- Shandong Provincial Key Laboratory of Applied Microbiology, Ecology Institute of Shandong Academy of Sciences, Qilu University of Technology (Shandong Academy of Sciences), Ji’nan, China
With the increasing demand for diesel in various countries, the ecological pollution caused by the improper use, storage, and accidental leakage of diesel needs to be addressed urgently. As an environmentally friendly and cost-effective method, bioremediation generally uses various microorganisms to remove pollutants from the environment. Here, the strain KJ-1, obtained through an enrichment culture using n-dodecane from oil-contaminated soil near a gas station as the substrate, was identified as Acinetobacter vivianii according to its morphology, biochemistry, and molecular biology. The isolate KJ-1 was able to use diesel as a sole carbon source and more than 40% of diesel was degraded after 12 days of incubation with strain KJ-1 in mineral salts medium. The most suitable diesel concentration and nitrogen source concentrations were 4,140 mg/L and 350–700 mg/L, respectively, for diesel degradation and bacterial growth. The optimal initial pH and temperature for strain KJ-1 growth and diesel degradation were 6.5–8.0 and 20–37°C, respectively. To investigate the diesel-degrading mechanisms of this strain, the complete genome was sequenced and annotated. The complete genome consists of one chromosome with a total length of 3,927,757 base pairs and a G + C content of 41.5%. The genes related to the two-component regulatory system and alkane degradation were analyzed. In addition, two putative alkane monooxygenases were analyzed, and the protein sequences were characterized and compared with other AlkBs in Acinetobacter spp. using sequences downloaded from NCBI. The results demonstrated that A. vivianii KJ-1 may be particularly useful for future bioremediation of diesel-polluted soil.
Introduction
As an important energy and fuel for the development of countries around the world, and due to rapid social economic development, the demand for such petroleum-derived products as diesel and gasoline is increasing. However, soil and groundwater contamination due to improper usage, storage, and disposal of petroleum-derived products, in particular diesel oil, as well as accidental leakage, is a major environmental problem that has attracted the attention of experts in various countries (Bunger et al., 2012; Xu et al., 2019). Diesel is a complex group of compounds consisting mainly of alkanes, cycloalkanes, olefins, and mono- to polycyclic aromatic hydrocarbons, which have caused great damage to the natural environment and even to human health, and are currently classified as hazardous wastes (Atlas and Hazen, 2011; Steiner et al., 2016). Fortunately, there are many indigenous microorganisms in the natural environment that can respond to environmental changes rapidly and use hydrocarbons as the sole source of carbon and energy, which plays an important role in pollutant degradation and environmental bioremediation (Xu et al., 2018; Keramea et al., 2021). At present, more than 90 genera and 200 species of bacteria have been reported to possess the ability to degrade diesel, including Bacillus (Gao et al., 2021; Khandelwal et al., 2022), Pseudomonas (Phulpoto et al., 2021), Alcanivorax (Kadri et al., 2018b), and Acinetobacter (Akinde and Obire, 2008; Zhang et al., 2014; Ho et al., 2020). Ultimately, diesel degradation by microorganisms varies and depends highly on the microbe’s ability to utilize the hydrocarbon components or byproducts of diesel degradation (Palanisamy et al., 2014; Liu et al., 2016). It is an effective way to find novel and efficient diesel-degrading microorganism to tackle the current severe pattern of diesel pollution.
The Acinetobacter spp. are aerobic gram-negative bacteria, which are rod-shaped and belong to g-proteobacteria in the family Moraxellaceae (Kostka et al., 2011). This Genus is widely distributed in a variety of environments and can survive on different types of surfaces, indicating that it is well adapted to different environments and shows considerable metabolic diversity (Jiao et al., 2016). It has been shown that the degradation of diesel compounds with Acinetobacter bacteria usually takes place under aerobic conditions, with three possible n-alkane degradation pathways involving different hydroxylases, commonly encoded by the genes alkB, almA, p450, and ladA (Varjani and Upasani, 2017). Among these hydroxylases, alkane-1 monooxygenase (AlkB) is the most common and most widely distributed of the alkane hydroxylases, acting mainly on medium and long chain alkanes (Park et al., 2017). It is engaged in the first step of the alkane terminal oxidative degradation pathway, and catalyzes the degradation of alkanes by oxidation of their terminal methylated carbons to 1-alkanols under aerobic conditions (Pal and Sengupta, 2020).
According to the latest database (July of 2022) on NCBI (http://www.ncbi.nlm.nih.gov), 86 Acinetobacter species have their genomes sequenced and about 70 of them carry genes involved in alkane degradation. However, there is little information about the genome of Acinetobacter vivianii. In this study, an Acinetobacter vivianii strain was isolated from diesel-contaminated soil from a gas station in Jinan, Shandong Province, China, and identified and characterized. To further understand its genetic traits for alkane degradation, the complete genome was sequenced and reported, and genes related to alkane degradation were analyzed.
Material and methods
Chemicals and media
The 0# diesel was purchased from a gas station owned by Shandong Kexin Oil Products Co., Ltd. in Jinan, China. Its cetane index was 56, and the density was 827.9 g/cm3 (20°C). The total pollutant content and the total sulfur content were 4.5 mg/kg and 4.0 mg/kg, respectively. n-Hexane, anhydrous sodium sulfate, and other chemicals and solvents were obtained from Sinopharm Chemical Reagent Co., Ltd. and Tianjin Guangfu Technology Development Co., Ltd., at the high est analytical reagent grade.
NB medium contained 10.0 g of tryptone, 3.0 g of beef extract, and 5.0 g of NaCl in 1 L of water at pH 7.0–7.2.
Mineral salts medium (MSM) contained 1 g of NH4NO3, 1 g of NaCl, 1.5 g of K2HPO4, 0.5 g of KH2PO4, and 0.2 g of MgSO4·7H2O in 1 L of water, with the pH adjusted to 7.2.
To prepare the diesel mineral medium, diesel was syringe filtered for sterilization with a 0.2-μm organic filter membrane (Tianjin Branch Billion Lung Experimental Equipment Co., Ltd.), and according to the experiment, some diesel was added to the sterile mineral medium as the sole carbon source.
