
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Sci. , 10 May 2021
Sec. Toxicology, Pollution and the Environment
Volume 9 - 2021 | https://doi.org/10.3389/fenvs.2021.554490
This article is part of the Research Topic The Environmental Hazards of Toxic Metals Pollution View all 10 articles
Textile wastewater (TWW) contains toxic metals that are inimical to microbiome, aesthetic quality, and the health of the receiving freshwater. TWW-impacted freshwater (L2) was assessed for metals eco-toxicity and the consequent impact on microbiome taxonomic profile (MTP) compared to a pristine environment (L1). The conductivity (1750 μS/cm), chemical oxygen demand (2,110 mg/L), biochemical oxygen demand (850 mg/L), and salinity (5,250 mg/L) of L2 were far above the permissible limits. Mercury posed very high ecological risks in the water column of L2 as lead, arsenic, and copper exerted high risk in the sediment. The MTP of L2 revealed the dominance of Euryarchaeota (48.6%) and Bathyarchaeota (45.9%) among the Archaea. The relative abundances of Proteobacteria and Bacteroidetes increased from 38.3 to 2.0%, respectively, in the L1 ecosystem to 42.1 and 12.9%, correspondingly, in L2. Unclassified Eukarya_uc_p (50.4%) and Fungi_uc (16.0%) were key players among the fungi kingdom in L2. The impact of the TWW on the microbiome was evident with the extinction of 6,249, 32,272, and 10,029 species of archaea, bacteria, and fungi, respectively. Whereas, 35,157, 32,394, and 7,291 species of archaea, bacteria, and fungi, correspondingly, exclusively found in L2 were assumed to be invading resident communities that combined with dominant autochthonous strains in shaping the ecophysiology dynamics in TWW-impacted freshwater. While the sensitive microorganisms in L2 are suggested bio-indicators of TWW ecotoxicity, the emergent and dominant taxa are pivotal to natural attenuation processes in the contaminated ecosystem that could be adopted for biotechnological strategy in decommissioning the TWW-impacted freshwater.
The textile industry is one of the most important sectors of the global economy as it contributes mass employment and contributes the gross domestic products (GDP) of many developing countries. Textile operations use a large volume of water and produce a large amount of wastewater that is rich in organic and inorganic pollutants (Dung et al., 2013; Bilinska et al., 2016). Textile dyeing and finishing treatments meted on fabric generate approximately 17–20% of textile wastewater (TWW) (Holkar et al., 2016). Worldwide, dye wastewater remains a major source of severe pollution snag due to industrialization, the huge demand for textile products, and the proportional humongous volume of production wastewaters discharged to the environment (Ali et al., 2008). Among the pollutants in TWW, are the biodegradables that may sometimes be recalcitrant while others, mainly heavy metals (HMs) and metalloids, are non-biodegradable and exert a toxicological effect on the receiving ecosystem (Dung et al., 2013).
The biodegradables contained in TWW are residues of reactive dyes and chemicals that enrich the chemical oxygen demand (COD) and biochemical oxygen demand (BOD) of the hydrosphere, leading to eutrophication (Bilinska et al., 2016). However, dyes used during the fabric dyeing process introduce, in addition to thick color that increases the turbidity of the water body, a diverse range of chemicals and HMs/metalloids that are hazardous to biota in the environment (Holkar et al., 2016). The commonly reported HMs and metalloids found in TWW include arsenic (As), Mercury (Hg), hexavalent chromium (Cr6+), iron (Fe), zinc (Zn), copper (Cu), lead (Pb), cadmium (Cd), and the mixtures of these HMs toxicants. The toxicity and persistence of HMs and metalloids in the receiving environment are of great concern with the resultant effect of biomagnifications along trophic levels (Gao and Chen, 2012). Bioaccumulation of wastewaters’ HMs/metalloids in the environmental matrixes has been reported to occur through physical, chemical, and biological processes (Rezaei and Sayadi, 2015).
The ecotoxicity of HMs/metalloids on the microbiome of the receiving environments varies from receptor interaction to membrane toxicity, narcosis, the disturbance of cell homeostasis, and enzyme inhibition (Worms et al., 2006). The degree of HMs/metalloids toxicity at the molecular level depends on the types of HMs/metalloids, dosage/bioavailable concentration at autochthonous microbiome, and other physico-chemical factors prevalent in such ecosystems. Consequently, the observable effects on the receiving biome include lethality, growth reduction, functional impairment, reduced fertility and reproduction, mutagenicity, behavioral disturbances, and eventual adaptation (Worms et al., 2006). Hg, like other HMs/metalloids with no known metabolic function, for example, exerts toxicity to different microbial processes and enzyme activities in impacted environments (Mahbub et al., 2016). The microbial processes impaired by HMs/metalloids toxicity include, but are not limited to, the activities of dehydrogenase (Mahbub et al., 2016), urease (Yang et al., 2007), nitrification (Mahbub et al., 2016), arylsulphatase (Casucci et al., 2003), methane oxidation (Contin et al., 2012), phosphatase and respiration (Tazisong et al., 2012), among others.
Microorganisms are one of the most sensitive bio-indicators for monitoring the influence of HMs toxicants in the polluted hydrosphere. The indiscriminate discharge of effluents from textile industries with disregard for environmental pollution regulations and legislatures, mostly in developing countries, can lead to temporal and spatial shifts of microbial communities in aquatic environments (Zhang et al., 2016). Although microorganisms are key players in the bioremediation of any deteriorating environment (Oyetibo et al., 2017a, Oyetibo et al., 2017b, Egbe et al., 2020; Oyetibo et al., 2021), the ecological response to such continual discharge of TWW is primarily the extinction of sensitive microbial taxa. The microbiome taxonomic profile in the receiving environment is then skewed toward the evolution of: 1) microorganisms that sequester HMs/metalloids; 2) microbes that exhibit resistance to the HMs/metalloids and thus perform their metabolic activities in the presence of the toxicants; and 3) HMs/metalloid-sensitive strains that can tolerate such HMs/metalloids but only become metabolically active once the HMs/metalloid toxicants have been mitigated (Oyetibo et al., 2010; Egbe et al., 2020; Ogwugwa et al., 2020).
To date, studies on the ecological consequences of TWW on the microbiome of the receiving environment are scarce. Therefore, the eventual alteration of ecological balance due to the toxicants (HMs/metalloids) contained in wastewater discharged by a textile industry is worthy of being studied. The present work, therefore, aimed to assess the HMs pollution status and impact of the textile wastewater from the textile industry in Nigeria (Africa) on the receiving freshwater. Therefore, the determination of the ecological impact of TWW was based on profiling the microbiome of the impacted ecosystem in relation to the pristine environment, using a culture independent approach. Consequently, the geochemistry and microbiome taxonomic profile of the TWW-impacted and non-impacted freshwater were integrated to elucidate the ecotoxicity of the TWW toxicants in freshwater. The success of this study would therefore suggest microbial bio-indicators for TWW pollution in freshwater. The outcome of this work provides information on the pivotal microbial taxa required in the circular economy concept for sustainable bioremediation of TWW-impacted freshwater environments.
