- 1Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 2Research Center in Bioresources for Agriculture, Industry and Medicine, Chiang Mai University, Chiang Mai, Thailand
- 3Research Center of Microbial Diversity and Sustainable Utilization, Faculty of Science, Chiang Mai University, Chiang Mai, Thailand
- 4Joint BioEnergy Institute, Emeryville, CA, United States
- 5Biological Systems and Engineering Division, Lawrence Berkeley National Laboratory, Berkeley, CA, United States
- 6Department of Life Sciences, Imperial College London, Silwood Park Campus, Ascot, United Kingdom
Heavy metal polluted wastewater from industries is currently one of the major environmental concerns leading to insufficient supply of clean water. Several strategies have been implemented to overcome this challenge including the use of microalgae as heavy metal bio-removers. However, there are still limitations that prevent microalgae to function optimally. Synthetic biology is a new biological discipline developed to solve challenging problems via bioengineering approaches. To date, synthetic biology has no universally affirmed definitions; however, it is uncontroversial that synthetic biology utilizes a constructive library of genetic standardized parts to create new biological systems or to redesign existing ones with improved characteristics. In this mini-review, we present state-of-the-art synthetic biology-based approaches that can be used to enhance heavy metal bio-removal from wastewater effluents by microalgae with a narrative synthetic biology workflow (Design-Build-Test-Learn cycle) to guide future developments of more advanced systems. We also provide insights into potent genes and proteins responsible for the bio-removal processes for stepwise developments of more advanced systems. A total of 49 unique genes and proteins are listed based on their eight heavy metals (Mn, Fe, Cu, Zn, As, Cd, Hg, and Pb) bio-removal functions in transport system, cellular tolerance, synthesis of key players in heavy metal bio-removal, biotransformation of heavy metals, and gene expression regulation. Thus, with our library, genetic parts are ready to be recruited for any synthetic biology-based designs. Thereby, this mini-review identifies potential avenues of future research and maps opportunities to unleash more potential of microalgae as heavy metal bio-removers with synthetic biology.
Introduction: Overview of Synthetic Biology and Wastewater Treatment
With recent advances in biological field, it is relatively more feasible to create purposeful synthetic systems through careful designs and engineering. This rapidly emerging discipline is referred to as “synthetic biology” and, since its birth in the year 2000, has demonstrated a number of major achievements (Meng and Ellis, 2020). With its widely applicable scope, synthetic biology has been used to overcome several challenges that our world has been facing including the development of CO2 fixing heterotrophic bacteria, CRISPR-based systems including for rapid diagnostics and, particularly for algal engineering (Gleizer et al., 2019; Krishnan et al., 2020; Meng and Ellis, 2020; Kaminski et al., 2021). Synthetic biology adopts the philosophy of bioengineering and aims at creating new biological systems or redesigning existing ones for particular purposes (Deplazes, 2009). This discipline builds up on constructive libraries of genetic parts ready to be recruited for any designs.
Water pollution is one of the global concerns affecting people in certain parts of the world. To alleviate this problem, microalgae have been used and developed extensively as a biological system for wastewater treatment (Wollmann et al., 2019). Heavy metals (HMs) are often referred to metal elements that are potentially toxic to human and the environment (Duffus, 2002; Rahman and Singh, 2019). Sources of HM contamination are varied, yet mostly industrial-based (Selvi et al., 2019). Heavy metal-contaminated water is conventionally treated via several approaches including the use of microalgae as a bio-remover that takes away HMs from liquid phases (Cheng et al., 2019).
Interestingly, previous reviews on similar topics only focus on 1) Genetic engineering tools and techniques for microalgae 2) Synthetic biology approaches for HM bio-removal in all potential microorganisms 3) Synthetic biology in microalgae for biomass and chemical production and 4) The use of customized algae for HM adsorption. To give some examples, a recent review by Fajardo et al. (2020) focuses on genetic engineering techniques and tools for microalgae ranging from classical to advanced ones (Fajardo et al., 2020) without any discussions on HM bio-removal. This may indicate that genetic engineering in microalgae is still considered under development. There are a few reviews on the use of genetically modified microorganisms (GMOs) to bio-remove HMs; however, they focus on all potential microorganisms, not particularly microalgae as a host in question (Diep et al., 2018; Capeness and Horsfall, 2020). Many reviews on the development of synthetic biology toolkits for microalgae only focus on biomass and chemical production (Wang et al., 2012; Jagadevan et al., 2018; Fabris et al., 2020; Kumar et al., 2020; Naduthodi et al., 2021) but not from the HM bio-removal perspective. Although, one review aims to emphasize the use of customized algae for HM bio-removal and discusses several approaches used to enhance algal performances including biological/molecular and chemical strategies, the review only focuses on adsorption as a removal mechanism (Cheng et al., 2019). Altogether, understandings on synthetic biology-based approaches applied to all microalgal HM bio-removal mechanisms is still missing. Potential genes/proteins involved in the process and their potential are yet to be collectively reported. To fill the knowledge gap, here, synthetic biology-based strategies for microalgal HM bio-removal are comprehensively discussed based on a synthetic biology concept (Design-Build-Test-Learn cycle) and all possible mechanisms used by microalgae to bio-remove HMs 1) biosorption 2) bioaccumulation and 3) biotransformation). A collective genetic part library of genes/proteins involved in and/or have been engineered for such purposes is also constructed to enable future developments of synthetic microalgal systems for HM bio-removal. Therefore, our work fills up the missing aspect on this topic and serves as a guide for stepwise developments. For instance, if a highly effective microalga is of interest as a host to bio-remove HMs, one can utilize the synthetic biology workflow presented in this mini-review to develop such systems by which they can select a strategy and handpick the most suitable genes/proteins listed here to start off the development. Specifically, if HM transporters of a microalga are observed to be ineffective, which limits the bio-removing performances, potential HM transporters from the library provided here can be handpicked and engineered.
Microalgal Mechanisms to Remove Heavy Metals From Wastewater Effluents
There are several mechanisms that microalgae use to remove HMs from contaminated sources. Here, we classify these mechanisms into 1) biosorption, 2) bioaccumulation, and 3) biotransformation.
Microalgae can be used as biosorbents by which HMs are absorbed onto their cell surface, or to be specific, onto the functional groups (e.g., -COO−, -OH- and -PO43−) or associated proteins present on their cell walls (Monteiro et al., 2012; Kumar et al., 2015; Mantzorou et al., 2018; Salama et al., 2019). In general, these functional groups are present in negatively charged forms, thereby, attribute to the overall anionics and attract positively charged metal ions. This mechanism is called biosorption. In this regard, the components of cell wall are functionally important key players and the binding affinity of cell surface to HMs in question is a key parameter determining the effectiveness of this mechanism. Modification of cultivation conditions to alter surface properties has been demonstrated to improve Hg biosorption in Scenedesmus obtusus XJ-15 (Huang et al., 2019). Moreover, increased accumulation of extracellular polymeric substances (EPSs) such as exopolysaccharides (Xiao and Zheng, 2016; Naveed et al., 2019) have been reported as a HM stress response mechanism in Chlamydomonas reinhardtii (Li et al., 2021) presumably because HMs can immobilize onto these substances.
Some may refer to biosorption and the second mechanism presented here, bioaccumulation, as the same process since they both ultimately result in free HMs taken away from liquid phases. However, in this mini-review, we propose that they are two different mechanisms to simplify the principles and lay out plain understandings toward synthetic biology-based approaches. Bioaccumulation is defined as a process that cells uptake metal ions and accumulate them intracellularly in a so-called “import-storage system” (Diep et al., 2018). Thus, it is important to state that in biosorption, HMs can be retrieved via simple methods such as washing with chelators; however, bioaccumulation requires cell disruption (Diep et al., 2018). Bioaccumulation is divided into 2 sequential steps: 1) cellular uptake of HMs, and 2) binding of the HMs to metal-biding peptides or polyphosphates (polyP) and sequestering in inactive forms. Microalgae uptake HMs via various transporters (Blaby-Haas and Merchant, 2012). Once HMs enter the cells, they can bind to designated biomolecules. In this section, metal-binding peptides (phytochelatins and metallothioneins) and polyphosphate bodies are discussed. Phytochelatins (PCs) are one of the metal-binding peptides (Cobbett and Goldsbrough, 2002) synthesized by biosynthetic enzymes (glutamate-cysteine ligase (GCL), glutathione synthetase (GSHS), and PC-synthase (PCS)) (Balzano et al., 2020; García-García et al., 2020). Typically, HM-PC complexes are sequestered in compartmentalized vacuoles, chloroplasts, and cytosol (Nagel et al., 1996; Zitka et al., 2011). Interestingly, glutathione—the final intermediate in PC synthesis—is also involved in HM-PC interactions (Balzano et al., 2020). Thus, all PC biosynthetic enzymes play a major part in HM bio-removal as they can determine intracellular availability of PCs (see Section 3). Metallothioneins (MT) are another well-known group of metal-binding peptides allowing the same process (Balzano et al., 2020). The main difference between PCs and MTs is that MTs are proteins translated directly from encoded gene, unlike PCs, which are enzymatically synthesized from intracellularly available precursors (Cobbett and Goldsbrough, 2002). Lastly, HMs can also bind to intracellular polyphosphates (polyP) because of the anionic nature of the phosphate groups (PO43−) making up the polymer. PolyP accumulation was shown to increase upon HM stress conditions (Sanz-Luque et al., 2020) emphasizing its unrealized potential in HM-bio-removal.
