
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Environ. Chem., 14 June 2024
Sec. Environmental Analytical Methods
Volume 5 - 2024 | https://doi.org/10.3389/fenvc.2024.1334207
 Kendra G. Selby
Kendra G. Selby Claire E. Korte
Claire E. Korte Lauren H. Phan
Lauren H. Phan Gabriel A. Bressendorff
Gabriel A. Bressendorff Ashley R. Chirchirillo
Ashley R. Chirchirillo Kevin R. Tucker*
Kevin R. Tucker*Matrix selection and application is a crucial step in obtaining meaningful results with matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI). Most instruments have a large spatial, and data with adequate spatial resolution can frequently be obtained on a benchtop instrument. The matrix application workflow has been optimized for the imaging of the earthworm (Eisenia hortensis), after exposure to various statins, a class of blood lipid-lowering agents. Lipids are nonpolar, often neutral molecules, making them difficult to ionize, and heightening the need for matrix optimization. The matrices 2,5-dihydroxybenzoic acid (DHB), α-cyano-4-hydroxycinammic acid (CHCA), 1,5-diaminonaphthalene (DAN), and 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid; SA) were studied. Samples were also washed in the ammonium salts of acetate, bicarbonate, formate, sulfate, or water as a control to enhance ionization and improve spatial resolution. A successful matrix for MSI is one that demonstrates homogenous tissue coverage, ionization of the analytes of interest, and does not require excessive laser power for ionization. All matrices showed sufficient tissue coverage; however, CHCA yielded unambiguous images of cholesterol and yielded sufficient signal over the lipid mass range (400–1,000 m/z), indicating that it successfully ionized endogenous lipids. Following additional optimization, the application of 50 mL of 10 mg/mL CHCA following a 5 s salt ammonium sulfate salt wash proved most successful for improving lipid ionization and enhancing spatial resolution.
Pharmaceuticals continue to be detected in the environment and are known to have toxic effects on the ecosystem, earning them the title of “contaminants of emerging concern.” Statins are a class of blood lipid-lowering agents that have been increasingly prescribed throughout the last decade (Matyori et al., 2023), which has led to their existence in the environment (Pino et al., 2015). The growing pharmaceutical industry and the difficulty removing these contaminants from wastewater treatment plants (Ternes et al., 2004; Yu et al., 2011) creates the urgent need for analysis of affected organisms including earthworms (Eisenia hortensis), an effective indicator of biotoxicity (Van-Gestel, Dirven-van Breemen and Baerselman, 1992; Fründ et al., 2010; Ciğerci et al., 2016). Dosing a model terrestrial environment with pharmaceuticals allows the uptake and distribution into biological tissue to be studied via mass spectrometry imaging (MSI). This technique is unique from bulk analytical methods in its ability to provide information regarding the spatial distribution of analytes within a sample, often with relatively simple sample preparation. Although MALDI MSI is a powerful technique, its application in environmental contexts is in the early stages of development (Selby et al., 2024). Moreover, such instruments are large, with time-of-flight (TOF) mass analyzers spanning from floor to ceiling. Instruments of this nature are expensive to purchase, cumbersome to care for, and have a large spatial footprint, thereby making benchtop MALDI TOF instruments more practical for some researchers, including those at primarily undergraduate institutions or labs with a small sample capacity.
The most important portion of the sample preparation workflow is the choice of matrix. Matrices are typically conjugated organic acids or bases; the conjugated organic nature allows for absorption of laser irradiation, whereas the acidic or basic properties of the matrix allow for proton exchange between the analyte and matrix. There are hundreds of matrices to choose from (Calvano et al., 2018; Leopold et al., 2018), and in selecting one, the analytes of interest, the tissue to which matrix is being applied, and the spatial resolution of the instrument must be considered. Although some matrices interact favorably with only a few classes compounds (e.g., 2,4,6-trihydroxyacetophenone (THAP) for nucleic acid and peptide/protein analysis (Patil et al., 2024), others span a wide range of assorted compounds (e.g., trans-2-[3-(4-tert-Butylphenyl)-2-methyl-2-propenylidene]malononitrile (DCTB) for analysis of oligomers, polymers, dendrimers, and small molecules (Ulmer et al., 2000; Brown et al., 2001)). However, the road to finding a suitable matrix for a given application is often considered a “trial-and-error” process because matrices do not always interact with analytes in the expected manner (Leopold et al., 2018). This is likely a consequence of an incomplete understanding of the exact mechanism of any given matrix. This necessitates matrix optimization, during which several matrices are examined for the most favorable interaction with the analytes of interest and the tissue being ionized from. A successful matrix for MSI is one that demonstrates homogenous tissue coverage, ionization of the analytes of interest, and does not require excessive laser power for ionization. The matrices tested in this study were 2,5-dihydroxybenzoic acid (DHB), α-cyano-4-hydroxycinammic acid (CHCA), 1,5-diaminonaphthalene (DAN), and 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid; SA) for the ionization of lipids and statins from earthworms. These matrices were chosen due to their ability to ionize lipids and statins (Jirásko et al., 2014; Leopold et al., 2018; Zhu et al., 2019; Angerer et al., 2022; Yin et al., 2022; Leopold et al., 2023).
