- 1School of Science Engineering and Technology, Abertay University, Dundee, United Kingdom
- 2Soil Microbial Ecology, FB 2 (Biology/Chemistry), University of Bremen, Bremen, Germany
- 3UMR ECOSYS, AgroParisTech, Université Paris-Saclay, Thiverval-Grignon, France
- 4School of Water, Energy and Environment, Cranfield University, Cranfield, United Kingdom
Simplified experimental systems, often referred to as microcosms, have played a central role in the development of modern ecological thinking on issues ranging from competitive exclusion to examination of spatial resources and competition mechanisms, with important model-driven insights to the field. It is widely recognized that soil architecture is the key driver of biological and physical processes underpinning ecosystem services, and the role of soil architecture and soil physical conditions is receiving growing interest. The difficulty to capture the architectural heterogeneity in microcosms means that we typically disrupt physical architecture when collecting soils. We then use surrogate measures of soil architecture such as aggregate size distribution and bulk-density, in an attempt to recreate conditions encountered in the field. These bulk-measures are too crude and do not describe the heterogeneity at microscopic scales where microorganisms operate. In the current paper we therefore ask the following questions: (i) To what extent can we control the pore geometry at microscopic scales in microcosm studies through manipulation of common variables such as density and aggregate size?; (ii) What is the effect of pore geometry on the growth and spread dynamics of bacteria following introduction into soil? To answer these questions, we focus on Pseudomonas sp. and Bacillus sp. We study the growth of populations introduced in replicated microcosms packed at densities ranging from 1.2 to 1.6 g cm−3, as well as packed with different aggregate sizes at identical bulk-density. We use X-ray CT and show how pore geometrical properties at microbial scales such as connectivity and solid-pore interface area, are affected by the way we prepare microcosms. At a bulk-density of 1.6 g cm−3 the average number of Pseudomonas was 63% lower than at a bulk-density of 1.3 g cm−3. For Bacillus this reduction was 66%. Depending on the physical conditions, bacteria in half the samples took between 1.62 and 9.22 days to spread 1.5 cm. Bacillus did spread faster than Pseudomonas and both did spread faster at a lower bulk-density. Our results highlight the importance that soil physical properties be considered in greater detail in soil microbiological studies than is currently the case.
Introduction
Simplified experimental systems, often referred to as microcosms, have played a central role in the development of modern ecological thinking on issues ranging from competitive exclusion to examination of spatial resources and competitive mechanisms, with important model-driven insights to the field (Drake et al., 1996; Jessup et al., 2004). In soil science, the complexity of soil ecosystems with interacting communities and their associated physico-chemical and biological processes has necessitated the development of simplified systems, with, for example, microcosms often used in transport studies and in studies quantifying dynamics of organic matter in soil. Microcosms help overcome problems associated with field studies that include difficulties in manipulative experiments and uncontrollability of temperature, wetness, and spatial heterogeneity. Other benefits include speed, repeatability, statistical power, and mechanistic insights (Carpenter, 1996). For the same reason, microcosms are often criticized based on the risks of investigating artifacts of the system and the absence of sound hypotheses that relate to real ecosystem functioning (Verhoef, 1996).
It is widely recognized that soil architecture is the key driver of biological and physical processes underpinning ecosystem services, and that the role of soil architecture and soil physical conditions is receiving growing interest (Nunan et al., 2001; Or et al., 2007; Tecon and Or, 2017). Nevertheless, the difficulty to capture the architectural heterogeneity in microcosms means that we typically disrupt physical architecture when collecting soils. Often this process is followed by drying and sieving, thereby exerting physical forces upon soil to disrupt its architecture. We then use surrogate measures of soil architecture such as aggregate size distribution and bulk-density, in an attempt to recreate conditions encountered in the field. These bulk measures are too crude and do not describe the heterogeneity at microscopic scales where microorganisms operate.
Recent years have seen a shift in soil science research toward non-destructive and explicit characterization of pore volumes. The complex pore geometry can offer refuge for microbes (Young et al., 2008), determine pathways of interaction, preferential pathways for fungal spread (Otten et al., 1999), and water flow, as well as provide surfaces for bacterial attachments, access to food sources, and nutrient adsorption (Young et al., 2008). Recent advances in the use of X-ray CT in research on soils enable these characteristics to be readily quantified, and various papers in the last few years have described the impact of management strategies and physical forces on soil architectural characteristics (e.g., Kravchenko et al., 2011). Soil characteristics that can be quantified using X-ray CT include the porosity, which quantifies the total volume available to microbial interactions and growth, the connectivity, which indicates how accessible the pore volume is for organisms to interact and find food sources, and the pore-solid interface area, which effectively defines the surface area accessible to microorganism in soils. Nevertheless, soil architecture and soil physical characteristics are poorly described in the majority of soil biological studies (Baveye et al., 2016), which often only give account of wetness without consideration of packing of the solid phase. Therefore we can identify 3 shortcomings in our current use of soil microcosms: (1) we have little insight in the loss of naturally-occurring architectural characteristics when we prepare soil microcosms, hampering extrapolations to field research, (2) we are unaware to what extend we can control soil architecture in a pre-described manner, and (3) we still have little insight into the effect of soil architecture on the growth and activity of micro-organisms when studied in microcosms.
In the current paper we therefore ask the following questions:
- To what extent can we control the pore geometry in microcosm studies through manipulation of common variables such as density and aggregate size? Are replicated microcosms really replicated at the microscale?
- What is the effect of pore geometry on the growth and spread of bacteria following introduction into soil?
We focus on Pseudomonas sp. and Bacillus sp. Both species are abundantly present in the rhizosphere and bulk soils in many locations and are frequently studied for their growth-promoting ability, yet there is still very little knowledge available on how their growth and spread is affected by soil physical conditions such as pore geometry.
Materials and Methods
Soil Sample Preparation
Samples were obtained from a sandy loam soil from an experimental site, Bullion Field, situated at the James Hutton Institute, Invergowrie, Scotland. Further description of the soil can be found in Sun et al. (2011). The soil was air-dried, sieved to size 1–2 and 2–4 mm, and stored in a cold room. Before usage, the soil was sterilized by autoclaving twice at 121°C at 100 kPa for 20 min within a 24 h interval time.
Bacteria and Preparation of Inoculum
Pseudomonas fluorescens SBW25-GFP (SBW25::mini-Tn7(Gm) P PrrnBP1 gfp.ASV-a, GmR (unpublished, A. Spiers), and Bacillus subtilis NRS1473 (NCIB3610 sacA::Phy−spank-GFPmut2, KmR; Hobley et al., 2013) cells were used as bacterial inoculum. Pseudomonas was grown on King's B medium (KB, 10 g Glycerol, 1.5 g K2HPO4, 1.5 g MgSO4.7H2O, 20 g Proteose peptone No.3 (Becton, Dickinson & Company, UK), 15 g Technical agar (1.5% w/v) per liter) (King et al., 1954). Bacillus was grown on Luria-Bertani medium (LB, 10 g NaCl, 10 g Tryptone, 10 g Yeast extract, 15 g Technical agar (1.5% w/v) per liter). Kanamycin (50 μg/ml) and Gentamycin (50 μg/ml) were added to the culture media.
