- 1CAS Key Laboratory of Coastal Environmental Processes and Ecological Remediation, Shandong Research Centre for Coastal Environmental Engineering and Technology, Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 2Laboratory for Marine Biology and Biotechnology, Pilot National Laboratory for Marine Science and Technology, Qingdao, China
- 3School of Pharmacy, Yantai University, Yantai, China
- 4School of Pharmacy, Binzhou Medical University, Yantai, China
- 5Center for Ocean Mega-Science, Chinese Academy of Sciences, Qingdao, China
Quinolone antibiotics are a common class of antibiotics in the environment and have received considerable attention. In this study, three groups of mixed degradation strains targeting mixed quinolone antibiotics, norfloxacin (NOR), and enrofloxacin (ENR) were selected through screening, enrichment, and microbial diversity detection experiments. The strains screened in this study are divided into two categories through degradation efficiency experiments, community composition detection and functional enrichment analysis. In groups mix and ENR, the resistant bacteria are the main microorganisms and the degrading bacteria are the secondary ones, while in group NOR, the strains with degradation effects are the main ones, and the strains with resistance effects are the secondary ones. What’s more, that carbon sources have little effect on the community composition of the quinolone antibiotic degrading and tolerant bacteria, the difference between groups is mainly controlled by the type of antibiotics. On this basis, we found the key to NOR degradation is the cleavage of carbon nitrogen bonds on the piperazine ring, followed by oxygenation and deethylation. Preliminary studies have confirmed that the optimal degradation conditions for NOR degrading strains, and also found that environmental factors did not significantly affect the degradation efficiency of the Mix and NOR degrading strains, which indicating that the mixed bacteria can degrade NOR in different real environments effectively such as tap water, seawater, river water, and lake water. This manuscript is the first report on a mixed strain of quinolone antibiotic microbial degradation, and it is also the study with the highest NOR degradation efficiency among known reports. It has great research value for the co-metabolism and biodegradation of quinolone antibiotics in the environment.
1 Introduction
Quinolone antibiotics are a common class of antibiotics discovered in the early 1960s, which contain a bicyclic core structure related to compound 4-quinolone. It has broad-spectrum activity against both Gram positive and Gram negative bacteria. Its function is to inhibit bacterial nucleic acid and cause damage to its chromosomes (Han et al., 2021; Jiwanti et al., 2022). Since 1980, antibiotics have been used in the aquaculture industry, and quinolones (QNs) are also one of the most commonly used antibiotics in aquaculture. Due to the low bioavailability, high polarity, and strong adsorption capacity of QNs, they have been widely detected in freshwater and seawater environments, and have caused cardiac developmental toxicity, neurotoxicity, immunotoxicity, neurotoxicity and other toxic effects to aquatic organisms such as zebrafish embryos (Danio rerio) (Xiao et al., 2018; Han et al., 2021), Chinese shrimp (Fenneropenaeus chinensis) (Zhang et al., 2011), peacock fish (Poecilia reticulata), which seriously threaten the development of aquaculture and human health, becoming one of the most important emerging pollutants in the world (Kovalakova et al., 2020).
Currently, there are multiple methods and pathways for studying the degradation of antibiotic pollutants in the environment, such as physical adsorption (activated carbon (Fu et al., 2017), Biochar-Chitosan Composite Microspheres (Liu et al., 2018)), chemical degradation (microbial fuel cell (MFC) (Li et al., 2017; Xu et al., 2021), Metal-organic polyhedron catalyzed photodegradation (Huang et al., 2022), UV and persulfate (Huang et al., 2022) and so on), microbial degradation (Kim et al., 2011; Wang and Wang, 2018) and other types of methods to remove residual antibiotics from the environment. Compared with other methods, microbial decomposition method has lower cost, higher decomposition efficiency, and less environmental pollution. It is considered one of the main methods for removing QNs in the natural environment and wastewater treatment at this stage.
Microbial degradation refers to the degradation of QNs antibiotics into various other degradation products under the action of microbial degrading bacteria. It is known that Lactobacillus reuteri WQ-Y1 can degrade fluoroquinolone antibiotics with a piperazine ring structure, with the highest degradation rate of 65.1% in 4 mg/mL ciprofloxacin (CIP) medium. Additionally, 1 mg/mL of glucose in Minimal Salt Medium (MSM) can enhance the degradation effect of CIP (Qu and Xiao, 2021). In addition, Labrys portucalensis F11 and Rhodococcus sp. FP1 have also been identified to have the ability to degrade ciprofloxacin and OFLX (Amorim et al., 2014; Maia et al., 2018). Some fungi can also degrade CIP through n acetylation, deethylene n acetylation, and/or n formylation reactions under the catalysis of functional enzymes (Parshikov et al., 2001). Most studies on the biodegradation of QNs focus on individual bacteria in pure cultures, while there are few reports on the degradation of QNs by mixed strains. Some studies have also shown that multiple strains typically have higher antibiotic degradation efficiency compared to a single strain, making mixed bacteria an effective method for removing antibiotics from the environment (Kassotaki et al., 2016).
In this study, the degradation efficiency of 20 mg/L norfloxacin (NOR) by composite microbial communities such as Acinetobacter bereziniae, Aeromonas caviae, Comamonas testosteroni, Pseudomonas putida, etc. reached 96.76% through screening, enrichment, and microbial diversity detection experiments. On this basis, we explored the molecular mechanism of QNs degradation by mixed bacteria, revealed the resistance mechanism related to protein outer membrane protein and peroxidase overexpression, and screened the key proteins for QNs degradation. Preliminary studies have confirmed that salinity has a small impact on the degradation efficiency of mixed bacterial communities, indicating that the mixed bacteria can effectively degrade QNs in different real environments such as tap water, seawater, river water, and lake water. This manuscript is currently known to have the highest NOR degradation efficiency in the report. The study of the biodegradation and degradation mechanism of QNs in the environment has important reference value.
2 Materials and methods
2.1 Chemicals and media
Quinolone antibiotics were purchased from J&K Chemical Inc. (Beijing, China), including Ofloxacin (OFLX), norfloxacin (NOR), ciprofloxacin (CIP), enrofloxacin (ENR). Acetonitrile (high performance liquid chromatography (HPLC) - grade) was purchased from CNW Technologies (Duesseldorf, Germany). Other chemical reagents were purchased from Sinopharm Chemical Reagent Co., Ltd. (Beijing, China).
The minimal medium (MM medium) had all the essential nutrients for bacteria except carbon, and QNs-degrading bacteria were enriched in the MM medium adding 20 mg/L QNs (OFLX:NOR:CIP:ENR = 1:1:1:1), 20 mg/L OFLX, 20 mg/L CIP, 20 mg/L NOR or 20 mg/L ENR as the only carbon namely the enrichment medium (Mix-MM, OFLX-MM, CIP-MM, NOR-MM, ENR-MM medium). The enriched QNs-degrading bacteria were isolated on the isolation medium which was the solid medium of the enrichment medium by adding 1.5% wt/vol agar powder. All bacteria were recovered and cultured in the growth medium (2216E seawater medium) which had adequate nutrients for bacteria (Wang et al., 2020). All of the above mediums and buffer solutions were autoclave sterilized. The formulation of all mediums is described in Supplementary Table S1.
