2010040
Liu, Q., Fan, Y., Zhang, J., Liu, H., Rahman, S., Islam, S., et al. (2021). Application of CRISPR/Cas9 in Crop Quality Improvement. Int. J. Mol. Sci. 22 (8), 4206. doi:10.3390/ijms22084206
Ma, J. X., Yan, Z., Zhang, J, Feng, Q., Wang, T., and Lu, X. (2018). Analysis of Genetic Diversity of 20 Wild Alfalfa Germplasms in Xinjiang with RAPD and ISSR Markers. J. Yunnan Agric. Univ. 33 (4), 577–587.
Mamirova, А., Nurzhanova, A., and Pidlisnyuk, V. (2019). Pop Pesticides and Reclamation Methods (Review). REPORTS. 6, 21–34. doi:10.32014/2019.2518-1483.164
Massot, F., Gkorezis, P., Van Hamme, J., Marino, D., Trifunovic, B., Vukovic, G., et al. (2021). Isolation, Biochemical and Genomic Characterization of Glyphosate Tolerant Bacteria to Perform Microbe-Assisted Phytoremediation. Front. Microbiol. 11, 3506. doi:10.3389/fmicb.2020.598507
Mello, I, Targanski, S., and Pietro-Souza, W. (2020). Endophytic Bacteria Stimulate Mercury Phytoremediation by Modulating its Bioaccumulation and Volatilization. Ecotoxicology Environ. Saf. 202, 110818. doi:10.1016/j.ecoenv.2020.110818
Mesa-Marín, J, Pérez-Romero, J. A., Redondo-Gómez, S., Pajuelo, E., Rodríguez-Llorente, I. D., and Mateos-Naranjo, E. (2020). Impact of Plant Growth Promoting Bacteria on Salicornia ramosissima Ecophysiology and Heavy Metal Phytoremediation Capacity in Estuarine Soils. Front. Microbiol. 11, 553018. doi:10.3389/fmicb.2020.553018
Mishra, S., Kumar, P., and Singh, R. (2020). “3 - Transgenic Plants in Phytoremediation of Organic Pollutants,” in Bioremediation of Pollutants. Editors V. C. Pandey, and V. Singh (Elsevier), 39–56. doi:10.1016/b978-0-12-819025-8.00003-x
Mizutani, M. (2012). Impacts of Diversification of Cytochrome P450 on Plant Metabolism. Biol. Pharm. Bull. 35 (6), 824–832. doi:10.1248/bpb.35.824
Montagnolli, R. N., Lopes, P. R., and Bidoia, E. D. (2015). Assessing Bacillus Subtilis Biosurfactant Effects on the Biodegradation of Petroleum Products. Environ. Monit. Assess. 187 (1), 4116. doi:10.1007/s10661-014-4116-8
Mouhamad, R., Ghanem, I., AlOrfi, M., Ibrahim, K., Ali, N., and Al-Daoude, A. (2012). Phytoremediation of Trichloroethylene and Dichlorodiphenyltrichloroethane-Polluted Water Using Transgenic Sesbania Grandiflora and Arabidopsis thaliana Plants Harboring Rabbit Cytochrome P450 2E1. Int. J. Phytoremediation. 14 (7), 656–668. doi:10.1080/15226514.2011.619232
Nebert, D. W., and Vasiliou, V. (2004). Analysis of the Glutathione S-Transferase (GST) Gene Family. Hum. Genomics. 1 (6), 460–464. doi:10.1186/1479-7364-1-6-460
Nedjimi, B. (2021). Phytoremediation: a Sustainable Environmental Technology for Heavy Metals Decontamination. SN Appl. Sci. 3 (3), 286. doi:10.1007/s42452-021-04301-4
Nelson, D. R. (2009). The Cytochrome P450 Homepage. Hum. Genomics. 4 (1), 59–65. doi:10.1186/1479-7364-4-1-59
Nelson, D. R. (2018). Cytochrome P450 Diversity in the Tree of Life. Biochim. Biophys. Acta Proteins Proteom. 1866 (1), 141–154. doi:10.1016/j.bbapap.2017.05.003
Novakova, M., Mackova, M., Antosova, Z., Viktorova, J., Szekeres, M., Demnerova, K., et al. (2010). Cloning the Bacterial bphC Gene into Nicotiana Tabacum to Improve the Efficiency of Phytoremediation of Polychlorinated Biphenyls. Bioeng. Bugs. 1 (6), 419–423. doi:10.4161/bbug.1.6.12723