For each experiment, an overnight culture was prepared by transferring a loop-full of colony in 10 ml of sterile broth and incubated at 28°C on a shaker at 200 rpm for 24 hr. The cells were harvested by centrifugation (4,000 × g) for 5 min and re-suspended in 10 ml PBS solution to a final concentration of OD600 = 0.95. The cell density of the solution used to inoculate was 6.46E+08 cells/ml for Pseudomonas sp. and 7.85E+08 cells/ml for Bacillus sp. The method of inoculation of the microcosms is described below.
To study the spread from localized sources, a colonized agarose pellet was used to provide a reproducible source of inoculum. A small 1 ml aliquot of inoculum of washed cells with densities as described above was mixed with 30 ml of LMP agarose solution in a centrifuge tube. The mixture was poured onto a petri dish that was left in a laminar flow cabinet at room temperature to solidify. The solidified agarose was then cut into small circular pellets using the circular end of a 1 ml pipette tip. Each pellet was of a size of 2.5 mm in diameter and 5 mm in height. Control pellet without bacteria were prepared in a similar way.
Microcosms to Study Spread of Bacteria as Affected by Soil Physical Conditions
Following Otten et al.'s (2001) approach to monitor the spread of fungi through soil, placement experiments were used where the probability of colonizing a target placed at distances from a source of inoculum is quantified over time. In these experiments, replicated microcosms of various thicknesses are prepared and a source of inoculum is placed on one side. On the other side a target is placed, which can be replaced on a daily basis and assessed for colonization. A colonized agarose bead is placed at the bottom of the sample. One autoclaved aggregate (2–4 mm in size) is placed on top of each sample. Aggregates are removed from time to time and assessed for colonization as described below. Each sample is placed in upright position in a closed centrifuge tube to reduce evaporation, and is incubated at 23°C. Each microcosm (distance) is replicated 10 times and a control series is set up using an agarose bead without bacteria.
The effect of aggregate size on the rate of spread was quantified in microcosms with a height of 1.5 cm prepared by repacking aggregates sized 0.5–1, 1–2, or 2–4 mm. In a similar way the effect of bulk-density (BD) was quantified by comparing microcosms packed at a density of 1.3 or 1.5 g cm−3 with an aggregate size of 1–2 mm. A wetness equivalent of 60% of the pores filled with water was maintained for all samples. For all experiments the target aggregate was replaced daily with a fresh aggregate till the aggregate was tested positive for colonization after which the sample was removed from the series. The removed aggregates were placed on KB media plates for detection of Pseudomonas and on LB media for Bacillus. Plates were incubated at 28°C for 48 h after which colonies were clearly visible on the plates for aggregates that had been colonized. This was taken as positive colonization and evidence that bacteria had traveled through the soil from the source of inoculum. Absence of colonization for the control samples confirmed the validity of this assumption.
Microcosms to Study Growth of Bacteria as Affected by Soil Architecture
Growth dynamics were determined in microcosms packed at different bulk-densities and aggregate-sizes. Soil microcosms were prepared in PE rings of size 3.40 cm3 (1.7 cm diameter and 1.5 cm height). The soil was wetted with sterile distilled water to achieve a moisture content so that 40% of the pores were water-filled. The gravimetric water content therefor differs per treatments, ranging from 0.13 to 0.06 g/g, and the amounts added to each sample are listed in Table 1. Two experiments were conducted, one looking at the effect of bulk-density, and a second looking at the effect of aggregate-size. In the first experiment, sterilized, sieved 1–2 mm aggregates were packed at a range of bulk-densities. The amount of soil required to obtain each bulk-density was inoculated with 500 μl of the bacterial suspension, mixed well, and packed in PE rings using a push rod. Bulk-densities of 1.2, 1.3, 1.4, 1.5, and 1.6 g cm−3 were obtained. This way the density of bacteria per volume soil (or microcosm) was identical for all bulk-densities. Control samples were packed in a similar manner except that sterile distilled water was used instead of a cell suspension. Three replicates per treatment for each sampling day were prepared, and the microcosms were sampled destructively 4 times. In the second experiment, sieved 1–2 and 2–4 mm aggregates were used. They were wetted to the same moisture content as above and packed in a similar way in PE rings at a bulk-density of 1.3 g cm−3. Soil in each ring was mixed with 500 μl of the bacterial suspension described above. The experiment was replicated 3 times and sampled 4 days after inoculation of the soil. All the microcosms were incubated at 23°C in the dark and sampled on 1, 5, 9, and 13 days after inoculation as described below.
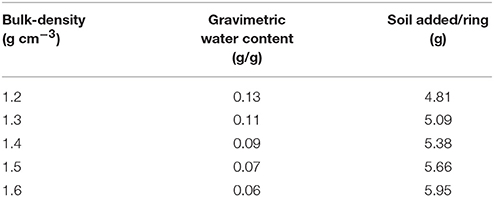
Table 1. The gravimetric water content that results in a moisture content of 40% water filled pores, and the amount of soil per ring/microcosm to pack at a particular bulk-density.
Preparation of Samples for in Situ Hybridization
On sampling day, each microcosm was mixed with 10 ml of sterile 1 × PBS solution and shaken for 15 min at room temperature. CARD-FISH was applied on soil suspensions according to the protocol described by Eickhorst and Tippkötter (2008). Briefly, 500 μl of soil suspension prepared as described above was fixed in 4% formaldehyde solution (216 μl of 37% formaldehyde and 2 × 642 μl 1 × PBS) at 4°C for 2.5 hr. The fixed samples were then washed thrice with 1 × PBS solution, centrifuged at 10,000 g for 5 min at 4°C and stored in 1 × PBS/ethanol (1:1) solution at −20°C. These fixed samples were sonicated (Sonopuls HD2200, Bandelin, Berlin, Germany) twice at 10% power for 30 s and then filtered on white polycarbonate filter (0.2 μm pores, 25 mm diameter; Sartorious, Germany) by applying vaccuum of 800 mbar. The filter membranes were then dipped in 0.2% low-melting-point agarose (Invitrogen Life Technologies) and dried at 46°C. To permeabilize cell walls, filters were incubated with 85 μl of lysozyme solution at 37°C for 60 min. The filters were then washed in H2OMQ and dehydrated in ethanol. For in-situ hybridization the membrane filters were cut into small sections.
Catalyzed Reporter Deposition (CARD) on Filter Sections
For in-situ hybridization, filter sections were incubated in 400 μl of hybridization buffer [100 mg ml−1 dextran sulfate (Sigma-Aldrich), 5M NaCl, 1M Tris-HCl (v/v), 35% Formamide (Fluka), 10% (v/v) SDS, blocking reagent (Roche, Germany) and H2OMQ] and 1.5 μl of 50 ng μl−1 horseradish peroxidase-labeled oligonucleotide probe working solution for 2 h in a rotating incubator at 35°C.
