
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Environ. Archaeol., 06 February 2025
Sec. Landscape and Geological Processes
Volume 4 - 2025 | https://doi.org/10.3389/fearc.2025.1544307
Reconstructing past environments can be challenging when archaeological materials are missing. The study of organic molecules, which remain as traces in the environment over millennia, represents one way to overcome this drawback. Fecal lipid markers (steroids and bile acids) and ancient sedimentary DNA offer a complementary and cross-validating analytical tool to broaden the range of methods used in environmental archaeology. However, little is known about the benefits of combining these two approaches. We present a brief overview of the current state of knowledge on fecal lipid markers and ancient sedimentary DNA. We identify scientific and methodological gaps and suggest their potential relevance for a better understanding of dynamic, human-animal relationships of the past. With this review, we aim to facilitate new research avenues, both in established disciplines and in conjunction with analytical approaches that have rarely been combined to date.
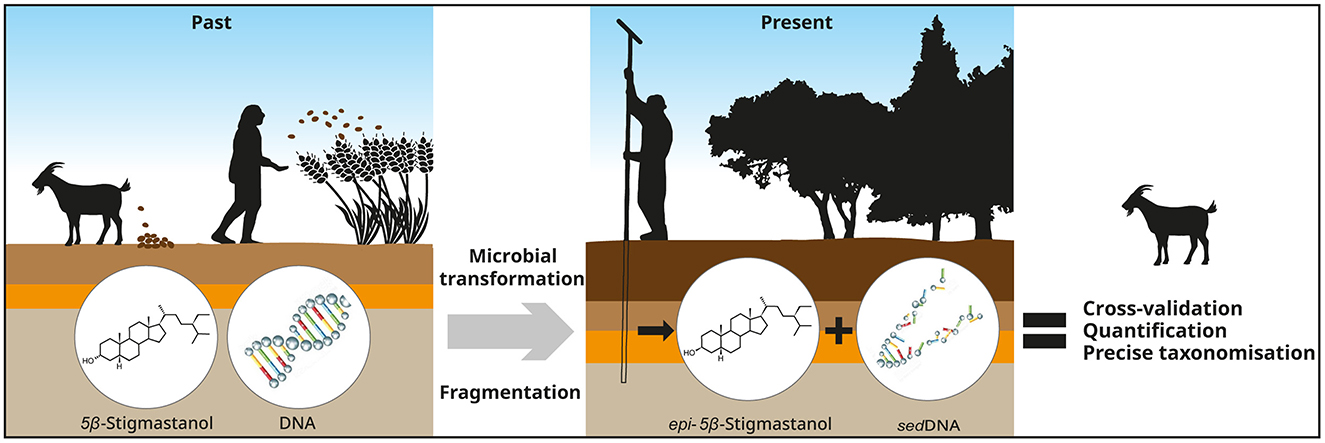
Graphical Abstract. Beneficial combination of fecal lipid marker and ancient sedimentary DNA to study past livestock farming.
Advances in analytical approaches for studying past environments have strengthened interdisciplinary collaboration between archaeology and geosciences. However, this can be challenging when archaeological materials are degraded or absent. Analyzing specific organic molecules that are recalcitrant against (e.g., aromatics) and protected from (encapsulated in aggregates) microbial turnover and therefore remain in the environment as traces over millennia, offers one approach to overcome this limitation (Curtin et al., 2021; Kjær et al., 2022).
The analysis of fecal lipid markers (FLM) such as steroids and bile acids has attracted considerable interest as complement to traditional approaches (e.g., coprophilous fungal spores, coprolites) to investigate the implications of past fecal inputs (Davies et al., 2022; Sistiaga et al., 2014). FLM in sediments have been used to reconstruct the onset of human occupation (Argiriadis et al., 2018; Ossendorf et al., 2019) and population fluctuations (Birkett et al., 2023; Keenan et al., 2021). They can provide in-depth insights into past land use, particularly livestock farming and manuring (Bull et al., 2001; Lavrieux et al., 2012), but also inform on functional areas of past settlements (Hjulström and Isaksson, 2009; Scherer et al., 2021).
Ancient sedimentary DNA (sedDNA) analysis is revolutionizing archaeology (Özdogan et al., 2024). It enables deciphering ancient ecosystems, biodiversity, and even human activity through the study of genetic traces preserved in soils and sediments. By analyzing sedDNA, precise information on the presence, taxonomy and diet of animals can be collected, allowing for unraveling past livestock farming strategies (Giguet-Covex et al., 2014; Zampirolo et al., 2024). Also, sedDNA can provide new insights into the composition and functions of human and animal gut microbiota (Borry et al., 2020).
In this mini review, we address the analysis of FLM and sedDNA from soils and sediments used to study past livestock farming. A brief summary of the current state of knowledge is followed by the identification of future research avenues for a joint application of FLM and sedDNA analysis.
The study of FLM has a long tradition, beginning in the 1980s (Knights et al., 1983; Pepe et al., 1989) with pioneering analytical work by Evershed's group (Evershed, 1993; Evershed et al., 1997; Evershed and Bethell, 1996). Many excellent reviews followed, summarizing the transformation processes of fecal lipids in the vertebrate gut and the environment, documenting their relevance in addressing research gaps in archaeology (Cramp et al., 2023; Evershed, 2008; Harrault et al., 2019).
Each type of diet has an individual steroidal profile with different proportions of steroidal precursors entering the digestive tract (Tyagi et al., 2009a). While cholesterol is omnipresent in eukaryotic cells (Gerber et al., 2015), β-sitosterol is predominant in plant biomass (Christie and Han, 2010). In the vertebrate gastrointestinal tract, Δ5-sterols are biosynthesised to 5β-stanols (coprostanol, 5β-stigmastanol) (Cuevas-Tena et al., 2018). 5β-stanols are useful not only for detecting fecal input in general, but also for distinguishing between omnivorous and herbivorous lifestyles (Leeming et al., 1996; Zocatelli et al., 2017). In a prominent example, Vachula et al. (2019) analyzed coprostanol from lake sediments in northern Alaska and provided evidence that eastern Beringia was inhabited before the formation of the land bridge across the Bering Strait during the last glaciation.
