- 1State Key Laboratory of Environmental Geochemistry, Institute of Geochemistry, Chinese Academy of Sciences, Guiyang, China
- 2University of Chinese Academy of Sciences, Beijing, China
- 3Department of Epidemiology and Biostatistics, School of Public Health, Peking University, Beijing, China
- 4Yunnan Provincial Key Laboratory of Soil Carbon Sequestration and Pollution Control, Faculty of Environmental Science and Engineering, Kunming University of Science and Technology, Kunming, China
- 5Guizhou Provincial People’s Hospital, Guiyang, China
In non-open environments, pathogenic microorganisms are more likely to invade the human respiratory tract due to their limited diffusion in the environment, which has received little attention. In this study, we explored the distribution characteristics of particulate matter (PM) in non-open environments, and included sewage treatment plants and farms, which are occupational exposure risks, and G-series high-speed trains and waiting rooms, which are crowded. The results showed orders of magnitude differences in PM and microbial concentrations and the DNA/PM values of adsorption in the different non-open spaces. The concentration of PM with a size in the 4.7–10.0 μm range was higher than those of PM in the 1.1–4.7 μm and 0.43–1.1 μm ranges in all three types of places, accounting for 74.64%, 46.59%, and 51.49%, respectively. The DNA/PM value for the 1.1–4.7 μm range was higher than those for PM in the other two ranges in all three types of places at 0.175, 3.78 × 10−3, and 9.98 ng/μg, respectively. Although the relative abundances of Class II potentially pathogenic bacteria with sizes ranging from 1.1 to 4.7 μm were higher in all three types of places, the total abundance and the relative abundance of identified pathogenic microorganisms with sizes ranging from 4.7 to 10.0 μm were higher in all three types of places. Here, in non-open spaces, the pathogen exposure risk associated with PM10, particularly the coarse fraction of PM10, deserves special attention. Infectious diseases caused by aerosol transmission of pathogens in non-open environments should receive more attention and require further investigation in the future.
1 Introduction
Aerosols are an important medium for the transmission of many pathogens that may cause respiratory infections. Pathogens may be absorbed into aerosols that are sufficiently small to be suspended in the air for a long time (even particles with a diameter larger than 100 μm may remain in still air for over 5 s) and be inhaled at both short and long ranges. Additionally, some pathogens have a long half-life, providing them sufficient time to infect people via the transmission of aerosols (Reche et al., 2018; Wu et al., 2020; Li et al., 2021a) and subsequently causing diseases such as pneumonia and allergic rhinitis (Booth, 2008; Warfel et al., 2012; Cao et al., 2014; Niu et al., 2021). Inhalable pathogenic microorganisms may also exert synergistic effects with fine particles in aerosols, increasing the risks of morbidity and toxicity (Horne et al., 2018; Liu et al., 2019; Matus and Oyarzún, 2019; Li et al., 2020), and the virus replication level increases with the concentration of fine particles (Dolci et al., 2018). Therefore, microorganisms may be maintained on, diffuse among and accumulate on aerosols (Wang Chia et al., 2021). For example, in Hong Kong, China, in 2003, severe acute respiratory syndrome (SARS) was transmitted to adjacent households through aerosols containing SARS-CoV (Yu et al., 2004). Since the outbreak of COVID-19, SARS-CoV-2 has been detected in aerosols in many sensitive places and atmospheric particles in epidemic areas, with a certain infectious ability (Liu et al., 2020; Setti et al., 2020; van Doremalen et al., 2020). These studies indicate that aerosols play an important role in the transmission of pathogenic microorganisms.
The microbial community structure and diversity in indoor aerosols are strongly correlated with those in outdoor aerosols, but indoor concentrations are higher than outdoor concentrations, and the content of human-related bacteria in aerosols in non-open spaces is higher than that in outdoor aerosols (Adhikari et al., 2014; Meadow et al., 2014; Guo et al., 2020; Kumar et al., 2022). Aerosols carrying pathogens are more likely to accumulate in non-open spaces than in open places due to the high crowd density and poor ventilation and thus are more likely to facilitate pathogen transmission (Lee et al., 2019; Chen et al., 2020; Li et al., 2021b; Grydaki et al., 2021). Therefore, the risk of aerosol transmission in non-open spaces must be studied. Current studies on pathogenic aerosols in closed places tend to focus on the community characteristics of pathogenic microorganisms or the effectiveness of various infection control measures in specific environments, such as medical settings (Lednicky et al., 2020; Wilson et al., 2020; Guo et al., 2022). However, microbial aerosols generated in the processes used in sewage treatment plants (STPs) may spread to the upper respiratory tract of employees (Gholipour et al., 2021; Ginn et al., 2021; Zieliński et al., 2021; Ma et al., 2022). Microbial aerosols present on public transportation may infect the human respiratory tract, with varying degrees of impact on passengers and staff (Wilson et al., 2020; Buitrago et al., 2021). In the aquaculture industry, zoonoses transmitted by inhalation and other transmission routes occur in farms, with varying degrees of health effects on farm employees (de la Rua-Domenech, 2006; Douglas et al., 2018; Kaikiti et al., 2022). At present, few studies have examined the characteristics of pathogenic microorganisms in aerosols, but these characteristics must be studied for the implementation of measures to protect the health of people in non-open environments.
We selected typical non-open spaces, including sewage treatment plants and farms, which represent occupational exposure risks, and G-series high-speed trains and waiting rooms which are crowded, to evaluate the potential risk of exposure to pathogenic aerosols with different particle sizes in different places. An Andersen multistage impaction sampler was used to capture aerosols of different particle size ranges and simulate the respiratory deposition of aerosols by the human respiratory tract. By analysing the captured particulate matter (PM) content, the PM concentration, microbial concentration, and distribution characteristics of microorganisms in different places were investigated. We explored the distribution characteristics of bacterial and fungal communities in different places based on the results of high-throughput sequencing and the adhesion characteristics of potential pathogens to aerosols with different particle sizes. This work provides a theoretical basis for the assessment of the risk of exposure to respiratory pathogenic microorganisms in non-open spaces, including population health risks in crowded places and health risks arising from occupational exposure.
2 Materials and methods
2.1 Sample collection
Samples were collected from typical non-open spaces in southwest China (Guiyang city, Chengdu city and Chongqing city) from August to October 2021 (Supplementary Figure S1). Sampling sites were selected by considering the health issues of people in crowded places and caused by occupational exposure. For crowded places, a G-series high-speed train (GT) and waiting room (WR) during rush hour were selected for sampling. Considering the average sitting height, the sampling height was set at 1.0 m. Considering the occupational exposure risks of employees, a sewage treatment plant (STP), a small-scale pig farm (SSPF), a large-scale pig farm (LSPF) and a cattle farm of similar size (CF) in Guiyang were selected for sampling. Based on the height of the human respiratory tract while standing, the sampling height was set at 1.5 m for the STP, LSPF, SSPF, and CF.
The carriage of GT has a volume of 100–120 m3 with a height of approximately 4 m, and 85 people are loaded in one carriage. The waiting room of the GT station is approximately 2.2 × 105 m2 with a height of 30–40 m and was very crowded during sampling. The inlet of the sewage plant is built underground, covering an area of approximately 10 m2, with a height of 5–6 m. Only a few staff work in the site for a short time. The cattle farm covers an area of 6,200 m2, with a height of 3–4 m, and only a few staff are present in the breeding area. For the breeding area of a large pig farm, its height is approximately 4 m, and only the workers were able to enter the site, while the small was built in a family yard with a height of 2.5 m, and only 3-4 farmers work here. The sampling sites were all comparable due to their similar sampling times and areas, and sampling was conducted at all sites in Southwest China in autumn (Supplementary Figure S5).