After the hybridization step, filter sections were subsequently washed in a pre-warmed washing buffer (1M Tris-HCl, 0.5M EDTA, 10% SDS, 5M NaCl and H2OMQ, 5 min at 37°C), H2OMQ (2 min at RT) and with TXP [Triton-X 100 (Bio-Rad), 1 × PBS) for 10 min at RT. For amplification of tyramide signals, filter sections were incubated with the amplification buffer [100 mg ml−1 dextran sulfate (Sigma-Aldrich), blocking reagent, 5M NaCl, 1 × PBS] along with 0.15% H2O2 solution and 1 μl of fluorescein-labeled tyramide solution for 20 min in a rotating incubator at 35°C. Afterwards, filter sections were washed in Triton-X-PBS (0.05% v/v) and dH20 for 10 min each at RT and dehydrated with ethanol.
Enumeration of Bacterial Cells With Epifluorescence Microscopy
For evaluation of CARD-FISH signals, air-dried filter sections were placed on glass slides, mounted with VectaShield H-1200 containing DAPI (4′, 6-diamino-2-phenylindole) stain and covered with coverslips. A ZEISS Axioskop 2 microscope equipped with an HBO 100 W Hg vapor lamp and a 63x objective (Carl Zeiss) was used for evaluating the filter sections. The tyramide stained cells signal was examined under a double excitation filter (Filter set 24, Carl Zeiss) and total cells were enumerated under UV excitation and a DAPI filter (F46-000, AHF, Tübingen, Germany). Bacterial cells were counted using a counting grid (10 × 10, 1.25 mm2; Carl Zeiss) integrated in the ocular of the microscope. The cells were counted at 15 random microscopic fields of views on each filter sections. Cell counts were extrapolated to obtain the number of cells per gram of soil.
Quantification of Soil Architecture With X-ray CT
An X-ray micro-tomography system, HMX225, was used to characterize and visualize the internal soil architecture (NIKON, Tring, UK). A series of samples packed at densities 1.2, 1.3, 1.4, 1.5, and 1.6 g cm−3 and with an aggregate size of 1–2 mm were prepared in triplicate as described above and scanned to quantify the effect of packing on pore geometry. In addition, samples with 1–2 or 2–4 mm aggregates (triplicate) were prepared to assess how aggregate-size affects pore geometry at a bulk-density of 1.3 g cm−3. All soil samples were scanned at 105 kV, 96 μA, and 2,000 angular projections with 2 frames per second. A molybdenum target was used with a 0.5 mm aluminum filter to minimize beam hardening effects. Radiographs were reconstructed into 3-D volume using CT-Pro at a resolution of 24 μm for the series looking at bulk-density; the samples comparing the effect of aggregate size at a single bulk-density value were scanned and reconstructed at 13.4 μm. Data were imported into VGStudiomax (Volumegraphics, Heidelberg, Germany), and converted into stacks of voxel-thick, 8-bit gray scale bmp images. Image stacks were cropped around a fixed central point to a cuboid sized 512 × 512 × 512 voxels. Segmentation of solid and pore phases was performed with an Indicator Kriging method (Houston et al., 2013) and in-house developed software was used to calculate porosity, connectivity and interface-surface area of the visible pore space in the samples. The connectivity corresponds to the volume fraction of visible pore space that is connected with the external surface of the image volume (Houston et al., 2013; Figure 1). It is noted that these properties are dependent on the resolution of the obtained scans.
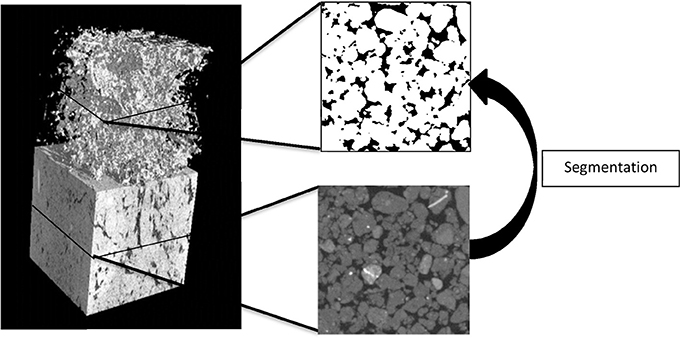
Figure 1. An example of a section of an X-ray CT scan of a repacked sieved soil sample (left) showing the solid and pore volumes and the 3D pore volume). An example of a transect is shown with the solid phase (gray) and pores (black). The pore volume is identified through segmentation which produces a binary image with pores (black) and the solid phase (white).
Data Analysis
Statistical analysis was performed with the statistical package SPSS version 2.1. An independent t-test with a 5% confidence interval was used to investigate architectural differences in mean porosity, connectivity and surface area across different bulk-densities and aggregate sizes. A generalized mixed effect Poisson model with the log link function was used to investigate significant differences in cell numbers between sampling days with day as a fixed factor. In different treatments, the significant difference between sampling days was investigated with treatments and days as fixed factor.
The rate and extent of spread was captured by 4-parameter sigmoidal curves following Otten et al. (2001). Curves were fitted to the data using Sigmaplot 11th Edition with the fraction of replicates with positive colonization, Y, given by: Y = Yo + a/(1 + exp –((x-x0)/b)), where a is the maximum fraction of replicates with successful colonization in all replicates (1.0), x0 is the point of inflection (when the fraction of replicates with positive colonization equals 0.5), and b is the steepness of the curve and reflects the variation in the rate of spread. The parameter Y0 reflects the number of positive colonizations in the control samples and was equal to 0 in all our experiments. The fitted relationship means that the rate and extent of spread can be captured by a relatively small set of parameters and the effect of treatments on parameter values can be compared.
Results
Effect of Bulk Soil Density and Aggregate Size on Pore Geometry
The effect of bulk-density on pore geometry is immediately apparent from the 2D slices selected from the 3D volumes with visibly less pore volume in the more compacted soil samples (Figure 2). In addition, the pore space looks more fragmented when the soil is packed at a higher density. This visual observation is confirmed by analysis of the thresholded 3D volumes, which showed a significant (P < 0.05) 57% decline in porosity with increasing bulk-density from 20.0% for BD = 1.2 g cm−3 to 8.7% for BD = 1.6 g cm−3. The connectivity of pores reduced from 98% (s.e. 0.5) for loosely packed soil (1.2 g cm−3) to 58% (s.e 6.1) for densely packed soil (1.6 g cm−3). The mean surface area of soil pores ranged from 43 (s.e 1.7) cm2 cm−3 for soil with a bulk-density of 1.2 g cm−3 to 35 (s.e. 5.1) cm2 cm−3 for soil with a bulk-density of 1.6 g cm−3, but this effect was not significant (Table 2).