Biotransformation of toxic HMs into less or non-toxic forms is another strategy used by microorganisms (Chaturvedi et al., 2015). Evidently, arsenic (As) is most toxic in the forms of As (III) and As (V), respectively. These forms can be detoxified by means of oxidation [for As (III)], reduction [for As(V)], methylation and conversion to arsenosugars/arsenolipids in different diatom species (Wang et al., 2015; Papry et al., 2019). Similarly, Hg and Cr reduction was reported to be catalyzed by intracellular reductases in microalgae (Kelly et al., 2007; Lee et al., 2017; Leong and Chang, 2020), which facilitates the bio-removal of these metals. Previously with synthetic biology, Sattayawat and colleagues described a novel concept “bioderivatization” in cyanobacteria where a toxic compound produced by the cells was biotransformed into a less toxic form. This successfully alleviated the deleterious effects on microbial cells and resulted in higher production yields (Sattayawat et al., 2020).
Intriguingly, we also reported that Pediastrum duplex AARLG060, a HM bio-removing microalga, performed a combination of mechanisms to remediate Mn (Thongpitak et al., 2019). This observation suggests that several engineering strategies could be implemented concurrently to achieve more impact.
Synthetic Biology-Based Approaches as Strategies to Enhance Heavy Metal Bio-Removal From Wastewater Effluents
Although the native bio-removal mechanisms in microalgae are promising, there is still room for improvement. For instance, the use of fast-growing microalgae is preferable as this could increase total surface area for biosorption and metal uptake. However, such fast-growing microalgae are usually sensitive to high concentrations of HMs.
Several synthetic biology toolkits for microalgae have been developed over the last few years (Gimpel et al., 2016; Crozet et al., 2018; Poliner et al., 2018, 2020; de Grahl et al., 2020; Mehrshahi et al., 2020; Südfeld et al., 2021; Windhagauer et al., 2021) with a recent comprehensive review on synthetic biology-based approaches for enhancement of biomass accumulation and carbon flux partitioning (Naduthodi et al., 2021). Construction of biosynthetic pathways for attractive chemicals has also been reported (Wichmann et al., 2018, 2021; Yunus et al., 2018). Recently, a work on ground-breaking CRISPR technology in Picochlorum celeri (Krishnan et al., 2020) emphasizes the rise of toolkit development even more.
Synthetic Biology Workflow—Design
Identification of desirable characteristics is a fundamental key toward engineering success that can facilitate the first step in synthetic biology workflow—design (Figure 1). Firstly, different species or even strains of microalgae exhibit different performances as biosorbents (Kumar et al., 2015; Salama et al., 2019). This suggests that the degree of anionics plays an important role in determining microalgal performances. From this observation, modification of anionic moieties on cell walls could be the first strategy. Physically, acid treatment could alter surface functional groups and increased HM bio-removal efficiency (Almomani and Bohsale, 2021). However, this strategy also requires an extra acid treatment step. Certainly, an alternative approach is to address this through synthetic biology. The next question is, how can microalgae be genetically modified to increase the binding affinity between microalgal cell surface and HMs? A recent work by Ma and colleagues suggested that biofilm formation facilitated Scenedesmus Cd bio-removal efficiency (Ma et al., 2021). Given that this process is a stress response mechanism toward HMs (Li et al., 2021; Ma et al., 2021) and extracellular polymeric substances (EPSs) forming up the biofilm are generally negatively charged, engineering microalgae for enhanced biofilm production can be implemented to facilitate HM bio-removal. The genetic control of biofilm formation in microorganisms has been studied and several reports on manipulation of biofilm formation in bacteria have been discussed elsewhere (Perni et al., 2013; Benedetti et al., 2016). Mutagenesis studies of cyanobacteria for enhanced EPS production have also been investigated (Pereira et al., 2019; Yadav et al., 2020). However, these remain to be explored in microalgae.
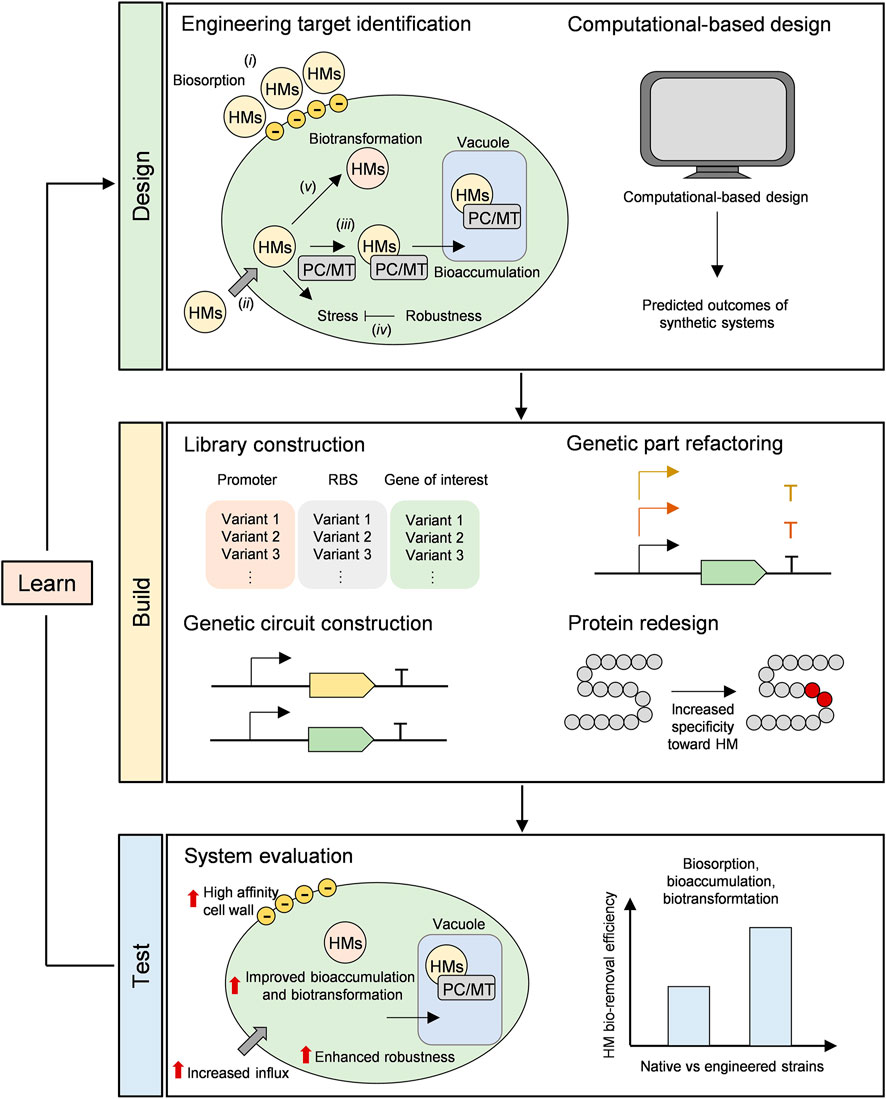
FIGURE 1. Design-Build-Test-Learn (DBTL) cycle of synthetic biology for HM bio-removal. Design: (i) Biosorption (ii) Cellular uptake of HMs (iii) Bioaccumulation (iv) Native cellular responses to HM-induced stress (v) Biotransformation.