Novakova, M., Mackova, M., Chrastilova, Z., Viktorova, J., Szekeres, M., Demnerova, K., et al. (2009). Cloning the Bacterial bphC Gene into Nicotiana Tabacum to Improve the Efficiency of PCB Phytoremediation. Biotechnol. Bioeng. 102 (1), 29–37. doi:10.1002/bit.22038
Nurzhanova, A., Kalugin, S., and Zhambakin, K. (2013). Obsolete Pesticides and Application of Colonizing Plant Species for Remediation of Contaminated Soil in Kazakhstan. Environ. Sci. Pollut. Res. 20 (4), 2054–2063. doi:10.1007/s11356-012-1111-x
Nurzhanova, A., Mukasheva, T., Berzhanova, R., Kalugin, S., Omirbekova, A., and Mikolasch, A. (2021). Optimization of Microbial Assisted Phytoremediation of Soils Contaminated with Pesticides. Int. J. Phytoremediation. 23 (5), 482–491. doi:10.1080/15226514.2020.1825330
Nurzhanova, A., Zhambakin, K., Rakhimbayev, I., Sedlovskiy, A., and Kalugin, S. (2011). Obsolete Pesticides and Phytoremediation of Polluted Soil in Kazakhstan. J. Life Sci. 5 (7), 12.
Nwoko, C. O. (2010). Trends in Phytoremediation of Toxic Elemental and Organic Pollutants. Afr. J. Biotechnol. 9 (37), 6010–6016. doi:10.5897/AJB09.061
Ochsner, U. A., Koch, A. K., Fiechter, A., and Reiser, J. (1994). Isolation and Characterization of a Regulatory Gene Affecting Rhamnolipid Biosurfactant Synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176 (7), 2044–2054. doi:10.1128/jb.176.7.2044-2054.1994
Olaniran, A. O., Balgobind, A., and Balakrishna, P. (2013). Bioavailability of Heavy Metals in Soil: Impact on Microbial Biodegradation of Organic Compounds and Possible Improvement Strategies. Int. J. Mol. Sci. 14 (5), 10197–10228. doi:10.3390/ijms140510197
Paz-Alberto, A. (2013). Phytoremediation: A Green Technology to Remove Environmental Pollutants. Am. J. Clim. Change 2, 71–86. doi:10.4236/ajcc.2013.21008
Pidlisnyuk, V., Mamirova, A., Kumar, P., Shapoval, P., Trögl, J., and Nurzhanova, A. (2020). Potential Role of Plant Growth-Promoting Bacteria in Miscanthus X Giganteus Phytotechnology Applied to the Trace Elements Contaminated Soils. Int. Biodeterioration Biodegradation 155, 105103. doi:10.1016/j.ibiod.2020.105103
Pilon-Smits, E. A., Hwang, S., Mel Lytle, C., Zhu, Y., Tai, J. C., Bravo, R. C., et al. (1999). Overexpression of ATP Sulfurylase in Indian Mustard Leads to Increased Selenate Uptake, Reduction, and Tolerance. Plant Physiol. 119 (1), 123–132. doi:10.1104/pp.119.1.123
Raskin, I., Nanda Kumar, P. B. A., Dushenkov, S., and Salt, D. E. (1994). Bioconcentration of Heavy Metals by Plants. Curr. Opin. Biotechnol. 5 (3), 285–290. doi:10.1016/0958-1669(94)90030-2
Ren, H., Wan, Y., and Zhao, Y. (2018). “Phytoremediation of Polychlorinated Biphenyl-Contaminated Soil by Transgenic Alfalfa Associated Bioemulsifier AlnA,” in Twenty Years of Research and Development on Soil Pollution and Remediation in China. Editors Y. Luo, and C. Tu (Singapore: Springer Singapore), 645–653. doi:10.1007/978-981-10-6029-8_39