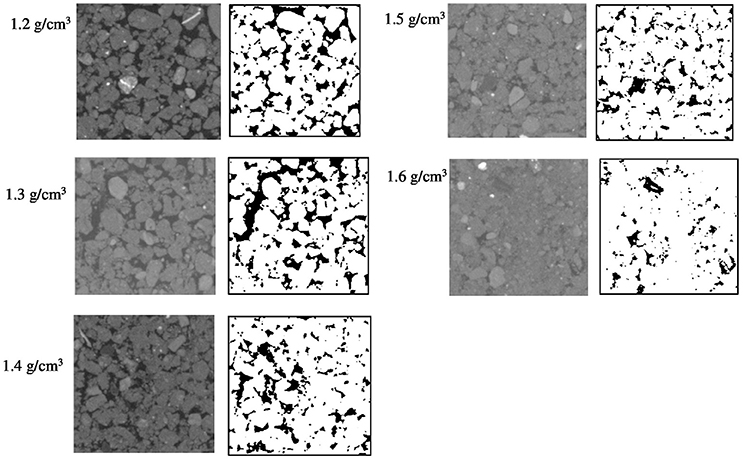
Figure 2. Examples of segmented two-dimensional images of samples packed with 1–2 mm sieved soil at BD ranging from 1.2 to 1.6 g cm−3. The solid phase is represented by different gray-scales and through thresholding transformed into binary images with black representing the pores and white representing the solid phases.
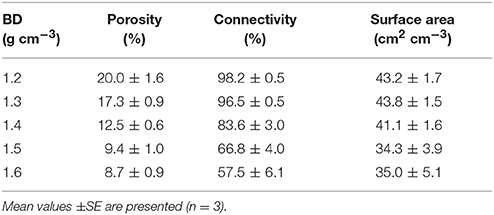
Table 2. Mean values of soil pore characteristics packed at bulk-densities (BD) of 1.2, 1.3, 1.4, 1.5, 1.6 g cm−3.
Representative 2D slices selected from the 3D volumes for soil packed with different aggregate sizes are presented in Figure 3. For the larger aggregate sizes (2–4 mm) the original aggregation of the soil is clearly visible in the resulting soil architecture. Smaller but still recognizable aggregates can also be seen in the other treatments. Overall, aggregate size distribution has a clear effect on pore geometry with wider pores in samples prepared with larger aggregate sizes. No significant difference is found for porosity and connectivity, and the only noticeable change is a minor decline in pore-solid interface with increasing aggregate size (Table 3). This is consistent with expectations, given that the soils were packed at the same bulk-density.
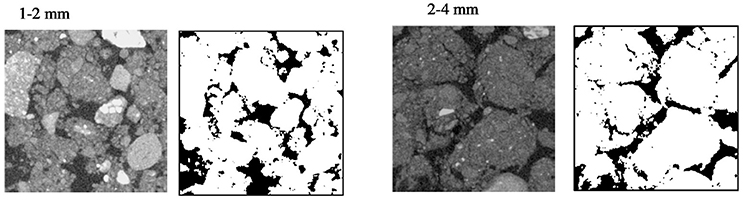
Figure 3. Examples of segmented two-dimensional images of microcosms packed at BD 1.3 g cm−3 with aggregates sizes 1–2 and 2–4 mm. The solid phase is represented by different gray-scales and through thresholding transformed into binary images with black representing the pores and white representing the solid phases.
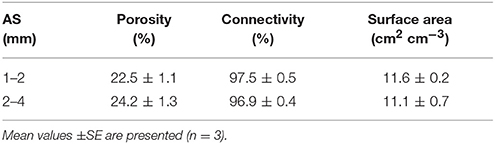
Table 3. Mean values of pore characteristics in soil of aggregate sizes (AS) 1–2 and 2–4 mm packed at a bulk-density of 1.3 g cm−3.
Visualization of Bacterial Cells in Soil Samples
The expression of GFP signals were detected on filter sections under double excitation filter (465–505 and 564–892 nm). GFP-tagged cells appear green in color against reddish color soil background, however the intensity of GFP signals appears very weak (Figure 4a). The counterstain DAPI shows that few GFP-tagged cells are not detected. The filter sections treated with CARD-FISH show brighter green signals compared to GFP signals (Figure 4b) against the soil background. Therefore, CARD-FISH is a logic choice to apply for enumeration of bacterial cells in all treatments.
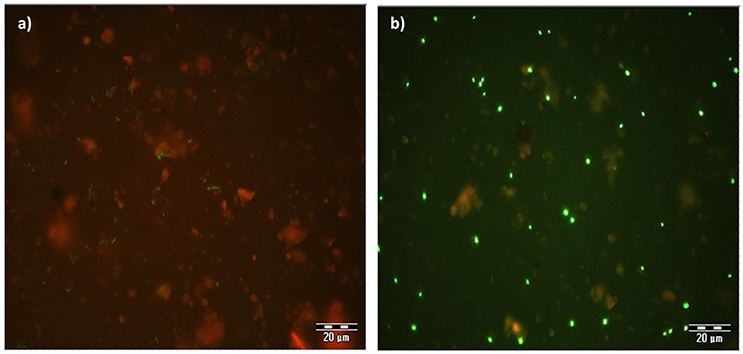
Figure 4. An example of microscopic images GFP-tagged (a) and CARD-FISH stained (b) Bacillus subtilis cells in soil filter sections under double excitation filter (465–505 and 564–892 nm). Scale bar 20 μm.
The Effect of Different Bulk Soil Densities on Growth of Bacteria
Average number of cell counts of Pseudomonas sp. and Bacillus sp. bacteria determined in different bulk densities of soil are presented in Figure 5. The growth of Pseudomonas and Bacillus cells in soil is significantly affected by the bulk-density of soil, with the increase in the cumulative number of cell dependent on bulk-density (Figure 5). For example, from days 1 to 13 at a bulk-density of 1.3 g cm−3 cell counts increase 3.56 times for Pseudomonas and 5 times for Bacillus with cell densities of 9.37E+08 (s.e 2.80E+07) cells g−1 soil, and 5.12E+08 (s.e 2.61E+07) cells g−1 soil for Pseudomonas sp. and Bacillus sp. at day 13 and 2.66E+08 (s.e 1.42E+07) cells g−1 (Pseudomonas), and 1.01E+08 (s.e 5.65E+06) cells g−1 soil (Bacillus) at day 1. This trend is expected due to the growth of bacteria in soil. For all bulk-densities and at all sampling times, the number of cell counts for Pseudomonas cells is significantly higher than Bacillus cells (P < 0.05).
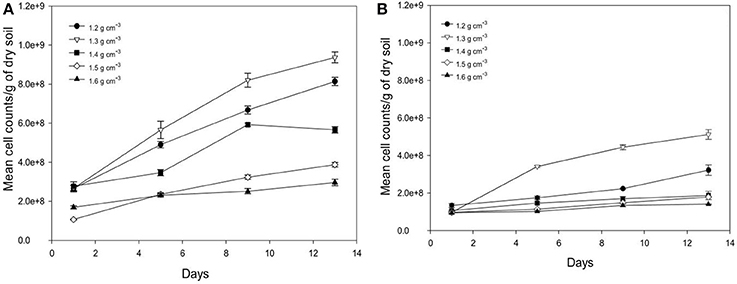
Figure 5. Change in number of cell counts per gram soil with time after inoculation with P. fluorescens (A) and B. subtilis (B) in soil at bulk densities of 1.2 g cm−3 (•),1.3 g cm−3(▿),1.4 g cm−3(■), 1.5 g cm−3(♢), and 1.6 g cm−3(▴). Data are means ±SE (n = 3).
