- 1Department of Entomology, University of Maryland, College Park, MD, United States
- 2National Socio-Environmental Synthesis Center, University of Maryland, College Park, MD, United States
- 3Department of Biological Science, University of Alabama, Tuscaloosa, AL, United States
Seasonally saturated wetlands are hydrologically dynamic ecosystems that provide various ecosystem services; however, their variable hydrologic conditions may promote greenhouse gas emissions. The extent to which wetlands produce and emit greenhouse gases is intimately tied to the underlying microbial community. We established a linear transect spanning a hydrologic gradient on the margins adjacent to five seasonally saturated wetlands to characterize how water level, saturation duration, and frequency of wet-dry cycles influenced the soil and methane-cycling microbial community composition. We found that the soil and methane-cycling microbial community diversity and structure were strongly related to water level changes and saturation duration but not saturation frequency. Soils that experienced inundation or saturation for more than 50% of the year harbored a soil microbial composition distinct from soils that were rarely saturated or inundated. While soils closer to the wetland basin supported a higher relative abundance of methanogens, methane-oxidizing microbes were present across the entire topographic gradient; however, the composition shifted from a primarily anaerobic to aerobic methanotroph community. Findings suggest that soil and methane cycling microorganisms are influenced by the soil’s hydrologic conditions, with methane producers being restricted to specific ranges and methane oxidizers able to occur under a variety of hydrologic conditions. The mechanisms driving methane oxidation by the latter, however, may depend on the hydrologic conditions.
Introduction
Seasonally flooded or saturated wetlands (e.g., vernal pools, wet prairies, and Carolina and Delmarva bays) temporarily hold water from fall to late spring and dry up periodically when evapotranspiration exceeds surface and groundwater inputs (Calhoun et al., 2017). Such dynamic hydroperiods influence every aspect of these systems – from the ecology and breeding success of amphibian species (Chandler et al., 2017) to soil nutrient cycling (Marton et al., 2015; Hansen et al., 2018). These variable hydrologic conditions also stimulate the aerobic and anaerobic heterotrophic decomposition of organic C, resulting in the production and emission of two important greenhouse gases: carbon dioxide (CO2) and methane (CH4) (Bansal et al., 2016; Kifner et al., 2018; Bledsoe and Peralta, 2020). The extent to which wetlands produce and emit greenhouse gases is intimately tied to the underlying soil microbial population; however, insights into seasonal wetland microbial communities are poorly understood.
Net CH4 emissions are ultimately controlled by two groups of microorganisms: methanogens and methanotrophs. Methanogens live in oxygen-poor environments and decompose simple organic and inorganic C compounds (e.g., acetate, formate, and carbon dioxide) into the potent greenhouse gas, CH4 (Kotsyurbenko et al., 2019; Conrad, 2020). The production of CH4, or methanogenesis, is a conserved metabolic process with only seven archaeal families in the phylum Euryarchaeota possessing the genes to carry out this activity. In contrast, consumers of CH4, or methanotrophs, that play a pivotal role in reducing wetland CH4 emissions (Chowdhury and Dick, 2013; Segarra et al., 2015) are not restricted to a single phylogenic group; instead, methane oxidation can be performed by many different bacterial and archaeal species. Until recently, methane oxidation was believed to be restricted to oxygen-rich soil environments; however, new evidence suggests that anaerobic oxidation of methane (AOM) may consume a much larger fraction of CH4 produced in freshwater environments, including wetland soils, than initially expected (Smemo and Yavitt, 2011; Cui et al., 2015; Wang et al., 2018).
While the effects of hydrologic conditions and variability on the associated soil and CH4-cycling microbial community are not well characterized for seasonally saturated wetlands, several studies have investigated microbial populations in ephemeral stream banks (Zeglin et al., 2011; Lee et al., 2018) and hydrologically variable regions adjacent to lakes (Siljanen et al., 2011), rivers (Kemnitz et al., 2004), and wetlands (Balasooriya et al., 2007; Yu and Ehrenfeld, 2010; Roy Chowdhury et al., 2014). Collectively, these studies suggest distinct microbial populations form between “wet” (predominantly anoxic) and “dry” (predominantly oxic) soil conditions. Contrary to traditional belief, researchers have found that CH4-cycling microorganisms are able to colonize a wide range of soil hydrologic conditions and that patterns in their relative abundance, diversity, and activity have not always been as expected. For example, permanently saturated soils were found to support a more extensive but less diverse methanogen population, and wetter littoral wetland soils were found to harbor a higher abundance and more diverse methanotroph population (Siljanen et al., 2011; Christiansen et al., 2016). Additionally, soils that experienced frequent wet-dry cycles were found to harbor more diverse, distinct, and highly adaptable soil microbial communities than soils experiencing stable, less dynamic hydrologic conditions (Foulquier et al., 2013; Peralta et al., 2014). Because soil hydrological conditions along the margins of seasonally saturated wetlands vary throughout the year, we expected seasonally saturated wetland habitats will support a diverse soil- and CH4-cycling microbial population.
Given this backdrop, our goals were to describe the soil and CH4-cycling microbial population associated with seasonally saturated wetlands and determine the relationship between soil hydrologic conditions and the distribution and composition of these microbial populations. Previous research has shown that the duration of inundation or saturation and the frequency of wetting and drying events affect the overall biodiversity and the composition of the microbial community (Foulquier et al., 2013; Peralta et al., 2014). Here, we expected soils undergoing more frequent wet-dry cycles to have a higher microbial richness (total OTUs) and diversity (Shannon’s diversity index) than soils with static hydrologic conditions. Furthermore, we expected microbial diversity would be reduced in soils saturated or inundated for prolonged periods. In addition, we hypothesized that water table position, saturation duration, and frequency of wet-dry cycles would result in overall differences in the relative abundance of major bacterial and archaeal phyla, including the distribution of methanogens and methanotrophs across a hydrologic gradient. To test these hypotheses, we characterized the water level and the soil microbial community across a wetland to an upland topographic gradient in five seasonally saturated wetland sites located on the Delmarva Peninsula (Maryland, United States).
Materials and Methods
Site History and Description
Five seasonally saturated wetlands located on lands owned by The Nature Conservancy in the Upper Choptank watershed of the Delmarva Peninsula (Maryland, United States) were selected for this study (Table 1). Estimates suggest that nearly 17,000 wetlands are located on the Delmarva Peninsula and that ∼80% of these non-tidal freshwater mineral wetlands range in size from ∼0.5 to 6.0 ha (Fenstermacher et al., 2014). Historical ditching and agricultural activity once impacted many of the wetlands located in this region; however, forests have regrown in areas where agricultural activities ceased several decades ago. Wetlands degraded by agricultural activity and other land use changes have also been restored as part of an initiative to reestablish native plant diversity and improve the water quality of the Chesapeake Bay.
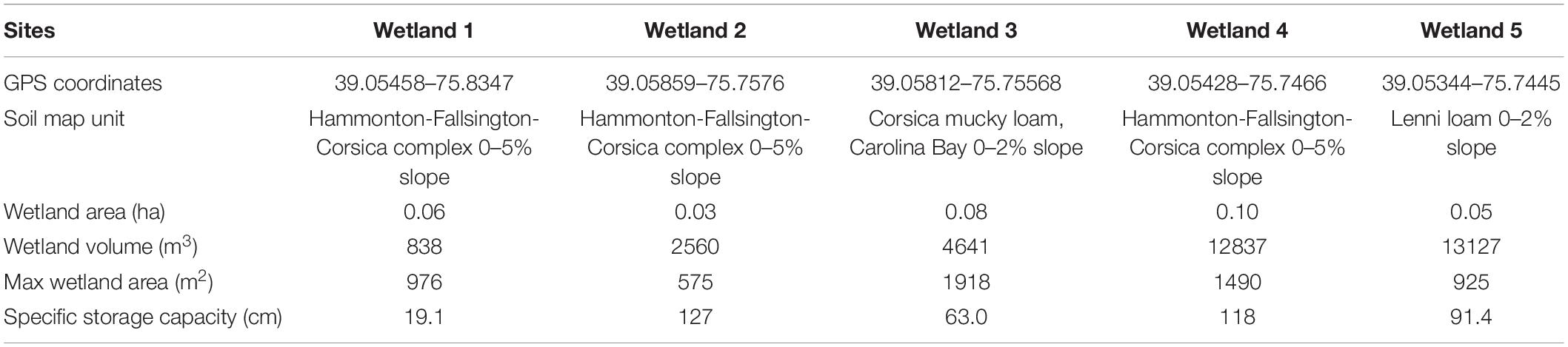
Table 1. Location and morphological description of the five sampled Delmarva wetland sites (Maryland, United States).
Photographs supplied by The Nature Conservancy suggest the lands surrounding our study sites (hereafter called “natural” wetlands) have not been disturbed for at least 70 years (data not shown). Based on USDA’s soil survey, wetlands in our study region have little topographic relief (<5% slope) and are composed of soils from the Hammonton-Fallsington-Corsica complex, Corsica mucky loam, or Lenni loam soil series. Previous studies have characterized wetland soil structure, carbon composition, hydrology, and plant communities in this region (Yepsen et al., 2014; McDonough et al., 2015; Epting et al., 2018; Jones et al., 2018; Lee et al., 2020). Whereas the exact nature of the hydroperiod varies among wetlands based on soil structure, vegetation, and topography, these wetlands generally experience a seasonal hydrologic cycle with increasing and higher water levels in fall and winter (October through March) and decreasing and lower water levels through spring and summer (April through September) (Phillips and Shedlock, 1993; Lee et al., 2020). Ongoing work includes extensive hydrologic monitoring within and among wetlands located on property owned by The Nature Conservancy. Thus, the wetlands we selected for this study offer an ideal opportunity to test our hypotheses.
