- 1Division of Agricultural Chemicals, ICAR-Indian Agricultural Research Institute, New Delhi, India
- 2Division of Environment Science, ICAR-Indian Agricultural Research Institute, New Delhi, India
- 3Division of Microbiology, ICAR-Indian Agricultural Research Institute, New Delhi, India
- 4Fishery Resources Assessment Division, ICAR-Central Marine Fisheries Research Institute, Kochi, India
Crude oil contamination of soil and water resources is a widespread issue. The present study evaluated the degradation of aliphatic hydrocarbons (C11–C36) in crude oil by 17 bacteria isolated from a crude oil–contaminated soil. The results suggested that Pseudomonas sp. and Bacillus amyloliquefaciens were the best hydrocarbon-degrading bacteria in the presence of surfactant Tween-80 (0.1% w/v). Based on the present investigation and a previous study, Pseudomonas sp. + B. amyloliquefaciens and fungus Aspergillus sydowii were identified as best oil degraders and were immobilized in alginate–bentonite beads, guargum–nanobenonite water dispersible granules (WDGs), and carboxy methyl cellulose (CMC)–bentonite composite. Sandy loam soil was fortified with 1, 2, and 5% crude oil, and total petroleum hydrocarbon (TPH) degradation efficiency of free cultures and bio-formulations was evaluated in sandy loam soils. Compared to a half-life (t1/2) of 69.7 days in the control soil (1% oil), free cultures of Pseudomonas sp. + B. amyloliquefaciens and A. sydowii degraded TPH with t1/2 of 10.8 and 19.4 days, respectively. Increasing the oil content slowed down degradation, and the t1/2 in the control and soils inoculated with Pseudomonas sp. + B. amyloliquefaciens and A. sydowii was 72.9, 14.7, and 22.2 days (2%) and 87.0, 23.4, and 30.8 days (5%), respectively. Supplementing soil with ammonium sulfate (1%) enhanced TPH degradation by Pseudomonas sp. + B. amyloliquefaciens (t1/2–10 days) and A. sydowii (t1/2–12.7 days). All three bio-formulations were effective in degrading TPH (1%), and the t1/2 was 10.7–11.9 days (Pseudomonas sp. + B. amyloliquefaciens and 14–20.2 days (A. sydowii) and were at par with free cultures. Microbial diversity analysis based on taxonomic markers and functional markers suggested that the bioaugmentation process helped keep soil in the active stage and restored the original microbial population to some extent. The present study concluded that bio-formulations of crude oil–degrading microbes can be exploited for its degradation in the contaminated environment.
Introduction
Crude oil is a rich source of hydrocarbons and a major source of energy to the ever growing automobile industry. Dependency on crude oil and products derived from it has greatly increased in the past few decades. At the same time, the number of incidences of accidental discharges of crude oil during exploration, dumping of industrial waste, damaged oil pipelines, petroleum refineries discharge, illegal theft, and accidents during transportation processes have been reported. Crude oil and petroleum products are carcinogenic and neurotoxic in nature (Deivakumari et al., 2020; Gangadhari et al., 2020). Therefore, it is of utmost importance to treat the exposure of crude oil contaminated soil in an effective way. India is not spared from such environmental threats, and several incidences of crude oil spillage have caused serious threat to the environment. It included destruction of more than 300 ha of mangrove across 100 km stretch of coastline in Mumbai due to collision of ships, four sq km spread of crude oil due to rupture of Mumbai–Uran trunk pipeline, and 25 miles of coastlines affected by ship collision in Chennai (THE ECONOMIC TIMES, 2011; THE HINDU, 2011; THE TIMES OF INDIA, 2017; Han et al., 2018). Several acres of paddy fields were ruined following irrigation by the oil slick–laden water from the channel, and few farmers got a compensation of INR 5000–80000 from Oil and Natural Gas Corporation Limited (ONGC) (THE HINDU, 2011). A one-acre Samba paddy fertile field was destroyed due to leakage of ONGC underground pipelines, and ONGC gave INR 75000 acre−1 compensation to farmers. A similar incidence happened again in 2018, and a 5-acre field was destroyed (THE TIMES OF INDIA, 2020). The 2020 Assam gas and oil leak, also referred to as Baghjan gas leak, occurred in Oil India Limited’s oil well in Tinsukia district, Assam, India, on May 27, 2020 due to a blowout of natural gas. The Baghjan oil Field has 21 active wells, of which 4 produce natural gas and 17 produce oil. The leak of gases and open fire incidences affected agricultural crops, plants including bamboos, tea, bananas, and betel nuts. Biodiversity were seriously affected, and condensate oil was found in the dead organism as well as on the agricultural land. Oil India Limited provided INR 2.5 million as immediate compensation to each of 12 families that completely lost their homes and continue to give INR 50000 per month as livelihood support to each family which are now forced to go away from their homes (BBC NEWS, 2020; WIKIPEDIA, 2020).
Both physical and chemical methods are used for treating/remediating oil contaminated sites, but they are neither cost-effective nor ecofriendly. Sometimes, by-products of treatment processes may pose a threat to public health and have a negative impact on the environment. Bioremediation is considered to be a safe and cost-effective approach, which leads to the complete mineralization of pollutants. However, it mainly depends on the biodegrading ability of native microbial populations or addition of exogenous microorganism(s) into the environment. Bioremediation of the contaminated site is mainly done either through bioaugmentation or bio-stimulation (Ali et al., 2020). Several studies reported the capabilities of indigenous microorganisms such as bacteria and fungus to degrade the hydrocarbons (Dean-Ross et al., 2002; Bundy et al., 2004; Maiti et al., 2008; Wang et al., 2011; Badr El-Din et al., 2014; Spini et al., 2018). Degradation of crude oil is limited due to low aqueous solubility; therefore, the use of non-toxic surfactant is suggested to enhance solubility and bioavailability. Cho et al. (1997) used eight different kinds of surfactants (alkylbenzene sulfonate, olefin sulfonate, alkyl sulfonate ester, alkylphenyl ether sulfonate ester, alkylphenyl ether sulfonate, dioctyl sulfosuccinic acid, alkylphenyl ether, and alkyl) and reported reclamation of 15–33% of the contaminated soil. Tween-80 (polyoxyethylene sorbitan monooleate) is a hydrophilic non-ionic surfactant and is widely used due to its non-toxic nature (Kachieng’a and Momba, 2017).
Microbiological cultures, enzyme additives, or nutrient additives that significantly increase the rate of biodegradation to mitigate the effect of contamination/discharges were defined as bioremediation agents by USEPA (Das and Chandran, 2011). Sometimes, addition of an inoculum at contaminated sites may lead to failure to show their maximum potential. It might be due to following reasons: 1) failure to adapt the prevailing environmental conditions (fluctuation or extremes in temperature, water content, pH, and/or nutrient availability); 2) tolerate co-contaminants like heavy metals; 3) competition with indigenous microbiota in cases of limited nutrients; and/or 4) unable to survive the predators (protozoa and bacteriophages) or antibiotic produced by competing microorganisms (Juhanson et al., 2009; Siles and García-Sánchez, 2018). To overcome such issues, bioaugmented products are typically recommended, either in liquid slurries or freeze-dried forms that may contain inorganic nutrients. Immobilization of microbes on natural (dextran, agar, agarose, alginate, chitosan, etc.) or synthetic (polyacrylamide, polyvinyl alcohol, etc.) carrier protects them from predatory and suboptimal environmental conditions (Zawierucha and Malina, 2011; Nwankwegu and Onwosi, 2017; Partovinia and Rasekh, 2018; Mehrotra et al., 2021).
Identification of suitable region-specific microbes/consortium microorganism is a very important task for the success of bioremediation. Studies on the effect of crude oil and bioaugmentation on microbial diversity in soil are very limited. The present study reports isolation of bacteria from oil-contaminated soils and their potential to degrade aliphatic hydrocarbon fractions and the effect of surfactant (Tween-80) on degradation. Microbes exhibiting best degradation were formulated for effective and successful degradation in oil-fortified sandy loam soil. A metagenomics study was performed to account variation among diversity and functional genes in control (without oil, uninoculated) and oil-fortified and -treated sandy loam soils.
Materials and Methods
Chemicals
Certified reference standard (1,000 μg ml−1) of C7–C40 saturated alkanes (49452-U) was purchased from Merck India. Crude oil was extracted from a crude oil–contaminated soil collected from a site adjacent to an oil well in Gujarat, India. It was characterized for total carbon (Elementar), heavy metals (atomic absorption spectrophotometer, Shimazdu Model AA 7000) (Supplementary Table S1), and aliphatic hydrocarbon using an analytical standard (C7–C40) mixture using gas chromatography. The molecular identification (16S rRNA) of isolated microbes was performed using Zymo Research Fungal/Bacterial Quick DNA MicroPrep™ D6005 following the manufacturer’s standard protocol. The soil DNA isolation kit (Nucleopore GDNA), was purchased from Genetix, New Delhi, India, and DNA was extracted as per standard protocol.
Soil
An alluvial soil from the experimental farm of ICAR-Indian Agricultural Research Institute (IARI), New Delhi, was collected for use in degradation studies. Sampling was done up to 15 cm depth of soil profile. The soil sample was air-dried at room temperature in shade, grounded using a mortar and pestle, sieved through a 2-mm sieve, and stored in plastic bags at room temperature. The pH of soil sample was measured using a Control Dynamics pH meter (APX 175 E/C) fitted with a calomel glass electrode (Jackson, 1967). The organic carbon content of soil was determined using the Walkley and Black method. The soil texture (sand, silt, and clay) was determined using the Bouyoucos hydrometer method (Day, 1965). The electrical conductivity (EC) of the soil sample was measured using a conductivity meter (Supplementary Table S1).
Isolation of Microbes Using Enrichment Technique and Identification
Bacteria were isolated from the crude oil–contaminated soil (Ahmedabad, Gujarat) using serial dilution methods. Crude oil–contaminated soil (1 g) was added in 10 ml minimal salt medium (MSM) and incubated at 27°C on a rotary shaker (120 rpm) for 7 days. Afterward, 1 ml of culture was serially diluted up to 109-fold, and 100 μl of each dilution was plated on 0.1% oil-enriched MSM agar plates. The bacteria grown on MSM agar were further purified and maintained on nutrient agar. They were identified using a standard molecular characterization approach based on the amplification of 16S rRNA gene sequencing. The partial sequences of the isolates were compared with sequences available in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov) through BLAST search to identify the nearest taxa and were submitted to NCBI GenBank, and accession number was obtained. The phylogenetic analysis for bacterial strains was performed by MEGAX software.
Degradation of Crude Oil in Media
The degradation of petroleum hydrocarbons in crude oil was studied using 17 isolated bacteria. Crude oil (250 mg, dissolved in 1 ml hexane) was added to a sterilized 100-ml Erlenmeyer flask, and hexane was allowed to evaporate under aseptic conditions. After evaporation of hexane, sterilized 25 ml slightly modified Reese’s minimal medium (RMM) (Reese and Mandels, 1963), without Tween-80 (−T80) and with 0.1% w/v Tween-80 (+T80), were added to two sets of flasks (Supplementary Table S2). Both sets of media were inoculated with bacteria (10% v/v culture, OD600∼1.0). Uninoculated samples served as control. Bacterial samples were incubated at 27°C in a rotary incubator maintained at 120 rpm for 30 days. After 30 days, flasks in triplicate were removed from each treatment for enzymatic studies and extraction/analysis of hydrocarbons.