2.2 Enrichment of QNs-degrading bacteria
In this study, we selected 20 mg/L Mix-MM, OFLX-MM, CIP-MM, NOR-MM, ENR-MM medium as the only carbon source to screen QNs degrading bacteria in the environment and we collected sediment from the mouth of the Guangdang River in Yantai City, Shandong Province as a source for screening degradation strains of QNs. Firstly, 5 ± 0.5 g of sediment were taken and placed in 200 mL of the Mix-MM, OFLX-MM, CIP-MM, NOR-MM, ENR-MM medium. The liquid culture medium containing sediment was placed in an air bath constant temperature oscillator and shaken for 1 week at 30 °C and 180 rpm. To ensure that the screened mixed strains have QNs degradation and resistance characteristics, the he mixed culture medium was divided into two parts for subsequent experiments: (1) accurately suck 1.5 mL of mixed culture solution using a pipette and place it in a 1.5 mL centrifuge tube. Then place it in a low speed centrifuge at 10,000 rpm for 2 min. Discard the sediment and filtered with 0.22 μm pore size polyethersulfone membrane (Jinteng experimental equipment Co., Ltd., Tianjin, China), the filtered filtrate is placed in a liquid phase injection bottle and detected for QNs by high-performance liquid chromatography- Ultraviolet Spectroscopy (HPLC-UV). (2) when a significant decrease in the concentration of QNs was detected in experimental step (1), 200 mL of the Mix-MM, OFLX-MM, CIP-MM, NOR-MM, ENR-MM medium was recalibrated, and 10 mL of the above mixed culture medium was accurately aspirated using a pipette and added to a new culture medium. This experimental step was repeated 5 times. When no changes in the concentration of QNs were detected in step (1) of the experiment, a new sampling site was selected and sediment samples were taken again, repeating the above experimental process.
In this experiment, HPLC-UV was used to detect the concentration of QNs. The experimental conditions were: column temperature 35 °C, mobile phase 0.4% acetic acid solution: acetonitrile = 80:20, and injection volume 10 μ L. The injection time is 6 min, the flow rate is 1 mL/min, and the detection wavelength is 294 nm. The degradation efficiency of QNs is calculated according to formula 1.
R is defined as the degradation rate (%) of QNs,
2.3 Isolation of mixed QNs-degrading bacteria
The selected mixed QNs-degrading bacteria (in Mix-MM, NOR-MM, ENR-MM) was has been proven to have significant degradation effects. 1mL mixed Mix, NOR, ENR-degrading bacteria were incubated in 50 mL Mix-MM, NOR-MM, ENR-MM medium and Mix-lysogeny broth (LB), NOR-LB, ENR-LB medium as the experiment group at 30°C, 160 rpm for 3 days to simulate the actual environment with sufficient and insufficient carbon sources, which were randomly divided into six groups: Mix-MM group, NOR-MM group, ENR-MM group, Mix-LB group, NOR-LB group, ENR-LB group. 1mL bacterial suspension was collected after 7 days, and then detected by HPLC-UV after filtering using 0.22 μm polyethersulfone membrane. Meanwhile, the bacterial cells of the experiment group were collected after 7 days by centrifugation at 10,000 rpm for 2 min, and DNA was extracted using Bacterial DNA Kit D3350 (Omega, Shanghai, China). Then the composition of microbial community diversity was analyzed by the Illumina MiSeq platform of Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China) for sequencing according to standard protocols.
The PE reads obtained from Illumina sequencing are first concatenated based on the overlap relationship, while quality control and filtering are performed on the sequence quality. After distinguishing the samples, operational taxonomic units (OTU) clustering analysis and species classification analysis are performed. Based on OTU, multiple diversity index analysis can be performed. Based on the results of OTU clustering analysis, multiple diversity index analysis can be performed on OTU, as well as detection of sequencing depth; Based on taxonomic information, statistical analysis of community structure can be conducted at various classification levels.
Raw fastq files were quality filtered by Trimmomatic and merged by Flash version 1.2.7. To detect the species richness in each sample, the effective tags in all samples is classified as operational taxonomic units (OTUs) based on 97% sequence similarity using UPARSE (version 7.0.1090 http://drive5.com/uparse/). The 16s rRNA gene sequence was classified and annotated by Silva database (Release138 http://www.arb-silva.de). Alpha-diversity (α) was assessed using the species richness indices (ace, chao and sobs) and diversity indices (shannon) by Mothur version 1.30.2 (https://www.mothur.org/wiki/Download_mothur). The KEGG and COG function prediction of 16s sequence is completed by PICRUSt version 1.1.0.
2.4 Biodegradation experiments
2.4.1 Growth curve of mixed QNs degrading bacteria
The mixed strains of Mix, NOR, and ENR degradation selected in 2.2 were resuspended in LB medium at 30°C and shaken at 180 r/min for 8–12 h to achieve a bacterial concentration of 1 OD. The activated bacterial solution was added to 200 mL of Mix-LB, NOR-LB, and ENR-LB medium, with an OD value of 0.05. The experimental conical flask was placed in a 30°C constant temperature incubator and shaken at 180 rpm for 24 h. Take 1 mL of LB every 2 h for cultivation. Based on measuring the UV absorbance at a wavelength of 600 nm using a UV visible spectrophotometer, plot the OD-t growth curve of the mixed strain in LB culture medium.
2.4.2 Detection of NOR degradation products
The mixed strains of NOR degradation selected in 2.2 were resuspended and added to 200 mL of NOR-MM medium, with an OD value of 0.05. The experimental conical flask was placed in a 30°C constant temperature incubator and shaken at 180 rpm for 7 days. Then samples were collected to detect the NOR degradation products of mixed bacteria by HPLC-ESI-MS/MS.
2.5 Effects of initial concentration, pH, and salinity on QNs degradation
QNs initial concentration, pH, and salinity were selected to investigate their effects on QNs biodegradation by mixed bacteria. Mix, NOR, and ENR initial concentrations were set to 1, 5, 10, 20, 50 mg/L. The pH values were set to 5, 6, 7, 8, 9 (shown in Supplementary Table S2), the seawater proportions were set to 0%, 20%, 40%, 60%, 80%, 100% with PBS buffer. Both QNs initial concentrations were 20 mg/L, the inoculum amount of the bacteria cell was 108 CFU/mL, and bacterial suspensions were detected after 7 days.
2.6 Data analysis
All data presented were the mean values of three independent sets of experiments. Each value was expressed as means ± standard deviation (S.D.). All data were tested for normality (Kolmogorov-Smirnov) and homogeneity of variance (Levene). Statistical analysis was carried out by one-way ANOVA using the Duncan’s test to evaluate whether the means were significantly different (p < 0.05). The statistical software SPSS 19.0 was used for all statistical calculations.
