
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Environ. Chem., 19 March 2025
Sec. Inorganic Pollutants
Volume 6 - 2025 | https://doi.org/10.3389/fenvc.2025.1559968
This article is part of the Research TopicEnvironmental Chemistry of Mercury: Sources, Pathways, Transformations and Impact Vol IIView all 3 articles
Mercury methylation by anaerobic microorganisms, including sulfate-reducing bacteria (SRB), is a key process in the production of neurotoxic methylmercury (MeHg). The chemical speciation of mercury (Hg) strongly influences its bioavailability as well as its potential for methylation and demethylation, with sulfur-containing ligands playing a critical role in these processes. In this study, we used isotopically enriched mercury species (199Hg(II), Me202Hg) to investigate how molecular speciation of mercury affects both methylation and demethylation processes by the sulfate-reducer Pseudodesulfovibrio hydrargyri BerOc1. Experimental assays were carried out: (i) without external addition of S-ligands, (ii) with the addition of increasing concentrations of exogenous cysteine (Cys) (0.01, 0.1, and 0.5 mM), or (iii) with the addition of exogenous sulfide (0.1 mM). We showed that the highest methylation rate (Kmeth) was obtained without the external addition of S-ligands, whereas the addition of Cys or sulfide decreased Hg methylation regardless of Cys concentration. By quantitatively determining Hg(II) speciation in extracellular fractions, we demonstrated that Hg(II) was mostly present in the form of Hg(Cys)2, when Cys was added. However, metabolically sulfide production from Cys degradation shifted the chemical speciation of Hg(II) from Hg(Cys)2 to a more insoluble fraction (HgS(S)). In the assay without externally added ligands (Cys or sulfide), speciation models were generated by taking in account the metabolically produced thiols. These models established the predominance of Hg(II) complexes with a mixed ligation involving biosynthesized thiols, OH−, and Cl− ions. Our results suggest that these complexes with lower thermodynamic stabilities enhance the MeHg formation rate compared to the more stable Hg(Cys)2 or HgS(s) species. Unlike Hg(II) methylation, the addition of S-ligands did not affect the rates of demethylation (Kdemeth) of MeHg, even though it caused a shift in the chemical speciation of MeHg (from MeHgCl to MeHgCys and MeHgSH). These findings contribute to our understanding of the potential role of specific S-ligands and chemical speciation in governing the environmental fate and toxicity of mercury.
The methylation of inorganic mercury [Hg(II)] by diverse anaerobic microorganisms, including sulfate reducers (SRB), iron reducers (IRB), and methanogens is driven by the hgcAB gene cluster. This process is a crucial step in the production of MeHg (Gilmour et al., 2013; Parks et al., 2013; Gilmour et al., 2018; Bravo et al., 2018; Goñi-Urriza et al., 2020). Microbial methylation occurs primarily in anoxic environments such as sediments and soils (Compeau and Bartha, 1985), where Hg speciation is expected to be regulated by thiol-containing ligands present in natural organic matter (NOM) or sulfides (Liem-Nguyen et al., 2017a). However, while sulfur-containing ligands exhibit the highest affinity for Hg compared to other ligands (Liem-Nguyen et al., 2017a; Liem-Nguyen et al., 2017b), our understanding of the mechanism by which these S-ligands affect the speciation and bioavailability of Hg(II) remains limited.
Experimental addition of exogenous S-ligands has been one approach to elucidate the potential impact of complexing ligands present in the environment. In summary, the addition of approximately 1–10 μM of specific low molecular mass thiols (LMM-RSH), particularly Cysteine (Cys), has been demonstrated to significantly increase the rates of methylation of Hg(II) in methylating strains such as the iron reducer Geobacter sulfurreducens (Schaefer and Morel, 2009; Schaefer et al., 2011; Lin et al., 2015; Lu et al., 2016), the sulfate reducer Desulfovibrio desulfuricans ND132 (Graham et al., 2012a), and non-methylating strains like Escherichia coli (Ndu et al., 2012; Thomas et al., 2014). Other thiols such as glutathione (GSH) or penicillamine (PEN) were reported to either increase or suppress Hg methylation depending on bacterial strain (Schaefer et al., 2011). This suggests that the concentration of thiols and their specific binding to Hg in the extracellular environment can regulate Hg methylation, and it is also strain-dependent. The increase in Hg(II) methylation in the presence of micromolar thiol addition has been generally attributed to a change in the chemical speciation of Hg(II) from HgClnOH(1-n) species to the predominance of Hg (LMM-RS)2 complexes (Schaefer and Morel, 2009; Lin et al., 2015), which were hypothesized to be more bioavailable to the model strains studied.
In addition to the role of thiols, the implication of sulfide, another Hg complexing ligand has likewise been investigated. Optimum exogenous addition of sulfides up to 500 μM and 200 µM enhanced Hg (II) methylation potential in the sulfate reducers Pseudodesulfovibrio hydrargyri BerOc1 (40%) (Barrouilhet et al., 2022), and ND132 (from ∼2 to >20-fold) (Graham et al., 2013), respectively. Maximal methylation was also observed at sulfide concentrations below 100 µM for three strains of methanogenic archaea (Gilmour et al., 2018). It has been suggested that the enhancement in Hg methylation was the consequence of the formation of nanoscale β-HgS(s) particles (Graham et al., 2013; Deonarine and Hsu-Kim, 2009; Gerbig et al., 2011; Graham et al., 2012b; Zhang et al., 2012; Pham et al., 2014).
The addition of exogenous S-ligands in excess (as discussed above) has been one approach for investigating their roles in Hg(II) methylation. However, thiols and/or sulfide may also be metabolically produced and exported by microorganisms (Barrouilhet et al., 2022; Hand and Honek, 2005; Adediran et al., 2019; Thomas et al., 2019; Bakour et al., 2023; Gutensohn et al., 2023) at concentrations that are sufficient to control Hg speciation and bioavailability (Adediran et al., 2019). Several microorganisms have demonstrated the ability to degrade Cys and produce biogenic sulfide. The degradation of Cys to sulfides has been observed in E. coli strains (Awano et al., 2005; Shimada et al., 2016; Thomas and Gaillard, 2017; Thomas et al., 2019; Stenzler et al., 2022), Desulfovibrio species (Graham et al., 2012a), including the model strain P. hydrargyri BerOc1 (Barrouilhet et al., 2022; Bakour et al., 2023). Consequently, previous studies with exogenous Cys addition are likely to involve a mixture of Hg(II)-thiol and Hg(II)-sulfide species, introducing uncertainties in the interpretation of the results. Moreover, for the methylating strains that are both able to methylate and demethylate Hg, these S-ligands may control both processes (methylation and demethylation) and impact the final net production of MeHg. The implications of S-ligands in the demethylation process are still unclear. In this study, we investigated the role of S-ligands (Cys and sulfide), metabolically produced and/or exogenously present in the external environment, in controlling Hg molecular speciation and transformations (methylation and demethylation) in the sulfate-reducing bacterium P. hydrargyri BerOc1, a well-established SRB model capable of methylating Hg(II) and demethylating MeHg (Barrouilhet et al., 2022; Ranchou-Peyruse et al., 2009; Pedrero et al., 2012; Goñi-Urriza et al., 2015; Isaure et al., 2020). We utilized isotopically enriched Hg species (199Hg(II), Me202Hg) to simultaneously track Hg species and transformations, which could lead to a more precise interpretation of the impact of S-ligands on these processes. Experiments were conducted (i) without the external addition of S-ligands, to investigate the effect of metabolically produced ones (ii) with increasing concentrations of Cys, or (iii) with the addition of sulfide. Subsequently, the potentials and rates of Hg methylation and demethylation, partitioning of Hg species, molecular speciation of Hg(II) and MeHg, microbial Cys degradation, and production of sulfide and other thiols were investigated in BerOc1. By quantifying and considering the microbially produced thiol compounds, we conducted experiments at environmentally relevant concentrations of such compounds and generated refined speciation models for Hg(II) and MeHg in assay media.
Pseudodesulfovibrio hydrargyri BerOc1, a sulfate-reducing strain isolated from Berre Lagoon sediment, is a well-known SRB model able to both methylate inorganic mercury Hg(II) and to demethylate methylmercury (MeHg) (Ranchou-Peyruse et al., 2009; Pedrero et al., 2012; Goñi-Urriza et al., 2015; Isaure et al., 2020; Barrouilhet et al., 2022). BerOc1 was grown anaerobically in the dark at 37 °C and pH 7.0–7.1 in a multipurpose medium (Widdel et al., 1992) under fumarate respiration with 40 mM pyruvate as the electron donor and 40 mM fumarate as the electron acceptor (Supplementary Text S2). Cells were harvested in the early stationary phase (optical density OD600 nm at ∼0.4–0.5). The growing cell cultures were carefully transferred into 250-mL PTFE centrifuge flasks under anoxic conditions and centrifuged (at 8,000 g, 15 min, 25°C). Cell pellets were collected, pooled, and sequentially washed three times with mineral base medium. The final washed cells were gently homogenized in 50 mL of fresh anoxic medium.