Biochemical Analysis
Microbial enzymatic parameters were measured during the degradation of crude oil in microbial culture in the RMM. The soluble protein content was estimated using an alkaline copper reagent (ACR) and Folin–Ciocalteu reagent (FCR) as per the procedure developed by Lowry et al. (1951). Fluorescein diacetate (FDA) hydrolase activity was analyzed following the method described by Green et al. (2006).
Preparation and Characterization of Microbial Formulations
Based on the ability to degrade crude oil/aliphatic hydrocarbons in the crude oil, non-pathogenicity and mutual compatibility studies, a consortium of Pseudomonas sp. + B. amyloliquefaciens and from our previous laboratory experiment, fungus A. sydowii exhibiting good crude oil–degrading ability (Khandelwal et al., 2021), were formulated. For preparing bio-formulations, bacterial cultures were grown in nutrient broth (250 ml), and after 1–2 days (till OD600 ∼ 1 achieved), cells were harvested by centrifugation at 4°C. Harvested cells were finally suspended in fresh nutrient broth to obtain 1010–1011 CFU ml−1. Similarly, fungal cultures were grown in potato dextrose agar plates (92 mm) for 30 days at 30°C for spore harvesting. After 30 days, fungal spores were scratched and transferred into 60 mM phosphate buffer so as to obtain a spore count of ∼107 ml−1. These bacterial cells and fungal spores were used for preparing three variants of solid formulations.
Bentonite–Alginate Composite Bead Formulation
Composite bead formulations of 1) Pseudomonas sp. + B. amyloliquefaciens and 2) Aspergillus sydowii were prepared. Initially, bentonite clay (1 g) was dispersed in 100 ml of microbial culture(s), followed by gradual mixing of sodium alginate (10 g) with continuous stirring for 30 min. After 30 min, the prepared mixture was dropwise added with a dropper in an aqueous solution of CaCl2 (2%) for bead formation. The beads were cured in CaCl2 solution overnight. The procedure was performed under aseptic conditions in laminar flow. The next day, beads were separated and washed 2–3 times with distilled water to remove CaCl2. The beads were stored at room temperature (Kumar et al., 2017). The size of wet beads was observed on a scale with an average of 20 beads.
Guargum–Nanobentonite Water-Dispersible Granule Formulation
Water-dispersible granules of 1) Pseudomonas sp. + B. amyloliquefaciens and 2) A. sydowii were prepared using the wet granulation method (Chumthong et al., 2008). All synthesis procedures were performed under aseptic conditions in laminar flow. Initially, 47 g nanobentonite clay (inert carrier) and 0.5 g guargum (binding agent) were mixed with 20 ml of distilled water and autoclaved for 15 min for three consecutive days. Then approximately 80 ml of microbial consortia/fungal spores and 2.5 g Tween-80 (dispersing agent) were mixed thoroughly with it to knead into loose dough. The kneaded dough was extruded through a hand extruder having a pore diameter of 1 mm. The developed formulations were dried in shade to attain <4% moisture content (Yadav et al., 2020).
The particle size of suspended WDGs was measured by using a particle size analyzer (Zetatrac, Microtrac, PA, United States) using the laser scattering principle. The polydispersity index (PDI) was calculated by the following formula (Eq. 1):
Carboxymethyl Cellulose–Bentonite Composite Formulation
CMC–bentonite composite formulations of 1) Pseudomonas sp. + B. amyloliquefaciens and 2) A. sydowii were prepared. Bentonite clay (1 g) was dispersed with 100 ml bacterial consortia/fungal spore solution and stirred for 2 h. After that, 1.75 g carboxymethyl cellulose (1,500–3,000 cP) was added to it and allowed to dissolve for another 2 h. The procedure was conducted aseptically in laminar flow. The viscous mass was lyophilized. First, viscous mass was kept at −20°C until it was completely frozen. Afterward, it was transferred to a high-vacuum lyophilizer at −80°C until the complete removal of water; the composite material was scratched and turned into powder using a mortar and pestle, and the dried powder was stored at room temperature.
Bacteria and Fungus Population Counts in Developed Bio-Formulations
The population of bacteria, in terms of colony forming unit (CFU), in the developed microbial formulation was performed by the using serial dilution method after storage at 4 ± 1°C (0, 1 day), 54 ± 1°C (0, 14 days), and 27 ± 1°C (0, 30, 90, 120, 150, and 180 days, and fungal spores were counted by using a hemocytometer.
Degradation of Crude Oil by Free and Formulated Cultures in the Sandy Loam Soil
Free cultures of 1) Pseudomonas sp. + B. amyloliquefaciens consortium and 2) A. sydowii fungus and their formulations were used to study crude oil degradation in the sandy loam soil. Air-dried soil (90) g in a 250-ml sterilized conical flask was supplemented with the sterile distilled water to attain 15% field capacity. Samples were incubated in a biological oxygen demand (BOD) incubator at 27 ± 1°C for 10 days for conditioning. Due to better degradation in the presence of Tween-80 [0.1% (w/w)], it was added. After 10 days, samples were fortified with the crude oil at 1, 2, and 5% fortification levels. This was done by fortifying 10 g soil with the required amount of crude oil in 5 ml hexane and mixing it with conditioned (90 g) soil. A total of 19 different treatments, in triplicate, were maintained as per details mentioned in Supplementary Table S3. Among different treatments, soils in T3 and T9 treatments were supplemented with ammonium sulfate (1%), while soils in treatments T4 and T10 were supplemented with compost (0.5%) to evaluate the effect of these amendments on the crude oil degradation. Soil samples in different treatments were inoculated with the free culture or formulated cultures. After inoculation, the samples were incubated at 27 ± 1°C. At regular intervals (0, 5, 10, 20, 30, and 60 days), the samples (5 g) were aseptically removed from each treatment for extraction of crude oil, and percent degradation was calculated by the difference in the weight of oil recovered on 0 days.
Extraction and Analysis
An analytical method for quantification of aliphatic hydrocarbons in the crude oil was standardized using analytical standard (C7–C40). Gas chromatography (GC-17 A, Shimadzu) equipped with an SG 5 column (30 m length, 0.53 mm ID, and 0.50 µm film thickness) and a flame ionization detector (FID) was used. Method conditions were: injection temperature 250°C (split mode), detector temperature 300°C, column temperature 100°C for 5 min, and a temperature increase at a rate of 10°C min−1 until 290°C and maintained till end of run.
The method for extraction of aliphatic fraction/crude oil from oil medium or soil was standardized. Aliphatic hydrocarbons from crude oil–fortified medium was extracted using 100, 70, and 50 ml hexane:methylene chloride (1:1). Organic fractions after three extractions were pooled, solvent was evaporated, and residues were redissolved in 10 ml hexane. Aliphatic hydrocarbons in samples were quantified using gas chromatography (GC), and percent degradation of individual hydrocarbon was quantified by the difference in the amount/area of hydrocarbon in the uninoculated control and bacteria-inoculated samples.
Total petroleum hydrocarbons (TPHs) from soil samples were extracted by taking the sample (5 g) in a 50-ml oak ridge centrifuge tube was mixed with 2.5 ml distilled water and left for 30 min. Subsequently, 5 ml of ethyl acetate was added and vortexed for 2 min. Then 2 g anhydrous magnesium sulfate was added and vortexed for 1 min. Subsequently, 0.75 g sodium chloride was added and shaken vigorously. The tube was vortexed for 1 min, and contents centrifuged at 5,000 rpm for 5 min. After centrifugation, 2.5 ml ethyl acetate fraction was transferred to a weighed test tube, and after solvent evaporation, gravimetrically weight of crude oil was measured.
Diversity of Microbial Groups Based on Taxonomic and Functional Gene Copies
The total soil genomic DNA, in triplicate, was extracted using the isolation kit (Nucleopore GDNA, Genetix, New Delhi, India) after the 10th day. The concentrations of total soil genomic DNA were determined using Nanodrop 3,300 spectrofluorometry (Waltham, Massachusetts, United States). The extracted DNA samples were stored at −20°C until further analysis. The abundance of 16S rRNA gene copies in different treatments was quantified from the total genomic DNA extracted from the sandy loam soil (Delhi) fortified with crude oil and the control soil. Similarly, the abundances of functional genes related to N-cycling were quantified using the quantitative PCR (qPCR) method. The extracted total genomic DNA was used as a template (4 µl) in the 20 µl reaction volume with primers specific for each gene tested, the SYBR Green I Master Mix (10 µl), bovine serum albumin (1 µl of 10 mg ml−1), and the nuclease-free water (5 µl). The qPCR assays were performed in the LightCycler 96 Real-Time PCR System (Roche Diagnostics Corp., Indianapolis, Indiana, United States).
Abundances of the 16S rRNA genes specific for Alphaproteobacteria (Eub338F- 5′ACTCCTACGGGAGGCAGCAG3′, Alf685R-5′TCTACGRATTTCACCYCTAC3′), Betaproteobacteria (Eub338F-5′ACTCCTACGGGAGGCAGCAG3′, Bet680R- 5′TCACTGCTACACGYG3′), Firmicutes (Lgc353F-5′GCAGTAGGGAATCTTCCG3′, Eub518R-5′ATTACCGCGGCTGCTGG3′), Bacteroidetes (Cfb319F- 5′GTACTGAGACACGGACCA3′, Eub518R- 5′ATTACCGCGGCTGCTGG3′), Bacteria (Eub338F-5′ACTCCTACGGGAGGCAGCAG3′, Eub518R-5′ATTACCGCGGCTGCTGG3′) (Fierer et al., 2005), and Archaea (Arch1F-5′ CGGTGAATACGTCCCTGC3′, Arch2R- 5′CGGTGAATATGCCCCTGC3′) (Suzuki et al., 2000) and the functional genes such as amoA (amoA1F-5′GGGGTTTCTACTGGTGGT3′, amoA2R-5′CCCCTCKGSAAAGCCTTCTTC3′) (Rotthauwe et al., 1997), nifH (nifH1F-5′TGCGAYCCSAARGCBGACTC3′, nifH2R- 5′ATSGCCATCATYTCRCCGGA3′) (Poly et al., 2001), nirS (nirS1F- 5′CCTA(C/T)TGGCC(A/G)CA(A/G)T3′, nirS6R-5′CGTTGAACTT(A/G)CCGGT3′), nirK (nirK1F-5′GG(A/C)ATGGT(G/T)CC(C/G)TGGCA3′, Amx1066R-nirK3R-5′GAACTTGCCGGT(A/C/G)G(C/T)CCAGAC3′) (Braker et al., 1998), and the anammox (Amx818F-5′ATGGGCACTMRGTAGAGGGGTTT3′,Amx1066R-5′AACGTCTCACGACACGAGCTG3′) (Li and Gu, 2011) were quantified. The fluorescence was detected during the extension step of each cycle while the melt-curve analysis confirmed the specificity of amplification. The calibration curves were generated using 10-fold serial dilutions of plasmid containing targeted genes and the CT values were plotted against the gene copies. The concentrations of genomic DNA and cloned plasmids were determined using Nanodrop 3300. The gene copy numbers were then calculated and expressed in terms of per gram soil.