Experimental Design
For each of the five selected wetland sites, we established a single transect along a topographic gradient from the wetland basin’s water edge to an upland point. The wetland basin edge was visually identified as the minimum extent of ponded water during August and September 2017. Within each transect, five evenly spaced sampling points (hereafter referred to as “hydrologic zones”) were identified (Figure 1). The first hydrologic zone (“A”) was positioned outside the wetland center near the basin water’s edge (hereafter, the “Basin Edge Zone”). The fifth hydrologic zone (“E”) was situated in the adjacent upland area (hereafter referred to as the “Upland Area”). The three remaining hydrologic zones (“B,” “C,” and “D”) were equally spaced across the topographic gradient. Hydrologic conditions were expected to vary along the topographic gradient, with waterlogged soil conditions decreasing with increasing distance from the wetland basin edge area (A) to the adjacent upland area (E).
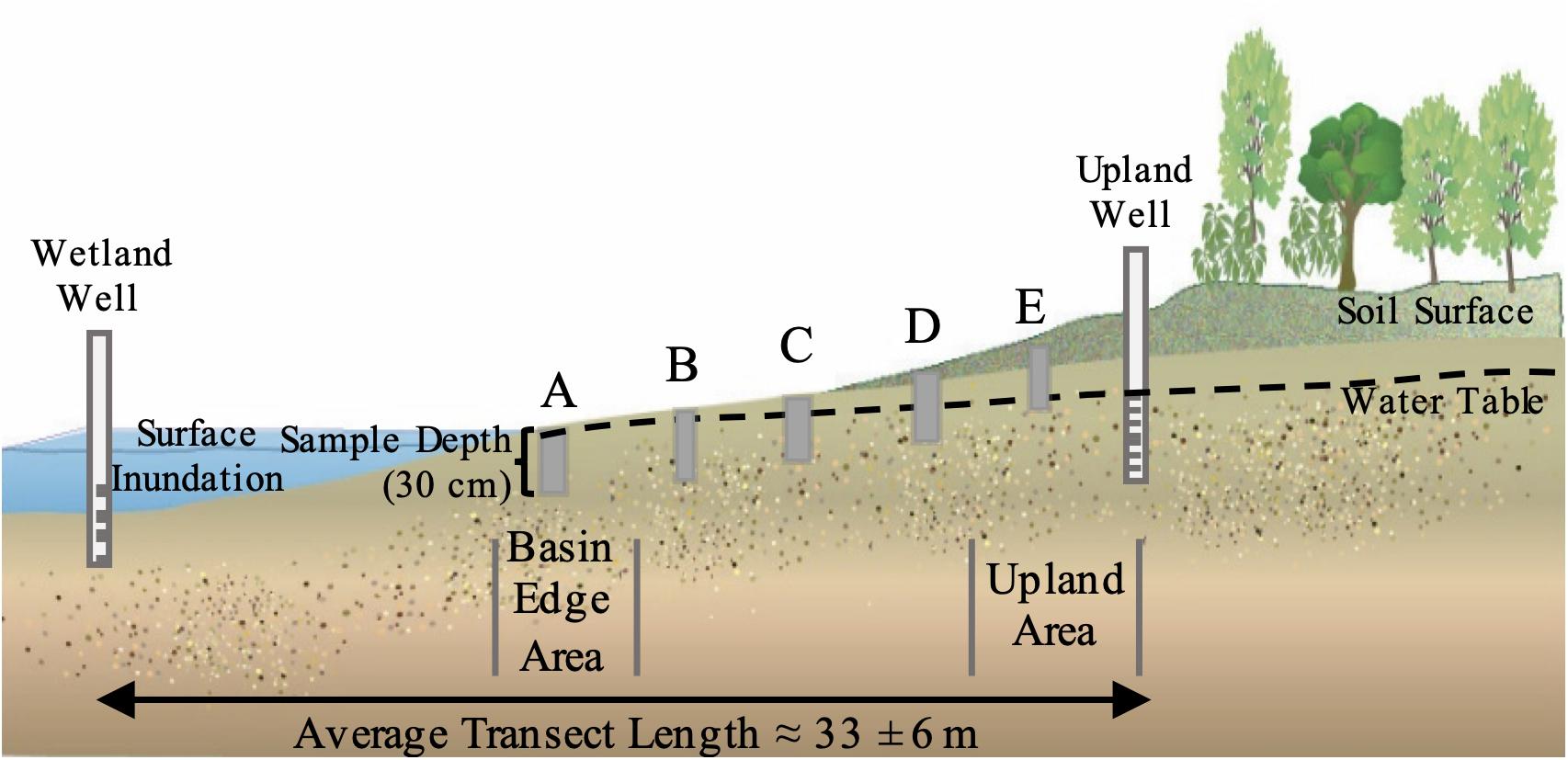
Figure 1. Approximate location and depth of groundwater well installation, as well as the approximate location of the five hydrologic zones (A–E) along the topographic gradient.
The study design and hydrologic data (described next) allowed us to evaluate average microbial diversity and composition as a function of average hydrologic conditions of each zone (i.e., a categorical analysis) and as a function of the hydrologic conditions at each specific sampling point along the gradient (i.e., using continuous hydrologic data).
Hydrological Monitoring and Characterization
We used a combination of water level, elevation, and a simple interpolation procedure to characterize the hydrologic variation from the wetland basin edge area (A) to the upland area (E). Surface water stilling wells and ∼2-m deep piezometers were installed near the center of the wetland basin (“Wetland Well”) and in the adjacent upland area (“Upland Well”), respectively. These bracketed each transect; however, due to differences in each wetland’s microtopography, well installation location and transect length varied by site (∼20 to 55 m). Surface water stilling wells and piezometers outfitted with a HOBO® U20 data loggers recorded water table position every 15-min for the 2018 water year (October 2017 – October 2018). Sampling points and well elevations were measured using a Topcon RL-H5A Laser Level (Topcon Positioning Systems, Livermore, CA, United States). Water table depth (cm) for each sampling point (i.e., Zones A–E) along each transect was determined by interpolating the water level between the Wetland and Upland wells. Notably, this approach considers the dynamic expansion and contraction of the ponded edge, and it assumes linearity between the wetland basin and the upland area. While seasonally flooded wetlands may experience temporary groundwater mounding (Phillips and Shedlock, 1993; Rosenberry and Winter, 1997), linearity is a reasonable assumption given the relatively small spatial scale and low-relief landscape. See the data availability statement for more details about these data.
Given the proximity of the study sites to each other, we assumed that all five wetland sites experienced the same local temperature and precipitation regimes. Notably, the 2018 water year was wetter than average. Based on the 72-year flow record from the Choptank River gauge located near our wetland sites (USGS Gage 01491000; nwis.usgs.gov), the 2018 water year was in the 77th percentile of total runoff.
Hydrologic variation was characterized for each sampling point along the topographic gradient. First, we summarized the 15-min water level readings into daily mean timestep intervals (n = 365 days). Daily water level values were then used to calculate the annual minimum, mean, and maximum water table depth (cm). Positive water level values signified the water table was present above the soil surface, and negative water level values indicate the water table fell below the soil surface. Daily water levels were then used to calculate the annual duration and frequency of saturation. While both of these metrics allow us to infer potential soil moisture conditions, neither of these metrics directly measure soil water content or redox conditions. The annual saturation duration (# of days) represents the total number of days the water table was predicted to be present in the soil horizon sampled (i.e., the top 30 cm), and the annual changes in the frequency of wet-dry cycles (hereafter referred to as “saturation frequency”) describes the total number of occasions the water table was present in the upper 30 cm of the soil horizon. Hydrological data for Wetland 1A was excluded from our analysis.
Soil Collection and Edaphic Analysis
In November 2017, we used a 12″ (30.5 cm) soil probe (Oakfield Apparatus, Wisconsin) to collect two soil cores just lateral to each established sampling zone (i.e., A–E) associated with each transect. Duplicate cores from each zone were placed into a sterile 24-oz sampling bag and gently homogenized into a single representative sample. At the time of the collection, high water levels at one wetland site obstructed sample collection from the basin edge area (i.e., Wetland 1A); therefore, a total of 24 samples were collected instead of 25 (5 wetlands × 1 transect per wetland × 5 hydrologic zones per transect × 1 homogenous soil core per hydrologic zone). All 24 samples were placed on ice and transferred back to the lab, where they were immediately subsampled and stored at 4°C or -40°C.
For each soil sample, we measured soil pH(CaCl2), gravimetric water content (Wm), and soil organic matter content (SOM, g OM g–1 soil). Soil pH was determined by mixing one-part field-moist soil with two parts 0.01 M calcium chloride (CaCl2). After a 10-min settling period, soil pH was recorded using a pH electrode probe (Fisher Scientific®, Waltham, MA, United States). To determine gravimetric water content, approximately ∼30 ± 10 g of wet soil was oven dried at 70°C for 72 h or until weights stabilized. Afterward, SOM was determined by combusting two oven-dried subsamples at 400°C for 16 h (Nelson and Sommers, 1996); duplicates values were averaged and used for statistical analysis.