For each experimental condition, 500 mL of multipurpose medium containing 1 mM pyruvate and 1 mM fumarate was inoculated with 8 mL aliquot of washed cell suspensions to a final optical density of 0.2, equivalent to final assay with ∼108 cells. mL−1, a cells density typical of pure culture experiments with P. hydrargyri BerOc1 (Pedrero et al., 2012; Isaure et al., 2020). For conditions with external S-ligand addition, Cys (0.01, 0.1, or 0.5 mM) or sulfide (0.1 mM) were added and allowed to equilibrate for 30 min before the addition of Hg. All stock solutions of Cys (≥99%, Acros Organics) and Na2S, 9H2O (98%, Alfa Aesar) were prepared in deoxygenated Milli-Q water (>18 MΩ cm) inside a glove box amended with N2.
All methylation assays were conducted in autoclaved penicillin flasks prewashed by ultrasonication in 10% HNO3 and HCl baths and rinsed with ultrapure Milli-Q water. For Hg exposure, 50 µL of 0.5 µM of 199HgCl2 enriched standard solution (ISC science), and 50 µL of 0.05 µM of Me202Hg chloride (LGC standards) were spiked into the cultures. The cultures under each condition were distributed into penicillin flasks and incubated at 37°C for 0, 4 h, 8 h, 20 h, 28 h, or 48 h in triplicates. Controls with inactive bacteria were performed with heated cells (80°C for 1 h) exposed to 0.05 µM of 199Hg(II), 0.05 µM of Me202Hg, either without the addition of S-ligands or with 0.1 mM Cys or 0.1 mM sulfide.
At the end of Hg exposure, an aliquot of 0.5 mL of the cell culture (bulk fraction) was sampled and digested in 50% (v/v) of 6 N nitric acid (HNO3) for Hg(II) and MeHg analysis. 1 mL of the remaining cell culture was then filtered at 0.2 µm (PVDF filter), and 0.5 mL was collected in 50% (v/v) 6 N HNO3 (extracellular fraction). The samples were stored at 4°C until further analysis. Isotopically enriched Hg(II) and MeHg concentrations were measured using capillary gas chromatography (GC TriPlus™ RSH™, Thermo Scientific) connected to an inductively coupled plasma mass spectrometer (ICPMS, X2-series, Thermo Electron) (Pedrero et al., 2012; Bridou et al., 2011). Quantification of isotopically enriched Hg(II) and MeHg was carried out by reverse isotope dilution analysis. To summarize, a known amount of fractions (bulk or extracellular) was buffered at pH 3.9 with a 0.1 M acetic acid/acetate buffer before being spiked with a known amount of isotopically enriched Hg species (198Hg(II) and Me201Hg) as internal standards. Both Hg(II) and MeHg were ethylated with 5% (v/v) NaBEt4 and extracted with isooctane by vigorous shaking for 20 min. The organic phase containing the Hg species was collected and analyzed using GC-ICPMS. The experimental data were mathematically processed by isotope pattern deconvolution approaches previously developed (Rodríguez-González et al., 2007). This technique allows the quantification of both Hg species concentrations and transformation factors (i.e., methylation and demethylation) affecting the two isotopic tracers during the analytical procedure. The analytical setup and methodology of GC-ICPMS for mercury speciation analysis are described in detail elsewhere (Ranchou-Peyruse et al., 2009; Pedrero et al., 2012).
Hg methylation potential and Hg species partitioning were determined in the same batch culture. Methylation potentials were calculated in bulk cultures by dividing the MeHg concentration produced (Tfinal- Tinitial, Tf-Ti) by the total Hg concentration measured at Ti. The partitioning of each Hg species (Hg(II) and MeHg) in the extracellular fractions was calculated by dividing the Hg species concentrations in the supernatant by the Hg species concentrations in the bulk fraction ×100. The percentage of Hg species in the pellet (i.e., cell-associated fraction including sorbed and intracellular Hg and particles in the medium) was determined by subtracting the percentage of Hg species in the supernatant fraction from 100%.
Specific methylation and demethylation rate constants (Kmeth and Kdemeth) were calculated for time series experiments using a first-order reversible reaction kinetic model. The mathematical steps for assessing the rate constants were discussed in detail by (Rodrı́guez Martı́n-Doimeadios et al., 2004). Assuming a pseudo-first-order reversible reaction, the net MeHg formation can be expressed as follows:
By applying the kinetic model, it was possible to obtain both rate constants from species concentrations obtained from time-series experiments using the BoxLucas1 fitting model (Origin Lab software).
With the assumption that [Me199Hg]0 = 0 at the onset of the reaction, Equation 1 can thus be integrated to obtain [Me199Hg] as a function of time.
The [Me199Hg]/[199Hg (II)]0 ratios were then plotted against incubation time (in hours) using Origin 6.1 software. The Box Lucas one exponential fitting model was used to resolve Equation 2 and calculate the methylation (Km) and demethylation (Kd) rate constants.
The concentrations of 199Hg(Cys)2, Me199Hg-Cys, and Me202Hg-Cys complexes were determined using an Acquity ultra performance liquid chromatography (UPLC) system (Waters, Milford, MA) equipped with a binary solvent pump and a cooled autosampler connected to a Xevo TQ mass spectrometer (MS). The complexes were separated using an Acquity UPLC HSS T3 C18 column (2.1 × 50 mm, 1.8 μm, Waters) with a matching Vanguard precolumn. Aliquots of 10 μL were injected at 0.35 mL·min−1 for the mobile phase consisting of 0.1% formic acid in water (A) and 0.1% formic acid in MeOH (B). We applied offline solid-phase extraction (SPE) using a mixed-mode reversed-phase ion-exchanger sorbent (Oasis Mcx 3 Cc Vac Cartridge, 60 mg, 30 µm particle Size) for selective preconcentration of the Hg complexes with an eluting solvent consisting of MeOH/water (60:40 v/v). Detection was performed in both positive and negative electrospray ionization modes (ESI). The method for MeHg complexes was adapted from Liem-Nguyen et al. (Liem-Nguyen et al., 2020), incorporating an offline SPE preconcentration. The limits of detection in this study were 8 nM for MeHg-Cys and 4 nM for Hg(Cys)2. The general operating conditions of the instrument are listed in Supplementary Table S1. The complexes were determined in extracellular fractions of BerOc1 at each kinetic point after filtration through 0.2 μm syringe filters.
To investigate the other possible Hg species that can be formed in P. hydrargyri BerOc1 culture assays, we used Visual Minteq software to perform thermodynamic modeling. The ranges of Log K and possible reactions were adopted from Compeau and Bartha (1985). Our thermodynamic calculations incorporated the total sulfide, cysteine, metabolically produced thiols, and total Hg and MeHg concentrations measured in the exposure medium. The complete speciation model is presented in Supplementary Table S2.
Sulfide was measured in the assay medium using a sulfide-reactive coumarin fluorescent probe, known as α, β-unsaturated ethanoylcoumarin (DHC) (Yang et al., 2019), after adapting the method for sulfide detection in P. hydrargyri BerOc1 cultures assays (Bakour et al., 2023). The detection limit using this approach was 20 nM. For analysis, 1 mL of the bulk fraction was collected at each kinetic point and analyzed using the DHC probe.
The concentrations of specific LMM-RSH thiol compounds including Cys, 2-Mercaptoethanesulfonic acid (SULF), PEN, N-acetyl-L-cysteine (NacCys), mercaptosuccinic acid (SUC), 2-Mercaptopropionic acid (2-MPA), GSH, cysteamine (Cyst),1-Thiolglycerol (Glyc), mercaptoacetic acid (Mac), N-acetyl-D-penicillamine (NacPen), L-homocysteine (HCys), γ-L-glutamyl-L-cysteine (γ-Glu-Cys), and cysteine-glycine (Cys-Gly) were determined according to (Liem-Nguyen et al., 2015), after adapting the method for UPLC-ESI-MS instrument and using offline SPE preconcentration (details cited above and in Supplementary Table S1). We applied offline SPE using an oasis hydrophilic-lipophilic balanced reversed-phase sorbent (HLB: 3 cc Vac Cartridge, 60 mg, 30 µm) for the selective preconcentration of thiol compounds with an eluting solvent consisting of MeOH/water (10:90 v/v). The limit of detection was 3–6 nM for all the LMM thiols investigated. The compounds were determined in extracellular fractions of BerOc1 at each kinetic point after filtration through 0.2 μm syringe filters.
We used isotopically enriched Hg species to conduct experimental assays with and without the addition of S-containing ligands (Cys and sulfide) to assess their roles in Hg transformations. The availability of isotopically enriched Hg species for the transformation processes was evaluated. The concentrations at the end of the incubation period were always recovered for both 199Hg(II) and Me202Hg (∼106 and 90%, respectively). A few Me202Hg losses were observed, which could be attributed to either sorption to the bottle walls or to particles in the culture medium. The production of Me199Hg resulting from 199Hg(II) methylation increased exponentially within the first few hours and reached a steady state after 28 h of incubation (Figure 1C). The transformation rate constants obtained using the kinetic model approaches applied to Hg species showed a high methylation rate (Kmeth) in the experiments without the external addition of S-ligands (2.1 ± 0.2 × 10−10 mL cell−1 h−1) (Figure 1A). The addition of exogenous Cys decreased the methylation rate and no significant difference was observed in Kmeth with increasing Cys concentrations from 0.01 to 0.5 mM (∼1.1 × 10−10 mL cell−1 h−1, p > 0.05, Kruskal–Wallis). The addition of 0.1 mM sulfide resulted in the lowest methylation rate (0.4 ± 0.2 × 10−10 mL cell−1 h-1) (Figure 1A). In another study, higher methylation potentials were observed when BerOc1 was exposed to 0.1 mM sulfide compared to 0.1 mM Cys (Barrouilhet et al., 2022). Here, we demonstrated a higher methylation potential upon the addition of 0.1 mM Cys compared to 0.1 mM of sulfide. However, it is important to note that, despite using the same strain, BerOc1, the experimental conditions varied. The previous study investigated BerOc1 under growth conditions, while in this study, we used resting cell conditions. Additionally, the concentrations of Hg(II) were not identical, leading to variations in the Hg(II)/Cys or Hg(II)/sulfide ratios in the exposure medium.