Statistical Analysis
Data were transformed using appropriate transformations, namely, square-root, and logarithmic to make it amenable for statistical analysis, and the comparison among various sources and their interactions was assessed based on the transformed values.
Degradation of Crude Oil by Bacteria in Medium
Crude oil degradation data were subjected to two-way ANOVA taking the two different sources of variability, namely, bacterial species and surfactant (−/+T80). Analysis of variance (ANOVA) was performed using SAS 9.4 software, and significant effects/interactions were noted at 5% level of significance. Tukey’s HSD test was done for pairwise comparison among the various effects and interactions. Clustering of the extracted compounds was done using R software, and the homogeneous groups of compounds were represented with the help of a dendrogram. The physicochemical properties of hydrocarbon that affected percent hydrocarbon degradation was identified using a principal component–based regression analysis which was performed using PROC PRINCOMP and PROC REG in SAS 9.4, and the adequacy of the model was assessed by the R2 values. Besides, a heat map has been prepared using an R package “pheatmap” (Kolde, 2019) using R-studio (R Core Team, 2020) to visualize the homogeneity among different bacteria in the presence/absence of surfactants and also among various compounds. The hierarchical clustering approach was used to prepare the dendrogram in the heat map based on the similarities calculated through the Euclidean distance.
Degradation of Total Petroleum Hydrocarbon in Crude Oil by Bacteria/Fungus in Soil
Crude oil degradation data in soil were subjected to one-way ANOVA, and the significance was noted at p < 0.05. Furthermore, Tukey’s HSD test was performed for multiple comparisons among treatment effects.
Abundance of Diverse Microbial Groups
One-way ANOVA was done to compare the effect of treatments on abundances of 16S rRNA gene copies after the 10th day of crude oil fortification in the sandy loam soil. The mean values of the treatments are depicted in the graph along with the error bars. The letter grouping was presented based on Tukey’s HSD test on the logarithmic values of abundances of gene copies g−1 of soil. The principal component analysis was performed using XLSTAT 2014.5.03.
Half-Life Calculation
The degradation data were fitted to the first-order kinetic Eq. 2:
where C0 is the apparent initial concentration (μg g−1), Ct is the concentration (μg g−1) after a lapse of time t (days), and k is the degradation rate constant. The half-life (t1/2) values were calculated from the k value using following formula (Eq. 3):
Results and Discussion
Crude Oil
Crude oil was characterized for aliphatic hydrocarbons and chromatograms of certified reference material and crude oil constituents are shown in Supplementary Figure S1. Crude oil components were identified based on the certified reference standards, and their concentration in oil were calculated (Supplementary Table S4). Results suggest that out of 33 peaks obtained in the Gujarat oil sample, 26 peaks could be identified, while the identity of seven peaks was uncertain. Twenty-six compounds, which could be identified, were C11 (undecane) to C36 (hexatriacontane) hydrocarbon. The total concentration of these aliphatic hydrocarbon compounds in the crude oil (equivalent to 10,000 mg kg−1) was found to be 1755.6 mg kg−1. Earlier, Mandal et al. (2012) reported C14–C34 aliphatic hydrocarbons in IOCL Panipat refinery, India, where initial total petroleum hydrocarbons were reported as 206.50–231.00 g kg−1 oily waste.
Isolation of Microbes From Oil Contaminated Soil and Their Identification
Seventeen bacteria were isolated from the enrichment culture (Supplementary Table S5). Based on rRNA gene sequences, the bacterial strains were identified as Lysinibacillus sp. AK1, Arthrobacter pascens AK2, Bacillus aryabhattai AK3, Kocuria rosea AK4, Arthrobacter sp. AK5, Kocuria flava AK6, Pseudomonas aeruginosa AK7, Bacillus megaterium AK8, Nocardioides sp. AK9, Bacillus sp. AK11, Pseudomonas sp. AK12, Sporosarcina luteola AK13, Bacillus niacin AK14, Bacillus siamensis AK15, Pseudomonas sp. A3, Bacillus amyloliquefaciens A9, and Staphylococcus sp. A14. The phylogenetic analysis was done using MEGAX, and a phylogenetic tree was constructed. All bacterial (Supplementary Figure S2) strains were showing approximately sequence similarity with their close relatives. Earlier, researchers reported about the potential of isolated microbes such as Bacillus sp., Rhodococcus sp., and Pseudomonas sp. (Pathak et al., 2017; Bilen Ozyurek and Bilkay, 2020; Viesser et al., 2020).
Degradation of Crude Oil by Isolated Bacteria in Medium
Degradation of aliphatic hydrocarbon fraction of crude oil was studied using 17 isolated bacteria, without (−T80) and with (+T80) Tween-80 (0.1%) and percent degradation of individual hydrocarbon was calculated with respect to its concentration/area in the uninoculated control sample at zero time. The control (uninoculated) medium showed 6–10% degradation of crude oil on the 30th day. It might be due to some abiotic factors like temperature, shaking over different days, and losses of moisture. However, it was very less than added inoculum in the medium. Therefore, the result of heat map was compared on the basis of added inoculum to assess the effect of bacterial strain in an effective way. The heat map (scale 0–100) was prepared using percent degradation of individual hydrocarbon by a bacterium in −T80 and +T80 treatments after 30 days (Figure 1). Aliphatic hydrocarbons (C11–C36) were clustered into different groups based on their degradation similarity, calculated using the Euclidean distance. Results suggested that C11–C15 hydrocarbons were degraded faster by all the isolated bacteria than aliphatic hydrocarbon >C16 and were grouped together. Probably, the physicochemical properties of hydrocarbons played an important role in their degradation. Among all bacteria, B. amyloliquefaciens, P. aeruginosa (a pathogenic bacteria), and Pseudomonas sp., showed fairly good degradation ability for aliphatic hydrocarbons in the crude oil. Tween-80 affected degradation of aliphatic hydrocarbons, but the effect varied for different bacteria (Table 1). Tween-80 increased degradation by P. aeruginosa (20.5%) K. flava (15.7%), B. niacini (14.8%), B. aryabhattai (12.3%), Pseudomonas sp. (11.7%), and A. pascens (9.2%), but the increase was non-significant. More than 80% degradation was observed by B. amyloliquefaciens, Pseudomonas species, and P. aeruginosa in the presence of Tween-80; however, the effect was non-significant. Thus, results of this study suggested that few of the isolated bacteria have fairly good capability to degrade aliphatic hydrocarbon fraction in the crude oil. Previously, Mittal and Singh (2009) reported that in 60 days, Pseudomonas Strain-I and Acinetobacter calcoaceticus showed 70.69 and 75.09% alkane and 45.37 and 47.7% aromatic fraction of crude oil. Pathak et al. (2017) and Patel and Bhaskaran (2018) reported that P. aeruginosa IASST201 and P. nitroreducens could degrade 77% (168 h) and 70% (10 days) crude oil degradation. Balachandran et al. (2012) observed that Streptomyces sp. (ERI-CPDA-1), isolated from oil-contaminated soil of Chennai region, had potential to degrade 98.25% diesel oil in 7 days at 30 C. Similarly, Bhattacharya et al., 2019 found that the mixed culture of Ochrobactrum pseudintemedium C1 +Bacillus cereus K1 was able to degrade 42.67–70.25% crude oil (4%) within 72 h.
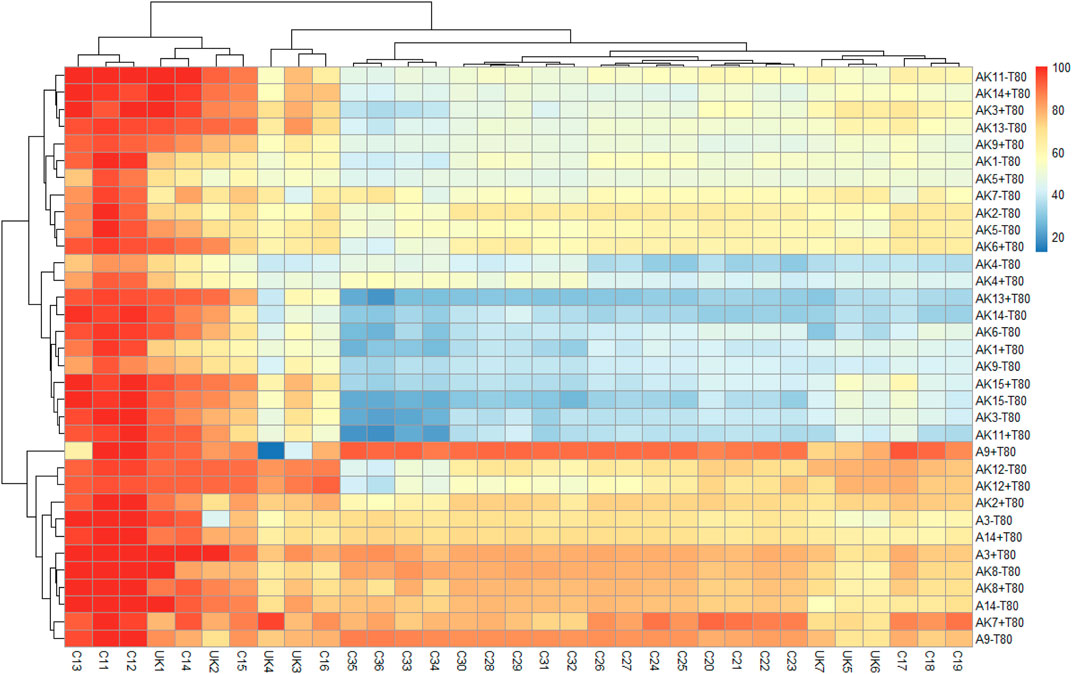
FIGURE 1. Heat map showing percent degradation of aliphatic hydrocarbons in the crude oil by different bacteria in mineral salts medium, without (−T80) and with Tween-80 (+T80). [AK1-Lysinibacillus sp.; AK2-Arthrobacter pascens; AK3-Bacillus aryabhattai; AK4-Kocuria rosea; AK5-Arthrobacter sp.; AK6-Kocuria flava; AK7-Pseudomonas aeruginosa; AK8-Bacillus megaterium; AK9-Nocardioides sp.; AK11-Bacillus sp.; AK12-Pseudomonas sp.; AK13-Sporosarcina luteola; AK14-Bacillus niacin; AK15-Bacillus siamensis; A3-Pseudomonas sp.; A9-Bacillus amyloliquefaciens; A14-Staphylococcus sp.; alkanes C11–C36 represents as undecane (C11H24), dodecane (C12H26), tridecane (C13H28), tetradecane (C14H30), pentadecane (C15H32), hexadecane (C16H34), heptadecane (C17H36), octadecane (C18H38), nonadecane (C19H40), eicosane (C20H42), heneicosane (C21H44), docosane (C22H46), tricosane (C23H48), tetracosane (C24H50), pentacosane (C25H52), hexacosane (C26H54), heptacosane (C27H56), octacosane (C28H58), nonacosane (C29H60), triacontane (C30H62), hentriacontane (C31H64), dotriacontane (C32H66), tritriacontane (C33H68), tetratriacontane (C34H70), pentatriacontane (C35H72), hexatriacontane (C36H74), and unknown compounds are represented as UK1-UK7].