3 Results and discussion
3.1 Screening of QNs degrading bacteria
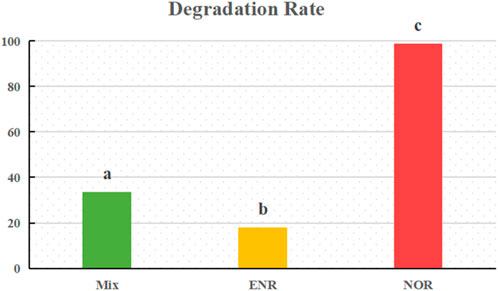
In this study, the mixed bacteria which could degrade ENR, NOR efficiently and possess OFLX, CIP resistance simultaneously was enriched after repeated screening. The degradation rate of NOR single antibiotic is the highest, reaching 98.66% (Figure 1). The highest degradation rate of norfloxacin by the degrading strain in this experiment at 20 mg/L was better than the currently known degradation rate of 34%–92.6% (Fu et al., 2017; Gu, 2023), indicating that the selected NOR degrading bacteria in this study have good application prospects. In repeated experiments, no significant decrease in antibiotics was observed in the experimental groups OFLX-MM and CIP-MM, confirming that no mixed strains capable of significantly degrading single antibiotics OFLX and CIP were screened in this batch of environmental samples. However, due to the high degradation efficiency of NOR, we choose to continue exploring the degradation mechanisms of NOR and ENR groups with degradation efficiency. Although the degradation strain of OFLX, CIP was not selected in this experiment, it was confirmed through the Mix-MM group experiment that the mixed strain has both OFLX, CIP resistance. Similar experimental results were observed in Dae Wi Kim’s research, Microbacterium sp. strain 4N2-2 did not grow on norfloxacin as a sole carbon source, but when adding other nutrients, it produced four possible metabolites from norfloxacin. Meanwhile, it also confirms that in this study some bacteria may degrade NOR and ENR to metabolites with less antibacterial activity that could be subject to further degradation by other microorganisms, in combination with these organic substances, the strain can exhibit degradation characteristics or resistance to OFLX and CIP (Kim et al., 2011).
3.2 Identification of QNs degrading bacteria
The raw data sets are available at the NCBI SRA with the accession number: SUB14112108. Complete diversity data analysis of 6 samples and obtain an optimized sequence of 450830193001211 bases, with an average sequence length of 428bp. The detailed information on each sample was shown in Supplrementary Material. The top 5 species at the phylum level include Proteobacteria, Bacteroidota, Actinobacteriota, and unclassified Bacteria. The top 5 species at the genus level include Klebsiella, Comamonas, Stenotrophomonas, Aeromonas, Dysgonomonas.
Klebsiella pneumoniae (K. Pneumoniae) is an important multidrug-resistant (MDR) pathogen affecting humans and a major source for hospital infections associated with high morbidity and mortality due to limited treatment options (Navon-Venezia et al., 2017) and has been extensively tested for its resistance to QNs (Martinez-Martinez et al., 2002; Paulraj et al., 2019). The QNs target bacterial topoisomerases to inhibit DNA replication. The mechanism of resistance of K. Pneumoniae to fluoroquinolones mainly includes mutations in the aminoglycoside acetyltransferase gene AAC (6)-Ib, production of MDR efflux pumps, and plasmid carrying resistance gene qnr (Redgrave et al., 2014). K. pneumoniae strains producing extended-spectrum β-lactamases (ESBL) and carbapenemases are more frequently resistant to fluoroquinolones than K. Pneumoniae strains lacking these enzymes (Martinez-Martinez et al., 2002; Wei et al., 2015), and multiple vitamins have an inhibitory effect on this resistance (Paulraj et al., 2019). Mechanisms of resistance were determined in 33 quinolone-resistant isolates of the species Aeromonas hydrophila, Aeromonas caviae, Aeromonas media, Aeromonas salmonicida, Aeromonas popoffii and Aeromonas veronii, recovered from humans, freshwater and eels by Elena Alcaide and Jordi Vila (Vila et al., 2002; Alcaide et al., 2010), and the mechanism of QNs resistance in the Aeromonas isolates studied was related to mutations in quinolone resistance-determining regions (QRDRs) of gyrA and parC genes.
In addition, it was also possible that antibiotics degradation by the mixed bacteria related to bacterial co-metabolism. Co-metabolism was an important mechanism of antibiotics biodegradation in the environment. The dominant strains detected in this study, Comamonas, Stenotrophomonas, and Dysgonomonas, have not yet been reported to be able to degrade QNs. However, the question of whether intermediate products in the degradation process of QNs can be utilized and whether they have resistance to QNs is still worth further research.
3.2.1 Species composition analysis
Through comparative analysis of the number of common and unique species (such as OTU) in 6 samples, it was found that 5 species coexist, mainly including Klebsiella pneumoniae, Stenotrophomonas acidaminiphila, Alcaligenes faecalis, Delftia tsuruhatensis, and Aeromonas Caviae (Figure 2A). In addition, the main degradation strains of NOR include: Methylorubrum extroquens, Leucobacter sp., Stenotrophomonas sp., Brevundimonas sp., Shinella sp., Castellaniella sp., Parapusillimonas sp., Phenylobacterium sp. and Flavobacterium. Among them, the community abundance of Leucobacter sp. account for 48.91% of the unique degradation bacteria of NOR-MM group, significantly higher than Shinella sp., which accounts for 14.13% (Figure 2B). Combined with the experimental results of shared strains, Shinella sp. and Comamonas testosteroni are the dominant strains in NOR-MM group.
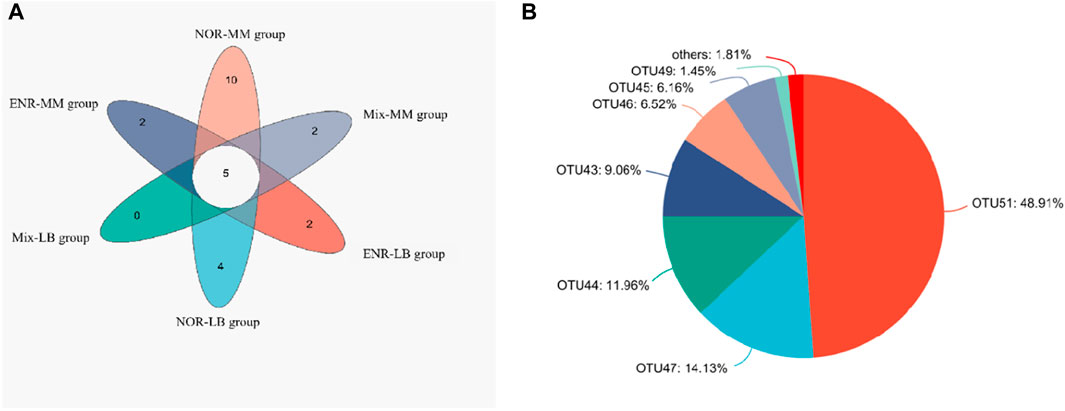
Figure 2. (A) Venn diagram of the comparative analysis of the common and unique species in 6 samples. (B) Pie diagram of ENR-MM group community composition.
By analyzing the community composition of 6 samples, the research results of the community barplot and community hotmap showed that Klebsiella pneumoniae was the main degrading bacteria of Mix-MM group, ENR-MM group, Mix-LB group and ENR-LB group. Comamonas was the main degrading bacteria of NOR-MM group and NOR-LB group. In addition, Comamonas also holds an important position in the ENR-MM group and Mix-MM group. In the inter group comparative analysis of species diversity, Beta diversity analysis also confirmed the results, with a stronger correlation between NOR-LB group and NOR-MM group, while the similarity in community composition between Mix-MM group, ENR-MM group, Mix-LB group, and ENR-LB group was stronger.