An eight-stage Andersen multistage impaction sampler (Qingdao Laoying Environmental Technology Co., Ltd., China) was used to collect aerosols (Supplementary Figure S6). The sampler simulates the anatomy and aerodynamics of the human respiratory system, with particle sizes ranging from 9.0–10, 5.8–9.0, 4.7–5.8, 3.3–4.7, 2.1–3.3, 1.1–2.1, 0.65–1.1, and 0.43–0.65 μm, as represented by STAGE 0–7. The sampling flow was 28.3 L/min. The flow rate of the sampler was calibrated before and after each sampling cycle. The detailed parameters are listed in Supplementary Table S1. Before sampling, the corresponding polycarbonate filter film, which had a diameter of 81 mm and an aperture size of 0.22 μm, was sterilized and weighed. In this study, the aerosol collection time was set to 48 h, except for on GT, for which the sampling time was 6 h due to the limitation of the train operating time.
2.2 Determination of environmental factors
Environmental parameters, including temperature (°C), atmospheric pressure (kPa) and relative humidity (%), were recorded when aerosolized microorganisms were sampled in non-open spaces. The sealing and ventilation degrees were recorded and scored according to the actual situation of the site. In addition, we noted whether the place was decontaminated during the sampling period. The results for environmental factors are shown in Supplementary Table S2.
2.3 DNA extraction and determination
PC membranes were cut into pieces with sterilized scissors and placed into 2 ml lysis tubes. A DNA extraction kit (Mega Gene, China) was used to extract the total DNA from the aerosol samples according to the manufacturer’s instructions. The total DNA concentration was determined using a Qubit 4.0 instrument (Thermo Fisher Scientific, United States). The Qubit dsDNA HS assay kit was used, and the detection limit was 0.01 ng/μL.
2.4 High-throughput 16S rRNA and ITS1 gene amplicon sequencing
The bacterial region used for analysis and identification was the 16S V3-V4 region. The primer sequences were 338F-5′-ACTCCTACGGGAGGCAGCA-3′ and 806R-5′-GGACTACHVGGGTWTCTAAT-3′. The fungal region analysed was the ITS1 region. The primer sequences were BD-ITS1F-5′-CTTGGTCATTTAGAGGAAGTAA-3′ and ITS2-2043R-5′-GCTGCGTTCTTCATCGATGC-3′. PCR amplification was performed, and the products were purified, quantified, and homogenized to form a sequencing library. The Illumina Nova 6,000 platform (Guangdong Magigene Biotechnology Co., Ltd., Guangzhou, China) was used for PE250 sequencing of the constructed amplicon library. Operational taxonomic units (OTUs) were clustered by UCLUST, with a 97% similarity level. The reagents and instruments used in the experiment are described in Supplementary Figure S3.
2.5 Data analysis
The species annotation and preliminary analysis of the data were completed using the Magigene Cloud platform. The parameters used in this paper are explained below. Usearch-sintax was used to compare the representative sequences of each OTU with SILVA (16S) and Unite (ITS) databases to obtain the annotated species information and understand the species origin of all sequences, and the default confidence threshold was 0.8. R 4.0.3 was used for statistical analyses of common and endemic species, community composition analysis, and species abundance cluster analysis. During principal coordinate analysis (PCoA) of the OTU abundance table, we used the vegan software package of R 4.0.3 combined with the Bray‒Curtis distance algorithm at the generic level. LEfse software was used to analyse the significance of differences in species between groups based on the homogenized OTU table for each species grade. The parametric Kruskal‒Wallis (KW) sum-rank test was used to determine the species with significant differences in abundance between two groups, and then the Wilcoxon rank sum test was used to determine the differences between two groups. Finally, linear discriminant analysis (LDA) was performed to reduce and evaluate the effect of species with significant differences (LDA score). The screening value of the LDA score was set to 3.0/4.0, and the result was the biomarker for each group. This study utilized the relative abundance to represent the risk of infection. A larger relative abundance represents a larger risk of infection.
3 Results and discussion
3.1 Characteristics of the PM distribution
The GT and WR were classified as crowded places, with relatively high risks of exposure to human-transmitted pathogens. The SSPF, LSPF, and CF were classified as animal gathering places where humans are at risk of contracting zoonoses. Because microbial concentrations in STPs are much higher than those in ordinary places, the STP was defined as a site with a potentially high risk of pathogen exposure. PM with different sizes is deposited in different parts of the human respiratory tract: 4.7–10 μm particles are deposited in the nose and pharynx, 1.1–4.7 μm particles are deposited in the trachea and bronchus, and 0.43–1.1 μm particles are deposited in alveoli (Guo et al., 2022). We defined PM4.7-10, PM1.1-4.7, and PM0.43-1.1 as the coarse part, fine part, and very fine part, respectively.
The concentration of PM4.7-10 was higher than those of PM1.1-4.7 and PM0.43-1.1 in all three types of places. PM4.7-10 accounted for 46.6%, 51.5%, and 74.64% of PM in the crowded places, animal gathering places, and STP, respectively. The proportion of PM4.7-10 on STP was higher, while the proportion of the PM concentration in different size ranges in the crowded places and animal gathering places were consistent (Figure 5C).
Specifically, the particle sizes measured at the GT and WR, with intensive human activities, were mainly in the 0.43–2.1 μm range. Similar results were also reported in previous studies (Du et al., 2021). However, the concentration on the GT (798 μg/m3) was higher than that in the WR (202 μg/m3) (Figures 1B,C,G). The explanation for the difference may be the higher crowd density (Lin et al., 2006) and poorer ventilation on the GT (Thomas, 2013). The STP (21.5 μg/m3) had a lower concentration of PM than crowded places (Figures 1A,G). In contrast to crowded places, relatively low PM concentrations were detected in animal gathering places, with orders of magnitude differences. The farm type and size had small effects on PM distributions but large effects on PM concentrations. The PM concentrations at the LSPF for all particle sizes were higher than those at the CF and SSPF on the same scale (Figures 1D–G). The PM concentration may be affected by the degree of sealing and ventilation and the sources of the farm aerosols themselves, including farming scales (Shao et al., 2021; Guo et al., 2022).
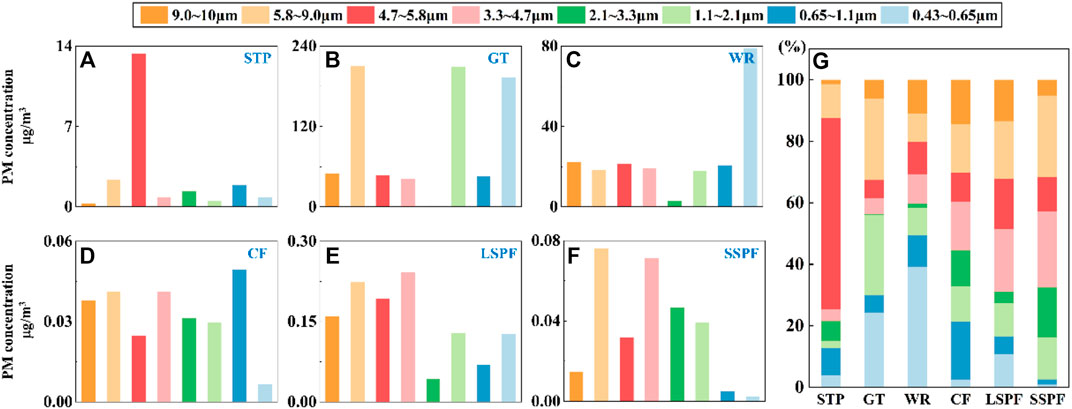
FIGURE 1. (A–G) The mass distribution characteristics of particles in non-open spaces. (A–F) The mass concentration of particles with different particle sizes at each site. (G) The distribution characteristics of PM in the different places. SSPF: small-scale pig farm, CF: cattle farm, LSPF: large-scale pig farm, GT: G-series high-speed train, WR: waiting room, STP: sewage treatment plant.