Isolation, screening and identification of alkane-degrading bacteria
Samples used for bacterial isolation were obtained from the oil-contaminated soil around the gas station of Shandong Kexin Oil Products Co., Ltd. (Jinan, China), which has been in business for 30 years. First, 10 g of contaminated soil was added to a 500-ml flask containing mineral media with n-dodecane (3,765 mg/L) as the sole carbon source. The flasks were incubated at 30°C and 160 rpm for 7 days, and then 2 ml of culture was transferred to another flask with fresh media for another 7 days of cultivation. After five sequential rounds were conducted, 1 ml of culture was used to isolate the alkane-degrading bacteria by a standard dilution plating technique. Alkane degradation was assessed by culturing in MSM broth supplemented with n-dodecane as the sole carbon source at 30°C and 160 rpm. The alkane-degrading bacteria were identified based on morphological, physiological, and biochemical characterizations according to Bergey’s Manual of Systematic Bacteriology. Molecular taxonomy was based on PCR amplification and sequencing of 16S rDNA with the universal primers 7F (5′-CAGAGTTTGATCCTGGCT-3′) and 1540R (5′-AGGAGGTGATCCAGCCGCA-3′).
Growth of KJ-1 in media supplemented with diesel
For analyzing the diesel degradation ability, strain KJ-1 was cultured and quantified by monitoring the concentration of the diesel in mineral salts media with diesel as the sole carbon source. Strain KJ-1 was grown in NB broth on a rotary shaker (160 rpm) for 12 h at 30°C, then cells were collected by centrifugation (5,000 rpm for 5 min), washed twice, and re-suspended to an OD600 = 0.7 (Lambda Bio Spectrophotometer, Perkin Elmer, United States) in sterile water. The cell suspension (approximately 1×108 cell/mL) was used to inoculate (2% v/v) 100 ml-flasks of MSM containing diesel as the sole carbon source and incubated at 30°C on a rotary shaker at 160 rpm. The cultures were taken out at different time points to measure the cell concentrations and the diesel residues. To confirm the individual effects of temperature (20°C, 25°C, 30°C, 31°C, 33°C, 35°C, 37°C, 39°C, and 40°C), pH (4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, and 9.0), initial concentration of diesel (2.07 g/L, 4.14 g/L, 8.28 g/L, 12.42 g/L, and 16.56 g/L), and initial concentration of nitrogen (200 mg/L, 350 mg/L, 700 mg/L, 1,000 mg/L, and 1,400 mg/L) on the degradation of diesel with strain KJ-1, four experiments were established. A non-inoculated control was set and each treatment was replicated three times.
The diesel in the liquid was extracted with n-hexane as a volume ratio of 1:1. The flasks were subjected to oscillation extraction in a shaker (25°C, 150 rpm) for 30 min, and then liquid-liquid extraction using a separating funnel. All steps were repeated three times. The concentration of the diesel fuel in the liquid was measured by gas chromatography using the method described in “Soil and Sediment—Determination of Petroleum Hydrocarbons (C10-C40)—Gas Chromatography (HJ 1021–2019)” and bacterial growth was monitored by UV spectrophotometer using an absorbance of 600 nm (Ho et al., 2020).
The degradation rate of diesel according to the following formula:
where: ƞ-diesel degradation rate; C1-residual amount of diesel fuel in the blank control; C2-remaining amount of diesel fuel in the sample.
In this study, the diesel concentration was determined through a gas chromatography-flame ionization detection equipped with an Agilent HP-5 capillary column (30 m × 0.32 mm × 0.25 μm). The analytical procedures were as follows: carrier gas (N2) flow rate of 1.5 ml/min, air flow rate of 300 ml/min, H2 flow rate of 30 ml/min, makeup gas flow rate of 28 ml/min, and injection volume of 1 μL. The oven temperature program was as follows: start at 40°C, held for 2 min, increase to 350°C at 10°C/min, and hold for 10 min. The inlet temperature and the detector temperature were both 300°C.
Genome sequencing analysis
A whole-genome shotgun strategy was adopted to construct the genome library, and sequencing was carried out using Illumina Hiseq and PacBio technology (Caporaso et al., 2012; Rhoads and Au, 2015). Data generated from the PacBio and Illumina platforms were used for bioinformatics analysis, all of which were performed using the free online platform Majorbio Cloud Platform (www.majorbio.com) from Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Ren et al., 2022). The software SOAPdenovo v2.04 was used for the assembly and the results were optimized using GapCloser v1.12. The coding sequences (CDS) in the genome were predicted using Glimmer (http://ccb.jhu.edu/software/glimmer/index.shtml), GeneMarkS, and Prodigal. The tRNA and rRNA sequences were predicted using tRNA-scan-SE and Barrnap, respectively. The nucleic acid and amino acid sequences of the functional genes were obtained by prediction. Using BLAST software, the amino acid sequences of the strains were compared using databases such as COG, GO, KEGG, NR and Swiss-Prot to obtain the corresponding functional annotation information (Sayers et al., 2011). Genomic annotation circles were mapped using the software Proksee (formerly CGView, https://proksee.ca/) and the secondary metabolite synthesis gene clusters were predicted using antiSMASH v6.1.0 (antiSMASH bacterial version (secondarymetabolites.org)).
Homology comparison of alkane monooxygenase
The sequences of alkane monooxygenase in different Acinetobacter spp. were obtained from NCBI database (http://www.ncbi.nlm.nih.gov). And phylogenetic tree of kinds of alkane monooxygenases was constructed by maximum likelihood method using MEGA 7.0 software. The sequences of alkane-1 monooxygenase in KJ-1 were aligned with other AlkB1 sequences downloaded from NCBI database by ClustalW online alignment tool (https://www.genome.jp/tools-bin/clustalw) and then plotted using ESPript3.0 online tool (https://espript.ibcp.fr/ESPript/ESPript/index.php).
Results and discussion
Isolation and identification of alkane-degrading strain
Seven bacterial strains (KJ-1, KJ-2, KJ-3, KJ-4, KJ-5, KJ-6 and KJ-7) were isolated from oil-contaminated soil with enrichment cultures. These strains showed good degradation ability on n-dodecane, and the degradation rate of n-dodecane was more than 60%. Among them, strain KJ-1 was the best degrader and was selected for further study.
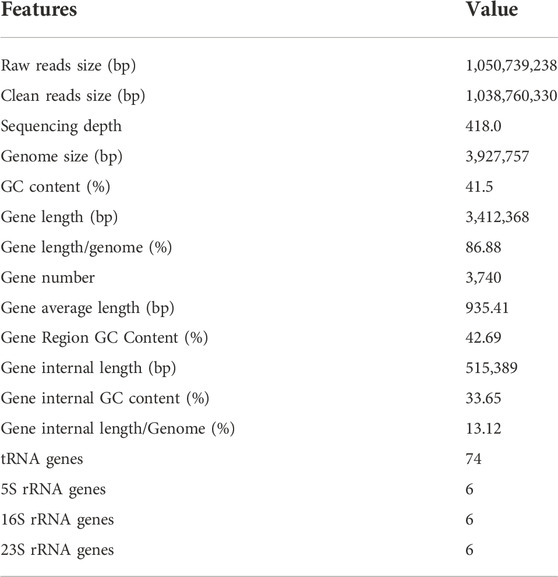
Strain KJ-1 was a gram-negative, aerobic, asporous, short-rod shaped bacterium with a size of 0.8 μm × 2.2 μm observed by scanning electron microscope (HITACHI SUB8011, Japan) (Figure 1A). The single colony of strain KJ-1 on NB agar plates was round, with smooth edge, light yellow and white, opaque and wet surface (Figure 1B). The strain KJ-1 showed negative oxidase, nitrate reductase, and positive for catalase and starch hydrolysis in biochemical test.