Ren, H., Su, Y., Zhang, J., Pan, H., Chen, B., and Wang, Y. (2016). Recombinant Protein, AlnA, Combined with Transgenic Alfalfa Remediates Polychlorinated Biphenyl-Contaminated Soils: Efficiency and Rhizosphere Microbial Community Response. Biotechnol. Lett. 38 (11), 1893–1901. doi:10.1007/s10529-016-2169-1
Rhouma, H., Taski-Ajdukovic, K., Zitouna, N., Dorra, S., Milic, D., and Trifi-Farah, N. (2017). Assessment of the Genetic Variation in Alfalfa Genotypes Using SRAP Markers for Breeding Purposes. Chilean J. Agric. Res. 77, 332–339. doi:10.4067/S0718-58392017000400332
Rylott, E. L., Johnston, E. J., and Bruce, N. C. (2015). Harnessing Microbial Gene Pools to Remediate Persistent Organic Pollutants Using Genetically Modified Plants—A Viable Technology. J. Exp. Bot. 66 (21), 6519–6533. doi:10.1093/jxb/erv384
Sadeghnia, H., Shahba, S., and Ebrahimzadeh-Bideskan, A. (2021). Atrazine Neural and Reproductive Toxicity. Toxin Rev., 1–14. doi:10.1080/15569543.2021.1966637
Salam, J. A., Hatha, M. A. A., and Das, N. (2017). Microbial-enhanced Lindane Removal by Sugarcane (Saccharum Officinarum) in Doped Soil-Applications in Phytoremediation and Bioaugmentation. J. Environ. Manage. 193, 394–399. doi:10.1016/j.jenvman.2017.02.006
Salt, D. E., Blaylock, M., Kumar, N. P., Dushenkov, V., Ensley, B. D., Chet, I., et al. (1995). Phytoremediation: A Novel Strategy for the Removal of Toxic Metals from the Environment Using Plants. Bio/Technology. 13 (5), 468–474. doi:10.1038/nbt0595-468
Schäffner, A., Messner, B., Langebartels, C., and Sandermann, H. (2002). Genes and Enzymes for In-Planta Phytoremediation of Air, Water and Soil. Acta Biotechnologica. 22, 141–151. doi:10.1002/1521-3846(200205)22:1/2<141::AID-ABIO141>3.0.CO;2-7
Shamustakimova, A. O., Mavlyutov, Y. M., and Klimenko, I. A. (2021). Application of SRAP Markers for DNA Identification of Russian Alfalfa Cultivars. Russ. J. Genet. 57 (5), 540–547. doi:10.1134/S1022795421050112
Shang, Yi., and Huang, S. (2020). Engineering Plant Cytochrome P450s for Enhanced Synthesis of Natural Products: Past Achievements and Future Perspectives. Plant Commun. 1 (1), 100012. doi:10.1016/j.xplc.2019.100012
Shankar, K., and Mehendale, H. M. (2014). “Cytochrome P450,” in Encyclopedia of Toxicology. Editor P. Wexler. Third Edition (Oxford: Academic Press), 1125–1127. doi:10.1016/b978-0-12-386454-3.00299-2
Sharma, P. (2021). Efficiency of Bacteria and Bacterial Assisted Phytoremediation of Heavy Metals: An Update. Bioresour. Techn. 328, 124835. doi:10.1016/j.biortech.2021.124835
Shen, C., Du, H., Chen, Z., Lu, H., Zhu, F., Chen, H., et al. (2020). The Chromosome-Level Genome Sequence of the Autotetraploid Alfalfa and Resequencing of Core Germplasms Provide Genomic Resources for Alfalfa Research. Mol. Plant 13 (9), 1250–1261. doi:10.1016/j.molp.2020.07.003
Shevtsov, A., Tarlykov, P., Zholdybayeva, E., Momynkulov, D., Sarsenova, A., Moldagulova, N., et al. (2013). Draft Genome Sequence of Rhodococcus Erythropolis DN1, a Crude Oil Biodegrader. Genome Announcements 1 (5), e00846-13. doi:10.1128/genomeA.00846-13
Silambarasan, S., Peter, L., Valentine, A., Cornejo, P., and Kannan, V. (2020). Pseudomonas Citronellolis Strain SLP6 Enhances the Phytoremediation Efficiency of Helianthus Annuus in Copper Contaminated Soils under Salinity Stress. Plant and Soil 457 (1), 241–253. doi:10.1007/s11104-020-04734-7