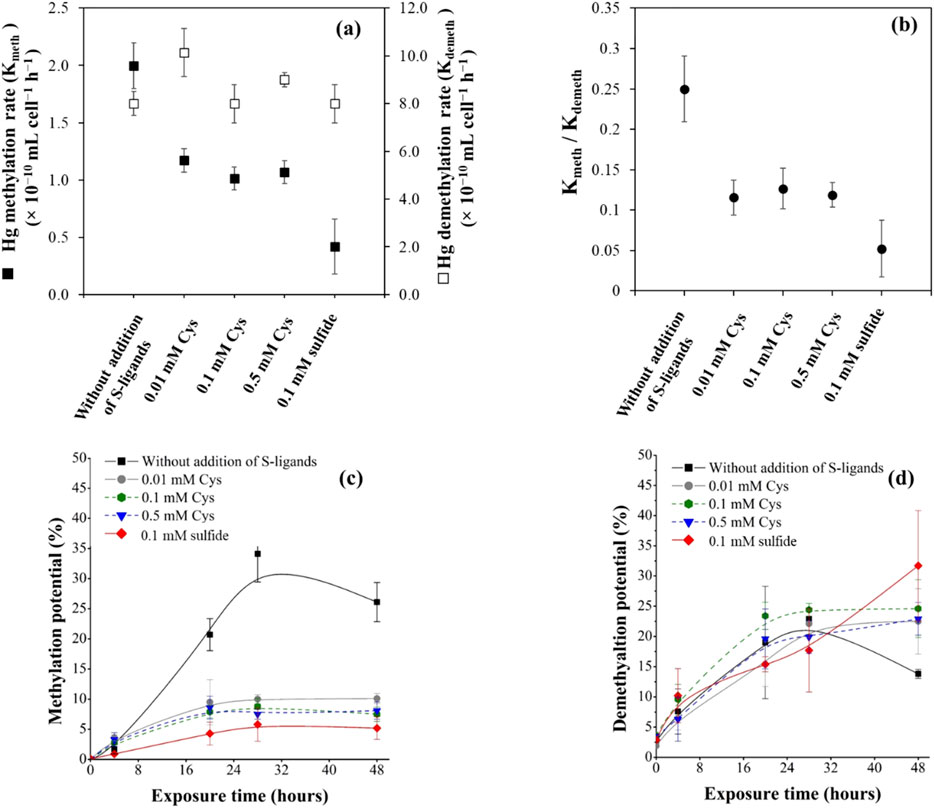
Figure 1. (a) Hg methylation rate Kmeth (black squares) and MeHg demethylation rate Kdemeth (white squares) determined for pure strain cultures of P. hydrargyri BerOc1 tested under different conditions (without external addition of S-ligands, with 0.01, 0.1, and 0.5 mM Cys, or with 0.1 mM of sulfide), after incubation with 0.5 µM 199Hg(II) and 0.05 µM of Me202Hg for 48 h. (b) Ratio of Kmeth/Kdemeth for the different conditions. (c) Hg methylation potentials (% of total recovered 199Hg(II) converted to Me199Hg) (d) MeHg demethylation potentials (% of total recovered Me202Hg converted to 202Hg(II)). Error bars represent the standard deviations of three independent replicates.
Unlike Hg(II) methylation, the results showed that the addition of Cys or sulfide had no effect on the MeHg demethylation rates by P. hydrargyri BerOc1. No significant difference has been found (p > 0.05, Kruskal-Wallis) (Figure 1A). The production of 202Hg(II) resulting from Me202Hg degradation increased exponentially within the first few hours and reached a steady state after 28 h of incubation (Figure 1D). In the absence of sulfur ligands, the concentration of 202Hg(II) decreased after 28 h (Figure 1D). This decrease is attributed to an enhanced methylation of Hg(II) under these conditions, accounting for the observed reduction.
The demethylation rates (Kdemeth) was 4–16 times higher than methylation rates (Kmeth), as previously reported for BerOc1 (Perrot et al., 2015), demonstrating that demethylation efficiency is greater than that of methylation. This ratio decreased with the addition of Cys or sulfide, highlighting the significant role of these S-complexing ligands in regulating the net MeHg production by bacterial cells. This decrease is attributed to the decrease in Hg(II) methylation observed in the presence of S-ligands, as demethylation was not affected.
To go deeper into the implications of S-ligands (Cys and sulfide) in Hg speciation and methylation/demethylation, we conducted further investigations into the molecular speciation and partitioning of Hg, as well as the metabolism of S-compounds.
We investigated Hg species partitioning between the extracellular and cell-associated/particulate (i.e., cell-sorbed, intracellular and particulate) fractions in BerOc1 incubated under different conditions (Figure 2; Figure 3).
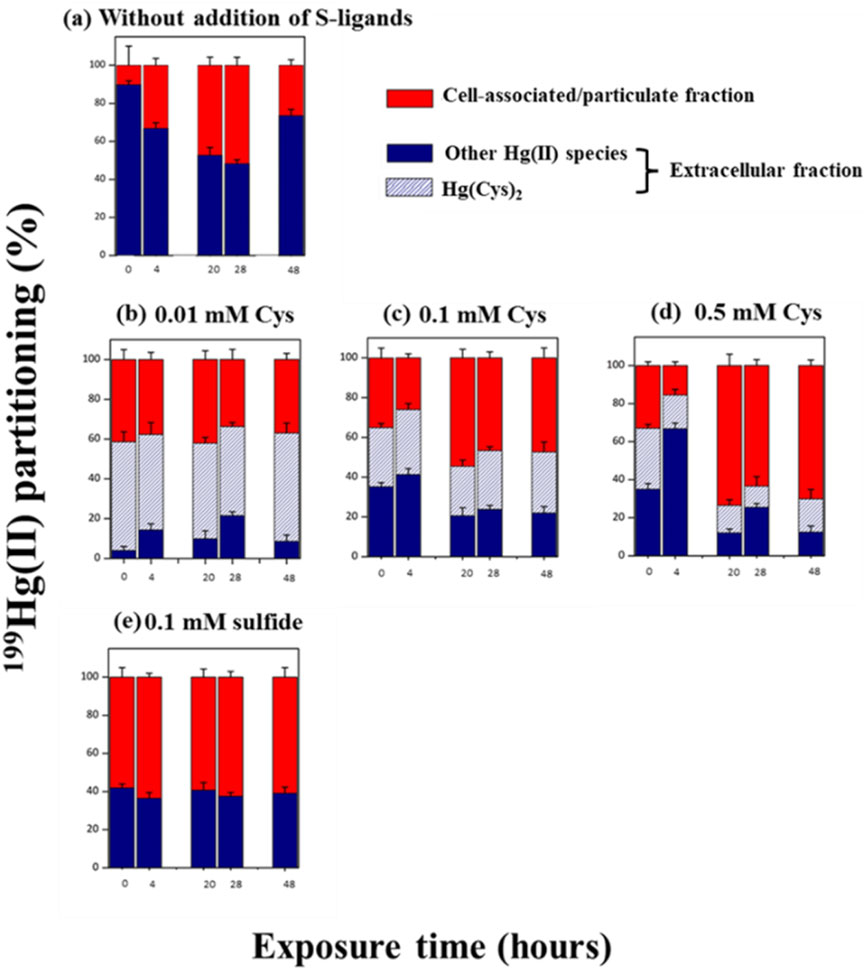
Figure 2. Partitioning (%) of added 199Hg(II) between extracellular (blue), the % of Hg(Cys)2 complex detected in the extracellular fraction (blue dash) and cell associated/particulate (red) fractions at the time = 0, 4, 20, 28 and 48 h during exposure to 0.5 µM of 199Hg(II) and 0.05 µM of Me202Hg. (a) Experimental without addition of external S-ligands (b) with the addition of 0.01 mM of Cys (c) 0.1 mM Cys (d) 0.5 mM Cys (e) 0.1 mM of sulfide. The percentage was calculated by considering the Hg concentration in the three studied fractions. The same culture batches were used to determine the Hg species in the bulk and extracellular fractions for each kinetics. After sampling for Hg species determination using GCICPMS (bulk + supernatant), cultures were filtered (0.2 µm), and the molecular speciation of Hg(II) was measured in the extracellular fraction using LCMSMS. Error bars represent the averages from three independent experiments, and the error bars are ±1 S.D.