Increasing cellular uptake of HMs is another strategy employed to enhance bio-removal efficiency. Mostly, this is carried out by increasing a number or activity of HM active transporters (Zagorski and Wilson, 2004; Deng et al., 2007; Deng and Jia, 2011; Sone et al., 2013). Heterologous expression and overexpression are one of the prime synthetic biology-based approaches leading to increased protein level and activity. For example, expression of a plant transporter for Zn and Cd (AtHMA4 C-terminal domain protein) in C. reinhardtii increases cellular tolerance and bioaccumulation of these HMs compared with the wild-type (Ibuot et al., 2020).
Expression and overexpression of genes involved in metal-binding peptide synthesis have also been reported to promote HM bio-removal. As to metal-binding peptides, PCs and MTs are the prime candidates. PCs are enzymatically synthesized from naturally available precursors (Hirata et al., 2001; Bräutigam et al., 2011; Filiz et al., 2019; Balzano et al., 2020); therefore, heterologous expression and overexpression of PC biosynthetic enzymes can increase their availability and capacity (Clemens et al., 1999; Bae et al., 2001; Pal and Rai, 2010; Singh et al., 2010). The expression of MT gene from chicken in C. reinhardtii was first reported in 1999 and showed to promote Cd bioaccumulation (Cai et al., 1999). Later, other works have also shown that the expression of MT enhanced binding affinity and cellular tolerance in the same host (Sihai et al., 2008; He et al., 2011). This is also the case when expressed in E. coli (Singh et al., 2008; Jafarian and Ghaffari, 2017; Shahpiri and Mohammadzadeh, 2018). Furthermore, overexpression of metal-tolerant proteins, CrMTP4, has shown to increase Cd tolerance in C. reinhardtii by 2.29 folds and resulted in a 3.09-fold increase in Cd bioaccumulation (Ibuot et al., 2017). In a more advanced level, synthetic phytochelatins were expressed and showed to enhance the ability of HM bioaccumulation in bacteria (Bae et al., 2000, 2002). Metalloregulatory proteins also play an important role in determining the efficiency of HM bioaccumulation and homeostasis (Quinn et al., 2003; Silver and Phung, 2005; Sommer et al., 2010; Reyes-Caballero et al., 2011; Merchant et al., 2020) with examples of engineering in E. coli (Sato and Kobayashi, 1998; Yang et al., 2013).
Highly tolerant microalgae tend to remove HMs more effectively due to their ability to withstand HMs at high concentrations. Several enzymes and proteins have been reported to play a role in cellular tolerance toward HMs. For example, increased superoxide dismutase (SOD) expression was observed during Hg exposure as a defense system in Halamphora veneta (Mu et al., 2017). Glutathione peroxidase (GPX), an enzyme responsible for cellular responses to oxidative stress from HM exposure, has also been identified in C. reinhardtii (Fu et al., 2002; Dayer et al., 2008; Ma et al., 2020). Another example, thioredoxins in C. reinhardtii are also induced upon Cd exposure (Lemaire et al., 1999). On an engineering point of view, heterologous expression of C. reinhardtii Fe-assimilating protein 1 (FEA1) was shown to alleviate HM toxicity in yeast and plants (Narayanan et al., 2011). Sequestration also plays a role in HM tolerance. For instance, CrMRP2 (a member of the ATP-binding cassette (ABC) transporter subfamily) in C. reinhardtii functions in cellular sequestration of PC-Cd complex. It was shown that Cd bioaccumulation and sequestration were higher in the wild-type compared with the CrMRP2 mutant. In the same study, expression of CrMRP2 could complement Cd-sensitive yeast mutant lacking a vacuolar glutathione (GSH)-conjugate ABC transporter (Wang and Wu, 2006).
As mentioned in the previous section, reductases have been shown to facilitate HM bio-removal. Expression of mercuric reductase from Bacillus megaterium MB1 in eukaryotic microalga Chlorella sp. DT was previously studied and shown to promote Hg bio-removal (Huang et al., 2006). For more exploratory features, construction of biotransformation pathways could be an option and has previously been implemented in bacteria (Dvořák et al., 2017).
Computational-based analysis to predict the outcomes is another approach facilitating the design step. Though, computational-based approaches for HM bio-removal have not been widely used in microalgal engineering. Molecular dynamics of PCs have been studied (Pochodylo and Aristilde, 2017) to gain better understandings on the HM-PC complexes and computational approaches for redesigning metal-binding sites were reviewed (Akcapinar and Sezerman, 2017). A web tool to facilitate the design of intron-containing transgene expression in microalgae has also been established (Jaeger et al., 2019). Note that gene expression or overexpression could burden overall cellular performances; hence, in silico modelling that can predict a balance state between protein expression and energy production (or metabolic flux) could be beneficial. Recently, this principle has been applied to microalgae for bioproduction purposes (Tibocha-Bonilla et al., 2018; Zuñiga et al., 2018), but not yet for HM bio-removal.
Synthetic Biology Workflow—Build
Variant libraries as a collection of genetic parts ready to be recruited are helpful in the second step of synthetic biology workflow—build (Figure 1). With Next Generation Sequencing (NGS) technology, more whole genome sequences are available for putative gene discovery. Previously, a list of metal transporters in microalgae has been reported based on a whole-genome analysis (Hanikenne et al., 2005). Similarly, metagenomic analysis allowed the discovery of a new MT family with confirmed ability to chelate Cu(I), Zn(II) and Cd(II) (Ziller et al., 2017). Moreover, putative HM tolerance genes from C. acidophila—an extremophilic green alga—are also reported based upon transcriptomic analysis (Puente-Sánchez et al., 2018).
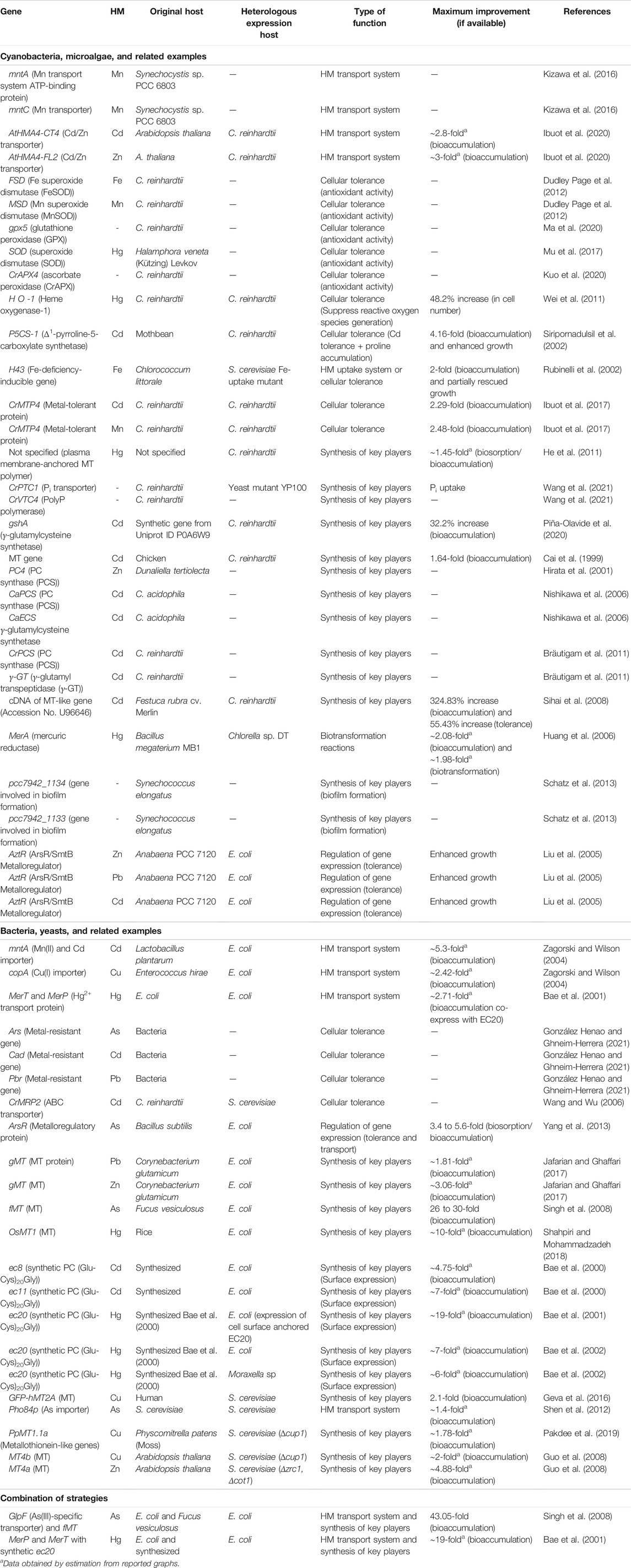
To emphasize the potentials of employing synthetic biology in microalgal bio-removal of HMs and facilitate future designs of such systems, we accumulatively search for genes taking part in HM bio-removal and summarized in Table 1. Subsequently, we group the genes based on their functions: 1) HM transport system, 2) cellular tolerance, 3) synthesis of key players, 4) biotransformation reactions of HMs, and 5) regulation of gene expression. However, it should be noted that these genes are listed based on their reported potential in HM bio-removal mechanisms, some of them have never been confirmed to enhance the process experimentally.