There is a significant effect of bulk-density on the growth of bacteria in soil. As the bulk-density increases, the number of cell counts decreases for both bacterial species (P < 0.05) at all sampling times, except for soil packed at bulk-density of 1.2 g cm−3 where the average cell counts is lower than for soil packed at 1.3 g cm−3 as observed in Figure 5. This is a striking result found for both bacteria suggesting there is an optimum density for bacterial growth. At a bulk-density of 1.6 g cm−3, the average number of Pseudomonas cells is 63% lower compared to that at a bulk-density of 1.3 g cm−3 (Figure 5A). A similar trend is observed for Bacillus cells where the cumulative number of cell counts is 66% lower at a bulk-density of 1.6 g cm−3 (P < 0.05, Figure 5B). As all cell densities are expressed per gram, these reductions are beyond those one might expect (81%) from an increase in bulk-density alone.
Effect of Aggregate Sizes on Growth of Bacteria
Over time, the growth of Pseudomonas and Bacillus is significantly increased (P < 0.05) for both aggregate size classes (Figure 6). For example, from days 1 to 13 in aggregates of size 2–4 mm, cumulative cell counts increase 3.3 times for Pseudomonas and 3.0 times for Bacillus with cell densities of 9.17E+08 (s.e 4.77E+07) cells g−1 soil and 3.71E+08 (s.e 9.55E+06) cells g−1 soil respectively for Pseudomonas and Bacillus at day 13 and 2.73E+08 (s.e 2.32E+07) cells g−1 soil (Pseudomonas), and 1.23E+08 (s.e 1.98E+07) cells g−1 soil (Bacillus) at day 1. The number of cell counts of Pseudomonas is significantly higher than that of Bacillus on all sampling days (Figure 6).
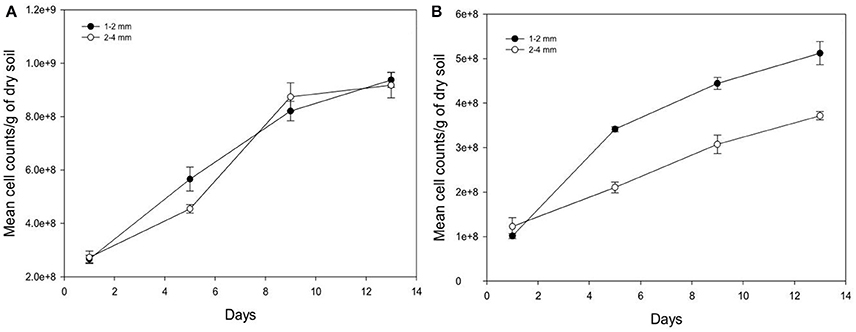
Figure 6. Change in the number of cells per gram soil with time after inoculation with P. fluorescens (A) and B. subtilis (B) detected at different sampling time in soil of aggregate size classes 1–2 mm (•) and 2–4 mm (◦). Data are means ±SE (n = 3).
Between the different aggregate size treatments, the number of cell counts of Pseudomonas is unaffected by aggregate size but the cell counts for Bacillus is higher in the 1–2 mm size aggregates class compared to 2–4 mm size aggregates (Figure 6). For example, on day 13 cell counts in smaller aggregates (1–2 mm) are 1.4 times higher for Bacillus than in larger aggregates (2–4 mm), with cell densities of 5.12E+08 (s.e 2.61E+07) cells g−1 soil in smaller aggregates (1–2 mm), and 3.71E+08 (s.e 9.55E+06) cells g−1 soil in larger aggregates (2–4 mm).
Effect of Bulk-Density on Spread Through Soil
Spread is quantified by a likelihood of spreading, expressed as the number of successful colonizations over time through a layer of soil with a thickness of 15 mm. In all replicates, all baits eventually become colonized irrespective of the bulk-density. However, the time it takes for replicates to become colonized is affected by the bulk-density for both bacterial strains. Increasing bulk-density decreases the movement of Pseudomonas and Bacillus in soil (Figure 7). In Bacillus-inoculated samples, the colonization day (Xo) is 1.62 for soil packed at a lower bulk-density, and 8.70 for soil packed at a higher bulk-density (Table 4). The colonization day (Xo) of Pseudomonas-inoculated samples is 3.00 in soil packed at lower bulk-density compared to soil packed at higher bulk-density where it is 9.22. In both bulk-density treatments, the spread of Bacillus was faster than that of Pseudomonas.
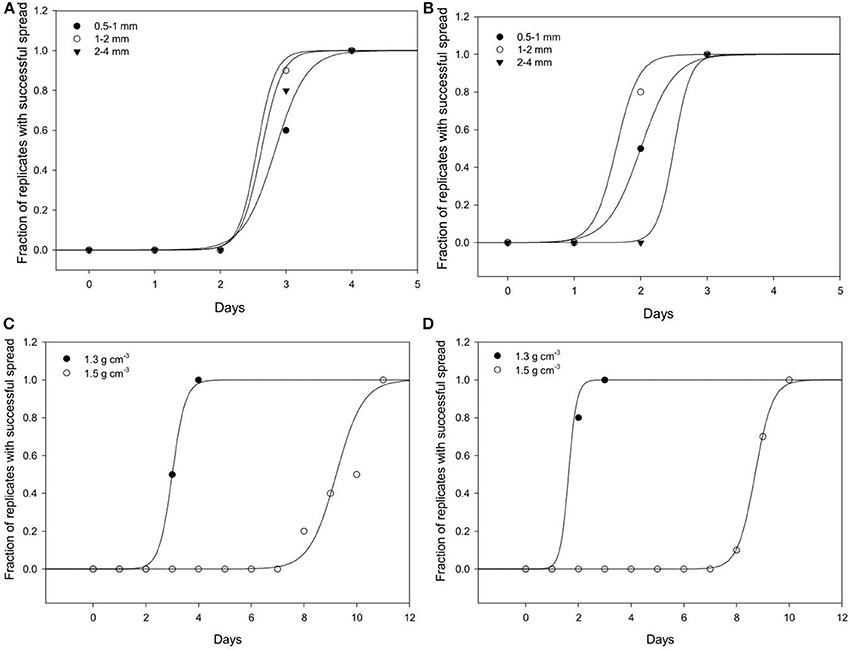
Figure 7. Dynamics of spread of Pseudomonas sp. and Bacillus sp. through soil. The number of replicates in which Pseudomonas (A) and Bacillus (B) successfully spread through soil of aggregates 0.5–1 (•); 1–2 (◦); 2–4 (▴) mm packed at a bulk-density of 1.3 g cm−3. The effect of soils packed at a BD of 1.3 or 1.5 g cm−3 (AS 1–2 mm and wetness 60%) on the spread of Pseudomonas (C) and Bacillus (D). For all treatments successful colonization was quantified as the number of successful colonizations of target bait placed at specified distances from a source of inoculum.