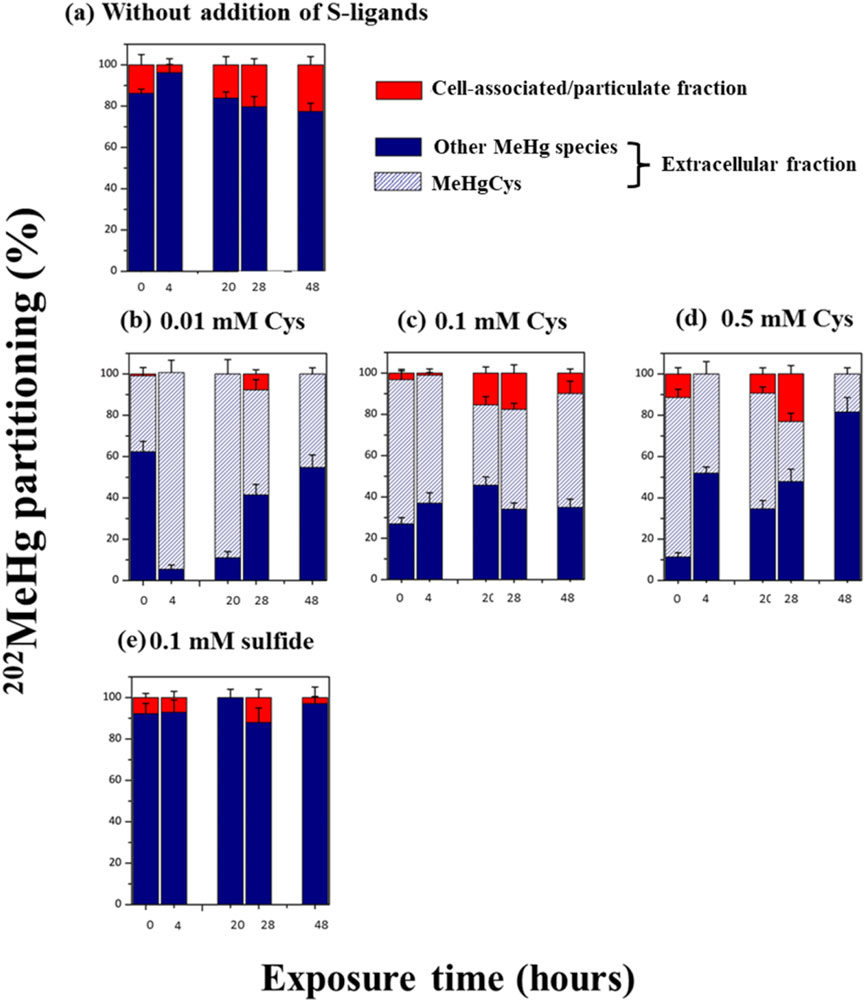
Figure 3. Partitioning (%) of added Me202Hg between extracellular (blue), the % of MeHgCys complex detected in the extracellular fraction (blue dash) and cell associated/particulate (red) fractions at the time = 0, 4, 20, 28 and 48 h during exposure to 0.5 µM of 199Hg(II) and 0.05 µM of Me202Hg. (a) Experimental without addition of external S-ligands (b) with the addition of 0.01 mM of Cys (c) 0.1 mM Cys (d) 0.5 mM Cys (e) 0.1 mM of sulfide. The percentage was calculated by considering the Hg concentration in the three studied fractions. The same culture batches were used to determine the Hg species in the bulk and extracellular fractions for each kinetics. After sampling for Hg species determination using GCICPMS (bulk + supernatant), cultures were filtered (0.2 µm), and the molecular speciation of MeHg was measured in the extracellular fraction using LCMSMS. Error bars represent the standard deviation of three independent replicates, each measured three times.
199Hg(II) added to the culture and 202Hg(II) formed by biotic demethylation showed dissimilar partitioning. In general, 199Hg(II) added to the culture was in the extracellular fraction, but also associated with cells/particulate fractions (20%–60%) (Figure 2), as previously observed for BerOc1(Pedrero et al., 2012; Barrouilhet et al., 2022). Conversely, only ∼20% of the 202Hg(II) resulting from demethylation is associated with cells, while the major fraction is found in the extracellular medium (Supplementary Figure S1).
Varying concentrations of Cys (0.01, 0.1, and 0.5 mM) or sulfide (0.1 mM) do not influence the distribution of 202Hg(II) produced through biotic demethylation (Supplementary Figure S1). Nevertheless, for 199Hg(II) introduced to the culture, the cellular/particulate fraction association increased with rising Cys levels after 20 h of exposure (Figures 2B–D). When 0.5 mM Cys was added, BerOc1 cells showed approximately 80% of Hg(II) associated with cellular/particulate fractions at 48 h (Figure 2D). In the case of sulfide addition, about 60% of 199Hg(II) was primarily associated with cellular/particulate fractions from the initial hours of exposure (Figure 2E).
The distribution of Me202Hg introduced to the culture and Me199Hg produced through biotic processes was found to be similar, with approximately 95% located in the extracellular fraction (Figure 3; Supplementary Figure S2), confirming previous findings. The formed MeHg was immediately exported. Earlier research noted no significant lags in the export of synthesized Hg(II) and MeHg, suggesting that the excreted Hg had minimal affinity or low selectivity for cell surface receptors and transporters (Schaefer and Morel, 2009; Gilmour et al., 2011; Schaefer et al., 2011; Graham et al., 2012a). Introducing increasing concentrations of Cys (0.01, 0.1, and 0.5 mM) or sulfide (0.1 mM) did not substantially impact the partitioning of either added Me202Hg or biotically methylated Me199Hg (Figure 3; Supplementary Figure S2). Nevertheless, it was observed that in the experiment with 0.1 mM sulfide addition, a higher proportion of Me199Hg (100%) was exported to the extracellular fraction compared to other conditions (∼80%) (Supplementary Figure S2). For BerOc1, a strong positive correlation was observed between MeHg in the extracellular medium and increased sulfide concentrations (Barrouilhet et al., 2022), indicating that sulfide may facilitate MeHg export and/or enhance desorption from the cell.
To control the absence of S-ligands in the medium at t = 0 in the assay, the cells were harvested, washed, and transferred to the experimental medium. When no exogenous Cys was added, sulfide remained undetectable (LOD 20 nM) in the medium for 48 h. Upon introducing exogenous Cys at 0.01, 0.1, and 0.5-mM concentrations, a notable reduction in Cys levels (Figure 4A) and an increase in sulfide production were observed over time (Figure 4B). After 48 h, the sulfide concentrations in the medium peaked at 4.6 ± 0.2 µM, 42.3 ± 1.9 µM, and 159.3 ± 2.1 µM, corresponding to initial exogenous Cys measurements of 8.2 ± 0.2 µM, 85.5 ± 7.4 µM and 471.4 ± 38.0 µM, respectively (Figures 4A, B). Faster sulfide production and Cys degradation occurred during the first 28 h of exposure, and then the rate decreased, leading to a plateau. Controls with heat-killed cells confirmed that the observed sulfide production from Cys degradation is a fully biotic process, as no sulfide production was observed when heated cells were exposed to 0.1 mM Cys (Figure 4B). Moreover, nanomolar concentration of Cys was detected in the assay without externally added ligands (Figure 4A insert). Increasing Cys concentration led to a higher sulfide concentration in the assay medium; however, the conversion rate of Cys to sulfide did not increase linearly with increasing external Cys (Figure 4C). Previous observations have shown that Cys degradation to sulfide reaches a threshold at 0.5 mM added Cys and above in E. coli (Thomas and Gaillard, 2017). Therefore, while increasing the concentration of external Cys may initially increase the conversion rate of Cys to sulfide, there may be a limit to the amount of additional Cys that can be converted, beyond which the conversion rate remains constant or even decreases due to various factors influencing microbial metabolism.
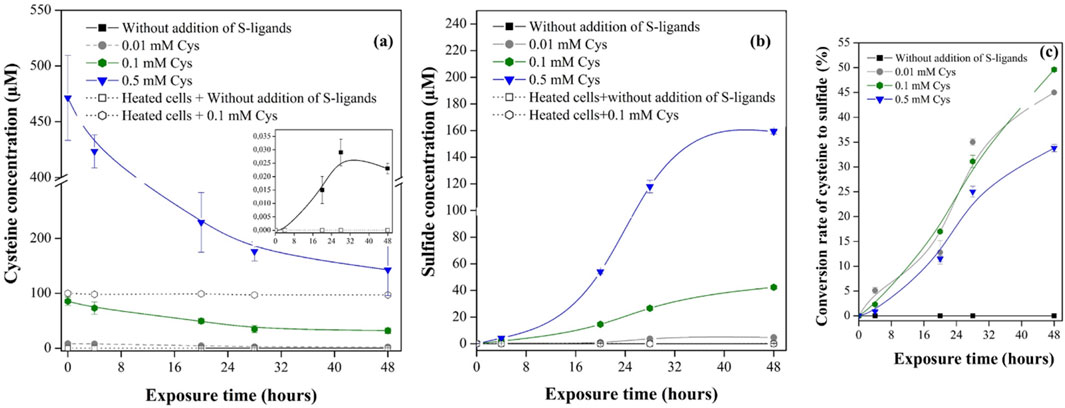
Figure 4. (a) Concentration of Cys measured in the extracellular fraction of BerOc1 culture at time = 0, 4, 20, 28 and 48 h during exposure to 0.05 µM of 199Hg(II), 0.05 µM of Me202Hg. The zoom represents the concentrations of Cys detected in the assay without externally added S-ligands. (b) Concentration of sulfide in the bulk fraction (c) The conversion rate of Cys to sulfide calculated by dividing the sulfide concentration detected at each kinetic point by the initial exogenous Cys concentration. The points are the averages from three independent experiments. Controls were performed with heated cells (80°C for 1 h) exposed to 0.05 µM of 199Hg(II), 0.05 µM of Me202Hg.