Matrix choice and application is of paramount importance because a matrix that is incompatible with the tissue or analytes of interest will result in unsuccessful ionization. Some analytes may be difficult to ionize regardless of matrix choice due to low concentration relative to other endogenous compounds. This may be addressed by washing the tissue prior to matrix application with a solution that will remove interfering endogenous compounds while allowing the tissue to retain the low concentration analytes of interest. The analytes of interest for this study included lipids—a typically neutral nonpolar species that can be difficult to positively ionize. Washes with ammoniated salts prior to matrix deposition have been shown to enhance biological lipid signal (Wang et al., 2011; Trim and Snel, 2016; Leopold et al., 2018), and amines are also known to interact favorably with lovastatin and simvastatin (Miao and Metcalfe, 2003). As a result, four different ammonium salts—acetate, bicarbonate, formate, and sulfate—and water as a control at varying submersion times were examined.
The aim of this research was to optimize the matrix application workflow for effectively analyzing the impact of exposure to atorvastatin, lovastatin, and simvastatin, along with endogenous lipids, in earthworms (E. hortensis). Images were generated using a linear, positive polarity MALDI TOF following matrix application via an artist airbrush sprayer (Zhang et al., 2020). Four matrices (CHCA, DAN, DHB, and SA) were examined for their ability to cocrystallize with the tissue to allow for analyte ionization. The amount of matrix was then standardized for matrices that demonstrated cocrystallization and analyte ionization before proceeding with an assessment of the effects of an ammonium-based salt wash prior to matrix deposition. The most successful workflow will then be utilized in a complementary qualitative and quantitative analysis of the effect of environmentally relevant concentrations of statins on earthworms.
For pharmaceutical exposures, atorvastatin calcium (certified reference material), lovastatin (>98%), and simvastatin (98%) were purchased from Sigma Aldrich (St. Louis, MO), TCI Co. (Kita-Ku, Tokyo, Japan), and Fisher Scientific (Hampton, NH), respectively. The MALDI matrices α-cyano-4-hydroxycinnamic acid (CHCA; 98%), 2,5-diaminonaphthalene (DAN; >98.0%), 2,5-dihydroxybenzoic acid (DHB; 99%), and 3,5-dimethoxy-4-hydroxycinnamic acid (sinapinic acid (SA); >98%) were purchased from Fisher Scientific, TCI Co., Thermo Fisher Scientific (Waltham, MA), and TCI Co., respectively. Standards and matrices were dissolved in methanol (HPLC grade; Fisher Scientific), ethanol (200 proof, Pharmco (Brookfield, CT)), or acetonitrile (HPLC grade; Fisher Scientific) depending on solubility. The protonating aid trifluoroacetic acid (TFA) (>99.5%) was purchased from VWR (Radnor, PA). For the assessment of salt washes, ammonium acetate (ACS, Reagent grade), bicarbonate (Baker Analyzed Reagent grade), formate (>99.0%), and sulfate (Certified ACS) were purchased from RICCA (Arlington, TX), Avantor (Radnor, PA), Honeywell (Charlotte, NC), and Thermo Fisher Scientific, respectively. Type I (≥18.2 MΩ) filtered water was used for any instance that water was needed.
The studies involving animals were reviewed and approved by the Southern Illinois University Edwardsville Institutional Animal Care and Use Committee under Protocol #11723-KT1. Earthworms (E. hortensis) were purchased from Speedy Worm (Alexandria, MN) and housed in 20-gal fish tanks containing locally purchased organic potting soil. The worms were acclimated for at least 2 weeks prior to exposure to the statins. Care measures were taken weekly, during which the worms were fed spent coffee grounds, and the soil moisture was adjusted via Type I water to fall within the range of 20%–30% moisture.
Twelve 2.5-gal tanks were prepared with 1.8 kg of potting soil and 20 acclimated worms were added for exposure to statins. To nine tanks, 400 mL either 1,000% of the environmentally relevant concentration of atorvastatin (2.0 ppb (Ottmar et al., 2012)), lovastatin (1.0 ppb (Conley et al., 2008)), or simvastatin (15 ppb (Pereira et al., 2016)) was added. The remaining three tanks received 400 mL of Type I water as control. The worms were kept in the treated soil for 2 weeks, and the soil was re-dosed with statin solution on day 7 maintaining the 20%–30% moisture level. The soil mass and volume of dose solution were designed to model the field irrigation and related water runoff (Shukla and Boman, 2006; Perdue and Hamer, 2019). Following the exposure period, three worms per tank were selected for MALDI MSI analysis, and the remaining were reserved for future studies.
The three worms designated for MALDI MSI analysis were rinsed with Type I water, coiled in a metal bottle cap, and covered with aluminum foil before flash freezing in liquid nitrogen. Worms were mounted in Type I water and cryosectioned (Lecia CM 1850; Deer Park, IL) at −20°C to obtain 20 μm longitudinal sections. Each section was thaw-mounted to an indium tin oxide (ITO)-coated glass slide (Shimadzu Scientific Instruments, Columbia, MD) and stored at −80°C. Slides were dried in a vacuum desiccator prior to matrix application using an artist’s airbrush sprayer (Paasche VL 0123, Kenosha, WI). Specific matrix application parameters varied depending on the purpose of the given step in matrix optimization, as described below. First, 10 mg/mL CHCA, DHB, DAN, and SA were dissolved in either methanol or acetonitrile according to solubility and received 0.1% (v/v) TFA to aid in ionization. The crystal sizes for these matrices vary, thus matrix was applied to each slide until there was consistent coverage of the tissue observed via optical microscopy. Several trials were performed at varying matrix concentrations to determine the average mass applied to the slide. This information was then used to determine the average volume of 10 mg/mL matrix that should be applied in matrix optimization. Next, comparatively low, middle, and high volumes of the successful 10 mg/mL matrices were examined. Finally, washes of four ammonium salts (acetate, formate, bicarbonate, and sulfate) prior to matrix deposition were examined prior to matrix application. All salt solutions were 50 mM; water was used as a control. Variable wash times (5, 10, and 30 s) were examined. For each acquisition, a white marker was used to indicate the corners of the imaging window to increase ease in defining this space. Additionally, 1 μL of 1:1 5 mg/mL CHCA:TOFMix standard (LaserBio Labs, Valbonne, France) was applied via the dried droplet method for mass spectral calibration.
