- 1Bigelow Laboratory for Ocean Sciences, East Boothbay, ME, United States
- 2Department of Soil, Water and Climate, University of Minnesota, Minneapolis, MN, United States
The crustal sub-seafloor covers a large portion of the Earth’s surface but is poorly understood as a habitat for life. It is unclear what metabolisms support the microscopic cells that have been observed, and how they survive under resource limitation. As the deep crustal subsurface represents a significant portion of the Earth’s surface, microbially mediated reactions may therefore be significant contributors to biogeochemical cycling. In the present study, we used electrochemical techniques to investigate the possibility that crustal subsurface microbial groups can use the solid rock matrix (basalts, etc.) as a source of electrons for redox reactions via extracellular electron transfer (EET). Subsurface crustal fluids and mineral colonization experiments from the cool and oxic basaltic crustal subsurface at the North Pond site on the western flank of the Mid-Atlantic Ridge were used as inocula in cathodic poised potential experiments. Electrodes in oxic microbial fuel cells (MFCs) were poised at −200 mV versus a standard hydrogen electrode to mimic the delivery of electrons in an energy range equivalent to iron oxidation. In this way, microbes that use reduced iron in solid minerals for energy were selected for from the general community onto the electrode surface for interrogation of EET activity, and potential identification by scanning electron microscopy (SEM) and DNA sequencing. The results document that there are cathodic EET-capable microbial groups in the low biomass crustal subsurface at this site. The patterns of current generation in the experiments indicate that these microbial groups are active but likely not growing under the low-resource condition of the experiments, consistent with other studies of activity versus growth in the deep biosphere. Lack of growth stymied attempts to determine the phylogeny of EET-capable microbial groups from this habitat, but the results indicate that these microbial groups are a small part of the overall crustal deep biosphere community. This first demonstration of using electromicrobiology techniques to investigate microbial metabolic potential in the crustal deep biosphere reveals the challenges and opportunities for studying EET in the crustal deep biosphere.
Introduction
Oceanic crust and fluids represent a significant portion of the Earth’s habitable volume (Edwards et al., 2005; Orcutt et al., 2011b), and may be an important contributor to biogeochemical cycling based on volume alone (Shah Walter et al., 2018). There are microbial cells present in this environment, often in very low cell densities (Orcutt et al., 2011b; Smith et al., 2011; Jungbluth et al., 2013, 2016; Jørgensen and Zhao, 2016; Meyer et al., 2016; Labonté et al., 2017; Früh-Green et al., 2018). Studies documenting the potential for microbial activity in the crustal subsurface (Torsvik et al., 1998; Lever et al., 2013; Orcutt et al., 2013a; Meyer et al., 2016; Robador et al., 2016; Zhang et al., 2016; D’Hondt et al., 2019; Jangir et al., 2019) indicate relatively low rates of activity consistent with the limiting resource and energy conditions in the subsurface (LaRowe and Amend, 2015; Bradley et al., 2018; Jones et al., 2018).
Oxidation of reduced iron bound in cool, oxic crustal basalts may be an important source of chemolithotrophic energy for supporting a crustal biosphere (Orcutt et al., 2013b; Barco et al., 2017). Theoretical calculations suggest that the oxidation of iron and sulfur can support 41 ± 21 × 1010 g C per year of autotrophic biomass carbon formation (Bach and Edwards, 2003), approximating recent estimates of a carbon fixation rate of 109–1012 g C per year based on incubation experiments with globally distributed basalts (Orcutt et al., 2015). Cultivation-based studies indicate the presence of autotrophic and heterotrophic iron oxidation metabolisms in oceanic crust (Edwards et al., 2003a, b, 2004; Meyer et al., 2016; Russell et al., 2016). If microbial oxidation of iron in the solid crustal matrix is occurring in subseafloor ocean crust, then this could have implications for the redox alteration state of oceanic crust over time, and on in-filling of vein networks with secondary alteration products that might affect fluid flow.
Crustal iron oxidation processes require capturing energy/electrons from a solid substrate and bringing them into the cell for use in an electron transport chain, a process often referred to as extracellular electron transfer (EET) (Lovley, 2011; Karbelkar et al., 2016; Hinks et al., 2017; Rowe et al., 2018). EET metabolisms rely on electron shuttle molecules, external structures such as nanowires, or direct contact of conductive structures in the cell wall (Lovley, 2008b; Tremblay and Zhang, 2015; Nealson and Rowe, 2016). One way to search for these EET microorganisms is by electrochemical techniques (Lovley, 2008a; Nealson and Rowe, 2016; Martin et al., 2018). In these techniques, the sites of reduction and oxidation reactions are physically separated, using metal wires and the movement of ions in fluid to set up a complete circuit of charge flow between the redox reaction. The voltage (i.e., the energy level of the electrons flowing through the circuit) of the system is set to mimic the redox couple energy output. This relies on the principle that EET microorganisms with the cellular machinery to use electrons at that potential can either take electrons from the electrode as if it were the electron donor (referred to as cathodic) or transfer electrons to an electrode as if it were the electron acceptor (referred to as anodic). Most work concerning EET concentrates on a few model microorganisms (Lovley, 2012; Nealson and Rowe, 2016; Rowe et al., 2018) and their use in microbial fuel cells (MFCs) (Reimers et al., 2006; Nielsen et al., 2008; Rittmann, 2017; Santoro et al., 2017). There are a growing number of anodic electrochemical studies in environmental samples to identify electrode-reducing EET-capable microorganisms (e.g., Bond et al., 2002; Williams et al., 2010; Reimers et al., 2017; Lam et al., 2019a). However, there are far fewer cathodic or electrode-oxidizing studies to identify environmental microorganisms capable of solid substrates as electron donors (Gregory et al., 2004; Strycharz-Glaven et al., 2013; Rowe et al., 2015, 2018; Jangir et al., 2019; Lam et al., 2019b). While EET-capable microbes are confirmed in the deep terrestrial subsurface (Summers et al., 2013; Badalamenti et al., 2016; Jangir et al., 2016, 2019; Karbelkar et al., 2016) and deep marine hydrothermal vents (Yamamoto et al., 2017; Pillot et al., 2018), to our knowledge there have been no studies in oceanic crust.
Here, we used cathodic poised potential MFC experiments to examine the capacity for microbial communities from the cool and oxic crustal subsurface to acquire energy from solid substrates. Source materials for the experiments originated from mineral colonization experiments called FLOCS (Orcutt et al., 2010) that had been deployed at North Pond on the western flank of the Mid-Atlantic Ridge as part of Integrated Ocean Drilling Program (IODP) Expedition 336 (Expedition 336 Scientists, 2012) and on subsequent follow up cruises (Figure 1). The mineral colonization experiments were incubated in subseafloor crustal fluids that are relatively cool and oxic (Meyer et al., 2016; Shah Walter et al., 2018; Tully et al., 2018) for periods of 3–6 years following approaches described elsewhere (Baquiran et al., 2016; Ramírez et al., 2019). In the MFCs, the colonized substrates were incubated under aerobic conditions in sterilized crustal fluid from the same environment, with voltages applied to mimic Fe2/O2 redox coupling and encourage EET-capable cells to use electrons from the working electrodes (WEs) (Figure 2). In brief, our results indicate the potential for EET-capable microbial groups in the crustal subsurface, although the identity of the specific microbial members enriched by this process is unclear due to low biomass limitations of the experiments.
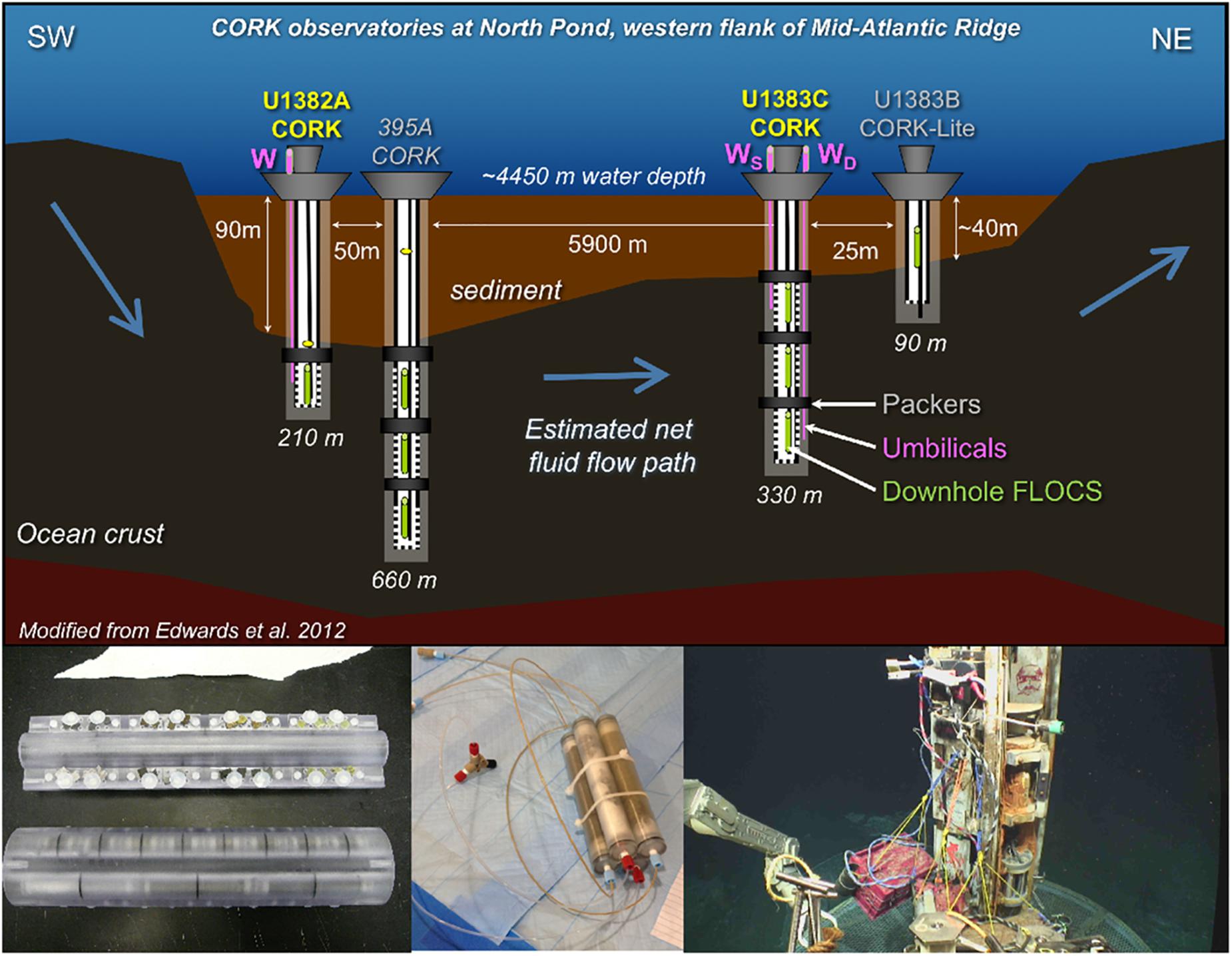
Figure 1. Overview of sampling from the North Pond crustal subsurface for this study. (Upper): Schematic of FLOCS colonization experiments and fluid sampling from CORK observatories installed during IODP Expedition 336. The primary locations targeted in this study – Holes U1382A and U1383C – are highlighted in yellow. FLOCS colonization experiments were deployed both downhole (green) and at the wellheads of the CORKs and accessing crustal fluid sourced from depth via stainless steel umbilical tubing (pink, with WS representing a FLOCS sourcing the shallow sealed horizon at WD sourcing the deeper sealed horizon). Figure modified from Edwards et al. (2012) with permission. (Lower) Left: Photograph of downhole FLOCS colonization experiment. Reprinted with permission from Edwards et al. (2012). Middle: Photograph of wellhead FLOCS experiment. Right: Deployment of wellhead FLOCS experiments in a milkcrate with OsmoSamplers connected to CORK wellhead umbilical. Photo courtesy RV Atlantis cruise AT39-01 ROV Jason dive J2-1032, 2017, chief scientist Geoff Wheat, University of Alaska Fairbanks, US National Science Foundation, ©Woods Hole Oceanographic Institution.
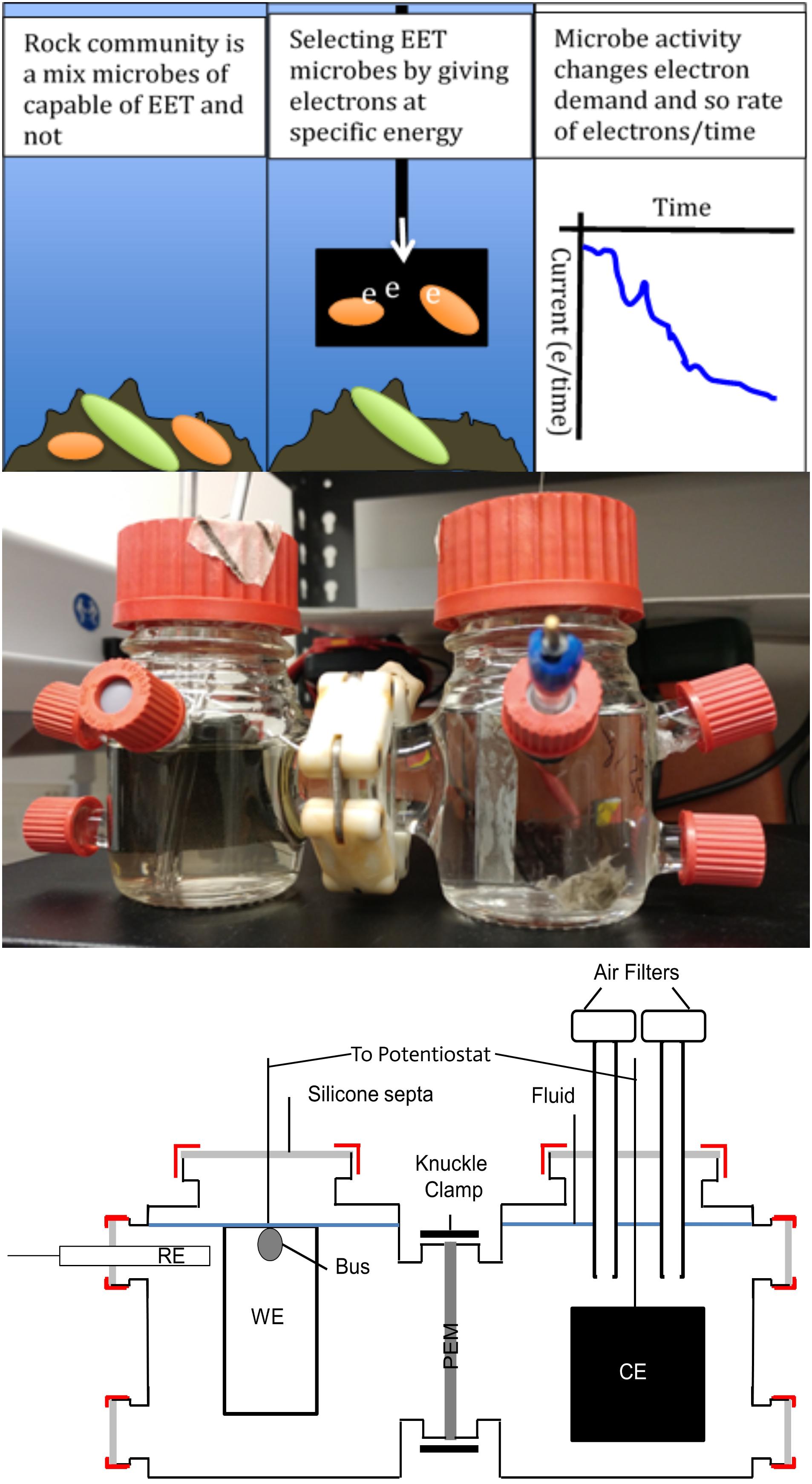
Figure 2. Overview of cathodic poised potential experiments for enriching microorganisms from North Pond subsurface crustal samples capable of extracellular electron transport (EET) when supplied with –200 mV (versus standard hydrogen electrode) and oxygen. Top: Conceptional cartoons of experiment principles. Middle: Photograph of a complete two-cell unit, with reference electrode (RE) and ITO-coated glass working electrode (WE) in the cell on the right and the counter electrode (CE) in the cell on the left separated by a Nafion proton exchange membrane (PEM). Bottom: Schematic of a complete two-cell unit.