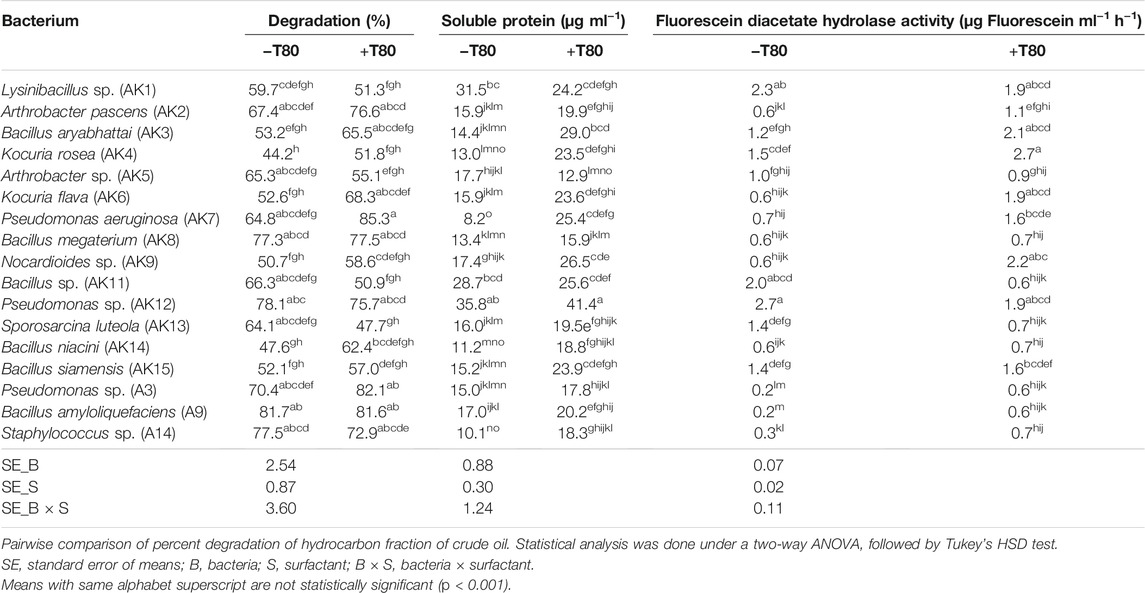
TABLE 1. Percent degradation of aliphatic fraction of crude and enzymatic activity during crude oil degradation by different bacteria, without (−T80) and with (+T80) Tween-80.
Percent aliphatic hydrocarbon degradation by different bacteria in −T80 and +T80 was correlated with physicochemical properties of hydrocarbons, so that parameters having a maximum effect on degradation can be identified. Due to high correlation among the various physicochemical properties of hydrocarbons (Supplementary Table S6), regression analysis, based on principal components, was performed. The first two extracted principal components explain 99% of the variability present in the data. Boiling point (BP) showed maximum contribution to the first principal component (PC1), followed by the melting point (MP), octanol–water partition coefficient (Kow), molecular weight (MW), and complexity, without much variability, while the aqueous solubility was having the least contribution to PC1. Regression analysis showed a significant negative correlation with all the physicochemical properties through PC1 for all the treatments. There are no available reports on correlating physicochemical properties of oil hydrocarbons with their degradation by bacteria.
Biochemical Analysis
Microbial parameters, soluble protein, and fluorescein diacetate (FDA) hydrolase activity were estimated in the medium (Table 1). The soluble protein content, an important parameter for assessing total enzymatic activity, varied from 8.2 to 35.8 μg ml−1 (−T80) and from 12.9 to 41.4 μg ml−1 (+T80). The significant increase in the soluble protein content by Tween-80 in B. aryabhattai, K. rosea, K. flava, P. aeruginosa, Nocardioides sp., B. niacini, B. siamensis, and Staphylococcus sp. were observed. Screened bacteria showed a variation in terms of soluble protein content in the presence of crude oil in the medium. It might be due to differences in the genetic potential of microbes to catabolize the oil fractions. In general, the increase/decrease in total soluble protein content corresponded with the increase/decrease in aliphatic hydrocarbon degradation.
The microbial enzyme FDA hydrolase, an extracellular enzyme, is mainly involved in the cleavage of ester linkage. The FDA hydrolase activity varied between 0.2 and 2.7 µg fluorescein ml−1 h−1 (−T80) and 0.7–2.7 µg fluorescein ml−1 h−1 (+T80). The results showed that Tween-80 significantly increased the FDA activity in medium inoculated with A. pascens, B. aryabhattai, K. rosea, K. flava, P. aeruginosa, Nocardioides sp., Bacillus sp., S. luteola, Pseudomonas sp., B. amyloliquefaciens, and Staphylococcus sp.. An increase in the FDA activity corroborated with the increase in percent aliphatic hydrocarbon degradation in certain cases.
Characterization of Bio-Formulations and Shelf Life Study
Six bio-formulations (alginate–bentonite beads, guargum–nanobentonite WDGs, and CMC-bentonite composite) of Pseudomonas sp. + B. amyloliquefaciens and A. sydowii were developed and characterized. The size of wet beads varied between 4 and 5 mm (Figure 2) and was measured by the average value of the size of 20 beads (Kumar et al., 2017). The particle size of WDGs (intensity-weighted mean hydrodynamic diameter) dispersed particles was 443 nm (Pseudomonas sp. + B. amyloliquefaciens) and 463 nm (A. sydowii). Results showed the unimodal distribution of A. sydowii WDGs, while the bimodal distribution was observed for WDGs of Pseudomonas sp. + B. amyloliquefaciens. Polydispersity indices (PDIs) of Pseudomonas sp. + B. amyloliquefaciens (0.35) and A. sydowii (0.19) formulations were less than those of nanobentonite (0.59), indicating homogeneous distribution of particles. Results revealed that the particle diameters at 50th percentile (D50) for different samples ranged between 282.4 and 433.0 nm. The average diameter of the dispersed particles of all the formulations was below 0.5 µ, suggesting their suitability to qualify for good preparations of WDGs.

FIGURE 2. Three types of bio-formulations of Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9 and Aspergillus sydowii AK20. (1) Alginate–bentonite beads (A,B). (2) Guargum–nanobentonite water-dispersible granules (WDG). (C,D) Carboxymethyl cellulose (CMC)–bentonite (E,F).
Bacterial CFU and the fungal spore count were expressed in the form of a logarithmic scale (Supplementary Table S7). Results suggested that the following storage at 27°C, bacterial population varied between 1011 and 1012 CFU g−1 of the carrier material. A similar trend was observed when samples were stored at 4 and 54°C. The fungal spore count was varied between 107 and 108 spore g−1 carriers over different days across different temperatures. It shows that formulated bacterial formulations were stable for a longer period of time. Compared to formulated products, storage of bacterial cultures beyond 30 days resulted in a decrease in counts, and after 180 days, 106 and 104 CFU/spore ml−1 were observed. However, storage at 4°C (1 day) and 54°C (14 days) showed no significant change in counts.
Degradation of Crude Oil Using Bio-Formulations in the Sandy Loam Soil
Degradation of total petroleum hydrocarbon (TPH) in crude oil was studied using free cultures and bio-formulations of Pseudomonas sp. + B. amyloliquefaciens and A. sydowii (Supplementary Table S8). Results using free cultures clearly indicated that compared to the uninoculated control (T1), percent TPH degradation (1% crude oil) was significantly higher in sandy loam soils inoculated with bacterial consortium (T2) and fungus (T8). At the 60th day, compared to 45.3% TPH degradation in T1 treatment, 97.5 and 87.5% degradation was observed in the T2 and T8 treatments, respectively, indicating that bacterial consortium was better in degrading TPH than the fungus. Addition of ammonium sulfate (1%, T3 and T9) as a nitrogen (N) source enhanced TPH degradation by bacterial consortium (T3) and fungus (T9) than respective T2 and T8 treatments, and at the 60th day, 100% (T3) and 96% (T9) degradations were observed. However, compost did not show significant enhancement in TPH degradation, and at the 60th day, 95.6% (T4) and 84.7% (T10) degradations were observed in compost-supplemented soils.
Increasing the crude oil content to 2 and 5% slowed down TPH degradation in soils. At the 60th day, compared to 43.6% degradation in the uninoculated control (T14), 93.4 and 83.5% degradations were observed in free bacterial consortium (T15) and fungus (T16) treatments, respectively. Increasing the crude oil content to 5% further slowed down degradation, and only 38, 81.6, and 72.6% degradations were observed in the control (T17), bacterial consortium (T18), and fungus (T19) treatments, respectively.
Bio-formulations of Pseudomonas sp. + B. amyloliquefaciens and A. sydowii, namely, bentonite–alginate beads (T5, T11), guargum–nanobentonite WDGs (T6, T12), and CMC–bentonite composite (T7, T13) were used to degrade TPH in 1% crude oil–fortified sandy loam soils (Supplementary Table S8). Results suggested that bio-formulations were effective in degrading TPH, and at the 60th day, 100.0% (T5), 100.0% (T6), 97.7% (T7), 88.9% (T11), 94.3% (T12), and 86.2% (T13) degradations were observed. Percent degradation observed using bio-formulations was slightly better or at par with the degradation by free bacterial consortium (T2-97.5%) and fungus (T8-87.5%) cultures.
Total petroleum hydrocarbon degradation data from different treatments (T1–T19) were fitted into the first-order kinetics (Table 2, Figure 3). Data fitted well to first-order kinetics as in most of the cases, the values of correlation coefficient (r) were >0.970. The half-life (t1/2) of TPH in 1% (T1), 2% (T14), and 5% (T17) uninoculated soils was 69.7, 72.9, and 87.0 days, respectively. Free cultures significantly enhanced degradation, and respective t1/2 values were 10.8 (T2), 14.7 (T15), and 23.4 (T18) days (bacterial consortium) and 19.4 (T8), 22.2 (T16), and 30.8 (T19) days (fungus). Thus, microbial cultures increased TPH degradation in soils, but higher concentrations were slightly toxic to microbes; and compared to 1% crude oil fortified soil (T2), the t1/2 values in the bacterial consortium 2% (T15) and 5% (T18) treatments increased by 36.1 and 116.7%, respectively. Similarly, the t1/2 in fungus treatments, compared to T8 (1%), increased by 14.4% (T16) and 58.8% (T19) in respective treatments. These results suggested that bacterial consortium was better in degrading the TPH content in crude oil–fortified soils, but fungi were more resilient at higher crude oil content and less strained to higher oil content. Supplementing soils with ammonium sulfate enhanced degradation by free bacterial consortium (t1/2-10.0 days) and fungus (t1/2-12.7 days), while no effect of compost was observed on respective t1/2 (T4-12.8 days; T10-21.4 days. The t1/2 values of TPH in bio-formulation–inoculated soils varied between 10.7 and 11.9 days (bacterial consortium) and 14.0–20.2 days (fungus). Among bacterial formulations, CMC–bentonite composite formulation (T7) was the best bacterial formulation (t1/2–10.7 days) and showed degradation at par with free consortium (T2, t1/2–10.8 days). Among fungus formulations, guargum–nanobentonite WDG formulation (T12) was better (t1/2–14.0 days) than the free culture (T8, t1/2–19.4 days).