Spectral and spatial data were acquired using MALDI Solutions Data Acquisition software (Shimadzu Scientific Instruments, Columbia, MD) on the Shimadzu MALDI-8020 (Columbia, MD): a linear, positive polarity, benchtop MALDI TOF MS with imaging capabilities. There were four assessment parameters used throughout matrix optimization summarized in Table 1. During Step 1, instrument acquisition conditions (laser power, pulsed extract, and number of shots) were optimized for each matrix to obtain high mass spectral signal-to-noise ratio (S/N), mass resolution, and on-tissue signal (mV). Final instrument operation conditions for remaining analyses were decided in Step 2 in accordance with the most successful amount of matrix. The mass range for all acquisitions was designed in such a way that the matrix ions (<400 m/z) and lipid signals could be captured (typically ∼400–1,000 m/z). The blanking mass was set to be the lower limit of the mass range. The number of profiles was automatically calculated according to the specified resolution (100 × 100 μm) and defined imaging area, which is directly correlated with the amount of time required to acquire data.
Images were generated and processed using IonView software (Shimadzu Scientific Instruments, Columbia, MD). Processing included normalizing the signal to the total ion count and the scales of each image were internally adjusted to maximize image contrast. Additionally, post hoc calibration was performed using a one-point calibration adjustment based on the signal from the CHCA dimer.
In identifying the most suitable matrix for ionization of lipids in earthworms, the spectral and spatial data were judged according to parameters 1, 2, and 3 (Table 1). CHCA, DAN, DHB, or SA was applied until the slide and tissue were uniformly coated as observed via optical microscopy due to varying crystal sizes. All matrices generated signal that was widespread across the entire imaging window, indicating consistent slide and tissue coverage (Figure 1A). Notably, the signal intensities of DAN and SA matrix were higher than that of the others. DAN and DHB poorly ionized cholesterol, as shown by the low signal intensity of cholesterol and remarkable similarity of the cholesterol and matrix images, respectively. Cholesterol was selected as an indicator of success because it is prevalent in biological tissue and has a well-established MALDI MS signal ([M+H–H2O]+ (369 m/z)) (McAvey et al., 2011). CHCA and SA were both successful in their ionization of cholesterol given the notable intensity and that the cholesterol signal can be easily distinguished from that of the matrix. Unsurprisingly given its poor ionization of cholesterol, DHB was unsuccessful in ionizing in the lipid region with at least threefold less signal intensity than the other matrices (Figure 1B). CHCA, DAN, and SA demonstrated sufficient signal intensity in the lipid region, with the most signal being generated by SA (Figure 1B). Both CHCA and SA demonstrated marked success compared to DAN and DHB in their tissue coverage, ionization of endogenous analytes, and ionization of lipids in earthworm tissue; therefore, both matrices were examined further.
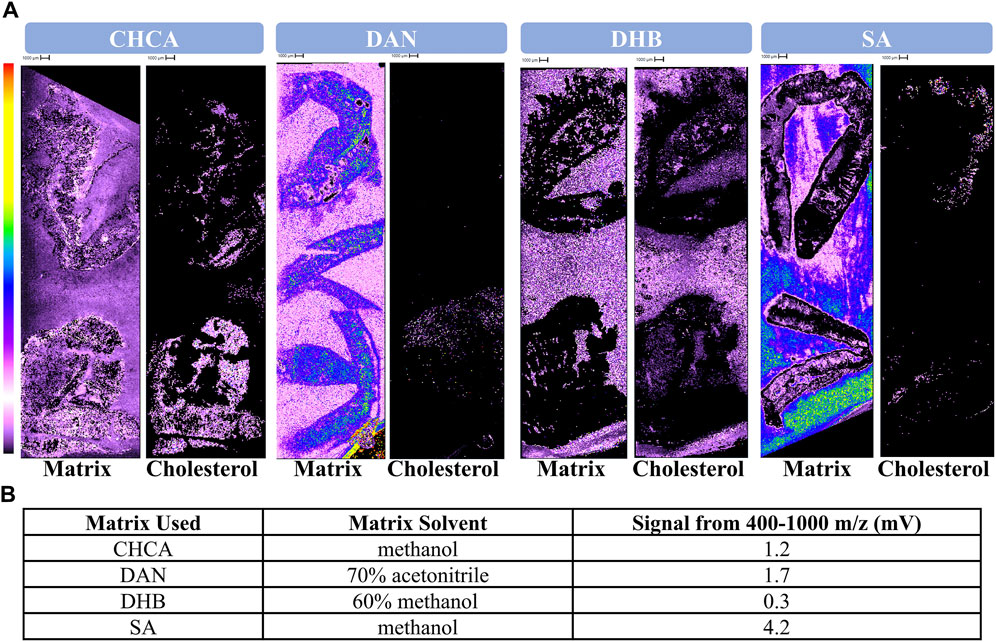
Figure 1. Comparison of CHCA, DAN, DHB, and SA. (A) Protonated matrix, cholesterol images, (B) and quantitative lipid signal following the application of four different matrices. 10 mg/mL CHCA, DAN, DHB, or SA were deposited onto each slide via airbrush sprayer until sufficient crystallization and tissue coverage was observed via optical microscope. The left-side image for each matrix header depicts the signal generated by protonated matrix (CHCA dimer 379.09, DAN 159.09, DHB 155.03, SA 225.08 m/z). The right-side image for each matrix header depicts the signal generated by cholesterol occurring at 369 m/z.