Through microbial reduction in soils, sterols are often converted to 5α-stanols (5α-cholestanol, 5α-stigmastanol) and epi-5β-stanols (epi-coprostanol; epi-5β-stigmastanol), mainly when fecal deposition has occurred long ago (Bull et al., 1999; Isaksson, 1998). Stanones are intermediates formed microbially during the conversion of Δ5-sterols to 5β/5α-stanols, and their epimers in the intestines of higher animals and the environment (Grimalt et al., 1990). They are thought to have potential as biomarkers comparable to 5β-stanols but have rarely been used to study past lifeworlds (Knights et al., 1983). Since not all steroidal compounds are exclusively attributed to feces, numerous ratios of distinct fecal lipids are calculated to interpret soil-specific degradation effects and to differentiate between omnivorous, herbivorous and plant-derived steroidal inputs (Bull et al., 2002; Lerch et al., 2022; Vázquez et al., 2021).
Bile acids (BAs) are the end products of cholesterol catabolism and are found exclusively in vertebrate feces. Individual patterns of BAs are the result of evolutionary variation and, to a lesser degree, diet (Hagey et al., 1993). Their analysis permits the differentiation between pig and human feces, which is not applicable with 5β-stanols (Prost et al., 2017; Zocatelli et al., 2017). The dominant secondary BA in pig feces is hyodeoxycholic acid (HDCA), which is not present in human or ruminant feces (Elhmmali et al., 1997; Tyagi et al., 2009b). Lithocholic acid (LCA) and deoxycholic acid (DCA), among others, are typical of human biochemistry (Perwaiz et al., 2002). A reliable source identification of fecal inputs combines both the analysis of (epi-)5β-stanols and bile acids (Prost et al., 2017).
The ability to decode species presence and biodiversity dynamics has been revolutionized by sedDNA analysis. Organic-rich sediments, such as those found in permafrost soils and caves, have proven to be excellent sources for recovering animal DNA (Boessenkool et al., 2014; Ficetola et al., 2008). Early studies demonstrated that sedDNA could identify extinct species in soils and sediments, offering new insights into past ecosystems (Pedersen et al., 2016; Rawlence et al., 2016; Willerslev et al., 2014). The long-term preservation of sedDNA in stratified archaeological layers allows scientists to track changes in animal populations in response to climate shifts, farming activities, and human migration (Pedersen et al., 2016; Willerslev et al., 2003). sedDNA has been used to trace animal domestication, revealing early evidence of domesticated species such as cattle, pigs, and sheep at prehistoric sites (Giguet-Covex et al., 2014). Haile et al. (2007) found that sedDNA could identify species, like moa and sheep, even without macrofossils, but they also revealed challenges in non-frozen environments where DNA leaching between layers can compromise chronological accuracy. This issue was illustrated by the presence of sheep DNA in older layers, likely due to downward migration. While sedDNA remains a valuable tool for reconstructing past ecosystems, non-frozen conditions pose significant risks for contamination.
Zampirolo et al. (2024) demonstrated the utility of sedDNA in tracing the introduction of pastoralism in Central Europe. By analyzing sediment samples from the Velký Mamuták rock shelter, modern-day Czech Republic, they reconstructed animal and plant DNA profiles across stratigraphic layers, revealing a gradual shift in herding practices. Their findings suggest herding began in the Late Neolithic with sheep, while pigs began to appear in the Late Bronze Age. This gradual transition, challenging to capture from fossil records alone, highlights sedDNA's potential to reveal human impacts on ecosystems.
By combining the analysis of FLM and sedDNA, researchers continue to broaden their understanding of human-animal relationships in archaeological contexts, helping to answer long-standing questions about past biodiversity and animal domestication (Slon et al., 2017; Zeder, 2015). To our knowledge, there are only a few studies that combine the analysis of FLM and sedDNA, but they are pioneering.
There is still much debate about the route taken by the Carthaginian commander Hannibal in the 2nd Punic War. To address this debate, Mahaney et al. (2017) examined a significant accumulation of mammalian dung in the upper Guil Valley near Mt. Viso dating to 2,168 cal 14C year. They were able to extract DCA, 5β-stigmastanol and genes of Clostridia, which were interpreted as evidence of the passage of the Hannibalian army.
Another remarkable study addresses the hitherto mystery as to whether the Vikings first colonized the Faroe Islands in the mid-ninth century. Apart from a few charred grains of barley dating to 351–543 BCE, there is no archaeological evidence of human activity before the Viking Age. Curtin et al. (2021) investigated lake sediments by combining FLM and sedDNA analyses and demonstrated that the Faroe Islands were colonized by humans and their livestock 300 years earlier than the Vikings' arrival.
In another study, geochemical and palynological indicators were used in combination with 5β-stanols, BAs and sedDNA from lake sediments to reconstruct human activity at medieval Celtic crannog settlements in Ireland (Brown et al., 2021) and Scotland (Brown et al., 2022). The results show a direct link between human presence, the processing of specific crops, livestock farming and local slaughters.
Chen et al. (2024) showed that the presence of coprostanol in Antarctic lacustrine sediments does not necessarily indicate human waste. In conjunction with high-throughput sequencing of the 16S rRNA gene, the conversion of seal feces-based cholesterol to coprostanol by anaerobic bacteria (Eubacterium coprostanoligenes) was illustrated. This example reveals that analyzing FLM and sedDNA in tandem enables for a better understanding of natural processes and helps avoiding misinterpretation.
Individually, FLM and sedDNA analyses already provide valuable insights in different cultural-historical contexts, but both methods have interpretive limitations. FLMs provide information on past fecal input and its quantification, whereas FLM cannot differentiate between non-/domesticated animal species. While sedDNA cannot quantify animal stocking, it gives precise data on animal phenotypes, physical traits and adaptations of past populations. The combination of both groups of molecular biomarkers as a complementary and a cross-validating analytical tool opens new windows into the past for reconstructing human-animal interactions.