Materials and Methods
Site Description and Sample Collection
North Pond is a sediment-covered depression overlying basalt crust of the western flank of the Mid-Atlantic Ridge at ∼4,450 m water depth (Figure 1). Details of the crustal composition may be found elsewhere (Expedition 336 Scientists, 2012). Fluids circulating within the crust at this site are oxic and 2–4°C with very low dissolved organic carbon and dissolved metal concentrations (Meyer et al., 2016).
Fluid and biofilm-containing mineral samples for this study come from CORK observatories installed at IODP Holes U1382A and U1383C as described elsewhere (Edwards et al., 2012). In brief, instrument strings containing OsmoSampler systems (Wheat et al., 2011) were deployed at different depths within the holes from 2011 to 2017. Additional OsmoSampler systems were deployed at the wellhead of the CORKs in 2014 (Table 1 and Figure 1) and were inoculated with bottom seawater, making them useful for identifying differences between crustal subsurface and bottom seawater inoculated microbial communities. OsmoSampler systems included Flow-through Osmo Colonization Systems (FLOCS) for mineral colonization experiments, as described elsewhere (Orcutt et al., 2010; Wheat et al., 2011; Edwards et al., 2012; Orcutt and Edwards, 2014; Ramírez et al., 2019). Each FLOCS contained sterile (autoclaved and ethanol-rinsed) substrates including crushed basalts, pyrite, pyrrhotite, or inert glass beads housed within polycarbonate cassettes and sleeves. Fluids were introduced into the FLOCS via the OsmoSampler pumps, allowing fluid microbial communities to colonize the substrates. Effluent of the FLOCS experiments was captured in Teflon coils in such a way to allow a time series of dissolved ion chemical composition to be constructed, following methods described in detail elsewhere (Orcutt et al., 2011a; Wheat et al., 2011; Edwards et al., 2012).
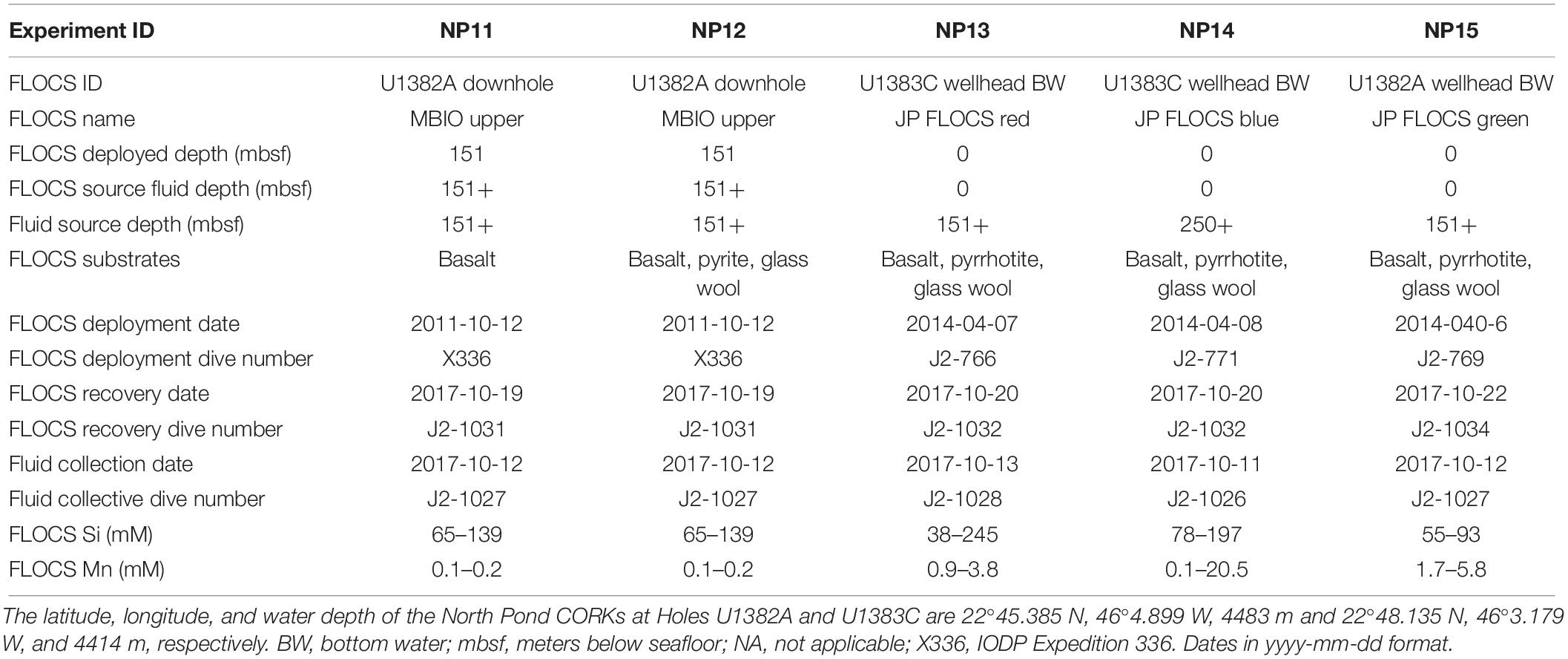
Table 1. Summary of mineral colonization experiments (i.e., FLOCS) and inoculating fluids used in the poised potential experiments, including deployment and recovery information about the FLOCS experiments and the source fluids used as inoculum and/or media, and the concentrations of dissolved silica and manganese within the FLOCS at the time of recovery.
All FLOCS were recovered in October 2017 during cruise AT39-01 of the RV Atlantis with the ROV Jason (Woods Hole Oceanographic Institution) following methods described elsewhere (Ramírez et al., 2019). In brief, the instrument strings with the downhole FLOCS were pulled up to the ship on a wire, then immediately disassembled to store the FLOCS in a cold room. Wellhead FLOCS incubated in milkcrates attached to crustal fluid umbilicals at the seafloor were also collected on this cruise with the ROV. FLOCS contents were distributed inside an ethanol-rinsed and UV-irradiated HEPA-filtered hood using sterile instruments. Substrates for the experiment were stored cold (2–4°C) and in the dark in sterile centrifuge tubes with ultrafiltered (0.22 μm-mesh Millipore Sterivex and 0.02 μm-mesh Whatman Anotop 25 filters) crustal fluid until incubation at the shore-based laboratory. Parallel samples for initial characterization of the substrate biofilms were transferred to sterile cyrovials, flash frozen with liquid nitrogen, and stored at −80°C until analysis. In addition to the FLOCS samples, 4–10 L samples of raw and unfiltered crustal fluids was collected into ethanol-rinsed cubitainers after collection into gamma-irradiated bags on the seafloor using a Mobile Pumping System as described in detail elsewhere (Cowen et al., 2012; Jungbluth et al., 2016; Meyer et al., 2016). Cubitainers were stored cold (2–4°C) and in the dark for ∼1 year before beginning the experiments.
The Teflon coils with fluid samples from the FLOCS were sub-sectioned at sea and analyzed for total silica and manganese following established protocols (Wheat et al., 2011). Prior work at North Pond has documented crustal fluid concentrations of 157 μM silica and 0.1 μM manganese and bottom seawater concentrations of 48 μM silica and <0.05 μM manganese (Meyer et al., 2016). The similarity of the FLOCS silica and manganese concentrations to those of crustal fluids (Table 1) is indicator that the FLOCS chambers were flushed with crustal fluid.
Cathodic Poised Potential Experiments
A detailed description of the protocol used is available elsewhere (Jones and Orcutt, 2019). In brief, glass two-cell, three-electrode MFC systems (Adams and Chittenden, CA, United States) were used as incubations chambers, run in parallel with a multichannel potentiostat (model CHI1030C, CHI Instruments, TX, United States). Nafion 117 proton exchange membrane (Fuel Cell Store, TX, United States) separated the half-cells. Cells were filled with distilled water and autoclaved prior to filling with media and sample inocula.
Ag/AgCl reference electrodes (Gamry Instruments, PA, United States part 932-00018 and/or Analytical Instrument Systems, NY, United States) were calibrated before each run by immersion in 3 M NaCl and compared against a known electrode kept only for that purpose (max ±20 mV drift from the value of lab master electrode). WEs were 2 × 4 cm2 indium tin oxide (ITO)-coated glass slides (Delta Technologies Ltd., CO, United States part CB-50IN-1111), constructed for MFC as described elsewhere (Rowe et al., 2015). Counter electrodes (CEs) were carbon cloth with a 4 × 4 cm2 surface area. Electrodes were sterilized by rinsing with 80% ethanol, air-drying, then exposing to UV radiation for 15 min per side.
Each experiment consisted of five treatments (Table 2): Three MFCs filled with buffered and double autoclaved crustal fluid (cool and oxic) and inoculated with colonized mineral samples. Colonized minerals were not pretreated to loosen cells from the mineral surfaces, as preserving biomass was considered more important. Chronoamperometric (CA) technique with a WE continuously poised at −200 mV versus a standard hydrogen electrode (SHE) was used as electron donor to incubate these cells. These are referred to as the “Echem” treatments. One MFC filled with the same fluid and WE but without any sample inoculum – referred to as the “Fluid” treatment. And finally, one glass bottle with a mixed sample inoculum and ITO electrode without any voltage applied – referred to as the “Offline” treatment. Samples were transferred into the MFCs in HEPA-filtered, UV-irradiated biosafety cabinet, or hood using sterile tools. An aliquot of each sample was also collected into a sterile plastic tube for microbial community analysis representing a “time zero” (T0) condition that may have changed from the Shipboard (SB) initial condition. For the Echem and Fluid treatments, the counter cell redox couple was H2O/O2. Testing (data not shown) determined that a strong kill control method was necessary to achieve sterility of the crustal fluid, consisting of 1 h autoclaving at 121°C, incubation at room temperature in the dark for ∼24 h to allow for spore germination, and then another 1 h autoclaving at 121°C before cooling down to 4°C for the addition of substrate. Sodium bicarbonate was added to the media (0.1 M) as a buffer. MFCs were incubated at 4°C in the dark.
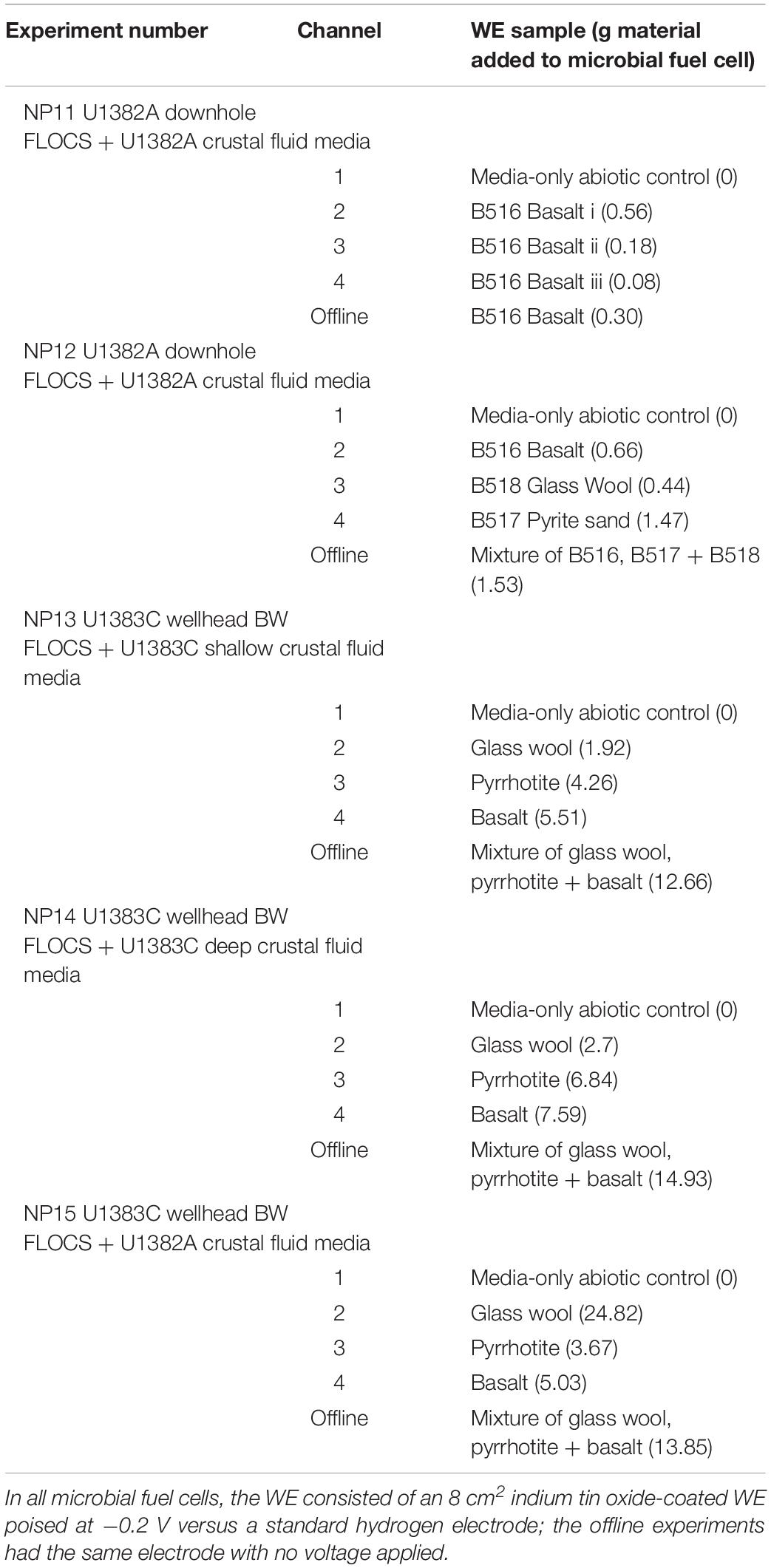
Table 2. Summary of poised potential experimental set-ups, indicating contents of the working electrode (WE) cell, the media used in the cells (ultra-filtered and double autoclaved fluid from either CORKs U1382A or U1383C supplemented with 10 mM sodium bicarbonate as a buffer).