However, it has not yet been seen to have a degradation effect or resistance to antibiotics. Although there is no literature confirming the biodegradability of QNs in Leucobacter sp. currently, Actinobacteria, such as Corynebacterium and Leukobacterium, are often the main carriers of sulfonamide resistance (SR) genes (sul1 and intI1) (Lin et al., 2017; Xia et al., 2019).In previous studies, Comamonas testosteroni has been confirmed to be able to degrade organic substances such as hexachlorobenzene (Dimova et al., 2022), phenanthrene, naphthalene (Goyal and Zylstra, 1997), and 4-chlorophenol (Bae et al., 1997; Wang et al., 2020). What’s more, it can be found in Figure 3 that carbon sources have little effect on the community composition of the QNs degrading and tolerant bacteria, the difference between groups is mainly controlled by the type of antibiotics. Based on the results of degradation efficiency experiments, it has been confirmed that the strains screened in this study are divided into two categories. In groups Mix and ENR, the resistant bacteria are the main microorganisms and the degrading bacteria are the secondary ones, while in group NOR, the strains with degradation effects are the main ones, and the strains with resistance effects are the secondary ones.
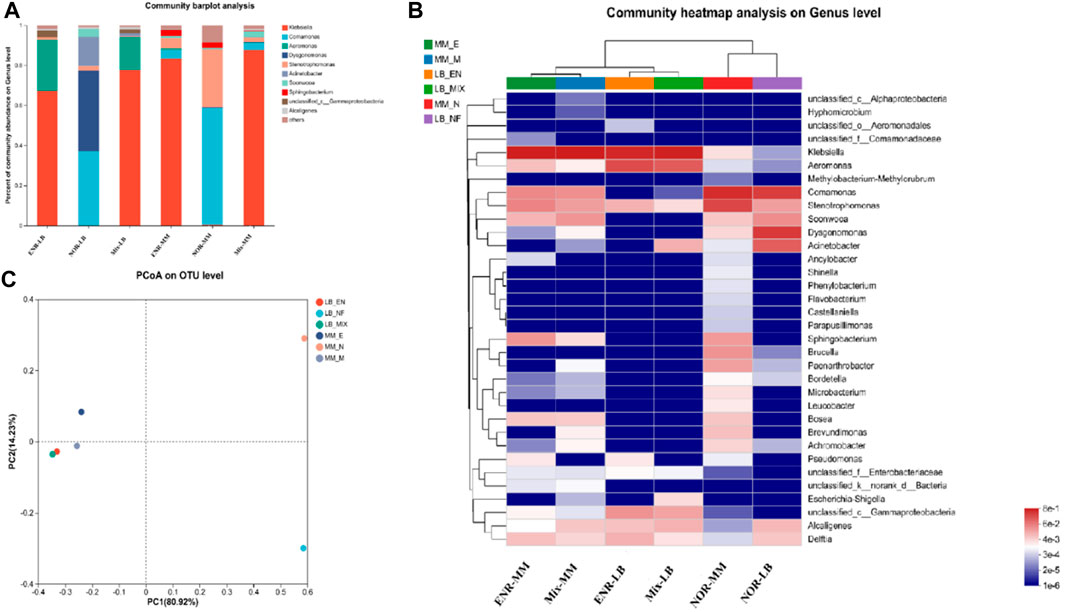
Figure 3. (A) The community barplot analysis. (B) The community heatmap analysis. (C) Sample Beta Diversity Analysis.
3.2.2 Functional prediction analysis
In this study, we obtained the COG family information and KEGG Ortholog (KO) information corresponding to each OTU through the greengene id corresponding to each OUT. And calculate the abundance of each COG and the abundance of KO. Based on the information in the COG database (Figure 4A), the description information and functional information of each COG can be parsed from the eggNOG database to obtain a functional abundance spectrum. We observed that the most frequently represented functional clusters in our transcriptome on the COG database were “Amino acid transport and metabolism”, “Inorganic ion transport and metabolism.” Through clustering analysis of the COG function in the NOR-MM group (Figure 4B), it was found that there was a significant enrichment of functions such as “Amino acid transport and metabolism”, “Inorganic ion transport and metabolism,” “Energy production and conversion” and “Translation, ribosomal structure and biogenesis.” In the study of the functional annotation of GOG, it was found that the 6 groups of samples were mainly involved in functional processes such as “amino acid transport and metabolism” and “inorganic ion transport and metabolism,” supporting the previous conclusion that carbon source is not the main factor affecting the composition of degradation strain communities proposed by the author.
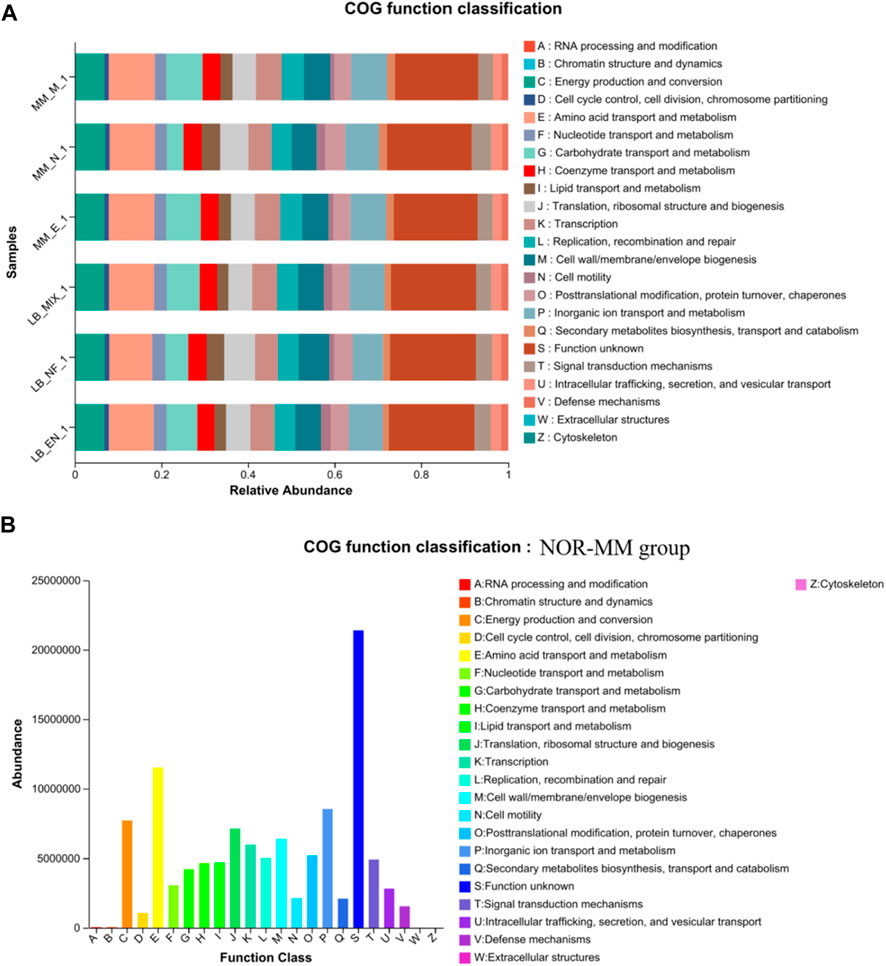
Figure 4. (A) The COG functional classification analysis in 6 samples. (B) The COG functional classification analysis in NOR-MM group.