Furthermore, FLMs are less frequently faced with cross-layer leakage, a common problem with sedDNA. Therefore, FLM analysis may serve as an effective pre-screening tool, allowing sedDNA analysis to focus on samples with a higher chance of DNA recovery. This reduces the risk of false positive signals and improves the detection of modern contaminants. Consequently, FLM and sedDNA analysis in tandem offer quantitatively interpretable, more precise and reliable datasets. This synergy permits the detection of subtle environmental changes and anthropogenic influences that may be less discernible when each approach is applied separately.
Understanding the interaction between soils and organic marker molecules is crucial for assessing the explanatory power of the latter. Ideally, unambiguous compounds that entered the soil simultaneously and are sufficiently preserved over millennia are used (Salisbury et al., 2022; van Mourik and Jansen, 2013). The following substance-specific and analytical challenges highlight potential pitfalls and research avenues for a combined application of FLM and sedDNA in soils and sediments.
Both FLM and sedDNA behave differently under varying soil conditions (e.g., particle size distribution, mineral composition, organic matter, redox and pH state, hydrology), indicating a high mutual dependency (Barnes and Turner, 2016; Islam et al., 2023; Jia et al., 2022; Thomsen and Willerslev, 2015). Organic molecules in soils are stabilized by highly reactive surfaces (organic matter, oxides, clay minerals) (Freeman et al., 2023; Gande et al., 2024; Islam et al., 2023; Lejzerowicz et al., 2013; Lloyd et al., 2012). Although our understanding of organic molecule behavior in soils still needs to be improved, some characteristics make such molecules promising candidates for advanced marker analysis.
FLMs are highly resistant against microbial turnover (Bull et al., 2002; Dubois and Jacob, 2016) primarily due to their aromatic ring structures and strong bonding to organic and mineral matter (Islam et al., 2023). Their hydrophobic nature inhibits large-scale translocation across stratigraphic layers, whereas a coarse soil texture, lower abundance of soil organic matter and co-transport by particulates are reported to have influence of FLM migration (von der Lühe et al., 2018; Lloyd et al., 2012).
sedDNA degradation occurs over time due to microbial enzymatic activity, mechanical damage, and spontaneous chemical reactions like hydrolysis and oxidation. Preservation is strongly influenced by environmental factors: Extreme pH (acidic or basic) and optimal conditions for microbial turnover (temperature, oxygen exposure and moisture) accelerate degradation. Cold and anoxic environments significantly enhance preservation, enabling sedDNA to persist for millennia (Kjær et al., 2022; Murchie et al., 2023; Parducci et al., 2017). In aquatic environments and through percolating water, sedDNA can be leached over considerable distances and across stratigraphic layers (Deiner et al., 2017; Haile et al., 2007). This transport is critical for interpreting genetic traces in chronological contexts. Giguet-Covex et al. (2019) emphasized the role of erosion in the preservation and transport of sedDNA within lacustrine environments. Their findings demonstrated how different soil horizons, vegetation cover and hydrographic connectivity influence sedDNA deposition and quality. Additionally, they highlighted the role of soil organic substance and soil milieu on sedDNA degradation, offering crucial insights into improving the reliability of datasets.
Future research is needed to refine our understanding of how environmental conditions affect the preservation, stabilization and transport of FLM and sedDNA in soils and sediments (Taberlet et al., 2018). In this context, column experiments with both FLM and sedDNA could help to identify differences in their transport sensibility to estimate their complementary and cross-validating character (Poté et al., 2007).
FLMs are leached from soil samples using a mixture of organic solvents (commonly used: dichloromethane:methanol, 2:1, v/v) by various extraction methods (Birk et al., 2012; Manley et al., 2020). Scherer et al. (2024) demonstrated that Soxhlet (SOX) extraction achieves significantly higher yields and better reproducibility of FLM than microwave-assisted and accelerated-solvent extraction. However, traditional SOX extraction methods are time-consuming (>24 h extraction time) and consume high volumes of solvent. Therefore, optimisation and automatisation of SOX extraction is crucial to achieve higher sample throughput, which enables larger data volumes with a higher reliability (by using replicates). Furthermore, methodological approaches from related disciplines could be promising to integrate into studies on past environments. Can the fecal BAs assay developed for analyzing medical plasma also be used for soil samples? Is it possible to develop non-destructive and extraction-free methods to pre-screen samples for certain substances (e.g., coprostanol)? One approach could be the use of high-resolution mass spectrometry (e.g., Orbitrap) combined with direct ionization technology (ambient ionization mass spectrometry) from flowing atmospheric pressure afterglow ion sources, allowing organic molecules to be desorbed and analyzed directly from surfaces.
The extraction of sedDNA from soil samples is particularly challenging due to soil heterogeneity, affecting the effectiveness of DNA extraction and amplification. Current DNA extraction methods for soil rely on a combination of chemical lysis buffers and mechanical disruption techniques, such as bead-beating, to release DNA from soil particles. These methods are implemented using both commercial kits and customs and self-made protocols. However, the effectiveness of these approaches can vary significantly depending on factors such as soil type, sample volume, and environmental conditions, warranting further investigation (Deiner et al., 2017). In this context, soil fractionation can help improve DNA recovery, particularly for specific plant or microbial DNA targets. This involves separating soil samples into size and density fractions to isolate finer particles where sedDNA may be better preserved (Lejzerowicz et al., 2013). Furthermore, the use of chloroform-isoamyl alcohol extraction and alternative lysis buffers that target specific organisms or DNA types, such as pollen or microbial DNA, are conceivable approaches to optimize DNA extraction from soil samples (Birks and Birks, 2016).