Ikorodu is one of the fastest-growing towns in the Lagos metropolis, owing to massive migration from other towns within Lagos State (Nigeria) and industrialization. A textile industry situated on the outskirts of Ikorodu has been operating for decades, discharging its effluents directly to a freshwater stream, forming a tributary that finally empties into Lagos lagoon. The sampling locations were a freshwater stream that daily receives wastewaters from the textile industry (L2: N 6.5719444°, 3.485833° E) and a pristine environment, Nigerian Conservation Foundation (NCF) with no known history of anthropogenic pollution (L1: N 6.43722°, 3.5359° E). Ten composite samples of water filtered through 0.45 µm syringe filters into sterile screw-capped bottles on-site were collected. Similarly, sediments (0–5 cm depth) via composite sampling technique were collected using a Ven Veen Grab into a sterile glass beaker and covered with sterile aluminum foil. Sampling in triplicates was between February to March 2017 at 3 weeks intervals. The composite samples were thoroughly mixed to represent each replicate of the triplicate analyses. All samples in triplicate were concealed in Ziploc bags and transported on ice to the laboratory and stored at −20°C for subsequent analyses.
The physicochemical analysis of wastewater and sediments was done as earlier reported (Oyetibo et al., 2019). The parameters including pH, appearance, and temperature were determined in situ, while others were determined ex situ. Eleven HMs/metalloids were assayed and quantified using Atomic Absorption Spectrophotometry (Perkin-Elmer Analyst 200; Pelkin-Elmer, Canada) after acidic digestion as explained previously (Oyetibo et al., 2010). The normalization, validation, operational conditions, and wavelengths of the analytical lines of the AAS used for the detection of HMs in samples were undertaken as earlier reported (Ogwugwa et al., 2020; Oyetibo et al., 2021). The geochemical indexes of the measured HMs/metalloids were determined to evaluate the degrees of contamination, accumulation, pollution, and eco-toxicity of the HMs/metalloids as previously described (Oyetibo et al., 2019).
where Cn is the concentration of the metal n and Bn is the natural local background concentration of metal n.
where Cn is the concentration of metal n and Bn is as indicated above. The factor K is the background matrix correction factor due to lithospheric effects, which is usually defined as 1.5 according to Muller (1969).
where CF is the contamination factor as described before.
where Tr is the toxic response factor for a given substance (see Supplementary Table SA1)
Genomic DNA extraction from 0.5 g (approx.) of sediment sample from each location was achieved with Fast DNA® Spin Kit for Soil (MP Biomedicals) using FastPrep® Cell Distruptor FP120 (Qbiogene, Heidelberg, Germany) at 6.5 speed for 30 s following the manufacturer’s instruction. The possible interference of humic substances in the DNA was removed based on the recommendation of Takada and Matsumoto (2005). DNA was purified and visualized in an ethidium bromide-stained 1% (w/v) agarose gel using UV trans-illumination, while quantification was via UV-Vis photo-spectrometry using the Epoch™ Spectrometer system (BioTek, Winooski, VT, United States).
The genomic DNA was amplified at the V3-V4 of the 16S rRNA using the primer set 341F and 805R; V4-V5 region of 16S rRNA gene using primer set A519F-Mi and A958R-Mi; and ITS2 region using primer set ITS3-Mi (forward) and ITS4-Mi (reverse) for bacteria, archaea, and eukarya, respectively. The first and second PCR recipes and conditions were presented in the supplementary materials (Supplementary Table SA2). The purified amplicons of 1st PCR were tagged with Illumina indices and adapters from a Nextera® XT Index Kit (Illumina, San Diego, CA, United States). Libraries were constructed at ChunLab Inc. Seoul, South Korea using the Illumina MiSeq platform, and the qualities of the constructed libraries were checked with Agilent 2,100 Bioanalyzer System (Agilent Technologies, Palo Alto, CA, United States) using a DNA 7500 chip, and thereafter quantified using Quanti-iT™ PicoGreen™ dsDNA Assay kit (Invitrogen) according to the manufacturer’s instructions. A short DNA fragment was removed using CleanPCR™ (CleanNA, Netherlands), and sequencing was performed using Illumina, MiSeq Reagent Kit v2 (500-cycles) of Illumina MiSeq platform at ChunLab Inc. Seoul, Korea.
Metagenome raw reads were processed beginning from the quality check and filtering of low quality (<Q25) reads using Trimmomatic 0.32 software (Bolger et al., 2014). The pair-end sequence of the same strand of PCR amplicon was merged based on overlapping sequence information using PANDAseq software (Masella et al., 2012). ChunLab’s pipeline in-house algorithms were used to remove 16S rRNA PCR primer sequences, and UNITE (https://unite.ut.ee) was used to analyze the ITS2 gene. Non-specific amplicons were identified and removed using the HMMER program-based search to exclude Singleton sequences (Eddy, 2011), while sequences denoizing were performed with DUDE-Seq software (Lee et al., 2017), sequences were de-replicated and non-redundant reads were extracted via UCLUST-clustering (Edgar, 2010). UCHIME (Edgar et al., 2011) was used for the detection and removal of chimera against BIOiPLUG’s chimera-free reference database, while the remaining non-chimeric sequences were clustered into operational taxonomic units (OTUs) using CD-HIT (Fu et al., 2012) and UCLUST (Edgar, 2010) as discussed by Lee et al. (2019). Query sequences that were matched with the reference sequences in the EzBioCloud database (https://www.ezbiocloud.net/) by ≥97% similarity were considered to be at the species level while <97% similarity cut-offs were used for the genus, family, order, class, and phylum. The sequencing metadata obtained and used in this study have been deposited in the NCBI’s sequence read archive (SRA) database under BioProject and SRA accession number PRJNA545318.
Statistical analyses including column/row statistics and regression analyses, unless otherwise stated, and bar charts in this study were performed using the prism 5 software program (GraphPad Software, San Diego, CA, United States). The estimated coverage of the constructed gene libraries was calculated as C = 1−(
The physico-chemistry of the water and sediment samples from the freshwater receiving textile wastewater (L2) revealed marked degrees of pollution, as shown in Table 1. In comparison with the pristine environment, the pH of the water from the impacted environment was alkaline (pH = 9.85) with characteristic high conductivity (1750 μS cm−1), COD (2,110 mg L−1), BOD (850 mg L−1), and salinity (5,250 mg L−1). The water of the impacted L2 contained a huge chloride level and appeared deep-bluish unlike that of the pristine environment (L1). It is noteworthy that the physico-chemical values of the water and sediment samples of the impacted L2 exceeded the recommended limits of the local and international regulatory bodies (US EPA, 1998; NESREA, 2010).
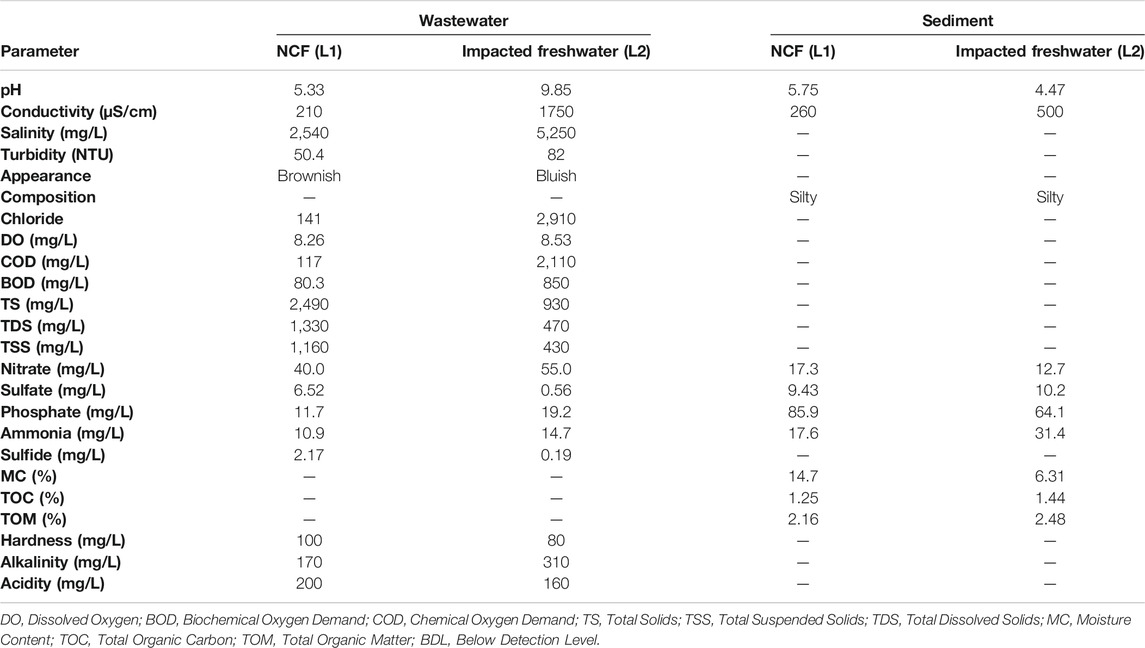
TABLE 1. Physicochemical status of the impacted freshwater receiving textile wastewater and a pristine environment.