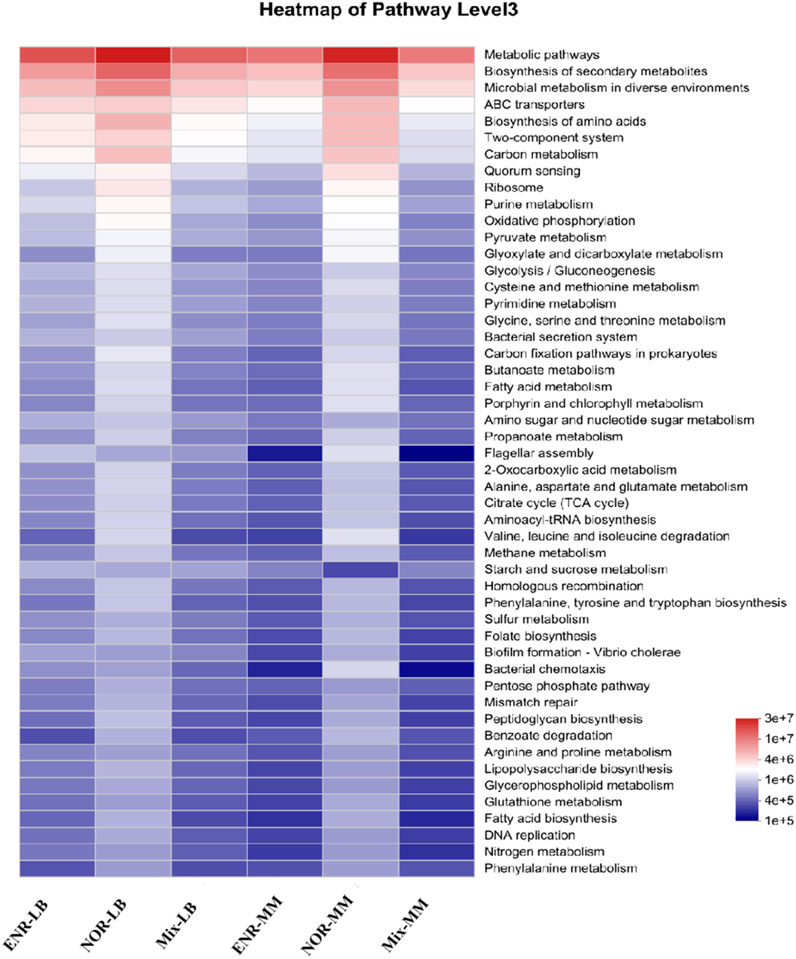
KEGG pathway analysis performed functional prediction analysis on 16S sequencing data from 6 groups of samples (Figure 5), and the results showed that the 6 groups of samples were significantly enriched in multiple pathways such as “metabolic paths,” “biosynthensis of secondary metabolites,” and “microbial metabolism” in diverse environments. In addition, the NOR-LB group and NOR-MM group also showed significant enrichment in signaling pathways such as “ABC transporters,” “Biosynthesis of amino acids,” “Two component system,” “Carbon metabolism,” and “Quorum sensing” and so on.
The 6 groups of samples were significantly enriched in various pathways such as “metabolic pathway,” “biosynthesis of secondary metabolites,” and “microbial metabolism” which confirmed that the selected degradation strains have the degradation effect of NQs, which can be degraded by metabolizing NQs or metabolic intermediate metabolites in the environment. At the same time, signal pathways such as “ABC transporters” and “Biosynthesis of amino acids” in the NOR-MM group were significantly enriched, confirming that NOR may increase drug resistance by passive diffusion through the cell membrane into the next metabolic process or enhancing the antioxidant system.
3.3 Growth curve of mixed QNs degrading bacteria
As shown in Figure 6A, the mixed strain showed a rapid growth effect after 4 h with sufficient carbon source. However, in the pre experiment, MM culture medium did not show a similar effect, confirms that the addition of carbon sources can promote the sustained and stable growth of mixed strains, but the impact on community composition is not significant. At 22 h, the growth of the Mix-LB group reached its peak for the first time, while the first peak of the ENR-LB group appeared at 24 h. The mixed bacteria in the NOR-LB group showed sustained and stable growth in the presence of NOR.
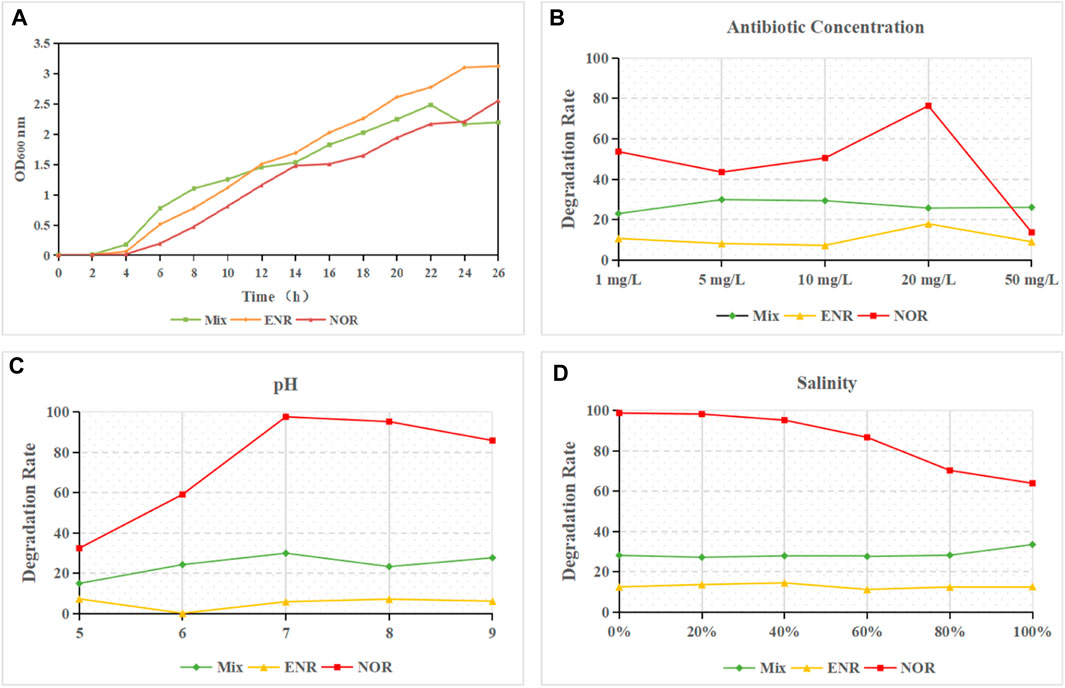
Figure 6. (A) Growth curve of mixed QNs degrading bacteria. (B) The Effect of initial concentration on the Degradation of QNs. (C) The Effect of pH on the Degradation of QNs. (D) The Effect of salinity on the Degradation of QNs.
3.4 Effects of initial concentration, pH, and salinity on QNs degradation
3.4.1 The effect of initial concentration on the degradation of QNs
In this experiment, concentration gradients of QNs (Mix, NOR, ENR) were set at 1, 5, 10, 20, and 50 mg/L, respectively. Samples before cultivation were taken as the control group and samples after 7 days of degradation were taken as the experimental group. The conversion rate of NQs was detected and calculated by HPLC-UV. The results are shown in Figure 6B. Firstly, the degradation rate of Mix-MM group by the mixed strains remained at 20%–30% under various concentration conditions, and there was no significant linear relationship between the degradation rate and the initial concentration. Secondly, the degradation rate of ENR-MM group remained at 5%–20% under various concentration conditions, and there was no significant linear relationship between the degradation rate and the initial concentration. And it is lower than the 80% degradation rate obtained by the strain Thermos sp.C419 (Pan et al., 2020), indicating that compared to other studies, the degradation rate of enrofloxacin by this mixed strain is not ideal. Finally, the degradation rate of NOR-MM group at a concentration of 20 mg/L reached 76.36%, approaching the 78% (14 days) degradation rate obtained from the Microbacterium sp. 4N2-2 (Kim et al., 2011) and the possible linear relationship needs conditioning in the future. Compared to the 20 mg/L group, the degradation efficiency of 50 mg/L was significantly reduced, indicating that the mixed strains had insufficient tolerance to ultra-high concentrations of NOR. Therefore, it indicates that excessive concentration (50 mg/L) of NOR still has an inhibitory effect on the growth of degrading strains. The 20 mg/L group showed a higher degradation effect, which indirectly confirmed the presence of NOR provided energy for the inventory of the strain. In the study on the degradation effect of antibiotics by Aeromonas caviae and mixed strains, it was found that mixed strains have more advantages in antibiotic degradation at high concentrations compared to single strains.