Analyzing FLM is biased toward material from humans and domestic animals, while associated (e.g., mice, rats) and co-associated animals (e.g., hyena, wolf, fox) are less frequently examined at archaeological sites. To improve fecal sourcing, the expansion of reference libraries that characterize steroid and bile acid profiles of domesticated and non-domesticated animals should be pursued. Further differentiation of organism-specific feces could be achieved by broaden the analytical spectrum of substances analyzed. According to Porru et al. (2022), mono/di/trioxo-BAs are able to distinguish between feces of wild boar and domestic pigs, while Vallejo et al. (2024) showed that the analysis of hormones such as progesterone provide insights into livestock husbandry favoring female pigs.
sedDNA studies often exhibit a bias toward bacterial communities and prominent taxa like domestic animals and megafauna, especially when using targeted sequencing methods. This focus can lead to an overrepresentation of specific groups while overlooking broader biodiversity. Shotgun metagenomic sequencing offers a more comprehensive, unbiased approach by capturing all DNA present, revealing greater ecological complexity and diversity (Armbrecht, 2020). However, it is resource-intensive and generates large datasets, which can complicate interpretation. Combining targeted and shotgun methods may balance the need for specificity and breadth, providing a fuller view of past environments and communities.
A promising approach to interpretating FLMs in modern and ancient contexts has been successfully demonstrated by Harrault et al. (2019) in which the distributions of eleven 5β-stanols of 90 fecal samples from 10 different species were used to conduct principal component analysis and hierarchical clustering. Extending this approach to include whole lipidomes and even sedDNA would allow the application of this approach to an increasingly large and diverse mixture of potential fecal sources. In order to detect fecal contamination, Grimalt et al. (1990) introduced the ratio coprostanol/(coprostanol/cholestanol). Human fecal input to the environment is argued to be reliably detected at a ratio ≥0.7, with no fecal contamination at a ratio < 0.3. However, strict application of these thresholds is debated (Isobe et al., 2002; Nakagawa et al., 2021), especially their suitability in archaeological contexts (Birk et al., 2011, 2022; Mackay et al., 2020). A combined analysis of FLM and sedDNA may prove an effective means of determining whether lower threshold values can also be indicative of fecal presence. The ongoing debate surrounding the imposition of these thresholds for samples from archaeological sites may soon reach a conclusion.
For both FLM and sedDNA analysis, sterile sampling and laboratory conditions (sterile tools, wearing gloves, strict protocols) are required to prevent modern contamination. Specifically, for sedDNA analysis, highest quality assurance is additionally obligatory (e.g., cleanroom conditions, negative controls) (Cooper and Poinar, 2000).
Authentication of sedDNA involves independent replications, analysis of molecular damage patterns, and assessment of DNA fragment sizes to distinguish ancient DNA from modern contaminants (Kjær et al., 2022). Computational tools further enhance reliability by filtering contaminants based on reference databases and damage signatures (Kjær et al., 2022; Capo et al., 2021). Integrating sedDNA data with chemical proxies, such as lipid biomarkers or isotopic signatures, offers a promising interdisciplinary approach to improve the differentiation between authentic ancient DNA and modern contamination, advancing the field of environmental archaeology.
In our mini review we have demonstrated that a combined application of FLM and sedDNA gives a significant contribution to the growing field of environmental archaeology. Both approaches in tandem represent a powerful tool in the ongoing effort to understand the complexities of past human ecosystem transformation across temporal and spatial scales, offering an innovative perspective on how ancient communities shaped, and were shaped by, the landscapes they inhabited. We hope that our review will stimulate new avenues of research in well-established disciplines, as well as inspire the combination of analytical approaches that have rarely been used in tandem to date.
SF: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. SS: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. BK-K: Conceptualization, Visualization, Writing – original draft, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was completed with the financial support of the Publication Fund of the Johannes Gutenberrg-University.
We are grateful to G. Janzen and M. C. A. Torbenson for critically proofreading the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Argiriadis, E., Battistel, D., McWethy, D. B., Vecchiato, M., Kirchgeorg, T., Kehrwald, N. M., et al. (2018). Lake sediment fecal and biomass burning biomarkers provide direct evidence for prehistoric human-lit fires in New Zealand. Sci. Rep. 8, 12113. doi: 10.1038/s41598-018-30606-3
Armbrecht, L. H. (2020). The potential of sedimentary ancient DNA to reconstruct past ocean ecosystems. Oceanography 33, 116–123. doi: 10.5670/oceanog.2020.211
Barnes, M. A., and Turner, C. R. (2016). The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 17, 1–17. doi: 10.1007/s10592-015-0775-4
Birk, J. J., Dippold, M., Wiesenberg, G. L. B., and Glaser, B. (2012). Combined quantification of faecal sterols, stanols, stanones and bile acids in soils and terrestrial sediments by gas chromatography–mass spectrometry. J. Chromatogr. A 1242, 1–10. doi: 10.1016/j.chroma.2012.04.027