Singh, A., and Cameotra, S. (2013). Rhamnolipids Production by Multi-Metal-Resistant and Plant-Growth-Promoting Rhizobacteria. Appl. Biochem. Biotechnol. 170, 1038–1056. doi:10.1007/s12010-013-0244-9
Song, Y., Shahir, S, and Abd Manan, F. (2021). Bacterial Inoculant-Assisted Phytoremediation of Heavy Metal-Contaminated Soil: Inoculant Development and the Inoculation Effects. Biologia 76 (9), 2675–2685. doi:10.1007/s11756-021-00804-y
Souza, M., Sadowsky, M., and Lawrence, W. (1996). Atrazine Chlorohydrolase from Pseudomonas Sp. Strain ADP: Gene Sequence, Enzyme Purification, and Protein Characterization. J. Bacteriol. 178, 4894–4900. doi:10.1128/jb.178.16.4894-4900.1996
Stepanova, A. Yu., Orlova, E. V., Teteshonok, D. V., and Dolgikh, Y. I. (2016). Obtaining Transgenic Alfalfa Plants for Improved Phytoremediation of Petroleum-Contaminated Soils. Russ. J. Genet. Appl. Res. 6 (6), 705–711. doi:10.1134/S2079059716060083
Su, Z. H., Xu, Z. S., Peng, R. H., Tian, Y. S., Zhao, W., Han, H. J., et al. (2012). Phytoremediation of Trichlorophenol by Phase II Metabolism in Transgenic Arabidopsis Overexpressing a Populus Glucosyltransferase. Environ. Sci. Technol. 46 (7), 4016–4024. doi:10.1021/es203753b
Sylvestre, M., Sirois, M., Hurtubise, Y., Bergeron, J., Ahmad, D., Shareck, F., et al. (1996). Sequencing of Comamonas Testosteroni Strain B-356-Biphenyl/chlorobiphenyl Dioxygenase Genes: Evolutionary Relationships Among Gram-Negative Bacterial Biphenyl Dioxygenases. Gene 174 (2), 195–202. doi:10.1016/0378-1119(96)00039-x
Taira, K., Hirose, J., Hayashida, S., and Furukawa, K. (1992). Analysis of Bph Operon from the Polychlorinated Biphenyl-Degrading Strain of Pseudomonas pseudoalcaligenes KF707. J. Biol. Chem. 267 (7), 4844–4853. doi:10.1016/s0021-9258(18)42908-0
Tang, L., Mao, B., Li, Y., Lv, Q., Zhang, L., Chen, C., et al. (2017). Knockout of OsNramp5 Using the CRISPR/Cas9 System Produces Low Cd-Accumulating Indica rice without Compromising Yield. Scientific Rep. 7 (1), 1–12. doi:10.1038/s41598-017-14832-9
Tarla, D. N., Davis, L. C., Nurzhanova, A., and Pidlisnyuk, V. (2020). Phytoremediation and Bioremediation of Pesticide-Contaminated Soil. Appl. Sci. 10 (4), 1217. doi:10.3390/app10041217
Tussipkan, D., and Manabayeva, S. A. (2021). Employing CRISPR/Cas Technology for the Improvement of Potato and Other Tuber Crops. Front. Plant Sci. 12, 747476. doi:10.3389/fpls.2021.747476
Varjani, S., and Upasani, V. N. (2021). Bioaugmentation of Pseudomonas aeruginosa NCIM 5514 – A Novel Oily Waste Degrader for Treatment of Petroleum Hydrocarbons. Bioresour. Techn. 319, 124240. doi:10.1016/j.biortech.2020.124240
Varjani, S., Upasani, V. N., and Pandey, A. (2020). Bioremediation of Oily Sludge Polluted Soil Employing a Novel Strain of Pseudomonas aeruginosa and Phytotoxicity of Petroleum Hydrocarbons for Seed Germination. Sci. Total Environ. 737, 139766. doi:10.1016/j.scitotenv.2020.139766
Vasil'eva, G. K., and Strizhakova, E. R. (2007). Bioremediation of Soils and Sediments Polluted by Polychlorinated Biphenyls. Mikrobiologiia 76 (6), 725–741.
