- Department of Biotechnology, Guru Nanak Dev University, Amritsar, India
Rice is an important cereal crop responsible for world's food security. The sensitivity of rice plants toward a range of abiotic stresses is a prime challenge for its overall growth and productivity. Among these, salinity is a major stress which results in a significant loss of global rice yield annually. For finding straightforward and strict future solutions in order to assure the food security to growing world population, understanding of the various mechanisms responsible for salt stress tolerance in rice is of paramount importance. In classical studies, identification of salt tolerant cultivars and the genetic markers linked to salt tolerance and breeding approaches have been given emphasis. It further affirmed on the identification of various pathways regulating the complex process of salt stress adaptation. However, only limited success has been achieved in these approaches as salt tolerance is a complex process and is governed by multiple factors. Hence, for better understanding of salt tolerance mechanisms, a comprehensive approach involving physiological, biochemical and molecular studies is much warranted. Modern experimental and genetic resources have provided a momentum in this direction and have provided molecular insights into different salt stress responsive pathways at the signaling and regulatory level. The integrative knowledge of classical and modern research of the understanding of salt stress adaptive pathways can help the researchers for designing effective strategies to fight against salt stress. Hence, the present review is focused on the understanding of the salt stress tolerance mechanisms in rice through the consolidative knowledge of classical and modern concepts. It further highlights the emerging new trends of salt stress adaptive pathways in rice.
Introduction
Rice as a crop has significant contribution in world food security. It feeds half of the world population and meets almost 80% of their food calorie requirements (Agarwal et al., 2016; Fan et al., 2016). However, the rice productivity is severely affected by biotic as well as abiotic stresses. Among the abiotic stresses, salinity stress has a significant impact on rice growth, development and its yield (Golldack et al., 2014). It affects more than 20% of total irrigated lands leading to forfeiture of 12 billion USD in world food economy (Fita et al., 2015; Volkov, 2015; Kaur et al., 2016a; Negrão et al., 2017). Therefore, to fulfill the ever-increasing demand of rice, conscious efforts to mitigate the adverse effects of salt stress are much warranted (Agarwal et al., 2016).
Traditional research tools have provided a wealth of information on the identification of salt tolerant cultivars and on the role of different physiological, biochemical mechanisms responsible for salinity tolerance in rice (Horie et al., 2012; Deinlein et al., 2014). Classical studies affirmed on the identification of different genetic markers and traits associated with salinity tolerance which were explored using breeding approach for the introgression of useful traits related to salt stress adaptation in rice (Fita et al., 2015; Mickelbart et al., 2015). However, little success has been achieved with this as salt stress tolerance is a complex phenomenon and controlled by a number of factors. Further plant breeding is a time consuming and a laborious process (Gregorio et al., 2002). Hence, for developing salt tolerant rice, designing alternative strategies which could help in comprehensive understanding of different mechanisms associated with salt stress adaptation is of paramount importance. The development of modern experimental resources and research tools like sequencing, microarray, imaging, recombinant DNA technology, proteomics etc. have given deeper insights into these mechanisms at the molecular and regulatory level. It is also realized that conventional research has provided appropriate platform for modern research and conceptual development for achieving salt tolerance in rice. There is a high degree of interdependency of wealth of information received and capitalizing this information for developing salinity tolerance in rice cultivars. Hence, integrating the classical and modern approaches will provide a better insight to salt tolerance mechanisms in rice and will provide future solution to increase the rice productivity in saline environment.
Physiological and Biochemical Mechanisms of Salt Tolerance
Plants respond to salinity stress through different mechanisms that operate at the various physiological and biochemical levels (Negrão et al., 2017). Salinity tolerance is governed by a combination of these different mechanisms in a synergistic manner. Classical studies emphasized on the identification of these pathways and modern research tools are helping in dissecting these pathways for their better understanding.
Ion Homeostasis and Compartmentalization
Plants cannot endure high concentration of salts in the cytoplasm both under the normal and stressful conditions. They get rid of these excessive salts either by their extrusion from the cell or their sequesterization into the vacuole or the older tissues (Gupta and Huang, 2014; Maathuis et al., 2014). Understanding of the mechanism behind ions transport and sequesterization has always remained a prime focus of plant science research. Potassium (K+) ions are the most predominant ions present in the plant cytosol which plays critical role in plant enzyme activation, photosynthesis, stomatal activity and sugar transport, etc. (Wang et al., 2013). Plants maintain high potassium (K+)/sodium (Na+) cytosolic ratio for maintaining their normal functioning.
Sodium chloride (NaCl) present in the soil during salt stress ionizes to form Na+ and Cl− ions. These excessive Na+ ions inhibit K+ ion uptake by interfering in the plasma membrane K+ transporters of root cells. During salinity, high affinity K+ ion transporters behave as low affinity Na+ ion transporters and act as a potential pathway for influx of Na+ ions in the cells (Brini and Masmoudi, 2012; Bojórquez-Quintal et al., 2014). These Na+ ions compete with K+ ions for binding to the catalytic sites of essential enzymes being physiochemically similar to each other (Brini and Masmoudi, 2012). Therefore, reestablishment of high K+/Na+ ions ratio is one of the critical aspect of salinity tolerance in plants (Brini and Masmoudi, 2012). This is achieved with the help of Na+/H+ antiporters that either export the excessive ions out of the cell or transport them to the vacuole (Zhao et al., 2017). Na+/H+ antiporters are broadly categorized into three types viz salt overly sensitive (SOS), NHX and high-affinity potassium transporters (HKT). Among these, the SOS ion transporters are the most extensively studied and well-characterized in plants (Ma et al., 2014). There are three genes that encode for SOS proteins in rice: SOS1, SOS2 and SOS3. These three SOS proteins work in a co-ordinate manner to transport excess of Na+ ions from the cytoplasm (Brini and Masmoudi, 2012). With the help of modern research tools, biologists have deciphered the detailed cascade of signaling events involved in SOS pathway. This pathway is triggered by the high Na+ concentration that is perceived by the intracellular calcium (Ca2+) ion signals. High concentration of NaCl perturbes intracellular Ca2+ level via hypothetical plasma membrane sensors (Türkan and Demiral, 2009). This unique and specific Ca2+ signal is recognized and decoded by SOS3 protein that belongs to calcineurin B-like protein (CBLs) family. SOS 3 then interacts with SOS 2 to form a SOS3/SOS2 protein kinase complex and these together activate SOS1. SOS1 encodes for a plasma membrane Na+/H+ antiporter that maintains Na+ ion homeostasis by effluxing excess of Na+ ions during salinity. SOS2 also differentially regulates vacuolar Na+/H+ and H+/Ca2+ antiporters contributing to enhanced sequesterization of Na+ ions in vacuole and thereby confer salinity tolerance. Further, the SOS2/SOS3 kinase complex downregulates the activity of Na+ ion transporters that mediates entry of these ions into root cells during salinity (Türkan and Demiral, 2009). In a recent report, a OsSOS1 gene isolated from salt tolerant rice variety Pokkali has been shown to confer salinity tolerance when expressed constitutively using CaMV35S promoter in rice salt sensitive cultivar BRRI dhan 28 (Yasmin et al., 2016). Apart from well-established role in Na+ ion homeostasis, SOS proteins have also been reported to play novel roles during stress like regulating the dynamics of cytoskeleton (Yasmin et al., 2016).
Along with SOS pathway, Ca2+ signal initiated by salinity is also perceived and decoded by calmoudlin like proteins (CMLs) (Zeng et al., 2015). CMLs bind to the Ca2+ ions and regulate a plethora of target proteins including ion transporters, protein kinases, transcription factors (TFs) and various metabolic proteins (Zeng et al., 2015). Recently, the interaction of vacuolar Na+/H+ antiporters with CML18 protein has been found to regulate the process of ion sequestration in the vacuole. Under the control conditions, these two proteins are found to interact strongly that in turn modifies the Na+/K+ antiporters ion selectivity resulting in reduced Na+/H+ exchange process. Under the effect of salt stress, due to the change in vacuolar pH, repression of Na+/H+ activity by CML18 is inhibited resulting in the sequesterization of Na+ ions in the vacuole (Yamaguchi et al., 2005). Further, rice CML proteins are reported to confer salt stress tolerance in an ABA dependent manner and in association with MYB TFs (Zeng et al., 2015).
NHX antiporters are localized in the tonoplast and are involved in sequesterization of excessive Na+ ions in the vacuole. Along with this; they have been lately reported to be involved in cell growth, osmoregulation and plant development (Deinlein et al., 2014). Constitutive overexpression of rice OsNHX1 gene isolated from well-known salt tolerant cultivar Pokkali has been reported to enhance the salt tolerance of rice salt sensitive cultivar (Amin et al., 2016). NHX antiporters also substantially contribute in the regulation of cellular pH and stomatal functioning of plants by K+ ion sequesterization in the vacuole (Barragán et al., 2012).
HKT family of ion transporters specifically transports Na+ ions although they were initially described as high affinity K+ ion transporters (Brini and Masmoudi, 2012). They can either transport Na+ ions prefentially or symport Na+/K+ ions depending upon the presence of serine or glycine residues in the protein structure. HKT transporters are of particular importance in regulating the influx of Na+ ions into the roots (Brini and Masmoudi, 2012). They are considered as the most widely known genes that confer salt tolerance in plants. Seven functional OsHKT genes have been reported to exist in japonica rice cultivar and are considered as a potential marker for salinity tolerance (Hamamoto et al., 2015). However, serious efforts are further required to explore the role of these genes in conferring salt stress adaptation.