Birk, J. J., Reetz, K., Sirocko, F., Wright, D. K., and Fiedler, S. (2022). Faecal biomarkers as tools to reconstruct land-use history in maar sediments in the Westeifel Volcanic Field, Germany. Boreas 51, 637–650. doi: 10.1111/bor.12576
Birk, J. J., Teixeira, W. G., Neves, E. G., and Glaser, B. (2011). Faeces deposition on Amazonian Anthrosols as assessed from 5β-stanols. J. Archaeol. Sci. 38, 1209–1220. doi: 10.1016/j.jas.2010.12.015
Birkett, B. A., Obrist-Farner, J., Rice, P. M., Parker, W. G., Douglas, P. M. J., Berke, M. A., et al. (2023). Preclassic environmental degradation of Lake Petén Itzá, Guatemala, by the early Maya of Nixtun-Ch'ich'. Commun. Earth Environ. 4, 59. doi: 10.1038/s43247-023-00726-4
Birks, H. J. B., and Birks, H. H. (2016). How have studies of ancient DNA from sediments contributed to the reconstruction of Quaternary floras? New Phytol. 209, 499–506. doi: 10.1111/nph.13657
Boessenkool, S., McGlynn, G., Epp, L. S., Taylor, D., Pimentel, M., Gizaw, A., et al. (2014). Use of ancient sedimentary DNA as a novel conservation tool for high-altitude tropical biodiversity. Conserv. Biol. J. Soc. Conserv. Biol. 28, 446–455. doi: 10.1111/cobi.12195
Borry, M., Cordova, B., Perri, A., Wibowo, M., Prasad Honap, T., Ko, J., et al. (2020). CoproID predicts the source of coprolites and paleofeces using microbiome composition and host DNA content. PeerJ 8, e9001. doi: 10.7717/peerj.9001
Brown, A. G., Fonville, T., van Hardenbroek, M., Cavers, G., Crone, A., McCormick, F., et al. (2022). New integrated molecular approaches for investigating lake settlements in north-western Europe. Antiquity 96, 1179–1199. doi: 10.15184/aqy.2022.70
Brown, A. G., van Hardenbroek, M., Fonville, T., Davies, K., Mackay, H., Murray, E., et al. (2021). Ancient DNA, lipid biomarkers and palaeoecological evidence reveals construction and life on early medieval lake settlements. Sci. Rep. 11, 11807. doi: 10.1038/s41598-021-91057-x
Bull, I. D., Evershed, R. P., and Betancourt, P. P. (2001). An organic geochemical investigation of the practice of manuring at a Minoan site on Pseira Island, Crete. Geoarchaeology 16, 223–242. doi: 10.1002/1520-6548(200102)16:2<223::AID-GEA1002>3.0.CO;2-7
Bull, I. D., Lockheart, M. J., Elhmmali, M. M., Roberts, D. J., and Evershed, R. P. (2002). The origin of faeces by means of biomarker detection. Environ. Int. 27, 647–654. doi: 10.1016/S0160-4120(01)00124-6
Bull, I. D., Simpson, I. A., Dockrill, S. J., and Evershed, R. P. (1999). Organic geochemical evidence for the origin of ancient anthropogenic soil deposits at Tofts Ness, Sanday, Orkney. Org. Geochem. 30, 535–556. doi: 10.1016/S0146-6380(99)00020-0
Capo, E., Giguet-Covex, C., Rouillard, A., Nota, K., Heintzman, P. D., Vuillemin, A., et al. (2021). Lake SEDIMENTARY DNA research on past terrestrial and aquatic biodiversity: overview and recommendations. J. Quat. 4, 6. doi: 10.3390/quat4010006
Chen, X., Wang, J., Jin, J., Nie, Y., Zheng, Z., Xue, Y., et al. (2024). Microbial hydrogenation of cholesterol to coprostanol by anaerobic bacteria: evidence from Antarctic lacustrine sediment. Biogeochemistry. 167, 1107–1122 doi: 10.1007/s10533-024-01121-7
Christie, W. W., and Han, H. (2010). Lipid Analysis, 5th Edn. Bridgwater: Oily Press Lipid Library Series.
Cooper, A., and Poinar, H. N. (2000). Ancient DNA: do it right or not at all. Science, 289, 1139–1139. doi: 10.1126/science.289.5482.1139b
Cramp, L. J. E., Bull, I. D., Casanova, E., Dunne, J., Roffet-Salque, M., Whelton, H.L., et al. (2023). “Lipids in archaeology,” in Handbook of Archaeological Sciences, ed. A. M. Pollard, R. A. Armitage, C., Makarewicz (Hoboken, NJ: Wiley), 529–556.
Cuevas-Tena, M., Alegría, A., and Lagarda, M. J. (2018). Relationship between dietary sterols and gut microbiota: a review. Eur. J. Lipid Sci. Technol. 120, 1800054. doi: 10.1002/ejlt.201800054
Curtin, L., D'Andrea, W. J., Balascio, N. L., Shirazi, S., Shapiro, B., Wet, D., et al. (2021). Sedimentary DNA and molecular evidence for early human occupation of the Faroe Islands. Commun. Earth Environ. 2, 253. doi: 10.1038/s43247-021-00318-0
Davies, A. L., Harrault, L., Milek, K., McClymont, E. L., Dallimer, M., Hamilton, A., et al. (2022). A multiproxy approach to long-term herbivore grazing dynamics in peatlands based on pollen, coprophilous fungi and faecal biomarkers. Palaeogeogr. Palaeoclimatol. Palaeoecol. 598, 111032. doi: 10.1016/j.palaeo.2022.111032
Deiner, K., Bik, H. M., Mächler, E., Seymour, M., Lacoursière-Roussel, A., Altermatt, F., et al. (2017). Environmental DNA metabarcoding: transforming how we survey animal and plant communities. Mol. Ecol. 26, 5872–5895. doi: 10.1111/mec.14350
Dubois, N., and Jacob, J. (2016). Molecular biomarkers of anthropic impacts in natural archives: a review. Front. Ecol. Evol. 4:92. doi: 10.3389/fevo.2016.00092
Elhmmali, M. M., Roberts, D. J., and Evershed, R. P. (1997). Bile acids as a new class of sewage pollution indicator. Environ. Sci. Technol. 31, 3663–3668. doi: 10.1021/es9704040
Evershed, R. P. (1993). Biomolecular archaeology and lipids. World Archaeol. 25, 74–93. doi: 10.1080/00438243.1993.9980229
Evershed, R. P. (2008). Organic residue analysis in archaeology: the archaeological biomarker revolution. Archaeometry 50, 895–924. doi: 10.1111/j.1475-4754.2008.00446.x
Evershed, R. P., and Bethell, P. H. (1996). “Application of multimolecular biomarker techniques to the identification of fecal material in archaeological soils and sediments,” in Molecular Markers in Environmental Geochemistry. Developed from a Symposium, ACS Symposium Series, ed. R. P. Eganhouse (Washington, DC: American Chemical Soc), 157–172.