Wackett, L. M., Sadowsky, B. M., and Shapir, N. (2002). Biodegradation of Atrazine and Related s-triazine Compounds: From Enzymes to Field Studies. Appl. Microbiol. Biotechnol. 58 (11), 39–45. doi:10.1007/s00253-001-0862-y
Wang, D., Lin, J., Lin, J., Wang, W., and Li, S. (2019). Biodegradation of Petroleum Hydrocarbons by Bacillus Subtilis BL-27, a Strain with Weak Hydrophobicity. Molecules 24 (17), 3021. doi:10.3390/molecules24173021
Wang, H., Chen, X., Xing, X., Hao, X., and Chen, D. (2010). Transgenic Tobacco Plants Expressing atzA Exhibit Resistance and strong Ability to Degrade Atrazine. Plant Cel Rep 29 (12), 1391–1399. doi:10.1007/s00299-010-0924-7
Wang, J. D., Li, X. X., and Qu, C. T. (2017). Exploration of Up-Regulated Key Proteins in Pseudomonas Aeruginosa for High-Efficiency Petroleum Degradation by Proteomic Analysis. Curr. Microbiol. 74 (10), 1178–1184. doi:10.1007/s00284-017-1302-2
Wang, L., Samac, D. A., Shapir, N., Wackett, L. P., Vance, C. P., Olszewski, N. E., et al. (2005). Biodegradation of Atrazine in Transgenic Plants Expressing a Modified Bacterial Atrazine Chlorohydrolase (atzA) Gene. Plant Biotechnol. J. 3 (5), 475–486. doi:10.1111/j.1467-7652.2005.00138.x
Wang, H., Wang, Q., Liu, Y., Liao, X., Chu, H., Chang, H., et al. (2021). PCPD: Plant Cytochrome P450 Database and Web-Based Tools for Structural Construction and Ligand Docking. Synth. Syst. Biotechnol. 6 (2), 102–109. doi:10.1016/j.synbio.2021.04.004
Wang, X., de Souza, M. F., Li, H., Filip, M., Tack, G., Yong Sik, O., et al. (2021). Zn Phytoextraction and Recycling of Alfalfa Biomass as Potential Zn-Biofortified Feed Crop. Sci. Total Environ. 760, 143424. doi:10.1016/j.scitotenv.2020.143424
Wang, Y., Ren, H., Pan, H., Liu, J., and Zhang, L. (2015). Enhanced Tolerance and Remediation to Mixed Contaminates of PCBs and 2,4-DCP by Transgenic Alfalfa Plants Expressing the 2,3-Dihydroxybiphenyl-1,2-Dioxygenase. J. Hazard. Mater. 286, 269–275. doi:10.1016/j.jhazmat.2014.12.049
Wang, Y., Wang, C., Ren, H., Jia, B., and Zhang, L. (2014). Effectiveness of Recombinant Protein AlnA in Enhancing the Extractability of Polychlorinated Biphenyls from Contaminated Soils. J. Hazard. Mater. 279, 67–74. doi:10.1016/j.jhazmat.2014.06.063
Wang, Z., and Şakiroğlu, M. (2021). “The Direct Diploid Progenitor of Autotetraploid Alfalfa,” in The Alfalfa GenomeLong-Xi Yu and Chittaranjan Kole (Cham: Springer International Publishing), 29–42. doi:10.1007/978-3-030-74466-3_3
Wangeline, A., Burkhead, J., Hale, K., Lindblom, S., Terry, N., and Pilon-Smits, E. (2004). Overexpression of ATP Sulfurylase in Indian Mustard: Effects on Tolerance and Accumulation of Twelve Metals. J. Environ. Qual. 33, 54–60. doi:10.2134/jeq2004.5400