Soil DNA Extraction and 16S rRNA Gene Sequencing
Soil microbial community composition was characterized by sequencing the V4 region of the 16S ribosomal RNA gene. Genomic DNA was extracted using the PowerSoil® DNA isolation kit (MoBio Laboratories, Inc.) and then quantified using a Qubit Fluorometer (Life Technologies). After extraction and quantification, we prepared 16S rRNA gene amplicons following Illumina’s 16S metagenomic sequencing library protocol, except we targeted the gene-specific sequences for the V4 region using the F515 and R806 primer set (Caporaso et al., 2011). PCR reactions (total volume = 35 μl) were set up using 3.5 μl of 5.0 ng μl–1 DNA, 7.0 μl of 1.0 μM forward (F515) and 7.0 μl of 1.0 μM reverse (R806) primers with adapters, and 17.5 μl of 2× Phusion Flash High-Fidelity PCR Master Mix (ThermoFisher Scientific). Briefly, 16S rRNA genes were amplified using the following thermocycling conditions: 95°C heat activation step for 3 min, followed by 25 cycles at 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s, and ended with a final extension step at 72°C for 5 min. After verifying the product band size on a 1.2% agarose gel, 25 μl of PCR product was purified using AMPure XP beads, indexed (Nextera XT Index v2 Set C), and then pooled into a single environmental sample. The purified environmental sample was quantified using a Bioanalyzer and the KAPA Library Quantification Kit (KAPA Biosystems). PhiX was added to a final concentration of 5%. Before sequencing, the pooled sample was diluted, denatured, and heat-shocked according to the manufacturer’s instructions. Pair-end reads were generated on a MiniSeq platform using a 300-cycle MiniSeq High Output Reagent Kit (FC-420-1003; Illumina, Penzberg, Germany).
Paired-end sequences were processed in R (v. 3.5.1) using the DADA2 bioinformatics (Callahan et al., 2016a) and the Phyloseq (McMurdie and Holmes, 2013) packages. The DADA2 workflow was followed to complete the initial filtering and taxonomic assignment (Callahan et al., 2016b). Forward and reverse sequences were not truncated before merging because the overlap between paired-end reads was only 7 bp. Merged sequences with a total length of 250 to 256 bp were clustered into amplicon sequence variants (ASV), more commonly referred to as operational taxonomic units (OTUs), using Silva version 132 (Quast et al., 2012). Following classification, singleton and doubleton OTUs, low abundance and prevalent phyla (<5 total abundance and <1% median prevalence across samples), and unclassified sequences at the phyla level were removed from the dataset. We rarified the number of sequences to create even read depth among samples (Supplementary Figure 1). Finally, taxa with a mean absolute abundance value less than or equal to 0.1% across all samples were removed before relativizing abundance to 100%.
Alpha diversity was measured by the number of observed OTUs (“Richness”) and Shannon’s diversity index, which combines estimates of microbial richness and evenness. We also assessed beta-diversity, or the variation in soil microbial community composition among the five hydrological zones, by generating a Bray-Curtis dissimilarity community matrix. We examined potential differences among the a priori groups using permutation multivariate analysis of variance (PERMANOVA). A non-metric multidimensional scaling (NMDS) plot was used to visualize patterns in the microbial communities, and environmental vector fitting was used to determine which, if any, edaphic and hydrologic properties significantly correlated with the ordination space defined by the composition of the microbial community. Individual microbial taxa identified as methanogen or methanotroph were summed into corresponding functional groups and used for statistical analysis.
Statistical Analysis
We used JMP Statistical Software (v. 14.1.0) to summarize data, evaluate assumptions of normality (i.e., distribution and variance structure), and perform the univariate statistical analysis. A one-way analysis of variance (ANOVA) was used to assess mean differences in edaphic properties (i.e., soil pH and SOM), hydrologic variability [i.e., annual mean water level depth (cm) and saturation duration (# of days)], and alpha diversity metrics (i.e., Richness and Shannon’s diversity index) across the five hydrologic zones. The relative abundance of the CH4-cycling microbial communities as well as the saturation frequency (# of events) were analyzed using the non-parametric Kruskal-Wallis Rank Sums test. Datasets with non-normal distributions were log10 or log10 (1+ relative abundance) transformed before statistical analysis. For statistically significant univariate models (alpha <0.05), differences among the mean were examined using Tukey’s Honest Significance Difference (HSD) or Wilcoxon Rank Sum test. In addition to our categorical analysis, we calculated the Pearson correlation coefficient (r) at a 95% confidence interval to assess the strength of associations among the measured edaphic, hydrologic, and microbial community parameters. The R (v. 4.0.1) vegan package (Oksanen et al., 2019) in RStudio was used to examine beta-diversity and visualize variation among the soil microbial communities.
Results
Hydrologic Variation Along the Transect
The water level for the 2018 water year (October 2017 – 2018) varied seasonally along the topographic gradient (Figure 2 and Supplementary Table 1). Overall, we observed a decrease in the annual mean water level and an increase in saturation frequency with increasing distance from the wetland’s basin edge. The water level measured in the wetlands’ basin edge areas (A) was at or above the soil surface for more than 300 days, suggesting that these soils were likely saturated for more than 80% of the year. In comparison, the soil horizons we sampled in the upland area (E) were rarely inundated or saturated, as the water level was seldom present in the top 30 cm of the soil column (<5% of the 2018 water year). The length of time the water table was present in the upper soil horizons of zones B through D varied from >30 to <80%. The water level in November was lower than the annual mean and less frequently saturated in the upper soil horizons (Supplementary Table 1).
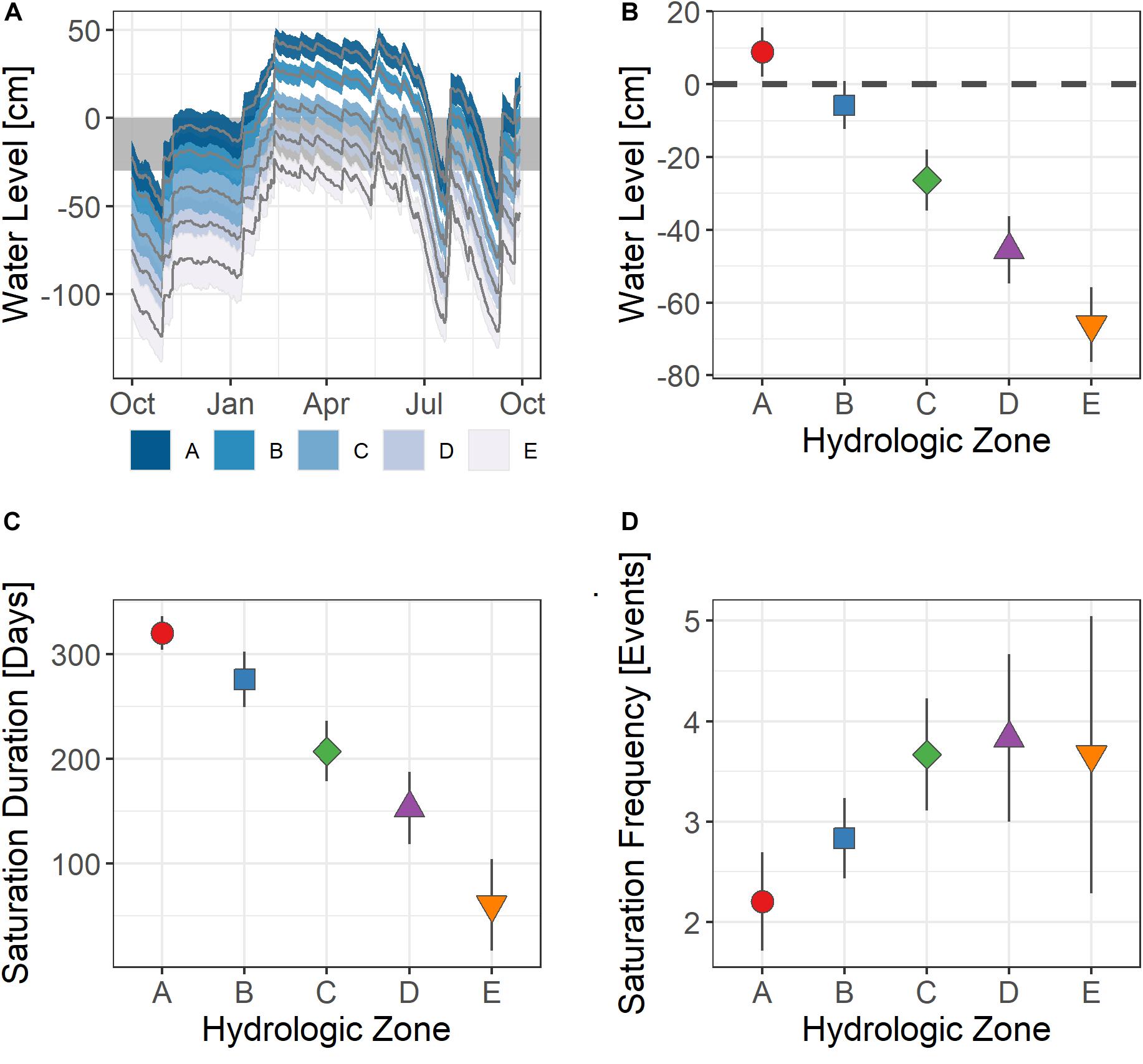
Figure 2. (A) Wetland hydrograph showing the mean and standard error of the mean (SEM) of wetland water level experienced across the 2018 water year. (B) Mean water level, (C) inundation duration, and (D) saturation frequency experienced at each transect point across the study wetlands. Here, water level is relative to the ground surface, where positive values indicate surface inundation and negative values indicate water level is below ground surface.