Evershed, R. P., Bethell, P. H., Reynolds, P. J., and Walsh, N. J. (1997). 5β-Stigmastanol and Related 5β-stanols as biomarkers of manuring: analysis of modern experimental material and assessment of the archaeological potential. J. Archaeol. Sci. 24, 485–495. doi: 10.1006/jasc.1996.0132
Ficetola, G. F., Miaud, C., Pompanon, F., and Taberlet, P. (2008). Species detection using environmental DNA from water samples. Biol. Lett. 4, 423–425. doi: 10.1098/rsbl.2008.0118
Freeman, C. L., Dieudonné, L., Agbaje, O. B. A., Žure, M., Sanz, J. Q., Collins, M., et al. (2023). Survival of environmental DNA in sediments: mineralogic control on DNA taphonomy. Environmental DNA 5, 1691–1705. doi: 10.1002/edn3.482
Gande, D., Hassenrück, C., Žure, M., Richter-Heitmann, T., Willerslev, E., and Friedrich, M.W. (2024). Recovering short DNA fragments from minerals and marine sediments: a comparative study evaluating lysis and isolation approaches. Environ. DNA 6, e547. doi: 10.1002/edn3.547
Gerber, A., Kleser, M., Biedendieck, R., Bernhardt, R., and Hannemann, F. (2015). Functionalized PHB granules provide the basis for the efficient side-chain cleavage of cholesterol and analogs in recombinant Bacillus megaterium. Microb. Cell Fact. 14, 107. doi: 10.1186/s12934-015-0300-y
Giguet-Covex, C., Ficetola, G. F., Walsh, K., Poulenard, J., Bajard, M., Fouinat, L., et al. (2019). New insights on lake sediment DNA from the catchment: importance of taphonomic and analytical issues on the record quality. Sci. Rep. 9, 14676. doi: 10.1038/s41598-019-50339-1
Giguet-Covex, C., Pansu, J., Arnaud, F., Rey, P-. J., Griggo, C., Gielly, L., et al. (2014). Long livestock farming history and human landscape shaping revealed by lake sediment DNA. Nat. Commun. 5, 3211. doi: 10.1038/ncomms4211
Grimalt, J. O., Fernández, P., Bayona, J. M., and Albalgés, J. (1990). Assessment of fecal sterols and ketones as indicators of urban sewage inputs to coastal waters. Environ. Sel. Technol. 24. doi: 10.1021/es00073a011
Hagey, L. R., Crombie, D. L., Espinosa, E., and Carey, M. C. (1993). Ursodeoxycholic acid in the Ursidae: Billarybile acids of bears, pandas, and related carnivores. JLR 34, 1911–1917. doi: 10.1016/S0022-2275(20)35109-9
Haile, J., Holdaway, R., Oliver, K., Bunce, M., Gilbert, M. T. P., Nielsen, R., et al. (2007). Ancient DNA chronology within sediment deposits: are paleobiological reconstructions possible and is DNA leaching a factor? Mol. Biol. Evol. 24, 982–989. doi: 10.1093/molbev/msm016
Harrault, L., Milek, K., Jardé, E., Jeanneau, L., Derrien, M., Anderson, D. G., et al. (2019). Faecal biomarkers can distinguish specific mammalian species in modern and past environments. PLoS ONE 14, e0211119. doi: 10.1371/journal.pone.0211119
Hjulström, B., and Isaksson, S. (2009). Identification of activity area signatures in a reconstructed Iron Age house by combining element and lipid analyses of sediments. J. Archaeol. Sci. 36, 174–183. doi: 10.1016/j.jas.2008.08.005
Isaksson, S. (1998). A kitchen entrance to the aristocracy—analysis of lipid biomarkers in cultural layers. Laborativ Arkeologi 10–11, 43–53.
Islam, M. S., Nakagawa, K. Yu, Z-. Q., Takao, Y., and Berndtsson, R. (2023). Coprostanol adsorption behavior in agricultural soil, riverbed sediment, and sand. J. Environ. Chem. Eng. 11, 110029. doi: 10.1016/j.jece.2023.110029
Isobe, K. O., Tarao, M., Zakaria, M. P., Chiem, N. H., Minh, L. Y., Takada, H., et al. (2002). Quantitative application of fecal sterols using gas chromatography–mass spectrometry to investigate fecal pollution in tropical waters: western Malaysia and Mekong Delta, Vietnam. Environ. Sci. Technol. 36, 4497–4507. doi: 10.1021/es020556h
Jia, W., Liu, X., Stoof-Leichsenring, K. R., Liu, S., Li, K., and Herzschuh, U. (2022). Preservation of sedimentary plant DNA is related to lake water chemistry. Environ. DNA 4, 425–439. doi: 10.1002/edn3.259
Keenan, B., Imfeld, A., Johnston, K., Breckenridge, A., Gélinas, Y., Douglas, P. M. J., et al. (2021). Molecular evidence for human population change associated with climate events in the Maya lowlands. Quat. Sci. Rev. 258, 106904. doi: 10.1016/j.quascirev.2021.106904
Kjær, K. H., Winther Pedersen, M., Sanctis, B., Cahsan, B., Korneliussen, T. S., Michelsen, C.S., et al. (2022). A 2-million-year-old ecosystem in Greenland uncovered by environmental DNA. Nature 612, 283–291. doi: 10.1038/s41586-022-05453-y
Knights, B. A., Dickson, C. A., Dickson, J. H., and Breeze, D. J. (1983). Evidence concerning the Roman military diet at Bearsden, Scotland, in the 2nd century AD. J. Archaeol. Sci. 10, 139–152. doi: 10.1016/0305-4403(83)90048-1
Lavrieux, M., Bréheret, J-. G., Disnar, J-. R., Jacob, J., Le Milbeau, C., Zocatelli, R., et al. (2012). Preservation of an ancient grassland biomarker signature in a forest soil from the French Massif Central. Org. Geochem. 51, 1–10. doi: 10.1016/j.orggeochem.2012.07.003
Leeming, R., Ball, A., Ashbolt, N., and Nichols, P. (1996). Using faecal sterols from humans and animals to distinguish faecal pollution in receiving waters. Water Res. 30, 2893–2900. doi: 10.1016/S0043-1354(96)00011-5
Lejzerowicz, F., Esling, P., Majewski, W., Szczuciński, W., Decelle, J., Obadia, C., et al. (2013). Ancient DNA complements microfossil record in deep-sea subsurface sediments. Biol. Lett. 9, 20130283. doi: 10.1098/rsbl.2013.0283