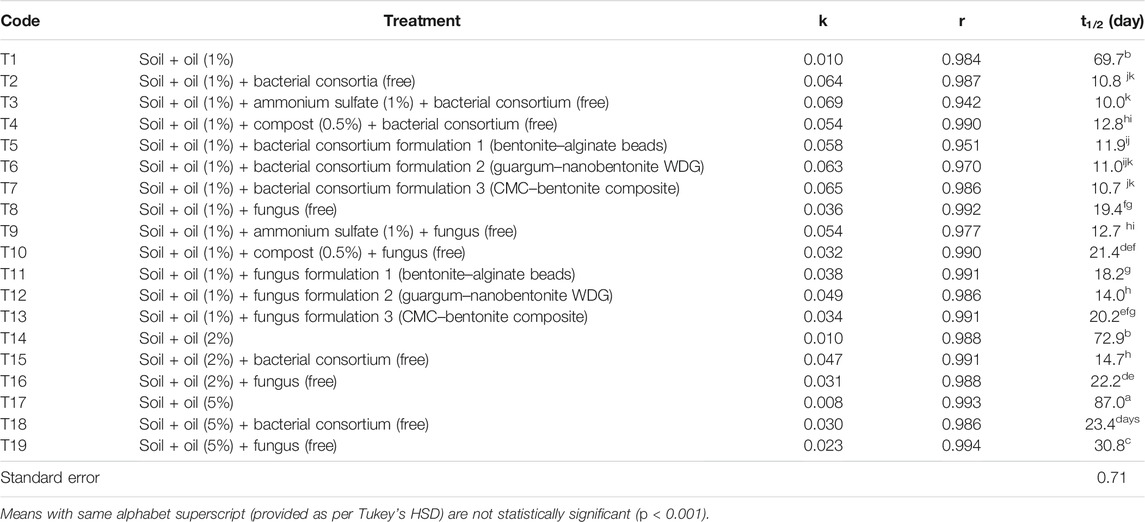
TABLE 2. Degradation parameters, rate constant (k), coefficient of correlation (r), and half-life (t1/2) for total petroleum hydrocarbons in the sandy loam soil.
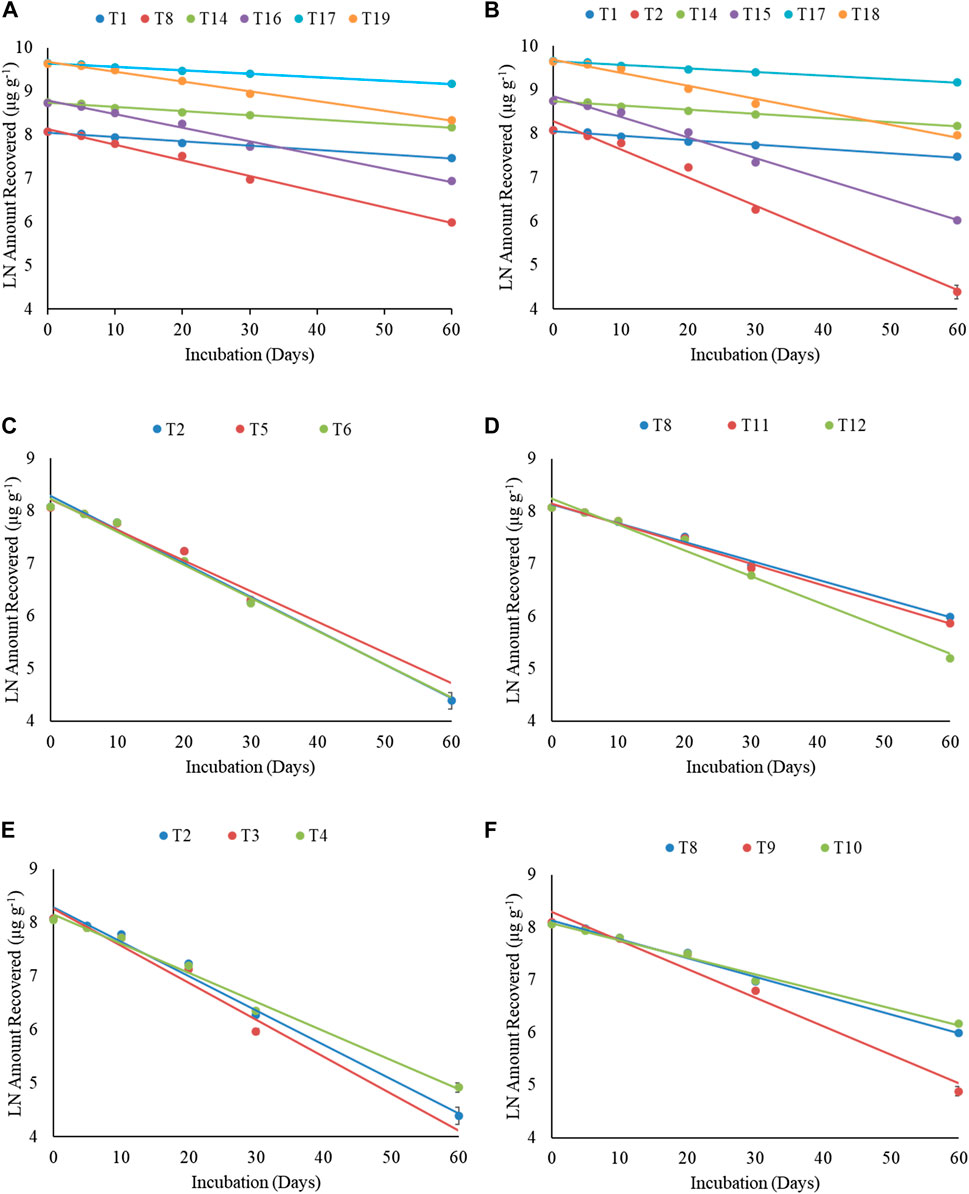
FIGURE 3. First-order degradation graphs for total petroleum hydrocarbons. (A) Effect of microbial consortia (free form) vis-a-vis crude oil concentration. (B) Effect of fungus (free form) vis-a-vis crude oil concentration. (C) Effect of microbial consortia based formulation vis-a-vis microbial consortia (free form) and control. (D) Effect of fungal spore–based formulation vis-a-vis fungus (free form) and control. (E) Effect of nutrient amendment vis-a-vis microbial consortia (free form) and control. (F) Effect of nutrient amendment vis-a-vis fungus (free form) and control in sandy loam soil.
The result of this study indicated that 1) degradation by free bacterial consortium was nearly two times faster than degradation by free fungus; 2) addition of ammonium sulfate slightly increased TPH degradation by free bacterial consortium and fungi; 3) formulating bacterial consortium and fungus did not affect their oil-degrading capability and; 4) degradation slowed down with increasing crude oil content in soil. Previously, Wang et al. (2012) studied the effect of addition of cotton stalk on TPH degradation and observed that after 220 days, compared to the control pile (20.44%), 49.62% of TPH was degraded in the cotton stalk mixed pile. Similarly, Beskoski et al. (2011) developed a consortium of microbes isolated from oil-contaminated soil (zymogenous microorganisms) of mazut (heavy fuel residual oil) and inoculated them in contaminated sites augmented with nutrients (N, P, and K). Mandal et al. (2012) reported that the microbial consortium was able to degrade initial total petroleum hydrocarbon from 206.50–231.00 g kg−1 oil waste to 10 g kg−1 within 3–7 months. Bio-stimulation and inoculation with consortium, compared to the control pile, increased the population of hydrocarbon-degrading microbes by more than 20 times after 50-day inoculation. The concentration of TPH was reduced from 5.2 to 0.3 g kg−1 in treated soil (6% of the initial concentration), while it was 96% of the initial concentration in the control pile. After 150 days, 96, 97, and 83% reduction in aliphatic, aromatic, and NSO asphaltene contents was observed. Lin et al. (2010) studied oil (C10–C28) and fuel oil (C10–C40) degradation in naturally contaminated soil. They observed that bioaugmentation (Gordonia alkanivorans CC-JG39, Rhodococcus erythropolis CC-BC11, Acinetobacter junii CC-FH2, and Exiguobacterium aurantiacum CC-LSH4-1) and biostimulation (rhamonolipid) resulted in 70 and 63% of diesel and fuel oil, respectively, in 28 days. Kim et al. (2005) reported that 3 or 6% (w/w) Arabian light crude oil and Tween-80 (0.075 or 0.75%) spiked Korean sandy soil was inoculated with a mixture of three oil-degrading microbes (Corynebacterium sp. IC-10, Sphingomonas sp. KH3-2, and Yarrowia sp. 180). After 1 week, 62 and 69% degradation of aliphatic and aromatic hydrocarbon fractions, respectively, was observed in 3% oil-fortified soil.
Diversity of Microbial Groups in the Crude Oil-Fortified Sandy Loam Soil
Microbial diversity in soil samples treated with different levels of crude oil (1, 2, and 5%) and inoculated with free cultures of bacterial consortium (Pseudomonas sp. + B. amyloliquefaciens) and fungus (A. sydowii) was analyzed by qPCR assays. The composition of microbial communities, based on the specific groups (16S rRNA genes of Bacteria, Archaea, Alphaproteobacteria, Betaproteobacteria, Bacteroidetes, and Firmicutes), varied. The gene copies of total bacteria (2.8 × 108 to 2.8 × 1010) were more than those of Archaea (5.0 × 103 to 1.4 × 107). The gene copies specific to different phyla followed the order given as follows: Betaproteobacteria (6.5 × 108 to 1.3 × 109) > Firmicutes (5.3 × 106 to 3.9 × 108) > Alphaproteobacteria (9.6 × 105 to 6.3 × 106) > Bacteroidetes (1.2 × 105 to 9.0 × 105). The abundances of 16S rRNA genes of Bacteria under different treatments (T1–T8) varied from 2.8 × 108 to 2.8 × 1010 g−1 soil (Figure 4). The highest abundance of these genes was observed in T3, significantly higher than abundance in other treatments. The abundances of this gene in different treatments followed the order: T3 > T4 > T1 > T5 ≥ T8 > T2 > T7 > T6. The 16S rRNA gene copies of Archaea under different treatments followed the order given as follows: T2 ≥ T3 ≥ T5 > T1 > T8 > T4 > T7 > T6. The abundances of 16S rRNA gene copies of Alphaproteobacteria (105–106), Betaproteobacteria (108–109), Bacteroidetes (105), and Firmicutes (106–108) are presented in Figure 4. Their abundances in different treatments followed the order T1, T6 ≥ T7 ≥ T8, T2 ≥ T4, and T1, respectively. The effect of bacterial consortia or fungus inoculation was observed in different treatments. Pseudomonas sp. + B. amyloliquefaciens inoculation in crude oil–fortified soil (1%) resulted in increased gene copies representing Bacteria, Alphaproteobacteria, and Firmicutes. Similarly, A. sydowii inoculation increased the population of Betaproteobacteria. The increases in gene copies of Betaproteobacteria following the A. sydowii inoculation in crude oil–fortified soils suggested that the members of Betaproteobacteria might have contributed to crude oil degradation. These results corroborated with the reports of Xu et al. (2014) and Benedek et al. (2013) who demonstrated the microbial diversity changes in soil, after the crude oil contamination. Likewise, Kuang et al. (2018) showed that the percentage of Proteobacteria increased in oil-contaminated soil (Shengli oil field, China) compared to unpolluted soil. Nevertheless, Deng et al. (2020) reported that after bioaugmentation of Bacillus sp. in crude oil–contaminated soil (Shannxi Province, Yanchang), the original microbial diversity was recovered at the phylum level. The present study suggested that microbial diversity changes were largely due to increase in the abundance of Betaproteobacteria. Moreover, consortium of Pseudomonas sp. + B. amyloliquefaciens and fungus A. sydowii established the microbial populations in the oil-contaminated soils, somewhat to the original levels of the uncontaminated soil. Hence, the bioaugmentation is required to enhance the resilience of contaminated soils.