Edaphic Properties
Acidic soils (pH < 4) were present across all five hydrologic zones (Table 2). Mean soil pH and organic matter content did not statistically differ by hydrologic zone even though the SOM content was nearly twice as high in the wetland basin’s edge area (hydrologic zone A) than in the adjacent upland area (hydrologic zone E). SOM content was significantly correlated with annual mean water level (r = +0.58) and saturation duration (r = +0.63) (Table 3).
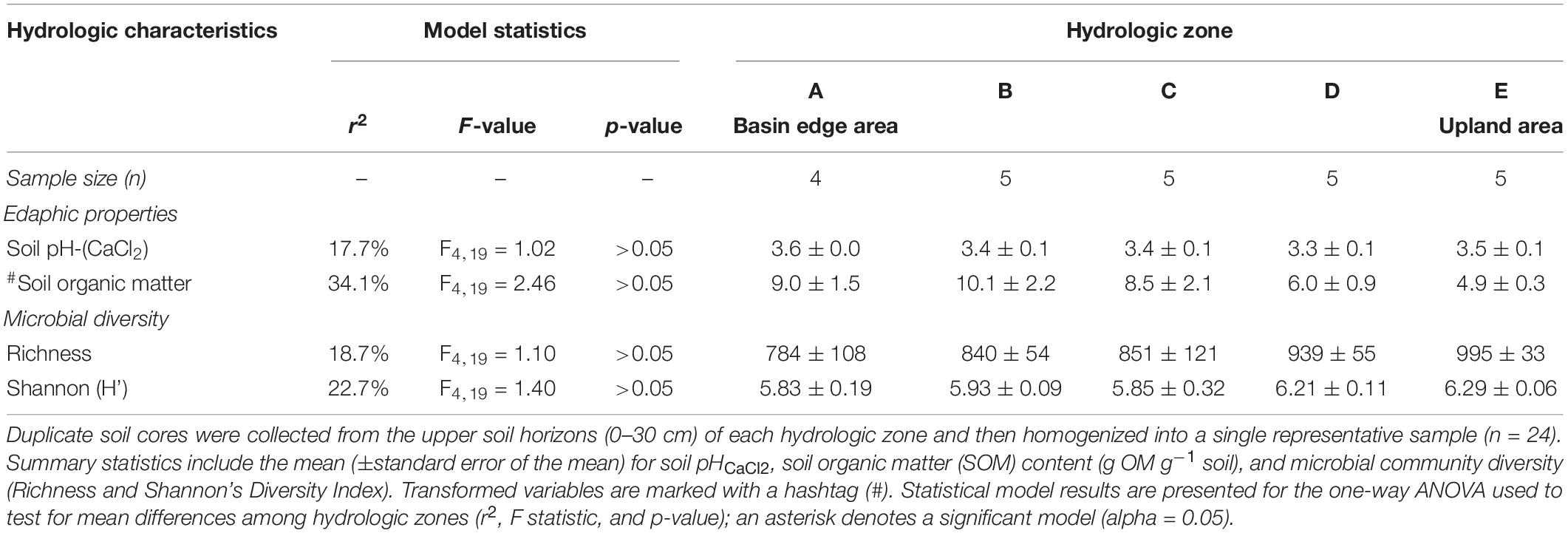
Table 2. Edaphic and microbial community diversity properties for each hydrologic zone (A–E) established along the topographic gradient.
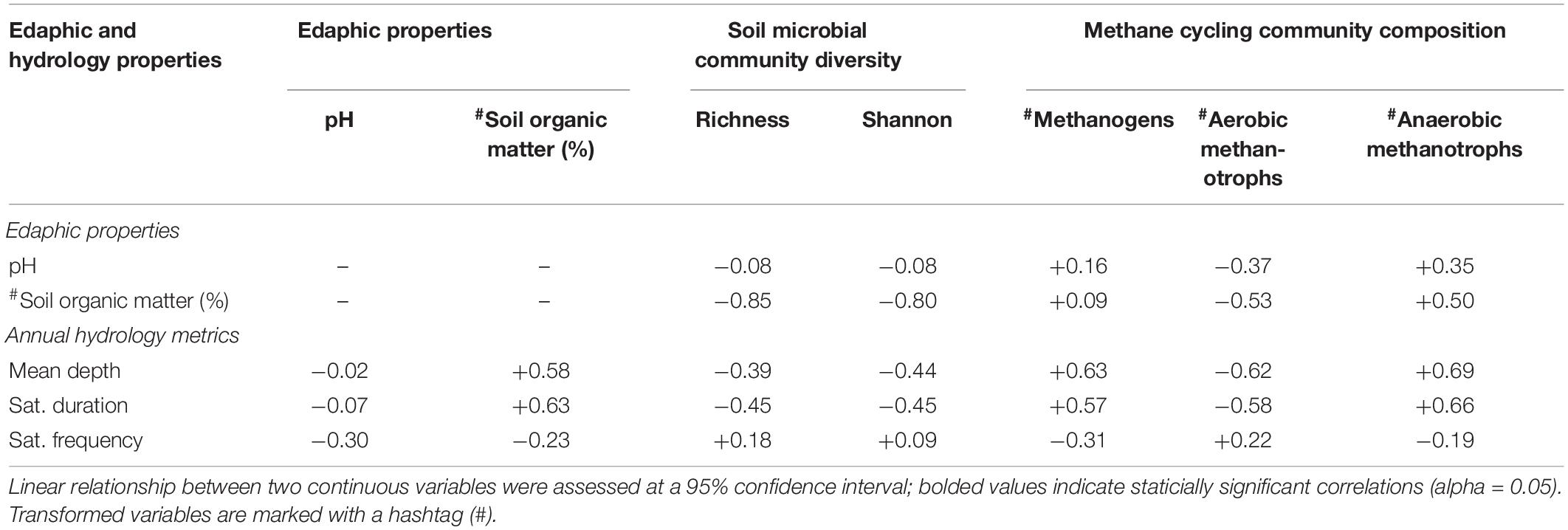
Table 3. Pearson correlation coefficient (r) among annual hydrology metrics, soil microbial community diversity measures, and relative abundance (%) of methane cycling microbes.
Soil Microbial Community Composition and Diversity
Paired-end sequencing of the 16S rRNA V4 gene region generated a total of 1,311,757 raw reads across 24 samples. Rarefaction curves for all 24 samples reached asymptote, suggesting that we sufficiently captured soil microbial richness across all locations (Supplementary Figure 1). The resulting quality-filtered sequences were clustered into 7,437 taxonomic groups, of which approximately 94.8% of these reads were clustered into 24 bacteria phyla and 5.2% clustered with five archaeal phyla (Supplementary Table 2). More than half of the taxa were classified into four bacterial phyla: Acidobacteria (17.5%), Chloroflexi (17.5%), Proteobacteria (16.4%), and Planctomycetes (13.4%). Most of the archaeal sequences were associated with Thaumarchaeota (2.9%) and Euryarchaeota (1.0%).
Alpha diversity (i.e., Richness and Shannon’s diversity index) mean differences were not statistically significant by hydrologic zone (Table 2) or site (Supplementary Table 3); however, lower microbial richness and Shannon diversity values were observed in sampling areas where soils were saturated or inundated for prolonged periods and possessed high concentrations of SOM (Table 3). Unlike alpha diversity, we found that the soil microbial community composition varied by hydrologic zone (PERMANOVA: F4,19 = 2.14, p < 0.05) but not by site (Supplementary Figure 2). These results suggest that soils experiencing inundation or saturation for more than 50% of the year (hydrologic zones A–C) shared a more similar microbial community composition than soils that were rarely saturated or inundated (hydrologic zones D and E) (Figure 3). We observed a significant correlation (p < 0.05) between soil microbial community composition and SOM content (r = +0.55), mean water depth (+0.67), and saturation duration (+0.70); saturation frequency was not significantly correlated with microbial community composition (+0.03) (Figure 3).