Classical studies mainly focused on the role of Na+ ion homeostasis in salt tolerance, however with the advent of modern research methodology, researchers have suggested that along with Na+ ion transporters, a thorough understanding of Cl− ion transporters is also essential for improving the salt tolerance in plants. The accumulation of Cl− ions is considered even more toxic for the plant vital processes like photosynthesis (Brini and Masmoudi, 2012; Li et al., 2017). However, the physiological and molecular mechanisms underlying chloride ion toxicity are not well-understood in plants (Khare et al., 2015). The fine tune balance of Cl− ions in the cytosol is achieved through their sequesterization in the vacuole with the help of ion transporters and voltage gated ion channels (Brini and Masmoudi, 2012; Li et al., 2017). As the net charge in the vacuole is slightly positive with respect to the negatively charged cytoplasm, a voltage gradient is maintained between the vacuole and the cytoplasm. Majority of the Cl− ions are sequestered in the vacuole through voltage–gated anion channels belonging to CLC family present on the tonoplast. In rice, the significant upregulation of a OsCLCa gene has been reported in response to salt stress. Further, in a comparative studies, the expression level of an OsCLCc gene was observed to be low in case of salt sensitive IR29 and high in the salt tolerant Pokkali cultivar of rice (Brini and Masmoudi, 2012). Cl− ion signal in the salt effected soil has also been linked to stomata closure in an ABA dependent manner as an early salt stress response (Geilfus et al., 2015). These studies indicate the importance of Cl− ion signaling and transporters in regulating the different pathways conferring salt tolerance.
Recently, researchers have identified that along with extrusion of excessive detrimental ions out of the cell cytosol, efficient uptake of K+ ions and their long distance distribution also plays critical role in deciding the salt tolerance level of a plant (Himabindu et al., 2016; Garriga et al., 2017). Three types of transporter families are involved in K+ ions transport namely HKT (High affinity K Transporter), HAK (High Affinity K)/KUP (K UP take)/KT (K Transporter) and Shaker K+ channel transporters (Shabala and Pottosin, 2014; Véry et al., 2014). But these transporters have not been well-studied in context to salt tolerance in rice. Hence, along with the conventional understanding of Na+ ion homeostasis exploring various mechanisms associated with Cl− ion and K+ ion mediated salinity tolerance can give a new hope to the researchers for developing salt tolerant rice varieties.
Production of Compatible Solutes and Osmoprotectants
Osmotic stress is another detrimental consequence of salinity on plant cells apart from ionic stress (Gupta and Huang, 2014). It occurs because of the intracellular water loss from the plant cells when the NaCl concentration increases above threshold level in the immediate environment of the cells (Türkan and Demiral, 2009). An instant response to this stress in plants is stomatal closure (Carillo et al., 2011). But this response is temporary and untenable for long duration because stomatal opening is required for carbon fixation (Carillo et al., 2011). So for long term strategy, plant cells tend to increase their osmotic potential by accumulating various metabolites better known as compatible (organic) solutes in the cytosol in order to reduce the water loss from the cells (Türkan and Demiral, 2009; Cardi et al., 2015). It is a key adaptation at the cellular level to combat the effect of osmotic stress (Hussain et al., 2016). Compatible solutes are water soluble, small polar, uncharged compounds and their high concenteration does not affect the plant's vital processes and enzymatic activities. They are mainly categorized into amino acid derivatives, sugars, sugar alcohols, complex sugars, sulfonium compounds, and tertiary amines (Flowers and Colmer, 2008). These solutes counterbalance the higher concenteration of salts outside the cells in one direction and accumulated Na+ and Cl− ions in the vacuole on the another direction (Türkan and Demiral, 2009). Along with osmoregulation, they also play key role in ROS detoxification and stabilization of membranes and proteins (Hussain et al., 2016).
The amino acid proline is one of the osmotically most active compound that substantially contributes to the osmotic adjustment (Hussain et al., 2016). It is also reported to act as a signaling/ regulatory molecule that can activate various pathways involved in salt stress adaptation process (Hussain et al., 2016). Moreover, it serves as a nitrogen reservoir when the plant recovers from stress (Hadi et al., 2014). The level of proline increases to many folds in response to salt stress in rice (Sharma et al., 2015). Modern molecular biology tools have given insights into the identification of different genes responsible for metabolic level of proline in the cell. This increased level of proline is consequence of the tight regulation of its anabolism and catabolism processes which are controlled by the two key enzymes viz biosynthetic enzyme pyrroline-5-carboxylate synthetase1 (P5CS1) and catabolic enzyme proline dehydrogenase (PDH1) (Verbruggen and Hermans, 2008; Kaur et al., 2016b). The expression of OsP5CS1 gene along with other biosynthetic genes like Δ1, -Pyrrolline-5-carboxylate reductase, and Ornithine-δ- aminotransferase increases whereas, the expression of catabolic gene OsPDH decreases in response to salt stress in rice (Bagdi et al., 2015). Its content has also been observed to be constitutively high in salt tolerant cultivars of rice (Kaur et al., 2016a). Further, the expression of biosynthetic gene OsP5CS was observed to be high in these tolerant cultivars (Kaur et al., 2016b). Therefore, OsP5CS and OsPDH can be considered as the potential candidates for developing transgenic rice cultivars with better salt tolerance ability in future.
Soluble, non-structural carbohydrates (sucrose, hexoses) are one of the key players in osmoregulation during salt stress. Their increase in the cell during stress provides optimism for enhancing the plant salt tolerance (Rosa et al., 2009). The level of these soluble sugars increases in plants when exposed to saline conditions. Trehalose is a disaccharide sugar synthesized from two glucose units linked by an α-α-1, 1 glycosidic bond that protects the plant cells from osmotic stress injuries (Redillas et al., 2012; Reina-Bueno et al., 2012). The level of trehalose increases significantly in response to salt stress in plants (Nounjan et al., 2012; Henry et al., 2015). Further the exogenous application of trehalose has been shown to be involved in better recovery of rice plants under salinity stress (Nounjan et al., 2012). Trehalose is synthesized in the plants via a conserved pathway that involves two steps catalyzed by trehalose-6-phosphate synthesis (TPS) and trehalose-6-phosphate phosphatases (TPP). The expression of these TPS and TPP genes increases in response to osmotic stress in plants (Henry et al., 2015). Furthermore, rice transgenic plants expressing TPSP gene that encodes for a bifunctional fusion enzyme TPSP were found to accumulate trehalose that resulted in enhanced salt stress tolerance. These plants were also observed to have more content of soluble sugars (sucrose, glucose, and fructose) (Redillas et al., 2012). Hence, trehalose can be considered as an absolute marker for salt stress tolerance in rice and genes involved in its biosynthesis are the vital candidates for crop improvement using genetic engineering. Along with this, trehalose-6- phosphate (T6P) that is an intermediate in biosynthesis of trehalose has been identified as an important signaling molecule that regulates plant growth and development (Gazzarrini and Tsai, 2014). It globally controls the process of plant metabolism and transcription by inhibiting the sucrose-non-fermenting-1 related kinase-1 (SnRk1) activity (Gazzarrini and Tsai, 2014; Figueroa and Lunn, 2016). T6P and SnRk1 also interact in an antagonistic manner under stressful conditions and thus maintains energy homeostasis required for the survival of plants (Gazzarrini and Tsai, 2014). However, the detailed pathway through which T6P regulates these vital processes is still required to be elucidated in detail.
Sugar alcohols also play diverse roles during salt stress. They are involved in osmoregulation, ROS scavenging and at the same time acts as molecular chaperons (Gupta and Huang, 2014). Mannitol is the chemically reduced form of mannose and is of utmost importance among sugar alcohols during osmoregulation (Trontin et al., 2014). It accumulates in plant cells in response to low water potential that occurs due to salt stress. Along with osmoregulation, it also acts as an alternative carbon and energy source during stress. In a comparative metabolic analysis in rice salt sensitive and tolerant cultivars, the level of mannitol was found to consistenly increase under the effect of salt stress hinting its role in providing salt stress adaptation in rice halophytes (Zhao et al., 2014). Mannitol level in the plant cell is tightly regulated at the level of its biosynthesis and degradation. Modern research resources have given deeper insights into the identification of critical genes involved in these processes. It is synthesized from fructose-6-phosphate by the activity of two enzymes mannitol-1-phosphate dehydrogenase (M1PDH) and a mannitol-1-phosphatase (M1Pase) (Groisillier et al., 2014). Higher level of mannitol is transported to the sink tissues of the plants where it is metabolized by the action of enzyme mannitol dehydrogenase (MTD). Overexpression of MTLD gene that encodes for M1PDH protein has been reported to accumulate mannitol in the transgenic rice plants thus improving their salt tolerance (Iwamoto and Shiraiwa, 2005). On the contarary, only meager information is available on exploitation of M1Pase and MTD genes in improving stress tolerance in plants. Hence, modulation of these key anabolic and catabolic genes regulating the level of mannitol in plants can be considered as an effective strategy for improving salt tolerance in rice.
Glycine betaine (GB) is a water soluble, non-toxic ammonium compound that plays critical role in salt stress adaptation by enhancing the content of cellular osmolytes (Gupta and Huang, 2014). Its implication in osmotic adjustment, lowering of ROS, stabilization of proteins and photosynthetic apparatus has been well-demonstrated by various workers (Cha-um and Kirdmanee, 2010; Gupta and Huang, 2014). Within a cell, it is synthesized either from glycine or choline that involves the participation of many enzymes including glycine sarcosine N-methyl transferase (GSMT), sarcosine dimethylglycine N-methyl transferase (SDMT), choline monooxygenase (CMO) and betaine aldehyde dehydrogenase (BADH). In rice, the biosynthesis of GB is not reported till date due to the formation of non-functional proteins involved in its biosynthetic pathway. However, transgenic rice plants raised with overexpression of choline oxidase (cod A) gene isolated from bacteria Arthrobacter glabiformis were found to highly tolerate the salinity (Mohanty et al., 2002; Giri, 2011). Therefore, raising the transgenic rice plants using the genes that encode for functional proteins involved in GB biosynthesis can act as an effective strategy for enhancing the salt stress adaptation in rice.