The contamination indexes of HMs/metalloids in L2 when the pristine environment was used as the background threshold are shown in Table 2. Based on the calculated contamination factor (CF), the very high contamination of Hg was observed in the water while a similar degree of contamination in the sediment was recorded for As and Pb. The water was also considerably contaminated with Cd and As while moderate contamination of the water was associated with Pb, Ni, Co., and Sb. On the contrary, Hg and Cd considerably contaminated the sediment. The potential geological accumulation of the HM/metalloid toxicants revealed moderate to strong pollution of Fe, Cd, Zn, Mg, and As in the water unlike in the sediment where such levels of pollution were found with Hg. However, the water of L2 was extremely polluted with Hg while such pollution in sediment was associated with Pb, Cu, and As. A very high toxic response of Hg (Eri = 410) was observed with the impacted water, but a lower Hg toxic response was associated with the sediment. Nevertheless, the very high toxic responses of Cu (Eri = 70,000), Pb (Eri = 67,500) and As (Eri = 540) were linked to the sediment in the impacted environment. Overall, the receiving environment is greatly impacted with higher HMs/metalloids pollution occurring at the sediment as exhibited with a higher degree of contamination (Cd = 110, water; 28,000, sediment), pollution load index (PLI = 3.2, water; 9.2, sediment), and potential ecological risk index (RI = 610, water; 140,000, sediment).
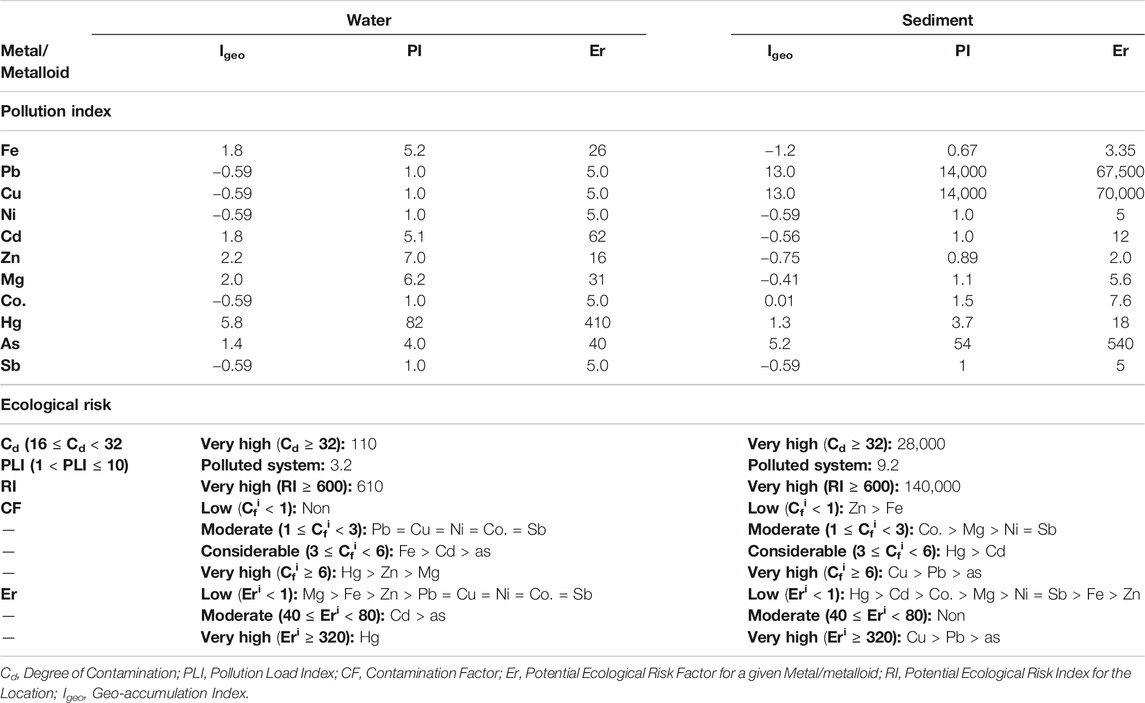
TABLE 2. Heavy metal and metalloid pollution indexes and ecological risks of the textile wastewater impacted freshwater.
The total valid reads of bacteria, archaea, and fungi after quality filtering, trimming and removing all chimeric reads are presented in Table 3. More sequence reads of the bacterial 16S rRNA genes were observed in the sediments of the pristine L1 environment (83,019) unlike fewer sequence reads of archaea 16S rRNA (113,810) and fungal ITS (67,121) genes than the impacted L2 sediment. After the removal of Singletons, more bacterial OTUs (9,950) were found in the impacted sediment than the pristine sediment (8,693). Figure 1 depicted the microbial composition and the relative abundance of the phyla taxa of the archaea (Figure 1A), bacteria (Figure 1B), and fungi (Figure 1C). A total of seven archaeal phyla were identified in all the sediments, where the impacted L2 ecosystem was mostly composed of Bathyarchaeota (45.9%), Euryarchaeota (48.6%), and not-yet-identified MBGB_p (4.3%) in comparison with the pristine sediments containing Bathyarchaeota (53.5%), Euryarchaeota (34.0%), and Thaumarchaeota (11.2%) as dominant phyla (Figure 1A). Proteobacteria (L1, 38.3; L2, 42.1%) and Chloroflex (L1, 16.9; L2, 14.4%) dominated the sediments from both ecosystems in addition to the visible dominance of Bacteroidetes (12.9%) in the textile impacted L2 ecosystem in contrast to the pristine L1 sediment that contained just 2.0% (Figure 1B). The relative abundance of Actinobacteria (L1, 5.57; L2, 4.66%) appeared relatively unaffected by the TWW unlike populations of Acidobacteria (11.2%) in the pristine L1 sediment that skewed down to 2.46% in the impacted L2 ecosystem (Figure 1B). The fungal community in the two ecosystems were dominantly Eukarya_uc_p, Fungi_uc, Ascomycota, Basidiomycota, and Chtridiomycota. The TWW appeared to have shifted the populations of not-yet-identified Eukarya_uc_p (15.4%), and Fungi_uc (9.6%) in L1 upward to 50.4 and 16.0%, respectively, in the TWW-impacted L2 (Figure 1C). Interestingly, Chitridiomycota (28.0%), which was the second-most dominant fungi in the pristine sediments, had become extinct with less than 0.3% relative abundance in the TWW-impacted L2.