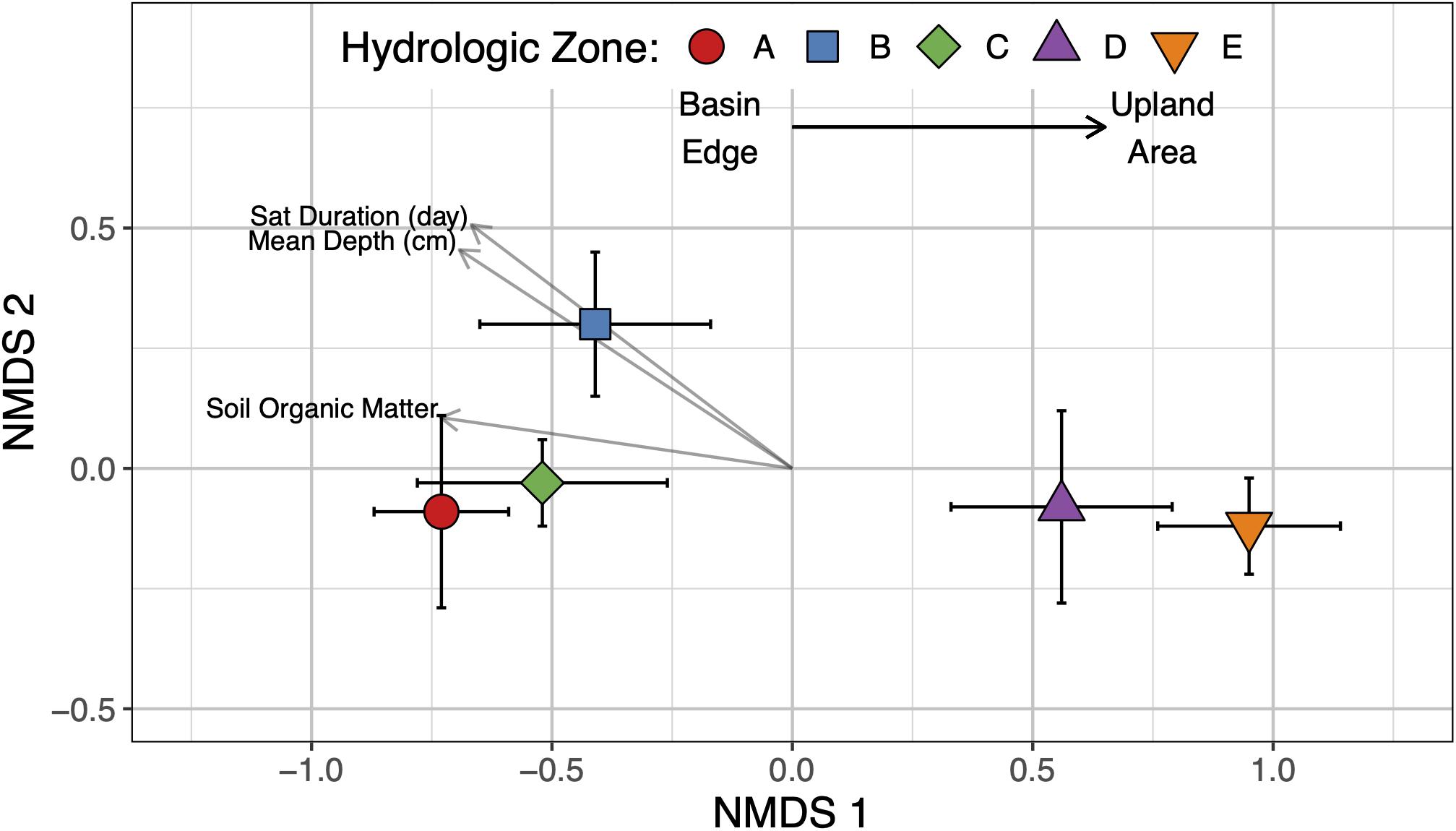
Figure 3. Non-metric multidimensional scaling plot showing separation of the soil microbiome based on Bray-Curtis dissimilarities. Environmental variables significantly correlating with the ordination space are included as vectors in the ordination plot (alpha = 0.05).
Methane Cycling Microbial Community Composition
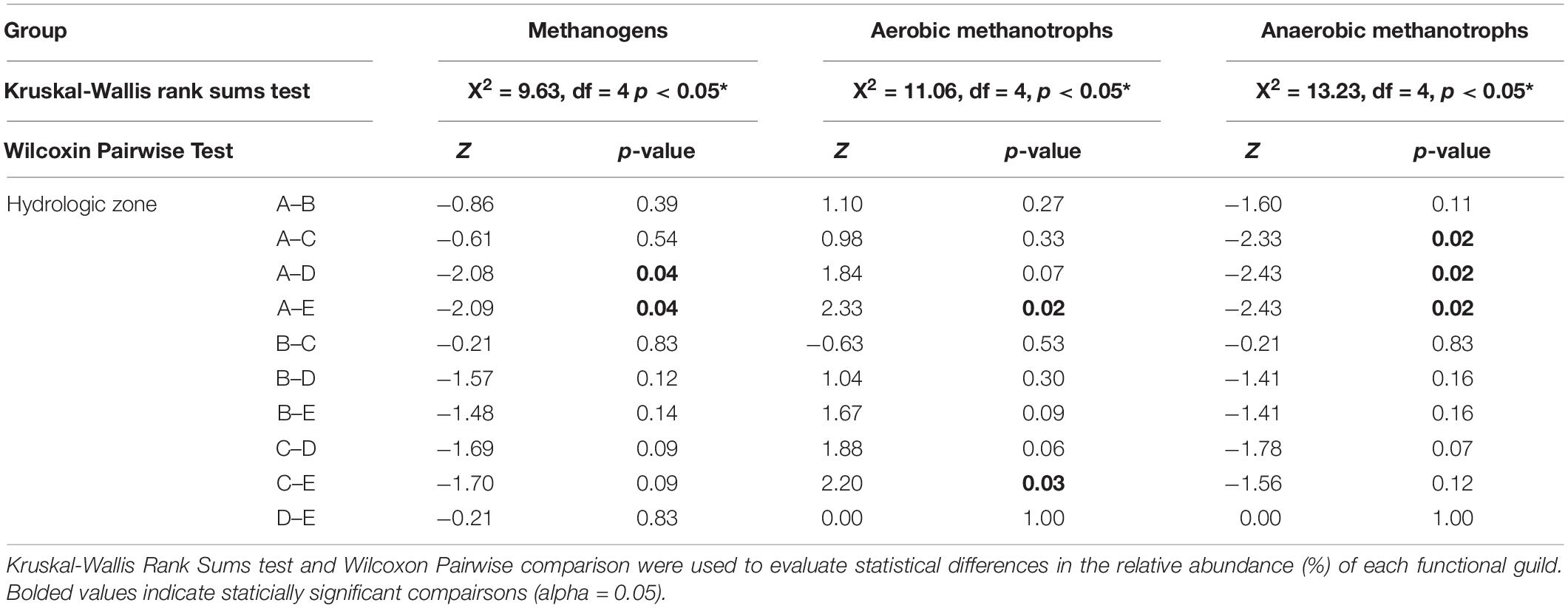
We identified a distinct assemblage of CH4-cycling microorganisms across the topographic gradient (Figure 4). Sequences putatively identified as methanogens were classified into six families: Methanobacteriaceae (Order Methanobacteriales), Methanocellaceae (Order Methanocellales), Methanoregulaceae (Order Methanomicrobiales, Methanosaetaceae (Order Methanosarcinales), Methanosarcinaceae (Order Methanosarcinales), Unclassified (Order Methanomassilii coccales). The composition and relative abundance of the methanogen community were similar among soils that were saturated for more than 50% of the year (hydrologic zones A–C); only a small fraction of methanogen sequences (∼0.25%) were identified in predominantly oxic soil conditions (hydrologic zones D,E). The methanotroph community, which was comprised of both aerobic and anaerobic methane oxidizing bacteria and archaea, differed by hydrologic zone (Figure 4 and Table 4). We identified a higher relative abundance of anaerobic methanotrophs (i.e., Methanoperedenaceae and Methylomirabilaceae) in “wet” zones (hydrologic zones A–C), whereas aerobic methanotrophs (i.e., Beijerinckiaceae and Methylacidiphilaceae) were dominant in “dry” zones (hydrologic zones D,E).
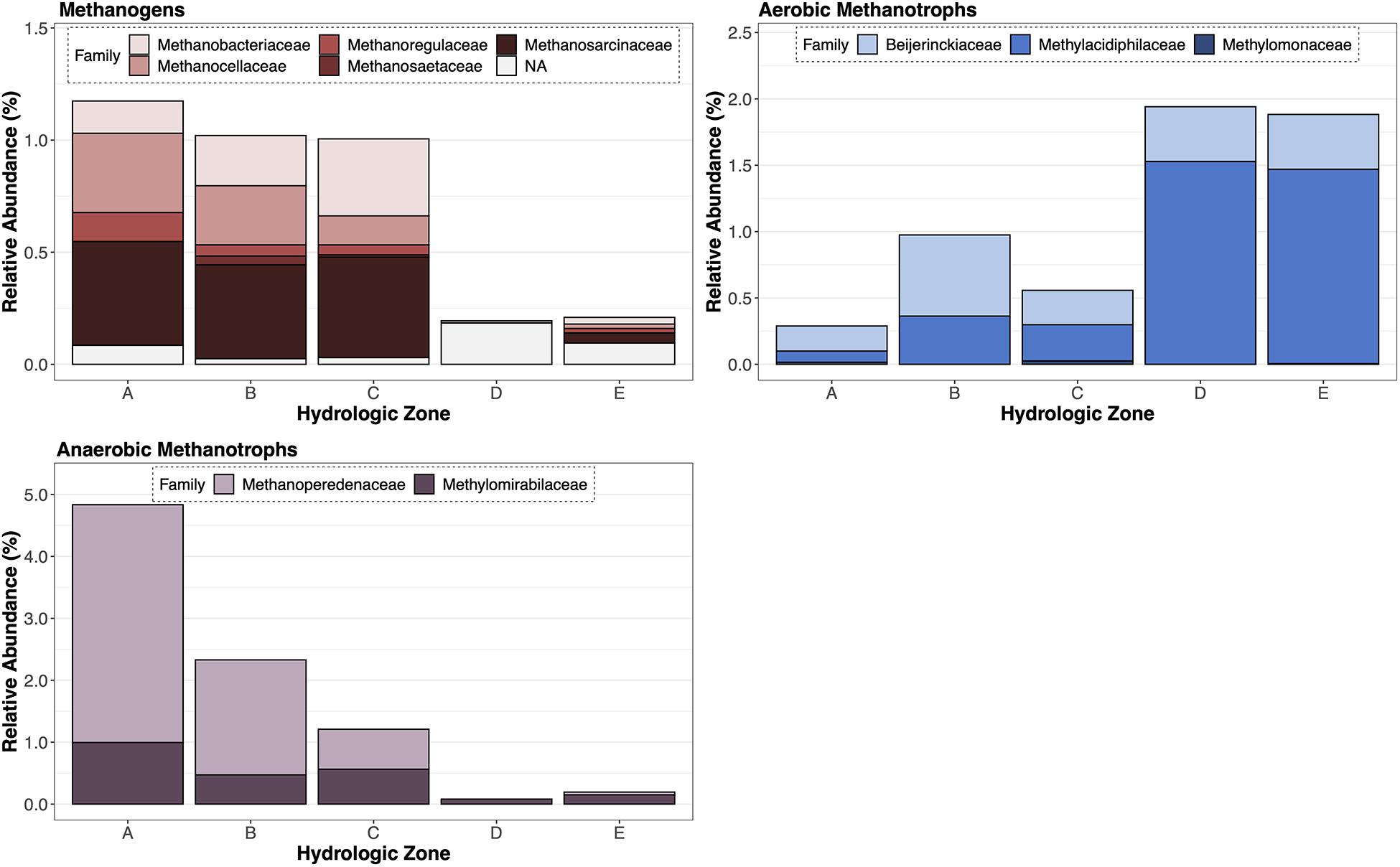
Figure 4. Total relative abundance of methanogens (red), aerobic methanotrophs (blue), and anaerobic methanotrophs (purple) by family for each hydrologic zone (A–E).