The amount of CHCA or SA applied to the slide was then optimized. Parameters 2, 3, and 4 were used to assess the data’s spectral and spatial quality, as well as the feasibility of instrument operation used for the optimized amount of matrix (Table 1). Comparatively low, middle, and high amounts of CHCA and SA were determined to be 20, 30, and 50 mL and 10, 20, and 40 mL, respectively. Cholesterol was well ionized by 50 mL CHCA and 30 mL SA given the distinct signal arising from the tissue, while other amounts generated cholesterol signal that was easily confused with matrix or produced low signal intensity (Figure 2A). 50 mL CHCA was likewise successful in lipid ionization whereas lower volumes generated insufficient signal intensity (Figure 2B). All volumes of SA yielded sufficient signal intensity for lipids, with 40 mL SA generating the highest signal intensity (Figure 2B). Uniquely considered during this portion of optimization was the laser power required for sufficient ionization to determine whether prolonged operation was feasible. Regardless of matrix amount, SA required a higher laser power than any volume of CHCA (Figure 2B). 50 mL CHCA successfully yielded meaningful results at a more reasonable laser power, and therefore this volume of matrix and instrument acquisition conditions were employed in subsequent analyses.
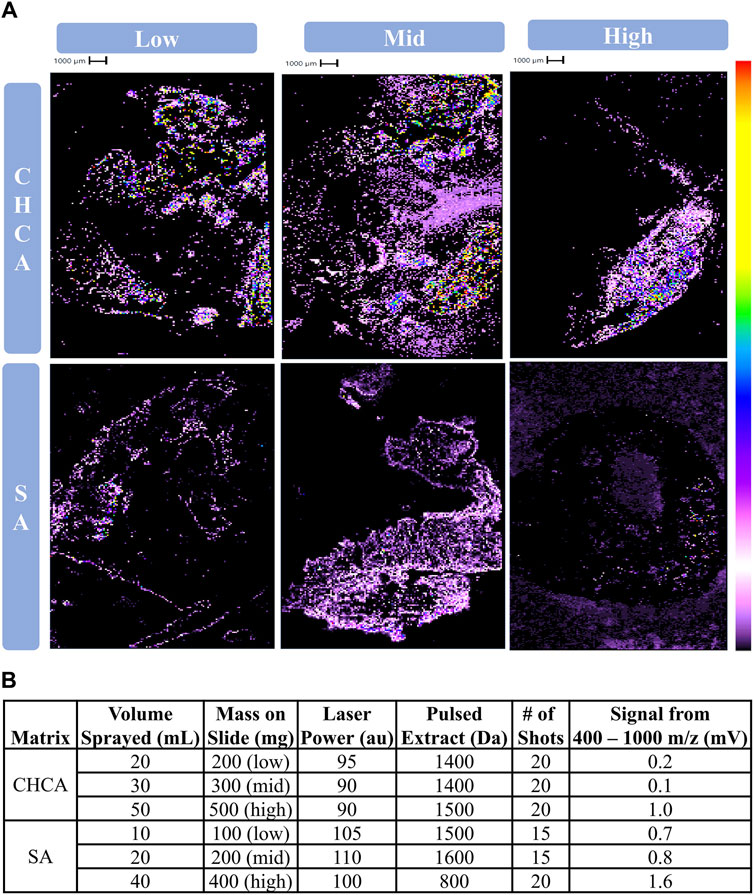
Figure 2. Determining the of amount of matrix applied to slide for CHCA and SA. (A) Cholesterol images (369 m/z), (B) lipid signal, and instrument operation conditions following the application of varying amounts of CHCA or SA. 10 mg/mL CHCA or SA were deposited on slides via airbrush sprayer at specified volumes. These masses deposited on the slides represent low, middle, and high amounts relative to the average applied mass of both CHCA and SA.
A salt wash prior to application of 50 mL CHCA was examined for its potential to aid in lipid ionization. According to parameters determined in Step 2, all salt wash data was acquired at a laser power of 90, pulsed extract of 1,500, and 20 shots. The quality of Step 3 data was assessed via parameters 2 and 3 (Table 1). Regardless of wash solution, 5 and 10 s washes generated the most distinct of cholesterol and most reasonable intensity for endogenous lipids, suggesting greater success at lipid ionization. A longer wash time enhances noise, causing cholesterol to be visually indistinguishable from off-tissue matrix signal (e.g., 30 s water) or from background signal (e.g., 30 s ammonium bicarbonate wash) (Figure 3A). When considering the 5 and 10 s washes, ammonium sulfate, ammonium bicarbonate, and water were all qualitatively successful in generating ionization of endogenous analytes with meaningful images. Short wash times of ammonium sulfate were the most successful both qualitatively (Figure 3A) and quantitatively (Figure 3B), and therefore this salt was identified as being the superior for ionization of lipids from statin-exposed earthworms. At this point in optimization, the worms were also assessed for ionization of statins; unfortunately, none were successfully ionized.