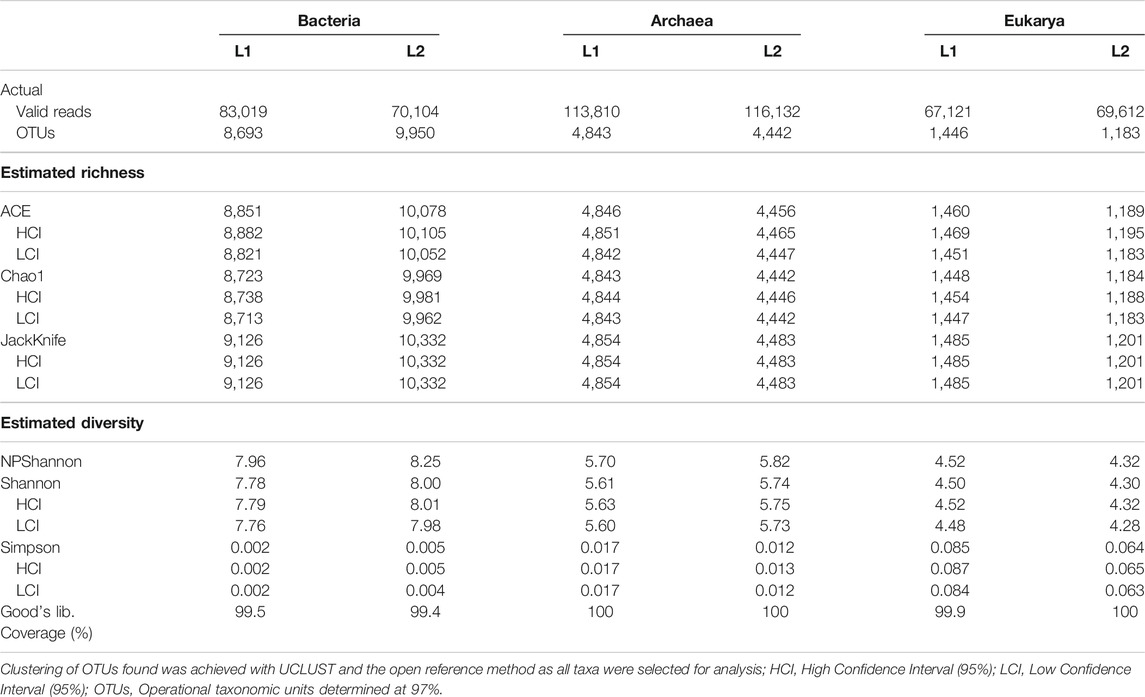
TABLE 3. Alpha diversity of microbiome evenness, richness, and varieties of species in the sediments.
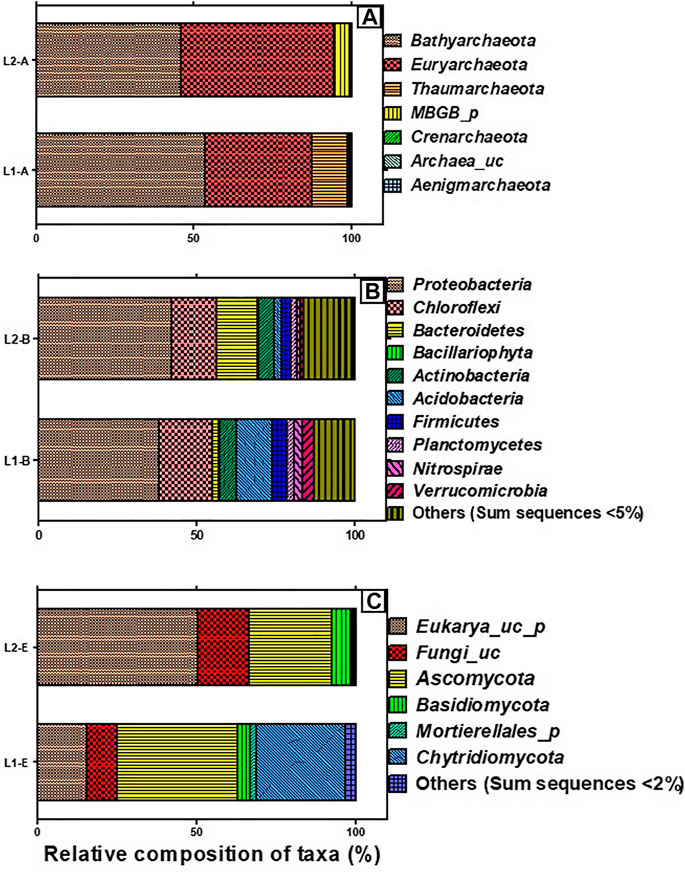
FIGURE 1. Taxonomic composition of microbiome present in the sediments of the pristine environment (L1) and textile wastewater impacted freshwater (L2), showing the archaeal phyla (A), bacterial phyla (B), and fungal phyla (C). Others represent the aggregation of phyla whose sum sequences are less than 5 and 2% in bacteria and fungi, respectively.
Analysis of the sequencing data of the microbial communities in the pristine and textile wastewater impacted ecosystems were revealed by a heat map based on the delineation of OTUs by the OrthoAni scale (Figure 2). The most abundant phyla of the three microbial domains in the ecosystems include Bathyarchaeota, Euryarchaeota, Proteobacteria, Eukarya_uc_p, Ascomycota, Chloroflexi, Chytridiomycota, and Fungi_uc, which is in tandem with the earlier discussed taxonomic compositions. Based on the Fast UniFrac metric and the OrthoAni values, the unweighted pair group method and arithmetic mean (UPGMA) dendrogram depicts the expected relatedness of the OTUs in the microbial domains with the highest OTUs richness among the Archaea and the least among the Fungi. Taxon Exclusive Or (XOR) analysis revealed that there was no archaeal phylum exclusively present in L1, except for two Class taxa (Thaumarchaeota_uc, and AF523942_c) that are not yet identified. Rather, 6,249 archaeal species that are mostly not yet identified were present in the sediments of L1 but found missing in those of the impacted L2. Whereas, 35,157 archaeal species were exclusively found in L2 (see Supplementary Table SA3 for the 20 most abundant taxa exclusively present in the ecosystems). Moreover, 12 bacterial phyla that were mostly yet to be identified, were only found in the pristine sediment contrary to the 30 bacterial phyla found in the impacted sediment but not found in L1. As such, 32,272 bacterial species were solely found in L1, while 32,394 bacterial species found in L2 were missing in L1. Nevertheless, the two fungal phyla (Crytomycota, n = 59; and Kickxellales_p, n = 1) present in L1 were missing in L2, while there was no fungal phylum uniquely found in L2 other than the 11 fungal Class taxa. Intrinsically, 10,029 fungal species were exclusively found in the pristine sediment while 7,291 fungal species were solely found in the TWW-impacted sediment.