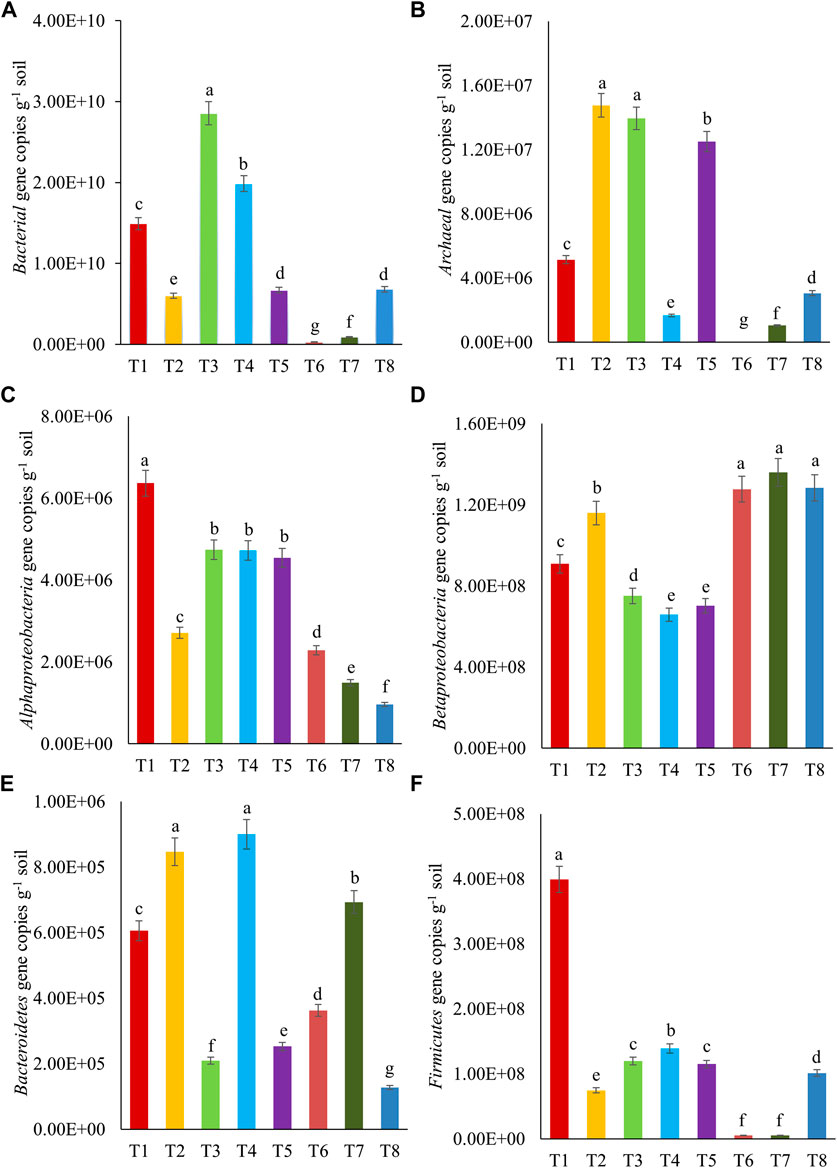
FIGURE 4. Abundances of 16S rRNA gene copies of (A) Bacteria, (B) Archaea, (C) Alphaproteobacteria, (D) Betaproteobacteria, (E) Bacteroidetes, and (F) Firmicutes after 10th day of crude oil fortification in sandy loam soil [T1–soil; T2–soil + crude oil (1%); T3–soil + crude oil (1%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T4–soil + crude oil (2%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T5–soil + crude oil (5%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T6–soil + crude oil (1%) + fungus (Aspergillus sydowii AK20); T7–soil + crude oil (2%) + fungus (Aspergillus sydowii AK20); T8–soil + crude oil (5%) + fungus (Aspergillus sydowii AK20)].
The functional gene copies related to the N-cycling, namely, bacterial amoA, nifH, nirK, nirS, and anammox, that were detected and quantified in the sandy loam soil under different treatments, are presented in Figure 5. The variable abundances of functional gene copies were more in nifH (105–108) than in nirK (105–107), amoA (103–107), nirS (103–105), and anammox (103–105). The functional gene copies of amoA, nifH, nirK, nirS, and anammox were found to be higher in T7, T1, T3, T2 ≥ T4, and T5 treatments, respectively. In general, the nitrogen-assimilating gene (nifH-encoding nitrogenase enzyme), those encoding nitrogen oxide reductase enzymes (nirK and nirS), and even the genes specific for anammox bacteria were found to be significantly higher after the inoculation of bacterial consortium in crude oil fortified–sandy loam soil. Increases in the gene copies suggested that the microbial inoculation enhanced certain nitrogen cycling enzymatic activities in crude oil–fortified soils. The regulation of nitrogen cycling in the contaminated soil is vital to keep the soil microbial communities active and improve their potential to use the contaminant(s) as the sources of energy. Therefore, the status of ammonia oxygenase enzyme (encoded by amoA), nitrogenase enzyme (encoded by nifH, nifD, and nifK), nitrogen oxide reductase (nirK, nirS, and nosZ), and hydrazine synthase (hzsA mediated by annamox) can be the significant indices to determine the soil condition (Rubio and Ludden, 2002; Khanal and Lee, 2020). The present study indicated that nitrogen regulation by the genes related to amoA and nifH was effective after the bioaugmentation of crude oil–degrading bacterial consortium (Pseudomonas sp. + B. amyloliquefaciens) and fungus (A. sydowii).
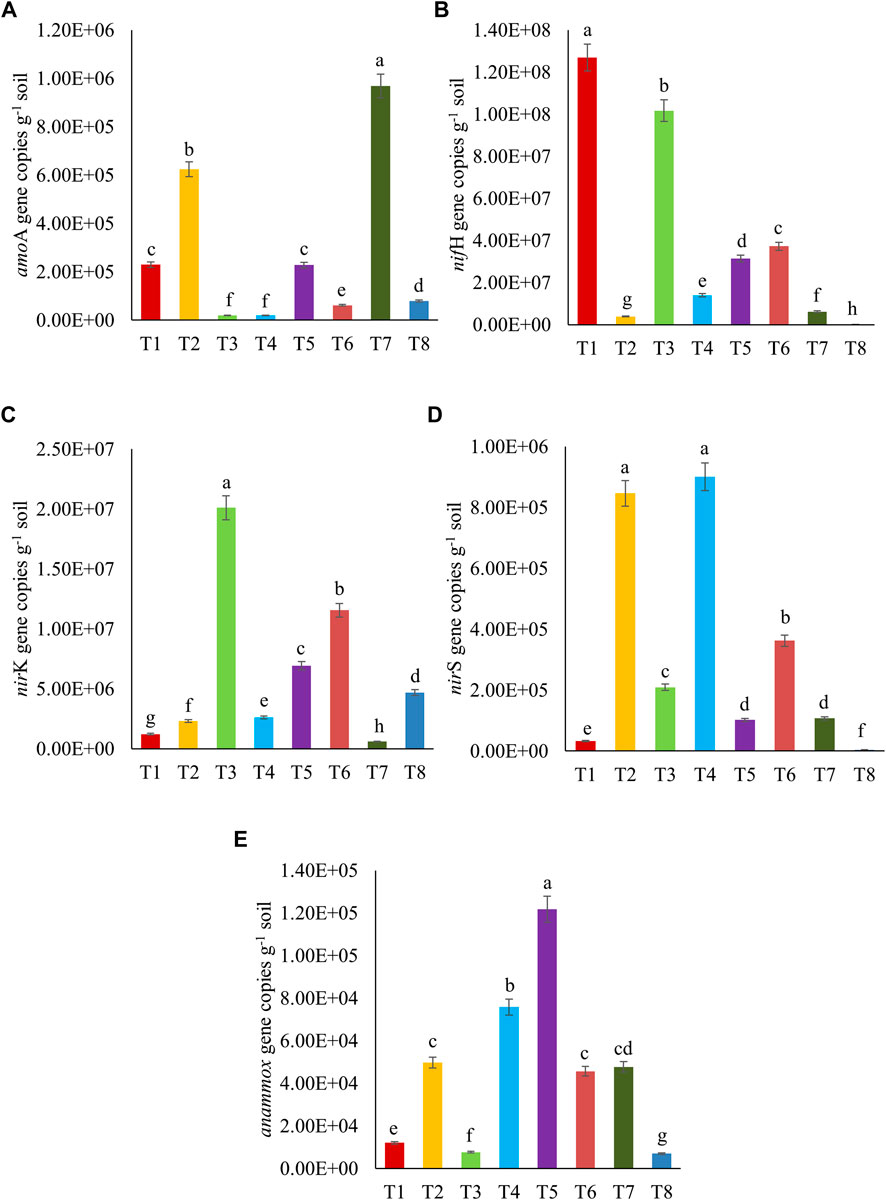
FIGURE 5. Abundances of microbial functional gene copies of (A) amoA, (B) nifH, (C) nirK, (D) nirS, and (E) anammox related to nitrogen cycling after 10th day of crude oil fortification in the sandy loam soil [T1–soil; T2–soil + crude oil (1%); T3–soil + crude oil (1%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T4–soil + crude oil (2%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T5–soil + crude oil (5%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T6–soil + crude oil (1%) + fungus (Aspergillus sydowii AK20); T7–soil + crude oil (2%) + fungus (Aspergillus sydowii AK20); T8–soil + crude oil (5%) + fungus (Aspergillus sydowii AK20)].
All the taxonomical and functional genes were subjected to PCA (Figure 6). To relate the clusters obtained with the treatments accountable for various genes, the dataset showed the contribution rate of principal components 1 (F1) and 2 (F2) was 38.41 and 21.75%, respectively. This suggested that two principal components represented 60.16% of gene variation among the treatments. Among 11 genes quantified, Alphaproteobacteria, Archaeal 16S, Firmicutes, Bacterial 16S, and nifH were positively associated. These genes were negatively correlated with Betaproteobacteria and amoA, while independent of Bacteroidetes, anammox, nirS, and nirK. These relationships suggest that it is possible to combine higher quantification of genes such as Alphaproteobacteria, Archaeal 16S, Firmicutes, Bacterial 16S, and nifH with lower quantification of Betaproteobacteria and amoA in each treatment. The treatments T1, T4, and T5 had the highest number of Alphaproteobacteria, Archaeal 16S, Firmicutes, Bacterial 16S, and nifH genes, and lower Betaproteobacteria and amoA gene numbers. The treatments T6, T7, and T8 constituted a cluster with similar quantification profiles.