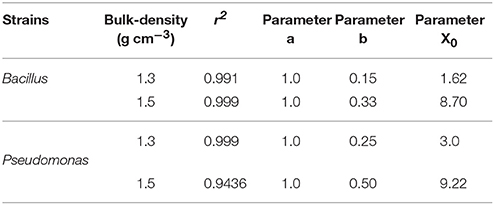
Table 4. Parameters of sigmoidal curve fitting between fraction of replicates with successful spread and different sampling days for Pseudomonas and Bacillus inoculated in soil with 60% moisture content packed to bulk densities 1.3 and 1.5 g cm−3.
Effect of Aggregate Size on Spread of Bacteria Trough Soil
Regardless of aggregate size, all replicates become colonized within 5 days, demonstrating rapid spread for all treatments. There is however an effect of aggregate size, with a different response for the two bacterial strains (Figure 7). In samples inoculated with Bacillus, colonization occurs within 2.0 days for packings of 0.5–1 mm aggregates, 1.62 days with 1–2 mm aggregates, and 2.50 days for soil with 2–4 mm aggregate sizes (Table 5). The colonization time of Pseudomonas-inoculated samples is 2.83 days for soil with 0.5–1 mm aggregates, 2.55 days with 1–2 mm aggregates, and 2.62 days for soil with 2–4 mm aggregate sizes. The spread of Pseudomonas and Bacillus is faster in soil with 1–2 mm compared to 0.5–1 mm aggregate-sizes. The spread of Bacillus was faster than that of Pseudomonas.
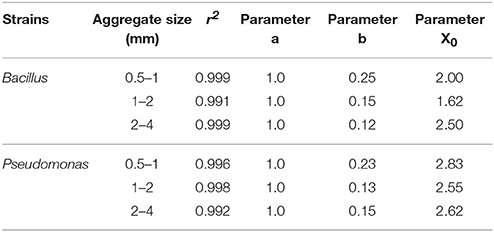
Table 5. Parameters of sigmoidal curve fitting between fraction of replicates with successful spread and sampling days for Pseudomonas and Bacillus inoculated in soil with aggregate sizes 0.5–1 mm; 1–2 mm and 2–4 mm with packed to bulk-density 1.3 g cm−3.
Discussion
Most laboratory studies of soil processes involving micro-organisms tend to be carried out in microcosms with often little consideration of the way soil is packed or what physical conditions are maintained. In the majority of cases, the water content of the soil is mentioned, often as gravimetric water content, but the density at which soil is packed is generally not provided. At best, when these characteristics are provided, they describe bulk-properties summarizing soil over scales that are much larger than those at which microorganisms operate in soil (Ettema and Wardle, 2002). In this paper we use bulk-density and aggregate size as experimental variables often encountered in soil studies and, via X-ray CT, we demonstrate that significant differences are generated at scales relevant to microorganisms. Our results show that it is possible to alter characteristics of pore geometry with the use of various initial conditions. Increasing aggregate size at the same density leads to formation of pore networks with a majority of macropores and also decreases the surface area of solid-pore interfaces. On the other hand, increasing the bulk-density of soil consisting of aggregates with the same size reduces the volume of pore space, its connectivity, and the pore-solid interface area.
The volume of available pore space and its characteristics have a major impact on a wide range of biological, chemical, and physical processes. Well-connected macro-pores are the preferential paths of fungal colony spread, followed by exploration of smaller connected pores and thin valleys (Otten et al., 2004; Pajor et al., 2010). On the other hand, meso- and micro-pores, where water menisci hold under larger negative pressures, are more suited for organisms that require instant access to water like bacteria (Young and Ritz, 2005). Pore size is a key determinant of the shape of the water retention curve, termed the “curve of life” by Young et al. (2008) since, at the scale relevant to microbial activity, it regulates the abundance of water and air. Macro-pores are the main pathways for the flow of soil water, which has a direct impact on transport of water soluble nutrients (Luo et al., 2010). By applying X-ray CT to a range of microcosms prepared in standard ways often applied in laboratory research, our results show for the first time how we can manipulate soil architecture to assess their impact on microbial dynamics in soil. It is noteworthy that we observed maximum growth at a bulk-density of 1.3, which may indicate that an optimal bulk-density exists for both species. Such an optimum might result from the interplay of contrasting effects. For example, with increasing bulk-density, the OM content per volume soil will increase. From this one might expect an increase in growth. On the other hand, there may also be a reduction in the pore-space and the water content in the sample. This will reduce the volume within which bacteria can grow to a smaller fraction of the soil volume, leading to reduced access to C, and therefore eventually to reduced growth. It is possible that a trade-off between such contrasting processes results in the optimal density for growth, in our case at a density of 1.3 g cm−3.
It is well documented that the pore volume that can be seen by X-ray CT is only a part of the pore volume as any pores smaller than the resolutions remain undetectable (e.g., Baveye et al., 2017). For meaningful samples for microcosm studies this can mean that pores less than 30 μm are not seen. For most soils this is a significant portion of the pore volume. In some soils, it may even represent all of the porosity. Perhaps equally importantly, pores from a few microns to 30 μm in size are in principle accessible to bacteria and archaea. We stress however, that for conditions under which most microcosm studies are conducted, the water content is such that sub-resolution are filled with water. This is certainly the case for our experimental set-up. As anaerobic conditions can occur at very small distances within saturated aggregates and we are here reporting on aerobic growth, these smaller pores may not play such a major part in the results as one might have assumed. The larger pores will however affect the distribution of water and the air-water interface, the diffusion pathways of dissolved organic carbon and the diffusion pathways of oxygen and hence can be expected to affect bacterial growth and spread. From the discussion above it follows that it is not just the pore geometry that should be considered, but that the water-air ratio within these pores is perhaps of greater relevance. It is clear from the results that the dominant characteristic affected by bulk-density is still the porosity, with connectivity only affected for the more densely packed soils. As expected there is a decrease of porosity values with increasing density of soil, consistent with other studies using thin sections (e.g., Harris et al., 2003) and general theory of porous media. Given that the samples were prepared from repacked sieved aggregates, it is no surprise that high values are reported for connected pore space, with only noticeable declines at higher densities when aggregates are broken during the packing and the pore space collapses. It should be noted that the values reported are for the specific resolutions obtained with X-ray CT, and different values might be expected at higher or lower resolutions as at the current resolution a significant amount of smaller pores is not considered.
We demonstrate clearly that the way microcosms are packed affects the growth and spread of bacteria, demonstrating the importance of reporting physical characteristics in microbial studies involving soil in the laboratory. Experimental results suggest that the rate of growth decreases with increasing bulk-density. These results are consistent with several other studies that report a reduction in the microbial community and its activity at higher bulk-density compared to the soil packed at lower bulk-density (Dick et al., 1988; Li et al., 2002; Tan et al., 2008; Frey et al., 2009; Pupin et al., 2009). For example, Pupin et al. (2009) report a reduction of 22–30% in the number of bacteria at a bulk-density of 1.7 g cm−3 compared to the control (1.3 g cm−3). Li et al. (2002) also report a negative relationship of microbial numbers with the bulk-density of soil. A reduction in the microbial biomass carbon and nitrogen was reported due to 13–36% decrease in air-filled porosity caused by compaction of soil. An increase in the bulk-density of soil reduces the number of large pores and the connectivity between the pores. These changes could result in reduced accessibility of organic substances, water movement, and gas exchange. A reduction in O2 diffusion through soil changes the soil environment into an anaerobic state, thus one of the factor in inhibiting the growth of aerobic microorganisms and its activity (Beylich et al., 2010).