Once the MFCs were constructed, a cyclic voltammetry (CV) sweep was performed for each cell with the potentiostat before each poised potential experiment at a scan rate of 0.1 V s–1 and sample interval of 0.001 V, across −1 to 1 V range. Then the chronoamperometry experiments began by applying a constant voltage (−200 mV versus SHE) to the Echem and Fluid MFCs. Current generation was monitored and recorded until current changes began to decline after a period of approximately 15 days. At the end of each CA experiment, two further CV sweeps were performed: one with the incubated ITO electrode (Tend i) and one with a fresh ITO electrode dipped into the incubated media (Tend ii).
Post-processing Cathodic Poised Potential Samples
After CV scanning, the ITO electrodes were carefully removed from the MFCs and split into two sections using a diamond scribe and tweezers for microscopy and DNA analysis. One piece was serially dehydrated in ethanol solutions (10 min steps each at 50:50, 60:40, 70:30, 80:20, 90:10, and 100:0 ratios of ethanol:distilled water) and hexamethyldisilizane (10 min) and air dried for 3 days in a HEPA-filtered biosafety cabinet in preparation for scanning electron microscopy (SEM), following methods available elsewhere (Braet et al., 1997; Hazrin-Chong and Manefield, 2012; D’Angelo and Orcutt, 2019b). Dehydrated slide pieces were then mounted onto 12-mm-diameter SEM stubs with double-sided carbon tape, sputter coated with gold with a Denton Vacuum Desk IV cold sputter instrument (Denton Vacuum LLC, Moorestown, NJ, United States), and imaged with a Zeiss Supra25 SEM at between 5 and 10 kV and a working distance between 3 and 9 mm. The second ITO piece was vigorously rubbed with a distilled water-soaked sterile cotton swab (sterile phosphate-buffered saline solution-soaked swab for NP14) to remove cells for DNA extraction.
In addition, aliquots of the residue FLOCS sample material in the MFCs were collected at the end of the experiment for “time final” (Tend) microbial community composition analysis. As with T0 samples, the aliquots were transferred into sterile plastic tubes and stored frozen at −80°C until analysis.
DNA Extraction, Quantification, Sequencing, and Bioinformatic Processing
The FastDNA kit from MP Biomedicals (Irvine, CA, United States) was used for all DNA extractions following manufacturer’s instructions with the exception of using a Retsch MM 400 shaker (frequency 20 r/S for 5 min) instead of vortexing as described in detail elsewhere (D’Angelo and Orcutt, 2019a). From the Echem and Fluid samples (Tables 2, 3), the ITO swab end was cut into a sterile plastic tube and subjected to a freeze-thaw cycle (8 min at −80°C then thawing at room temperature for 10 min or until ice barely melted into water) before transferring into a FastDNA kit tube for DNA extraction. For the SB and NP12 samples, 0.232 μg/μl polyadenine (polyA, final concentration) was added to the sample at the first step to increase DNA yield from low biomass samples. For each batch of samples extracted, a no template control (NTC) was included as a check for contamination. DNA concentrations were measured by Q-bit Fluorometer 3.0 (Thermo Fisher Scientific, Waltham, MA, United States) using the Qubit HS dsDNA kit (Thermo Fisher Scientific, Waltham, MA, United States) according to manufacturer instructions. Note that inclusion of polyA interferes with DNA quantification following this approach (Table 3).
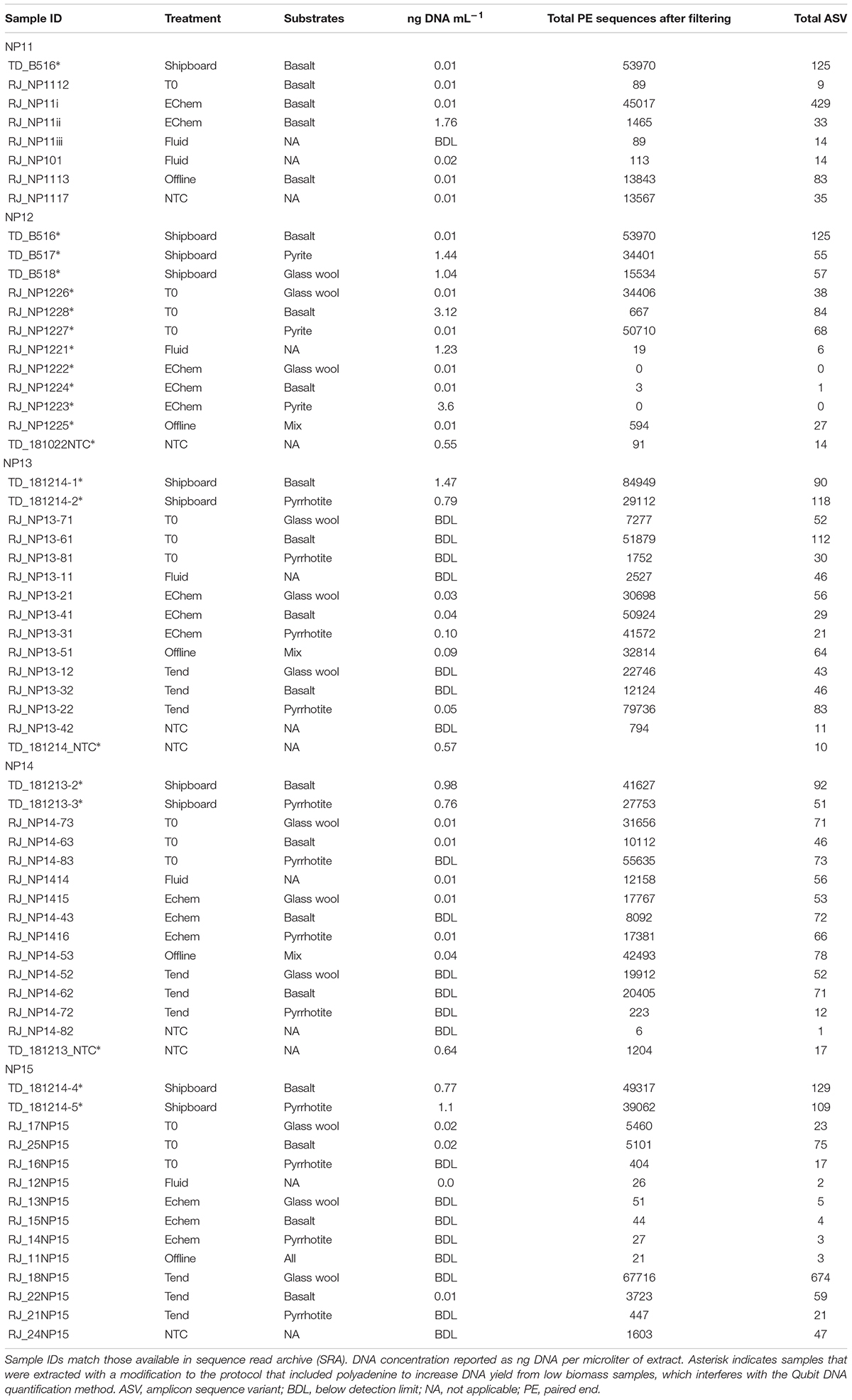
Table 3. Summary of DNA extractions and 16S rRNA gene amplicon sequence analysis (after quality control filtering) for each experiment and treatment, including in the original samples after collection and direct preservation on the ship (shipboard or SB), at the start of the experiment after 1+ year of sample storage (T0), on the ITO electrode at the end of the experiment (EChem), on the sample material in the WE cell at the end of the experiment (Tend), in an electrochemical cell with only sterile fluid (Fluid), in an electrochemical cell without voltage applied (Offline), and in a no template negative DNA extraction controls run with the same batch of samples (NTC).
Aliquots of DNA extracts were sent to the Integrated Microbiome Resource facility at Dalhousie University (Halifax, Canada) for sequencing of the V4–V5 hypervariable region of the 16S rRNA gene via Illumina Mi-Seq technology using the 515F/926R primer pair (Walters et al., 2015; Parada et al., 2016). Sequencing was performed on an Illumina MiSeq using 300 × 300 bp paired-end V3 chemistry.
Raw sequenced reads were processed at the same time as sequenced reads from mineral incubation experiments taken from the same environments, as well as additional NTC samples (data not shown, but full data available at the NCBI Sequence Read Archive BioProject PRJNA564565 samples SAMN12723399-489), using modifications of the standard DADA2 pipeline Version 1.12 (Callahan et al., 2016) to construct unique amplicon sequence variants (ASVs). This was done to directly link ASVs between the two datasets and to give the DADA2 algorithm the most possible data to use for sequence inference. Thirty base-pairs were trimmed off of the start of each read, forward reads were truncated at 250 base-pairs, and reverse reads were truncated at 200 base-pairs. This allowed for 50 BP of overlap for the forward and reverse reads to be merged after sequence inference. Approximately 100 million bases from ∼500K reads were used by DADA2 to infer the base-call error rates in the data. Inference of ASVs, merging of forward and reverse reads, and chimera removal resulted in 2,011 ASVs ranging from 345 to 360 BP in length. These sequences were assigned taxonomy by the naïve Bayesian classifier built into the DADA2 package using the Silva v132 database. Scripts used to process the data are available via https://github.com/orcuttlab/northpond.
Results and Discussion
Evidence for Fe2/O2 Cathodic EET in North Pond Crustal Microbial Communities
Only a few of the cathodic continuously poised potential experiments with colonized substrate samples from North Pond incubated in MFCs set at −200 mV vs. SHE (Table 2) resulted in an increase in current density relative to the double-autoclaved crustal fluid only treatment (Figure 3). Current responses with values greater than the fluid only treatments were in basalt or glass wool treatments in the NP11, NP12, and NP14 experiments. While the current responses between the samples and the fluid only treatments only varied by a few μA cm–2, we note that other environmental cathodic EET experiments have observed similar patterns between biotic and abiotic treatments (Li and Nealson, 2015; Rowe et al., 2015; Jangir et al., 2016; Karbelkar et al., 2016; Lam et al., 2019b). In NP11, which was three basalt replicates from the Hole U1382A downhole incubation (Table 2), there was considerable variation between the signals. This may be because of founder effects or from variation in viable cell biomass between sample inoculum. The basalt signal for NP12, which was the same substrate inoculum, is most similar to NP11 replicate iii, suggesting EET-capable biomass is the driver of the observed signals, as these samples came from a similar section of substrate. In some experiments (i.e., NP13 and NP15), the fluid treatment had an increase in current density compared to the samples (Figure 3), suggesting that some hardy microbial groups that survived the extreme pressure/temperature changes of double autoclaving are also capable of EET. However, NP13 and NP15 curves indicated possible electrode interference, so this interpretation should be viewed with caution. Specifically, the NP13 Fluid reference electrode drifted out of calibration during the course of the incubation (data not shown), and the NP15 Echem basalt treatment exhibited a very noisy profile until day 5. The MFCs incubated with metal sulfide substrates (i.e., pyrite in NP12 and pyrrhotite in NP13-15) barely displayed any current response. Cumulatively, these results indicate that: (1) microbial groups capable of cathodic EET at −200 mV versus SHE (roughly the Fe2/O2 redox coupling) are present in the crustal subsurface, but at heterogeneous density; and (2) that cathodic EET-capable microbial groups may still exist in cold and oxic crustal fluids that are subjected to intense heating (i.e., double autoclaving).
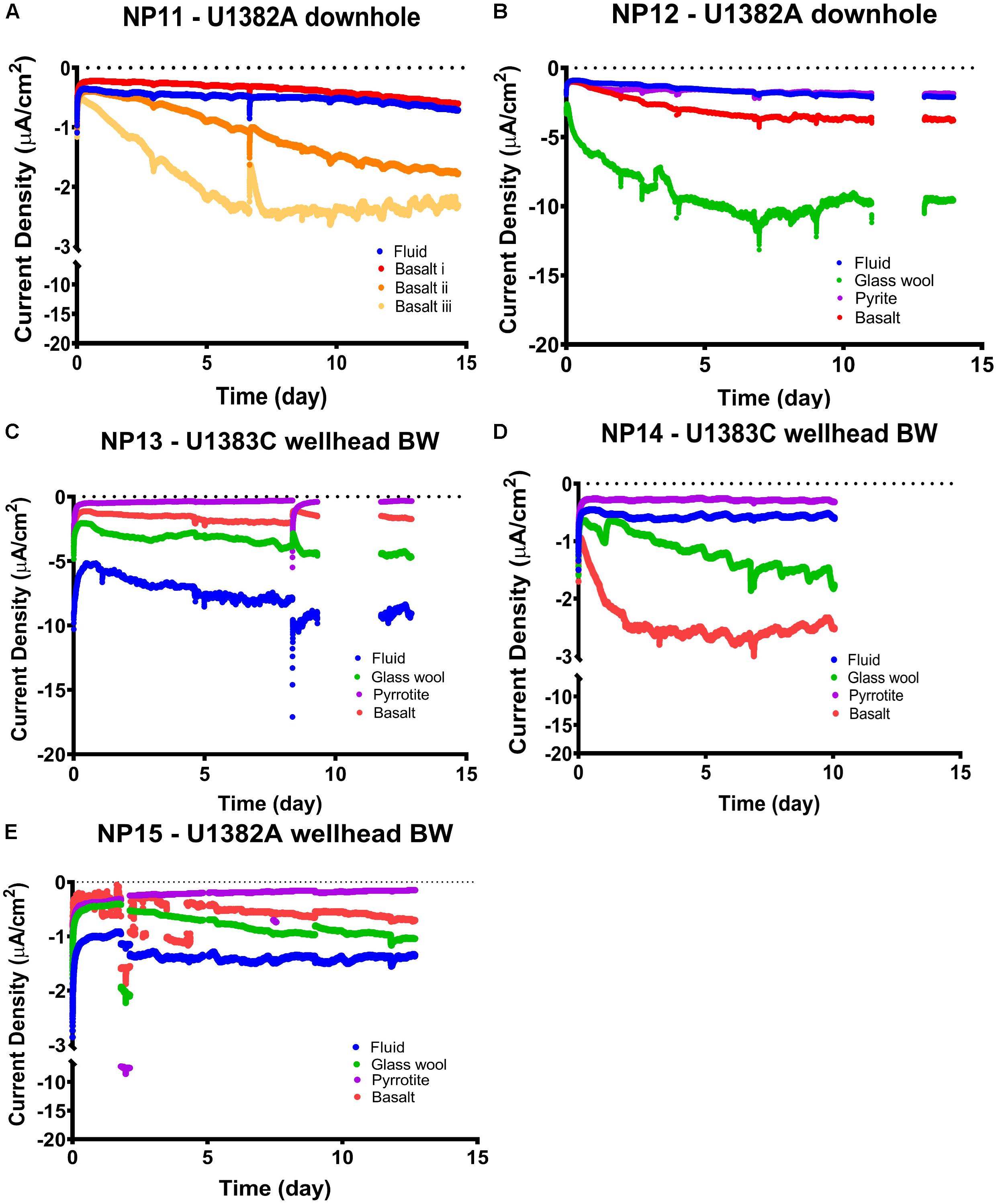
Figure 3. Chronoamperometry profiles of current (normalized to electrode surface area) versus incubation time in cathodic poised potential experiments at –200 mV (versus standard hydrogen electrode), with channels and conditions as listed as in Tables 1, 2. (A) NP11, (B) NP12, (C) NP13, (D) NP14, and (E) NP15. Breaks in data records indicate periods when data were not recorded due to glitches in software. Note the breaks in the y-axes in the NP11, NP14, and NP15 experiments, and that ‘-’ here denotes current direction and not mathematical negative so increasingly negative values represent increasing current density of the electron flow direction out of the electrode.