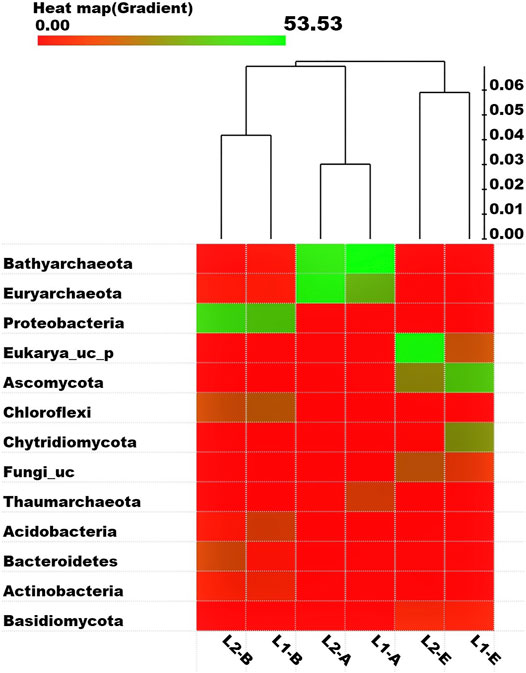
FIGURE 2. Heat map (gradient) representing the abundance of eight bacterial families, above the cut off (phylotypes that are over 5% from an individual sample). Each taxon and its proportion are represented by a square and relativity of samples based on operational taxonomic units (OTUs) delineated by the OrthoAni scale is shown as a dendrogram. The full green square equals 53.53%.
The plot of the correlation between the size of the sediment data and the number of OTUs delineated are depicted in asymptotic rarefaction curves (Figure 3). The highest numbers of OTUs were observed with bacteria while the least OTUs were associated with fungi, corresponding to species diversities in the ecosystems. The textile wastewater impacted sediment contained more bacterial OTUs than the pristine sediment. By contrast, less diverse OTUs of Archaea and Fungi domains were associated with the textile impacted sediment. The evenness, richness, and diversity estimations are summarized in Table 3.
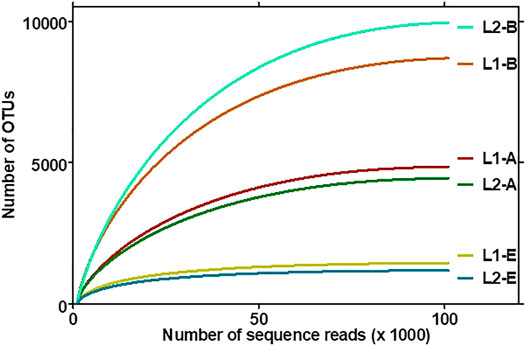
FIGURE 3. Rarefaction curve of the operational taxonomic units (OTUs) of the archaeal (L1-A, L2-A), bacterial (L1-B, L2-B), and fungal (L1-E, L2-E) sequence reads associated with the sediments. Clustering of OTUs in sediments based on taxonomy-dependent clustering and taxonomy-based clustering (TDC-TBC), which first identified sequences at species level using a similarity-based identification method that hits against the EzBioCloud database (https://www.ezbiocloud.net/), and the sequences that cannot be identified (sequences with <97% similarity) were then subjected to a TBC to be assigned OTUs. The confidence intervals of the curves were 95%.
More than 99% of the sequences in both environments represented the bacteria, archaea, and fungi present in the sediments, signifying the low probability of finding new species, should the number of sequence reads be increased based on Goods library coverage. The values of ACE, Chao 1, and JackKnife were proportional to microbiome richness, whereby the impacted sediment contained richer bacterial OTUs than the pristine sediment. Thus, the TWW-impacted sediment was more diverse, with respect to the sequence reads picked as OTUs by UCLUST and the open reference method. However, the pristine sediments contained higher varieties of Archaea and Fungi than the TWW-impacted sediment. NPShannon, Shannon, and Simpson indexes further revealed the diversity of microbiome in the sediments. While the values of NPShannon and Shannon were proportional to the degree of species diversity, the values of the Simpson index were inversely proportional to species diversity among sediments. It might therefore be adduced that the biodiversity of the microbiome in the textile wastewater impacted sediment and that of the pristine was not marginal. The impacted ecosystem appeared more diverse than the pristine ecosystem but in reality, as revealed by the actual OTUs pick by UCLUST, the pristine sediment was more diverse than the textile wastewater impacted sediment.
Industrialization is a quest of every nation to drive the economic growth of its citizens. The textile industry, unlike other industries, remains one of the major generators of effluent wastewater due to the large volume of water used for its different wet processing operations. The components of textile wastewaters are considered important pollutants that hamper the usability of receiving freshwater and thus damage the environment and public health (Holkar et al., 2016). The nature of pollution that accompanies the textile industry is such that the physico-chemistry of the receiving environment is compromised as evident with parameters of such affected ecosystem fall beyond the recommended limits. As observed in this study, the pH, conductivity, salinity, COD, BOD, and chloride among others of the TWW-impacted system (L2) were unacceptably at variance with the parameters of the pristine environment (L1) and consequently above the recommended limit. The physico-chemical parameters exceeding the recommended limits of regulatory bodies have variously been reported as an indicator of environmental pollution (Gupta et al., 2017; Obijiofor et al., 2018). For example, the Chloride levels in unpolluted waters are often below 10 mg L−1 unlike the 2,907 mg L−1 recorded for the TWW-impacted L2 that might have been contributed during the textile production step involving bleaching with hypochlorites and chlorites in the wet processing of fabrics. Similarly, the high pH recorded for the TWW-impacted L2 is connected to the alkali treatment of fabrics at a temperature of around 90°C during de-sizing, scouring, and mercerization.
The sources of the high values of COD and BOD in the TWW-impacted L2 must be from impurities in cotton and hemicellulose, and the residual dyes used in textile production. Consequently, the organic compounds in TWW as reflected in the high values of COD and BOD in the water column, and total organic matter of sediments in the impacted ecosystem against the unpolluted ecosystem must have contributed immensely to actual toxicity from the wastewater.
Previous studies have affirmed the relevance of the organic compounds of TWW to the ecotoxicity of the receiving environments (Khan and Malik, 2018). The high conductivity values of both water and the sediment of TWW-impacted L2 suggested that dissolved solids are mostly mineral salts (Ado et al., 2015). Although the microbial degradation of textile dyes is possible (Chen and Yien Ting, 2015; Ajaz et al., 2019), the eco-toxicological impact of textile dyes on autochthonous microorganisms has been reported (Bilinska et al., 2016). The high ecological risk of Hg in the water of the TWW-impacted L2 based on contamination factor (CF), revealed highly mobile Hg in the aqua system driving the bioavailability of toxic Hg in the ecosystem. Other toxic metals of note that variously contaminated the TWW-impacted L2, including Cd, As, Pb, and Ni, do not have any metabolic relevance to microbiota other than toxicity. Hg toxicity along with those of other HMs/metalloids to various ecosystems has been reported (Mahbub et al., 2017; Oyetibo et al., 2019; Ogwugwa et al., 2020; Oyetibo et al., 2021). Interestingly, the higher ecological risk of Hg in the water column (than the sediment) of impacted L2 is evidence that many mercuric ions (Hg2+) were bioavailable to the indigenous microbiota of the freshwater. However, the humongous sum of pollution indexes of the TWW-impacted L2 sediment is an indication of overtime accumulation of the HMs/metalloid toxicants from the continuous discharge of wastewaters from the textile industry. The consequences of these eco-toxicants have been variously reported (Ado et al., 2015; Obijiofor et al., 2018).