In synthetic biology, genetic circuits can be designed and constructed in a manner that would maximize system performances. In this sense, refactoring native genetic parts—promoters, terminators—is a useful approach to enhance protein expression. Promoter and terminator engineering in C. reinhardtii have been studied and reported (Einhaus et al., 2020). To boost up the engineered constructs even further, fine-tuning of regulatory systems could be implemented. This strategy has been illustrated in microalgae, though in the area not related to HM bio-removal (Sun et al., 2018). Additionally, protein redesign based on computational predictions could be implemented experimentally in this step.
Synthetic Biology Workflow—Test and Learn
The final two steps in synthetic biology workflow are to test and to learn from the newly designed system (Figure 1). In these steps, system performances are assessed and factors affecting the system are taken into consideration for the second round of the Design-Build-Test-Learn (DBTL) cycle. The goal of each cycle is to achieve a more optimal system, thus observations made from the previous cycle essentially put each piece of jigsaws together toward better performances. For instance, a bacterial system expressing synthetic phytochelatins showed enhanced HM bioaccumulation (Bae et al., 2000); however, it was learnt that HM uptake was a rate-limiting step of the process. Thus, the same system was improved in the second round of DBLT cycle by expressing anchored synthetic phytochelatins on the cell surface or expressing the synthetic phytochelatins together with HM transporters to overcome the limitation (Bae et al., 2001).
Prospects of Synthetic Biology-Based Approaches for Microalgal Heavy Metal Bio-Removal
Synthetic biology builds solely on genetic manipulations. Thus, legislations that are in effect to control the use of genetically modified organisms (GMOs) in each country are the major restrictions. Moreover, relatively few numbers of genetic parts and tools compared with that of other microorganisms also limit the development of microalgal chassis for synthetic biology, though the knowledge is building up (Tran and Kaldenhoff, 2020; Sproles et al., 2021). Omics information is also required for accurate computational predictions. This allows us to foresee cellular states after modifications and facilitates the system design. Altogether, despite such limitations, we expected that synthetic biology will gain more attention as a tool to improve microalgal HM bio-removal from wastewater effluents in the near future.
Conclusion
In microalgae, synthetic biology is often investigated as a tool to improve biomass and chemical production. Here, we discuss synthetic biology from another point of view—for heavy metal bio-removal from wastewater. A descriptive Design-Build-Test-Learn synthetic biology cycle is also presented for the first time to serve as a direction for straightforward implementations of synthetic biology for microalgal HM bio-removal. To facilitate this even further, we construct a genetic part library of genes and proteins involved in the process and/or reported to be engineered in a synthetic biology manner that in future, each part can be handpicked directly from our library.
Author Contributions
PS conceptualized and drafted the manuscript. IY, NN, NM, WP-A, JP ,and CP revised the manuscript critically and approved the final draft.
Funding
This research work was partially supported by Chiang Mai University.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Akcapinar, G. B., and Sezerman, O. U. (2017). Computational Approaches for De Novo Design and Redesign of Metal-Binding Sites on Proteins. Biosci. Rep. 37, 1–12. doi:10.1042/BSR20160179
Almomani, F., and Bhosale, R. R. (2021). Bio-sorption of Toxic Metals from Industrial Wastewater by Algae Strains Spirulina Platensis and Chlorella Vulgaris: Application of Isotherm, Kinetic Models and Process Optimization. Sci. Total Environ. 755, 142654. doi:10.1016/j.scitotenv.2020.142654
Bae, W., Mehra, R. K., Mulchandani, A., and Chen, W. (2001). Genetic Engineering of Escherichia coli for Enhanced Uptake and Bioaccumulation of Mercury. Appl. Environ. Microbiol. 67, 5335–5338. doi:10.1128/aem.67.11.5335-5338.2001
Bae, W., Mulchandani, A., and Chen, W. (2002). Cell Surface Display of Synthetic Phytochelatins Using Ice Nucleation Protein for Enhanced Heavy Metal Bioaccumulation. J. Inorg. Biochem. 88, 223–227. doi:10.1016/S0162-0134(01)00392-0
Balzano, S., Sardo, A., Blasio, M., Chahine, T. B., Dell’Anno, F., Sansone, C., et al. (2020). Microalgal Metallothioneins and Phytochelatins and Their Potential Use in Bioremediation. Front. Microbiol. 11, 1–16. doi:10.3389/fmicb.2020.00517
Benedetti, I., de Lorenzo, V., and Nikel, P. I. (2016). Genetic Programming of Catalytic Pseudomonas Putida Biofilms for Boosting Biodegradation of Haloalkanes. Metab. Eng. 33, 109–118. doi:10.1016/j.ymben.2015.11.004
Blaby-Haas, C. E., and Merchant, S. S. (2012). The Ins and Outs of Algal Metal Transport. Biochim. Biophys. Acta (Bba) - Mol. Cell Res. 1823, 1531–1552. doi:10.1016/j.bbamcr.2012.04.010
Bräutigam, A., Schaumlöffel, D., Preud'homme, H., Thondorf, I., and Wesenberg, D. (2011). Physiological Characterization of Cadmium-Exposed Chlamydomonas Reinhardtii. Plant Cell Environ. 34, 2071–2082. doi:10.1111/j.1365-3040.2011.02404.x
Cai, X.-H., Brown, C., Adhiya, J., Traina, S. J., Sayre, R. T., Brown, C., et al. (1999). Growth and Heavy Metal Binding Properties of Transgenic Chlamydomonas Expressing a Foreign Metallothionein Gene. Int. J. Phytoremediation 1, 53–65. doi:10.1080/15226519908500004
Capeness, M. J., and Horsfall, L. E. (2020). Synthetic Biology Approaches Towards The Recycling of Metals From the Environment. Biochem. Soc. Trans. 48, 1367–1378. doi:10.1042/BST20190837
Chaturvedi, A. D., Pal, D., Penta, S., and Kumar, A. (2015). Ecotoxic Heavy Metals Transformation by Bacteria and Fungi in Aquatic Ecosystem. World J. Microbiol. Biotechnol. 31, 1595–1603. doi:10.1007/s11274-015-1911-5
Chen, W., Bae, W., Mehra, R., and Mulchandani, A. (2002). Enhanced Bioaccumulation of Heavy Metals by Bacterial Cells with Surface-Displayed Synthetic Phytochelatins. ACS Symp. Ser. 806, 411–418. doi:10.1021/bk-2002-0806.ch024
Cheng, S. Y., Show, P.-L., Lau, B. F., Chang, J.-S., and Ling, T. C. (2019). New Prospects for Modified Algae in Heavy Metal Adsorption. Trends Biotechnol. 37, 1255–1268. doi:10.1016/j.tibtech.2019.04.007
Clemens, S., Kim, E. J., Neumann, D., and Schroeder, J. I. (1999). Tolerance to Toxic Metals by a Gene Family of Phytochelatin Synthases from Plants and Yeast. EMBO 18, 3325–3333. doi:10.1093/emboj/18.12.3325