FIGURE 1. Transmission electron micrograph of strain KJ-1 (A) and physiological and biochemical morphology of strain KJ-1 on MSM agar medium (B)
A 16S rRNA fragment was amplified from strain KJ-1 and sequenced. Then, a phylogenetic tree was constructed based on the 16S rRNA gene sequence (GenBank accession: MZ994727). The 16S rRNA gene sequence of strain KJ-1 showed that strain KJ-1 was very similar to Acinetobacter vivianii NIPH 2168, and the corresponding phylogenetic tree analysis supported a strong relationship between KJ-1 and members of Acinetobacter spp. (Figure 2). According to the morphological, physiological, and biochemical characterization, and the 16S rRNA sequence analysis, strain KJ-1 was identified as Acinetobacter vivianii. Acinetobacter bacteria are widely found in nature and can survive on wet and dry surfaces, and it is the most common genera that can degrade hydrocarbons. Many strains of this genus were reported to be involved in alkane degradation in previous studies, such as A. junii (Ohadi et al., 2017), A. mesopotamicus (Acer et al., 2020), A. venetianus (Fondi et al., 2016), A. calcoaceticus (Singkham-In and Chatsuwan, 2022), and A. baumannii (Hossain et al., 2022). In this study, we isolated a strain, KJ-1, from a gas station from Shandong Kexin Oil Products Co., Ltd., Jinan, China. This strain was able to grow in MSM with n-dodecane as the sole carbon source and degrade n-dodecane very efficiently. It is closely homologous to Acinetobacter vivianii NIPH 2168.
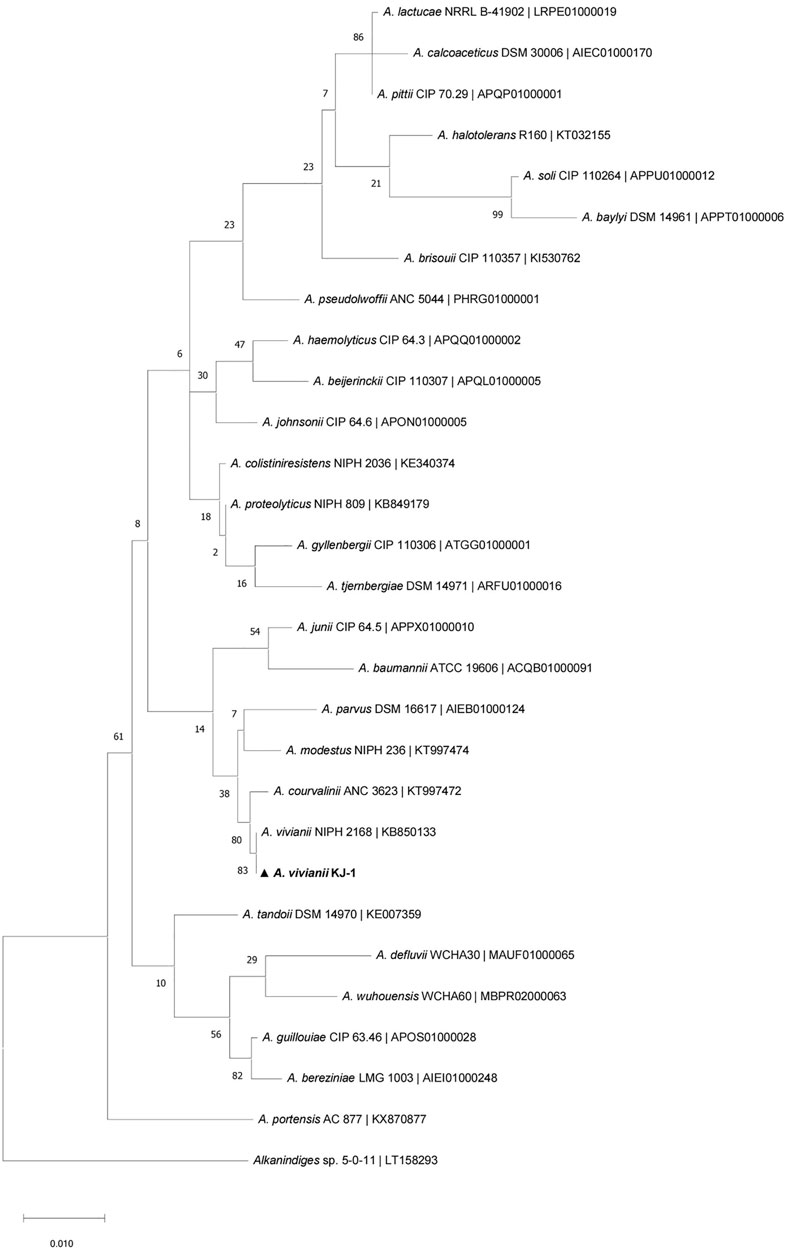
FIGURE 2. Phylogenetic tree of Acinetobacter sp. KJ-1 and its closest related species based on 16S rRNA gene sequence similarity using maximum likelihood method. Data for all 16S rRNA genes were downloaded from NCBI (http://www.ncbi.nlm.nih.gov). The outgroup used was Alkanindiges sp. 5-0-11. Bootstrap value represents 1,000 replicates and random seeding. Tree was built using Mega 7.0.26.