ROS Homeostasis
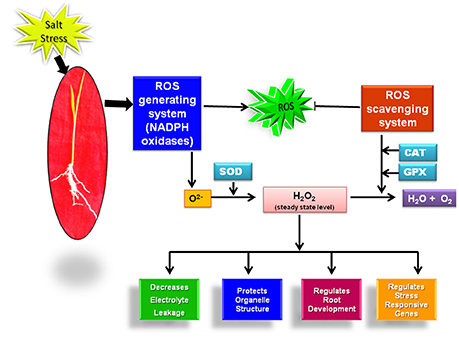
One of the pronounced effect of salinity stress on plants is the oxidative damage to the biomolecules like proteins, lipids triggered by reactive oxygen species (ROS) (AbdElgawad et al., 2016). ROS including superoxide radical (O2−), hydrogen peroxide (H2O2) and hydroxyl radical (OH−) are generated in the plant organelle mitochondria, chloroplast and cytoplasm in minute concentration when the electrons escaped from the electron transport chains are accepted by oxygen leading to the formation of O2− in the absence of other acceptors (Türkan and Demiral, 2009; Kaur et al., 2016a). At this low concenteration, ROS are involved in regulation of plant growth, development and stress adaptation (Kaur et al., 2016a). But under the effect of salt stress, the disruption of ROS homeostasis occurs which harms the plant (Baxter et al., 2014). Plants acclimatize under the salt stress conditions by restricting their ROS level to a threshold value with the help of ROS generating and scavenging machinery.
The utmost and extensively studied enzyme system involved in O2− generation is NADPH oxidase that catalyzes the shifting of electrons from NADPH to molecular oxygen (O2). In plants, NADPH oxidase genes are known as respiratory burst oxidase homologs (Rbohs) (Kaur et al., 2014). In rice, nine genes have been proposed to encode for Rbohs (Kaur et al., 2014). The expression of two rice genes OsRbohA and OsRbohI has been shown to be up-regulated at the transcript level under the effect of salt stress (Wang et al., 2013). Moreover, the enzymatic activity of NADPH oxidase has been reported to be more in salt tolerant cultivars of rice than the sensitive cultivars (Kaur et al., 2016a). These findings clearly hint toward the possible role of OsRboh genes in salt stress acclimization in rice.
Superoxide ions (O2−) generated by the action of NADPH oxidase is further converted into H2O2 by virtue of enzyme superoxide dismutase (SOD). H2O2 acts as an important signaling molecule and activates an array of stress responsive pathways in plants (Kaur et al., 2016a). Initially, H2O2 was considered as a harmful reactive molecule that damages the plants. But in the recent years, with the advancement of research tools, it has emerged as a vital molecule for plant growth and survival as it regulates a number of stress adaptive pathways (Elizabeth and Alison, 2011; Orabi et al., 2015). The level of H2O2 has been reported to increase in rice under the effect of NaCl. Its level is also observed to be constitutively high in salt tolerant cultivars of rice (Luna Sankhi and Luna Suvarna) (Kaur et al., 2016a). Although, H2O2 plays immensely important role in regulation of different plant processes, its level above threshold value can result in serious harm to the plant. So, its level in plants is tightly regulated with the help of SOD and H2O2 scavenging enzymes. The enzymatic activity of SOD increases in response to salt stress in rice plants (Sharma et al., 2015). Further, at the transcript level, higher gene expression level of two isoforms of SOD viz. Fe-SOD and Cu/Zn-SOD has been reported under the effect of salt stress (Sharma et al., 2013). Furthermore, overexpression studies conducted with cytosolic Cu/Zn-SOD and Fe-SOD have shown improved tolerance towards oxidative and salt stress in rice (Prashanth et al., 2008; Bose et al., 2013). A comparative study done on rice glycophytes and halophytes have also shown the higher expression level of Fe-SOD and Cu/Zn SOD genes in halophytic rice (Kaur et al., 2016a). Once the signaling cascade has been initiated, excessive H2O2 is scavenged by downstream antioxidant enzymes ascorbate oxidase (APX), catalase (CAT) and guaicol peroxidase (GR) (You and Chan, 2015). The expression level of H2O2 scavenging enzymes also shows interesting trend in response to salt stress in rice. The expression level of CAT, APX and GPX has been reported to increase under the effect of 100 mM of NaCl in rice cultivar Pusa basmati-1 (Sharma et al., 2015). The higher activity of these enzymes is even considered as a well-known marker for salinity tolerance in plants (Kaur et al., 2016a).
Till now; the majority of literature has linked the salt stress tolerance to the level of these antioxidant enzymes. But, in the past few years with the advancement of biochemical, microscopic and molecular tools, ROS generating NADPH oxidase enzyme system has also proved its candidature as a potential marker for salinity tolerance. It clearly indicates that along with our existing knowledge on the role of ROS scavenging system in rice salt stress acclimization, we should give equal emphasis to the ROS generating system for getting better insights into the mechanism of salt stress adaptation in rice as these two pathways may work in a coordinated manner for achieving salt stress tolerance (Figure 1).
Hormonal Regulation of Salt Stress
Phytohormones are well-known as the key players regulating plant growth and development. With the advent of modern research tools, their critical role in modulation of different abiotic stress responses has attained huge attention (Kohli et al., 2013; Wani et al., 2016). They act as a mediator for controlling the interaction between the environmental stresses and plant responses (Tao et al., 2008).
The plant hormone abscisic acid (ABA) was originally discovered in early 1960s and was known as an inhibitory rather than stimulatory hormone. It is reported to be involved in the process of stomata closure, bud dormancy, seed dormancy and inhibition of shoot growth (Rodríguez-Gacio et al., 2009). The critical role of ABA in regulation of salt stress adaptation is also well-established. The level of ABA increases in response to salt stress in rice and lead to the activation of cascade of responses regulating salt stress tolerance (Kumar et al., 2013). The most predominant ABA mediated response that regulates salt stress tolerance is stomatal closure, thus preventing the loss of water as a stress adaptive feature (Sah et al., 2016). Recently, it is found that ABA also modulates root architecture in response to stress. The increased ABA level increases the root biomass leading to higher root to shoot ratio (Sah et al., 2016). Modern research tools and genetic resources have helped the researchers in deciphering the detailed pathway and identification of key genes involved in ABA biosynthesis and catabolism that could be targeted for achieving salt tolerance. NCED (9-cis-epoxycarotenoid dioxygenase) is the critical gene that encodes for the rate limiting enzyme involved in ABA biosynthesis in plants (Jisha et al., 2015). Recently, the overexpression of a rice transcription factor OsEREBP1 has been shown to increase the expression of NCED that in turn confers salt tolerance in rice (Jisha et al., 2015). These studies show that manipulating the expression levels of key genes involved in ABA biosynthesis and degradation can be considered as an effective approach for achieving salt tolerance in rice.
The phytohormone auxin is classically known as a master regulator of plant root development (Saini et al., 2013). But, in the past few years, the role of auxins in salt stress tolerance has also been well-realized (Du et al., 2013; Zörb et al., 2013). The level of different types of auxins like IAA and IBA increases significantly in response to salt stress. They are known to modulate the root architecture of the plant under stress conditions for achieving salt tolerance (Kazan, 2013). Auxins also increase the photosynthetic pigment concentration, alter the activities of key antioxidant enzymes and reduces the membrane permeability for conferring salt tolerance (Kaya et al., 2013). However, the specific role of auxin in providing salt stress tolerance in rice is still not well-understood. The advanced research tools have played a pivotal role in the identification of the key genes involved in auxin biosynthesis, transport and degradation that constitutes an essential component of the plant's abiotic stress tolerance mechanisms. Further, exploration of the role of these genes using the current research techniques can ultimately lead to the better understanding of role of auxins in development of crops which are better adapted to stressful environments.
Brassinosteroids are the steroidal plant hormones that are indispensible for plant growth and development as they regulate a plethora of vital plant processes including growth, flowering, seed germination, senescence, rhizogenesis, abscission and maturation (Gudesblat and Russinova, 2011; Fridman and Savaldi-Goldstein, 2013). In the past decade, the involvement of brassinosteroids has been well-established in the amelioration of different abiotic stresses in rice (Bajguz and Hayat, 2009). Exogenous treatment of brassinosteroids leads to salt stress adaptation in rice through the modulation of ROS machinery (Sharma et al., 2015). Brassinosteroids promote the transcript level expression and protein activity of different ROS detoxification enzymes such as superoxide dismutase, ascorbate peroxidase and catalase (Bajguz and Hayat, 2009; Choudhary et al., 2012; Sharma et al., 2015). Brassinosteroids also prevent the water loss during salt stress by inactivating the YDA-mediated signaling cascades of stomata development which is triggered by BIN2 (Kumar et al., 2013).
Gibberellic acid (GA) has been well-known as a growth stimulating hormones in plants (Gupta and Chakrabarty, 2013). It also mitigates the drastic effects of salt stress in different crops by enhancing the carbohydrates, proline, protein and amino acid content (Shaddad et al., 2013). The increased endogenous level of GA has been linked to salt stress tolerance in rice (Sahoo et al., 2014). However, the detailed mechanism how GA regulates salt tolerance is still not well-explored in rice. Modern research tools have enhanced our understanding of the detailed pathway involved in GA biosynthesis, signaling and degradation. Recently, the GA signaling genes (ga3ox1-3, rga28, gai-2, della) have been linked to endoreduplication during cell cycle in response to mild osmotic stress (Colebrook et al., 2014). Exploring the role of these genes using transgenics approach can help further in deduction of the probable mechanism of action of gibberellic acid in salt stress amelioration in rice.