Cobbett, C., and Goldsbrough, P. (2002). Phytochelatins and Metallothioneins: Roles in Heavy Metal Detoxification and Homeostasis. Annu. Rev. Plant Biol. 53, 159–182. doi:10.1146/annurev.arplant.53.100301.135154
Crozet, P., Navarro, F. J., Willmund, F., Mehrshahi, P., Bakowski, K., Lauersen, K. J., et al. (2018). Birth of a Photosynthetic Chassis: A MoClo Toolkit Enabling Synthetic Biology in the Microalga Chlamydomonas Reinhardtii. ACS Synth. Biol. 7, 2074–2086. doi:10.1021/acssynbio.8b00251
Dayer, R., Fischer, B. B., Eggen, R. I. L., and Lemaire, S. D. (2008). The Peroxiredoxin and Glutathione Peroxidase Families in Chlamydomonas Reinhardtii. Genetics 179, 41–57. doi:10.1534/genetics.107.086041
de Grahl, I., Rout, S. S., Maple-Grødem, J., and Reumann, S. (2020). Development of a Constitutive and an Auto-Inducible High-Yield Expression System for Recombinant Protein Production in the Microalga Nannochloropsis Oceanica. Appl. Microbiol. Biotechnol. 104, 8747–8760. doi:10.1007/s00253-020-10789-4
Deng, X., and Jia, P. (2011). Construction and Characterization of a Photosynthetic Bacterium Genetically Engineered for Hg2+ Uptake. Bioresour. Tech. 102, 3083–3088. doi:10.1016/j.biortech.2010.10.051
Deng, X., Yi, X. E., and Liu, G. (2007). Cadmium Removal from Aqueous Solution by Gene-Modified Escherichia coli JM109. J. Hazard. Mater. 139, 340–344. doi:10.1016/j.jhazmat.2006.06.043
Diep, P., Mahadevan, R., and Yakunin, A. F. (2018). Heavy Metal Removal by Bioaccumulation Using Genetically Engineered Microorganisms. Front. Bioeng. Biotechnol. 6, 1–20. doi:10.3389/fbioe.2018.00157
Duffus, J. H. (2002). "Heavy Metals" a Meaningless Term? (IUPAC Technical Report). Pure Appl. Chem. 74, 793–807. doi:10.1351/pac200274050793
Dvořák, P., Nikel, P. I., Damborský, J., and de Lorenzo, V. (2017). Bioremediation 3.0 : Engineering Pollutant-Removing Bacteria in the Times of Systemic Biology. Biotechnol. Adv. 35, 845–866. doi:10.1016/j.biotechadv.2017.08.001
Einhaus, A., Baier, T., Rosenstengel, M., Freudenberg, R. A., and Kruse, O. (2021). Rational Promoter Engineering Enables Robust Terpene Production in Microalgae. ACS Synth. Biol. 10, 847–856. doi:10.1021/acssynbio.0c00632
Fabris, M., Abbriano, R. M., Pernice, M., Sutherland, D. L., Commault, A. S., Hall, C. C., et al. (2020). Emerging Technologies in Algal Biotechnology: Toward the Establishment of a Sustainable, Algae-Based Bioeconomy. Front. Plant Sci. 11, 1–22. doi:10.3389/fpls.2020.00279
Fajardo, C., Donato, M., Carrasco, R., Martínez‐Rodríguez, G., Mancera, J. M., and Fernández‐Acero, F. J. (2020). Advances and Challenges in Genetic Engineering of Microalgae. Rev. Aquacult 12, 365–381. doi:10.1111/raq.12322
Filiz, E., Saracoglu, I. A., Ozyigit, I. I., and Yalcin, B. (2019). Comparative Analyses of Phytochelatin Synthase (PCS) Genes in Higher Plants. Biotechnol. Biotechnological Equipment 33, 178–194. doi:10.1080/13102818.2018.1559096
Fu, L.-H., Wang, X.-F., Eyal, Y., She, Y.-M., Donald, L. J., Standing, K. G., et al. (2002). A Selenoprotein in the Plant Kingdom.Mass Spectrometry Confirms that an Opal Codon(UGA) Encodes Selenocysteine in Chlamydomonas Reinhardtii Glutathione Peroxidase. J. Biol. Chem. 277, 25983–25991. doi:10.1074/jbc.M202912200
García-García, J. D., Sánchez-Thomas, R., Saavedra, E., Fernández-Velasco, D. A., Romero-Romero, S., Casanova-Figueroa, K. I., et al. (2020). Mapping the Metal-Catalytic Site of a Zinc-Activated Phytochelatin Synthase. Algal Res. 47, 101890. doi:10.1016/j.algal.2020.101890
Geva, P., Kahta, R., Nakonechny, F., Aronov, S., and Nisnevitch, M. (2016). Increased Copper Bioremediation Ability of New Transgenic and Adapted Saccharomyces cerevisiae Strains. Environ. Sci. Pollut. Res. 23, 19613–19625. doi:10.1007/s11356-016-7157-4
Gimpel, J. A., Nour-Eldin, H. H., Scranton, M. A., Li, D., and Mayfield, S. P. (2016). Refactoring the Six-Gene Photosystem II Core in the Chloroplast of the Green Algae Chlamydomonas Reinhardtii. ACS Synth. Biol. 5, 589–596. doi:10.1021/acssynbio.5b00076
Gleizer, S., Ben-Nissan, R., Bar-On, Y. M., Antonovsky, N., Noor, E., Zohar, Y., et al. (2019). Conversion of Escherichia coli to Generate All Biomass Carbon from CO2. Cell 179, 1255–1263.e12. doi:10.1016/j.cell.2019.11.009
GonzálezHenao, S., and Ghneim-Herrera, T. (2021). Heavy Metals in Soils and the Remediation Potential of Bacteria Associated with the Plant Microbiome. Front. Environ. Sci. 9, 1–17. doi:10.3389/fenvs.2021.604216
Guo, W.-J., Meetam, M., and Goldsbrough, P. B. (2008). Examining the Specific Contributions of Individual Arabidopsis Metallothioneins to Copper Distribution and Metal Tolerance. Plant Physiol. 146, 1697–1706. doi:10.1104/pp.108.115782
Han, S., Hu, Z., and Lei, A. (2008). Expression and Function Analysis of the Metallothionein-like (MT-like) Gene from Festuca Rubra in Chlamydomonas Reinhardtii Chloroplast. Sci. China Ser. C 51, 1076–1081. doi:10.1007/s11427-008-0136-3
Hanikenne, M., Krämer, U., Demoulin, V., and Baurain, D. (2005). A Comparative Inventory of Metal Transporters in the Green Alga Chlamydomonas Reinhardtii and the Red Alga Cyanidioschizon Merolae. Plant Phys. 137, 428–446. doi:10.1104/pp.104.054189.428
He, Z., Siripornadulsil, S., Sayre, R. T., Traina, S. J., and Weavers, L. K. (2011). Removal of Mercury from Sediment by Ultrasound Combined with Biomass (Transgenic Chlamydomonas Reinhardtii). Chemosphere 83, 1249–1254. doi:10.1016/j.chemosphere.2011.03.004
Hirata, K., Tsujimoto, Y., Namba, T., Ohta, T., Hirayanagi, N., Miyasaka, H., et al. (2001). Strong Induction of Phytochelatin Synthesis by Zinc in marine green Alga, Dunaliella Tertiolecta. J. Biosci. Bioeng. 92, 24–29. doi:10.1016/s1389-1723(01)80193-6
Huang, C.-C., Chen, M.-W., Hsieh, J.-L., Lin, W.-H., Chen, P.-C., and Chien, L.-F. (2006). Expression of Mercuric Reductase from Bacillus Megaterium MB1 in Eukaryotic Microalga Chlorella Sp. DT: An Approach for Mercury Phytoremediation. Appl. Microbiol. Biotechnol. 72, 197–205. doi:10.1007/s00253-005-0250-0
Huang, R., Huo, G., Song, S., Li, Y., Xia, L., and Gaillard, J.-F. (2019). Immobilization of Mercury Using High-Phosphate Culture-Modified Microalgae. Environ. Pollut. 254, 112966. doi:10.1016/j.envpol.2019.112966
Ibuot, A., Dean, A. P., McIntosh, O. A., and Pittman, J. K. (2017). Metal Bioremediation by CrMTP4 Over-expressing Chlamydomonas Reinhardtii in Comparison to Natural Wastewater-Tolerant Microalgae Strains. Algal Res. 24, 89–96. doi:10.1016/j.algal.2017.03.002