3.4.2 The effect of pH on the degradation of QNs
In this experiment, QNs (Mix, NOR, ENR, 20 mg/L) were added to a pH buffer, and three replicates were set up. After 7 days of cultivation at 30°C and 160 r/min, the conversion rate of NQs was detected and calculated by HPLC-UV. The results are shown in Figure 6C. Firstly, the degradation rate of Mix-MM group by mixed strains remained between 15% and 30% under various pH conditions, with low degradation rate and no significant linear relationship with pH. Secondly, the degradation rate of ENR-MM group remained below 10% under various pH conditions, with low degradation rate and no significant linear relationship with pH. Thirdly, under the condition of pH 7, the degradation rate of NOR-MM group can reach to the peak as high as 97.55%, and as the pH decreases, the NOR degradation efficiency significantly decreases. Overall, the degradation strains of Mix and ENR groups were not strongly affected by pH, while the mixed strains of NDR group were more significantly affected by environmental pH. Adamek et al. (2016) found that in rivers with severe antibiotic pollution, there is a significant positive correlation between the biological removal rate of antibiotics and pH, that is, the higher the pH, the higher the antibiotic removal rate. In the process of photodegradation, the degradation rate of antibiotics is negatively correlated with pH, meaning that the degradation rate of antibiotics is higher at low pH (Lester et al., 2012). Some studies have shown that pH can affect the degradation efficiency of antibiotics and other pollutants by affecting bacterial growth, reproduction, and enzyme activity in the bacterial (Wang and Wang, 2018), which explains that the degradation efficiency of the NOR experimental group with high degradation efficiency in this manuscript is greatly affected by pH in the environment.
3.4.3 The effect of salinity on the degradation of QNs
In this study, 20 mg/L QNs (Mix, NOR, ENR) were added to different seawater ratios of culture medium. Three replicates were set up and cultured under the same conditions. Samples were taken before and 7 days after cultivation, and the conversion rate of QNs was measured and calculated by HPLC-UV. The results are shown in Figure 6D. As shown in the figure, the degradation rate of Mix-MM group by the mixed strains remained around 30% under various salinity conditions, with low degradation rate and no significant linear relationship with salinity. The degradation rate of ENR-MM group remained around 10% under various salinity conditions, with low degradation rate and no significant linear relationship with salinity. The response of Mix-MM and ENR-MM group to salinity is not significant, which may be related to the widespread presence of monarch K. pneumoniae in the environment (Navon-Venezia et al., 2017). Therefore, the impact of environmental factors on its effectiveness is not significant.
The degradation efficiency of NOR-MM group decreases with increasing salinity. Under the condition of pure seawater with 100% salinity, the degradation rate is about 60%, while under the condition of zero, the degradation rate can reach as high as 98.66%. The degradation rate of NOR in seawater is significantly lower than that in freshwater environment, but it also reaches over 60%. This result confirms the NOR degrading mixed bacteria in the NOR-MM group can simultaneously degrade in multiple environmental media such as rivers, lakes, and oceans, and has strong application value.
3.5 Analysis of NOR degradation products
According to the mass charge ratio of each product fragment in the first order mass spectrometry (Figure 7A), it can be inferred that two products were obtained after the degradation of NOR, with m/z of 243.88 and 278.80, respectively. It is preliminarily speculated that the molecular formula of product 1 is C16H18FN3O4, product 2 is C14H16FN3O3. The co-metabolic mechanism of norfloxacin degradation by mixed bacteria suggests that its degradation starting point is the cleavage of carbon nitrogen bonds on the piperazine ring, followed by oxidation and deethylation reactions of the ring opening piperazine ring, and the structure of the product is confirmed (Figure 7B).
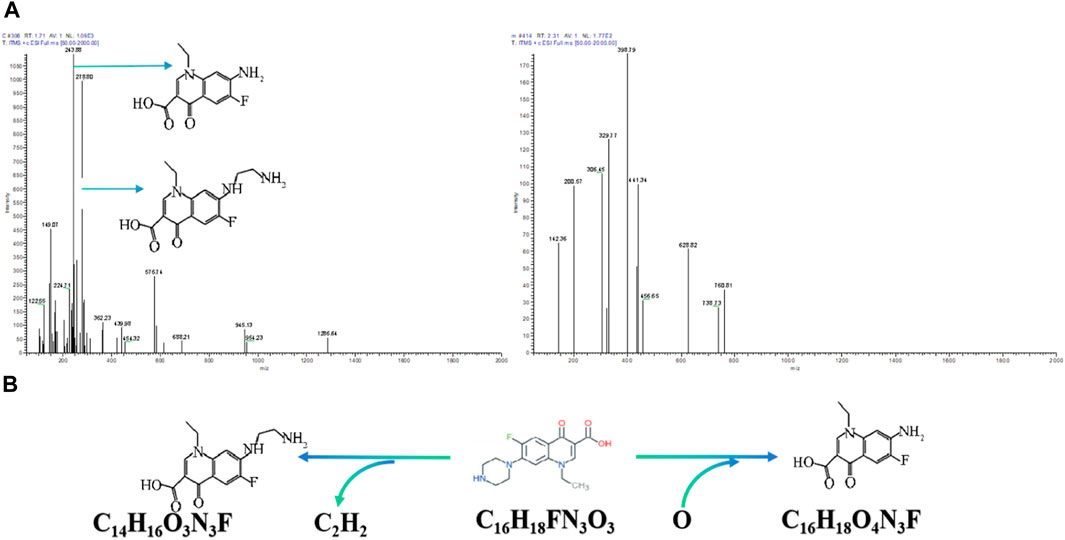
Figure 7. (A) The mass spectrogram of Norfloxacin. (B) The proposed Degradation pathway of Norfloxacin.
In the study of the degradation pathways of QNs in existing wastewater treated with ozone, more than 20 products were detected. Among them, the analysis results of NOR degradation products 1 and 9 are similar to those of our degradation products 2 and 1, which with the same molecular formula (Liu et al., 2012). The degradation of these species occurs by the attack of either molecular ozone or hydroxyl radicals on both the piperazinyl moieties. Transformations of the piperazinyl ring appear to occur via both the ring cleavage and the addition of oxygen atom to the ring. Loss of an ethylene group from the piperazine ring produces desethylene norfloxacin (product 2). This pathway and the formation of product 2 are in agreement with the results of Kim et al. (2011) who examined the modification of norfloxacin by a Microbacterium sp. strain isolated from a wastewater treatment plant. The formation of another direct product of NOR oxidation (product 1) appears to take place via the addition of an oxygen atom that produces the hydroxy-derivative of piperazine ring, indicating that the degradation pathway of NOR in this article is similar to it (Liu et al., 2012; Cai et al., 2022).