In experiments without the exogenous addition of S-ligands, six different thiol compounds were detected in the extracellular assay medium of metabolically active P. hydrargyri BerOc1 (Figure 5A). After 48 h of incubation, the detected LMM-RSH thiols were 2-MPA (35 ± 4 nM), NacCys (30 ± 6 nM), Cys (20 ± 3 nM), HCys (16 ± 2 nM), γ-Glu-Cys (12 ± 3), and Cys-Gly (10 ± 2 nM). The total concentration of thiol compounds reached 123 nM in the assay medium at a cell concentration of 1.1 × 108 cells mL−1. During the first 28 h of incubation, the thiol compounds were rapidly produced and exported (Figure 5A). The assay with heat-killed cells revealed no detectable formation of thiol compounds, indicating that thiols were biosynthesized by BerOc1 via active metabolism and were not produced by dead disrupted cells. The iron-reducer G. sulfurreducens PCA has been also shown to produce and export nanomolar concentrations of thiol compounds including Cys, PEN, NacCys, γ-GluCys, Cyst, Glyc, Mac, and HCys (Adediran et al., 2019).
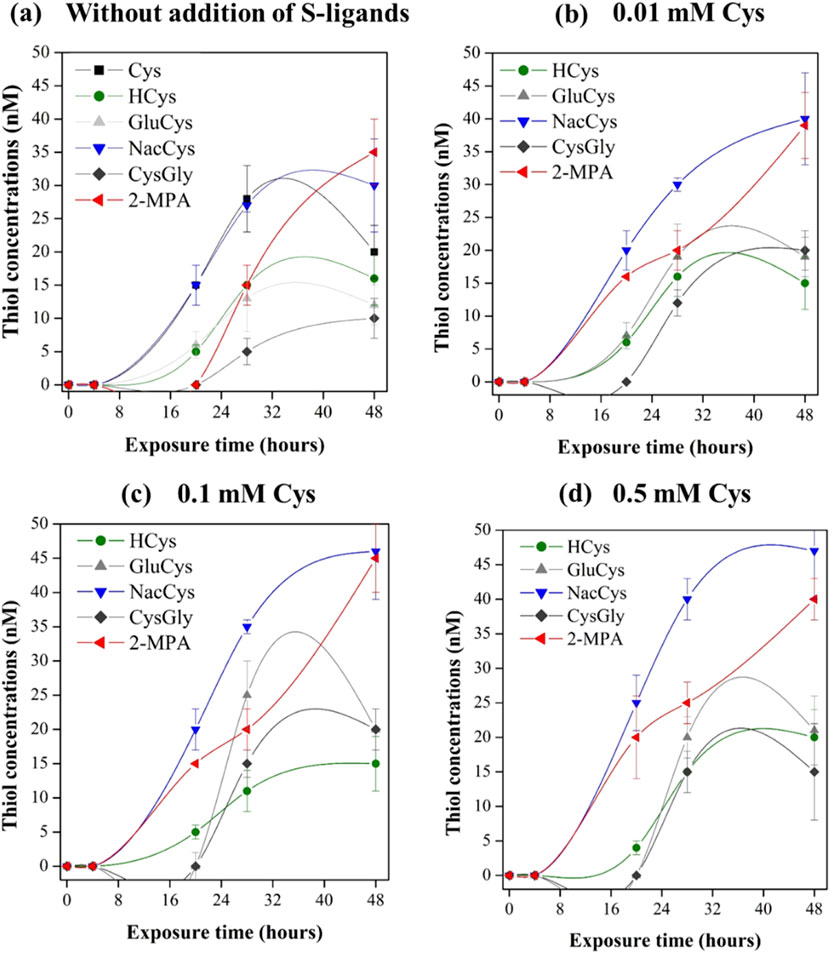
Figure 5. Concentrations of biosynthesized LMM-RSH compounds in extracellular assay medium at the time = 0, 4, 20, 28 and 48 h during exposure to 0.5 µM of 199Hg(II) and 0.05 µM of Me202Hg with metabolically active P. hydrargyri BerOc1 (∼108 cells mL-1) for (a) Experimental without addition of external S-ligands (b) with the addition of 0.01 mM Cys (c) 0.1 mM Cys (d) 0.5 mM Cys. The thiols detected were Cys, HCys, NacCys, Cys-Gly, γ-Glu-Cys, and 2-MPA. Total biosynthesized thiol concentrations were as follows: (a) 123 ± 5 nM, (b) 135 ± 6 nM, (c) 141 ± 7 nM, and (d) 146 ± 5 nM.
With the addition of exogenous Cys at increasing concentrations, the same thiols were detected in the medium (Figures 5B–D). The total concentration of produced thiol compounds reached 135, 141, and 146 nM for Cys additions of 0.01, 0.1, and 0.5 mM, respectively. The addition of increasing Cys concentration do not impact the production of the other thiols. This suggest that the production of these thiols may be not strongly influenced by changes in Cys levels. Recently, Gutensohn et al., 2023 demonstrated that external exposure of G. sulfurreducens PCA to Cys resulted in increased formation of PEN. Experiments using isotopically labeled Cys confirmed that PEN was formed directly from Cys via dimethylation of the C-3 atom. Moreover, PEN was found to not accumulate intracellularly, which could imply that it is part of G. sulfurreducens PCA metabolic strategy to maintain Cys homeostasis. In contrast to PCA, PEN was not detected in our experiments with BerOc1. However, BerOc1 degrade Cys and produce sulfide to maintain a low intracellular Cys concentration.
The addition of exogenous Cys led to sulfide production by P. hydrargyri BerOc1. To determine whether Hg(II)-Cys or Hg(II)-sulfide species were favored, we used liquid chromatography-tandem mass spectrometry to measure Hg(Cys)2 formation in the extracellular environment over time. The results, shown as percentages of Hg(Cys)2 relative to total extracellular Hg(II), are displayed in Figure 2. When 0.01 mM Cys was added, about 80% of the introduced 199Hg(II) formed Hg(Cys)2 complexes (Figure 2B). However, with increasing Cys concentrations, the proportion of 199Hg(Cys)2 decreased significantly, reaching 40% and 20% for 0.1 and 0.5 mM Cys, respectively (Figures 2C, D). Chemical modeling predicted HgS(s) formation in all Cys exposure experiments (Supplementary Figure S4). HgS(s) precipitation occurred rapidly, with 100% of Hg(II) forming HgS(s) after 4 h with 0.1 and 0.5 mM Cys, and 20 h with 0.01 mM Cys (Supplementary Figure S4). This indicates that sulfide production from Cys degradation shifted Hg(II) speciation from Hg(Cys)2 to HgS(s), explaining the observed decrease in Hg(Cys)2. Previous research has shown cell-associated HgS(s) formation in P. hydrargyri BerOc1 (Isaure et al., 2020) and biomediated HgS(s) formation from Hg(II)-Cys complexes with Cys addition (0.1–1 mM) (Thomas et al., 2018). However, our results do not fully align with thermodynamic predictions, which suggest 100% Hg(II) as HgS(s) at measured sulfide levels. We detected Hg(Cys)2 even with sulfide concentrations up to 0.16 mM, indicating the coexistence of Hg(Cys)2 and HgS(s) in the medium.
Without exogenous S-ligands introduction, BerOc1 cells exported six different thiols, including Cys, at nanomolar levels, and 199Hg(Cys)2 (LOD 4 nM) was not detected (Figure 2A). Thermodynamic modeling predicted that HgCl2 (aq), HgCl3− (aq), and HgCl42- (aq) were the dominant Hg(II) species during the initial incubation hours (Supplementary Figure S4). The metabolically active synthesis and export of thiols by BerOc1 shifted Hg(II) speciation in the medium to HgOH(LMM-RS), HgCl(LMM-RS), and Hg(II)-thiol complexes with a 1:1 stoichiometry.