Diesel degradation by KJ-1
Alkanes are the main components of diesel oil. To detect the degradation ability of strain KJ-1 on diesel oil, the cells were cultured in mineral salts media with diesel as the sole carbon source at 30°C. According to the growth curve in MSM with diesel as the carbon source, 4–6 days were required to reach the maximum value of the specific growth rate, after which time the culture grew slowly (Figure 3). More than 45% of the diesel in the media (12,418.5 mg/L) was degraded, which suggests the strong diesel degradation ability of strain KJ-1 under the culture conditions compared to A. calcoaceticus CA16 and A. haemolyticus strain 2SA (Ho et al., 2020; Diallo et al., 2021). As shown in Figure 3, the growth of strain KJ-1 was positively correlated with the degradation of diesel during days 2–20. From days 22–28, the OD600 tended to a steady state, and although its value decreased from days 26–28, there was no significant difference, and a balance may have been struck between bacterial growth and cell autolysis. According to the diesel degradation rate from day 22–28, it was speculated that there was no significant change in the growth of strain KJ-1; however, various enzymes related to diesel degradation were still functioning. Because C12 is one of the components in diesel, the concentration of C12 and other alkanes in diesel oil at different time points were analyzed (Supplementary Figure S1). In the treatment group, the concentration of C12 decreased gradually in the first 4 days and increased on the sixth day. Therefore, it was speculated that the increase of C12 was due to the degradation of long-chain alkanes. This phenomenon also pertained to C10, C11, and C13. The different long chain alkanes (C14 to C26 in diesel oil) decreased compared to the control culture without strain KJ-1 (data unpublished). It was speculated that this bacterium has genes involved in long chain alkane degradation. This was confirmed through genomic analysis. The putative genes involved in alkane degradation are shown in Supplementary Table S2.
With diesel as the sole carbon source, its concentration has an important effect on the growth and metabolism of microorganisms. If the concentration is too low, the strain cannot obtain enough carbon and its growth and reproduction are affected; if the concentration is too high, the requirements for sufficient oxygen and nutrients are increased, thus affecting normal growth. Here, the effect of different initial diesel concentrations on the degradation ability of strain KJ-1 was analyzed (Figure 4A). When the initial concentration of diesel was 2,070 mg/L to 12,418.5 mg/L, there was no significant difference in the growth or degradation of diesel of strain KJ-1. The OD600 value increased slowly when the initial concentration was between 24,837 mg/L to 33,116 mg/L. At 28,976.5 mg/L, the OD600 was the highest; however, the degradation rate decreased compared to 24,837 mg/L. Therefore, there is a point between 24,837 mg/L to 33,116 mg/L at which the growth rate of the strain decreased, while the initial concentration increased. When the concentration was 37,273.5 mg/L, the degradation rate was the lowest, at only 22.89%, suggesting that high concentrations of diesel had a certain toxicity to cells.
FIGURE 4. The growth of the strain KJ-1 under different conditions and its degradation effect on diesel. (A), the initial diesel concentration (mg/L); (B), the initial concentration of nitrogen source (mg/L); (C), incubation temperature; (D), the initial pH in MSM media.
Nitrogen, as an essential element of proteins, nucleic acids, and other nitrogen compounds of living organisms, plays a vital role in microbial growth and product synthesis (Kahraman et al., 2017). Ammonium nitrate (NH4NO3) includes two forms of nitrogen, ammonium and nitric acid, which can supply two different nitrogen sources at the same time. Here, different concentrations of ammonium nitrate (NH4NO3) were chosen as the inorganic nitrogen source to analyze the degradation of diesel by A. vivianii KJ-1 (Figure 4B). Under the influence of different concentrations of ammonium nitrate, the degradation rate of diesel was above 50% in all cultures of strain KJ-1. Moreover, there was no difference in the diesel degradation rate when the concentration of the nitrogen source was 350–700 mg/L. However, when the concentration of the nitrogen source exceeded 700 mg/L, the growth of strain KJ-1 increased and the degradation rate of diesel decreased.
The culture temperature affects the growth, reproduction, and metabolic activity of microorganisms. In this experiment, temperature had a significant effect on growth and diesel degradation of strain KJ-1. The optimal growth and degradation temperature was analyzed in the 20°C–37°C range. In this range, 35°C was the best incubation temperature for both the growth of strain KJ-1 and the degradation of diesel oil. Strain KJ-1 grew and degraded diesel in a wide range of temperatures. However, when the culture temperature was higher than 39°C, the growth of strain KJ-1 and the degradation of diesel were inhibited (Figure 4C).
The pH value is closely related to the essential activities and metabolism of microorganisms. The effect of the initial pH (pH range 4.0–9.0) was investigated in MSM with 0.5% diesel as the sole carbon source. As indicated in Figure 4D, strain KJ-1 grew well on MSM with diesel at an initial pH in the range of 4.0–9.0. The pH had a significant effect on biodegradation of diesel by strain KJ-1 (one-way ANOVA, F10, 22 = 5.986, p < 0.0001). With an initial pH value of 6.5–8.0, the diesel degradation rate was above 60%. According to the results, A. vivianii. KJ-1 could grow and use diesel as a carbon source in a wide pH range.
Genomic characteristics of the isolate KJ-1
The genomic properties of strain KJ-1 are shown in Table 1 and Figure 5. The complete genome consists of one chromosome with a total length of 3,927,757 base pairs (bp) and a G + C content of 41.5%. The genome contained 3,740 predicted coding sequences, with 3,648 coding for protein sequences and 92 coding for RNAs. Of the entire 3,648 CDSs, 1,899 (52.56%) of the CDSs were identified as participating in 212 pathways, according to the KEGG database (http://www.genome.jp/kegg/) (Supplementary Figure S2). A total of 635 genes were found to focus on biosynthesis of secondary metabolites, fatty acids, amino acids, antibiotics, and similar pathways. Among these, the synthesis of various antibiotics, including streptomycin, penicillin, cephalosporin, novobiocin, prodigiosin, and vancomycin, indicated that the strain has a certain advantage in competition with other microorganisms in the environment. There were 235 CDSs involved in the degradation of various substances, particularly in the pathway of xenobiotics, such as aromatic compounds, benzoates, naphthalenes, alkanes, atrazine, and polycyclic aromatic hydrocarbons, indicating that this strain could be used as a degrader for various organic pollutants. In addition, according to the clusters of orthologous genes (Huerta-Cepas et al., 2019), 503 genes are responsible for cellular processes and signaling, 499 genes for information storage and processing, and 1,132 genes for metabolism, while the remaining 916 (30.61%) sequences were found to have unknown functions designated as hypothetical proteins (Supplementary Figure S3).