Particulate inhalation alone may cause respiratory tract damage, and thus a high PM concentration is also a key concern (Grydaki et al., 2021). A high PM concentration also provides many attachment sites for microorganisms. Dust should be removed at some places, such as the GT, WR, STP and others, especially the coarse part of PM10, to reduce the risk of exposure.
3.2 Characteristics of microorganism distributions
We tested the DNA concentrations of PM in different sizes, which represented the microbial concentration, to further evaluate the distributions of microorganisms on PM of different sizes. The DNA concentration in some samples was lower than the detection limit (0.01 ng/μl). In general, in the comparison of the distribution of DNA among the different places (Figure 2G), the total DNA concentration measured in aerosols from the STP was 1.19 ng/m3, and the DNA concentrations in samples of different particle size ranges were similar. The DNA concentration in the sample from the GT (1.64 ng/m3) was much higher than that in the sample from the WR (0.065 ng/m3). In all stages, the DNA concentrations on the GT were greater than those in the WR. The explanations for these differences may be different crowd densities (Lin et al., 2006) and ventilation conditions (Liu et al., 2020). Among the animal gathering places, DNA concentrations in samples from the LSPF (9.73 ng/m3) for all sizes of PM were much higher than those from the CF (0.522 ng/m3) and SSPF (0.287 ng/m3) of the same size. The DNA distributions in animal gathering places were completely different. The DNA concentrations in samples of PM of all sizes from the LSPF were higher than those in samples from the SSPF and CF (Figures 2A–F).
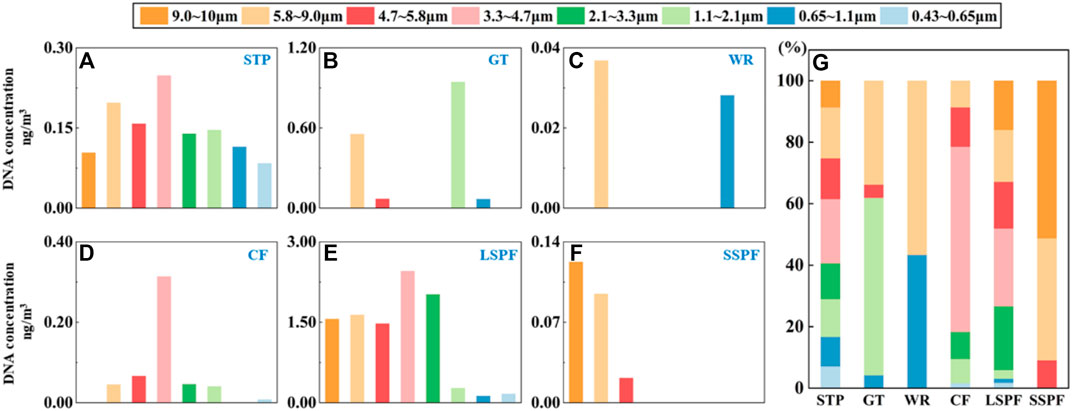
FIGURE 2. Characteristics of the distribution of the DNA concentration on particles from non-open spaces. (A–G) Concentrations of DNA on particles with different sizes from each place. (G) Distribution characteristics of DNA in the different places.
Considering the risk of microbial adsorption (Thomas, 2013), the values of DNA/PM adsorption from all places were calculated (Supplementary Table S4). In all three types of places, PM with sizes ranging from 1.1 to 4.7 μm had the highest DNA/PM adsorption value (Figure 5D). The DNA/PM values were 0.175, 3.78 × 10−3, and 9.98 ng/μg in crowded places, animal gathering places, and STP, respectively. The DNA/PM values were inconsistent among the different places. For the animal gathering places (1.99, 8.20, and 0.833 ng/μg), the DNA/PM values were much higher than those for the crowded places and STP, and were more than 100 times higher than that for the STP (0.055 ng/μg) and 103 times higher than those for the crowded places (2.05 × 10−3 ng/mg and 3.22 × 10−4 ng/mg).
3.3 Characteristics of the distribution of microbial communities
3.3.1 Overall distribution characteristics
We conducted PCoA at the OTU level and common and endemic genera levels and a linear discriminant analysis effect size (LEfSe) at the genus level to explore the effects of the different places and particle size ranges on the characteristics of microbial population distributions.
For the different places, PCoA showed that principal components PC1 and PC2 explained 25.2% and 23.1% of the total variance in bacterial communities in the samples at the OTU level, respectively (Figure 3A). The bacterial compositions from each place were separated from the samples from other places, indicating differences in aerosol bacterial community compositions between the different places (p < 0.05), except GT and WR (Supplementary Table S5). Based on these results, the community composition of bacteria of the same species in aerosol samples was relatively similar among the different places. At the OTU level, PC1 and PC2 explained 17.6% and 11% of the total variance in the fungal communities in the samples, respectively. A remarkable difference was observed among various sampling places (p < 0.05, Supplementary Table S5; Figure 3E). However, the distance between the SSPF and CF was close (p = 0.075), and both WR and CF (p = 0.124) and WR and LSPF (p = 0.121) were close.