The strongest current response of ∼10 μA cm–2 increase compared to the control was observed in the glass wool treatment from the U1382A downhole samples (i.e., experiments NP11 and NP12; Figure 3). This result was unexpected, given the inert nature of the colonized substrate compared with basalt or metal sulfides. However, the glass wool samples in the FLOCS were located downstream of the cassettes containing basalts and metal sulfides, possibly functioning as a sieve that concentrated biomass exiting those samples. Supporting this assessment, the lower starting current for this sample may be an indicator of higher starting biomass for this treatment.
It is notable that the current response from the two experiments with downhole-incubated FLOCS samples (i.e., experiments NP11 and NP12) had similar or increased current density responses to those observed in the wellhead bottom-seawater-incubated FLOCS (Figure 3); despite the wellhead samples having roughly one order of magnitude more starting material in the MFCs compared with the downhole samples (Table 2). This indicates a lower amount of EET-capable microbial groups in the bottom seawater inoculated samples versus the downhole samples. We attribute this difference to a presumed selection for EET-capable microbial groups in the crustal subsurface compared to bottom seawater. There also may be an effect from the different incubation times on the colonizing biofilms in the FLOCS (i.e., 4 years or bottom water FLOCS versus 6 years for downhole FLOCS; Table 2), which allowed more EET-capable groups to colonize in the subsurface samples.
Considering the probable low starting biomass on crustal subsurface FLOCS [i.e., crustal fluid at this site has cell densities of 103–104 cells ml–1 (Meyer et al., 2016; Tully et al., 2018)] and rock core samples are estimated at 104 cells per gram rock (Jørgensen and Zhao, 2016), it is remarkable that any current response was detected from these crustal subsurface samples, and at current densities comparable to much shallower and higher cell density marine environments (Lam et al., 2019b). This is particularly so given the presumed low-energy, low-activity nature of the deep marine crustal subsurface microbial community. Current response was most rapid in the first 5 days of the experiment, leveling off for the remainder of the experiment (Figure 3). However, previous work (Meyer et al., 2016) also noted a quick response to incubation conditions by the North Pond crustal fluid microbial community, proposing this as evidence that the microbes were poised to quickly take advantage of transient resources as an adaptive advantage in their resource limiting environment. This suggests that metabolic activity, rather than growth of cells, explains this current response, similar to prior experiments (Deng and Okamoto, 2018). Activity rather than growth in these experiments makes sense given that no additional resources (i.e., vitamins, minerals, and carbon) were provided in the MFC media. EET-capable microorganisms are known to need additional organic carbon (Koch and Harnisch, 2016).
In each experiment, metal sulfides had the weakest current increase, often almost indistinguishable from the control (Figure 3). This is contrary to expectation, as pyrite and pyrrhotite are more reactive than basalt, providing a more favorable substrate for iron oxidation. For comparison, in an anoxic crustal subsurface environment, metal sulfide samples had higher cell densities than basalt samples (Orcutt et al., 2011a), and sulfides have also been observed to be preferentially colonized at the seafloor (Edwards et al., 2003b; Orcutt et al., 2011a). The result of our experiment suggests that: (1) replicating the Fe2/O2 redox couple with poised potential at −200 mV may be impacted by sulfide from the metal sulfides, (2) microbial communities colonizing the metal sulfides preferred other redox reactions than those involving Fe2/O2, (3) in situ EET-capable microbial communities preferentially colonize the dominant substrate versus minor components of the crust at this site (i.e., basalt is more prevalent than metal sulfides at North Pond: Expedition 336 Scientists, 2012), or (4) that EET-capable microbes on pyrrhotite may have remained attached to the mineral and did not migrate to the electrode. Additionally, when pyrrhotite oxidizes, it preferentially loses iron in dissolved form (Belzile et al., 2004), making it less attractive to iron oxidizers that require surface-bound iron.
Evidence of Redox Reactions in Cathodic EET-Capable Microbial Communities From Cyclic Voltammetry
Cyclic voltammetry scans of attached biomass were performed immediately after CA scans to gain insight into conductive pathways of attached biomass and/or in shuttle molecules released into the fluid (Figure 4). Scan configurations were characteristic of voltammetry in the absence of substrate (LaBelle and Bond, 2009), adding to evidence that these communities were resource limited in this experiment. In this configuration, peaks in a scan correspond to an EET mechanism. Peaks in these curves represent the mid-point potential of a given mechanism, essentially the optimum voltage at which an EET mechanism functions (LaBelle and Bond, 2009).
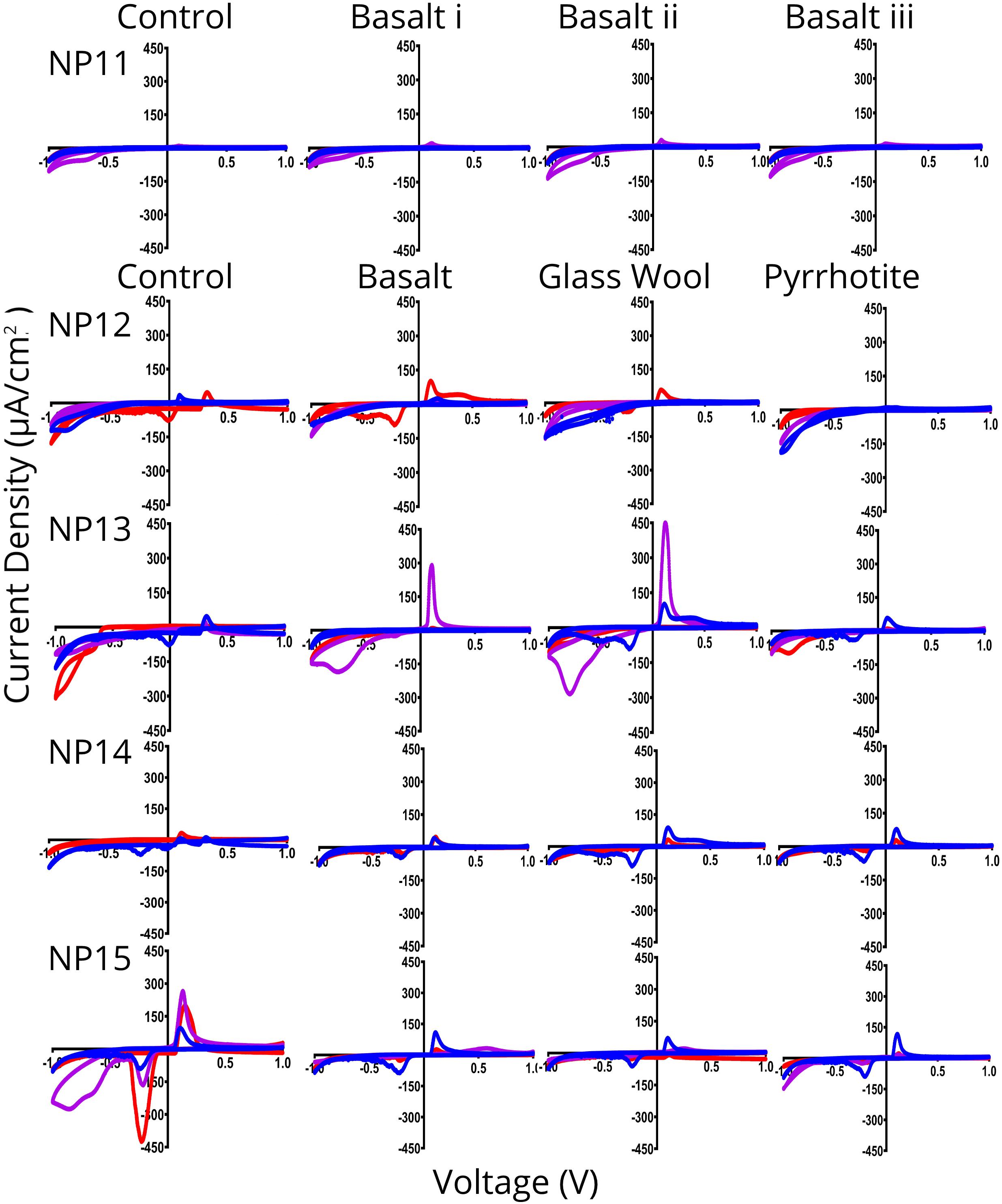
Figure 4. Cyclic voltammetry (CV) scans of indium tin oxide (ITO) electrodes from cathodic poised potential experiments. In each plot, red lines are scans of electrodes immediately before incubation, blue lines are scans of electrodes immediately after incubation, and purple scans are from fresh ITO electrodes inserted into incubated media. Overall, electrodes have current peaks at –0.8, –0.35, and 0.2 V vs. Ag/AgCl on both the incubated ITO electrode at the end of experiment (blue) and on a fresh ITO electrode inserted into incubated media (purple), indicating redox couples likely equivalent to H2/O, Fe2/O2, and an as yet unknown mechanism, respectively. Note that ‘-’ here indicates current and voltage direction and not mathematical negative so increasingly negative values represent increasing current density in the opposite electron flow direction. Plots are average of triplicate CV scans at 0.1 V/s (duplicate for NP11 Tend i, NP11 Tend ii, NP12 Tend i, and NP13 Tend i).
Cyclic voltammetry scans show similar patterns across the experiments, with current density peaks at approximately −0.8, −0.25, and +0.1 V vs. Ag/AgCl except for NP12 and NP13 fluid, which had peaks at −0.65 and +0.35 V in the Tend i and −0.8 and +0.1 in the Tend ii with fresh electrode (Figure 4). This indicates that there is some EET activity in the fluid control despite double-autoclaving and taking measures to ensure sterility. It is less likely that abiotic reactions are responsible for these peaks, as these would be more similar to each other if there were no biology involved. The CV-observed EET activity in the controls is particularly strong in the NP12, NP13, and NP15 fluid scans, which correlates with higher CA current density in those same samples (Figure 3).
In some instances – the NP11, NP2, and NP15 curves particularly – initial experiment scans have a peak at +0.1 V that disappears or is overreached in Tend curves, indicating that there was an abiotic reaction present in the initial seawater that changes by the end of the experiments. These changes are likely due to changes in chemistry in response to the microbial communities present.
NP11, NP12 basalt, NP13 pyrrhotite, and NP15 curves, with the exception of the fluid control, show a Tend i CV curve with the old electrode that has higher peaks than that of the Tend ii CV curves with fresh electrode. This indicates that biofilm organisms with direct contact are responsible for the EET in these cells. All other samples showed a higher current response at the end of an experiment with a fresh electrode. This indicates that planktonic cells and extracellular shuttle mechanisms were more EET capable in these experiments. These differences in capability between fractions of a single environment are noted elsewhere, but remain an understudied part of microbe-EET understanding (LaBelle and Bond, 2009; Lam et al., 2019b). This result is perhaps surprising given that releasing electron shuttles would seem an expensive strategy in the low-resource deep marine crustal subsurface (Jones et al., 2018).
The CV peak occurring at approximately −0.25 V vs. Ag/AgCl is likely indicating the mechanism that is giving the electrode community energy, as the CA experiment voltage is set to −0.4 V vs. Ag/AgCl (−0.2 V vs. SHE). From the CV scans, it appears that the set voltage is not optimum for this community, however. Overpotential may play a role in this and having a peak close to but not a match for a CA voltage is also seen with isolation of EET capable microbes from other environmental samples (Rowe et al., 2015; Jangir et al., 2016; Lam et al., 2019b). This peak, in combination with that of the +0.1 V vs. Ag/AgCl, may also be indicative of cytochrome-based EET (Bewley et al., 2013).
There is a diffuse peak between −0.9 and −0.7 V vs. Ag/AgCl. This peak covers the range at which hydrogen is abiotically generated but is in addition to the abiotic signal (Deng and Okamoto, 2018). This may indicate a capacity for using hydrogen as a source of electrons. The capacity to use hydrogen as an electron donor is widespread among acidophilic iron-oxidizers (Hedrich and Johnson, 2013) and there is also some evidence that this metabolism is present in neutrophilic iron-oxidizers (Mori et al., 2017), indicating a possible alternative explanation. This peak is also associated with some environmental iron-oxidizers (Lam et al., 2019b), though these appear to have a wide range of potential redox values they can use (Summers et al., 2013).