In this study, both Pseudomonas sp. and Bacillus sp. are aerobic microorganisms and they were both shown to be negatively affected by the increase in bulk-density of soil. We tried to mitigate this effect by choosing a wetness equivalent to 40–60% of the pore space filled with water (and hence 60–40% with air) and maintain this ratio between our treatments. Inevitably, other factors were also altered. For example, as more soil is packed in a microcosm at a higher bulk-density, and the number of cells at inoculation is constant per volume, the cell count expressed per gram of soil is lower in soil with a higher density. This is inherent to quantifying microorganisms in soil and is the main difference between comparing numbers or densities expressed gravimetrically or volumetrically. However, the differences we found are larger than could be explained by such a simple dilution effect. On the other hand, all other parameters being equal, soil with a higher bulk-density has larger organic matter content per volume of soil. So each microcosm contains more organic matter at a higher bulk-density. The fact that this may also have affected the growth highlights the complex web of interactions that take place between physical space and other conditions. Disentangling this through targeted experimentation is not easy and the way forward would be to develop mathematical models that consider the impact of microscopic heterogeneities on microbial dynamics. Examples of such an approach are given for bacteria in Monga et al. (2014) and for fungi in Falconer et al. (2015) where it was shown how biological, physical, and chemical characteristics interact at the microscale to influence emerging processes at larger scales.
A significant effect of aggregate size on the growth is observed only for samples inoculated with Bacillus. The numbers of Bacillus cell counts are higher in smaller aggregates of 1–2 mm in size. The possibility of active growth in smaller size aggregates could be due to the availability of more nutrients in smaller sized aggregates. A non-significant effect of aggregate size on Pseudomonas cells counts is observed. This result agrees with the finding of Drazkiewicz (1994) who found that soil type had more influence on the number of Pseudomonas than the aggregate size.
Spread of microorganism is a critical trait that affects their ability to find food sources, and to interact with other species. Yet data on mobility of bacteria through soil are limited to studies under conditions of convective flow. We developed a simple experimental system that enables the spread of bacteria through soil, following the concept of dispersal kernels commonly used in ecology. Our results show that both species spread significant distances in relatively short timescales even in absence of convective flow. Our results demonstrate spread beyond 1.5 cm in the absence of convective flow and that the spread of bacteria is species-dependent and determined by soil physical conditions. Interestingly, whereas the growth of Bacillus is slower than that of Pseudomonas, the spread is faster. Potentially this could indicate that energy devoted to spread is diverged from energy devoted to growth. In any case our results show a differential effect of soil physical conditions on the ability of bacteria to grow and spread, which are likely to be significant in relation to the way species explore soil and interact with each other. The results showed a different response to soil architecture on spread than the results we found on growth. Whereas one might expect some similarities to occur, it is also not surprising that factors like connectivity of the water-filled pore space will have a greater effect on spread than it will have on the growth. This is expected as in the growth experiment, bacteria were mixed through the soil, and growth can therefore be initiated in disconnected parts of the pore volume. This in contrast to the spread, where a single source was used, and a connected pathway will be required for spread. The spread would therefore be expected to show a greater dependency on water content that the growth data within the range of water content tested in this study.
Our results have implications for the way soil microbiological studies are reported. We demonstrate in this study that the physical composition of soil has a significant effect on the growth of bacteria in soil. Cell counts of both bacterial strains selected for this study show a significant influence of bulk-density on their growth in soil whereas aggregate size only affected Bacillus. We also show that bacterial strains respond differentially to soil physical conditions. This highlights the need to include detailed reporting on soil physical conditions in soil microbiological studies. This is true whether the characteristics are measured in terms of bulk properties, such as bulk-density and aggregate size, or in terms of the microscopic heterogeneity of the pore geometry. We also demonstrate how specific characteristics of the pore volume, such as connectivity of the pore space or the pore-solid interface can be manipulated through bulk properties of soil microcosms.
Author Contributions
All authors contributed to original ideas and design, analysis and writing, AJ performed experiments.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
WO and RF are grateful for funding received from the National Environment and Research Council (NE/P014208/1). AJ acknowledges support from SORSAS and DAAD (A/12/76235). TE received funding from the University of Bremen (ZF/02/600/10). The authors acknowledge the contributions made by Dr Hannes Schmidt for assistance in the application of CARD-FISH, by Dr Radoslaw Pajor in the production of part of some images used in the paper, and Dr Simona Hapca for assistance with data analysis.
References
Baveye, P., Berthelin, J., and Munch, J. (2016). Too much or not enough: reflection on two contrasting perspectives on soil biodiversity. Soil Biol. Biochem. 103, 320–326. doi: 10.1016/j.soilbio.2016.09.008
Baveye, P. C., Pot, V., and Garnier, P. (2017). Accounting for sub-resolution pores in models of water and solute transport in soils based on computed tomography images: are we there yet? J. Hydrol. 555, 253–256. doi: 10.1016/j.jhydrol.2017.10.021
Beylich, A., Oberholzer, H.-R., Schrader, S., Hoper, H., and Wilke, B.-M. (2010). Evaluation of soil compaction effects on soil biota and soil biological processes in soils. Soil Tillage Res. 109, 133–143. doi: 10.1016/j.still.2010.05.010
Carpenter, S. R. (1996). Microcosm experiments have limited relevance for community and ecosystem ecology. Ecology 77, 677–680 doi: 10.2307/2265490
Dick, R. P., Rasmussen, P. E., and Kerle, E. A. (1988). Influence of long-term residue management on soil enzyme activities in relation to soil chemical properties of a wheat-fallow system. Biol. Fertil. Soils 6, 159–164. doi: 10.1007/BF00257667
Drake, J. A., Huxel, G. R., and Hewitt, C. L. (1996). Microcosms as models for generating and testing community theory. Ecology 77, 670–677. doi: 10.2307/2265489
Drazkiewicz, M. (1994). Distribution of microorganisms in soil aggregates: effect of aggregate size. Folia Microbiol. 39, 276–282. doi: 10.1007/BF02814312
Eickhorst, T., and Tippkötter, R. (2008). Improved detection of soil microorganisms using fluorescence in situ hybridization (FISH) and catalyzed reporter deposition (CARD-FISH). Soil Biol. Biochem. 40, 1883–1891. doi: 10.1016/j.soilbio.2008.03.024
Ettema, C. H., and Wardle, D. A. (2002). Spatial soil ecology. Trends Ecol. Evol. 17 177–183. doi: 10.1016/S0169-5347(02)02496-5
Falconer, R. E., Battaia, G., Schmidt, S., Baveye, P., Chenu, C., and andOtten, W. (2015). Microscale heterogeneity explains experimental variability and non-linearity in mineralisation of soil organic matter. PLoS ONE 10:e0123774 doi: 10.1371/journal.pone.0123774
Frey, B., Kremer, J., Rudt, A., Sciacca, S., Matthies, D., and Luscher, P. (2009). Compaction of forest soils with heavy logging machinery affects soil bacterial community structure. Eur. J. Soil Biol. 45, 312–320. doi: 10.1016/j.ejsobi.2009.05.006
Harris, K., Young, I. M., Gilligan, C. A., Otten, W., and Ritz, K. (2003). Effect of bulk density on the spatial organisation of the fungus Rhizoctonia solani in soil. FEMS Microb. Ecol. 44, 45–56. doi: 10.1111/j.1574-6941.2003.tb01089.x
Hobley, L., Ostrowski, A., Rao, F. V., Bromley, K. M., Porter, M., Prescott, A. R., et al. (2013). BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc. Natl. Acad. Sci. U.S.A. 110, 13600–13605. doi: 10.1073/pnas.1306390110
Houston, A. N., Otten, W., Baveye, P. C., and Hapca, S. (2013). Adaptive-window indicator kriging: a thresholding method for computed tomography images of porous media. Comput. Geosci. 54, 239–248. doi: 10.1016/j.cageo.2012.11.016
Jessup, C. M., Kassen, R., Forde, S. E., Kerr, B., Buckling, A., Rainey, P. B. J., et al. (2004). Big questions, small worlds: microbial model systems in ecology. Trends Ecol. Evol. 19, 189–197.doi: 10.1016/j.tree.2004.01.008
King, E. O., Ward, M. K., and Raney, D. E. (1954). Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med.44, 301–307.