In addition to these categorical differences, we found that the relative abundance of methanogens and anaerobic methanotrophs increased with increasing annual mean water level and saturation duration at sampling points, whereas the relative abundance of aerobic methanotroph decreased (Table 3). We found no relationship between the relative abundance of methanogen and edaphic properties; however, we found a positive association between SOM content and the relative abundance of aerobic (r = −0.51) and anaerobic (r = +0.48) methanotrophs (Table 3).
Discussion
Hydrological conditions and variability are well-known to influence soil microbial community composition; however, the response of microbial communities in ephemeral wetlands is still poorly understood. This study aimed to characterize the soil microbiome associated with the lateral margins of seasonally saturated wetlands and understand how different hydrologic conditions (i.e., water level, saturation duration, and frequency of wet-dry cycles) influenced microbial community structure, particularly the microorganisms responsible for regulating soil CH4 cycling (i.e., methanogens and methanotrophs). The results of this study partially support our hypotheses. First, we found that soil and CH4-cycling microbial communities associated with the upper soil horizons were strongly linked to changes in water level, saturation duration, and SOM content but not to the saturation frequency. Second, the microbial and the CH4-cycling community composition differed across the hydrologic gradient. Specifically, we found that the methanogen community was relatively restricted to saturated zones, but the CH4-oxidizing community was distributed across the entire hydrologic gradient and shifted from a predominantly aerobic to an anaerobic community. In addition to changes in the soil hydrologic conditions, other environmental factors, such as extreme soil acidity (pH < 5), likely influenced the microbial population, particularly the composition of aerobic methanotrophs. The presence of these highly adapted but metabolically diverse methanotroph communities suggests that these wetland-to-upland interfaces could be important and dynamic CH4 sinks.
Microbial Richness, Diversity, and Composition Strongly Related to Hydrologic and Soil Conditions
The relatively even distribution and abundance of dominant bacterial phyla across the hydrologic gradient suggest wetland soils in this region maintain a core microbiome (Supplementary Figure 3), but the varying hydrologic conditions influence the composition and overall relative abundance of different microbial taxa (Figure 3). Overall microbial richness (total OTUs) and diversity (Shannon’s index) decreased with increasing annual mean water level and saturation duration (Table 3) but did not differ among the hydrologic zones, even between the two opposite ends of the transect (Table 2). In terms of overall community composition though, soils that were inundated or saturated for more than 50% of the year were distinct from soils that were rarely saturated or inundated (Figure 3). Here, we found that soils adjacent to the wetland basin were colonized by a microbial population better adapted to sustained anaerobic conditions (e.g., obligate and facultative anaerobes such as iron reducers, methanogens, and anaerobic methane oxidizers). In contrast, drier, well-aerated soils had a more diverse community, comprised of both obligate aerobes (e.g., nitrifiers and methanotrophs) and facultative anaerobes (e.g., iron oxidizers and denitrifiers), as well as having a “seed bank” of dormant or inactive CH4-cycling microbes (Figure 4). We suspect that environmental variables, such as O2 availability and redox conditions, are important conditions affecting the distribution and relative abundance of various bacterial and archaeal taxa. Our analysis also suggests that soil microbial community diversity, richness, and composition were strongly related to SOM content; however, because SOM and hydrology are typically related, it is difficult to discern the link between hydrology, organic matter and microbial populations (Fierer et al., 2003; Bossio et al., 2006; Balasooriya et al., 2007).
Unlike other studies (Kemnitz et al., 2004; Peralta et al., 2014), we did not observe a relationship between hydrologic variability (i.e., saturation frequency) and microbial community structure (Table 3). The 2018 water year was wetter than average for the region (see section “Hydrological Monitoring and Characterization”), which may explain why we observed a similar magnitude of static saturation frequency across the wetland-to-upland gradient (Figure 2) and did not find a relationship between frequency and microbial communities. We believe that the patterns we observed in the soil microbial population resulted from long-term seasonal changes in the wetland hydrologic regime and not short-term changes. Previously published literature suggests that relatively short-term changes in hydrology (i.e., hours to 6 mo.) impact soil function (Foulquier et al., 2013), but the underlying structure of the microbial composition is shaped by former and long-term historical trends in hydrology (Banerjee et al., 2016; Bledsoe and Peralta, 2020; Veach and Zeglin, 2020). That being said, small but sustained changes in hydrology could surpass thresholds and lead to fundamental changes in the structure and function of the microbial community (Lee et al., 2018). Land use and restoration efforts are already impacting the current hydrologic regime of these wetlands (Fenstermacher et al., 2014; Jones et al., 2018); therefore, it is imperative that we continue to research how microbial populations are changing and responding to hydrologic modification over different timescales. These new insights will help us better understand the potential outcomes of restoration efforts and how to effectively evaluate and monitor their progress.
Oxygen-Tolerant Hydrogenotrophic Methanogens Restricted to “Wet” Zones, but Methane Oxidizers Flourish Across a Hydrologic Gradient
Methanogens and methanotrophs are capable of colonizing a range of moisture conditions (Angel et al., 2011; Siljanen et al., 2011), but the relative abundances of these communities, their composition, and activity are expected to vary between well and poorly oxygenated conditions (Miller et al., 2004; Angel et al., 2012) and as a function of available substrate (Macalady et al., 2002; Sun et al., 2012; Gauthier et al., 2015). Our findings suggest that the distribution and composition of the CH4-cycling community were strongly influenced by the hydrologic and soil conditions.
The relative abundance of methanogens was significantly positively related to water level and saturation duration (Table 3). Of the six identified methanogen families (Figure 4), three families made up more than 66% of the sequences: Methanosarcinaceae, Methanocellaceae, and Methanoregulaceae. Methanosarcinaceae and Methanocellaceae tend to be more tolerant of desiccation and able to activate rapidly under ideal conditions (Liu and Whitman, 2008; Sakai et al., 2010). In contrast, Methanoregulaceae sp. tends to be more sensitive to O2 and more likely to associate with bulk peat soil than with microaerophilic conditions or oxidized rhizospheres (He et al., 2015). This sensitivity to O2 may explain why we found a slightly higher presence of Methanoregulaceae in soils with the shallowest water levels and the longest durations of saturation than any other location sampled. Surprisingly, Methanosaetaceae, a common methanogen identified in temperate wetlands, lakes, and minerotrophic fens (Bridgham et al., 2013), made up a minor fraction of sequences in our dataset (Figure 4). Given that a relatively large population of methanogens is maintained in soils adjacent to the wetland basin, these regions are likely the main producers of CH4; however, rates of CH4 production are dependent on additional restrictions, such as the presence of competing terminal electron acceptors (Segers, 1998; Dalcin Martins et al., 2017) and the chemical composition and quantity of the dissolved organic pool (Schipper and Reddy, 1994; Boye et al., 2018). Looking more closely at the three families of methanogens we identified, we find that they are all hydrogenotrophic; however, species associated with these families are metabolically versatile and can utilize a variety of dissolved inorganic (e.g., CO2) and organic C (e.g., acetate, formate, and other methylated organic C) compounds. The metabolic versatility of these three methanogen families will thus allow methanogenesis to proceed even as the composition of the dissolved organic and inorganic C substrate pool varies.
Methane-oxidizing microorganisms were distributed across the hydrologic gradient we sampled, but the relative abundance of aerobic and anaerobic methanotrophs was quite different. For aerobic methanotrophs, the relative abundance decreased with increasing periods of inundation or saturation (Table 3). While these results align with previous research, it is not uncommon for conditions that favor methanogenesis to also support a diverse methanotroph population (Miller et al., 2004; Siljanen et al., 2011; Christiansen et al., 2016). Therefore, we were surprised that a larger, more diverse population of aerobic methanotrophs were not present across all hydrologic conditions. Not only was the spatial distribution of aerobic methanotrophs limited, only two families, Beijerinckiaceae and Methylacidiphilaceae, were identified in our sequence dataset. The relatively high abundance of Methylacidiphilaceae was unexpected because these microorganisms are more commonly associated with geothermal environments than wetlands (Sharp et al., 2014). Notably, the optimal pH range for both aerobic methanotroph families is reported to be less than 5. The extremely acidic soil conditions associated with these wetland sites (pH < 4) may be restricting the colonization of other aerobic methane-oxidizing bacteria commonly found in wetlands.