The high ecological risk indices observed in the TWW-impacted L2 were evident in the taxonomic composition and alpha diversities of the indigenous microbiome. The impact of TWW was more pronounced on the fungal taxa, where most OTUs that were dominant in the pristine ecosystem had gone extinct due to toxicities of HMs/metalloid toxicants present in the wastewater. Similar observations have been reported from other environments that were either treated with HMs/metalloid toxicants or had been naturally exposed to toxic HMs/metalloids (Catania et al., 2018; Lee et al., 2019). It should also be noted that archaea and bacteria can generally better adapt to extreme environments than eukarya, as represented by fungi in this study. Oyetibo et al., 2015 had earlier reported the low capabilities of fungi, except for a few strains such as Yarrowia sp Idd1 and Idd2, to resist/tolerate a toxic dosage of Hg in the environment. Similar eco-toxicity of HMs to the growth or survival of some of the archaea, leading to the observance of a significant reduction in the relative abundance and qualitative difference in the Archaean community (Sandaa et al., 1999; Guo et al., 2019). Moreover, salinity is a key factor in controlling the diversity and composition of the archaeal community in freshwater (Liu et al., 2016) as observed in the TWW-impacted L2 sediment where archaeal richness and evenness were reduced.
The dominance of Bathyarchaeota (53.5%), Euryarchaeota (34.0%), and Thaumarchaeota in the pristine L1 were in tandem with earlier reported archaeal composition in freshwater sediments (Yang et al., 2016; Guo et al., 2019). However, the skewness of archaeal composition in the TWW-impacted L2, as depicted with a lowered relative abundance of Bathyarchaeota (45.9%), increase in abundance of Euryarchaeota (48.6%), and relegation of Thaumarchaeota to <1% sequence reads was due to the influence of the HMs/metalloid toxicants present in the TWW discharged into the ecosystem. It can, therefore, be adduced that members of Thaumarchaeota were sensitive to HMs and chemicals contained in the TWW that were consistently released into L2. This observation agrees with the conclusions of Liu et al. (2018) and Guo et al. (2019) but negates the suggestion of He and colleagues (2018) that there may be HM-resistant Thaumarchaeota organisms in metal polluted freshwater. In agreement with the findings of Guo and colleagues (2019), Bathyarchaeota remained abundant in both pristine L1 and TWW-impacted L2, only that the relative abundance became reduced in the TWW-impacted L2 ecosystem, possibly due to the toxic HMs/metalloids in the TWW, and some members might exhibit resistance to HMs/metalloids. The total organic carbon was reportedly correlated with the abundance of Bathyarchaeota (Pan et al., 2019). Members of Bathyarchaeota might have been active in carbon cycling within the ecosystems despite the impact of HMs toxicants on their diversity and abundance in the TWW-impacted L2 ecosystem. Low abundances of Crenarchaeota (<1% sequence reads) in both pristine L1 and TWW-impacted L2 may be due to the low availability of nitrogen and CO2 in the two ecosystems since members of this archaeal phylum are greatly involved in ammonia oxidation to obtain energy that is used for carbon fixation (Lu et al., 2019). Furthermore, an increase in the relative abundance of Euryarchaeota in the TWW-impacted L2 indicated possible biogeochemical activities that drive the natural attenuation in the polluted hydrosphere as earlier reported by Yang and colleagues (2016).
Bacteria are usually numerically dominant in comparison with Archaea in aquatic environments. Bacterial abundance and diversity are regulated by the physico-chemistry of sediments, playing important roles in the transformation of organic matter and the biogeochemical cycling of primary elements including nitrogen, sulphur, metals, and phosphorus (Cheng et al., 2014). The influence of environmental factors such as salinity, organic matter, and various degrees of pollution on the ecology of the bacterial community has been extensively studied (Oyetibo et al., 2019). Currently, the impact of TWW on the diversity and community of bacteria as found in TWW-impacted L2 is not quite different from previous reports of Oyetibo et al. (2019). Of note, is the increased dominance of Proteobacteria and Bacteroidetes, and lesser relative abundance of Acidobacteria in the TWW-impacted L2, which is not unusual. Members of Proteobacteria are known to be key players in the amelioration of the TWW-impacted aquatic ecosystems. Their dominance in the present study suggested that they must have been actively involved in the functions and processes of the TWW-impacted L2. Furthermore, Bacteroidetes remain pivotal in converting complex molecules into simpler compounds in freshwater sediment. Thus, it is assumed that the increased composition of Bacteroidetes in TWW-impacted L2 could be linked to the increased mineralization of the chemicals contained in the TWW.
The fungal taxonomic profile of the TWW-impacted L2 defied the usual dominance of Ascomycota, Basidiomycota, Cryptomycota, and Chytridiomycota in a freshwater ecosystem (Lepere et al., 2019). On the contrary, the vast majority of ITS sequence reads taxonomy in the TWW-impacted L2 was uncharacterized Eukarya_uc_p (50.4%) and Fungi_uc (16.0%) indicating the plausible toxic consequences of the components of TWW consistently discharged into L2. The dominance of Ascomycota after the uncharacterized phyla in the two ecosystems has been reported in freshwater and is possibly due to the introduction of spores and strands of mycelia during rainwater, runoff, and wind events. As such, it is difficult to categorically conclude that the dominant fungal groups were in-dwellers but rather they might be periodic immigrants in the ecosystems (Grossart et al., 2019). The high sensitivity of Crytomycota to toxic HMs/metalloids may be connected to a lack of chitin in their cell wall, while the posterior whiplash flagella motility of Chytridiomycota due to TWW chemotaxis may explain the low abundance of the phyla in the TWW-impacted L2 (Grossart et al., 2016; Lepere et al., 2019). Moreover, the direct trophic antagonism between fungi and bacteria seemingly explains the low numbers of fungal OTUs in the two ecosystems with fewer OTUs occurring in TWW-impacted L2.
The effects of HMs/metalloid toxicants in textile wastewater on the microbiome of receiving freshwater ecosystems were toward the extinction of some species and emergence of some others. The sensitive microbial taxa in form of 6,028 OTUs present in pristine LI but absent in TWW-impacted L2 are considered efficient bio-indicators of TWW pollution. These efficient bio-indicators are cogent for the early and consistent environmental monitoring of TWW toxicity in the receiving ecosystem in attempts to safely guide the biota in such ecosystems. The gradual decrease and/or eventual loss of the sensitive taxa would indicate the toxic level of the TWW compounds, thereby signifying the need for mitigating TWW discharges. Nevertheless, the activities of the dominant autochthonous microorganisms present in TWW-impacted freshwater may provide solutions for the effective stimulations required for sustainable bioremediation processes. Hence, the 5,271 OTUs exclusively present in the TWW-impacted L2 are assumed to be functional in the impacted ecosystem and may be crucial to the transformational processes of the HMs/metalloid toxicants. As such, the exclusive dominant OTUs would be prospective biotechnological tools for the circular economy concept to protect environments from the ecotoxicity of TWW discharges and the decommissioning strategies applicable to TWW-impacted freshwater.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA545318.
GOd participated in sample collection, experimentation, data collation and manuscript preparation; GOy participated in experimental design, supervising experimentation, interpretation of data, and manuscript preparation; MI participated in experimental design, and supervising experimentation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2021.554490/full#supplementary-material
Ado, A., Tukur, A. I., Ladan, M., Gumel, S. M., Muhammad, A. A., Habibu, S., et al. (2015). A Review on Industrial Effluents as Major Sources of Water Pollution in Nigeria. Chem. J. 1, 159–164.