Cytokinin is a well-known plant growth regulator that play a pivotal role in many of the plant processes including cell division, photomorphogenetic differentiation, photosynthesis promotion and inhibition of leaf senescence (Wang et al., 2015; Zwack and Rashotte, 2015). The level of cytokinins increases during salt stress thus improving the salt stress adaptation by reprogramming the transcript level expression of genes related to plant growth and development. It helps in retaining the expression of these genes which otherwise is masked under the effect of salt stress (Golan et al., 2016). The exogenous application of cytokinins increases the grain yield of rice under the salt stress conditions (Javid et al., 2011) However, a complete understanding of the events that regulate cytokinin mediated salt stress tolerance is still lacking. This could be due to the complex communication between stress signaling and cytokinins (Zwack and Rashotte, 2015). Further, overexpression of cytokinin biosynthetic gene AtlPT8 (Adenosine phosphate-isopentenyl transferase 8) decreases the salt stress tolerance in Arabidopsis by increasing the level of ROS, inhibition of primary root growth and decreasing the chlorophyll content (Wang et al., 2015). Therefore, the role of cytokinins in regulation of salt stress adaptation is contradictory as there are equal number of evidences supporting the positive as well as negative role of these in salt stress tolerance (Wang et al., 2015; Zwack and Rashotte, 2015). Hence, further comprehensive study is required to solve the mystery that how cytokinins regulate salt stress adaptation pathways.
Polyamines are a class of aliphatic nitrogen containing plant growth regulator that plays a significant role in cellular growth and development in plants (Gill and Tuteja, 2010). They are also well-known to regulate salt stress adaptation in plants. They confer salt stress tolerance mainly by modulation of ROS homeostasis (Liu et al., 2015; Saha et al., 2015). Recently, they have been hypothesized to regulate the process of ion transport during salt stress adaptation (Saha et al., 2015). However, the crosstalk that exists between polyamines, ROS and ion transporters is complex and more research is required to decipher this. Further, there is scarcity of information available on the signaling cascade of how polyamine genes are linked to salt stress responses (Liu et al., 2015).
Strigolactones are the plant signaling molecules which have been added in the growing list of plant growth regulators in the past decade (Smith and Li, 2014; Pandey et al., 2016). They are well-known for their role in plant-microbe symbiotic interactions, root growth, internode elongation and senescence (Van Ha et al., 2014; Pandey et al., 2016). The role of strogolactones has also been well-established in salt stress adaptation using loss-of-function mutation. The loss of function of strigolactone biosynthetic genes (MAX) increases the salt stress susceptivity establishing a direct positive link between strigolactone and salt stress adaptation (Van Ha et al., 2014). However, serious research efforts are still required to uncover the detailed signaling mechanism of strigolactone mediated salt stress tolerance particularly in rice.
Molecular Regulation of Salinity Tolerance
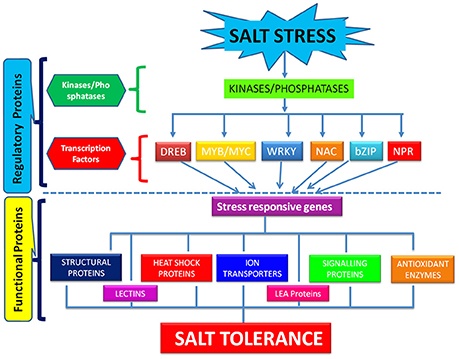
The process of salt tolerance is regulated by an array of genes at the molecular level. The identification and characterization of these stress responsive genes is a prerequisite for engineering stress tolerant crops (Himabindu et al., 2016). Till date, a number of genes have been identified and validated using modern molecular biology and transgenics approach (Table 1). These are further divided into two categories: regulatory genes and functional genes (Figure 2). Regulatory genes including transcription factors, protein phosphatases and protein kinases encode for the proteins which controls the various stress responsive signaling pathways (Yoshida et al., 2014). On the other hand functional proteins include important enzymes and metabolic proteins that directly protect the plants under stress (Kosová et al., 2013).
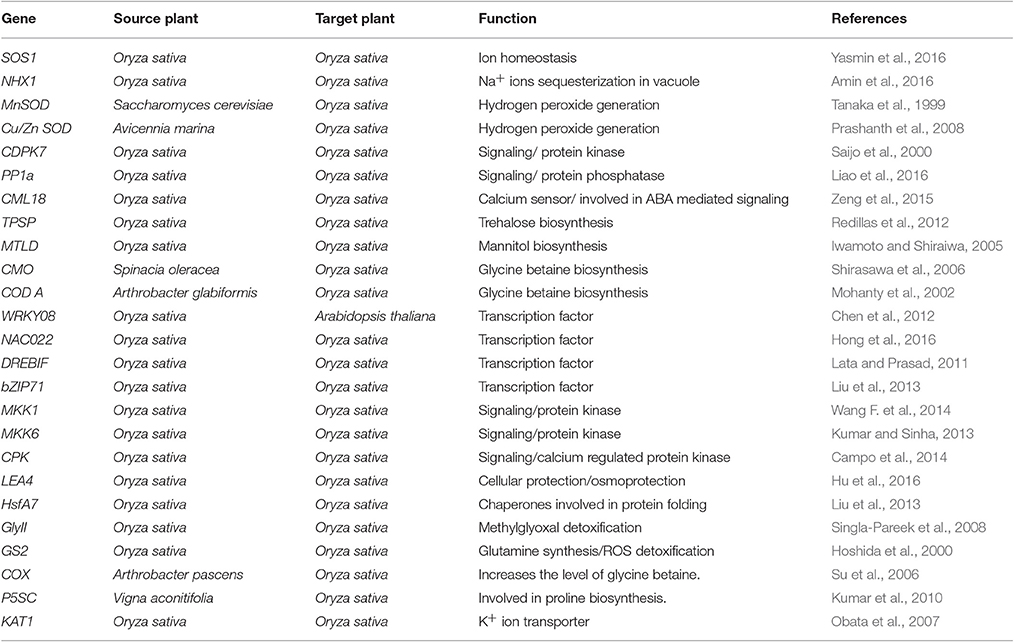
Table 1. Summary of the genes used for improvement of salinity tolerance in rice using transgenics approach.
Regulatory Proteins
All the mechanisms involved in salinity tolerance are tightly regulated by different transcription factors and genes that are either up-regulated or down-regulated in response to salt stress and thereby serve distinct functions in stress adaptation (Yoshida et al., 2014). The major transcription factors playing critical role in salt stress tolerance comprises WRKY, NAC, DREB, bZIP, AP2, C2H2 zinc finger families (Gupta and Huang, 2014). All these bind to the regulatory regions of the stress responsive genes and hence modulate their expression for conferring salt tolerance. WRKY belongs to one of the largest family of transcription factors involved in regulation of salt stress signaling pathways in rice (Banerjee and Roychoudhury, 2015). Overexpression of rice OsWRKY08 gene has been reported to enhance the salt tolerance in Arabidopsis through regulation of stress responsive COR and RD genes in an ABA independent manner (Chen et al., 2012). NAC family of transcription factors are also the key players in plant's fight against salt stress (Hong et al., 2016). But the detailed function of many of the members of this family in abiotic stress tolerance is not well-understood in rice. Recently, a rice OsNAC022 gene was overexpressed in rice that lead to salt and drought tolerance by increasing the gene expression of functional LEA and ERD genes (Hong et al., 2016). DREB is another one of the most critical transcription factors to synchronize the plants response toward salinity. They recognize and bind to the DRE motif in the promoter region of stress responsive genes. Among the major DRE binding proteins, the overexpression of rice DREB genes like OsDREBIF confers salt tolerance (Lata and Prasad, 2011). It confers salt tolerance in rice in a ABA dependent and ABA independent manner. They regulate the expression of an array of genes like ERD, KIN, and RD for conferring salt tolerance in rice. GRAS belongs to the plant specific class of transcription factors that play a significant role in achieving salt stress tolerance (Xu et al., 2015). Overexpression of GRAS transcription factors in plants like Arabidopsis thaliana has proved its role in salt tolerance but very less information is available in literature upon its role in regulation of salt stress in rice (Ma et al., 2010). MYB/MYC transcription factors are well-known for their role in abiotic stress tolerance (Dubos et al., 2010). They regulate the expression of salt stress signaling genes like those encoding ion transporters, proline synthase etc. in an ABA dependent manner for conferring salt tolerance (Yang et al., 2012). bZIP belongs to the largest family of transcription factors in plants. They play significant role in salt tolerance of rice in an ABA dependent manner (Liu et al., 2013). Overexpression of OsbZIP71gene increased the expression of stress regulated RD and NHX1 genes. RD genes encode for cell protective LEA/Dehydrin proteins and NHX1 plays significant role in ion compartmentalization. Moreover, it increased the transcript level of OsMYB4 which is involved in salt tolerance (Liu et al., 2014). Although, the role of NPR1 in biotic stress tolerance is well-established, its role in abiotic stress tolerance also cannot be ignored (Srinivasan et al., 2009). It interacts with bZIP transcription factor and leads to salt and drought tolerance in rice by regulating the expression of PR proteins (Alves et al., 2013).
Protein kinases and phosphates are the key enzymatic proteins that play a pivotal role in salt stress adaptation. They are involved in the regulation of the key components of stress signaling pathways by their phosphorylation and dephosphorylation, respectively. They act as on/off switch for controlling the salt stress adaptive pathways (País et al., 2009). The most widely studied protein kinases that play immensely important role in salt stress adaptation are mitogen activated protein kinases (MAPKs). Their involvement has been reported in response to salt stress in rice (De Zelicourt et al., 2016). They are considered as a central hub in different salt stress adaptation pathways like ROS signaling, ion homeostasis and hormonal regulated salt tolerance (Golldack et al., 2014). In the context of abiotic stress tolerance in plants, a MAP kinase core composed of 3 MAPK kinases viz.MEKK1→MKK2→MPK4/MPK6 has been identified. But still a complete cascade of MAPK involved in salt stress adaptation is missing. Researchers have established the role of different members of MAPKs in rice in response to salt stress by hunting a transgenic circuit approach (Moustafa, 2014). Overexpression of MKK1 that regulates the activity of MPK4 confers salt stress tolerance in rice through a cascade of signaling events (Wang F. et al., 2014). Further, the constitutive activation of OsMKK6 achieved through site directed mutagenesis of serine and threonine residues into glutamic acid improved salt tolerance in rice cultivar Pusa Basmati-1 suggesting the definite role of MKK6 in salt stress adaptation pathways in rice (Kumar and Sinha, 2013). Apart from MAPKs, calcium dependent protein kinases (CDPKs) are also critical for salt stress adaptation. They belong to the serine/threonine kinases family and translate the cytosolic Ca2+ stimuli generated by salinity into phosphorylation signals (Latz et al., 2013). The overexpression of rice OsCPK4 gene is reported to confer salt stress tolerance in plants by inhibiting the lipid peroxidation of plasma membrane (Campo et al., 2014). Although, CDPK genes are known to behave as an integral part of salt stress adaptive pathways, the function of only a few members has been deduced in rice. Hence, functional characterization of various salt stress responsive CDPKs is of vital importance for achieving salt stress tolerance in rice.