In many of our experiments, there was a prominent peak at approximately +0.1 V vs. Ag/AgCl, indicating that this community has the capacity to directly use electrodes as a terminal electron acceptor. Anaerobic metabolisms should be unfavorable in the bulk oxic conditions of North Pond, but this result may indicate anaerobic microniche in this habitat that support this type of metabolism. It may be that microbial groups with anaerobic metabolic potential are more well adapted to take advantage of anaerobic micro-niches within the cracks and crevices of the fluid flow system, or that the conditions within the FLOCS facilitated micro-anaerobic pockets within the sample chambers. This evidence corroborates other work on the microbial community at this site, which observed taxa and metabolisms connected to anaerobic processes (Tully et al., 2018). As the ability to perform both anodic and cathodic electron transfer is identified in as many as 20 EET species to date (Koch and Harnisch, 2016; Rowe et al., 2018), our results suggest that the co-occurrence of solid substrate oxidation and reaction metabolisms in the same microbial communities, as we observe in our experiments, is not out of the ordinary. Future anodic experiments could explore these metabolisms in the crustal subsurface in more detail.
Morphology of Cathode-Attached Microbial Groups
Viewed under SEM, the morphology of particles on the Echem sample ITO electrodes is consistently ultra-small coccoids of 100–300 nm diameter (example provided in Figure 5, all data available in BCO-DMO dataset). These ultrasmall coccoids are not seen in the fluid-only treatments, though spheres of <100 nm are occasionally present on those. A total of 100–300 nm coccoids without observable attachment structures are also present on polished chips of basalt and pyrrhotite/pyrite from the downhole experiments at this same site (data available at https://www.bco-dmo.org/dataset/756152). These polished mineral coupons are not directly comparable to the poised potential experiments described here, as the bioelectrochemical samples were crushed mineral sands and not polished chips. They are from the same locations and are comparable minerals, however, so give some idea of morphological diversity.
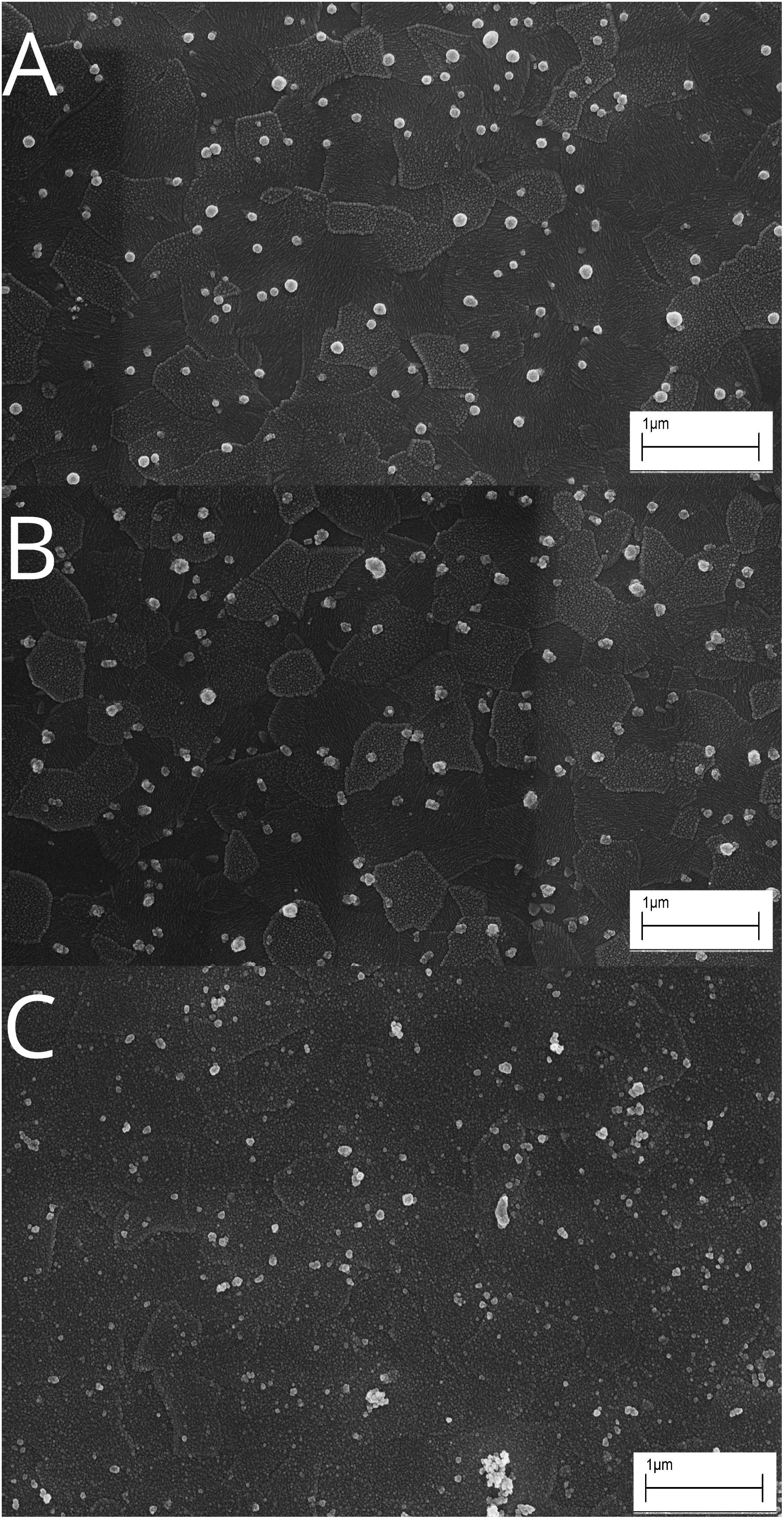
Figure 5. Representative scanning electron microscopy (SEM) images of particle morphology observed on indium tin oxide (ITO) electrodes in NP14 Glass Wool (A), NP14 Basalt (B), and NP14 fluid control (C) on termination of incubations indicates very small coccoid-shaped particles, possibly cells, adhering to the electrodes in the live samples. Scale bars 1 mm.
We presume that these small coccoids are cells that have colonized the electrodes. If correct, the size of these cells would classify them as ultra-small bacteria (Ghuneim et al., 2018). If these are cells, this very small size may indicate adaption to resource limited conditions of the deep crustal environment, as suggested for other resource limited environments (Schut et al., 1993; Kuhn et al., 2014), or it may indicate cells in a starvation configuration (Álvarez et al., 2008). A starvation response is consistent with the chronoamperometry profiles showing limited current response after the first few days, which seem to indicate activity rather than growth.
No pili or attachment structures were detected in any SEM image, indicating that electron transport does not happen through conductive structures. Electron transport is therefore likely through direct contact of some mechanism embedded in the cell wall or via electron shuttle mechanisms. There is currently too little information to speculate on what these cathode-oxidizing mechanisms might be, beyond the CV peaks showing little similarity to flavin-mediated mechanisms (Marsili et al., 2008).
Microbial Groups in North Pond Crustal Subsurface Sample Capable of Cathodic EET
The low biomass expected from the FLOCS samples (Ramírez et al., 2019), as well as on the ITO electrodes, necessitated a stringent protocol of including numerous no template extraction controls (NTCs) for bulk DNA extraction and sequencing (Labonté et al., 2017; Sheik et al., 2018; Ramírez et al., 2019). Instead of simply removing any sequence groups that appeared in any NTC from sample sequence libraries, we present the full results of all taxonomic groups in all samples including NTCs (Figures 6, 7), with samples grouped by treatment type rather than by experiment, to show trends more clearly.
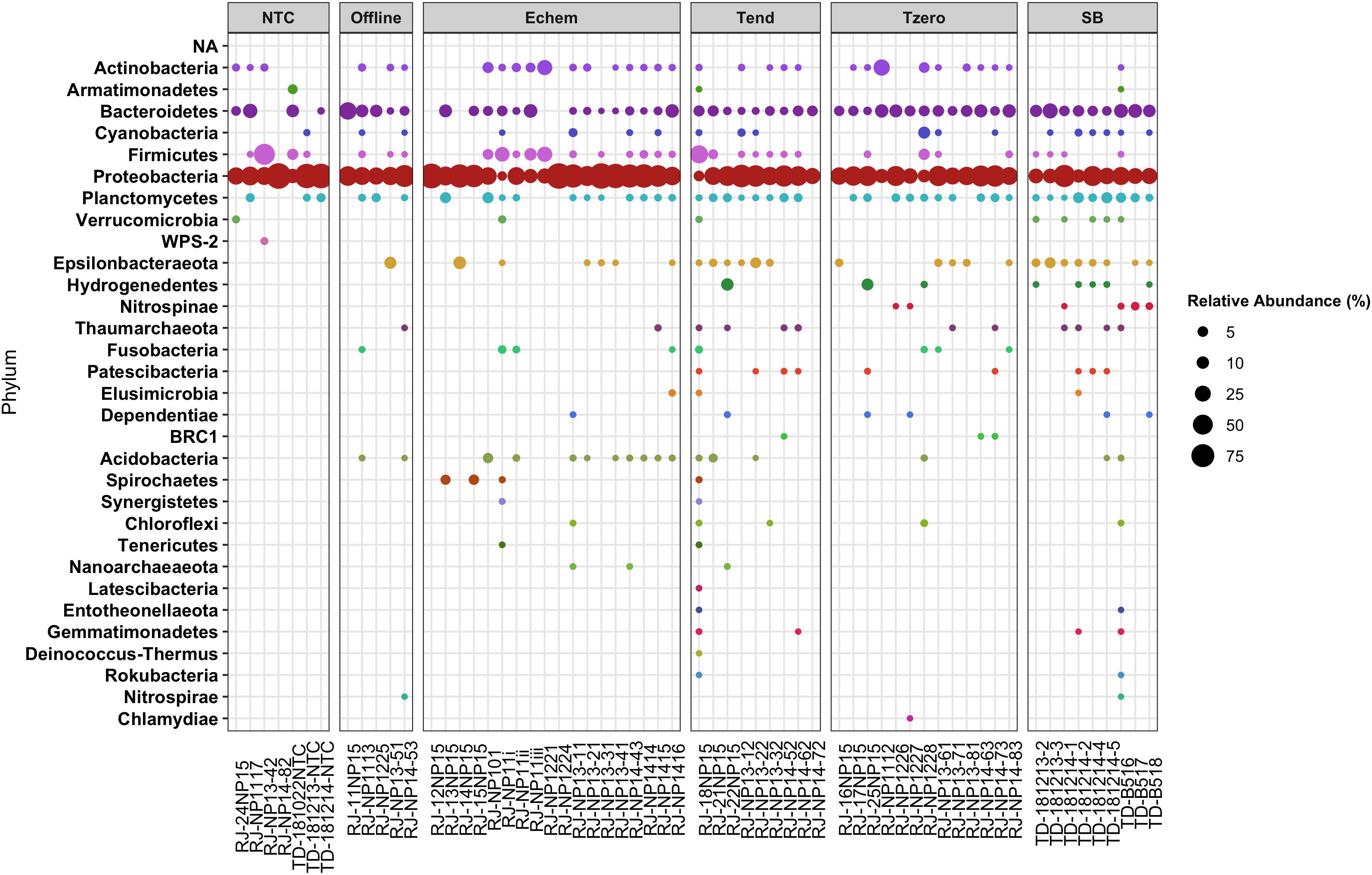
Figure 6. Relative abundance of taxonomic groups at the Phyla level in poised potential experiment samples, based on 16S rRNA gene (V4–V5 hypervariable region) Illumina sequence data, with sample names matching those provided in Table 2. Sample order grouped by Treatment type, and taxonomic groups sorted as taxa that appear at least once in a no-template-control blank DNA extraction versus those that do not.
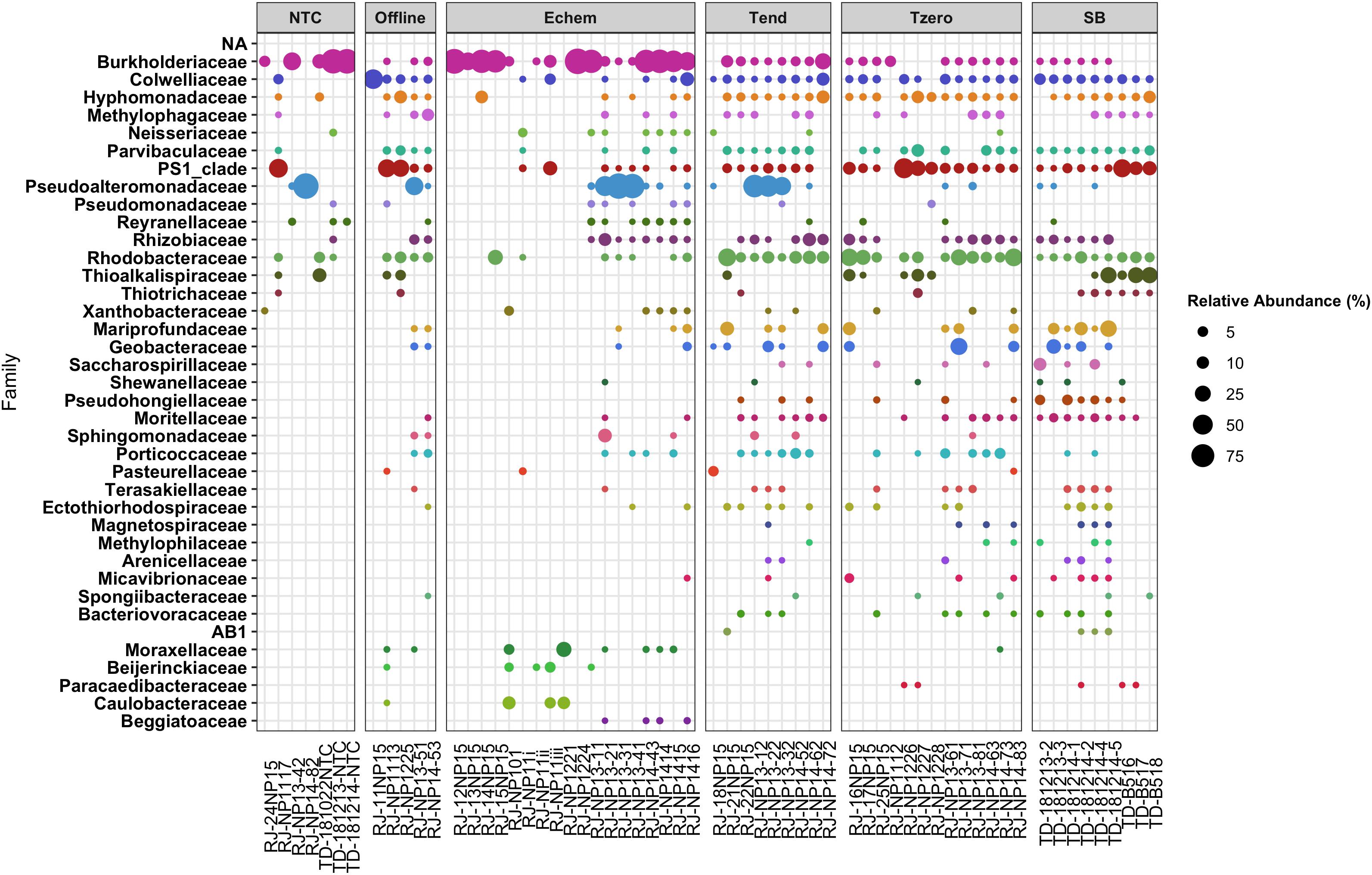
Figure 7. Relative abundance of Proteobacteria taxonomic families as a subset of data shown in Figure 6.