Hg bioavailability for methylation was highest in the absence of externally added S-ligands (only metabolically produced S-ligands were present). Under these conditions, Hg(II) predominantly forms inorganic complexes (HgOHnCl2−n) and mixed ligands like HgOH(LMM-RS), HgCl(LMM-RS), and Hg(II)-thiol complexes with 1:1 stoichiometry. This observation suggests that complexes with lower thermodynamic stability can enhance the MeHg formation rate compared to more stable species like Hg(Cys)2 or HgS(s) (Supplementary Table S2). Adediran et al., 2019 (24) determined Hg(II) methylation rate constants for various Hg(II) species in G. sulfurreducens PCA, revealing species-specific k′meth values (×10–13 L cell−1 h−1) in the following order: HgOHnCl2−n (k′meth = 45) > HgOHnCl1−n (LMM-RS), Hg (LMM-RORS) (k′meth = 8.8) > Hg(Cysteamine)2 (k′meth = 7.3) > Hg(Cys)2 (k′meth = 3.4) > Hg(NacCys)2 (k′meth = 0.49). These findings imply that Hg(II) complexes with lower thermodynamic stability are more bioavailable for bacterial methylation, reinforcing the notion that the specific nature of metal-ligand interactions plays a crucial role in methylation efficiency. In contrast, previous studies have shown excess Cys (∼1–100 μM) promotes Hg(II) uptake and methylation (Schaefer and Morel, 2009; Schaefer et al., 2011; Ndu et al., 2012; Thomas et al., 2014; Szczuka et al., 2015), while nanomolar levels of Cys (<200 nM) decrease Hg(II) methylation (Lu et al., 2016). The increase in Hg(II) methylation with micromolar Cys additions, compared to nanomolar levels, was attributed to a concentration-dependent shift in Hg(II) speciation from HgOHnCl2−n to Hg(Cys)2, presumed to be more bioavailable (Schaefer and Morel, 2009; Lin et al., 2015). However, Adediran et al., 2019 found that in the iron-reducing G. sulfurreducens PCA, the enhancement in Hg(II) methylation with micromolar Cys (0–10 µM) was not solely due to changes in Hg(II) speciation, suggesting that other biochemical pathways regulating Hg(II) methylation warrant further investigation.
Our results align with the framework that indicates as the thermodynamic stability of an aqueous metal complex increases, the bioavailability of the metal decreases (Morel et al., 1991; Slaveykova et al., 2005; Xu et al., 2012). We observed a higher Kmeth for less stable Hg(II) complexes, underscoring that biotic ligand models explain much of the data, yet some aspects of Hg(II) uptake and methylation remain inadequately addressed by ligand affinity alone. This is crucial, as less stable mixed ligand Hg(II) species, while not predominant in thermodynamic equilibrium, may be kinetically favored under specific conditions (Miller et al., 2009; Jiang et al., 2015), Our study demonstrates that these species can significantly contribute to MeHg formation when present.
The mechanisms of Hg(II) uptake by methylating microorganisms remain to be fully elucidated. Proposed mechanisms include the uptake of thermodynamically stable Hg(II) species via passive diffusion of neutral HgS0(aq) and Hg(SH)20 (aq) (Benoit et al., 1999), and active uptake processes for the Hg(Cys)2 complex (Schaefer and Morel, 2009). Additionally, metal uptake likely involves binding to cell surface functional groups (Hudson, 1998; Campbell et al., 2002), with mechanisms such as ligand exchange reactions, either through complex dissociation in solution (Slaveykova et al., 2005; Di Toro et al., 2001) or transient ternary complex formation (Aristilde et al., 2012; Flynn et al., 2014; Fein, 2017). Regardless of the uptake mechanism, the metal transfer rate from solution to cell surface depends on the metal’s affinity for aqueous and cell surface ligands and their relative concentration (Zhao et al., 2016). As the thermodynamic stability of an aqueous metal complex increases, the bioavailability of the metal decreases (Morel et al., 1991; Slaveykova et al., 2005; Xu et al., 2012). Our results, showing a higher Kmeth for less stable Hg(II) complexes, align with this framework. Although biotic ligand models explain most data, some aspects of Hg(II) uptake and methylation remain unexplained by ligand affinity alone.
The formation of MeHg-Cys in the extracellular medium was quantified using liquid chromatography tandem mass spectrometry. Figure 3 displays the percentage of Me202Hg-Cys species relative to total extracellular Me202Hg. The experiments involving Cys addition demonstrated that Me202Hg formed complexes with Cys to produce Me202Hg-Cys (Figure 3). The proportion of Me202Hg-Cys in the extracellular medium remained relatively stable (approximately 60%–80% of total added Me202Hg) as Cys concentrations increased from 0.01 to 0.1 mM. However, at the highest Cys exposure (0.5 mM), the concentration of Me202HgCys significantly decreased over time, reaching only 10% of the added Me202Hg after 48 h (Figure 3). To investigate the formation of MeHg-sulfide species under the experimental conditions, thermodynamic modeling was conducted. Chemical modeling of MeHg revealed the presence of MeHgSH and S (MeHg)2 in all experiments with Cys exposure (Supplementary Figure S5). The formation of MeHgSH increased with higher Cys concentrations, reaching 100% of MeHg at 0.5 mM addition. This explains the observed decrease in Me202HgCys, which can be attributed to a shift in MeHg chemical speciation from MeHgCys to MeHgSH and S (MeHg)2. In experiments without externally added S-ligands, MeHg202Cys was not detected. Thermodynamic modeling was performed to examine the species formed in the absence of excess S ligands (Supplementary Figure S5). MeHgCl was found to be the dominant MeHg species throughout the exposure period. The metabolically active synthesis and cellular export of thiol compounds (LMM-RSH) by P. hydrargyri BerOc1 did not alter the chemical speciation of MeHg in the assay media. While extensive literature exists on the thermodynamics of Hg(II) complexation (Dyrssen and Wedborg, 1991; Ravichandran, 2004; Smith et al., 2004; Cardiano et al., 2011; Drott et al., 2013; Warner and Jalilehvand, 2016; Liem-Nguyen et al., 2017b), there is less data available on the thermodynamic formation constant for MeHg, particularly regarding its complexation with thiol compounds. This lack of data may lead to an underestimation of MeHg-thiol compound formation. This study demonstrated that the addition of S-ligands (Cys and sulfide) induced a change in the chemical speciation of MeHg. However, the shift in MeHg chemical speciation from MeHgCl to MeHgCys, MeHgSH, and S (MeHg)2 did not impact demethylation rates (Kdemeth).
In this work, Hg(II) and MeHg speciation and molecular transformations were investigated using the model strain P. hydrargyri BerOc1. The use of isotopic enriched mercury species allowed precise tracking of mercury species and their transformations. We showed that the highest methylation rate (Kmeth) was obtained under conditions without addition of ligands. Under such conditions, by quantifying and taking in account microbial produced thiols, refined speciation models were generated. These models revealed that Hg(II) formed complexes with mixed binding involving biosynthesized thiols, OH− ions and Cl− ions. Our results suggest that these complexes increase the rate of MeHg formation compared to the more stable Hg(Cys)2 or HgS(s) species. Unlike Hg(II) methylation, the addition of S-ligands did not affect the rates of demethylation of MeHg, even though it caused a shift in the chemical speciation of MeHg. The results of this study provide valuable insights into the underlying mechanisms of mercury methylation and demethylation.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
IK: Conceptualization, Data curation, Formal analysis, Validation, Vizualization, Writing–original draft, Writing–review and editing. M-PI: Conceptualization, Validation, Vizualization, Writing–original draft, Writing–review and editing, Supervision, Funding acquisition, SB: Conceptualization, Data curation, Formal analysis, Writing-review and editing. MS-G: Conceptualization, Validation, Vizualization, Writing–original draft, Writing–review and editing, Supervision, Funding acquisition. MM: Conceptualization, Data curation, Formal analysis, Validation, Vizualization, Writing–original draft, Writing–review and editing, Supervision, Funding acquisition
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Go-Beam project, funded by the Agence Nationale de la Recherche, through the E2S-UPPA call Key Scientific challenges.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvc.2025.1559968/full#supplementary-material