In contrast to the plenty of information available on the protein kinase enzymes, the knowledge on the role of protein phosphatases in salt stress adaptation is very limited. They are usually considered as unexciting enzymes as they are involved in the turning off of the signaling cascades triggered by protein kinases (País et al., 2009). But, recently it has been documented that it positively regulate the salt stress responsive pathways (Liao et al., 2016). In a latest research, overexpression of protein phosphatase 1a (OsPP1a) has been reported to enhance salinity tolerance in rice (Liao et al., 2016). In an another study, a rice phosphatase 2C has been found to confer salt tolerance in Arabidopsis (Singh et al., 2015). Thus, protein phosphatases can also be useful along with transcription factors and protein kinases in enhancing the salt tolerance in rice.
Functional Proteins
The identification and characterization of different proteins playing critical roles in salt stress signaling acts as a fascinating approach for understanding the complex process of salt tolerance (Himabindu et al., 2016). The major functional proteins involved in the process of salt tolerance are heat shock proteins (HSPs), late embryogenesis abundant (LEA) proteins, ion transporters, ROS detoxification enzymes, etc. (Kosová et al., 2013). LEA proteins are the low molecular weight hydrophilic proteins that belong to a huge and diverse gene family present in different plant species (Bhardwaj et al., 2013; Gao and Lan, 2016). They are rich in glycine, glutamic acid, lysine, and threoinine amino acids. The high content of glycine enables them to adopt random- coiled structures (Hu et al., 2016). The role of LEA proteins in conferring salt and drought tolerance in various plants is well-established. LEA proteins directly accumulate in plant cells in response to salt stress and thereof confers tolerance by mainly protecting the mitochondrial membranes. They also function as enzyme protectants, molecular chaperones, multi-functional water-binding molecules, membrane stabilizers and space fillers, thus inhibiting cellular collapse during salt stress (Liang et al., 2013; Hu et al., 2016). In a recent, study overexpression of a OsLEA4 gene that encodes for a LEA protein has been reported to confer salt tolerance in rice (Hu et al., 2016). These studies indicate that LEA family members are the absolutely potential candidates for genetic modifications in rice for achieving salt tolerance.
HS Ps are stress inducible proteins having chaperone activity and are synthesized in response to almost all kind of stresses in plants (Wang et al., 2004; Wang Y. et al., 2014; Hu et al., 2017). According to their molecular weight, major categories of HSPs are small heat shock proteins, Hsp60, Hsp70, Hsp90 and Hsp100 (Wang Y. et al., 2014). The expression of HSPs in response to different stresses is regulated by the transcription factors known as heat shock transcription factors (Hsfs; Liu et al., 2013). Overexpression rice OsHsfA7 gene encoding HSP transcription factor has been reported to confer salt stress tolerance in rice (Liu et al., 2013). Further, the overexpression of LimHSP16.45 which is a small heat shock protein has been shown to confer multiple abiotic stress tolerance including salinity in Arabidopsis (Mu et al., 2013). These reports emphasize on the prospective role of HSPs in achieving salt stress tolerance in rice.
Lectins are the carbohydrate binding proteins that possesses at least one non catalytic domain that binds reversibly and specifically to carbohydrates and their derivatives, without altering their structure (De Hoff et al., 2009). They are found to have a ubiquitous distribution in all kingdoms of life playing a diverse array of roles (De Hoff et al., 2009). The role of plant lectins have been well-explored in relation to regulation of innate immune responses against predators and pathogens (De Hoff et al., 2009). However, from the past few years, they are in limelight for their potential role in regulation of abiotic stress responses in plants. A plethora of uncharacterized abiotic stress responsive lectins have been identified at the genomic scale in plants suggesting their novel and unexplored roles (Komath et al., 2006; De Hoff et al., 2009). A study conducted on rice suggests that mannose binding lectins are involved in regulation of salt stress tolerance (Sharma et al., 2013). But the exact role of lectins in salt stress adaptation in rice is completely unrevealed till date. As these proteins accumulate in response to salt stress, their functional characterization can open a new chapter in the area of rice salt stress management in future.
Conclusion
Rice is considered an important cereal crop and greatly contributes toward world's food security. Hence, deciphering strategies to mitigate major abiotic stresses such as salinity which drastically affect rice production is in the top of agenda of scientists. The scientific inputs on the subject suggest that salinity stress is a complex problem and it cannot be addressed through a singular approach. Hence, it is imperative to understand the various levels through which it operates and develop suitable strategies by integrating various approaches. The initial efforts in this direction focused on identifying salt tolerant cultivars and thus developing useful markers for breeding purposes to obtain salt tolerant varieties. Further, information obtained on salt stress induced responses and critical insights to the adaptive stress mechanisms including ion homeostasis, osmoregulation, ROS homeostasis of rice plants provides necessary clues for developing plants which could efficiently withstand salt stress. Besides this, the detailed understanding of the different processes at the physiological, biochemical and molecular levels has provided a suitable platform to formulate the future strategies to solve this complex problem which is linked to food security of the world. In conclusion, our effort in integrating knowledge of classical studies and modern approaches will definitely provide a better understanding and facilitate appropriate remedial strategies for salinity stress tolerance in rice.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
NK is thankful to the Department of Biotechnology (Govt. of India) for Senior Research Fellowship.
References
AbdElgawad, H., Zinta, G., Hegab, M. M., Pandey, R., Asard, H., and Abuelsoud, W. (2016). High salinity induces different oxidative stress and antioxidant responses in maize seedlings organs. Front. Plant Sci. 7:276. doi: 10.3389/fpls.2016.00276
Agarwal, P., Parida, S. K., Raghuvanshi, S., Kapoor, S., Khurana, P., Khurana, J. P., et al. (2016). Rice improvement through genome-based functional analysis and molecular breeding in India. Rice 9:1. doi: 10.1186/s12284-015-0073-2
Alves, M. S., Dadalto, S. P., Gonçalves, A. B., De Souza, G. B., Barros, V. A., and Fietto, L. G. (2013). Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14, 7815–7828. doi: 10.3390/ijms14047815
Amin, U. S. M., Biswas, S., Elias, S. M., Razzaque, S., Haque, T., Malo, R., et al. (2016). Enhanced salt tolerance conferred by the complete 2.3 kb cDNA of the rice vacuolar Na+/H+ antiporter gene compared to 1.9 kb coding region with 5′-UTR in transgenic lines of rice. Front. Plant Sci. 7:14. doi: 10.3389/fpls.2016.00014
Bagdi, D. L., Shaw, B. P., Sahu, B. B., and Purohit, G. K. (2015). Real time PCR expression analysis of gene encoding p5cs enzyme and proline metabolism under NaCI salinity in rice. J. Environ. Biol. 36, 955–961.