Ibuot, A., Webster, R. E., Williams, L. E., and Pittman, J. K. (2020). Increased Metal Tolerance and Bioaccumulation of Zinc and Cadmium inChlamydomonas Reinhardtiiexpressing a AtHMA4 C‐terminal Domain Protein. Biotechnol. Bioeng. 117, 2996–3005. doi:10.1002/bit.27476
Jaeger, D., Baier, T., and Lauersen, K. J. (2019). Intronserter, an Advanced Online Tool for Design of Intron Containing Transgenes. Algal Res. 42, 101588. doi:10.1016/j.algal.2019.101588
Jafarian, V., and Ghaffari, F. (2017). A Unique Metallothionein-Engineered in Escherichia coli for Biosorption of Lead, Zinc, and Cadmium; Absorption or Adsorption? Microbiology 86, 73–81. doi:10.1134/S0026261717010064
Jagadevan, S., Banerjee, A., Banerjee, C., Guria, C., Tiwari, R., Baweja, M., et al. (2018). Recent Developments in Synthetic Biology and Metabolic Engineering in Microalgae towards Biofuel Production. Biotechnol. Biofuels 11, 1–21. doi:10.1186/s13068-018-1181-1
Kaminski, M. M., Abudayyeh, O. O., Gootenberg, J. S., Zhang, F., and Collins, J. J. (2021). CRISPR-based Diagnostics. Nat. Biomed. Eng. 5, 643–656. doi:10.1038/s41551-021-00760-7
Kelly, D. J. A., Budd, K., and Lefebvre, D. D. (2006). Biotransformation of Mercury in pH-Stat Cultures of Eukaryotic Freshwater Algae. Arch. Microbiol. 187, 45–53. doi:10.1007/s00203-006-0170-0
Kizawa, A., Kawahara, A., Takimura, Y., Nishiyama, Y., and Hihara, Y. (2016). RNA-seq Profiling Reveals Novel Target Genes of LexA in the Cyanobacterium Synechocystis Sp. PCC 6803. Front. Microbiol. 7, 1–14. doi:10.3389/fmicb.2016.00193
Krishnan, A., Cano, M., Burch, T. A., Weissman, J. C., and Posewitz, M. C. (2020). Genome Editing Using Cas9-RNA Ribonucleoprotein Complexes in the High-Productivity marine Alga Picochlorum Celeri. Algal Res. 49, 101944. doi:10.1016/j.algal.2020.101944
Kumar, G., Shekh, A., Jakhu, S., Sharma, Y., Kapoor, R., and Sharma, T. R. (2020). Bioengineering of Microalgae: Recent Advances, Perspectives, and Regulatory Challenges for Industrial Application. Front. Bioeng. Biotechnol. 8, 914. doi:10.3389/fbioe.2020.00914
Kumar, K. S., Dahms, H.-U., Won, E.-J., Lee, J.-S., and Shin, K.-H. (2015). Microalgae - A Promising Tool for Heavy Metal Remediation. Ecotoxicology Environ. Saf. 113, 329–352. doi:10.1016/j.ecoenv.2014.12.019
Kuo, E. Y., Cai, M.-S., and Lee, T.-M. (2020). Ascorbate Peroxidase 4 Plays a Role in the Tolerance of Chlamydomonas Reinhardtii to Photo-Oxidative Stress. Sci. Rep. 10, 1–12. doi:10.1038/s41598-020-70247-z
Lee, L., Hsu, C.-Y., and Yen, H.-W. (2017). The Effects of Hydraulic Retention Time (HRT) on Chromium(VI) Reduction Using Autotrophic Cultivation of Chlorella Vulgaris. Bioproc. Biosyst. Eng. 40, 1725–1731. doi:10.1007/s00449-017-1827-6
Lemaire, S., Keryer, E., Stein, M., Schepens, I., Issakidis-Bourguet, E., Gérard-Hirne, C., et al. (1999). Heavy-Metal Regulation of Thioredoxin Gene Expression inChlamydomonas Reinhardtii. Plant Physiol. 120, 773–778. doi:10.1104/pp.120.3.773
Leong, Y. K., and Chang, J.-S. (2020). Bioremediation of Heavy Metals Using Microalgae: Recent Advances and Mechanisms. Bioresour. Tech. 303, 122886. doi:10.1016/j.biortech.2020.122886
Li, C., Zheng, C., Fu, H., Zhai, S., Hu, F., Naveed, S., et al. (2021). Contrasting Detoxification Mechanisms of Chlamydomonas Reinhardtii under Cd and Pb Stress. Chemosphere 274, 129771. doi:10.1016/j.chemosphere.2021.129771
Liu, T., Golden, J. W., and Giedroc, D. P. (2005). A Zinc(II)/Lead(II)/Cadmium(II)-Inducible Operon from the CyanobacteriumAnabaenaIs Regulated by AztR, an α3N ArsR/SmtB Metalloregulator. Biochemistry 44, 8673–8683. doi:10.1021/bi050450+
Ma, X., Chen, Y., Liu, F., Zhang, S., and Wei, Q. (2021). Enhanced Tolerance and Resistance Characteristics of Scenedesmus Obliquus FACHB-12 with K3 Carrier in Cadmium Polluted Water. Algal Res. 55, 102267. doi:10.1016/j.algal.2021.102267
Ma, X., Zhang, B., Miao, R., Deng, X., Duan, Y., Cheng, Y., et al. (2020). Transcriptomic and Physiological Responses to Oxidative Stress in a Chlamydomonas Reinhardtii Glutathione Peroxidase Mutant. Genes 11, 1–26. doi:10.3390/genes11040463
Mantzorou, A., Navakoudis, E., Paschalidis, K., and Ververidis, F. (2018). Microalgae: a Potential Tool for Remediating Aquatic Environments from Toxic Metals. Int. J. Environ. Sci. Technol. 15, 1815–1830. doi:10.1007/s13762-018-1783-y
Mehrshahi, P., Nguyen, G. T. D. T., Gorchs Rovira, A., Sayer, A., Llavero-Pasquina, M., Lim Huei Sin, M., et al. (2020). Development of Novel Riboswitches for Synthetic Biology in the Green Alga Chlamydomonas. ACS Synth. Biol. 9, 1406–1417. doi:10.1021/acssynbio.0c00082
Meng, F., and Ellis, T. (2020). The Second Decade of Synthetic Biology: 2010-2020. Nat. Commun. 11, 1–4. doi:10.1038/s41467-020-19092-2
Merchant, S. S., Schmollinger, S., Strenkert, D., Moseley, J. L., and Blaby-Haas, C. E. (2020). From Economy to Luxury: Copper Homeostasis in Chlamydomonas and Other Algae. Biochim. Biophys. Acta (Bba) - Mol. Cell Res. 1867, 118822. doi:10.1016/j.bbamcr.2020.118822
Monteiro, C. M., Castro, P. M. L., and Malcata, F. X. (2012). Metal Uptake by Microalgae: Underlying Mechanisms and Practical Applications. Biotechnol. Prog. 28, 299–311. doi:10.1002/btpr.1504
Mu, W., Jia, K., Liu, Y., Pan, X., and Fan, Y. (2017). Response of the Freshwater Diatom Halamphora veneta (Kützing) Levkov to Copper and Mercury and its Potential for Bioassessment of Heavy Metal Toxicity in Aquatic Habitats. Environ. Sci. Pollut. Res. 24, 26375–26386. doi:10.1007/s11356-017-0225-6
Naduthodi, M. I. S., Claassens, N. J., D’Adamo, S., van der Oost, J., and Barbosa, M. J. (2021). Synthetic Biology Approaches to Enhance Microalgal Productivity. Trends Biotechnol. 39, 1019–1036. doi:10.1016/j.tibtech.2020.12.010
Nagel, K., Adelmeier, U., and Voigt, J. (1996). Subcellular Distribution of Cadmium in the Unicellular green Alga Chlamydomonas Reinhardtii. J. Plant Physiol. 149, 86–90. doi:10.1016/S0176-1617(96)80178-7
Narayanan, N. N., Ihemere, U., Chiu, W. T., Siritunga, D., Rajamani, S., Singh, S., et al. (2011). The Iron Assimilatory Protein, FEA1, from Chlamydomonas Reinhardtii Facilitates Iron-specific Metal Uptake in Yeast and Plants. Front. Plant Sci. 2, 1–13. doi:10.3389/fpls.2011.00067
Naveed, S., Li, C., Lu, X., Chen, S., Yin, B., Zhang, C., et al. (2019). Microalgal Extracellular Polymeric Substances and Their Interactions with Metal(loid)s: A Review. Crit. Rev. Environ. Sci. Tech. 49, 1769–1802. doi:10.1080/10643389.2019.1583052
Nishikawa, K., Onodera, A., and Tominaga, N. (2006). Phytochelatins Do Not Correlate with the Level of Cd Accumulation in Chlamydomonas Spp. Chemosphere 63, 1553–1559. doi:10.1016/j.chemosphere.2005.09.056