Lerch, M., Bromm, T., Geitner, C., Haas, J. N., Schäfer, D., Glaser, B., et al. (2022). Human and livestock faecal biomarkers at the prehistorical encampment site of Ullafelsen in the Fotsch Valley, Stubai Alps, Austria—potential and limitations. Biogeosciences 19, 1135–1150. doi: 10.5194/bg-19-1135-2022
Lloyd, C. E. M., Michaelides, K., Chadwick, D. R., Dungait, J. A. J., and Evershed, R. P. (2012). Tracing the flow-driven vertical transport of livestock-derived organic matter through soil using biomarkers. Organ. Geochem. 43, 56–66. doi: 10.1016/j.orggeochem.2011.11.001
Mackay, H., Davies, K. L., Robertson, J., Roy, L., Bull, I. D., Whitehouse, N. J., et al. (2020). Characterising life in settlements and structures: incorporating faecal lipid biomarkers within a multiproxy case study of a wetland village. J. Archaeol. Sci. 121, 105202. doi: 10.1016/j.jas.2020.105202
Mahaney, W. C., Allen, C. C. R., Pentlavalli, P., Kulakova, A., Young, J. M., Dirszowsky, R. W., et al. (2017). Biostratigraphic evidence relating to the age-old question of Hannibal's invasion of Italy. Archaeometry 59, 179–190. doi: 10.1111/arcm.12228
Manley, A., Collins, A. L., Joynes, A. Mellander, P-. E., and Jordan, P. (2020). Comparing extraction methods for biomarker steroid characterisation from soil and slurry. Water Air Soil Pollut. 231, 524. doi: 10.1007/s11270-020-04871-w
Murchie, T. J., Giguet-Covex, C., Heintzman, P. D., Slon, V., and Wang, Y. (2023). “Terrestrial fauna and hominin DNA from sedimentary archives,” in Tracking Environmental Change Using Lake Sediments. Volume 6: Sedimentary DNA, Developments in Paleoenvironmental Research, ed. E. Capo, C. Barouillet, J. P. Smol (Cham: Springer International Publishing), 299–378. doi: 10.1007/978-3-031-43799-1_11
Nakagawa, K., Amano, H., Persson, M., and Berndtsson, R. (2021). Spatiotemporal variation of nitrate concentrations in soil and groundwater of an intensely polluted agricultural area. Sci. Rep. 11, 2598. doi: 10.1038/s41598-021-82188-2
Ossendorf, G., Groos, A. R., Bromm, T., Tekelemariam, M. G., Glaser, B., Lesur, J., et al. (2019). Middle Stone Age foragers resided in high elevations of the glaciated Bale Mountains, Ethiopia. Science 365, 583–587. doi: 10.1126/science.aaw8942
Özdogan, K. T., Gelabert, P., Hammers, N., Altinişik, N.E., De Groot, A., and Plets, G. (2024). Archaeology meets environmental genomics: implementing sedaDNA in the study of the human past. Archaeol. Anthropol. Sci. 16, 108. doi: 10.1007/s12520-024-01999-2
Parducci, L., Bennett, K. D., Ficetola, G. F., Alsos, I. G., Suyama, Y., Wood, J. R., et al. (2017). Ancient plant DNA in lake sediments. New Phytol. 214, 924–942. doi: 10.1111/nph.14470
Pedersen, M. W., Ruter, A., Schweger, C., Friebe, H., Staff, R. A., Kjeldsen, K. K., et al. (2016). Postglacial viability and colonization in North America's ice-free corridor. Nature 537, 45–49. doi: 10.1038/nature19085
Pepe, C., Dizabo, P., Scribe, P., Dagaut, J., Fillaux, J., Saliot, A., et al. (1989). Les marqueurs biogéochimiques: application à l'archéologie. Revue d' Archéométrie 13:1e12. doi: 10.3406/arsci.1989.866
Perwaiz, S., Mignault, D., Tuchweber, B., and Yousef, I. M. (2002). Rapid and improved method for the determination of bile acids in human feces using MS. Lipids 37, 1093–1100. doi: 10.1007/s11745-002-1005-0
Porru, E., Scicchitano, D., Interino, N., Tavella, T., Candela, M., Roda, A., et al. (2022). Analysis of fecal bile acids and metabolites by high resolution mass spectrometry in farm animals and correlation with microbiota. Sci. Rep. 12, 2866. doi: 10.1038/s41598-022-06692-9
Poté, J., Rosselli, W., Wigger, A., and Wildi, W. (2007). Release and leaching of plant DNA in unsaturated soil column. Ecotoxicol. Environ. Saf. 68, 293–298. doi: 10.1016/j.ecoenv.2006.11.004
Prost, K., Birk, J. J., Lehndorff, E., Gerlach, R., and Amelung, W. (2017). Steroid biomarkers revisited—improved source identification of faecal remains in archaeological soil material. PLoS ONE 12, e0164882. doi: 10.1371/journal.pone.0164882
Rawlence, N. J., Wood, J. R., Bocherens, H. R., and Karyne, M. (2016). Dietary interpretations for extinct megafauna using coprolites, intestinal contents and stable isotopes: complimentary or contradictory? Quat. Sci. Rev. 142, 173–178. doi: 10.1016/j.quascirev.2016.05.017
Salisbury, R. B., Bull, I. D., Cereda, S., Draganits, E., Dulias, K., Kowarik, K., et al. (2022). Making the most of soils in archaeology. a review. Archaeologia Band 106, 319–334. doi: 10.1553/archaeologia106s319
Scherer, S., Birk, J. J., Klassen, S., and Fiedler, S. (2024). A matter of extraction—extraction yields and ratios of faecal lipid biomarker from archaeological soils using Soxhlet, microwave-assisted and accelerated-solvent extraction. Org. Geochem. 197, 104863. doi: 10.1016/j.orggeochem.2024.104863
Scherer, S., Höpfer, B., Deckers, K., Fischer, E., Fuchs, M., Kandeler, E.et al. (2021). Middle Bronze Age land use practices in the northwestern Alpine foreland—a multi-proxy study of colluvial deposits, archaeological features and peat bogs. Soil 7, 269–304. doi: 10.5194/soil-7-269-2021
Sistiaga, A., Mallol, C., Galván, B., and Summons, R. E. (2014). The neanderthal meal: a new perspective using faecal biomarkers. PLoS ONE 9, e101045. doi: 10.1371/journal.pone.0101045
Slon, V., Hopfe, C., Weiß, C. L., Mafessoni, F., La Rasilla, M., Lalueza-Fox, C., et al. (2017). Neandertal and denisovan DNA from pleistocene sediments. Science 356, 605–608. doi: 10.1126/science.aam9695
Taberlet, P., Bonin, A., Zinger, L., and Coissac, E. (2018). Environmental DNA, First edition. Oxford: Oxford University Press.