Bajguz, A., and Hayat, S. (2009). Effects of brassinosteroids on the plant responses to environmental stresses. Plant Physiol. Biochem. 47, 1–8. doi: 10.1016/j.plaphy.2008.10.002
Banerjee, A., and Roychoudhury, A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci. World J. 17, 1127–1142. doi: 10.1155/2015/807560
Barragán, V., Leidi, E. O., Andrés, Z., Rubio, L., De Luca, A., Fernández, J. A., et al. (2012). Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell. 24, 1127–1142. doi: 10.1105/tpc.111.095273
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bhardwaj, R., Sharma, I., Kanwar, M., Sharma, R., Handa, N., Kaur, H., et al. (2013). “LEA proteins in salt stress tolerance,” in Salt Stress in Plants (New York, NY: Springer), 79–112. doi: 10.1007/978-1-4614-6108-1_5
Bojórquez-Quintal, E., Velarde-Buendía, A., Ku-González, A., Carillo-Pech, M., Ortega-Camacho, D., Echevarría-Machado, I., et al. (2014). Mechanisms of salt tolerance in habanero pepper plants (Capsicum chinense Jacq.): Proline accumulation, ions dynamics and sodium root-shoot partition and compartmentation. Front. Plant Sci. 5:605. doi: 10.3389/fpls.2014.00605
Bose, J., Rodrigo-Moreno, A., and Shabala, S. (2013). ROS homeostasis in halophytes in the context of salinity stress tolerance. J. Exp. Bot. 65, 1241–1257. doi: 10.1093/jxb/ert430
Brini, F., and Masmoudi, K. (2012). Ion transporters and abiotic stress tolerance in plants. ISRN Mol. Biol. 2012:927436. doi: 10.5402/2012/927436
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., and San Segundo, B. (2014). Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. doi: 10.1104/pp.113.230268
Cardi, M., Castiglia, D., Ferrara, M., Guerriero, G., Chiurazzi, M., and Esposito, S. (2015). The effects of salt stress cause a diversion of basal metabolism in barley roots: possible different roles for glucose-6-phosphate dehydrogenase isoforms. Plant Physiol. Biochem. 86, 44–54. doi: 10.1016/j.plaphy.2014.11.001
Carillo, P., Annunziata, M. G., Pontecorvo, G., Fuggi, A., and Woodrow, P. (2011). “Salinity stress and salt tolerance,” in Abiotic Stress in Plants-Mechanisms and Adaptations, ed Arun Shanker (InTech). doi: 10.5772/22331
Cha-um, S., and Kirdmanee, C. (2010). Effect of glycinebetaine on proline, water use, and photosynthetic efficiencies, and growth of rice seedlings under salt stress. Turk. J. Agric. Forest. 34, 517–527. doi: 10.3906/tar-0906-34
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., and Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Choudhary, S. P., Yu, J. Q., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2012). Benefits of brassinosteroid crosstalk. Trends Plant Sci. 17, 594–605. doi: 10.1016/j.tplants.2012.05.012
Colebrook, E. H., Thomas, S. G., Phillips, A. L., and Hedden, P. (2014). The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 217, 67–75. doi: 10.1242/jeb.089938
De Hoff, P. L., Brill, L. M., and Hirsch, A. M. (2009). Plant lectins: the ties that bind in root symbiosis and plant defense. Mol. Genet. Genomics 282, 1–15. doi: 10.1007/s00438-009-0460-8
De Zelicourt, A., Colcombet, J., and Hirt, H. (2016). The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 21, 677–685. doi: 10.1016/j.tplants.2016.04.004
Deinlein, U., Stephan, A. B., Horie, T., Luo, W., Xu, G., and Schroeder, J. I. (2014). Plant salt-tolerance mechanisms. Trends Plant Sci. 19, 371–379. doi: 10.1016/j.tplants.2014.02.001
Du, H., Liu, H., and Xiong, L. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4:397. doi: 10.3389/fpls.2013.00397
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Elizabeth, V., and Alison, D. (2011). Hydrogen peroxide as a signaling molecule. Antioxid. Redox Signal. 15, 147–151. doi: 10.1089/ars.2011.3968
Fan, X., Tang, Z., Tan, Y., Zhang, Y., Luo, B., Yang, M., et al. (2016). Overexpression of a pH-sensitive nitrate transporter in rice increases crop yields. Proc. Natl. Acad. Sci. 113, 7118–7123. doi: 10.1073/pnas.1525184113
Figueroa, C. M., and Lunn, J. E. (2016). A tale of two sugars: trehalose 6-phosphate and sucrose. Plant Physiol. 172, 7–27. doi: 10.1104/pp.16.00417
Fita, A., Rodríguez-Burruezo, A., Boscaiu, M., Prohens, J., and Vicente, O. (2015). Breeding and domesticating crops adapted to drought and salinity: a new paradigm for increasing food production. Front. Plant Sci. 6:978. doi: 10.3389/fpls.2015.00978
Flowers, T. J., and Colmer, T. D. (2008). Salinity tolerance in halophytes. New Phytologist. 179, 945–963. doi: 10.1111/j.1469-8137.2008.02531.x
Fridman, Y., and Savaldi-Goldstein, S. (2013). Brassinosteroids in growth control: how, when and where. Plant Sci. 209, 24–31. doi: 10.1016/j.plantsci.2013.04.002
Gao, J., and Lan, T. (2016). Functional characterization of the late embryogenesis abundant (LEA) protein gene family from Pinus tabuliformis (Pinaceae) in Escherichia coli. Sci. Rep. 6:19467. doi: 10.1038/srep19467
Garriga, M., Raddatz, N., Véry, A. A., Sentenac, H., Rubio-Meléndez, M. E., González, W., et al. (2017). Cloning and functional characterization of HKT1 and AKT1 genes of Fragaria spp.—Relationship to plant response to salt stress. J. Plant Physiol. 210, 9–17. doi: 10.1016/j.jplph.2016.12.007
Gazzarrini, S., and Tsai, A. Y. L. (2014). Trehalose-6-phosphate and SnRK1 kinases in plant development and signaling: the emerging picture. Front. Plant Sci. 5:119. doi: 10.3389/fpls.2014.00119
Geilfus, C. M., Mithöfer, A., Ludwig-Müller, J., Zörb, C., and Muehling, K. H. (2015). Chloride-inducible transient apoplastic alkalinizations induce stomata closure by controlling abscisic acid distribution between leaf apoplast and guard cells in salt-stressed Vicia faba. New Phytologist. 208, 803–816. doi: 10.1111/nph.13507
Gill, S. S., and Tuteja, N. (2010). Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 5, 26–33. doi: 10.4161/psb.5.1.10291
Giri, J. (2011). Glycinebetaine and abiotic stress tolerance in plants. Plant Signal. Behav. 6, 1746–1751. doi: 10.4161/psb.6.11.17801
Golan, Y., Shirron, N., Avni, A., Shmoish, M., and Gepstein, S. (2016). Cytokinins induce transcriptional reprograming and improve arabidopsis plant performance under drought and salt stress conditions. Front. Environ. Sci. 4:63. doi: 10.3389/fenvs.2016.00063
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. doi: 10.3389/fpls.2014.00151
Gregorio, G. B., Senadhira, D., Mendoza, R. D., Manigbas, N. L., Roxas, J. P., and Guerta, C. Q. (2002). Progress in breeding for salinity tolerance and associated abiotic stresses in rice. Field Crops Res. 76, 91–101. doi: 10.1016/S0378-4290(02)00031-X
Groisillier, A., Shao, Z., Michel, G., Goulitquer, S., Bonin, P., Krahulec, S., et al. (2014). Mannitol metabolism in brown algae involves a new phosphatase family. J. Exp. Bot. 65, 559–570. doi: 10.1093/jxb/ert405
Gudesblat, G. E., and Russinova, E. (2011). Plants grow on brassinosteroids. Curr. Opin. Plant Biol. 14, 530–537. doi: 10.1016/j.pbi.2011.05.004
Gupta, B., and Huang, B. (2014). Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int. J. Genomics 2014:701596. doi: 10.1155/2014/701596
Gupta, R., and Chakrabarty, S. K. (2013). Gibberellic acid in plant: still a mystery unresolved. Plant Signal. Behav. 8:e25504. doi: 10.4161/psb.25504
Hadi, S. M., Ibrahim, K. M., and Yousif, S. I. (2014). Effect of shock and elevated levels of mannitol on callus growth, regeneration and proline accumulation in Ruta graveolens cultures. Int. J. Curr. Microbiol. Appl. Sci. 3, 479–488.
Hamamoto, S., Horie, T., Hauser, F., Deinlein, U., Schroeder, J. I., and Uozumi, N. (2015). HKT transporters mediate salt stress resistance in plants: From structure and function to the field. Curr. Opin. Biotechnol. 32, 113–120. doi: 10.1016/j.copbio.2014.11.025
Henry, C., Griffiths, C. A., Paul, M. J., Kollman, A., Soulaiman, S. A. K. R., and Lagrimini, M. (2015). Differential role for trehalose metabolism in salt stressed maize. Plant Physiol. 169, 1072–1089. doi: 10.1104/pp.15.00729
Himabindu, Y., Chakradhar, T., Reddy, M. C., Kanygin, A., Redding, K. E., and Chandrasekhar, T. (2016). Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 124, 39–63. doi: 10.1016/j.envexpbot.2015.11.010
Hong, Y., Zhang, H., Huang, L., Li, D., and Song, F. (2016). Overexpression of a stress-responsive NAC transcription factor gene ONAC022 improves drought and salt tolerance in rice. Front. Plant Sci. 7:4. doi: 10.3389/fpls.2016.00004
Horie, T., Karahara, I., and Katsuhara, M. (2012). Salinity tolerance mechanisms in glycophytes: an overview with the central focus on rice plants. Rice 5:11. doi: 10.1186/1939-8433-5-11
Hoshida, H., Tanaka, Y., Hibino, T., Hayashi, Y., Tanaka, A., Takabe, T., et al. (2000). Enhanced tolerance to salt stress in transgenic rice that overexpresses chloroplast glutamine synthetase. Plant Mol. Biol. 43, 103–111. doi: 10.1023/A:1006408712416
Hu, T., Zhu, S., Tan, L., Qi, W., He, S., and Wang, G. (2016). Overexpression of OsLEA4 enhances drought, high salt and heavy metal stress tolerance in transgenic rice (Oryza sativa L.). Environ. Exp. Bot. 123, 68–77. doi: 10.1016/j.envexpbot.2015.10.002
Hu, W., Ding, Z., Tie, W., Yan, Y., Liu, Y., Wu, C., et al. (2017). Comparative physiological and transcriptomic analyses provide integrated insight into osmotic, cold, and salt stress tolerance mechanisms in banana. Sci. Rep. 7:43007. doi: 10.1038/srep43007
Hussain, M. I., Lyra, D. A., Farooq, M., Nikoloudakis, N., and Khalid, N. (2016). Salt and drought stresses in safflower: a review. Agron. Sustain. Dev. 36, 1–31. doi: 10.1007/s13593-015-0344-8
Iwamoto, K., and Shiraiwa, Y. (2005). Salt-regulated mannitol metabolism in algae. Mar. Biotechnol. 7, 407–415. doi: 10.1007/s10126-005-0029-4
Javid, M. G., Sorooshzadeh, A., Moradi, F., Sanavy, S. A. M. M., and Allahdadi, I. (2011). The role of phytohormones in alleviating salt stress in crop plants. Aust. J. Crop Sci. 5:726.