Overall, the Echem ITO electrode swabs revealed that the dominant taxonomic groups in the sequence data overlapped with the dominant groups in the NTC’s sequence libraries (Figures 6, 7). This suggests overamplification of DNA contaminants in these extremely low biomass samples, as is known to occur (Chandler et al., 1997; Sheik et al., 2018; Ramírez et al., 2019), swamping out any signal of any cells that may have attached to the ITO electrodes. It may also indicate cross-contamination from the samples into the NTCs during sample handling, as some of the taxonomic groups that appear in the NTC samples are not “typical” sequence contaminant groups, such as Thioalkalispiraceae (Figure 7). Because of these uncertainties in provenance, taxonomic groups that appear in the NTCs are not discussed further.
Of the remaining taxonomic groups not observed in the NTCs, the majority of these groups observed in the Echem samples were also observed on the ITO electrodes incubated in the Offline controls without voltage applied (Figures 6, 7). This indicates cathodic EET did not enrich these taxa.
Only a small fraction of the total sequences in this study are found on the Echem samples but not in the NTCs or Offline treatments (Figures 6, 7), which may indicate that they are involved in the cathodic EET observed in the experiments. One of the basalt treatments from the U1382A downhole NP11 experiment (RJ_NP11i from Table 3) had a few sequences grouping within the Spirochaetes, Synergistetes, and Tenericutes phyla (Figure 6). The Spirochaetes phyla was also observed in the sequence libraries from the bottom seawater NP15 experiment Echem samples from basalt and glass wool (RJ_15NP15 and RJ_13NP15), and it was also enriched in a NP15 Tend sample (RJ_18NP15) compared to SB or T0 samples where it is absent or very rare. The NP15 experiment had current response that was inconclusive for indicating EET activity, so it is not clear if this phylum is responding to stimulating conditions for cathodic EET or to some other factor. Spirochaetes members have been identified elsewhere in crust-associated subsurface sediment (Liao et al., 2011) and can perform acetogenesis (Leadbetter et al., 1999; Miura et al., 2016), which is a known EET-associated metabolism (Yang et al., 2012; Igarashi and Kato, 2017). They were also detected in a shallow subsurface sediments EET study (Lam et al., 2019b). Flavins are noted as potentially metabolically important in some Spirochaetes (Gutiérrez-Preciado et al., 2015; Radolf et al., 2016) which are a known EET mechanism in other phyla (Yang et al., 2012; Xu et al., 2016; Zhao et al., 2019). However, characteristic flavin-based peaks, as described elsewhere (Marsili et al., 2008; Fuller et al., 2014; Light et al., 2018), are absent from in our data. There are also marine Synergistetes that can metabolize hydrogen (Jumas-Bilak and Marchandin, 2014; Miura et al., 2016), but these are described as strict anaerobes and are not described as acetogens specifically (Jumas-Bilak and Marchandin, 2014). Tenericutes members occur in a bioelectrochemical study, though little information is available on their specific role (Liu et al., 2019). These groups have yet to be investigated for EET capability directly.
The Fluid treatment from the NP13 experiment (RJ-NP13-11), which had the strongest current response of all samples in that experiment (Figure 3) despite going through double autoclaving, had sequences falling into this non-NTC/non-Offline category that grouped within the Chloroflexi and Nanoarchaeota phyla (Figure 6). Chloroflexi strain Ardentica maritima 110 is EET capable (Kawaichi et al., 2018). Nanoarchaeota-associated sequences were also detected in the NP14 Echem sample from basalt (RJ_NP14_43), which also had the strongest current response in that experiment (Figure 3). There are no specific EET studies involving Nanoarchaeota, though they have a gene for cytochrome bd-I ubiquinol oxidase subunit I, a protein that shuttles electrons from the host’s membrane coupled to O2 reduction (Jarett et al., 2018). A syntrophic quinone electron transfer mechanism has been shown to support Nanoarchaeota in co-culture (Smith et al., 2015) and interspecies electron transfer is a known mechanism (Lovley, 2017) as are using cytochromes as an EET mechanisms (Rosenbaum et al., 2011; Santos et al., 2015). CV peaks that may indicate a cytochrome mechanism (Bewley et al., 2013) are present in our data. Other archaea show some evidence of EET capability (Uria et al., 2017). Particle size and coccoid morphology observed by SEM are also consistent with Nanoarchaeota members (Huber et al., 2003). This may indicate that Nanoarchaeota are involved in the cathodic EET activity observed in those experiments.
The NP13 glass wool sample (RJ_NP13-21), which showed the second strongest current response in that sample (Figure 3), had a small fraction of Shewanellaceae-associated sequences which were not observed in the NTCs or offline samples (Figure 7). This taxon was also observed in a few SB, T0, and Tend samples, indicating that this group is a crustal generalist. Shewanellaceaea are well known for their ability for anodic and cathodic EET across multiple environments (Lovley, 2012; Rowe et al., 2018; Zou et al., 2019). Our results suggest that Shewanellacaea can also perform similar electron transfer reactions in the crustal subsurface.
Several Echem samples had Beggiatoaceae-associated sequences that did not appear in any other samples (Figure 7). This taxon is commonly detected on sediment surfaces and at shallow depths rather than the subsurface (Amend and Teske, 2005; Jørgensen et al., 2010; Teske et al., 2016), so it is a rare member of the crustal deep biosphere that may have responded to the cathodic EET stimulating conditions of this experiment. No EET study has specifically looked at Beggiatoaceae, and it is very rarely mentioned in a bioelectrochemical context. Cultured members of this Proteobacteria family are sulfide oxidizers that can use oxygen or nitrate as a terminal electron acceptor. Beggiatoecae also metabolize acetate (Burton et al., 1966; Güde et al., 1981; Strohl et al., 1981), which may be being generated on the cathode as a product of acetogenesis (Min et al., 2013) or possibly as a waste product from EET-capable microbes.
Finally, the NP14 Echem sample with pyrite (RJ_NP1416), which did not show any current response (Figure 3), had non-NTC/non-Offline sequences grouping within the Elusimicrobia phylum (Figure 6) and the Micavibrionaceae family of Proteobacteria (Figure 7). As this experiment did not indicate EET current response, it is not likely that these taxa are involved in EET.
Conclusion
These results indicate that solid–substrate-utilizing EET-capable microbes are present in the oxic crustal subsurface and the deep sea, but those that potentially oxidize iron with this mechanism are likely minor overall contributors to the general community despite the energetic advantages to do so. It may also be that this mechanism is one of many of these microbes are capable of, reflective of a strategy of taking advantage of resources as they become available in North Pond, as suggested elsewhere (Tully et al., 2018). This may also be an example of an available niche that is not well filled (Cockell et al., 2016). Moreover, if the cathodic EET-capable microbial groups require organic carbon, then the extreme carbon limitation of this oligotrophic basalt environment (Mason et al., 2009) is a limiting factor to this subset of the North Pond community, as suggested for other oligotrophic environments (Morono et al., 2011; Ziebis et al., 2012). Despite the evidence for cathodic EET reactions, this incubation approach was not successful for conclusively determining for which members of the low biomass crustal subsurface are responsible for these reactions, although there are indications that Nanoarchaeota and Shewanellaeceae taxa may be involved. Overall, this study shows that cathodic EET-capable microbial groups may be more widespread in the general environment than previously considered, and that this enrichment approach to searching for them can be useful to quantify their activity and metabolic potential in challenging environments such as the cool, oxic basalt of North Pond.
Data Availability Statement
Chronoamperometry, CV, and SEM data are available from the Biological and Chemical Oceanography Data Management Office (BCO-DMO) under project 707762: https://www.bco-dmo.org/dataset/780261 (SEM), https://www.bco-dmo.org/dataset/780127 (CA profiles), https://www.bco-dmo.org/dataset/780248 (CV profiles), and https://www.bco-dmo.org/dataset/780225 (experimental metadata). 16S rRNA gene sequence data are available at the National Center for Biotechnology’s Sequence Read Archive BioProject PRJNA564565 samples 274 SAMN12723345–SAMN12723415.
Author Contributions
RJ and BO formulated the original hypotheses, designed the project, and wrote the manuscript with contributions from TD’A. All authors participated in the sample collection, data analysis, and interpretation. RJ performed the experiments. RJ and TD’A performed the DNA extractions and bioinformatic analysis.
Funding
This work was supported by the U.S. National Science Foundation (NSF, awards OCE-1536539 and OCE-1737017 to BO and funding for IODP Expedition 336, involvement in cruise MSM37, and funding support for cruise AT39-01), the NSF-funded Center for Dark Energy Biosphere Investigations Science and Technology Center (C-DEBI, OCE-0939564), and the NASA Astrobiology Institute Life Underground program (NNA13AA92A). This is C-DEBI contribution number 503.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the AT39-01 science party, the ROV Jason party, and the ship’s crew for their assistance in recovering the experiments, including Geoff Wheat as a chief scientist, Kelle Freel and Clarisse Sullivan for operating the mobile pumping system, Olivia Nigro for ultrafiltered crustal fluids, and Jackie Goordial for sample assistance. Silica and manganese concentration data were kindly provided by Geoff Wheat. This project would not have been possible without the Integrated Ocean Discovery Program (IODP) Expedition 336, the Gordon and Betty Moore Foundation supporting the CORK borehole observatory infrastructure installed in 2011, the help of Amanda Haddad and Samuel Hulme in deploying the downhole and wellhead experiments, and Michael Rappé for provisioning the Mobile Pumping System. We also thank Heiner Villinger, members of the MSM37 science party, and the DFG German Science Foundation for supporting the deployment of the wellhead experiments in 2014, aided by Jean Paul Baquiran and Stephanie Carr. Finally, we thank Peter Countway for use of laboratory equipment and André Comeau for assistance with sequencing our low biomass DNA samples.
References
Álvarez, B., López, M. M., and Biosca, E. G. (2008). Survival strategies and pathogenicity of Ralstonia solanacearum phylotype II subjected to prolonged starvation in environmental water microcosms. Microbiology 154, 3590–3598. doi: 10.1099/mic.0.2008/019448-0
Amend, J. P., and Teske, A. P. (2005). Expanding frontiers in deep subsurface microbiology. Palaeogeogr. Palaeoclimatol. Palaeoecol. 219, 131–155. doi: 10.1016/j.palaeo.2004.10.018
Bach, W., and Edwards, K. J. (2003). Iron and sulfide oxidation within the basaltic ocean crust: implications for chemolithoautotrophic microbial biomass production. Geochim. Cosmochim. Acta 67, 3871–3887. doi: 10.1016/S0016-7037(03)00304-1
Badalamenti, J. P., Summers, Z. M., Chan, C. H., Gralnick, J. A., and Bond, D. R. (2016). Isolation and genomic characterization of ‘desulfuromonas soudanensis WTL’, a metal- and electrode-respiring bacterium from anoxic deep subsurface brine. Front. Microbiol. 7:913. doi: 10.3389/fmicb.2016.00913
Baquiran, J. P. M., Ramírez, G. A., Haddad, A. G., Toner, B. M., Hulme, S. M., Wheat, C. G., et al. (2016). Temperature and redox effect on mineral colonization in Juan de Fuca ridge flank subsurface crustal fluids. Front. Microbiol. 7:396. doi: 10.3389/fmicb.2016.00396
Barco, R. A., Hoffman, C. L., Ramírez, G. A., Toner, B. M., Edwards, K. J., and Sylvan, J. B. (2017). In-situ incubation of iron-sulfur mineral reveals a diverse chemolithoautotrophic community and a new biogeochemical role for thiomicrospira. Environ. Microbiol. 19, 1322–1337. doi: 10.1111/1462-2920.13666
Belzile, N., Chen, Y.-W., Cai, M.-F., and Li, Y. (2004). A review on pyrrhotite oxidation. J. Geochem. Explor. 84, 65–76. doi: 10.1016/j.gexplo.2004.03.003
Bewley, K. D., Ellis, K. E., Firer-Sherwood, M. A., and Elliott, S. J. (2013). Multi-heme proteins: nature’s electronic multi-purpose tool. Biochim. Biophys. Acta Bioenerg. 1827, 938–948. doi: 10.1016/j.bbabio.2013.03.010
Bond, D. R., Holmes, D. E., Tender, L. M., and Lovley, D. R. (2002). Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295, 483–485. doi: 10.1126/science.1066771