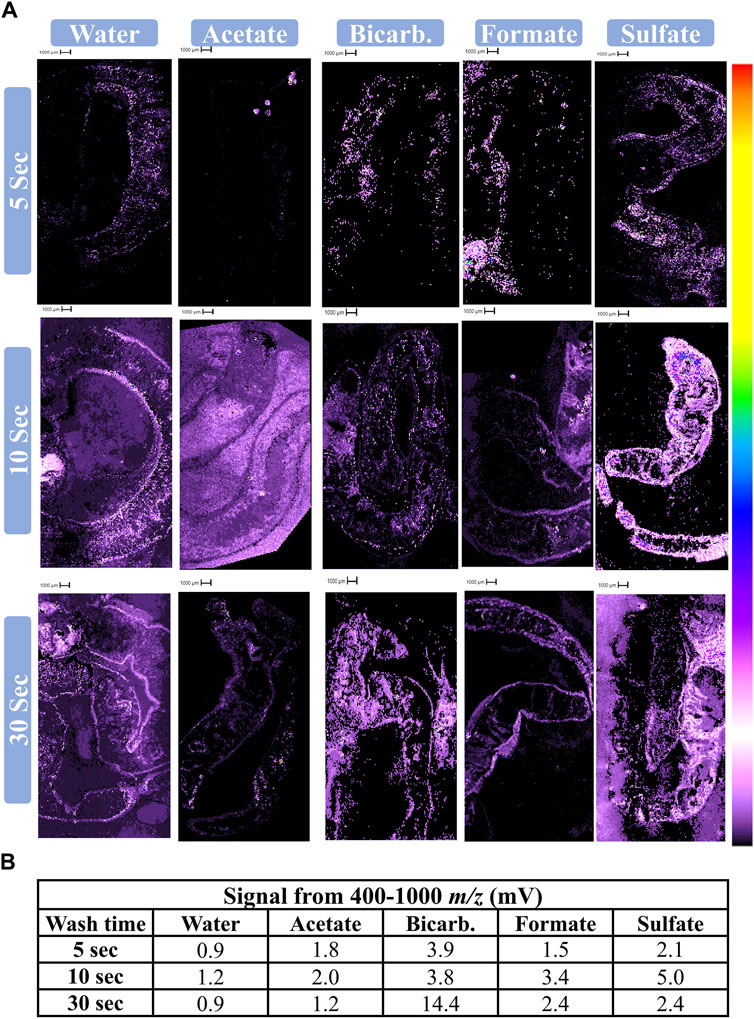
Figure 3. Assessment of ammonium salt wash before applying CHCA. (A) Cholesterol images (369 m/z) and (B) lipid signal following various salt washes. 50 mL of 10 mg/mL CHCA was deposited on slides via an artist’s airbrush following a wash (50 mM specified salt) for a given length of time. The instrument was operated at a laser power of 90, pulsed extract of 1,500 Da, and 20 shots.
DAN was ruled out as a probable matrix for ionization of lipids in earthworm tissue due to the high intensity of matrix signal and minimal signal for cholesterol (Figure 1A). The high intensity of the matrix signal is likely a consequence of it being a basic rather than acidic matrix like CHCA, DHB, and SA. As a result, DAN will accept a proton rather than donating one to the analytes of interest, thereby allowing for easy matrix detection in positive mode and suppressing ionization of the analytes. The Shimadzu MALDI-8020 can only perform positive ionization, so DAN and other basic matrices are not suitable for applications on this instrument.
DHB was ruled out as a probable matrix for ionization lipids in earthworm tissue due to images from matrix and cholesterol signal being indistinguishable from one another (Figure 1A). This may seem surprising, as DHB is a well-known matrix applied to a variety of different analytes including proteins, oligosaccharides, and perhaps most relevant, lipids (Hillenkamp et al., 1991; Strupat et al., 1991; Leopold et al., 2023). DHB frequently generates large crystals (Li et al., 2016) when compared to other matrices used in this study. Larger crystals are more difficult to apply in a homogenous and reproducible manner, especially using an artist’s airbrush. Due to the difficulty in successful homogenous matrix application to the tissue, it is unlikely that co-crystallization of the endogenous analytes and matrix occurred. This thereby prevents the matrix from assisting in desorption and ionization of the analytes, effectively rendering the matrix ineffective. The disadvantage of large crystal size is supported by the success of CHCA and SA, which both have smaller crystal sizes. It is possible that DHB would have been successful for this application had it been applied with an automated sprayer and therefore well-controlled matrix application system.
SA was initially promising as a potential matrix as it generated sufficient matrix coverage and successful ionization of endogenous analytes (Figure 1). SA required a higher laser power regardless of the amount of matrix applied to the slide (Figure 2), indicating that the matrix is ill-suited for lipids (Leopold et al., 2023). Other studies suggest that SA would be more suitable for ionization of higher mass analytes such as proteins (Patil et al., 2024). Moreover, laser power and ionization are positively related regardless of matrix choice; however, there is no way to confidently ensure that only the desired analytes are being ionized. Instead, additional classes of compounds compete for ionization with the analytes of interest, thus reducing the signal from analytes of interest. Additionally, enhancing overall signal can lead to earlier burnout of the detector. The over-enhancement of signal is supported by the higher signal within the lipid region for those slides receiving SA (Figures 1B, 2B). Furthermore, the laser itself may suffer early burnout, as earthworms are relatively large, and typically require anywhere from 2 to 4 h to image at a 100 μm resolution. Repeatedly operating the instrument for longer run times at an excessive laser power to obtain replicates and sufficient data would be impractical and harmful to the instrument’s long-term performance. As a result, SA was deemed an unsuitable matrix for detecting lipids from biological tissue on this linear benchtop MALDI TOF.