In addition to identifying aerobic methanotrophs, we found that the relative abundance of anaerobic methane oxidizers exceeded aerobic methanotrophs and was approximately 2–5 times greater than the methanogen population (Figure 4). Recent evidence suggests that anaerobic methanotrophs can significantly reduce CH4 emissions from wetland soils (Smemo and Yavitt, 2011; Segarra et al., 2015; Wang et al., 2018); however, most models and studies do not include these processes or microorganisms in their analysis (Gauthier et al., 2015). While it is unclear how active these communities are in these soils, the phylotypes that we identified, the anaerobic methane-oxidizing archaea (ANME), Candidatus Methanoperedens (formerly classified as ANME-2d), and the anaerobic methane-oxidizing bacterial species, Methylomirabilaceae, suggest that CH4 consumption and subsequent emissions is linked to and controlled by inorganic N (i.e., nitrite or nitrate) availability. However, a study recently identified two new species, Candidatus Methanoperedens manganicus and Candidatus Methanoperedens manganireducens, and found that CH4 oxidation is coupled to manganese oxide reduction (Leu et al., 2020). This emerging area of research highlights an important gap in our understanding about microbial populations and their influence on soil biogeochemical cycles. While the evidence suggests that aerobic methane oxidizers are not the only microorganisms consuming CH4 in freshwater wetlands, additional information about the anaerobic methane oxidizing population and their activity is needed to more accurately assess their role in regulating soil CH4 cycling and emissions from freshwater mineral wetlands.
Future Research
The results of this study solely rely on 16S rRNA gene abundances from a limited number of wetland sites. While we can infer functional potential and population size, it does not quantify the rates of CH4 production or oxidation or allow us to make inferences about where and how CH4-cycling microbial communities assemble and interact across the gradient. However, understanding the distribution of these communities and their composition does help us understand potential pathways that methanogens and methanotrophs use, particularly in the context of hydrology, which will help inform mechanistic models needed to predict and reduce future wetland emissions (Bridgham et al., 2013).
Conclusion
This research provides insight into the composition and metabolic diversity of the soil microbial community, particularly the microorganisms responsible for regulating soil CH4 cycling, in the margins of seasonally saturated wetlands. We found that the overall richness and diversity of the microbial community declined, and the composition converged in areas where soils were inundated or saturated for more than 50% of the year. Additionally, soils that experienced prolonged periods of inundation and saturation support a relatively high abundance of methanogens and thus these areas could serve as a significant source area of CH4 flux. However, we also found that hydrologic conditions that favored methanogenesis also maintained a fairly large population of anaerobic methanotrophs. Regardless of water level and saturation duration, CH4-oxidizing microorganisms were present across the hydrologic gradient. While the activity of these communities is unknown, we cannot assume that CH4 oxidation is merely compartmentalized to specific soil hydrologic conditions. Rather CH4 consumption can occur across short spatial gradients and a wide range of hydrologic conditions.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/664757; www.doi.org/10.5281/zenodo.3879223.
Author Contributions
CM and KH conceived this study. CM collected soils and processed samples and wrote the manuscript. CNJ collected the hydrology data and completed the analysis. MP provided feedback on the experimental design and assisted with laboratory work. MP created images in Figure 1 using Adobe Illustrator. All authors commented on the draft manuscript.
Funding
This research benefited by funding from the University of Maryland and partial salary support from the National Socio-Environmental Synthesis Center (NSF DBI-1052875).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to The Nature Conservancy for providing permission to access the sites, and to Dr. Stephanie Yarwood for allowing us to use lab space and equipment to process and sequence the soil samples. Additionally, Michael Williams provided important help with field deployment of wells and downloads of hydrologic data.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2020.593942/full#supplementary-material
References
Angel, R., Claus, P., and Conrad, R. (2012). Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 6, 847–862.
Angel, R., Matthies, D., and Conrad, R. (2011). Activation of methanogenesis in arid biological soil crusts despite the presence of oxygen. PLoS One 6:e20453. doi: 10.1371/journal.pone.0020453
Balasooriya, W. K., Denef, K., Peters, J., Verhoest, N. E. C., and Boeckx, P. (2007). Vegetation composition and soil microbial community structural changes along a wetland hydrological gradient. Hydrol. Earth Syst. Sci. 12, 277–291.
Banerjee, S., Helgason, B., Wang, L., Winsley, T., Ferrari, B. C., and Siciliano, S. D. (2016). Legacy effects of soil moisture on microbial community structure and N 2 O emissions. Soil Biol. Biochem. 95, 40–50. doi: 10.1016/j.soilbio.2015.12.004
Bansal, S., Tangen, B., and Finocchiaro, R. (2016). Temperature and hydrology affect methane emissions from prairie pothole Wetlands. Wetlands 36, 371–381. doi: 10.1007/s13157-016-0826-8
Bledsoe, R. B., and Peralta, A. L. (2020). Dynamic hydrology and plant-mediated effects reduce greenhouse gas emissions and alter wetland microbial communities. bioRxiv doi: 10.1101/2020.06.29.178533
Bossio, D. A., Fleck, J. A., Scow, K. M., and Fujii, R. (2006). Alteration of soil microbial communities and water quality in restored wetlands. Soil Biol. Biochem. 38, 1223–1233. doi: 10.1016/j.soilbio.2005.09.027
Boye, K., Herrmann, A. M., Schaefer, M. V., Tfaily, M. M., and Fendorf, S. (2018). Discerning microbially mediated processes during redox transitions in flooded soils using carbon and energy balances. Front. Environ. Sci. 6:15. doi: 10.3389/fenvs.2018.00015
Bridgham, S. D., Cadillo-Quiroz, H., Keller, J. K., and Zhuang, Q. (2013). Methane emissions from wetlands: biogeochemical, microbial, and modeling perspectives from local to global scales. Glob. Change Biol. 19, 1325–1346. doi: 10.1111/gcb.12131
Calhoun, A. J. K., Mushet, D. M., Bell, K. P., Boix, D., Fitzsimons, J. A., and Isselin-Nondedeu, F. (2017). Temporary wetlands: challenges and solutions to conserving a ‘disappearing’ ecosystem. Biol. Conserv. 211, 3–11. doi: 10.1016/j.biocon.2016.11.024
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016a). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Callahan, B. J., Sankaran, K., Fukuyama, J. A., McMurdie, P. J., and Holmes, S. P. (2016b). Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. F1000Research 5:1492. doi: 10.12688/f1000research.8986.1
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chandler, H. C., McLaughlin, D. L., Gorman, T. A., McGuire, K. J., Feaga, J. B., and Haas, C. A. (2017). Drying rates of ephemeral wetlands: implications for breeding amphibians. Wetlands 37, 545–557. doi: 10.1007/s13157-017-0889-1
Chowdhury, T. R., and Dick, R. P. (2013). Ecology of aerobic methanotrophs in controlling methane fluxes from wetlands. Appl. Soil Ecol. 65, 8–22. doi: 10.1016/j.apsoil.2012.12.014
Christiansen, J. R., Levy-Booth, D., Prescott, C. E., and Grayston, S. J. (2016). Microbial and environmental controls of methane fluxes along a soil moisture gradient in a pacific coastal temperate rainforest. Ecosystems 19, 1255–1270. doi: 10.1007/s10021-016-0003-1
Conrad, R. (2020). Importance of hydrogenotrophic, aceticlastic and methylotrophic methanogenesis for methane production in terrestrial, aquatic and other anoxic environments: a mini review. Pedosphere 30, 25–39. doi: 10.1016/S1002-0160(18)60052-9
Cui, M., Ma, A., Qi, H., Zhuang, X., and Zhuang, G. (2015). Anaerobic oxidation of methane: an “active” microbial process. MicrobiologyOpen 4, 1–11. doi: 10.1002/mbo3.232
Dalcin Martins, P., Hoyt, D. W., Bansal, S., Mills, C. T., Tfaily, M., Tangen, B. A., et al. (2017). Abundant carbon substrates drive extremely high sulfate reduction rates and methane fluxes in prairie pothole Wetlands. Glob. Chang. Biol. 23, 3107–3120. doi: 10.1111/gcb.13633
Epting, S. M., Hosen, J. D., Alexander, L. C., Lang, M. W., Armstrong, A. W., and Palmer, M. A. (2018). Landscape metrics as predictors of hydrologic connectivity between Coastal Plain forested wetlands and streams. Hydrol. Process. 32, 516–532. doi: 10.1002/hyp.11433
Fenstermacher, D. E., Rabenhorst, M. C., Lang, M. W., McCarty, G. W., and Needelman, B. A. (2014). Distribution, morphometry, and land use of delmarva bays. Wetlands 34, 1219–1228. doi: 10.1007/s13157-014-0583-5
Fierer, N., Schimel, J. P., and Holden, P. A. (2003). Variations in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35, 167–176. doi: 10.1016/S0038-0717(02)00251-1
Foulquier, A., Volat, B., Neyra, M., Bornette, G., and Montuelle, B. (2013). Long-term impact of hydrological regime on structure and functions of microbial communities in riverine wetland sediments. FEMS Microbiol. Ecol. 85, 211–226. doi: 10.1111/1574-6941.12112
Gauthier, M., Bradley, R. L., and Šimek, M. (2015). More evidence that anaerobic oxidation of methane is prevalent in soils: is it time to upgrade our biogeochemical models? Soil Biol. Biochem. 80, 167–174. doi: 10.1016/j.soilbio.2014.10.009
Hansen, A. T., Dolph, C. L., Foufoula-Georgiou, E., and Finlay, J. C. (2018). Contribution of wetlands to nitrate removal at the watershed scale. Nat. Geosci. 11, 127–132. doi: 10.1038/s41561-017-0056-6
He, S., Malfatti, S. A., McFarland, J. W., Anderson, F. E., Pati, A., Huntemann, M., et al. (2015). Patterns in wetland microbial community composition and functional gene repertoire associated with methane emissions. mBio 6:e0066-15. doi: 10.1128/mBio.00066-15
Jones, C. N., Evenson, G. R., McLaughlin, D. L., Vanderhoof, M. K., Lang, M. W., McCarty, G. W., et al. (2018). Estimating restorable wetland water storage at landscape scales. Hydrol. Process. 32, 305–313. doi: 10.1002/hyp.11405
Kemnitz, D., Chin, K.-J., Bodelier, P., and Conrad, R. (2004). Community analysis of methanogenic archaea within a riparian flooding gradient. Environ. Microbiol. 6, 449–461. doi: 10.1111/j.1462-2920.2004.00573.x
Kifner, L. H., Calhoun, A. J. K., Norton, S. A., Hoffmann, K. E., and Amirbahman, A. (2018). Methane and carbon dioxide dynamics within four vernal pools in Maine, USA. Biogeochemistry 139, 275–291. doi: 10.1007/s10533-018-0467-5
Kotsyurbenko, O. R., Glagolev, M. V., Merkel, A. Y., Sabrekov, A. F., and Terentieva, I. E. (2019). “Methanogenesis in Soils, Wetlands, and peat,” in Biogenesis of Hydrocarbons, eds A. J. M. Stams and D. Sousa, (Cham: Springer International Publishing), 1–18.