Kravchenko, S., Falconer, R., Grinev, D., and Otten, W. (2011). Fungal colonization in soils of contrasting management: modeling growth in 3D pore volumes of undisturbed soil samples. Ecol. Appl. 21, 1202–1210. doi: 10.1890/10-0525.1
Li, C. H., Ma, B. L., and Zhang, T. Q. (2002). Soil bulk density effects on soil microbial populations and enzyme activities during the growth of maize (Zea mays L.) planted in large pots under field exposure. Can. J. Soil Sci. 82, 147–154. doi: 10.4141/S01-026
Luo, L., Lin, H., and Li, S. (2010). Quantification of 3-D soil macropore networks in different soil types and land uses using computed tomography. J.Hydrol. 393, 53–64.doi: 10.1016/j.jhydrol.2010.03.031
Monga, O., Garnier, P., Pot, V., Coucheney, E., Nunan, N., Otten, W., et al. (2014). Simulating microbial degradation of organic matter in a simple porous system using the 3-D diffusion-based model MOSAIC. Biogeosciences 11, 2201–2209. doi: 10.5194/bg-11-2201-2014
Nunan, N., Ritz, K., Crabb, D., Harris, K., Wu, K., Crawford, J. W., et al. (2001). Quantification of the in situ distribution of soil bacteria by large-scale imaging of thin sections of undisturbed soil. FEMS Microbiol. Ecol. 37, 67–77. doi: 10.1111/j.1574-6941.2001.tb00854.x
Or, D., Smets, B. F., Wraith, J. M., Dechesne, A., and Friedman, S. P. (2007). Physical constraints affecting bacterial habitats and activity in unsaturated porous media–a review. Adv. Water Resour.30, 1505–1527. doi: 10.1016/j.advwatres.2006.05.025
Otten, W., Gilligan, C. A., Hall, D., Harris, K., Ritz, K., and Young, I. M. (2001). Soil physics, fungal epidemiology and the spread of Rhizoctonia solani. N. Phytol. 151, 459–468. doi: 10.1046/j.0028-646x.2001.00190.x
Otten, W., Gilligan, C. A., Watts, C., Dexter, A. R., and Hall, D. (1999). Continuity of air-filled pores and invasion thresholds for a soil-borne fungal plant pathogen, Rhizoctonia solani. Soil Biol. Biochem. 31, 1803–1810.
Otten, W., Harris, K., Young, I. M., Ritz, K., and Gilligan, C. A. (2004). Preferential spread of the pathogenic fungus Rhizoctonia solani through structured soil. Soil Biol. Biochem. 36, 203–210. doi: 10.1016/j.soilbio.2003.09.006
Pajor, R., Falconer, R., Hapca, S., and Otten, W. (2010). Modelling and quantifying the effect of heterogeneity in soil physical conditions on fungal growth. Biogeosciences 7, 3731–3740. doi: 10.5194/bg-7-3731-2010
Pupin, B., Freddi, S., and Nahas, E. (2009). Microbial alterations of the soil influenced compaction. Rev. Bras. Ciênc. 33, 1207–1213. doi: 10.1590/S0100-06832009000500014
Sun, B., Hallett, P. D., Caul, S., Daniell, T. J., and Hopkins, D. W. (2011). Distribution of soil carbon and microbial biomass in arable soils under different tillage regimes. Plant Soil 338, 17–25. doi: 10.1007/s11104-010-0459-2
Tan, X., Chang, S. X., and Kabzems, R. (2008). Soil compaction and forest floor removal reduced microbial biomass and enzyme activities in a boreal aspen forest soil. Biol. Fertil. Soils. 44, 471–479. doi: 10.1007/s00374-007-0229-3
Tecon, R., and Or, D. (2017). Biophysical processes supporting the diversity of microbial life in soil FEMS Microbiol. Rev. 41, 599–623. doi: 10.1093/femsre/fux039
Verhoef, H. A. (1996). The role of microcosms in the study of ecosystem processes. Ecology 77, 685–690 doi: 10.2307/2265492
Keywords: X-ray CT scanning, bacterial growth, bacterial spread, CARD-FISH, microcosm experiment, pseudomonas, Bacillus subtilis
Citation: Juyal A, Eickhorst T, Falconer R, Baveye PC, Spiers A and Otten W (2018) Control of Pore Geometry in Soil Microcosms and Its Effect on the Growth and Spread of Pseudomonas and Bacillus sp. Front. Environ. Sci. 6:73. doi: 10.3389/fenvs.2018.00073
Received: 19 March 2018; Accepted: 20 June 2018;
Published: 13 July 2018.
Edited by:
Denis Angers, Agriculture and Agri-Food Canada (AAFC), CanadaReviewed by:
Andrea Carminati, University of Bayreuth, GermanyPascal Benard, University of Bayreuth, Germany
Copyright © 2018 Juyal, Eickhorst, Falconer, Baveye, Spiers and Otten. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thilo Eickhorst, ZWlja2hvcnN0QHVuaS1icmVtZW4uZGU=
†Present Address: Ruth Falconer, School of Arts, Media and Computer Games, Abertay University, Dundee, United Kingdom
 Archana Juyal
Archana Juyal Thilo Eickhorst
Thilo Eickhorst Ruth Falconer1†
Ruth Falconer1† Philippe C. Baveye
Philippe C. Baveye Andrew Spiers
Andrew Spiers Wilfred Otten
Wilfred Otten