CHCA yielded consistent tissue coverage and successful ionization of endogenous analytes (Figure 1). Following an assessment of the amount of matrix to be applied, a relatively high amount (50 mL) was deemed to be the most successful as it generated the most quantitative lipid ionization while also maintaining a more reasonable laser power than SA (Figure 2). CHCA has been employed for the ionization of numerous compounds such as peptides, smaller proteins, and lipids, among others (Lou et al., 2015; Itonori et al., 2018; Høiem et al., 2022). The ability of CHCA to ionize smaller molecules compared to the other matrices is the most probable reason as to why it is the most suitable matrix for ionization within the lipid region on a linear MALDI TOF. Moreover, CHCA crystals are relatively small, making them more compatible for use with a handheld sprayer. The success of CHCA led to its use in the assessment of ammonium-based salt washes.
Washing the slide with an ammonium salt prior to deposition of 50 mL 10 mg/mL CHCA led to enhanced signal within the lipid region for most of the salts examined (Figure 3). This is likely because the wash solution removes polar interfering compounds, thereby enabling the detection of less polar analytes, including lipids. Longer washes reduced spatial quality (Figure 3A), likely because the compounds of interest and perhaps the tissue itself were also being washed away, as well as analyte redistribution on the surface. This is supported by the decreased spatial quality regardless of which salt or water was used (Figure 3A). Additionally, the lipid signal for four of the five conditions at the 30 s wash time more closely resembles that of the 5 s wash, with a peak in signal intensity at 10 s (Figure 3B). Ammonium sulfate was most successful at enhancing signal within the lipid region while also rendering spatially resolved images of endogenous analytes. This may be the result of a favorable interaction between sulfate and the earthworm tissue and/or CHCA that allows for more homogenous crystal distribution and analyte/matrix co-crystallization.
Despite successful ionization of lipids and related analytes, neither water nor salt wash enhanced detection of statins. It may have been the case that the exposure concentrations may have been too low, leading to minimal uptake by the earthworms resulting in the concentrations being below the limit of detection. It is also possible that the wash removed the statins from the tissue as they are more polar and smaller than lipids. Lastly, the endogenous lipids are known to exist in the tissue at greater concentrations than the statins, and therefore enhancing their signal using a salt wash may have masked any signal created by the drugs.
This project aimed to enhance meaningful lipid signal and spatial resolution of statins and lipids from earthworm tissue analyzed by MSI on linear benchtop MALDI TOF MS. A successful matrix for MSI is one that demonstrates homogenous tissue coverage, ionization of the analytes of interest, and does not require excessive laser power for ionization. A 5 s wash with 50 mM ammonium sulfate prior to application of 50 mL of 10 mg/mL CHCA (with 0.1% v/v TFA) via artist airbrush sprayer proved superior for lipid ionization compared to DAN, DHB, and SA; however, these conditions failed to ionize statins. As a result, variable ammonium sulfate concentrations should be investigated to optimize the ionization of statins. Additionally, earthworms may be exposed to higher concentrations of statins with the aim of ensuring statin detection. Ammonium or amine-based salts should continue to be examined including the use of ionic liquid matrices (ILM) including ammonium-CHCA salts. These solutions have recently been found to be useful for small molecule detection due to the smaller crystal size and reduced lipid fragmentation (Abdelhamid, 2018). The inclusion of salt into the CHCA matrix solution rather than washing the slide with the salt prior to matrix deposition could also improve the quality of lipid images.
It is important to note that regardless of the matrix used, the high laser power (90) and pulsed extract (1,500 Da) were required due to the difficulty in consistently ionizing analytes within a thick longitudinal section of tissue. Analyte ionization is heavily dependent on the tissue, section thickness, the matrix of choice, and the instrument being used. Therefore, matrix optimization should be performed with each new tissue type for analysis. This study serves as the first MALDI MSI optimization study for endogenous compounds within model invertebrates exposed to pharmaceuticals as environmental xenobiotics. Because of this as well as the fact that matrix optimization should be performed for each combination of tissue types and analytes of interest, this work should be considered an extension to the present literature regarding MSI of biological tissues featuring a novel application. These results are proposed as a starting point for those seeking to ionize lipids in heterogenous biological tissue using a positive polarity, linear MALDI TOF for MSI analysis and a painter’s airbrush sprayer for matrix application. The workflow outlined here will make MSI more accessible to those employing handheld sprayers and linear MALDI TOF instruments with imaging capabilities.
The raw data supporting the conclusion of this article will be made available by the authors, without undue reservation.
The manuscript presents research on animals that do not require ethical approval for their study.
KS: Conceptualization, Data curation, Funding acquisition, Investigation, Methodology, Visualization, Writing–original draft. CK: Investigation, Writing–original draft. LP: Investigation, Writing–review and editing. GB: Investigation, Writing–review and editing. AC: Investigation, Writing–review and editing. KT: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Writing–original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was partly supported by funding to KS from the National Science Foundation under Grant No. 2130471. Funding was also provided by Southern Illinois University Edwardsville through the Research Grants for Graduate Students, Competitive Graduate Awards, and Undergraduate Research and Creative Activities programs.