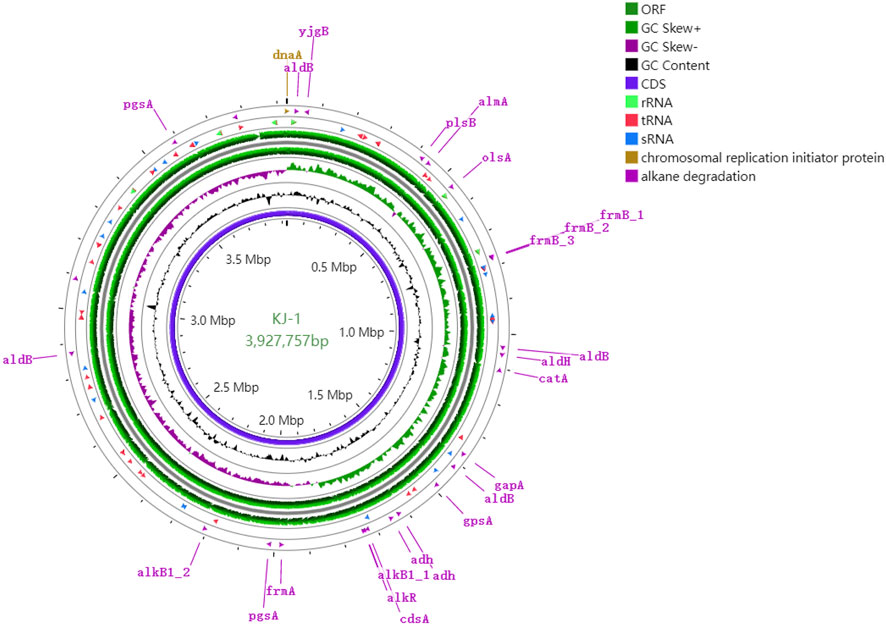
FIGURE 5. Circular representation of genome and features of the KJ-1. The contents of the featured rings (starting with the outermost ring to the center) are as follows. Ring 1: genes involved in alkane degradation, combined forward and reverse strand; Ring 2: RNA (including tRNA, sRNA and rRNA) combined forward and reverse strands; Ring 3 and 4: CDS in forward and reverse strands; Ring 5: GC skew plot, values above average is depicted in green and below average in purple; Ring 6: plot of GC content; Ring 7: Combined ORFs in forward and reverse strands; Ring 8: sequence ruler. The figure was produced using Proksee (https://proksee.ca/).
The genes associated with the two-component regulatory system were environmental stress factors, which are of great significance for enabling bacterial strains to respond to changes in environmental conditions (Bhagirath et al., 2019). When specific changes in the extracellular environment are detected, histidine kinases perform autophosphorylation, transferring the phosphate groups from adenosine triphosphate to specific histidine residues. The corresponding reaction regulator then catalyzes the transfer of the phosphate group to aspartic acid residues in the reception domain of the reaction regulator, resulting in a series of regulatory reactions (Bhagirath et al., 2019). Here, 59 genes in strain KJ-1 genome were involved in two-component regulatory systems, including 34 genes as regulators, 19 genes for sensors, and 6 genes for hybrid types. The genes encoding parts of histidine kinases and response regulatory proteins are shown in Supplementary Table S1. These systems include EnzC/OmpR to maintain osmotic pressure of cells, Pho/PhoB to regulate the phosphate levels and quorum sensing systems, CusS/CusR to respond to heavy metal ions (Cu2+), KdpE/KdpD for potassium transport, GlnG/GlnL for nitrogen regulation, PilS/PilR to regulate the production of antibacterial substance, and EvgA/EvgS and RstA/RstB for drug resistance. Therefore, the strain KJ-1 adapted to the changing environment by using the two-component signal transduction system as a degrader to move alkane and other organic substance into different environments.
Homology comparison of alkane monooxygenase
Because of the presence of multiple alkane degradable related enzymes, the hydrocarbon-consuming bacteria are capable to utilize kinds of alkanes (Nie et al., 2014; Williams et al., 2022). The genome of A. vivianii KJ-1 contains multiple hydrocarbon degrading genes, therefore it can grow on MSM media with n-dodecane or diesel as the sole carbon source (Supplementary Table S2 and Figure 5). Here, the genome of A. vivianii KJ-1 encodes 25 enzymes that may be involved in alkane degradation. Among these putative enzymes, two AlkB sequences, named AlkB1_1 (encoded with gene1663) and AlkB1_2 (encoded with gene2103), were found based on genome-wide annotation analysis. Here, gene1663 (alkB1_1) and gene2103 (alkB1_2) encode the putative AlkB monooxygenases, comprising 414 and 397 amino acids with the estimated molecular weight of 47.1 kDa and 45.3 kDa, separately. The clustering analysis of alkane monooxygenases in KJ-1 with other same class of enzymes downloaded from NCBI (http://www.ncbi.nlm.nih.gov) were performed (Figure 6). According to the phylogenetic tree of amino acid sequence, AlkB1_1 was most homologous to alkane monooxygenase from Acinetobacter sp. (GeneBank No. WP_047427165.1), while AlkB1_2, was close to sequence from Acinetobacter sp. YK3 (GeneBank No. WP_069578407.1).

FIGURE 6. Phylogenetic relationship of Alkane monooxygenase of A. vivianii KJ-1 with other closely related species using maximum likelihood method. The outgroup used was AlkB protein from Alkanindiges illinoisensis (WP 134243320.1). The bootstrap value represents 1,000 replicates, and tree was built using Mega 7.0.26.
The sequences of alkane-1 monooxygenase in KJ-1 were aligned with other AlkB1 sequences downloaded from NCBI database by ClustalW online alignment tool (https://www.genome.jp/tools-bin/clustalw) and then plotted using ESPript3.0 online tool (https://espript.ibcp.fr/ESPript/ESPript/index.php) (Figure 7). The alignment results showed that the overall similarity of the 10 AlkB protein sequences was 76.67%, and the region, located at 57-371aa and 52-365aa of the two AlkB sequences of KJ-1, has relatively high consistency, suggesting a feature that is relatively evolutionarily conserved. This region is consistent with the cl00615 (PSSMID 294412) superfamily domain described in a previous report (Lu et al., 2020). This conserved domain encodes an alkane hydroxylase responsible for the initial oxidation of inactivated alkanes, using the oxygen rebound mechanism to hydroxylate alkanes (Kadri et al., 2022). This mechanism involves the homolytic cleavage of the C-H bond by an electrophilic metal-oxo intermediate to generate a substrate-based radical (Naing et al., 2013). This domain family has a broad hydrophobic region, and is therefore able to span the membrane bilayer at least twice, which enables bacteria to make efficient use of petroleum hydrocarbons (Kadri et al., 2018a).