Page, M. D., Allen, M. D., Kropat, J., Urzica, E. I., Karpowicz, S. J., Hsieh, S. I., et al. (2012). Fe Sparing and Fe Recycling Contribute to Increased Superoxide Dismutase Capacity in Iron-Starved Chlamydomonas Reinhardtii. Plant Cell 24, 2649–2665. doi:10.1105/tpc.112.098962
Pakdee, O., Songnuan, W., Panvisavas, N., Pokethitiyook, P., Yokthongwattana, K., and Meetam, M. (2019). Functional Characterization of Metallothionein-like Genes from Physcomitrella Patens: Expression Profiling, Yeast Heterologous Expression, and Disruption of PpMT1.2a Gene. Planta 250, 427–443. doi:10.1007/s00425-019-03173-8
Pal, R., and Rai, J. P. N. (2010). Phytochelatins: Peptides Involved in Heavy Metal Detoxification. Appl. Biochem. Biotechnol. 160, 945–963. doi:10.1007/s12010-009-8565-4
Papry, R. I., Ishii, K., Mamun, M. A. A., Miah, S., Naito, K., Mashio, A. S., et al. (2019). Arsenic Biotransformation Potential of Six marine Diatom Species: Effect of Temperature and Salinity. Sci. Rep. 9, 1–16. doi:10.1038/s41598-019-46551-8
Pereira, S. B., Sousa, A., Santos, M., Araújo, M., Serôdio, F., Granja, P., et al. (2019). Strategies to Obtain Designer Polymers Based on Cyanobacterial Extracellular Polymeric Substances (EPS). Ijms 20, 1–18. doi:10.3390/ijms20225693
Perni, S., Hackett, L., Goss, R. J., Simmons, M. J., and Overton, T. W. (2013). Optimisation of Engineered Escherichia coli Biofilms for Enzymatic Biosynthesis of L-Halotryptophans. AMB Expr. 3, 1–10. doi:10.1186/2191-0855-3-66
Piña-Olavide, R., Paz-Maldonado, L. M. T., Alfaro-De La Torre, M. C., García-Soto, M. J., Ramírez-Rodríguez, A. E., Rosales-Mendoza, S., et al. (2020). Increased Removal of Cadmium by Chlamydomonas Reinhardtii Modified with a Synthetic Gene for γ-glutamylcysteine Synthetase. Int. J. Phytoremediation 22, 1269–1277. doi:10.1080/15226514.2020.1765138
Pochodylo, A. L., and Aristilde, L. (2017). Molecular Dynamics of Stability and Structures in Phytochelatin Complexes with Zn, Cu, Fe, Mg, and Ca: Implications for Metal Detoxification. Environ. Chem. Lett. 15, 495–500. doi:10.1007/s10311-017-0609-3
Poliner, E., Clark, E., Cummings, C., Benning, C., and Farre, E. M. (2020). A High-Capacity Gene Stacking Toolkit for the Oleaginous Microalga, Nannochloropsis Oceanica CCMP1779. Algal Res. 45, 101664. doi:10.1016/j.algal.2019.101664
Poliner, E., Farré, E. M., and Benning, C. (2018). Advanced Genetic Tools Enable Synthetic Biology in the Oleaginous Microalgae Nannochloropsis Sp. Plant Cell Rep 37, 1383–1399. doi:10.1007/s00299-018-2270-0
Puente-Sánchez, F., Díaz, S., Penacho, V., Aguilera, A., and Olsson, S. (2018). Basis of Genetic Adaptation to Heavy Metal Stress in the Acidophilic green Alga Chlamydomonas Acidophila. Aquat. Toxicol. 200, 62–72. doi:10.1016/j.aquatox.2018.04.020
Quinn, J. M., Kropat, J., and Merchant, S. (2003). Copper Response Element and Crr1-dependent Ni 2+ -Responsive Promoter for Induced, Reversible Gene Expression in Chlamydomonas Reinhardtii. Eukaryot. Cell 2, 995–1002. doi:10.1128/EC.2.5.995-1002.2003
Rahman, Z., and Singh, V. P. (2019). The Relative Impact of Toxic Heavy Metals (THMs) (Arsenic (As), Cadmium (Cd), Chromium (Cr)(VI), Mercury (Hg), and lead (Pb)) on the Total Environment: an Overview. Environ. Monit. Assess. 191, 419. doi:10.1007/s10661-019-7528-7
Reyes-Caballero, H., Campanello, G. C., and Giedroc, D. P. (2011). Metalloregulatory Proteins: Metal Selectivity and Allosteric Switching. Biophysical Chem. 156, 103–114. doi:10.1016/j.bpc.2011.03.010
Rubinelli, P., Siripornadulsil, S., Gao-Rubinelli, F., and Sayre, R. (2002). Cadmium- and Iron-Stress-Inducible Gene Expression in the green Alga Chlamydomonas Reinhardtii : Evidence for H43 Protein Function in Iron Assimilation. Planta 215, 1–13. doi:10.1007/s00425-001-0711-3
Salama, E.-S., Roh, H.-S., Dev, S., Khan, M. A., Abou-Shanab, R. A. I., Chang, S. W., et al. (2019). Algae as a green Technology for Heavy Metals Removal from Various Wastewater. World J. Microbiol. Biotechnol. 35, 1–19. doi:10.1007/s11274-019-2648-3
Sanz-Luque, E., Bhaya, D., and Grossman, A. R. (2020). Polyphosphate: A Multifunctional Metabolite in Cyanobacteria and Algae. Front. Plant Sci. 11, 1–21. doi:10.3389/fpls.2020.00938
Sato, T., and Kobayashi, Y. (1998). The Ars Operon in the Skin Element of Bacillus Subtilis Confers Resistance to Arsenate and Arsenite. J. Bacteriol. 180, 1655–1661. doi:10.1128/jb.180.7.1655-1661.1998
Sattayawat, P., Yunus, I. S., and Jones, P. R. (2020). Bioderivatization as a Concept for Renewable Production of Chemicals that Are Toxic or Poorly Soluble in the Liquid Phase. Proc. Natl. Acad. Sci. USA 117, 1404–1413. doi:10.1073/pnas.1914069117
Schatz, D., Nagar, E., Sendersky, E., Parnasa, R., Zilberman, S., Carmeli, S., et al. (2013). Self-suppression of Biofilm Formation in the cyanobacteriumSynechococcus Elongatus. Environ. Microbiol. 15, 1786–1794. doi:10.1111/1462-2920.12070
Selvi, A., Rajasekar, A., Theerthagiri, J., Ananthaselvam, A., Sathishkumar, K., Madhavan, J., et al. (2019). Integrated Remediation Processes toward Heavy Metal Removal/recovery from Various Environments-A Review. Front. Environ. Sci. 7, 66. doi:10.3389/fenvs.2019.00066
Shahpiri, A., and Mohammadzadeh, A. (2018). Mercury Removal by Engineered Escherichia coli Cells Expressing Different rice Metallothionein Isoforms. Ann. Microbiol. 68, 145–152. doi:10.1007/s13213-018-1326-2
Shen, M. W. Y., Shah, D., Chen, W., and Da Silva, N. (2012). Enhanced Arsenate Uptake in Saccharomyces cerevisiae Overexpressing the Pho84 Phosphate Transporter. Biotechnol. Prog. 28, 654–661. doi:10.1002/btpr.1531
Silver, S., and Phung, L. T. (2005). A Bacterial View of the Periodic Table: Genes and Proteins for Toxic Inorganic Ions. J. Ind. Microbiol. Biotechnol. 32, 587–605. doi:10.1007/s10295-005-0019-6
Singh, S., Kang, S. H., Lee, W., Mulchandani, A., and Chen, W. (2009). Systematic Engineering of Phytochelatin Synthesis and Arsenic Transport for Enhanced Arsenic Accumulation in E. Coli. Biotechnol. Bioeng. 105, 780–785. doi:10.1002/bit.22585
Singh, S., Mulchandani, A., and Chen, W. (2008). Highly Selective and Rapid Arsenic Removal by Metabolically Engineered Escherichia coli Cells Expressing Fucus Vesiculosus Metallothionein. Appl. Environ. Microbiol. 74, 2924–2927. doi:10.1128/AEM.02871-07
Siripornadulsil, S., Traina, S., Verma, D. P. S., and Sayre, R. T. (2002). Molecular Mechanisms of Proline-Mediated Tolerance to Toxic Heavy Metals in Transgenic Microalgae. Plant Cell 14, 2837–2847. doi:10.1105/tpc.004853
Sommer, F., Kropat, J., Malasarn, D., Grossoehme, N. E., Chen, X., Giedroc, D. P., et al. (2010). The CRR1 Nutritional Copper Sensor in Chlamydomonas Contains Two Distinct Metal-Responsive Domains. Plant Cell 22, 4098–4113. doi:10.1105/tpc.110.080069