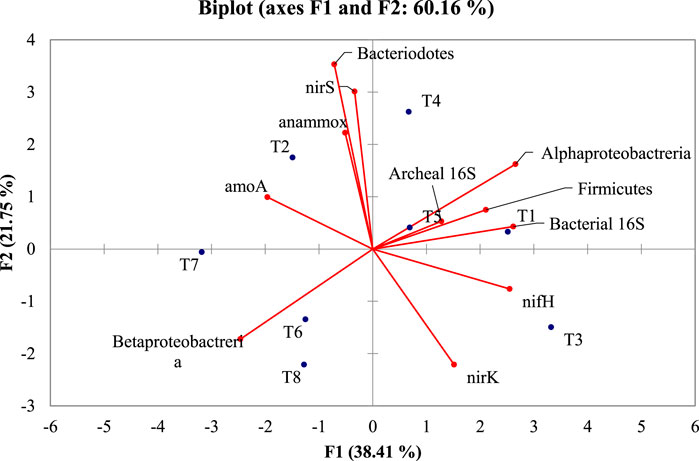
FIGURE 6. PCA biplot depicting the taxonomic and functional gene markers of diverse microbial groups and treatments in crude oil fortified sandy loam soil [T1–soil; T2–soil + crude oil (1%); T3–soil + crude oil (1%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T4–soil + crude oil (2%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T5–soil + crude oil (5%) + bacterial consortia (Pseudomonas sp. A3 + Bacillus amyloliquefaciens A9); T6–soil + crude oil (1%) + fungus (Aspergillus sydowii AK20); T7–soil + crude oil (2%) + fungus (Aspergillus sydowii AK20); T8–soil + crude oil (5%) + fungus (Aspergillus sydowii AK20)].
Conclusion
Microbes isolated from crude oil–contaminated soils were identified and evaluated for aliphatic hydrocarbon (C11–C36) degradation in Reese’s minimal medium, without (−T80) and with Tween-80 (+T80). Isolated microbes showed 44.2–81.7% (−T80) and 47.7–85.3% (+T80) degradations of aliphatic hydrocarbon fraction. Among all bacteria, Pseudomonas sp. A3, Bacillus amyloliquefaciens A9, and Pseudomonas aeruginosa AK7 were identified as best aliphatic hydrocarbon degraders. Based on non-pathogenecity and compatibility studies, consortium of Pseudomonas sp. + B. amyloliquefaciens and Aspergillus sydowii, separately, was formulated in three types of formulations. Both free cultures and formulations were effective in degrading TPH in 1–5% oil-fortified sandy loam soil. Increasing the crude oil content slowed down degradation. The addition of ammonium sulfate (1%) enhanced TPH degradation by free microbial cultures. Moreover, the consortium of Pseudomonas sp. + B. amyloliquefaciens and fungus A. sydowii established the microbial populations in the oil-contaminated soils, somewhat to the original levels of the uncontaminated soil. In addition, amoA and nifH were also effective after the bioaugmentation of crude oil. Hence, the bioaugmentation is required to enhance the resilience of contaminated soils. Microbes identified in the present study can be exploited for remediating the contaminated natural environment.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
AK conducted experiment studies, analyzed data, and wrote the manuscript. RS analyzed metagenomics data. BR contributed to the planning of metagenomics analysis and manuscript correction. AD helped with conceptualization for development of bio-formulations. EV assisted with statistical analysis. LN contributed to microbe isolation and identification. TB developed the method of analysis. NS contributed to concept, experiment planning, coordination, and manuscript correction.
Funding
The work related to Ph.D. was supported by INSPIRE Fellowship and IARI Fellowship in the form of contingency.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors, and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
AK is thankful to the Department of Science and Technology, Government of India, New Delhi, and the Indian Agricultural Research Institute (ICAR) for giving INSPIRE Fellowship and IARI Senior Fellowship during the Ph.D. program, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.794303/full#supplementary-material
References
Ali, N., Dashti, N., Khanafer, M., Al-Awadhi, H., and Radwan, S. (2020). Bioremediation of Soils Saturated with Spilled Crude Oil. Sci. Rep. 10, 1–9. doi:10.1038/s41598-019-57224-x
Badr El-Din, S. M., Moussa, T. A., Moawad, H., and Sharaf, O. A. (2014). Isolation and Characterization of Polyaromatic Hydrocarbons Degrading Bacteria from Compost Leachate. J. Adv. Biol. 5, 651–660. doi:10.24297/jab.v5i2.3772
Balachandran, C., Duraipandiyan, V., Balakrishna, K., and Ignacimuthu, S. (2012). Petroleum and Polycyclic Aromatic Hydrocarbons (PAHs) Degradation and Naphthalene Metabolism in Streptomyces Sp. (ERI-CPDA-1) Isolated from Oil Contaminated Soil. Bioresour. Technol. 112, 83–90. doi:10.1016/j.biortech.2012.02.059
BBC NEWS (2020). Assam Fire: India's Longest Burning Gas Blaze Is Destroying Lives. Available at: https://www.bbc.com/news/world-asia-india54719286#:∼:text=The%20blaze%20that%20started%20on,longest%20such%20fire%20in%20India.2020/ (Accessed May 27, 2020).
Benedek, T., Vajna, B., Táncsics, A., Márialigeti, K., Lányi, S., and Máthé, I. (2013). Remarkable Impact of PAHs and TPHs on the Richness and Diversity of Bacterial Species in Surface Soils Exposed to Long-Term Hydrocarbon Pollution. World J. Microbiol. Biotechnol. 29, 1989–2002. doi:10.1007/s11274-013-1362-9
Beskoski, V. P., Gojgic-Cvijovic, G., Milic, J., Ilic, M., Miletic, S., Solevic, T., et al. (2011). Ex Situ Bioremediation of a Soil Contaminated by Mazut (Heavy Residual Fuel Oil)-A Field experiment. Chemosphere 83, 34–40. doi:10.1016/j.chemosphere.2011.01.020
Bhattacharya, M., Guchhait, S., Biswas, D., and Singh, R. (2019). Evaluation of a Microbial Consortium for Crude Oil Spill Bioremediation and its Potential Uses in Enhanced Oil Recovery. Biocatal. Agric. Biotechnol. 18, 101034. doi:10.1016/j.bcab.2019.101034
Bilen Ozyurek, S., and Bilkay, S. I. (2020). Comparison of Petroleum Biodegradation Efficiencies of Three Different Bacterial Consortia Determined in Petroleum-Contaminated Waste Mud Pit. SN Appl. Sci. 2, 272. doi:10.1007/s42452-020-2044-5
Braker, G., Fesefeldt, A., and Witzel, K.-P. (1998). Development of PCR Primer Systems for Amplification of Nitrite Reductase Genes ( nirK and nirS ) to Detect Denitrifying Bacteria in Environmental Samples. Appl. Environ. Microbiol. 64, 3769–3775. doi:10.1128/aem.64.10.3769-3775.1998
Bundy, J. G., Paton, G. I., and Campbell, C. D. (2004). Combined Microbial Community Level and Single Species Biosensor Responses to Monitor Recovery of Oil Polluted Soil. Soil Biol. Biochem. 36, 1149–1159. doi:10.1016/j.soilbio.2004.02.025
Cho, B.-H., Chino, H., Tsuji, H., Kunito, T., Nagaoka, K., Otsuka, S., et al. (1997). Laboratory-scale Bioremediation of Oil-Contaminated Soil of Kuwait with Soil Amendment Materials. Chemosphere 35, 1599–1611. doi:10.1016/s0045-6535(97)00220-8
Chumthong, A., Kanjanamaneesathian, M., Pengnoo, A., and Wiwattanapatapee, R. (2008). Water-Soluble Granules Containing Bacillus Megaterium for Biological Control of rice Sheath Blight: Formulation, Bacterial Viability and Efficacy Testing. World J. Microbiol. Biotechnol. 24, 2499–2507. doi:10.1007/s11274-008-9774-7
Das, N., and Chandran, P. (2011). Microbial Degradation of Petroleum Hydrocarbon Contaminants: An Overview. Biotechnol. Res. Int. 2011, 1–13. doi:10.4061/2011/941810
Day, P. R. (1965). “Particle Fractionation and Particle-Size Analysis,” in Methods of Soil Analysis, Part I. Editor C. A. Black (Madison, Wisconsin: Agronomy Society of America), 545–567.
Dean-Ross, D., Moody, J., and Cerniglia, C. E. (2002). Utilization of Mixtures of Polycyclic Aromatic Hydrocarbons by Bacteria Isolated from Contaminated Sediment. FEMS Microbiol. Ecol. 41, 1–7. doi:10.1111/j.1574-6941.2002.tb00960.x
Deivakumari, M., Sanjivkumar, M., Suganya, A. M., Prabakaran, J. R., Palavesam, A., and Immanuel, G. (2020). Studies on Reclamation of Crude Oil Polluted Soil by Biosurfactant Producing Pseudomonas aeruginosa (DKB1). Biocatal. Agric. Biotechnol. 29, 101773. doi:10.1016/j.bcab.2020.101773
Deng, Z., Jiang, Y., Chen, K., Gao, F., and Liu, X. (2020). Petroleum Depletion Property and Microbial Community Shift after Bioremediation Using Bacillus Halotolerans T-04 and Bacillus Cereus 1-1. Front. Microbiol. 11, 353. doi:10.3389/fmicb.2020.00353
Fierer, N., Jackson, J. A., Vilgalys, R., and Jackson, R. B. (2005). Assessment of Soil Microbial Community Structure by Use of Taxon-specific Quantitative PCR Assays. Appl. Environ. Microbiol. 71, 4117–4120. doi:10.1128/AEM.71.7.4117-4120.2005
Gangadhari, R. K., Murty, S., and Khanzode, V. (2020). Analysis of Accidents Involving Petroleum Tankers and Their Consequences in India. Proc. Saf. Prog. 40, e12154. doi:10.1002/prs.12154
Green, V. S., Stott, D. E., and Diack, M. (2006). Assay for Fluorescein Diacetate Hydrolytic Activity: Optimization for Soil Samples. Soil Biol. Biochem. 38, 693–701. doi:10.1016/j.soilbio.2005.06.020
Han, Y., Nambi, I. M., and Prabhakar Clement, T. (2018). Environmental Impacts of the Chennai Oil Spill Accident - A Case Study. Sci. Total Environ. 626, 795–806. doi:10.1016/j.scitotenv.2018.01.128
Juhanson, J., Truu, J., Heinaru, E., and Heinaru, A. (2009). Survival and Catabolic Performance of introducedPseudomonasstrains during Phytoremediation and Bioaugmentation Field experiment. FEMS Microbiol. Ecol. 70, 446–455. doi:10.1111/j.1574-6941.2009.00754.x
Kachieng’a, L., and Momba, M. (2017). Kinetics of Petroleum Oil Biodegradation by a Consortium of Three Protozoan Isolates (Aspidisca sp., Trachelophyllum Sp. And Peranema sp.). Biotechnol. Rep. 15, 125–131. doi:10.1016/j.btre.2017.07.001
Khanal, A., and Lee, J.-H. (2020). Functional Diversity and Abundance of Nitrogen Cycle-Related Genes in Paddy Soil. Appl. Biol. Chem. 63, 1–13. doi:10.1186/s13765-020-00500-6
Khandelwal, A., Singh, S. B., Sharma, A., Nain, L., Varghese, E., and Singh, N. (2021). Effect of Surfactant on Degradation of Aspergillus Sp. And Trichoderma Sp. Mediated Crude Oil. Int. J. Environ. Anal. Chem. 1-14, 1–14. doi:10.1080/03067319.2021.1879800
Kim, S.-J., Choi, D. H., Sim, D. S., and Oh, Y.-S. (2005). Evaluation of Bioremediation Effectiveness on Crude Oil-Contaminated Sand. Chemosphere 59, 845–852. doi:10.1016/j.chemosphere.2004.10.058
Kolde, R. (2019). Pheatmap: Implementation of Heatmaps that Offers More Control over Dimensions and Appearance. R package version 1.0.12. Available at: https://CRAN.R-project.org/package=pheatmap/ (Accessed June 14, 2021).