Bradley, J. A., Amend, J. P., and LaRowe, D. E. (2018). Necromass as a limited source of energy for microorganisms in marine sediments. J. Geophys. Res. Biogeosci. 123, 577–590. doi: 10.1002/2017JG004186
Braet, F., De Zanger, R., and Wisse, E. (1997). Drying cells for SEM, AFM and TEM by hexamethyldisilazane: a study on hepatic endothelial cells. J. Microsci. 186, 84–87. doi: 10.1046/j.1365-2818.1997.1940755.x
Burton, S. D., Morita, R. Y., and Miller, W. (1966). Utilization of acetate by Beggiatoa. J. Bacteriol. 91, 1192–1200.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chandler, D. P., Fredrickson, J. K., and Brockman, F. J. (1997). Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol. Ecol. 6, 475–482. doi: 10.1046/j.1365-294X.1997.00205.x
Cockell, C. S., Bush, T., Bryce, C., Direito, S. O. L., Fox-Powell, M. G., Harrison, J. P., et al. (2016). Habitability: a review. Astrobiology 16, 89–117. doi: 10.1089/ast.2015.1295
Cowen, J. P., Copson, D. A., Jolly, J., Hsieh, C.-C., Lin, H. T., Glazer, B. T., et al. (2012). Advanced instrument system for real-time and time-series microbial geochemical sampling of the deep (basaltic) crustal biosphere. Deep. Res. Part I Oceanogr. Res. Pap. 61, 43–56. doi: 10.1016/j.dsr.2011.11.004
D’Angelo, T., and Orcutt, B. N. (2019a). DNA extraction using FastDNA spinkit for soil with PolyA modifictaion. Protocols.io. doi: 10.17504/protocols.io.ur8ev9w
D’Angelo, T., and Orcutt, B. N. (2019b). SEM sample preparation. Protocols.io. doi: 10.17504/protocols.io.w9ufh6w
Deng, X., and Okamoto, A. (2018). Electrode potential dependency of single-cell activity identifies the energetics of slow microbial electron uptake process. Front. Microbiol. 9:2744. doi: 10.3389/fmicb.2018.02744
D’Hondt, S., Pockalny, R., Fulfer, V. M., and Spivack, A. J. (2019). Subseafloor life and its biogeochemical impacts. Nat. Commun. 10, 1–13. doi: 10.1038/s41467-019-11450-z
Edwards, K. J., Bach, W., and McCollom, T. M. (2005). Geomicrobiology in oceanography: microbe-mineral interactions at and below the seafloor. Trends Microbiol. 13, 449–456. doi: 10.1016/j.tim.2005.07.005
Edwards, K. J., Bach, W., McCollom, T. M., and Rogers, D. R. (2004). Neutrophilic iron-oxidizing bacteria in the ocean: their habitats, diversity, and roles in mineral deposition, rock alteration, and biomass production in the deep-sea. Geomicrobiol. J. 21, 393–404. doi: 10.1080/01490450490485863
Edwards, K. J., Bach, W., and Rogers, D. R. (2003a). Geomicrobiology of the ocean crust: a role for chemoautotrophic Fe-bacteria. Biol. Bull. 204, 180–185. doi: 10.2307/1543555
Edwards, K. J., Rogers, D. R., Wirsen, C. O., and McCollom, T. M. (2003b). Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing, chemolithoautotrophic - and Proteobacteria from the Deep Sea. Appl. Environ. Microbiol. 69, 2906–2913. doi: 10.1128/AEM.69.5.2906-2913.2003
Edwards, K. J., Wheat, C. G., Orcutt, B. N., Hulme, S. M., Becker, K., Jannasch, H. W., et al. (2012). “Design and deployment of borehole observatories and experiments during IODP expedition 336 Mid-atlantic ridge flank at north nond,” in Proceedings of the Integrated Ocean Drilling Program, eds K. J. Edwards, W. Bach, and A. Klaus (Tokyo: Integrated Ocean Drilling Program Management International Inc.), doi: 10.2204/iodp.proc.336.109.2012
Expedition 336 Scientists (2012). “Site U1383,” in Proceedings of the Integrated Ocean Drilling Program, eds K. J. Edwards, W. Bach, and A. Klaus (Tokyo: Integrated Ocean Drilling Program Management International Inc.), doi: 10.2204/iodp.proc.336.105.2012
Früh-Green, G. L., Orcutt, B. N., Rouméjon, S., Lilley, M. D., Morono, Y., Cotterill, C., et al. (2018). Magmatism, serpentinization and life: insights through drilling the Atlantis massif (IODP expedition 357). Lithos 323, 137–155. doi: 10.1016/j.lithos.2018.09.012
Fuller, S. J., McMillan, D. G. G., Renz, M. B., Schmidt, M., Burke, I. T., and Stewart, D. I. (2014). Extracellular electron transport-mediated Fe(iii) reduction by a community of alkaliphilic bacteria that use flavins as electron shuttles. Appl. Environ. Microbiol. 80, 128–137. doi: 10.1128/AEM.02282-13
Ghuneim, L.-A. J., Jones, D. L., Golyshin, P. N., and Golyshina, O. V. (2018). Nano-sized and filterable bacteria and archaea: biodiversity and function. Front. Microbiol. 9:1971. doi: 10.3389/fmicb.2018.01971
Gregory, K. B., Bond, D. R., and Lovley, D. R. (2004). Graphite electrodes as electron donors for anaerobic respiration. Environ. Microbiol. 6, 596–604. doi: 10.1111/j.1462-2920.2004.00593.x
Güde, H., Strohl, W. R., and Larkin, J. M. (1981). Mixotrophic and heterotrophic growth of Beggiatoa alba in continuous culture. Arch. Microbiol. 129, 357–360. doi: 10.1007/BF00406462
Gutiérrez-Preciado, A., Torres, A. G., Merino, E., Bonomi, H. R., Goldbaum, F. A., and García-Angulo, V. A. (2015). Extensive identification of bacterial riboflavin transporters and their distribution across bacterial species. PLoS One 10:e0126124. doi: 10.1371/journal.pone.0126124
Hazrin-Chong, N. H., and Manefield, M. (2012). An alternative SEM drying method using hexamethyldisilazane (HMDS) for microbial cell attachment studies on sub-bituminous coal. J. Microbiol. Methods 90, 96–99. doi: 10.1016/j.mimet.2012.04.014
Hedrich, S., and Johnson, D. B. (2013). Aerobic and anaerobic oxidation of hydrogen by acidophilic bacteria. FEMS Microbiol. Lett. 349, 40–45. doi: 10.1111/1574-6968.12290
Hinks, J., Zhou, M., and Dolfing, J. (2017). “Microbial Electron Transport in the Deep Subsurface,” in Microbial Ecology of Extreme Environments. Cham: Springer International Publishing, 81–102. doi: 10.1007/978-3-319-51686-8_4
Huber, H., Hohn, M. J., Stetter, K. O., and Rachel, R. (2003). The phylum Nanoarchaeota: present knowledge and future perspectives of a unique form of life. Res. Microbiol. 154, 165–171. doi: 10.1016/S0923-2508(03)00035-4
Igarashi, K., and Kato, S. (2017). Extracellular electron transfer in acetogenic bacteria and its application for conversion of carbon dioxide into organic compounds. Appl. Microbiol. Biotechnol. 101, 6301–6307. doi: 10.1007/s00253-017-8421-3
Jangir, Y., French, S., Momper, L. M., Moser, D. P., Amend, J. P., and El-Naggar, M. Y. (2016). Isolation and characterization of electrochemically active subsurface Delftia and Azonexus species. Front. Microbiol. 7:756. doi: 10.3389/fmicb.2016.00756
Jangir, Y., Karbelkar, A. A., Beedle, N. M., Zinke, L. A., Wanger, G., Anderson, C. M., et al. (2019). In situ electrochemical studies of the terrestrial deep subsurface biosphere at the sanford underground research facility. South Dakota, USA. Front. Energy Res. 7:121. doi: 10.3389/fenrg.2019.00121
Jarett, J. K., Nayfach, S., Podar, M., Inskeep, W., Ivanova, N. N., Munson-Mcgee, J., et al. (2018). Single-cell genomics of co-sorted Nanoarchaeota suggests novel putative host associations and diversification of proteins involved in symbiosis. Microbiome 6, 1–14. doi: 10.1186/s40168-018-0539-8
Jones, R. M., Goordial, J. M., and Orcutt, B. N. (2018). Low energy subsurface environments as extraterrestrial analogs. Front. Microbiol. 9:1605. doi: 10.3389/fmicb.2018.01605
Jones, R. M., and Orcutt, B. N. (2019). Bioelectrochemistry protocol for CHI potentiostat. Protocols.io doi: 10.17504/protocols.io.xihfkb6
Jørgensen, B. B., Dunker, R., Grünke, S., and Røy, H. (2010). Filamentous sulfur bacteria, Beggiatoa spp., in arctic marine sediments (Svalbard, 79°N). FEMS Microbiol. Ecol. 73, 500–513. doi: 10.1111/j.1574-6941.2010.00918.x
Jørgensen, S. L., and Zhao, R. (2016). Microbial inventory of deeply buried oceanic crust from a young ridge flank. Front. Microbiol. 7:820. doi: 10.3389/fmicb.2016.00820
Jumas-Bilak, E., and Marchandin, H. (2014). “The phylum synergistetes,” in The Prokaryotes, eds E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer). doi: 10.1007/978-3-642-38954-2_384
Jungbluth, S. P., Bowers, R. M., Lin, H. T., Cowen, J. P., and Rappé, M. S. (2016). Novel microbial assemblages inhabiting crustal fluids within mid-ocean ridge flank subsurface basalt. ISME J. 10, 2033–2047. doi: 10.1038/ismej.2015.248
Jungbluth, S. P., Grote, J., Lin, H. T., Cowen, J. P., and Rappé, M. S. (2013). Microbial diversity within basement fluids of the sediment-buried Juan de Fuca Ridge flank. ISME J. 7, 161–172. doi: 10.1038/ismej.2012.73
Karbelkar, A. A., Jangir, Y., Reese, B. K., Wanger, G., Anderson, C., El-Naggar, M. Y., et al. (2016). Electrode Cultivation and Interfacial Electron Transport in Subsurface Microorganisms in American Geophysical Union, Fall General Assembly 2016. Available at: https://ui.adsabs.harvard.edu/abs/2016AGUFM.B53C0546K/abstract (accessed January 30, 2018).
Kawaichi, S., Yamada, T., Umezawa, A., McGlynn, S. E., Suzuki, T., Dohmae, N., et al. (2018). Anodic and cathodic extracellular electron transfer by the filamentous bacterium Ardenticatena maritima 110S. Front. Microbiol. 9:68. doi: 10.3389/fmicb.2018.00068
Koch, C., and Harnisch, F. (2016). Is there a specific ecological niche for electroactive microorganisms? Chemelectrochem 3, 1282–1295. doi: 10.1002/celc.201600079
Kuhn, E., Ichimura, A. S., Peng, V., Fritsen, C. H., Trubl, G., Doran, P. T., et al. (2014). Brine assemblages of ultrasmall microbial cells within the ice cover of Lake Vida Antarctica. Appl. Environ. Microbiol. 80, 3687–3698. doi: 10.1128/AEM.00276-14
LaBelle, E., and Bond, D. R. (2009). “Cyclic voltammetry for the study of microbial electron transfer at electrodes,” in Bioelectrochemical Systems: From Extracellular Electron Transfer to Biotechnological Application, eds K. Rabaey, L. Angenent, U. Schröder, and J. Keller (London: IWA Publishing), 137–152.
Labonté, J. M., Lever, M. A., Edwards, K. J., and Orcutt, B. N. (2017). Influence of igneous basement on deep sediment microbial diversity on the eastern Juan de Fuca Ridge flank. Front. Microbiol. 8:1434. doi: 10.3389/fmicb.2017.01434
Lam, B. R., Barge, L. M., Noell, A. C., and Nealson, K. H. (2019a). Detecting endogenous microbial metabolism and differentiating between abiotic and biotic signals observed by bioelectrochemical systems in soils. Astrobiology 20:ast.2018.1892. doi: 10.1089/ast.2018.1892
Lam, B. R., Barr, C. R., Rowe, A. R., and Nealson, K. H. (2019b). Differences in applied redox potential on cathodes enrich for diverse electrochemically active microbial isolates from a marine sediment. Front. Microbiol. 10:1979. doi: 10.3389/fmicb.2019.01979
LaRowe, D. E., and Amend, J. P. (2015). Power limits for microbial life. Front. Microbiol. 6:718. doi: 10.3389/fmicb.2015.00718
Leadbetter, J. R., Schmidt, T. M., Graber, J. R., and Breznak, J. A. (1999). Acetogenesis from H2 plus CO2 by spirochetes from termite Guts. Science 283, 686–689. doi: 10.1126/science.283.5402.686
Lever, M. A., Rouxel, O., Alt, J. C., Shimizu, N., Ono, S., Coggon, R. M., et al. (2013). Evidence for microbial carbon and sulfur cycling in deeply buried ridge flank basalt. Science 339, 1305–1308. doi: 10.1126/science.1229240
Li, S. L., and Nealson, K. H. (2015). Enriching distinctive microbial communities from marine sediments via an electrochemical-sulfide-oxidizing process on carbon electrodes. Front. Microbiol. 6:111. doi: 10.3389/fmicb.2015.00111
Liao, L., Xu, X. W., Jiang, X. W., Wang, C. S., Zhang, D. S., Ni, J. Y., et al. (2011). Microbial diversity in deep-sea sediment from the cobalt-rich crust deposit region in the Pacific Ocean. FEMS Microbiol. Ecol. 78, 565–585. doi: 10.1111/j.1574-6941.2011.01186.x
Light, S. H., Su, L., Rivera-Lugo, R., Cornejo, J. A., Louie, A., Iavarone, A. T., et al. (2018). A flavin-based extracellular electron transfer mechanism in diverse Gram-positive bacteria. Nature 562, 140–144. doi: 10.1038/s41586-018-0498-z
Liu, W., Wang, L., Gao, L., and Wang, A.-J. (2019). “Hydrogen and methane production in bioelectrochemical system: biocathode structure and material upgrading,” in Microbial Electrochemical Technology: Sustainable Platform for Fuels, Chemicals and Remediation, eds S. V. Mohan, S. Varjani, and A. Pandey (Amsterdam: Elsevier), 921–953. doi: 10.1016/B978-0-444-64052-9.00038-8
Lovley, D. R. (2008a). Extracellular electron transfer: wires, capacitors, iron lungs, and more. Geobiology 6, 225–231. doi: 10.1111/j.1472-4669.2008.00148.x
Lovley, D. R. (2008b). The microbe electric: conversion of organic matter to electricity. Curr. Opin. Biotechnol. 19, 564–571. doi: 10.1016/j.copbio.2008.10.005
Lovley, D. R. (2011). Powering microbes with electricity: direct electron transfer from electrodes to microbes. Environ. Microbiol. Rep. 3, 27–35. doi: 10.1111/j.1758-2229.2010.00211.x
Lovley, D. R. (2012). Electromicrobiology. Annu. Rev. Microbiol. 66, 391–409. doi: 10.1146/annurev-micro-092611-150104
Lovley, D. R. (2017). Happy together: microbial communities that hook up to swap electrons. ISME J. 11, 327–336. doi: 10.1038/ismej.2016.136
Marsili, E., Baron, D. B., Shikhare, I. D., Coursolle, D., Gralnick, J. A., and Bond, D. R. (2008). Shewanella secretes flavins that mediate extracellular electron transfer. Proc. Natl. Acad. Sci. U.S.A. 105, 3968–3973. doi: 10.1073/pnas.0710525105