The authors acknowledge M. Nazim Boutaghou at Shimadzu Scientific Instruments, Matthew Openshaw, and Tom Abban at Kratos Analytical, as well as the rest of the Shimadzu family for their continued support.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdelhamid, H. N. (2018). Ionic liquid-assisted laser desorption/ionization–mass spectrometry: matrices, microextraction, and separation. Methods Protoc. 1 (2), 23. doi:10.3390/mps1020023
Angerer, T. B., Bour, J., Biagi, J. L., Moskovets, E., and Frache, G. (2022). Evaluation of 6 MALDI-matrices for 10 μm lipid imaging and on-tissue MSn with AP-MALDI-orbitrap. J. Am. Soc. Mass Spectrom. 33 (5), 760–771. doi:10.1021/jasms.1c00327
Brown, T., Clipston, N. L., Simjee, N., Luftmann, H., Hungerbühler, H., and Drewello, T. (2001). Matrix-assisted laser desorption/ionization of amphiphilic fullerene derivatives. Int. J. Mass Spectrom. 210–211, 249–263. doi:10.1016/S1387-3806(01)00429-8
Calvano, C. D., Monopoli, A., Cataldi, T. R. I., and Palmisano, F. (2018). MALDI matrices for low molecular weight compounds: an endless story? Anal. Bioanal. Chem. 410 (17), 4015–4038. doi:10.1007/s00216-018-1014-x
Ciğerci, İ. H., Ali, M. M., Kaygısız, Ş. Y., and Liman, R. (2016). Genotoxicity assessment of cobalt chloride in Eisenia hortensis earthworms coelomocytes by comet assay and micronucleus test. Chemosphere 144, 754–757. doi:10.1016/j.chemosphere.2015.09.053
Conley, J. M., Symes, S. J., Schorr, M. S., and Richards, S. M. (2008). Spatial and temporal analysis of pharmaceutical concentrations in the upper Tennessee River basin. Chemosphere 73 (8), 1178–1187. doi:10.1016/j.chemosphere.2008.07.062
Fründ, H.-C., Butt, K., Capowiez, Y., Eisenhauer, N., Emmerling, C., Ernst, G., et al. (2010). Using earthworms as model organisms in the laboratory: recommendations for experimental implementations. Pedobiologia 53, 119–125. doi:10.1016/j.pedobi.2009.07.002
Hillenkamp, F., Karas, M., Beavis, R. C., and Chait, B. T. (1991). Matrix-assisted laser desorption/ionization mass spectrometry of biopolymers. Anal. Chem. 63 (24), 1193A–1203A. doi:10.1021/ac00024a716
Høiem, T. S., Andersen, M. K., Martin-Lorenzo, M., Longuespée, R., Claes, B. S., Nordborg, A., et al. (2022). An optimized MALDI MSI protocol for spatial detection of tryptic peptides in fresh frozen prostate tissue. Proteomics 22 (10), 2100223. doi:10.1002/pmic.202100223
Itonori, S., Hashimoto, K., Nakagawa, M., Harada, M., Suzuki, T., Kojima, H., et al. (2018). Structural analysis of neutral glycosphingolipids from the silkworm Bombyx mori and the difference in ceramide composition between larvae and pupae. J. Biochem. 163 (3), 201–214. doi:10.1093/jb/mvx072
Jirásko, R., Holčapek, M., Kuneš, M., and Svatoš, A. (2014). Distribution study of atorvastatin and its metabolites in rat tissues using combined information from UHPLC/MS and MALDI-Orbitrap-MS imaging. Anal. Bioanal. Chem. 406 (19), 4601–4610. doi:10.1007/s00216-014-7880-y
Leopold, J., Popkova, Y., Engel, K., and Schiller, J. (2018). Recent developments of useful MALDI matrices for the mass spectrometric characterization of lipids. Biomolecules 8 (4), 173. doi:10.3390/biom8040173
Leopold, J., Prabutzki, P., Engel, K. M., and Schiller, J. (2023). A five-year update on matrix compounds for MALDI-MS analysis of lipids. Biomolecules 13 (3), 546. doi:10.3390/biom13030546
Li, S., Zhang, Y., Liu, J., Han, J., Guan, M., Yang, H., et al. (2016). Electrospray deposition device used to precisely control the matrix crystal to improve the performance of MALDI MSI. Sci. Rep. 6 (1), 37903. doi:10.1038/srep37903
Lou, X., de Waal, B. F. M., Milroy, L., and van Dongen, J. L. J. (2015). A sample preparation method for recovering suppressed analyte ions in MALDI TOF MS. J. Mass Spectrom. 50 (5), 766–770. doi:10.1002/jms.3587
Matyori, A., Brown, C. P., Ali, A., and Sherbeny, F. (2023). Statins utilization trends and expenditures in the U.S. before and after the implementation of the 2013 ACC/AHA guidelines. Saudi Pharm. J. SPJ 31 (6), 795–800. doi:10.1016/j.jsps.2023.04.002
McAvey, K. M., Guan, B., Fortier, C. A., Tarr, M. A., and Cole, R. B. (2011). Laser-induced oxidation of cholesterol observed during MALDI-TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 22 (4), 659–669. doi:10.1007/s13361-011-0074-3
Miao, X.-S., and Metcalfe, C. D. (2003). Determination of cholesterol-lowering statin drugs in aqueous samples using liquid chromatography–electrospray ionization tandem mass spectrometry. J. Chromatogr. A 998 (1), 133–141. doi:10.1016/S0021-9673(03)00645-9