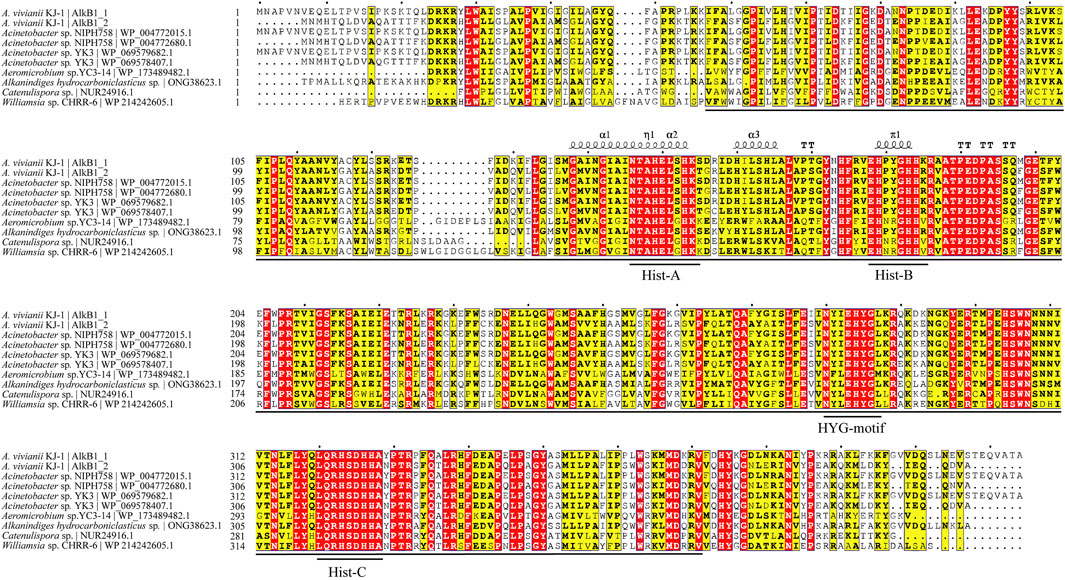
FIGURE 7. Sequence alignment of homologous proteins of AlkB. Fully conserved amino acids are highlighted in red, and semi-conserved amino acids are highlighted in yellow. Conserved domains are marked with horizontal lines. The η symbol refers to a 310-helix. α-helices, 310-helices and π-helices are displayed as medium, small and large squiggles, respectively. β-strands are rendered as arrows, strict β-turns as TT letters and strict α-turns as TTT. The amino acid sequences were aligned by the ClustalW online alignment tool (https://www.genome.jp/tools-bin/clustalw) and then plotted using ESPript3.0 online tool (https://espript.ibcp.fr/ESPript/ESPript/index.php).
The amino acid identity of the more closely related genus Acinetobacter is higher than that of the other bacteria, suggesting that the more closely related proteins are also more functionally conserved (Guo et al., 2021). The sequence alignments of AlkB1_1 and AlkB1_2 with other AlkB sequences indicated that all of these alkane monooxygenase share one conserved HYG motif (N[Y/F][I/L]EHYG) and three highly conserved regions of the Hist boxes (Hist-A: HEL[S/G]HK; Hist-B: EH[P/N][Y/R] GHH; Hist-C: LQRHSDHHA), which were similar to alkane hydroxylase from other strains, such as Alcanivorax sp. 2B5(Liu et al., 2010), Pseudomonas putida GPo1 (Van-Beilen et al., 2005), Dietzia sp. DQ12-45-1b (Nie et al., 2011). The order and position of motifs are roughly the same, indicating that these four motifs and their functions are relatively conserved in the evolution process. However, in addition to these four motifs, AlkB1_1 also contains two other conserved motifs at the front and end of the amino acid sequence, which are the same as AlkB1 sequences in Acinetobacter sp. NIPH 758 and YK3. About HYG-motif, its histidine sequence is also conservative in related hydrocarbon monooxygenase. It is of great significance to study these conserved sequences because they can be one of the important identification marks of alkB. It can be used as probes to detect the activities of related degradation enzymes in the process of alkane degradation and has been reported (Bagi et al., 2022)
Conclusion
In this study, an n-dodecane-degrading bacterium strain KJ-1 was isolated from oil-contaminated soil around a gas station and identified as Acinetobacter vivianii based on morphological, physiological, biochemical, and molecular biology analyses. A. vivianii grew well in MSM with diesel as the sole carbon source, and the diesel degradation rate reached more than 40% after incubation for 12 days. Diesel degradation by this isolate was not significantly affected by the initial concentration of diesel (2070 mg/L to 33,116 mg/L). There was no difference in the degradation rate and bacterial growth when the concentration of ammonium nitrate (NH4NO3) was between 350 and 700 mg/L. The optimal growth and degradation was found to be in the range of 20°C–37°C and at pH 6.5–8.0, respectively. Through these experiments, it was demonstrated that A. vivianii KJ-1 was capable of utilizing diesel as a sole carbon source.
The genomic analysis of the isolate KJ-1 (NCBI GenBank No. CP085083.1) verified its genetic basis for these properties. The gene annotation analysis revealed potential clusters of orthologous genes encoding putative oxygenases and reductases, including alkane monooxygenases, NAD(P)/FAD-dependent oxidoreductases, alcohol dehydrogenases, catechol dioxygenases, and other genes related to hydrocarbon degradation. Based on the annotation results, the genes associated with the two-component regulatory system related to environmental stress factors were analyzed, and there were 59 genes in the strain KJ-1 genome involved in two-component regulatory systems responsible for such functions as potassium transport, nitrogen regulation responses, and drug resistance. In addition, two putative alkane monooxygenases were analyzed, and the amino acid sequences were characterized and aligned with other published AlkB sequences. Since A. vivianii KJ-1 was isolated from soil near a gas station in Jinan, China, it may be particularly useful for future bioremediation for diesel-polluted soil.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/nuccore/MZ994727.
Author contributions
YZ and YH designed the study; YZ, YH, FS and ZW analyzed samples; JW, YH, and XZ designed the graphics; YZ and YH wrote the manuscript; LZ, YG and GC supervised the study; and all authors contributed to previous versions and approved the final manuscript.
Funding
This work was supported by Shandong Provincial Natural Science Foundation (ZR2021MD124), National Key Research and Development Program of China (2019YFC1804103), the Twenty Policies for University in Jinan Project (2020GXRC059) and Science and Education Fusion Projects of Qilu University of Technology (2022PYI009, 2022PY016).