Ajaz, M., Rehman, A., Khan, Z., Nisar, M. A., and Hussain, S. (2019). Degradation of Azo Dyes by Alcaligenes Aquatilis 3c and its Potential Use in the Wastewater Treatment. AMB Expr. 9, 64–75. doi:10.1186/s13568-019-0788-3
Ali, N., Ikramullah, G., Lutfullah, G., Hameed, A., and Ahmed, S. (2008). Decolorization of Acid Red 151 by Aspergillus niger SA1 under Different Physicochemical Conditions. World J. Microbiol. Biotechnol. 24, 1099–1105. doi:10.1007/s11274-007-9581-6
Bilinska, L., Gmurek, M., and Ledakowicz, S. (2016). Comparison between Industrial and Simulated Textile Wastewater Treatment by AOPs – Biodegadability, Toxicity and Cost Assessment. Chem. Eng. J. 306, 550–559. doi:10.1016/j.cej.2016.07.100
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 30, 2114–2120. doi:10.1093/bioinformatics/btu170
Casucci, C., Okeke, B. C., and Frankenberger, W. T. (2003). Effects of Mercury on Microbial Biomass and Enzyme Activities in Soil. Bter 94, 179–192. doi:10.1385/bter:94:2:179
Catania, V., Cappello, S., Di Giorgi, V., Santisi, S., Di Maria, R., Mazzola, A., et al. (2018). Microbial Communities of Polluted Sub-surface Marine Sediments. Mar. Pollut. Bull. 131, 396–406. doi:10.1016/j.marpolbul.2018.04.015
Chen, S. H., and Yien Ting, A. S. (2015). Biodecolorization and Biodegradation Potential of Recalcitrant Triphenylmethane Dyes by Coriolopsis Sp. Isolated from Compost. J. Environ. Manage. 150, 274–280. doi:10.1016/j.jenvman.2014.09.014
Cheng, W., Zhang, J., Wang, Z., Wang, M., and Xie, S. (2014). Bacterial Communities in Sediments of a Drinking Water Reservoir. Ann. Microbiol. 64, 875–878. doi:10.1007/s13213-013-0712-z
Contin, M., Rizzardini, C. B., Catalano, L., and De Nobili, M. (2012). Contamination by Mercury Affects Methane Oxidation Capacity of Aerobic Arable Soils. Geoderma 189–190, 250–256. doi:10.1016/j.geoderma.2012.06.031
Dung, T. T. T., Cappuyns, V., Swennen, R., and Phung, N. K. (2013). From Geochemical Background Determination to Pollution Assessment of Heavy Metals in Sediments and Soils. Rev. Environ. Sci. Biotechnol. 12, 335–353. doi:10.1007/s11157-013-9315-1
Eddy, S. R. (2011). Accelerated Profile HMM Searches. Plos Comput. Biol. 7, c1002195. doi:10.1371/journal.pcbi.1002195
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 27, 2194–2200. doi:10.1093/bioinformatics/btr381
Edgar, R. C. (2010). Search and Clustering Orders of Magnitude Faster Than BLAST. Bioinformatics 26, 2460–2461. doi:10.1093/bioinformatics/btq461
Egbe, C. C., Oyetibo, G. O., and Ilori, M. O. (2020). Ecological Impact of Organochlorine Pesticides Consortium on Autochthonous Microbial Community in Agricultural Soil. Ecotoxicol Environ. Saf. 207, 111319. doi:10.1016/j.ecoenv.2020.111319
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: Accelerated for Clustering the Next-Generation Sequencing Data. Bioinformatics 28, 3150–3152. doi:10.1093/bioinformatics/bts565
Gao, X., and Chen, C.-T. A. (2012). Heavy Metal Pollution Status in Surface Sediments of the Coastal Bohai Bay. Water Res. 46, 1901–1911. doi:10.1016/j.watres.2012.01.007
Grossart, H-P., Van den Wyngaert, S., Kagami, M., Wurzbacher, C., Cunliffe, M., and Rojas-Jimenez, K. (2019). Fungi in Aquatic Ecosystems. Nat. Rev. Microbiol. 17(6). 339–354.doi:10.1038/s41579-019-0175-8
Grossart, H.-P., Wurzbacher, C., James, T. Y., and Kagami, M. (2016). Discovery of Dark Matter Fungi in Aquatic Ecosystems Demands a Reappraisal of the Phylogeny and Ecology of Zoosporic Fungi. Fungal Ecol. 19, 28–38. doi:10.1016/j.funeco.2015.06.004
Guo, Q., Li, N., Chen, S., Chen, Y., and Xie, S. (2019). Response of Freshwater Sediment Archaeal Community to Metal Spill. Chemosphere 217, 584–590. doi:10.1016/j.chemosphere.2018.11.054
Gupta, B. G., Biswas, J. K., and Agrawal, K. M. (2017). Physico-chemical Parameters, Water Quality Index and Statistical Analysis of Surface Water Contamination by Bleaching and Dyeing Effluents at Kalikapur, West Bengal, India. J. Environ. Sci. Pollut. Res. 3, 177–180.
Holkar, C. R., Jadhav, A. J., Pinjari, D. V., Mahamuni, N. M., and Pandit, A. B. (2016). A Critical Review on Textile Wastewater Treatments: Possible Approaches. J. Environ. Manage. 182, 351–366. doi:10.1016/j.jenvman.2016.07.090
Kemp, P. F., and Aller, J. Y. (2004). Estimating Prokaryotic Diversity: When Are 16S rDNA Libraries Large Enough? Limnol. Oceanogr. Methods 2, 114–125. doi:10.4319/lom.2004.2.114
Khan, S., and Malik, A. (2018). Toxicity Evaluation of Textile Effluents and Role of Native Soil Bacterium in Biodegradation of a Textile Dye. Environ. Sci. Pollut. Res. 25, 4446–4458. doi:10.1007/s11356-017-0783-7
Lee, B., Moon, T., and Yoon, S. (2017). DUDE-seq: Fast, Flexible, and Robust Denoising for Targeted Amplicon Sequencing. PLoS One 12, e0181463. doi:10.1371/journal.pone.0181463
Lee, S. M., Kim, N., Nam, R. H., Park, J. H., Choi, S. I., Park, Y-T., et al. (2019). Gut Microbiota and Butyrate Level Changes Associated with the Long-Term Administration of Proton Pump Inhibitors to Old Rats. Sci. Rep. 9, 6626. doi:10.1038/s41598-019-43112-x
Lepere, C., Domaizon, I., Humbert, J-F., Jardillier, L., Hugo, M., and Debroas, D. (2019). Diversity, Spacial Distribution and Activity of Fungi in Freshwater Ecosystems. PeerJ. 7, e6247. doi:10.7717/peerj.6247
Liu, J., Cao, W., Jiang, H., Cui, J., Shi, C., Qiao, X., et al. (2018). Impact of Heavy Metal Pollution on Ammonia Oxidizers in Soils in the Vicinity of a Tailings Dam, Baotou, China. Bull. Environ. Contam. Toxicol. 101, 110–116. doi:10.1007/s00128-018-2345-1
Liu, Y., Priscu, J. C., Xiong, J., Conrad, R., Vick-Majors, T., Chu, H., et al. (2016). Salinity Drives Archaeal Distribution Patterns in High Altitude Lake Sediments on the Tibetan Plateau. FEMS Microbiol. Ecol. 92. doi:10.1093/femsec/fiw033
Lu, S., Liu, X., Liu, C., Wang, X., and Cheng, G. (2019). Review of Ammonia-Oxidizing Bacteria and Archaea in Freshwater Ponds. Rev. Environ. Sci. Biotechnol. 18, 1–10. doi:10.1007/s11157-018-9486-x
Mahbub, K. R., Krishnan, K., Megharaj, M., and Naidu, R. (2016). Mercury Inhibits Soil Enzyme Activity in a Lower Concentration Than the Guideline Value. Bull. Environ. Contam. Toxicol. 96, 76–82. doi:10.1007/s00128-015-1664-8
Mahbub, K. R., Krishnan, K., Naidu, R., Andrews, S., and Megharaj, M. (2017). Mercury Toxicity to Terrestrial Biota. Ecol. Indicators 74, 451–462. doi:10.1016/j.ecolind.2016.12.004
Masella, A. P., Bartram, A. K., Truszkowski, J. M., Brown, D. G., and Neufeld, J. D. (2012). PANDAseq: Paired-End Assembler for Illumina Sequences. BMC Bioinformatics 13, 31. doi:10.1186/1471-2105-13-31
Muller, G. (1969). Index of Geoaccumulation in Sediments of the Rhine River. Geol. J. 2, 109–118. doi:10.1016/j.envint.2020.106032
NESREA (National Environmental Standards and Regulations Enforcement Agency) (2010). National Environmental (Surface and Groundwater Quality Control) Regulations. Abuja, Nigeria: Federal Ministry of Environment, FCT, 32.