Adediran, G. A., Liem-Nguyen, V., Song, Y., Schaefer, J. K., Skyllberg, U., and Björn, E. (2019). Microbial biosynthesis of thiol compounds: implications for speciation, cellular uptake, and methylation of Hg(II). Environ. Sci. Technol. 53 (14), 8187–8196. doi:10.1021/acs.est.9b01502
Aristilde, L., Xu, Y., and Morel, F. M. M. (2012). Weak organic ligands enhance zinc uptake in marine phytoplankton. Environ. Sci. Technol. 46 (10), 5438–5445. doi:10.1021/es300335u
Awano, N., Wada, M., Mori, H., Nakamori, S., and Takagi, H. (2005). Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl. Environ. Microbiol. 71 (7), 4149–4152. doi:10.1128/aem.71.7.4149-4152.2005
Bakour, I., Isaure, M. P., Barrouilhet, S., Goñi-Urriza, M., and Monperrus, M. (2023). Coupling fluorescent probes to characterize S-containing compounds in a sulfate reducing bacteria involved in Hg methylation. Talanta Open 7, 100228. doi:10.1016/j.talo.2023.100228
Barrouilhet, S., Monperrus, M., Tessier, E., Khalfaoui-Hassani, B., Guyoneaud, R., Isaure, M. P., et al. (2022). Effect of exogenous and endogenous sulfide on the production and the export of methylmercury by sulfate-reducing bacteria. Environ. Sci. Pollut. Res. 30, 3835–3846. doi:10.1007/s11356-022-22173-y
Benoit, J. M., Gilmour, C. C., Mason, R. P., and Heyes, A. (1999). Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters. Environ. Sci. Technol. 33 (6), 951–957. doi:10.1021/es9808200
Bravo, A. G., Zopfi, J., Buck, M., Xu, J., Bertilsson, S., Schaefer, J. K., et al. (2018). Geobacteraceae are important members of mercury-methylating microbial communities of sediments impacted by waste water releases. ISME J. 12 (3), 802–812. doi:10.1038/s41396-017-0007-7
Bridou, R., Monperrus, M., Gonzalez, P. R., Guyoneaud, R., and Amouroux, D. (2011). Simultaneous determination of mercury methylation and demethylation capacities of various sulfate-reducing bacteria using species-specific isotopic tracers. Environ. Toxicol. Chem. 30 (2), 337–344. doi:10.1002/etc.395
Campbell, P. G. C., Errécalde, O., Fortin, C., Hiriart-Baer, V. P., and Vigneault, B. (2002). Metal bioavailability to phytoplankton—applicability of the biotic ligand model. Comp. Biochem. Physiology Part C Toxicol. and Pharmacol. 133 (1), 189–206. doi:10.1016/s1532-0456(02)00104-7
Cardiano, P., Falcone, G., Foti, C., and Sammartano, S. (2011). Sequestration of Hg2+ by some biologically important thiols. J. Chem. and Eng. Data, 56. doi:10.1021/je200735r
Compeau, G. C., and Bartha, R. (1985). Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl. Environ. Microbiol. 50 (2), 498–502. doi:10.1128/aem.50.2.498-502.1985
Deonarine, A., and Hsu-Kim, H. (2009). Precipitation of mercuric sulfide nanoparticles in NOM-containing water: implications for the natural environment. Environ. Sci. Technol. 43 (7), 2368–2373. doi:10.1021/es803130h
Di Toro, D. M., Allen, H. E., Bergman, H. L., Meyer, J. S., Paquin, P. R., and Santore, R. C. (2001). Biotic ligand model of the acute toxicity of metals. 1. Technical basis. Environ. Toxicol. Chem. 20 (10), 2383–2396. doi:10.1897/1551-5028(2001)020<2383:blmota>2.0.co;2
Drott, A., Björn, E., Bouchet, S., and Skyllberg, U. (2013). Refining thermodynamic constants for mercury(II)-Sulfides in equilibrium with metacinnabar at sub-micromolar aqueous sulfide concentrations. Environ. Sci. Technol. 47 (9), 4197–4203. doi:10.1021/es304824n
Dyrssen, D., and Wedborg, M. (1991). The sulphur-mercury(II) system in natural waters. Water, Air, Soil Pollut. 56 (1), 507–519. doi:10.1007/bf00342295
Fein, J. B. (2017). Advanced biotic ligand models: using surface complexation modeling to quantify metal bioavailability to bacteria in geologic systems. Chem. Geol. 464, 127–136. doi:10.1016/j.chemgeo.2016.10.001
Flynn, S. L., Szymanowski, J. E. S., and Fein, J. B. (2014). Modeling bacterial metal toxicity using a surface complexation approach. Chem. Geol. 374–375, 110–116. doi:10.1016/j.chemgeo.2014.03.010
Gerbig, C., Kim, C., Stegemeier, J., Ryan, J., and Aiken, G. (2011). Formation of nanocolloidal metacinnabar in mercury-DOM-sulfide systems. Environ. Sci. and Technol. 45, 9180–9187. doi:10.1021/es201837h
Gilmour, C. C., Bullock, A. L., McBurney, A., Podar, M., and Elias, D. A. (2018). Robust mercury methylation across diverse methanogenic archaea. Lovley DR. mBio 9 (2), e02403. doi:10.1128/mBio.02403-17
Gilmour, C. C., Elias, D. A., Kucken, A. M., Brown, S. D., Palumbo, A. V., Schadt, C. W., et al. (2011). Sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 as a model for understanding bacterial mercury methylation. Appl. Environ. Microbiol. 77 (12), 3938–3951. doi:10.1128/aem.02993-10
Gilmour, C. C., Podar, M., Bullock, A. L., Graham, A. M., Brown, S. D., Somenahally, A. C., et al. (2013). Mercury methylation by novel microorganisms from new environments. Environ. Sci. Technol. 47 (20), 11810–11820. doi:10.1021/es403075t
Goñi-Urriza, M., Corsellis, Y., Lanceleur, L., Tessier, E., Gury, J., Monperrus, M., et al. (2015). Relationships between bacterial energetic metabolism, mercury methylation potential, and hgcA/hgcB gene expression in Desulfovibrio dechloroacetivorans BerOc1. Environ. Sci. Pollut. Res. Int. 22 (18), 13764–13771. doi:10.1007/s11356-015-4273-5
Goñi-Urriza, M., Klopp, C., Ranchou-Peyruse, M., Ranchou-Peyruse, A., Monperrus, M., Khalfaoui-Hassani, B., et al. (2020). Genome insights of mercury methylation among Desulfovibrio and Pseudodesulfovibrio strains. Res. Microbiol. 171 (1), 3–12. doi:10.1016/j.resmic.2019.10.003
Graham, A. M., Aiken, G. R., and Gilmour, C. C. (2012b). Dissolved organic matter enhances microbial mercury methylation under sulfidic conditions. Environ. Sci. Technol. 46 (5), 2715–2723. doi:10.1021/es203658f
Graham, A. M., Aiken, G. R., and Gilmour, C. C. (2013). Effect of dissolved organic matter source and character on microbial Hg methylation in Hg–S–dom solutions. Environ. Sci. Technol. 47 (11), 5746–5754. doi:10.1021/es400414a
Graham, A. M., Bullock, A. L., Maizel, A. C., Elias, D. A., and Gilmour, C. C. (2012a). Detailed assessment of the kinetics of Hg-cell association, Hg methylation, and methylmercury degradation in several Desulfovibrio species. Appl. Environ. Microbiol. 78 (20), 7337–7346. doi:10.1128/aem.01792-12
Gutensohn, M., Schaefer, J. K., Maas, T. J., Skyllberg, U., and Björn, E. (2023). Metabolic turnover of cysteine-related thiol compounds at environmentally relevant concentrations by Geobacter sulfurreducens. Front. Microbiol. 13, 1085214. doi:10.3389/fmicb.2022.1085214
Hand, C. E., and Honek, J. F. (2005). Biological chemistry of naturally occurring thiols of microbial and marine Origin. J. Nat. Prod. 68 (2), 293–308. doi:10.1021/np049685x
Hudson, R. J. M. (1998). Which aqueous species control the rates of trace metal uptake by aquatic biota? Observations and predictions of non-equilibrium effects. Sci. Total Environ. 219 (2), 95–115. doi:10.1016/s0048-9697(98)00230-7
Isaure, M. P., Albertelli, M., Kieffer, I., Tucoulou, R., Petrel, M., Gontier, E., et al. (2020). Relationship between Hg speciation and Hg methylation/demethylation processes in the sulfate-reducing bacterium Pseudodesulfovibrio hydrargyri: evidences from HERFD-XANES and nano-XRF. Front. Microbiol. 11, 584715. doi:10.3389/fmicb.2020.584715
Jiang, T., Skyllberg, U., Wei, S., Wang, D., Lu, S., Jiang, Z., et al. (2015). Modeling of the structure-specific kinetics of abiotic, dark reduction of Hg(II) complexed by O/N and S functional groups in humic acids while accounting for time-dependent structural rearrangement. Geochimica Cosmochimica Acta 154, 151–167. doi:10.1016/j.gca.2015.01.011
Liem-Nguyen, V., Bouchet, S., and Björn, E. (2015). Determination of sub-nanomolar levels of low molecular mass thiols in natural waters by liquid chromatography tandem mass spectrometry after derivatization with p-(hydroxymercuri) benzoate and online preconcentration. Anal. Chem. 87 (2), 1089–1096. doi:10.1021/ac503679y
Liem-Nguyen, V., Nguyen-Ngoc, H. T., Adediran, G. A., and Björn, E. (2020). Determination of picomolar levels of methylmercury complexes with low molecular mass thiols by liquid chromatography tandem mass spectrometry and online preconcentration. Anal. Bioanal. Chem. 412 (7), 1619–1628. doi:10.1007/s00216-020-02389-y
Liem-Nguyen, V., Skyllberg, U., and Björn, E. (2017a). Thermodynamic modeling of the solubility and chemical speciation of mercury and methylmercury driven by organic thiols and micromolar sulfide concentrations in boreal wetland soils. Environ. Sci. Technol. 51 (7), 3678–3686. doi:10.1021/acs.est.6b04622