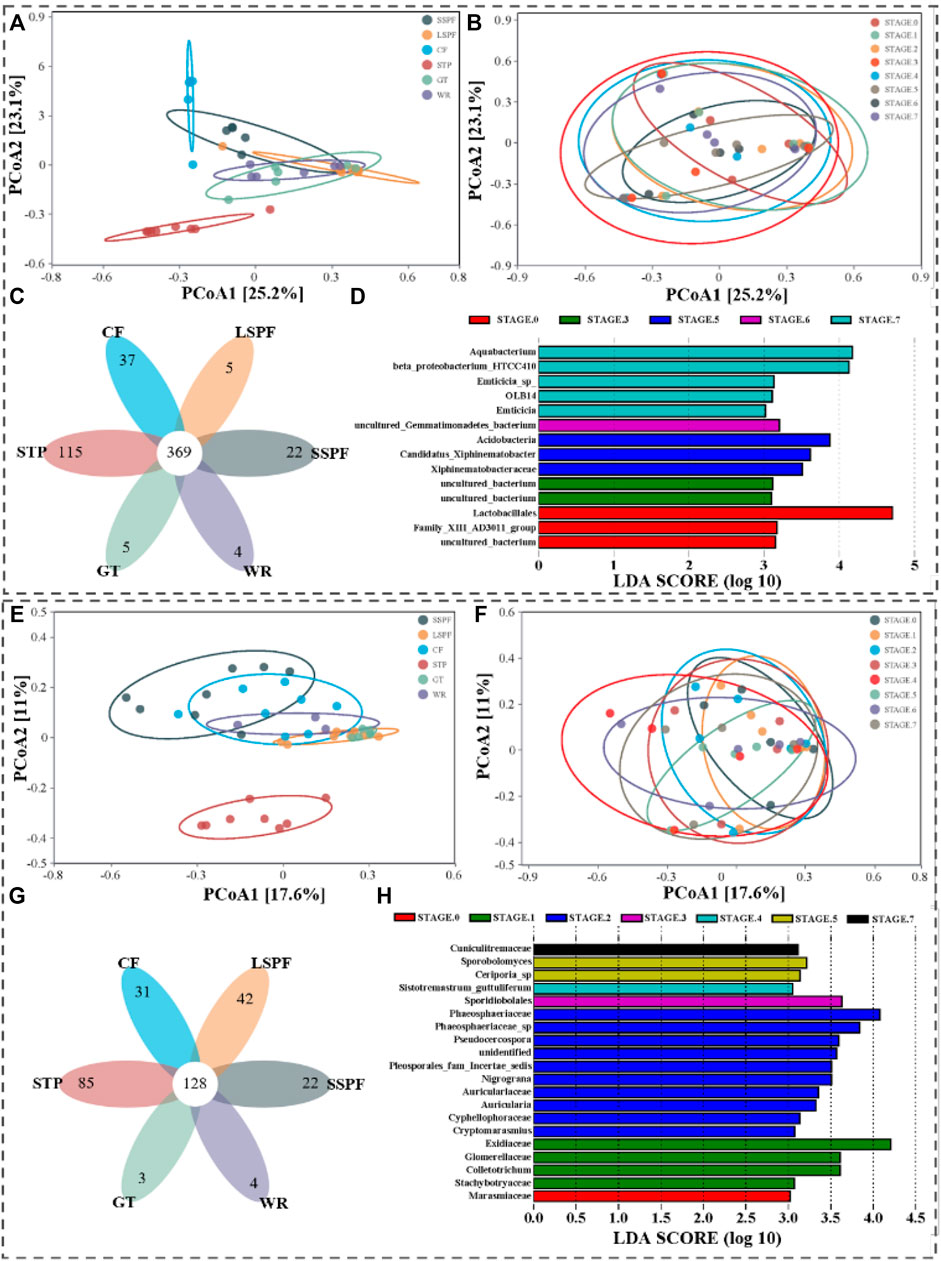
FIGURE 3. Overall differences in microbial population distributions among the different places. (A–D) and (E–G) Differences in bacterial and fungal population distributions, respectively. (A,E) Coordinate maps of the principal components of bacterial classification by place. (B,F) Coordinate diagrams of principal component classifications by particle size; (C,G) Venn diagrams of common and endemic genera by place. (G,H) Histograms showing the LDA values for the distribution of microorganisms by particle size (LDA>3.0).
For bacteria, 369 genera were identified in aerosols collected from the six places, and all the genera had relative abundances greater than 2% (Figure 3C). For fungi, 128 genera were identified in aerosols collected from the six places, including Phlebia, Aspergillus, Trechispora, Erysiphe, Schizophyllum, Amphinema, Periconia, Coprinopsis, Steccherinum, and Bjerkandera. All the genera had relative abundances higher than 2% (Figure 3G).
Although the species compositions of bacteria and fungi distributed on particles with different sizes were generally similar (p > 0.05, Figures 3B,F), some microorganisms still showed differences (Figures 3D,H). When the LDA value was >3.0, significant differences were observed among taxa at all levels.
3.3.2 Characteristics of the microbial distributions at the different places
We found that the place affected the population distribution characteristics of microorganisms in aerosols, and thus we further compared differences in bacterial and fungal populations in aerosols from places with similar characteristics, including crowded places (GT and WR) and animal gathering places (LSPF, CF, and SSPF).
In the crowded places, unassigned and uncultured bacteria accounted for 12.3% on the GT and 24.1% in the WR. At the genus level, 523 common genera were identified in aerosols collected from the GT and WR. 59 endemic genera were detected in samples from the GT and 25 in samples from the WR (Figure 4A). PCoA showed that PC1 and PC2 accounted for 26.3% and 19.8% of the OTUs, respectively. A significant difference in the bacterial community composition was not observed between the aerosols collected from the GT and WR (p = 0.216, Figure 4B). LEfSe (LDA>4.0) showed that groups with significant differences, such as Clostridia, Clostridiales, and Deinococcus, were identified in samples from only the GT (Figures 4C,D). Unassigned and uncultured fungi accounted for 76.8% in samples from the GT and 81.3% in samples from the WR. At the genus level, 164 common genera were identified in samples from the GT and WR. 39 endemic genera were detected in samples from the GT and 44 in samples from the WR (Figure 4E). PCoA showed that PC1 and PC2 accounted for 45.8% and 20.8% of the variance at the OTU level, respectively. Significant differences in fungal community compositions were observed between aerosol samples collected from the GT and WR (p < 0.05, Figure 4F). LEfSe (LDA>4.0) showed significant differences in Erysiphe, Leotiomycetes, Erysiphaceae, Erysiphales and other fungi in samples from the GT. Significant differences in Phaeosphaeriaceae_sp, Sordariomycetes, Xylariales, Phaeosphaeriaceae, and other fungi were observed among the samples from the WR (Figures 4G,H).
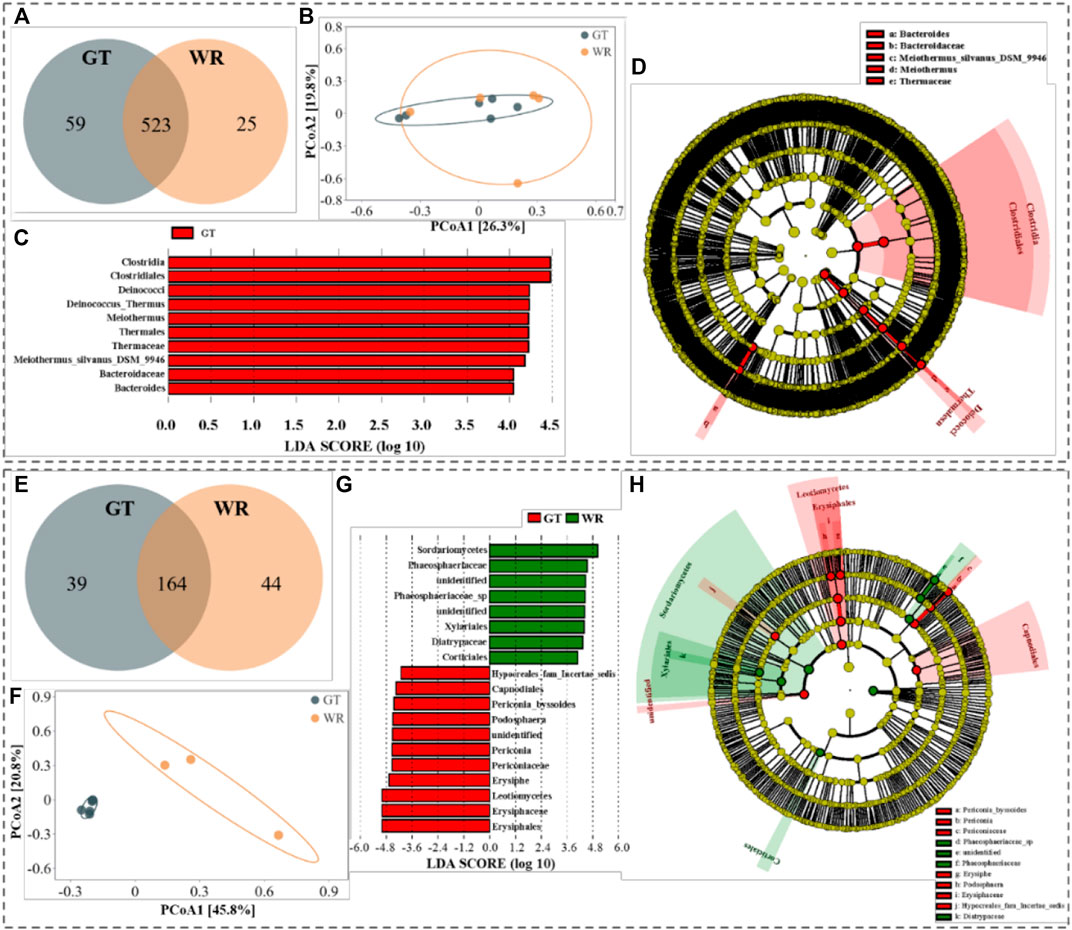
FIGURE 4. Comparison of bacteria and fungi in aerosols collected from GT and WR. (A,E) Statistical analyses of common and endemic genera; b and f Principal component coordinate diagrams. (C,D,G,H) Diagrams of the LEfSe analysis, c and d histograms of the LDA value distribution, and (D,H) phylogenetic clades. Among the figures, (A–D) and (E–H) show bacteria and fungi detected in the samples, respectively.