Wu, Z., Bañuelos, G. S., Lin, Z-Q., Liu, Y., Yuan, L., Yin, X., et al. (2015). Biofortification and Phytoremediation of Selenium in China. Front. Plant Sci. 6, 136. doi:10.3389/fpls.2015.00136
Yamada, T., Ishige, T., Shiota, N., Inui, H., Ohkawa, H., and Ohkawa, Y. (2002). Enhancement of Metabolizing Herbicides in Young Tubers of Transgenic Potato Plants with the Rat CYP1A1 Gene. Theor. Appl. Genet. 105 (4), 515–520. doi:10.1007/s00122-002-0961-x
Yang, C-h., Zhang, Y., and Huang, C-f. (2019). Reduction in Cadmium Accumulation in Japonica rice Grains by CRISPR/Cas9-mediated Editing of OsNRAMP5. J. Integr. Agric. 18 (3), 688–697. doi:10.1016/s2095-3119(18)61904-5
Yang, Y., Ding, J., Chi, Y., and Yuan, J. (2020). Characterization of Bacterial Communities Associated with the Exotic and Heavy Metal Tolerant Wetland Plant Spartina Alterniflora. Scientific Rep. 10 (1), 1–11. doi:10.1038/s41598-020-75041-5
Ye, J. Y., Zhang, J. B., Gao, J. G., Li, H. T., Liang, D., and Liu, R. M. (2016). Isolation and Characterization of Atrazine-Degrading Strain Shewanella Sp. YJY4 from Cornfield Soil. Lett. Appl. Microbiol. 63 (1), 45–52. doi:10.1111/lam.12584
Yin, S., Wang, Y., and Nan, Z. (2018). Genetic Diversity Studies of Alfalfa Germplasm (Medicago Sativa L. Subsp Sativa) of United States Origin Using Microsatellite Analysis. Legume Res. 41 (2), 202–207. doi:10.18805/LR-358
Zhang, Y., Liu, J., Zhou, Y., Gong, T., Wang, J., and Ge, Y. (2013). Enhanced Phytoremediation of Mixed Heavy Metal (Mercury)-organic Pollutants (Trichloroethylene) with Transgenic Alfalfa Co-expressing Glutathione S-Transferase and Human P450 2E1. J. Hazard. Mater. 260, 1100–1107. doi:10.1016/j.jhazmat.2013.06.065
Zhang, Y., and Liu, J. (2011). Transgenic Alfalfa Plants Co-expressing Glutathione S-Transferase (GST) and Human CYP2E1 Show Enhanced Resistance to Mixed Contaminates of Heavy Metals and Organic Pollutants. J. Hazard. Mater. 189 (1), 357–362. doi:10.1016/j.jhazmat.2011.02.042
Keywords: alfalfa (Medicago sativa L.), genotypic diversity, phytoremediation, POPs, petroleum, heavy metals
Citation: Tussipkan D and Manabayeva SA (2022) Alfalfa (Medicago Sativa L.): Genotypic Diversity and Transgenic Alfalfa for Phytoremediation. Front. Environ. Sci. 10:828257. doi: 10.3389/fenvs.2022.828257
Received: 03 December 2021; Accepted: 25 January 2022;
Published: 04 March 2022.
Edited by:
Liangang Mao, Institute of Plant Protection (CAAS), ChinaReviewed by:
Vineet Kumar, National Environmental Engineering Research Institute (CSIR), IndiaNeerja Srivastava, Chhatrapati Shahu Ji Maharaj University, India
Copyright © 2022 Tussipkan and Manabayeva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuga A. Manabayeva, bWFuYWJheWV2YUBiaW9jZW50ZXIua3o=
 Dilnur Tussipkan
Dilnur Tussipkan Shuga A. Manabayeva
Shuga A. Manabayeva