Liem-Nguyen, V., Skyllberg, U., Nam, K., Björn, E., Liem-Nguyen, V., Skyllberg, U., et al. (2017b). Thermodynamic stability of mercury(II) complexes formed with environmentally relevant low-molecular-mass thiols studied by competing ligand exchange and density functional theory. Environ. Chem. 14 (4), 243–253. doi:10.1071/en17062
Lin, H., Lu, X., Liang, L., and Gu, B. (2015). Cysteine inhibits mercury methylation by Geobacter sulfurreducens PCA mutant Δ omcBESTZ. Environ. Sci. and Technol. Lett. 2 (5), 144–148. doi:10.1021/acs.estlett.5b00068
Lu, X., Liu, Y., Johs, A., Zhao, L., Wang, T., Yang, Z., et al. (2016). Anaerobic mercury methylation and demethylation by Geobacter bemidjiensis bem. Environ. Sci. and Technol. 50 (8), 4366–4373. doi:10.1021/acs.est.6b00401
Miller, C. L., Southworth, G., Brooks, S., Liang, L., and Gu, B. (2009). Kinetic controls on the complexation between mercury and dissolved organic matter in a contaminated environment. Environ. Sci. Technol. 43 (22), 8548–8553. doi:10.1021/es901891t
Morel, F. M. M., Hudson, R. J. M., and Price, N. M. (1991). Limitation of productivity by trace metals in the sea. Limnol. Oceanogr. 36 (8), 1742–1755. doi:10.4319/lo.1991.36.8.1742
Ndu, U., Mason, R. P., Zhang, H., Lin, S., and Visscher, P. T. (2012). Effect of inorganic and organic ligands on the bioavailability of methylmercury as determined by using a mer-lux bioreporter. Appl. Environ. Microbiol. 78 (20), 7276–7282. doi:10.1128/aem.00362-12
Parks, J. M., Johs, A., Podar, M., Bridou, R., Hurt, R. A., Smith, S. D., et al. (2013). The genetic basis for bacterial mercury methylation. Science. 339 (6125), 1332–1335. doi:10.1126/science.1230667
Pedrero, Z., Bridou, R., Mounicou, S., Guyoneaud, R., Monperrus, M., and Amouroux, D. (2012). Transformation, localization, and biomolecular binding of Hg species at subcellular level in methylating and nonmethylating sulfate-reducing bacteria. Environ. Sci. Technol. 46 (21), 11744–11751. doi:10.1021/es302412q
Perrot, V., Bridou, R., Pedrero, Z., Guyoneaud, R., Monperrus, M., and Amouroux, D. (2015). Identical Hg isotope mass dependent fractionation signature during methylation by sulfate-reducing bacteria in sulfate and sulfate-free environment. Environ. Sci. Technol. 49 (3), 1365–1373. doi:10.1021/es5033376
Pham, A. L. T., Morris, A., Zhang, T., Ticknor, J., Levard, C., and Hsu-Kim, H. (2014). Precipitation of nanoscale mercuric sulfides in the presence of natural organic matter: structural properties, aggregation, and biotransformation. Geochimica Cosmochimica Acta 133, 204–215. doi:10.1016/j.gca.2014.02.027
Ranchou-Peyruse, M., Monperrus, M., Bridou, R., Duran, R., Amouroux, D., Salvado, J. C., et al. (2009). Overview of mercury methylation capacities among anaerobic bacteria including representatives of the sulphate-reducers: implications for environmental studies. Geomicrobiol. J. 26 (1), 1–8. doi:10.1080/01490450802599227
Ravichandran, M. (2004). Interactions between mercury and dissolved organic matter–a review. Chemosphere 55 (3), 319–331. doi:10.1016/j.chemosphere.2003.11.011
Rodríguez-González, P., Monperrus, M., Alonso, J. I. G., Amouroux, D., and Donard Of, X. (2007). Comparison of different numerical approaches for multiple spiking species-specific isotope dilution analysis exemplified by the determination of butyltin species in sediments. J. Anal. Atomic Spectrom. 22 (11), 1373–1382. doi:10.1039/b706542f
Rodrı́guez Martı́n-Doimeadios, R. C., Tessier, E., Amouroux, D., Guyoneaud, R., Duran, R., Caumette, P., et al. (2004). Mercury methylation/demethylation and volatilization pathways in estuarine sediment slurries using species-specific enriched stable isotopes. Mar. Chem. 90 (1), 107–123. doi:10.1016/j.marchem.2004.02.022
Schaefer, J. K., and Morel, F. M. M. (2009). High methylation rates of mercury bound to cysteine by Geobacter sulfurreducens. Nat. Geosci. 2 (2), 123–126. doi:10.1038/ngeo412
Schaefer, J. K., Rocks, S. S., Zheng, W., Liang, L., Gu, B., and Morel, F. M. M. (2011). Active transport, substrate specificity, and methylation of Hg(II) in anaerobic bacteria. PNAS 108 (21), 8714–8719. doi:10.1073/pnas.1105781108
Shimada, T., Tanaka, K., and Ishihama, A. (2016). Transcription factor DecR (YbaO) controls detoxification of L-cysteine in Escherichia coli. Microbiol. Read. 162 (9), 1698–1707. doi:10.1099/mic.0.000337
Slaveykova, V. I., Wilkinson, K. J., Slaveykova, V. I., and Wilkinson, K. J. (2005). Predicting the bioavailability of metals and metal complexes: critical review of the biotic ligand model. Environ. Chem. 2 (1), 9–24. doi:10.1071/en04076
Smith, R. M., Martell, A., Motekaitis, R., Smith, R., and Motekaitis, R. (2004). NIST critically selected stability constants of metal complexes database. Available online at: https://www.semanticscholar.org/paper/NIST-Critically-Selected-Stability-Constants-of-Smith-Martell/4d1e936ecb2cd7c641ada4f9db589ad5b50072ba.
Stenzler, B. R., Zhang, R., Semrau, J. D., DiSpirito, A. A., and Poulain, A. J. (2022). Diffusion of H 2 S from anaerobic thiolated ligand biodegradation rapidly generates bioavailable mercury. Environ. Microbiol. 24 (7), 3212–3228. doi:10.1111/1462-2920.16078
Szczuka, A., Morel, F. M. M., and Schaefer, J. K. (2015). Effect of thiols, zinc, and redox conditions on Hg uptake in shewanella oneidensis. Environ. Sci. Technol. 49 (12), 7432–7438. doi:10.1021/acs.est.5b00676
Thomas, S. A., Catty, P., Hazemann, J. L., Michaud-Soret, I., and Gaillard, J. F. (2019). The role of cysteine and sulfide in the interplay between microbial Hg(II) uptake and sulfur metabolism. Metallomics. 11 (7), 1219–1229. doi:10.1039/c9mt00077a
Thomas, S. A., and Gaillard, J. F. (2017). Cysteine addition promotes sulfide production and 4-fold Hg(II)–S coordination in actively metabolizing Escherichia coli. Environ. Sci. Technol. 51 (8), 4642–4651. doi:10.1021/acs.est.6b06400
Thomas, S. A., Rodby, K. E., Roth, E. W., Wu, J., and Gaillard, J. F. (2018). Spectroscopic and microscopic evidence of biomediated HgS species formation from Hg(II)–Cysteine complexes: implications for Hg(II) bioavailability. Environ. Sci. Technol. 52 (17), 10030–10039. doi:10.1021/acs.est.8b01305
Thomas, S. A., Tong, T., and Gaillard, J. F. (2014). Hg(ii) bacterial biouptake: the role of anthropogenic and biogenic ligands present in solution and spectroscopic evidence of ligand exchange reactions at the cell surface. Metallomics 6 (12), 2213–2222. doi:10.1039/c4mt00172a
Warner, T., and Jalilehvand, F. (2016). Formation of Hg(II) tetrathiolate complexes with cysteine at neutral pH. Can. J. Chem. 94 (4), 373–379. doi:10.1139/cjc-2015-0375
Widdel, F., and Bak, F. (1992). “Gram-negative mesophilic sulfate-reducing bacteria,” in The prokaryotes: a handbook on the biology of bacteria: ecophysiology, isolation, identification, applications. Editors A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (New York, NY: Springer), 3352–3378. doi:10.1007/978-1-4757-2191-1_21
Xu, Y., Shi, D., Aristilde, L., and Morel, F. M. M. (2012). The effect of pH on the uptake of zinc and cadmium in marine phytoplankton: possible role of weak complexes. Limnol. Oceanogr. 57 (1), 293–304. doi:10.4319/lo.2012.57.1.0293
Yang, Y., Feng, Y., Jiang, Y., Qiu, F., Wang, Y., Song, X., et al. (2019). A coumarin-based colorimetric fluorescent probe for rapid response and highly sensitive detection of hydrogen sulfide in living cells. Talanta 197, 122–129. doi:10.1016/j.talanta.2018.12.081
Zhang, T., Kim, B., Levard, C., Reinsch, B. C., Lowry, G. V., Deshusses, M. A., et al. (2012). Methylation of mercury by bacteria exposed to dissolved, nanoparticulate, and microparticulate mercuric sulfides. Environ. Sci. Technol. 46 (13), 6950–6958. doi:10.1021/es203181m
Keywords: speciation, bioavailability, thiols, sulfides, Hg transformations, pure strain culture, isotopic tracers, hyphenated mass spectrometry techniques
Citation: Bakour I, Isaure M-P, Barrouilhet S, Goñi-Urriza M and Monperrus M (2025) Mercury interaction with S-containing molecules: implications for methylation and demethylation processes in a sulfate reducing bacteria. Front. Environ. Chem. 6:1559968. doi: 10.3389/fenvc.2025.1559968
Received: 13 January 2025; Accepted: 05 March 2025;
Published: 19 March 2025.
Edited by:
Robert Peter Mason, University of Connecticut, United StatesCopyright © 2025 Bakour, Isaure, Barrouilhet, Goñi-Urriza and Monperrus. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mathilde Monperrus, bWF0aGlsZGUubW9ucGVycnVzQHVuaXYtcGF1LmZy
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.