Jisha, V., Dampanaboina, L., Vadassery, J., Mithöfer, A., Kappara, S., and Ramanan, R. (2015). Overexpression of an AP2/ERF type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 10:e0127831. doi: 10.1371/journal.pone.0127831
Kaur, G., Sharma, A., Guruprasad, K., and Pati, P. K. (2014). Versatile roles of plant NADPH oxidases and emerging concepts. Biotechnol. Adv. 32, 551–563. doi: 10.1016/j.biotechadv.2014.02.002
Kaur, N., Dhawan, M., Sharma, I., and Pati, P. K. (2016a). Interdependency of reactive oxygen species generating and scavenging system in salt sensitive and salt tolerant cultivars of rice. BMC Plant Biol. 16:1. doi: 10.1186/s12870-016-0824-2
Kaur, N., Kirat, K., Saini, S., Sharma, I., Gantet, P., and Pati, P. K. (2016b). Reactive oxygen species generating system and brassinosteroids are linked to salt stress adaptation mechanisms in rice. Plant Signal. Behav. 11:e1247136 doi: 10.1080/15592324.2016.1247136
Kaya, C., Ashraf, M., Dikilitas, M., and Tuna, A. L. (2013). Alleviation of salt stress-induced adverse effects on maize plants by exogenous application of indoleacetic acid (IAA) and inorganic nutrients-A field trial. Aust. J. Crop Sci. 7:249.
Kazan, K. (2013). Auxin and the integration of environmental signals into plant root development. Ann. Bot. 112, 655–665. doi: 10.1093/aob/mct229
Khare, T., Kumar, V., and Kishor, P. K. (2015). Na+ and Cl− ions show additive effects under NaCl stress on induction of oxidative stress and the responsive antioxidative defense in rice. Protoplasma 252, 1149–1165. doi: 10.1007/s00709-014-0749-2
Kohli, A., Sreenivasulu, N., Lakshmanan, P., and Kumar, P. P. (2013). The phytohormone crosstalk paradigm takes center stage in understanding how plants respond to abioticstresses. Plant Cell Rep. 32, 945–957. doi: 10.1007/s00299-013-1461-y
Komath, S. S., Kavitha, M., and Swamy, M. J. (2006). Beyond carbohydrate binding: new directions in plant lectin research. Organ. Biomol. Chem. 4, 973–988. doi: 10.1039/b515446d
Kosová, K., Prášil, I. T., and Vítámvás, P. (2013). Protein contribution to plant salinity response and tolerance acquisition. Int. J. Mol. Sci. 14, 6757–6789. doi: 10.3390/ijms14046757
Kumar, K., and Sinha, A. K. (2013). Overexpression of constitutively active mitogen activated protein kinase kinase 6 enhances tolerance to salt stress in rice. Rice 6:25. doi: 10.1186/1939-8433-6-25
Kumar, K., Kumar, M., Kim, S. R., Ryu, H., and Cho, Y. G. (2013). Insights into genomics of salt stress response in rice. Rice 6:27. doi: 10.1186/1939-8433-6-27
Kumar, V., Shriram, V., Kishor, P. B. K., Jawali, N., and Shitole, M. G. (2010). Enhanced proline accumulation and salt stress tolerance of transgenic indica rice by over-expressing P5CSF129A gene. Plant Biotechnol. Rep. 4, 37–48. doi: 10.1007/s11816-009-0118-3
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Latz, A., Mehlmer, N., Zapf, S., Mueller, T. D., Wurzinger, B., Pfister, B., et al. (2013). Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 6, 1274–1289. doi: 10.1093/mp/sss158
Li, B., Tester, M., and Gilliham, M. (2017). Chloride on the move. Trends Plant Sci. 22, 236–248. doi: 10.1016/j.tplants.2016.12.004
Liang, J., Zhou, M., Zhou, X., Jin, Y., Xu, M., and Lin, J. (2013). JcLEA, a novel LEA-like protein from Jatropha curcas, confers a high level of tolerance to dehydration and salinity in Arabidopsis thaliana. PLoS ONE 8:e83056. doi: 10.1371/journal.pone.0083056
Liao, Y.-D., Lin, K.-H., Chen, C.-C., and Chiang, C.-M. (2016). Oryza sativa protein phosphatase 1a (OsPP1a) involved in salt stress tolerance in transgenic rice. Mol. Breed. 36:22. doi: 10.1007/s11032-016-0446-2
Liu, A.-L., Zou, J., Liu, C.-F., Zhou, X.-Y., Zhang, X.-W., Luo, G.-Y., et al. (2013). Over-expression of OsHsfA7 enhanced salt and drought tolerance in transgenic rice. BMB Rep. 46, 31–36. doi: 10.5483/BMBRep.2013.46.1.090
Liu, C., Mao, B., Ou, S., Wang, W., Liu, L., Wu, Y., et al. (2014). OsbZIP71, a bZIP transcription factor, confers salinity and drought tolerance in rice. Plant Mol. Biol. 84, 19–36. doi: 10.1007/s11103-013-0115-3
Liu, J. H., Wang, W., Wu, H., Gong, X., and Moriguchi, T. (2015). Polyamines function in stress tolerance: from synthesis to regulation. Front. Plant Sci. 6:827. doi: 10.3389/fpls.2015.00827
Ma, D. M., Xu, W. R., Li, H. W., Jin, F. X., Guo, L. N., Wang, J., et al. (2014). Co-expression of the Arabidopsis SOS genes enhances salt tolerance in transgenic tall fescue (Festuca arundinacea Schreb.). Protoplasma 251, 219–231. doi: 10.1007/s00709-013-0540-9
Ma, H. S., Liang, D., Shuai, P., Xia, X. L., and Yin, W. L. (2010). The salt-and drought-inducible poplar GRAS protein SCL7 confers salt and drought tolerance in Arabidopsis thaliana. J. Exp. Bot. 61, 4011–4019. doi: 10.1093/jxb/erq217
Maathuis, F. J. M., Ahmad, I., and Patishtan, J. (2014). Regulation of Na+ fluxes in plants. Front. Plant Sci. 5:467. doi: 10.3389/fpls.2014.00467
Mickelbart, M. V., Hasegawa, P. M., and Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251. doi: 10.1038/nrg3901
Mohanty, A., Kathuria, H., Ferjani, A., Sakamoto, A., Mohanty, P., Murata, N., et al. (2002). Transgenics of an elite indica rice variety Pusa Basmati 1 harbouring the codA gene are highly tolerant to salt stress. Theor. Appl. Genet. 106, 51–57. doi: 10.1007/s00122-002-1063-5
Moustafa, K. (2014). MAPK transgenic circuit to improve plant stress-tolerance? Plant Signal. Behav. 9:e970101. doi: 10.4161/15592316.2014.970101
Mu, C., Zhang, S., Yu, G., Chen, N., Li, X., and Liu, H. (2013). Overexpression of small heat shock protein LimHSP16.45 in Arabidopsis enhances tolerance to abiotic stresses. PLoS ONE 8:e82264. doi: 10.1371/journal.pone.0082264
Negrão, S., Schmöckel, S. M., and Tester, M. (2017). Evaluating physiological responses of plants to salinity stress. Ann. Bot. 119, 1–11. doi: 10.1093/aob/mcw191
Nounjan, N., Nghia, P. T., and Theerakulpisut, P. (2012). Exogenous proline and trehalose promote recovery of rice seedlings from salt-stress and differentially modulate antioxidant enzymes and expression of related genes. J. Plant Physiol. 169, 596–604. doi: 10.1016/j.jplph.2012.01.004
Obata, T., Kitamoto, H. K., Nakamura, A., Fukuda, A., and Tanaka, Y. (2007). Rice shaker potassium channel OsKAT1 confers tolerance to salinity stress on yeast and rice cells. Plant Physiol. 144, 1978–1985. doi: 10.1104/pp.107.101154
Orabi, S. A., Dawood, M. G., and Salman, S. R. (2015). Comparative study between the physiological role of hydrogen peroxide and salicylic acid in alleviating the harmful effect of low temperature on tomato plants grown under sand-ponic culture. Sci. Agric. 9, 49–59. doi: 10.15192/PSCP.SA.2015.1.9.4959
País, S. M., Téllez-iñón, M. T., Capiati, D. A., Pais, S. M., Tellez-Inon, M. T., Capiati, D. A., et al. (2009). Serine/threonine protein phosphatases type 2A and their roles in stress signaling. Plant Signal. Behav. 4, 1013–1015. doi: 10.4161/psb.4.11.9783
Pandey, A., Sharma, M., and Pandey, G. K. (2016). Emerging roles of strigolactones in plant responses to stress and development. Front. Plant Sci. 7:434. doi: 10.3389/fpls.2016.00434
Prashanth, S. R., Sadhasivam, V., and Parida, A. (2008). Over expression of cytosolic copper/zinc superoxide dismutase from a mangrove plant Avicennia marina in indica Rice var Pusa Basmati-1 confers abiotic stress tolerance. Transgenic Res. 17, 281–291. doi: 10.1007/s11248-007-9099-6
Redillas, M. C., Park, S. H., Lee, J. W., Kim, Y. S., Jeong, J. S., Jung, H., et al. (2012). Accumulation of trehalose increases soluble sugar contents in rice plants conferring tolerance to drought and salt stress. Plant Biotechnol. Rep. 6, 89–96. doi: 10.1007/s11816-011-0210-3
Reina-Bueno, M., Argandoña, M., Salvador, M., Rodríguez-Moya, J., Iglesias-Guerra, F., Csonka, L. N., et al. (2012). Role of trehalose in salinity and temperature tolerance in the model halophilic bacterium Chromohalobacter salexigens. PLoS ONE 7:e33587. doi: 10.1371/journal.pone.0033587
Rodríguez-Gacio, M. D. C., Matilla-Vázquez, M. A., and Matilla, A. J. (2009). Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signal. Behav. 4, 1035–1048. doi: 10.4161/psb.4.11.9902
Rosa, M., Prado, C., Podazza, G., Interdonato, R., González, J. A., Hilal, M., et al. (2009). Soluble sugars–metabolism, sensing and abiotic stress: a complex network in the life of plants. Plant Signal. Behav. 4, 388–393. doi: 10.4161/psb.4.5.8294
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:e33587. doi: 10.3389/fpls.2016.00571
Saha, J., Brauer, E. K., Sengupta, A., Popescu, S. C., Gupta, K., and Gupta, B. (2015). Polyamines as redox homeostasis regulators during salt stress in plants. Front. Environ. Sci. 3:21. doi: 10.3389/fenvs.2015.00021