A comparison of differences in aerosol microbial populations among the animal gathering places yielded the conclusions described below (Supplementary Figure S2). Significant differences in bacterial community compositions were observed between the aerosol samples collected from the LSPF and SSPF and CF (p < 0.05), consistent with previous findings (Amin et al., 2022). Significant differences in fungal community compositions were observed between samples collected from the LSPF and SSPF and CF (p < 0.05), but not between SSPF and CF (p > 0.05).
3.4 Pathogen exposure risk in a non-open environment
Based on the results of high-throughput sequencing, we compared our findings to the List of Pathogenic Microorganisms Transmitted to Humans (Bulletin, 2006). Four identified pathogenic bacterial genera were detected, including Enterobacter, Erysipelothrix, Streptococcus, and Prevotella. A few species of potentially pathogenic bacteria that are classified as Class II pathogens were detected, but the vast majority were classified as Class III. 5 genera containing Class II potentially pathogenic bacteria, including Bacillus, Burkholderia, Coxiella, Mycobacterium, and Seleniivibrio, were identified. 34 genera containing Class III potentially pathogenic bacteria were identified, and the bacteria detected in all six places were Acinetobacter, Actinobacillus, Actinomyces, Aeromonas, Bacteroides, Campylobacter, Clostridium_sensu_stricto_1, Corynebacterium, Dermatophilus, Flavobacterium, Fusobacterium, Legionella, Neisseria, Peptostreptococcus, Proteus, Pseudomonas, Serratia, Staphylococcus, and Treponema. 19 common bacterial genera were detected in all sites. Moreover, seven pathogenic fungal genera, including Alternaria, Arthrinium, Geotrichum, Mucor, Stachybotrys, Trichoderma, and Trichothecium, were identified. Eight genera containing Class III potentially pathogenic fungi, including Aspergillus, Candida, Cladosporium, Cryptococcus, Exophiala, Fusarium, Penicillium, and Trichophyton, were detected. All the potential pathogenic fungi were classified as Class III pathogens, which are harmless to the human body under general conditions. These results are generally consistent with previous publications. For example, Bacillus and Aspergillus have been reported to have the highest contributions to the environment (Hernández-Castillo et al., 2014; Alananbeh et al., 2017; Madhwal et al., 2020). The relative abundances of pathogenic bacteria and fungi of all sizes from all places are shown in Figures 5A,B, respectively. The proportion of pathogenic bacteria in animal gathering places is relatively high. The reason may be that pathogenic bacteria from farm breeding wastewater (including animal excreta) enter the aerosol, increasing the concentration of pathogenic bacteria (Matjuda and Aiyegoro, 2019). The proportion of pathogens in crowded places is also higher because humans act as the direct source of bacterial emissions (from skin, body fluids and breathing) in crowded public spaces (Abdel Hameed et al., 2009). In addition, in the sewage treatment plant, the proportions of pathogenic bacteria and fungi are relatively high, potentially due to the abundant source of the aerosol and the strong water flow in the water inlet, enabling the bacteria and fungi to enter the aerosol.
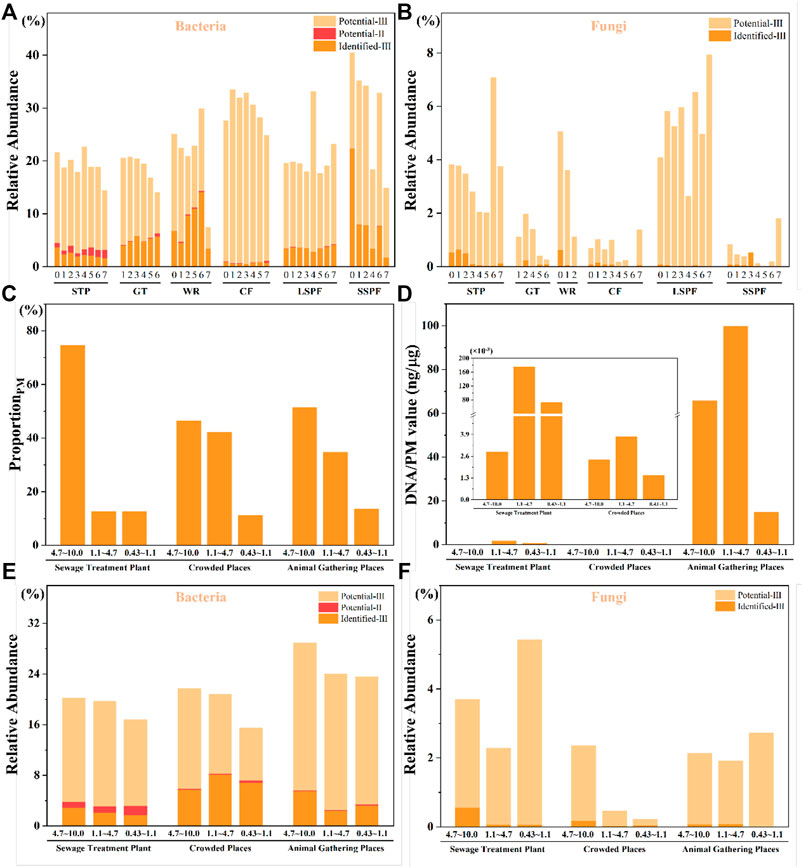
FIGURE 5. Risk assessment in the different places. (A) The relative abundance of pathogenic bacteria, and (B) the relative abundance of pathogenic fungi. The term “identified” refers to identified pathogenic microorganisms, and Ⅱ and Ⅲ represent Class Ⅱ and Class Ⅲ pathogens, respectively. Because amplicon sequencing can be performed at only the genus level and only some sequences are detected at the species level, the term “potential” indicates that the organisms have not been determined at the species level, but the genus includes pathogenic species, which are defined as microorganisms that may be pathogenic. (C–F) The proportion of PM, the DNA/PM values and the pathogens detected in PM with sizes ranging from 4.7–10.0 μm, 1.1–4.7 μm and 0.43–1.1 μm.