4 Conclusion
In this study, the mixed bacteria which could degrade ENR, NOR efficiently and possess OFLX, CIP resistance simultaneously was enriched after repeated screening. Through degradation efficiency experiments, community composition detection and functional enrichment analysis, it was found that, the strains screened in this study are divided into two categories. In groups Mix and ENR, the resistant bacteria are the main microorganisms and the degrading bacteria are the secondary ones, while in group NOR, the strains with degradation effects are the main ones, and the strains with resistance effects are the secondary ones. What’s more, that carbon sources have little effect on the community composition of QNs, the difference between groups is mainly controlled by the type of antibiotics. The ring opening oxidation and deethylation of piperazine ring are key factors in NOR degradation. What’s more, the optimal degradation conditions for NOR degrading strains which indicating that the mixed bacteria can degrade NOR in different real environments effectively such as tap water, seawater, river water, and lake water. It has great research value for the co-metabolism and biodegradation of QNs in the environment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI, accession number PRJNA1056513.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
HW: Conceptualization, Formal Analysis, Investigation, Methodology, Writing–original draft. QW: Investigation, Writing–review and editing. ML: Investigation, Writing–review and editing. ZS: Investigation, Writing–review and editing. JY: Conceptualization, Investigation, Writing–review and editing. XW: Investigation, Writing–review and editing. JL: Supervision, Writing–review and editing. LC: Funding acquisition, Supervision, Visualization, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the Natural Science Foundation of Shandong Province of China (ZR2020QC011, ZR2022QD058, ZR2020QB105, ZR2020KC032, ZR2021MD039), the Youth Innovation Promotion Association CAS (2021212), the National Nature Science Foundation of China (22176210, 22376216, 42276168), and the Taishan Scholars Program (ts20190962).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvc.2024.1326206/full#supplementary-material
Abbreviations
ENR, Enrofloxacin; HPLC, high performance liquid chromatography; K.Pneumoniae, Klebsiella pneumoniae; MDR, multidrug-resistant; MFC, microbial fuel cell; MM, minimal medium; MSM, Minimal Salt Medium; NOR, Norfloxacin; QNs, Quinolones; QRDRs, quinolone resistance-determining regions; OFLX, Ofloxacin; OTU, operational taxonomic units; SR, sulfonamide resistance.
References
Adamek, E., Baran, W., and Sobczak, A. (2016). Assessment of the biodegradability of selected sulfa drugs in two polluted rivers in Poland: effects of seasonal variations, accidental contamination, turbidity and salinity. J. Hazard. Mater. 313, 147–158. doi:10.1016/j.jhazmat.2016.03.064
Alcaide, E., Blasco, M. D., and Esteve, C. (2010). Mechanisms of quinolone resistance in Aeromonas species isolated from humans, water and eels. Res. Microbiol. 161, 40–45. doi:10.1016/j.resmic.2009.10.006
Amorim, C. L., Moreira, I. S., Maia, A. S., Tiritan, M. E., and Castro, P. M. L. (2014). Biodegradation of ofloxacin, norfloxacin, and ciprofloxacin as single and mixed substrates by Labrys portucalensis F11. Appl. Microbiol. Biotechnol. 98, 3181–3190. doi:10.1007/s00253-013-5333-8
Bae, H. S., Lee, J. M., Kim, Y. B., and Lee, S. T. (1997). Biodegradation of the mixtures of 4-chlorophenol and phenol by Comamonas testosteroni CPW301. Biodegradation 7, 463–469. doi:10.1007/bf00115293
Cai, H., Ma, Y., Li, J., Jin, Y., Zhu, P., and Chen, M. (2022). Norfloxacin degradation by persulfate activated with Cu2O@WO3 composites: efficiency, stability, mechanism, and degradation pathway. Industrial Eng. Chem. Res. 61 (30), 11237–11248. doi:10.1021/acs.iecr.1c04918
Dimova, M., Iutynska, G., Yamborko, N., Dordevi, D., and Kushkevych, I. (2022). Possible processes and mechanisms of hexachlorobenzene decomposition by the selected Comamonas testosteroni bacterial strains. Processes 10 (11), 2170. doi:10.3390/pr10112170
Fu, B. M., Chen, L. W., and Cai, T. M. (2017a). Isolation, screening, and degradation characteristics of norfloxacin degrading bacterium NOR-36. J. Environ. Sci. 37 (2), 576–584. doi:10.13671/j.hjkxxb.2016.0245
Fu, H., Li, X., Wang, J., Lin, P., Chen, C., Zhang, X., et al. (2017b). Activated carbon adsorption of quinolone antibiotics in water: performance, mechanism, and modeling. J. Environ. Sci. 56, 145–152. doi:10.1016/j.jes.2016.09.010
Goyal, A. K., and Zylstra, G. J. (1997). Genetics of naphthalene and phenanthrene degradation by Comamonas testosteroni. J. Industrial Microbiol. Biotechnol. 19, 401–407. doi:10.1038/sj.jim.2900476
Gu, C. Q. (2023). Research progress in microbial degradation of quinolone antibiotics. Shandong Chem. Ind. 52, 100–103. doi:10.19319/j.cnki.issn.1008-021x.2023.04.002
Han, Y., Ma, Y., Yao, S., Zhang, J., and Hu, C. (2021). In vivo and in silico evaluations of survival and cardiac developmental toxicity of quinolone antibiotics in zebrafish embryos (Danio rerio). Environ. Pollut. 277, 116779. doi:10.1016/j.envpol.2021.116779
Huang, H., Liu, Z. Y., Li, S. B., Zhu, J., Jiang, B. X., and Zhang, Y. T. (2022). Amino-functionalized Zr (IV) metal-organic polyhedron as water-stable catalyst for the photocatalytic degradation of tetracycline. J. Solid State Chem. 307, 122821. doi:10.1016/j.jssc.2021.122821
Jiwanti, P. K., Wardhana, B. Y., Sutanto, L. G., and Chanif, M. F. (2022). A review on carbon-based electrodes for electrochemical sensor of quinolone antibiotics. ChemistrySelect 7 (15), e202103997. doi:10.1002/slct.202103997
Kassotaki, E., Gianluigi, B., Laura, F. C., Rodriguez-Roda, I., and Pijuan, M. (2016). Enhanced sulfamethoxazole degradation through ammonia oxidizing bacteria co-metabolism and fate of transformation products. Water Res. A J. Int. Water Assoc. Water Res. 94 (1), 111–119. doi:10.1016/j.watres.2016.02.022
Kim, D. W., Heinze, T. M., Kim, B. S., Schnackenberg, L. K., Woodling, K. A., and Sutherland, J. B. (2011). Modification of norfloxacin by a Microbacterium sp. strain isolated from a wastewater treatment plant. Appl. Environ. Microbiol. 77, 6100–6108. doi:10.1128/aem.00545-11