Kuang, S., Su, Y., Wang, H., Yu, W., Lang, Q., and Matangi, R. (2018). Soil Microbial Community Structure and Diversity Around the Aging Oil Sludge in Yellow River delta as Determined by High-Throughput Sequencing. Archaea 2018, 1–10. doi:10.1155/2018/7861805
Kumar, A., Nain, L., and Singh, N. (2017). Alginate Immobilized Enrichment Culture for Atrazine Degradation in Soil and Water System. J. Environ. Sci. Health B 52, 229–236. doi:10.1080/03601234.2016.1270680
Li, M., and Gu, J.-D. (2011). Advances in Methods for Detection of Anaerobic Ammonium Oxidizing (Anammox) Bacteria. Appl. Microbiol. Biotechnol. 90, 1241–1252. doi:10.1007/s00253-011-3230-6
Lin, T.-C., Pan, P.-T., and Cheng, S.-S. (2010). Ex Situ Bioremediation of Oil-Contaminated Soil. J. Hazard. Mater. 176, 27–34. doi:10.1016/j.jhazmat.2009.10.080
Lowry, O., Rosebrough, N., Farr, A. L., and Randall, R. (1951). Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 193, 265–275. PMID: 14907713. doi:10.1016/s0021-9258(19)52451-6
Maiti, D., Chandra, K., Mondal, S., Ojha, A. K., Das, D., Roy, S. K., et al. (2008). Isolation and Characterization of a Heteroglycan from the Fruits of Astraeus Hygrometricus. Carbohydr. Res. 343, 817–824. doi:10.1016/j.carres.2007.12.003
Mandal, A. K., Sarma, P. M., Jeyaseelan, C. P., Channashettar, V. A., Singh, B., Agnihotri, A., et al. (2012). Large Scale Bioremediation of Petroleum Hydrocarbon Contaminated Waste at Various Installations of ONGC. India: Case Studies. Erem 68, L114–L128. doi:10.5755/J01.EREM.68.2.5632
Mehrotra, T., Dev, S., Banerjee, A., Chatterjee, A., Singh, R., and Aggarwal, S. (2021). Use of Immobilized Bacteria for Environmental Bioremediation: A Review. J. Environ. Chem. Eng. 9, 105920. doi:10.1016/j.jece.2021.105920
Mittal, A., and Singh, P. (2009). Isolation of Hydrocarbon Degrading Bacteria from Soils Contaminated with Crude Oil Spills. Indian J. Exp. Biol. 47, 760–765.
Nwankwegu, A. S., and Onwosi, C. O. (2017). Microbial Cell Immobilization: A Renaissance to Bioaugmentation Inadequacies. A Review. Environ. Technol. Rev. 6, 186–198. doi:10.1080/21622515.2017.1356877
Partovinia, A., and Rasekh, B. (2018). Review of the Immobilized Microbial Cell Systems for Bioremediation of Petroleum Hydrocarbons Polluted Environments. Crit. Rev. Environ. Sci. Technol. 48, 1–38. doi:10.1080/10643389.2018.1439652
Patel, D. D., and Bhaskaran, L. (2018). Study on Paraffin Wax Degrading Ability of Pseudomonas Nitroreducens Isolated from Oil wells of Gujarat, India. Pet. Sci. Technol. 36, 583–590. doi:10.1080/10916466.2018.1437634
Pathak, M., Sarma, H. K., Bhattacharyya, K. G., Subudhi, S., Bisht, V., Lal, B., et al. (2017). Characterization of a Novel Polymeric Bioflocculant Produced from Bacterial Utilization of N-Hexadecane and its Application in Removal of Heavy Metals. Front. Microbiol. 8, 1–15. doi:10.3389/fmicb.2017.00170
Poly, F., Ranjard, L., Nazaret, S., Gourbière, F., and Monrozier, L. J. (2001). Comparison of nifH Gene Pools in Soils and Soil Microenvironments with Contrasting Properties. Appl. Environ. Microbiol. 67, 2255–2262. doi:10.1128/aem.67.5.2255-2262.2001
R Core Team (2020). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.R-project.org/ (Accessed March 31, 2021).
Reese, E. T., and Mandels, M. (1963). Methods Carbohydrate Chemistry. 3rd Edn.. London: Academic Press, 139–143.
Rotthauwe, J. H., Witzel, K. P., and Liesack, W. (1997). The Ammonia Monooxygenase Structural Gene amoA as a Functional Marker: Molecular fine-scale Analysis of Natural Ammonia-Oxidizing Populations. Appl. Environ. Microbiol. 63, 4704–4712. doi:10.1128/aem.63.12.4704-4712.1997
Rubio, L. M., and Ludden, P. W. (2002). “The Gene Products of the Nif Regulon,” in Nitrogen Fixation at the Millenium. Editor G. J. Leigh (Amsterdam: Elsevier), 101–136. doi:10.1016/B978-044450965-9/50004-5
Siles, J. A., and García-Sánchez, M. (2018). “Microbial Dynamics during the Bioremediation of Petroleum Hydrocarbon-Contaminated Soils through Biostimulation: An Overview,” in Approaches in Bioremediation: The New Era of Environmental Microbiology and Nanobiotechnology. Editors R. Prasad, and E. Aranda (Cham: Springer), 115–134. doi:10.1007/978-3-030-02369-0_7
Spini, G., Spina, F., Poli, A., Blieux, A.-L., Regnier, T., Gramellini, C., et al. (2018). Molecular and Microbiological Insights on the Enrichment Procedures for the Isolation of Petroleum Degrading Bacteria and Fungi. Front. Microbiol. 9, 2543. doi:10.3389/fmicb.2018.02543
Suzuki, M. T., Taylor, L. T., and DeLong, E. F. (2000). Quantitative Analysis of Small-Subunit rRNA Genes in Mixed Microbial Populations via 5′-Nuclease Assays. Appl. Environ. Microbiol. 66, 4605–4614. doi:10.1128/AEM.66.11.4605-4614.2000
THE ECONOMIC TIMES (2011). Oil Spill at Uran; ONGC Fixes Leak in Pipeline. Available at: https://economictimes.indiatimes.com/industry/energy/oil-gas/oil-spill-at-uran-ongc-fixes-leak-in-pipeline/articleshow/23657251.cms?from=mdr/ (Accessed July 21, 2020).
THE HINDU (2011). Oil Spill on fields Leaves Farmers on Edge. https://www.thehindu.com/news/national/tamil-nadu/oil-spill-on-fields-leaves-farmers-on-edge/article4062601.ece/ (Accessed October 12, 2020).
THE TIMES OF INDIA (2017). Chennai Oil Spill. Available at: https://timesofindia.indiatimes.com/topic/chennai-oil-spill/ (Accessed July 21, 2020).
THE TIMES OF INDIA (2020). Crude Oil Flows into Paddy Field as ONGC. Available at: https://timesofindia.indiatimes.com/city/trichy/crude-oil-flows-into-paddy-field-as-ongc-pipe-develops-leak/articleshow/78285275.cms/ (Accessed October 12, 2020).
Viesser, J. A., Sugai-Guerios, M. H., Malucelli, L. C., Pincerati, M. R., Karp, S. G., and Maranho, L. T. (2020). Petroleum-Tolerant Rhizospheric Bacteria: Isolation, Characterization and Bioremediation Potential. Sci. Rep. 10, 2060. doi:10.1038/s41598-020-59029-9
Wang, X.-B., Chi, C.-Q., Nie, Y., Tang, Y.-Q., Tan, Y., Wu, G., et al. (2011). Degradation of Petroleum Hydrocarbons (C6-C40) and Crude Oil by a Novel Dietzia Strain. Bioresour. Technol. 102, 7755–7761. doi:10.1016/j.biortech.2011.06.009
Wang, X., Wang, Q., Wang, S., Li, F., and Guo, G. (2012). Effect of Biostimulation on Community Level Physiological Profiles of Microorganisms in Field-Scale Biopiles Composed of Aged Oil Sludge. Bioresour. Technol. 111, 308–315. doi:10.1016/j.biortech.2012.01.158
WIKIPEDIA (2020). Assam Gas and Oil Leak. Available at: https://en.wikipedia.org/wiki/2020_Assam_gas_and_oil_leak#cite_note-:0-2 2020a/ (Accessed May 27, 2021).
Xu, Y., Sun, G.-D., Jin, J.-H., Liu, Y., Luo, M., Zhong, Z.-P., et al. (2014). Successful Bioremediation of an Aged and Heavily Contaminated Soil Using a Microbial/plant Combination Strategy. J. Hazard. Mater. 264, 430–438. doi:10.1016/j.jhazmat.2013.10.071
Yadav, S., Sharma, A., Khan, M. A., Sharma, R., Celin, M., Malik, A., et al. (2020). Enhancing Hexahydro-1, 3, 5-trinitro-1, 3, 5-triazine (RDX) Remediation through Water-Dispersible Microbacterium Esteraromaticum Granules. J. Environ. Manage. 264, 110446. doi:10.1016/j.jenvman.2020.110446
Zawierucha, I., and Malina, G. (2011). “Bioremediation of Contaminated Soils: Effects of Bioaugmentation and Biostimulation on Enhancing Biodegradation of Oil Hydrocarbons,” in Microbial Action on Hydrocarbons. Editors V. Kumar, M. Kumar, and R. Prasad (Singapore: Springer Singapore), 187–201. doi:10.1007/978-3-642-19769-7_8
Keywords: total petroleum hydrocarbon, half-life, bio-formulation, taxonomic marker, functional marker
Citation: Khandelwal A, Sugavanam R, Ramakrishnan B, Dutta A, Varghese E, Nain L, Banerjee T and Singh N (2022) Free and Immobilized Microbial Culture–Mediated Crude Oil Degradation and Microbial Diversity Changes Through Taxonomic and Functional Markers in a Sandy Loam Soil. Front. Environ. Sci. 9:794303. doi: 10.3389/fenvs.2021.794303
Received: 13 October 2021; Accepted: 08 December 2021;
Published: 12 January 2022.
Edited by:
Eric D. van Hullebusch, Université de Paris, FranceReviewed by:
Sharmila Jayasena, University of Colombo, Sri LankaAli Partovinia, Shahid Beheshti University, Iran
Copyright © 2022 Khandelwal, Sugavanam, Ramakrishnan, Dutta, Varghese, Nain, Banerjee and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Neera Singh, ZHJuZWVyYXNpbmdoQHlhaG9vLmNvbQ==; Ashish Khandelwal, YXNoaXNoLmlhc2JodUBnbWFpbC5jb20=
 Ashish Khandelwal
Ashish Khandelwal Ramya Sugavanam
Ramya Sugavanam B. Ramakrishnan
B. Ramakrishnan Anirban Dutta
Anirban Dutta Eldho Varghese
Eldho Varghese Lata Nain
Lata Nain Tirthankar Banerjee
Tirthankar Banerjee Neera Singh
Neera Singh