Sahoo, R. K., Ansari, M. W., Tuteja, R., and Tuteja, N. (2014). OsSUV3 transgenic rice maintains higher endogenous levels of plant hormones that mitigates adverse effects of salinity and sustains crop productivity. Rice 7:17. doi: 10.1186/s12284-014-0017-2
Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., and Izui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 23, 319–327. doi: 10.1046/j.1365-313x.2000.00787.x
Saini, S., Sharma, I., Kaur, N., and Pati, P. K. (2013). Auxin: a master regulator in plant root development. Plant Cell Rep. 32, 741–757. doi: 10.1007/s00299-013-1430-5
Shabala, S., and Pottosin, I. (2014). Regulation of potassium transport in plants under hostile conditions: implications for abiotic and biotic stress tolerance. Physiol. Plant. 151, 257–279. doi: 10.1111/ppl.12165
Shaddad, M. A. K., Abd El-Samad, H. M., and Mostafa, D. (2013). Role of gibberellic acid (GA3) in improving salt stress tolerance of two wheat cultivars. Int. J. Plant Physiol. Biochem. 5, 50–57. doi: 10.5897/IJPPB11.055
Sharma, I., Bhardwaj, R., and Pati, P. K. (2015). Exogenous application of 28-homobrassinolide modulates the dynamics of salt and pesticides induced stress responses in an elite rice variety Pusa Basmati-1. J. Plant Growth Regul. 34, 509–518. doi: 10.1007/s00344-015-9486-9
Sharma, I., Ching, E., Saini, S., Bhardwaj, R., and Kumar, P. (2013). Plant physiology and biochemistry exogenous application of brassinosteroid offers tolerance to salinity by altering stress responses in rice variety Pusa Basmati-1. Plant Physiol. Biochem. 69, 17–26. doi: 10.1016/j.plaphy.2013.04.013
Shirasawa, K., Takabe, T., Takabe, T., and Kishitani, S. (2006). Accumulation of glycinebetaine in rice plants that overexpress choline monooxygenase from spinach and evaluation of their tolerance to abiotic stress. Ann. Bot. 98, 565–571. doi: 10.1093/aob/mcl126
Singh, A., Jha, S. K., Bagri, J., and Pandey, G. K. (2015). ABA inducible rice protein phosphatase 2C confers ABA insensitivity and abiotic stress tolerance in Arabidopsis. PLoS ONE 10:e0125168. doi: 10.1371/journal.pone.0125168
Singla-Pareek, S. L., Yadav, S. K., Pareek, A., Reddy, M. K., and Sopory, S. K. (2008). Enhancing salt tolerance in a crop plant by overexpression of glyoxalase II. Transgenic Res. 17, 171–180. doi: 10.1007/s11248-007-9082-2
Smith, S. M., and Li, J. (2014). Signalling and responses to strigolactones and karrikins. Curr. Opin. Plant Biol. 21, 23–29. doi: 10.1016/j.pbi.2014.06.003
Srinivasan, T., Kumar, K. R. R., Meur, G., and Kirti, P. B. (2009). Heterologous expression of Arabidopsis NPR1 (AtNPR1) enhances oxidative stress tolerance in transgenic tobacco plants. Biotechnol. Lett. 31, 1343–1351. doi: 10.1007/s10529-009-0022-5
Su, J., Hirji, R., Zhang, L., He, C., Selvaraj, G., and Wu, R. (2006). Evaluation of the stress-inducible production of choline oxidase in transgenic rice as a strategy for producing the stress-protectant glycine betaine. J. Exp. Bot. 57, 1129–1135. doi: 10.1093/jxb/erj133
Tanaka, Y., Hibino, T., Hayashi, Y., Tanaka, A., Kishitani, S., Takabe, T., et al. (1999). Salt tolerance of transgenic rice overexpressing yeast mitochondrial Mn-SOD in chloroplasts. Plant Sci. 148, 131–138. doi: 10.1016/s0168-9452(99)00133-8
Tao, Y., Ferrer, J. L., Ljung, K., Pojer, F., Hong, F., Long, J. A., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. doi: 10.1016/j.cell.2008.01.049
Trontin, C., Kiani, S., Corwin, J. A., Hematy, K., Yansouni, J., Kliebenstein, D. J., et al. (2014). A pair of receptor-like kinases is responsible for natural variation in shoot growth response to mannitol treatment in Arabidopsis thaliana. Plant J. 78, 121–133. doi: 10.1111/tpj.12454
Türkan, I., and Demiral, T. (2009). Recent developments in understanding salinity tolerance. Environ. Exp. Bot. 67, 2–9. doi: 10.1016/j.envexpbot.2009.05.008
Van Ha, C., Leyva-González, M. A., Osakabe, Y., Tran, U. T., Nishiyama, R., Watanabe, Y., et al. (2014). Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. 111, 851–856. doi: 10.1073/pnas.1322135111
Verbruggen, N., and Hermans, C. (2008). Proline accumulation in plants: a review. 35, 753–759. doi: 10.1007/s00726-008-0061-6
Véry, A. A., Nieves-Cordones, M., Daly, M., Khan, I., Fizames, C., and Sentenac, H. (2014). Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J. Plant Physiol. 171, 748–769. doi: 10.1016/j.jplph.2014.01.011
Volkov, V. (2015). Salinity tolerance in plants. Quantitative approach to ion transport starting from halophytes and stepping to genetic and protein engineering for manipulating ion fluxes. Front. Plant Sci. 6:873. doi: 10.3389/fpls.2015.00873
Wang, F., Jing, W., and Zhang, W. (2014). The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci. 227, 181–189. doi: 10.1016/j.plantsci.2014.08.007
Wang, M., Zheng, Q., Shen, Q., and Guo, S. (2013). The critical role of potassium in plant stress response. Int. J. Mol. Sci. 14, 7370–7390. doi: 10.3390/ijms14047370
Wang, W., Vinocur, B., Shoseyov, O., and Altman, A. (2004). Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 9, 244–252. doi: 10.1016/j.tplants.2004.03.006
Wang, Y., Lin, S., Song, Q., Li, K., Tao, H., Huang, J., et al. (2014). Genome-wide identification of heat shock proteins (Hsps) and Hsp interactors in rice: Hsp70s as a case study. BMC Genomics 15:344. doi: 10.1186/1471-2164-15-344
Wang, Y., Shen, W., Chan, Z., and Wu, Y. (2015). Endogenous cytokinin overproduction modulates ROS homeostasis and decreases salt stress resistance in Arabidopsis thaliana. Front. Plant Sci. 6:1004. doi: 10.3389/fpls.2015.01004
Wani, S. H., Kumar, V., Shriram, V., and Sah, S. K. (2016). Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 4, 162–176. doi: 10.1016/j.cj.2016.01.010
Xu, K., Chen, S., Li, T., Ma, X., Liang, X., Ding, X., et al. (2015). OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 15:141. doi: 10.1186/s12870-015-0532-3
Yamaguchi, T., Aharon, G. S., Sottosanto, J. B., and Blumwald, E. (2005). Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+-and pH-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 102, 16107–16112. doi: 10.1073/pnas.0504437102
Yang, A., Dai, X., and Zhang, W. H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 63, 2541–2556. doi: 10.1093/jxb/err431
Yasmin, F., Biswas, S., Jewel, G. N. A., Elias, S. M., and Seraj, Z. I. (2016). Constitutive overexpression of the plasma membrane Na+/H+ antiporter for conferring salinity tolerance in rice. Plant Tissue Cult. Biotechnol. 25, 257–272. doi: 10.3329/ptcb.v25i2.26259
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2014). ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 21, 133–139. doi: 10.1016/j.pbi.2014.07.009
You, J., and Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Front. Plant Sci. 6:1092. doi: 10.3389/fpls.2015.01092
Zeng, H., Xu, L., Singh, A., Wang, H., Du, L., and Poovaiah, B. W. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6:600. doi: 10.3389/fpls.2015.00600
Zhao, X., Wang, W., Zhang, F., Deng, J., Li, Z., and Fu, B. (2014). Comparative metabolite profiling of two rice genotypes with contrasting salt stress tolerance at the seedling stage. PLoS ONE 9:e108020. doi: 10.1371/journal.pone.0108020
Zhao, X., Wei, P., Liu, Z., Yu, B., and Shi, H. (2017). Soybean Na+/H+ antiporter GmsSOS1 enhances antioxidant enzyme activity and reduces Na+ accumulation in Arabidopsis and yeast cells under salt stress. Acta Physiol. Plant. 39:19. doi: 10.1007/s11738-016-2323-3
Zörb, C., Geilfus, C. M., Mühling, K. H., and Ludwig-Müller, J. (2013). The influence of salt stress on ABA and auxin concentrations in two maize cultivars differing in salt resistance. J. Plant Physiol. 170, 220–224. doi: 10.1016/j.jplph.2012.09.012
Keywords: rice, abiotic stress, salinity, ion homeostasis, osmoregulation, reactive oxygen species
Citation: Kaur N and Pati PK (2017) Integrating Classical with Emerging Concepts for Better Understanding of Salinity Stress Tolerance Mechanisms in Rice. Front. Environ. Sci. 5:42. doi: 10.3389/fenvs.2017.00042
Received: 30 January 2017; Accepted: 20 June 2017;
Published: 05 July 2017.
Edited by:
David Delmail, University of Rennes 1, FranceReviewed by:
Suriyan Cha-um, National Science and Technology Development Agency, ThailandSergio Esposito, University of Naples Federico II, Italy
Michael Riemann, Karlsruhe Institute of Technology, Germany
Copyright © 2017 Kaur and Pati. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pratap Kumar Pati, cGtwYXRpQHlhaG9vLmNvbQ==
 Navdeep Kaur
Navdeep Kaur Pratap Kumar Pati
Pratap Kumar Pati