Thomsen, P. F., and Willerslev, E. (2015). Environmental DNA—An emerging tool in conservation for monitoring past and present biodiversity. Biol. Conserv. 183, 4–18. doi: 10.1016/j.biocon.2014.11.019
Tyagi, P., Edwards, D. R., and Coyne, M. S. (2009a). Distinguishing between human and animal sources of fecal pollution in waters: a review. IJW 5, 15. doi: 10.1504/IJW.2009.023080
Tyagi, P., Edwards, D. R., and Coyne, M. S. (2009b). Fecal sterol and bile acid biomarkers. Water Air Soil Pollut. 198, 45–54. doi: 10.1007/s11270-008-9824-7
Vachula, R. S., Huang, Y., Longo, W. M., Dee, S. G., Daniels, W. C., Russell, J. M., et al. (2019). Evidence of Ice Age humans in eastern Beringia suggests early migration to North America. Q. Sci. Rev. 205, 35–44. doi: 10.1016/j.quascirev.2018.12.003
Vallejo, A., Forgia, V., Vergès, J. M., Gorostizu-Orkaiztegi, A., Alday-Izaguirre, A., Elejaga-Jimeno, A., et al. (2024). Identification of animal species housed and herding practices in ancient sediments from the Vallone Inferno rock-shelter (Scillato, Sicily, Italy) using faecal biomarkers, hormones, and their metabolites. Quat. Int. 683–684, 123–134. doi: 10.1016/j.quaint.2023.08.003
van Mourik, J. M., and Jansen, B. (2013). The added value of biomarker analysis in palaeopedology; reconstruction of the vegetation during stable periods in a polycyclic driftsand sequence in SE-Netherlands. Quat. Int. 306, 14–23. doi: 10.1016/j.quaint.2013.05.034
Vázquez, C., Vallejo, A., Vergès, J. M., and Barrio, R. J. (2021). Livestock activity biomarkers: Estimating domestication and diet of livestock in ancient samples. J. Archaeol. Sci. Rep. 40, 103220. doi: 10.1016/j.jasrep.2021.103220
von der Lühe, B., Birk, J. J., Dawson, L., Mayes, R. W., and Fiedler, S. (2018). Steroid fingerprints: efficient biomarkers of human decomposition fluids in soil. Org. Geochem. 124, 228–237. doi: 10.1016/j.orggeochem.2018.07.016
Willerslev, E., Davison, J., Moora, M., Zobel, M., Coissac, E., Edwards, M. E., et al. (2014). Fifty thousand years of Arctic vegetation and megafaunal diet. Nature 506, 47–51. doi: 10.1038/nature12921
Willerslev, E., Hansen, A. J., Binladen, J., Brand, T. B., Gilbert, M. T. P., Shapiro, B., et al. (2003). Diverse plant and animal genetic records from Holocene and Pleistocene sediments. Science 300, 791–795. doi: 10.1126/science.1084114
Zampirolo, G., Holman, L. E., Sawafuji, R., Ptáková, M., Kovačiková, L., Šída, P., et al. (2024). Tracing early pastoralism in Central Europe using sedimentary ancient DNA. Curr. Biol. 34, 4650–4661.e4. doi: 10.1016/j.cub.2024.08.047
Zeder, M. A. (2015). Core questions in domestication research. Proc. Natl. Acad. Sci. U.S.A. 112, 3191–3198. doi: 10.1073/pnas.1501711112
Zocatelli, R., Lavrieux, M., Guillemot, T., Chassiot, L., Le Milbeau, C., Jacob, J., et al. (2017). Fecal biomarker imprints as indicators of past human land uses: source distinction and preservation potential in archaeological and natural archives. J. Archaeol. Sci. 81, 79–89. doi: 10.1016/j.jas.2017.03.010
Keywords: past livestock farming, archaeology, steroids, bile acids, sedDNA
Citation: Fiedler S, Scherer S and Krause-Kyora B (2025) Fecal lipid markers in tandem with ancient sedimentary DNA as a tool for tracing past livestock farming from soils and sediments. Front. Environ. Archaeol. 4:1544307. doi: 10.3389/fearc.2025.1544307
Received: 12 December 2024; Accepted: 17 January 2025;
Published: 06 February 2025.
Edited by:
Aron L. Crowell, Smithsonian National Museum of Natural History (SI), United StatesReviewed by:
Anna Irto, University of Messina, ItalyCopyright © 2025 Fiedler, Scherer and Krause-Kyora. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sabine Fiedler, ZmllZGxlcnNAdW5pLW1haW56LmRl
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.