Acknowledgments
Thanks to Leilei Wang for assisting the author to complete the gas chromatography analysis. Thanks to Tengfei Zhang for preparation work. Thanks to Qingchen Meng for his help in data processing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.1044754/full#supplementary-material
References
Acer, O., Guven, K., Poli, A., Di-Donato, P., Leone, L., Buono, L., et al. (2020). Acinetobacter mesopotamicus sp. nov., petroleum-degrading bacterium, isolated from petroleum-contaminated soil in diyarbakir, in the southeast of Turkey. Curr. Microbiol. 77, 3192–3200. doi:10.1007/s00284-020-02134-9
Akinde, S. B., and Obire, O. (2008). Aerobic heterotrophic bacteria and petroleum-utilizing bacteria from cow dung and poultry manure. World J. Microbiol. Biotechnol. 24, 1999–2002. doi:10.1007/s11274-008-9700-z
Atlas, R. M., and Hazen, T. C. (2011). Oil biodegradation and bioremediation: A tale of the two worst spills in U.S History. Environ. Sci. Technol. 45, 6709–6715. doi:10.1021/es2013227
Bagi, A., Knapik, K., and Baussant, T. (2022). Abundance and diversity of n-alkane and PAH-degrading bacteria and their functional genes – potential for use in detection of marine oil pollution. Sci. Total Environ. 810, 152238. doi:10.1016/j.scitotenv.2021.152238
Bhagirath, A. Y., Li, Y., Patidar, R., Yerex, K., Ma, X., Kumar, A., et al. (2019). Two component regulatory systems and antibiotic resistance in gram-negative pathogens. Int. J. Mol. Sci. 20, 1781. doi:10.3390/ijms20071781
Bunger, J., Krahl, J., Schroder, O., Schmidt, L., and Westphal, G. A. (2012). Potential hazards associated with combustion of bio-derived versus petroleum-derived diesel fuel. Crit. Rev. Toxicol. 42 (9), 732–750. doi:10.3109/10408444.2012.710194
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Huntley, J., Fierer, N., et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 6, 1621–1624. doi:10.1038/ismej.2012.8
Fondi, M., Maida, I., Perrin, E., Orlandini, V., Torre, L.-L., Bosi, E., et al. (2016). Genomic and phenotypic characterization of the species Acinetobacter venetianus. Sci. Rep. 6, 21985. doi:10.1038/srep21985
Gao, J., Ming, J., Xu, M., Fu, X., Duan, L.-F., Xu, C.-C., et al. (2021). Isolation and characterization of a high-efficiency marine diesel oil-degrading bacterium. Pet. Sci. 18, 641–653. doi:10.1007/s12182-020-00540-z
Guo, M., Wang, J., Zhang, Y., and Zhang, L. (2021). Increased WD40 motifs in Planctomycete bacteria and their evolutionary relevance. Mol. Phylogenet. Evol. 155, 107018. doi:10.1016/j.ympev.2020.107018
Ho, M. T., Li, M. S. M., Mcdowell, T., Macdonald, J., and Yuan, Z.- C. (2020). Characterization and genomic analysis of a diesel-degrading bacterium, Acinetobacter calcoaceticus CA16, isolated from Canadian soil. BMC Biotechnol. 20 (1), 39. doi:10.1186/s12896-020-00632-z
Hossain, M. F., Akter, M. A., Rahman-Sohan, M. S., Sultana, D. N., Reza, M. A., and Faisal-Hoque, K. M. (2022). Bioremediation potential of hydrocarbon degrading bacteria: Isolation, characterization, and assessment. Saudi J. Biol. Sci. 29 (1), 211–216. doi:10.1016/j.sjbs.2021.08.069
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernandez-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47 (1), 309–314. doi:10.1093/nar/gky1085
Jiao, S., Liu, Z., Lin, Y., Yang, J., Chen, W., and Wei, G. (2016). Bacterial communities in oil contaminated soils: Biogeography and co-occurrence patterns. Soil Biol. Biochem. 98, 64–73. doi:10.1016/j.soilbio.2016.04.005
Kadri, T., Cuprys, A., Rouissi, T., Brar, S. K., Daghrir, R., and Lauzon, J.-M. (2018a). Nanoencapsulation and release study of enzymes from Alkanivorax borkumensis in chitosan-tripolyphosphate formulation. Biochem. Eng. J. 137, 1–10. doi:10.1016/j.bej.2018.05.013
Kadri, T., Magdouli, S., Rouissi, T., and Brar, S. K. (2018b). Ex-situ biodegradation of petroleum hydrocarbons using Alcanivorax borkumensis enzymes. Biochem. Eng. J. 132, 279–287. doi:10.1016/j.bej.2018.01.014
Kadri, T., Miri, S., Robert, T., Brar, K. S., Rouissi, T., Laxman-pachapur, V., et al. (2022). Pilot-scale production and in-situ application of petroleum-degrading enzyme cocktail from Alcanivorax borkumensis. Chemosphere 295, 133840. doi:10.1016/j.chemosphere.2022.133840
Kahraman, B. F., Altin, A., Altin, S., and Bayik, G. D. (2017). Biostimulation of n-alkane degradation in diesel fuel-spiked soils. Soil Sediment Contam. Int. J. 26 (5), 486–500. doi:10.1080/15320383.2017.1355351
Keramea, P., Spanoudaki, K., Zodiatis, G., Gikas, G., and Sylaios, G. (2021). Oil spill modeling: A critical review on current trends, perspectives, and challenges. J. Mar. Sci. Eng. 9, 181. doi:10.3390/jmse9020181
Khandelwal, A., Sugavanam, R., Ramakrishnan, B., Dutta, A., Varghese, E., Nain, L., et al. (2022). Free and immobilized microbial culture–mediated crude oil degradation and microbial diversity changes through taxonomic and functional markers in a sandy loam soil. Front. Environ. Sci. 9, 794303. doi:10.3389/fenvs.2021.794303
Kostka, J. E., Prakash, O., Overholt, W. A., Green, S. J., Freyer, G., Canion, A., et al. (2011). Hydrocarbon-degrading bacteria and the bacterial community response in gulf of Mexico beach sands impacted by the deepwater horizon oil spill. Appl. Environ. Microbiol. 77 (22), 7962–7974. doi:10.1128/AEM.05402-11
Liu, Y. C., Li, L. Z., Wu, Y., Tian, W., Zhang, L. P., Xu, L., et al. (2010). Isolation of an alkane-degrading Alcanivorax sp. strain 2B5 and cloning of the alkB gene. Bioresour. Technol. 101 (1), 310–316. doi:10.1016/j.biortech.2009.08.028
Liu, Z., Xie, W., Li, D., Peng, Y., Li, Z., and Liu, S. (2016). Biodegradation of phenol by bacteria strain acinetobacter calcoaceticus PA isolated from phenolic wastewater. Int. J. Environ. Res. Public Health 13, 300. doi:10.3390/ijerph13030300