Lee, K. C., Caruso, T., Archer, S. D. J., Gillman, L. N., Lau, M. C. Y., Cary, S. C., et al. (2018). Stochastic and deterministic effects of a moisture gradient on soil microbial communities in the mcmurdo Dry Valleys of Antarctica. Front. Microbiol. 9:2619. doi: 10.3389/fmicb.2018.02619
Lee, S., McCarty, G. W., Moglen, G. E., Lang, M. W., Nathan Jones, C., Palmer, M., et al. (2020). Seasonal drivers of geographically isolated wetland hydrology in a low-gradient, Coastal Plain landscape. J. Hydrol. 583:124608. doi: 10.1016/j.jhydrol.2020.124608
Leu, A. O., Cai, C., McIlroy, S. J., Southam, G., Orphan, V. J., Yuan, Z., et al. (2020). Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J. 14, 1030–1041. doi: 10.1038/s41396-020-0590-x
Liu, Y., and Whitman, W. B. (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 1125, 171–189. doi: 10.1196/annals.1419.019
Macalady, J. L., McMillan, A., Dickens, A. F., Tyler, S. C., and Scow, K. M. (2002). Population dynamics of type I and II methanotrophic bacteria in rice soils. Environ. Microbiol. 4, 148–157.
Marton, J. M., Creed, I. F., Lewis, D. B., Lane, C. R., Basu, N. B., Cohen, M. J., et al. (2015). Geographically isolated wetlands are important biogeochemical reactors on the landscape. BioScience 65, 408–418. doi: 10.1093/biosci/biv009
McDonough, O. T., Lang, M. W., Hosen, J. D., and Palmer, M. A. (2015). Surface hydrologic connectivity between delmarva bay wetlands and nearby streams along a gradient of agricultural alteration. Wetlands 35, 41–53. doi: 10.1007/s13157-014-0591-5
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8:e61217. doi: 10.1371/journal.pone.0061217
Miller, D. N., Yavitt, J. B., Madsen, E. L., and Ghiorse, W. C. (2004). Methanotrophic activity, abundance, and diversity in forested swamp pools: spatiotemporal dynamics and influences on methane fluxes. Geomicrobiol. J. 21, 257–271. doi: 10.1080/01490450490438766
Nelson, D., and Sommers, L. (1996). “Chapter 34: total carbon, organic carbon, and organic matter,” in Methods of Soil Analysis Part 3—Chemical Methods, eds D.L. Sparks, A.L. Page, P.A. Helmke, R.H. Loeppert, and P. N. Soltanpour, (Madison: Soil Science Society of America), 961–1010.
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2019). vegan: Community Ecology Package. R package version 2.5-6. Available online at: https://CRAN.R-project.org/package=vegan
Peralta, A. L., Ludmer, S., Matthews, J. W., and Kent, A. D. (2014). Bacterial community response to changes in soil redox potential along a moisture gradient in restored wetlands. Ecol. Eng. 73, 246–253. doi: 10.1016/j.ecoleng.2014.09.047
Phillips, P. J., and Shedlock, R. J. (1993). Hydrology and chemistry of groundwater and seasonal ponds in the Atlantic Coastal Plain in Delaware, USA. J. Hydrol. 141, 157–178.
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Rosenberry, D. O., and Winter, T. C. (1997). Dynamics of water-table fluctuations in an upland between two prairie-pothole wetlands in North Dakota. J. Hydrol. 191, 266–289. doi: 10.1016/S0022-1694(96)03050-8
Roy Chowdhury, T., Mitsch, W. J., and Dick, R. P. (2014). Seasonal methanotrophy across a hydrological gradient in a freshwater wetland. Ecol. Eng. 72, 116–124. doi: 10.1016/j.ecoleng.2014.08.015
Sakai, S., Conrad, R., Liesack, W., and Imachi, H. (2010). Methanocella arvoryzae sp. nov., a hydrogenotrophic methanogen isolated from rice field soil. Int. J. Syst. Evol. Microbiol. 60, 2918–2923. doi: 10.1099/ijs.0.020883-0
Schipper, L. A., and Reddy, K. R. (1994). Methane production and emissions from four reclaimed and pristine wetlands of southeastern United States. Soil Sci. Soc. Am. J. 58, 1270–1275.
Segarra, K. E. A., Schubotz, F., Samarkin, V., Yoshinaga, M. Y., Hinrichs, K.-U., and Joye, S. B. (2015). High rates of anaerobic methane oxidation in freshwater wetlands reduce potential atmospheric methane emissions. Nat. Commun. 6:7477. doi: 10.1038/ncomms8477
Segers, R. (1998). Methane production and methane consumption: a review of processes underlying wetland methane fluxes. Biogeochemistry 41, 23–51. doi: 10.1023/A:1005929032764
Sharp, C. E., Smirnova, A. V., Graham, J. M., Stott, M. B., Khadka, R., Moore, T. R., et al. (2014). Distribution and diversity of V errucomicrobia methanotrophs in geothermal and acidic environments: diversity of verrucomicrobial methanotrophs. Environ. Microbiol. 16, 1867–1878. doi: 10.1111/1462-2920.12454
Siljanen, H. M. P., Saari, A., Krause, S., Lensu, A., Abell, G. C. J., Bodrossy, L., et al. (2011). Hydrology is reflected in the functioning and community composition of methanotrophs in the littoral wetland of a boreal lake: spatial aspects of methanotrophs in littoral wetland. FEMS Microbiol. Ecol. 75, 430–445. doi: 10.1111/j.1574-6941.2010.01015.x
Smemo, K. A., and Yavitt, J. B. (2011). Anaerobic oxidation of methane: an underappreciated aspect of methane cycling in peatland ecosystems? Biogeosciences 8, 779–793. doi: 10.5194/bg-8-779-2011
Sun, C. L., Brauer, S. L., Cadillo-Quiroz, H., Zinder, S. H., and Yavitt, J. B. (2012). Seasonal changes in methanogenesis and methanogenic community in three peatlands, New York State. Front. Microbiol. 3:81. doi: 10.3389/fmicb.2012.00081
Veach, A. M., and Zeglin, L. H. (2020). Historical drought affects microbial population dynamics and activity during soil drying and re-wet. Microb. Ecol. 79, 662–674. doi: 10.1007/s00248-019-01432-5
Wang, W., Zeng, C., Sardans, J., Wang, C., Tong, C., and Peñuelas, J. (2018). Soil methane production, anaerobic and aerobic oxidation in porewater of wetland soils of the minjiang river estuarine, China. Wetlands 38, 627–640. doi: 10.1007/s13157-018-1006-9
Yepsen, M., Baldwin, A. H., Whigham, D. F., McFarland, E., LaForgia, M., and Lang, M. (2014). Agricultural wetland restorations on the USA Atlantic Coastal Plain achieve diverse native wetland plant communities but differ from natural wetlands. Agric. Ecosyst. Environ. 197, 11–20. doi: 10.1016/j.agee.2014.07.007
Yu, S., and Ehrenfeld, J. G. (2010). Relationships among plants, soils and microbial communities along a hydrological gradient in the New Jersey Pinelands, USA. Ann. Bot. 105, 185–196. doi: 10.1093/aob/mcp183
Keywords: freshwater mineral wetlands, methanogens, methanotrophs, methane emissions, microbial diversity
Citation: Maietta CE, Hondula KL, Jones CN and Palmer MA (2020) Hydrological Conditions Influence Soil and Methane-Cycling Microbial Populations in Seasonally Saturated Wetlands. Front. Environ. Sci. 8:593942. doi: 10.3389/fenvs.2020.593942
Received: 11 August 2020; Accepted: 12 October 2020;
Published: 04 November 2020.
Edited by:
Vinicius Fortes Farjalla, Federal University of Rio de Janeiro, BrazilReviewed by:
Jean Remy Davee Guimaraes, Federal University of Rio de Janeiro, BrazilDaniele Kasper, Federal University of Rio de Janeiro, Brazil
Copyright © 2020 Maietta, Hondula, Jones and Palmer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christine E. Maietta, Y3ByYXNzZUB1bWQuZWR1
 Christine E. Maietta
Christine E. Maietta Kelly L. Hondula
Kelly L. Hondula C. Nathan Jones
C. Nathan Jones Margaret A. Palmer
Margaret A. Palmer