The deposition of pathogenic microorganisms in different parts of the respiratory tract exerts different effects on respiratory tract infection, and if pathogenic microorganisms do not settle on the active parts, the threat to human health is low. Therefore, studies exploring the characteristics of pathogenic microorganisms distributed in different places and on particles of different sizes can also be used as one to evaluate potentially pathogenicity. By exploring the distribution characteristics of identified pathogenic microorganisms in the six places, we observed certain differences, but the results for the distribution characteristics of different pathogenic microorganisms were still preliminary. The relative abundances of identified pathogenic bacteria in the different places and on PM of different sizes are shown in Supplementary Figures S3,S4. Regarding the identified pathogenic bacteria, Enterobacter, Erysipelothrix, Streptococcus, and Prevotella were detected. These four bacterial genera were found in all types of places and in all stages. Regarding the identified pathogenic fungi, seven genera were detected, including Alternaria, Arthrinium, Geotrichum, Mucor, Stachybotrys, Trichoderma, and Trichothecium. Among them, Alternaria, Arthrinium, Trichoderma, and Trichothecium were found in all types of places.
Overall, the relative abundances of pathogenic bacteria in the six places ranged from 7.50% to 40.60%. The relative abundances of pathogenic fungi ranged from 0.87‱ to 7.95%. Class II potentially pathogenic bacteria must be identified at a lower taxonomic level to determine whether they might exert a direct harmful effect on the human body. However, the proportion of Class III pathogens that were identified was very large. They may pose a low threat to healthy people but a high threat to vulnerable people with weakened immunity due to other diseases or long-term occupational exposure (Tsay et al., 2020).
We further compared the three PM size ranges from three types of places. For pathogenic bacteria, the total abundances in PM with sizes ranging from 4.7–10.0 μm (21.78%) and 1.1–4.7 μm (20.84%) were higher than those in PM with sizes ranging from 0.43–1.1 μm (15.53%) in crowded places. However, the relative abundance of Class II potentially pathogenic bacteria in PM with sizes ranging from 1.1–4.7 μm range (8.07%) was higher than those in PM with other size ranges, and the relative abundance of identified pathogenic bacteria in PM with sizes ranging from 0.43–1.1 μm (0.35%) was higher than those in the other ranges. For pathogenic fungi, the total abundance in PM with sizes ranging from 4.7–10.0 μm (2.36%) was much higher than those in PM with sizes ranging from 1.1–4.7 μm (0.46%) and 0.43–1.1 μm (0.23%). A similar result was obtained for identified pathogenic fungi (0.17%, 0.02%, and 0.04%, respectively). Considering the total abundance and the relative abundance of Class II potentially pathogenic microorganisms and identified pathogenic microorganisms, we must focus on PM of all sizes in crowded places. For pathogenic bacteria, the total abundances in PM of all 3 size ranges were similar in the animal gathering places. However, the relative abundance of identified pathogenic bacteria in PM with sizes ranging from 4.7–10.0 μm (5.53%) was higher than those in PM of the other size ranges, and the relative abundance of Class II potentially pathogenic bacteria in PM with sizes ranging from 0.43–1.1 μm (0.21%) was higher than those in PM of the other size ranges. For pathogenic fungi, the total abundance in PM with sizes ranging from 0.43–1.1 μm (2.74%) was higher than those in PM with sizes ranging from 4.7–10.0 μm (2.15%) and 1.1–4.7 μm (1.92%). However, the relative abundances of identified pathogenic fungi in PM from the 1.1–4.7 μm size range (0.08%) and in the 4.7–10.0 μm size range (0.07%) were higher than those in the other size ranges. For the STP, the total abundances of pathogenic bacteria in PM with sizes ranging from 4.7–10.0 μm (20.22%) and 1.1–4.7 μm (19.74%) were higher than those in PM with sizes ranging from 0.43–1.1 μm (16.81%). For pathogenic fungi, the total abundance in PM with sizes ranging from 0.43–1.1 μm (5.44%) was higher than those in PM with sizes ranging from 4.7–10.0 μm (3.70%) and 1.1–4.7 μm (2.29%). Additionally, the relative abundances of identified pathogenic bacteria (2.85%) and fungi (0.55%) in PM with the 4.7–10.0 μm size range were the highest. The relative abundance of Class II potentially pathogenic bacteria in PM with the 0.43–1.1 μm size range (1.45%) was the highest (Figures 5E,F).
In general, the total abundances of pathogenic microorganisms in PM of all 3 size ranges were similar. However, because of the more serious harm caused by Class II potentially pathogenic bacteria and identified pathogenic microorganisms, we should factor in these two categories for a higher reference value. The relative abundances of identified pathogenic microorganisms in PM with the 4.7–10.0 μm size range were higher in the animal gathering places and the STP than in the crowded places, and the relative abundance of identified pathogenic microorganisms in PM with the 1.1–4.7 μm size range was higher in crowded places, although the abundance values for all particle sizes were greater than 5% in the crowded places. The relative abundance of Class II potentially pathogenic bacteria in PM with sizes ranging from 1.1–4.7 μm was highest in all three types of places.
This study did not consider the survival time, probability of microorganisms leaving the human body or the effects of other factors, such as respiratory settlement, air turbulence, ventilation rate, or exposure time (Tellier, 2006; To and Chao, 2010; Kumar et al., 2022). Future studies consider these factors and combine them in models such as the MA and Wells-Riley models to predict potential health risks in susceptible populations (Wang et al., 2020). In addition, appropriately extending the sampling time to ensure that the microbial concentration reaches the detection limit or using other methods, such as qPCR, to accurately quantify the microbial concentration will aid in the exploration of the distribution characteristics of microorganisms with different particle sizes.
4 Conclusion
In this study, systematic and detailed analysis of PM and microbes in the air along with their relationships was conducted using a combination of eight-stage sampling and 16S rRNA sequencing. STP, crowded places including WR and GT, and farms such as SSPF and LSPF were selected to represent sites of different types. Microbes were widely distributed in all types of locations, while the farms presented much higher abundances than STP and crowded places. Moreover, the pathogen exposure risk of PM10 should not be ignored in a non-open environment, while the risk of exposure to PM10 with coarse particles was higher than exposure to other particles. In addition, although the size of PM did not affect the diversity of microbes, it exerted a significant effect on the contents of microorganisms. These results verified the wide existence of pathogens in PM in non-open environments, especially the coarse fraction of PM10. Furthermore, the health of staff who work in relatively non-open spaces, particularly STPs and farms, should receive more attention. In summary, this work provides an understanding of the pathogen exposure risk associated with different sizes of PM10 in non-open environments.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author contributions
YZ: conceptualization, methodology, software, data curation, writing- original draft preparation. KM: supervision, conceptualization, methodology, writing, reviewing, and editing. HC: writing- reviewing and editing. BW: writing, reviewing, and editing. XZ: writing, reviewing, and editing. WD: writing, reviewing, and editing. XiZ: writing, reviewing, and editing. HZ: conceptualization, methodology, writing, reviewing, and editing.