Ottmar, K. J., Colosi, L. M., and Smith, J. A. (2012). Fate and transport of atorvastatin and simvastatin drugs during conventional wastewater treatment. Chemosphere 88 (10), 1184–1189. doi:10.1016/j.chemosphere.2012.03.066
Patil, A. A., Descanzo, M. J. N., Dhisale, V. B., and Peng, W. P. (2024). MALDI sample preparation methods: a mini review. Int. J. Mass Spectrom. 498, 117219. doi:10.1016/j.ijms.2024.117219
Pereira, A. M. P. T., Silva, L. J., Lino, C. M., Meisel, L. M., and Pena, A. (2016). Assessing environmental risk of pharmaceuticals in Portugal: an approach for the selection of the Portuguese monitoring stations in line with Directive 2013/39/EU. Chemosphere 144, 2507–2515. doi:10.1016/j.chemosphere.2015.10.100
Pino, M. R., Val, J., Mainar, A. M., Zuriaga, E., Español, C., and Langa, E. (2015). Acute toxicological effects on the earthworm Eisenia fetida of 18 common pharmaceuticals in artificial soil. Sci. Total Environ. 518–519, 225–237. doi:10.1016/j.scitotenv.2015.02.080
Selby, K. G., Hubecky, E. M., Zerda-Pinto, V., Korte, C. E., Bressendorff, G. A., and Tucker, K. R. (2024). Mass spectrometry imaging for environmental sciences: a review of current and future applications. Trends Environ. Anal. Chem. 42, e00232. doi:10.1016/j.teac.2024.e00232
Strupat, K., Karas, M., and Hillenkamp, F. (1991). 2,5-Dihydroxybenzoic acid: a new matrix for laser desorption—ionization mass spectrometry. Int. J. Mass Spectrom. Ion Process. 111, 89–102. doi:10.1016/0168-1176(91)85050-V
Ternes, T. A., Joss, A., and Siegrist, H. (2004). Peer reviewed: scrutinizing pharmaceuticals and personal care products in wastewater treatment. Environ. Sci. Technol. 38 (20), 392A–399A. doi:10.1021/es040639t
Trim, P. J., and Snel, M. F. (2016). Small molecule MALDI MS imaging: current technologies and future challenges. Methods 104, 127–141. doi:10.1016/j.ymeth.2016.01.011
Ulmer, L., Mattay, J., Torres-Garcia, H. G., and Luftmann, H. (2000). Letter: the use of 2-[(2E)-3-(4-tert-butylphenyl)-2-methylprop-2-Enylidene]Malononitrile as a matrix for matrix-assisted laser desorption/ionization mass spectrometry. Eur. J. Mass Spectrom. 6 (1), 49–52. doi:10.1255/ejms.329
Van-Gestel, C. A. M., Dirven-van Breemen, E. M., and Baerselman, R. (1992). Influence of environmental conditions on the growth and reproduction of the earthworm Eisenia andrei in an artificial soil substrate. Pedobiol. (Jena) 36 (2), 109–120. doi:10.1016/s0031-4056(24)00779-0
Wang, H.-Y. J., Liu, C. B., and Wu, H.-W. (2011). A simple desalting method for direct MALDI mass spectrometry profiling of tissue lipids. J. Lipid Res. 52 (4), 840–849. doi:10.1194/jlr.D013060
Yin, W., Al-Wabli, R. I., Attwa, M. W., Rahman, A. F. M. M., and Kadi, A. A. (2022). Detection and characterization of simvastatin and its metabolites in rat tissues and biological fluids using MALDI high resolution mass spectrometry approach. Sci. Rep. 12 (1), 4757. doi:10.1038/s41598-022-08804-x
Yu, Y., Huang, Q., Wang, Z., Zhang, K., Tang, C., Cui, J., et al. (2011). Occurrence and behavior of pharmaceuticals, steroid hormones, and endocrine-disrupting personal care products in wastewater and the recipient river water of the Pearl River Delta, South China. J. Environ. Monit. 13 (4), 871–878. doi:10.1039/C0EM00602E
Zhang, Y., Qin, L., Sun, J., Chen, L., Jia, L., Zhao, J., et al. (2020). Metabolite changes associated with earthworms (Eisenia fetida) graphene exposure revealed by matrix-assisted laser desorption/ionization mass spectrometry imaging. Ecotoxicol. Environ. Saf. 205, 111102. doi:10.1016/j.ecoenv.2020.111102
Zhu, Z., Shen, J., Wang, D., Chen, C., Xu, Y., Guo, H., et al. (2019). An auxiliary matrix for routine analysis of small molecules and biological macromolecules using matrix-assisted laser desorption ionization mass spectrometry. Anal. Bioanal. Chem. 411 (5), 1041–1052. doi:10.1007/s00216-018-1532-6
Keywords: esenia, statin, MALDI (matrix-assisted laser desorption/ionisation), mass spectrometry imaging (MSI), matrix, alphacyano- 4-hydroxcinnamic acid, lipid, cholesterol
Citation: Selby KG, Korte CE, Phan LH, Bressendorff GA, Chirchirillo AR and Tucker KR (2024) Method optimization for benchtop mass spectrometry imaging of lipids in Eisenia hortensis. Front. Environ. Chem. 5:1334207. doi: 10.3389/fenvc.2024.1334207
Received: 06 November 2023; Accepted: 30 May 2024;
Published: 14 June 2024.
Edited by:
Gary Fones, University of Portsmouth, United KingdomReviewed by:
Elizabeth Neumann, University of California, Davis, United StatesCopyright © 2024 Selby, Korte, Phan, Bressendorff, Chirchirillo and Tucker. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kevin R. Tucker, a2V2dHVja0BzaXVlLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.