Martin, A. L., Satjaritanun, P., Shimpalee, S., Devivo, B. A., Weidner, J., Greenway, S., et al. (2018). In-situ electrochemical analysis of microbial activity. AMB Express 8:162. doi: 10.1186/s13568-018-0692-2
Mason, O. U., Di Meo-Savoie, C. A., Van Nostrand, J. D., Zhou, J., Fisk, M. R., and Giovannoni, S. J. (2009). Prokaryotic diversity, distribution, and insights into their role in biogeochemical cycling in marine basalts. ISME J. 3, 231–242. doi: 10.1038/ismej.2008.92
Meyer, J. L., Jaekel, U., Tully, B. J., Glazer, B. T., Wheat, C. G., Lin, H.-T., et al. (2016). A distinct and active bacterial community in cold oxygenated fluids circulating beneath the western flank of the Mid-Atlantic ridge. Sci. Rep. 6:22541. doi: 10.1038/srep22541
Min, S., Jiang, Y., and Li, D. (2013). Production of acetate from carbon dioxide in bioelectrochemical systems based on autotrophic mixed culture. J. Microbiol. Biotechnol. 23, 1140–1146. doi: 10.4014/jmb.1304.04039
Miura, T., Kita, A., Okamura, Y., Aki, T., Matsumura, Y., Tajima, T., et al. (2016). Semi-continuous methane production from undiluted brown algae using a halophilic marine microbial community. Bioresour. Technol. 200, 616–623. doi: 10.1016/j.biortech.2015.10.090
Mori, J. F., Scott, J. J., Hager, K. W., Moyer, C. L., Küsel, K., and Emerson, D. (2017). Physiological and ecological implications of an iron- or hydrogen-oxidizing member of the Zeta proteobacteria, Ghiorsea bivora, gen. nov., sp. Nov. ISME J. 11, 2624–2636. doi: 10.1038/ismej.2017.132
Morono, Y., Terada, T., Nishizawa, M., Ito, M., Hillion, F., Takahata, N., et al. (2011). Carbon and nitrogen assimilation in deep subseafloor microbial cells. Proc. Natl. Acad. Sci. U.S.A. 108, 18295–18300. doi: 10.1073/pnas.1107763108
Nealson, K. H., and Rowe, A. R. (2016). Electromicrobiology: realities, grand challenges, goals and predictions. Microb. Biotechnol. 9, 595–600. doi: 10.1111/1751-7915.12400
Nielsen, M. E., Reimers, C. E., White, H. K., Sharma, S., and Girguis, P. R. (2008). Sustainable energy from deep ocean cold seeps. Energy Environ. Sci. 1:584. doi: 10.1039/b811899j
Orcutt, B. N., Bach, W., Becker, K., Fisher, A. T., Hentscher, M., Toner, B. M., et al. (2011a). Colonization of subsurface microbial observatories deployed in young ocean crust. ISME J. 5, 692–703. doi: 10.1038/ismej.2010.157
Orcutt, B. N., and Edwards, K. J. (2014). Life in the ocean crust: lessons from subseafloor laboratories. Dev. Mar. Geol. 7, 175–196. doi: 10.1016/B978-0-444-62617-2.00007-4
Orcutt, B. N., LaRowe, D. E., Biddle, J. F., Colwell, F. S., Glazer, B. T., Reese, B. K., et al. (2013a). Microbial activity in the marine deep biosphere: progress and prospects. Front. Microbiol. 4:189. doi: 10.3389/fmicb.2013.00189
Orcutt, B. N., Sylvan, J. B., Knab, N. J., and Edwards, K. J. (2011b). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiol. Mol. Biol. Rev. 75, 361–422. doi: 10.1128/MMBR.00039-10
Orcutt, B. N., Sylvan, J. B., Rogers, D. R., Delaney, J., Lee, R. W., and Girguis, P. R. (2015). Carbon fixation by basalt-hosted microbial communities. Front. Microbiol. 6:904. doi: 10.3389/fmicb.2015.00904
Orcutt, B. N., Wheat, C. G., and Edwards, K. J. (2010). Subseafloor ocean crust microbial observatories: development of FLOCS (FLow-through Osmo colonization system) and Evaluation of Borehole Construction Materials. Geomicrobiol. J. 27, 143–157. doi: 10.1080/01490450903456772
Orcutt, B. N., Wheat, C. G., Rouxel, O. J., Hulme, S. M., Edwards, K. J., and Bach, W. (2013b). Oxygen consumption rates in subseafloor basaltic crust derived from a reaction transport model. Nat. Commun. 4, 1–8. doi: 10.1038/ncomms3539
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403–1414. doi: 10.1111/1462-2920.13023
Pillot, G., Frouin, E., Pasero, E., Godfroy, A., Combet-Blanc, Y., Davidson, S., et al. (2018). Specific enrichment of hyperthermophilic electroactive Archaea from deep-sea hydrothermal vent on electrically conductive support. Bioresour. Technol. 259, 304–311. doi: 10.1016/j.biortech.2018.03.053
Radolf, J. D., Deka, R. K., Anand, A., Šmajs, D., Norgard, M. V., and Yang, X. F. (2016). Treponema pallidum, the syphilis spirochete: making a living as a stealth pathogen. Nat. Rev. Microbiol. 14, 744–759. doi: 10.1038/nrmicro.2016.141
Ramírez, G. A., Garber, A. I., Lecoeuvre, A., D’Angelo, T., Wheat, C. G., and Orcutt, B. N. (2019). Ecology of subseafloor crustal biofilms. Front. Microbiol. 10:1983. doi: 10.3389/fmicb.2019.01983
Reimers, C. E., Girguis, P. R., Stecher, H. A., Tender, L. M., Ryckelynck, N., and Whaling, P. (2006). Microbial fuel cell energy from an ocean cold seep. Geobiology 4, 123–136. doi: 10.1111/j.1472-4669.2006.00071.x
Reimers, C. E., Li, C., Graw, M. F., Schrader, P. S., and Wolf, M. (2017). The identification of cable bacteria attached to the anode of a benthic microbial fuel cell: evidence of long distance extracellular electron transport to electrodes. Front. Microbiol. 8:2055. doi: 10.3389/fmicb.2017.02055
Rittmann, B. E. (2017). Ironies in microbial electrochemistry. J. Environ. Eng. 143:03117001. doi: 10.1061/(ASCE)EE.1943-7870.0001202
Robador, A., LaRowe, D. E., Jungbluth, S. P., Lin, H. T., Rappé, M. S., Nealson, K. H., et al. (2016). Nanocalorimetric characterization of microbial activity in deep subsurface oceanic crustal fluids. Front. Microbiol. 7:454. doi: 10.3389/fmicb.2016.00454
Rosenbaum, M., Aulenta, F., Villano, M., and Angenent, L. T. (2011). Cathodes as electron donors for microbial metabolism: which extracellular electron transfer mechanisms are involved? Bioresour. Technol. 102, 324–333. doi: 10.1016/j.biortech.2010.07.008
Rowe, A. R., Chellamuthu, P., Lam, B. R., Okamoto, A., and Nealson, K. H. (2015). Marine sediments microbes capable of electrode oxidation as a surrogate for lithotrophic insoluble substrate metabolism. Front. Microbiol. 6:784. doi: 10.3389/fmicb.2014.00784
Rowe, A. R., Rajeev, P., Jain, A., Pirbadian, S., Okamoto, A., Gralnick, J. A., et al. (2018). Tracking electron uptake from a cathode into Shewanella cells: implications for energy acquisition from solid-substrate electron donors. mBio 9:e002203-17. doi: 10.1128/mBio.02203-17
Russell, J. A., Leon-Zayas, R., Wrighton, K. C., and Biddle, J. F. (2016). Deep subsurface life from North Pond: enrichment, isolation, characterization and genomes of heterotrophic bacteria. Front. Microbiol. 7:678. doi: 10.3389/fmicb.2016.00678
Santoro, C., Arbizzani, C., Erable, B., and Ieropoulos, I. (2017). Microbial fuel cells: from fundamentals to applications. A review. J. Power Sources 356, 225–244. doi: 10.1016/j.jpowsour.2017.03.109
Santos, T. C., Silva, M. A., Morgado, L., Dantas, J. M., and Salgueiro, C. A. (2015). Diving into the redox properties of Geobacter sulfurreducens cytochromes: a model for extracellular electron transfer. Dalt. Trans. 44, 9335–9344. doi: 10.1039/c5dt00556f
Schut, F., de Vries, E. J., Gottschal, J. C., Robertson, B. R., Harder, W., Prins, R. A., et al. (1993). Isolation of typical marine bacteria by dilution culture: growth, maintenance, and characteristics of isolates under laboratory conditions. Appl. Environ. Microbiol. 59:2150.
Shah Walter, S. R., Jaekel, U., Osterholz, H., Fisher, A. T., Huber, J. A., Pearson, A., et al. (2018). Microbial decomposition of marine dissolved organic matter in cool oceanic crust. Nat. Geosci. 11, 334–339. doi: 10.1038/s41561-018-0109-5
Sheik, C. S., Reese, B. K., Twing, K. I., Sylvan, J. B., Grim, S. L., Schrenk, M. O., et al. (2018). Identification and removal of contaminant sequences from ribosomal gene databases: lessons from the census of deep life. Front. Microbiol. 9:840. doi: 10.3389/FMICB.2018.00840
Smith, A. R., Popa, R., Fisk, M. R., Nielsen, M. E., Wheat, C. G., Jannasch, H. W., et al. (2011). In situ enrichment of Ocean crust microbes on igneous minerals and glasses using an osmotic flow-through device. Geochem. Geophys. Geosyst. 12:Q06007. doi: 10.1029/2010GC003424
Smith, J. A., Nevin, K. P., and Lovley, D. R. (2015). Syntrophic growth via quinone-mediated interspecies electron transfer. Front. Microbiol. 6:121. doi: 10.3389/fmicb.2015.00121
Strohl, W. R., Cannon, G. C., Shively, J. M., Güde, H., Hook, L. A., Lane, C. M., et al. (1981). Heterotrophic carbon metabolism by Beggiatoa alba. J. Bacteriol. 148, 572–583.
Strycharz-Glaven, S. M., Glaven, R. H., Wang, Z., Zhou, J., Vora, G. J., and Tender, L. M. (2013). Electrochemical investigation of a microbial solar cell reveals a nonphotosynthetic biocathode catalyst. Appl. Environ. Microbiol. 79, 3933–3942. doi: 10.1128/AEM.00431-13
Summers, Z. M., Gralnick, J. A., and Bond, D. R. (2013). Cultivation of an obligate Fe(II)-oxidizing lithoautotrophic bacterium using electrodes. mBio 4:e00420-12. doi: 10.1128/mBio.00420-12
Teske, A. P., de Beer, D., McKay, L. J., Tivey, M. K., Biddle, J. F., Hoer, D., et al. (2016). The guaymas basin hiking guide to hydrothermal mounds, chimneys, and microbial mats: complex seafloor expressions of subsurface hydrothermal circulation. Front. Microbiol. 7:75. doi: 10.3389/fmicb.2016.00075
Torsvik, T., Furnes, H., Muehlenbachs, K., Thorseth, I. H., and Tumyr, O. (1998). Evidence for microbial activity at the glass-alteration interface in oceanic basalts. Earth Planet. Sci. Lett. 162, 165–176. doi: 10.1016/S0012-821X(98)00164-2
Tremblay, P.-L., and Zhang, T. (2015). Electrifying microbes for the production of chemicals. Front. Microbiol. 6:201. doi: 10.3389/fmicb.2015.00201
Tully, B. J., Wheat, C. G., Glazer, B. T., and Huber, J. A. (2018). A dynamic microbial community with high functional redundancy inhabits the cold, oxic subseafloor aquifer. ISME J. 12, 1–16. doi: 10.1038/ismej.2017.187
Uria, N., Ferrera, I., and Mas, J. (2017). Electrochemical performance and microbial community profiles in microbial fuel cells in relation to electron transfer mechanisms. BMC Microbiol. 17:2. doi: 10.1186/s12866-017-1115-2
Walters, W., Hyde, E. R., Berg-lyons, D., Ackermann, G., Humphrey, G., Parada, A., et al. (2015). Improved bacterial 16S rRNA Gene (V4 abd V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e0009-15. doi: 10.1128/mSystems.00009-15
Wheat, C. G., Jannasch, H. W., Kastner, M., Hulme, S. M., Cowen, J. P., Edwards, K. J., et al. (2011). “Fluid sampling from oceanic borehole observatories: design and methods for CORK activities (1990-2010),” in Proceedings of the Integrated Ocean Drilling Program, eds A. T. Fisher, T. Tsuji, and K. Petronotis (Toky: Integrated Ocean Drilling Program Management International Inc.), doi: 10.2204/iodp.proc.327.109.2011
Williams, K. H., Nevin, K. P., Franks, A. E., Englert, A., Long, P. E., and Lovley, D. R. (2010). Electrode-based approach for monitoring in situ microbial activity during subsurface bioremediation. Environ. Sci. Technol. 44, 47–54. doi: 10.1021/es9017464
Xu, S., Jangir, Y., and El-Naggar, M. Y. (2016). Disentangling the roles of free and cytochrome-bound flavins in extracellular electron transport from Shewanella oneidensis MR-1. Electrochim. Acta 198, 49–55. doi: 10.1016/j.electacta.2016.03.074
Yamamoto, M., Nakamura, R., Kasaya, T., Kumagai, H., Suzuki, K., and Takai, K. (2017). Spontaneous and widespread electricity generation in natural deep-sea hydrothermal fields. Angew. Chem. 56, 5725–5728. doi: 10.1002/anie.201701768
Yang, Y., Xu, M., Guo, J., and Sun, G. (2012). Bacterial extracellular electron transfer in bioelectrochemical systems. Process Biochem. 47, 1707–1714. doi: 10.1016/j.procbio.2012.07.032
Zhang, X., Feng, X., and Wang, F. (2016). Diversity and metabolic potentials of subsurface crustal microorganisms from the western flank of the mid-atlantic ridge. Front. Microbiol. 7:363. doi: 10.3389/fmicb.2016.00363
Zhao, G., Li, E., Li, J., Liu, F., Yang, X., and Xu, M. (2019). Effects of flavin-goethite interaction on goethite reduction by Shewanella decolorationis S12. Front. Microbiol. 10:1623. doi: 10.3389/fmicb.2019.01623
Ziebis, W., McManus, J., Ferdelman, T. G., Schmidt-Schierhorn, F., Bach, W., Muratli, J., et al. (2012). Interstitial fluid chemistry of sediments underlying the North Atlantic gyre and the influence of subsurface fluid flow. Earth Planet. Sci. Lett. 323–324, 79–91. doi: 10.1016/j.epsl.2012.01.018
Keywords: deep biosphere, ocean crust, electromicrobiology, extracellular electron transfer, microbial fuel cells, iron oxidation, North Pond
Citation: Jones RM, D’Angelo T and Orcutt BN (2020) Using Cathodic Poised Potential Experiments to Investigate Extracellular Electron Transport in the Crustal Deep Biosphere of North Pond, Mid-Atlantic Ridge. Front. Environ. Sci. 8:11. doi: 10.3389/fenvs.2020.00011
Received: 30 September 2019; Accepted: 16 January 2020;
Published: 11 February 2020.
Edited by:
Amelia-Elena Rotaru, University of Southern Denmark, DenmarkReviewed by:
Enrico Marsili, Nazarbayev University, KazakhstanMichael J. Wilkins, Colorado State University, United States
Copyright © 2020 Jones, D’Angelo and Orcutt. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beth N. Orcutt, Ym9yY3V0dEBiaWdlbG93Lm9yZw==
 Rose M. Jones
Rose M. Jones Timothy D’Angelo
Timothy D’Angelo Beth N. Orcutt
Beth N. Orcutt