Acknowledgments
The authors acknowledge support from the Guizhou Provincial Science and Technology Projects (Qiankehe Zhicheng [2020] 4Y190 and Qiankehe Jichu-ZK [2022] Yiban 565), and Youth Cross Team Project of CAS (JCTD-2021–17).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2022.1006209/full#supplementary-material
References
Abdel Hameed, A. A., Khoder, M. I., Yuosra, S., Osman, A. M., and Ghanem, S. (2009). Diurnal distribution of airborne bacteria and fungi in the atmosphere of helwan area, Egypt. Sci. Total Environ. 407, 6217–6222. doi:10.1016/j.scitotenv.2009.08.028
Adhikari, A., Kettleson, E. M., Vesper, S., Kumar, S., Popham, D. L., Schaffer, C., et al. (2014). Dustborne and airborne gram-positive and gram-negative bacteria in high versus low ermi homes. Sci. Total Environ. 482-483, 92–99. doi:10.1016/j.scitotenv.2014.02.110
Alananbeh, K. M., Boquellah, N., Kaff, N. A., and Ahmadi, M. A. (2017). Evaluation of aerial microbial pollutants in al-haram al-nabawi during pilgrimage of 2013. Saudi J. Biol. Sci. 24, 217–225. doi:10.1016/j.sjbs.2015.08.003
Amin, H., Šantl-Temkiv, T., Cramer, C., Vestergaard, D. V., Holst, G. J., Elholm, G., et al. (2022). Cow farmers’ homes host more diverse airborne bacterial communities than pig farmers’ homes and suburban homes. Front. Microbiol. 13, 883991. doi:10.3389/fmicb.2022.883991
Booth, B. (2008). Microbes in the air near swine farms—More or less. Environ. Sci. Technol. 42, 8618–8619. doi:10.1021/es802620k
Buitrago, N. D., Savdie, J., Almeida, S. M., and Verde, S. C. (2021). Factors affecting the exposure to physicochemical and microbiological pollutants in vehicle cabins while commuting in lisbon. Environ. Pollut. 270, 116062. doi:10.1016/j.envpol.2020.116062
Bulletin, P. (2006). List of pathogenic microorganisms transmitted from human to human. Available at: https://www.chinacdc.cn/lac/gzzd/gwfgbz/202003/t20200320_214605.htm.
Cao, C., Jiang, W., Wang, B., Fang, J., Lang, J., Tian, G., et al. (2014). Inhalable microorganisms in beijing's pm2.5 and pm10 pollutants during a severe smog event. Environ. Sci. Technol. 48, 1499–1507. doi:10.1021/es4048472
Chen, W., Zhang, N., Wei, J., Yen, H.-L., and Li, Y. (2020). Short-range airborne route dominates exposure of respiratory infection during close contact. Build. Environ. 176, 106859. doi:10.1016/j.buildenv.2020.106859
de la Rua-Domenech, R. (2006). Human mycobacterium bovis infection in the United Kingdom: Incidence, risks, control measures and review of the zoonotic aspects of bovine tuberculosis. Tuberculosis 86, 77–109. doi:10.1016/j.tube.2005.05.002
Dolci, M., Favero, C., Bollati, V., Campo, L., Cattaneo, A., Bonzini, M., et al. (2018). Particulate matter exposure increases jc polyomavirus replication in the human host. Environ. Pollut. 241, 234–239. doi:10.1016/j.envpol.2018.05.044
Douglas, P., Robertson, S., Gay, R., Hansell, A. L., and Gant, T. W. (2018). A systematic review of the public health risks of bioaerosols from intensive farming. Int. J. Hyg. Environ. Health 221, 134–173. doi:10.1016/j.ijheh.2017.10.019
Du, W., Wang, J., Wang, Z., Lei, Y., Huang, Y., Liu, S., et al. (2021). Influence of Covid-19 lockdown overlapping Chinese spring festival on household pm2.5 in rural Chinese homes. Chemosphere 278, 130406. doi:10.1016/j.chemosphere.2021.130406
Gholipour, S., Mohammadi, F., Nikaeen, M., Shamsizadeh, Z., Khazeni, A., Sahbaei, Z., et al. (2021). Covid-19 infection risk from exposure to aerosols of wastewater treatment plants. Chemosphere 273, 129701. doi:10.1016/j.chemosphere.2021.129701
Ginn, O., Rocha-Melogno, L., Bivins, A., Lowry, S., Cardelino, M., Nichols, D., et al. (2021). Detection and quantification of enteric pathogens in aerosols near open wastewater canals in cities with poor sanitation. Environ. Sci. Technol. 55, 14758–14771. doi:10.1021/acs.est.1c05060
Grydaki, N., Colbeck, I., Mendes, L., Eleftheriadis, K., and Whitby, C. (2021). Bioaerosols in the athens metro: Metagenetic insights into the pm10 microbiome in a naturally ventilated subway station. Environ. Int. 146, 106186. doi:10.1016/j.envint.2020.106186
Guo, J., Xiong, Y., Shi, C., Liu, C., Li, H., Qian, H., et al. (2020). Characteristics of airborne bacterial communities in indoor and outdoor environments during continuous haze events in beijing: Implications for health care. Environ. Int. 139, 105721. doi:10.1016/j.envint.2020.105721
Guo, W., Fu, Y., Jia, R., Guo, Z., Su, C., Li, J., et al. (2022). Visualization of the infection risk assessment of sars-cov-2 through aerosol and surface transmission in a negative-pressure ward. Environ. Int. 162, 107153. doi:10.1016/j.envint.2022.107153
Hernández-Castillo, O., Mugica-Álvarez, V., Castañeda-Briones, M. T., Murcia, J. M., García-Franco, F., and Falcón Briseño, Y. (2014). Aerobiological study in the Mexico city subway system. Aerobiologia 30, 357–367. doi:10.1007/s10453-014-9334-6
Horne, B. D., Joy, E. A., Hofmann, M. G., Gesteland, P. H., Cannon, J. B., Lefler, J. S., et al. (2018). Short-term elevation of fine particulate matter air pollution and acute lower respiratory infection. Am. J. Respir. Crit. Care Med. 198, 759–766. doi:10.1164/rccm.201709-1883OC
Kaikiti, C., Stylianou, M., and Agapiou, A. (2022). Td-gc/ms analysis of indoor air pollutants (vocs, pm) in hair salons. Chemosphere 294, 133691. doi:10.1016/j.chemosphere.2022.133691
Kumar, P., Hama, S., Abbass, R. A., Nogueira, T., Brand, V. S., Wu, H.-W., et al. (2022). In-kitchen aerosol exposure in twelve cities across the globe. Environ. Int. 162, 107155. doi:10.1016/j.envint.2022.107155
Lednicky, J. A., Lauzardo, M., Fan, Z. H., Jutla, A., Tilly, T. B., Gangwar, M., et al. (2020). Viable sars-cov-2 in the air of a hospital room with Covid-19 patients. Int. J. Infect. Dis. 100, 476–482. doi:10.1016/j.ijid.2020.09.025
Lee, J., Yoo, D., Ryu, S., Ham, S., Lee, K., Yeo, M., et al. (2019). Quantity, size distribution, and characteristics of cough-generated aerosol produced by patients with an upper respiratory tract infection. Aerosol Air Qual. Res. 19, 840–853. doi:10.4209/aaqr.2018.01.0031
Li, L., Liu, H., Wang, Y., Han, X., Ge, T., and Pan, L. (2020). Construction of a nomogram for predicting the risk of allergic rhinitis among employees of long-distance bus stations in China. Indoor Air 30, 1178–1188. doi:10.1111/ina.12694
Li, Y. G., Qian, H., Hang, J., Chen, X. G., Cheng, P., Ling, H., et al. (2021b). Probable airborne transmission of sars-cov-2 in a poorly ventilated restaurant. Build. Environ. 196, 107788. doi:10.1016/j.buildenv.2021.107788