Sone, Y., Nakamura, R., Pan-Hou, H., Itoh, T., and Kiyono, M. (2013). Role of MerC, MerE, MerF, MerT, And/or MerP in Resistance to Mercurials and the Transport of Mercurials in Escherichia coli. Biol. Pharm. Bull. 36, 1835–1841. doi:10.1248/bpb.b13-00554
Sproles, A. E., Fields, F. J., Smalley, T. N., Le, C. H., Badary, A., and Mayfield, S. P. (2021). Recent Advancements in the Genetic Engineering of Microalgae. Algal Res. 53, 102158. doi:10.1016/j.algal.2020.102158
Südfeld, C., Hubáček, M., Figueiredo, D., Naduthodi, M. I. S., van der Oost, J., Wijffels, R. H., et al. (2021). High-throughput Insertional Mutagenesis Reveals Novel Targets for Enhancing Lipid Accumulation in Nannochloropsis Oceanica. Metab. Eng. 66, 239–258. doi:10.1016/j.ymben.2021.04.012
Sun, X.-M., Ren, L.-J., Zhao, Q.-Y., Ji, X.-J., and Huang, H. (2018). Microalgae for the Production of Lipid and Carotenoids: A Review with Focus on Stress Regulation and Adaptation. Biotechnol. Biofuels 11, 1–16. doi:10.1186/s13068-018-1275-9
Thongpitak, J., Pekkoh, J., and Pumas, C. (2019). Remediation of Manganese-Contaminated Coal-Mine Water Using Bio-Sorption and Bio-Oxidation by the Microalga Pediastrum Duplex (AARLG060): A Laboratory-Scale Feasibility Study. Front. Microbiol. 10, 1–14. doi:10.3389/fmicb.2019.02605
Tibocha-Bonilla, J. D., Zuñiga, C., Godoy-Silva, R. D., and Zengler, K. (2018). Advances in Metabolic Modeling of Oleaginous Microalgae. Biotechnol. Biofuels 11, 1–15. doi:10.1186/s13068-018-1244-3
Tran, N. T., and Kaldenhoff, R. (2020). Achievements and Challenges of Genetic Engineering of the Model green Alga Chlamydomonas Reinhardtii. Algal Res. 50, 101986. doi:10.1016/j.algal.2020.101986
Wang, B., Wang, J., Zhang, W., and Meldrum, D. R. (2012). Application of Synthetic Biology in Cyanobacteria and Algae. Front. Microbio. 3, 1–15. doi:10.3389/fmicb.2012.00344
Wang, L., Jia, X., Zhang, Y., Xu, L., Menand, B., Zhao, H., et al. (2021). Loss of Two Families of SPX Domain-Containing Proteins Required for Vacuolar Polyphosphate Accumulation Coincides with the Transition to Phosphate Storage in green Plants. Mol. Plant 14, 838–846. doi:10.1016/j.molp.2021.01.015
Wang, T., and Wu, M. (2006). An ATP-Binding Cassette Transporter Related to Yeast Vacuolar ScYCF1 Is Important for Cd Sequestration in Chlamydomonas Reinhardtii. Plant Cell Environ 29, 1901–1912. doi:10.1111/j.1365-3040.2006.01566.x
Wang, Y., Wang, S., Xu, P., Liu, C., Liu, M., Wang, Y., et al. (2015). Review of Arsenic Speciation, Toxicity and Metabolism in Microalgae. Rev. Environ. Sci. Biotechnol. 14, 427–451. doi:10.1007/s11157-015-9371-9
Wei, Y. Y., Zheng, Q., Liu, Z. P., and Yang, Z. M. (2011). Regulation of Tolerance of Chlamydomonas Reinhardtii to Heavy Metal Toxicity by Heme Oxygenase-1 and Carbon Monoxide. Plant Cell Physiol. 52, 1665–1675. doi:10.1093/pcp/pcr102
Wichmann, J., Baier, T., Wentnagel, E., Lauersen, K. J., and Kruse, O. (2018). Tailored Carbon Partitioning for Phototrophic Production of (E)-α-bisabolene from the green Microalga Chlamydomonas Reinhardtii. Metab. Eng. 45, 211–222. doi:10.1016/j.ymben.2017.12.010
Wichmann, J., Lauersen, K. J., Biondi, N., Christensen, M., Guerra, T., Hellgardt, K., et al. (2021). Engineering Biocatalytic Solar Fuel Production: The PHOTOFUEL Consortium. Trends Biotechnol. 39, 323–327. doi:10.1016/j.tibtech.2021.01.003
Windhagauer, M., Abbriano, R. M., Ashworth, J., Barolo, L., Jaramillo-Madrid, A. C., Pernice, M., et al. (2021). Characterisation of Novel Regulatory Sequences Compatible with Modular Assembly in the Diatom Phaeodactylum Tricornutum. Algal Res. 53, 102159. doi:10.1016/j.algal.2020.102159
Wollmann, F., Dietze, S., Ackermann, J. U., Bley, T., Walther, T., Steingroewer, J., et al. (2019). Microalgae Wastewater Treatment: Biological and Technological Approaches. Eng. Life Sci. 19, 860–871. doi:10.1002/elsc.201900071
Xiao, R., and Zheng, Y. (2016). Overview of Microalgal Extracellular Polymeric Substances (EPS) and Their Applications. Biotechnol. Adv. 34, 1225–1244. doi:10.1016/j.biotechadv.2016.08.004
Yadav, A. P. S., Dwivedi, V., Kumar, S., Kushwaha, A., Goswami, L., and Reddy, B. S. (2020). Cyanobacterial Extracellular Polymeric Substances for Heavy Metal Removal: A Mini Review. J. Compos. Sci. 5, 1. doi:10.3390/jcs5010001
Yang, T., Liu, J.-W., Gu, C., Chen, M.-L., and Wang, J.-H. (2013). Expression of Arsenic Regulatory Protein in escherichia Coli for Selective Accumulation of Methylated Arsenic Species. ACS Appl. Mater. Inter. 5, 2767–2772. doi:10.1021/am400578y
Yunus, I. S., Wichmann, J., Wördenweber, R., Lauersen, K. J., Kruse, O., and Jones, P. R. (2018). Synthetic Metabolic Pathways for Photobiological Conversion of CO2 into Hydrocarbon Fuel. Metab. Eng. 49, 201–211. doi:10.1016/j.ymben.2018.08.008
Zagorski, N., and Wilson, D. B. (2004). Characterization and Comparison of Metal Accumulation in Two Escherichia coli Strains Expressing Either CopA or MntA, Heavy Metal-Transporting Bacterial P-type Adenosine Triphosphatases. Abab 117, 33–48. doi:10.1385/abab:117:1:33
Ziller, A., Yadav, R. K., Capdevila, M., Reddy, M. S., Vallon, L., Marmeisse, R., et al. (2017). Metagenomics Analysis Reveals a New Metallothionein Family: Sequence and Metal-Binding Features of New Environmental Cysteine-Rich Proteins. J. Inorg. Biochem. 167, 1–11. doi:10.1016/j.jinorgbio.2016.11.017
Zitka, O., Krystofova, O., Sobrova, P., Adam, V., Zehnalek, J., Beklova, M., et al. (2011). Phytochelatin Synthase Activity as a Marker of Metal Pollution. J. Hazard. Mater. 192, 794–800. doi:10.1016/j.jhazmat.2011.05.088
Keywords: synthetic biology, heavy metal, microalgae, bio-removal, wastewater treament
Citation: Sattayawat P, Yunus IS, Noirungsee N, Mukjang N, Pathom-Aree W, Pekkoh J and Pumas C (2021) Synthetic Biology-Based Approaches for Microalgal Bio-Removal of Heavy Metals From Wastewater Effluents. Front. Environ. Sci. 9:778260. doi: 10.3389/fenvs.2021.778260
Received: 16 September 2021; Accepted: 08 November 2021;
Published: 24 November 2021.
Edited by:
Yongjun Wei, Zhengzhou University, ChinaReviewed by:
Adel Al-Gheethi, Universiti Tun Hussein Onn Malaysia, MalaysiaDillirani Nagarajan, National Cheng Kung University, Taiwan
Copyright © 2021 Sattayawat, Yunus, Noirungsee, Mukjang, Pathom-Aree, Pekkoh and Pumas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pachara Sattayawat, cGFjaGFyYS5zYXR0YXlhd2F0QGNtdS5hYy50aA==
 Pachara Sattayawat
Pachara Sattayawat Ian S. Yunus
Ian S. Yunus Nuttapol Noirungsee
Nuttapol Noirungsee Nilita Mukjang6
Nilita Mukjang6 Wasu Pathom-Aree
Wasu Pathom-Aree Chayakorn Pumas
Chayakorn Pumas