Obijiofor, O. C., Okoye, P. A. C., and Ekejiuba, I. O. C. (2018). Assessment of Surface Water Contamination and Effect of Textile Effluents on Ibeshe River, Ikorodu, Lagos State, Nigeria. J. Chem. Soc. Nigeria 43, 69–79.
Ogwugwa, V. H., Oyetibo, G. O., and Amund, O. O. (2020). Taxonomic Profiling of Bacteria and Fungi in Freshwater Sewer Receiving Hospital Wastewater. Environ. Res. 192, 110319. doi:10.1016/j.envres.2020.110319
Oyetibo, G. O., Chien, M.-F., Ikeda-Ohtsubo, W., Suzuki, H., Obayori, O. S., Adebusoye, S. A., et al. (2017b). Biodegradation of Crude Oil and Phenanthrene by Heavy Metal Resistant Bacillus Subtilis Isolated from a Multi-Polluted Industrial Wastewater Creek. Int. Biodeterioration Biodegradation 120, 143–151. doi:10.1016/j.ibiod.2017.02.021
Oyetibo, G. O., Ige, O. O., Obinani, P. K., and Amund, O. O. (2021). Ecological Risk Potentials of Petroleum Hydrocarbons and Heavy Metals Shape the Bacterial Communities of Marine Hydrosphere at Atlantic Ocean, Atlas Cove, Nigeria. J. Environ. Manag. 289, 112563. doi:10.1016/j.jenvman.2021.112563
Oyetibo, G. O., Ilori, M. O., Adebusoye, S. A., Obayori, O. S., and Amund, O. O. (2010). Bacteria with Dual Resistance to Elevated Concentrations of Heavy Metals and Antibiotics in Nigerian Contaminated Systems. Environ. Monit. Assess. 168, 305–314. doi:10.1007/s10661-009-1114-3
Oyetibo, G. O., Ishola, S. T., Ikeda-Ohtsubo, W., Miyauchi, K., Ilori, M. O., and Endo, G. (2015). Mercury Bioremoval by Yarrowia Strains Isolated from Sediments of Mercury-Polluted Estuarine Water. Appl. Microbiol. Biotechnol. 99, 3651–3657. doi:10.1007/s00253-014-6279-1
Oyetibo, G. O., Miyauchi, K., Huang, Y., Chien, M.-F., Ilori, M. O., Amund, O. O., et al. (2017a). Biotechnological Remedies for the Estuarine Environment Polluted with Heavy Metals and Persistent Organic Pollutants. Int. Biodeterioration Biodegradation 119, 614–625. doi:10.1016/j.ibiod.2016.10.005
Oyetibo, G. O., Miyauchi, K., Huang, Y., Ikeda-Ohtsubo, W., Chien, M.-F., Ilori, M. O., et al. (2019). Comparative Geochemical Evaluation of Toxic Metals Pollution and Bacterial Communities of Industrial Effluent Tributary and a Receiving Estuary in Nigeria. Chemosphere 227, 638–646. doi:10.1016/j.chemosphere.2019.04.048
Pan, J., Chen, Y., Wang, Y., Zhou, Z., and Li, M. (2019). Vertical Distribution of Bathyarchaeotal Communities in Mangrove Wetlands Suggests Distinct Niche Preference of Bathyarchaeota Subgroup 6. Microb. Ecol 77 (2), 417–428. doi:10.1007/s00248-018-1309-7
Rezaei, A., and Sayadi, M. H. (2015). Long-term Evolution of the Composition of Surface Water from the River Gharasoo, Iran: a Case Study Using Multivariate Statistical Techniques. Environ. Geochem. Health 37, 251–261. doi:10.1007/s10653-014-9643-2
Sandaa, R.-A., Enger, Ø., and Torsvik, V. (1999). Abundance and Diversity of Archaea in Heavy-Metal-Contaminated Soils. Appl. Environ. Microbiol. 65, 3293–3297. doi:10.1128/aem.65.8.3293-3297.1999
Takada, Y., and Matsumoto, N. (2005). Skim Milk Drastically Improves the Efficacy of DNA Extraction from Andisol, a Volcanic Ash Soil. Jpn. Agric. Res. Quart. 39, 247–252. doi:10.6090/jarq.39.247
Tazisong, I. A., Senwo, Z. N., and Williams, M. I. (2012). Mercury Speciation and Effects on Soil Microbial Activities. J. Environ. Sci. Health A 47, 854–862. doi:10.1080/10934529.2012.665000
U.S EPA (United States Environmental Protection Agency) (1998). Guidelines for Ecological Risk Assessment, Risk Assessment Forum. Washington, DC: U.S Environmental Agency, 114.
Worms, I., Simon, D. F., Hassler, C. S., and Wilkinson, K. J. (2006). Bioavailability of Trace Metals to Aquatic Microorganisms: Importance of Chemical, Biological and Physical Processes on Biouptake. Biochimie 88, 1721–1731. doi:10.1016/j.biochi.2006.09.008
Yang, C.-l., Sun, T.-h., He, W.-x., Zhou, Q.-x., and Chen, S. (2007). Single and Joint Effects of Pesticides and Mercury on Soil Urease. J. Environ. Sci. 19, 210–216. doi:10.1016/s1001-0742(07)60034-5
Yang, Y. Y., Dai, Y., Wu, Z., Xie, S. G., and Liu, Y. (2016). Temporal and Spatial Dynamics of Archaeal Communities in Two Freshwater Lakes at Different Trophic Status. Front. Microbiol. 7, 451. doi:10.3389/fmicb.2016.00451
Keywords: ecotoxicology, textile wastewater, heavy metals, microbiome, freshwater, pollution
Citation: Odubanjo GO, Oyetibo GO and Ilori MO (2021) Ecological Risks of Heavy Metals and Microbiome Taxonomic Profile of a Freshwater Stream Receiving Wastewater of Textile Industry. Front. Environ. Sci. 9:554490. doi: 10.3389/fenvs.2021.554490
Received: 22 April 2020; Accepted: 12 April 2021;
Published: 10 May 2021.
Edited by:
Senthil Kumar Ponnusamy, SSN College of Engineering, IndiaReviewed by:
Jennifer Mesa-Marín, Sevilla University, SpainCopyright © 2021 Odubanjo, Oyetibo and Ilori. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ganiyu Oladunjoye Oyetibo, Z295ZXRpYm9AdW5pbGFnLmVkdS5uZw==
†Present address: Grace Olunike Odubanjo, Doctoral student at the Department of Microbiology, Faculty of Science, University of Lagos, Lagos, Nigeria
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.