Li, Z., Zheng, W., Wang, Y., Li, B., and Wang, Y. (2021a). Spatiotemporal variations in the association between particulate matter and airborne bacteria based on the size-resolved respiratory tract deposition in concentrated layer feeding operations. Environ. Int. 150, 106413. doi:10.1016/j.envint.2021.106413
Lin, K. U. N., Yee-Tak Fong, D., Zhu, B., and Karlberg, J. (2006). Environmental factors on the sars epidemic: Air temperature, passage of time and multiplicative effect of hospital infection. Epidemiol. Infect. 134, 223–230. doi:10.1017/S0950268805005054
Liu, X. X., Li, Y., Qin, G., Zhu, Y., Li, X., Zhang, J., et al. (2019). Effects of air pollutants on occurrences of influenza-like illness and laboratory-confirmed influenza in hefei, China. Int. J. Biometeorol. 63, 51–60. doi:10.1007/s00484-018-1633-0
Liu, Y., Ning, Z., Chen, Y., Guo, M., Liu, Y., Gali, N. K., et al. (2020). Aerodynamic analysis of sars-cov-2 in two wuhan hospitals. Nature 582, 557–560. doi:10.1038/s41586-020-2271-3
Ma, J., Chen, Z., Wang, J., Wang, Y., and Li, L. (2022). Diffusion simulation, health risks, ozone and secondary organic aerosol formation potential of gaseous pollutants from rural comprehensive waste treatment plant. Chemosphere 286, 131857. doi:10.1016/j.chemosphere.2021.131857
Madhwal, S., Prabhu, V., Sundriyal, S., and Shridhar, V. (2020). Ambient bioaerosol distribution and associated health risks at a high traffic density junction at dehradun city, India. Environ. Monit. Assess. 192, 196. doi:10.1007/s10661-020-8158-9
Matjuda, D. S.-m., and Aiyegoro, O. A. (2019). Analysis of bacteriological pollution and the detection of antibiotic resistance genes of prevailing bacteria emanating from pig farm seepage. MicrobiologyOpen 8, e00737. doi:10.1002/mbo3.737
Matus, C. P., and Oyarzún, G. M. (2019). Impacto del material particulado aéreo (mp2.5) sobre las hospitalizaciones por enfermedades respiratorias en niños: Estudio caso-control alterno. Rev. Chil. Pediatr. 90, 166–174. doi:10.32641/rchped.v90i2.750
Meadow, J. F., Altrichter, A. E., Kembel, S. W., Kline, J., Mhuireach, G., Moriyama, M., et al. (2014). Indoor airborne bacterial communities are influenced by ventilation, occupancy, and outdoor air source. Indoor Air 24, 41–48. doi:10.1111/ina.12047
Niu, X., Wang, Y., Ho, S. S. H., Chuang, H.-C., Sun, J., Qu, L., et al. (2021). Characterization of organic aerosols in pm1 and their cytotoxicity in an urban roadside area in Hong Kong. Chemosphere 263, 128239. doi:10.1016/j.chemosphere.2020.128239
Reche, I., D’Orta, G., Mladenov, N., Winget, D. M., and Suttle, C. A. (2018). Deposition rates of viruses and bacteria above the atmospheric boundary layer. ISME J. 12, 1154–1162. doi:10.1038/s41396-017-0042-4
Setti, L., Passarini, F., De Gennaro, G., Barbieri, P., Perrone, M. G., Borelli, M., et al. (2020). Sars-cov-2rna found on particulate matter of bergamo in northern Italy: First evidence. Environ. Res. 188, 109754. doi:10.1016/j.envres.2020.109754
Shao, S., Zhou, D., He, R., Li, J., Zou, S., Mallery, K., et al. (2021). Risk assessment of airborne transmission of Covid-19 by asymptomatic individuals under different practical settings. J. Aerosol Sci. 151, 105661. doi:10.1016/j.jaerosci.2020.105661
Tellier, R. (2006). Review of aerosol transmission of influenza a virus. Emerg. Infect. Dis. 12, 1657–1662. doi:10.3201/eid1211.060426
Thomas, R. J. (2013). Particle size and pathogenicity in the respiratory tract. Virulence 4, 847–858. doi:10.4161/viru.27172
To, G. N. S., and Chao, C. Y. H. (2010). Review and comparison between the Wells–Riley and dose-response approaches to risk assessment of infectious respiratory diseases. Indoor Air 20, 2–16. doi:10.1111/j.1600-0668.2009.00621.x
Tsay, M.-D., Tseng, C.-C., Wu, N.-X., and Lai, C.-Y. (2020). Size distribution and antibiotic-resistant characteristics of bacterial bioaerosol in intensive care unit before and during visits to patients. Environ. Int. 144, 106024. doi:10.1016/j.envint.2020.106024
van Doremalen, N., Bushmaker, T., Morris, D. H., Holbrook, M. G., Gamble, A., Williamson, B. N., et al. (2020). Aerosol and surface stability of sars-cov-2 as compared with sars-cov-1. N. Engl. J. Med. 382, 1564–1567. doi:10.1056/NEJMc2004973
Wang Chia, C., Prather Kimberly, A., Sznitman, J., Jimenez Jose, L., Lakdawala Seema, S., Tufekci, Z., et al. (2021). Airborne transmission of respiratory viruses. Science 373, eabd9149. doi:10.1126/science.abd9149
Wang, P., Qiao, X., and Zhang, H. (2020). Modeling pm2.5 and o3 with aerosol feedbacks using wrf/chem over the sichuan basin, southwestern China. Chemosphere 254, 126735. doi:10.1016/j.chemosphere.2020.126735
Warfel, J. M., Beren, J., and Merkel, T. J. (2012). Airborne transmission of bordetella pertussis. J. Infect. Dis. 206, 902–906. doi:10.1093/infdis/jis443
Wilson, N. M., Norton, A., Young, F. P., and Collins, D. W. (2020). Airborne transmission of severe acute respiratory syndrome coronavirus-2 to healthcare workers: A narrative review. Anaesthesia 75, 1086–1095. doi:10.1111/anae.15093
Wu, B., Qi, C., Wang, L., Yang, W., Zhou, D., Wang, M., et al. (2020). Detection of microbial aerosols in hospital wards and molecular identification and dissemination of drug resistance of escherichia coli. Environ. Int. 137, 105479. doi:10.1016/j.envint.2020.105479
Yu, I. T. S., Li, Y., Wong, T. W., Tam, W., Chan, A. T., Lee, J. H. W., et al. (2004). Evidence of airborne transmission of the severe acute respiratory syndrome virus. N. Engl. J. Med. 350, 1731–1739. doi:10.1056/nejmoa032867
Keywords: non-open environment, pathogens, health risk, aerosol, particulate matter
Citation: Zhao Y, Mao K, Cao H, Wang B, Zheng X, Du W, Zhang X and Zhang H (2022) Nonnegligible pathogenic exposure risk of coarse part of PM10 in non-open environments. Front. Environ. Sci. 10:1006209. doi: 10.3389/fenvs.2022.1006209
Received: 01 August 2022; Accepted: 25 October 2022;
Published: 11 November 2022.
Edited by:
Nsikak U. Benson, Covenant University, NigeriaReviewed by:
Mikalai Filonchyk, Lanzhou Jiaotong University, ChinaMagdalena Reizer, Warsaw University of Technology, Poland
Yikun Yang, Peking University, China
Copyright © 2022 Zhao, Mao, Cao, Wang, Zheng, Du, Zhang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kang Mao, bWFva2FuZ0BtYWlsLmd5aWcuYWMuY24=; Hua Zhang, emhhbmdodWFAbWFpbC5neWlnLmFjLmNu
 Yirong Zhao
Yirong Zhao Kang Mao
Kang Mao Haorui Cao1,2
Haorui Cao1,2 Wei Du
Wei Du Hua Zhang
Hua Zhang