Lu, S., Wang, J., Chitsaz, F., Derbyshire, M. K., Geer, R. C., Gonzales, N. R., et al. (2020). CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48 (1), 265–268. doi:10.1093/nar/gkz991
Naing, S. H., Parvez, S., Pender-Cudlip, M., Groves, J. T., and Austin, R. N. (2013). Substrate specificity and reaction mechanism of purified alkane hydroxylase from the hydrocarbonoclastic bacterium Alcanivorax borkumensis (AbAlkB). J. Inorg. Biochem. 121, 46–52. doi:10.1016/j.jinorgbio.2012.12.012
Nie, Y., Liang, J., Fang, H., Tang, Y. Q., and Wu, X. L. (2011). Two novel alkane hydroxylase-rubredoxin fusion genes isolated from a Dietzia bacterium and the functions of fused rubredoxin domains in long-chain n-alkane degradation. Appl. Environ. Microbiol. 77 (20), 7279–7288. doi:10.1128/AEM.00203-11
Nie, Y., Liang, J. L., Fang, H., Tang, Y. Q., and Wu, X. L. (2014). Characterization of a CYP153 alkane hydroxylase gene in a Gram-positive Dietzia sp. DQ12-45-1b and its "team role" with alkW1 in alkane degradation. Appl. Microbiol. Biotechnol. 98 (1), 163–173. doi:10.1007/s00253-013-4821-1
Ohadi, M., Dehghannoudeh, G., Shakibaie, M., Banat, I. M., Pournamdari, M., and Forootanfar, H. (2017). Isolation, characterization, and optimization of biosurfactant production by an oil-degrading Acinetobacter junii B6 isolated from an Iranian oil excavation site. Biocatal. Agric. Biotechnol. 12, 1–9. doi:10.1016/j.bcab.2017.08.007
Pal, S., and Sengupta, K. (2020). Computational-based insights into the phylogeny, structure, and function of Rhodococcus alkane-1-monooxygenase. 3 Biotech. 10, 391. doi:10.1007/s13205-020-02388-x
Palanisamy, N., Ramya, J., Kumar, S., Vasanthi, N. S., Chandran, P., and Khan, S. (2014). Diesel biodegradation capacities of indigenous bacterial species isolated from diesel contaminated soil. J. Environ. Health Sci. Eng. 12, 142. doi:10.1186/s40201-014-0142-2
Park, C., Shin, B., Jung, J., Lee, Y., and Park, W. (2017). Metabolic and stress responses of Acinetobacter oleivorans DR1 during long-chain alkane degradation. Microb. Biotechnol. 10 (6), 1809–1823. doi:10.1111/1751-7915.12852
Phulpoto, I. A., Hu, B., Wang, Y., Ndayisenga, F., Li, J., and Yu, Z. (2021). Effect of natural microbiome and culturable biosurfactants-producing bacterial consortia of freshwater lake on petroleum-hydrocarbon degradation. Sci. Total Environ. 751, 141720. doi:10.1016/j.scitotenv.2020.141720
Ren, Y., Yu, G., Shi, C., Liu, L., Guo, Q., Han, C., et al. (2022). Majorbio Cloud: A one-stop, comprehensive bioinformatic platform for multiomics analyses. iMeta 1, 12. doi:10.1002/imt2.12
Rhoads, A., and Au, K. F. (2015). PacBio sequencing and its applications. Genomics Proteomics Bioinforma. 13 (5), 278–289. doi:10.1016/j.gpb.2015.08.002
Sayers, E. W., Barrett, T., Benson, D. A., Bolton, E., Bryant, S. H., Canese, K., et al. (2011). Database resources of the national center for biotechnology information. Nucleic Acids Res. 39, 38–51. doi:10.1093/nar/gkq1172
Singkham-In, U., and Chatsuwan, T. (2022). Synergism of imipenem with fosfomycin associated with the active cell wall recycling and heteroresistance in Acinetobacter calcoaceticus-baumannii complex. Sci. Rep. 12 (1), 230. doi:10.1038/s41598-021-04303-7
Steiner, S., Bisig, C., Petri-Fink, A., and Rothen-Rutishauser, B. (2016). Diesel exhaust: Current knowledge of adverse effects and underlying cellular mechanisms. Arch. Toxicol. 90 (7), 1541–1553. doi:10.1007/s00204-016-1736-5
Van-Beilen, J. B., Smits, T. H., Roos, F. F., Brunner, T., Balada, S. B., Rothlisberger, M., et al. (2005). Identification of an amino acid position that determines the substrate range of integral membrane alkane hydroxylases. J. Bacteriol. 187 (1), 85–91. doi:10.1128/JB.187.1.85–91.2005
Varjani, S. J., and Upasani, V. N. (2017). A new look on factors affecting microbial degradation of petroleum hydrocarbon pollutants. Int. Biodeterior. Biodegrad. 120, 71–83. doi:10.1016/j.ibiod.2017.02.006
Williams, S. C., Luongo, D., Orman, M., Vizcarra, C. L., and Austin, R. N. (2022). An alkane monooxygenase (AlkB) family in which all electron transfer partners are covalently bound to the oxygen-activating hydroxylase. J. Inorg. Biochem. 228, 111707. doi:10.1016/j.jinorgbio.2021.111707
Xu, B., Sun, Q. J., Lan, C. W., Chen, W. M., Hsueh, C. C., and Chen, B. Y. (2019). Exploring the glyphosate-degrading characteristics of a newly isolated, highly adapted indigenous bacterial strain, Providencia rettgeri GDB 1. J. Biosci. Bioeng. 128 (1), 80–87. doi:10.1016/j.jbiosc.2019.01.012
Xu, X., Liu, W., Tian, S., Wang, W., Qi, Q., Jiang, P., et al. (2018). Petroleum hydrocarbon-degrading bacteria for the remediation of oil pollution under aerobic conditions: A perspective analysis. Front. Microbiol. 9, 2885. doi:10.3389/fmicb.2018.02885
Keywords: Acinetobacter vivianii, diesel degradation, genome analysis, alkane monooxygenase, two-component regulatory system
Citation: Zhang Y, Song F, Wang J, Zhao Q, Zheng L, Wang Z, Zhang X, Gao Y, Chen G and Huang Y (2022) Complete genome sequence analysis of a novel alkane-degrading bacterial strain, Acinetobacter vivianii KJ-1, and its diesel degradation ability. Front. Environ. Sci. 10:1044754. doi: 10.3389/fenvs.2022.1044754
Received: 15 September 2022; Accepted: 01 November 2022;
Published: 18 November 2022.
Edited by:
Aiju Liu, Shandong University of Technology, ChinaReviewed by:
Mezbaul Bahar, The University of Newcastle, AustraliaShiqiang Gao, University of Wuerzburg, Germany
Tongtong Li, Zhejiang University of Technology, China
Copyright © 2022 Zhang, Song, Wang, Zhao, Zheng, Wang, Zhang, Gao, Chen and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yujie Huang, aHVhbmd5akBzZGFzLm9yZw==
 Yali Zhang
Yali Zhang Fanyong Song
Fanyong Song Jianing Wang
Jianing Wang Qingqing Zhao
Qingqing Zhao Xinjian Zhang
Xinjian Zhang Yongchao Gao
Yongchao Gao Yujie Huang
Yujie Huang