Kovalakova, P., Cizmas, L., Mcdonald, T. J., Marsalek, B., Feng, M., and Sharma, V. K. (2020). Occurrence and toxicity of antibiotics in the aquatic environment: a review. Chemosphere 251, 126351. doi:10.1016/j.chemosphere.2020.126351
Lester, Y., Mamane, H., and Avisar, D. (2012). Enhanced removal of micropollutants from groundwater, using pH modification coupled with photolysis. Water air soil Pollut. 223 (4), 1639–1647. doi:10.1007/s11270-011-0971-x
Li, H., Yang, X., and Song, H. (2017). Study on antibiotics removal by microbial fuel cell coupled constructed wetland. Dongnan Daxue Xuebao (Ziran Kexue Ban)/Journal Southeast Univ. Nat. Sci. Ed. 47, 410–415. doi:10.3969/j.issn.1001-0505.2017.02.034
Lin, H., Zhang, J., Chen, H., Wang, J., Sun, W., Zhang, X., et al. (2017). Effect of temperature on sulfonamide antibiotics degradation, and on antibiotic resistance determinants and hosts in animal manures. Sci. Total Environ. 607–608 (31), 725–732. doi:10.1016/j.scitotenv.2017.07.057
Liu, B., Lang, Y., Zhu, C., and Zhao, H. (2018). Optimal preparation of biochar-chitosan composite Microspheres for highly efficient adsorption of norfloxacin. Res. Environ. Sci. 31, 1138–1143. doi:10.13198/j.issn.1001-6929.2018.03.22
Liu, C. V., Nanaboina, G. V. K., and Jiang, W. (2012). Spectroscopic study of degradation products of ciprofloxacin, norfloxacin and lomefloxacin formed in ozonated wastewater. Water Res. 46, 5235–5246. doi:10.1016/j.watres.2012.07.005
Maia, A. S., Tiritan, M. E., and Castro, P. M. L. (2018). Enantioselective degradation of ofloxacin and levofloxacin by the bacterial strains Labrys portucalensis F11 and Rhodococcus sp. FP1. Ecotoxicol. Environ. Saf. 155, 144–151. doi:10.1016/j.ecoenv.2018.02.067
Martinez-Martinez, L., Pascual, A., Conejo, M., Garcia, I., Joyanes, P., Domenech-Sanchez, A., et al. (2002). Energy-dependent accumulation of norfloxacin and porin expression in clinical isolates of Klebsiella pneumoniae and relationship to extended-spectrum β-lactamase production. Antimicrob. Agents Chemother. 46, 3926–3932. doi:10.1128/aac.46.12.3926-3932.2002
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41 (3), 252–275. doi:10.1093/femsre/fux013
Pan, L. J., Li, J., and Li, C. X. (2020). Thermophilic thermophilic bacteria degrade fluoroquinolone antibiotics. J. Environ. Eng. 14, 11. doi:10.12030/j.cjee.201907008
Parshikov, I., Heinze, T., Moody, J., Freeman, J., Williams, A., and Sutherland, J. (2001). The fungus Pestalotiopsis guepini as a model for biotransformation of ciprofloxacin and norfloxacin. General Meet. Am. Soc. Microbiol. Appl. Microbiol. Biotechnol. 56 (3-4), 474–477. doi:10.1007/s002530100672
Paulraj, P., Ozogbuda, P., Sajna, K., Chandramohan, M., Jenifer, S. A., Javad, P., et al. (2019). Effect of glutathione, ascorbic acid and multivitamins on sensitivity of norfloxacin againstPseudomonas aeruginosaandKlebsiella pneumoniae. J. Pure Appl. Microbiol. 13, 835–840. doi:10.22207/jpam.13.2.18
Qu, C. X., Wu, Z., Pan, D. D., Cai, Z. D., and Liu, X. T. (2021). Characterization of Lactobacillus reuteri WQ-Y1 with the ciprofloxacin degradation ability. Biotechnol. Lett. 43 (4), 855–864. doi:10.1007/s10529-020-03068-9
Redgrave, L. S., Sutton, S. B., Webber, M. A., and Piddock, L. J. V. (2014). Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 22, 438–445. doi:10.1016/j.tim.2014.04.007
Vila, J., Marco, F., Soler, L., Chacon, M., and Figueras, M. J. (2002). In vitro antimicrobial susceptibility of clinical isolates of Aeromonas caviae, Aeromonas hydrophila and Aeromonas veronii biotype sobria. J. Antimicrob. Chemother. 49 (4), 701–702. doi:10.1093/jac/49.4.701
Wang, J., and Wang, S. (2018). Microbial degradation of sulfamethoxazole in the environment. Appl. Microbiol. Biotechnol. 102 (8), 3573–3582. doi:10.1007/s00253-018-8845-4
Wang, Q., Zheng, R., and Sun, X. T. (2020b). Effects of testosterone pleuromonas on PAHs degradation and bacterial community structure in leymus chinensis rhizosphere. Soil J. Biotechnol. 36 (12), 2657–3267. doi:10.13345/j.cjb.200381
Wang, Q. N., Jiang, Y. R., Wang, H. D., Chang, X., Lv, M., and Chen, L. (2020a). Isolation and characterization of a marine bacterium Vibrio diabolicus strain L2-2 capable of biotransforming sulfonamides. Environ. Res. 188, 109718. doi:10.1016/j.envres.2020.109718
Wei, D. D., Wan, L. G., Yu, Y., Xu, Q. F., Deng, Q., Cao, X. W., et al. (2015). Characterization of extended-spectrum beta-lactamase, carbapenemase, and plasmid quinolone determinants in Klebsiella pneumoniae isolates carrying distinct types of 16S rRNA methylase genes, and their association with mobile genetic elements. Microb. drug Resist. Larchmt. N.Y.) 21, 186–193. doi:10.1089/mdr.2014.0073
Xia, J., Sun, H., Zhang, X. X., Zhang, T., and Ye, L. (2019). Aromatic compounds lead to increased abundance of antibiotic resistance genes in wastewater treatment bioreactors. Water Res. 166, 115073. doi:10.1016/j.watres.2019.115073
Xiao, C. Q., Han, Y., Liu, Y., Zhang, J., and Hu, C. Q. (2018). Relationship between fluoroquinolone structure and neurotoxicity revealed by zebrafish neurobehavior. Chem. Res. Toxicol. 31, 238–250. doi:10.1021/acs.chemrestox.7b00300
Xu, H., Hai, L. S., Yang, R. P., XuY, L., Xu, J. Y., and Yang, X. L. (2021). Simultaneous reduction of antibiotics leakage and methane emission from constructed wetland by integrating microbial fuel cell. Bioresour. Technol. Biomass, Bioenergy, Biowastes, Convers. Technol. Biotransformations, Prod. Technol. 320, 124285. doi:10.1016/j.biortech.2020.124285
Keywords: quinolone antibiotic, norfloxacin, enrofloxacin, microbial community diversity, microbial degradation
Citation: Wang H, Wang Q, Lv M, Song Z, Yu J, Wang X, Li J and Chen L (2024) Study on the degradation and metabolic mechanism of four quinolone antibiotics by mixed strains. Front. Environ. Chem. 5:1326206. doi: 10.3389/fenvc.2024.1326206
Received: 23 October 2023; Accepted: 17 January 2024;
Published: 02 February 2024.
Edited by:
Pedro Neves Carvalho, Aarhus University, DenmarkReviewed by:
Thomas Thiebault, Université Paris Sciences et Lettres, FranceTu Binh Minh, VNU University of Science, Vietnam
Copyright © 2024 Wang, Wang, Lv, Song, Yu, Wang, Li and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingxin Chen, bHhjaGVuQHlpYy5hYy5jbg==
 Hongdan Wang
Hongdan Wang Qiaoning Wang
Qiaoning Wang Min Lv
Min Lv Zhihua Song3
Zhihua Song3 Jinhua Li
Jinhua Li Lingxin Chen
Lingxin Chen